
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 19 July 2021
Sec. Gastroenterology
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.655474
This article is part of the Research Topic Gut Microbiota and Gastrointestinal Disorders View all 13 articles
 Antonio De Vincentis1*
Antonio De Vincentis1* Marco Santonico2
Marco Santonico2 Federica Del Chierico3
Federica Del Chierico3 Annamaria Altomare4
Annamaria Altomare4 Benedetta Marigliano5
Benedetta Marigliano5 Alice Laudisio6
Alice Laudisio6 Sofia Reddel3
Sofia Reddel3 Simone Grasso2
Simone Grasso2 Alessandro Zompanti7
Alessandro Zompanti7 Giorgio Pennazza7
Giorgio Pennazza7 Lorenza Putignani3,8
Lorenza Putignani3,8 Michele Pier Luca Guarino4
Michele Pier Luca Guarino4 Michele Cicala4
Michele Cicala4 Raffaele Antonelli Incalzi1
Raffaele Antonelli Incalzi1Background: Intestinal dysbiosis might play a pathogenetic role in subjects with symptomatic uncomplicated diverticular disease (SUDD), but the effect of rifaximin therapy has been scantly explored with regard to gut microbiota variations in patients with SUDD.
Aims: To verify to which extent rifaximin treatment affects the gut microbiota and whether an electronic multisensorial assessment of stools and breath has the potential for detecting these changes.
Methods: Breath and stool samples were collected from consecutive patients with SUDD before and after a 7 days' therapy with rifaximin. Stool microbiota was assessed, and the electronic multisensorial assessment was carried out by means of the BIONOTE electronic (e-)tongue in stools and (e-)nose in breath.
Results: Forty-three subjects (female 60%, median age 66 years) were included, and 20 (47%) reported clinical improvement after rifaximin therapy. Alpha and beta diversity of stool microbiota did not significantly change after treatment, while a significant variation of selected taxa was shown (i.e., Citrobacter, Coprococcus, Anaerotruncus, Blautia, Eggerthella lenta, Dehalobacterium, SMB53, and Haemophilus parainfluenzae). Overall, the electronic multisensorial system suboptimally mirrored microbiota changes, but it was able to efficiently predict patients' clinical improvement after rifaximin with accuracies ranging from 0.81 to 0.98.
Conclusions: In patients with SUDD, rifaximin administration is associated with significant variation of selected taxa. While inaccurate in predicting gut microbiota change, an electronic multisensorial system, made up of e-tongue and e-nose, was able to predict clinical improvement, thus potentially qualifying as an easy and cheap tool to forecast subjects taking most likely benefit from rifaximin therapy.
Colonic diverticulosis is a complex multifactorial disorder, in which dysbiosis could play a key role. It affects up to one third of people over the age of 60 years and causes symptoms including abdominal pain or bloating and changes in bowel habit (condition termed symptomatic uncomplicated diverticular disease—SUDD) in 20% of cases (1). Approximately 10–25% of patients with SUDD may develop acute diverticulitis (2). The fact that the small bowel diverticula are associated with bacterial overgrowth and most complications of colonic diverticular disease are bacterial in nature and may benefit from antibiotic therapy or fecal stream diversion further supports the importance of gut microbiota in diverticular disease pathogenesis (3). Fiber deficiency, attributed to a Western diet, plays a crucial role, considering that DNA sequencing confirms that fecal microbiota composition is affected by consumption of supplemental fibers (4). A low-fiber diet not only affects colonic motility but can account for the microbiota composition bending toward a prevalence of Bacteroides (5). Low-grade inflammation, altered intestinal microbiota, visceral hypersensitivity, and an abnormal colonic motility are likely to play a variable pathogenetic role (6). The presence of inflammation and dysbiosis in SUDD validates the responsiveness to anti-inflammatory medications like non-absorbable enteral antibiotics such as rifaximin, a semisynthetic antibiotic synthesized in 1982 from rifamycin (7). Rifaximin has a broad spectrum of antibacterial action, is unlikely to induce bacterial resistance (7), and decreases the metabolic activity of the intestinal bacterial flora and the degradation of dietary fiber (8). Rifaximin acts by binding to the beta subunit of bacterial DNA-dependent RNA polymerase resulting in the inhibition of bacterial RNA synthesis (9). It has in vitro bactericidal and bacteriostatic activity against aerobic and anaerobic gram-positive and gram-negative species, being also able to reduce bacterial virulence and translocation and to inhibit bacterial adherence to gut mucosa (10). In fact, cyclic administration of rifaximin with dietary fiber supplementation outperforms simple dietary fiber supplementation in reducing both symptoms and complication frequency of SUDD (11). In conclusion, the better comprehension of the inflammatory patterns and the gut microbiota has increased the therapeutic options: current evidence enhances the therapeutic role of rifaximin (as well as mesalazine that acts directly on the gastrointestinal epithelium) in the treatment of SUDD symptoms, and the fiber supplementation is still recommended in SUDD by much of the international guidelines even if its use is not supported by the recent evidence. Unfortunately, the only available study of rifaximin effects on gut microbiota refers to only four women, being thus less than exploratory in nature (12). Thus, we purposed to verify to which extent rifaximin treatment affects the gut microbiota and whether an electronic multisensorial assessment of stools and breath has the potential for detecting these changes. Indeed, the genetic study of microbiota is cumbersome and requires both time and money. Instead, both a gas sensor array (e-nose) and the liquid sensor array (e-tongue) qualify as very simple and inexpensive methods for assessing, respectively, the spectrum of volatile organic compounds and the electrical impedance of a given liquid, which clearly reflect its physical properties and chemical composition (13). Both e-nose and e-tongue have displayed a wide spectrum of diagnostic and classificatory properties in different conditions, e.g., in liver diseases, chronic obstructive pulmonary disease, asthma, and selected tumors for the e-nose (14–19), in ascites, pleural effusion, urinary tract infections, and wound infections for e-tongue (20–23). Thus, e-nose and e-tongue might be able to catch the rifaximin-induced changes in gut microbiota, allowing either to interpret or to monitor the response to rifaximin in SUDD patients.
In this prospective longitudinal study, consecutive subjects with a diagnosis of SUDD were recruited at the Campus Bio-Medico University Hospital of Rome (Italy) from January to July 2017. Both design and size of this study are consistent with the intention of performing a proof-of-concept study testing the diagnostic properties of e-nose and e-tongue toward genetically proved changes in gut microbioma induced by rifaximin. SUDD was defined as the presence of abdominal pain, bloating, and/or bowel habit changes that include diarrhea, constipation, or a mixed bowel habit, in patients with diverticulosis in the absence of macroscopic inflammation and of any complications (stenosis, abscesses, fistula) (2, 24). Subjects referring allergy to rifaximin; or taking medication with a potential modifying role on microbiota (i.e., antibiotics, prebiotics, probiotics, proton pump inhibitors) in the previous month; or with active cancer, COPD exacerbation, end stage of liver, or kidney disease; or suffering from inflammatory bowel disease were excluded. Finally, considering the well-known influence of diet on intestinal microbiota, only subjects following a Mediterranean normocaloric omnivorous diet (2,000–2,100 and 2,500–2,800 kcal/diet for women and men, respectively) were included.
Since a reduction of potentially pathogenic components of the intestinal microbiota was expected after rifaximin therapy in at least the 70% of participants, the enrollment of 20 subjects was considered adequate to guarantee a statistical power of 80% assuming an alpha error of 0.05. Based on previous experience with multisensorial systems (14, 15, 20–23), this sample size was increased to at least 40 subjects in order to allow the investigation of the discriminative and classificatory capacities of these systems toward rifaximin-induced microbiome changes.
All the main socio-demographic and clinical variables along with blood tests were collected for each participant. Nutritional assessment was performed through the Mini Nutritional Assessment (MNA) (25), abdominal symptoms (i.e., pain and bloating) were scored as mild, moderate, or severe, and bowel habits were assessed through the Bristol Stool Scale. Eligible subjects were evaluated at baseline and, then, were prescribed with a cycle of 7 days of rifaximin 800 mg/diet. A follow-up evaluation was performed after the completion of the antibiotic therapy. Breath and fecal samples were collected for each participant at baseline and at follow-up for the multisensorial system analysis and for the gut microbiota analysis. The study protocol was approved by the local Ethical Committee (n 47/2016), and all participants signed an informed consent.
Fecal samples were collected by each subject in the same morning of the outpatient visit. DNA was extracted from 200 mg of stools using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany), following the manufacturer's instructions. The 16S rRNA V3–V4 variable region (~460 bp) was amplified by using the primer pairs described in the MiSeq rRNA Amplicon Sequencing protocol (Illumina, San Diego, CA, USA). The PCR reactions were set up using a 2 × KAPA HiFi HotStart ReadyMix (KAPA Biosystems Inc., Wilmington, MA, USA) following the manufacturer's protocol. AMPure XP beads (Beckman Coulter Inc., Beverly, MA, USA) were employed to clean DNA amplicons from primers and dimer primers. A unique combination of Illumina Nextera adaptor-primers for each sample was incorporated in amplicons by a second amplification step. The final library was cleaned up and quantified using Quant-iT™ PicoGreen® dsDNA Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Samples were pooled together before the sequencing on an Illumina MiSeq™ platform according to the manufacturer's specifications to generate paired-end reads of 300 base-length. QIIME v1.9 software (26) was used to filter raw reads for quality, read length, and chimera presence. Cleaned sequences were then clustered into operational taxonomic units (OTUs) with a 97% clustering threshold of pairwise identity. OTUs' representative sequences were aligned using PyNAST v.0.1. software (27) against the Greengenes 13_08 database with a 97% similarity for bacterial sequences. All raw sequences have been archived in the NCBI database: PRJNA731467 (https://www.ncbi.nlm.nih.gov/bioproject).
The multisensorial system was made up of the BIONOTE e-Tongue for liquids and of the BIONOTE e-Nose for gases (13). Breath collection was performed in the morning by means of the Pneumopipe® (European Patent n 12425057.2), with all participants who have been fasting and smoke-free for at least 8 h. The breath samples were stored in adsorbent cartridges (Tenax GR by Supelco) at −20°C after collection. Before analysis, the VOC mixture was desorbed with a thermal procedure at four different temperatures (50, 100, 150, and 200°C) and, then, analyzed by the BIONOTE e-Nose (13). This instrument is composed of eight transducers consisting of quartz crystals with a resonance frequency of 20 MHz in thickness shear mode, functionalized with a combination of anthocyanins extracted from three different plant tissues: red rose, red cabbage, and blue hortensia. Each VOC in the mixture binds to the different anthocyanins in the measure cell, thus causing a frequency shift of the respective quartz forming the reference value: this frequency shift is acquired by the system as the sensor response. The final dataset is composed of a fingerprint of 32 responses for each sample, which derive from the registration of the eight-sensor behavior at the four different temperatures of VOC desorption from the cartridge.
Fecal samples were collected by each subject in the same morning of the outpatient visit and stored at +4°C. Before the analysis, feces were prepared for the analysis by diluting 50 mg in 5 ml of distilled water and subsequently centrifuged at 10°C at 1,000 relative centrifugal forces for 10 min. The supernatant was taken with a pipette and analyzed by the BIONOTE e-Tongue (13). This instrument consists of voltammetric screen-printed electrodes controlled by a high-stability electronic interface. The sensor probe is made up of three electrodes: a silver working, a platinum reference, and a gold counter electrode. The applied input signal consists of a triangular waveform with a working range from −1 to 1 V with 500 input voltages and 500 corresponding output current values. The time duration of the complete measurement process is of about 90 s, repeated five times for reproducibility assessment. Data array is made up by the characteristic fingerprints extracted by each voltammogram registered for the measured samples. The setup of the parameters for the acquisition, the number of samples, and the sampling interval are controlled by a dedicated software interface.
Data were presented by means of descriptive statistics and compared using non-parametric tests. Alpha diversity of the gut microbiota was measured on the raw data by the Good's coverage, Chao-1, and Shannon indices, using the Wilcoxon signed-rank test to assess differences after rifaximin therapy. For further analyses, OTUs not seen in at least 20% of the samples or with a relative abundance <1% in the total dataset were removed. Principal component analysis (PCoA) on unweighted and weighted UniFrac distance matrices was used as ordination method to compare gut microbiota of subjects pre- and post-rifaximin therapy by permutational multivariate analysis of variance (PERMANOVA). In addition, the differential abundance analysis of gut bacteria was conducted through the negative binomial distribution on raw counts normalized by “size factors,” taking into account the sequencing depth between the samples. The differences in bacterial abundance were expressed as log2 fold change (log2FC). Analyses of gut microbiota changes were stratified by gender and by presence of abdominal pain, and linear mixed models were applied to verify the related impact in the amount of alpha diversity change after rifaximin therapy.
The ability of e-tongue and e-nose to predict gut microbiota changes or clinical improvement after rifaximin therapy was verified using partial-least-squared discriminant analysis (PLS-DA) with 10-fold cross-validation. Predictive capacities were expressed with the root-mean-square-error cross validation (RMSECV) to aggregate in a single measure of predictive power the magnitudes of the machine errors in prediction of continuous variables (e.g., Shannon and Chao-1 indices). Since RMSECV expresses the prediction error in the same unit of the original measurement, it cannot be compared between different variables. To allow comparability, RMSECV% was computed standardizing RMSECV by the 95% interval of the variable-specific distribution. Conversely, for dichotomous outcomes (i.e., clinical improvement), overall accuracy, sensitivity, specificity, and positive (PPV) and negative (NPV) predictive values were computed. All the analyses were performed using R version 4.0.2 (The R Foundation for Statistical Computing, Vienna, Austria).
Forty-three subjects with SUDD were included in the study (Table 1). The median age was 66 years, and 26 (60%) were women, with a median BMI of 24.5 kg/m2. Sixteen subjects (37%) were at risk of malnutrition, and no malnourished subjects were identified. At baseline, all subjects complained about abdominal bloating [mild: 29 (67%); moderate: 4 (9.3%); and severe: 10 (23%)] and 18 (42%) referred also abdominal pain [mild: 13 (72.2%); moderate: 5 (27.8%)]. According to the Bristol Stool Scale, 12 participants (28%) had diarrheal stool, while only 2 (4.7%) were constipated.
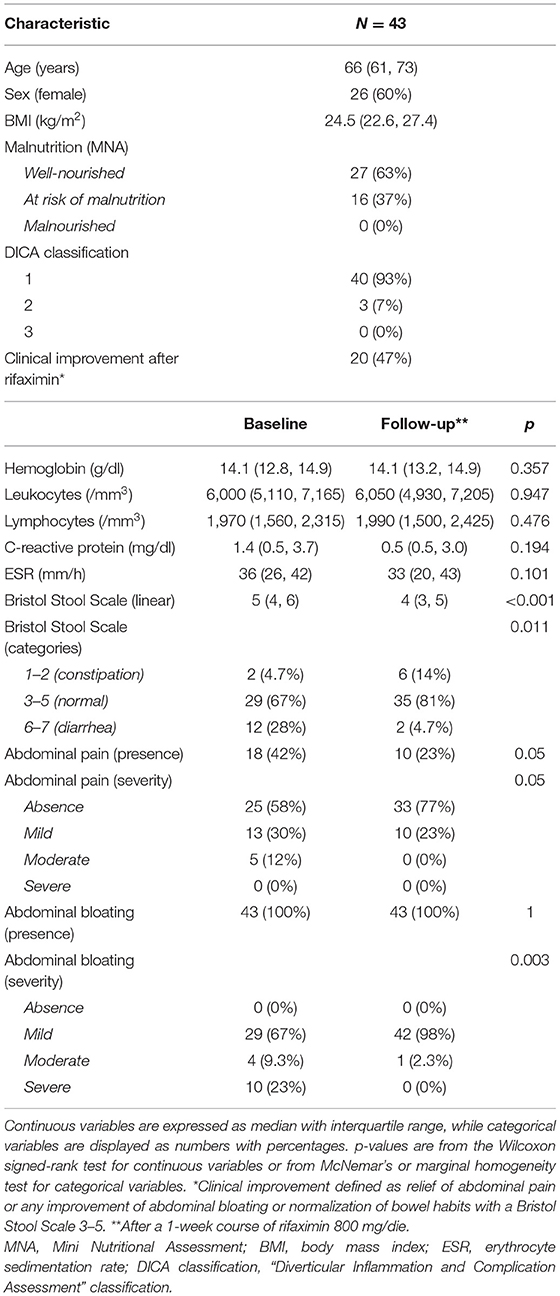
Table 1. Main clinical and biochemical characteristics of the study cohort at baseline and after rifaximin therapy (follow-up).
After rifaximin therapy, no significant differences were observed regarding the main inflammatory markers (leukocytes, C-reactive protein, and erythrocyte sedimentation rate). Conversely, patients reported a significant improvement of symptoms in terms of abdominal pain (p = 0.05), bloating (p = 0.003), and bowel habits (p = 0.011—Table 1). Overall, 20 subjects (47%) experienced a clinical improvement after therapy, defined as relief of abdominal pain or any improvement of abdominal bloating or normalization of bowel habits.
Good's coverage averaged 96% in baseline samples and 97% in follow-up, indicating that most of the OTUs in the samples were detected. Alpha diversity measures did not significantly change after rifaximin therapy [Shannon index: baseline 4.5 (4.0–4.8) vs. follow-up 4.1 (3.6–4.6), p 0.06; Chao-1 index: baseline 6,246.1 (3,796.5–8,239.8) vs. follow-up 5,615.7 (3,634.5–7,741), p 0.77; Figure 1, upper panels]. Similarly, the UniFrac PCoA plot did not show a shift in the overall gut microbiota composition from baseline to post-rifaximin treatment (PERMANOVA p 0.08 for unweighted and 0.06 for weighted analysis; Figure 1, lower panels). Similar findings were observed after stratification according to gender and to the presence of abdominal pain (Supplementary Figure 1), and no difference was found in the amount of alpha diversity change after rifaximin therapy across gender (p for linear mixed model 0.40 and 0.92 for Shannon and Chao-1 indices, respectively) and across patients with/out abdominal pain (p for linear mixed model 0.26 and 0.42 for Shannon and Chao-1 indices, respectively).
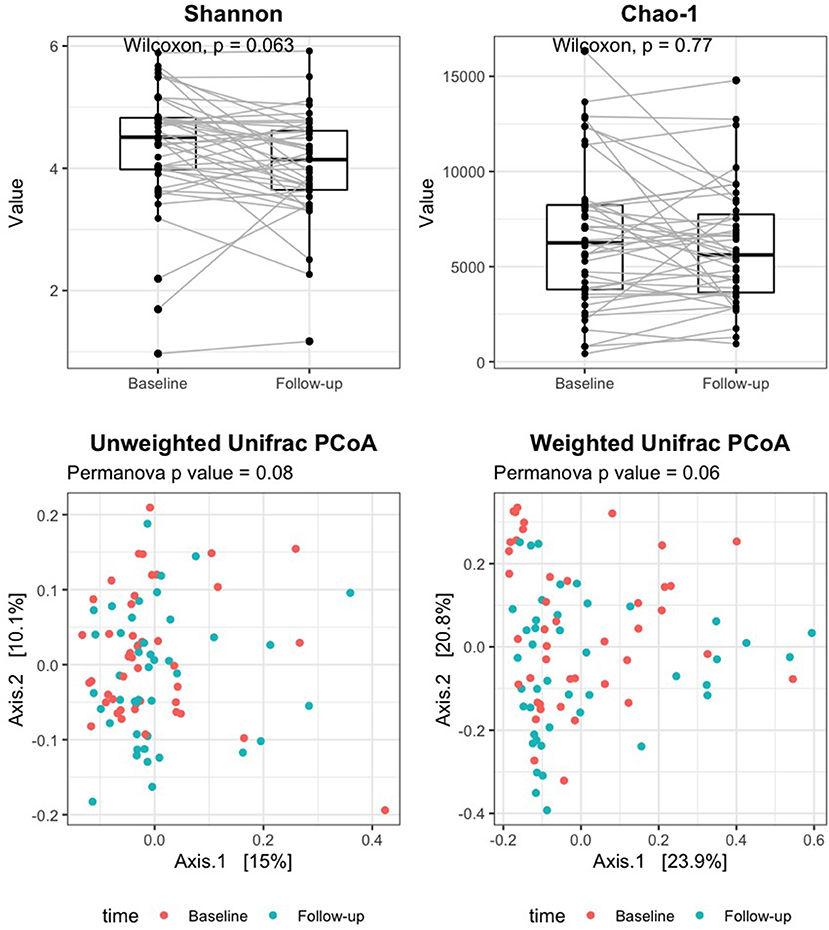
Figure 1. Change of alpha and beta diversity measures after rifaximin therapy in SUDD patients. Alpha diversity of the gut microbiota was measured on the raw data by the Shannon and Chao-1 indices (upper panels). Beta diversity was assessed by principal component analysis (PCoA) on unweighted and weighted UniFrac distance matrices (lower panels). The Wilcoxon signed-rank test and permutational multivariate analysis of variance (PERMANOVA) were applied to assess differences in alpha and beta diversity measures after rifaximin therapy, respectively.
The relative abundance analysis revealed significant changes in selected families and genera (Figure 2). In particular, after rifaximin therapy the gut microbiota was enriched in Bacteroidaceae, Citrobacter, and Coprococcus and deficient in Mogibacteriaceae, Christensenellaceae, Dehalobacteriaceae, Pasteurellaceae, Anaerotruncus, Blautia, Eggerthella lenta, Dehalobacterium, SMB53, and Haemophilus parainfluenzae (p-adj <0.05) at the family and genus levels, respectively (Figure 2). Other selected families and genera showed large log2FC (>2 or <-2), but without reaching statistical significance: Peptostreptococcaceae, EtOH8, Leuconostocaceae, Eubacteriaceae, Clostridium, Bifidobacterium, and Klebsiella (Figure 2). Subgroup analyses revealed a significant decrease of Pasteurellaceae, Clostridiaceae, Blautia, Veillonella dispar, and Haemophilus parainfluenzae in men and of Christensenellaceae in women. Parabacteroides were significantly increased in men (Supplementary Figures 2A,B). Selected variations at the phylum, family, and genus levels according to the presence of abdominal pain have been also evidenced and are reported in Supplementary Figures 2C,D.
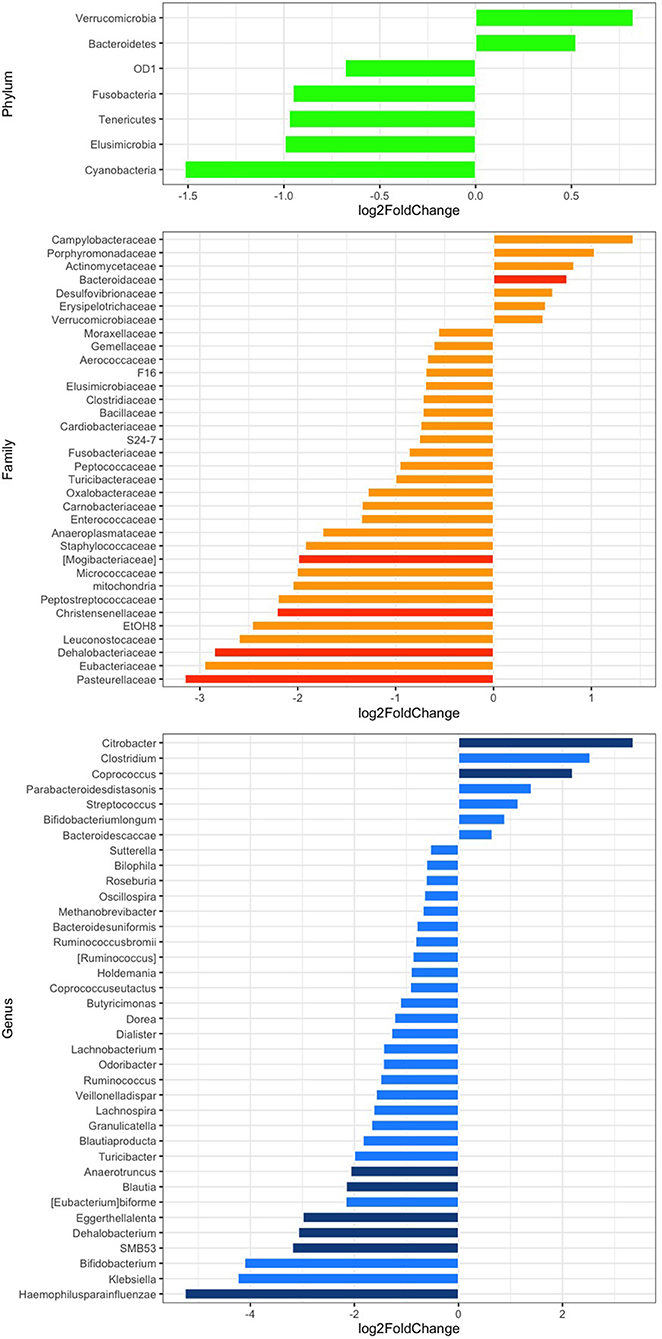
Figure 2. Differential abundance analysis of the gut microbiota composition at the phylum (green), family (orange), and genus (blue) levels after rifaximin therapy. Differential bacterial abundance is expressed as log2 fold change (log2FC); positive or negative values indicate an increase or decrease proportional to the absolute value of log2FC. Comparisons with a log2FC higher or lower than 0.5 are displayed. p < 0.05 adjusted for multiple comparisons with the Benjamini-Hochberg method (p-adj) are considered significant and represented by a darker color.
Predictive performances of the electronic multisensorial system toward alpha diversity measures of gut microbiota in pre–post rifaximin samples are reported in Table 2. E-tongue and e-nose predicted Shannon index and Shannon index variation after rifaximin with a RMSECV% between 20 and 25 (RMSECV 0.62–0.95). A similar range of RMSECV% was observed for the prediction of Chao-1 index and Chao-1 index variation after rifaximin (19–26) with RMSECV between 2,292 and 3,314. The integration of e-tongue and e-nose data only minimally improved predictive performances (Table 2). The abilities of both e-tongue and e-nose to predict the main phyla, families, and genera variation were generally suboptimal, with exception made for a few selected cases (Supplementary Table 1).
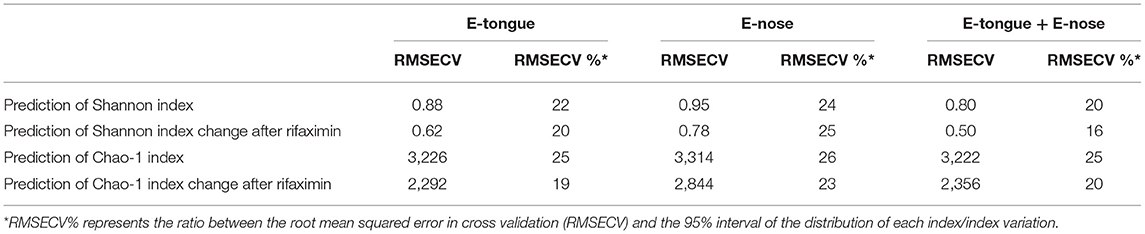
Table 2. E-tongue and e-nose prediction of alpha measure variation in patients with SUDD undergoing rifaximin therapy.
Conversely, the accuracy for the discrimination of pre/post rifaximin samples was 0.81 (0.72–0.89) for e-tongue, 0.9 (0.81–0.95) for e-nose, and 0.87 (0.78–0.93) for the integration of both techniques (Table 3). Both e-tongue and e-nose evidenced good to excellent capacities to predict clinical outcome after rifaximin therapy. Two different potential predictors were tested to this purpose, i.e., the sensors' responses obtained from the analysis of only pre-rifaximin samples, or the change in sensors' responses obtained from the analysis of both pre- and post-rifaximin samples. Indeed, the analysis of only pre-rifaximin samples could correctly predict the occurrence of clinical improvement after treatment in 36/43 subjects with e-tongue [accuracy 0.84 (0.69–0.93), sensitivity 0.91, specificity 0.75, PPV 0.81, NPV 0.88] and in 41/43 subjects with e-nose [accuracy 0.95 (0.84–0.99), sensitivity 1.00, specificity 0.90, PPV 0.92, NPV 1.00]. The integration of both techniques did not lead to an improvement of the predictive performances [accuracy 0.81 (0.67–0.92), sensitivity 0.87, specificity 0.75, PPV 0.80, NPV 0.83]. Similarly, the change in sensors' responses after rifaximin could efficiently predict clinical improvement [accuracy 0.84 (0.69–0.93) for e-tongue; accuracy 0.81 (0.67–0.92) for e-nose]. In this case, the integration of both techniques could further increase the accuracy to 0.98 (0.88–1) (sensitivity 1.00, specificity 0.95, PPV 0.96, NPV 1.00).
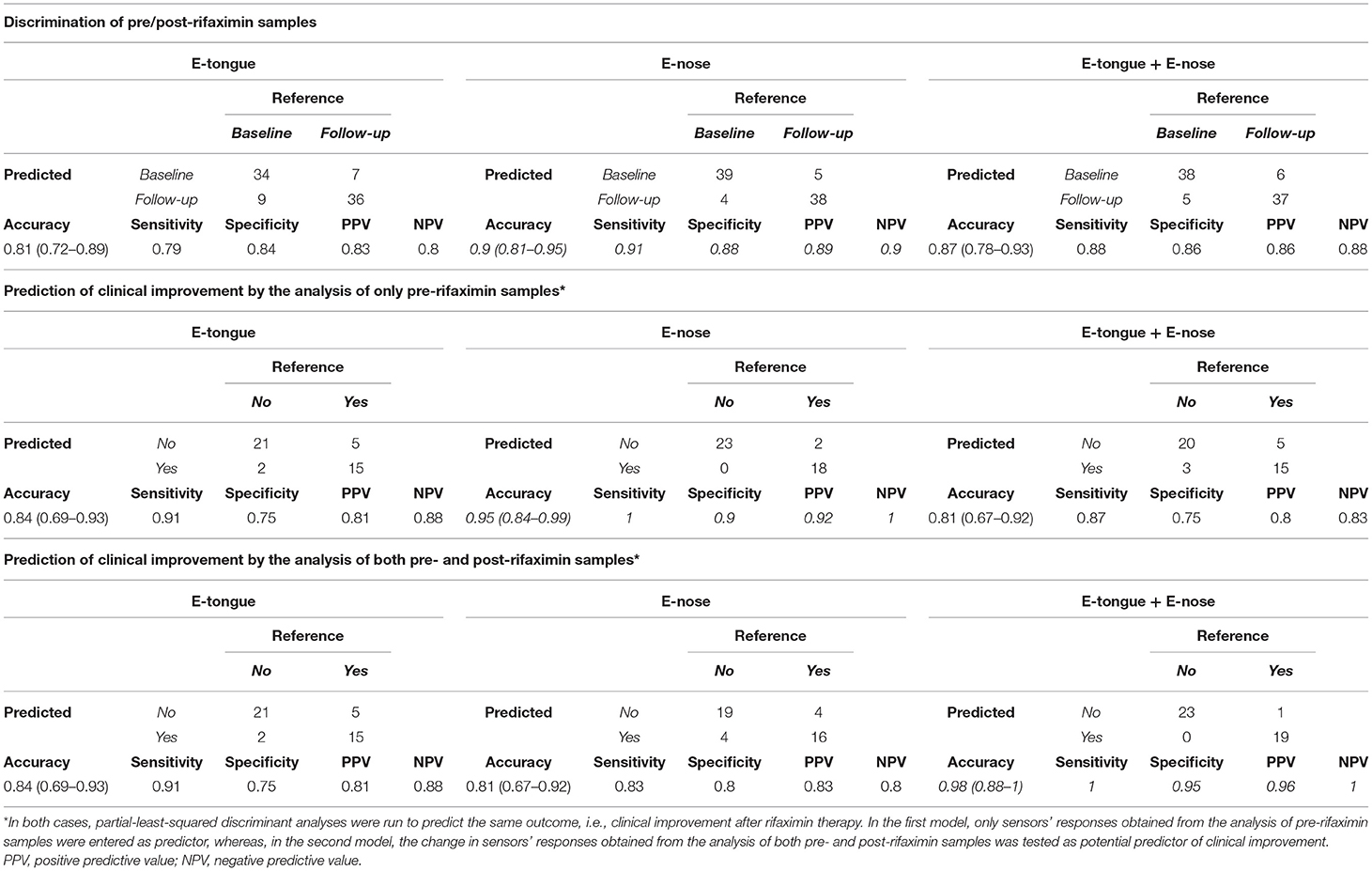
Table 3. E-tongue and e-nose discrimination of pre/post-rifaximin sample and prediction of clinical improvement.
In this study, we comprehensively explored the effects of rifaximin on fecal microbiota in SUDD patients, correlating them with clinical and laboratory data and assessing whether sensor-based methods can gain insight into microbiota status and its rifaximin-related changes. We showed that rifaximin significantly affects the relative abundance of selected bacteria and that an electronic multisensorial system (e-nose and e-tongue) has the potential for predicting and discriminating rifaximin-induced clinical response.
Several studies have demonstrated that rifaximin, a poorly absorbed oral antibiotic with an activity against anaerobic, gram-positive, and gram-negative bacteria, generates an “eubiotic” effect, also promoting the growth of beneficial bacteria such as Lactobacillus and Bifidobacteria (28, 29). Moreover, it has been reported that rifaximin decreases the metabolic activity of intestinal microbiota, increases fecal mass, and reduces bacterial overgrowth (30), and its very low absorption rates imply a low level of bacterial resistance onset (31). However, the effects of rifaximin on the intestinal microbiota are largely unknown. In fact, it seems to not affect the overall microbiome composition (32) but to induce selective depletion of a few taxa involved in the regulation of inflammation and mucosal barrier functionality (33).
In the present study, after treatment of rifaximin, an increase of Bacteroidaceae was observed in fecal microbiota of SUDD patients; even if the impact of these bacteria on SUDD pathogenesis is not known, in several studies using animal models of colitis, Bacteroidaceae or their metabolites seem to exert a protective role against inflammation (34, 35). Actually, a significant increase of Bacteroidetes spp., which usually represent a significant and stable part of the GI microbiota, plays an important metabolic role with the production of succinic acid, acetic acid, and in some cases propionic acid. Interestingly, propionic acid is mainly produced by the fermentation of indigested food by the microbiota in the colon, but can reach the blood compartment and the adipose tissue, where it reduces fatty acid levels in plasma via inhibition of lipolysis and induction of lipogenesis in adipose tissue and suppression of fatty acid production in liver (36). Moreover, after rifaximin treatment, we observed a significant reduction of Christensenellaceae, which are usually present, in a large amount, in patients with a previous history of diverticulitis, suggesting a possible role of this bacterial family in the pathogenesis of SUDD (37). Moreover, after rifaximin treatment a significant decrease of several microbial species was also observed such as Eggerthella lenta, which is an emerging pathogen responsible for bacteremia in several pathological conditions, among which is diverticular disease (38).
Overall, we did not observe a significant variation of serum inflammatory markers, and only less than half of the participants reported a clinical improvement after rifaximin. This could be likely due to the fact that the participants were not naïve to rifaximin. Furthermore, nearly two-thirds of enrolled subjects had normal bowel habits or absence of abdominal pain or only mild bloating at baseline. As such, the impact of rifaximin therapy in this specific cohort of not-naïve and pauci-symptomatic subjects could have been less evident.
Both e-nose and e-tongue were able to distinguish patients who ultimately will benefit from rifaximin from those who will not. However, the integration of the two methods did not improve the discrimination (prediction), as if the information inherent to (in) each technique was in itself complete and could not benefit from any integration. We remind that e-nose assesses the pattern of VOCs, while e-tongue assesses the bioelectric properties of a liquid, which reflects the chemical composition. It is noteworthy that all the components of the liquid contribute to shape its e-tongue pattern, whereas only volatile compounds contribute to the e-nose pattern. Accordingly, it is not surprising that adding information based on selected components (e-nose) to that derived from the whole set of liquid components (e-tongue) does not significantly improve the prognostic and discriminatory properties of the latter. The same consideration likely applies to the lack of improvement in the prediction of alpha diversity measures of gut microbiota by integrating e-nose and e-tongue. Unfortunately, the available literature does not provide any example of integration of e-tongue and e-nose, making our hypothesis worthy of validation in other settings. Instead, the integration of the two methods significantly improved the discrimination based on both baseline and post-rifaximin data. This seemingly contradictory (variant) finding is likely due to the distinctive effects of rifaximin on bacterial phyla. Indeed, the observed changes in loads of individual phyla might account for changing proportions of volatile compounds after rifaximin, making thus a “repeated information,” the one on volatile compounds collected by e-nose and, in their liquid form, by e-tongue, more representative of the biological changes underlying clinical changes. Finally, both e-nose and e-tongue were weakly correlated with changes in the vast majority of bacterial taxa, with only a few exceptions. Given, we found relevant changes in 7 phyla, 34 families, and 37 genera; the few significant correlations might be chance findings. However, it is of interest that they pertain to bacteria with plausible biological importance.
The present study has some limitations. First, the limited sample size makes the obtained results exploratory in nature, thus requiring confirmation in largest studies. Subgroup analyses did not reveal differences in diversity measures according to gender and to the presence of abdominal pain, and selected families and genera were found increased or decreased across strata. However, the reduced statistical power hampers sensible speculations on gender- and symptom-specific findings, and a wider cohort should be enrolled to highlight such subtle microbiological differences at the family and genus levels. Secondly, the studied cohort of SUDD subjects was not naïve to rifaximin therapy, and this might have smoothed the discriminative properties of multisensory systems. However, the fact that these properties were evident even in this “difficult” population testifies to the potentialities of the proposed method. Then, we did not assess intestinal permeability or inflammatory status, which might have helped our understanding of observed changes in microbioma. However, interpreting changes was out of the scope of our study, which was designed to test whether e-nose and e-tongue could assess rifaximin-induced changes in microbioma. Finally, the lack of concomitant stool metabolomic data prevents a more comprehensive understanding of the impact of the observed microbiota changes after therapy.
However, strengths also are worthy of mention. This is the first study characterizing the microbiota in SUDD subjects before and after rifaximin, and, as such, its results should be regarded with attention. In particular, the time by dose cumulative exposition to rifaximin chosen for treatment corresponds to the minimal one known to be clinically effective. Thus, the multisensorial system showed discriminative properties in a difficult experimental condition. It is likely that expanding the research toward stronger therapeutic regimens will further disclose the diagnostic potential of this method. Moreover, it tests a very innovative approach based on different sensor methods (electronic multisensorial system made up of e-nose and e-tongue) in the search for an easy and inexpensive method to surrogate microbiota genetic study. Finally, the e-tongue method can be easily standardized, given its dependence upon the operating electrical field, which guarantees for its reproducibility.
In conclusion, the present study highlights specific rifaximin-related changes in stool microbiota of SUDD patients in the selected bacterial population. Moreover, it shows that an electronic multisensorial system is able to efficiently predict rifaximin-induced clinical response. These preliminary results need to be confirmed and expanded in other SUDD and not SUDD populations. If confirmed, they might open the way to a fast and low-cost stool characterization with many potential perspectives of use not only in gastroenteric diseases.
All sequences have been archived in NCBI database: PRJNA731467 (https://www.ncbi.nlm.nih.gov/bioproject).
The studies involving human participants were reviewed and approved by Campus Bio-Medico. The patients/participants provided their written informed consent to participate in this study.
RA, BM, MC, LP, AD, MS, and GP conceived the study. BM, MG, and AA recruited participants. AD, FD, SR, LP, and RA conducted the analysis. SG, GP, MS, and AZ analyzed breath and fecal samples. AD, RA, AA, and AL wrote the article. All authors revised and approved the written manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.655474/full#supplementary-material
1. Barbara G, Scaioli E, Barbaro MR, Biagi E, Laghi L, Cremon C, et al. Gut microbiota, metabolome and immune signatures in patients with uncomplicated diverticular disease. Gut. (2017) 66:1252–61. doi: 10.1136/gutjnl-2016-312377
2. Cuomo R, Barbara G, Pace F, Annese V, Bassotti G, Binda GA, et al. Italian consensus conference for colonic diverticulosis and diverticular disease. United Euro Gastroenterol J. (2014) 2:413–42. doi: 10.1177/2050640614547068
3. Jacobs DO. Clinical practice. Diverticulitis. N Engl J Med. (2007) 357:2057–66. doi: 10.1056/NEJMcp073228
4. Hooda S, Boler BMV, Serao MCR, Brulc JM, Staeger MA, Boileau TW, et al. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutr. (2012) 142:1259–65. doi: 10.3945/jn.112.158766
5. Tap J, Furet J-P, Bensaada M, Philippe C, Roth H, Rabot S, et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol. (2015) 17:4954–64. doi: 10.1111/1462-2920.13006
6. Seth A, Yan F, Polk DB, Rao RK. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am J Physiol Gastrointest Liver Physiol. (2008) 294:G1060–9. doi: 10.1152/ajpgi.00202.2007
7. Calanni F, Renzulli C, Barbanti M, Viscomi GC. Rifaximin: beyond the traditional antibiotic activity. J Antibiot. (2014) 67:667–70. doi: 10.1038/ja.2014.106
8. Scaioli E, Colecchia A, Marasco G, Schiumerini R, Festi D. Pathophysiology and therapeutic strategies for symptomatic uncomplicated diverticular disease of the colon. Dig Dis Sci. (2016) 61:673–83. doi: 10.1007/s10620-015-3925-0
9. Kruis W, Germer C-T, Leifeld L, German Society for Gastroenterology, Digestive and Metabolic Diseases, and the German Society for General and Visceral Surgery. Diverticular disease: guidelines of the german society for gastroenterology, digestive and metabolic diseases and the german society for general and visceral surgery. Digestion. (2014) 90:190–207. doi: 10.1159/000367625
10. Bielaszewska M, Idelevich EA, Zhang W, Bauwens A, Schaumburg F, Mellmann A, et al. Effects of antibiotics on Shiga toxin 2 production and bacteriophage induction by epidemic Escherichia coli O104:H4 strain. Antimicrob Agents Chemother. (2012) 56:3277–82. doi: 10.1128/AAC.06315-11
11. Bianchi M, Festa V, Moretti A, Ciaco A, Mangone M, Tornatore V, et al. Meta-analysis: long-term therapy with rifaximin in the management of uncomplicated diverticular disease. Aliment Pharmacol Ther. (2011) 33:902–10. doi: 10.1111/j.1365-2036.2011.04606.x
12. Laghi L, Mastromarino P, Elisei W, Capobianco D, Zhu CL, Picchio M, et al. Impact of treatments on fecal microbiota and fecal metabolome in symptomatic uncomplicated diverticular disease of the colon: a pilot study. J Biol Regul Homeost Agents. (2018) 32:1421–32.
13. Santonico M, Pennazza G, Grasso S, D'Amico A, Bizzarri M. Design and test of a biosensor-based multisensorial system: a proof of concept study. Sensors. (2013) 13:16625–40. doi: 10.3390/s131216625
14. De Vincentis A, Pennazza G, Santonico M, Vespasiani-Gentilucci U, Galati G, Gallo P, et al. Breath-print analysis by e-nose for classifying and monitoring chronic liver disease: a proof-of-concept study. Sci Rep. (2016) 6:25337. doi: 10.1038/srep25337
15. De Vincentis A, Pennazza G, Santonico M, Vespasiani-Gentilucci U, Galati G, Gallo P, et al. Breath-print analysis by e-nose may refine risk stratification for adverse outcomes in cirrhotic patients. Liver Int. (2017) 37:242–50. doi: 10.1111/liv.13214
16. De Vincentis A, Vespasiani-Gentilucci U, Sabatini A, Antonelli-Incalzi R, Picardi A. Exhaled breath analysis in hepatology: state-of-the-art and perspectives. World J Gastroenterol. (2019) 25:4043–50. doi: 10.3748/wjg.v25.i30.4043
17. Scarlata S, Pennazza G, Santonico M, Santangelo S, Rossi Bartoli I, Rivera C, et al. Screening of obstructive sleep apnea syndrome by electronic-nose analysis of volatile organic compounds. Sci Rep. (2017) 7:11938. doi: 10.1038/s41598-017-12108-w
18. Finamore P, Pedone C, Lelli D, Costanzo L, Bartoli IR, De Vincentis A, et al. Analysis of volatile organic compounds: an innovative approach to heart failure characterization in older patients. J Breath Res. (2018) 12:026007. doi: 10.1088/1752-7163/aa8cd4
19. Rocco R, Incalzi RA, Pennazza G, Santonico M, Pedone C, Bartoli IR, et al. BIONOTE e-nose technology may reduce false positives in lung cancer screening programmes†. Eur J Cardiothorac Surg. (2016) 49:1112–7; discussion: 1117. doi: 10.1093/ejcts/ezv328
20. Muley M, Vespasiani-Gentilucci U, De Vincentis A, Santonico M, Pennazza G, Sanguedolce S, et al. Voltammetric analysis for distinguishing portal hypertension-related from malignancy-related ascites: a proof of concept study. PLoS ONE. (2020) 15:e0233350. doi: 10.1371/journal.pone.0233350
21. Pipita ME, Santonico M, Pennazza G, Zompanti A, Fazzina S, Cavalieri D, et al. Integration of voltammetric analysis, protein electrophoresis and pH measurement for diagnosis of pleural effusions: a non-conventional diagnostic approach. Sci Rep. (2020) 10:15222. doi: 10.1038/s41598-020-71542-5
22. Lelli D, Pedone C, Alemanno P, Bertini A, Di Gioia C, Fazzina S, et al. Voltammetric analysis for fast and inexpensive diagnosis of urinary tract infection: a diagnostic study. J Transl Med. (2018) 16:17. doi: 10.1186/s12967-018-1393-y
23. Santonico M, Frezzotti E, Antonelli Incalzi R, Pedone C, Lelli D, Zompanti A, et al. Non-invasive monitoring of lower-limb ulcers via exudate fingerprinting using BIONOTE. Sens Actuators B Chem. (2016) 232:68–74. doi: 10.1016/j.snb.2016.03.101
24. Maconi G. Diagnosis of symptomatic uncomplicated diverticular disease and the role of Rifaximin in management. Acta Biomed. (2017) 88:25–32. doi: 10.23750/abm.v88i1.6360
25. Rubenstein LZ, Harker JO, Salvà A, Guigoz Y, Vellas B. Screening for undernutrition in geriatric practice: developing the short-form mini-nutritional assessment (MNA-SF). J Gerontol A Biol Sci Med Sci. (2001) 56:M366–72. doi: 10.1093/gerona/56.6.M366
26. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. (2010) 7:335–6. doi: 10.1038/nmeth.f.303
27. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. (2010) 26:266–7. doi: 10.1093/bioinformatics/btp636
28. Lopetuso LR, Petito V, Scaldaferri F, Gasbarrini A. Gut microbiota modulation and mucosal immunity: focus on rifaximin. Mini Rev Med Chem. (2015) 16:179–85. doi: 10.2174/138955751603151126121633
29. Ponziani FR, Scaldaferri F, Petito V, Paroni Sterbini F, Pecere S, Lopetuso LR, et al. The role of antibiotics in gut microbiota modulation: the eubiotic effects of rifaximin. Dig Dis. (2016) 34:269–78. doi: 10.1159/000443361
30. Lanas A, Abad-Baroja D, Lanas-Gimeno A. Progress and challenges in the management of diverticular disease: which treatment? Therap Adv Gastroenterol. (2018) 11:1756284818789055. doi: 10.1177/1756284818789055
31. Scarpignato C, Barbara G, Lanas A, Strate LL. Management of colonic diverticular disease in the third millennium: highlights from a symposium held during the United European Gastroenterology week 2017. Therap Adv Gastroenterol. (2018) 11:1756284818771305. doi: 10.1177/1756284818771305
32. Kaji K, Takaya H, Saikawa S, Furukawa M, Sato S, Kawaratani H, et al. Rifaximin ameliorates hepatic encephalopathy and endotoxemia without affecting the gut microbiome diversity. World J Gastroenterol. (2017) 23:8355–66. doi: 10.3748/wjg.v23.i47.8355
33. Kawaguchi T, Suzuki F, Imamura M, Murashima N, Yanase M, Mine T, et al. Rifaximin-altered gut microbiota components associated with liver/neuropsychological functions in patients with hepatic encephalopathy: an exploratory data analysis of phase II/III clinical trials. Hepatol Res. (2019) 49:404–18. doi: 10.1111/hepr.13300
34. Delday M, Mulder I, Logan ET, Grant G. Bacteroides thetaiotaomicron ameliorates colon inflammation in preclinical models of Crohn's disease. Inflamm Bowel Dis. (2019) 25:85–96. doi: 10.1093/ibd/izy281
35. Brown EM, Ke X, Hitchcock D, Jeanfavre S, Avila-Pacheco J, Nakata T, et al. Bacteroides-derived sphingolipids are critical for maintaining intestinal homeostasis and symbiosis. Cell Host Microbe. (2019) 25:668–80.e7. doi: 10.1016/j.chom.2019.04.002
36. Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. (2010) 1801:1175–83. doi: 10.1016/j.bbalip.2010.07.007
37. Kvasnovsky CL, Leong LEX, Choo JM, Abell GCJ, Papagrigoriadis S, Bruce KD, et al. Clinical and symptom scores are significantly correlated with fecal microbiota features in patients with symptomatic uncomplicated diverticular disease: a pilot study. Eur J Gastroenterol Hepatol. (2018) 30:107–12. doi: 10.1097/MEG.0000000000000995
Keywords: microbiota, e-tongue, e-nose, diverticular disease, rifaximin
Citation: De Vincentis A, Santonico M, Del Chierico F, Altomare A, Marigliano B, Laudisio A, Reddel S, Grasso S, Zompanti A, Pennazza G, Putignani L, Guarino MPL, Cicala M and Antonelli Incalzi R (2021) Gut Microbiota and Related Electronic Multisensorial System Changes in Subjects With Symptomatic Uncomplicated Diverticular Disease Undergoing Rifaximin Therapy. Front. Med. 8:655474. doi: 10.3389/fmed.2021.655474
Received: 27 January 2021; Accepted: 11 June 2021;
Published: 19 July 2021.
Edited by:
Giovanni Marasco, University of Bologna, ItalyCopyright © 2021 De Vincentis, Santonico, Del Chierico, Altomare, Marigliano, Laudisio, Reddel, Grasso, Zompanti, Pennazza, Putignani, Guarino, Cicala and Antonelli Incalzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio De Vincentis, YS5kZXZpbmNlbnRpc0B1bmljYW1wdXMuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.