
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med., 27 September 2021
Sec. Gastroenterology
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.704328
Background and Aims: Biliary atresia (BA) is the most common cholestatic liver disease in neonates. Although the Kasai procedure can improve temporary biliary drainage in some cases, complications and liver fibrosis still develop. Liver transplantation is the ultimate treatment. The current study aimed to investigate the effect of previous Kasai surgery on gut microbiota and bile acid in BA with end-stage liver disease.
Methods: Patients with BA with end-stage liver disease were divided into two groups according to whether they had previously undergone Kasai surgery (non-Kasai: n = 8, post-Kasai: n = 8). Metagenomic sequencing and ultraperformance liquid chromatography/tandem mass spectrometry were performed to identify the gut microbiota and bile acid.
Results: Previous Kasai surgery had some effects on gut microbiota and bile acid in BA with end-stage liver disease. In the gut microbiome, the differential species were mainly distributed at the species level. Veillonella atypica had a significant increase in the non-Kasai group (P < 0.05). Bacteroides spp., Prevotella spp., Barnesiella spp., Parabacteroides spp., Heliobacterium spp., Erysipelatoclostridium spp. and Diaporthe spp. were increased in the post-Kasai group (P < 0.05). Concerning functional profiles, methionine biosynthesis was enriched in the non-Kasai group, while pyridoxal biosynthesis and riboflavin biosynthesis were enriched in the post-Kasai group (linear discriminant analysis > 2, P < 0.05). In stools, 17 bile acids were distinctly elevated in the post-Kasai group, such as cholic acid, chenodeoxycholic acid, β-muricholic acid and tauro α-muricholate (P < 0.05). Spearman correlation test showed that V. atypica had an enormously positive correlation with liver enzymes. Faecalibacterium prausnitzii and Escherichia coli were associated with derivatives of the alternative pathway of bile acid metabolism.
Conclusion: Previous Kasai surgery can improve the gut microbiota and bile acid in patients with BA with end-stage liver disease. This improvement contributes to maintaining the intestinal barrier.
Biliary atresia (BA) is the most common cause of cholestatic liver disease in children. It is characterized by intrahepatic and extrahepatic bile duct occlusion and bile drainage obstruction (1, 2). It is fatal if left untreated, with a reported survival rate of <10% at 3 years of age (3). Kasai portoenterostomy (KPE) is considered the primary treatment of BA, but its outcome is still unsatisfactory (4). Although this approach can improve the short-term work, most patients develop fibrosis and progress to end-stage liver disease (5). Liver transplantation is still the only available salvage treatment. It is indicated when: (1) the Kasai procedure fails; (2) patients develop progressive deterioration of liver function despite an initially successful Kasai operation; or (3) end-stage liver disease develops in children who have not undergone Kasai surgery (6).
BA is a rare disease with an unclear etiology. There is strong evidence that viruses and toxins contribute to BA. Cytomegalovirus, human papillomavirus, human herpesvirus 6, Epstein–Barr virus, reovirus, and rotavirus have been detected directly in injured liver and biliary remnants, or indirectly by the presence of serological markers of infection in patients with BA (7). Viruses trigger an inflammatory response that injures the duct epithelium and produces rapidly progressive cholangiopathy. As for disease progression, Isaacs-Ten et al. have demonstrated that exposure to bacterial endotoxin sensitizes liver cells to bile-acid-induced cell death in cholestatic liver disease (8). When the intestinal barrier is damaged, translocated bacteria and microbial toxins can enter the portal circulation and access the liver (9). Therefore, there is a special relationship between the gut microbiota and liver injury.
The human gut microbiota composition can be altered by multiple factors such as diet, age, antibiotics, and various diseases. Among these, bile acids appear to be significant regulators. Bile acids interact with gut microbiota through the gut–liver axis (10). Bile acids can affect the composition of gut microbes by controlling the pH of the gut environment, inhibiting the growth of pathogens, and maintaining the balance of gut microbes (11). Decreased bile acids in the gut contribute to the overgrowth of potential pathogens, many of which produce lipopolysaccharide (LPS). Conversely, increased bile acids favor growth of Gram-positive Firmicutes (12).
The gut microbiota participates in and influences the enterohepatic circulation of bile acids. The conversion of primary to secondary bile acids depends on the bile salt hydrolase on the surface of certain bacteria in the gut (13–15). Previous research has reported that bacterial dysbiosis is linked to low bile acid levels entering the intestine in cirrhosis (16). There is still some unknown crosstalk between specific microbiota and bile acid metabolism that needs to be explored.
In the current study, we aimed to investigate the effect of previous Kasai surgery on the gut microbiome and bile acids in patients with BA with end-stage liver disease, then explore their relationship and its impact on health. High-throughput techniques, such as 16S rRNA genes and metagenomic sequencing, compared to traditional culture-dependent methods, have significantly improved the ability to rapidly determine the composition of the gut microbiome and its functions (17). The present study combined 16S rRNA genes with subsequently metagenomic sequencing to make the results more reliable.
We recruited patients with BA listed for liver transplantation at Beijing Friendship Hospital, Capital Medical University, between September 2017 and December 2018. The diagnosis in these patients was previously confirmed by laparotomy or operative cholangiography. Enrolled patients had to meet the following criteria: (1) age <3 years; (2) diagnosed with type III BA; (3) no antibiotics or probiotics within 1 month; (4) no digestive diseases such as diarrhea or constipation; and (5) similar dietary habits. Differential diagnoses were excluded, including bile duct dysplasia, progressive familial intrahepatic cholestasis, citrin deficiency disease, tyrosinemia type 1, and α-1 antitrypsin deficiency. The patients were divided into two groups based on whether they had previously undergone Kasai surgery. Each group consisted of eight patients (the non-Kasai and post-Kasai groups). The detailed demographic data and clinical indicators are shown in Table 1.
All samples were stored at −80°C within 4 h of collection. Bacterial DNA was extracted using the QIAamp Fast DNA Stool Mini Kit (51604; Qiagen, Hilden, Germany). Ten micrograms of stool sample were weighed in a centrifuge tube, approximately 25 mg of precooled submerged beads were added, and 200 μl acetonitrile/methanol (v/v = 8:2) solvent containing 10 μl internal standard for homogeneous mixing was added and centrifuged at 13,500 rpm and 4°C for 20 min to remove proteins. After centrifugation, 10 μl supernatant was obtained, diluted with 90 μl 1:1 acetonitrile/methanol (v/v = 80/20) and ultrapure water mixed solvent, shaken and centrifuged for analysis. The injection volume was 5 μL. The DNA concentration was measured with a NanoDrop (Thermo Scientific, MA, USA) and Qubit®2.0 (Invitrogen, Carlsbad, CA, USA), and the molecular size was estimated by agarose gel electrophoresis.
Following the Illumina TruSeq DNA Sample Prep v2 Guide (San Diego, CA, USA), we constructed the DNA paired-end libraries with an insert size of 500 bp for the 40 stool samples (16 from pre-LT, 14 from post-LT, and 10 from controls). The quality of all libraries was evaluated using an Agilent 2100 bioAnalyzer (Agilent Technologies, Wokingham, UK) and an Agilent 2100 DNA 1000 kit. All samples were subjected to 150-bp paired-end sequencing on an Hiseq X-ten platform (Illumina). Illumina raw reads were screened according to the following criteria: (1) adaptor contamination reads were removed; (2) reads containing more than three ambiguous N bases were removed; (3) reads containing low quality (Q <20) bases were trimmed; and (4) reads containing <60% of high-quality bases (Phred score ≥20) were deleted.
Clean reads were subjected to bacterial genomes from the National Center for Biotechnology Information GenBank with SOAPaligner (version 2.21), and reads mapped to the host genome were abandoned.
For species classification, the NCBI database (http://www.ncbi.nlm.nih.gov) was used to align the clean reads with known bacteria, fungi, viruses, and archaea by SOAPaligner 2.21. As to the functional profiles, the non-redundant genes were annotated against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using BLAST (v. 2.2.28+). When the assembled protein sequence was similar (score ≥60 and E <1 × 10−5) to a protein sequence in the database, the assembled protein was considered to play the same role as the database protein. The relative abundance of all orthologous genes was accumulated to generate the close lot of each KEGG ortholog. The results of metagenomic sequencing and assembly data in each sample are listed in Supplementary Table 1.
Bile acid standards (Steraloids, USA), six stable isotopes labeled standards (C/D/NIsotopes, Canada/Steraloids), ammonium acetate (analytical grade, Sigma–Aldrich, St Louis, MO, USA), methanol (Optima LC-MS), acetonitrile (Optima LC-MS), isopropanol (Optima LC-MS), glacial acetic acid and formic acid (Optima LC-MS) were all purchased from ThermoFisher Scientific (Fairlawn, NJ, USA). The following equipment was used: ACQUITYUPLC-XevoTQ-S liquid-mass spectrometer (WatersCorp., Milford, MA, USA); Mill-Q ultrapure water system (Millipore, Billerica, MA, USA); homogenizer (BB24; NextAdvance, Averill Park, NY, USA); microcentrifuge (Microfuge20R; Beckman Coulter, Indianapolis, IN, USA); and lyophilizer (Labconco, Kansas City, MO, USA).
Seventy-three bile acid standards were used, and six representative isotope bile acids were used as internal standards for calibration. Standards and isotope markers were accurately weighed and prepared with methanol to a concentration of 5.0 mM. We mixed the standards in serum matrix without bile acids and set seven concentrations of 2000, 1000, 400, 100, 25, 10 and 5 nM. We weighed 10 mg stool sample in a centrifuge tube, added ~25 mg of precooled submerged beads, and 200 μl acetonitrile/methanol (v/v = 8:2) solvent containing 10 μl internal standard for homogeneous mixing, centrifuged at 13,500 rpm and 4°C for 20 min to remove protein. After centrifugation, 10μl supernatant was diluted with 90 μl 1:1 acetonitrile/methanol (80/20) and ultrapure water mixed solvent, shaken and centrifuged before injection analysis. The injection volume was 5μl. Ultra-high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) system (ACQUITYUPLC-XevoTQ-S; Waters) was used for quantification of metabolites (18).
The non-parametric Wilcoxon test (Wilcox. test in R) was performed to analyze the statistical significance of the different taxonomic levels between the different cohorts. Differences were considered significant at P < 0.05 or false discovery rate (FDR) <0.1. Linear discriminant analysis (LDA) effect size (LEfSe) analysis was used to identify the taxa most likely to explain differences between the post-Kasai and non-Kasai groups. The LDA score cut-off of 2.0 indicated a significant difference. Orthogonal partial least squares discriminate analysis (OPLS-DA) was used for statistical analysis to determine stool bile acid changes between the two groups. All the metabolite variables were scaled to pareto scaling prior to conducting the OPLS-DA. The model validity was evaluated from model parameters R2 and Q2, which provided information for the interpretability and predictability, respectively, of the model and avoided the risk of overfitting. Variable importance in the projection (VIP) was calculated in the OPLS-DA model. The VIP score cut-off of 1.0 indicated a significant difference. The Spearman correlation test was conducted to investigate the relationship between the clinical parameters, bile acid, and microbial composition. A heat map was drawn using the R software corrplot package/gplots package to illustrate the results.
16S rRNA gene sequencing was performed to determine the alterations in the gut microbiota between the two groups. It showed no significant difference at the phylum, order or genus level (Figures 1A–C). Shigella, Streptococcus and Enterococcus abundances were higher in the non-Kasai group although they did not reach statistical significance (P > 0.05, Supplementary Table 2). However, Veillonella atypica had a noticeable increase in the non-Kasai group at the species level (Figure 1D, P < 0.05) (Supplementary Table 3).
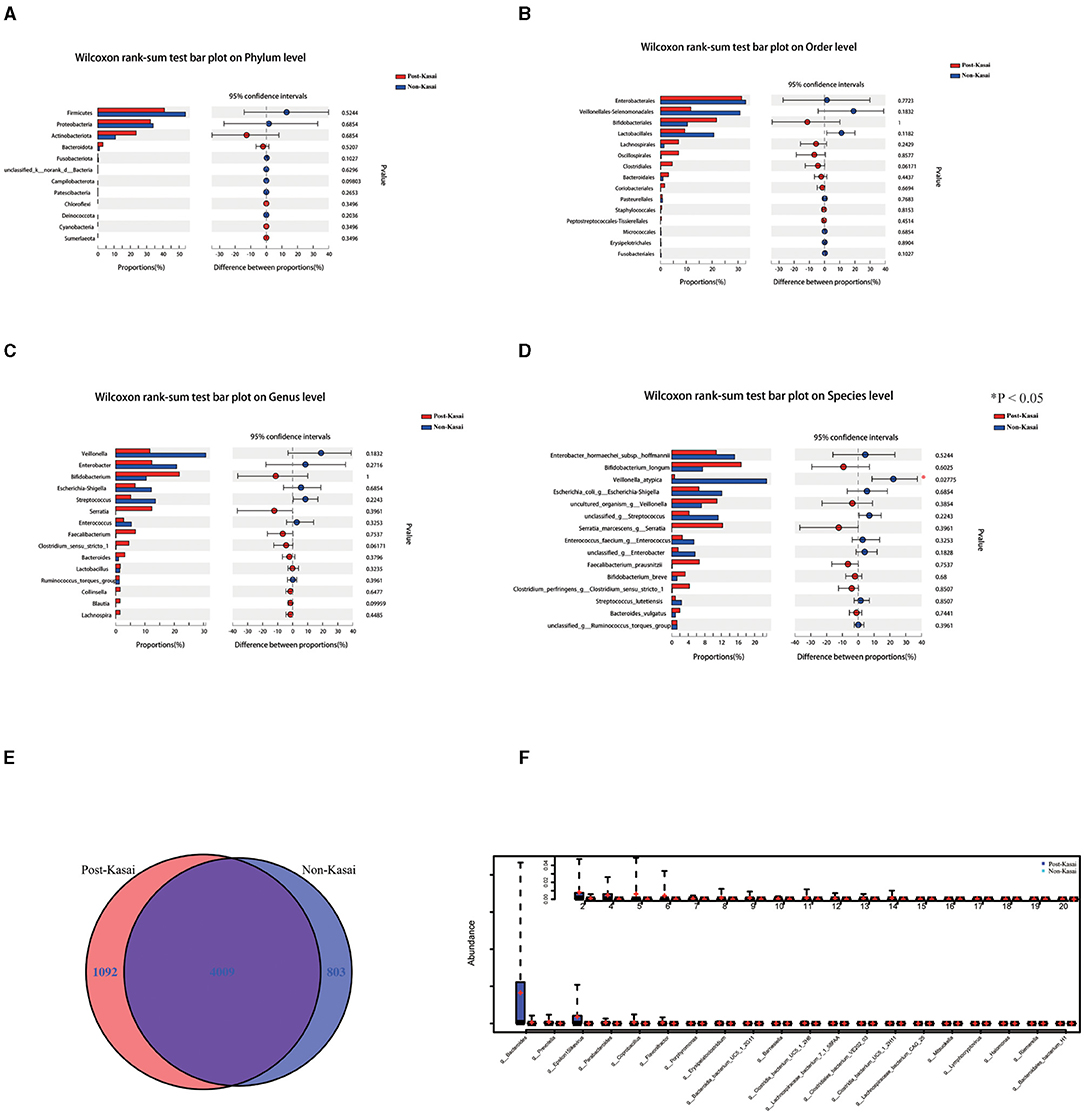
Figure 1. (A–D) Differential gut microbiota between the post-Kasai and non-Kasai groups at the phylum, order, genus and species levels (*P < 0.05, 16S rRNA genes). (E) Venn diagram of the post-Kasai and non-Kasai groups at the species level. (F) Differential gut microbiota between the post-Kasai and non-Kasai groups performed by metagenomic sequencing.
Metagenomic sequencing was used further to identify the differential species between the two groups. There were 803 and 1,092 species enriched in the non-Kasai and post-Kasai groups, respectively (Figure 1E). We concluded that Kasai surgery increased the diversity of species in BA. Bacteroides, Prevotella, Barnesiella, Parabacteroides, Heliobacterium, Erysipelatoclostridium and Diaporthe were increased in the post-Kasai group (Figure 1F, Supplementary Table 4). Spearman correlation test showed that the abundance of Veillonella spp. (e.g., V. atypica) was strongly positively correlated with liver enzyme alanine aminotransferase (ALT) and aspartate aminotransferase (AST), but had no significant correlation with total bile acid (Figure 2G, Supplementary Table 5). Therefore, we speculated that V. atypica contributed to the liver injury in BA.
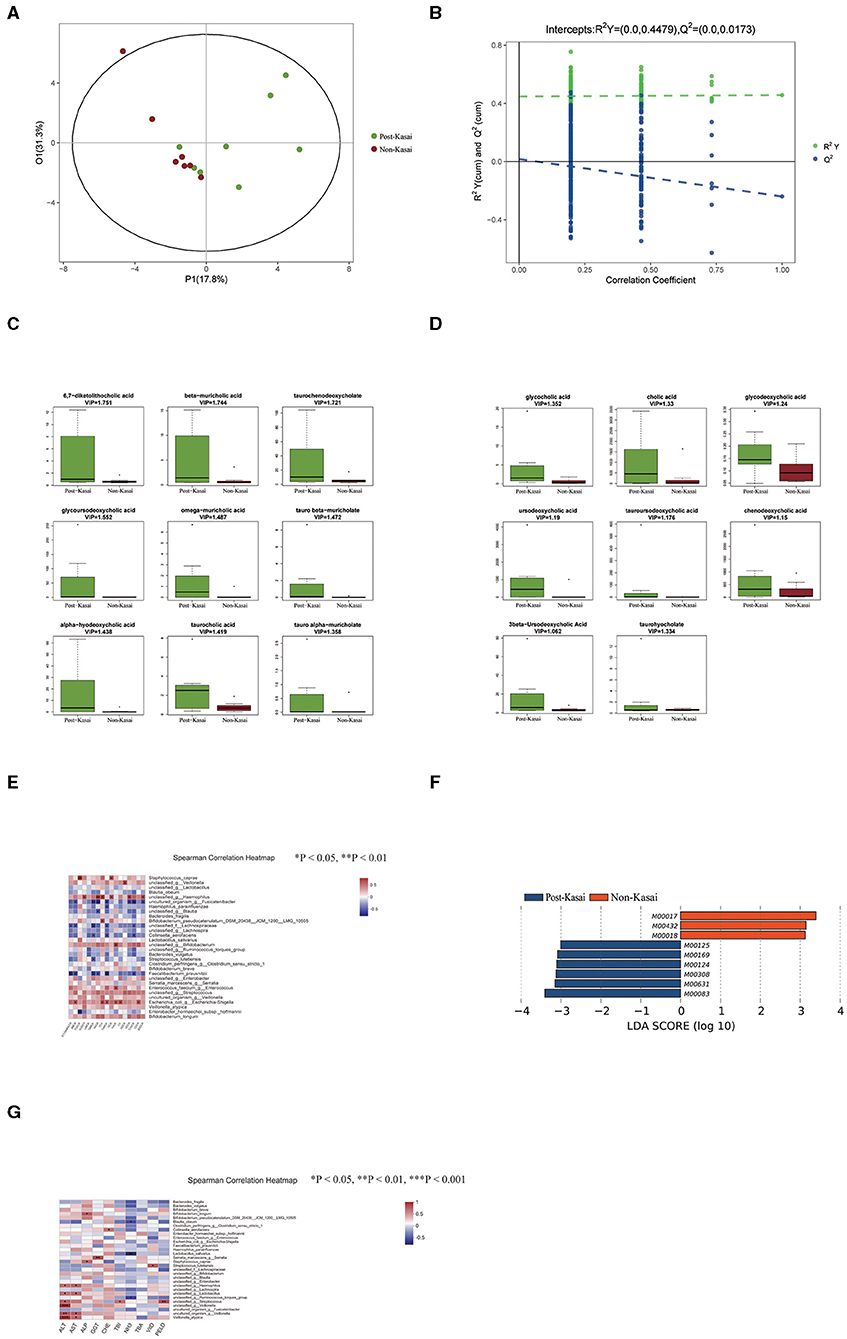
Figure 2. (A,B) Significant differences in stool bile acid composition between the post-Kasai and non-Kasai groups identified by OPLS-DA (left: OPLS-DA map; right: model verification map OPLS-DA). Model verification map of OPLS-DA: the x axis represents the replacement retention of the replacement test; the y axis represents the R2 (green dot) and Q2 (blue dot) replacement test values. The two dashes represent the regression lines of R2 and Q2, respectively. (C,D) Differential bile acids between the two groups. The x axis represents the different groups, and the y axis expression of screened differential bile acids. Different colors represent different groups, and the boxplot shows five statistical values (minimum, first quartile, median, third quartile, and maximum, namely five lines from bottom to top). (E) Spearman correlations between gut species and bile acids. The x axis represents the differential bile acids, and the y axis the species (*P < 0.05, **P < 0.01, ***P < 0.001). Blue denotes a negative correlation and red a positive correlation. (F) Differential functional profiles between the two groups. (G) Spearman correlations between gut species and clinical indicators (*P < 0.05, **P < 0.01, ***P < 0.001). The x axis represents the environmental factors, and the y axis the species. Blue denotes a negative correlation and red a positive correlation.
A total of 46 fecal bile acids were detected, and OPLS-DA was used to screen for differential metabolites between the two groups (Figures 2A,B, permutation test: R2Y = 0.0–0.4479, Q2 = 0.0–0.0173). Seventeen bile acid levels were significantly elevated in the post-Kasai group (Figures 2C,D, VIP > 1) (Supplementary Table 6).
In the increased bile acid, β-muricholic acid (βMCA), ω-muricholic acid (ωMCA), tauro α-muricholate (TαMCA), tauro β-muricholate (TβMCA), taurohyocholate (THCA), and α-hyodeoxycholic acid (HDCA) belonged to the products of the alternative pathway, and the remaining bile acids were the products of the classical pathway. Spearman correlation test was subsequently conducted to investigate the relationship between the differential bile acids and species (Figure 2E, Supplementary Table 7). The level of βMCA, TαMCA, TβMCA and HDCA was strongly negatively correlated with the abundance of Faecalibacterium prausnitzii but positively correlated with Escherichia coli. Importantly, these bile acids were all derived from the alternative pathway. Previous research has shown that the classical pathway of bile acid metabolism is impaired, while the alternative pathway is preserved in infantile cholestasis (19). We inferred that the altered abundance of F. prausnitzii and E. coli contributed to the changed bile acid metabolism in BA.
We annotated the catalogs using the KEGG database to investigate the gut microbiome's functional profiles (http://www.genome.jp/kegg/). There were nine differential functional modules between the post-Kasai and non-Kasai groups (Figure 2F). Six were enriched in the post-Kasai group and the remaining three were in the non-Kasai group (Supplementary Table 8, P < 0.05). Methionine biosynthesis (M00017) was enriched in the non-Kasai group, while pyridoxal biosynthesis and riboflavin biosynthesis (M00124 and M00125) were enriched the post-Kasai group.
As the largest immune organ in the human body, intestinal microbes play an essential role in multiple physiological functions and are called the second human genome (20). Mounting evidence supports an association between liver disease and changes in the microbiome composition (16, 21–25). However, the role of previous Kasai surgery in the gut microbiota and bile acids in patients with BA with the end-stage liver disease remains largely undefined. An in-depth understanding of the gut microbiota is essential for exploring the relationship between the microbiome and disease pathogenesis. We, therefore, used 16S rRNA and metagenomic sequencing to analyze and identify fecal microorganisms.
Compared to 16S rRNA sequencing, metagenomic sequencing identified more differential species between the non-Kasai and post-Kasai groups. Overall, the number of species was greater in the post-Kasai group, attributed to bile acid drainage. Differences in microbiota composition are mainly at the species level, and in particular, V. atypica showed a significant decrease in the post-Kasai group. Veillonella is reported to be associated with autoimmune liver disease (22). Correlation analysis showed that V. atypica was positively correlated with liver enzymes. The above data suggested that the abundance of V. atypica contributed to liver injury in BA. Besides, Bacteroides, Prevotella, Clostridium spp., Barnesiella, Parabacteroides, Heliobacterium, Erysipelatoclostridium and Diaporthe were also enriched in the post-Kasai group. Clostridium spp., Barnesiella and Prevotellaceae contain genes that produce the short-chain fatty acids, propionate and butyrate, which play important roles in maintaining the intestinal barrier and anti-inflammation (26). Bacteroides spp. have been associated with health (9). Consistent with these results, methionine biosynthesis was decreased in the post-Kasai group. Previous research has demonstrated that dietary methionine restriction improves the gut microbiota and reduces intestinal permeability and inflammation (27). We concluded that the gut microbiota, intestinal permeability, and inflammation were improved in the post-Kasai group.
Bile acids are synthesized in the liver by multistep reactions catalyzed via two distinct routes, the classical and alternative pathways (28). The classical pathway is initiated by the rate-limiting enzyme cholesterol 7α-hydroxylase (CYP7A1) and results in the formation of the primary BAs, CA and CDCA. The alternative pathway is initiated with the oxidation of the cholesterol side-chain by the mitochondrial cytochrome p450 sterol 27-hydroxylase (CYP27A1) followed by 25-hydroxycholesterol 7-alpha-hydroxylase (CYP7B1) (29). HCA, αMCA, βMCA, and their conjugated bile acids are the products of this pathway. The classical pathway accounts for about 75% of bile acid production. The gut microbiome harbors hundreds of pathways, many of which modulate host biology. In the intestine, bile acids are subject to extensive metabolism by gut microbes, namely deconjugation of glycine or taurine and biotransformation of the unconjugated primary bile acids to secondary bile acids (30). Deoxycholic acid, lithocholic acid (LCA) and its derivatives are major components of the recirculating bile acid pool (31). Consistently, 6,7-diketolithocholic acid (6,7-DiketoLCA), one derivative of LCA, was increased in the post-Kasai group. Previous research has demonstrated that disorder of bile acid metabolism is related to inflammatory bowel disease (32). We observed that the abundance of F. prausnitzii and E. coli was related to the alternative pathway of bile acid metabolism.
As for functional profiles, it was observed that the pathway of pyridoxal and riboflavin biosynthesis was higher in the post-Kasai group. Pyridoxal is one of the pyridine derivatives from vitamin B6. Vitamin B6 deficiency affects cell-mediated immunity in both animal and human studies (33). Riboflavin (vitamin B2) is unique among water-soluble vitamins. There are reports of various congenital malformations associated with riboflavin deficiency in rats and mice. Besides, riboflavin synthesized by bacterial metabolism in the colon might be a more critical source (34). Based on functional results, it appeared that the post-Kasai group was healthier although it still needs verification by microbial metabolomics.
This study had some limitations. (1) The number of patients was small, and a greater number of patients should be enrolled. We will expand the sample size in future research to verify these results. (2) Alterations in the functional microbiome profiles and the correlations between the gut microbiome, bile acid, and clinical indicators need to be verified. Microbial metabolomics will be performed to understand their functions and correlations deeply.
In conclusion, 16S rRNA and metagenomic sequencing revealed that previous Kasai surgery can improve the gut microbiota composition in patients with BA with end-stage liver disease. V. atypica was decreased while Bacteroides, Prevotella, Barnesiella, Parabacteroides, Heliobacterium, Erysipelatoclostridium and Diaporthe were increased in the post-Kasai group. V. atypica might contribute to liver injury in BA. UPLC-MS/MS was performed to detect characteristic changes in stool bile acids. We conclude that the abundance of F. prausnitzii and E. coli is related to the alternative pathway of bile acid metabolism.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI SRA (https://www.ncbi.nlm.nih.gov/bioproject PRJNA730640), it can be accessed with the BioProject identifier PRJNA730640.
The studies involving human participants were reviewed and approved by Beijing Friendship Hospital, Capital Medical University (Approval ID: 2019-P2–131-02). Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
WS: study design, data collection, analysis and interpretation of the data, and writing of the report. L-YS and Z-JZ: study design, study supervision, and critical revision of the manuscript for important intellectual content. All authors have read and approved the final manuscript to be submitted.
This study was supported by the National Natural Science Foundation of China (Grant No. 81570586).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.704328/full#supplementary-material
BA, biliary atresia; ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; GGT, γ-glutamyltransferase; TBA, total bile acids; TBIL, total bilirubin; PELD, pediatric end-stage liver disease; KEGG, Kyoto encyclopedia of genes and genomes; LEfSe, LDA effect size; LDA, linear discriminant analysis; βMCA, β-muricholic acid; ωMCA, ω-muricholic acid; TαMCA, Tauro α-muricholate; TβMCA, Tauro β-muricholate; THCA, taurohyocholate; HDCA, alpha-hyodeoxycholic acid; CDCA, chenodeoxycholic acid; CA, cholic acid; GCA, glycocholic acid; TCDCA, taurochenodeoxycholic acid; GUDCA, glycoursodeoxycholic acid; TCA, taurocholic acid; UDCA, ursodeoxycholic acid; GDCA, glycodeoxycholic acid; TUDCA, tauroursodeoxycholic acid; 6, 7-DiketoLCA, 6, 7-diketolithocholic acid; βUDCA, 3β-Ursodeoxycholic Acid; OPLS-DA, orthogonal partial least squares discrimination analysis; VIP, variable importance in the projection.
1. Lakshminarayanan B, Davenport M. Biliary atresia: a comprehensive review. J Autoimmun. (2016) 73:1–9. doi: 10.1016/j.jaut.2016.06.005
2. Vij M, Rela M. Biliary atresia: pathology, etiology and pathogenesis. Future Sci OA. (2020) 6:FSO466. doi: 10.2144/fsoa-2019-0153
4. Chung PHY, Zheng S, Tam PKH. Biliary atresia: east versus west. Semin Pediatr Surg. (2020) 29:150950. doi: 10.1016/j.sempedsurg.2020.150950
5. Sundaram SS, Mack CL, Feldman AG, Sokol RJ. Biliary atresia: Indications and timing of liver transplantation and optimization of pretransplant care. Liver Transpl. (2017) 23:96–109. doi: 10.1002/lt.24640
6. Wang SH, Chen CL, Concejero A, Wang CC, Lin CC, Liu YW, et al. Living donor liver transplantation for biliary atresia. Chang Gung Med J. (2007) 30:103–8. doi: 10.1016/j.transproceed.2010.11.002
7. Asai A, Miethke A, Bezerra JA. Pathogenesis of biliary atresia: defining biology to understand clinical phenotypes. Nat Rev Gastroenterol Hepatol. (2015) 12:342–52. doi: 10.1038/nrgastro.2015.74
8. Isaacs-Ten A, Echeandia M, Moreno-Gonzalez M, Brion A, Goldson A, Philo M, et al. Intestinal microbiome-macrophage crosstalk contributes to cholestatic liver disease by promoting intestinal permeability in mice. Hepatology. (2020) 72:2090–108. doi: 10.1002/hep.31228
9. Chopyk DM, Grakoui A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology. (2020) 159:849–63. doi: 10.1053/j.gastro.2020.04.077
10. Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. (2018) 15:397–411. doi: 10.1038/s41575-018-0011-z
11. Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol Aspects Med. (2017) 56:54–65. doi: 10.1016/j.mam.2017.06.002
12. Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. (2014) 30:332–8. doi: 10.1097/MOG.0000000000000057
13. Yang X, Lu D, Zhuo J, Lin Z, Yang M, Xu X. The gut-liver axis in immune remodeling: new insight into liver diseases. Int J Biol Sci. (2020) 16:2357–66. doi: 10.7150/ijbs.46405
14. Woodhouse C, Singanayagam A, Patel VC. Modulating the gut-liver axis and the pivotal role of the faecal microbiome in cirrhosis. Clin Med. (2020) 20:493–500. doi: 10.7861/clinmed.2020-0676
15. Wang F, Cui Q, Zeng Y, Chen P. [Gut microbiota-an important contributor to liver diseases]. Nan Fang Yi Ke Da Xue Xue Bao. (2020) 40:595–600. doi: 10.12122/j.issn.1673-4254.2020.04.23
16. Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. (2014) 513:59–64. doi: 10.1038/nature13568
17. Human Microbiome Project C. A framework for human microbiome research. Nature. (2012) 486:215–21. doi: 10.1038/nature11209
18. Xie G, Zhong W, Li H, Li Q, Qiu Y, Zheng X, et al. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. (2013) 27:3583–93. doi: 10.1096/fj.13-231860
19. Wang Y, Gao X, Zhang X, Xiao Y, Huang J, Yu D, et al. Gut Microbiota dysbiosis is associated with altered bile acid metabolism in infantile cholestasis. mSystems. (2019) 4. doi: 10.1128/mSystems.00463-19
20. Zhu B, Wang X, Li L. Human gut microbiome: the second genome of human body. Protein Cell. (2010) 1:718–25. doi: 10.1007/s13238-010-0093-z
21. Abe K, Fujita M, Hayashi M, Okai K, Takahashi A, Ohira H. Gut and oral microbiota in autoimmune liver disease. Fukushima J Med Sci. (2020) 65:71–5. doi: 10.5387/fms.2019-21
22. Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut. (2019) 69:569–77. doi: 10.1136/gutjnl-2018-317836
23. Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. (2018) 67:534–41. doi: 10.1136/gutjnl-2016-313332
24. Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. (2017) 14:527–39. doi: 10.1038/nrgastro.2017.72
25. Dubinkina VB, Tyakht AV, Odintsova VY, Yarygin KS, Kovarsky BA, Pavlenko AV, et al. Links of gut microbiota composition with alcohol dependence syndrome and alcoholic liver disease. Microbiome. (2017) 5:141. doi: 10.1186/s40168-017-0359-2
26. Holota Y, Dovbynchuk T, Kaji I, Vareniuk I, Dzyubenko N, Chervinska T, et al. The long-term consequences of antibiotic therapy: Role of colonic short-chain fatty acids (SCFA) system and intestinal barrier integrity. PLoS ONE. (2019) 14:e0220642. doi: 10.1371/journal.pone.0220642
27. Yang Y, Zhang Y, Xu Y, Luo T, Ge Y, Jiang Y, et al. Dietary methionine restriction improves the gut microbiota and reduces intestinal permeability and inflammation in high-fat-fed mice. Food Funct. (2019) 10:5952–68. doi: 10.1039/C9FO00766K
28. Jia W, Wei M, Rajani C, Zheng X. Targeting the alternative bile acid synthetic pathway for metabolic diseases. Protein Cell. (2021) 12:411–25. doi: 10.1007/s13238-020-00804-9
29. Rizzolo D, Buckley K, Kong B, Zhan L, Shen J, Stofan M, et al. Bile acid homeostasis in a cholesterol 7alpha-hydroxylase and sterol 27-hydroxylase double knockout mouse model. Hepatology. (2019) 70:389–402. doi: 10.1002/hep.30612
30. Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome. (2019) 7:75. doi: 10.1186/s40168-019-0689-3
31. Funabashi M, Grove TL, Wang M, Varma Y, McFadden ME, Brown LC, et al. A metabolic pathway for bile acid dehydroxylation by the gut microbiome. Nature. (2020) 582:566–70. doi: 10.1038/s41586-020-2396-4
32. Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. (2013) 62:531–9. doi: 10.1136/gutjnl-2012-302578
33. Ueland PM, McCann A, Midttun O, Ulvik A. Inflammation, vitamin B6 and related pathways. Mol Aspects Med. (2017) 53:10–27. doi: 10.1016/j.mam.2016.08.001
Keywords: biliary atresia, Kasai surgery, bile acids, gut microbiota (GM), metagenomic sequencing
Citation: Song W, Sun L-Y and Zhu Z-J (2021) Effects of Previous Kasai Surgery on Gut Microbiota and Bile Acid in Biliary Atresia With End-Stage Liver Disease. Front. Med. 8:704328. doi: 10.3389/fmed.2021.704328
Received: 02 May 2021; Accepted: 31 August 2021;
Published: 27 September 2021.
Edited by:
Li Ang, First Affiliated Hospital of Zhengzhou University, ChinaReviewed by:
Fabio Sallustio, University of Bari Aldo Moro, ItalyCopyright © 2021 Song, Sun and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Ying Sun, c3VueGx4QG91dGxvb2suY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.