
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 13 January 2022
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.759648
This article is part of the Research Topic "Source-tracking”, molecular epidemiology and antigenic diversity of SARS-CoV-2 infections causing coronavirus disease 2019, COVID-19. View all 37 articles
 Herbert F. Jelinek1,2,3
Herbert F. Jelinek1,2,3 Mira Mousa1,4
Mira Mousa1,4 Nawal Alkaabi5
Nawal Alkaabi5 Eman Alefishat1,6
Eman Alefishat1,6 Gihan Daw Elbait1
Gihan Daw Elbait1 Hussein Kannout1
Hussein Kannout1 Hiba AlHumaidan7
Hiba AlHumaidan7 Francis Amirtharaj Selvaraj7
Francis Amirtharaj Selvaraj7 Hala Imambaccus7
Hala Imambaccus7 Stefan Weber7
Stefan Weber7 Maimunah Uddin5
Maimunah Uddin5 Fatema Abdulkarim8
Fatema Abdulkarim8 Bassam Mahboub8
Bassam Mahboub8 Guan Tay1,9,10
Guan Tay1,9,10 Habiba Alsafar1,2,11* on behalf of the UAE COVID-19 Collaborative Partnership
Habiba Alsafar1,2,11* on behalf of the UAE COVID-19 Collaborative PartnershipIntroduction: Coronavirus disease 2019 (COVID-19) disease severity differs widely due to numerous factors including ABO gene-derived susceptibility or resistance. The objective of this study was to investigate the association of the ABO blood group and genetic variations of the ABO gene with COVID-19 severity in a heterogeneous hospital population sample from the United Arab Emirates, with the use of an epidemiological and candidate gene approach from a genome-wide association study (GWAS).
Methods: In this cross-sectional study, a total of 646 participants who tested positive for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) were recruited from multiple hospitals and population-based (quarantine camps) recruitment sites from March 2020 to February 2021. The participants were divided into two groups based on the severity of COVID-19: noncritical (n = 453) and critical [intensive care unit (ICU) patients] (n = 193), as per the COVID-19 Reporting and Data System (CO-RADS) classification. The multivariate logistic regression analysis demonstrated the association of ABO blood type as well as circulating anti-A antibodies and anti-B antibodies as well as A and B antigens, in association with critical COVID-19 hospital presentation. A candidate gene analysis approach was conducted from a GWAS where we examined 240 single nucleotide polymorphisms (SNPs) (position in chr9: 136125788-136150617) in the ABO gene, in association with critical COVID-19 hospital presentation.
Results: Patients with blood group O [odds ratio (OR): 0.51 (0.33, 0.79); p = 0.003] were less likely to develop critical COVID-19 symptoms. Eight alleles have been identified to be associated with a protective effect of blood group O in ABO 3'untranslated region (UTR): rs199969472 (p = 0.0052), rs34266669 (p = 0.0052), rs76700116 (p = 0.0052), rs7849280 (p = 0.0052), rs34039247 (p = 0.0104), rs10901251 (p = 0.0165), rs9411475 (p = 0.0377), and rs13291798 (p = 0.0377).
Conclusion: Our findings suggest that there are novel allelic variants that link genetic variants of the ABO gene and ABO blood groups contributing to the reduced risk of critical COVID-19 disease. This study is the first study to combine genetic and serological evidence of the involvement of the ABO blood groups and the ABO gene allelic associations with COVID-19 severity within the Middle Eastern population.
Coronavirus disease (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread rapidly worldwide (1). Disease severity differs widely due to numerous factors including SARS-CoV-2 variants and viral load, presence of chronic disease comorbidity, genetic factors, and age among others as well as oxidative stress and inflammation (2–6). The ABO gene-derived susceptibility or resistance and ABO blood group antigens have also been associated to COVID-19 disease severity (7). Age has been well described as a risk factor with the 0–4 years age group being 4 times less likely and the 85+ age group being 13 times more likely to be hospitalized compared to the 18–29 age groups (3). Comorbidities including obesity, diabetes, cardiovascular disease, chronic kidney disease, hypertension, tuberculosis, respiratory disease, inflammatory disease, coagulation dysfunction, and cancer have been reported to play a role in disease severity (4). Timing to hospital admission, challenges on health systems and expertise in healthcare delivery are also associated with the risk of increased progression in noncommunicable and communicable diseases including progression to severe COVID-19 presentation requiring hospitalization (8).
The ABO blood typing and the ABO gene variant analysis have been a central component of the immune response in transplantation and transfusion medicine, but has also been of importance in the immune response and progression of viral infections (9). The ABO blood system consists of different molecular determinants leading to different antigenic structures that play varying roles in the immune response. The A and B alleles encode different glycosyltransferases that add N-acetylgalactosamine and D-galactose to the common precursor H determinant side chain, which converts to A or B antigen. As O alleles do not encode a functional enzyme due to a premature stop codon within the glycosyltransferase gene yielding a truncated gene, it results in a complete loss of enzymatic activity and unchanged H-antigen (10, 11).
The genetic basis of the human ABO blood groups is located on chromosome 9 (9q34.1) and is associated with the synthesis of specific ABO glycosyltransferases (12). The ABO blood groups have been included in disease risk and severity of COVID-19, although with mixed results (13). The first link between coronavirus infection and ABO was reported in 2005 following the SARS-CoV-2 pandemic (14). These authors reported that health professionals with type O blood group were less likely to be infected compared to those with other blood types. More recent studies on SARS-CoV-2 reported similar associations between viral infection and the ABO blood groups (15, 16). However, divergent results are still coming forth with respect to the role of the ABO blood groups as a risk marker for COVID-19 severity linked to possible comorbidities and methodological differences as well as population differences in tissue receptors associated with SARS-CoV-2 infection (4, 13, 17–19).
Determining any link between the ABO blood groups and disease is complicated due to the highly polymorphic nature of the ABO blood group system and, hence, more often related to ABO phenotype rather than ABO genotype (20, 21). The risk of disease associated with a specific blood group has been shown for coronary artery disease, ischemic stroke, cancer, dementia, hypertension, hyperlipidemia, and diabetes (22–25). The ABO blood groups modulate both the hemostasis and endothelial function, leading to interactions with inflammatory cells and a positive association to metabolic and cardiovascular conditions (26, 27). Therefore, when investigating the role of ABO serology and association with COVID-19 severity, the role of ABO blood groups in nonviral pathology is an important factor because severe COVID-19 is characterized as an inflammatory state that damages the alveolar capillary barrier and compromises gas exchange, leading to intracapillary thrombosis and endothelial dysfunction. This is further highlighted by recent findings of increased morbidity and mortality of patients with COVID-19 with comorbidities (28).
Similar to the link between the risk of nonviral diseases, the ABO blood groups have been associated with diverse bacterial and viral infections. Several enteric organisms, including Vibrio cholera are linked to the risk of disease and disease severity, where individuals with blood group O have a more severe disease after infection (29). Other infections include mumps, tuberculosis, plague, malaria, and norovirus among others (20, 21, 30, 31). Type O blood has been linked to cholera bacteria and greater susceptibility to infection by Helicobacter pylori, plague, and mumps compared to people with either the A or B variants (22, 32, 33). Blood group A increases the risk of smallpox, while blood group B is associated with an increased risk of tuberculosis, Streptococcus pneumoniae, Escherichia coli, and Salmonella infections (21). Viral infections and association with the ABO blood groups have been reported for influenza, picornaviruses, hepatitis B virus, norovirus, and HIV infection (34–38). The association between the ABO blood group and infectious diseases may be due to modifications of a key target cell surface glycoprotein or glycolipid, thereby affecting important cellular functions, such as endocytosis, phagocytosis, and signal transduction, in response to infection (16).
Coto et al. conducted a large meta-analysis investigating the susceptibility or protection of the ABO blood groups associated with SARS-CoV-2 of over 30,000 cases. This study was not able to determine a link between the ABO blood group and severity or mortality associated with COVID-19 (39). Although a 45% higher risk of developing COVID-19, if the A blood type was present and a 35% lower risk with type O blood reported by others (2). More recent studies of available data have drawn similar conclusions highlighting the necessity to account for potential confounders (16).
The gene locus for the human ABO blood groups is located on chromosome 9 (9q34.1) and is associated with the synthesis of specific ABO glycosyltransferases (12). The ABO glycosyltransferase gene also presents with common polymorphisms including a one nucleotide deletion in exon 6 (codon 87) determining the O allele and four single nucleotide polymorphisms (SNPs) at residues 176, 235, 266, and 268, which switch enzyme function from A transferase to B transferase activity (12, 40). Preliminary results from the 23andMe consortium of genome sequencing have indicated a protective effect of group O and reported that the rs505922 SNP in the ABO gene with a T substitution at that location is associated with a lower risk of SARS-CoV-2 infection. Similar findings were reported for individuals with the type O blood group who were less likely to be infected, whereas those individuals with the type A blood group were more likely to be infected (13, 41). Genome data from Europe and Australia confirmed these findings. In COVID-19 genome-wide association study (GWAS), the authors studied a cohort in Italy and Spain, suggesting a correlation between the ABO blood groups and SARS-CoV-2 susceptibility (2). This cohort included 835 patients with severe COVID-19 who were hospitalized with respiratory failure and 1,255 control participants from Italy and 775 patients and 950 control participants from Spain. The final analysis showed significant associations with rs657152 at locus 9q34.2, which is in almost complete linkage disequilibrium (LD) with rs8176719 of the ABO blood group locus, the main determinant of the group O, allele ABO*O.01.01 (2). In contrast, a Danish study reported no association of rs657152 with COVID-19 infection or outcome (42). Data from other studies participating in the COVID-19 Host Genetics Initiative combined with the data from COVID-19 GWAS also found no association with the ABO blood group locus (43).
Therefore, further investigation and study are warranted to clarify the relationship between COVID-19 and the ABO gene variants, especially with respect to regional differences. Khalil et al. (44) analyzing the ABO blood group associations with COVID-19 in a Middle East and North African cohort could not corroborate previous studies that indicated a link between the ABO blood groups and susceptibility to or severity of SARS-CoV-2 infection. Only a limited number of genetic studies have addressed COVID-19 severity and comparing critical to noncritical in hospital patients with conflicting results and not with respect to ABO gene polymorphisms (15, 45, 46). One study provided results for single polymorphism and COVID-19 infection (2). Therefore, the aim of this study is to ascertain whether the specific ABO blood group is associated with COVID-19 severity and identify allelic variants on the ABO gene that is associated with disease severity in a heterogeneous population sample from the United Arab Emirates (UAE).
Patients with COVID-19 were recruited from multiple recruitment sites (hospitals and facility quarantine sites) across the UAE. Only patients who tested positive for SARS-CoV-2 by real-time PCR (RT-PCR) were included in this study cohort. The participants were divided into two groups based on the severity of COVID-19, which was indicated by the treating physician as noncritical (n = 453) or critical (n = 193) based on the COVID-19 Reporting and Data System (CO-RADS) classification (47). In brief, participants were defined as critical COVID-19 cases, if they are admitted to the intensive care unit (ICU) with the use of oxygen supplementation or mechanical ventilation. An informed written consent form was obtained in accordance with the Declaration of Helsinki. This study was approved by the Abu Dhabi Health COVID-19 Research Ethics Committee (DOH/DQD/2020/538), the Dubai Scientific Research Ethics Committee (DSREC-04/2020_09), and the SEHA Research Ethics Committee (SEHA-IRB-005). Consent was obtained from a family member of the patients who were on ventilators, with the approval of a supervising physician.
A total of 2 ml blood samples of 646 patients with COVID-19 were collected in a sterile 5-ml sample tube supplemented with ethylenediaminetetraacetic acid (EDTA) from the cubital vein by experienced venipuncture nurses. Samples were transported in a sealed biohazard bag using a cool transport container to Khalifa University Center for Biotechnology in Abu Dhabi. COVID-19 infection was confirmed by a RT-PCR test of SARS-CoV-2.
Deoxyribonucleic acid was extracted using the automated MagPurix 12 system according to the protocol of the manufacturer. DNA was quantified using the DS-11 Series of Spectrophotometer/Fluorometer (DeNovix, Wilmington, Delaware, USA). Genotyping was performed using the Infinium Global Screening Array (Illumina Incorporation, San Diego, California, USA), which contained around 654,027 genetic markers and developed by Avera Institute for Huma Genetics (Sioux Falls, South Dakota, USA). Quality control (QC) on the data was performed using the PLINK software (version 1.07) to exclude SNPs with a low minor allele frequency (<0.01), low genotyping rate (<95%), and deviation from Hardy–Weinberg equilibrium (p < 10−4) significance level. Samples that failed to reach 98.5% call rate were reanalyzed. After quality control, 417,263 variants passed filters. Participant genotype data were phased and imputed using the Phase 3 1000 Genomes Projects panel (https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html). Variants with low imputation quality (r2 < 0.5 averaged across the batch) were removed. A total of 240 SNPs (position in chr9: 136,125,788–136,150,617) in the ABO gene located at chromosome 9q34.2 were extracted for the association of this study for candidate gene analyses.
The statistical analysis was conducted as a case–control panel, with controls characterized as noncritical symptoms and cases characterized as critical symptoms, with the use of the PLINK software (version 1.9), R software (version 3.4), and the SPSS software (version 16.0). The chi-squared test was used to study categorical variables by cross-tabulation. The bivariate and multivariate logistic regression analyses were used to estimate the odds ratio (OR) and p-values of the association between blood type and COVID-19 severity phenotypes. All the regression models accounted for gender (male and female), age (continuous), current body mass index (BMI) (continuous), and presence of comorbidities (yes and no). Comorbidities were defined as a previous medical diagnosis of diabetes mellitus, hypertension, cardiac disease, lung disease, liver disease, kidney disease, metabolic disorder, and/or an autoimmune disease. For candidate gene analysis, we examined 240 SNPs (position in chr9: 136125788–136150617) in the ABO gene. Two candidate gene association tests were conducted that included unadjusted analysis and adjustment on the top ten eigenvectors for population stratification, age, and gender. We tested for association using logistic regression, assuming additive allelic effects. The significance level adopted for all the analyses was p ≤ 0.05. For the significant SNPs, we evaluated the frequency of genotype and alleles by stratifying based on the ABO blood group.
A total of 646 participants were included in the cohort, with 453 participants admitted to hospital for noncritical COVID-19 and 193 participants admitted for critical COVID-19 (Table 1). The average age was 44.70 years (SD ± 15.53) and average BMI was 28.11 kg/m2 (SD ± 15.53). Males (78.6%) and Asian nationality (57.0%) made up the majority of the overall population. The noncritical COVID-19 group was significantly younger (p < 0.001) and contained the lower BMI group (p = 0.001). Gender was not associated with a critical presentation of COVID-19 disease (p = 0.640).
Table 2 demonstrates the prevalence and logistic regression analysis of the ABO blood type characteristics of a subset of the participants (n = 527). The multivariate regression analysis adjusted for age, gender, BMI, and presence of comorbid conditions demonstrated that patients with a blood group B [OR: 0.97 (0.57, 1.65); p = 0.975] and blood group AB [OR: 0.61 (0.25, 1.46): p = 0.273] had no association with critical COVID-19. However, there was a protective effect for blood group O [OR: 0.51 (0.31, 0.84); p = 0.008] from developing critical COVID-19, which is consistent with previous findings. After adjustment, circulating anti-A antibodies and anti-B antibodies, as well as A and B antigens, in the plasma had no association to COVID-19 critical phenotype. Given that age and BMI play a significant role on COVID-19 severity, stratification of the blood group was conducted (Supplementary Table 1), demonstrating that age plays a significant role in COVID-19 severity.
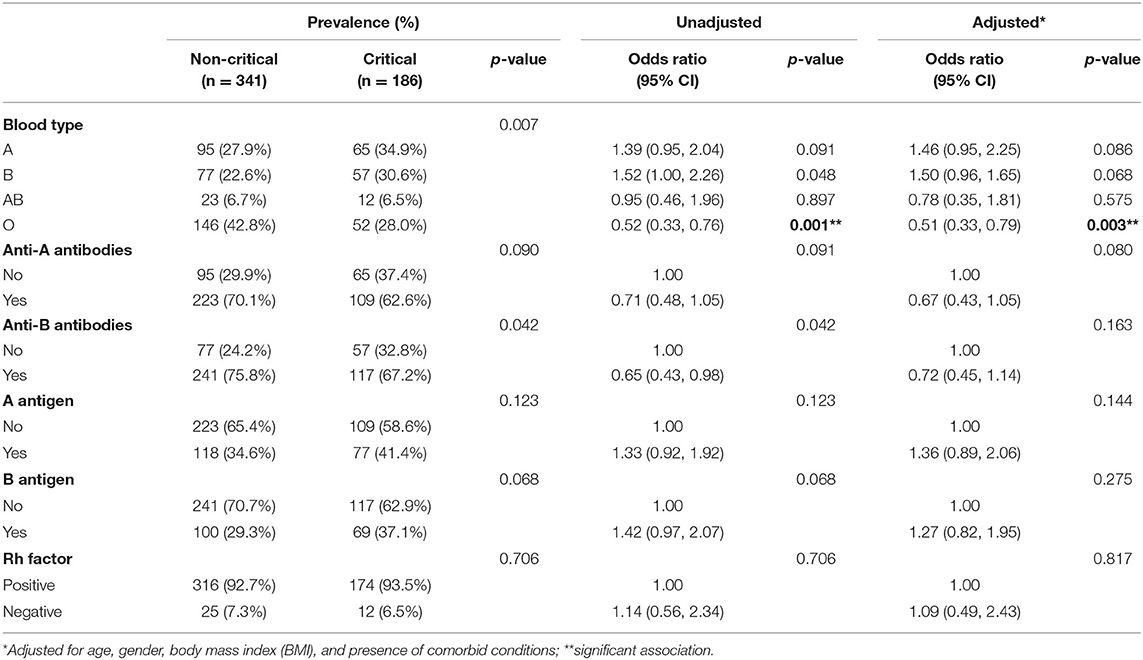
Table 2. Association of blood type characteristics to noncritical vs. critical coronavirus disease 2019 (COVID-19) hospital presentation.
To investigate if COVID-19 severity was associated with specific SNPs located on the ABO locus, all the 240 SNPs located in the ABO candidate gene were extracted from a GWAS and analyzed in this study. The ABO candidate gene consists of a 5′-untranslated region (UTR), seven exons, and 3′-UTR. Supplementary Table 1 demonstrates the unadjusted and adjusted OR and p-value of the SNPs at the ABO locus. As blood type frequencies vary across the ancestry groups, the confounding effect of ancestry was evaluated by adjusting for ethnicity. The first ten eigenvectors were used in subsequent analyses for adjustment to population stratification. Following these adjustments, eight SNPs were identified on the ABO gene locus that was associated with COVID-19 severity, all located in the 3'-UTR region (Table 3).
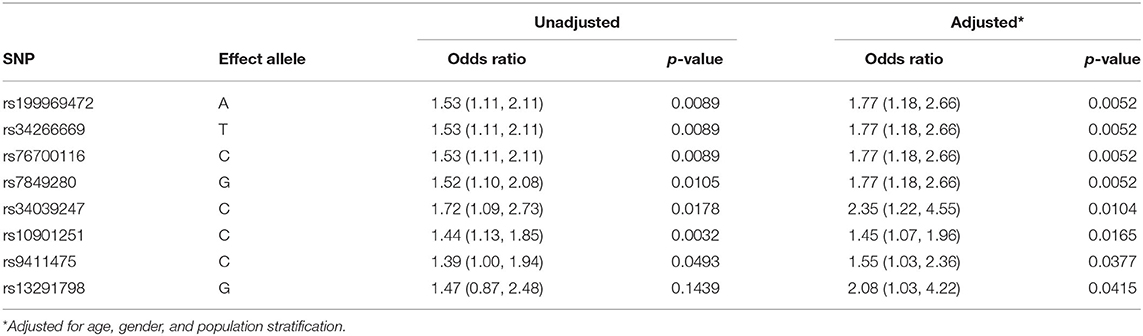
Table 3. Unadjusted and adjusted effect size and p-value of the eight significant single nucleotide polymorphisms (SNPs) in chromosome 9 on the ABO gene.
An upregulation of the following SNPs, located in the same haplotype block (D′ = 1.0; as given in Figure 1), is associated with the critical COVID-19 phenotype: rs199969472 [OR: 1.77 (95% CI: 1.18, 2.66); p = 0.0052], rs10901251 [OR: 1.45 (95% CI: 1.07, 1.96), p = 0.0165], rs34266669 [OR: 1.77 (95% CI: 1.18, 2.66); p = 0.0052], rs76700116 [OR: 1.77 (95% CI: 1.18, 2.66); p = 0.0052], and rs7849280 [OR: 1.77 (95% CI: 1.18, 2.66); p = 0.0052]. Haplotype analyses (Figure 1 and Table 4) of the 5 SNPs were performed to estimate the genetic contribution of haplotypes to critical COVID-19 phenotype, demonstrating a significant association to two haplotype blocks (ACGAA, p = 0.0028; CTACG, p = 0.0079). SNP rs34039247 [OR: 2.35 (95% CI: 1.22, 4.55); p = 0.0104], SNP rs9411475 [OR: 1.55 (95% CI: 1.03, 2.36); p = 0.0377], and SNP rs13291798 [OR: 1.55 (1.03, 2.36); p = 0.0377] have also been identified to be associated with critical COVID-19 phenotype.
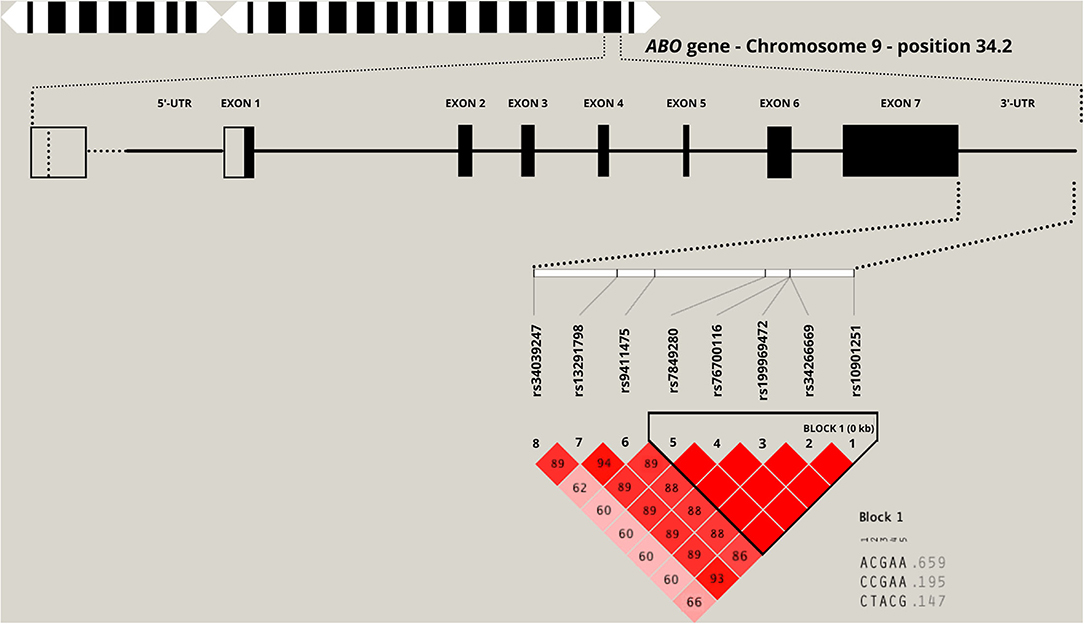
Figure 1. Location of the top eight significant single nucleotide polymorphisms (SNPs) in the 3-untranslated region (3'-UTR) of the ABO candidate gene. The numbers inside each square and the square shading indicate the degree of linkage disequilibrium (LD) between SNPs (complete LD, D′ = 1.0 and no LD, D′ = 0.0).
To determine which of these alleles contribute to the protective effect found for blood group O in the current cohort, the genotype and allele frequencies of the eight significant SNPs were stratified according to blood group (Table 5). The effect alleles of the eight SNPs associated with higher risk of COVID-19 severity (Table 3) had a significantly lower frequency in patients with blood group O in noncritical vs. critical patients. Of interest is that this is the first report of an allele being identified that is associated with a protective effect of blood group O. For the SNP rs19996947, the A allele was found to be present in 28.8% of critical cases vs. the G allele, which was present in 71.2% of the cases, indicating a significant association (p = 0.002).
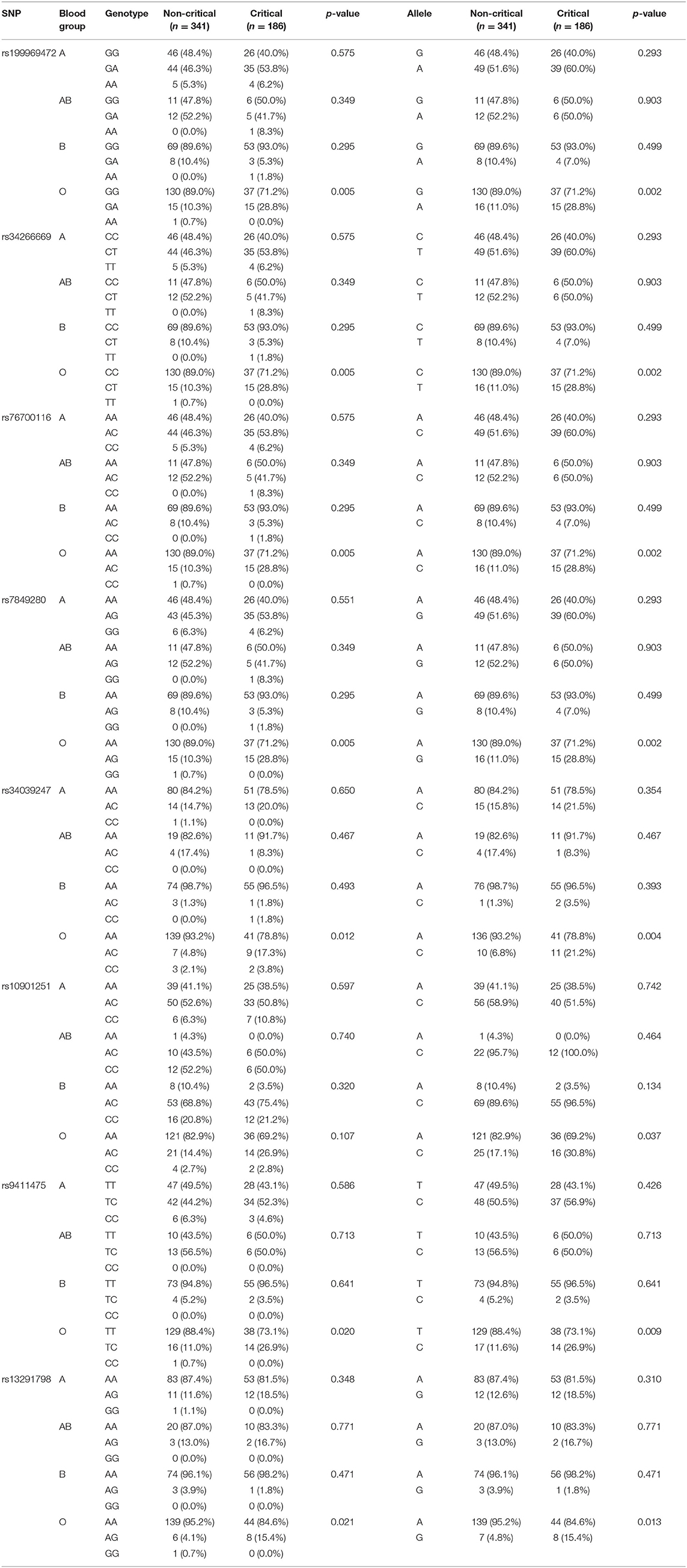
Table 5. Genotype and allelic stratification of eight significant SNPs in chromosome 9 on the ABO gene into the ABO blood group.
Five SNPs are in moderate-strong LD with the SNP associated with the blood type O allele (rs8176719; rs199969472, D′ = 0.62; rs10901251, D′ = 0.53; rs76700116, D′ = 0.62; rs7849280, D′ = 0.62; rs9411475, D′ = 0.55). Only two SNPs are in moderate-strong LD with the SNP associated with the blood type A allele (rs8176746; rs10901251, D′ = 0.66; rs34039247, D′ = 0.96) and blood type B allele (rs8176747; rs10901251, D′ = 0.66; rs34039247, D′ = 0.96). Although our data did not demonstrate significant association to the blood type O allele (rs8176719), the SNPs in moderate and strong LD was associated with a significantly reduced risk of critical phenotype presentation with COVID-19.
This study is the first study to combine genetic and serological evidence of the involvement of the ABO blood groups and ABO gene allelic associations with COVID-19 severity within a Middle Eastern population investigated in the UAE, a country that is characterized by the convergence of multiple cultural and socioeconomic factors.
Patients with blood group O [OR: 0.51 (0.31, 0.84); p = 0.008] were less likely to present with critical COVID-19 to the hospitals included in this study, which is consistent with previous findings. Studies have demonstrated that the adhesion of S protein and angiotensin-converting enzyme 2 (ACE2), a receptor for SARS-CoV-2, can be inhibited by anti-A natural antibody (11). Therefore, the anti-A and anti-B natural antibodies produced by patients with blood group O may decrease the risk of developing critical symptoms by blocking the interaction with ACE2 receptor and prevent viral entry into the lung epithelium. Our data strengthen the evidence for a role for ABO in COVID-19 susceptibility and severity and is notable given the reported links between COVID-19 and blood clotting complications that has now been associated with C5a activity (48).
With respect to the ABO locus, eight SNPs showed an association signal in a candidate gene level and play a role in dysregulating the antibody, natural killer cell, and immune mediator profiles: rs199969472 (p = 0.0052), rs34266669 (p = 0.0052), rs76700116 (p = 0.0052), rs7849280 (p = 0.0052), rs34039247 (p = 0.0104), rs10901251 (p = 0.0165), rs9411475 (p = 0.0377), and rs13291798 (p = 0.0377). Stratification of the SNPs with respect to the ABO blood group indicated significant associations between blood group O and the allelic polymorphism that provided protection to the current cohort for critical presentation of COVID-19. All the SNPs that have showed an association signal are located at 3'-UTR, which play an important role in the ABO gene expression and transcriptional signaling in association with COVID-19 symptom presentation (49).
The eight SNPs identified in this study, however, did not include rs657152 and rs8176719, which have been previously observed to be associated with COVID-19 (2). Several reasons can be proposed for this finding including a yet unidentified interaction between different genome variants with minor allele frequencies below 5% and very weak effects (50). Geographical distribution of ancestral risk alleles and population characteristics such as consanguinity, environmental pressures, and incorrect reporting of phenotype may also have contributed to the differences to previous observed results (51). However, the differences in association between the ABO blood group and SNPs may also be explained by the study of Valenti et al. (48) who reported that the association between impact of rs11385949 and the ABO blood group on complement activation disappeared at 30 days after admission during remission of symptoms in surviving patients.
Conflicting findings are present in studies that have investigated the association of the ABO blood group and COVID-19 susceptibility and severity (16, 41, 52–58). Most systematic reviews and meta-analysis studies have demonstrated a decreased risk in susceptibility and severity among patients with COVID-19 in O group (10, 13, 28, 54, 59), whereas other reviews and manuscripts demonstrated no significant association (39, 53, 60–62). These differences may be due to ascertainment bias, multiple confounding effects, diverse studies populations and their geographics locations, presence and adjustment to comorbidities, and case and control selection criteria. These factors may lead to inaccurate estimation of the relative risk. Therefore, appropriately designed observational case–control studies would be more suited to minimize bias and investigate the association of the ABO blood group and COVID-19 severity. In addition, given that the population ancestries in Asian and Middle Eastern countries are relatively homogeneous, relative frequencies of ABO phenotypes may affect the findings of this study. In this study, even with the adjustment of stratification for ancestry, 8 SNPs were identified on the ABO gene locus to be significantly associated with COVID-19 severity, with a lower frequency (protective factor) of the effect allele in patients with blood group O.
Direct susceptibility or resistance of disease may be a function of disease severity, as shown by a meta-analysis reported by Pourali et al. (28). In addition, host protein levels may play a role in disease severity in terms of virulence, as blood group O is associated with lower ACE levels and the converse has been found for blood group A (27). Associated with this finding is the binding probability of SARS-CoV-2. ACE2, being the main receptor, binds to cells expressing blood group moieties most reported in the mucous membrane of the respiratory tract. Thus, blood group AB has the most contact and blood group O the least contact with SARS-CoV-2 (27, 63). This is further demonstrated in this study, where patients with blood type O were less likely to present to hospital with critical COVID-19. The association between COVID-19 severity and the ABO group may be due to the development of neutralizing antibodies against N-linked glycans that extensively cover SARS-CoV-2 spike protein or due to the stabilization of the von Willebrand factor including a one nucleotide deletion in exon 6 (codon 87) determining O allele. Four SNPs at residues 176, 235, 266, and 268, which switch enzyme function from A transferase to B transferase activity, may also contribute to the current findings (12, 40, 64).
Blood groups are also linked to several other diseases, including cardiovascular diseases and pulmonary thromboembolism, which have been identified as one of the main complications in COVID-19 disease progression (21, 65). The ABO blood type trait reflects polymorphisms within the ABO gene. The ABO gene is associated with a number of other traits including risk factors for COVID-19 morbidity and mortality. The genetic locus located within the ABO gene plays a role in hemoglobin concentration, hematocrit (66, 67), von Willebrand factor (68), myocardial infection (69), coronary artery disease (70), ischemic stroke (71), type 2 diabetes (72), and venous thromboembolism (73). In fact, one study recently reported an overall 24% cumulative incidence of pulmonary embolism in patients with COVID-19 pneumonia, of which 50% were the ICU patients (74, 75). Hence, this study does not limit itself to identify ABO links with COVID-19, but investigated comorbidities reported in a population sample that were admitted to hospitals in the UAE with confirmed SARS-CoV-2 infection. However, a similar finding has been demonstrated in this study, where specific risk alleles of SNPs were upregulated in the critical COVID-19 group.
Potential limitations of this study also need to be considered. Due to the pandemic associated lockdown over several months, we were unable to collect data on a large number of patients. Selection bias is a fundamental limitation of this study, so the association estimates are conditional on presentation to the hospital. Nonetheless, the methodology of the cross-sectional study minimized additional selection bias, with respect to the outcome of interest. Due to the geographical location of the Middle East region, there is substantial genetic admixture present within the population. However, to limit population stratification, 10 eigenvectors were adjusted for. An important limitation was the use of a GWAS arrays that were manufactured based on the Caucasian population and this limits the possibility of including targeted SNPs in the genome. To limit this error, imputation of genotypes for genetic variants that are untyped in the arrays increases the information provided by each microarray by accurately evaluating the evidence for association at genetic markers that are not directly genotyped. However, the imputed SNPs from previous studies did not reach significance suggesting that there may be specific population characteristics associated with COVID-19 severity. The inclusion of multiple data collection sites from across the country may have included cases that were not entirely homogeneous. It is possible that the criteria for hospitalization of patients with COVID-19 are different across the collection sites, thus measurement errors may exist in this study. Misclassification and ascertainment biases in the control group were limited by only selecting patients with COVID positive with the noncritical or critical cases.
There is much to be done to understand the role of ABO and its association to COVID-19 severe phenotypes. The combination of genetic and serological evidence of the involvement of the ABO blood groups and ABO gene allelic associations with COVID-19 severity provides a unique opportunity to study host genomics to the interindividual phenotypic variability. By conducting a GWAS and extracting the SNPs in the ABO gene, we have provided further insight into the genetic mechanisms associated with COVID-19 disease severity and the possible link to allelic variants and COVID-19 critical phenotypes. Future study will address linking genomic data to electronic health records that can be leveraged to improve clinical management and lead to better patient outcomes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
This study was approved by the Abu Dhabi Health COVID-19 Research Ethics Committee (DOH/DQD/2020/538), Dubai Scientific Research Ethics Committee (DSREC-04/2020_09), and SEHA Research Ethics Committee (SEHA-IRB-005). The patients/participants provided their written informed consent to participate in this study.
Juan Acuna, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates; Eman Alefishat, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates; Ernesto Damiani, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates; Samuel F. Feng, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates; Andreas Henschel, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates; Abdulrahim Sajini, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates; Ahmed Yousef, Khalifa University of Science and Technology, Abu Dhabi, United Arab Emirates; Bassam Ali, United Arab Emirates University, Al Ain, United Arab Emirates; Hiba Alhumaidan, Sheikh Khalifa Medical City and SEHA, Abu Dhabi; Hala Imambabaccus, Sheikh Khalifa Medical City and SEHA, Abu Dhabi, United Arab Emirates; Amirtharaj Francis, Sheikh Khalifa Medical City and SEHA, Abu Dhabi, United Arab Emirates; Stefan Weber, Sheikh Khalifa Medical City and SEHA, Abu Dhabi, United Arab Emirates; Mohammad Tahseen Al Bataineh, University of Sharjah, Sharjah, United Arab Emirates; Rabih Halwani, University of Sharjah, Sharjah, United Arab Emirates; Rifat Akram Hamoudi, University of Sharjah, Sharjah, United Arab Emirates; Abdulmajeed Al Khajeh, Dubai Health Authority, Dubai, United Arab Emirates; Laila Salameh, Dubai Health Authority, Dubai, United Arab Emirates.
HAls and GT conceived the project to study the role of the virus and host in COVID-19 in the United Arab Emirates. HJ and HAlH conceived the central research questions for the ABO data and initiated the first draft of the manuscript. MM, GD, and HJ analyzed and constructed the figures. BM, FA, MU, and NA were responsible for the recruitment of the patients and collecting data for the study. HJ, GT, EA, and HAls provided critical review during manuscript preparation. All authors on the primary list contributed to the data interpretation and critically reviewed the manuscript and approved the final manuscript for submission.
This study was commissioned as part of a project to study the host cell receptors of coronaviruses funded by Khalifa University's CPRA grant (Reference Number 2020-004). The project was funded by internal funds provided by Khalifa University awarded to HAls.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the participants of this study for their generosity in providing samples to advance our understanding of COVID-19 infection. We acknowledge the assistance of the healthcare workers at the frontline of COVID-19 pandemic, specially Ms. Laila Salameh, Dr. Amna Saeed, Dr. Mawada Hussein, Dr. Nimmi Kurien, Dr. Mona Elhassan, Dr. Eman Elzain, Dr. Salma Rahma, Dr. Sarrosh Khan, Dr. Omar Al-Assaf, Dr. Muneeba Moin, Dr. Hend Alsaidi, Dr. Rufaida Osman, Dr. Khalid Hassan, and Dr. Afrah Joher without their assistance, this study would not have been possible. We are also grateful to Hema Vurivi, who assisted in processing the samples in the laboratory.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.759648/full#supplementary-material
1. Team EE. Latest updates on COVID-19 from the European Centre for Disease Prevention and Control. Eurosurveillance. (2020) 25:2002131. doi: 10.2807/1560-7917.ES.2020.25.6.2002131
2. Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, et al. Genomewide association study of severe Covid-19 with respiratory failure. NEJM. (2020) 383:1522–34. doi: 10.1056/NEJMoa2020283
3. Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin. Immunol. (2020) 146:110–8. doi: 10.1016/j.jaci.2020.04.006
4. Wolff D, Nee S, Hickey NS, Marschollek M. Risk factors for Covid-19 severity and fatality: a structured literature review. Infection. (2020) 1–14. doi: 10.1007/s15010-020-01509-1
5. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
6. Mousa M, Vurivi H, Kannout H, Uddin M, Alkaabi N, Mahboub B, et al. Genome-wide association study of hospitalized COVID-19 patients in the United Arab Emirates. EBioMedicine. (2021) 74:103695. doi: 10.1016/j.ebiom.2021.103695
7. Novaes RD, Teixeira AL, De Miranda AS. Oxidative stress in microbial diseases: pathogen, host, and therapeutics. Hindawi. (2019). doi: 10.1155/2019/8159562
8. Faes C, Abrams S, Van Beckhoven D, Meyfroidt G, Vlieghe E, Hens N. Time between symptom onset, hospitalisation and recovery or death: statistical analysis of Belgian COVID-19 patients. Int J Environ Res Public Health. (2020) 17:7560. doi: 10.3390/ijerph17207560
9. Landsteiner K. Ueber Agglutinationserscheinungen normalen menschlichen Blutes. Wien Klin Wschr. (1901) 14:1132.
10. Franchini M, Capra F, Targher G, Montagnana M, Lippi G. Relationship between ABO blood group and von Willebrand factor levels: from biology to clinical implications. Thromb J. (2007) 5:1–5. doi: 10.1186/1477-9560-5-14
11. Guillon P, Clément M, Sébille V, Rivain J-G, Chou C-F, Ruvoën-Clouet N, et al. Inhibition of the interaction between the SARS-CoV spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology. (2008) 18:1085–93. doi: 10.1093/glycob/cwn093
12. Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. (1990) 345:229–33. doi: 10.1038/345229a0
13. Wu B-B, Gu D-Z, Yu J-N, Yang J, Shen W-Q. Association between ABO blood groups and COVID-19 infection, severity and demise: A systematic review and meta-analysis. Infect Genet Evol. (2020) 84:104485–104485. doi: 10.1016/j.meegid.2020.104485
14. Cheng Y, Cheng G, Chui CH, Lau FY, Chan PK, Ng MH, et al. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA. (2005) 293:1450–1. doi: 10.1001/jama.293.12.1450-c
15. Mizuno-Matsumoto Y, Inoguchi Y, Carpels SMA, Muramatsu A, Yamamoto Y. Cerebral cortex and autonomic nervous system responses during emotional memory processing. PLoS ONE. (2020) 15:e0229890. doi: 10.1371/journal.pone.0229890
16. Goel R, Bloch EM, Pirenne F, Al-Riyami AZ, Crowe E, Dau L, et al. ABO blood group and COVID-19: a review on behalf of the ISBT COVID-19 working group. Vox Sang. (2021) 116:849–61. doi: 10.1111/vox.13076
17. Bajgain KT, Badal S, Bajgain BB, Santana MJ. Prevalence of comorbidities among individuals with COVID-19: A rapid review of current literature. Am J Infect Control. (2020) 49:238–46. doi: 10.1016/j.ajic.2020.06.213
18. Flegel, H. COVID-19: risk of infection is high, independently of ABO blood group. Haematologica. (2020) 105. doi: 10.3324/haematol.2020.266593
19. Latz CA, Decarlo C, Boitano L, Png CYM, Patell R, Conrad MF, et al. Blood type and outcomes in patients with COVID-19. Ann Hematol. (2020) 99:2113–8. doi: 10.1007/s00277-020-04169-1
20. Yamamoto F, Cid E, Yamamoto M, Blancher A. ABO research in the modern era of genomics. Transfus Med Rev. (2012) 26:103–18. doi: 10.1016/j.tmrv.2011.08.002
21. Ewald DR, Sumner SCJ. Blood type biochemistry and human disease. Wiley Interdiscip Rev Syst Biol Med. (2016) 8:517–35. doi: 10.1002/wsbm.1355
22. Garratty G. Blood groups and disease: a historical perspective. Transfus Med Rev. (2000) 14:291–301. doi: 10.1053/tmrv.2000.16228
23. Sari I, Ozer O, Davutoglu V, Gorgulu S, Eren M, Aksoy M. ABO blood group distribution and major cardiovascular risk factors in patients with acute myocardial infarction. Blood Coagul Fibrinolysis. (2008) 19:231–4. doi: 10.1097/MBC.0b013e3282f54522
24. Zakai NA, Judd SE, Alexander K, Mcclure LA, Kissela BM, Howard G, et al. ABO blood type and stroke risk: the REasons for Geographic And Racial Differences in Stroke Study. J Thromb Haemost. (2014) 12:564–70. doi: 10.1111/jth.12507
25. Fagherazzi G, Gusto G, Clavel-Chapelon F, Balkau B, Bonnet F. ABO and Rhesus blood groups and risk of type 2 diabetes: evidence from the large E3N cohort study. Diabetologia. (2015) 58:519–22. doi: 10.1007/s00125-014-3472-9
26. Abdelmassih AF, Mahrous R, Taha A, Saud A, Osman A, Kamel B, et al. The potential use of ABO blood group system for risk stratification of COVID-19. Med Hypotheses. (2020) 145:110343. doi: 10.1016/j.mehy.2020.110343
27. Dai X. ABO blood group predisposes to COVID-19 severity and cardiovascular diseases. Eur J Prev Cardiol. (2020) 27:1436–7. doi: 10.1177/2047487320922370
28. Pourali F, Afshari M, Alizadeh-Navaei R, Javidnia J, Moosazadeh M, Hessami A. Relationship between blood group and risk of infection and death in COVID-19: a live meta-analysis. New Microbes and New Infections. (2020) 37:100743. doi: 10.1016/j.nmni.2020.100743
29. Kuhlmann FM, Santhanam S, Kumar P, Luo Q, Ciorba MA, Fleckenstein JM. Blood group O-dependent cellular responses to cholera toxin: parallel clinical and epidemiological links to severe cholera. Am J Trop Med Hyg. (2016) 95:440–3. doi: 10.4269/ajtmh.16-0161
30. Moulds JM, Moulds JJ. Blood group associations with parasites, bacteria, and viruses. Transfus Med Rev. (2000) 14:302–11. doi: 10.1053/tmrv.2000.16227
31. Cooling L. Blood groups in infection and host susceptibility. Clin Microbiol Rev. (2015) 28:801–70. doi: 10.1128/CMR.00109-14
32. Barua D, Paguio AS. ABO blood groups and cholera. Ann Hum Biol. (1977) 4:489–92. doi: 10.1080/03014467700002481
33. Boren T, Falk P, Roth K, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. (1993) 262:1892–5. doi: 10.1126/science.8018146
34. Tyrrell DA, Sparrow P, Beare AS. Relation between blood groups and resistance to infection with influenza and spome picornaviruses. Nature. (1968) 220:819–20. doi: 10.1038/220819a0
35. Evans AS, Shepard DA, Richards VA. ABO blood groups and viral diseases. Yale J Biol Med. (1972) 45:81–92.
36. Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, Lindblad L, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. (2003) 9:548–53. doi: 10.1038/nm860
37. Siransy LK, Nanga ZY, Zaba FS, Tufa NY, Dasse SR. ABO/Rh blood groups and risk of HIV infection and Hepatitis B among blood donors of Abidjan, Côte D'ivoire. Eur J Microbiol Immunol. (2015) 5:205–9. doi: 10.1556/1886.2015.00029
38. Jing W, Zhao S, Liu J, Liu M. ABO blood groups and hepatitis B virus infection: a systematic review and meta-analysis. BMJ Open. (2020) 10:e034114–e034114. doi: 10.1136/bmjopen-2019-034114
39. Coto, E., Albaiceta, G. M., Clemente, M. G., and Gómez, J. (2020). Lack of association between SNPsrs8176719 (O blood group) and COVID-19: Data from Spanish age matched patients and controls. Transfusion, 10.1111/trf.16206. doi: 10.1111/trf.16206
40. Fry AE, Griffiths MJ, Auburn S, Diakite M, Forton JT, Green A, et al. Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum Mol Genet. (2007) 17:567–76. doi: 10.1093/hmg/ddm331
41. Zhao J, Yang Y, Huang H, Li D, Gu D, Lu X, et al. Relationship between the ABO Blood Group and the COVID-19 Susceptibility. medRxiv. (2020) 2020.2003.2011.20031096. doi: 10.1101/2020.03.11.20031096
42. Lehrer S, Rheinstein PH. ABO blood groups, COVID-19 infection and mortality. Blood Cells, Molecules, and Diseases. (2021) 89:102571. doi: 10.1016/j.bcmd.2021.102571
43. COVID-19 Host Genetics Initiative. The COVID-19 Host Genetics Initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. Eur J Hum Genet. (2020) 28:715–8. doi: 10.1038/s41431-020-0636-6
44. Khalil A, Feghali R, Hassoun M. The Lebanese COVID-19 Cohort; A challenge for the ABO blood group system. Front Med. (2020) 7:585341–585341. doi: 10.3389/fmed.2020.585341
45. Anastassopoulou C, Gkizarioti Z, Patrinos GP, Tsakris A. Human genetic factors associated with susceptibility to SARS-CoV-2 infection and COVID-19 disease severity. Hum Genomics. (2020) 14. doi: 10.1186/s40246-020-00290-4
46. ASM. American Society of Mammalogist. Chiroptera (ASM Mammal Diversity Database). (2020). Available online at: https://mammaldiversity.org/#Y2hpcm9wdGVyYSZnbG9iYWxfc2VhcmNoPXRydWUmbG9vc2U9dHJ1ZQ (accessed June 22, 2021).
47. Fujioka T, Takahashi M, Mori M, Tsuchiya J, Yamaga E, Horii T, et al. Evaluation of the usefulness of CO-RADS for chest CT in patients suspected of having COVID-19. Diagnostics. (2020) 10:608. doi: 10.3390/diagnostics10090608
48. Valenti L, Griffini S, Lamorte G, Grovetti E, Renteria SCU, Malvestiti F, et al. Chromosome 3 cluster rs11385942 variant links complement activation with severe COVID-19. J Autoimmun. (2021) 117:102595. doi: 10.1016/j.jaut.2021.102595
49. Heit JA, Armasu SM, Asmann YW, Cunningham JM, Matsumoto ME, Petterson TM, et al. A genome-wide association study of venous thromboembolism identifies risk variants in chromosomes 1q24. 2 and 9q. J Thromb Haemost. (2012) 10:1521–31. doi: 10.1111/j.1538-7836.2012.04810.x
50. Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. (2009) 461:747–53. doi: 10.1038/nature08494
51. Bergström A, Mccarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. (2020) 367. doi: 10.1126/science.aay5012
52. Focosi D, Carla IM, Lanza M. ABO blood group correlations with COVID-19: Cohort choice makes a difference. Clin Infect Dis. (2021) 72:e919–e919. doi: 10.1093/cid/ciaa1495
53. Gutiérrez-Valencia M, Leache L, Librero J, Jericó C, Enguita Germán M, García-Erce JA. ABO blood group and risk of COVID-19 infection and complications: A systematic review and meta-analysis. Transfusion. (2021) 10.1111/trf.16748. doi: 10.1111/trf.16748
54. Liu N, Zhang T, Ma L, Zhang H, Wang H, Wei W, et al. The impact of ABO blood group on COVID-19 infection risk and mortality: A systematic review and meta-analysis. Blood Rev. (2021) 48:100785. doi: 10.1016/j.blre.2020.100785
55. Muñiz-Diaz E, Llopis J, Parra R, Roig I, Ferrer G, Grifols J, et al. Relationship between the ABO blood group and COVID-19 susceptibility, severity and mortality in two cohorts of patients. Blood Transfusion. (2021) 19:54. doi: 10.2450/2020.0256-20
56. Pendu JL, Breiman A, Rocher J, Dion M, Ruvoën-Clouet N. ABO blood types and COVID-19: spurious, anecdotal, or truly important relationships? A reasoned review of available data. Viruses. (2021) 13:160. doi: 10.3390/v13020160
57. Ray JG, Schull MJ, Vermeulen MJ, Park AL. Association between ABO and Rh blood groups and SARS-CoV-2 infection or severe COVID-19 illness: a population-based cohort study. Ann Intern Med. (2021) 174:308–15. doi: 10.7326/M20-4511
58. Zhang Y, Garner R, Salehi S, La Rocca M, Duncan D. Association between ABO blood types and coronavirus disease 2019 (COVID-19), genetic associations, and underlying molecular mechanisms: a literature review of 23 studies. Ann Hematol. (2021) 100:1123-1132. doi: 10.1007/s00277-021-04489-w
59. Golinelli D, Boetto E, Maietti E, Fantini MP. The association between ABO blood group and SARS-CoV-2 infection: A meta-analysis. PLoS ONE. (2020) 15:e0239508. doi: 10.1371/journal.pone.0239508
60. Levi JE, Telles PR, Scrivani H, Oliveira F, Vieira L, Dias C, et al. Lack of association between ABO blood groups and susceptibility to SARS-CoV-2 infection. Hematol Transfus Cell Ther. (2020) 42:541. doi: 10.1016/j.htct.2020.10.914
61. Roberts GH, Park DS, Coignet MV, Mccurdy SR, Knight SC, Partha R, et al. AncestryDNA COVID-19 host genetic study identifies three novel loci. medRxiv. (2020) doi: 10.1101/2020.10.06.20205864
62. Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in Covid-19. Nature. (2021) 591:92–8. doi: 10.1038/s41586-020-03065-y
63. Mattos LCD, Moreira HW. Genetic of the ABO blood system and its link with the immune system. Rev Bras Hematol Hemoter. (2004) 26:60–3. doi: 10.1590/S1516-84842004000100012
64. Walls AC, Xiong X, Park Y-J, Tortorici MA, Snijder J, Quispe J, et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell. (2020) 183:1732. doi: 10.1016/j.cell.2020.11.031
65. Groot HE, Sierra LEV, Said MA, Lipsic E, Karper JC, Harst PVD. Genetically determined ABO blood group and its associations with health and disease. Arterioscler Thromb Vasc Biol. (2020) 40:830–8. doi: 10.1161/ATVBAHA.119.313658
66. Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. (2016) 167:1415–29. e1419. doi: 10.1016/j.cell.2016.10.042
67. Yasumizu Y, Sakaue S, Konuma T, Suzuki K, Matsuda K, Murakami Y, et al. Genome-wide natural selection signatures are linked to genetic risk of modern phenotypes in the Japanese population. Mol Biol Evol. (2020) 37:1306–16. doi: 10.1093/molbev/msaa005
68. Smith NL, Chen M-H, Dehghan A, Strachan DP, Basu S, Soranzo N, et al. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: The CHARGE Consortium. Circulation. (2010) 121:1382. doi: 10.1161/CIRCULATIONAHA.109.869156
69. Nikpay M, Goel A, Won H-H, Hall LM, Willenborg C, Kanoni S, et al. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nat Genet. (2015) 47:1121. doi: 10.1038/ng.3396
70. Dichgans M, Malik R, König IR, Rosand J, Clarke R, Gretarsdottir S, et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke. (2014) 45:24–36. doi: 10.1161/STROKEAHA.113.002707
71. Williams FM, Carter AM, Hysi PG, Surdulescu G, Hodgkiss D, Soranzo N, et al. Ischemic stroke is associated with the ABO locus: the EuroCLOT study. Ann Neurol. (2013) 73:16–31. doi: 10.1002/ana.23838
72. Scott RA, Scott LJ, Mägi R, Marullo L, Gaulton KJ, Kaakinen M, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. (2017) 66:2888–902. doi: 10.2337/db16-1253
73. Dentali F, Sironi AP, Ageno W, Turato S, Bonfanti C, Frattini F, et al. Non-O blood type is the commonest genetic risk factor for VTE: results from a meta-analysis of the literature. Semin Thromb Hemost. (2012) 38:535–48. doi: 10.1055/s-0032-1315758
74. Bompard F, Monnier H, Saab I, Tordjman M, Abdoul H, Fournier L, et al. Pulmonary embolism in patients with COVID-19 pneumonia. Eur Respir J. (2020) 56:2001365. doi: 10.1183/13993003.01365-2020
Keywords: ABO blood group, SARS-CoV-2, disease severity, UAE, COVID-19 pandemic, infection–immunology, Middle East
Citation: Jelinek HF, Mousa M, Alkaabi N, Alefishat E, Daw Elbait G, Kannout H, AlHumaidan H, Selvaraj FA, Imambaccus H, Weber S, Uddin M, Abdulkarim F, Mahboub B, Tay G and Alsafar H (2022) Allelic Variants Within the ABO Blood Group Phenotype Confer Protection Against Critical COVID-19 Hospital Presentation. Front. Med. 8:759648. doi: 10.3389/fmed.2021.759648
Received: 16 August 2021; Accepted: 13 December 2021;
Published: 13 January 2022.
Edited by:
Bathri Narayan Vajravelu, MCPHS University, United StatesReviewed by:
Paul Simonson, Weill Cornell Medical Center, United StatesCopyright © 2022 Jelinek, Mousa, Alkaabi, Alefishat, Daw Elbait, Kannout, AlHumaidan, Selvaraj, Imambaccus, Weber, Uddin, Abdulkarim, Mahboub, Tay and Alsafar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Habiba Alsafar, aGFiaWJhLmFsc2FmYXJAa3UuYWMuYWU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.