- 1Beijing Institute of Radiation Medicine, Academy of Military Medical Sciences (AMMS), Beijing, China
- 2Department of Basic Medicine, Chengde Medical College, Chengde, China
- 3Academy of Life Sciences, Anhui Medical University, Hefei, China
Diabetic wounds are recalcitrant to healing. One of the important characteristics of diabetic trauma is impaired macrophage polarization with an excessive inflammatory response. Many studies have described the important regulatory roles of microRNAs (miRNAs) in macrophage differentiation and polarization. However, the differentially expressed miRNAs involved in wound healing and their effects on diabetic wounds remain to be further explored. In this study, we first identified differentially expressed miRNAs in the inflammation, tissue formation and reconstruction phases in wound healing using Illumina sequencing and RT-qPCR techniques. Thereafter, the expression of musculus (mmu)-miR-145a-5p (“miR-145a-5p” for short) in excisional wounds of diabetic mice was identified. Finally, expression of miR-145a-5p was measured to determine its effects on macrophage polarization in murine RAW 264.7 macrophage cells and wound healing in diabetic mice. We identified differentially expressed miRNAs at different stages of wound healing, ten of which were further confirmed by RT-qPCR. Expression of miR-145a-5p in diabetic wounds was downregulated during the tissue formation stage. Furthermore, we observed that miR-145a-5p blocked M1 macrophage polarization while promoting M2 phenotype activation in vitro. Administration of miR-145a-5p mimics during initiation of the repair phase significantly accelerated wound healing in db/db diabetic mice. In conclusion, our findings suggest that rectifying macrophage function using miR-145a-5p overexpression accelerates diabetic chronic wound healing.
Introduction
Wound healing is a complex process involving a variety of cell types and the coordination of various cytokines to regulate wound repair. Macrophages tightly participate in and regulate the entire process of wound healing, and their numbers increase during the inflammation phase, peak during the tissue formation phase, and gradually decline during the maturation phase (1, 2). Generally, macrophages are divided into M1 (classical activation) and M2 (alternative activation) phenotypes. M1 macrophages phagocytize pathogens and necrotic tissues and secrete inflammatory factors, while M2 macrophages help to relieve inflammation and promote angiogenesis by releasing anti-inflammatory factors and growth factors. Diabetic patients experience wounds characterized by a prolonged inflammatory state, with overactivated M1-like macrophages and elevated levels of proinflammatory factors with inadequately activated M2 phenotypes and impaired expression of growth factors (3, 4). Because macrophages are an important target for the treatment of chronic diabetic wounds, studying the regulation of macrophage polarization may uncover therapeutic avenues to accelerate wound repair in diabetes.
miRNAs, 20–24 nucleotide (nt) non-coding RNAs, have the ability to regulate gene expression and the network of cellular processes. One miRNA can regulate the expression of multiple target genes, while genes involved in the same physiological or pathological process may be regulated by a single or a few miRNAs. Accordingly, miRNAs have advantages over conventional genes when used as therapeutic targets in diseases (5). Many studies have determined that miRNAs are important in regulating macrophage polarization (6, 7). Furthermore, M1 and M2 phenotypes play distinctive roles in the progression of refractory diabetic wounds. Thus, miRNAs might affect the repair of diabetic wounds by regulating the conversion between M1 and M2 macrophages.
In the present study, we determined the miRNA expression profiles in different stages of wound healing using Illumina sequencing and real-time quantitative reverse transcript polymerase chain reaction (RT-qPCR) techniques. Furthermore, overexpression of mmu-miR-145a-5p using mimics inhibited M1 polarization and promoted M2 polarization in vitro. Administration of miR-145a-5p mimics during the initiation of the repair phase also significantly accelerated wound healing in diabetic mice.
Materials and Methods
Animals
BKS-Leprem2Cd479/Nju (db/db) diabetic mice (male, 8–10-week-old) were purchased from the Peking University Experimental Animal Center (Beijing, China). The measured blood glucose value of the db/db mice was 23.86 ± 2.07 mmol/L while the normal was ~6 mmol/L, indicating that the diabetic mouse models were successfully established. Normal C57BL/6J mice (male, 8–10-week-old) were purchased from Weitonglihua Co., Ltd. (Beijing, China). All animals were maintained in an animal facility, where the temperature was 22 ± 2°C and the humidity 55 ± 10% on a 12 h light-dark cycle. Food and water were freely available.
Wound Healing Model
Mice were anesthetized with pentobarbital (i.p., 80 mg/kg). The dorsal surface was then shaved and cleaned with 75% ethanol. Two round wounds with a diameter of 5 mm were made on each side of the midline of the shaved dorsum using a punch biopsy tool (Miltex, USA). The wounds were left undressed, and all mice were housed separately thereafter. The wounds were imaged using a high-resolution camera, the sizes of which were calculated using ImageJ software (National Institutes of Health, USA). Biopsies were harvested on postoperative days 0, 1, 3, 5, 7, 15, 19, and 21.
RNA Isolation and Library Preparation
During the inflammation, tissue formation, and tissue reconstruction phases of skin wound healing in control mice, namely, at 0, 1, 3, and 7 days post-operation, the wounds and a 5 mm unwounded skin border were harvested and stored at −80°C until further processing. Total RNA was isolated using the miRNeasy Serum/Plasma Kit (Qiagen, USA). The A (260/280) absorbance ratio of isolated RNA was 1.8–2.0, while the A (260/230) absorbance ratio was >1.6. Integrity of total RNA was determined using formaldehyde denaturing gel electrophoresis. The libraries were constructed from total RNA using the Illumina TruSeq Small RNA Sample Preparation kit (Illumina, USA). Single-stranded cDNA was created by reverse transcription reactions with the ligation products. The cDNA was amplified by PCR. Finally, Illumina sequencing technology was used to sequence these prepared samples.
Illumina Sequencing and RT-QPCR Validation
Raw sequences were processed using the Illumina pipeline program. Clean reads were filtered, and contaminated reads were removed. Secondary structure prediction of individual miRNAs was performed using Mfold software. Clean sequence reads were mapped using miRBase 20.0, and a mismatch of 1–2 nucleotide bases was allowed. All data were transformed to log base 2. Differences between the samples were analyzed using chi-square and Fisher's exact tests. miRNAs with a fold difference N2.0 and P < 0.05 were considered statistically significant. miRNAs with high expression abundance and significant differences (fold change >4) in each phase of skin trauma were verified using RT-qPCR with Applied Biosystems TaqManTM MGB (minor groove binder) probes (Thermo Fisher Scientific, Waltham, MA, USA). The experiment was performed in strict accordance with the reagent instructions. All samples were analyzed in triplicate. The specificity of each PCR product was validated by melting curve analysis at the end of PCR cycles, and the cycle threshold (Ct) value was defined as the number of cycles required for the fluorescent signal to reach the threshold. Relative expression levels of miRNAs in wound samples were calculated using the 2–ΔΔCt formula, where ΔΔCt = [Ct (target, test) – Ct (ref, test)] – [Ct (target, calibrator) – Ct (ref, calibrator)]. All primers used were obtained from Invitrogen (Carlsbad, CA, USA).
Transfection of miR-145a-5p Mimics and Negative Control
RAW 264.7 macrophage cells were grown in Roswell Park Memorial Institute medium (RPMI) 1640 (Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37°C in a 5% CO2 atmosphere. Macrophage polarization was induced with 1 μg/ml lipopolysaccharide (LPS, Sigma, L2880) or 20 ng/ml interleukin-4 (IL-4, Proteintech, 214-14). The cells were then transfected with 4 nM miR-145a-5p mimics (4464066, MC11480, Thermo Fisher Scientific) and negative control (4464059, Thermo Fisher Scientific) using Lipofectamine RNAiMAX (Invitrogen) according to the instructions and were harvested 48 h later. Three repeats were performed for each group.
For the in vivo study, miR-145a-5p mimics and negative control were diluted to 4 nM in autoclaved phosphate-buffered saline (PBS) and injected subcutaneously around the wound at a volume of 4 μL in each quadrant. The injections were repeated every 3 days for 2 weeks.
Quantitative Real-Time PCR
At predetermined time points, total RNA was isolated from skin tissues and RAW 264.7 macrophage cells using TRIzol reagent (Invitrogen). Thereafter, complementary DNAs (cDNAs) were synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The resulting cDNAs were amplified with 40 cycles by RT-qPCR using Power SYBR@ Green PCR Master Mix (Thermo Fisher Scientific). Each sample was analyzed three times and normalized to β-actin as the internal control. Primer sequences for RT-qPCR are listed in Table 1.
Histological Analysis and Immunohistochemistry
The wounds and a 5 mm unwounded skin border were harvested from mice and processed for histological analysis. Wound widths were measured as the distance between wound margins. Wound areas were determined using image analysis (measured below the clot and above the panniculus muscle). Re-epithelialization was defined as the percentage of distance migrated by the neoepidermis compared to the upper wound width. Immunohistochemistry analysis was performed on formalin-fixed, paraffin-embedded 5-μm sections. The primary antibodies used in this study were against keratin 14 (K14, 1:1000, Covance, Princeton, NJ, USA), Ki67 (1:1000, Covance) and CD206 (1:500, Novus Biologicals, CO, USA). To quantify CD206 expression, ImageJ 1.50d (USA) was used to analyze the integrated optical density.
Bioinformatics Analysis of Downstream Target Genes of mmu-miR-145a-5p
To predict target genes of mmu-miR-145a-5p, TargetScan (5.0), miRanda (3.3a), and PITA (1.0) analyses were conducted and the default thresholds were targetscan_score ≥50, miranda_Energy < −10, and ddG < −5. Thereafter, the intersections of the three databases were selected as candidate downstream miRNAs of mmu-miR-145a-5p for further analysis. Functional enrichment analysis of the target genes of miR-145a-5p was conducted using clusterProfiler with Gene Ontology.db. (GO) and the Kyoto Encyclopedia of Genes and Genomes.db. (KEGG). Significance was assessed using hypergeometric tests and Benjamini-Hochberg correction, with a significance threshold of p < 0.05 (8, 9).
Statistical Analysis
All results are presented as the mean ± SD. All experiments were performed with a minimum of three independent replicates. One-way ANOVA followed by Bonferroni post-hoc tests were performed to analyze multiple groups and unpaired Student's t-test was performed to analyze two parallel groups, using SPSS software (IBM, Armonk, NY, USA). Differences were considered significant at two-sided p < 0.05.
Results
Differentially Expressed miRNAs During Wound Healing in Mice
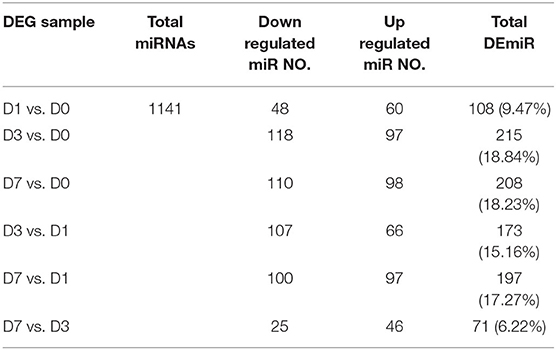
High-throughput miRNA sequencing (miRNA-Seq, 10 million reads/sample) was performed on unharmed skin tissues (D0) and during inflammation (postinjury day 1, D1), tissue formation (postinjury day 3, D3) and tissue reconstruction (postinjury day 7, D7) phases during wound healing in normal male C57BL/6J mice. Table 2 shows the total number of differentially expressed miRNAs between different groups (fold change ≥2 and P ≤ 0.01). The miRNAs with high expression abundance and significant differences (a fold change >4) in each phase of skin trauma were verified using real-time PCR with the probe method. The verification results for 10 miRNAs were consistent with the sequencing results, and expression levels of mmu-miR-183a-5p, mmu-miR-143-3p, mmu-miR-145a-5p, mmu-miR-let-7c, mmu-miR-26a-5p, mmu-miR-27a-3p, mmu-miR-30e-5p, mmu-miR-1a-3p, mmu-miR-1b-5p, and mmu-miR-let-7b were significantly downregulated 1–7 days after trauma (Figure 1). These results suggest that miRNAs participate in the regulation of different stages of wound healing and might be potential therapeutic targets in diseases related to abnormal wound repair.
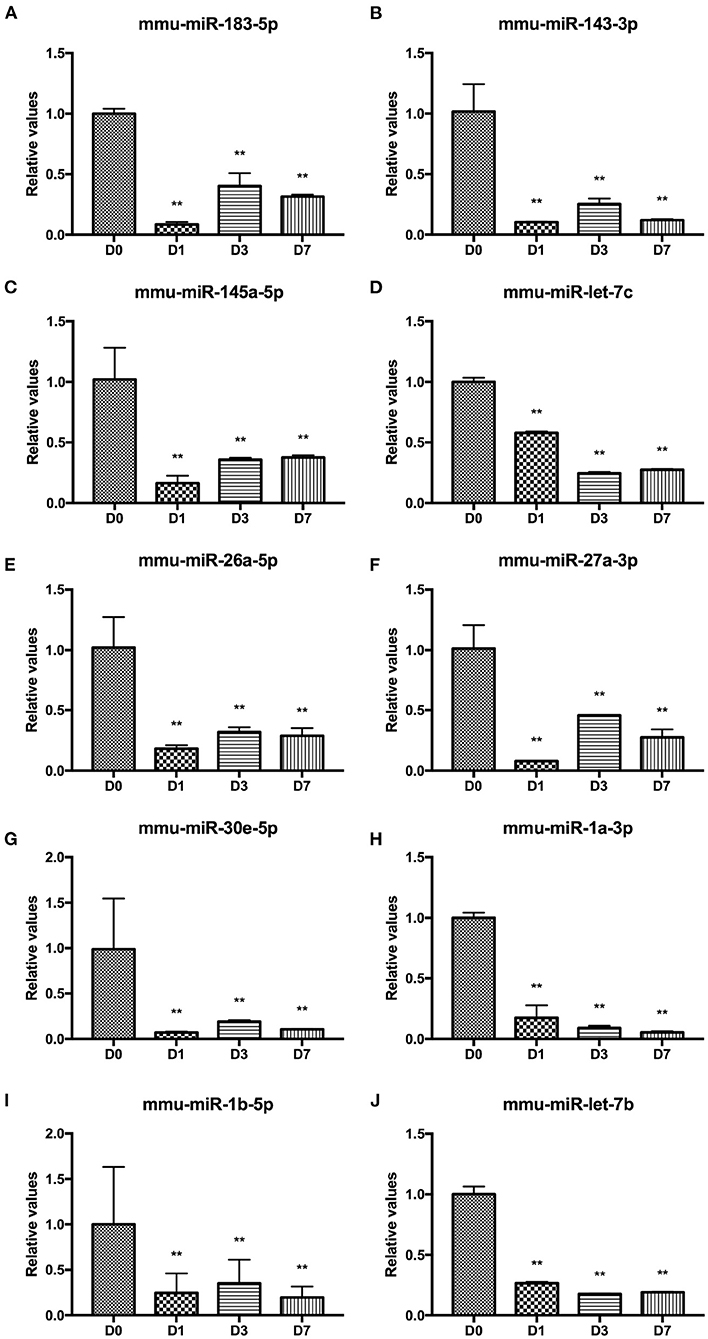
Figure 1. Differentially expressed miRNAs at different stages of wound healing. (A) mmu-miR-183a-5p. (B) mmu-miR-143-3p, (C) mmu-miR-145a-5p. (D) mmu-miR-let-7c. (E) mmu-miR-26a-5p. (F) mmu-miR-27a-3p. (G) mmu-miR-30e-5p. (H) mmu-miR-1a-3p. (I) mmu-miR-1b-5p. (J) mmu-miR-let-7b. **P < 0.01 vs. control (D0).
An Excessive Inflammatory Response and Insufficient Growth Factor Secretion During the Formation Phase of Skin Trauma Causes Delayed Wound Healing in Diabetic Mice
Next, we examined the changes in wound repair and the inflammatory state in diabetic mice. The wound closure speed in the db/db mice was dramatically lower than that in the control group (Figure 2A). We also determined cytokine expression in wound tissues and found that expression levels of the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-1alpha (IL-1α) in db/db mice were downregulated on days 1 and 3 and elevated on day 7 post-injury, suggesting insufficient activation of M1 macrophages during the inflammatory stage of the wounds but overreaction during the tissue formation stage (Figures 2B,C). Quite a number of studies have reported the abnormal inflammation in chronic diabetic wound, which is characterized by the upregulation of pro-inflammatory cytokines, such as TNF-a, IL-1α, IL-1β, and IL-6 (10–13). Shang et al. found the increased M1-like macrophages on d7 in diabetic mice, which were believed to be closely related to the expression of pro-inflammatory factors (14). In addition, the growth factors transforming growth factor beta 2 (TGF-β2) and Smad5 in the db/db mice decreased significantly at different time points post-injury, indicating reduced polarization of M2 macrophages in all stages of diabetic wound healing (Figures 2D,E). Collectively, the failure of transformation from M1 macrophages to M2 phenotypes during the tissue formation stage might be an important mechanism of delayed wound healing in diabetes mellitus.
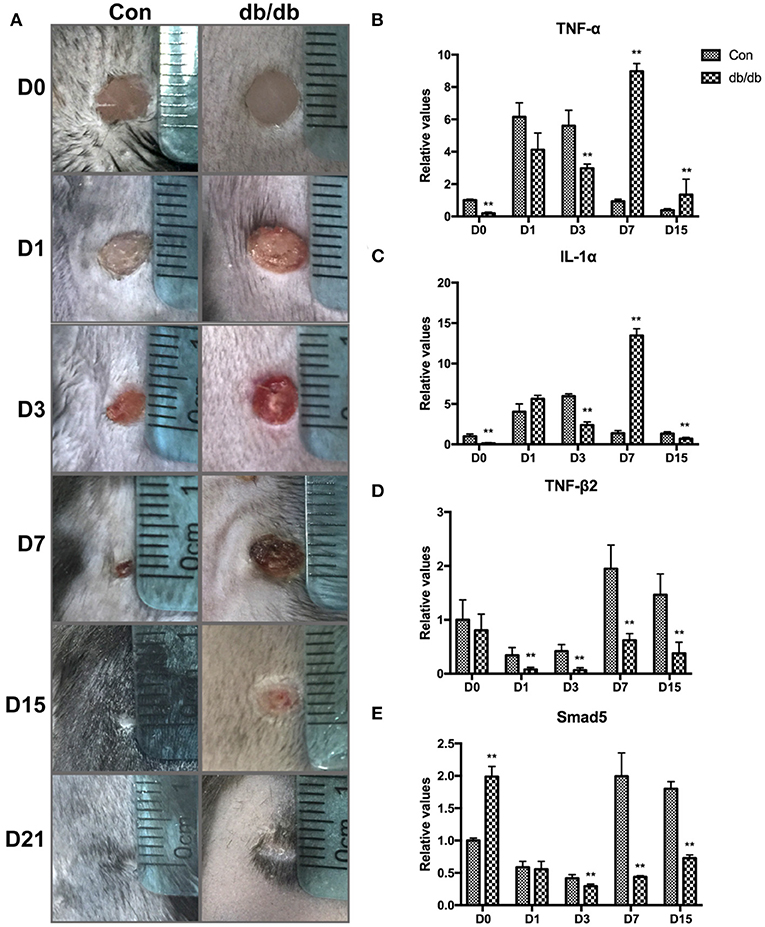
Figure 2. Changes in wound size and the expression of proinflammatory and growth factors in diabetic wound healing. (A) Representative images of excisional wounds in db/db and control mice on days 0, 1, 3, 7, 15, and 21. (B–E) Expression of proinflammatory and growth cytokines in the db/db and control mice on days 0, 1, 3, 7, and 15. **P < 0.01 vs. the corresponding control (Con) group at the same time points.
miR-145a-5p Promotes Macrophage Polarization Toward M2 in vitro
miR-145a-5p was one of the differentially expressed miRNAs during wound healing identified by microarray and RT-qPCR. Many studies have reported the regulatory effects of miR-145a-5p on cell differentiation, such as myoblasts, adipocytes, and microglia (15–17). Whether miR-145a-5p affects the polarization of macrophages in diabetic wound healing greatly excited our interest. First, we studied the expression of miR-145a-5p in wound tissues of db/db mice and found that expression levels of miR-145a-5p increased on days 0, 1, and 15 but were significantly decreased on day 7 after the operation (Figure 3A). miR-145a-5p was involved in the regulation of inflammation (18), and the higher expression level of miR-145a-5p in db/db mice than in control mice on day 0 might be explained by diabetic microangiopathy and inflammation. These results suggest that miR-145a-5p is involved in the pathophysiological process of diabetic wound healing.
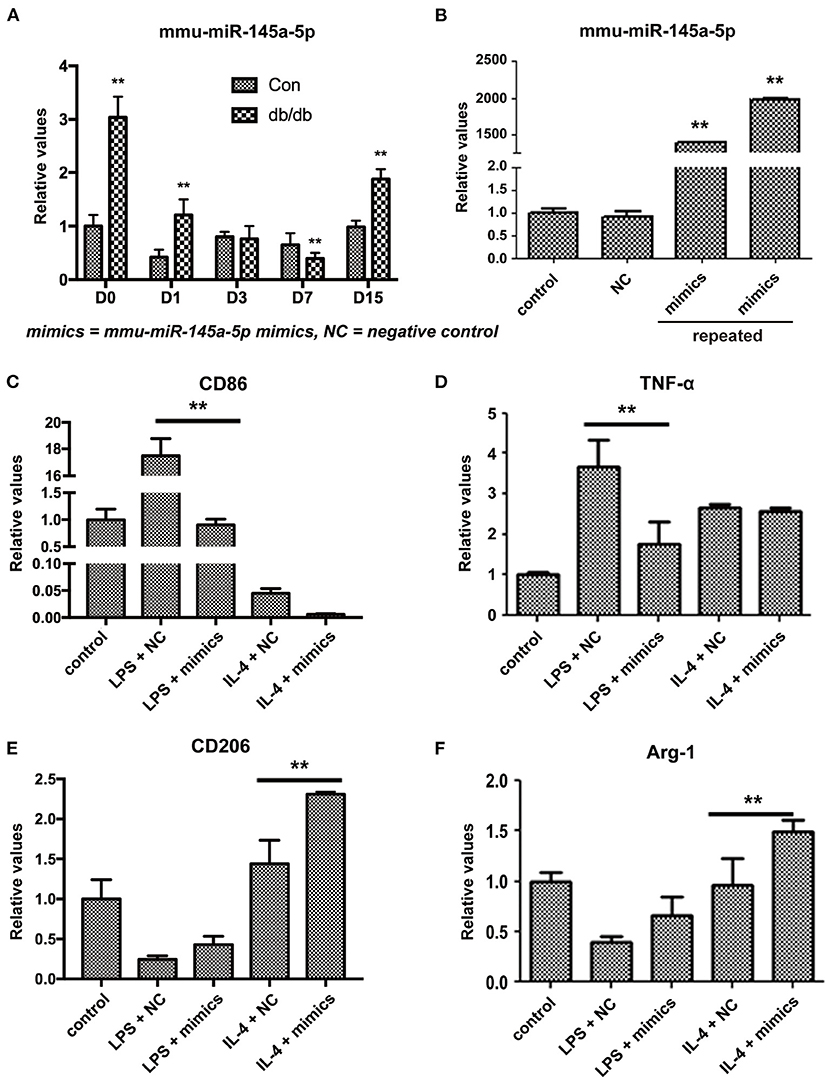
Figure 3. Influence of miR-145a-5p on macrophage polarization in RAW 264.7 cells. (A) Expression of miR-145a-5p in db/db and control mice on postoperative days 0, 1, 3, 7, and 15. (B) Expression of miR-145a-5p in RAW 264.7 macrophages 24 h after the addition of miR-145a-5p mimics or negative control miRNA. (C–F) Expression of CD86, TNF-α, CD206 and Arg-1 in RAW 264.7 macrophages pretreated with LPS or IL-4 in combination with miR-145a-5p mimics/negative control miRNA. **P < 0.01 vs. the corresponding control group.
To further clarify the effects of miR-145a-5p on macrophage polarization, we transfected miR-145a mimics or the related negative control (NC) in RAW 264.7 macrophage cells pretreated with LPS or IL-4. Transfection of miR-145a-5p mimics into RAW 264.7 cells increased the expression levels of miR-145a-5p > 1000-fold compared to that of the NC group (Figure 3B). Generally, CD86 and tumor necrotic factor-α (TNF-α) are used as biomarkers generated by M1 macrophages, while CD206 and arginase 1 (Arg-1) are accepted markers of the M2 phenotype (19, 20). miR-145a-5p mimic transfection dramatically downregulated expression of CD86 and TNF-α in RAW 264.7 macrophages pretreated with LPS, suggesting that miR-145a-5p abolished M1 polarization induced by LPS (Figures 3C,D). On the other hand, miR-145a-5p overexpression caused increased expression of CD206 and Arg-1 in macrophages pretreated with LPS or IL-4, indicating the roles played by miR-145a-5p in facilitating M2 polarization (Figures 3E,F). These results demonstrate that miR-145a-5p promotes macrophage polarization toward M2.
miR-145a-5p Mimics Accelerate Wound Healing in db/db Mice
miR-145a-5p mimics were administered locally to assess their therapeutic effect on diabetic trauma. The wound closure rate in db/db mice treated with miR-145a-5p mimics was dramatically higher than that in the control group (Figures 4A,B). Histological analysis revealed that wounds in the miR-145a-5p mimic group healed at a faster rate than those in the control group (Figure 5A). Immunohistochemistry of K14 showed that the neoepithelial lengths were longer in the healing skin of db/db mice transfected with miR-145a-5p mimics than in those treated with negative control miRNAs, and enhanced expression of Ki67 on postoperative day 7 suggested an increased number of proliferative cells during the tissue formation stage when miR-145a-5p was overexpressed (Figures 5B,C). Importantly, miR-145a-5p mimics increased the expression of CD206 in diabetic wounds, suggesting the ability of miR-145a-5p to promote M2 polarization in vivo (Figures 5D,E). These results show that overexpression of miR-145a-5p accelerates diabetic wound healing and increases M2 phenotypes in diabetic wounds.
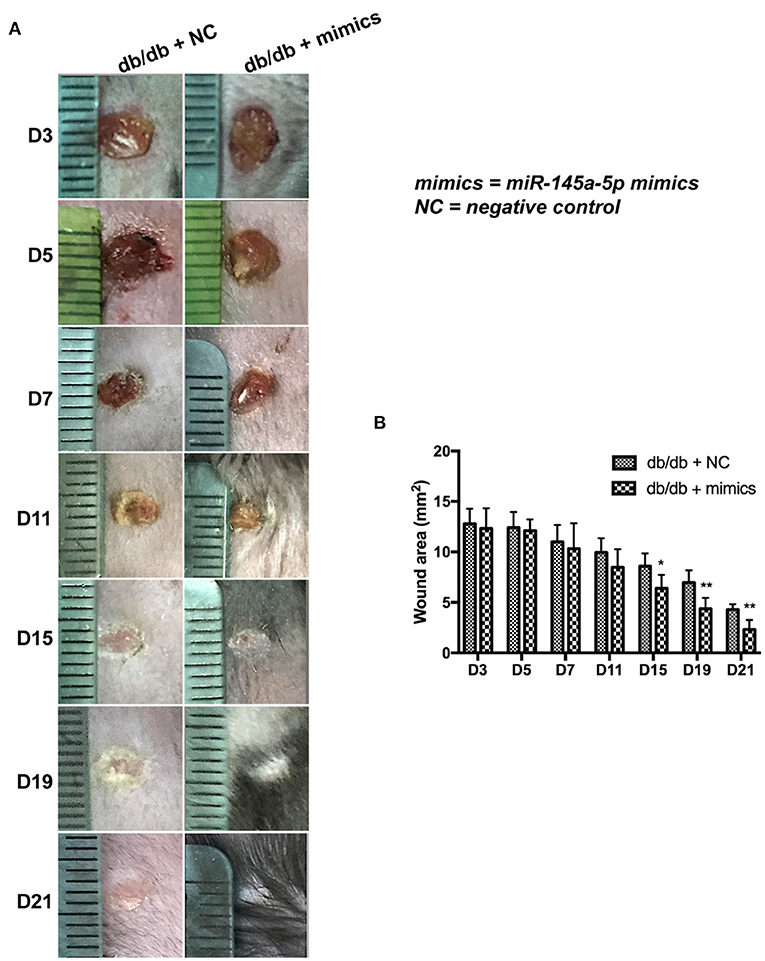
Figure 4. Effect of miR-145a-5p on wound healing size in diabetic mice. (A) Representative images of excisional wounds of db/db mice treated with miR-145a-5p mimics or the corresponding negative control. (B) Wound size measurement. *P < 0.05, **P < 0.01 vs. the corresponding control group.
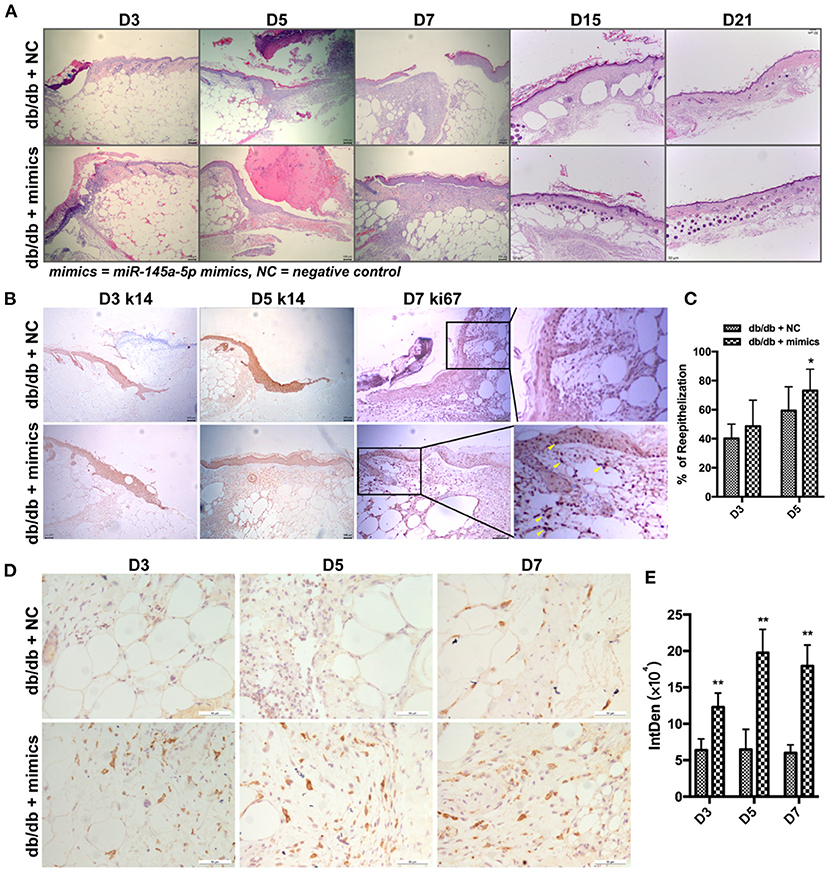
Figure 5. Effect of miR-145a-5p on keratinocyte reepithelialization of wound healing in diabetic mice. (A) Representative H&E stained images of excisional wounds in db/db mice treated with miR-145a-5p mimics or negative control on days 3, 5, 7, 15, and 21. (B) Representative immunohistochemistry (IHC) stained images of K14 and Ki67 in wounds of db/db mice treated with miR-145a-5p mimics or negative control on days 3, 5, and 7. Ki67+ cells are indicated with yellow arrows. (C) Statistical analysis of epithelialization ratio. (D) Representative IHC images of CD206 in diabetic wounds treated with miR-145a-5p mimics or negative control on days 3, 5, and 7. (E) Statistical analysis of the integrated optical density of CD206 from D. *P < 0.05 and **P < 0.01 vs. the corresponding control group at the same time points.
Go and KEGG Functional Enrichment Analysis of miR-145a-5p Downstream Genes
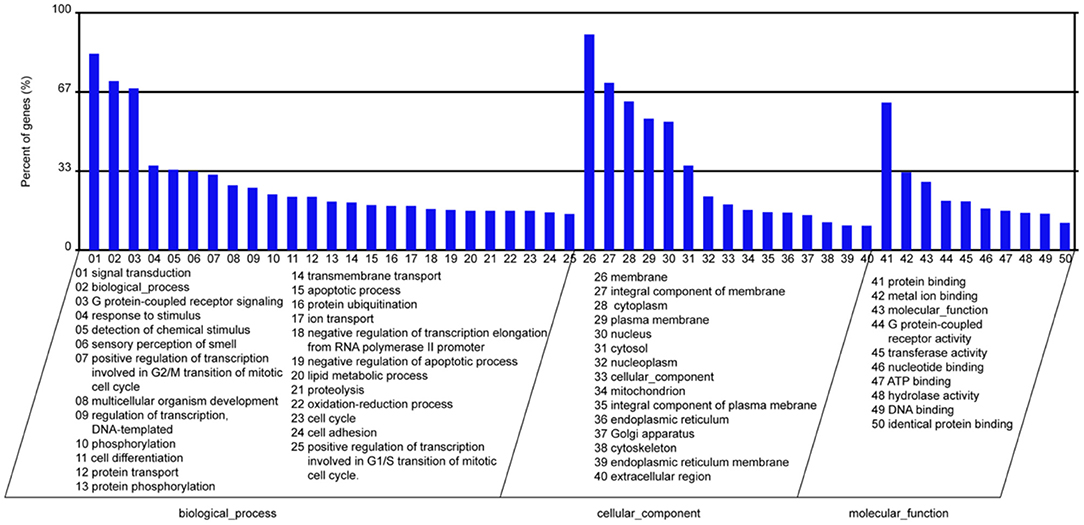
In the present study, we screened the downstream genes of miR-145a-5p using 3 databases, including TargetScan, miRanda, and PITA. Subsequently, 2,473 target genes of miR-145a-5p predicted by all three databases were selected. GO and KEGG functional enrichment analyses revealed that miR-145a-5p is involved in regulating many biological processes, including signal transduction, cell differentiation, apoptotic processes, etc. (Figure 6).
Discussion
The normal wound healing response is characterized by progression from clot formation to an inflammatory phase, a repair phase, and finally remodeling. Macrophages have been verified to be important in the inflammatory phase of tissue repair, where their dynamic plasticity allows these cells to mediate both tissue-destructive and tissue-reparative functions (10). In diabetic chronic wounds, an extended inflammatory phase stemming from macrophage functional disturbance halts this progression (21, 22). The phagocytosis rate of M1 macrophages in the diabetic state is >5 times slower than normal, which may lead to slower clearance of wound necrotic tissue and apoptotic cells and sustained release of inflammatory factors. The blocked transformation from the M1 to M2 phenotype and the insufficient secretion of anti-inflammatory factors and growth factors lead to failure of normal tissue formation and eventually prolonged healing in diabetic mellitus (23, 24). Some studies have attempted to treat diabetic trauma by regulating the macrophage-mediated inflammatory response and have achieved positive effects, suggesting macrophages as a potential therapeutic target in refractory wounds in diabetic mellitus (25–30). Our research group has been committed to investigating wound healing mechanisms and therapy and previously found that knockdown of casein kinase 2-interacting protein 1 (CKIP-1) in macrophages inhibited M1-type activation and slowed the wound healing rate (31). However, innovative methods still need to be explored to regulate the macrophage response during wound healing. In this study, we identified miR-145a-5p as one of the differentially expressed miRNAs between db/db diabetic mice and controls. miR-145a-5p has been demonstrated to play important regulatory roles in cell differentiation and polarization, such as myoblasts, adipocytes, and microglia (15–17). Therefore, the regulation of miR-145a-5p on the macrophage response and its effect on diabetic wound healing aroused our interest.
miRNAs are a class of endogenous RNAs of ~22 nucleotides in length produced by dicer 1, ribonuclease III (Dicer) processing single-stranded RNA precursors of ~70–90 bases (32). miRNAs bind to the corresponding 3′ untranslated region (3′UTR) of the target mRNA through complete or incomplete pairing, preventing its translation or disrupting its stability to regulate gene expression (33). miRNAs regulate 30% of all human genes and have extensive regulatory functions in life activities and profound effects on gene expression, growth, and behavior (34, 35). miRNAs are widely involved in various skin wound healing processes (36). miR-125b regulates leukocyte aggregation and activation by inhibiting TNF-α expression (37). miR-125b inhibits keratinocyte proliferation by downregulating fibroblast growth factor receptor 2 (FGFR2) (38). In ischemic trauma, hypoxia in the wound site and high miR-210 expression reduce keratinocyte proliferation by inhibiting E2F transcription factor 3 (E2F3) expression (39). miR-483-3p depresses keratinocyte proliferation and migration by downregulating mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2, or MK2), Ki67, and Yes-associated protein 1 (YAP1) (40). miR-203 regulates keratinocyte proliferation and migration at the wound margin by affecting RAN and Ras association (RalGDS/AF-6) and pleckstrin homology domain 1 (RAPH1) (41). miR-205 promotes keratinocyte migration by inhibiting inositol polyphosphate phosphatase-like 1 (SHIP2) (42, 43). miR-21 downregulates tissue metalloproteinase inhibitor 3 (TIMP3) and T-cell lymphoma invasion and metastasis 1 (TIAM1) to promote keratinocyte migration (44). miR-21 is elevated in the skin of diabetic mice and regulates wound contraction and collagen deposition (45, 46). miR-27b inhibits mouse mesenchymal stem cell migration to burn sites by silencing C-X-C motif chemokine ligand 12 (SDF-1A), which slows wound healing (47). Members of the miR-199 family are involved in the regulation of wound healing through the AKT/mammalian target of rapamycin (mTOR) pathway (48). let-7c overexpression inhibits the M1 macrophage phenotype and promotes the M2 phenotype (49). LPS-pretreated mesenchymal cells secrete let-7b from exosomes, promoting macrophage secretion of anti-inflammatory factors and targeting M2 differentiation, while miR-130b inhibits M2 macrophage activation by targeting peroxisome proliferator-activated receptor gamma (PPARγ) (50). In conclusion, miRNAs are intimately involved in the regulation of wound healing. In this study, we screened out miRNAs related to the traumatic inflammatory response, tissue formation, and tissue remodeling by chip and finally verified 10 differentially expressed miRNAs (high expression and significant difference) using RT-qPCR. Among these miRNAs, miR-145a-5p was selected for further study due to its potential to regulate macrophage polarization.
In this study, we found that local application of miR-145a-5p dramatically promoted wound healing in db/db mice. K14 is an intermediate filament protein that is mainly expressed in epithelial cells, which are generally used as a research biomarker of epidermal skin (51). Ki67 is universally expressed among proliferating cells, but is absent from quiescent cells (52). miR-145a-5p was found to facilitate diabetic wound healing, of which the indicators were chiefly the accelerated repair, the larger K14 positive area, as well as the increased Ki67 positive cells. Macrophage dysfunction is an important characteristic of delayed wound healing in diabetes mellitus (21, 22). In this study, functional experiments revealed that miR-145-5p overexpression inhibited M1 macrophage polarization while promoting M2 polarization in RAW 264.7 macrophage cells. Importantly, the overexpression of miR-145-5p increased the number of M2 macrophages (CD206+) in diabetic wound. It has been demonstrated that macrophages of M2 facilitate the resolution of inflammation and promote tissue remodeling by releasing growth cytokines and anti-inflammatory cytokines (10, 11). Therefore, the polarization of macrophage toward the M2 phenotype mediated by miR-145a-5p might be an important mechanism responsible for its therapeutic effects on diabetic wound repair. Additionally, the bioinformatics analysis of the downstream miRNAs of miR-145a-5p suggested that miR-145a-5p was involved in the regulation of signal transduction, cell differentiation, apoptosis, cell cycle, lipid metabolic process, etc. Wang et al. found that miR-145a-5p regulated microglial polarization and the production of inflammatory cytokines, in which the NF-κB pathway might be involved (17). Chen et al. reported that miR-145a-5p modulated the polarization of M2 macrophage by targeting PAK7 and regulating β-catenin signaling in hyperlipidemia (53). Moreover, miR-145a-5p regulates the differentiation of myoblasts, adipocytes, and T cells (15, 16, 54). However, the underlying mechanisms by which miR-145a-5p regulates macrophage polarization have not been elucidated. More work is needed to verify the downstream genes and related signaling pathways of miR-145a-5p in regulating macrophage polarization.
The other differentially expressed miRNAs identified in our study, such as miR-183-5p, miR-143-3p, miR-let-7c, miR-26a-5p, miR-27a-3p, miR-30e-5p, and miR-let-7b, were mainly involved in the regulation of cellular proliferation and inflammation. The functions of these miRNAs were found to be chiefly as follows: First, miR-183-5p transferred to macrophages by exosomes promoted the secretion of proinflammatory cytokines (55); second, miR-143-3p participated in the regulation of LPS induced inflammation (56); third, miR-let-7c inhibited proinflammatory cytokine production in osteoarthritis and rheumatoid arthritis synovial fibroblasts (57); fourth, miR-26a-5p was capable of protecting against retinal neuronal impairment in diabetic mice by down-regulating phosphatase and tensin homolog (PTEN) (58); fifth, miR-27a-3p attenuated LPS-induced HK-2 cell apoptosis by downregulating the protein levels of TLR4 and NF-kappaB (59, 60); sixth, miR-30e-5p alleviated inflammation and cardiac dysfunction after myocardial infarction through targeting PTEN (61, 62); and seventh, miR-let-7b regulated the expression of inflammation-associated genes in monocytes, macrophages, and neutrophil (63, 64). The literature on the biological roles of miR-1a-3p and miR-1b-5p has been scanty. While these miRNAs might play important roles in the process of wound healing through the regulation of inflammation and cell proliferation, the effects of these identified miRNAs on wound healing as well as the underlying mechanisms remain to be further explored, where potential therapeutic targets, for instance, diabetic trauma, are expected to be pinpointed for chronic wound healing.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://datadryad.org/stash/dataset/, https://doi.org/10.5061/dryad.n5tb2rbwx.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Beijing Institute of Radiation Medicine.
Author Contributions
YLi and LY: conceptualization. YH, HZ, YLiu, YY, CY, HY, YX, ZJ, and CH: methodology. YH and JW: formal analysis and writing—original draft preparation. YLi: writing—review and editing and supervision. LY: project administration and funding acquisition. All authors have read and agreed to the final version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 81272104.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.775523/full#supplementary-material
References
1. Martin P, Leibovich SJ. Inflammatory cells during wound repair: the good, the bad and the ugly. Trends Cell Biol. (2005) 15:599–607. doi: 10.1016/j.tcb.2005.09.002
2. Eming SA, Hammerschmidt M, Krieg T, Roers A. Interrelation of immunity and tissue repair or regeneration. Semin Cell Dev Biol. (2009) 20:517–27. doi: 10.1016/j.semcdb.2009.04.009
3. Loots MA, Lamme EN, Zeegelaar J, Mekkes JR, Bos JD, Middelkoop E. Differences in cellular infiltrate and extracellular matrix of chronic diabetic and venous ulcers versus acute wounds. J Invest Dermatol. (1998) 111:850–7. doi: 10.1046/j.1523-1747.1998.00381.x
4. Eming SA, Krieg T, Davidson JM. Inflammation in wound repair: molecular and cellular mechanisms. J Invest Dermatol. (2007) 127:514–25. doi: 10.1038/sj.jid.5700701
5. Choi PJ, Oskouian RJ, Tubbs RS. The current understanding of microRNA's therapeutic, diagnostic, and prognostic role in chordomas: a review of the literature. Cureus. (2018) 10:e3772. doi: 10.7759/cureus.3772
6. Essandoh K, Li Y, Huo J, Fan GC. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. (2016) 46:122–31. doi: 10.1097/SHK.0000000000000604
7. Roy S. miRNA in macrophage development and function. Antioxid Redox Signal. (2016) 25:795–804. doi: 10.1089/ars.2016.6728
8. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
9. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. (2000) 28:27–30. doi: 10.1093/nar/28.1.27
10. Boniakowski AE, Kimball AS, Jacobs BN, Kunkel SL, Gallagher KA. Macrophage-mediated inflammation in normal and diabetic wound healing. J Immunol. (2017) 199:17–24. doi: 10.4049/jimmunol.1700223
11. Aitcheson SM, Frentiu FD, Hurn SE, Edwards K, Murray RZ. Skin wound healing: normal macrophage function and macrophage dysfunction in diabetic wounds. Molecules. (2021) 26:4917. doi: 10.3390/molecules26164917
12. Kim SY, Nair MG. Macrophages in wound healing: activation and plasticity. Immunol Cell Biol. (2019) 97:258–67. doi: 10.1111/imcb.12236
13. Zhao R, Liang H, Clarke E, Jackson C, Xue M. Inflammation in chronic wounds. Int J Mol Sci. (2016) 17:2085. doi: 10.3390/ijms17122085
14. Shang W, Chen G, Li Y, Zhuo Y, Wang Y, Fang Z, et al. Static magnetic field accelerates diabetic wound healing by facilitating resolution of inflammation. J Diabetes Res. (2019) 2019:5641271. doi: 10.1155/2019/5641271
15. Du J, Li Q, Shen L, Lei H, Luo J, Liu Y, et al. miR-145a-5p promotes myoblast differentiation. Biomed Res Int. (2016) 2016:5276271. doi: 10.1155/2016/5276271
16. Du J, Cheng X, Shen L, Tan Z, Luo J, Wu X, et al. Methylation of miR-145a-5p promoter mediates adipocytes differentiation. Biochem Biophys Res Commun. (2016) 475:140–8. doi: 10.1016/j.bbrc.2016.05.057
17. Wang H, Liao S, Li H, Chen Y, Yu J. Long non-coding RNA TUG1 sponges mir-145a-5p to regulate microglial polarization after oxygen-glucose deprivation. Front Mol Neurosci. (2019) 12:215. doi: 10.3389/fnmol.2019.00215
18. Ramelli SC, Comer BS, McLendon JM, Sandy LL, Ferretti AP, Barrington R, et al. Nanoparticle delivery of anti-inflammatory LNA oligonucleotides prevents airway inflammation in a HDM model of asthma. Mol Ther Nucleic Acids. (2020) 19:1000–14. doi: 10.1016/j.omtn.2019.12.033
19. Lisi L, Ciotti GM, Braun D, Kalinin S, Curro D, Dello Russo C, et al. Expression of iNOS, CD163 and ARG-1 taken as M1 and M2 markers of microglial polarization in human glioblastoma and the surrounding normal parenchyma. Neurosci Lett. (2017) 645:106–12. doi: 10.1016/j.neulet.2017.02.076
20. Xu HT, Lee CW Li MY, Wang YF, Yung PS, Lee OK. The shift in macrophages polarisation after tendon injury: a systematic review. J Orthop Translat. (2020) 21:24–34. doi: 10.1016/j.jot.2019.11.009
21. Jawerbaum A, Gonzalez E. Diabetic pregnancies: the challenge of developing in a pro-inflammatory environment. Curr Med Chem. (2006) 13:2127–38. doi: 10.2174/092986706777935302
22. Acosta JB, del Barco DG, Vera DC, Savigne W, Lopez-Saura P, Guillen Nieto G, et al. The pro-inflammatory environment in recalcitrant diabetic foot wounds. Int Wound J. (2008) 5:530–9. doi: 10.1111/j.1742-481X.2008.00457.x
23. Arya AK, Tripathi R, Kumar S, Tripathi K. Recent advances on the association of apoptosis in chronic non healing diabetic wound. World J Diabetes. (2014) 5:756–62. doi: 10.4239/wjd.v5.i6.756
24. Mirza RE, Koh TJ. Contributions of cell subsets to cytokine production during normal and impaired wound healing. Cytokine. (2015) 71:409–12. doi: 10.1016/j.cyto.2014.09.005
25. Zhang Q, Oh JH, Park CH, Baek JH, Ryoo HM, Woo KM. Effects of dimethyloxalylglycine-embedded poly(epsilon-caprolactone) fiber meshes on wound healing in diabetic rats. ACS Appl Mater Interfaces. (2017) 9:7950–63. doi: 10.1021/acsami.6b15815
26. Yu T, Gao M, Yang P, Liu D, Wang D, Song F, et al. Insulin promotes macrophage phenotype transition through PI3K/Akt and PPAR-gamma signaling during diabetic wound healing. J Cell Physiol. (2019) 234:4217–31. doi: 10.1002/jcp.27185
27. Lee J, Rodero MP, Patel J, Moi D, Mazzieri R, Khosrotehrani K. Interleukin-23 regulates interleukin-17 expression in wounds, and its inhibition accelerates diabetic wound healing through the alteration of macrophage polarization. FASEB J. (2018) 32:2086–94. doi: 10.1096/fj.201700773R
28. Di G, Du X, Qi X, Zhao X, Duan H, Li S, et al. Mesenchymal stem cells promote diabetic corneal epithelial wound healing through TSG-6-dependent stem cell activation and macrophage switch. Invest Ophthalmol Vis Sci. (2017) 58:4344–54. doi: 10.1167/iovs.17-21506
29. Boniakowski AM, denDekker AD, Davis FM, Joshi A, Kimball AS, Schaller M, et al. SIRT3 regulates macrophage-mediated inflammation in diabetic wound repair. J Invest Dermatol. (2019) 139:2528–37 e2. doi: 10.1016/j.jid.2019.05.017
30. Ishida Y, Kuninaka Y, Nosaka M, Furuta M, Kimura A, Taruya A, et al. CCL2-mediated reversal of impaired skin wound healing in diabetic mice by normalization of neovascularization and collagen accumulation. J Invest Dermatol. (2019) 139:2517–27 e5. doi: 10.1016/j.jid.2019.05.022
31. Zhang L, Xia X, Zhang M, Wang Y, Xing G, Yin X, et al. Integrated analysis of genomics and proteomics reveals that CKIP-1 is a novel macrophage migration regulator. Biochem Biophys Res Commun. (2013) 436:382–7. doi: 10.1016/j.bbrc.2013.05.109
32. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-y
33. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. (2000) 403:901–6. doi: 10.1038/35002607
34. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-867400045-5
35. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. (2008) 9:102–14. doi: 10.1038/nrg2290
36. Banerjee J, Chan YC, Sen CK. MicroRNAs in skin and wound healing. Physiol Genomics. (2011) 43:543–56. doi: 10.1152/physiolgenomics.00157.2010
37. Tili E, Michaille JJ, Gandhi V, Plunkett W, Sampath D, Calin GA. miRNAs and their potential for use against cancer and other diseases. Future Oncol. (2007) 3:521–37. doi: 10.2217/14796694.3.5.521
38. Xu N, Brodin P, Wei T, Meisgen F, Eidsmo L, Nagy N, et al. MiR-125b, a microRNA downregulated in psoriasis, modulates keratinocyte proliferation by targeting FGFR2. J Invest Dermatol. (2011) 131:1521–9. doi: 10.1038/jid.2011.55
39. Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, et al. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci U S A. (2010) 107:6976–81. doi: 10.1073/pnas.1001653107
40. Bertero T, Gastaldi C, Bourget-Ponzio I, Imbert V, Loubat A, Selva E, et al. miR-483-3p controls proliferation in wounded epithelial cells. FASEB J. (2011) 25:3092–105. doi: 10.1096/fj.10-168401
41. Viticchie G, Lena AM, Cianfarani F, Odorisio T, Annicchiarico-Petruzzelli M, Melino G, et al. MicroRNA-203 contributes to skin re-epithelialization. Cell Death Dis. (2012) 3:e435. doi: 10.1038/cddis.2012.174
42. Yu J, Ryan DG, Getsios S, Oliveira-Fernandes M, Fatima A, Lavker RM. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci U S A. (2008) 105:19300–5. doi: 10.1073/pnas.0803992105
43. Yu J, Peng H, Ruan Q, Fatima A, Getsios S, Lavker RM. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. (2010) 24:3950–9. doi: 10.1096/fj.10-157404
44. Yang X, Wang J, Guo SL, Fan KJ, Li J, Wang YL, et al. miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int J Biol Sci. (2011) 7:685–90. doi: 10.7150/ijbs.7.685
45. Madhyastha R, Madhyastha H, Nakajima Y, Omura S, Maruyama M. MicroRNA signature in diabetic wound healing: promotive role of miR-21 in fibroblast migration. Int Wound J. (2012) 9:355–61. doi: 10.1111/j.1742-481X.2011.00890.x
46. Wang T, Feng Y, Sun H, Zhang L, Hao L, Shi C, et al. miR-21 regulates skin wound healing by targeting multiple aspects of the healing process. Am J Pathol. (2012) 181:1911–20. doi: 10.1016/j.ajpath.2012.08.022
47. Lu MH, Hu CJ, Chen L, Peng X, Chen J, Hu JY, et al. miR-27b represses migration of mouse MSCs to burned margins and prolongs wound repair through silencing SDF-1a. PLoS ONE. (2013) 8:e68972. doi: 10.1371/journal.pone.0068972
48. Jin Y, Tymen SD, Chen D, Fang ZJ, Zhao Y, Dragas D, et al. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS ONE. (2013) 8:e64434. doi: 10.1371/journal.pone.0064434
49. Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. (2013) 190:6542–9. doi: 10.4049/jimmunol.1202496
50. Ti D, Hao H, Tong C, Liu J, Dong L, Zheng J, et al. LPS-preconditioned mesenchymal stromal cells modify macrophage polarization for resolution of chronic inflammation via exosome-shuttled let-7b. J Transl Med. (2015) 13:308. doi: 10.1186/s12967-015-0642-6
51. Toivola DM, Boor P, Alam C, Strnad P. Keratins in health and disease. Curr Opin Cell Biol. (2015) 32:73–81. doi: 10.1016/j.ceb.2014.12.008
52. Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer. (2010) 17:R245–62. doi: 10.1677/ERC-10-0136
53. Chen H, Gao J, Xu Q, Wan D, Zhai W, Deng L, et al. MiR-145-5p modulates lipid metabolism and M2 macrophage polarization by targeting PAK7 and regulating β-catenin signaling in hyperlipidemia. Can J Physiol Pharmacol. (2021) 99:857–63. doi: 10.1139/cjpp-2020-0539
54. Zhong W, Yan J, Wang Z, Liang Y. [CD137 signaling regulates the expression of nuclear factor of activated T cells c1 through miR-145a-5p in ApoE(-)/(-) mice]. Zhonghua Xin Xue Guan Bing Za Zhi. (2015) 43:887–93. doi: 10.3760/cma.j.issn.0253-3758.2015.10.010
55. Guo J, Duan Z, Zhang C, Wang W, He H, Liu Y, et al. Mouse 4T1 breast cancer cell-derived exosomes induce proinflammatory cytokine production in macrophages via miR-183. J Immunol. (2020) 205:2916–25. doi: 10.4049/jimmunol.1901104
56. Wang S, Tan Y, Yang T, Liu C, Li R. Pulmonary AngII promotes LPS-induced lung inflammation by regulating microRNA-143. Cytotechnology. (2021) 73:1–10. doi: 10.1007/s10616-021-00493-0
57. Law YY, Lee WF, Hsu CJ, Lin YY, Tsai CH, Huang CC, et al. miR-let-7c-5p and miR-149-5p inhibit proinflammatory cytokine production in osteoarthritis and rheumatoid arthritis synovial fibroblasts. Aging. (2021) 13:17227–36. doi: 10.18632/aging.203201
58. Shi R, Liu DD, Cao Y, Xue YS. microRNA-26a-5p prevents retinal neuronal cell death in diabetic mice by targeting PTEN. Curr Eye Res. (2021):1–9. doi: 10.1080/02713683.2021.1975760
59. Hu M, Wei J, Yang L, Xu J, He Z, Li H, et al. Linc-KIAA1737-2 promoted LPS-induced HK-2 cell apoptosis by regulating miR-27a-3p/TLR4/NF-κB axis. J Bioenerg Biomembr. (2021) 53:393–403. doi: 10.1007/s10863-021-09897-1
60. Chen L, Lu Q, Chen J, Feng R, Yang C. Upregulating miR-27a-3p inhibits cell proliferation and inflammation of rheumatoid arthritis synovial fibroblasts through targeting toll-like receptor 5. Exp Ther Med. (2021) 22:1227. doi: 10.3892/etm.2021.10661
61. Raga-Cervera J, Bolarin JM, Millan JM, Garcia-Medina JJ, Pedrola L, Abellán-Abenza J, et al. miRNAs and genes involved in the interplay between ocular hypertension and primary open-angle glaucoma. Oxidative stress, inflammation, and apoptosis networks. J Clin Med. (2021) 10:2227. doi: 10.3390/jcm10112227
62. Chen Y, Yin Y, Jiang H. miR-30e-5p alleviates inflammation and cardiac dysfunction after myocardial infarction through targeting PTEN. Inflammation. (2021) 44:769–79. doi: 10.1007/s10753-020-01376-w
63. Chen B, Han J, Chen S, Xie R, Yang J, Zhou T, et al. MicroLet-7b regulates neutrophil function and dampens neutrophilic inflammation by suppressing the canonical TLR4/NF-κB pathway. Front Immunol. (2021) 12:653344. doi: 10.3389/fimmu.2021.653344
Keywords: mmu-miR-145a-5p, diabetes, wound healing, macrophage, inflammation
Citation: Hao Y, Yang L, Liu Y, Ye Y, Wang J, Yu C, Yan H, Xing Y, Jia Z, Hu C, Zuo H and Li Y (2021) mmu-miR-145a-5p Accelerates Diabetic Wound Healing by Promoting Macrophage Polarization Toward the M2 Phenotype. Front. Med. 8:775523. doi: 10.3389/fmed.2021.775523
Received: 14 September 2021; Accepted: 22 November 2021;
Published: 21 December 2021.
Edited by:
Ralf J. Ludwig, University of Lübeck, GermanyReviewed by:
Christoph G. S. Nabzdyk, Mayo Clinic, United StatesGiuseppe Cappellano, University of Eastern Piedmont, Italy
Copyright © 2021 Hao, Yang, Liu, Ye, Wang, Yu, Yan, Xing, Jia, Hu, Zuo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyan Zuo, enVvaHkyMDA1QDEyNi5jb20=; Yang Li, bGVleW91bmcxMDlAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Yanhui Hao
Yanhui Hao Leilei Yang1†
Leilei Yang1† Hua Yan
Hua Yan Yuan Xing
Yuan Xing Hongyan Zuo
Hongyan Zuo Yang Li
Yang Li