- 1Department of Cellular and Molecular Pathology, Stefan S. Nicolau Institute of Virology, Bucharest, Romania
- 2Faculty of Medicine, Titu Maiorescu University, Bucharest, Romania
Liquid biopsy represents an exciting new area in the field of cancer diagnosis and management, offering a less invasive and more convenient approach to obtain a time-point image of the tumor burden and its genomic profile. Samples collected from several body fluids, mostly blood, can be used to gain access to circulating tumor cells and DNA, non-coding RNAs, microRNAs, and exosomes, at any moment, offering a dynamic picture of the tumor. For patients with GC, the use of blood-based biopsies may be particularly beneficial since tissue biopsies are difficult to obtain and cause real distress to the patient. With advantages such as repeatability and minimal invasion, it is no wonder that the field of liquid biopsy has received tremendous attention. However, the abundance of studies, involving a wide range of assays with different principles, prevented for the moment the reproducibility of the results and therefore the translation into the clinic of liquid biopsy. In this review, we present the latest technical development and data on circulating biomarkers available through liquid biopsy in gastric cancer with an emphasis on their clinical utility in areas such as cancer screening, prognostic stratification, and therapeutic management.
Introduction
Gastric cancer (GC) is the fifth most common cancer worldwide, with 1.089.103 new cases being registered in 2020 (5.6%), after female breast cancer (11.7%), lung (11.4%), colorectal (10.0 %), and prostate (7.3%). Compared with other cancers, morbidity rates are quite high. With 768.793 (7.7%) deaths each year, GC represents the fourth cause of cancer-related death in the world (1). To overcome this burden, significant efforts are being made by clinicians and researchers around the world.
To date, several serum biomarkers have been identified and widely used for GC diagnosis and prognosis, including carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA19-9), cancer antigen 125 (CA125), and cancer antigen 72-4 (CA72-4). However, those biomarkers have low sensitivity (<40%), and their specificities are modest. Only when used as a triple marker, CEA, CA19-9, and CA72.4, the sensitivity was 62.0% (2). Moreover, they may offer a certain value only in determining peritoneal metastasis, estimating poor prognosis and a higher risk for recurrence of GC rather than in setting up a diagnosis (3). Preoperative levels of CEA, CA19-9, and CA125 were analyzed on 768 patients with GC and found above the cut-off levels in only 15.4, 8.7, and 5.7% of all cases, respectively (4). In a comprehensive meta-analysis of literature published on GC serum markers, the Japanese Gastric Cancer Association concluded that they are not useful for early cancer diagnosis, but they are useful for detecting recurrence and distant metastasis, predicting patient survival, and monitoring after surgery (5).
In this regard, liquid biopsy emerged as a promising tool for early detection, treatment selection, and real-time progression information. In the last decade, it became clear that closer monitoring of disease progression during second line and salvage chemotherapy proved to be extremely important for overall survival. In that aspect, novel technologies, like a liquid biopsy and new biomarkers offer guidance on the proper timing for therapy and impact assessment. Another major advantage of liquid biopsies consists in offering a rapid and precise, less invasive and more convenient approach to obtain cancer information. It may be particularly beneficial since biopsies are difficult to obtain and cause real distress to the patient.
Samples can be collected from several biological fluids such as blood, saliva, breast milk, cerebrospinal fluid, urine, gastric juice, semen, etc., but the most used and characterized type is blood (Figure 1).
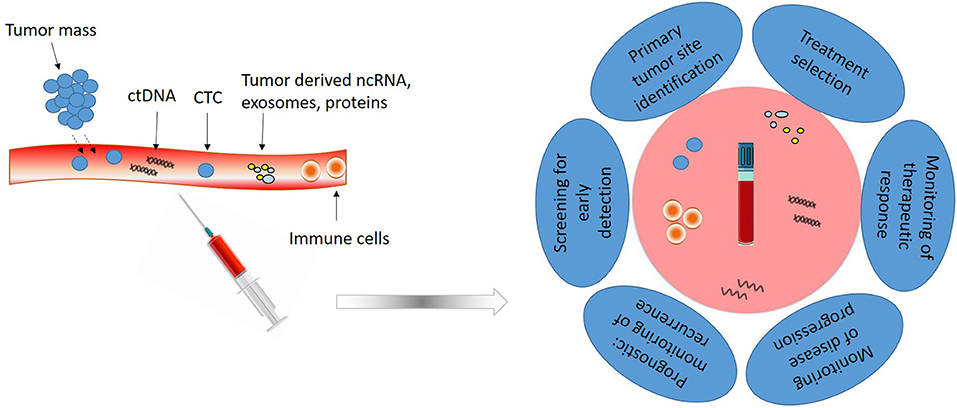
Figure 1. Liquid biopsy composition and utility. Liquid biopsy obtained from peripheral blood is composed of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), non-coding RNAs (ncRNAs) and different tumor derived proteins and vesicles. Analysis of these elements has clinical utility for early detection, treatment selection, real-time progression information and prognosis.
Nowadays, liquid biopsy cover for the investigation of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating non-coding RNAs (ncRNAs), microRNAs, and circulating exosomes, in the blood of patients with cancer. It can assess more accurately the tumor dynamics and heterogeneity, monitor treatment responses and potential emergence of drug resistance, determine the minimal residual disease and indicate a personalized cancer management strategy. Liquid biopsies provide access to CTCs, DNA, and ncRNA, at any moment, with minimal discomfort for the patients, offering a dynamic picture of the tumor. However, the technology to obtain enough material for monitoring disease progression and drug resistance is labor-intensive, must have high sensitivity and specificity, and should permit multiplex determination of panels of relevant markers.
As today, the FDA has approved several technologies for monitoring cancer patients using liquid biopsy: CELLSEARCH CTC test for quantification of circulating tumor cells from Veridex, Guardant360 CDx and FoundationOne Liquid CDx, both using circulating cell-free DNA and next-generation sequencing (NGS) technology to provide tumor mutation profiling. These platforms have received clearance for malignancies such as metastatic lung, breast, prostate, or colorectal cancer. For GC clinical management, they have not yet been introduced in daily practice, although a great benefit for patients and clinicians could be anticipated, by overcoming limitations of traditional tissue biopsies.
Circulating Tumor Cells (CTCs)
CTCs are cells derived from the primary tumor that infiltrate into the bloodstream, and from there on to other body fluids. CTCs are extremely rare events, with about one to 10 cells in 10 ml of peripheral blood (6). Since they represent the phenotypic and genotypic image of the primary tumor, identification and analysis of CTCs emerged as a promising assay to screen for early carcinogenesis, and to monitor cancer progression and treatment effectiveness in a real-time manner. CTCs are currently approved by the FDA as a prognostic biomarker and used in monitoring cancer evolution in patients with breast, prostate, and colorectal cancer (7). In GC, CTCs are very rare and difficult to identify, the best method of detection and specific markers remaining controversial.
Currently used methods for CTC isolation include a primary step of enrichment consisting on depletion of red and white blood cells known as negative selection, and a positive selection based on their biological and physical properties, with subsequent identification supported by immunological, molecular, and functional markers. There are several enrichment technologies known, such as Cell Search, Adna Test, CTC-chip, MACS, or MagSweeper, all of them using epithelial cell adhesion molecule (EpCAM) as a surface marker for enrichment (8). The main issue is that during the metastasis process, involving migration, intravasation, circulation into the bloodstream, and extravasation with new organ colonization, CTCs undergo epithelial to mesenchymal transition (EMT) and lose their epithelial markers becoming more difficult to identify. However, with EMT comes the overexpression of extracellular matrix (ECM) proteins such as vimentin, fibronectin, twist, ZEB1, ZEB2, snail, slug, and N-cadherin (9, 10). These could help more accurately identify CTCs if they are included in the existing enrichment technologies.
Other methodologies for the enrichment of CTCs are based on physical properties: size (isolation by size of epithelial tumor cells ISET) or density (OncoQuick, RosetteSep), but these can result in losses in the number of isolated CTCs.
After enrichment, follows the CTC identification step which is performed by immunocytological methods (detection of other specific surface antigens like cytokeratins (CK), 8, 18, 19, 20, CD44), molecular (amplification by qRT-PCR of specific CTC markers like surviving, CKs, VEGFR), or functional [epithelial immunospot EPISPOT that detects protein secretion (e.g., CK19)]. Of these methods, the one that has the highest specificity in identifying CTC is molecular testing, with the limitation related to the fact that cell viability is lost.
To date, CellSearch System (Veridex) was approved by FDA for clinical use in breast, colorectal, and prostate cancers (11–13). The system immunomagnetically enriches CTCs expressing EpCAM from 7.5 mL of blood using ferrofluids beads coated with anti-EpCAM antibodies and then selects fluorescently cells based on DAPI, CD45-APC, and 8, 18 and 19 CK-PE. Other system for positive selection of CTC is cytokeratin (CK)-dependent immunomagnetic separation system from Miltenyi based on anti-pan CK antibody that recognizes CK 7, 8, 18, and 19.
Clinical Utility of CTCs
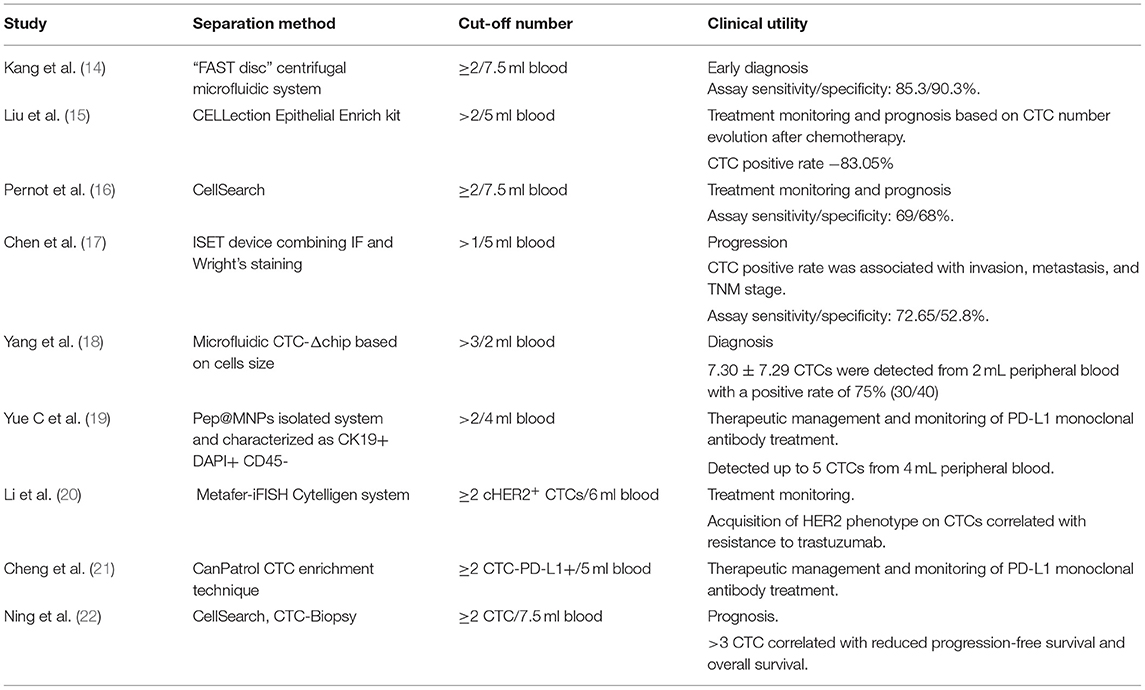
Several studies have shown that CTCs have significant values in the diagnosis, prognosis, and treatment management of GC at various stages (Table 1).
CTCs detection has been suggested to be a useful biomarker for diagnosis. In a study conducted by Kang HM el al. using a CTC level of ≥2 per 7.5 mL of blood, the authors successfully differentiate patients with GC from healthy controls. Sensitivity and specificity were 85.3 and 90.3%, respectively (14). In addition, CTCs were identified in more than 80% of early-stage (T1 or N0) cases, suggesting that CTCs could be an early diagnostic biomarker for GC with high sensitivity and specificity.
As bloodstream circulating cells detached from the primary tumor, CTCs are significantly involved in cancer metastasis and recurrence. As a result, it is not surprising that many studies have shown that the presence of CTC is associated with advanced stages, poor survival, and progression-free survival in GC (23). In a meta-analysis performed on 3,814 GC patients, Gao Y et al. concludes that the presence of CTCs identifies a group of patients with poor overall survival (OS) (CTCs: HR = 1.84, 95% CI 1.50–2.26, p < 0.001) (24).
In addition to diagnosis and prediction of prognosis, several studies reported that monitoring changes in the number of CTCs during treatment may be a predictive marker of treatment response. Li et al. (25) showed that high CTCs numbers (≥3) during treatment and post-therapy correlates with ineffective therapeutic response and reduced PFS and OS. In another study (26), on 100 patients with metastatic GC, was found that a CTCs number ≥5 during palliative chemotherapy is associated with limited response to the treatment and poor prognosis. Similar results were obtained in another study (27), in which quantification of CTCs in patients with advanced GC at 2 or 4 weeks after initiation of the treatment (S-1-based, with or without cisplatin, or base on paclitaxel) demonstrated that patients with ≥4 CTCs at 4 weeks had 3.5 months inferior median OS than the patient with <4 CTCs.
Besides CTC counting, molecular and phenotypic characterization of CTCs may provide additional information on the mechanism of therapy resistance and recurrence risk in patients with GC. This date is sustained by Szczepanik et al. (28), who demonstrated that only a subpopulation of CTCs expressing cytokeratins (8, 18, and 19) and CD44 marker (CK+CD44+) are significantly associated with distant metastases and with reduced survival.
New biomarkers such as the HER2 and PD-L1 positivity or amplification on CTCs are nowadays considered useful in predicting a positive response to trastuzumab and checkpoint therapy. For instance, HER2-positive CTCs were detected in nonmetastatic gastric adenocarcinoma patients with disease progression but HER2-negative primary tumors. Mishima et al. (29) demonstrated that a re-evaluation of HER-2 status on CTCs in advanced GC patients (n = 15) whose primary tumors were HER2-, but HER2 positive CTCs, may have clinical implications, allowing a better selection of patients for trastuzumab therapies (29, 30).
Moreover, two studies based on immune-checkpoints detection on CTC PD-L1 demonstrated that the abundance of PD-L1-positive CTCs compared with baseline might be used to screen patients with GC that will most likely benefit from PD-1/PD-L1 blockade therapies (19, 21). Moreover, dynamic changes of PD-L1 positive CTC count might be used to monitor treatment response as a non-invasive strategy.
Circulating Tumor DNA (ctDNA)
ctDNA originates as a result of different physiological events like apoptosis, necroptosis, tumor secretion, and micro-metastasis, and can be detected in different body fluids like blood serum or plasma, synovial fluid, cerebrospinal fluid, etc. In cancer patients, circulating tumor DNA (ctDNA) can have up to four- or five-times greater levels than circulating free DNA (cfDNA) in healthy controls and harbor tumor-associated molecular alterations (31). It offers the possibility to explore alterations in tumor DNA at the genetic and epigenetic level through assays such as copy number variations (CNVs), gene integrity, gene mutations, DNA methylation, and therefore proved to be essential to study tumor genomics distinguishing molecular subtypes, screening of EBV-associated cancers, treatment selection based on detected mutations in ctDNA, and to follow tumor progression and clonal evolution of tumors, at regular bases without difficulties in obtaining serial biopsies (32, 33).
Nevertheless, there are several limitations in performing ctDNA analyses: the detection and quantification of ctDNA are limited by its instability and kinetics, and also ctDNA concentration is highly variable, being very low especially in early stage cancers, and requires highly sensitive detection methods. The detection of ctDNA is facilitated if the primary tumor genetic abnormalities are already known.
Currently developed methods for ctDNA-based assays are varying widely between detection of a single-point mutation and the analysis of the entire genome; techniques such as PCR or PCR-based assays (qPCR, ddPCR, ARMS, MS-PCR, etc.) or targeted sequencing are used. Nowadays, novel high throughput sequencing techniques such as NGS or WES can identify mutations in multiple genetic regions (34, 35). Thus, two platforms based on NGS have already been approved by the FDA for the analysis of genomic profile in cancer patients, by liquid biopsy. Guardant360 CDx received FDA approval for clinical use in August 2020. The analysis facilitates the detection of single nucleotide variants (SNVs), insertions and deletions (indels) in 55 genes, copy number amplifications (CNAs) in two genes, and fusions in four genes. The panel is indicated for the clinical management of patients with non-small cell long cancer (NSCL) (36, 37). Also in August 2020, FDA has approved the FoundationOne Liquid CDx, which uses NGS technology optimized for cfDNA to investigate >300 genes. It detects major types of genomic alterations, microsatellite instability, blood tumor mutational burden, and tumor fraction values. FoundationOne Liquid CDx system was validated on several types of solid cancers, including mostly NSCLC, prostate, ovarian and breast cancer samples. The assay performance studies were conducted on cfDNA isolated from plasma of cancer patients representing 37 cancer types, including gastric cancer (38).
Clinical Utility of ctDNA
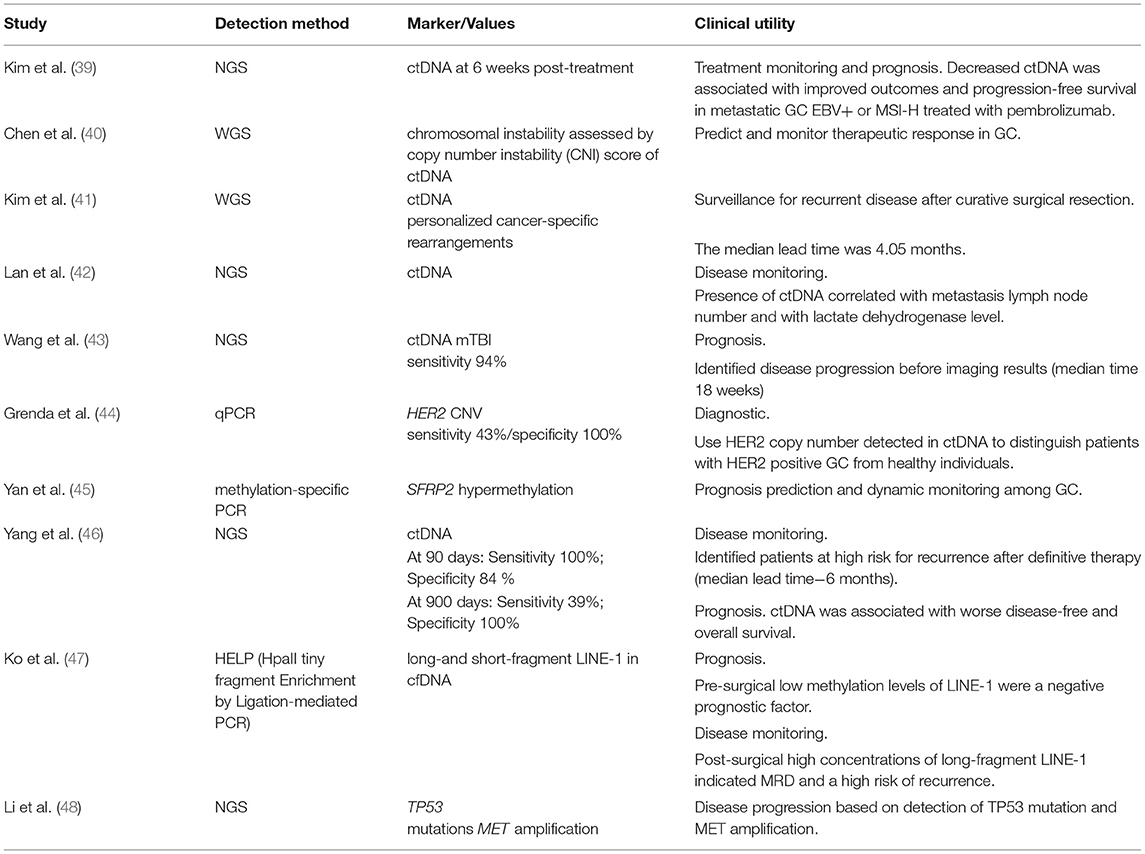
The analysis of ctDNA can be used in the detection of different genetic and epigenetic alterations, microsatellite instability, deletion, amplification, chromosome translocation, and loss of heterozygosity. The results of some recent studies in the GC field being reported in Table 2.
Usually, ctDNA can be detected before therapy and disappears after complete surgical resection. Its presence after or its reappearance at follow-up indicates minimal residual disease (MRD), which is a cause of recurrence (41, 42, 46, 47, 49, 50).
The analysis of ctDNA can be used to guide treatment decisions and evaluate clinical response. Thus, Zhang M et al. evaluated the ctDNA genomic profile of Chinese advanced GC patients by NGS and determined genomic alterations (like del-alterations in TP53, LRP1B, MYC, ERBB2, and KRAS genes), blood tumor mutation burden, and blood microsatellite instability status that can advise the clinical decision in advanced GC (51). Analyzing the profile of ctDNA genetic alterations in a cohort of 46 patients with GC, Iqbal M et al.established that only a few genes are altered, in the top 11 being TP53, KRAS, PIK3CA, ARID1A, EGFR, APC, ERBB2/HER2, CDK6, MET, PTEN, and MYC, presenting single nucleotide variations, CNV, and indels (32).
Kim ST et al.used Guardant360 ctDNA NGS assay to assess tumor mutational load and to determine the microsatellite instability (MSI) status in pretreatment tissue aiming to identify patients with metastatic gastric cancer who are most likely to benefit from pembrolizumab treatment. In addition, patients were followed with a serial collection of plasma-derived ctDNA. The obtained results showed a decrease in ctDNA level at six weeks post-treatment, which predicted response to immune chechpoint therapy and was associated with improved outcomes and progression-free survival (39).
ctDNA can be used for HER2-targeted population screening, since there is a high concordance of HER2 amplification between ctDNA and tumor tissues (52, 53). Also, HER2 CNV detected in ctDNA could be used to monitor trastuzumab efficacy (52, 54, 55), as well as to predict innate trastuzumab resistance (54) and the developed one (43).
In the VIKTORY trial (NCT 02299648), high MET copy number in ctDNA correlated with response to savolitinib, an experimental small-molecule inhibitor of c-Met. The concordance rate between tumor and ctDNA for MET amplification was 89.5%, with 100% specificity and 83.3% sensitivity relative to tissue testing, which increased to 100% when patients without detectable ctDNA were excluded (56). Du J et al.also reported the potential of ctDNA profiling for treatment decision and prognosis in a case of a stage IV GC patient with high levels of MET amplification.
Periodic mutation profiling of ctDNA by NGS revealed the appearance of several genetic alterations including re-occurrence of MET amplification, multiple secondary MET mutations, a dramatic increase of FGFR2 gene relative copy number as well as mutations in other downstream and bypassing elements that were correlated with the patient's cancer progression, transient response, and resistance development to crizotinib treatment, a receptor tyrosine kinase inhibitor (57). Furthermore, the utility of ctDNA monitoring in resistance development during treatment and progression was reported by Frigault MM et al. who identified the mechanisms of acquired resistance to savolitinib in three patients with GC and MET-amplified tumors that showed a clinical response followed by cancer progression (58).
Circulating Non-coding RNAs (ncRNAs)
The development of RNAseq techniques highlighted the circulating transcriptome, including coding and non-coding RNA, as an important source of potential diagnostic, prognostic, or predictive cancer biomarkers (59). Aberrant expression of circulating non-coding RNAs (ncRNAs), mostly microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), has been reported in several cancers, including GC, where these molecules seem to act as tumor suppressors or oncogenes, mediating important intracellular processes. Therefore, many studies suggested that an increased/decreased of ncRNA expression in blood could guide the therapeutic approach being associated with disease grade, malignant progression, or treatment response (60).
ncRNAs are usually secreted in the blood, urine, or other body fluids as a result of cell necrosis, apoptosis, or due to an active secretion of the tumor cells and these molecules are stable enough to be manipulated since they are protected by exosomes and microvesicles. Therefore, analyzing the expression of circulating ncRNAs could bring important information about the expression profile of the primary tumor (61).
miRNAs could be considered promising GC biomarkers since many studies reported an aberrant expression of these molecules in gastric tissue in preneoplastic events, such as Helicobacter pylori infection, chronic gastritis, preneoplastic conditions (atrophic gastritis, intestinal metaplasia), and also in early and advanced cancer (62). Circulating lncRNAs expression was also correlated with diagnosis, prognosis, and therapy monitoring of GC patients (60, 63).
Clinical Utility of Non-coding RNAs (ncRNAs)
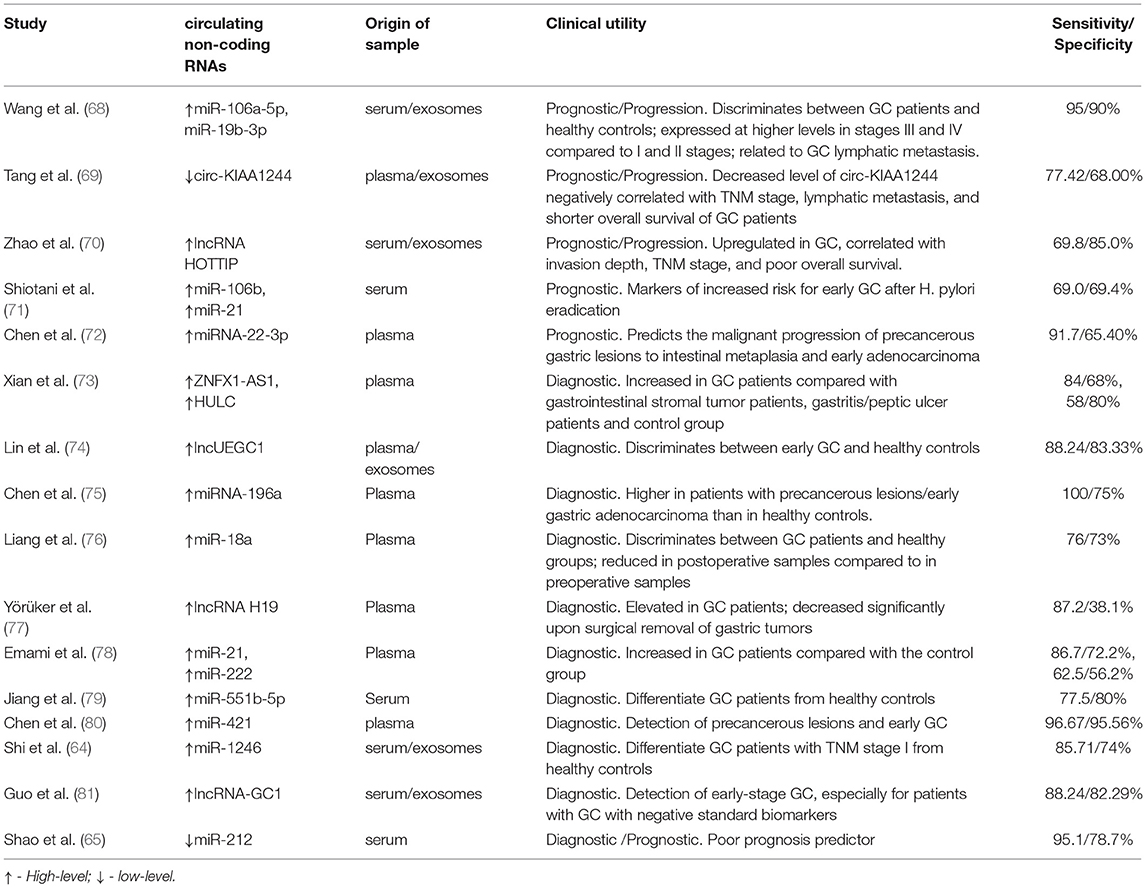
A recent study showed that an increased expression of miR-1246 in serum could distinguish between GC patients with TNM stage I, healthy controls, and patients with benign diseases, highlighting the utility of this molecule as a biomarker for early diagnosis of GC. This elevated serum level of miR-1246 seems to be tumor-derived and it is packaged into exosomes (64). miR-212, a molecule that is epigenetically downregulated in GC showed a decreased expression in serum of GC patients compared with healthy donors, being negatively correlated with tumor stages (65). Low serum levels of miR-203 are associated with lymph node, peritoneal, and distant metastases in GC patients (66) while a high level of miR-21-5p could be detected in urine samples of GC patients and this level is reduced after the surgical removal of the tumors (67).
In Table 3 are presented several ncRNAs that are found to be differentially expressed in GC patients and are associated with prognostic and therapy monitoring. However, more studies are needed to validate a panel of ncRNA biomarkers for GC.
Exosomes
Exosomes are small membrane vesicles of endocytic origin, with a diameter between 30 and 150 nm. They are secreted by cells into the environment in response to various physiological or pathological processes (82). The majority of the circulating transcriptome secreted by tumor cells is assisted by exosomes that protect cargos to travel along with the bloodstream. Through them, tumor cells can alter the microenvironment, influence the anti-tumor immune response, and modulate invasion and angiogenesis (83, 84). As a result, these regulatory properties of tumor cell-derived exosomes are essential in promoting tumor growth. Once released into the bloodstream or tumor microenvironment, the exosomes can interact with adjacent cells producing varied biological effects: direct exosome-cell stimulation or via transferred exosome cargo (85).
Due to their endosomal origin, they can be identified based on surface proteins such as CD81, CD62, and CD9 (86). In addition, they may have cell-specific or tumor-associated antigens on the surface reflecting the identity of the cells they come from (67, 87, 88).
This specific identity of the exomes makes them important candidates as reliable biomarkers. So, exosomes are macrovesicles rich in transcripts specific to tumors (89) that could be used as a biomarker of cancer diagnosis, progression, and metastasis.
It was shown that based on their composition GC exomes promote proliferation in an autocrine manner (90) or by activation of MAPK/ERK pathways (91) and PI3K/Akt (91). Exosomes are involved in mesothelial invasion and tumor dissemination within the abdominal wall and diaphragm (92), inducing apoptosis and mesothelial-to-mesenchymal transition (MMT), with mesothelial barrier destruction and peritoneal fibrosis (93) by enhancing the expression of fibronectin 1 and laminin gamma 1 (94).
GC-derived exosomes are involved in the modulation of tumor immunity. Thus, circulating exosomal PD-L1 was shown to be an independent prognostic factor in GC, associated with the immunosuppressive status of GC patients and decrease in CD4+ T cell count, CD8+ T-cell count, and granzyme B (95). Moreover, exosomal PD-L1 significantly decreased T-cell surface CD69 by increasing PD-1+ tumor-associated macrophages (TAMs), which by interacting with PD-L1+ cells can increase IL-10 production and CD8+ T cell dysfunction (96).
We can conclude that exosomal cargos are implicated in GC progression and modulation of tumor immunity via cellular communication and interactions in the tumor microenvironment.
Conclusions
There is currently an abundance of work both in terms of clinical studies on different types of biomarkers and as a study methodology used to track these biomarkers in liquid biopsies. So much diversity in assays and principles of work has made liquid biopsy difficult to translate to the clinic, mostly due to the lack of reproducibility.
Before introducing liquid biopsies in current clinical use for surveillance, the methodologies should be standardized to ensure reproducibility, and proper controls should be developed and used. Quantification of CTCs, ctDNA, ncRNAs, cell heterogeneity analysis, and molecular modifications resulted from liquid biopsies assessment should be included in designing surveillance protocols based on international guidelines outlined from consistent evidence.
In this regard, an international consortium The International Liquid Biopsy Standardization Alliance (ILSA) was founded in December 2020 (97), with the role of promoting liquid biopsy. The organization aims to standardize work protocols, implement them and evaluate their performance and clinical utility.
However, several biomarkers obtained by liquid biopsy already proved their usefulness. Thus, CTC cut-off number (≥2), HER2 and PD-L1 expression on their surface showed to have clinical utility in therapeutic management, allowing a better selection of patients for specific therapies like trastuzumab or immune-checkpoints inhibitors. CtDNA offers the possibility to explore alterations in tumor DNA at the genetic and epigenetic level through assays such as CNVs, gene integrity, gene mutations, DNA methylation, and therefore proved to be essential for distinguishing tumor subtypes, and guide personalized therapy. Regarding ncRNA, a lot of studies highlighted their utility in early diagnosis and surveillance of cancer progression but it is difficult to select keys ncRNAs from a large number of candidates, however, their combination may shape-up the basis for the development of an early diagnostic or prognostic panel.
Author Contributions
MC-E wrote the circulating tumor cells section and reviewed and edited the manuscript. LN wrote the circulating non-coding RNAs section. LM wrote the circulating tumor DNA section. CB wrote on the exosomes. DD wrote on the role of liquid biopsy in monitoring treatment response and CD wrote its role in surveillance for recurrent disease. All authors approved the submitted version.
Funding
This work was supported by Grants of the Ministry of Research, Innovation and Digitization, CNCS/CCCDI – UEFISCDI, Project Number PN-III-P4-ID-PCCF-2016-0158 (contract PCCF17/2018), within PNCDI III and Project Number TE 36/2020 (PN-III-P1-1.1-TE-2019-1864) within PNCDI III.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Li Y, Yang Y, Lu M, Shen L. Predictive value of serum CEA, CA19-9 and CA72. 4 in early diagnosis of recurrence after radical resection of gastric cancer. Hepatogastroenterology. (2011) 58:2166–70. doi: 10.5754/hge11753
3. Kim JH, Jun KH, Jung H, Park IS, Chin HM. Prognostic value of preoperative serum levels of five tumor markers (carcinoembryonic antigen, CA19-9, Alpha-fetoprotein, CA72-4, and CA125) in gastric cancer. Hepatogastroenterology. (2014) 61:863–9
4. Hwang GI, Yoo CH, Sohn BH, Shin JH, Park YL, Kim HD, et al. Predictive value of preoperative serum CEA, CA19-9 and CA125 levels for peritoneal metastasis in patients with gastric carcinoma. Cancer Res Treat. (2004) 36:178–81. doi: 10.4143/crt.2004.36.3.178
5. Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: a systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. (2014) 17:26–33. doi: 10.1007/s10120-013-0259-5
6. Ross AA, Cooper BW, Lazarus HM, Mackay W, Moss TJ, Ciobanu N, et al. Detection and viability of tumor cells in peripheral blood stem cell collections from breast cancer patients using immunocytochemical and clonogenic assay techniques. Blood. (1993) 82:2605–10. doi: 10.1182/blood.V82.9.2605.2605
7. Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. (2010) 16:398–406. doi: 10.1016/j.molmed.2010.07.001
8. Nakamura K, Iwatsuki M, Kurashige J, Ishimoto T, Baba Y, Miyamoto Y, et al. Circulating tumor cells in gastric cancer. J Cancer Metastasis and Treatment. (2018) 4:32. doi: 10.20517/2394-4722.2017.78
9. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. (2009) 119:1420–8. doi: 10.1172/JCI39104
10. Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. (2011) 9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490
11. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. (2004) 351:781–91. doi: 10.1056/NEJMoa040766
12. Negin BP, Cohen SJ. Circulating tumor cells in colorectal cancer: past, present, and future challenges. Curr Treat Options Oncol. (2010) 11:1–13. doi: 10.1007/s11864-010-0115-3
13. Resel Folkersma L, Olivier Gomez C, San Jose Manso L, Veganzones de, Castro S, Galante Romo I, Vidaurreta Lazaro M, et al. Immunomagnetic quantification of circulating tumoral cells in patients with prostate cancer: clinical and pathological correlation. Arch Esp Urol. (2010) 63:23–31. doi: 10.4321/S0004-06142010000100004
14. Kang HM, Kim GH, Jeon HK, Kim DH, Jeon TY, Park DY, et al. Circulating tumor cells detected by lab-on-a-disc: Role in early diagnosis of gastric cancer. PLoS ONE. (2017) 12:e0180251. doi: 10.1371/journal.pone.0180251
15. Liu Y, Ling Y, Qi Q, Lan F, Zhu M, Zhang Y, et al. Prognostic value of circulating tumor cells in advanced gastric cancer patients receiving chemotherapy. Mol Clin Oncol. (2017) 6:235–42. doi: 10.3892/mco.2017.1125
16. Pernot S, Badoual C, Terme M, Castan F, Cazes A, Bouche O, et al. Dynamic evaluation of circulating tumour cells in patients with advanced gastric and oesogastric junction adenocarcinoma: Prognostic value and early assessment of therapeutic effects. Eur J Cancer. (2017) 79:15–22. doi: 10.1016/j.ejca.2017.03.036
17. Chen F, Wang S, Fang Y, Zheng L, Zhi X, Cheng B, et al. Feasibility of a novel one-stop ISET device to capture CTCs and its clinical application. Oncotarget. (2017) 8:3029–41. doi: 10.18632/oncotarget.13823
18. Yang C, Zhang N, Wang S, Shi D, Zhang C, Liu K, et al. Wedge-shaped microfluidic chip for circulating tumor cells isolation and its clinical significance in gastric cancer. J Transl Med. (2018) 16:139. doi: 10.1186/s12967-018-1521-8
19. Yue C, Jiang Y, Li P, Wang Y, Xue J, Li N, et al. Dynamic change of PD-L1 expression on circulating tumor cells in advanced solid tumor patients undergoing PD-1 blockade therapy. Oncoimmunology. (2018) 7:e1438111. doi: 10.1080/2162402X.2018.1438111
20. Li Y, Zhang X, Liu D, Gong J, Wang DD Li S, et al. Evolutionary expression of HER2 conferred by chromosome aneuploidy on circulating gastric cancer cells contributes to developing targeted and chemotherapeutic resistance. Clin Cancer Res. (2018) 24:5261–71. doi: 10.1158/1078-0432.CCR-18-1205
21. Cheng B, Tong G, Wu X, Cai W, Li Z, Tong Z, et al. Enumeration and characterization of circulating tumor cells and its application in advanced gastric cancer. Onco Targets Ther. (2019) 12:7887–96. doi: 10.2147/OTT.S223222
22. Ning D, Cui K, Liu M, Ou Y, Wang Z, Zou B, et al. Comparison of CellSearch and Circulating Tumor Cells (CTC)-biopsy systems in detecting peripheral blood circulating tumor cells in patients with gastric cancer. Med Sci Monit. (2021) 27:e926565. doi: 10.12659/MSM.926565
23. Wang S, Zheng G, Cheng B, Chen F, Wang Z, Chen Y, et al. Circulating tumor cells (CTCs) detected by RT-PCR and its prognostic role in gastric cancer: a meta-analysis of published literature. PLoS ONE. (2014) 9:e99259. doi: 10.1371/journal.pone.0099259
24. Gao Y, Xi H, Wei B, Cui J, Zhang K, Li H, et al. Association between liquid biopsy and prognosis of gastric cancer patients: a systematic review and meta-analysis. Front Oncol. (2019) 9:1222. doi: 10.3389/fonc.2019.01222
25. Li Y, Gong J, Zhang Q, Lu Z, Gao J, Li Y, et al. Dynamic monitoring of circulating tumour cells to evaluate therapeutic efficacy in advanced gastric cancer. Br J Cancer. (2016) 114:138–45. doi: 10.1038/bjc.2015.417
26. Lee SJ, Lee J, Kim ST, Park SH, Park JO, Park YS, et al. Circulating tumor cells are predictive of poor response to chemotherapy in metastatic gastric cancer. Int J Biol Markers. (2015) 30:e382–6. doi: 10.5301/jbm.5000151
27. Matsusaka S, Chin K, Ogura M, Suenaga M, Shinozaki E, Mishima Y, et al. Circulating tumor cells as a surrogate marker for determining response to chemotherapy in patients with advanced gastric cancer. Cancer Sci. (2010) 101:1067–71. doi: 10.1111/j.1349-7006.2010.01492.x
28. Szczepanik A, Sierzega M, Drabik G, Pituch-Noworolska A, Kolodziejczyk P, Zembala M. CD44(+) cytokeratin-positive tumor cells in blood and bone marrow are associated with poor prognosis of patients with gastric cancer. Gastric Cancer. (2019) 22:264–72. doi: 10.1007/s10120-018-0858-2
29. Mishima Y, Matsusaka S, Chin K, Mikuniya M, Minowa S, Takayama T, et al. Detection of HER2 amplification in circulating tumor cells of her2-negative gastric cancer patients. Target Oncol. (2017) 12:341–51. doi: 10.1007/s11523-017-0493-6
30. Abdallah EA, Braun AC, Flores B, Senda L, Urvanegia AC, Calsavara V, et al. The potential clinical implications of circulating tumor cells and circulating tumor microemboli in gastric cancer. Oncologist. (2019) 24:e854–e63. doi: 10.1634/theoncologist.2018-0741
31. De Mattos-Arruda L, Olmos D, Tabernero J. Prognostic and predictive roles for circulating biomarkers in gastrointestinal cancer. Future Oncol. (2011) 7:1385–97. doi: 10.2217/fon.11.122
32. Iqbal M, Roberts A, Starr J, Mody K, Kasi PM. Feasibility and clinical value of circulating tumor DNA testing in patients with gastric adenocarcinomas. J Gastrointest Oncol. (2019) 10:400–6. doi: 10.21037/jgo.2019.01.14
33. Cheung AH, Chow C, To KF. Latest development of liquid biopsy. J Thorac Dis. (2018) 10:S1645–S51. doi: 10.21037/jtd.2018.04.68
34. Koldby KM, Mortensen MB, Detlefsen S, Pfeiffer P, Thomassen M, Kruse TA. Tumor-specific genetic aberrations in cell-free DNA of gastroesophageal cancer patients. J Gastroenterol. (2019) 54:108–21. doi: 10.1007/s00535-018-1508-5
35. Buono G, Gerratana L, Bulfoni M, Provinciali N, Basile D, Giuliano M, et al. Circulating tumor DNA analysis in breast cancer: Is it ready for prime-time? Cancer Treat Rev. (2019) 73:73–83. doi: 10.1016/j.ctrv.2019.01.004
36. Richardson AO, Hardin A, Shukla S, Zhao J, Barbacioru C, Beas H, et al. Abstract 572: The liquid biopsy Guardant360 CDx has robust performance at low inputs allowing for high rate of returning patient results. Cancer Research. (2021) 81:572. doi: 10.1158/1538-7445.AM2021-572
37. Leighl NB, Page RD, Raymond VM, Daniel DB, Divers SG, Reckamp KL, et al. Clinical utility of comprehensive cell-free DNA analysis to identify genomic biomarkers in patients with newly diagnosed metastatic non-small cell lung cancer. Clin Cancer Res. (2019) 25:4691–700. doi: 10.1158/1078-0432.CCR-19-0624
38. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS ONE. (2020) 15:e0237802. doi: 10.1371/journal.pone.0237802
39. Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. (2018) 24:1449–58. doi: 10.1038/s41591-018-0101-z
40. Chen Z, Zhang C, Zhang M, Li B, Niu Y, Chen L, et al. Chromosomal instability of circulating tumor DNA reflect therapeutic responses in advanced gastric cancer. Cell Death Dis. (2019) 10:697. doi: 10.1038/s41419-019-1907-4
41. Kim YW, Kim YH, Song Y, Kim HS, Sim HW, Poojan S, et al. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp Mol Med. (2019) 51:1–10. doi: 10.1038/s12276-019-0292-5
42. Lan J, Lu Y, Guan Y, Chang L, Yu Z, Qian H. Identification of circulating tumor DNA using a targeted 545-gene next generation sequencing panel in patients with gastric cancer. Oncol Lett. (2020) 19:2251–7. doi: 10.3892/ol.2020.11305
43. Wang Y, Zhao C, Chang L, Jia R, Liu R, Zhang Y, et al. Circulating tumor DNA analyses predict progressive disease and indicate trastuzumab-resistant mechanism in advanced gastric cancer. EBioMedicine. (2019) 43:261–9. doi: 10.1016/j.ebiom.2019.04.003
44. Grenda A, Wojas-Krawczyk K, Skoczylas T, Krawczyk P, Sierocinska-Sawa J, Wallner G, et al. HER2 gene assessment in liquid biopsy of gastric and esophagogastric junction cancer patients qualified for surgery. BMC Gastroenterol. (2020) 20:382. doi: 10.1186/s12876-020-01531-5
45. Yan H, Chen W, Ge K, Mao X, Li X, Liu W, et al. Value of plasma methylated SFRP2 in prognosis of gastric cancer. Dig Dis Sci. (2020) 9:1449–58. doi: 10.1007/s10620-020-06710-8
46. Yang J, Gong Y, Lam VK, Shi Y, Guan Y, Zhang Y, et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. (2020) 11:346. doi: 10.1038/s41419-020-2531-z
47. Ko K, Kananazawa Y, Yamada T, Kakinuma D, Matsuno K, Ando F, et al. Methylation status and long-fragment cell-free DNA are prognostic biomarkers for gastric cancer. Cancer Med. (2021) 10:2003–12. doi: 10.1002/cam4.3755
48. Li J, Li Z, Ding Y, Xu Y, Zhu X, Cao N, et al. TP53 mutation and MET amplification in circulating tumor DNA analysis predict disease progression in patients with advanced gastric cancer. PeerJ. (2021) 9:e11146. doi: 10.7717/peerj.11146
49. Leal A, van Grieken NCT, Palsgrove DN, Phallen J, Medina JE, Hruban C, et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun. (2020) 11:525. doi: 10.1038/s41467-020-14310-3
50. Wu R, Shi C, Chen Q, Wu F, Li Q. Detection of circulating tumor cell DNA for monitoring advanced gastric cancer. Int J Clin Exp Pathol. (2020) 13:203–11
51. Zhang M, Qi C, Wang Z, Chen H, Zhao X, Zhang X, et al. Molecular characterization of ctDNA from Chinese patients with advanced gastric adenocarcinoma reveals actionable alterations for targeted and immune therapy. J Mol Med (Berl). (2021). doi: 10.1007/s00109-021-02093-z
52. Wang H, Li B, Liu Z, Gong J, Shao L, Ren J, et al. HER2 copy number of circulating tumour DNA functions as a biomarker to predict and monitor trastuzumab efficacy in advanced gastric cancer. Eur J Cancer. (2018) 88:92–100. doi: 10.1016/j.ejca.2017.10.032
53. Zhang C, Chen Z, Chong X, Chen Y, Wang Z, Yu R, et al. Clinical implications of plasma ctDNA features and dynamics in gastric cancer treated with HER2-targeted therapies. Clin Transl Med. (2020) 10:e254. doi: 10.1002/ctm2.254
54. Wang DS, Liu ZX, Lu YX, Bao H, Wu X, Zeng ZL, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut. (2019) 68:1152–61. doi: 10.1136/gutjnl-2018-316522
55. Aguilar-Mahecha A, Joseph S, Cavallone L, Buchanan M, Krzemien U, Batist G, et al. Precision medicine tools to guide therapy and monitor response to treatment in a HER-2+ gastric cancer patient: case report. Front Oncol. (2019) 9:698. doi: 10.3389/fonc.2019.00698
56. Lee J, Kim ST, Kim K, Lee H, Kozarewa I, Mortimer PGS, et al. Tumor genomic profiling guides patients with metastatic gastric cancer to targeted treatment: the VIKTORY umbrella trial. Cancer Discov. (2019) 9:1388–405. doi: 10.1158/2159-8290.CD-19-0442
57. Du J, Wu X, Tong X, Wang X, Wei J, Yang Y, et al. Circulating tumor DNA profiling by next generation sequencing reveals heterogeneity of crizotinib resistance mechanisms in a gastric cancer patient with MET amplification. Oncotarget. (2017) 8:26281–7. doi: 10.18632/oncotarget.15457
58. Frigault MM, Markovets A, Nuttall B, Kim KM, Park SH, Gangolli EA, et al. Mechanisms of Acquired Resistance to Savolitinib, a Selective MET Inhibitor in MET-Amplified Gastric Cancer. JCO Precis Oncol. (2020) 4:222–32. doi: 10.1200/PO.19.00386
59. Sole C, Arnaiz E, Manterola L, Otaegui D, Lawrie CH. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin Cancer Biol. (2019) 58:100–8. doi: 10.1016/j.semcancer.2019.01.003
60. Uchoa Guimaraes CT, Ferreira Martins NN, Cristina da, Silva Oliveira K, Almeida CM, Pinheiro TM, Gigek CO, et al. Liquid biopsy provides new insights into gastric cancer. Oncotarget. (2018) 9:15144–56. doi: 10.18632/oncotarget.24540
61. Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. (2014) 11:145–56. doi: 10.1038/nrclinonc.2014.5
62. Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: Current insights and future perspectives. World J Gastroenterol. (2018) 24:3313–29. doi: 10.3748/wjg.v24.i30.3313
63. Gao Y, Wang JW, Ren JY, Guo M, Guo CW, Ning SW, et al. Long noncoding RNAs in gastric cancer: From molecular dissection to clinical application. World J Gastroenterol. (2020) 26:3401–12. doi: 10.3748/wjg.v26.i24.3401
64. Shi Y, Wang Z, Zhu X, Chen L, Ma Y, Wang J, et al. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int J Clin Oncol. (2020) 25:89–99. doi: 10.1007/s10147-019-01532-9
65. Shao JP, Su F, Zhang SP, Chen HK Li ZJ, Xing GQ, et al. miR-212 as potential biomarker suppresses the proliferation of gastric cancer via targeting SOX4. J Clin Lab Anal. (2020) 34:e23511. doi: 10.1002/jcla.23511
66. Imaoka H, Toiyama Y, Okigami M, Yasuda H, Saigusa S, Ohi M, et al. Circulating microRNA-203 predicts metastases, early recurrence, and poor prognosis in human gastric cancer. Gastric Cancer. (2016) 19:744–53. doi: 10.1007/s10120-015-0521-0
67. Kao HW, Pan CY, Lai CH, Wu CW, Fang WL, Huang KH, et al. Urine miR-21-5p as a potential non-invasive biomarker for gastric cancer. Oncotarget. (2017) 8:56389–97. doi: 10.18632/oncotarget.16916
68. Wang N, Wang L, Yang Y, Gong L, Xiao B, Liu X, et al. serum exosomal microRNA panel as a potential biomarker test for gastric cancer. Biochem Biophys Res Commun. (2017) 493:1322–8. doi: 10.1016/j.bbrc.2017.10.003
69. Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. (2018) 17:137. doi: 10.1186/s12943-018-0888-8
70. Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. (2018) 17:68. doi: 10.1186/s12943-018-0817-x
71. Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, Kusunoki H, et al. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer. (2013) 109:2323–30. doi: 10.1038/bjc.2013.596
72. Chen TH, Chiu CT, Lee C, Chu YY, Cheng HT, Hsu JT, et al. Circulating microRNA-22-3p predicts the malignant progression of precancerous gastric lesions from intestinal metaplasia to early adenocarcinoma. Dig Dis Sci. (2018) 63:2301–8. doi: 10.1007/s10620-018-5106-4
73. Xian HP, Zhuo ZL, Sun YJ, Liang B, Zhao XT. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. (2018) 16:4689–98. doi: 10.3892/ol.2018.9199
74. Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH, et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. (2018) 17:84. doi: 10.1186/s12943-018-0834-9
75. Chen TH, Lee C, Chiu CT, Chu YY, Cheng HT, Hsu JT, et al. Circulating microRNA-196a is an early gastric cancer biomarker. Oncotarget. (2018) 9:10317–23. doi: 10.18632/oncotarget.23126
76. Liang Q, Zhang G, Wang J, Sheng S. Diagnostic value of MicroRNA-18a for gastric cancer: a meta-analysis. Clin Lab. (2018) 64:177–84. doi: 10.7754/Clin.Lab.2017.170827
77. Yoruker EE, Keskin M, Kulle CB, Holdenrieder S, Gezer U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed Rep. (2018) 9:181–6. doi: 10.3892/br.2018.1116
78. Emami SS, Nekouian R, Akbari A, Faraji A, Abbasi V, Agah S. Evaluation of circulating miR-21 and miR-222 as diagnostic biomarkers for gastric cancer. J Cancer Res Ther. (2019) 15:115–9. doi: 10.4103/jcrt.JCRT_592_17
79. Jiang X, Jiang M, Xu M, Xu J, Li Y. Identification of diagnostic utility and molecular mechanisms of circulating miR-551b-5p in gastric cancer. Pathol Res Pract. (2019) 215:900–4. doi: 10.1016/j.prp.2019.01.035
80. Chen J, Wu L, Sun Y, Yin Q, Chen X, Liang S, et al. Mir-421 in plasma as a potential diagnostic biomarker for precancerous gastric lesions and early gastric cancer. PeerJ. (2019) 7:e7002. doi: 10.7717/peerj.7002
81. Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H, et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: a multiphase study. JAMA Surg. (2020) 155:572–9. doi: 10.1001/jamasurg.2020.1133
82. Al-Nedawi K, Meehan B, Rak J. Microvesicles: messengers and mediators of tumor progression. Cell Cycle. (2009) 8:2014–8. doi: 10.4161/cc.8.13.8988
83. Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. (2008) 15:80–8. doi: 10.1038/sj.cdd.4402237
84. Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. (2007) 67:2912–5. doi: 10.1158/0008-5472.CAN-07-0520
85. Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. (2006) 20:1487–95. doi: 10.1038/sj.leu.2404296
86. Xue H, Lu B, Zhang J, Wu M, Huang Q, Wu Q, et al. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J Proteome Res. (2010) 9:545–55. doi: 10.1021/pr9008817
87. Zhang X, Shi H, Yuan X, Jiang P, Qian H, Xu W. Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol Cancer. (2018) 17:146. doi: 10.1186/s12943-018-0898-6
88. Fu M, Gu J, Jiang P, Qian H, Xu W, Zhang X. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer. (2019) 18:41. doi: 10.1186/s12943-019-1001-7
89. Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol Ther. (2020) 207:107458. doi: 10.1016/j.pharmthera.2019.107458
90. Filipazzi P, Burdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. (2012) 22:342–9. doi: 10.1016/j.semcancer.2012.02.005
91. Qu JL, Qu XJ, Zhao MF, Teng YE, Zhang Y, Hou KZ, et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. (2009) 41:875–80. doi: 10.1016/j.dld.2009.04.006
92. Tanaka M, Kuriyama S, Itoh G, Maeda D, Goto A, Tamiya Y, et al. Mesothelial cells create a novel tissue niche that facilitates gastric cancer invasion. Cancer Res. (2017) 77:684–95. doi: 10.1158/0008-5472.CAN-16-0964
93. Deng G, Qu J, Zhang Y, Che X, Cheng Y, Fan Y, et al. Gastric cancer-derived exosomes promote peritoneal metastasis by destroying the mesothelial barrier. FEBS Lett. (2017) 591:2167–79. doi: 10.1002/1873-3468.12722
94. Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Ogino S, et al. Tumor exosome-mediated promotion of adhesion to mesothelial cells in gastric cancer cells. Oncotarget. (2016) 7:56855–63. doi: 10.18632/oncotarget.10869
95. Fan Y, Che X, Qu J, Hou K, Wen T, Li Z, et al. Exosomal PD-L1 Retains immunosuppressive activity and is associated with gastric cancer prognosis. Ann Surg Oncol. (2019) 26:3745–55. doi: 10.1245/s10434-019-07431-7
96. Wang F, Li B, Wei Y, Zhao Y, Wang L, Zhang P, et al. Tumor-derived exosomes induce PD1(+) macrophage population in human gastric cancer that promotes disease progression. Oncogenesis. (2018) 7:41. doi: 10.1038/s41389-018-0049-3
Keywords: liquid biopsy, gastric cancer, screening, prognosis, circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), circulating non-coding RNAs (ncRNA), exosomes
Citation: Chivu-Economescu M, Necula L, Matei L, Dragu D, Bleotu C and Diaconu CC (2021) Clinical Applications of Liquid Biopsy in Gastric Cancer. Front. Med. 8:749250. doi: 10.3389/fmed.2021.749250
Received: 29 July 2021; Accepted: 31 August 2021;
Published: 28 September 2021.
Edited by:
Razvan Iacob, Fundeni Clinical Institute, RomaniaReviewed by:
Bianca Galateanu, University of Bucharest, RomaniaThomas Aparicio, Assistance Publique Hopitaux De Paris, France
Copyright © 2021 Chivu-Economescu, Necula, Matei, Dragu, Bleotu and Diaconu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mihaela Chivu-Economescu, bWloYWVsYS5lY29ub21lc2N1QHZpcm9sb2d5LnJv; bWloYWVsYS5jaGl2dUBnbWFpbC5jb20=
 Mihaela Chivu-Economescu
Mihaela Chivu-Economescu Laura Necula
Laura Necula Lilia Matei
Lilia Matei Denisa Dragu1
Denisa Dragu1 Coralia Bleotu
Coralia Bleotu Carmen C. Diaconu
Carmen C. Diaconu