
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med., 31 May 2021
Sec. Gastroenterology
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.664045
This article is part of the Research TopicLoss of Epithelial Barrier Integrity in Inflammatory Diseases: Cellular Mediators and Therapeutic TargetsView all 13 articles
 Miguel Gonzalez Acera1,2†
Miguel Gonzalez Acera1,2† Jay V. Patankar1,2†
Jay V. Patankar1,2† Leonard Diemand1,2
Leonard Diemand1,2 Britta Siegmund3
Britta Siegmund3 Markus F. Neurath1,2
Markus F. Neurath1,2 Stefan Wirtz1,2*‡
Stefan Wirtz1,2*‡ Christoph Becker1,2*‡
Christoph Becker1,2*‡Inflammatory cytokines initiate and sustain the perpetuation of processes leading to chronic inflammatory conditions such as inflammatory bowel diseases (IBD). The nature of the trigger causing an inflammatory reaction decides whether type 1, type 17, or type 2 immune responses, typically characterized by the respective T- helper cell subsets, come into effect. In the intestine, Type 2 responses have been linked with mucosal healing and resolution upon an immune challenge involving parasitic infections. However, type 2 cytokines are frequently elevated in certain types of IBD in particular ulcerative colitis (UC) leading to the assumption that Th2 cells might critically support the pathogenesis of UC raising the question of whether such elevated type 2 responses in IBD are beneficial or detrimental. In line with this, previous studies showed that suppression of IL-13 and other type 2 related molecules in murine models could improve the outcomes of intestinal inflammation. However, therapeutic attempts of neutralizing IL-13 in ulcerative colitis patients have yielded no benefits. Thus, a better understanding of the role of type 2 cytokines in regulating intestinal inflammation is required. Here, we took a comparative transcriptomic approach to address how Th2 responses evolve in different mouse models of colitis and human IBD datasets. Our data show that type 2 immune-related transcripts are induced in the inflamed gut of IBD patients in both Crohn's disease and UC and across widely used mouse models of IBD. Collectively our data implicate that the presence of a type 2 signature rather defines a distinct state of intestinal inflammation than a disease-specific pathomechanism.
Immune responses that detect and eliminate multicellular metazoan parasites and allergic reactions have evolved fundamentally differently than those that tackle other immune challenges. The tissue destruction caused by such parasitic infections and allergic reactions, signals an “eliminate and repair” type of innate immune response, characterized by the emergence of the type 2 helper T cells (Th2) (1). The “type 2” nomenclature was initially coined after Th2 lymphocytes, thought of as being the sole cellular sources capable of producing the core type 2 cytokines interleukin (IL)-4, IL-5, and IL-13 (2). However, key discoveries in the field of innate immunity revealed the presence of new cellular sources of type 2 cytokines (3). For e.g., the type 2 innate lymphoid cells, which also express the polarizing transcription factor GATA3 are rich sources of IL-4 and express IL-5 and IL-13 (4). The effector cells of type 2 responses are epithelia, myeloid cells and granulocytes including dendritic cells, macrophages, neutrophils, basophils, eosinophils, and mast cells and the various secretory and absorptive lineages of epithelial cells. Multiple, differentially polarized subsets of T cells are involved in disease pathogenesis in inflammatory conditions, for e.g., Th1, Th2, Th9, and Th17 (5, 6).
It has been speculated that a breakdown in type 2-mediated processes is the underlying cause of the failure of tissue repair and healing in chronic debilitating conditions affecting mucosal tissues (1). However, our understanding of whether this hypothesis holds true is limited. Studies on chronic diseases affecting the gut such as inflammatory bowel diseases (IBD) show a dichotomous relationship between type 2 responses and the two predominant types of IBD, namely Crohn's disease (CD) and ulcerative colitis (UC). Both CD and UC are chronic mucosal inflammatory diseases, an elevated expression of type 2 cytokines may be detected in the tissues and serum of UC, but not of CD patients, however, this concept has recently been challenged (7, 8). Therefore, elevation in type 2 cytokines does not establish a causal connection between UC and type 2 immune responses and crucial questions on when and why such an elevation may be observed remain poorly addressed. Administration of neutralizing antibodies against IL-13 failed to induce any clinical response in UC patients, which begs the question whether defective signaling via respective receptors could cause a compensatory elevation in the expression of the type 2 cytokines in UC (9–11). Indeed some of the well-documented downstream effects of type 2 cytokines such as increased smooth muscle proliferation and cholinergic activation, and elevated mucin production are not uniquely seen to occur in UC, indicating that there is, at least, a partial functional blockade in the mediation of type 2 responses despite the elevation in their levels. By contrast, some studies indicate that elevated type 2 responses in CD patients during parasite infections can in fact prove therapeutically beneficial (12). It is therefore complicated to dissect how type 2 cytokines influence IBD.
Strong type 2 responses are elicited in mucosal tissues during allergic conditions or helminth infections. Therefore, the most common mouse models to study type 2 immune responses are based on infecting mice with parasitic helminths. Some of the most common nematode worms used to provoke mucosal type 2 responses in mice are Nippostrongylus brasiliensis and Heligmosomoides polygyrus (13). Another mouse model, which is reported to show a predominant type 2 response, is the oxazolone colitis model (14, 15). Other common mouse models of colitis such as the chemically induced dextran sulfate sodium (DSS) colitis and tri-nitrobenzidium sulfate (TNBS) colitis are described to have a more mixed Th 1, Th 17 response (16). Whereas, the chronic DSS colitis model is described to involve more of a type 2 immune response (16). One of the most commonly employed models to study the contribution of T cells to the pathogenicity of colitis is the transfer colitis mouse model, which is more of a type 1 model (17). Although these generalized distinctions hold out, there has also been evidence that indicates the contrary. Therefore, the dogmatic view of UC as a predominantly type 2 and CD a type 1 immune disease has been challenged. This is important given that both therapeutic and diagnostic attempts in IBD have evolved around our understanding of the immune homeostasis.
In order to improve our understanding of the type 2 response in IBD, we present here a comparative transcriptomic analysis of various mouse models and human cohorts of IBD using a well-defined list of markers of type 2 immune responses. Our data show that an elevation in type 2 responses is a generalized phenomenon seen intestinal inflammation and identifies a core network of type 2 associated genes that is shared between preclinical models and patients of IBD.
The C57Bl6 strain has been widely used in colitis research and was purchased from Charles river laboratories GmbH (Sulzfeld, Germany). All mice included in this study were of both genders and aged between 8 and 18 weeks. Acute and chronic DSS, Oxazolone, and T-cell transfer colitis were induced as previously described (18, 19). At the end of the colitis induction period, animals were euthanized and tissues were harvested for further analysis. The housing, animal care and experimentation was performed according to the institutional guidelines that were preapproved by the ethics commission of Lower Franconia and Rhineland.
The RNA from the tissue samples were harvested using the peqGold total tissue RNA kit (peqlab GmbH) according to the manufacturer's instructions. The RNA degradation, contamination and quantification were checked using a combination of 1% agarose gels checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA), the Nanodrop (thermofischer), the Qbit (Thermo) and the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
For library preparation for transcriptome, sequencing a total amount of 1 μg RNA per sample was used as input material per sample. Sequencing libraries were generated using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer's recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5X). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3' ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150–200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, USA). Then 3 μl USER Enzyme (NEB, USA) was used with size-selected, adaptorligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina) according to the manufacturer's instructions. After cluster generation, the library preparations were sequenced on an Illumina platform and paired-end reads were generated. Raw data (raw reads) of FASTQ format were firstly processed through in-house scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter and poly-N sequences and reads with low quality from raw data. At the same time, Q20, Q30, and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality. Reference genome and gene model annotation files were downloaded from NCBI, UCSC and Ensembl directly. Paired-end clean reads were mapped to the reference mouse genome (GRCm38.p6) using STAR software (20) (2.7.0d).
Transcriptomic data from the duodenal samples of mice infected with the parasite H. polygyrus that has been previously published was obtained from NCBI's Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/gds (GSE102789). Raw RNAseq data was mapped against the mouse genome and processed and analyzed using the same procedures and tools described above for the mouse colitis models.
Transcriptomic data from the respective intestinal tissues from IBD patients from the previously published studies were obtained from –
a) NCBI's Gene Expression Omnibus (GEO) https://www.ncbi.nlm.nih.gov/gds and from
b) EBI's Array Express https://www.ebi.ac.uk/arrayexpress/.
The specifics of the cohorts and the respective accession numbers are provided here:
Where applicable, microarray data and its annotation was downloaded and processed using the limma R package [3.42.2] (25). Raw RNAseq data was mapped against the human genome [GRCh38.p13], processed and analyzed using the same procedures and tools described above for the mouse colitis models.
Network extension for detection of pathways and physical interactors for a subset type 2 identifier genes that were commonly regulated between mouse and human samples was determined using the freely available tool Genemania for Cytoscape 3.7 (26, 27).
FeatureCounts (v2.0.1) was used to count the read numbers mapped of each gene included in the Ensembl database 21. Then the median of ratios of each gene were calculated, based on the sequencing depth and the RNA composition. Although the Reads Per Kilobase of exon model per Million (RPKM) is currently one of the most commonly used methods for normalization of RNA seq data, the median of ratios used by DESeq2 is more accurate for the differential expression analysis22.
Differential expression analysis between two conditions/groups (three biological replicates per condition) was performed using DESeq2 (v 1.26.0) R package22. DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution22. The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the False Discovery Rate (FDR). Genes with an adjusted P < 0.05 found by DESeq2 were assigned as differentially expressed.
To generate a discovery gene set for type 2 immunity that would then be used to query transcriptomic datasets, we surveyed gene sets in the molecular signatures database (MSigDB, https://www.gsea-msigdb.org/gsea/msigdb) (28). The MSigDB database serves as a standard resource for gene sets used in gene set enrichment analyses. A list of all MSigDB gene sets used for this analysis is available as Supplemental Table 1. Surprisingly, an integrated list of the 15 gene sets that we investigated, failed to show the necessary attributes of a canonical type 2 signature. Therefore, based on reliable literature resources, we compiled a list of 39 genes, which are known to be classical markers for type 2 immune responses (Table 1). This list includes genes categorized into five major groups: cytokines, transcription factors, chemokines, receptors, and other genes, which served as a molecular identifier to interrogate transcriptomic datasets and associate a specific type 2 signature (Table 1).
The genes included in the cytokines group are involved in multiple types of signaling for the activation of the type 2 response. These include the classical type 2 cytokines Il4, Il5, IL9, and IL13 that are involved in the activation of the effector cells (30–33, 36, 37). The cytokines Il33, Il25, and TLSP are primarily produced by parenchymal cells in response to damage, thereby rapidly activating type 2 ILCs (ILC2s) and other immune cells in an early activation stage (2, 29, 40). The ILC2s and Treg cells secrete AREG in order to help repair the damaged epithelium (41, 42). Finally, IL10 is produced by macrophages and lymphocytes, and has an anti-inflammatory function, in order to maintain intestinal homeostasis (34, 35).
Among the key transcription factors (TF) that regulates type 2 polarization and expression of multiple cytokines is GATA3 (43). Another key regulatory TF assigned to type 2 responses is MAF, which controls the production of Il4, and attenuates type 1 signaling (44). The IL4 signaling is crucial in dictating type 2 responses and its actions are mediated by the activation of the TF STAT6, which regulates a plethora of type 2 related effects including polarization and recruitment of Th2 cells (45).
Among the chemokines that represent the type 2 signature were CCL1 and CCL8 that promote migration and activation of ILC2s and Treg cells. CCL17 and CCL22 have similar effects, dendritic cells (DC) produce both and they interact with T helper cells (46–49).
Key cytokine receptors, which influence the type 2 polarization and function are IL4R1 (lymphocytes, ILC, fibroblasts, and epithelium), IL9R (eosinophils, mast cells, ILC2s), IL13RA1 (ILC, granulocytes, epithelial cells, fibroblasts), and IL31RA (monocytes, epithelium) (56, 57, 59). Other cytokine receptors which influence type 2 behaviors include the IL-33 receptor subunit IL1RL1 (hematopoietic), which has been seen involved in allergic responses and IL17RB (ILC2s, monocyte, Tuft cells), which is receptor of IL25 (50, 52). The receptors for type 2-related chemokines are also included in this group, with the genes CCR4 (Th2) and CCR8 (ILCs, T cells) (54, 55). PTGDR2 is a prostaglandin D receptor expressed in Th2 cells, mediating allergic responses (51). The receptor of TSLP, CRLF2 (DC, hematopoietic) is also a key component determining the activation and initiation of Th2 responses (40, 53).
Finally, we identified 12 additional genes that are known to control type 2 responses, but do not fall into any of the above categories. Among these were DENND1B which is involved in the down-modulation of the T cell receptor, and its absence, malfunction or delay has been associated with asthma and allergic response (60). The Tec family tyrosine kinase ITK, also included in this group, is required for the production of type 2 cytokines and the differentiation of ILC2s and T cells (61). ARG1 and ARG2 are paralogues, both encoding for the metabolic enzyme arginase, which metabolizes L-arginine, and has been identified as a marker of ILC2s and alternatively activated macrophages; and an upstream regulator of these metabolic genes is ECM1 (62–64). PRKCZ encodes an atypical protein kinase C, which is involved in immune surveillance (65). Next, we included RETNLA and RETNLB that encode proteins of the Resistin family that are generally elevated upon type 2 immune activation via the actions of IL-13 (66, 67). During nematode driven intestinal type 2 responses, the upregulation of the chitinase CHIL3 is detectable (68). This enzyme controls the degradation of chitin, and is produced and released by intestinal epithelial cells promoting host cell survival and proliferation. Various scenarios of type 2 activation in the intestine have shown that MUC5AC is induced in the epithelium and is responsible for the elevated mucus production, characteristic of these models (69). MRC1 is a receptor induced by the type 2 cytokine IL4 and can bind high-mannose structures on parasite walls, aiding their neutralization and engulfment (70).
In an initial attempt to find discernable patterns of the involvement of type 2 immune responses in different mouse models of IBD, we screened colonic transcriptomes against the type 2 immune response identifier gene set. The mouse models included for this were a time course of dextran sulfate sodium (DSS) colitis representing mild inflammation at day 3, high inflammation at day 8, moderate recovery at day 12 and full recovery from colitis at day 19 of the experimental protocol, characterized by distinct changes in body weights during and after DSS challenge (Figures 1A,B). Apart from these 4 stages of DSS colitis we also included samples of acute DSS colitis, chronic DSS colitis, Oxazolone colitis, and adoptive T-cell transfer colitis, where inflammation was ascertained by histochemical staining (Figures 1C,D). Colonic transcriptomes from unchallenged Rag1 knockout mice and unchallenged C57BL/6 mice were used as control datasets for the T-cell transfer colitis and the rest of the mouse models, respectively. Our exploration yielded 25 genes representing 64% of the identifier gene set which met all requisite criteria for technical thresholds across all the mouse model datasets (Figures 1C,D). Intriguingly, while a type 2 immune signature was evident throughout the course of DSS-induced colitis, we failed to identify a clear type 2 discriminatory signature in specific mouse models and observed that surprisingly, DSS colitis had the most dynamic Th2 response followed by T-cell transfer colitis and lastly oxazolone colitis (Figure 1D). Oxazolone colitis showed the mildest regulatory shift to a type 2 response with only 5 genes of the identifier gene set being significantly regulated. These included Itk, Ccl17, Retnla, Areg, and Ccl8 (Figure 1D). Interestingly, oxazolone colitis was the only dataset in which Ccl17 was significantly regulated. There was an overall tendency toward upregulation of most of the transcripts that we assessed barring 3 transcripts, Dennd1b, Prkcz, and Il17rb which showed an overall downregulation and clustered together across all the datasets that we tested (Figure 1D). The protein products of these three genes functionally cooperate downstream of IL-17 signaling to modulate cellular stress response (71). Among all the genes and across all the datasets, the expression of Tslp showed the least significant variation in expression making it the only gene in the identifier that failed to show significant expression changes in at least one of the datasets (Figures 1C,D). The gene that showed high similarity across all the datasets was Ccl8, which was significantly upregulated in all the tested datasets (Figures 1C,D).
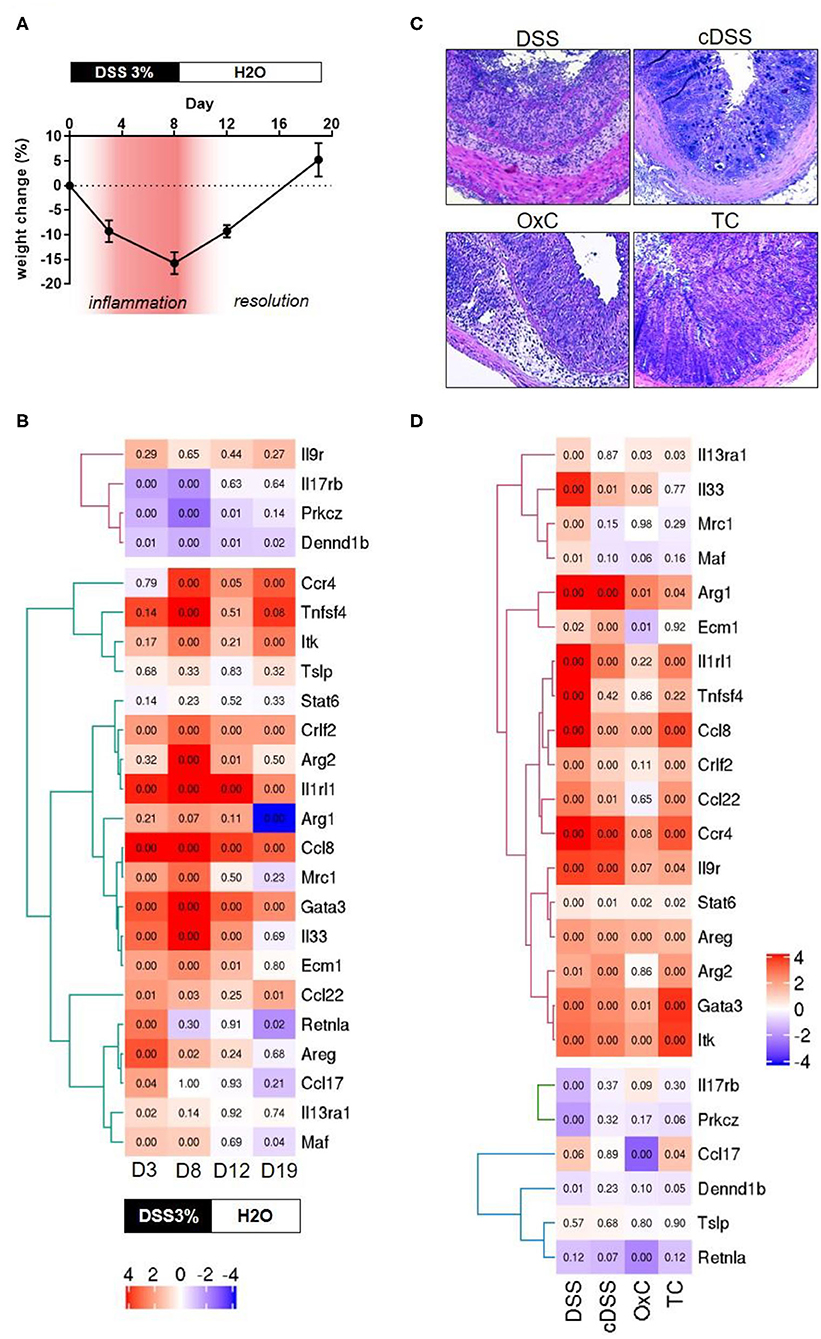
Figure 1. Transcriptomic analyses of type 2 identifiers in multiple mouse models of colitis. (A) percent change in body weight from baseline and DSS treatment paradigm. Time points representing inflammation are represented in red (B) Heatmap depicting the normalized expression of the indicated genes from four different time-points of dextran sulfate sodium (DSS) induced colitis D3 = day 3, D8 = day 8, D12 = day 12, and D19 = day 19. The text boxes below represent the treatment type and duration. Hierarchical clustering shows clear segregation of two distinct groups of genes. (C) Representative photomicrographs of H&E-stained tissue sections from the indicated colitis mouse models. DSS, acute dextran sulfate sodium colitis; cDSS, chronic dextran sulfate sodium colitis; OxC, Oxazolone induced colitis; TC, T-cell adoptive transfer colitis. (D) Heatmap showing normalized expression levels of the indicated genes the type of regimen used to induce colitis is depicted below. Numbers in each quadrant represent the p-value and the legend denotes the fold changes in expression ratios.
Several genes such as Gata3, Itk, Ccl8, Ccl22, Ccr4, Crlf2, Il9r, Il1rl1, and Arg2 are regulated concordantly in T-cell transfer colitis, acute DSS colitis and the time point representing high inflammation in the DSS time course (Figures 1C,D). Finally, some genes tended to show a greater proportion of upregulation only in the DSS colitis models. These included Il33, Ecm1, Arg1 and Mrc1, and Maf (Figures 1C,D). No gene was exclusively upregulated in the Transfer or Oxazolone colitis models. Thus, our analysis fails to identify a discriminatory type 2 signature in mouse models of IBD, which are known to have distinct immune phenotypes (14–17).
Classically, UC has been classified as having a dominant type 2 immune signature. This view has always been controversial and there is evidence in support of and against this view (72, 73). To address whether the type 2 gene set identifier may discriminate between human IBD subtypes, we screened five publicly available transcriptomic datasets representing 22 distinct comparisons against respective control samples from intestinal biopsies of human IBD patients. The datasets represented ileum (Figure 2A) as well as the colon and rectum (Figure 2B) at various degrees of inflammation from CD and UC patients of both genders. After applying the appropriate abundance thresholds our analysis yielded 22 genes representing 56% of the identifier gene set that were analyzed across all the cohorts (Figures 2A,B). A comparison of the overall magnitude of the most upregulated genes, showed that the rectal tissues of UC patients, on an average, had 11 out of the 22 genes upregulated (Figure 2B). These included ARG2, MAF, EVM1, MRC1, CCL22, GATA3, IL33, ITK, CCR4, and IL1RL1. However, their regulatory behavior was non-discriminatory when comparing disease subset or tissue of origin because several of them were also upregulated in CD patients and in inflamed ileal tissues (Figures 2A,B). The least informative genes vis à vis their significance across all the datasets were TNFSF4, DENND1B, STAT6, and TSLP with significant changes in expression detectable in just 1, 3, 3, and 4 comparisons, respectively (Figures 2A,B). Whereas, genes IL1RL1, IL33, ECM1, and PRKCZ were the most informative with significance reached in 15, 12, 12, 12, and 12 comparisons, respectively (Figures 2A,B).
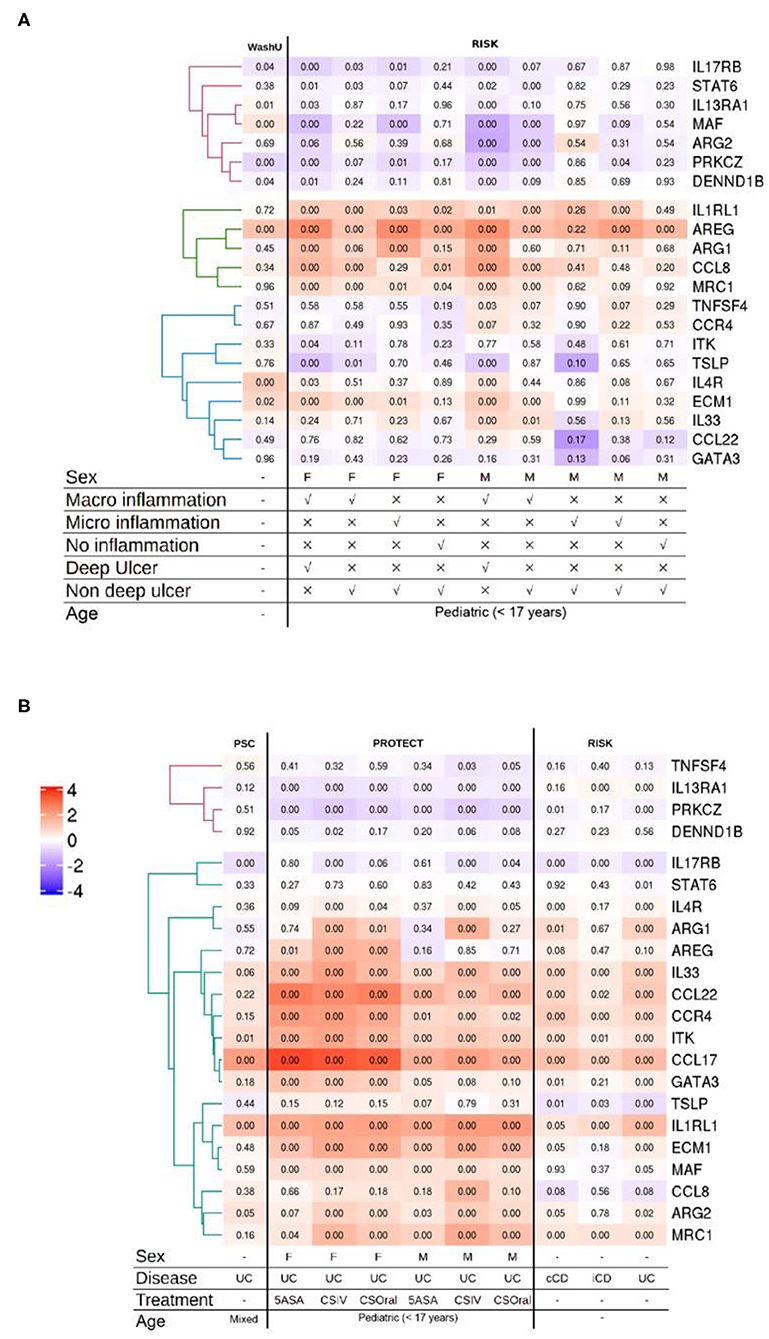
Figure 2. Analysis of multiple IBD cohorts using the type 2 identifier gene set. (A) Heatmap of the normalized ileal gene expression profiles from the indicated cohorts of CD patients. Where available, gender, inflammation type and ulceration status are depicted below. (B) Heatmap of the normalized gene expression profiles of the indicated genes in the colonic and rectal tissues from IBD patients. Where available, gender, disease diagnosis (UC, Ulcerative Colitis; iCD, ileal Crohn's disease; and cCD, colonic Crohn's disease) and treatments (5-ASA, 5-aminosalicylic acid; CSIV, Cyclosporin intravenous; and CSOral, Cyclosporin oral) are indicated below. Numbers in each quadrant represent the p-value and the legend denotes the fold changes in expression ratios.
In the analysis of the more severe cases of disease across all the cohorts, the gene IL1RL1 showed the most significant upregulation in all tissues including ileum, colon and rectum (Figures 2A,B). Remarkably, the upregulation in CCL22 and GATA3 was restricted to the colonic and rectal samples with no regulation detected in any of the ileal samples irrespective of IBD subtype and gender (Figures 2A,B). Among these, CCL22 afforded the most discriminatory power against ileal tissues, with none of the ileal datasets showing changes in regulation and only the rectal and colonic datasets showing an upregulation. We also detected a discordant expression pattern for two genes ARG2 and MAF between the ileum and the rectum with an overall tendency of being downregulated in the inflamed ilea and upregulated in the inflamed rectum (Figures 2A,B). Among the comparisons involving inflamed vs. control ileal tissues, the genes CCL8, ARG1 and AREG were upregulated in patients of both genders with endoscopic evidence of macroinflammation with deep ulcers (Figures 2A,B).
A generalized trend toward downregulation was observed for a cluster of 3 genes that included IL13RA1, PRKCZ, and DENND1B (Figures 2A,B). Among this cluster, IL13RA1 and PRKCZ were significantly downregulated across all the cohorts representing rectal tissues, whereas in ileal cohorts, significance was only reached for the most inflamed ileal tissues with evidence of macroinflammation and deep ulceration (Figures 2A,B).
By comparing the genes that showed significant regulation (up or down) in the mouse models and the human cohorts, we identified two clusters of genes the regulatory behavior of which was conserved. For both, up as well as down regulated genes that were commonly regulated, the greatest overlap was observed between the mouse models showing acute inflammation and human cohorts that were highly inflamed. The seven genes encoding for ITK, GATA3, CCL22, IL1RL1, CCR4, and ARG2 were upregulated in DSS and T-cell transfer colitis, and in the human colon and rectum cohorts (Figures 1C,D, 2A,B). An analysis of pathways and physical interactors of these genes yielded an extended network (Figure 3). Some of the members of this extended network such as IL33, CCL17, and MAF were among our type 2 identifier gene set; the others are downstream components, which may participate in regulating type 2 immune response (Figure 3).
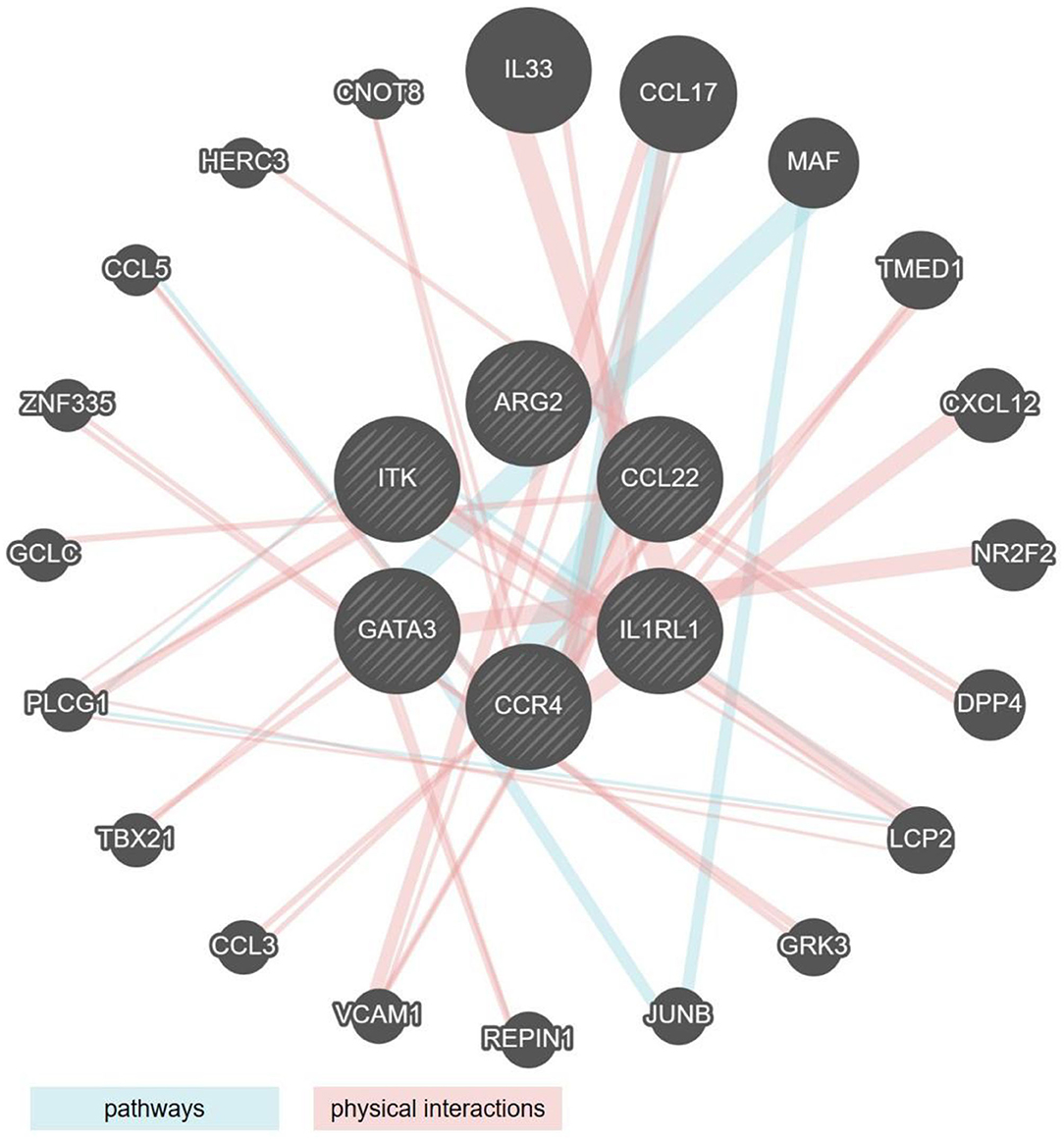
Figure 3. Network analysis of the six type 2 associated genes similarly regulated in human IBD and mouse models of colitis. Radial interaction map of type 2 associated marker genes common to both human IBD and mouse models of colitis (the six central circles, shaded gray). Each non-shaded gray circle represents a predicted pathway interactor (aqua lines) or physical interactor (mauve lines) of the six core factors. Size of each circle represents the rank generated by an automatically selected weighting method in Cytoscape.
Interestingly, and contrary to our expectation, except for ITK, the regulatory behavior of none of these seven shared genes was significantly altered in the Oxazolone colitis mouse model, a model previously associated with Th2-driven pathology (Figure 1D). We also identified another module of genes, including MRC1, ARG2, ECM1, MAF, and IL33 that was upregulated in the DSS models as well as in the human rectum cohorts with some degree of overlap with the most inflamed of the ileal cohorts (Figures 1C,D, 2A,B). Among these, the upregulation of only ARG2 and MAF was truly restricted solely to the rectal datasets (Figures 1C,D, 2A,B). Among the DSS mouse models, Maf expression was restricted to the mild, high and acute inflammation stages, returned to normal levels during the resolution stage, and remained unaltered in the chronic DSS model (Figures 1C,D).
Interestingly, a cognate behavior for the regulation of STAT6 and TSLP was identified among the mouse models and human cohorts that showed a lack of any significant regulation (Figures 1C,D, 2A,B). The upregulation of AREG was observed across all the mouse models except for controls and recovery cohorts and was shared predominantly among in the human ileal but not colonic and rectal ones (Figures 1C,D, 2A,B). Interestingly, the downregulation of the genes PRKCZ and DENND1B was common among the mouse models and the human cohorts (Figures 1C,D, 2A,B).
Research into the cytokine biology of IBD has nurtured the dogmatic view that different IBD subtypes are characterized by distinct immunotypes (6). This classical view has been repeatedly challenged and recent evidence calls for a revision of this point of view. Although several studies have identified an upregulation in type 2 markers in ulcerative colitis, there is no conclusive evidence that a diagnostic discrimination of UC vs. CD can be attained by measuring the markers of type 1 and type 2 immune responses (8, 74, 75). Thus, the type of mucosal immune response on its own cannot explain the differences in the clinical pathogenesis of CD vs. UC. Preclinical mouse models, which resemble these classical human IBD immunotypes, have been widely employed for gaining a better understanding of disease biology as well as for drug discovery purposes. However, due to a lack of consensus on whether certain archetypal immune responses are associated with a certain mouse model that reflects a specific human IBD subtype, we took advantage of a comparative transcriptomic approach. To our knowledge, such an approach has not been applied so far in gaining an understanding of type 2 immunity in preclinical and clinical IBD.
Classically, colitis induced using the adoptive transfer of CD4+CD45RBhigh T cells into Rag 1, 2 knockout mice that lack T and B cells has been considered to be Th1 -mediated (76). More recently, both Th1 and Th17 cells were broadly implicated in colitogenic disease mechanisms in this model. Another commonly used mouse model for IBD, DSS colitis, is also considered to be predominantly a Th1/Th17 type of colitis, although colitis can develop even in the absence of T cells (77). Conversely, the models of chronic DSS colitis and Oxazolone colitis are considered to be Th2-driven forms of colitis (78). Interestingly, our analysis showed a significant overlap in the expression of cognate type 2 gene set in the DSS as well as the T cell transfer colitis models, which as stated earlier are classically considered type 1-driven models. Interestingly, and contrary to our expectation, chronic DSS colitis and Oxazolone colitis showed the least regulation of the type 2 identifier gene set. Using a single cell sequencing approach, Kiner et al. recently also failed to identify classical T helper cell subsets and could not define distinct Th1, Th17, or Th2 restricted clusters but rather identified phenotypically flexible clusters that depended on the overarching microbial milieu (79). Notably, the lack of Th stereotypes in the gut mucosa was not only observed at steady state, but also when mice were infected with different pathogens including the metazoan parasite H. polygurus and N. brasiliensi (79). In line with this, we analyzed two transcriptomic datasets, one from the lung and the other from the small intestine of mice that were infected with the helminth parasites Nippostrongylus brasiliensis and Heligmosomoides polygyrus, respectively (80, 81). Similar to the findings of Kiner et al., we also failed to identify a significant proportion of canonical type 2 response signatures, which were shared between the two tissues.
One technical caveat of transcriptome sequencing methods used in our study is the cutoff for minimum sequence length that guarantees a good sequence read. In our method, this cutoff was 150 base pairs for a paired end sequencing, which inadvertently causes a loss of short length transcripts, which we were unable to include in our analysis. Nonetheless, our analysis contributes to the growing body of evidence that points to a reassessment of the classical immune subtyping in IBD.
In addition, our data reveal a conserved group of type 2 associated genes, which are regulated similarly in the commonly used mouse models of IBD and in human IBD. The analyses presented in our work shows that most of the classical markers of type 2 immune response do not behave in a presumed categorical pattern. We also show that the conception that in preclinical studies of IBD, specific types of immune responses can be modeled using specific mouse models needs to be revised.
The data for the mRNA sequencing datasets generated in this study for the time course of experimental DSS colitis are available via the array express service of EMBL-EBI through the accession number E-MTAB-9850. All other datasets analyzed in this study along with their accession numbers, references, and sources for the data can be found in the materials and methods section of this article.
The animal study was reviewed and approved by The ethics commission of Lower Franconia and Rhineland.
LD: experimentation and analysis. MG, JP, SW, and CB: study concept, design, literature search, experimentation, analysis, interpretation of data, and critical revision of the manuscript. BS: critical revision of the manuscript. All authors contributed to the article and approved the submitted version.
This work received funding from the DFG projects TRR241 (A02, A03, A08, B04, B05, C02, C04, and INF), and the SFB1181 (C02 and C05), KFO257 (TP01), FOR 2438 (TPZ). The project was further supported by the Interdisciplinary Center for Clinical Research (IZKF: J68, A76). LD holds a scholarship from the Transregio initiative TRR241 (A03) for a master's thesis in medicine.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Reyes Gamez Belmonte and Mousumi Mahapatro for their helpful discussions during the drafting of this manuscript. The authors also acknowledge the help provided by the TRR241 IBDome Project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.664045/full#supplementary-material
1. Gieseck RL, Wilson MS, Wynn TA. Type 2 immunity in tissue repair and fibrosis. Nat Rev Immunol. (2018) 18:62–76. doi: 10.1038/nri.2017.90
2. Fort MM, Cheung J, Yen D, Li J, Zurawski SM, Lo S, et al. IL-25 induces IL-4, IL-5, and IL-13 and Th2-associated pathologies in vivo. Immunity. (2001) 15:985–95. doi: 10.1016/S1074-7613(01)00243-6
3. Oliphant CJ, Barlow JL, McKenzie AN. Insights into the initiation of type 2 immune responses. Immunology. (2011) 134:378–85. doi: 10.1111/j.1365-2567.2011.03499.x
4. von Moltke J, Pepper M. Sentinels of the Type 2 immune response. Trends Immunol. (2018) 39:99–111. doi: 10.1016/j.it.2017.10.004
5. Chen ML, Sundrud MS. Cytokine networks and T-cell subsets in inflammatory bowel diseases. Inflamm Bowel Dis. (2016) 22:1157–67. doi: 10.1097/MIB.0000000000000714
6. Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. (2014) 14:329–42. doi: 10.1038/nri3661
7. Strober W, Fuss IJ. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology. (2011) 140:1756–67. doi: 10.1053/j.gastro.2011.02.016
8. Korolkova OY, Myers JN, Pellom ST, Wang L, M'Koma AE. Characterization of serum cytokine profile in predominantly colonic inflammatory bowel disease to delineate ulcerative and Crohn's colitides. Clin Med Insights Gastroenterol. (2015) 8:29–44. doi: 10.4137/CGast.S20612
9. Abraham C, Dulai PS, Vermeire S, Sandborn WJ. Lessons learned from trials targeting cytokine pathways in patients with inflammatory bowel diseases. Gastroenterology. (2017) 152:374–88.e4. doi: 10.1053/j.gastro.2016.10.018
10. Danese S, Rudzinski J, Brandt W, Dupas JL, Peyrin-Biroulet L, Bouhnik Y, et al. Tralokinumab for moderate-to-severe UC: a randomised, double-blind, placebo-controlled, phase IIa study. Gut. (2015) 64:243–9. doi: 10.1136/gutjnl-2014-308004
11. Reinisch W, Panes J, Khurana S, Toth G, Hua F, Comer GM, et al. Anrukinzumab, an anti-interleukin 13 monoclonal antibody, in active UC: efficacy and safety from a phase IIa randomised multicentre study. Gut. (2015) 64:894–900. doi: 10.1136/gutjnl-2014-308337
12. Broadhurst MJ, Leung JM, Kashyap V, McCune JM, Mahadevan U, McKerrow JH, et al. IL-22+ CD4+ T cells are associated with therapeutic trichuris trichiura infection in an ulcerative colitis patient. Sci Transl Med. (2010) 2:60ra88. doi: 10.1126/scitranslmed.3001500
13. Bouchery T, Volpe B, Shah K, Lebon L, Filbey K, LeGros G, et al. The study of host immune responses elicited by the model murine hookworms nippostrongylus brasiliensis and heligmosomoides polygyrus. Curr Protoc Mouse Biol. (2017) 7:236–86. doi: 10.1002/cpmo.34
14. Heller F, Fuss IJ, Nieuwenhuis EE, Blumberg RS, Strober W. Oxazolone colitis, a Th2 colitis model resembling ulcerative colitis, is mediated by IL-13-producing NK-T cells. Immunity. (2002) 17:629–38. doi: 10.1016/S1074-7613(02)00453-3
15. Weigmann B, Neurath MF. Oxazolone-induced colitis as a model of Th2 immune responses in the intestinal mucosa. Methods Mol Biol. (2016) 1422:253–61. doi: 10.1007/978-1-4939-3603-8_23
16. Alex P, Zachos NC, Nguyen T, Gonzales L, Chen TE, Conklin LS, et al. Distinct cytokine patterns identified from multiplex profiles of murine DSS and TNBS-induced colitis. Inflamm Bowel Dis. (2009) 15:341–52. doi: 10.1002/ibd.20753
17. Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, Coffman RL. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. (1994) 1:553–62. doi: 10.1016/1074-7613(94)90045-0
18. Wirtz S, Popp V, Kindermann M, Gerlach K, Weigmann B, Fichtner-Feigl S, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. (2017) 12:1295–309. doi: 10.1038/nprot.2017.044
19. Eden K. Adoptive transfer colitis. Methods Mol Biol. (2019) 1960:207–14. doi: 10.1007/978-1-4939-9167-9_18
20. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
21. VanDussen KL, Stojmirovic A, Li K, Liu TC, Kimes PK, Muegge BD, et al. Abnormal small intestinal epithelial microvilli in patients with Crohn's disease. Gastroenterology. (2018) 155:815–28. doi: 10.1053/j.gastro.2018.05.028
22. Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. (2014) 124:3617–33. doi: 10.1172/JCI75436
23. Quraishi MN, Acharjee A, Beggs AD, Horniblow R, Tselepis C, Gkoutos G, et al. A pilot integrative analysis of colonic gene expression, gut microbiota, and immune infiltration in primary sclerosing cholangitis-inflammatory bowel disease: association of disease with bile acid pathways. J Crohns Colitis. (2020) 14:935–47. doi: 10.1093/ecco-jcc/jjaa021
24. Haberman Y, Karns R, Dexheimer PJ, Schirmer M, Somekh J, Jurickova I, et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat Commun. (2019) 10:38. doi: 10.1038/s41467-018-07841-3
25. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
26. Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. (2008) 9(Suppl. 1):S4. doi: 10.1186/gb-2008-9-s1-s4
27. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303
28. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
29. Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. (2005) 23:479–90. doi: 10.1016/j.immuni.2005.09.015
30. Chatelain R, Varkila K, Coffman RL. IL-4 induces a Th2 response in Leishmania major-infected mice. J Immunol. (1992) 148:1182–7.
31. Takatsu K, Tominaga A, Hamaoka T. Antigen-induced T cell-replacing factor (TRF). I. Functional characterization of a TRF-producing helper T cell subset and genetic studies on TRF production. J Immunol. (1980) 124:2414–22.
32. Schmitt E, Germann T, Goedert S, Hoehn P, Huels C, Koelsch S, et al. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. (1994) 153:3989–96.
33. Renauld JC, Goethals A, Houssiau F, Merz H, Van Roost E, Van Snick J. Human P40/IL-9. Expression in activated CD4+ T cells, genomic organization, and comparison with the mouse gene. J Immunol. (1990) 144:4235–41.
34. Mosmann TR, Schumacher JH, Fiorentino DF, Leverah J, Moore KW, Bond MW. Isolation of monoclonal antibodies specific for IL-4, IL-5, IL-6, and a new Th2-specific cytokine (IL-10), cytokine synthesis inhibitory factor, by using a solid phase radioimmunoadsorbent assay. J Immunol. (1990) 145:2938–45.
35. Moore KW, Vieira P, Fiorentino DF, Trounstine ML, Khan TA, Mosmann TR. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science. (1990) 248:1230–4. doi: 10.1126/science.2161559
36. McKenzie AN, Li X, Largaespada DA, Sato A, Kaneda A, Zurawski SM, et al. Structural comparison and chromosomal localization of the human and mouse IL-13 genes. J Immunol. (1993) 150:5436–44.
37. Herbert JM, Savi P, Laplace MC, Lale A, Dol F, Dumas A, et al. IL-4 and IL-13 exhibit comparable abilities to reduce pyrogen-induced expression of procoagulant activity in endothelial cells and monocytes. FEBS Lett. (1993) 328:268–70. doi: 10.1016/0014-5793(93)80941-M
38. Takaoka A, Arai I, Sugimoto M, Yamaguchi A, Tanaka M, Nakaike S. Expression of IL-31 gene transcripts in NC/Nga mice with atopic dermatitis. Eur J Pharmacol. (2005) 516:180–1. doi: 10.1016/j.ejphar.2005.04.040
39. Stott B, Lavender P, Lehmann S, Pennino D, Durham S, Schmidt-Weber CB. Human IL-31 is induced by IL-4 and promotes TH2-driven inflammation. J Allergy Clin Immunol. (2013) 132:446–54.e5. doi: 10.1016/j.jaci.2013.03.050
40. Ray RJ, Furlonger C, Williams DE, Paige CJ. Characterization of thymic stromal-derived lymphopoietin (TSLP) in murine B cell development in vitro. Eur J Immunol. (1996) 26:10–6. doi: 10.1002/eji.1830260103
41. Zaiss DM, Yang L, Shah PR, Kobie JJ, Urban JF, Mosmann TR. Amphiregulin, a TH2 cytokine enhancing resistance to nematodes. Science. (2006) 314:1746. doi: 10.1126/science.1133715
42. Shoyab M, Plowman GD, McDonald VL, Bradley JG, Todaro GJ. Structure and function of human amphiregulin: a member of the epidermal growth factor family. Science. (1989) 243:1074–6. doi: 10.1126/science.2466334
43. Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, et al. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. (1991) 10:1187–92. doi: 10.1002/j.1460-2075.1991.tb08059.x
44. Ho IC, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. (1996) 85:973–83. doi: 10.1016/S0092-8674(00)81299-4
45. Quelle FW, Shimoda K, Thierfelder W, Fischer C, Kim A, Ruben SM, et al. Cloning of murine Stat6 and human Stat6, Stat proteins that are tyrosine phosphorylated in responses to IL-4 and IL-3 but are not required for mitogenesis. Mol Cell Biol. (1995) 15:3336–43. doi: 10.1128/MCB.15.6.3336
46. Rodriguez-Sosa M, Rosas LE, Terrazas LI, Lu B, Gerard C, Satoskar AR. CC chemokine receptor 1 enhances susceptibility to Leishmania major during early phase of infection. Immunol Cell Biol. (2003) 81:114–20. doi: 10.1046/j.0818-9641.2002.01132.x
47. Goya I, Gutierrez J, Varona R, Kremer L, Zaballos A, Marquez G. Identification of CCR8 as the specific receptor for the human beta-chemokine I-309: cloning and molecular characterization of murine CCR8 as the receptor for TCA-3. J Immunol. (1998) 160:1975–81.
48. Islam SA, Chang DS, Colvin RA, Byrne MH, McCully ML, Moser B, et al. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat Immunol. (2011) 12:167–77. doi: 10.1038/ni.1984
49. Oliveira SH, Lukacs NW. Stem cell factor and igE-stimulated murine mast cells produce chemokines (CCL2, CCL17, CCL22) and express chemokine receptors. Inflamm Res. (2001) 50:168–74. doi: 10.1007/s000110050741
50. Lohning M, Stroehmann A, Coyle AJ, Grogan JL, Lin S, Gutierrez-Ramos JC, et al. T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci USA. (1998) 95:6930–5. doi: 10.1073/pnas.95.12.6930
51. Nagata K, Hirai H, Tanaka K, Ogawa K, Aso T, Sugamura K, et al. CRTH2, an orphan receptor of T-helper-2-cells, is expressed on basophils and eosinophils and responds to mast cell-derived factor(s). FEBS Lett. (1999) 459:195–9. doi: 10.1016/S0014-5793(99)01251-X
52. Rickel EA, Siegel LA, Yoon BR, Rottman JB, Kugler DG, Swart DA, et al. Identification of functional roles for both IL-17RB and IL-17RA in mediating IL-25-induced activities. J Immunol. (2008) 181:4299–310. doi: 10.4049/jimmunol.181.6.4299
53. Park LS, Martin U, Garka K, Gliniak B, Di Santo JP, Muller W, et al. Cloning of the murine thymic stromal lymphopoietin (TSLP) receptor: Formation of a functional heteromeric complex requires interleukin 7 receptor. J Exp Med. (2000) 192:659–70. doi: 10.1084/jem.192.5.659
54. Annunziato F, Galli G, Cosmi L, Romagnani P, Manetti R, Maggi E, et al. Molecules associated with human Th1 or Th2 cells. Eur Cytokine Netw. (1998) 9:12–6. doi: 10.1016/S1286-4579(99)80021-5
55. Zingoni A, Soto H, Hedrick JA, Stoppacciaro A, Storlazzi CT, Sinigaglia F, et al. The chemokine receptor CCR8 is preferentially expressed in Th2 but not Th1 cells. J Immunol. (1998) 161:547–51.
56. Vella AT, Hulsebosch MD, Pearce EJ. Schistosoma mansoni eggs induce antigen-responsive CD44-hi T helper 2 cells and IL-4-secreting CD44-lo cells. Potential for T helper 2 subset differentiation is evident at the precursor level. J Immunol. (1992) 149:1714–22.
57. Umeshita-Suyama R, Sugimoto R, Akaiwa M, Arima K, Yu B, Wada M, et al. Characterization of IL-4 and IL-13 signals dependent on the human IL-13 receptor alpha chain 1: redundancy of requirement of tyrosine residue for STAT3 activation. Int Immunol. (2000) 12:1499–509. doi: 10.1093/intimm/12.11.1499
58. Saito S, Aoki A, Arai I, Takaishi S, Ito H, Akiyama N, et al. Regulation of Th2 responses by different cell types expressing the interleukin-31 receptor. Allergy Asthma Clin Immunol. (2017) 13:23. doi: 10.1186/s13223-017-0194-9
59. Demoulin JB, Renauld JC. Interleukin 9 and its receptor: an overview of structure and function. Int Rev Immunol. (1998) 16:345–64. doi: 10.3109/08830189809043001
60. Yang CW, Hojer CD, Zhou M, Wu X, Wuster A, Lee WP, et al. Regulation of T cell receptor signaling by DENND1B in TH2 cells and allergic disease. Cell. (2016) 164:141–55. doi: 10.1016/j.cell.2015.11.052
61. Miller AT, Wilcox HM, Lai Z, Berg LJ. Signaling through Itk promotes T helper 2 differentiation via negative regulation of T-bet. Immunity. (2004) 21:67–80. doi: 10.1016/j.immuni.2004.06.009
62. Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, et al. Differential regulation of nitric oxide synthase-2 and arginase-1 by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. (2001) 167:6533–44. doi: 10.4049/jimmunol.167.11.6533
63. Xu W, Ghosh S, Comhair SA, Asosingh K, Janocha AJ, Mavrakis DA, et al. Increased mitochondrial arginine metabolism supports bioenergetics in asthma. J Clin Invest. (2016) 126:2465–81. doi: 10.1172/JCI82925
64. Li Z, Zhang Y, Liu Z, Wu X, Zheng Y, Tao Z, et al. ECM1 controls T(H)2 cell egress from lymph nodes through re-expression of S1P(1). Nat Immunol. (2011) 12:178–85. doi: 10.1038/ni.1983
65. Wang Z, Yoo YJ, De La Torre R, Topham C, Hanifin J, Simpson E, et al. Inverse correlation of TRIM32 and protein kinase C zeta in T helper type 2-biased inflammation. J Invest Dermatol. (2020) 141:1297–307.e3. doi: 10.1016/j.jid.2020.09.021
66. Loke P, Nair MG, Parkinson J, Guiliano D, Blaxter M, Allen JE. IL-4 dependent alternatively-activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. (2002) 3:7. doi: 10.1186/1471-2172-3-7
67. Mishra A, Wang M, Schlotman J, Nikolaidis NM, DeBrosse CW, Karow ML, et al. Resistin-like molecule-beta is an allergen-induced cytokine with inflammatory and remodeling activity in the murine lung. Am J Physiol Lung Cell Mol Physiol. (2007) 293:L305–13. doi: 10.1152/ajplung.00147.2007
68. Low D, Subramaniam R, Lin L, Aomatsu T, Mizoguchi A, Ng A, et al. Chitinase 3-like 1 induces survival and proliferation of intestinal epithelial cells during chronic inflammation and colitis-associated cancer by regulating S100A9. Oncotarget. (2015) 6:36535–50. doi: 10.18632/oncotarget.5440
69. Dabbagh K, Takeyama K, Lee HM, Ueki IF, Lausier JA, Nadel JA. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J Immunol. (1999) 162:6233–7.
70. Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. (1992) 176:287–92. doi: 10.1084/jem.176.1.287
71. Bulek K, Chen X, Parron V, Sundaram A, Herjan T, Ouyang S, et al. IL-17A recruits Rab35 to IL-17R to mediate PKCalpha-dependent stress fiber formation and airway smooth muscle contractility. J Immunol. (2019) 202:1540–8. doi: 10.4049/jimmunol.1801025
72. Biancheri P, Di Sabatino A, Ammoscato F, Facciotti F, Caprioli F, Curciarello R, et al. Absence of a role for interleukin-13 in inflammatory bowel disease. Eur J Immunol. (2014) 44:370–85. doi: 10.1002/eji.201343524
73. Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, Strong SA, et al. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn's disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol. (1996) 157:1261–70. doi: 10.1097/00024382-199703001-00528
74. Nemeth ZH, Bogdanovski DA, Barratt-Stopper P, Paglinco SR, Antonioli L, Rolandelli RH. Crohn's disease and ulcerative colitis show unique cytokine profiles. Cureus. (2017) 9:e1177. doi: 10.7759/cureus.1177
75. Bamias G, Cominelli F. Role of type 2 immunity in intestinal inflammation. Curr Opin Gastroenterol. (2015) 31:471–6. doi: 10.1097/MOG.0000000000000212
76. Kanai T, Kawamura T, Dohi T, Makita S, Nemoto Y, Totsuka T, et al. TH1/TH2-mediated colitis induced by adoptive transfer of CD4+CD45RBhigh T lymphocytes into nude mice. Inflamm Bowel Dis. (2006) 12:89–99. doi: 10.1097/01.MIB.0000197237.21387.mL
77. Dieleman LA, Ridwan BU, Tennyson GS, Beagley KW, Bucy RP, Elson CO. Dextran sulfate sodium-induced colitis occurs in severe combined immunodeficient mice. Gastroenterology. (1994) 107:1643–52. doi: 10.1016/0016-5085(94)90803-6
78. Boirivant M, Fuss IJ, Chu A, Strober W. Oxazolone colitis: a murine model of T helper cell type 2 colitis treatable with antibodies to interleukin 4. J Exp Med. (1998) 188:1929–39. doi: 10.1084/jem.188.10.1929
79. Kiner E, Willie E, Vijaykumar B, Chowdhary K, Schmutz H, Chandler J, et al. Gut CD4(+) T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat Immunol. (2021) 22:216–28. doi: 10.1038/s41590-020-00836-7
80. Reece JJ, Siracusa MC, Scott AL. Innate immune responses to lung-stage helminth infection induce alternatively activated alveolar macrophages. Infect Immun. (2006) 74:4970–81. doi: 10.1128/IAI.00687-06
Keywords: IBD, type 2 immunity, intestine, chronic inflammation, ulcerative colitis
Citation: Gonzalez Acera M, Patankar JV, Diemand L, Siegmund B, Neurath MF, Wirtz S and Becker C (2021) Comparative Transcriptomics of IBD Patients Indicates Induction of Type 2 Immunity Irrespective of the Disease Ideotype. Front. Med. 8:664045. doi: 10.3389/fmed.2021.664045
Received: 04 February 2021; Accepted: 04 May 2021;
Published: 31 May 2021.
Edited by:
Alexander R. Moschen, Innsbruck Medical University, AustriaReviewed by:
Hiroshi Nakase, Sapporo Medical University, JapanCopyright © 2021 Gonzalez Acera, Patankar, Diemand, Siegmund, Neurath, Wirtz and Becker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Wirtz, c3RlZmFuLndpcnR6QHVrLWVybGFuZ2VuLmRl; Christoph Becker, Y2hyaXN0b3BoLmJlY2tlckB1ay1lcmxhbmdlbi5kZQ==
†These authors have contributed equally to this work
‡These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.