
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 24 June 2021
Sec. Precision Medicine
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.667525
This article is part of the Research Topic Nucleic Acids-Based Cancer Theranostics View all 29 articles
 Haipeng Jia1†
Haipeng Jia1† Xiaofen Zhang1†
Xiaofen Zhang1† Xinxin Liu1†
Xinxin Liu1† Ruifang Qiao2
Ruifang Qiao2 Yan Liu1
Yan Liu1 Sulong Lv1
Sulong Lv1 Hongbo Zhu1
Hongbo Zhu1 Jie Wang1
Jie Wang1 Qiuhong Kong1
Qiuhong Kong1 Hong Zhang1
Hong Zhang1 Zhirong Zhang1*
Zhirong Zhang1*Objective: Multiple myeloma is an incurable hematological malignancy. It is imperative to identify immune markers for early diagnosis and therapy. Here, this study analyzed immune-related mRNAs and assessed their prognostic value and therapeutic potential.
Methods: Abnormally expressed immune-related mRNAs were screened between multiple myeloma and normal bone marrow specimens in the GSE47552 and GSE6477 datasets. Their biological functions were then explored. Survival analysis was presented for assessing prognosis-related mRNAs. CIBERSORT was utilized for identifying 22 immune cell compositions of each bone marrow specimen. Correlation between FABP5 mRNA and immune cells was then analyzed in multiple myeloma.
Results: Thirty-one immune-related mRNAs were abnormally expressed in multiple myeloma, which were primarily enriched in B cells-related biological processes and pathways. Following validation, FABP5 mRNA was a key risk factor of multiple myeloma. Patients with its up-regulation usually experienced unfavorable outcomes. There were distinct differences in the infiltration levels of B cells naïve, B cells memory, plasma cells, T cells CD4 naïve, resting memory CD4 T cells, activated memory CD4 T cells, Tregs, resting NK cells, M0 macrophages, M1 macrophages, M2 macrophages, and neutrophils between multiple myeloma and normal samples. FABP5 mRNA had correlations to B cells memory, B cells naïve, dendritic cells activated, macrophages M0, macrophages M1, macrophages M2, neutrophils, activated NK cells, resting memory CD4 T cells, CD8 T cells and Tregs.
Conclusion: Collectively, our data showed that FABP5 mRNA was related to immune microenvironment, which could be a target of immunotherapy and prognostic marker for multiple myeloma.
Multiple myeloma is a heterogenous and incurable neoplasm of plasma cells, accounting for 1% of malignancies and around 10% of all hematological malignancies (1). Stem cell transplantation, proteasome inhibitors and immunosuppressive agents are the main therapeutic strategies, which have prolonged survival time of multiple myeloma subjects (2). Multiple myeloma is a multi-step disease, initially characterized by asymptomatic monoclonal gammopathy (MGUS). MGUS accounts for 1% of the adult population, and ~1% of MGUS patients turn into malignant multiple myeloma every year (3). Despite the remarkable clinical outcomes, nearly all subjects experience recurrence. Hence, it still requires novel therapeutic approaches.
The survival, growth as well as proliferation of myeloma cells depend upon the bone marrow microenvironment (4, 5). The microenvironment contains a variety of cell types, including hematopoietic cells (B cells, T cells, natural killer cells, bone marrow-derived suppressor cells, and osteoclasts) and non-hematopoietic cells (marrow stromal cells, osteoblasts, and endothelial cells) (6). These cells secrete various factors, thereby promoting the migrative as well as proliferative behaviors of multiple myeloma cells (7–9). Multiple myeloma progress closely associates with abnormal innate and adaptive immune systems (10). There is evidence that the residual immune cell dysfunction in the tumor microenvironment can lead to the suppression of the host's anti-tumor immune function (11–13). In the normal microenvironment, effector cells can produce a powerful anti-tumor response. Nevertheless, tumor cells often protect themselves from the host immune system by inhibiting the anti-tumor immunity of their surrounding microenvironment. The sophisticated interplays between immune microenvironment and this disease may affect the clinical responses to immuno-oncology therapies such as thalidomide, lenalidomide (14) as well as pomalidomide (15). Early studies have mainly established the role of the bone marrow microenvironment in the pathological process of multiple myeloma, but the immune components of the microenvironment have not received enough attention. The immune response is a dynamic and complex process. It is of importance for understanding the main factors that contribute to the immunosuppressive environment in multiple myeloma (16). Herein, we focused on abnormally expressed immune-related mRNAs in multiple myeloma. Among them, high expression of FABP5 mRNA displayed poor outcomes of subjects. FABP5 is a member of the FABP family, which is overexpressed in several cancers such as prostate cancer (17). Increasing evidence has demonstrated that FABP5 is involved in tumor progression (18). Nevertheless, the biological roles of FABP5 remain indistinct in multiple myeloma. Our further analysis demonstrated that FABP5 dysregulation was in relationship with immune microenvironment of multiple myeloma, indicating that FABP5 was an underlying immunotherapeutic target for multiple myeloma.
From the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/), microarray data of bone marrow specimens from 5 normal and 41 multiple myeloma subjects were retrieved from the GSE47552 dataset on the GPL6244 platform. Furthermore, microarray expression profiling of 15 normal and 103 multiple myeloma bone marrow samples was obtained from the GSE6477 dataset on the GPL96 platform. The gene expression profiles from the GSE47552 and GSE6477 datasets were utilized for differential expression analysis. Probe IDs were transformed to gene symbols, followed by log2 transformation. The GSE4452 dataset contained complete follow-up information of 65 multiple myeloma subjects. Moreover, the GSE4204 dataset comprised survival information of 538 multiple myeloma patients. The GSE4452 and GSE4204 datasets were used for survival analysis.
Two thousand four hundred and ninety-eight immune-related mRNAs were obtained from the Immunology Database and Analysis Portal (ImmPort) database (https://www.immport.org/home) (19). Supplementary Table 1 listed the list of immune-related mRNAs.
The differences in immune-related mRNAs between multiple myeloma and normal bone marrow specimens from the GSE47552 and GSE6477 datasets were analyzed through “limma” package (20). These mRNAs with false discovery rate (FDR) <0.05 and |log2fold change| > 1 displayed differential expression. Abnormally expressed immune-related mRNAs were intersected between the GSE47552 and GSE6477 datasets.
Gene Ontology (GO) as well as Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotation analyses for abnormally expressed immune-related mRNAs were carried out via “clusterProfiler” package (21). GO terms contained biological process (BP), molecular function (MF), and cellular component (CC). Terms with FDR <0.05 were significantly enriched. The enrichment results of the top 10 terms were depicted by “ggplot2” package.
Univariate cox regression analyses were presented for assessment of which mRNAs displayed significant relationships to outcomes of multiple myeloma in the GSE4204 dataset through “survival” package. Hazard ratio (HR), 95% confidence interval (CI) as well as p-value were separately determined. The mRNAs with p <0.05 were survival-related mRNAs. The mRNAs with HR > 1 were protective factors, while those with HR <1 were risk factors. Then, patients in the GSE4204 dataset were separated into high and low expression subgroups in line with the median value of each survival-related mRNA. Kaplan-Meier curves between two subgroups were conducted. Log-rank test was utilized for evaluating whether there were differences in overall survival between subgroups. The prognostic implications of survival-related mRNAs were verified in the GSE4452 dataset.
Twenty-two immune cell compositions of bone marrow specimens were characterized based on the mRNA expression profiling from the GSE4204 dataset through the CIBERSORT algorithm (http://cibersort.stanford.edu/) (22). The perm was set at 1000. Specimens with p < 0.05 were retained for the analyses. These immune cells contained B cells naïve, B cells memory, plasma cells, CD8 T cells, T cells CD4, naïve T cells, CD4 memory resting, T cells CD4 memory activated, T cells follicular helper, T cells regulatory (Tregs), T cells gamma delta, NK cells resting, activated NK cells, monocytes, macrophages M0, macrophages M1, macrophages M2, dendritic cells resting, dendritic cells activated, mast cells resting, mast cells activated, eosinophils, and neutrophils. Correlation analyses were carried out between different immune cell compositions in each sample. The differences in fractions of 22 immune cells between normal and multiple myeloma bone marrow specimens were assessed through Mann-Whitney U-test. Correlations between FABP5 expression and immune cell fractions were determined among bone marrow samples by Pearson correlation analysis.
R language (https://www.r-project.org/) and corresponding packages were carried out for statistical analyses.
In the GSE6477 dataset, 139 immune-related mRNAs displayed abnormal expression between multiple myeloma and normal bone marrow specimens (Supplementary Table 2). Among them, 96 mRNAs showed down-regulation and 43 mRNAs exhibited up-regulation in multiple myeloma than normal bone marrow specimens (Figure 1A). The expression patterns of these mRNAs possessed conspicuous differences between multiple myeloma and normal subgroups (Figure 1B). Table 1 listed the top 10 up- and down-regulated mRNAs in multiple myeloma. In the GSE47552 dataset, there were 33 up-regulated and 81 down-regulated immune-related mRNAs (Figure 1C). Supplementary Table 3 listed the detailed information of 114 immune-related mRNAs. These mRNAs were distinguished multiple myeloma from normal bone marrow specimens (Figure 1D). Table 2 displayed the top 10 up- and down-regulated mRNAs for multiple myeloma. Thus, immune-related mRNAs may participate in multiple myeloma progress.
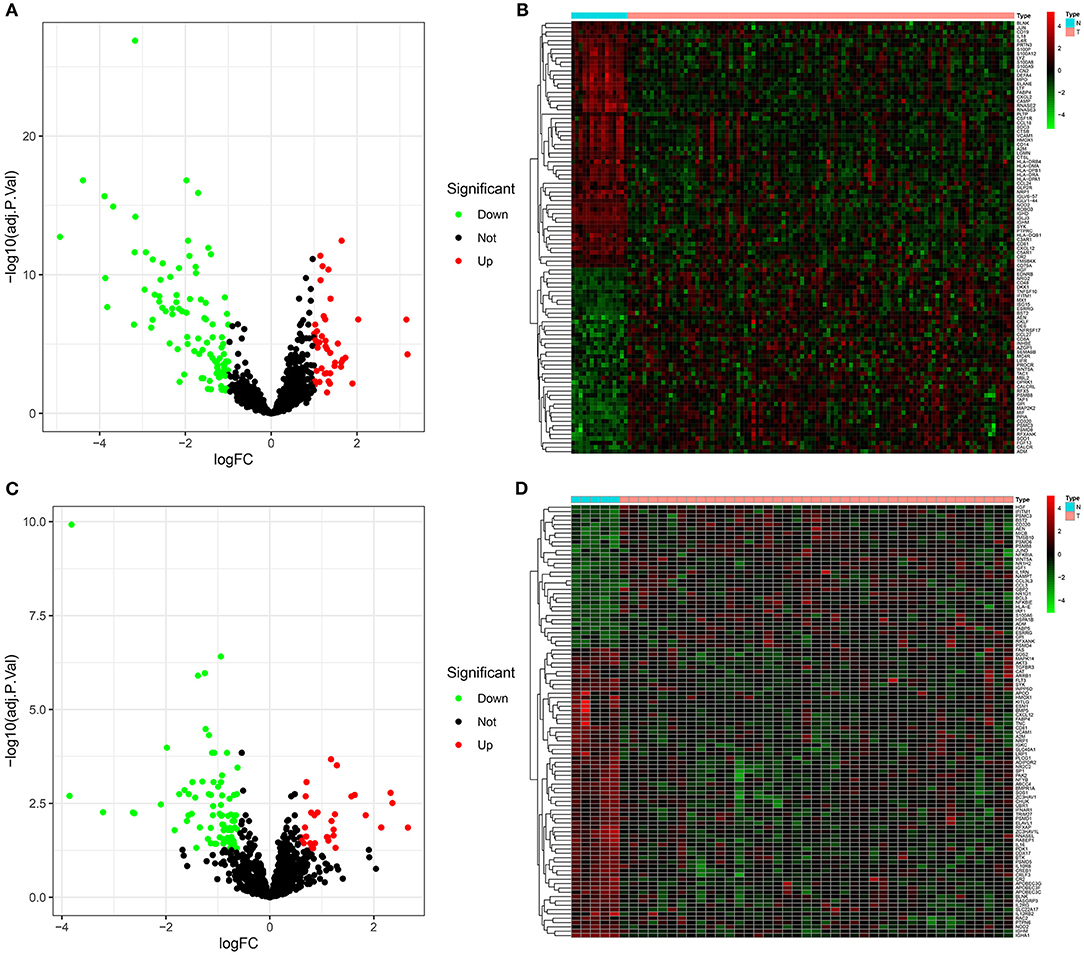
Figure 1. Abnormally expressed immune-related mRNAs between multiple myeloma and normal bone marrow specimens. (A) Volcano plots for up-regulated (red) and down-regulated (green) immune-related mRNAs in multiple myeloma than normal specimens in the GSE6477 dataset. (B) Hierarchical clustering analyses for differential expression patterns of immune-related mRNAs in multiple myeloma and normal samples in the GSE6477 dataset. N indicates normal samples and T indicates multiple myeloma samples. (C) Volcano plots for highly (red) and lowly (green) expressed immune-related mRNAs in multiple myeloma than normal specimens in the GSE47552 dataset. (D) Heat map for abnormal expression patterns of immune-related mRNAs in multiple myeloma and normal samples in the GSE47552 dataset.
Following intersection of abnormally expressed immune-related mRNAs in the GSE6477 and GSE47552 datasets, 31 common mRNAs were obtained, including CD81, PSMB8, CR2, ABCC4, ADM, IGHM, SYK, IFITM1, RASGRP3, IGKC, CXCL12, VCAM1, ESRRG, AEN, SLC22A17, NRP1, A2M, NOD2, BLNK, CD320, HGF, CCL8, BST2, RFXANK, HMOX1, PSMC3, FABP5, GPI, FABP4, WNT5A, and CD19 (Figure 2A). The biological functions that they were involved in were analyzed in depth. These mRNAs primarily participated in B cell-related biological processes such as B cell activation, regulation of immune effector process, immune response-activating cell surface receptor signaling pathway, immune response-activating signal transduction, leukocyte proliferation, antigen receptor-mediated signaling pathway, humoral immune response, regulation of B cell activation, B cell receptor signaling pathway, and regulation of production of molecular mediator of immune response (Figure 2B). They were mainly involved in regulating the cellular components of external side of plasma membrane, secretory granule lumen, cytoplasmic vesicle lumen, vesicle lumen, blood microparticle, immunoglobulin complex, proteasome complex, platelet alpha granule lumen, endopeptidase complex, and immunoglobulin complex, circulating and the like. Moreover, they possessed the molecular functions of receptor ligand activity, signaling receptor activator activity, integrin binding, growth factor activity, glycosaminoglycan binding, cytokine activity, G protein-coupled receptor binding, long-chain fatty acid transporter activity, peptidoglycan binding, and chemoattractant activity and the like. The pathways enriched by these common mRNAs were analyzed. In Figure 2C, they principally participated in B cell receptor signaling pathway, Epstein-Barr virus infection, NF-kappa B signaling pathway, primary immunodeficiency, malaria, hepatocellular carcinoma, tuberculosis, axon guidance, proteasome, PPAR signaling pathway, complement and coagulation cascades, hematopoietic cell lineage, viral protein interaction with cytokine and cytokine receptor, TNF signaling pathway and leukocyte transendothelial migration.

Figure 2. Functional annotation analyses for dysregulated immune-related mRNAs. (A) Venn diagram for common dysregulated immune-related mRNAs in the GSE6477 and GSE47552 datasets. (B) The top 10 GO results enriched by common mRNAs, composed of biological process (BP), cellular component (CC), and molecular function (MF) terms. (C) KEGG pathways involved in common mRNAs. The size of the bubble is proportional to the count of enriched genes. The more the color tends to red, the smaller the p-value.
As depicted by univariate cox regression analyses, among 31 common dysregulated immune-related mRNAs, AEN (HR: 1.35132218004432, 95% CI: 1.00336950593063–1.81993933788734, p-value: 0.0474670415899411), CD320 (HR: 1.2666572113788, 95% CI: 1.06872217896594–1.50125123508741, p-value: 0.0063988822159732), FABP5 (HR: 1.44824784649006, 95% CI: 1.25386429924306–1.67276620454803, p-value: 4.74125786831674e-07), and GPI (HR: 1.45171072863824, 95% CI: 1.06338050769041–1.98185317899107, p-value: 0.0189309122327046) were risk factors of multiple myeloma subjects in the GSE4204 dataset (Figure 3A). Meanwhile, CXCL12 (HR: 0.730888940033441, 95% CI: 0.568482755798023–0.939691903078592, p-value: 0.0144802428352221), IGKC (HR: 0.82233697739277, 95% CI: 0.704030330155922–0.960524107303318, p-value: 0.0135803713395964), NOD2 (HR: 0.842773450385212, 95% CI: 0.719652065331376–0.986959008235654, p-value: 0.0337646407355474), VCAM1 (HR: 0.761232879031875, 95% CI: 0.6455778262167–0.897607496086219, p-value: 0.00117564947725508) and WNT5A (HR: 0.891231234461453, 95% CI: 0.800514277560811–0.992228540507633, p-value: 0.035517571353358) were protective factors of multiple myeloma. In the GSE4204 dataset, our Kaplan-Meier survival analyses demonstrated that low expression of CXCL12 (p-value: 0.009800177; Figure 3B) and WNT5A (p-value: 0.012647237; Figure 3C) displayed more unfavorable outcomes for multiple myeloma patients. Meanwhile, subjects with high FABP5 (p-value: 5.22E-05; Figure 3D) and PSMB8 (p-value: 0.040988621; Figure 3E) expression often experienced shorter survival time. In the GSE4452 dataset, we found that subjects with high CD320 (p-value: 0.100794819; Figure 3F), FABP5 (p-value: 0.002496168; Figure 3G) and GPI (p-value: 0.087747724; Figure 3H) were indicative of poorer outcomes compared to those with their low expression. Combining above data, FABP5 mRNA was a key immune-related prognostic marker for multiple myeloma.
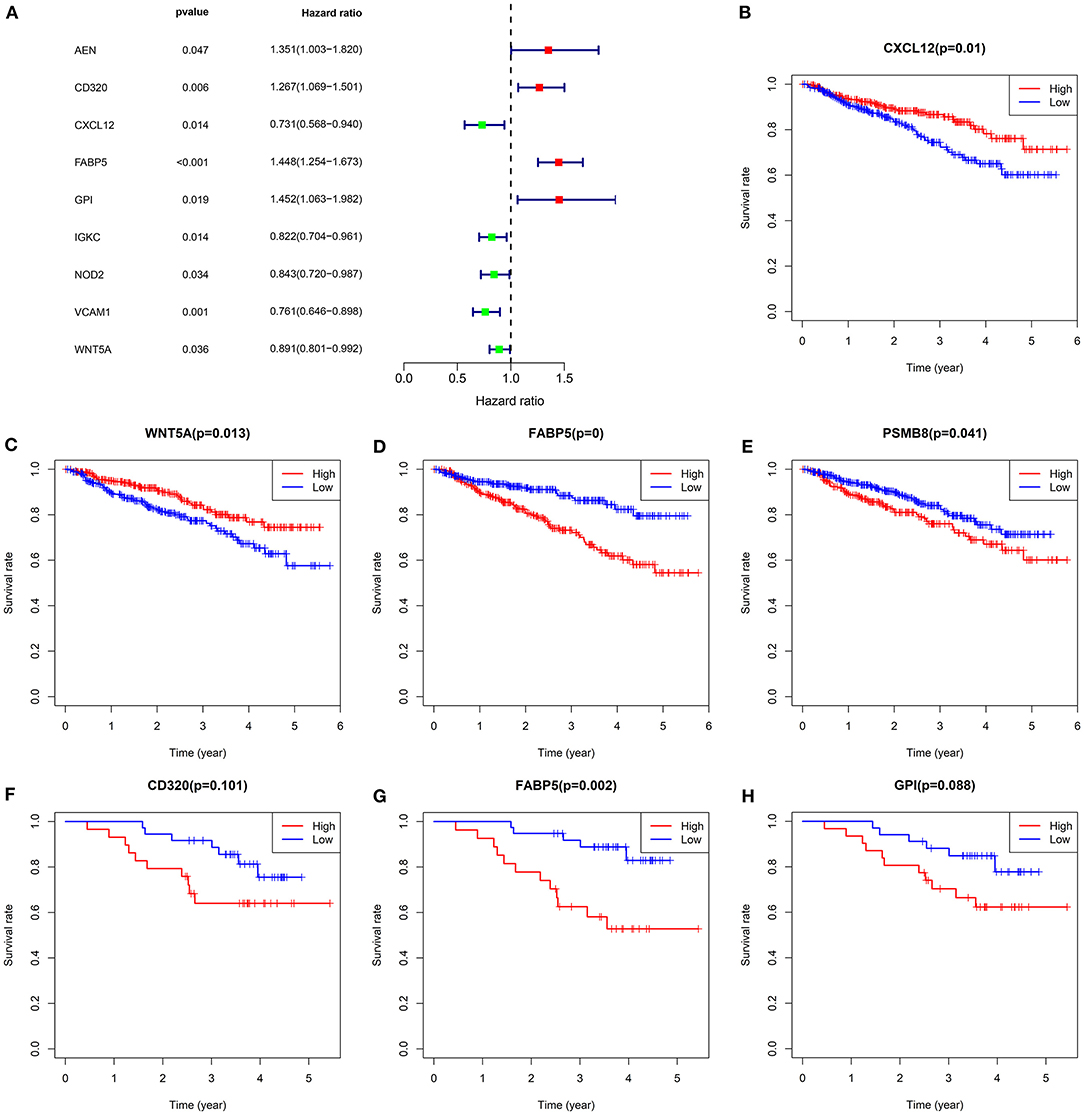
Figure 3. Survival analysis for common dysregulated immune-related mRNAs in multiple myeloma. (A) Forest plots for univariate cox regression analysis of common dysregulated immune-related mRNAs in the GSE4204 dataset. Kaplan-Meier survival curves of (B) CXCL12, (C) WNT5A, (D) FABP5, and (E) PSMB8 expression among multiple myeloma subjects in the GSE4204 dataset. Kaplan-Meier survival curves of (F) CD320, (G) FABP5, and (H) GPI expression among subjects in the GSE4452 dataset. P-values for log-rank test. Red indicates high expression group while blue indicates low expression group.
CIBERSORT was employed to identify the mixture of B cells naïve, B cells memory, plasma cells, CD8 T cells, T cells CD4, naïve T cells, CD4 memory resting, T cells CD4 memory activated, T cells follicular helper, Tregs, T cells gamma delta, NK cells resting, activated NK cells, monocytes, macrophages M0, macrophages M1, macrophages M2, dendritic cells resting, dendritic cells activated, mast cells resting, mast cells activated, eosinophils and neutrophils in multiple myeloma bone marrow specimens (Figure 4A). Correlations between the proportions of 22 immune cells in each multiple myeloma bone marrow sample were further analyzed. In Figure 4B, plasma cells displayed a strongly negative correlation to B cells memory (r = −0.84). Monocytes possessed a positive relationship with NK cells resting (r = 0.41).
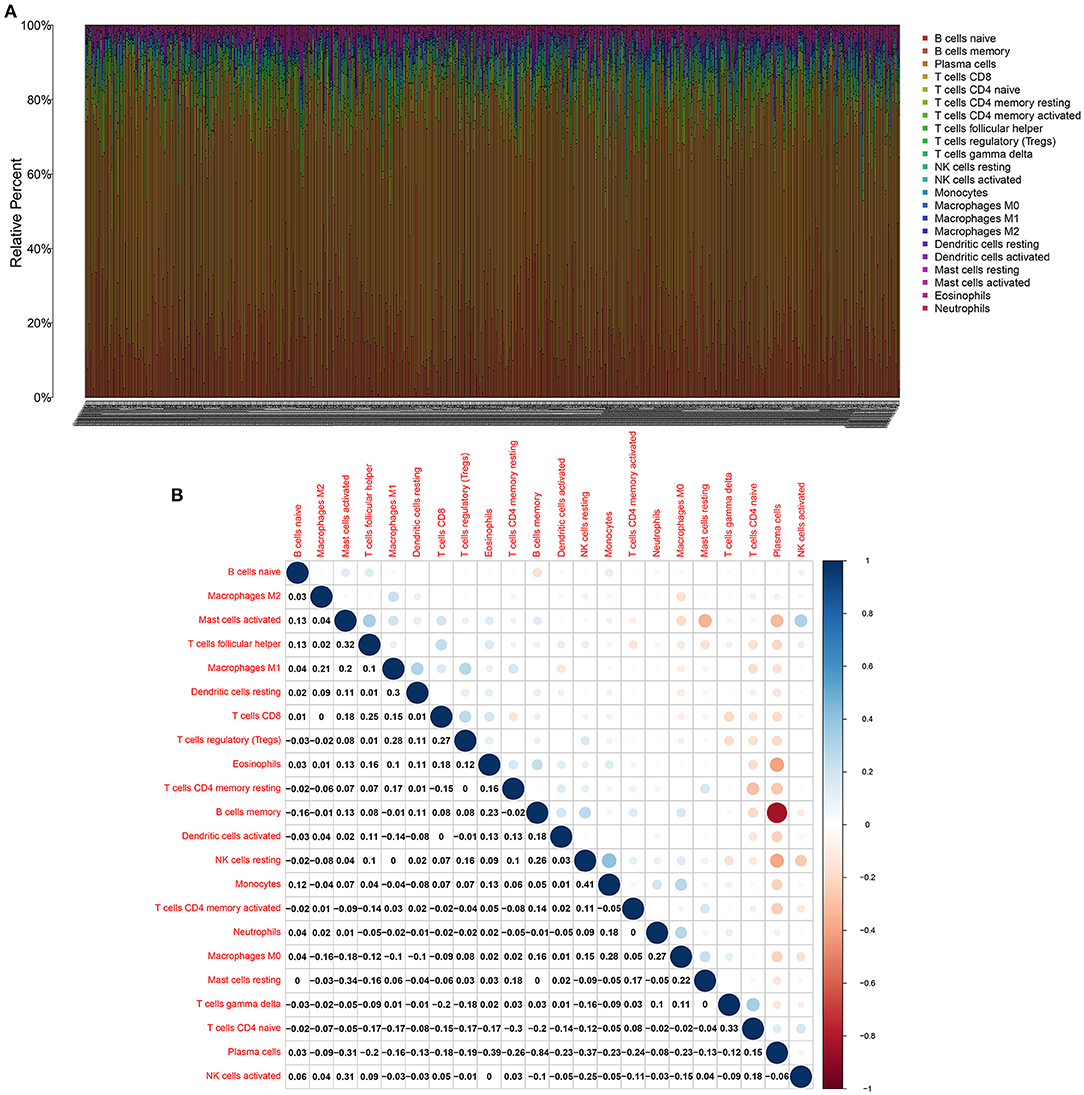
Figure 4. CIBERSORT identifies tumor immune cells in multiple myeloma bone marrow specimens. (A) Stacked bar chart for the proportions of 22 kinds of immune cells in each multiple myeloma bone marrow samples. Each type of immune cell is identified by a unique color. (B) The correlation between the proportions of immune cells in multiple myeloma bone marrow specimens. Blue indicates negative correlation and red indicates positive correlation. The larger the correlation coefficient, the bigger the bubble.
We compared the infiltrations of immune cells in multiple myeloma and normal bone marrow specimens in the GSE6477 dataset. Data showed that there were significant differences in the infiltration levels of B cells naïve (p < 0.001), B cells memory (p < 0.001), plasma cells (p < 0.001), T cells CD4 naïve (p = 0.028), resting memory CD4 T cells (p = 0.012), activated memory CD4 T cells (p = 0.016), Tregs (p < 0.001), resting NK cells (p = 0.007), M0 macrophages (p = 0.002), M1 macrophages (p < 0.001), M2 macrophages (p = 0.004), and neutrophils (p < 0.001) between multiple myeloma and normal bone marrow specimens (Figure 5).
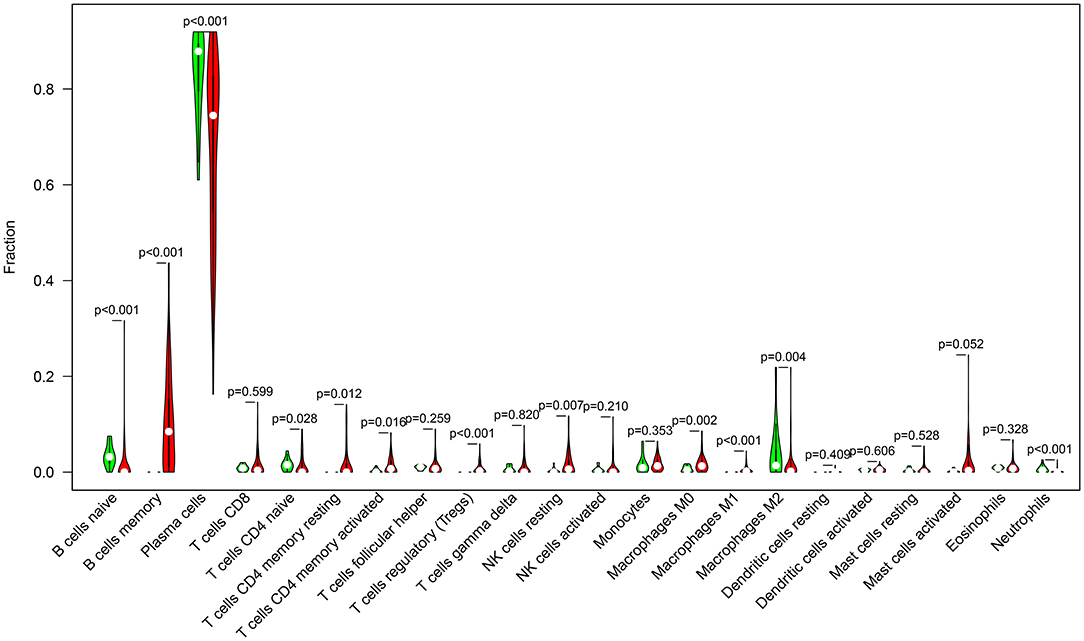
Figure 5. Violin diagram for abnormal expression of immune cells in multiple myeloma (red) and normal (green) bone marrow specimens in the GSE6477 dataset.
The relationships between FABP5 mRNA expression and infiltrations of immune cells were assessed in multiple myeloma bone marrow specimens. In Figure 6A, FABP5 mRNA displayed a negative association with infiltration of B cells memory (r = −0.16, p = 3e-04). A positive correlation between FABP5 mRNA and B cells naïve was detected in multiple myeloma bone marrow specimens (r = 0.11, p = 0.015; Figure 6B). FABP5 mRNA was negatively correlated to levels of dendritic cells activated (r = −0.098, p = 0.023; Figure 6C) and macrophages M0 (r = −0.11, p = 0.0091; Figure 6D). Furthermore, FABP5 mRNA exhibited positive correlations to infiltrations of macrophages M1 (r = 0.09, p = 0.036; Figure 6E), macrophages M2 (r = 0.093, p = 0.03; Figure 6F), neutrophils (r = 0.15, p = 6e-04; Figure 6G), activated NK cells (r = 0.095, p = 0.028; Figure 6H) and resting memory CD4 T cells (r = 0.18, p = 3.4e-05; Figure 6I). We also found that there were negative associations between FABP5 mRNA and infiltrations of CD8 T cells (r = −0.14, p = 0.0016; Figure 6J) and Tregs (r = −0.1, p = 0.016; Figure 6K). Both in the GSE6477 (Figure 7A) and GSE47552 (Figure 7B) datasets, FABP5 mRNA was up-regulated in multiple myeloma compared to normal bone marrow specimens (p = 0.042 and 0.002). There were higher levels of B cells naïve (p = 0.014), T cells CD4 memory resting (p < 0.001), macrophages M2 (p = 0.010) and neutrophils (p = 0.009) as well as lower levels of B cells memory (p = 0.004) and T cells CD8 (p = 0.002) in the high FABP5 expression group compared to its low expression group (Figure 7C).

Figure 6. Associations between FABP5 mRNA and infiltrations of immune cells in multiple myeloma bone marrow specimens. (A) B cells memory; (B) B cells naïve; (C) dendritic cells activated; (D) macrophages M0; (E) macrophages M1; (F) macrophages M2; (G) neutrophils; (H) activated NK cells; (I) resting memory CD4 T cells; (J) CD8 T cells; (K) Tregs.
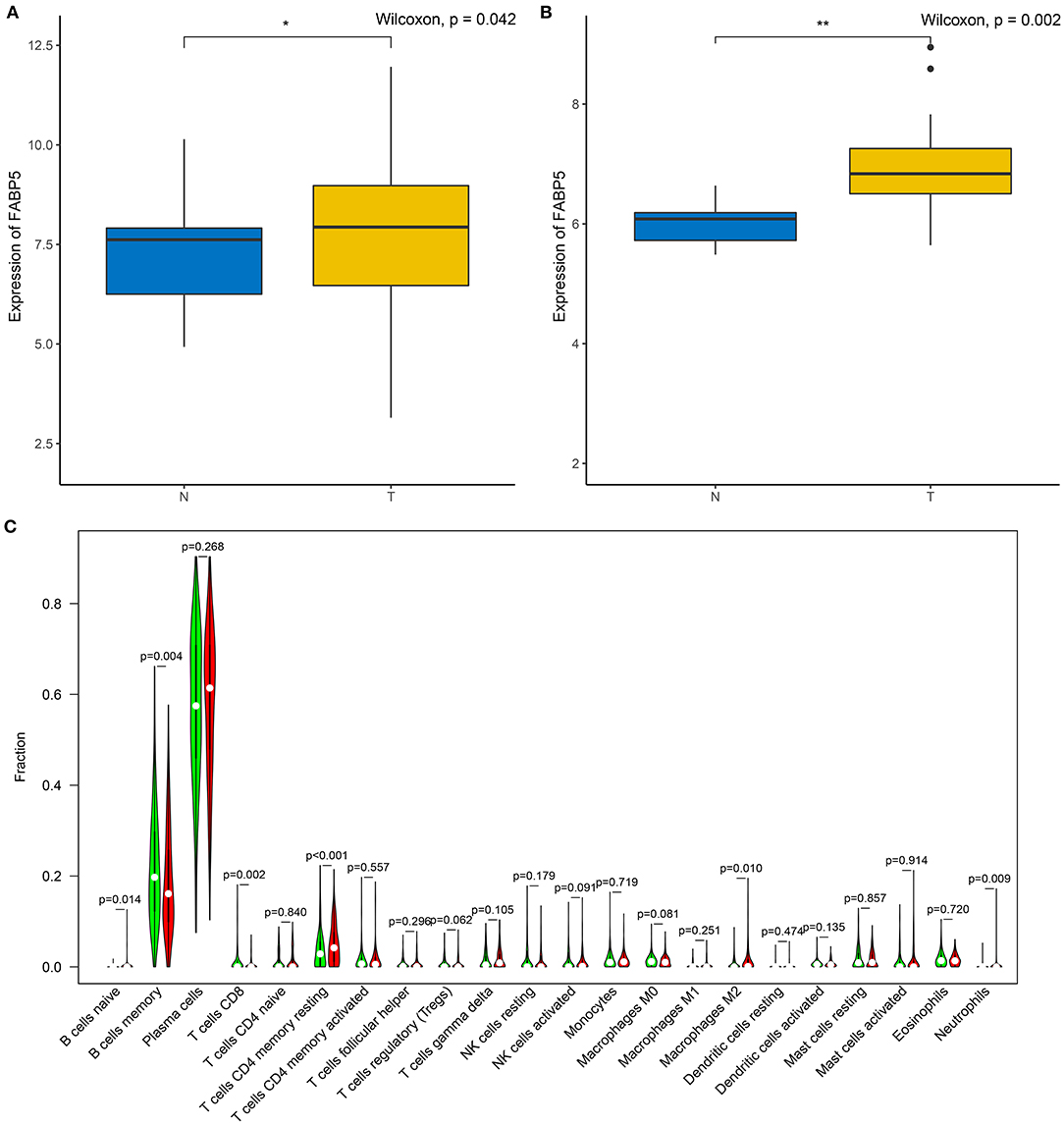
Figure 7. Association between FABP5 mRNA and immune microenvironment of multiple myeloma. (A,B) Expression of FABP5 mRNA in multiple myeloma and normal bone marrow specimens in the (A) GSE6477 and (B) GSE47552 datasets. (C) Infiltration levels of immune cells between high and low FABP5 expression groups. Red: high FABP5 expression group; green: low expression group. *P < 0.05; **P < 0.01.
Multiple myeloma is an aggressive and incurable hematological malignancy, manifested by the malignant proliferation of abnormal plasma cells (23). Despite the advancement of treatment strategies, more therapeutic targets are still required for multiple myeloma. Here, we identified FABP5 as a novel immune-related mRNA as well as a prognostic marker for this malignancy.
Through integrative analyses of the GSE6477 and GSE47552 datasets, this study screened 31 common dysregulated immune-related mRNAs in multiple myeloma. These mRNAs were primarily enriched in B cell-related biological functions such as B cell activation, B cell receptor signaling pathway, plasma membrane. These data were indicative that they participated in the pathogenesis of multiple myeloma. Different patients' outcomes vary greatly. Thus, the establishment of a precise prognostic evaluation system to identify patients with different risks, especially for high-risk patients, is essential for clinicians to formulate overall treatment strategies, and can provide an important reference for patients and their families to understand the disease and achieve better management. Among all dysregulated immune-related mRNAs, AEN, CD320, FABP5, and GPI were risk factors for multiple myeloma, while CXCL12, IGKC, NOD2, VCAM1, and WNT5A were protective factors for multiple myeloma. Among them, CXCL12 up-regulation could be induced by HIF-2α in multiple myeloma plasma cells, thereby reducing migration as well as adhesion to mesenchymal stromal cells (24). CXCL12 is a target for reversing cellular adhesion-induced drug resistance against multiple myeloma (25). NOD2/CARD15 variant is correlated to the sensitivity of multiple myeloma bone marrow cells to bortezomib (26). Sialyltransferase inhibitor may suppress relationships between E-selectin, VCAM1, and MADCAM1, thereby prolonging survival time of multiple myeloma patients (27). WNT5A is an abundant growth factor upon myeloma bone marrow specimens (28). After multiple dataset verification, FABP5 was a key prognostic factor for multiple myeloma. Subjects with FABP5 up-regulation were indicative of more unfavorable outcomes. Hence, it was a risk factor of this disease. Nevertheless, the malignant roles of FABP5 mRNA remain undefined in multiple myeloma. Liu et al. reported that FABP5 was a stage-related prognostic factor of this malignancy (29). Its roles have been expounded in other malignancies. For instance, FABP5 is a critical driving factor of metastatic prostate carcinoma (30). Furthermore, it may promote lymph node metastases for cervical carcinoma via reprogramming fatty acid metabolisms (17). Up-regulated FABP5 induced by fatty acid actuates hepatocellular carcinoma progress (31). FABP5 may elevate PI3K/AKT-mediated proliferation of renal carcinoma cells (18). Combining previous research, FABP5 mRNA might be a potential immunotherapeutic target of multiple myeloma. However, it remains unclear about which immunotherapy (dendritic cell vaccine, CAR T cells, or CAR NK cells) is good for targeting FABP5. Furthermore, whether FAPB5 targeted treatment causes any toxicity in patients with multiple myeloma requires further analysis. Thus, in our future studies, we will validate the therapeutic effects of FABP5 mRNA therapy in multiple myeloma by experiments.
There was a strongly negative correlation of plasma cells and B cells memory in multiple myeloma bone marrow specimens, indicating that there was interplay between plasma cells and B cells memory. There were distinct differences in the infiltration levels of B cells naïve, B cells memory, plasma cells, T cells CD4 naïve, resting memory CD4 T cells, activated memory CD4 T cells, Tregs, resting NK cells, M0 macrophages, M1 macrophages, M2 macrophages, and neutrophils between multiple myeloma and normal samples. Macrophages are innate immune cells that play a role in the host's own defense and maintenance of tissue homeostasis. Macrophages maintain the growth of myeloma cells through cell-to-cell contact-dependent behaviors and non-contact-mediated mechanisms in the bone marrow compartment, while enhancing the protective effect of mesenchymal stem cells on tumor cells (32). As an important part of the bone marrow microenvironment, through the connection between macrophages and tumor cell activation signal pathways, they can inhibit the protease pathway and block drug-induced apoptosis (33). Similar to solid tumors, multiple myeloma cells regulate immune molecules in the bone marrow microenvironment to adapt them to the growth of myeloma itself (34). The main immunosuppressive mechanisms during tumor progression include regulation of the expansion of immune cells, dysfunction of antigen presenting cells and suppression of immune effector cells (like effector T cells, NK cells) (35). Our data were indicative that FABP5 mRNA displayed correlations to B cells memory, B cells naïve, dendritic cells activated, macrophages M0, macrophages M1, macrophages M2, neutrophils, activated NK cells, resting memory CD4 T cells, CD8 T cells and Tregs, suggesting the relationships of FABP5 mRNA with immune microenvironment of multiple myeloma.
However, the prognostic value of FABP5 mRNA will be confirmed in a multicenter multiple myeloma cohort. Moreover, more experiments should be verified the functions of FABP5 in multiple myeloma progress and immune microenvironment.
This study screened 31 abnormally expressed immune-related mRNAs for multiple myeloma. These mRNAs were primarily involved in B cell-related pathways. After verification in multiple datasets, patients with FABP5 mRNA usually unfavorable outcomes. There was crosstalk between immune cells in multiple myeloma bone marrow specimens. FABP5 mRNA displayed significant correlations to infiltrations of immune cells. Taken together, FABP5 mRNA could be a promising therapeutic target as well as prognostic marker in multiple myeloma.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Materials.
ZZ conceived and designed the study. HJ, XZ, XL, RQ, and YL conducted most of the experiments, data analysis, and wrote the manuscript. SL, HZhu, JW, QK, and HZha participated in collecting data and helped to draft the manuscript. All authors reviewed and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.667525/full#supplementary-material
Supplementary Table 1. A list of 2,498 immune-related mRNAs.
Supplementary Table 2. One hundred and thirty-nine abnormally expressed immune-related mRNAs for multiple myeloma in the GSE6477 dataset.
Supplementary Table 3. One hundred and thirty-nine abnormally expressed immune-related mRNAs for multiple myeloma in the GSE47552 dataset.
GEO, Gene Expression Omnibus; ImmPort, Immunology Database and Analysis Portal; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; BP, biological process; MF, molecular function; CC, cellular component; HR, hazard ratio; CI, confidence interval; CIBERSORT, Cell-type Identification By Estimating Relative Subsets of RNA Transcripts; Tregs, T cells regulatory.
1. Luo SQ, Xiong DH, Li J, Li G, Wang Y, Zhang JM, et al. C1orf35 contributes to tumorigenesis by activating c-MYC transcription in multiple myeloma. Oncogene. (2020) 39:3354–66. doi: 10.1038/s41388-020-1222-7
2. Jung SH, Lee HJ, Vo MC, Kim HJ, Lee JJ. Immunotherapy for the treatment of multiple myeloma. Crit Rev Oncol Hematol. (2017) 111:87–93. doi: 10.1016/j.critrevonc.2017.01.011
3. Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. (2002) 2:175–87. doi: 10.1038/nrc746
4. Hou J, Wei R, Qian J, Wang R, Fan Z, Gu C, et al. The impact of the bone marrow microenvironment on multiple myeloma (review). Oncol Rep. (2019) 42:1272–282. doi: 10.3892/or.2019.7261
5. Ho M, Chen T, Liu J, Dowling P, Hideshima T, Zhang L, et al. Targeting histone deacetylase 3 (HDAC3) in the bone marrow microenvironment inhibits multiple myeloma proliferation by modulating exosomes and IL-6 trans-signaling. Leukemia. (2020) 34:196–209. doi: 10.1038/s41375-019-0493-x
6. Ryu D, Kim SJ, Hong Y, Jo A, Kim N, Kim HJ, et al. Alterations in the transcriptional programs of myeloma cells and the microenvironment during extramedullary progression affect proliferation and immune evasion. Clin Cancer Res. (2020) 26:935–44. doi: 10.1158/1078-0432.CCR-19-0694
7. Umezu T, Imanishi S, Yoshizawa S, Kawana C, Ohyashiki JH, Ohyashiki K. Induction of multiple myeloma bone marrow stromal cell apoptosis by inhibiting extracellular vesicle miR-10a secretion. Blood Adv. (2019) 3:3228–40. doi: 10.1182/bloodadvances.2019000403
8. Kawano Y, Zavidij O, Park J, Moschetta M, Kokubun K, Mouhieddine TH, et al. Blocking IFNAR1 inhibits multiple myeloma-driven Treg expansion and immunosuppression. J Clin Invest. (2018) 128:2487–99. doi: 10.1172/JCI88169
9. Chen T, Moscvin M, Bianchi G. Exosomes in the pathogenesis and treatment of multiple myeloma in the context of the bone marrow microenvironment. Front Oncol. (2020) 10:608815. doi: 10.3389/fonc.2020.608815
10. Venken K, Favreau M, Gaublomme D, Menu E, Vanderkerken K, Elewaut D. Checkpoint inhibition in the treatment of multiple myeloma: a way to boost innate-like T cell anti-tumor function? Mol Immunol. (2018) 101:521–6. doi: 10.1016/j.molimm.2018.08.019
11. Bailur JK, McCachren SS, Doxie DB, Shrestha M, Pendleton K, Nooka AK, et al. Early alterations in stem-like/resident T cells, innate and myeloid cells in the bone marrow in preneoplastic gammopathy. JCI Insight. (2019) 5:e127807. doi: 10.1172/jci.insight.127807
12. Bilotta MT, Abruzzese MP, Molfetta R, Scarno G, Fionda C, Zingoni A, et al. Activation of liver X receptor up-regulates the expression of the NKG2D ligands MICA and MICB in multiple myeloma through different molecular mechanisms. FASEB J. (2019) 33:9489–504. doi: 10.1096/fj.201900319R
13. Martín-Antonio B, Suñe G, Najjar A, Perez-Amill L, Antoñana-Vildosola A, Castella M, et al. Extracellular NK histones promote immune cell anti-tumor activity by inducing cell clusters through binding to CD138 receptor. J Immunother Cancer. (2019) 7:259. doi: 10.1186/s40425-019-0739-1
14. Vo MC, Anh-NguyenThi T, Lee HJ, Nguyen-Pham TN, Jaya Lakshmi T, Jung SH, et al. Lenalidomide enhances the function of dendritic cells generated from patients with multiple myeloma. Exp Hematol. (2017) 46:48–55. doi: 10.1016/j.exphem.2016.11.004
15. Chu TH, Vo MC, Park HS, Lakshmi TJ, Jung SH, Kim HJ, et al. Potent anti-myeloma efficacy of dendritic cell therapy in combination with pomalidomide and programmed death-ligand 1 blockade in a preclinical model of multiple myeloma. Cancer Immunol Immunother. (2021) 70:31–45. doi: 10.1007/s00262-020-02654-0
16. Vo MC, Yang S, Jung SH, Chu TH, Lee HJ, Lakshmi TJ, et al. Synergistic antimyeloma activity of dendritic cells and pomalidomide in a murine myeloma model. Front Immunol. (2018) 9:1798. doi: 10.3389/fimmu.2018.01798
17. Zhang C, Liao Y, Liu P, Du Q, Liang Y, Ooi S, et al. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics. (2020) 10:6561–80. doi: 10.7150/thno.44868
18. Lv Q, Wang G, Zhang Y, Han X, Li H, Le W, et al. FABP5 regulates the proliferation of clear cell renal cell carcinoma cells via the PI3K/AKT signaling pathway. Int J Oncol. (2019) 54:1221–32. doi: 10.3892/ijo.2019.4721
19. Bhattacharya S, Andorf S, Gomes L, Dunn P, Schaefer H, Pontius J, et al. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. (2014) 58:234–9. doi: 10.1007/s12026-014-8516-1
20. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
21. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. (2012) 16:284–7. doi: 10.1089/omi.2011.0118
22. Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. (2015) 12:453–7. doi: 10.1038/nmeth.3337
23. Teramachi J, Tenshin H, Hiasa M, Oda A, Bat-Erdene A, Harada T, et al. TAK1 is a pivotal therapeutic target for tumor progression and bone destruction in myeloma. Haematologica. (2020) 106:1401–13. doi: 10.3324/haematol.2019.234476
24. Vandyke K, Zeissig MN, Hewett DR, Martin SK, Mrozik KM, Cheong CM, et al. HIF-2α promotes dissemination of plasma cells in multiple myeloma by regulating CXCL12/CXCR4 and CCR1. Cancer Res. (2017) 77:5452–63. doi: 10.1158/0008-5472.CAN-17-0115
25. Waldschmidt JM, Simon A, Wider D, Müller SJ, Follo M, Ihorst G, et al. CXCL12 and CXCR7 are relevant targets to reverse cell adhesion-mediated drug resistance in multiple myeloma. Br J Haematol. (2017) 179:36–49. doi: 10.1111/bjh.14807
26. Zmorzyński S, Popek-Marciniec S, Styk W, Wojcierowska-Litwin M, Korszeń-Pilecka I, Szudy-Szczyrek A, et al. The impact of the NOD2/CARD15 variant (3020insC) and PSMA6 polymorphism (-8C>G) on the development and outcome of multiple myeloma. Biomed Res Int. (2020) 2020:7629456. doi: 10.1155/2020/7629456
27. Natoni A, Farrell ML, Harris S, Falank C, Kirkham-McCarthy L, Macauley MS, et al. Sialyltransferase inhibition leads to inhibition of tumor cell interactions with E-selectin, VCAM1, and MADCAM1, and improves survival in a human multiple myeloma mouse model. Haematologica. (2020) 105:457–67. doi: 10.3324/haematol.2018.212266
28. Frenquelli M, Caridi N, Antonini E, Storti F, Viganò V, Gaviraghi M, et al. The WNT receptor ROR2 drives the interaction of multiple myeloma cells with the microenvironment through AKT activation. Leukemia. (2020) 34:257–70. doi: 10.1038/s41375-019-0486-9
29. Liu XP, Yin XH, Meng XY, Yan XH, Wang F, He L. Development and validation of a 9-gene prognostic signature in patients with multiple myeloma. Front Oncol. (2018) 8:615. doi: 10.3389/fonc.2018.00615
30. Carbonetti G, Wilpshaar T, Kroonen J, Studholme K, Converso C, d'Oelsnitz S, et al. FABP5 coordinates lipid signaling that promotes prostate cancer metastasis. Sci Rep. (2019) 9:18944. doi: 10.1038/s41598-019-55418-x
31. Seo J, Jeong DW, Park JW, Lee KW, Fukuda J, Chun YS. Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun Biol. (2020) 3:638. doi: 10.1038/s42003-020-01367-5
32. Chen H, Li M, Sanchez E, Soof CM, Bujarski S, Ng N, et al. JAK1/2 pathway inhibition suppresses M2 polarization and overcomes resistance of myeloma to lenalidomide by reducing TRIB1, MUC1, CD44, CXCL12, and CXCR4 expression. Br J Haematol. (2020) 188:283–94. doi: 10.1111/bjh.16158
33. De Beule N, De Veirman K, Maes K, De Bruyne E, Menu E, Breckpot K, et al. Tumour-associated macrophage-mediated survival of myeloma cells through STAT3 activation. J Pathol. (2017) 241:534–46. doi: 10.1002/path.4860
34. Liu Z, Liu H, He J, Lin P, Tong Q, Yang J. Myeloma cells shift osteoblastogenesis to adipogenesis by inhibiting the ubiquitin ligase MURF1 in mesenchymal stem cells. Sci Signal. (2020) 13:eaay8203. doi: 10.1126/scisignal.aay8203
Keywords: multiple myeloma, FABP5, immunotherapy, prognosis, therapeutic target, immune microenvironment
Citation: Jia H, Zhang X, Liu X, Qiao R, Liu Y, Lv S, Zhu H, Wang J, Kong Q, Zhang H and Zhang Z (2021) FABP5, a Novel Immune-Related mRNA Prognostic Marker and a Target of Immunotherapy for Multiple Myeloma. Front. Med. 8:667525. doi: 10.3389/fmed.2021.667525
Received: 13 February 2021; Accepted: 29 April 2021;
Published: 24 June 2021.
Edited by:
Fu Wang, Xi'an Jiaotong University, ChinaReviewed by:
Manh-Cuong Vo, Chonnam National University Hwasun Hospital, South KoreaCopyright © 2021 Jia, Zhang, Liu, Qiao, Liu, Lv, Zhu, Wang, Kong, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhirong Zhang, emhhbmd6aGlyb25nQHNkZm11LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.