- 1Department of Orthopedics, The First Hospital of Jilin University, Changchun, China
- 2Department of Orthopedics, The First People's Hospital of Yunnan Province, Kunming, China
Background: Abnormal expression levels of microRNAs (miRNAs) were observed in ankylosing spondylitis (AS) in recent articles, suggesting that miRNAs may be used as biomarkers for AS diagnoses. In this paper, we conducted a meta-analysis to identify the overall diagnostic accuracy of miRNA biomarkers in AS patients.
Methods: An extensive search was undertaken in PubMed, Embase, Cochrane databases, and Wan Fang database up to 30 December 2020 using the following key words: (“microRNAs” or “microRNA” or “miRNA” or “miR” or “RNA, Micro” or “Primary MicroRNA”) and (“Spondylitis Ankylosing” or “Spondyloarthritis Ankylopoietica” or “Ankylosing Spondylarthritis” or “Ankylosing Spondylarthritides” or “Spondylarthritides Ankylosing” or “Ankylosing Spondylitis”) and (“blood” or “serum” or “plasma”). Statistical evaluation of dysregulated miRNAs using the sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and the area under the curve (AUC).
Results: Twenty-nine articles reporting on the miRNAs of AS were included. A total of 42 miRNAs were observed to be up-regulated and 45 miRNAs were down-regulated in the AS cases compared with the controls. Besides, 29 studies from nine articles were included in our meta-analysis. The pooled sensitivity, specificity, PLR, NLR, DOR, and AUC were 0. 76 (95% CI, 0.70–0.81), 0.80 (95% CI, 0.74–0.85), 3.75 (95% CI, 2.82–5.01), 0.30 (95% CI, 0.24–0.39), 12.32 (95% CI, 7.65–19.83), 0.85 (95% CI, 0.81–0.88), respectively, suggesting a good diagnostic accuracy of miRNAs for AS.
Conclusions: Circulating miRNAs are deregulated in AS patients. miRNAs may be used as a relatively non-invasive biomarkers for the detection of AS.
Introduction
Ankylosing spondylitis (AS) is a common chronic rheumatic disease. It is usually characterized by inflammatory back pain, structural and functional disorders, and restricted motion of spine (1). The prevalence of AS is ~0.32% in North America, 0.24% in Europe, 0.17% in Asia, 0.10% in Latin America, and 0.07% in Africa (2). Until now, AS remains difficult to diagnose before sacroiliac joint damage is visible on X-ray. In addition, the pathogenesis of AS is not fully understood yet. Thus, the diagnosis and treatment of this disease still remain difficult. Although AS is genetically closely related to human leukocyte antigen (HLA)-B27, in people of European descent, about 85% of AS patients are HLA-B27 positive (3). However, HLA-B27 has low specificity for AS, and only 5~6% of people with HLA-B27 will suffer from AS disease (4). Besides, HLA-B27 only accounts for <30% of the overall genetic risks of the pathology of AS (5). As a result, there is an urgent need for early biomarkers for AS diagnosis. In addition, there have also numerous other associated genetic polymorphisms been identified, including HLA class I and II alleles (6), the human major histocompatibility complex class I chain-related gene A (7), non-major histocompatibility complex (MHC) regions (8) and microRNAs gene (9), and so on.
MicroRNA (miRNA) is a highly conserved noncoding RNA oligonucleotide during evolution, which can regulate the expression of target genes at the post-transcriptional level. It was validated that miRNAs can regulate cell metabolism, differentiation, proliferation, apoptosis, etc. (10). The expressions of miRNAs can be changed under physiological and pathological processes or treatment (11). Moreover, miRNAs can be detected in a variety of sources, including blood, tissue and body fluids. miRNAs have a high accuracy for the diagnosis of osteosarcoma (12), cancers of central nervous system (13), colorectal cancer (14), and glioma (15). Over the past years, the diagnosis accuracy of circulating miRNAs for early detection of AS have been reported in multiple articles. However, some results were inconsistent due to the difference in sample size, study design, and type of specimen. To date, there is no comprehensive meta-analysis on this topic. Therefore, we conducted such meta-analysis to identify the overall diagnostic accuracy of miRNA biomarkers in AS patients.
Methods
Literature Search Strategy
PubMed, Embase databases, Cochrane databases, and Wan Fang database for relevant articles published before 30 December 2020 were searched using the following key words: (“microRNAs” or “microRNA” or “miRNA” or “miR” or “RNA, Micro” or “Primary MicroRNA”) and (“Spondylitis Ankylosing” or “Spondyloarthritis Ankylopoietica” or “Ankylosing Spondylarthritis” or “Ankylosing Spondylarthritides” or “Spondylarthritides Ankylosing” or “Ankylosing Spondylitis”) and (“blood” or “serum” or “plasma”). Only English and Chinese articles were involved.
Article Selection
All included articles had to meet the following criteria: (1) all the patients reported must be diagnosed as AS; (2) the articles are aimed at identifying the overall accuracy of miRNAs in AS patients; (3) miRNAs for the diagnosis of AS were compared with the diagnosis of AS according to the modified New York criteria or the AS classification criteria; (4) primary outcomes were the fold-change, p-value, and the sensitivity, specificity, PLR, NLR, DOR, and AUC of miRNAs for AS; (5) the articles are either a retrospective or prospective observational design (for prospective design, we used baseline data). The criteria for exclusion were as follows: (1) duplicated articles; (2) case reports, reviews, meta-analyses, editorials, and letters; (3) insufficient data; (4) papers conducted in animals or cell lines.
Data Extraction, Meta-Analysis, and Quality Assessment
Data extraction was performed as follows: last name of the first author, publication year; ethnicity of patients; miRNA expression profiling; sample numbers; sample types; direction of expression difference (up or down regulated); sensitivity and specificity data, fold-change from miRNA expression profile. Studies identified the sensitivity and specificity of individual miRNA about the diagnosis of AS were included in the meta-analysis, and the qualities of selected articles were further assessed using the modified version of the QUADAS-2 tool (16). Moreover, the expression analysis of a specific miRNA in articles is considered to be an independent study.
Statistical Analysis
All statistical analyses were performed using Review Manager 5.3 and Stata 12.0. P < 0.05 and fold change ≥ 2 or ≤ 0.5 indicated statistical significance. Studies identified the sensitivity and specificity of individual miRNA about the diagnosis of AS were included in the meta-analysis. Several common evaluation indicators, including sensitivity, specificity, PLR, NLR, DOR, and AUC value, were used to evaluate the diagnostic accuracy of miRNAs. The between-study heterogeneity was evaluated with the I2 and χ2 statistics, I2 > 50% indicated significant heterogeneity; hence, a random-effect model was subsequently used to pool these results. Subgroup analysis and meta-regression analysis were used to identify sources of heterogeneity. Finally, Deeks' funnel plot analysis was performed to explore the potential publication bias.
Results
Included Articles
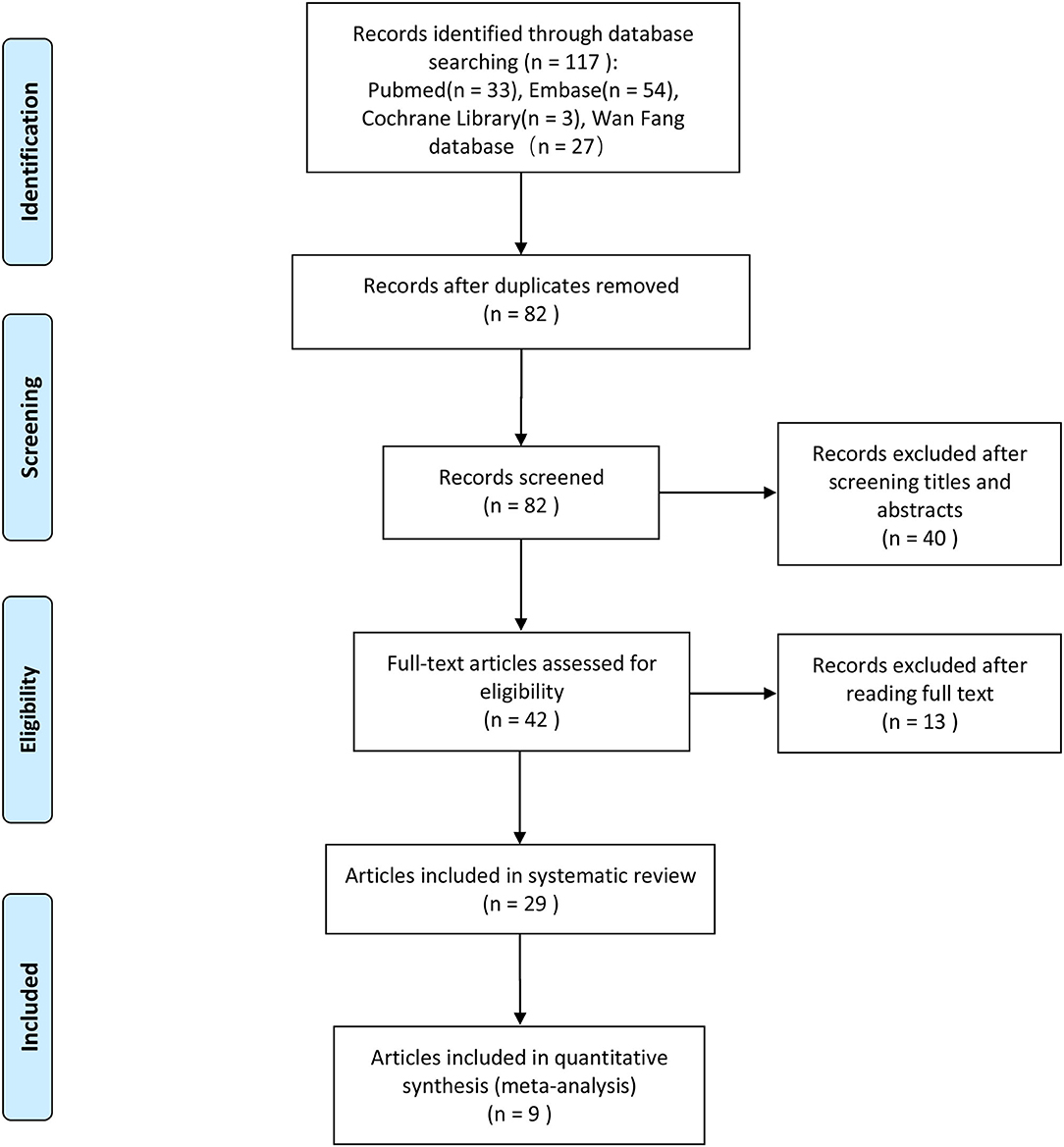
We initially identified 117 potentially eligible articles after the literature search process, and after removing 35 duplicated articles, 82 articles remained. In which, we excluded 40 articles after reading the titles and abstracts. And after full-text assessment, we eventually identified 29 articles that could be used for the systematic review of the miRNA expression profile. The full text of the 29 articles was assessed again in details, and nine of them (17–25) were included in our meta-analysis. A flowchart of the selection process of the articles is presented in Figure 1.
Systematic Review of miRNAs Expression Profiles
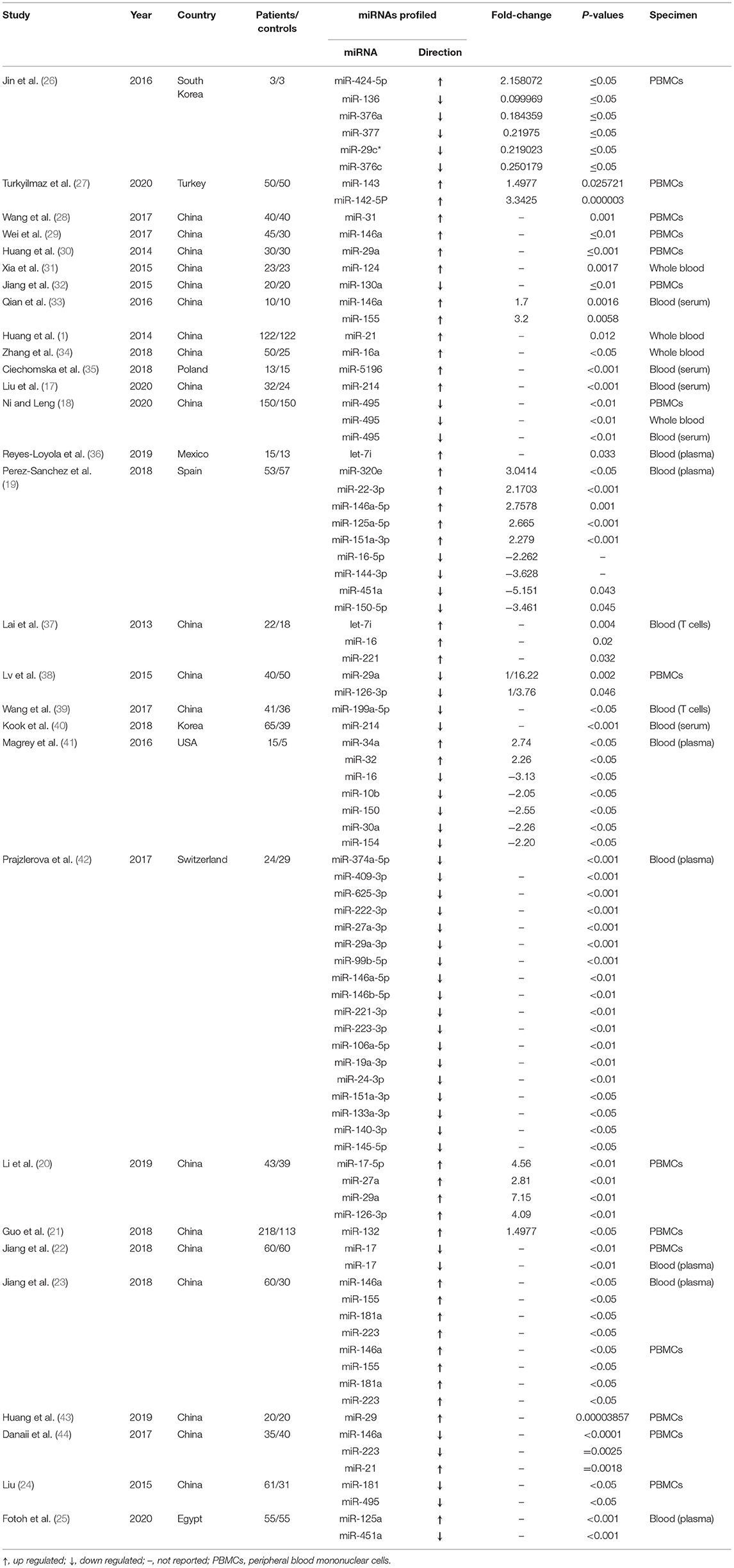
A summary of 29 articles reporting on the miRNA expression profiles is shown in Table 1. A total of 87 miRNAs were differentially expressed between AS patients and healthy controls in these screened articles, and of which, 42 miRNAs were found to be up-regulated and 45 miRNAs were down-regulated in the AS cases compared with the controls. Twelve up-regulated miRNAs (miR-29, miR-146a, miR-126-3p, miR-155, miR-21, miR-214, miR-16, miR-181a, miR-223, miR-146a-5p, miR-151a-3p, and let 7i) and two down-regulated miRNAs (miR-17, miR-495) were identified by more than one studies, miR-29 was the most frequently identified dysregulated miRNA. Seven miRNAs (miR-29, miR-214, miR-16, miR-146a-5p, miR-151a-3p, miR-146a, and miR-223) were found to be either up-regulated or down-regulated in the AS cases as compared with the controls.
Article Characteristics in the Meta-Analysis and Quality Assessment
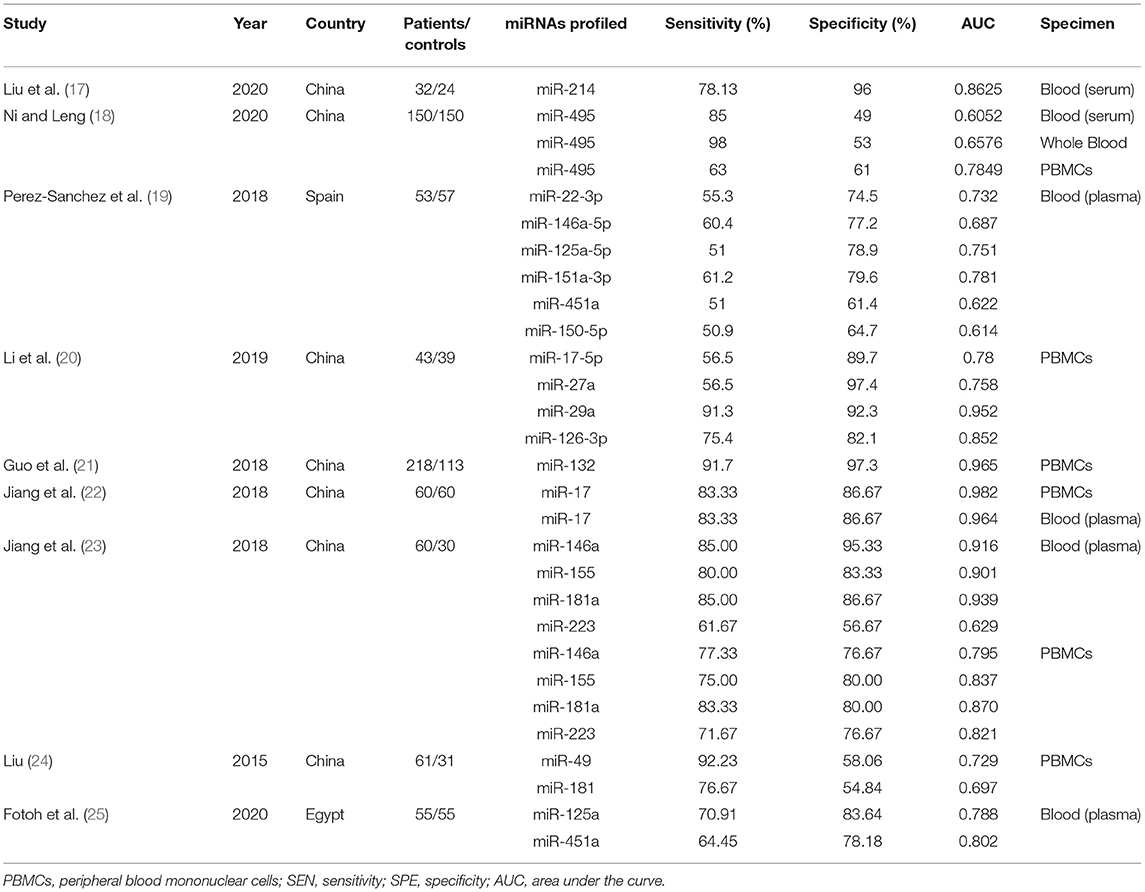
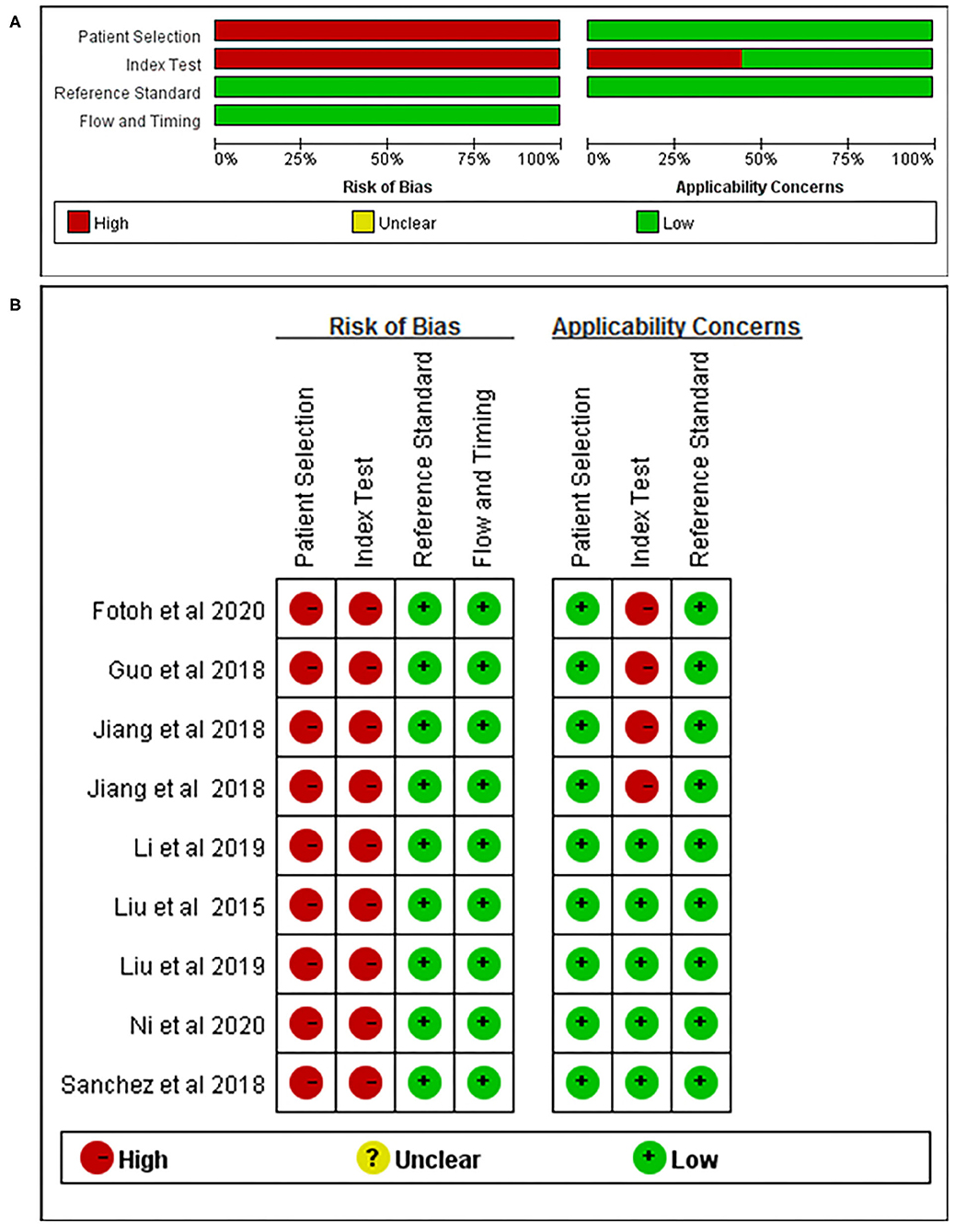
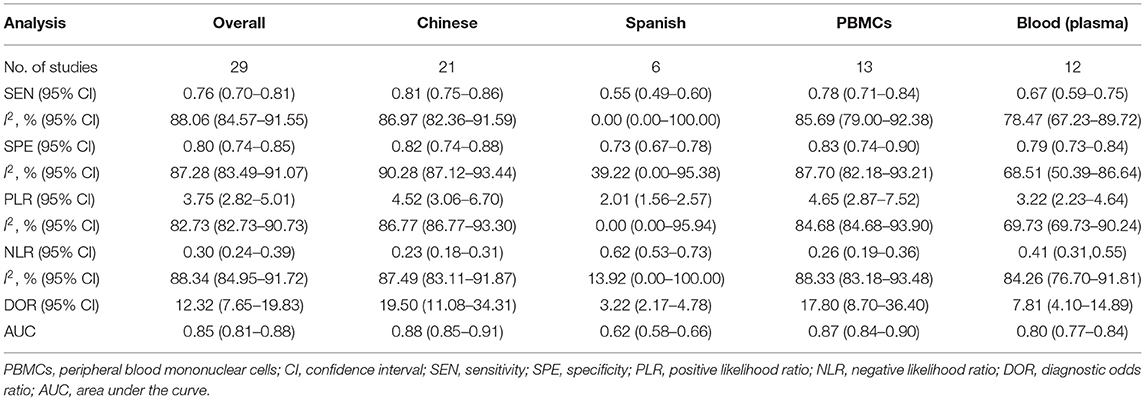
Nine articles that reported on the diagnostic accuracy of AS were included in the systematic review (Table 2). In total, the nine articles (time period ranging from years of 2015 to 2020) reported 29 studies, including 496 AS patients and 383 controls. Among the 29 included studies, one study detected miRNA in whole blood, two studies detected miRNA in blood serum, 13 studies researched on blood plasma, and 13 studies researched peripheral blood mononuclear cells (PBMCs). Among the 29 studies, 21 studies were conducted in Chinese populations, six studies were conducted in Spanish populations, and the remaining two studies focused on Egyptian populations. The quality of the included articles was assessed using the modified version of the QUADAS-2 tool. The risk of bias and applicability concerns graph for the included articles is presented in Figure 2.
Diagnostic Accuracy
Plots of sensitivities and specificities are shown in Figure 3. The results of the I2 test were 88.06% for sensitivity and 87.28% for specificity, which suggested that there was significant heterogeneity between the results of the included studies, and thus the random-effects model was selected. The sensitivity [±95% confidence intervals (CIs)] and specificity (±95% CIs) for each miRNA are shown in the corresponding forest plots (Figures 3A,B). The overall sensitivity and specificity of the 29 individual miRNAs in the diagnosis of AS were 0.76 (95% CI, 0.70–0.81), and 0.80 (95% CI, 0.74–0.85), respectively. The SROC curve is shown in Figure 4, with AUC of 0.85 (95% CI, 0.81–0.88). The overall PLR and NLR are shown in Table 3, the pooled PLR and NLR were 3.75 (95% CI, 2.82–5.01), 0.30 (95% CI, 0.24–0.39), These results indicate good discriminative ability of using miRNAs as biomarkers for the detection of AS.
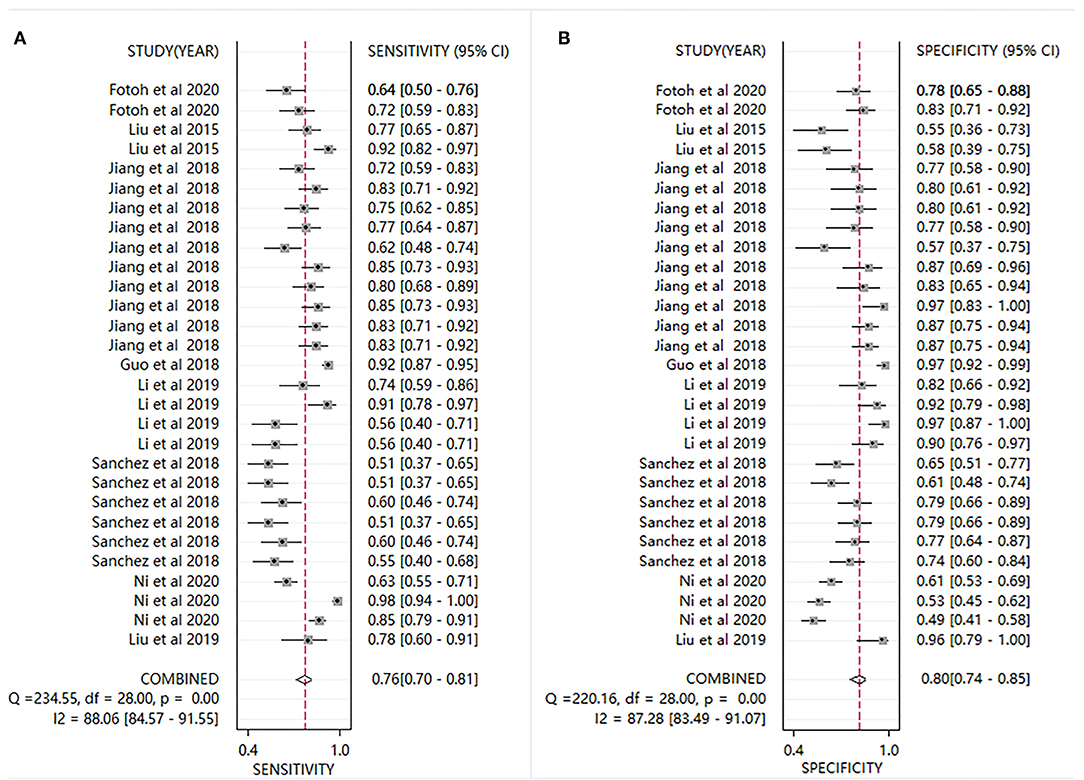
Figure 3. Forest plots for studies on individual miRNA used in the diagnosis of AS among the 29 studies included in the meta-analysis. (A) sensitivity; (B) specificity.
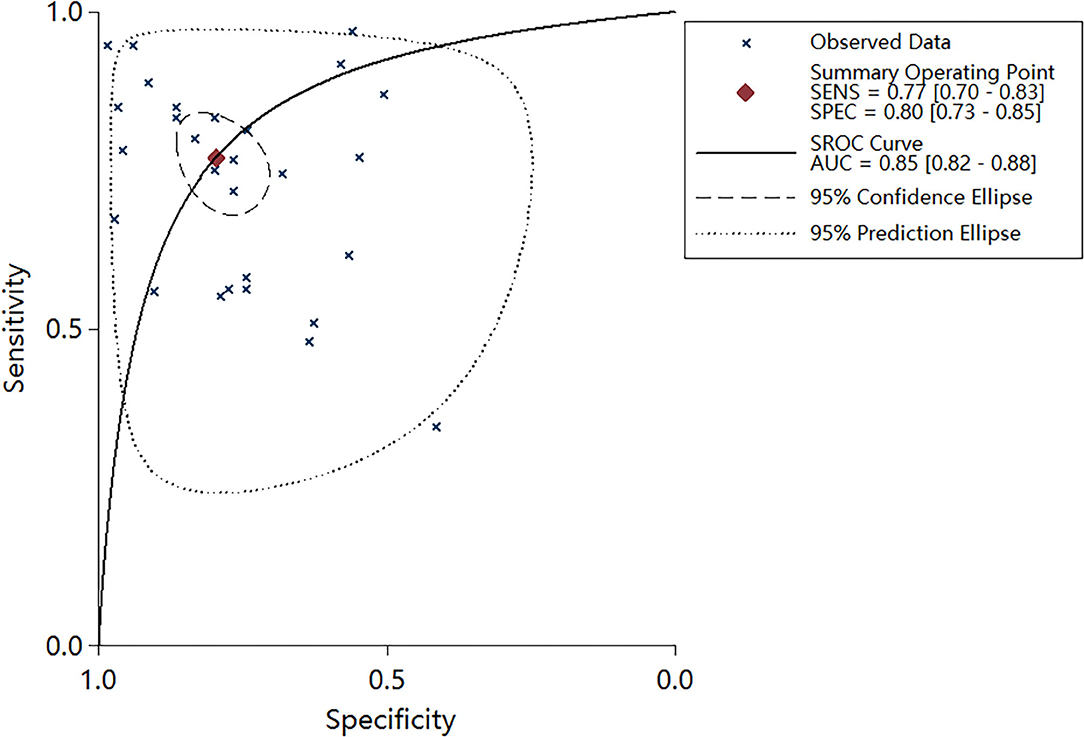
Figure 4. Summary receiver operator characteristic (SROC) curve with area under curve (AUC) based on miRNAs.
Subgroup Analysis and Meta-Regression Analysis
To explore the between-study heterogeneity in this meta-analysis, we carried out subgroup analyses and meta-regression analysis based on ethnicity and detected sample. As shown in Table 3, I2 > 50% of each parameter indicated significant heterogeneity and thus the random-effects model was selected. However, ethnicity and sample types aren't the sources of heterogeneity in sensitivity and specificity. The pooled sensitivity, specificity, DOR, and AUC were 0.76 (95% CI, 0.70–0.81), 0.80 (95% CI, 0.74–0.85), 12.316 (7.651–19.826), 0.85 (0.81–0.88), respectively. In detected samples of peripheral blood mononuclear cells (PBMCs), the results were 0.78 (0.71–0.84) for sensitivity, 0.83 (0.74–0.90) for specificity, 4.65 (2.87–7.52) for PLR, 0.26 (0.19–0.36) for PLR, 17.80 (8.70–36.40) for DOR, and 0.87 (0.84–0.90) for AUC. In the blood plasma samples, the sensitivity, specificity, PLR, NLR, DOR, and AUC was 0.67 (0.59–0.75), 0.79 (0.73–0.84), 3.22 (2.23–4.64), 0.41 (0.31–0.55), 7.81 (4.10–14.89), and 0.80 (0.77–0.84), suggesting that miRNAs in PBMCs, rather than in blood plasma, has a higher diagnostic accuracy. Furthermore, the sensitivity, specificity, PLR, and NLR for Chinese were 0.81 (95% CI, 0.75–0.86), 0.82 (95% CI, 0.74–0.88), 4.52 (95% CI, 3.06–6.70), and 0.23 (95% CI, 0.18–0.31), respectively, with a DOR of 19.50 (95% CI, 11.08–34.31) and AUC value of 0.88 (95% CI, 0.85–0.91). Compared with the Spanish, miRNAs have a higher overall diagnostic accuracy in the Chinese, and they have important diagnostic value for AS disease in the Chinese. Meta-regression was further conducted to explore the potential sources of the interstudy heterogeneity in sensitivity and specificity. Ethnicity were not the source of heterogeneity in sensitivity (P = 0.07) and specificity (P = 0.21). Besides, the specimen types were also not the source of heterogeneity in sensitivity (P = 0.57) and specificity (P = 0.71).
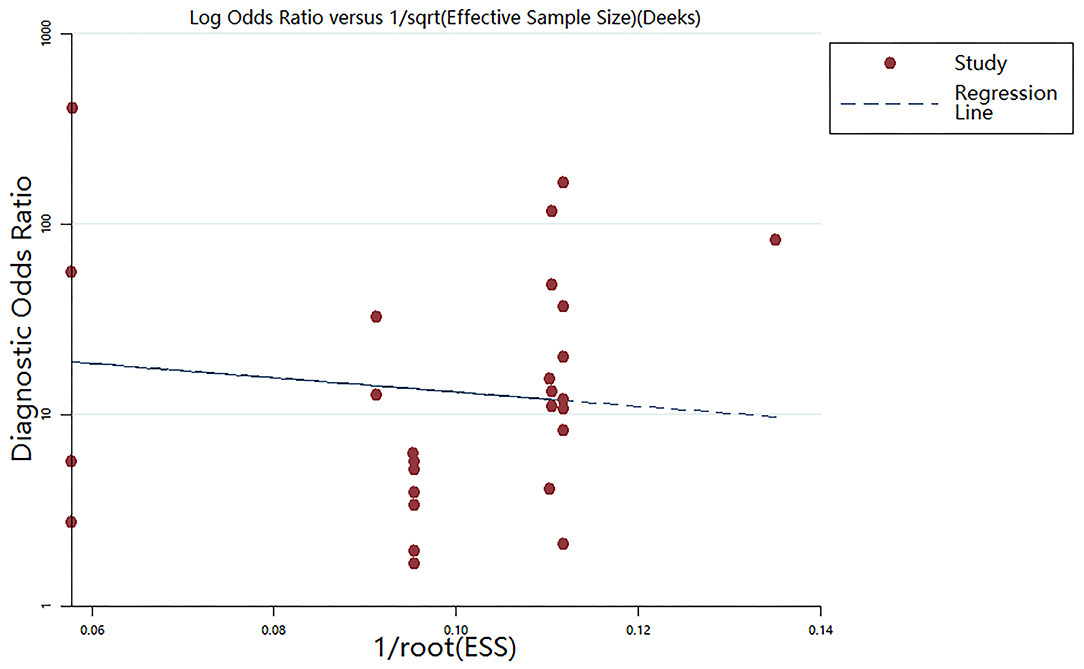
Publication Bias
The Deeks' funnel plot asymmetry test was performed (Figure 5). The P-value of 0.144 suggested that no bias existed in the included publications.
Discussion
To date, the early diagnosis of AS is still challenging, because the pathogenesis of AS remains unclear. Besides, X-ray fails to detect early radiological changes of AS. As a result, the diagnosis of AS is often delayed, even up to 7–11 years after the onset of the symptoms (45, 46). Currently the biomarkers such as HLA-B27, erythrocyte sedimentation rate, and C-reactive protein only have moderate diagnostic value, and their sensitivity and specificity are suboptimal (47). On the contrary, substantial progress has been made in improving the early diagnosis for other rheumatic diseases, such as rheumatoid arthritis (RA) (48). Novel biomarkers for the early diagnosis of AS are urgently needed. Over the past years, miRNAs have been observed an important role in the pathologies of systemic rheumatic diseases (49). miRNAs are actively involved in the deregulation of immune cell functions, regulation of T-cell survival signaling, and proinflammatory pathway. In addition, bone remodeling is controlled through miRNAs, such as miR-29a, miR-21, miR-34a, miR-17-5p, and miR-27b-3p. miRNAs, such as miR-214, also control T-cell autophagy, which might be involved in the pathological process of AS (50). Circulating miRNAs are promising diagnostic biomarkers for AS because of their high sensitivity and specificity for AS. Besides, they are relatively stable and can be easily detected in blood serum or plasma, which allows repeated sampling to monitor the development of the disease (51). Since the first AS-associated miRNAs was discovered in blood (37), there are increasing number of articles reporting on their expression in recent years. Thus, we conduct this research to evaluate the overall diagnostic accuracy of miRNAs in AS patients.
This systematic review has identified 87 miRNAs which were differentially expressed between AS and healthy controls in these screened articles, and of which, 42 miRNAs were observed to be up-regulated and 45 miRNAs were down-regulated in AS cases compared with the controls. Twelve up-regulated miRNAs (miR-29, miR-146a, miR-126-3p, miR-155, miR-21, miR-214, miR-16, miR-181a, miR-223, miR-146a-5p, miR-151a-3p, and let 7i) and two down-regulated miRNAs (miR-17, miR-495) were identified by more than one studies, and miR-29 was the most frequently identified dysregulated miRNA. Besides, miR-29 was found to be either up-regulated or down-regulated in AS cases compared with controls. Disease activity and therapy may be the sources of heterogeneity about the expression of miR-29. The expression of miR-29a was significantly lower in active AS patients than in controls. However, miR-29a was significantly upregulated in AS patients after 12-week of etanercept therapy than that in controls. When there was a good therapeutic effect, the expression of the miRNAs would get up-regulated and tend to be normal (38). On the contrary, plasma miR-22–3p, miR-146a-5p, and miR-125a-5p expression levels were significantly up-regulated in active AS patients when compared with non-active AS patients (19). miR-29a, miR-22–3p, miR-146a-5p, and miR-125a-5p will probably become the potential biomarkers, activity evaluation, and the curative effect monitoring of AS. miR-29a is part of the miR-29 family (miR-29a, miR-29b, and miR-29c), which are expressed in both T- and B-cells, as well as dendritic cells. It has been identified that miR-29a plays an important role in the regulator of immunity (52) and the bone metabolism, and miR-29a can modulate Wnt signaling to accelerate bone formation (53). Interestingly, miR-29c, another member of the miR-29 family, was observed to be down-regulated in AS patients compared with controls in two studies (26). Additionally, six miRNAs (miR-214, miR-223, miR-151a-3p, miR-146a-5p, miR-146a, and miR-16) were observed to be either up-regulated or down-regulated in the AS cases as compared with the controls. Inconsistent results about dysregulated miRNAs may be caused by a variety of factors, including ethnic differences, the severity of the disease, as well as the source of samples, and sample sizes. In the present meta-analysis, a pooled sensitivity of 0.76 (95% CI, 0.70–0.81), specificity of 0.80 (95% CI, 0.74–0.85), PLR of 3.75 (95% CI, 2.82–5.01), NLR of 0.30 (95% CI, 0.24–0.39), DOR of 12.32 (7.65–19.83) and AUC of 0.85 (95% CI, 0.81–0.88) demonstrate that as a diagnostic biomarker, circulating miRNAs achieved a relatively high overall accuracy in diagnosing AS. However, also as a diagnostic biomarker for AS, its serum CRP has only 50% sensitivity and 80% specificity (54). In addition, we carried out subgroup analyses to explore the sources of heterogeneity. Compared with the Spanish, miRNAs have a higher overall diagnostic accuracy in Chinese. This result may be related to the sample size of the reported articles, because we eventually only included 53 patients from one article about the miRNAs expression profile of Spanish. However, 624 patients from seven articles about the miRNAs expression profile of Chinese were included. Besides, miRNAs in PBMCs showed higher diagnostic value than in blood plasm. The expression of miRNAs in PBMCs is likely to be more stable than that in blood plasm. Thus, for a better blood-based biomarker for AS, miRNAs in PBMCs is preferred.
The current meta-analysis is the first ever to evaluate the overall diagnostic accuracy of miRNAs for AS as to our knowledge. We combined the results of 29 studies with 732 patients and 559 controls, which may enhance the statistical power and the reliability of the findings. Because the study's heterogeneity (I2 88.06% for sensitivity, and 87.28% for specificity) was significant, we adopted the random effects model to reduce the heterogeneity related bias. We found that miRNAs in PBMCs, rather than in blood plasma, had a higher diagnostic accuracy for AS. However, meta-regression and subgroup analysis didn't find the sources of heterogeneity, suggesting that complex factors influence the final results. There is almost no publication bias by the Deeks' funnel plot asymmetry test, strengthening the reliability of the results. We not only reviewed the pooled diagnostic accuracy of circulating miRNAs, but also systematically summarized the diagnostic effectiveness of every individual miRNA, which provide valuable reference data for further research. However, the present paper has some limitations. First, due to the different kinds of miRNAs, small sample size, and different cut-off values in the selected articles, significant heterogeneity was existed in the current meta-analysis, which is a common situation in other published meta-analyses of diagnostic accuracy. Moreover, the overall diagnostic accuracy may be amplified, because those articles with positive results are more likely to be published. In addition, the biological characteristics and mechanisms of the different miRNAs in AS may differ, which may limit the applicability of the pooled analysis. It is expected that in the future more studies from different genetics, geography, and ethnicity may be carried out to increase the validity of our conclusions and the reliability of miRNA analysis for AS.
Conclusion
In summary, we identified numerous circulating miRNAs associated with AS in this systematic review. The circulating miRNAs may be used as biomarkers for the detection of AS. It was also found that miRNA in PBMCs, rather than in blood plasma, has a higher diagnostic accuracy. The diagnosis of miRNAs for AS seems to be more sensitive in Chinese than that in Spanish. Further studies based on larger sample of patients and controls are still required.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
JL and XX designed the study and collected data. JL drafted the manuscript. WL, FG, KZ, ZSu, QW, ZSui, and PZ contributed to the writing. TY provided critical feedback and contributed to the review of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This paper was supported by the National Natural Science Foundation of China (Grant No. 31970090).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.701789/full#supplementary-material
References
1. Huang CH, Wei JC, Chang WC, Chiou SY, Chou CH, Lin YJ, et al. Higher expression of whole blood microRNA-21 in patients with ankylosing spondylitis associated with programmed cell death 4 mRNA expression and collagen cross-linked C-telopeptide concentration. J Rheumatol. (2014) 41:1104–11. doi: 10.3899/jrheum.130515
2. Dean LE, Jones GT, MacDonald AG, Downham C, Sturrock RD, Macfarlane GJ. Global prevalence of ankylosing spondylitis. Rheumatology (Oxford). (2014) 53:650–7. doi: 10.1093/rheumatology/ket387
3. Cortes A, Pulit SL, Leo PJ, Pointon JJ, Robinson PC, Weisman MH, et al. Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat Commun. (2015) 6:7146. doi: 10.1038/ncomms8146
4. Dougados M, Baeten D. Spondyloarthritis. Lancet. (2011) 377:2127–37. doi: 10.1016/S0140-6736(11)60071-8
5. Brown MA, Kennedy LG, Darke C, Gibson K, Pile KD, Shatford JL, et al. The effect of HLA-DR genes on susceptibility to and severity of ankylosing spondylitis. Arthritis Rheum. (1998) 41:460–5. doi: 10.1002/1529-0131(199803)41:3<460::AID-ART12>3.0.CO;2-X
6. Reveille JD, Zhou X, Lee M, Weisman MH, Yi L, Gensler LS, et al. HLA class I and II alleles in susceptibility to ankylosing spondylitis. Ann Rheum Dis. (2019) 78:66–73. doi: 10.1136/annrheumdis-2018-213779
7. Zhou X, Wang J, Zou H, Ward MM, Weisman MH, Espitia MG, et al. MICA, a gene contributing strong susceptibility to ankylosing spondylitis. Ann Rheum Dis. (2014) 73:1552–7. doi: 10.1136/annrheumdis-2013-203352
8. Liu J, Pu W, Li Y, Ma Y, Zhu Q, Wan W, et al. Genetic association of non-MHC region with ankylosing spondylitis in a Chinese population. Ann Rheum Dis. (2019) 78:852–3. doi: 10.1136/annrheumdis-2018-214625
9. Xu HY, Wang ZY, Chen JF, Wang TY, Wang LL, Tang LL, et al. Association between ankylosing spondylitis and the miR-146a and miR-499 polymorphisms. PLoS One. (2015) 10:e0122055. doi: 10.1371/journal.pone.0122055
10. Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. (2011) 67:129–39. doi: 10.1007/s13105-010-0050-6
11. Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. (2012) 148:1172–87. doi: 10.1016/j.cell.2012.02.005
12. Wang X, Ning Y, Yang L, Liu H, Wu C, Wang S, et al. Diagnostic value of circulating microRNAs for osteosarcoma in Asian populations: a meta-analysis. Clin Exp Med. (2017) 17:175–83. doi: 10.1007/s10238-016-0422-5
13. Wei D, Wan Q, Li L, Jin H, Liu Y, Wang Y, et al. MicroRNAs as potential biomarkers for diagnosing cancers of central nervous system: a meta-analysis. Mol Neurobiol. (2015) 51:1452–61. doi: 10.1007/s12035-014-8822-6
14. Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. (2017) 116:762–74. doi: 10.1038/bjc.2017.12
15. Zhou Q, Liu J, Quan J, Liu W, Tan H, Li W. MicroRNAs as potential biomarkers for the diagnosis of glioma: a systematic review and meta-analysis. Cancer Sci. (2018) 109:2651–9. doi: 10.1111/cas.13714
16. Zhelev Z, Hyde C, Youngman E, Rogers M, Fleming S, Slade T, et al. Diagnostic accuracy of single baseline measurement of Elecsys Troponin T high-sensitive assay for diagnosis of acute myocardial infarction in emergency department: systematic review and meta-analysis. BMJ. (2015) 350:h15. doi: 10.1136/bmj.h15
17. Liu Z, Huang F, Luo G, Wang Y, Du R, Sun W, et al. miR-214 stimulated by IL-17A regulates bone loss in patients with ankylosing spondylitis. Rheumatology (Oxford). (2020) 59:1159–69. doi: 10.1093/rheumatology/kez594
18. Ni WJ, Leng XM. Down-regulated miR-495 can target programmed cell death 10 in ankylosing spondylitis. Mol Med. (2020) 26:50. doi: 10.1186/s10020-020-00157-3
19. Perez-Sanchez C, Font-Ugalde P, Ruiz-Limon P, Lopez-Pedrera C, Castro-Villegas MC, Abalos-Aguilera MC, et al. Circulating microRNAs as potential biomarkers of disease activity and structural damage in ankylosing spondylitis patients. Hum Mol Genet. (2018) 27:875–90. doi: 10.1093/hmg/ddy008
20. Li X, Lv Q, Tu L, Zhao M, Zhang P, Li Q, et al. Aberrant expression of microRNAs in peripheral blood mononuclear cells as candidate biomarkers in patients with axial spondyloarthritis. Int J Rheum Dis. (2019) 22:1188–95. doi: 10.1111/1756-185X.13563
21. Guo TM, Yan Y, Cao WN, Liu Q, Zhu HY, Yang L, et al. Predictive value of microRNA-132 and its target gene NAG-1 in evaluating therapeutic efficacy of non-steroidal anti-inflammatory drugs treatment in patients with ankylosing spondylitis. Clin Rheumatol. (2018) 37:1281–93. doi: 10.1007/s10067-018-4017-2
22. Jiang F, Xu m, Zheng XM. Expression and clinical significance of miR-17 and MMP-3 in patients with ankylosing spondylitis. Chin J Health Lab Tec. (2018) 28:2701–4.
23. Jiang F, Xu M, Zheng XM. miR-146a, miR-155, miR-181a and miR-223 level differences in peripheral blood of patients with ankylosing spondylitis. Exp Lab Med. (2018) 36:766–9+72.
24. Liu X. Detection the Expression Levels of Five Kinds of MicroRNAs in Ankylosing Spondylitis Patients' Peripheral Blood Mononuclear Cells and the Regulatory Effect of miR 495 on DVL-2 Protein. NingXia Medical University (2015).
25. Fotoh DS, Noreldin RI, Rizk MS, Elsabaawy MM, Esaily HA. miRNA-451a and miRNA-125a expression levels in ankylosing spondylitis: impact on disease diagnosis, prognosis, and outcomes. J Immunol Res. (2020) 2020:2180913. doi: 10.1155/2020/2180913
26. Jin HM, Cho YN, Kee SJ, Lee SS, Park YW, Kim TJ. Micro-ribonucleic acid profiles from microarray in ankylosing spondylitis. Arch Rheumatol. (2016) 31:121–6. doi: 10.5606/ArchRheumatol.2016.5733
27. Turkyilmaz A, Ata P, Akbas F, Yagci I. The expression levels of microRNAs associated with T and B cell differentiation/stimulation in ankylosing spondylitis. Balkan J Med Genet. (2020) 23:25–32. doi: 10.2478/bjmg-2020-0006
28. Wang M, Wang L, Zhang X, Yang X, Li X, Xia Q, et al. Overexpression of miR-31 in peripheral blood mononuclear cells (PBMC) from patients with ankylosing spondylitis. Med Sci Monit. (2017) 23:5488–94. doi: 10.12659/MSM.905238
29. Wei C, Zhang H, Wei C, Mao Y. Correlation of the expression of miR-146a in peripheral blood mononuclear cells of patients with ankylosing spondylitis and inflammatory factors. Exp Ther Med. (2017) 14:5027–31. doi: 10.3892/etm.2017.5155
30. Huang J, Song G, Yin Z, Luo X, Ye Z. Elevated miR-29a expression is not correlated with disease activity index in PBMCs of patients with ankylosing spondylitis. Mod Rheumatol. (2014) 24:331–4. doi: 10.3109/14397595.2013.854077
31. Xia Y, Chen K, Zhang MH, Wang LC, Ma CY, Lin YL, et al. MicroRNA-124 involves in ankylosing spondylitis by targeting ANTXR2. Mod Rheumatol. (2015) 25:784–9. doi: 10.3109/14397595.2015.1023887
32. Jiang Y, Wang L. Role of histone deacetylase 3 in ankylosing spondylitis via negative feedback loop with microRNA-130a and enhancement of tumor necrosis factor-1alpha expression in peripheral blood mononuclear cells. Mol Med Rep. (2016) 13:35–40. doi: 10.3892/mmr.2015.4494
33. Qian BP, Ji ML, Qiu Y, Wang B, Yu Y, Shi W, et al. Identification of serum miR-146a and miR-155 as novel noninvasive complementary biomarkers for ankylosing spondylitis. Spine. (2016) 41:735–42. doi: 10.1097/BRS.0000000000001339
34. Zhang CL, Li YC, Wu JW, Zhu BL. Expression and function of peripheral blood miRNA16a in patients with ankylosing spondylitis. Eur Rev Med Pharmacol Sci. (2018) 22:5106–13. doi: 10.26355/eurrev_201808_15704
35. Ciechomska M, Bonek K, Merdas M, Zarecki P, Swierkot J, Gluszko P, et al. Changes in MiRNA-5196 expression as a potential biomarker of anti-TNF-alpha therapy in rheumatoid arthritis and ankylosing spondylitis patients. Arch Immunol Ther Exp (Warsz). (2018) 66:389–97. doi: 10.1007/s00005-018-0513-y
36. Reyes-Loyola P, Rodriguez-Henriquez P, Ballinas-Verdugo MA, Amezcua-Castillo LM, Juarez-Vicuna Y, Jimenez-Rojas V, et al. Plasma let-7i, miR-16, and miR-221 levels as candidate biomarkers for the assessment of ankylosing spondylitis in Mexican patients naive to anti-TNF therapy. Clin Rheumatol. (2019) 38:1367–73. doi: 10.1007/s10067-019-04509-1
37. Lai NS, Yu HC, Chen HC, Yu CL, Huang HB, Lu MC. Aberrant expression of microRNAs in T cells from patients with ankylosing spondylitis contributes to the immunopathogenesis. Clin Exp Immunol. (2013) 173:47–57. doi: 10.1111/cei.12089
38. Lv Q, Li Q, Zhang P, Jiang Y, Wang X, Wei Q, et al. Disorders of microRNAs in peripheral blood mononuclear cells: as novel biomarkers of ankylosing spondylitis and provocative therapeutic targets. Biomed Res Int. (2015) 2015:504208. doi: 10.1155/2015/504208
39. Wang Y, Luo J, Wang X, Yang B, Cui L. MicroRNA-199a-5p induced autophagy and inhibits the pathogenesis of ankylosing spondylitis by modulating the mTOR signaling via directly targeting ras homolog enriched in brain (Rheb). Cell Physiol Biochem. (2017) 42:2481–91. doi: 10.1159/000480211
40. Kook HY, Jin SH, Park PR, Lee SJ, Shin HJ, Kim TJ. Serum miR-214 as a novel biomarker for ankylosing spondylitis. Int J Rheum Dis. (2019) 22:1196–201. doi: 10.1111/1756-185X.13475
41. Magrey MN, Haqqi T, Haseeb A. Identification of plasma microRNA expression profile in radiographic axial spondyloarthritis-a pilot study. Clin Rheumatol. (2016) 35:1323–7. doi: 10.1007/s10067-015-3123-7
42. Prajzlerova K, Grobelna K, Husakova M, Forejtova S, Jungel A, Gay S, et al. Association between circulating miRNAs and spinal involvement in patients with axial spondyloarthritis. PLoS One. (2017) 12:e0185323. doi: 10.1371/journal.pone.0185323
43. Huang J, Song G, Yin Z, Fu Z, Zhang L. Altered expression of microRNAs targeting Dkk-1 in peripheral blood mononuclear cells of patients with ankylosing spondylitis. Cent Eur J Immunol. (2019) 44:59–64. doi: 10.5114/ceji.2019.84018
44. Danaii S, Abolhasani R, Soltani-Zangbar MS, Zamani M, Yousefi M. Oxidative stress and immunological biomarkers in Ankylosing spondylitis patients. Gene Rep. (2019) 18:100574. doi: 10.1016/j.genrep.2019.100574
45. Feldtkeller E, Khan MA, van der Heijde D, van der Linden S, Braun J. Age at disease onset and diagnosis delay in HLA-B27 negative vs. positive patients with ankylosing spondylitis. Rheumatol Int. (2003) 23:61–6. doi: 10.1007/s00296-002-0237-4
46. Garrido-Cumbrera M, Poddubnyy D, Gossec L, Galvez-Ruiz D, Bundy C, Mahapatra R, et al. The European map of axial spondyloarthritis: capturing the patient perspective-an analysis of 2846 patients across 13 countries. Curr Rheumatol Rep. (2019) 21:19. doi: 10.1007/s11926-019-0819-8
47. Brown MA, Li Z, Cao KL. Biomarker development for axial spondyloarthritis. Nat Rev Rheumatol. (2020) 16:448–63. doi: 10.1038/s41584-020-0450-0
48. Aletaha D, Smolen JS. Diagnosis and management of rheumatoid arthritis: a review. JAMA. (2018) 320:1360–72. doi: 10.1001/jama.2018.13103
49. Alevizos I, Illei GG. MicroRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol. (2010) 6:391–8. doi: 10.1038/nrrheum.2010.81
50. Mohammadi H, Hemmatzadeh M, Babaie F, Gowhari Shabgah A, Azizi G, Hosseini F, et al. MicroRNA implicationsin the etiopathogenesis of ankylosing spondylitis. J Cell Physiol. (2018) 233:5564–73. doi: 10.1002/jcp.26500
51. Poel D, Buffart TE, Oosterling-Jansen J, Verheul HM, Voortman J. Evaluation of several methodological challenges in circulating miRNA qPCR studies in patients with head and neck cancer. Exp Mol Med. (2018) 50:e454. doi: 10.1038/emm.2017.288
52. Liston A, Papadopoulou AS, Danso-Abeam D, Dooley J. MicroRNA-29 in the adaptive immune system: setting the threshold. Cell Mol Life Sci. (2012) 69:3533–41. doi: 10.1007/s00018-012-1124-0
53. Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. (2010) 285:25221–31. doi: 10.1074/jbc.M110.116137
Keywords: ankylosing spondylitis, microRNA, biomarkers, systematic review, meta-analysis
Citation: Li J, Xie X, Liu W, Gu F, Zhang K, Su Z, Wen Q, Sui Z, Zhou P and Yu T (2021) MicroRNAs as Biomarkers for the Diagnosis of Ankylosing Spondylitis: A Systematic Review and Meta-Analysis. Front. Med. 8:701789. doi: 10.3389/fmed.2021.701789
Received: 28 April 2021; Accepted: 15 July 2021;
Published: 10 August 2021.
Edited by:
Miroslav Harjaček, United Arab Emirates University, United Arab EmiratesReviewed by:
Fernando Manuel Pimentel-Santos, Universidade NOVA de Lisboa, PortugalÂngelo Calado, Faculdade de Medicina da Universidade de Lisboa, Portugal
Copyright © 2021 Li, Xie, Liu, Gu, Zhang, Su, Wen, Sui, Zhou and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiecheng Yu, eXV0Y0BqbHUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Jiangbi Li1†
Jiangbi Li1† Xiaoping Xie
Xiaoping Xie Tiecheng Yu
Tiecheng Yu