
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Med. , 24 January 2022
Sec. Ophthalmology
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.770138
This article is part of the Research Topic Reviews in: Ophthalmology 2022 View all 22 articles
Keratoconus (KC) is an etiologically heterogeneous corneal ectatic disorder. To systematically display the pathogenesis of keratoconus (KC), this study reviewed all the reported genes involved in KC, and performed an enrichment analysis of genes identified at the genome, transcription, and protein levels respectively. Combined analysis of multi-level results revealed their shared genes, gene ontology (GO), and pathway terms, to explore the possible pathogenesis of KC. After an initial search, 80 candidate genes, 2,933 transcriptional differential genes, and 947 differential proteins were collected. The candidate genes were significantly enriched in extracellular matrix (ECM) related terms, Wnt signaling pathway and cytokine activities. The enriched GO/pathway terms of transcription and protein levels highlight the importance of ECM, cell adhesion, and inflammatory once again. Combined analysis of multi-levels identified 13 genes, 43 GOs, and 12 pathways. The pathogenic relationships among these overlapping factors maybe as follows. The gene mutations/variants caused insufficient protein dosage or abnormal function, together with environmental stimulation, leading to the related functions and pathways changes in the corneal cells. These included response to the glucocorticoid and reactive oxygen species; regulation of various signaling (P13K-AKT, MAPK and NF-kappaB), apoptosis and aging; upregulation of cytokines and collagen-related enzymes; and downregulation of collagen and other ECM-related proteins. These undoubtedly lead to a reduction of extracellular components and induction of cell apoptosis, resulting in the loosening and thinning of corneal tissue structure. This study, in addition to providing information about the genes involved, also provides an integrated insight into the gene-based etiology and pathogenesis of KC.
Keratoconus (KC) is a complex multifactor degenerative disorder of the cornea, characterized by corneal ectasia, thinning, and cone-shaped protrusion, leading to reduced vision, irregular astigmatism, and corneal scarring (1–4). The worldwide prevalence of KC is approximately 1:2000 (4). KC usually manifests during puberty, and the clinical manifestation vary depending on disease severity (3). Myopia and astigmatism in one or both eyes were the main symptoms in the early stage. With disease progression, visual acuity of patients is progressive loss, and cannot be corrected with spectacles. KC at completion stage often has typical clinical sign, including Munson sign, a V-shape deformation of the lower eyelid in downward position; Fleischer ring, a hemosiderin arc or circle line around the cone base; Vogt's striae, fine vertical lines produced by Descemet's membrane compression (3, 4). In addition, the central or lower temporal part of the cornea shows obvious conical protrusion, and the central cornea becomes thinner obviously. In the completion stage, KC spontaneously or due to external forces such as eye rubbing, rupture of the posterior elastic layer of the cornea occurs, resulting in acute corneal edema and significant decline in visual acuity (3, 4). Because of the unclear pathogenesis and limited availability of medical treatments, KC has become a significant clinical problem worldwide and a leading indication for corneal transplantation (5).
Probing KC's etiology and pathogenesis and adopting effective control methods are the fundamentals of prevention and treatment of KC. KC has a clear genetic tendency. Genetic factors are involved in the development of KC (2, 6, 7). Until now, more than 70 candidate genes and regions have been screened and identified by genome-wide linkage analysis, whole-exome sequencing (WES), candidate gene sequencing, genome-wide association study (GWAS), or candidate gene association study (7–90). Due to the genetic heterogeneity and population differences among KC patients, the genetic cause of most cases has not been effectively identified, and the pathogenesis underlying the genetic mutation remains unclear. This represents the current bottleneck in KC etiology research, so it is very important to find a breakthrough point to explore the key related genes, and the common pathogenesis, of KC.
Traditional genetic studies have typically focused on high-quality families to map and identify new disease-causing genes or screen susceptible sites through population association analysis. However, the pathogenesis caused by mutations or susceptible sites is obscure, leading to the slow progress of pathogenesis studies of most genetic diseases, especially complex ones (91). The combined analysis of multi-levels can achieve the display of the candidate genes screened at the DNA level at the transcription level, the analysis of mutations in the genome at the RNA level for significantly differentially expressed genes, and the enrichment analysis of key genes in the pathway. Integrated genomics, transcription, and protein data can be leveraged to systematically analyze multiple consecutive events occurring in diseases, including changes in expression levels caused by gene mutations, and various forms of heterogeneity in transcriptional regulation, translation, and post-translational regulation body and feedback regulation. According to the changes in candidate factors at different levels, the candidate pathogenic factors can be thoroughly explored and the target of pathogenicity can be identified. Multiomics analysis can also be used to build a gene regulatory network in order to clarify the regulatory and causal relationships between various molecules, so as to gain a deeper understanding of the molecular mechanism and genetic basis of complex traits in genetic diseases.
In this analytical study, we examined all the reported genes involved in KC and performed an enrichment analysis of genes identified at the genome, transcription, and protein levels, respectively. In addition, by using gene set enrichment analysis, we attempted to explore the important mechanisms at different levels. Combined analysis of multi-level results revealed their shared genes, GOs, and pathways, allowing us to explore the possible pathogenesis of KC. The results of this study, in addition to providing information about the changed genes involved in the disease, provide an integrated insight into the common pathogenesis of KC.
To find genes associated with KC, the literature was reviewed and data were collected manually. All the studies describing genome changes (including pathogenic mutations or susceptible variants) and differentially expressed encoding genes at the transcription and protein levels were scrutinized using the following keywords in the PubMed and Web of Science databases: “keratoconus” AND (“gene” OR “expression” OR “transcriptome”). We limited our search to articles published up to search date that were written in English. The search was conducted in November 2020. Then, each article was read and classified carefully. For the genome level, only those studies on patients with keratoconus were collected, excluding those on other syndromes patients with keratoconus, central corneal thickness and corneal curvature of normal person unless their results were confirmed in keratoconus samples. For the transcription and protein levels, only those human studies that directly used in situ KC corneal tissue or primary KC corneal cells were selected. Finally, we identified and recorded all the reported genes with pathogenic mutations or susceptible variants, differentially expressed genes at the transcription level, and differentially expressed or abnormally distributed genes at the protein level. This study did not search other databases, such as clinical trials and so. The relevant genes collected just included the genes published in the PubMed and Web of Science databases.
Enrichment analysis is a statistics-based method for classifying genes that are overrepresented in a specific set of genes. All the genes associated with KC were classified into three groups according to changed levels, and gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed via the online Database for Annotation, Visualization, and Integrated Discovery (DAVID) software, version 6.8 (92, 93).
Combined analysis of multi-level enrichment revealed their shared GOs and pathways, allowing us to explore the possible pathogenesis of KC. To further identify the putative pathogenicity of gene mutations/susceptible variants and to detect the KC-related gene function and pathway changes, we conducted a combined analysis of the multi-level results. Online tools (http://bioinformatics.psb.ugent.be/webtools/Venn/) were used to calculate and draw custom Venn diagrams depict the genes, GOs, and KEGG pathways shared by the multi-level results.
After an extensive review of resources, more than 200 studies were selected from the published articles and were reviewed in greater detail. Our study confirmed the 80 candidate genes, 2,933 differentially expressed genes at the transcription level, and 947 differentially expressed genes at the protein level.
KC has a clear genetic tendency (2, 6, 7). In the past 20 years, scholars have extensively investigated the genetic cause of KC (7–90). Thus far, more than 40 candidate genes and regions have been located and screened via genome-wide linkage analysis, WES, or candidate gene sequencing. In addition, candidate gene association studies and GWAS have been carried out successively. Thus far, research has found that SNP (Single Nucleotide Polymorphism) loci of 30 genes are related to KC, which can increase or decrease the risk of KC. After an initial search, 80 candidate genes were collected for further analysis. As the results show, a few genes were identified by more than one type of analysis, which represents stronger evidence of their involvement in KC (Figure 1A). The top seven genes were COL5A1, MIR184, LOX, ZNF469, VSX1, COL4A3 and COL4A4 (Figure 1A). Detailed literature informations of reported KC associated genes at the genomic level were shown in Supplementary Table S1.
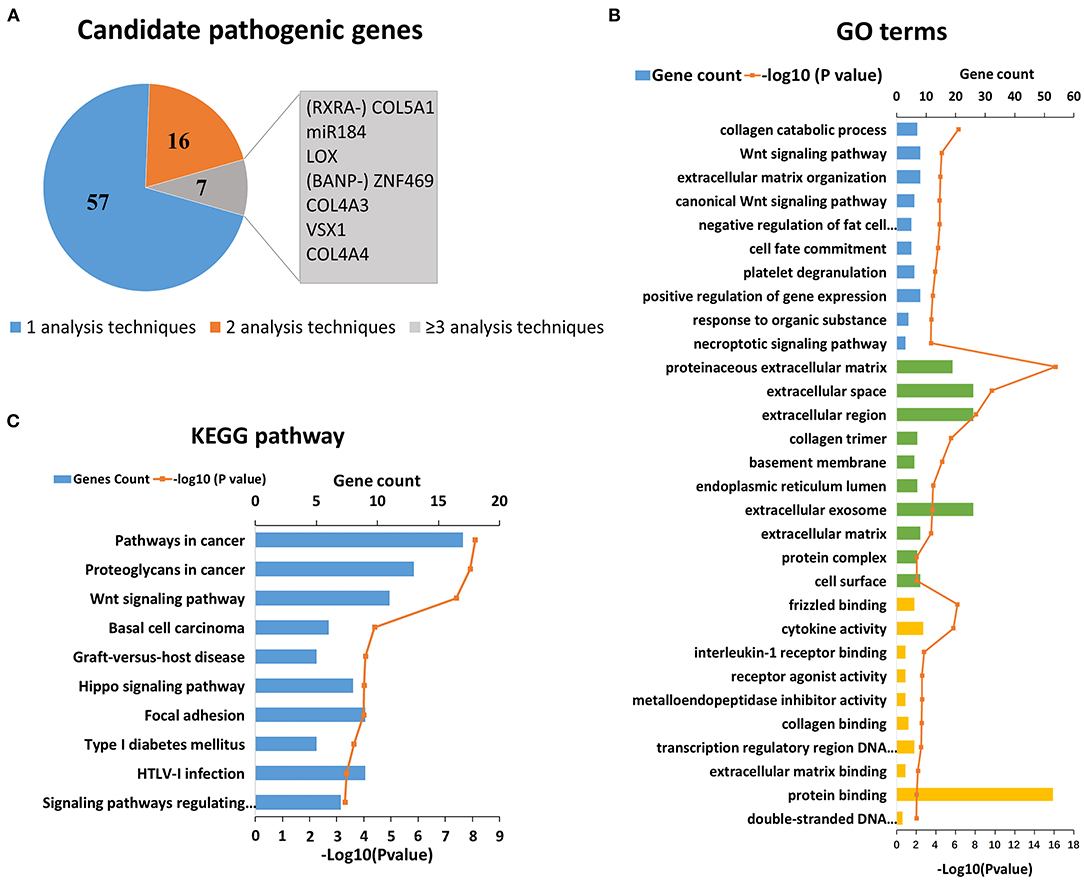
Figure 1. Reported candidate genes in keratoconus at the DNA level. (A) The associated genes identified by different analysis techniques. (B) Top ten enriched terms of each GO category at the DNA level. The three colors represent biological process (blue), cell component (green), and molecular function (yellow), respectively. (C) Top ten enriched KEGG pathways at the DNA level.
On the basis of the DAVID results, we tested whether the genes associated with KC clustered into certain GO terms and KEGG pathways. Figures 1B,C shows the significant GO terms and KEGG pathways with a p < 0.05. We found that the top-ranking GO terms were proteinaceous extracellular matrix (ECM) (GO: 0005578, p = 6.00 × 10−17), extracellular space (GO: 0005615, p = 2.10 × 10−10) and their related pathways in the cell component category, including extracellular region, collagen timer, basement membrane, extracellular exosome, as well as ECM (Figure 1B). The top five enriched biological processes were collagen catabolic process (GO: 0010033, p = 5.00 × 10−7), Wnt signaling pathway (GO: 0009611, p = 2.60 × 10−5), ECM organization (GO: 0033554, p = 3.50 × 10−5), canonical Wnt signaling pathway (GO: 0006916, p = 4.20 × 10−5), and negative regulation of fat cell differentiation (GO: 0010941, p = 4.20 × 10−5). The top molecular function terms most were involved in or related to various binding (frizzled binding, interleukin-1 receptor binding, collagen binding, transcription regulatory region DNA binding and ECM binding etc.), and activities of cytokine, receptor agonist, and metalloendopeptidase inhibitor.
The top-ranking KEGG pathways were associated with certain complex diseases, including cancer, type 1 diabetes mellitus, and graft-vs.-host disease (Figure 1C). These results suggested that KC might overlap with some of the pathogenesis of these diseases. The others top-ranking KEGG pathways were Wnt signaling pathway, Hippo signaling pathway, and Focal adhesion, which suggest that these pathways may play a role in the pathogenicity of KC.
Detailed literature information of differentially expressed encoding genes were shown in Supplementary Table S2 (49, 90, 94–136). In addition to many studies that focused on particular genes or gene families, other studies investigated the transcriptome. Quantitative real-time PCR (qRT-PCR), RNA-seq and microarray were the main methods used in the differential expression studies (49, 90, 94–136). All the significantly differentially expressed encoding genes were collected regardless of whether the different corneal tissue types including corneal tissue, corneal epithelia, corneal stroma and primary stromal fibroblast.
After an initial search, a total of 2,933 reported differentially expressed encoding genes between KC corneas and normal corneas at the transcription level were collected (Figure 2A). There are 1,948 downregulated genes, 802 upregulated genes, and 183 genes with opposite results in different studies (Supplementary Table S2). About two-thirds of differentially expressed encoding genes were reported only once in single studies and need to be verified by further research. The remaining 933 genes were reported more than twice in the same or different corneal tissue types (Figure 2A). Among them, 13 genes, including GPNMB, TIMP3, CA12, CTGF, ID3, IGFBP3, JUN, PRELP, RGS5, SFRP1, SOD2, TIMP1 and VEGFA were reported in five or more studies (Figure 2A). All of these genes were reported significantly down regulated in KC corneal tissues, except for a few opposite results in individual studies. The changes in these genes indicate the decrease of growth factor (CTGF, IGFBP3 and VEGFA), transcription factor (ID3 and JUN), superoxide dismutase (SOD2), and metalloproteinase inhibitor (TIMP3 and TIMP1) in KC corneas. These results highlight the importance of these changes in the pathogenicity of KC.
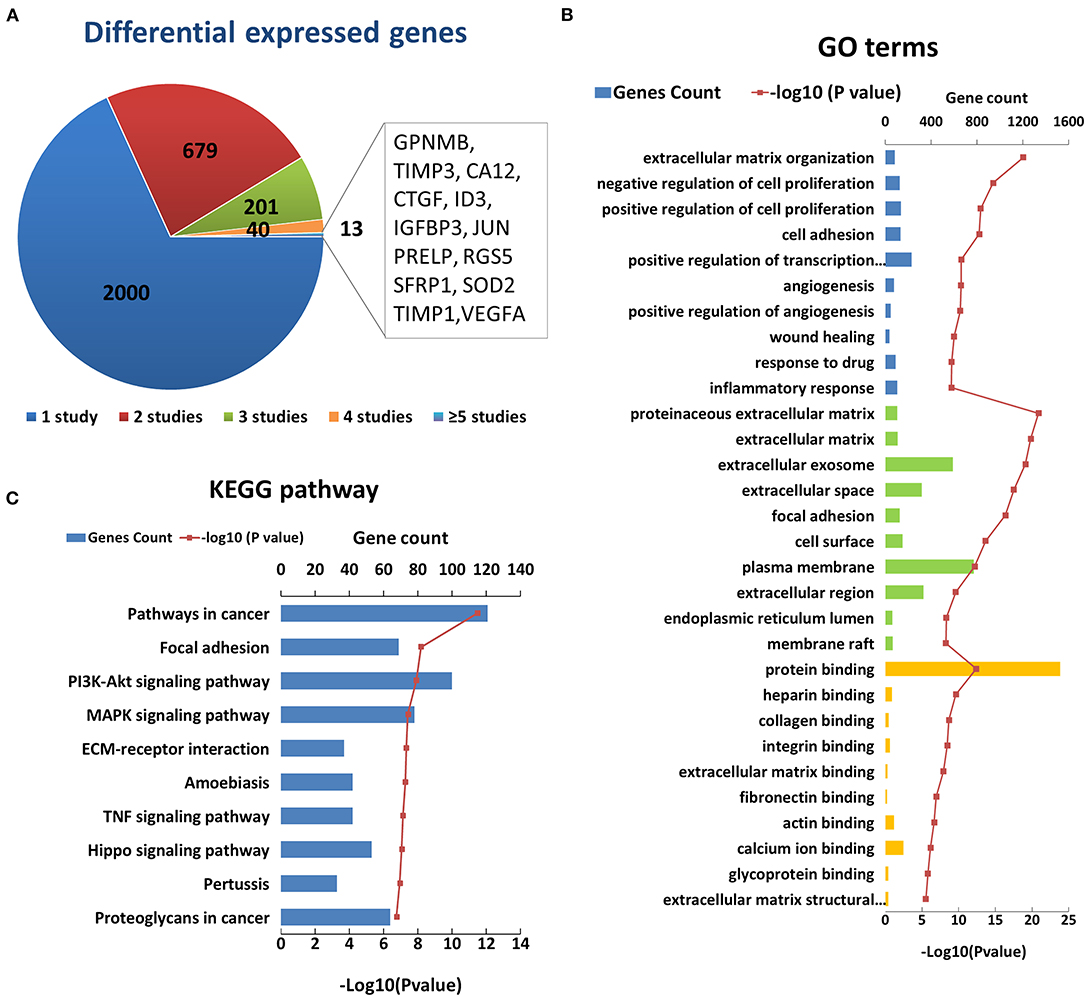
Figure 2. Enriched GO terms and KEGG pathways based on 2933 differential genes at the transcription level. (A) The classification of the 2933 differential genes according to the number and results of related studies. (B) Top 10 enriched terms of each GO category at the transcription level. The three colors represent biological process (blue), cell component (green), and molecular function (yellow), respectively. (C) Top ten enriched KEGG pathways at the transcription level.
The GO analysis of differentially expressed genes showed that there were 497 biological process, 84 cell component, and 127 molecular factor terms with a p < 0.05. The top ten significantly enriched biological process terms were involved in or related to ECM organization, regulation of cell proliferation, cell adhesion, angiogenesis, wound healing, response to drug and inflammatory (Figure 2B). The top ten significant cell component terms were involved in or related to proteinaceous ECM, ECM, extracellular exosome, extracellular space, focal adhesion, cell surface, plasma membrane, extracellular region, endoplasmic reticulum lumen, and membrane raft (Figure 2B). The top ten significant enriched molecular function terms were associated with various bindings, such as protein, heparin, collagen, integrin, extracellular matrix, fibronectin, actin, calcium ion, and glycoprotein bindings. These results further highlight the importance of extracellular region and ECM related proteins in the pathogenicity of KC. In addition, cell proliferation, cell adhesion angiogenesis and response to drug and inflammatory may be involved in the process of KC, which requires further study. The KEGG pathway analysis identified 86 enriched pathways. The top five significantly enriched KEGG pathways were pathways in cancer (hsa05200, p = 3.20 × 10−12), focal adhesion (hsa04510, p = 6.50 × 10−9), PI3K-Akt signaling pathway (hsa 04210, p = 1.20 × 10−8), MAPK signaling pathway (hsa04010, p = 3.7 × 10−8), and ECM-receptor interaction (hsa04512, p = 4.60 × 10−8) (Figure 2C). These results suggest that these pathways may play a role in the pathogenicity of KC.
After an initial search, a total of 946 reported differentially expressed proteins between KC corneas and normal corneas were collected (98–103, 109, 114, 117, 118, 120, 125, 126, 128, 130, 131, 133, 137–193). Immunohistochemistry, immunofluorescence, and western blot were used for candidate protein level detection, and proteomic analysis was performed via mass spectrometry. All the reported differentially expressed proteins were collected, including downregulated, upregulated, and abnormally distributed proteins. There are 427 downregulated genes, 398 upregulated genes, 14 abnormally distributed proteins, and 107 genes with opposite results in different studies (Supplementary Table S3). About half differentially expressed genes were reported only once. The remaining 466 genes were reported more than twice in the same or different corneal tissue types (Figure 3A). Among them, 30 genes were reported in four or more times, including five genes (HSPB1, VIM, HMGA1, TSTD1, RUVBL2) with upregulation and two genes (BZW1, LOX) with downregulation in at least four studies. The changes in these genes indicate the decrease of transcription factor (BZW1), lysyl oxidase (LOX), and increase of small heat shock protein (HSPB1), vimentins (VIM), thiosulfate sulfurtransferase (TSTD1) and ATPase (RUVBL2) in KC corneas. These results highlight the importance of these changes in the pathogenicity of KC. The other genes with conflicting results suggest that the pathogenesis of KC is very complex, and further research is needed to clarify the role of these genes.
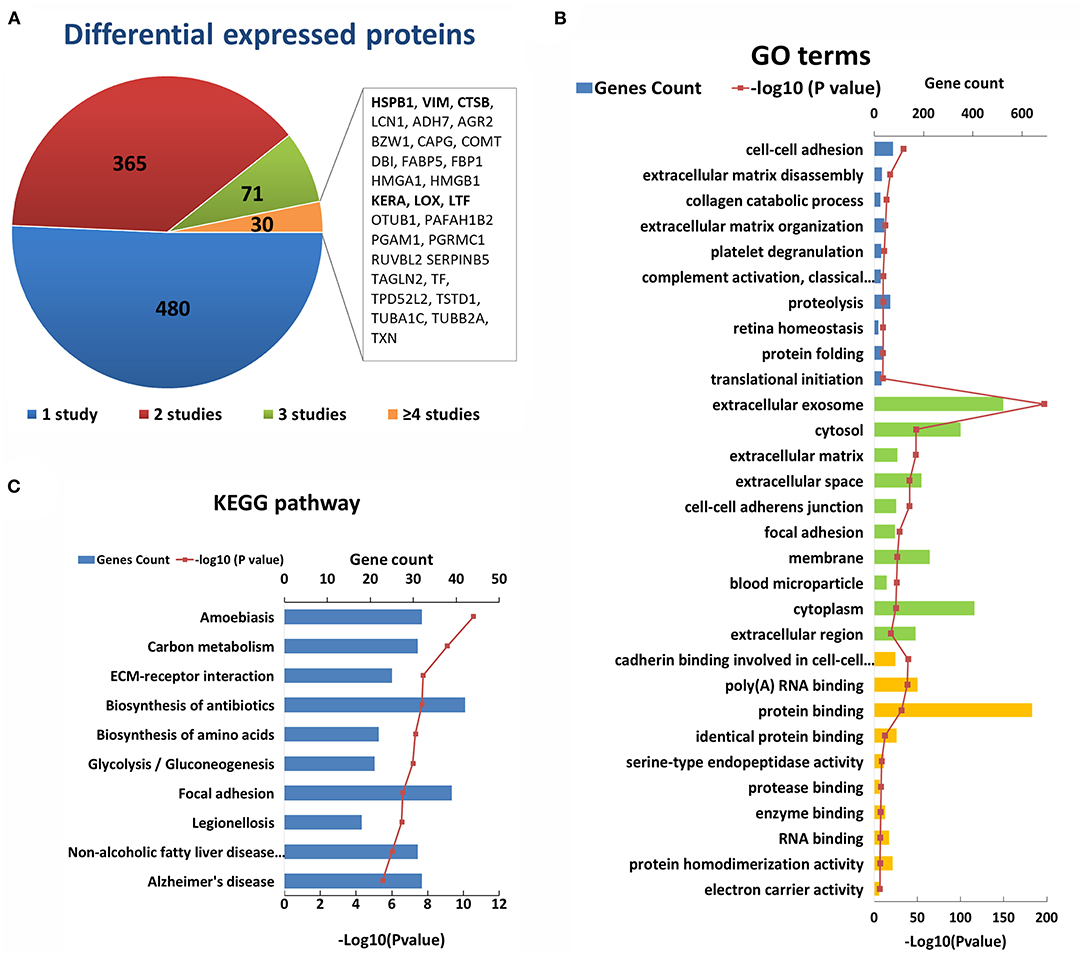
Figure 3. Enriched GO terms and KEGG pathways based on 947 differential genes at the protein level. (A) The classification of the 947 differential proteins according to the number and results of related studies. (B) Top 10 enriched terms of each GO category at the protein level. The three colors represent biological process (blue), cell component (green), and molecular function (yellow), respectively. (C) Top ten enriched KEGG pathways at the protein level.
The GO analysis of differentially expressed proteins showed that there were 344 biological process, 127 cell component, and 98 molecular factor terms with a p < 0.05. The top ten significant enriched biological process terms were involved in or related to ECM disassembly, ECM organization, collagen catabolic process, proteolysis, cell-cell adhesion, platelet degranulation, complement activation, retina homeostasis, protein folding and translational initiation (Figure 3B). The top ten significant cell component terms were involved in or related to extracellular exosome, ECM, extracellular space, extracellular region, cell-cell adherens junction, focal adhesion, cytosol, membrane, cytoplasm, and blood microparticle (Figure 3B). The top 10 significant enriched molecular function terms were associated various bindings (cadherin, RNA, protein, identical protein, protease and enzyme bindings), endopeptidase activity, protein homodimerization activity and electron carrier activity (Figure 3B). These results once again highlight the importance of collagen and ECM in the pathogenicity of KC at the protein level. Except for ECM and its related GO terms, the results showed that proteolysis, cell-cell adhesion, focal adhesion, protein binding, and endopeptidase may be involved in the process of KC (Figure 3B). The top five significant enriched KEGG pathways were Amoebiasis, Carbon metabolism, ECM-receptor interaction, Biosynthesis of antibiotics and Biosynthesis of amino acids (Figure 3C). These results provided further evidence of the important role of ECM pathways in the pathogenicity of KC at the protein level.
To further identify the putative pathogenicity of gene variants and detect the KC-related gene function and pathway changes, we conducted a combined analysis of DNA, RNA, and protein level results. First, we analyzed the genes shared between the different levels. The results showed that there were 13 overlapping genes between all three levels (Figure 4A, Table 1). They were COL4A3, COL6A2, MMP9, TIMP1, LOX, TGFBI, TNF, IL1A, IL1RN, SOD1, CAT, VSX1 and TF. All the genes except TGFBI and SOD1 were reported to have KC-susceptibility SNPs. Potential pathogenic mutations of seven overlapping genes (LOX, IL1RN, COL4A3, VSX1, TGFBI, SOD1 and COL6A2) were identified in KC patients. In addition, LOX and IL1RN were located in the susceptible loci detected by linkage analysis. Five of the 13 genes (LOX, IL1RN, VSX1, COL4A3, and TGFBI) were identified by multiple types of analysis at the DNA level. Three of the 13 genes (LOX, TIMP1, and TNF) were verified in multiple studies with consistent expression changes at the transcription level. Five of the 13 proteins (LOX, IL1RN, COL6A2, MMP9, and TNF) were verified in multiple studies with consistent changes at the protein level. These results suggested that these overlapping genes might be key genes of KC and might play an important role in the pathogenesis of KC. As the results show (Table 1), five genes (TNF, MMP9, IL1A, CAT, and VSX1) had significant upregulation, and six genes (TIMP1, COL6A2, SOD1, TGFBI, COL4A3, and LOX) had significant downregulation in KC at both the transcription and protein levels. The coincident changes of these genes also indicated the decrease of collagen (COL4A3 and COL6A2), metallopeptidase inhibitor (TIMP1), lysyl oxidase (LOX), superoxide dismutase (SOD1), and the increase of metallopeptidase (MMP9), antioxidant enzyme (CAT) and inflammatory cytokines (TNF and IL1A). These results highlight the importance of these changes in the pathogenicity of KC.
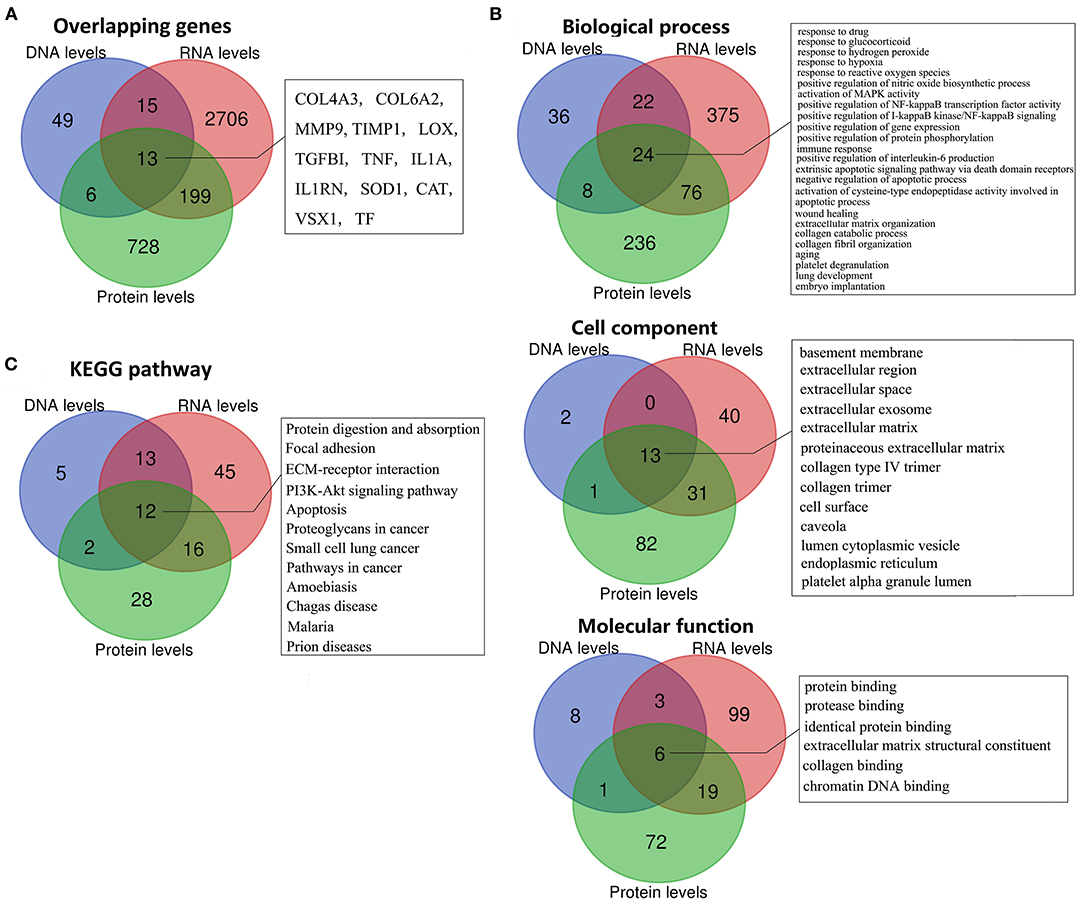
Figure 4. Overlapping genes, GO terms, and KEGG pathways between multi-levels. (A) Overlapping genes; (B) Overlapping GO terms; (C) Overlapping KEGG pathways.
Combined analysis of different levels' enrichment revealed their shared significant GOs and pathways, allowing the researchers to explore the possible pathogenesis of KC. The overlapping significant GOs between all three levels, including 24 biological process, 13 cell component, and six molecular function terms, are shown in Figure 4B. The shared biological process terms can divided into four groups, including various response-related GOs (response to drug, glucocorticoid, hydrogen peroxide, reactive oxygen species, and hypoxia), apoptosis-related GOs (activation of cysteine-type endopeptidase activity involved in apoptotic process, negative regulation of apoptotic process, and extrinsic apoptotic signaling pathway via death domain receptors), ECM-related GOs (ECM organization, collagen catabolic process, and collagen fibril organization) and activation and positive regulation of various signaling (MAPK activity, NF-kappaB activity, and I-kappaB kinase/NF-kappaB signaling). For the cell components, most of the overlapping GOs were ECM related (including basement membrane, ECM, proteinaceous ECM, collagen type IV trimer, and collagen trimer), extracellular related (extracellular region, extracellular space and extracellular exosome) and pinocytosis related (caveola and cytoplasmic vesicle). For the molecular functions, the overlapping GOs were ECM related (ECM structural constituent and collagen binding), and various binding-related GOs (protein, identical protein, protease and chromatin DNA binding). The combined analysis of the KEGG pathway showed that Protein digestion and absorption, Focal adhesion, ECM-receptor interaction, PI3K-Akt signaling pathway, apoptosis, and various diseases related pathways (including cancer, Prion diseases, Malaria, Amoebiasis and Chagas disease) were significantly enriched at DNA, RNA, and protein levels (Figure 4C). Most GO and pathway enrichments shared at DNA, RNA, and protein levels were related to collagen, ECM, extracellular, various responses and apoptosis, suggesting that these GO and pathway changes might have been etiological—serving as mechanisms of KC.
KC is an etiologically heterogeneous corneal ectatic disorder, and both environmental and genetic factors play a role in its etiopathogenesis (194). Based on results from studies that have investigated the genetic etiology, expression, and translation changes in the process of development, it is becoming increasingly clear that KC is a complex disease with a complex etiology or convergence of multiple disease pathways. However, the common pathogenesis underlying the different etiologies remains unclear. In this study, we reviewed all the studies of KC-related genes identified at the genome, transcription, and protein levels. Through multi-level related gene enrichment-based review, we systematically explored the schematic representing factors responsible for KC at different levels. The results of this study, in addition to providing information about the genes involved in the disease, clearly provide an integrated insight into the gene-based etiology and pathogenesis of KC.
Genetic changes play an important role in the etiopathogenesis of KC (2, 6, 7, 194). Many forms of gene variation, such as inheritance gene mutation, de novo mutation, and polymorphism, have been reported to be involved in the etiology of KC (7–90). More than half of the genes were reported in one type of study or in single studies. A few genes, encoded chains of collagens (COL5A1, COL4A3, and COL4A4) (57, 59, 61, 67, 72, 76, 78, 84, 87, 88, 90), collagen cross-linking enzyme (LOX) (18, 30, 90), factor for the synthesis or organization of collagen fibers (ZNF469) (27, 32, 34, 46, 47, 62, 65), and others (MIR184 and VSX1) (6, 7, 16, 19, 24, 40, 56, 82, 85) were identified in different types of studies, such as pathogenic mutation analysis, polymorphism association analysis, and family-based linkage analysis. However, the occurrence rate of these gene mutations in the population was relatively low, and in many populations, it could not even be verified (25, 195–202), which suggested that KC is genetically heterogeneous. Among the reported KC associated genetic changes, there were 11 genes responsible for apoptosis related process (FAS, FASLG, TNF, BIRC2, SMAD3, WNT5A, CAT, TIMP1, MMP9, FOXO1, and COL4A3), 14 reported corneal biomechanics loci (MPDZ, COL6A2, MYOF, LOX, ZNF469, SMAD3, NFIB, FNDC3B, COL5A1, WNT10A, TGFBI, SLC4A11, FOXO1 and COL4A3) (203), and five genes responsible for inflammatory processes (IL1A, IL1B, FAS, TNF and IL17B). Genetic changes in these genes might lead to the changes in related functions and pathways in the corneal cells, then lead to induction of apoptosis, inflammatory and altered biomechanics of cornea, which have been reported involved in the etiopathogenesis of KC (204–206), and lead to the occurrence of KC. The top candidate gene based enrichment, including ECM and their related pathways (76, 90), Wnt signaling pathway (60), and cytokine activities (204), have been reported involved in the etiopathogenesis of KC. The role of negative regulation of fat cell differentiation in the etiopathogenesis of KC has not been reported, but body mass index was reported associated with keratoconus before (207). So, more studies are needed on the relationship between this functional change in order to clarify its relationship and mechanism of action in KC.
Differences between the expression of genes in normal and KC corneas suggested disease pathways. The top 13 verified differentially expressed genes at RNA level indicated a decrease of growth factor, transcription factor, superoxide dismutase, and metalloproteinase inhibitor, which highlights the importance of these changes in the pathogenicity of KC. The top differentially expressed genes based enrichment were similar to the GOs at the genomic level, mainly including ECM and its related GO terms, response to inflammatory and various bindings. These results represent further confirmation of the importance of collagen (61, 67, 75, 78, 84, 87, 88, 113, 117, 125, 163–165), ECM (76, 90, 97, 158), cell adhesion (72, 97), and inflammatory (49, 101, 124, 128, 139, 204) in the pathogenicity of KC. In addition, cell proliferation, angiogenesis, and response to drug have not been reported before, which may be involved in the process of KC, and should be investigated further. For reported differentially expressed proteins, the genes with upregulation or downregulation in at least four studies highlight the importance of transcription factor (98, 106, 112, 128, 145, 160), lysyl oxidase (18, 30, 81, 125, 150, 151), small heat shock protein (156, 188, 193), vimentins (126, 156, 188, 193), thiosulfate sulfurtransferase and ATPase (188) in KC corneas. Gene-based enrichment analysis showed that the differentially expressed proteins were significantly enriched in ECM and its related GO terms, proteolysis, and various bindings. These results once again highlight the importance of these GO terms in the pathogenicity of KC at the protein level.
Different approaches have been used to investigate and define the phenotype, mechanisms, and causes of KC. Thousands of genes were identified at genomic, transcription, and protein levels. Observations of corneal changes that occur in KC often do not distinguish between primary changes and secondary inflammatory or degenerative effects. Although research has identified many differences that distinguish KC corneas from normal corneas, it has not been possible to trace these changes back to primary causes or to identify the triggers that precipitate the cascade of events that leads to the clinical picture of KC. The results at different levels were clearly similar. In order to explore the key points, combined analysis of multi-levels was performed. Integrated genomics, transcription, and protein data can be leveraged to systematically analyze multiple consecutive events occurring in diseases. According to the changes in candidate factors at different levels, the candidate pathogenic factors can be thoroughly explored and the target of pathogenicity can be identified. The consistent changes in these factors at different levels suggested that these factors play an important role in the pathogenesis of KC. The DNA, RNA, and protein changes represented the cause and process changes of KC, respectively.
Based on the results of multi-level combined analysis, we hypothesized that the pathogenic relationships among these related factors is as follows. The etiology of KC can be divided into environmental and genetic factors. The environmental factors may include endogenous and exogenous factors, such as glucocorticoid, hydrogen peroxide, and reactive oxygen species (ROS) (120). The gene mutations or variants involved in collagen (57, 61, 64, 67, 75, 76, 78, 88), metallopeptidase inhibitor (75, 81, 109, 157), lysyl oxidase (18, 30, 81, 90), metallopeptidase (64), antioxidant enzyme (16, 25, 81, 199), inflammatory cytokines (11, 45, 83, 201), and others cause insufficient protein dosage or abnormal function. These genetic changes, together with the aforementioned stimulation, lead to the changes in related functions and pathways in the corneal cells. The related functions include the response to the stimulation of hormones and reactive oxygen species (96, 106, 120, 121, 189), activation and positive regulation of various signaling (MAPK activity, NF-kappaB activity, and I-kappaB kinase/NF-kappaB signaling) (123, 128), upregulation of cytokines and collagen-related enzymes (49, 101, 122, 124, 147, 174, 187), and downregulation of collagen, collagen-crossing, and other ECM-related proteins (97, 103, 117, 163, 164), and regulation of apoptosis (36, 175, 186). These undoubtedly lead to the reduction of extracellular components and the induction of apoptosis and aging. The change in extracellular structure, decrease of extracellular composition, and apoptosis of corneal cells all lead to the loosening and thinning of corneal tissue structure, which leads to the occurrence of KC.
In addition to the different levels and combined analysis results of this paper, our hypothesis was supported by many other studies of molecular mechanisms and cell events of KC. A few hormones and substances have been reported to be associated with KC (208–214). However, the relationship between glucocorticoid and KC has not been studied before, and should be investigated further. Chronic inflammatory events were detected in the tears of KC patients (215–217). A significant increase in the apoptosis of KC cells has been reported in several studies (186, 218, 219). Moreover, a decrease of dulfated epitopes of keratan sulfate KC corneas was also reported (220). The electron microscope results of KC showed that the content of the stroma increases, whereas the fibril diameter is reduced, the mean diameter and interfibrillar spacing of collagen fibrils are reduced, and the collagen fibrils and proteoglycans number density and area fractions are significantly increased (138).
Our study has some limitations. All the results were obtained through multi-level related gene enrichment-based analysis. More studies are needed on the relationship between these functional changes in order to clarify their relationship and mechanism of action, which could provide a new direction for the treatment of KC. For the expression studies collected in this study, there were several different corneal tissue types. Most of the studies used the corneal tissue, and a few studies used the corneal epithelia, corneal stroma or primary stromal fibroblast (see Supplementary Tables S1–S3). In this analysis-based study, because of the limited space, instead of categorizing different genes detected in different tissues, we just conducted a unified analysis in different levels. Analysis that is more detailed needs to be carried out in the future to find the role of different corneal cells in KC pathogenesis. Furthermore, the interaction between genetic factors and environmental factors in the pathogenesis of KC has not been effectively solved, and further research is needed. Epigenetic mechanisms might help explain environmental contributions to the pathogenesis of KC (221). There are few studies on the relationship between epigenetic changes and KC (60). Recently, certain epigenetic changes, such as circle RNA, have been confirmed to play an important role in other diseases having overlapped pathogenesis pathway with KC (222–225), suggesting its potential role in KC pathogenesis. Study the role of these epigenetic changes might be a new research direction of KC in future.
Keratoconus is an etiologically heterogeneous corneal ectatic disorder, and both environmental and genetic factors play a role in its etiopathogenesis. Based on results from studies that have investigated the genetic etiology, expression, and translation changes in the process of development, it is becoming increasingly clear that KC is a complex disease with a complex etiology or convergence of multiple disease pathways. The common pathogenesis underlying the different etiologies remains unclear. In this study, we reviewed all the studies of KC-related genes identified at the genome, transcription, and protein levels. Through multi-level related gene enrichment-based review, we systematically explored the schematic representing factors responsible for KC at different levels. The results of this study, in addition to providing information about the genes involved in the disease, clearly provide an integrated insight into the gene-based etiology and pathogenesis of KC. Base on the results, we hypothesized that the pathogenic relationships among these related factors is as follows. The gene mutations/variants caused insufficient protein dosage or abnormal function, together with environmental stimulation, leading to the changes in the related functions and pathways in the corneal cells. These included response to the glucocorticoid and reactive oxygen species; regulation of various signaling (P13K-AKT, MAPK and NF-kappaB), apoptosis and aging; upregulation of cytokines and collagen-related enzymes; and downregulation of collagen and other ECM-related proteins. These undoubtedly lead to a reduction of extracellular components and induction of cell apoptosis, resulting in the loosening and thinning of corneal tissue structure. This hypothesis was supported by many other studies of molecular mechanisms and cell events of KC. More studies are needed on the relationship between these functional changes in order to clarify their relationship and mechanism of action, which could provide a new direction for the treatment of KC.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
X-DH and HG designed the research. W-HX, CS, YL, and Z-XZ performed the literature search. X-DH analyzed the data, participated in the discussion, and wrote and revised the paper. KW and P-FL participated in the revision of the paper.
This study was supported by National Natural Science Foundation of China (81500763 and 91849209), Shandong Provincial Natural Science Foundation, China (ZR2020MC059), China Postdoctoral Science Foundation (2019M652311), Special Support for Post-doc Creative Funding in Shandong Province, and Applied Research Program for Post-Doctoral in Qingdao.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are greatly indebted to authorship of involved reported literatures.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.770138/full#supplementary-material
Supplementary Table S1. Reported candidate genes in keratoconus at the DNA level.
Supplementary Table S2. The gene lists of reported differential expressed genes between KC and normal cornea at the transcription level.
Supplementary Table S3. The gene lists of reported differential expressed genes between KC and normal cornea at the protein level.
1. Rabinowitz YS. Keratoconus. Surv Ophthalmol. (1998) 42:297–319. doi: 10.1016/S0039-6257(97)00119-7
2. Mas Tur V, MacGregor C, Jayaswal R, O'Brart D, Maycock N. A review of keratoconus: Diagnosis, pathophysiology, and genetics. Surv Ophthalmol. (2017) 62:770–83. doi: 10.1016/j.survophthal.2017.06.009
3. Keratology Group OBoCA. Expert consensus on diagnosis and treatment of keratoconus in China (2019. Chinese Journal of Ophthalmology. (2019) 55:891–5.
4. Ferrari G, Rama P. The keratoconus enigma: A review with emphasis on pathogenesis. Ocul Surf. (2020) 18:363–73. doi: 10.1016/j.jtos.2020.03.006
5. Rahman I, Carley F, Hillarby C, Brahma A, Tullo AB. Penetrating keratoplasty: indications, outcomes, and complications. Eye (Lond). (2009) 23:1288–94. doi: 10.1038/eye.2008.305
6. Karolak JA, Gajecka M. Genomic strategies to understand causes of keratoconus. Mol Genet Genomics. (2017) 292:251–69. doi: 10.1007/s00438-016-1283-z
7. Valgaeren H, Koppen C, Van Camp G. A new perspective on the genetics of keratoconus: why have we not been more successful? Ophthalmic Genet. (2018) 39:158–74. doi: 10.1080/13816810.2017.1393831
8. Burdon KP, Vincent AL. Insights into keratoconus from a genetic perspective. Clin Exp Optom. (2013) 96:146–54. doi: 10.1111/cxo.12024
9. Nielsen K, Hjortdal J, Pihlmann M, Corydon TJ. Update on the keratoconus genetics. Acta Ophthalmol. (2013) 91:106–13. doi: 10.1111/j.1755-3768.2012.02400.x
10. Rabinowitz YS, Galvis V, Tello A, Rueda D, Garcia JD. Genetics vs chronic corneal mechanical trauma in the etiology of keratoconus. Exp Eye Res. (2021) 202:108328. doi: 10.1016/j.exer.2020.108328
11. Kim SH, Mok JW, Kim HS, Joo CK. Association of−31T>C and−511 C>T polymorphisms in the interleukin 1 beta (IL1B) promoter in Korean keratoconus patients. Mol Vis. (2008) 14:2109–16.
12. Guan T, Ma ZW, Ding SP. [Analyses of coding sequence point mutation and polymorphism of TGFBI gene in Chinese patients with keratoconus]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. (2011) 28:152–5. doi: 10.3760/cma.j.issn.1003-9406.2011.02.007
13. Pathak D, Nayak B, Singh M, Sharma N, Tandon R, Sinha R, et al. Mitochondrial complex 1 gene analysis in keratoconus. Mol Vis. (2011) 17:1514–25.
14. Li X, Bykhovskaya Y, Haritunians T, Siscovick D, Aldave A, Szczotka-Flynn L, et al. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum Mol Genet. (2012) 21:421–9. doi: 10.1093/hmg/ddr460
15. Czugala M, Karolak JA, Nowak DM, Polakowski P, Pitarque J, Molinari A, et al. Novel mutation and three other sequence variants segregating with phenotype at keratoconus 13q32 susceptibility locus. Eur J Hum Genet. (2012) 20:389–97. doi: 10.1038/ejhg.2011.203
16. Saee-Rad S, Hashemi H, Miraftab M, Noori-Daloii MR, Chaleshtori MH, Raoofian R, et al. Mutation analysis of VSX1 and SOD1 in Iranian patients with keratoconus. Mol Vis. (2011) 17:3128–36.
17. Guan T, Liu C, Ma Z, Ding S. The point mutation and polymorphism in keratoconus candidate gene TGFBI in Chinese population. Gene. (2012) 503:137–9. doi: 10.1016/j.gene.2012.04.061
18. Bykhovskaya Y, Li X, Epifantseva I, Haritunians T, Siscovick D, Aldave A, et al. Variation in the lysyl oxidase (LOX) gene is associated with keratoconus in family-based and case-control studies. Invest Ophthalmol Vis Sci. (2012) 53:4152–7. doi: 10.1167/iovs.11-9268
19. Wang Y, Jin T, Zhang X, Wei W, Cui Y, Geng T, et al. Common single nucleotide polymorphisms and keratoconus in the Han Chinese population. Ophthalmic Genet. (2013) 34:160–6. doi: 10.3109/13816810.2012.743569
20. Lu Y, Vitart V, Burdon KP, Khor CC, Bykhovskaya Y, Mirshahi A, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. (2013) 45:155–63.
21. Li X, Bykhovskaya Y, Tang YG, Picornell Y, Haritunians T, Aldave AJ, et al. An association between the calpastatin (CAST) gene and keratoconus. Cornea. (2013) 32:696–701. doi: 10.1097/ICO.0b013e3182821c1c
22. Mikami T, Meguro A, Teshigawara T, Takeuchi M, Uemoto R, Kawagoe T, et al. Interleukin 1 beta promoter polymorphism is associated with keratoconus in a Japanese population. Mol Vis. (2013) 19:845–51.
23. Bae HA, Mills RA, Lindsay RG, Phillips T, Coster DJ, Mitchell P, et al. Replication and meta-analysis of candidate loci identified variation at RAB3GAP1 associated with keratoconus. Invest Ophthalmol Vis Sci. (2013) 54:5132–5. doi: 10.1167/iovs.13-12377
24. Lechner J, Bae HA, Guduric-Fuchs J, Rice A, Govindarajan G, Siddiqui S, et al. Mutational analysis of MIR184 in sporadic keratoconus and myopia. Invest Ophthalmol Vis Sci. (2013) 54:5266–72. doi: 10.1167/iovs.13-12035
25. Moschos MM, Kokolakis N, Gazouli M, Chatziralli IP, Droutsas D, Anagnou NP, et al. Polymorphism Analysis of VSX1 and SOD1 Genes in Greek Patients with Keratoconus. Ophthalmic Genet. (2015) 36:213–7. doi: 10.3109/13816810.2013.843712
26. Synowiec E, Wojcik KA, Izdebska J, Binczyk E, Blasiak J, Szaflik J, et al. Polymorphisms of the homologous recombination gene RAD51 in keratoconus and Fuchs endothelial corneal dystrophy. Dis Markers. (2013) 35:353–62. doi: 10.1155/2013/851817
27. Sahebjada S, Schache M, Richardson AJ, Snibson G, MacGregor S, Daniell M, et al. Evaluating the association between keratoconus and the corneal thickness genes in an independent Australian population. Invest Ophthalmol Vis Sci. (2013) 54:8224–8. doi: 10.1167/iovs.13-12982
28. Wojcik KA, Synowiec E, Jimenez-Garcia MP, Kaminska A, Polakowski P, Blasiak J, et al. Polymorphism of the transferrin gene in eye diseases: keratoconus and Fuchs endothelial corneal dystrophy. Biomed Res Int. (2013) 2013:247438. doi: 10.1155/2013/247438
29. Sahebjada S, Schache M, Richardson AJ, Snibson G, Daniell M, Baird PN. Association of the hepatocyte growth factor gene with keratoconus in an Australian population. PLoS ONE. (2014) 9:e84067. doi: 10.1371/journal.pone.0084067
30. Hasanian-Langroudi F, Saravani R, Validad MH, Bahari G, Yari D. Association of Lysyl oxidase (LOX) Polymorphisms with the risk of keratoconus in an Iranian population. Ophthalmic Genet. (2015) 36:309–14. doi: 10.3109/13816810.2014.881507
31. Rho CR, Park JH, Jung YH, Kim MS. A case of concomitant keratoconus and granular corneal dystrophy type II. Cont Lens Anterior Eye. (2014) 37:314–6. doi: 10.1016/j.clae.2014.02.001
32. Lechner J, Porter LF, Rice A, Vitart V, Armstrong DJ, Schorderet DF, et al. Enrichment of pathogenic alleles in the brittle cornea gene, ZNF469, in keratoconus. Hum Mol Genet. (2014) 23:5527–35. doi: 10.1093/hmg/ddu253
33. Karolak JA, Polakowski P, Szaflik J, Szaflik JP, Gajecka M. Molecular Screening of Keratoconus Susceptibility Sequence Variants in VSX1, TGFBI, DOCK9, STK24, and IPO5 Genes in Polish Patients and Novel TGFBI Variant Identification. Ophthalmic Genet. (2016) 37:37–43. doi: 10.3109/13816810.2014.926375
34. Vincent AL, Jordan CA, Cadzow MJ, Merriman TR, McGhee CN. Mutations in the zinc finger protein gene, ZNF469, contribute to the pathogenesis of keratoconus. Invest Ophthalmol Vis Sci. (2014) 55:5629–35. doi: 10.1167/iovs.14-14532
35. Wojcik KA, Synowiec E, Polakowski P, Glowacki S, Izdebska J, Lloyd S, et al. Polymorphism of the flap endonuclease 1 gene in keratoconus and Fuchs endothelial corneal dystrophy. Int J Mol Sci. (2014) 15:14786–802. doi: 10.3390/ijms150814786
36. Synowiec E, Wojcik KA, Izdebska J, Blasiak J, Szaflik J, Szaflik JP. Polymorphisms of the apoptosis-related FAS and FAS ligand genes in keratoconus and Fuchs endothelial corneal dystrophy. Tohoku J Exp Med. (2014) 234:17–27. doi: 10.1620/tjem.234.17
37. Wojcik KA, Synowiec E, Sobierajczyk K, Izdebska J, Blasiak J, Szaflik J, et al. Polymorphism of the DNA base excision repair genes in keratoconus. Int J Mol Sci. (2014) 15:19682–99. doi: 10.3390/ijms151119682
38. Hao XD, Chen P, Chen ZL Li SX, Wang Y. Evaluating the association between keratoconus and reported genetic loci in a han Chinese population. Ophthalmic Genet. (2015) 36:132–6. doi: 10.3109/13816810.2015.1005317
39. Synowiec E, Wojcik KA, Izdebska J, Binczyk E, Szaflik J, Blasiak J, et al. Polymorphism of the LIG3 gene in keratoconus and Fuchs endothelial corneal dystrophy. Cell Mol Biol (Noisy-le-grand). (2015) 61:56–63.
40. Shetty R, Nuijts RM, Nanaiah SG, Anandula VR, Ghosh A, Jayadev C, et al. Two novel missense substitutions in the VSX1 gene: clinical and genetic analysis of families with Keratoconus from India. BMC Med Genet. (2015) 16:33. doi: 10.1186/s12881-015-0178-x
41. Cuellar-Partida G, Springelkamp H, Lucas SE, Yazar S, Hewitt AW, Iglesias AI, et al. WNT10A exonic variant increases the risk of keratoconus by decreasing corneal thickness. Hum Mol Genet. (2015) 24:5060–8. doi: 10.1093/hmg/ddv211
42. Wang Y, Wei W, Zhang C, Zhang X, Liu M, Zhu X, et al. Association of interleukin-1 gene single nucleotide polymorphisms with keratoconus in Chinese han population. Curr Eye Res. (2016) 41:630–5. doi: 10.3109/02713683.2015.1045083
43. Wojcik KA, Synowiec E, Kaminska A, Izdebska J, Polakowski P, Pawlowska E, et al. Polymorphism of the APEX nuclease 1 gene in keratoconus and Fuchs endothelial corneal dystrophy. Cell Mol Biol Lett. (2015) 20:48–65. doi: 10.1515/cmble-2015-0001
44. Karolak JA, Rydzanicz M, Ginter-Matuszewska B, Pitarque JA, Molinari A, Bejjani BA, et al. Variant c.2262A>C in DOCK9 Leads to Exon Skipping in Keratoconus Family. Invest Ophthalmol Vis Sci. (2015) 56:7687–90. doi: 10.1167/iovs.15-17538
45. Karolak JA, Gambin T, Pitarque JA, Molinari A, Jhangiani S, Stankiewicz P, et al. Variants in SKP1, PROB1, and IL17B genes at keratoconus 5q311-q353 susceptibility locus identified by whole-exome sequencing. Eur J Hum Genet. (2017) 25:73–8. doi: 10.1038/ejhg.2016.130
46. Yu X, Chen B, Zhang X, Shentu X. Identification of seven novel ZNF469 mutations in keratoconus patients in a Han Chinese population. Mol Vis. (2017) 23:296–305.
47. Yildiz E, Bardak H, Gunay M, Bardak Y, Imamoglu S, Ozbas H, et al. Novel Zinc Finger Protein Gene 469 (ZNF469) Variants in Advanced Keratoconus. Curr Eye Res. (2017) 42:1396–400. doi: 10.1080/02713683.2017.1325910
48. Rong SS, Ma STU Yu XT, Ma L, Chu WK, Chan TCY, et al. Genetic associations for keratoconus: a systematic review and meta-analysis. Sci Rep. (2017) 7:4620. doi: 10.1038/s41598-017-04393-2
49. Arbab M, Tahir S, Niazi MK, Ishaq M, Hussain A, Siddique PM, et al. TNF-alpha genetic predisposition and higher expression of inflammatory pathway components in keratoconus. Invest Ophthalmol Vis Sci. (2017) 58:3481–7. doi: 10.1167/iovs.16-21400
50. Guan T, Wang X, Zheng LB, Wu HJ, Yao YF. Analysis of the VSX1 gene in sporadic keratoconus patients from China. BMC Ophthalmol. (2017) 17:173. doi: 10.1186/s12886-017-0567-3
51. Hao XD, Chen P, Zhang YY Li SX, Shi WY, Gao H. De novo mutations of TUBA3D are associated with keratoconus. Sci Rep. (2017) 7:13570. doi: 10.1038/s41598-017-13162-0
52. Bykhovskaya Y, Fardaei M, Khaled ML, Nejabat M, Salouti R, Dastsooz H, et al. TSC1 mutations in keratoconus patients with or without tuberous sclerosis. Invest Ophthalmol Vis Sci. (2017) 58:6462–9. doi: 10.1167/iovs.17-22819
53. Zhang J, Wu D, Li Y, Fan Y, Chen H, Xu J. Evaluating the association between calpastatin (CAST) gene and keratoconus in the Han Chinese population. Gene. (2018) 653:10–3. doi: 10.1016/j.gene.2018.02.016
54. Guan T, Wu HJ, Zhang LJ, Xu DJ, Zheng LB, Yao YF. [A novel VSX1 gene mutation identified in a sporadic keratoconus patient from China]. Zhonghua Yan Ke Za Zhi. (2018) 54:212–7. doi: 10.3760/cma.j.issn.0412-4081.2018.03.012
55. Wang YM, Ma L, Lu SY, Chan TCY, Yam JCS, Tang SM, et al. Analysis of multiple genetic loci reveals MPDZ-NF1B rs1324183 as a putative genetic marker for keratoconus. Br J Ophthalmol. (2018) 102:1736–41. doi: 10.1136/bjophthalmol-2018-312218
56. da Silva DC, Gadelha BNB, Feitosa AFB, da Silva RG, Albuquerque T, Santos D, et al. Analysis of VSX1 variations in brazilian subjects with keratoconus. J Ophthalmic Vis Res. (2018) 13:266–73. doi: 10.4103/jovr.jovr_116_17
57. Zhang J, Wu D, Dai Y, Xu J. Functional relevance for central cornea thickness-associated genetic variants by using integrative analyses. BioData Min. (2018) 11:19. doi: 10.1186/s13040-018-0179-3
58. Yari D, Saravani R, Saravani S, Ebrahimian K, Galavi HR. Genetic polymorphisms of catalase and glutathione peroxidase-1 in keratoconus. Iran J Public Health. (2018) 47:1567–74.
59. Skorodumova LO, Belodedova AV, Sharova EI, Malyugin BE. [Search for genetic markers for precise diagnostics of keratoconus]. Biomed Khim. (2019) 65:9–20. doi: 10.18097/PBMC20196501009
60. Kabza M, Karolak JA, Rydzanicz M, Udziela M, Gasperowicz P, Ploski R, et al. Multiple Differentially Methylated Regions Specific to Keratoconus Explain Known Keratoconus Linkage Loci. Invest Ophthalmol Vis Sci. (2019) 60:1501–9. doi: 10.1167/iovs.18-25916
61. Sargazi S, Moudi M, Heidari Nia M, Saravani R, Malek Raisi H. Association of KIF26B and COL4A4 gene polymorphisms with the risk of keratoconus in a sample of Iranian population. Int Ophthalmol. (2019) 39:2621–8. doi: 10.1007/s10792-019-01111-x
62. Zhang W, Margines JB, Jacobs DS, Rabinowitz YS, Hanser EM, Chauhan T, et al. Corneal perforation after corneal cross-linking in keratoconus associated with potentially pathogenic ZNF469 mutations. Cornea. (2019) 38:1033–9. doi: 10.1097/ICO.0000000000002002
63. Froukh T, Hawwari A. Autosomal recessive non-syndromic keratoconus: homozygous frameshift variant in the candidate novel gene GALNT14. Curr Mol Med. (2019) 19:683–7. doi: 10.2174/1566524019666190730095630
64. Abdullah OA, El Gazzar WB, Salem TI, Al-Kamil EA. Role of extracellular matrix remodelling gene SNPs in keratoconus. Br J Biomed Sci. (2020) 77:13–8. doi: 10.1080/09674845.2019.1654346
65. Magalhaes OA, Kowalski TW, Wachholz GE, Schuler-Faccini L. Whole-exome sequencing in familial keratoconus: the challenges of a genetically complex disorder. Arq Bras Oftalmol. (2019) 82:453–9. doi: 10.5935/0004-2749.20190087
66. Ilhan A, Altun S, Durukan I, Yolcu U, Erdem U. The association between genetic polymorphism of glutathione peroxidase 1 (rs1050450) and keratoconus in a Turkish population. Arq Bras Oftalmol. (2019) 82:501–6. doi: 10.5935/0004-2749.20190102
67. Lin Q, Zheng L, Shen Z, Jie L. A Novel Splice-Site Variation in COL5A1 Causes Keratoconus in an Indian Family. J Ophthalmol. (2019) 2019:2851380. doi: 10.1155/2019/2851380
68. Cao K, Sahebjada S, Richardson AJ, Baird PN. Do age-related macular degeneration genes show association with keratoconus? Eye Vis (Lond). (2019) 6:38. doi: 10.1186/s40662-019-0164-z
69. Khaled ML, Bykhovskaya Y, Gu C, Liu A, Drewry MD, Chen Z, et al. PPIP5K2 and PCSK1 are candidate genetic contributors to familial keratoconus. Sci Rep. (2019) 9:19406. doi: 10.1038/s41598-019-55866-5
70. McComish BJ, Sahebjada S, Bykhovskaya Y, Willoughby CE, Richardson AJ, Tenen A, et al. Association of genetic variation with keratoconus. JAMA Ophthalmol. (2020) 138:174–81. doi: 10.1001/jamaophthalmol.2019.5293
71. Abdul-Maksoud RS, Fouad RA, Elsayed TG, Ibrahem RA, Badawi AE. The impact of catalase and glutathione peroxidase-1 genetic polymorphisms on their enzyme activities among Egyptian patients with keratoconus. J Gene Med. (2020) 22:e3192. doi: 10.1002/jgm.3192
72. Karolak JA, Gambin T, Rydzanicz M, Polakowski P, Ploski R, Szaflik JP, et al. Accumulation of sequence variants in genes of Wnt signaling and focal adhesion pathways in human corneas further explains their involvement in keratoconus. PeerJ. (2020) 8:e8982. doi: 10.7717/peerj.8982
73. Awd-Allah NA, Ismail SM, Salah El-Dine MM, Mohammed MM. Association between POLG and XRCC1 gene polymorphisms and keratoconus occurrence among Egyptian patients. Arch Soc Esp Oftalmol (Engl Ed). (2020) 95:439–46. doi: 10.1016/j.oftale.2020.03.002
74. Hosoda Y, Miyake M, Meguro A, Tabara Y, Iwai S, Ueda-Arakawa N, et al. Keratoconus-susceptibility gene identification by corneal thickness genome-wide association study and artificial intelligence IBM Watson. Commun Biol. (2020) 3:410. doi: 10.1038/s42003-020-01137-3
75. Yari D, Ehsanbakhsh Z, Validad MH, Langroudi FH. Association of TIMP-1 and COL4A4 gene polymorphisms with keratoconus in an Iranian population. J Ophthalmic Vis Res. (2020) 15:299–307. doi: 10.18502/jovr.v15i3.7448
76. Hao XD, Chen XN, Zhang YY, Chen P, Wei C, Shi WY, et al. Multi-level consistent changes of the ECM pathway identified in a typical keratoconus twin's family by multi-omics analysis. Orphanet J Rare Dis. (2020) 15:227. doi: 10.1186/s13023-020-01512-7
77. Froukh T, Hawwari A, Al Zubi K. Whole exome sequencing highlights variants in association with Keratoconus in Jordanian families. BMC Med Genet. (2020) 21:177. doi: 10.1186/s12881-020-01112-z
78. Abdelghany AA, Toraih EA, Abdelaziz EZ, El-Sherbeeny NA, Fawzy MS. Association of collagen gene (COL4A3) rs55703767 variant with response to riboflavin/ultraviolet a-induced collagen cross-linking in female patients with keratoconus. Cornea. (2021) 40:88–98. doi: 10.1097/ICO.0000000000002489
79. Droitcourt C, Touboul D, Ged C, Ezzedine K, Cario-Andre M, de Verneuil H, et al. A prospective study of filaggrin null mutations in keratoconus patients with or without atopic disorders. Dermatology. (2011) 222:336–41. doi: 10.1159/000328408
80. Rathi VM, Vemuganti GK, Sangwan VS, Kannabiran C. Late occurrence of granular dystrophy in bilateral keratoconus: penetrating keratoplasty and long-term follow-up. Indian J Ophthalmol. (2011) 59:398–400. doi: 10.4103/0301-4738.83624
81. De Bonis P, Laborante A, Pizzicoli C, Stallone R, Barbano R, Longo C, et al. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol Vis. (2011) 17:2482–94.
82. Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. (2011) 89:628–33. doi: 10.1016/j.ajhg.2011.09.014
83. Nowak DM, Karolak JA, Kubiak J, Gut M, Pitarque JA, Molinari A, et al. Substitution at IL1RN and deletion at SLC4A11 segregating with phenotype in familial keratoconus. Invest Ophthalmol Vis Sci. (2013) 54:2207–15. doi: 10.1167/iovs.13-11592
84. Li X, Bykhovskaya Y, Canedo AL, Haritunians T, Siscovick D, Aldave AJ, et al. Genetic association of COL5A1 variants in keratoconus patients suggests a complex connection between corneal thinning and keratoconus. Invest Ophthalmol Vis Sci. (2013) 54:2696–704. doi: 10.1167/iovs.13-11601
85. Vincent AL, Jordan C, Sheck L, Niederer R, Patel DV, McGhee CN. Screening the visual system homeobox 1 gene in keratoconus and posterior polymorphous dystrophy cohorts identifies a novel variant. Mol Vis. (2013) 19:852–60.
86. Dehkordi FA, Rashki A, Bagheri N, Chaleshtori MH, Memarzadeh E, Salehi A, et al. Study of VSX1 mutations in patients with keratoconus in southwest Iran using PCR-single-strand conformation polymorphism/heteroduplex analysis and sequencing method. Acta Cytol. (2013) 57:646–51. doi: 10.1159/000353297
87. Kokolakis NS, Gazouli M, Chatziralli IP, Koutsandrea C, Gatzioufas Z, Peponis VG, et al. Polymorphism analysis of COL4A3 and COL4A4 genes in Greek patients with keratoconus. Ophthalmic Genet. (2014) 35:226–8. doi: 10.3109/13816810.2014.946055
88. Saravani R, Hasanian-Langroudi F, Validad MH, Yari D, Bahari G, Faramarzi M, et al. Evaluation of possible relationship between COL4A4 gene polymorphisms and risk of keratoconus. Cornea. (2015) 34:318–22. doi: 10.1097/ICO.0000000000000356
89. Bardak H, Gunay M, Yildiz E, Bardak Y, Gunay B, Ozbas H, et al. Novel visual system homeobox 1 gene mutations in Turkish patients with keratoconus. Genet Mol Res. (2016) 15. doi: 10.4238/gmr15049024
90. Xu X, Zhang X, Cui Y, Yang H, Ping X, Wu J, et al. Three novel variants identified within ECM-related genes in Chinese Han keratoconus patients. Sci Rep. (2020) 10:5844. doi: 10.1038/s41598-020-62572-0
91. Jackson M, Marks L, May GHW, Wilson JB. The genetic basis of disease. Essays Biochem. (2018) 62:643–723. doi: 10.1042/EBC20170053
92. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. (2009) 4:44–57. doi: 10.1038/nprot.2008.211
93. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. (2009) 37:1–13. doi: 10.1093/nar/gkn923
94. Sarker-Nag A, Hutcheon AE, Karamichos D. Mitochondrial Profile and Responses to TGF-beta Ligands in Keratoconus. Curr Eye Res. (2016) 41:900–7. doi: 10.3109/02713683.2015.1078361
95. Atilano SR, Lee DH, Fukuhara PS, Chwa M, Nesburn AB, Udar N, et al. Corneal oxidative damage in keratoconus cells due to decreased oxidant elimination from modified expression levels of SOD enzymes, PRDX6, SCARA3, CPSF3, and FOXM1. J Ophthalmic Vis Res. (2019) 14:62–70. doi: 10.4103/jovr.jovr_80_18
96. Ayan B, Yuksel N, Carhan A, Gumuskaya Ocal B, Akcay E, Cagil N, et al. Evaluation estrogen, progesteron and androgen receptor expressions in corneal epithelium in keratoconus. Cont Lens Anterior Eye. (2019) 42:492–6. doi: 10.1016/j.clae.2018.11.015
97. Bykhovskaya Y, Gromova A, Makarenkova HP, Rabinowitz YS. Abnormal regulation of extracellular matrix and adhesion molecules in corneas of patients with keratoconus. Int J Keratoconus Ectatic Corneal Dis. (2016) 5:63–70. doi: 10.5005/jp-journals-10025-1123
98. Chiambaretta F, Nakamura H, De Graeve F, Sakai H, Marceau G, Maruyama Y, et al. Kruppel-like factor 6 (KLF6) affects the promoter activity of the alpha1-proteinase inhibitor gene. Invest Ophthalmol Vis Sci. (2006) 47:582–90. doi: 10.1167/iovs.05-0551
99. Chiplunkar S, Chamblis K, Chwa M, Rosenberg S, Kenney MC, Brown DJ. Enhanced expression of a transmembrane phosphotyrosine phosphatase (LAR) in keratoconus cultures and corneas. Exp Eye Res. (1999) 68:283–93. doi: 10.1006/exer.1998.0604
100. Chung ES, Lee KH, Kim M, Chang EJ, Chung TY, Kim EK, et al. Expression of neurotrophic factors and their receptors in keratoconic cornea. Curr Eye Res. (2013) 38:743–50. doi: 10.3109/02713683.2013.774421
101. Du G, Liu C, Li X, Chen W, He R, Wang X, et al. Induction of matrix metalloproteinase-1 by tumor necrosis factor-alpha is mediated by interleukin-6 in cultured fibroblasts of keratoconus. Exp Biol Med (Maywood). (2016) 241:2033–41. doi: 10.1177/1535370216650940
102. Engler C, Chakravarti S, Doyle J, Eberhart CG, Meng H, Stark WJ, et al. Transforming growth factor-beta signaling pathway activation in Keratoconus. Am J Ophthalmol. (2011) 151:752–9 e2. doi: 10.1016/j.ajo.2010.11.008
103. Garcia B, Garcia-Suarez O, Merayo-Lloves J, Alcalde I, Alfonso JF, Fernandez-Vega Cueto L, et al. Differential expression of proteoglycans by corneal stromal cells in keratoconus. Invest Ophthalmol Vis Sci. (2016) 57:2618–28. doi: 10.1167/iovs.15-16692
104. Garcia B, Garcia-Suarez O, Merayo-Lloves J, Ferrara G, Alcalde I, Gonzalez J, et al. Heparanase overexpresses in keratoconic cornea and tears depending on the pathologic grade. Dis Markers. (2017) 2017:3502386. doi: 10.1155/2017/3502386
105. Ha NT, Nakayasu K, Murakami A, Ishidoh K, Kanai A. Microarray analysis identified differentially expressed genes in keratocytes from keratoconus patients. Curr Eye Res. (2004) 28:373–9. doi: 10.1080/02713680490502201
106. Hao XD, Chen ZL, Qu ML, Zhao XW Li SX, Chen P. Decreased Integrity, Content, and Increased Transcript Level of Mitochondrial DNA Are Associated with Keratoconus. PLoS ONE. (2016) 11:e0165580. doi: 10.1371/journal.pone.0165580
107. Joseph R, Srivastava OP, Pfister RR. Downregulation of beta-actin gene and human antigen R in human keratoconus. Invest Ophthalmol Vis Sci. (2012) 53:4032–41. doi: 10.1167/iovs.11-9062
108. Kanai A. [The pathogenesis and treatment of corneal disorders]. Nippon Ganka Gakkai Zasshi. (2002) 106:757–76.
109. Kenney MC, Chwa M, Atilano SR, Tran A, Carballo M, Saghizadeh M, et al. Increased levels of catalase and cathepsin V/L2 but decreased TIMP-1 in keratoconus corneas: evidence that oxidative stress plays a role in this disorder. Invest Ophthalmol Vis Sci. (2005) 46:823–32. doi: 10.1167/iovs.04-0549
110. Khaled ML, Bykhovskaya Y, Yablonski SER Li H, Drewry MD, Aboobakar IF, et al. Differential Expression of Coding and Long Noncoding RNAs in Keratoconus-Affected Corneas. Invest Ophthalmol Vis Sci. (2018) 59:2717–28. doi: 10.1167/iovs.18-24267
111. Lee JE, Oum BS, Choi HY, Lee SU, Lee JS. Evaluation of differentially expressed genes identified in keratoconus. Mol Vis. (2009) 15:2480–7.
112. Mace M, Galiacy SD, Erraud A, Mejia JE, Etchevers H, Allouche M, et al. Comparative transcriptome and network biology analyses demonstrate antiproliferative and hyperapoptotic phenotypes in human keratoconus corneas. Invest Ophthalmol Vis Sci. (2011) 52:6181–91. doi: 10.1167/iovs.10-70981
113. Kabza M, Karolak JA, Rydzanicz M, Szczesniak MW, Nowak DM, Ginter-Matuszewska B, et al. Collagen synthesis disruption and downregulation of core elements of TGF-beta, Hippo, and Wnt pathways in keratoconus corneas. Eur J Hum Genet. (2017) 25:582–90. doi: 10.1038/ejhg.2017.4
114. Mootha VV, Kanoff JM, Shankardas J, Dimitrijevich S. Marked reduction of alcohol dehydrogenase in keratoconus corneal fibroblasts. Mol Vis. (2009) 15:706–12.
115. Nielsen K, Birkenkamp-Demtroder K, Ehlers N, Orntoft TF. Identification of differentially expressed genes in keratoconus epithelium analyzed on microarrays. Invest Ophthalmol Vis Sci. (2003) 44:2466–76. doi: 10.1167/iovs.02-0671
116. Pahuja N, Kumar NR, Shroff R, Shetty R, Nuijts RM, Ghosh A, et al. Differential molecular expression of extracellular matrix and inflammatory genes at the corneal cone apex drives focal weakening in keratoconus. Invest Ophthalmol Vis Sci. (2016) 57:5372–82. doi: 10.1167/iovs.16-19677
117. Peters DP, Harrison DA, Brandt CR. Heterogeneity of type I collagen expression in human corneal keratoconus fibroblasts. Ophthalmic Res. (1993) 25:273–9. doi: 10.1159/000267325
118. Priyadarsini S, McKay TB, Sarker-Nag A, Karamichos D. Keratoconus in vitro and the key players of the TGF-beta pathway. Mol Vis. (2015) 21:577–88.
119. Rabinowitz YS, Dong L, Wistow G. Gene expression profile studies of human keratoconus cornea for NEIBank: a novel cornea-expressed gene and the absence of transcripts for aquaporin 5. Invest Ophthalmol Vis Sci. (2005) 46:1239–46. doi: 10.1167/iovs.04-1148
120. Shetty R, Sharma A, Pahuja N, Chevour P, Padmajan N, Dhamodaran K, et al. Oxidative stress induces dysregulated autophagy in corneal epithelium of keratoconus patients. PLoS ONE. (2017) 12:e0184628. doi: 10.1371/journal.pone.0184628
121. Saee-Rad S, Raoofian R, Mahbod M, Miraftab M, Mojarrad M, Asgari S, et al. Analysis of superoxide dismutase 1, dual-specificity phosphatase 1, and transforming growth factor, beta 1 genes expression in keratoconic and non-keratoconic corneas. Mol Vis. (2013) 19:2501–7.
122. Saghizadeh M, Chwa M, Aoki A, Lin B, Pirouzmanesh A, Brown DJ, et al. Altered expression of growth factors and cytokines in keratoconus, bullous keratopathy and diabetic human corneas. Exp Eye Res. (2001) 73:179–89. doi: 10.1006/exer.2001.1028
123. Sharif R, Khaled ML, McKay TB, Liu Y, Karamichos D. Transcriptional profiling of corneal stromal cells derived from patients with keratoconus. Sci Rep. (2019) 9:12567. doi: 10.1038/s41598-019-48983-8
124. Shetty R, Ghosh A, Lim RR, Subramani M, Mihir K, Reshma AR, et al. Elevated expression of matrix metalloproteinase-9 and inflammatory cytokines in keratoconus patients is inhibited by cyclosporine A. Invest Ophthalmol Vis Sci. (2015) 56:738–50. doi: 10.1167/iovs.14-14831
125. Shetty R, Sathyanarayanamoorthy A, Ramachandra RA, Arora V, Ghosh A, Srivatsa PR, et al. Attenuation of lysyl oxidase and collagen gene expression in keratoconus patient corneal epithelium corresponds to disease severity. Mol Vis. (2015) 21:12–25.
126. Shetty R, Vunnava KP, Dhamodaran K, Matalia H, Murali S, Jayadev C, et al. Characterization of Corneal Epithelial Cells in Keratoconus. Transl Vis Sci Technol. (2019) 8:2. doi: 10.1167/tvst.8.1.2
127. Shinde V, Hu N, Mahale A, Maiti G, Daoud Y, Eberhart CG, et al. RNA sequencing of corneas from two keratoconus patient groups identifies potential biomarkers and decreased NRF2-antioxidant responses. Sci Rep. (2020) 10:9907. doi: 10.1038/s41598-020-66735-x
128. Stachon T, Latta L, Kolev K, Seitz B, Langenbucher A, Szentmary N. [Increased NF-kappaB and iNOS Expression in Keratoconus Keratocytes - Hints for an Inflammatory Component?]. Klin Monbl Augenheilkd. (2019) 238:1010–7. doi: 10.1055/a-1002-0100
129. Stachs O, Bochert A, Gerber T, Koczan D, Thiessen HJ, Guthoff RF. [The extracellular matrix structure in keratoconus]. Ophthalmologe. (2004) 101:384–9. doi: 10.1007/s00347-003-0902-3
130. Sutton G, Madigan M, Roufas A, McAvoy J. Secreted frizzled-related protein 1 (SFRP1) is highly upregulated in keratoconus epithelium: a novel finding highlighting a new potential focus for keratoconus research and treatment. Clin Exp Ophthalmol. (2010) 38:43–8. doi: 10.1111/j.1442-9071.2009.02216.x
131. Wang YN, Liu XN, Wang XD, Yin Y, Chen Y, Xiao XH, et al. Expression of visual system homeobox 1 in human keratoconus. Int J Ophthalmol. (2019) 12:201–6. doi: 10.18240/ijo.2019.02.03
132. Wentz-Hunter K, Cheng EL, Ueda J, Sugar J, Yue BY. Keratocan expression is increased in the stroma of keratoconus corneas. Mol Med. (2001) 7:470–7. doi: 10.1007/BF03401852
133. Whitelock RB, Fukuchi T, Zhou L, Twining SS, Sugar J, Feder RS, et al. Cathepsin G, acid phosphatase, and alpha 1-proteinase inhibitor messenger RNA levels in keratoconus corneas. Invest Ophthalmol Vis Sci. (1997) 38:529–34.
134. You J, Corley SM, Wen L, Hodge C, Hollhumer R, Madigan MC, et al. RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci Rep. (2018) 8:389. doi: 10.1038/s41598-017-18480-x
135. Zhang LY, Zou LH. [Study on enhanced leukocyte antigen-related tyrosine phosphatase in keratoconus]. Zhonghua Yan Ke Za Zhi. (2005) 41:234–8.
136. Zhao G, Wang C, Sun W, Zhang W, Li Y, Sheng H, et al. The expression of protein betaig-h3 inducible by transforming growth factor-beta in keratoconus and normal cornea. Zhonghua Yan Ke Za Zhi. (2002) 38:419–21.
137. Akhtar S, Bron AJ, Hayes AJ, Meek KM, Caterson B. Role of keratan sulphate (sulphated poly -N-acetyllactosamine repeats) in keratoconic cornea, histochemical, and ultrastructural analysis. Graefes Arch Clin Exp Ophthalmol. (2011) 249:413–20. doi: 10.1007/s00417-010-1512-9
138. Akhtar S, Bron AJ, Salvi SM, Hawksworth NR, Tuft SJ, Meek KM. Ultrastructural analysis of collagen fibrils and proteoglycans in keratoconus. Acta Ophthalmol. (2008) 86:764–72. doi: 10.1111/j.1755-3768.2007.01142.x
139. Bosnar D, Dekaris I, Gabric N, Markotic A, Lazic R, Spoljaric N. Influence of interleukin-1alpha and tumor necrosis factor-alpha production on corneal graft survival. Croat Med J. (2006) 47:59–66.
140. Brookes NH, Loh IP, Clover GM, Poole CA, Sherwin T. Involvement of corneal nerves in the progression of keratoconus. Exp Eye Res. (2003) 77:515–24. doi: 10.1016/S0014-4835(03)00148-9
141. Bureau J, Pouliquen Y, Lorans G. [Fibrocyte reaction to interleukin 1 stimulation in keratoconus. Original title: Fibrocyte response to interleukin 1 stimulation in keratoconus]. Klin Monbl Augenheilkd. (1993) 203:269–74. doi: 10.1055/s-2008-1045679
142. Bystrom B, Carracedo S, Behndig A, Gullberg D, Pedrosa-Domellof F. Alpha11 integrin in the human cornea: importance in development and disease. Invest Ophthalmol Vis Sci. (2009) 50:5044–53. doi: 10.1167/iovs.08-3261
143. Caglayan M, Kocamis SI, Sarac O, Tatli Dogan H, Kosekahya P, Ayan M, et al. Investigation of Heme Oxygenase 2 Enzyme Protein Expression in Keratoconus and Normal Human Corneal Epithelium: An Immunohistochemical Study. Curr Eye Res. (2019) 44:25–9. doi: 10.1080/02713683.2018.1521980
144. Chaerkady R, Shao H, Scott SG, Pandey A, Jun AS, Chakravarti S. The keratoconus corneal proteome: loss of epithelial integrity and stromal degeneration. J Proteomics. (2013) 87:122–31. doi: 10.1016/j.jprot.2013.05.023
145. Cheng EL Li Y, Sugar J, Yue BY. Cell density regulated expression of transcription factor Sp1 in corneal stromal cultures. Exp Eye Res. (2001) 73:17–24. doi: 10.1006/exer.2001.1014
146. Cheung IM, McGhee C, Sherwin T. Deficient repair regulatory response to injury in keratoconic stromal cells. Clin Exp Optom. (2014) 97:234–9. doi: 10.1111/cxo.12118
147. Collier SA, Madigan MC, Penfold PL. Expression of membrane-type 1 matrix metalloproteinase (MT1-MMP) and MMP-2 in normal and keratoconus corneas. Curr Eye Res. (2000) 21:662–8. doi: 10.1076/0271-3683(200008)2121-VFT662
148. Collier SA. Is the corneal degradation in keratoconus caused by matrix-metalloproteinases? Clin Exp Ophthalmol. (2001) 29:340–4. doi: 10.1046/j.1442-9071.2001.d01-17.x
149. Andrade FEC, Covre JL, Ramos L, Hazarbassanov RM, Santos MSD, Campos M, et al. Evaluation of galectin-1 and galectin-3 as prospective biomarkers in keratoconus. Br J Ophthalmol. (2018) 102:700–7. doi: 10.1136/bjophthalmol-2017-311495
150. Dudakova L, Liskova P, Trojek T, Palos M, Kalasova S, Jirsova K. Changes in lysyl oxidase (LOX) distribution and its decreased activity in keratoconus corneas. Exp Eye Res. (2012) 104:74–81. doi: 10.1016/j.exer.2012.09.005
151. Dudakova L, Sasaki T, Liskova P, Palos M, Jirsova K. The presence of lysyl oxidase-like enzymes in human control and keratoconic corneas. Histol Histopathol. (2016) 31:63–71.
152. Gatzioufas Z, Charalambous P, Thanos S. Reduced expression of the gap junction protein Connexin 43 in keratoconus. Eye (Lond). (2008) 22:294–9. doi: 10.1038/sj.eye.6702972
153. Greene CA, Kuo C, Sherwin T. Aberrant Patterns of Key Epithelial Basement Membrane Components in Keratoconus. Cornea. (2017) 36:1549–55. doi: 10.1097/ICO.0000000000001393
154. Hasby EA, Saad HA. Immunohistochemical expression of Fas ligand (FasL) and neprilysin (neutral endopeptidase/CD10) in keratoconus. Int Ophthalmol. (2013) 33:125–31. doi: 10.1007/s10792-012-9651-0
155. Iqbal O, Fisher G, Vira S, Syed D, Sadeghi N, Freeman D, et al. Increased expression of secreted frizzled-related protein-1 and microtubule-associated protein light chain 3 in keratoconus. Cornea. (2013) 32:702–7. doi: 10.1097/ICO.0b013e318282987a
156. Joseph R, Srivastava OP, Pfister RR. Differential epithelial and stromal protein profiles in keratoconus and normal human corneas. Exp Eye Res. (2011) 92:282–98. doi: 10.1016/j.exer.2011.01.008
157. Kenney MC, Chwa M, Alba A, Saghizadeh M, Huang ZS, Brown DJ. Localization of TIMP-1, TIMP-2, TIMP-3, gelatinase A and gelatinase B in pathological human corneas. Curr Eye Res. (1998) 17:238–46. doi: 10.1076/ceyr.17.3.238.5222
158. Kenney MC, Nesburn AB, Burgeson RE, Butkowski RJ, Ljubimov AV. Abnormalities of the extracellular matrix in keratoconus corneas. Cornea. (1997) 16:345–51. doi: 10.1097/00003226-199705000-00016
159. Lackner EM, Matthaei M, Meng H, Ardjomand N, Eberhart CG, Jun AS. Design and analysis of keratoconus tissue microarrays. Cornea. (2014) 33:49–55. doi: 10.1097/ICO.0000000000000012
160. Lambiase A, Merlo D, Mollinari C, Bonini P, Rinaldi AM, DA M, et al. Molecular basis for keratoconus: lack of TrkA expression and its transcriptional repression by Sp3. Proc Natl Acad Sci U S A. (2005) 102:16795–800. doi: 10.1073/pnas.0508516102
161. Li Y, Zhou L, Twining SS, Sugar J, Yue BY. Involvement of Sp1 elements in the promoter activity of the alpha1-proteinase inhibitor gene. J Biol Chem. (1998) 273:9959–65. doi: 10.1074/jbc.273.16.9959
162. Lyon D, McKay TB, Sarkar-Nag A, Priyadarsini S, Karamichos D. Human Keratoconus Cell Contractility is Mediated by Transforming Growth Factor-Beta Isoforms. J Funct Biomater. (2015) 6:422–38. doi: 10.3390/jfb6020422
163. Maatta M, Heljasvaara R, Sormunen R, Pihlajaniemi T, Autio-Harmainen H, Tervo T. Differential expression of collagen types XVIII/endostatin and XV in normal, keratoconus, and scarred human corneas. Cornea. (2006) 25:341–9. doi: 10.1097/01.ico.0000178729.57435.96
164. Maatta M, Vaisanen T, Vaisanen MR, Pihlajaniemi T, Tervo T. Altered expression of type XIII collagen in keratoconus and scarred human cornea - Increased expression in scarred cornea is associated with myofibroblast transformation. Cornea. (2006) 25:448–53. doi: 10.1097/01.ico.0000183537.45393.1f
165. Mackiewicz Z, Maatta M, Stenman M, Konttinen L, Tervo T, Konttinen YT. Collagenolytic proteinases in keratoconus. Cornea. (2006) 25:603–10. doi: 10.1097/01.ico.0000208820.32614.00
166. Malfeito M, Regueiro U, Perez-Mato M, Campos F, Sobrino T, Lema I. Innate Immunity Biomarkers for Early Detection of Keratoconus. Ocul Immunol Inflamm. (2019) 27:942–8. doi: 10.1080/09273948.2018.1511813
167. Marini M, Mencucci R, Rosa I, Favuzza E, Guasti D, Ibba-Manneschi L, et al. Telocytes in normal and keratoconic human cornea: an immunohistochemical and transmission electron microscopy study. J Cell Mol Med. (2017) 21:3602–11. doi: 10.1111/jcmm.13270
168. Nielsen K, Vorum H, Fagerholm P, Birkenkamp-Demtroder K, Honore B, Ehlers N, et al. Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study. Exp Eye Res. (2006) 82:201–9. doi: 10.1016/j.exer.2005.06.009
169. Olofsson EM, Marklund SL, Pedrosa-Domellof F, Behndig A. Interleukin-1alpha downregulates extracellular-superoxide dismutase in human corneal keratoconus stromal cells. Mol Vis. (2007) 13:1285–90.
170. Priyadarsini S, Hjortdal J, Sarker-Nag A, Sejersen H, Asara JM, Karamichos D. Gross cystic disease fluid protein-15/prolactin-inducible protein as a biomarker for keratoconus disease. PLoS ONE. (2014) 9:e113310. doi: 10.1371/journal.pone.0113310
171. Regueiro U, Perez-Mato M, Hervella P, Campos F, Sobrino T, Lema I. Toll-like receptors as diagnostic targets in pellucid marginal degeneration. Exp Eye Res. (2020) 200:108211. doi: 10.1016/j.exer.2020.108211
172. Sacchetti M, Scorcia V, Lambiase A, Bonini S. Preliminary evidence of neuropeptides involvement in keratoconus. Acta Ophthalmol. (2015) 93:e315–6. doi: 10.1111/aos.12483
173. Sawaguchi S, Yue BY, Sugar J, Gilboy JE. Lysosomal enzyme abnormalities in keratoconus. Arch Ophthalmol. (1989) 107:1507–10. doi: 10.1001/archopht.1989.01070020581044
174. Seppala HPS, Maatta M, Rautia M, Mackiewicz Z, Tuisku I, Tervo T, et al. EMMPRIN and MMP-1 in keratoconus. Cornea. (2006) 25:325–30. doi: 10.1097/01.ico.0000183534.22522.39
175. Sevost'ianov EN, Giniatullin RU, Gorskova EN, Teplova SN. Keratocyte apoptosis in keratoconus. Vestnik oftalmologii. (2002) 118:36–8.
176. Sherwin T, Brookes NH, Low IP, Poole CA, Clover GM. Cellular incursion into Bowman's membrane in the peripheral cone of the keratoconic cornea. Exp Eye Res. (2002) 74:473–82. doi: 10.1006/exer.2001.1157
177. Smith VA, Hoh HB, Littleton M, Easty DL. Over-expression of a gelatinase a activity in. keratoconus. Eye. (1995) 9:429–33. doi: 10.1038/eye.1995.100
178. Smith VA, Matthews FJ, Majid MA, Cook SD. Keratoconus: Matrix metalloproteinase-2 activation and TIMP modulation. Biochim Biophys Acta Mol Basis Dis. (2006) 1762:431–9. doi: 10.1016/j.bbadis.2006.01.010
179. Srivastava OP, Chandrasekaran D, Pfister RR. Molecular changes in selected epithelial proteins in human keratoconus corneas compared to normal corneas. Molecular Vision. (2006) 12:1615–25.
180. Tai TYT, Damani MR, Vo R, Rayner SA, Glasgow BJ, Hofbauer JD, et al. Keratoconus associated with corneal stromal amyloid deposition containing TGFBIp. Cornea. (2009) 28:589–93. doi: 10.1097/ICO.0b013e31818c9003
181. Takacs L, Csutak A, Balazs E, Modis L, Berta A. Expression of beta ig-h3 is lower than normal in keratoconus corneas but increases with scarring. Cornea. (1999) 18:599–605. doi: 10.1097/00003226-199909000-00014
182. Thanos S, Oellers P, Zu Horste MM, Prokosch V, Schlatt S, Seitz B, et al. Role of thyroxine in the development of keratoconus. Cornea. (2016) 35:1338–46. doi: 10.1097/ICO.0000000000000988
183. Toti P, Tosi GM, Traversi C, Schurfeld K, Cardone C, Caporossi A. CD-34 stromal expression pattern in normal and altered human corneas. Ophthalmology. (2002) 109:1167–71. doi: 10.1016/s0161-6420(02)01042-4
184. Tuori A, Virtanen I, Aine E, Uusitalo H. The expression of tenascin and fibronectin in keratoconus, scarred and normal human cornea. Graefes Arch Clin Exp Ophthalmol. (1997) 235:222–9. doi: 10.1007/BF00941763
185. Tuori AJ, Virtanen I, Aine E, Kalluri R, Miner JH, Uusitalo HM. The immunohistochemical composition of corneal basement membrane in keratoconus. Curr Eye Res. (1997) 16:792–801. doi: 10.1076/ceyr.16.8.792.8989
186. Wang X, Zou L, Jin T, Pan Z. Apoptosis in keratoconus and its relevance to the expression of Fas-L protein. Ophthalmic Research. (2008) 26:53–6.
187. Wolf M, Clay SM, Oldenburg CE, Rose-Nussbaumer J, Hwang DG, Chan MF. Overexpression of MMPs in corneas requiring penetrating and deep anterior lamellar keratoplasty. Invest Ophthalmol Vis Sci. (2019) 60:1734–47. doi: 10.1167/iovs.18-25961
188. Yam GH-F, Fuest M, Zhou L, Liu Y-C, Deng L, Chan AS-Y, et al. Differential epithelial and stromal protein profiles in cone and non-cone regions of keratoconus corneas. Scientific Reports. (2019) 9. doi: 10.1038/s41598-019-39182-6
189. Yin H, Luo C, Tian Y, Deng Y. Altered expression of sex hormone receptors in keratoconus corneas. Biomedical Research-India. (2017) 28:5089–92.
190. You J, Wen L, Roufas A, Madigan MC, Sutton G. Expression of SFRP Family Proteins in Human Keratoconus Corneas. Plos ONE. (2013) 8. doi: 10.1371/journal.pone.0066770
191. You J, Wen L, Roufas A, Hodge C, Sutton G, Madigan MC. Expression of HGF and c-met proteins in human keratoconus corneas. J Ophthalmol. (2015). doi: 10.1155/2015/852986
192. Zhou LL, Sawaguchi S, Twining SS, Sugar J, Feder RS, Yue B. Expression of degradative enzymes and protease inhibitors in corneas with keratoconus. Investigative Ophthalmology & Visual Science. (1998) 39:1117–24.
193. Zhou LL, Yue B, Twining SS, Sugar J, Feder RS. Expression of wound healing and stress-related proteins in keratoconus corneas. Curr Eye Res. (1996) 15:1124–31. doi: 10.3109/02713689608995144
194. Roy S, Yadav S, Dasgupta T, Chawla S, Tandon R, Ghosh S. Interplay between hereditary and environmental factors to establish an in vitro disease model of keratoconus. Drug Discov Today. (2019) 24:403–16. doi: 10.1016/j.drudis.2018.10.017
195. Farzadfard A, Nassiri N, Moghadam TN, Paylakhi SH, Elahi E. Screening for MIR184 Mutations in Iranian Patients with Keratoconus. J Ophthalmic Vis Res. (2016) 11:3–7. doi: 10.4103/2008-322X.180715
196. Lucas SEM, Zhou T, Blackburn NB, Mills RA, Ellis J, Leo P, et al. Rare, potentially pathogenic variants in ZNF469 are not enriched in keratoconus in a large australian cohort of european descent. Invest Ophthalmol Vis Sci. (2017) 58:6248–56. doi: 10.1167/iovs.17-22417
197. Kalantan H, Kondkar AA, Sultan T, Azad TA, Alsabaani NA, AlQahtani MA, et al. Polymorphism rs13334190 in zinc finger protein 469 (ZNF469) is not a risk factor for keratoconus in a Saudi cohort. BMC Res Notes. (2017) 10:652. doi: 10.1186/s13104-017-2996-8
198. Jeoung JW, Kim MK, Park SS, Kim SY, Ko HS, Wee WR, et al. VSX1 gene and keratoconus: genetic analysis in Korean patients. Cornea. (2012) 31:746–50. doi: 10.1097/ICO.0b013e3181e16dd0
199. Nejabat M, Naghash P, Dastsooz H, Mohammadi S, Alipour M, Fardaei M. VSX1 and SOD1 mutation screening in patients with keratoconus in the South of Iran. J Ophthalmic Vis Res. (2017) 12:135–40. doi: 10.4103/jovr.jovr_97_16
200. Verma A, Das M, Srinivasan M, Prajna NV, Sundaresan P. Investigation of VSX1 sequence variants in South Indian patients with sporadic cases of keratoconus. BMC Res Notes. (2013) 6:103. doi: 10.1186/1756-0500-6-103
201. Palamar M, Onay H, Ozdemir TR, Arslan E, Egrilmez S, Ozkinay F, et al. Relationship between IL1beta-511C>T and ILRN VNTR polymorphisms and keratoconus. Cornea. (2014) 33:145–7. doi: 10.1097/ICO.0000000000000027
202. Davidson AE, Borasio E, Liskova P, Khan AO, Hassan H, Cheetham ME, et al. Brittle cornea syndrome ZNF469 mutation carrier phenotype and segregation analysis of rare ZNF469 variants in familial keratoconus. Invest Ophthalmol Vis Sci. (2015) 56:578–86. doi: 10.1167/iovs.14-15792
203. Simcoe MJ, Khawaja AP, Hysi PG, Hammond CJ, Eye UKB, Vision C. Genome-wide association study of corneal biomechanical properties identifies over 200 loci providing insight into the genetic etiology of ocular diseases. Hum Mol Genet. (2020) 29:3154–64. doi: 10.1093/hmg/ddaa155
204. Loh IP, Sherwin T. Is Keratoconus an Inflammatory Disease? The Implication of Inflammatory Pathways. Ocul Immunol Inflamm. (2020):1–10. doi: 10.1080/09273948.2020.1780271
205. Kim WJ, Rabinowitz YS, Meisler DM, Wilson SE. Keratocyte apoptosis associated with keratoconus. Exp Eye Res. (1999) 69:475–81. doi: 10.1006/exer.1999.0719
206. Wolffsohn JS, Safeen S, Shah S, Laiquzzaman M. Changes of corneal biomechanics with keratoconus. Cornea. (2012) 31:849–54. doi: 10.1097/ICO.0b013e318243e42d
207. Eliasi E, Bez M, Megreli J, Avramovich E, Fischer N, Barak A, et al. The Association Between Keratoconus and Body Mass Index: A Population-Based Cross-Sectional Study Among Half a Million Adolescents. Am J Ophthalmol. (2021) 224:200–6. doi: 10.1016/j.ajo.2020.11.021
208. Gatzioufas Z, Thanos S. Acute keratoconus induced by hypothyroxinemia during pregnancy. J Endocrinol Invest. (2008) 31:262–6. doi: 10.1007/BF03345600
209. Bilgihan K, Hondur A, Sul S, Ozturk S. Pregnancy-induced Progression of Keratoconus. Cornea. (2011) 30:991–4. doi: 10.1097/ICO.0b013e3182068adc
210. Dutta D, Shivaprasad K, Ghosh S, Mukhopadhyay S, Chowdhury S. Adrenal myelolipoma with keratoconus: A novel clinical association. Indian J Endocrinol Metab. (2012) 16:S364–6. doi: 10.4103/2230-8210.104094
211. McKay TB, Hjortdal J, Sejersen H, Karamichos D. Differential Effects of Hormones on Cellular Metabolism in Keratoconus In Vitro. Scientific Reports. (2017) 7. doi: 10.1038/srep42896
212. Sharif R, Bak-Nielsen S, Sejersen H, Ding K, Hjortdal J, Karamichos D. Prolactin-induced protein is a novel biomarker for Keratoconus. Exp Eye Res. (2019) 179:55–63. doi: 10.1016/j.exer.2018.10.015
213. Dawczynski J, Franke S, Blum M, Kasper M, Stein G, Strobel J. Advanced glycation end-products in corneas of patients with keratoconus. Graefes Arch Clin Exp Ophthalmol. (2002) 240:296–301. doi: 10.1007/s00417-002-0445-3
214. Arnal E, Peris-Martinez C, Luis Menezo J, Johnsen-Soriano S, Javier Romero F. Oxidative stress in keratoconus? Invest Ophthalmol Vis Sci. (2011) 52:8592–7. doi: 10.1167/iovs.11-7732
215. Lema I, Duran JA. Inflammatory molecules in the tears of patients with keratoconus. Ophthalmology. (2005) 112:654–9. doi: 10.1016/j.ophtha.2004.11.050
216. Sorkhabi R, Ghorbanihaghjo A, Taheri N, Ahoor MH. Tear film inflammatory mediators in patients with keratoconus. Int Ophthalmol. (2015) 35:467–72. doi: 10.1007/s10792-014-9971-3
217. Ionescu C, Corbu CG, Tanase C, Jonescu-Cuypers C, Nicula C, Dascalescu D, et al. Inflammatory biomarkers profile as microenvironmental expression in keratoconus. Dis Markers. (2016). doi: 10.1155/2016/1243819
218. Uzunoglu E, Lortlar N, Erdogan D, Erdamar H, Akyol ES, Akata F. Immunohistochemical and ultrastructural presentation of apoptosis and aqueous humor's nitric oxide levels in keratoconus. Nobel Medicus. (2013) 9:5–9.
219. Kaldawy RM, Wagner J, Ching S, Seigel GM. Evidence of apoptotic cell death in keratoconus. Cornea. (2002) 21:206–9. doi: 10.1097/00003226-200203000-00017
220. Rodrigues M, Nirankari V, Rajagopalan S, Jones K, Funderburgh J. Clinical and histopathologic changes in the host cornea after epikeratoplasty for keratoconus. Am J Ophthalmol. (1992) 114:161–70. doi: 10.1016/S0002-9394(14)73980-7
221. McMonnies CW. Epigenetic mechanisms might help explain environmental contributions to the pathogenesis of keratoconus. Eye Contact Lens. (2014) 40:371–5. doi: 10.1097/ICL.0000000000000078
222. Hu W, Han Q, Zhao L, Wang L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-beta1. J Cell Physiol. (2019) 234:1469–76. doi: 10.1002/jcp.26959
223. Wen ZJ, Xin H, Wang YC, Liu HW, Gao YY, Zhang YF. Emerging roles of circRNAs in the pathological process of myocardial infarction. Mol Ther Nucleic Acids. (2021) 26:828–48. doi: 10.1016/j.omtn.2021.10.002
224. Zhang Y, Jia DD, Zhang YF, Cheng MD, Zhu WX Li PF, et al. The emerging function and clinical significance of circRNAs in Thyroid Cancer and Autoimmune Thyroid Diseases. Int J Biol Sci. (2021) 17:1731–41. doi: 10.7150/ijbs.55381
Keywords: keratoconus, candidate genes, multi-level combined analysis, gene enrichment, pathogenesis
Citation: Hao X-D, Gao H, Xu W-H, Shan C, Liu Y, Zhou Z-X, Wang K and Li P-F (2022) Systematically Displaying the Pathogenesis of Keratoconus via Multi-Level Related Gene Enrichment-Based Review. Front. Med. 8:770138. doi: 10.3389/fmed.2021.770138
Received: 03 September 2021; Accepted: 31 December 2021;
Published: 24 January 2022.
Edited by:
Michele Lanza, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Dimitrios Karamichos, University of North Texas Health Science Center, United StatesCopyright © 2022 Hao, Gao, Xu, Shan, Liu, Zhou, Wang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Dan Hao, aGFveGlhb2RhbjE5ODdAMTYzLmNvbQ==; Kun Wang, d2FuZ2s2OTZAcWR1LmVkdS5jbg==; Pei-Feng Li, cGVpZmxpQHFkdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.