- 1Department of Respiratory and Critical Care Medicine, West China Hospital/West China School of Medicine, Sichuan University, Chengdu, China
- 2Department of Periodical Press and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University, Chengdu, China
- 3Chinese Evidence-Based Medicine Center, West China Hospital, Sichuan University, Chengdu, China
- 4Department of Anesthesiology and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University and The Research Units of West China, Chinese Academy of Medical Sciences, Chengdu, China
- 5Northern Ireland Methodology Hub, Queen's University Belfast, Belfast, United Kingdom
Background: Recently, Coronavirus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome virus 2 (SARS-CoV-2), has affected more than 200 countries and lead to enormous losses. This study systematically reviews the application of Artificial Intelligence (AI) techniques in COVID-19, especially for diagnosis, estimation of epidemic trends, prognosis, and exploration of effective and safe drugs and vaccines; and discusses the potential limitations.
Methods: We report this systematic review following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. We searched PubMed, Embase and the Cochrane Library from inception to 19 September 2020 for published studies of AI applications in COVID-19. We used PROBAST (prediction model risk of bias assessment tool) to assess the quality of literature related to the diagnosis and prognosis of COVID-19. We registered the protocol (PROSPERO CRD42020211555).
Results: We included 78 studies: 46 articles discussed AI-assisted diagnosis for COVID-19 with total accuracy of 70.00 to 99.92%, sensitivity of 73.00 to 100.00%, specificity of 25 to 100.00%, and area under the curve of 0.732 to 1.000. Fourteen articles evaluated prognosis based on clinical characteristics at hospital admission, such as clinical, laboratory and radiological characteristics, reaching accuracy of 74.4 to 95.20%, sensitivity of 72.8 to 98.00%, specificity of 55 to 96.87% and AUC of 0.66 to 0.997 in predicting critical COVID-19. Nine articles used AI models to predict the epidemic of the COVID-19, such as epidemic peak, infection rate, number of infected cases, transmission laws, and development trend. Eight articles used AI to explore potential effective drugs, primarily through drug repurposing and drug development. Finally, 1 article predicted vaccine targets that have the potential to develop COVID-19 vaccines.
Conclusions: In this review, we have shown that AI achieved high performance in diagnosis, prognosis evaluation, epidemic prediction and drug discovery for COVID-19. AI has the potential to enhance significantly existing medical and healthcare system efficiency during the COVID-19 pandemic.
Introduction
Coronavirus Disease 2019 (COVID-19), caused by severe acute respiratory syndrome virus 2 (SARS-CoV-2) was first detected in December 2019, and spread rapidly to most cities and countries around the world (1–3). During face-to-face contact, SARS-CoV-2 is mainly transmitted through respiratory droplets (4). The infection may cause mild symptoms of upper respiratory tract infections, as well as extremely severe sepsis and shock. It may lead to serious and lethal complications in vulnerable populations, especially in the elderly with comorbidities (4–6). As of 16 March 2021, SARS-CoV-2 has affected more than 200 countries and led to enormous losses, causing more than 120 million confirmed cases and 2.6 million identified deaths. The rising incidence and massive casualties caused by COVID-19 exert considerable pressure on limited healthcare resources. Effective tools are needed to streamline the diagnosis, treatment and surveillance of COVID-19 and increase the clinical efficiency of healthcare systems (7). Recent studies have shown that artificial intelligence is a promising technology as they can achieve better scale-up, accelerate processing power, and even outperform humans in specific healthcare tasks (8).
Artificial intelligence (AI) is a field of algorithm-based applications that enable machines to solve knowledge problems and use algorithms to simulate human decision-making, and continuously improves performance by applying inputted data to perform specific tasks (9–11). The advantages of AI are reflected in high sensitivity and specificity in identifying the object, the speed of reporting and consistency of results (9). In recent years, AI has made significant progress, especially in predictive machine learning models for medical care. Deep learning is a method of ML, based on the complex architectures of Artificial Neural Networks (ANN). Deep learning reveals significant discriminative performance after providing sufficient training data sets and is essential for making predictions (12). In medicine, technologies based on Artificial intelligence and machine learning (AI/ML) aim to improve the quality of medical care, increase diagnostic accuracy and reduce potential errors and predict outcomes by discovering new insights from the enormous amount of data produced by the experience of many patients (10).
Researchers have made significant contributions to the campaign against COVID-19, and new COVID-19-related AI models in the literature are rapidly increasing. Well-trained artificial intelligence models can ensure accurate and rapid diagnosis or assist doctors to streamline the diagnosis and reduce manual labor (13, 14). AI models could early detect the patients at higher risk and characterize the epidemiology of COVID-19 and model disease transmission by training data (15, 16). Artificial intelligence-based methods could assist in the discovery of novel drugs and vaccines, such as repurpose exist drugs, screen targets as vaccines based on the potential mutation model to SARS-CoV-2, as well as screen compounds as potential adjuvants for vaccines (3, 17). AI-powered chatbots have been used with success in clinical scenarios and can advise many more people than a manned call center and ease the stress placed on medical hotlines (18). AI could manage the pandemic by using thermal imaging to scan public spaces for people potentially infected, and by enforcing social distancing and lockdown measures (3, 17).
Artificial intelligence has been widely used in COVID-19, including diagnosis, public health, clinical decision making, social control, therapeutics, vaccine development, surveillance, combination with big data, operation of other core clinical services, and management of patients with COVID-19 (3, 18, 19). In order to solve the significant pressure of the limited medical resources caused by the pandemic of COVID-19, rapid diagnosis, accurate prediction, enhanced monitoring, and effective treatments are the most important measures to control the spread of the pandemic. Many related review articles have been published. However, the results of these studies are inconsistent and there is little research systematically assessing the application of AI for COVID-19 in accordance with PRISMA, and most of them only discuss aspects such as diagnosis or treatment. Therefore, we conducted this review to assess the performance of AI for COVID-19 systematically, and to describe the main categories of AI use, the potential benefits and limitations and future directions for AI.
Methods
We registered the protocol for this review in advance (PROSPERO CRD42020211555, URL: https://www.crd.york.ac.uk/prospero/).
Search Strategy and Eligibility Criteria
This systematic review is reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (20) (Supplementary Material 2). We searched PubMed, Embase and the Cochrane Library for published studies from the inception of these resources to 19 September 2020 using the following terms related to artificial intelligence and COVID-19: “Artificial intelligence,” “Machine Intelligence,” “Machine learning,” “Deep learning,” “Predictive model,” “2019 novel coronavirus disease,” “COVID-19,” “2019 novel coronavirus infection,” “coronavirus disease-19,” and “2019-nCoV disease.” The details of the search strategy are in the Supplementary Material 1.
We included original studies fulfilling the following criteria: (I) research topic was focused on the application of AI for COVID-19, (II) participants had a confirmed diagnosis of COVID-19 by reverse transcription-polymerase chain reaction (RT-PCR) testing or other laboratory examination (where appropriate), and (III) article was published in English.
We excluded studies if: (I) insufficient data were available, (II) we were unable to access the full text or complete data, or (III) the report was a review, case-report or comment.
Two trained researchers (Lian Wang, Dongguang Wang) screened titles, abstracts and the full text of potentially eligible studies independently using Endnote X8.2 software, Thomson Reuters. Discrepancies were resolved through consultation with a third researcher (Xiang Tong, Tao Liu).
Data Abstraction and Quality Assessment
We extracted data and recorded the following information for each study: basic information for the article (title, first author, date of publication), experimental design (algorithm, sample size) and primary outcome (sensitivity, accuracy and specificity of AI for diagnosis and prognosis evaluation; prediction of epidemic; drug repurposing and development). If a study used multiple models, we extracted the most discriminative one.
Three researchers (SZ, JH, LZ) used PROBAST (prediction model risk of bias assessment tool) to assess the risk of bias in the included studies (21). The PROBAST statement was divided into four domains: participants, predictors, outcome, and analysis. These domains contain a total of 20 signal questions to help structure judgment of risk of bias for prediction models, such as the range of included patients, whether the same predictors and results were defined for all participants, whether the clinical decision rule was determined prospectively, and whether a relevant measure of accuracy was reported (22). The details of PROBAST are in the Supplementary Material 3.
We divided the included studies into four categories: diagnosis, prognosis, epidemic prediction, and drug discovery. In the diagnosis and prognosis domains, we evaluated the classification performance using AUC, accuracy, sensitivity, and specificity. For epidemic trends and drugs and vaccines discovery, we listed the results only because there are no suitable evaluation indicators.
Role of the Funding Source
This study was supported by National Key R&D Program of China (2017YFC1309703) and 1·3·5 project for disciplines of excellence–Clinical Research Incubation Project, West China Hospital, Sichuan University (2019HXFH008). The funders of this research did not contribute to the study design, data analysis, data interpretation or preparation of the manuscript.
Results
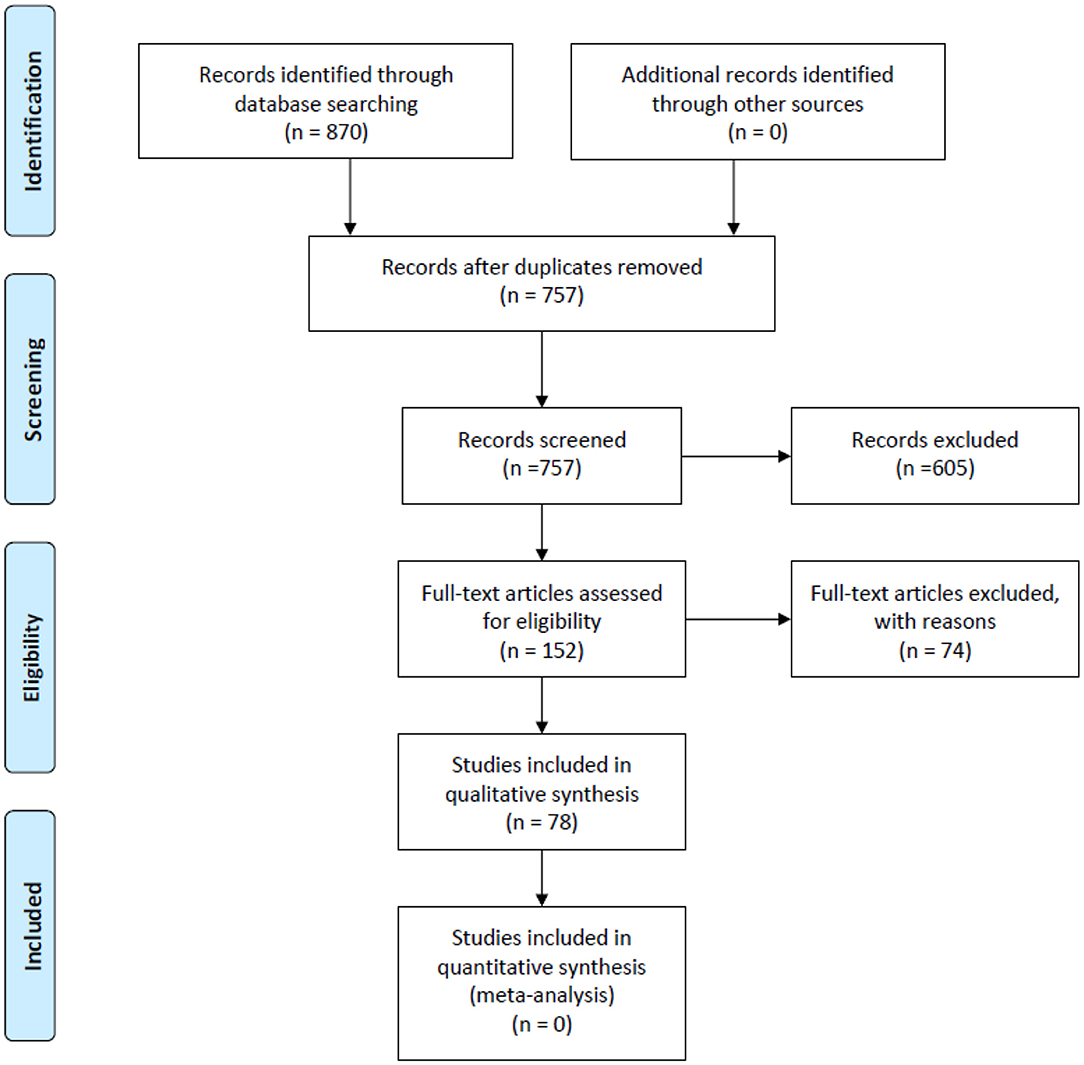
We retrieved a total of 870 records from PubMed, Embase and the Cochrane Library. Of these, 78 studies met our inclusion criteria. Details of the study selection process are shown in Figure 1. Among the 78 included studies (12, 14, 17, 23–97), 46 discussed AI-assisted diagnosis of COVID-19 (14, 23–67), 14 evaluated prognosis (12, 68–80), 9 estimated infected cases, infection rates and epidemic trends (81–89), 8 explored potential effective and safe drugs (90–97), primarily through drug repurposing and drug development and 1 article predicted vaccine targets that has the potential to develop COVID-19 vaccines (17).
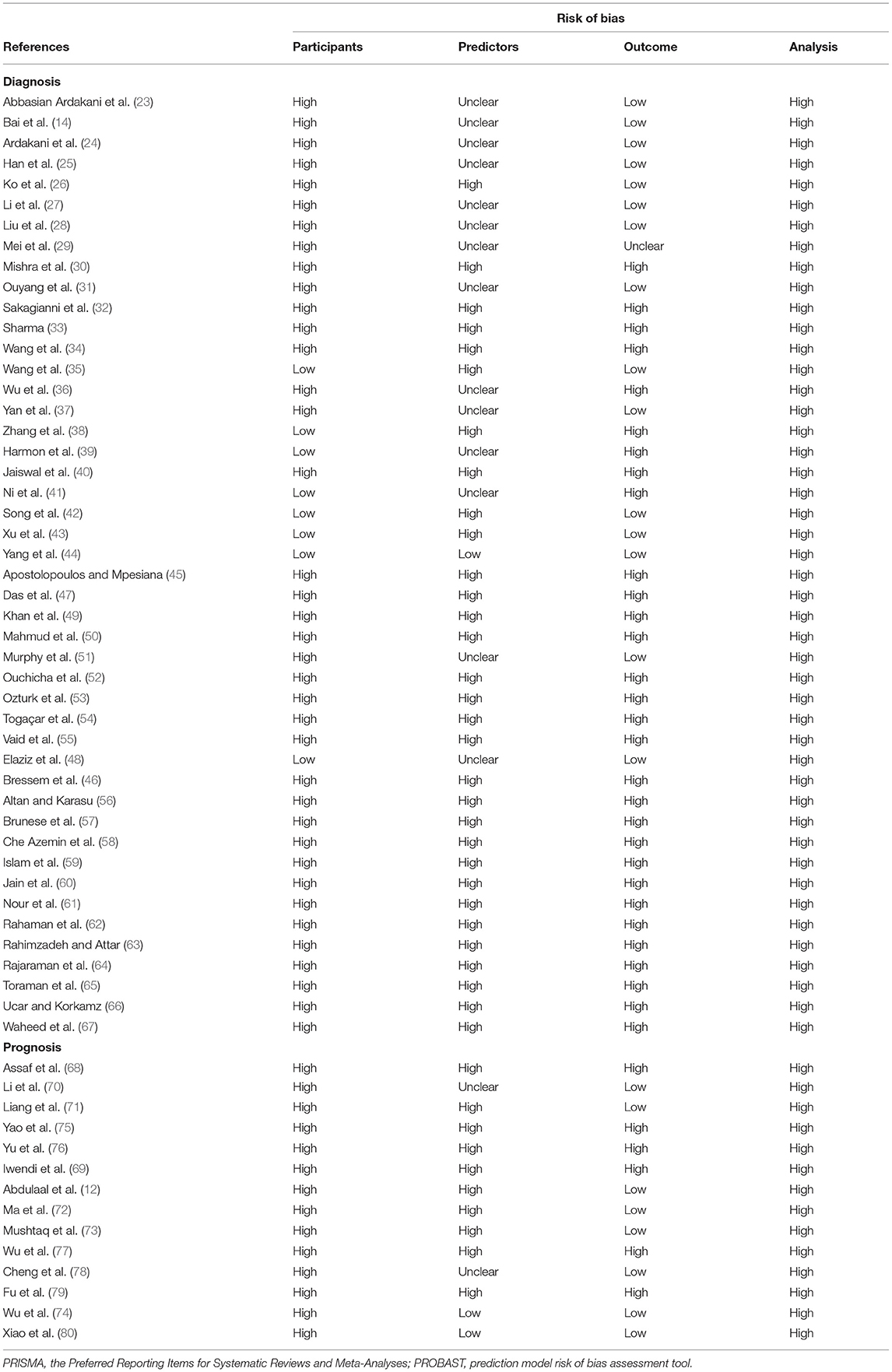
We used PROBAST to evaluate the quality of the 60 articles related to diagnosis or prognosis for COVID-19 (Table 1) (12, 14, 23–80). According to the assessment with PROBAST, all models had a high risk of bias. In the absence of appropriate evaluation tools for the other 18 articles, the quality of these was not assessed (17, 81–97).
AI-Assisted Diagnosis for COVID-19
We included 46 studies related to AI-assisted diagnosis through chest images for COVID-19 (14, 23–67). The findings of these studies ranged as follows, total accuracy: 70.00 to 99.92%, sensitivity: 73.00 to 100.00%, specificity: 25 to 100.00%, AUC: 0.732 to 1.000.
Chest CT Images
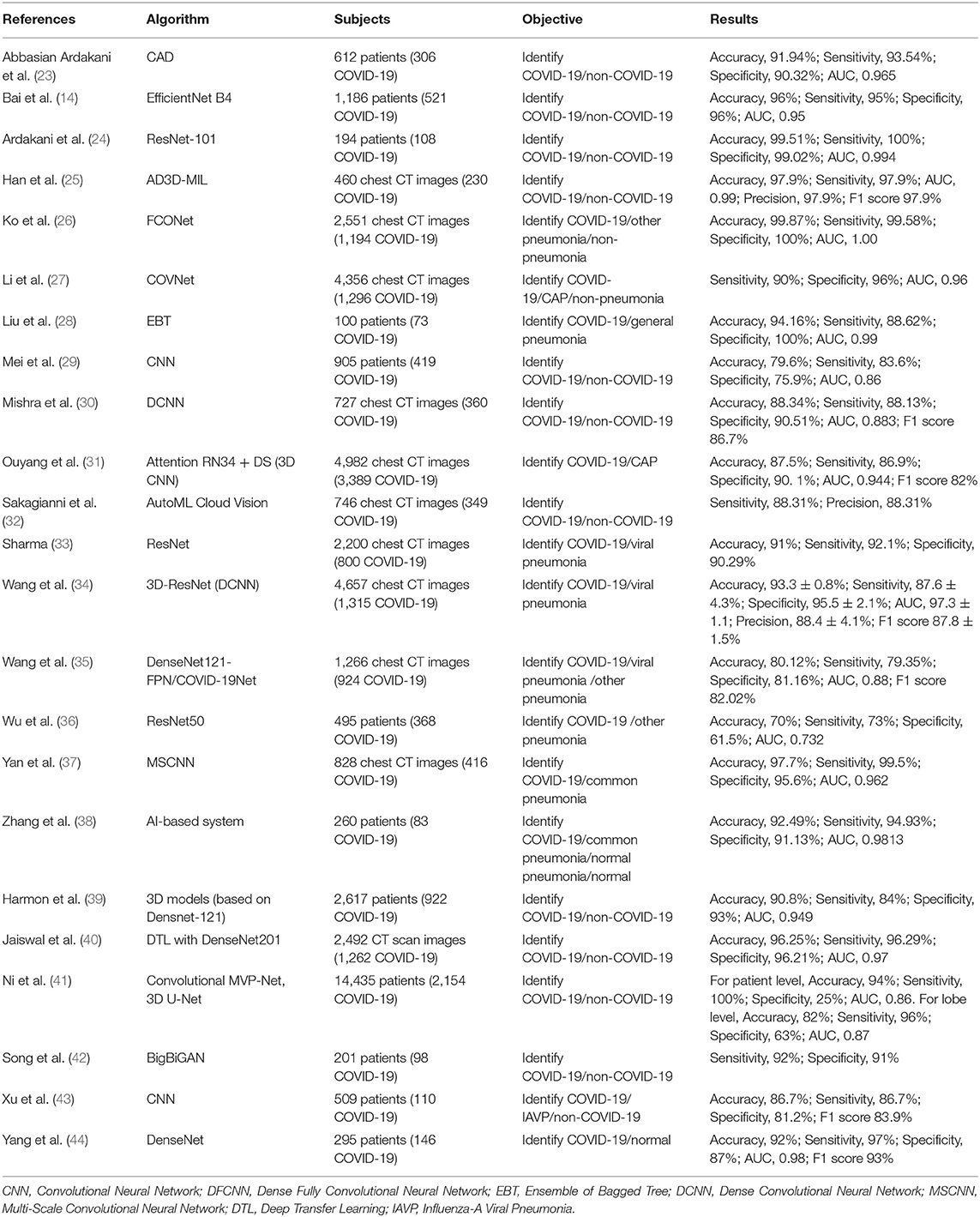
Deep learning with a convolutional neural network (CNN) has gained increasing attention for its outstanding image recognition performance (98). Several of the studies (n = 18) we identified had developed AI models based on CNN and these showed excellent ability to discriminate COVID-19 and non-COVID pneumonia by automatically detecting chest CT images with an accuracy of 70.00 to 99.87%, sensitivity of 73.00 to 100.00%, specificity of 25 to 100.00%, and AUC of 0.732 to 1.000 (Table 2) (14, 23, 24, 26, 27, 29–31, 33–40, 43, 44). Mei et al. (29) developed a joint CNN model that diagnoses COVID-19 patients rapidly by combining chest CT findings with clinical symptoms, exposure history, and laboratory tests. Moreover, Mishra et al. (30) proposed a decision-fusion approach, which combined the predictions of each Deep CNN model and achieved results above 86% for all the performance metrics under consideration. Three studies (14, 41, 42) found that AI models had higher test accuracy, sensitivity, specificity than radiologists, and with the assistance of AI, the radiologists made diagnosis with much faster speeds and achieved a higher diagnostic performance.
In addition, some studies reported applications distinguishing COVID-19 from non-COVID-19 or other pneumonia by using the AD3D-MIL model (25), Ensemble of Bagged Tree (28), AutoML Cloud Vision (32). All of these performed well.
Chest X-Ray Images
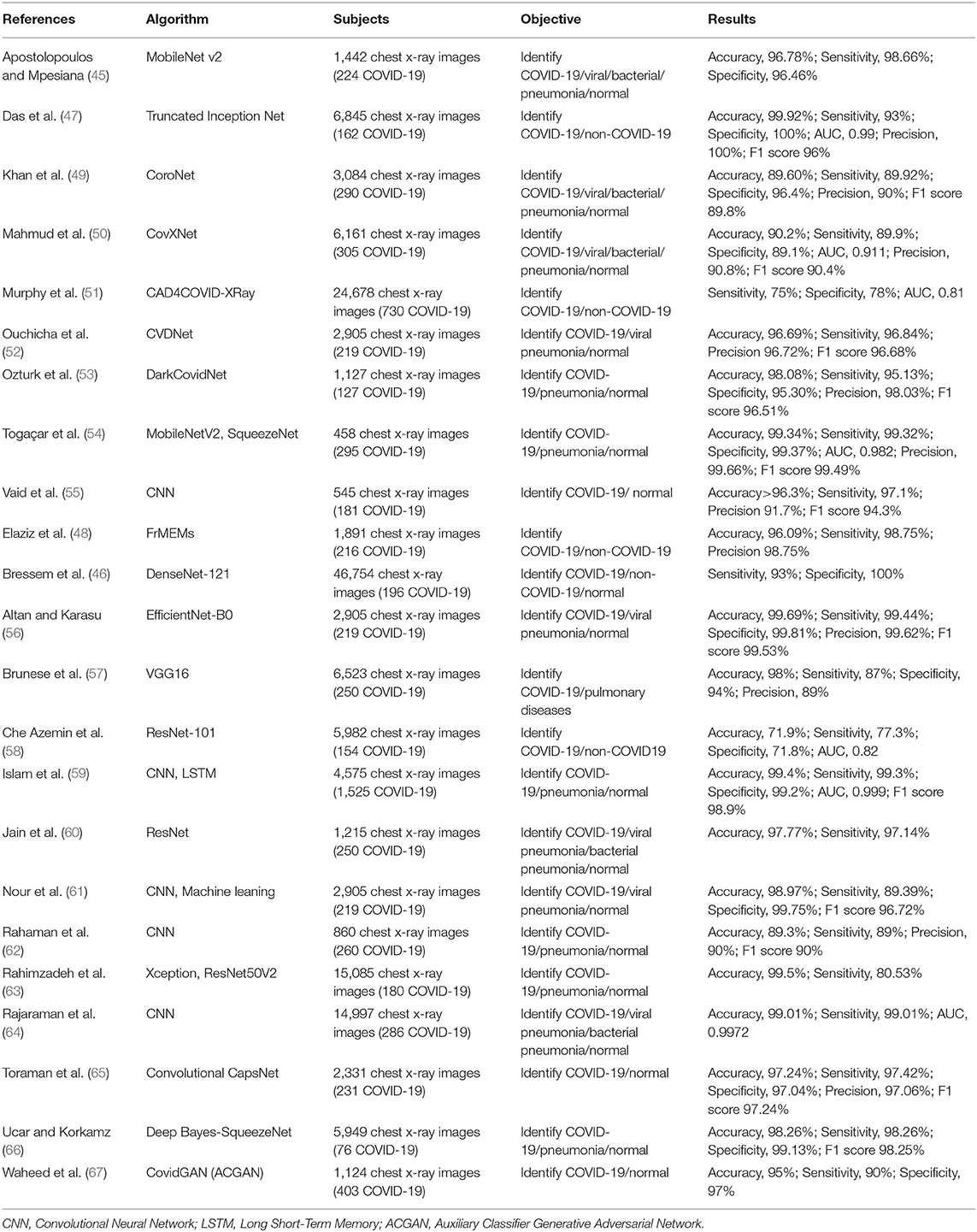
Although CT images have high sensitivity in detecting COVID-19, costs and radiation doses are relatively high. On the contrary, chest X-ray is a low-cost and rapid detection method, which might be used in initial screening of suspected cases of COVID-19 infection, supporting the timely application of quarantine measures in positive patients (45). Several studies have developed AI techniques to automatically detect and extract features from chest X-rays to assist in the diagnosis of COVID-19 with high accuracy (71.90 to 99.92%), sensitivity (75.00 to 99.44%), specificity (71.80 to 100.00%), and AUC (0.81 to 0.999) (45–67) (Table 3).
Predicting the Prognosis of COVID-19
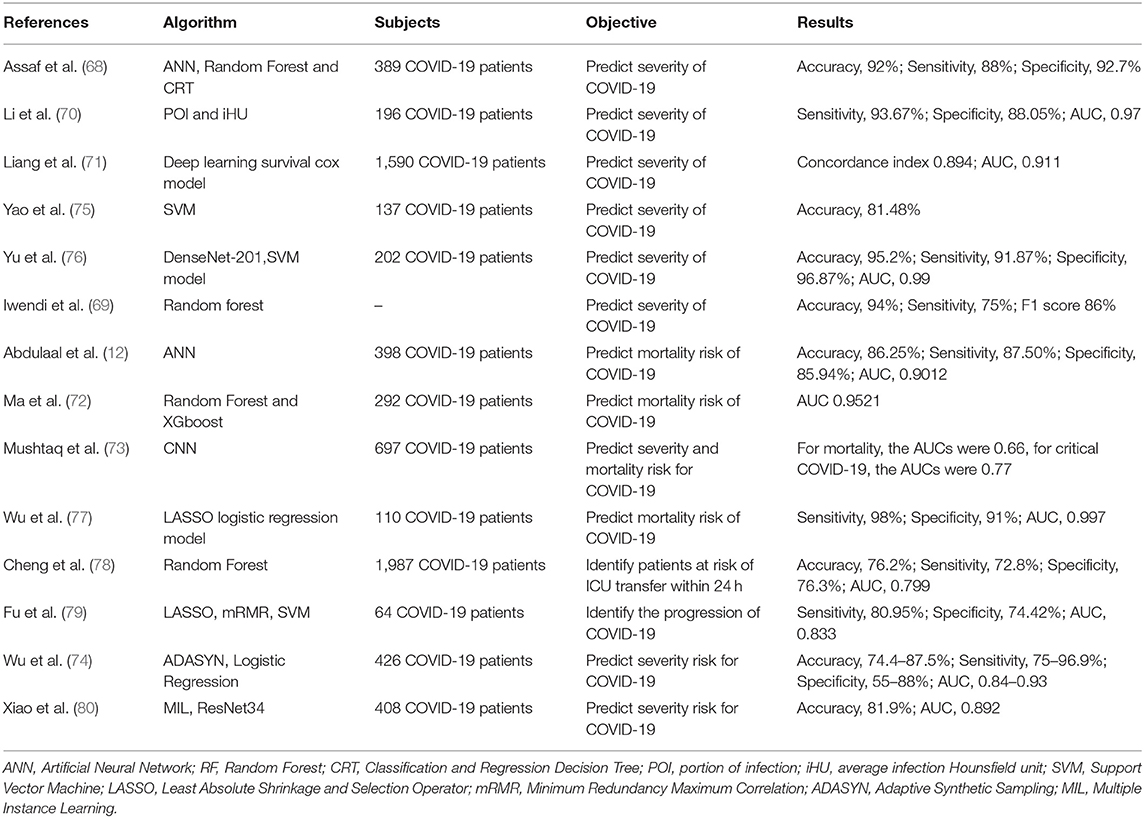
The ability to identify a patient's risk of deterioration during their hospitalization is critical for effective medical resource allocation and to ensure that patients receive appropriate management during the COVID-19 pandemic. We identified several AI models built on the chest CT images that accurately quantified lung abnormalities related to COVID-19 and evaluated the severity and prognosis of the disease (70, 76, 79, 80). Some studies showed that deep learning models could predict the risk of COVID-19 patients developing critical illness, based on clinical characteristics at hospital admission, such as clinical, laboratory and radiological characteristics (68, 71, 73–75, 77, 78). Iwendi et al. (69) developed a model using the geographical, traveling, health, and demographic data of COVID-19 patients to predict the severity and the possible outcomes of the cases. In general, AI models reached accuracy of 74.4 to 95.20%, sensitivity of 72.8 to 98.00%, specificity of 55 to 96.87% and AUC of 0.66 to 0.997 in predicting critical COVID-19 (Table 4). Accurately determining the prognosis of COVID-19 patients as early as possible and starting early treatment may improve their prognosis and reduce mortality from COVID-19.
Predicting the Epidemic Trend of COVID-19
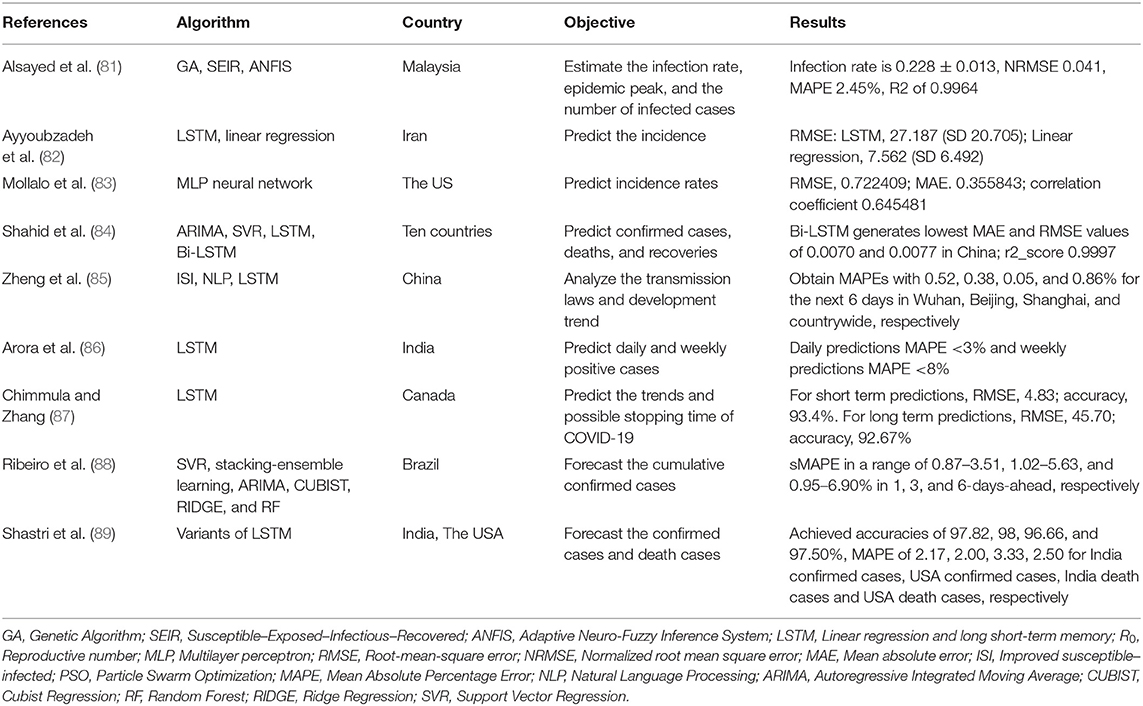
COVID-19 has spread globally and had a substantial impact. It was defined as a pandemic by the World Health Organization (WHO) in March 2020. As the COVID-19 pandemic evolves, it is vital to focus on building prediction models to help policymakers and health managers to allocate healthcare resources and prevent or limit outbreaks (82). We identified 9 studies that sought to predict the epidemic trend of COVID-19 (81–89) (Table 5). Of these, 6 studies used long short-term memory (LSTM) models with or without other models to predict the incidence, confirmed cases, deaths and recoveries, development trend and possible stopping time of COVID-19 (82, 84–86, 88, 89). Alsayed et al. (81) used the Susceptible–Exposed–Infectious–Recovered (SEIR) model combined with machine learning to predict the epidemic's evolution or estimate the unreported number of infections. Mollalo et al. (83) tested the applicability of multi-layer perceptron (MLP) artificial neural networks in simulating cumulative incidence of COVID-19 at the county-level across the continental USA. Shahid et al. (84) proposed prediction models including support vector regression (SVR), autoregressive integrated moving average (ARIMA), long short-term memory (LSTM) and Bi-directional long short-term memory (Bi-LSTM) to predict confirmed cases, deaths and recoveries in ten major countries affected by COVID-19. Zheng et al. (85) proposed an improved susceptible–infected (ISI) model to estimate the variety of the infection rates and to analyze the transmission laws and development trend. Ribeiro et al. (88) used several machine learning models to forecast the cumulative confirmed cases of COVID-19 in the ten Brazilian states with a high daily incidence, and rank the models based on their accuracy. The results of these studies may bring broad benefits by helping to control and prevent COVID-19.
Drug Discovery and Vaccine Development for COVID-19
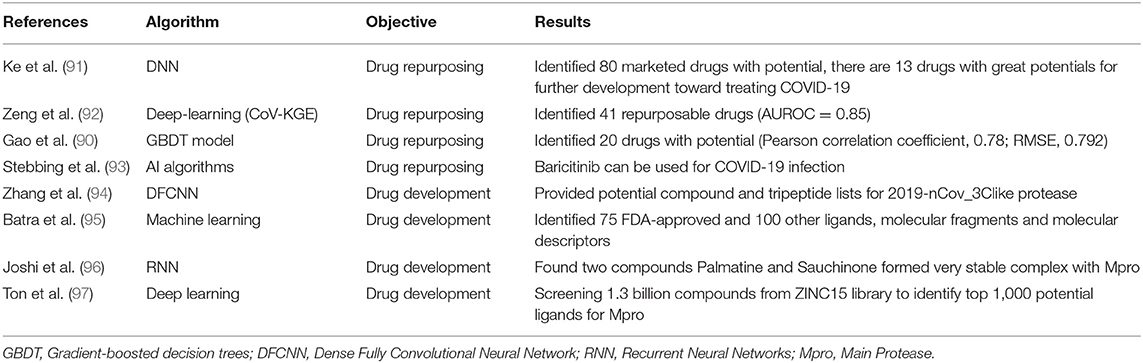
With the spread of COVID-19 showing no signs of slowing and there are few proven effective therapeutics for COVID-19, thousands of people continue to die from the disease every day. It is essential to develop antiviral drugs and vaccines against SARS-CoV-2. It usually needs a long time to develop a drug or vaccine using traditional methods but to try to accelerate this process, several studies have applied AI techniques to identify potential drugs and develop effective and safe vaccines for COVID-19. We identified 9 studies that developed models to find potential drugs and vaccines for COVID-19(17, 90–97) (Table 6).
Drug Repurposing
Drug repurposing refers to the application of approved drugs to new therapeutic indications, which has become a successful drug development strategy for reducing development costs and increasing the simplicity of drug approval procedures (99). AI algorithms could be trained and then be used to screen existing drugs that may prove effective in the treatment of COVID-19. Ke et al. (91) used AI to identify 13 drugs with activities against feline infectious peritonitis (FIP) coronavirus, and further studies proved their activities against SARS-CoV-2 in clinical applications. In another study, Zeng et al. (92) identified 41 high-confidence repurposed drug candidates with a higher area under the receiver operating characteristic (AUROC) of 0.85. Gao et al. (90) developed a gradient-boosted decision trees (GBDT) model for screening 8,565 drugs in DrugBank, finally finding 20 FDA-approved drugs and 20 investigational or off-market drugs that might be effective against SARS-CoV-2. Stebbing et al. (93) used AI prediction to identify Baricitinib, which is used to treat rheumatoid arthritis and myelofibrosis, can be used for COVID-19 infection through proposed anti-cytokine effects and as an inhibitor of host cell viral propagation.
Drug Development
Zhang et al. (94) built a protein 3D model of 3CLpro and used a deep learning method to identify protein-ligand interacting pairs, and finally provided potential compound and tripeptide lists for 3CLpro. Batra et al. (95) combined machine learning and high-fidelity ensemble docking to identify 75 FDA-approved and 100 other ligands from drug data sets as potential therapeutic agents against COVID-19. Joshi et al. (96) used deep-learning models to screen natural compounds and found that two compounds Palmatine and Sauchinone formed very stable complex with Mpro, which may be considered for therapeutic development against the SARS-CoV-2. Ton et al. (97) developed Deep Docking (DD) to screen 1.3 billion compounds from ZINC15 library and identify top 1,000 potential ligands for SARS-CoV-2 Mpro protein.
Vaccine Development
Without an existing effective medical therapy, the development of an effective and safe vaccine is an important method to deal with this highly infectious disease caused by the SARS-CoV-2 coronavirus. Ong et al. applied a ML tool to predict the S protein, nsp3, 3CL-pro, and nsp8-10 were crucial to the viral adhering and host invasion by investigating the entire proteome of SARS-CoV-2. SARS-CoV-2 S protein has the highest protective antigenicity score and was identified as the most favorable vaccine candidate, besides, the nsp3 protein was selected for further investigation (17). The predicted vaccine targets have the potential for COVID-19 vaccine developed, however, they need to be further evaluated in clinical studies.
Discussion
Our systematic review includes 78 articles on the application of AI for COVID-19. These spanned radiological diagnosis, prediction of prognosis, estimation of epidemic trends and drugs and vaccines discovery for COVID-19.
The gold standard for diagnostic tests for COVID-19 is real-time reverse-transcriptase polymerase chain reaction (RT-PCR). However, RT-PCR does produce false negatives or fluctuating results (100). A study compared the diagnostic performance of chest computed tomography (CT) scan with RT-PCR and found that the chest CT is more sensitive than RT-PCR (98 vs. 71%, respectively, P < 0·001) (101), suggesting that Chest CT could be a supplementary diagnostic measure to help physicians make faster and more accurate decisions. AI technique is used for identifying or classifying images, recognizing speech and processing natural language (102). It is well-suited to developing tools to assist with the use of chest CT to diagnose COVID-19 (103). Advanced AI-based algorithms can learn the typical CT image signs, such as bilateral and subpleural ground-glass opacities (GGO), vascular thickening, spider web, and even crazy-paving patterns (104). In addition, the algorithms can also learn some high-dimensional features that radiologists cannot handle, such as texture and wavelet information, thereby allowing pneumonia caused by SARS-CoV-2 to be distinguished from that caused by other pathogens, through advanced AI-based algorithms (36). Several studies have shown that deep learning can automatically differentiate COVID-19 from non-COVID-19 or other pneumonia diseases through extracting features from chest CT and X-rays images. As shown in Tables 2, 3, most of the studies achieved over 90% accuracy, sensitivity and specificity. The performance of AI-assisted diagnosis was comparable to radiologists with significant clinical experience and could assist and improve the performance of radiologists. This means that AI-assisted diagnosis is a useful screening tool, which might shorten patient waiting time, simplify the workflow, reduce the workload of radiologists and allow them to respond more quickly and effectively in emergencies (38). AI techniques have recently shown great potential in the real-time diagnosis of COVID-19 by using images. However, the severity of disease, comorbidities, and the proportion of asymptomatic patients have an impact on the diagnostic sensitivity of chest CT (105). Chest CT have a relatively high sensitivity in symptomatic COVID-19 patients, but low specificity (106). The Italian Society of Medicine and Interventional Radiology recommends that CT should be used as a screening tool only for symptomatic patients with specific indications (107). AI should be used to assist diagnosis, not an independent diagnostic tool. Second, the evaluation of patients based on a single data type may be biased, therefore, AI-assisted diagnosis needs to be used in combination with other laboratory tests and a multimodal AI framework was required to analyze different data types (108). Third, several studies used a relatively small amount of data to train the deep learning models, and the testing data set had the same sources as the training data set. This may cause the problem of overfitting of the models (108, 109). Fourth, there is little evidence to directly compare the performance of humans and machines or the performance of AI in actual clinical work. Only Bai, Song, Ni, and a latest research show that AI assistance improved radiologists' performance in identifying COVID-19(13, 14, 41, 42). In addition, confounding factors can influence the internal validity of researches and the accuracy of AI-based radiological interpretation, such as the variation of respiratory effort, image contrast, technique, and resolution of radiological images (110).
In regard to the prognosis of patients with COVID-19, information available at hospital admission, typically 6 days (median) before the patient developed severe COVID-19, can be used by AI for early detection of patients at higher risk, allowing adjustments to their in-hospital allocation and management (68). AI can evaluate the prognosis of COVID-19 patients by clinical manifestations, laboratory and radiological characteristics and identify potential predictive biomarkers related to the disease's severity. The significantly elevated LDH levels reflects the severity of pneumonia, and increased serum CRP predicts the risk of death in patients with severe COVID-19 (72). Age and comorbidities may be risk factors for severe COVID-19 after hospitalization, such as diabetes, hypertension and cardiovascular diseases (74). As well as chest CT images being a powerful tool to assist clinical diagnosis because of its high sensitivity and the ability to quantify the COVID-19 associated lung abnormalities, they also help assess the severity and prognosis of the disease and monitor the development of the disease (70, 76, 80). Accurate risk prediction of patients with critical COVID-19 may help to optimize patient triage and in-hospital allocation, monitor disease progression and treatment response, prioritize medical resources and improve the overall management of the COVID-19 pandemic (68). However, several of the studies we identified were retrospective single-center studies, which reduces their external validity. Therefore, the results in this review may not be generalizable to other environment and healthcare systems, especially considering the high variability of COVID-19 in different countries and populations (68, 72). In addition, prospective studies with a larger number of patients from multiple locations are required to verify the predictive ability of a model (81).
Many statistical and numerical models have been used to predict the trend of the COVID-19 pandemic, such as the epidemic peak, transmission and development trend, the SEIR model is one of the most popular models (81). Alsayed et al. combined the SEIR model with machine learning to characterize the epidemic dynamics and to predict possible contagion scenarios of COVID-19 in Malaysia (81). Long short-term memory (LSTM) is a recurrent neural network that is an effective model for the prediction of time series where data are sequential (82). Therefore, LSTM has been widely used to predict the confirmed cases, death and recovery, development trend of COVID-19 through time (82, 84–87, 89). Embed the NLP and LSTM into the Improved susceptible–infected (ISI) model is more accurate and reliable than the traditional epidemic model, providing a basis for estimating the law of virus transmission (85). In addition, AI models, such as SVR, stacking-ensemble learning, ARIMA, CUBIST, RIDGE, RF and MLP also play an important role in estimating the epidemic of COVID-19 (83, 88). AI techniques provide useful tools to help policy makers make decisions and take actions to prevent diffusion at the early stage of the epidemic and to minimize the subsequent impact of COVID-19. However, we cannot verify and validate the database, so it is difficult to compare and calibrate results with other studies.
Traditional drug repurposing design methods are based on repeated trials and there is no systematic way to screen the enormous drug-dose parameter space (111). AI is an effective approach to quickly detect potential drugs as antiviral therapeutics for COVID-19 (91). Deep learning, using the relationship between drug targets and diseases can be used as a helpful tool to assist drug repurposing and minimize the possibility of failure in clinical trials (92). Chymotrypsin-like protease is a major therapeutic target, and several studies used it to identify potential therapeutic drugs against COVID-19 by performing drug screening over protein-ligand or protein-peptide among existing drugs (90, 94, 112). In addition, using AI techniques to conduct virtual screening of biologically active compounds can support new drug discovery. However, all predicted drugs must be tested in randomized trials before being used in COVID-19 patients (92).
During the COVID-19 pandemic, an effective and safe vaccine is essential to prevent infection and reduce deaths. The development of vaccines was a complicated process with many difficulties, such as the complexity of the human immune system and the variability between different populations (113). Research organizations in many nations and multinational companies are developing various vaccines, including whole virus vaccine, subunit vaccine, nucleic acid vaccines. Researchers are trying to use AI techniques to explore the vaccine development. Ong et al. (17) predict 6 vaccine candidates, including S protein, nsp3, 3CL-pro, and nsp8-10, S protein was identified as the most favorable vaccine candidate. Nsp3 has a high antigen protection score and has not been used for vaccine development, therefore it was also selected for further investigation. Currently, S protein has been widely used in subunit vaccines, and other proteins are expected to be used in vaccine development. In addition, the latest research analyzed the entire SARS-CoV-2 proteome via AI and identified some of the epitope hotspots that can be used in vaccine formulations (114).
AI has the potential to be an important tool in the fight against COVID-19 and similar pandemics. However, there were many problems in using AI to predict and diagnose COVID-19, and rigorous clinical trials were required before drugs and vaccines developed by AI are approved, so the use of AI has so far been rather limited. It requires continuous efforts by researchers. But recent studies have shown that AI tools such as computer vision and robots have the potential to be widely promoted and used in the short term, such as infrared thermal cameras have been paired with AI-powered facial recognition systems to determine if the individual are wearing masks, using camera images to observe whether social distancing rules are complied, AI-based dialogue chatbots can complete symptom screening and patient education (19). Robotics, AI, and digital technology have been implemented in sanitation for hospitals and public areas, delivery in hospitals and public spaces, patrolling, screening, health consulting, and virus tracking (115).
Our systematic review has several limitations that should be noted. First, although we conducted a systematic search, we only included articles published in English, introducing the possibility of publication bias. Second, in the included studies, the models in 60 studies were at high risk of bias according to assessment with PROBAST (Table 1), and the remaining 18 studies were not evaluated due to lack of suitable evaluation tools. Therefore, the predictive performance of these AI models when used in practice is probably lower than that reported, which means that the predictions of these models may be unreliable. Third, many studies had small sample sizes, and the testing data set had the same sources as the training data set, which leads to an increased risk of overfitting. Fourth, over one fifth of the studies were retrospective single-center studies, which might limit their applicability to the specific center or the same geographical region. This means that results may not be generalizable to other settings and places.
Conclusions
Artificial intelligence has been widely explored in the medical field, especially for enhancing medical and healthcare capabilities. At present, many countries continue to struggle to contain the spread of COVID-19. Facing limited medical resource and increasing healthcare pressure, the use of AI techniques to assist with diagnosis, treatment, prediction of prognosis, evaluation of epidemic trends, surveillance and public health decision-making may improve the efficiency and ability of humans to fight the COVID-19 pandemic.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YZ and LW conceived of the study. LW and DW screened the literature for relevancy and did the data extraction. SZ, JH, and LZ did the quality appraisal. XT and TL resolved any disagreements in study relevancy, extraction, and quality appraisal. LW and LC drafted and revised the manuscript. HF, YZ, and MC directed and revised the manuscript. All authors participated in data interpretation and revised the manuscript for intellectual content.
Funding
This study was supported by National Key R&D Program of China (2017YFC1309703), 1·3·5 project for disciplines of excellence–Clinical Research Incubation Project, West China Hospital, Sichuan University (2019HXFH008), and Science & Technology Department of Sichuan Province (2020YFS0186). The funders of this research did not contribute to the study design, data analysis, data interpretation or preparation of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.704256/full#supplementary-material
References
1. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
2. Zhou T, Liu Q, Yang Z, Liao J, Yang K, Bai W, et al. Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. J Evid Based Med. (2020) 13:3–7. doi: 10.1111/jebm.12376
3. Ma X, Wang Y, Gao T, He Q, He Y, Yue R, et al. Challenges and strategies to research ethics in conducting COVID-19 research. J Evid Based Med. (2020) 13:173–177. doi: 10.1111/jebm.12388
4. Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. (2020) 324:782–93. doi: 10.1001/jama.2020.12839
5. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
6. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. (2020) 323:1061–9. doi: 10.1001/jama.2020.1585
7. Yassine HM, Shah Z. How could artificial intelligence aid in the fight against coronavirus? Expert Rev Anti Infect Ther. (2020) 18:493–7. doi: 10.1080/14787210.2020.1744275
8. Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Fut Healthc J. (2019) 6:94–8. doi: 10.7861/futurehosp.6-2-94
9. Shen J, Zhang CJP, Jiang B, Chen J, Song J, Liu Z, et al. Artificial intelligence versus clinicians in disease diagnosis: systematic review. JMIR Med Informat. (2019) 7:e10010. doi: 10.2196/10010
10. Hwang TJ, Kesselheim AS, Vokinger KN. Lifecycle regulation of artificial intelligence- and machine learning-based software devices in medicine. JAMA. (2019) 322:2285–6. doi: 10.1001/jama.2019.16842
11. Adamson AS, Smith A. Machine learning and health care disparities in dermatology. JAMA Dermatol. (2018) 154:1247–8. doi: 10.1001/jamadermatol.2018.2348
12. Abdulaal A, Patel A, Charani E, Denny S, Mughal N, Moore L. Prognostic modeling of COVID-19 using artificial intelligence in the united kingdom: model development and validation. J Med Internet Res. (2020) 22:e20259. doi: 10.2196/20259
13. Yousefzadeh M, Esfahanian P, Movahed SMS, Gorgin S, Rahmati D, Abedini A, et al. ai-corona: Radiologist-assistant deep learning framework for COVID-19 diagnosis in chest CT scans. PLoS ONE. (2021) 16:e0250952. doi: 10.1371/journal.pone.0250952
14. Bai HX, Wang R, Xiong Z, Hsieh B, Chang K, Halsey K, et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. (2020) 296:E156–65. doi: 10.1148/radiol.2020201491
15. Fang C, Bai S, Chen Q, Zhou Y, Xia L, Qin L, et al. Deep learning for predicting COVID-19 malignant progression. Med Image Anal. (2021) 72:102096. doi: 10.1016/j.media.2021.102096
16. Al-Qaness MAA, Saba AI, Elsheikh AH, Elaziz MA, Ibrahim RA, Lu S, et al. Efficient artificial intelligence forecasting models for COVID-19 outbreak in Russia and Brazil. Process Saf Environ Prot. (2021) 149:399–409. doi: 10.1016/j.psep.2020.11.007
17. Ong E, Wong MU, Huffman A, He Y. COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Front Immunol. (2020) 11:1581. doi: 10.3389/fimmu.2020.01581
18. Naudé W. Artificial intelligence vs COVID-19: limitations, constraints and pitfalls. AI Soc. (2020) 35:761–65. doi: 10.1007/s00146-020-00978-0
19. Chen J, See KC. Artificial intelligence for COVID-19: rapid review. J Med Internet Res. (2020) 22:e21476. doi: 10.2196/21476
20. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
21. Moons KGM, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. (2019) 170:W1–33. doi: 10.7326/M18-1377
22. Wolff RF, Moons KGM, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. (2019) 170:51–8. doi: 10.7326/M18-1376
23. Abbasian Ardakani A, Acharya UR, Habibollahi S, Mohammadi A. COVIDiag: a clinical CAD system to diagnose COVID-19 pneumonia based on CT findings. Eur Radiol. (2020) 31:121–30. doi: 10.1007/s00330-020-07087-y
24. Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput Biol Med. (2020) 121:103795. doi: 10.1016/j.compbiomed.2020.103795
25. Han Z, Wei B, Hong Y, Li T, Cong J, Zhu X, et al. Accurate screening of COVID-19 using attention-based deep 3D multiple instance learning. IEEE Trans Med Imaging. (2020) 39:2584–94. doi: 10.1109/TMI.2020.2996256
26. Ko H, Chung H, Kang WS, Kim KW, Shin Y, Kang SJ, et al. COVID-19 pneumonia diagnosis using a simple 2d deep learning framework with a single chest CT image: model development and validation. J Med Internet Res. (2020) 22:e19569. doi: 10.2196/19569
27. Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al. Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. (2020) 296:E65–71. doi: 10.1148/radiol.2020200905
28. Liu C, Wang X, Liu C, Sun Q, Peng W. Differentiating novel coronavirus pneumonia from general pneumonia based on machine learning. Biomed Eng Online. (2020) 19:66. doi: 10.1186/s12938-020-00809-9
29. Mei X, Lee HC, Diao KY, Huang M, Lin B, Liu C, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat Med. (2020) 26:1224–8. doi: 10.1038/s41591-020-0931-3
30. Mishra AK, Das SK, Roy P, Bandyopadhyay S. Identifying COVID19 from chest CT images: a deep convolutional neural networks based approach. J Healthc Eng. (2020) 2020:8843664. doi: 10.1155/2020/8843664
31. Ouyang X, Huo J, Xia L, Shan F, Liu J, Mo Z, et al. Dual-Sampling attention network for diagnosis of COVID-19 from community acquired pneumonia. IEEE Trans Med Imaging. (2020) 39:2595–605. doi: 10.1109/TMI.2020.2995508
32. Sakagianni A, Feretzakis G, Kalles D, Koufopoulou C, Kaldis V. Setting up an easy-to-use machine learning pipeline for medical decision support: a case study for COVID-19 diagnosis based on deep learning with CT scans. Stud Health Technol Inform. (2020) 272:13–6. doi: 10.3233/SHTI200481
33. Sharma S. Drawing insights from COVID-19-infected patients using CT scan images and machine learning techniques: a study on 200 patients. Environ Sci Pollut Res Int. (2020) 27:37155–63. doi: 10.1007/s11356-020-10133-3
34. Wang J, Bao Y, Wen Y, Lu H, Luo H, Xiang Y, et al. Prior-Attention residual learning for more discriminative COVID-19 screening in CT images. IEEE Trans Med Imaging. (2020) 39:2572–83. doi: 10.1109/TMI.2020.2994908
35. Wang S, Zha Y, Li W, Wu Q, Li X, Niu M, et al. A fully automatic deep learning system for COVID-19 diagnostic and prognostic analysis. Eur Respir J. (2020) 56:2000775. doi: 10.1183/13993003.00775-2020
36. Wu X, Hui H, Niu M, Li L, Wang L, He B, et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur J Radiol. (2020) 128:109041. doi: 10.1016/j.ejrad.2020.109041
37. Yan T, Wong PK, Ren H, Wang H, Wang J, Li Y. Automatic distinction between COVID-19 and common pneumonia using multi-scale convolutional neural network on chest CT scans. Chaos Solitons Fractals. (2020) 140:110153. doi: 10.1016/j.chaos.2020.110153
38. Zhang K, Liu X, Shen J, Li Z, Sang Y, Wu X, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. (2020) 181:1423–33.e11. doi: 10.1016/j.cell.2020.04.045
39. Harmon SA, Sanford TH, Xu S, Turkbey EB, Roth H, Xu Z, et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Commun. (2020) 11:4080. doi: 10.1038/s41467-020-17971-2
40. Jaiswal A, Gianchandani N, Singh D, Kumar V, Kaur M. Classification of the COVID-19 infected patients using DenseNet201 based deep transfer learning. J Biomol Struct Dyn. (2020) 1–8. doi: 10.1080/07391102.2020.1788642
41. Ni Q, Sun ZY, Qi L, Chen W, Yang Y, Wang L, et al. A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest CT images. Eur Radiol. (2020) 30:6517–27. doi: 10.1007/s00330-020-07044-9
42. Song J, Wang H, Liu Y, Wu W, Dai G, Wu Z, et al. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur J Nucl Med Mol Imaging. (2020) 47:2516–24. doi: 10.1007/s00259-020-04929-1
43. Xu X, Jiang X, Ma C, Du P, Li X, Lv S, et al. a deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. (2020) 6:1122–9. doi: 10.1016/j.eng.2020.04.010
44. Yang S, Jiang L, Cao Z, Wang L, Cao J, Feng R, et al. Deep learning for detecting corona virus disease 2019 (COVID-19) on high-resolution computed tomography: a pilot study. Ann Transl Med. (2020) 8:450. doi: 10.21037/atm.2020.03.132
45. Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. (2020) 43:635–40. doi: 10.1007/s13246-020-00865-4
46. Bressem KK, Adams LC, Erxleben C, Hamm B, Niehues SM, Vahldiek JL. Comparing different deep learning architectures for classification of chest radiographs. Sci Rep. (2020) 10:13590. doi: 10.1038/s41598-020-70479-z
47. Das D, Santosh KC, Pal U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys Eng Sci Med. (2020) 43:915–25. doi: 10.1007/s13246-020-00888-x
48. Elaziz MA, Hosny KM, Salah A, Darwish MM, Lu S, Sahlol AT. New machine learning method for image-based diagnosis of COVID-19. PLoS ONE. (2020) 15:e0235187. doi: 10.1371/journal.pone.0235187
49. Khan AI, Shah JL, Bhat MM. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed. (2020) 196:105581. doi: 10.1016/j.cmpb.2020.105581
50. Mahmud T, Rahman MA, Fattah SA. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput Biol Med. (2020) 122:103869. doi: 10.1016/j.compbiomed.2020.103869
51. Murphy K, Smits H, Knoops AJG, Korst M, Samson T, Scholten ET, et al. COVID-19 on chest radiographs: a multireader evaluation of an artificial intelligence system. Radiology. (2020) 296:E166–72. doi: 10.1148/radiol.2020201874
52. Ouchicha C, Ammor O, Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid-19) from chest x-ray images. Chaos Solitons Fractals. (2020) 140:110245. doi: 10.1016/j.chaos.2020.110245
53. Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. (2020) 121:103792. doi: 10.1016/j.compbiomed.2020.103792
54. Togaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. (2020) 121:103805. doi: 10.1016/j.compbiomed.2020.103805
55. Vaid S, Kalantar R, Bhandari M. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int Orthop. (2020) 44:1539–42. doi: 10.1007/s00264-020-04609-7
56. Altan A, Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos Solitons Fractals. (2020) 140:110071. doi: 10.1016/j.chaos.2020.110071
57. Brunese L, Mercaldo F, Reginelli A, Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput Methods Programs Biomed. (2020) 196:105608. doi: 10.1016/j.cmpb.2020.105608
58. Che Azemin MZ, Hassan R, Mohd Tamrin MI, Md Ali MA. COVID-19 deep learning prediction model using publicly available radiologist-adjudicated chest X-Ray images as training data: preliminary findings. Int J Biomed Imaging. (2020) 2020:8828855. doi: 10.1155/2020/8828855
59. Islam MZ, Islam MM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Informat Med Unlock. (2020) 20:100412. doi: 10.1016/j.imu.2020.100412
60. Jain G, Mittal D, Thakur D, Mittal MK. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybernet Biomed Eng. (2020) 40:1391–405. doi: 10.1016/j.bbe.2020.08.008
61. Nour M, Cömert Z, Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and bayesian optimization. Appl Soft Comput. (2020) 97:106580. doi: 10.1016/j.asoc.2020.106580
62. Rahaman MM, Li C, Yao Y, Kulwa F, Rahman MA, Wang Q, et al. Identification of COVID-19 samples from chest X-Ray images using deep learning: a comparison of transfer learning approaches. J Xray Sci Technol. (2020) 28:821–39. doi: 10.3233/XST-200715
63. Rahimzadeh M, Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Informatics in medicine unlocked. (2020) 19:100360. doi: 10.1016/j.imu.2020.100360
64. Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani SK. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. IEEE Access Pract Innovat Open Solut. (2020) 8:115041–50. doi: 10.1109/ACCESS.2020.3003810
65. Toraman S, Alakus TB, Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals. (2020) 140:110122. doi: 10.1016/j.chaos.2020.110122
66. Ucar F, Korkmaz D. COVIDiagnosis-Net: deep bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. (2020) 140:109761. doi: 10.1016/j.mehy.2020.109761
67. Waheed A, Goyal M, Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. CovidGAN: data augmentation using auxiliary classifier GAN for improved Covid-19 detection. IEEE Access Pract Innovat Open Solut. (2020) 8:91916–23. doi: 10.1109/ACCESS.2020.2994762
68. Assaf D, Gutman Y, Neuman Y, Segal G, Amit S, Gefen-Halevi S, et al. Utilization of machine-learning models to accurately predict the risk for critical COVID-19. Intern Emerg Med. (2020) 15:1435–43. doi: 10.1007/s11739-020-02475-0
69. Iwendi C, Bashir AK, Peshkar A, Sujatha R, Chatterjee JM, Pasupuleti S, et al. COVID-19 patient health prediction using boosted random forest algorithm. Front Public Health. (2020) 8:357. doi: 10.3389/fpubh.2020.00357
70. Li Z, Zhong Z, Li Y, Zhang T, Gao L, Jin D, et al. From community-acquired pneumonia to COVID-19: a deep learning–based method for quantitative analysis of COVID-19 on thick-section CT scans. Eur Radiol. (2020) 30:6828–37. doi: 10.1101/2020.04.17.20070219
71. Liang W, Yao J, Chen A, Lv Q, Zanin M, Liu J, et al. Early triage of critically ill COVID-19 patients using deep learning. Nat Commun. (2020) 11:3543. doi: 10.1038/s41467-020-17280-8
72. Ma X, Ng M, Xu S, Xu Z, Qiu H, Liu Y, et al. Development and validation of prognosis model of mortality risk in patients with COVID-19. Epidemiol Infect. (2020) 148:e168. doi: 10.1017/S0950268820001727
73. Mushtaq J, Pennella R, Lavalle S, Colarieti A, Steidler S, Martinenghi CMA, et al. Initial chest radiographs and artificial intelligence (AI) predict clinical outcomes in COVID-19 patients: analysis of 697 Italian patients. Eur Radiol. (2020) 31:1770–9. doi: 10.1007/s00330-020-07269-8
74. Wu G, Yang P, Xie Y, Woodruff HC, Rao X, Guiot J, et al. Development of a clinical decision support system for severity risk prediction and triage of COVID-19 patients at hospital admission: an international multicentre study. Eur Respir J. (2020) 56:2001104. doi: 10.1183/13993003.01104-2020
75. Yao H, Zhang N, Zhang R, Duan M, Xie T, Pan J, et al. Severity detection for the coronavirus disease 2019 (COVID-19) patients using a machine learning model based on the blood and urine tests. Front Cell Dev Biol. (2020) 8:683. doi: 10.3389/fcell.2020.00683
76. Yu Z, Li X, Sun H, Wang J, Zhao T, Chen H, et al. Rapid identification of COVID-19 severity in CT scans through classification of deep features. Biomed Eng Online. (2020) 19:63. doi: 10.1186/s12938-020-00807-x
77. Wu G, Zhou S, Wang Y, Lv W, Wang S, Wang T, et al. A prediction model of outcome of SARS-CoV-2 pneumonia based on laboratory findings. Sci Rep. (2020) 10:14042. doi: 10.1038/s41598-020-71114-7
78. Cheng FY, Joshi H, Tandon P, Freeman R, Reich DL, Mazumdar M, et al. Using machine learning to predict ICU transfer in hospitalized COVID-19 patients. J Clin Med. (2020) 9:1668. doi: 10.3390/jcm9061668
79. Fu L, Li Y, Cheng A, Pang P, Shu Z. A novel machine learning-derived radiomic signature of the whole lung differentiates stable from progressive COVID-19 infection: a retrospective cohort study. J Thorac Imaging. (2020) 35:361–8. doi: 10.1097/RTI.0000000000000544
80. Xiao LS, Li P, Sun F, Zhang Y, Xu C, Zhu H, et al. Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Front Bioeng Biotechnol. (2020) 8:898. doi: 10.3389/fbioe.2020.00898
81. Alsayed A, Sadir H, Kamil R, Sari H. Prediction of epidemic peak and infected cases for COVID-19 disease in Malaysia, 2020. Int J Environ Res Public Health. (2020) 17:1–15. doi: 10.3390/ijerph17114076
82. Ayyoubzadeh SM, Ayyoubzadeh SM, Zahedi H, Ahmadi M, S RNK. Predicting COVID-19 incidence through analysis of google trends data in iran: data mining and deep learning pilot study. JMIR Public Health Surveill. (2020) 6:e18828. doi: 10.2196/18828
83. Mollalo A, Rivera KM, Vahedi B. Artificial neural network modeling of novel coronavirus (COVID-19) incidence rates across the continental United States. Int J Environ Res Public Health. (2020) 17:4204. doi: 10.3390/ijerph17124204
84. Shahid F, Zameer A, Muneeb M. Predictions for COVID-19 with deep learning models of LSTM, GRU and Bi-LSTM. Chaos Solitons Fractals. (2020) 140:110212. doi: 10.1016/j.chaos.2020.110212
85. Zheng N, Du S, Wang J, Zhang H, Cui W, Kang Z, et al. Predicting COVID-19 in China using hybrid AI model. IEEE Trans Cybern. (2020) 50:2891–904. doi: 10.1109/TCYB.2020.2990162
86. Arora P, Kumar H, Panigrahi BK. Prediction and analysis of COVID-19 positive cases using deep learning models: a descriptive case study of India. Chaos Solitons Fractals. (2020) 139:110017. doi: 10.1016/j.chaos.2020.110017
87. Chimmula VKR, Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos Solitons Fractals. (2020) 135:109864. doi: 10.1016/j.chaos.2020.109864
88. Ribeiro M, da Silva RG, Mariani VC, Coelho LDS. Short-term forecasting COVID-19 cumulative confirmed cases: perspectives for Brazil. Chaos Solitons Fractals. (2020) 135:109853. doi: 10.1016/j.chaos.2020.109853
89. Shastri S, Singh K, Kumar S, Kour P, Mansotra V. Time series forecasting of Covid-19 using deep learning models: India-USA comparative case study. Chaos Solitons Fractals. (2020) 140:110227. doi: 10.1016/j.chaos.2020.110227
90. Gao K, Nguyen DD, Chen J, Wang R, Wei GW. Repositioning of 8565 existing drugs for COVID-19. J Phys Chem Lett. (2020) 11:5373–82. doi: 10.1021/acs.jpclett.0c01579
91. Ke YY, Peng TT, Yeh TK, Huang WZ, Chang SE, Wu SH, et al. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed J. (2020) 43:355–62. doi: 10.1016/j.bj.2020.05.001
92. Zeng X, Song X, Ma T, Pan X, Zhou Y, Hou Y, et al. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J Proteome Res. (2020) 19:4624–36. doi: 10.1021/acs.jproteome.0c00316
93. Stebbing J, Krishnan V, de Bono S, Ottaviani S, Casalini G, Richardson PJ, et al. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol Med. (2020) 12:e12697. doi: 10.15252/emmm.202012697
94. Zhang H, Saravanan KM, Yang Y, Hossain MT, Li J, Ren X, et al. Deep learning based drug screening for novel coronavirus 2019-nCov. Interdiscip Sci. (2020) 12:368–76. doi: 10.1007/s12539-020-00376-6
95. Batra R, Chan H, Kamath G, Ramprasad R, Cherukara MJ, Sankaranarayanan S. Screening of therapeutic agents for COVID-19 using machine learning and ensemble docking studies. J Phys Chem Lett. (2020) 11:7058–65. doi: 10.1021/acs.jpclett.0c02278
96. Joshi T, Joshi T, Pundir H, Sharma P, Mathpal S, Chandra S. Predictive modeling by deep learning, virtual screening and molecular dynamics study of natural compounds against SARS-CoV-2 main protease. J Biomol Struct Dyn. (2020) 1–19. doi: 10.1080/07391102.2020.1802341
97. Ton AT, Gentile F, Hsing M, Ban F, Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol Inform. (2020) 39:e2000028. doi: 10.1002/minf.202000028
98. Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Deep learning with convolutional neural network in radiology. Jpn J Radiol. (2018) 36:257–72. doi: 10.1007/s11604-018-0726-3
99. Amelio I, Gostev M, Knight RA, Willis AE, Melino G, Antonov AV. DRUGSURV: a resource for repositioning of approved and experimental drugs in oncology based on patient survival information. Cell Death Dis. (2014) 5:e1051. doi: 10.1038/cddis.2014.9
100. Li Y, Yao L, Li J, Chen L, Song Y, Cai Z, et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J Med Virol. (2020) 92:903–8. doi: 10.1002/jmv.25786
101. Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. (2020) 296:E115–7. doi: 10.1148/radiol.2020200432
102. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. (2015) 521:436–44. doi: 10.1038/nature14539
103. Miller DD, Brown EW. Artificial intelligence in medical practice: the question to the answer? Am J Med. (2018) 131:129–33. doi: 10.1016/j.amjmed.2017.10.035
104. Kanne JP. Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: key points for the radiologist. Radiology. (2020) 295:16–7. doi: 10.1148/radiol.2020200241
105. Kim H, Hong H, Yoon SH. Diagnostic performance of CT and reverse transcriptase polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. (2020) 296:E145–55. doi: 10.1148/radiol.2020201343
106. Adams HJA, Kwee TC, Yakar D, Hope MD, Kwee RM. Systematic review and meta-analysis on the value of chest ct in the diagnosis of coronavirus disease (COVID-19): sol scientiae, illustra nos. AJR Am J Roentgenol. (2020) 215:1342–50. doi: 10.2214/AJR.20.23391
107. Neri E, Miele V, Coppola F, Grassi R. Use of CT and artificial intelligence in suspected or COVID-19 positive patients: statement of the Italian society of medical and interventional radiology. Radiol Med. (2020) 125:505–8. doi: 10.1007/s11547-020-01197-9
108. Santosh KC. AI-Driven tools for coronavirus outbreak: need of active learning and cross-population train/test models on multitudinal/multimodal data. J Med Syst. (2020) 44:93. doi: 10.1007/s10916-020-01562-1
109. Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol. (2019) 20:405–10. doi: 10.3348/kjr.2019.0025
110. Chen J, See KC. Artificial intelligence for COVID-19: a rapid review. J Med Internet Res. (2020) 22:e21476. doi: 10.2196/preprints.21476
111. Abdulla A, Wang B, Qian F, Kee T, Blasiak A, Ong YH, et al. Project IDentif.AI: harnessing artificial intelligence to rapidly optimize combination therapy development for infectious disease intervention. Adv Ther. (2020) 2000034. doi: 10.1002/adtp.202000034
112. Liu S, Zheng Q, Wang Z. Potential covalent drugs targeting the main protease of the SARS-CoV-2 coronavirus. Bioinformatics. (2020) 36:3295–8. doi: 10.1093/bioinformatics/btaa224
113. Kaushal K, Sarma P, Rana SV, Medhi B, Naithani M. Emerging role of artificial intelligence in therapeutics for COVID-19: a systematic review. J Biomol Struct Dyn. (2020) 1–16. doi: 10.1080/07391102.2020.1855250
114. Malone B, Simovski B, Moliné C, Cheng J, Gheorghe M, Fontenelle H, et al. Artificial intelligence predicts the immunogenic landscape of SARS-CoV-2 leading to universal blueprints for vaccine designs. Sci Rep. (2020) 10:22375. doi: 10.1038/s41598-020-78758-5
Keywords: artificial intelligence, COVID-19, diagnosis, prognosis evaluation, epidemic prediction, drug discovery 2
Citation: Wang L, Zhang Y, Wang D, Tong X, Liu T, Zhang S, Huang J, Zhang L, Chen L, Fan H and Clarke M (2021) Artificial Intelligence for COVID-19: A Systematic Review. Front. Med. 8:704256. doi: 10.3389/fmed.2021.704256
Received: 02 May 2021; Accepted: 09 August 2021;
Published: 30 September 2021.
Edited by:
Reza Lashgari, Shahid Beheshti University, IranReviewed by:
Saeid Gorgin, Iranian Research Organization for Science and Technology, IranSeyed Mohammad Sadegh Movahed, Shahid Beheshti University, Iran
Hadi Choubdar, Shahid Beheshti University of Medical Sciences, Iran
Copyright © 2021 Wang, Zhang, Wang, Tong, Liu, Zhang, Huang, Zhang, Chen, Fan and Clarke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Fan, ZmFuaG9uZ2ZhbkBxcS5jb20=; Yonggang Zhang, amVibV96aGFuZ0B5YWhvby5jb20=; Mike Clarke, bS5jbGFya2VAcXViLmFjLnVr
†These authors have contributed equally to this work and share first authorship
 Lian Wang
Lian Wang Yonggang Zhang2,3*†
Yonggang Zhang2,3*† Dongguang Wang
Dongguang Wang Xiang Tong
Xiang Tong Tao Liu
Tao Liu Shijie Zhang
Shijie Zhang Jizhen Huang
Jizhen Huang Li Zhang
Li Zhang Hong Fan
Hong Fan