
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 04 June 2021
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.648005
This article is part of the Research Topic Coronavirus Disease (COVID-19): Pathophysiology, Epidemiology, Clinical Management and Public Health Response, Volume II View all 165 articles
The SARS-CoV-2 virus is causing devastating morbidity and mortality worldwide. Nanomedicine approaches have a high potential to enhance conventional diagnostics, drugs and vaccines. In fact, lipid nanoparticle/mRNA vaccines are already widely used to protect from COVID-19. In this review, we present an overview of the taxonomy, structure, variants of concern, epidemiology, pathophysiology and detection methods of SARS-CoV-2. The efforts of repurposing, tailoring, and adapting pre-existing medications to battle COVID-19 and the state of vaccine developments are presented. Next, we discuss the broad concepts and limitations of how nanomedicine could address the COVID-19 threat. Nanomaterials are particles in the nanometer scale (10–100 nm) which possess unique properties related to their size, polarity, structural and chemical composition. Nanoparticles can be composed of precious metals (copper, silver, gold), inorganic materials (graphene, silicon), proteins, carbohydrates, lipids, RNA/DNA, or conjugates, combinations and polymers of all of the aforementioned. The advanced biochemical features of these nanoscale particles allow them to directly interact with virions and irreversibly disrupt their structure, which can render a virus incapable of replicating within the host. Virus-neutralizing coats and surfaces impregnated with nanomaterials can enhance personal protective equipment, hand sanitizers and air filter systems. Nanoparticles can enhance drug-based therapies by optimizing uptake, stability, target cell-specific delivery, and magnetic properties. In fact, recent studies have highlighted the potential of nanoparticles in different aspects of the fight against SARS-CoV-2, such as enhancing biosensors and diagnostic tests, drug therapies, designing new delivery mechanisms, and optimizing vaccines. This article summarizes the ongoing research on diagnostic strategies, treatments, and vaccines for COVID-19, while emphasizing the potential of nanoparticle-based pharmaceuticals and vaccines.
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) infection causes the ongoing pandemic of Coronavirus Disease 2019 (COVID-19). In 2019, the first confirmed and documented cases of COVID-19 in China rapidly progressed to a worldwide state of emergency unparalleled since the outbreak of the Spanish Flu in 1918. The failure to control the spread of COVID-19 has highlighted the urgency of developing diagnostic and therapeutic approaches against highly contagious pathogens. A plethora of innovative treatments is being proposed which incorporate the use of traditional and futuristic methods to minimize the pathogenicity, morbidity and mortality of SARS-CoV-2. Nanotechnology is an emerging field that has branched into the world of medicine. Due to its progressive nature, nanomedicine can overcome difficulties facing conventional medicine. Most importantly, it will hopefully contribute to revolutionizing drug-based medicine in the twenty first century.
Nanomaterials have properties that, if exploited correctly, may improve treatments and vaccines, and provide alternative and safer ways to battle diseases (1). However, emergence of side effects of these nanoparticles, such as unwanted interactions with tissues or increased inflammation, could put a temporary hold on the utilization of nanotechnology (2). The COVID-19 crisis sets the stage to evolve the concepts of nanotechnology into reality. As its potential is revealed, it can offer innovative ways of protecting healthy and infected individuals, detecting SARS-CoV-2, and helping to end the pandemic.
In this review, we present an overview of SARS-CoV-2 pathophysiology, diagnostics, treatment and vaccines followed by discussing the current and future applications of nanomedicine aiming to mitigate the COVID-19 pandemic. The nanoparticle approaches presented here will help to win the fight against SARS-CoV-2 and other pathogens.
In the first week of January 2020, the Chinese Center for Disease Control and Prevention (CCDC) disclosed that 27 cases of pneumonia admitted during late December of 2019, were attributed to Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), later named COVID-19 by the World Health Organization (WHO) (3). The patients had visited one of the “wet markets” in Wuhan city, located in China's Hubei province, which are known for their considerable variety of wild animals for sale (4). Recent genomic analysis has revealed that the SARS-CoV-2 genome is 96% identical to a known bat coronavirus (BatCoV RaTG13) from Rhinolophus affinis, a species found in Yunnan province (5, 6). The WHO declared the viral outbreak a public health emergency of global proportions at the end of January, when there were approximately 10,000 diagnosed cases around the globe (7). It was estimated that SARS-CoV-2 has a Case Fatality Rate (CFR) of 2–4% (8, 9) with substantial variation between countries, as well as a higher basic reproduction number (median R0 range: 3.5–4.7) compared to other coronaviruses or influenza (Figure 1) (10–12). As of April 2021, more than 140 million people across the globe have contracted COVID-19, and more than 3 million of those cases resulted in fatalities.
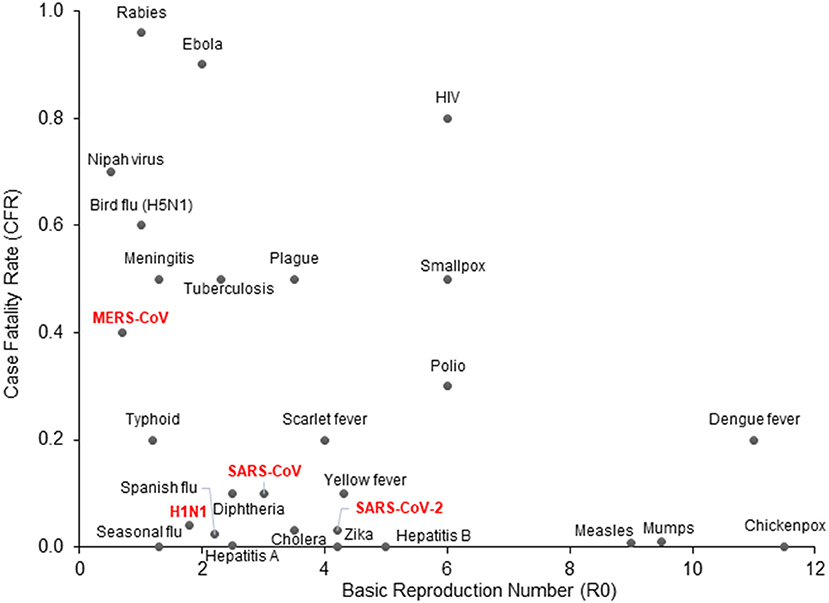
Figure 1. COVID-19 epidemiologic characteristics compared to other prevalent infections. The Case Fatality Rate (CFR) for COVID-19 is estimated around 2–4% with some variation and a recent decline due to optimized supportive care. The Basic Reproduction Number (R0) shown on the x-axis is also an estimate from epidemiological data. SARS-CoV-2 is more contagious than SARS-CoV and MERS-CoV, which may be attributed to longer incubation periods and asymptomatic carriers.
Coronaviruses (CoVs) belong to the Cornidovirineae suborder under the Coronaviridae family. CoVs are a predominant group of viruses, but of the 46 known CoVs only 7 have been confirmed to infect humans (13). Human coronavirus 229E (HCoV-229E) and human coronavirus NL63 (HCoV-NL63) are members of the genus Alphacoronavirus while human coronavirus OC43 (HCoV-OC43) and human coronavirus HKU1 (HCoV-HKU1) belong to the genus Betacoronavirus. These viruses are linked to mild upper respiratory tract diseases and can be attributed to 15–30% of common cold cases with regional/global and seasonal patterns (Table 1) (14, 22, 23). In contrast, SARS-CoV (sometimes referred to as SARS-CoV-1), MERS-CoV, and SARS-CoV-2 of the Betacoronavirus genus, are associated with severe disease pathophysiology, including respiratory disease, multi-organ failure, sepsis and death (14–16, 20, 21, 24–26).
SARS-CoV-2 is encoded by positive single-stranded RNA (ssRNA) bound to the nucleocapsid phosphoprotein (N). It is enclosed in a bilipid envelope surrounded by transmembrane proteins, such as the small envelope glycoprotein (E), membrane glycoprotein (M), and type-I trimeric spike protein (S) (Figure 2) (27–33). SARS-CoV-2 spike protein binds the Angiotensin Converting Enzyme 2 (ACE2) receptor located on type I and II pneumocytes and other epithelial and non-epithelial tissues, to enter host cells (34). More specifically, the spike protein monomers depend on host proteases for entry, such as the transmembrane serine protease 2 (TMPRSS2). TMPRSS2 can hydrolyze peptide bonds between the S1 and S2 subunits (35, 36). This process primes the spike protein and allows the S1 subunit, which contains the receptor binding domain (RBD) held together by several disulfide bonds, to bind with the N-terminal helix of ACE2 (17, 18, 37, 38). After internalization into the host cell, SARS-CoV-2 undergoes an uncoating process and initial viral transcription which requires supportive proteins and enzymes, including some rarely found in other RNA viruses such as (3'-to-5' exoribonuclease, 2'-O-ribose methyltransferase, ADP ribose 1'-phosphatase) (27). The viral transcripts can amass to 15–30% of the transcriptome in infected host cells (39). The translation of viral proteins occurs in the cytoplasm and viral proteins control the replication process. Viral proteins are inserted into the Golgi apparatus and are transported to the plasma membrane, where virions are released and begin infecting neighboring cells (19).
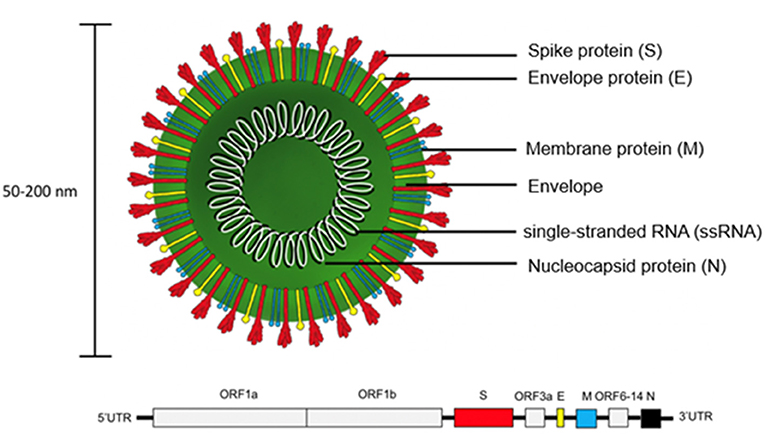
Figure 2. Structure of SARS-CoV-2 virion and genome. The trimeric spike protein (S) is required for docking to the hACE2 receptor. S protein is targeted by antibody-based therapies and is used as the immunogen for vaccine candidates. The single-stranded RNA (ssRNA) genome is bound to the Nucleocapsid phosphoprotein (N) which facilitates transcription after virus entry into host cells. The large viral genome (29.9 kb) is arranged as open reading frames (ORF) encoding for about 27 non-structural proteins (e.g., replicase, protease) and the four structural proteins (S, E, M, N).
The genome sequences by next-generation sequencing (NGS) indicated that SARS-CoV-2 is more closely related to bat coronaviruses (BatCoV RaTG13 [96%], SL-CoVZXC21 [88%], SL-CoVZC45 [88%] (5), than to SARS-CoV (79% similarity) and MERS-CoV (50% similarity) (27, 40). SARS-CoV-2 spike protein has ~75% sequence similarity to the amino acid sequence of SARS-CoV spike protein (41), including a mutation in the C-terminal RBD for enhanced binding to ACE2 (5, 42, 43). SARS-CoV-2 quasispecies have been described, although the mutation rate is slower than for influenza virus (44). Virus variants of concern with higher infectivity and pathogenicity and a risk for resistance against the first generation of vaccines have emerged (45). A variant encoding a D614G mutation (conversion of aspartic acid to glycine at position 614) in the spike protein, located in the S1 domain has become most prevalent (46). This new D614G variant is associated with increased replication and transmission when compared to other less common isolates, such as the USA-WA1/2020 variant, which contains an aspartic acid residue at this position (47, 48). There are several sub-variants, such as the D16 INMI1 isolated in Italy, the G614 PV08449/2020 isolated in New York and the G614 BavPat1/2020 isolated in Germany (49, 50). Three variants of concern each with 17 amino acid changes and all featuring a N501Y spike protein mutation have emerged in the end of 2020 (51): A VUI-202012/01 (B.1.1.7) variant was first detected in the United Kingdom (52). The 501Y.V2 (B.1.351) variant was first discovered in South Africa and the P.1 variant was initially reported in Brazil and Japan (53). The P.1 and B.1.351 variants contain an E484K spike mutation.
The clinical presentation of COVID-19 can be grouped in three categories based on disease severity and progression: the asymptomatic phase/stage, the mild symptomatic stage, and the severe respiratory infection stage (Table 2). Most individuals do not pass through all stages and asymptomatic or mild symptoms are most common (61). It is estimated that 15–30% of cases are asymptomatic, which may contribute to herd immunity (62). Individuals in the first category, also known as “stealth carriers,” do not present any symptoms and molecular testing can even be negative. If COVID-19 progresses to stage 2, mild infection symptoms are observed such as fever and coughing, and the patient typically tests positive in RT-PCR assays (59). It can take 1–3 weeks after the first symptoms for the production of antibodies against SARS-CoV-2. The third and most severe phase may present as a flu-like stage, a respiratory inflammation stage including pneumonia, acute respiratory distress syndrome (ARDS), pulmonary edema, and sometimes the complications of coagulopathy and fibrotic changes due to lung remodeling (54). This sequence of events can result in dramatically compromised gas exchange and respiratory failure (Table 2) (55, 63). Severe COVID-19 (stage 3) appears to be associated with a higher production of neutralizing antibodies. Additional symptoms include gastrointestinal dysfunction and secondary infections, as well as harmful tissue destruction due to pro-inflammatory leukocytes such as macrophages and granulocytes (56, 58–60).
Emerging evidence suggests that a previous infection with one of the four endemic coronaviruses that cause “common cold” (HCoV-OC43, -HKU1, -NL63, and−229E) is associated with mitigated SARS-CoV-2 illness, which may be explained by a better pre-existing immune response and heterotypic immunity to homologous viruses (64). In addition to neutralizing, cross-reactive antibodies, memory CD4+ T cells have been hypothesized to reduce lung viral burden, accelerate antibody production and to enable heterotypic immunity (65–67). On the other hand, anti-SARS-CoV-2 antibodies with cross-reactivity for host proteins may contribute to pathologies such as Kawasaki-like disease and Guillain-Barré syndrome (68–70).
Many factors can influence the severity and outcome of COVID-19 infection such as age, gender, pre-existing health conditions and comorbidities (71). In general, the rates for apparent infection, hospitalization and death are higher for individuals aged 65 and above. Men have a higher risk for severe disease, an observation that has not been fully explained (72, 73). One hypothesis is centered around the higher tobacco use in men (4:1) and that long-term smokers develop cardiovascular and respiratory conditions which correlate with rapid and severe progression of COVID-19 (57). Increased vulnerability to SARS-CoV-2 is also correlated with a variety of health factors such as severe obesity, type II diabetes mellitus, serious cardiovascular conditions and immunocompromised states such as autoimmune disease or recent chemotherapy (74, 75). Last but not least, susceptibility to COVID-19 has been linked to certain genetic traits including polymorphisms for IFNAR2, TYK2, TLR7, OAS1, DPP9, and CCR2, and the major histocompatibility complex loci (HLA) (76) which also provide susceptibility to other infections such as Influenza, Hepatitis B, and leprosy (77).
The WHO and the Center for Disease Control and Prevention (CDC) have established detailed protocols regarding the use, containment, culturing, and testing for SARS-CoV-2. The CDC has classified any research work with infective SARS-CoV-2 as Biosafety Level 3 (BSL-3) category, while protocols with inactivated forms of SARS-CoV-2 or pseudotyped viruses can be performed in a BSL-2 laboratory (78).
It is widely accepted that suitable cell lines to propagate SARS-CoV-2 must express sufficient numbers of ACE2 and TMPRSS2 on their surface. The Vero cell line is derived from kidney cells of the African green monkey and sublineages such as the Vero E6 and Vero CCL81 cell lines produce even higher SARS-CoV-2 titers. Other cell types such Calu-3 (a human lung cancer line), Caco-2 (a human colorectal adenocarcinoma line), HEK 293T (derived human embryonic kidney line) and Huh7 (a human hepatocellular carcinoma line) can also be used for infection studies, but are not suitable for generating high titer virus stocks (79). Genetically modified cell lines, such as an ACE2 overexpressing HEK 293T line and Air-Liquid Interface (ALI) epithelial cell models exist (80). Human induced Pluripotent Stem Cell (iPSC)-derived alveolar type 2 cells (iAT2) are susceptible to SARS-CoV-2 infection in ALI culture. SARS-CoV-2 infection of the iAT2 cells hijacks the transcriptomic machinery, deprograms host cell differentiation, while inducing the NFkB pathway and interferon-dependent host defense programs (39, 81).
SARS-CoV-2 infection can be investigated in animal models. Mice (Mus musculus) were genetically engineered to over-express human ACE2 because SARS-CoV-2 spike protein does not bind well to murine ACE2. In addition, non-modified Syrian hamsters (Mesocricetus auratus), ferrets (Mustela putorius furo), non-human primates (Cynomolgus macaques and Rhesus Macaques) and other mammalian species can be infected to study the pathobiology of COVID-19 (79, 82–86).
Molecular diagnostic test for SARS-CoV-2 were rapidly developed (Table 3). The first Reverse Transcription-Polymerase Chain Reaction (RT-PCR) assay was released by the WHO and targeted three regions of the SARS-CoV-2 genome: the N gene, the E gene, and a highly conserved gene for RNA-dependent RNA polymerase (RdRp) (91, 92). Meanwhile, multiple alternative RT-PCR primer sets are available, while additional methods such as SARS-CoV-2 nucleoprotein antigen tests and antibody detection kits have also been developed (90, 93–95). The SARS-CoV-2 specific antibodies can be detected by rapid diagnostic tests (90). CRISPR-Cas12 based assays, such as DETECTR, identify the presence of SARS-CoV-2 RNA (89, 95). Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) is a faster (30–40 min) and cheaper alternative for RT-PCR with the advantage of point-of-risk testing (87, 88). Another method is termed Specific High-sensitivity Enzymatic Reporter Unlocking (SHERLOCK) and utilizes Cas13a for the accurate and highly sensitive detection of viral RNA copies (96).
Chloroquine is a malaria drug, which passively diffuses into acidic lysosomes, endosomes, and Golgi vesicles. While initial reports and studies were promising for chloroquine/hydroxychloroquine in COVID-19 patients, these findings were not confirmed and the NIH discontinued clinical trials investigating the efficiency of chloroquine (3, 97). In 2020, the FDA had authorized the administration of chloroquine to certain COVID-19 patients, but shortly thereafter, the agency terminated its use due to the high ambiguity regarding its efficiency and side effects (Table 4). In fact, hydroxychloroquine did not improve 28 day mortality in hospitalized COVID-19 patients (1,561 patients, 27% non-survivors) as compared to standard care (3,155 patients, 25% non-survivors) (98). A meta-analysis of studies on the efficiency of chloroquine for treating COVID-19 has shown that there was no significant difference in patient outcome and that the side effects posed a larger threat (99, 107).
Azithromycin is classified as a broad-spectrum macrolide antibiotic. Azithromycin also amplifies antiviral immune recognition and interferon pathways in airway epithelial cells (108). A single-center study had suggested that a combination of hydroxychloroquine and azithromycin significantly reduced viral loads and time to a negative PCR test after SARS-CoV-2 infection (100). However, subsequent trials failed to reproduce these results, with no significant difference in viral burden as well as continuing positive PCR results (109). The latter study suggests that the antiviral properties of both medications have been overestimated and their side effect profiles might adversly manifest in COVID-19.
Remdesivir was originally designed to target hepatitis C virus, and later studied for effectiveness against Ebola virus. It is classified as an anti-viral adenosine-tri-phosphate analog, which is incorporated into the forming viral RNA chain by the RNA-dependent polymerase and disrupts viral replication (98). Remdesivir showed some efficacy in inhibiting infection of mammalian cells by human coronaviruses (110). In clinical trials, remdesivir tended to shorten the recovery time for adult patients and reduced symptoms of upper respiratory infection (101).
Remdesivir was one of the first drugs granted emergency use authorization, and it is now approved by the FDA for use in adults children (>12 years) for the treatment of COVID-19 requiring hospitalization (102). However, remdesivir only achieves modest benefits for subgroups of hospitalized COVID-19 patients.
Dexamethasone is a potent anti-inflammatory corticosteroid that binds to the glucocorticoid receptor and depending on the dosage either reduces the expression of certain pro-inflammatory genes or boosts the transcription of a subset of anti-inflammatory regulators (103). A meta-analysis of n = 1,703 severely ill COVID-19 patients found glucocorticoids to reduce 28 day mortality (32% vs. 40%) without an increased risk for severe adverse events (111). The RECOVERY trial (n = 2,104) showed that dexamethasone decreased COVID-19 mortality (29% vs. 41%) in patients on mechanical ventilation or receiving oxygen without mechanical ventilation (23 vs. 26%) (104). No difference in survival was found in patients who did not require respiratory support. Hence, dexamethasone is recommended for severe cases of SARS-CoV-2 infection and its best role could be as part of a combination therapy (22).
Tocilizumab is a humanized monoclonal antibody against the Interleukin-6 (IL-6) receptor used in autoimmune diseases and inflammatory disorders (112). Clinical trials suggest that Tocilizumab can reduce hyperinflammation during severe COVID-19. More specifically, one trial showed that Tocilizumab reduced mortality of COVID-19 when compared to standard care while increasing the risk of secondary infections (105). Tocilizumab relieves clinical symptoms, reduces the requirement for supplementary oxygen, reverses lymphopenia and decreases C-Reactive Protein (CRP) levels (106). A direct positive correlation was found between CRP levels, lung lesions and higher severity of COVID-19 (113). While not all studies have shown significant differences in disease severity or survival of infected patients treated with Tocilizumab compared to a placebo (114), a meta-analysis of n = 2,120 patients supported a reduction of mortality in severe cases of COVID-19 (115). In a more recent analysis of n = 4,116 adults, Tocilizumab reduced COVID-19-associated mortality (29% vs. 33%) and was more effective in combination with glucocorticoids (54% vs. 47%) (116). Patients receiving Tocilizumab were less likely to require mechanical ventilation and showed improved clinical outcomes (116). Sarilumab is another blocking anti-IL-6R antibody which is studied for COVID-19.
Neutralizing antibodies and passive immunization are a feasible approach to mitigate SARS-CoV-2 infection (117, 118). Passive immunization could be especially helpful for immunocompromised individuals at risk for severe clinical manifestations such as respiratory failure (119). Prophylaxis against infectious agents using purified polyclonal immunoglobulin (Ig), also known as polyvalent immunoglobulin, is not a new idea (120). Ranging from highly specific to very broad, neutralizing monoclonal antibodies have been designed against a variety of viral agents such as MERS-CoV (121, 122).
Recent work on the antibody repertoire produced by infected humanized mice and recovered patients has generated a large bank of antibodies that can be used against COVID-19. Anti-SARS-CoV-2 spike antibodies were generated by immunizing mice with a DNA plasmid encoding the RBD protein. In addition, B-cells were isolated from the peripheral blood of recovered patients (117).
The antibodies generated from both studies were reported to be highly similar in function and efficacy against many spike variants. However, four of them, utilized individually or in cocktails, showed promising results against newer strains that had originated from human populations (117). A cocktail therapy was proposed to limit viral resistance to therapy by using antibodies that target two distinct, non-overlapping regions of the RBD (123). Nevertheless, the antibodies were not effective in neutralizing SARS-CoV-2 when new spike mutations arose from in vitro passaging or when combinations of antibodies that target overlapping regions were administered (123). Other neutralizing antibodies (LY-CoV555 and LY-CoV016) have shown promising results in the BLAZE-2 clinical trial. Bamlanivimab (LY-CoV555) alone reduced the risk of symptomatic COVID-19 by 80% (124), while in a separate study the combination of Bamlanivimab (LY-CoV555) with Etesevimab (LY-CoV016) was found to decrease hospitalization and death from COVID-19 by 70% (124–127). Furthermore, Regeneron's REGN-COV2 neutralizing antibody cocktail (Casirivimab and Imdevimab) was effective in reducing the viral load in patients with delayed immune responses or with high initial virus titers (128).
Vaccines are the best approach for prevention of infection. There are five types of vaccines under development (Figure 3) (129):
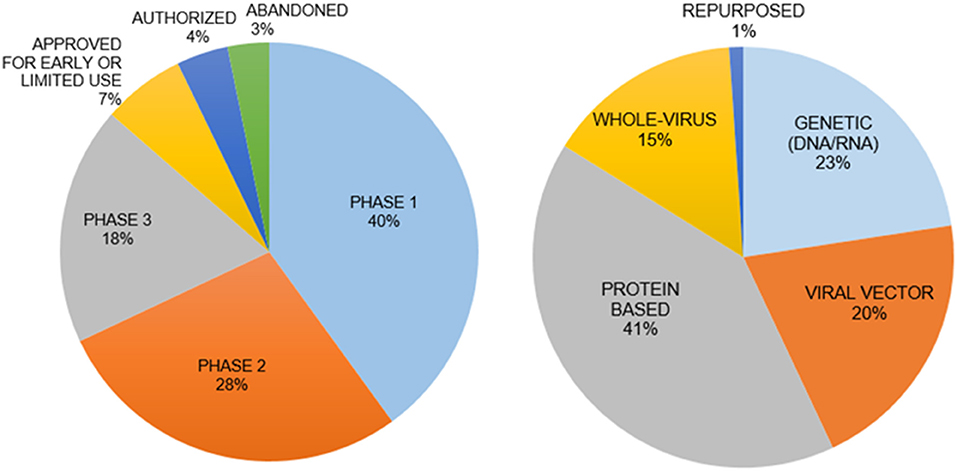
Figure 3. Vaccine types and clinical phases of their development. The genetic vaccines immunize with mRNA for SARS-CoV-2 and protein-based vaccines immunize with spike protein to induce immunity. The interim analysis of phase 3 clinical trials for both types of vaccines have been disclosed with promising results in November 2020. The mRNA vaccines are a new principle with little information on how long the induced immunity will last.
(i) Genetic Vaccines use SARS-CoV-2 specific DNA/RNA sequences to stimulate an immune response. (ii) Viral Vector Vaccines employ alternative viruses as “carriers” for SARS-CoV-2 genes. (iii) Whole Virus Vaccines present an inactivated form of the virus to the immune system. (iv) Protein-based Vaccines incorporate selected virus proteins such as the spike protein. (v) Repurposing the Bacillus Calmette-Guérin vaccine to stimulate the immune system. Recent discussions raised questions regarding the safety, long-term side effects, and social implications of a vaccine developed in a short period time without sufficient pre-clinical testing and adequate clinical trials. A major concern for a vaccine is the syndrome of acquired cellular immunopathology, a condition observed when the delivery platform for the viral proteins or genes leads to a violent pro-inflammatory response from T-cells. This results in the migration of white blood cells into target tissues, further deteriorating the health of a patient. Another concern is Antibody-Dependent Enhancement (ADE), when non-specific antibodies generated by the vaccine allow for enhanced viral internalization, thus potentially worsening the infection and pathophysiology of COVID-19 (130). A vaccine could also have low efficacy in terms of long-lasting protection from infection because of insufficient neutralizing antibodies, weak memory T cell responses or new SARS-CoV-2 variants.
In the pre-COVID-19 era, vaccine development lasted on average about a decade and required extensive funding, scientific diversity, and countless volunteers. The federal Center for Biologics Evaluation and Research is a branch of the FDA responsible for evaluating the safety and efficacy of novel medication and vaccines. The FDA and CDC have established a strict set of clinical trials (phase 1–3), through which the safety and efficacy are investigated. In the last year, there has been a race to develop the first vaccine to prevent COVID-19.
There are more than 110 potential vaccine candidates, with over 80 in human trials and almost another 80 vaccine candidates in preclinical testing. There are currently seven approved vaccines, developed mainly in the US, Russia, China, India, UK, Germany and Belgium. In some cases, the development of vaccine candidates came to a halt, such as for Merck, Imperial College London, Themis, Institut Pasteur and IAVI (131–136) (Figure 3).
The clinical trial of the mRNA-based vaccine (BNT162b2) from Pfizer/BioNTech enrolling 43,000 participants showed a reduced risk for SARS-CoV-2 infection by over 90%. This vaccine received FDA emergency approval in the US, while it has been fully approved in other countries (137). Further studies have shown that the BNT162b2 vaccine has 95% efficacy in preventing a COVID-19 infection 7 days after the second dose (137, 138). Of note, the nanoparticles that deliver the mRNA contain polyethylene glycol (PEG), a compound that has been linked to unwanted severe allergy-like symptoms. Similar concerns have been raised with the nanoparticles used in the mRNA vaccine from Moderna. It consists of mRNA-1273 encapsulated in lipid nanoparticles (139). The mRNA-1273 showed similar efficiency (94%) to the Pfizer vaccine and was granted FDA emergency use authorization. The protection by a protein-based adenovirus vector vaccine from AstraZeneca was around 70% with some uncertainties about optimal dosing and recent concerns about a risk for thrombotic complications. Regardless of these promising results, the duration of long-lasting immunity induced by these vaccines has yet to be determined. The Johnson & Johnson vaccine also uses an adenovirus vector to express SARS-CoV-2 spike protein in the host cells to induce immunity. This process yields SARS-CoV-2-specific antibodies in ~90% of individuals after the first dose (140). The Johnson & Johnson vaccine has been associate with a very rare risk for cerebral venous sinus thrombosis. Mild to moderate local (e.g., pain and swelling at injection site) and systemic (e.g., fever, chills) side effects are very common for the current COVID-19 vaccines. Seropositive participants develop higher antibody titers and experience higher rates of systemic side effects (141). It is expected that SARS-CoV-2 will eventually transition from a pandemic to an endemic disease, a change that is associated with the distribution of infected individuals. Endemic dynamics are characterized by a shift of primary infections to younger ages in the population, which for COVID-19 usually causes only mild disease or asymptomatic infection. The shift to mild endemic disease depends on the rate of virus transmission and may be accelerated by vaccination (142).
Nanomedicine approaches may provide new solutions in the fight against COVID-19. The hope is that nanotechnology can improve the effectiveness and specificity of drugs and vaccines. The nanomedical field utilizes nanomaterials: particles in the nanometer scale that possess unique chemo-physiological properties. Two key characteristics of nanoparticles are their size and polarity. Their size, ranging from 10 to 100 nm, allows them to easily interact with a biological target of similar size and pass through several types of membranes, such as the lung-blood vessel junction and the blood-brain barrier (143). In addition, the polarity of nanoparticles can be modified to facilitate a specific purpose such as binding other drugs, increasing the surface stability, or reducing aggregation and precipitation (144, 145). Specialized nanoparticles with a magnetic nature can be guided through the body via a system of external magnets and forced to increase their temperature by exposing them to an oscillating magnetic field, a technique currently used in oncology for tumor suppression (146–148). Moreover, these particles can be both organic and inorganic, used individually or aggregated, and combined with other medication or other nanoparticles. Due to the unique features of nanomaterials, widespread applications in both the prevention and treatment of SARS-CoV-2 are feasible. Nanotechnology could be applied for personal protective equipment, gene silencing, creating biosensors, developing pharmacologically active compounds and nano-vaccines, and for directly destroying SARS-CoV-2 particles (149–151).
Biosynthesis of nanoparticles by microorganisms has recently emerged as an alternative to conventional chemical and physical synthesis. Biosynthetic nanoparticles can have similar morphology and properties to their conventional counterparts (152, 153). There are several benefits of large-scale synthesis of microbe-derived nanoparticles such as avoiding hazardous chemicals, expensive reagents or toxic materials for stabilization and synthesis. Nanoparticles can bioconjugate, genetically engineer, infuse, mineralize or even assist in self-assembly of viral and bacterial particles. These techniques could be used as tools for vaccine design and production (152, 153).
There are several designs for nanoparticle-based peptide vaccines. Nanoparticles can be used to construct a multiple antigen-presenting platform. Self-assembling lipo-peptides, consisting of a lipid chain bound to an antigen, can form micelles with enhanced epitope presentation ability (154). Another safe and effective method of antigen delivery to antigen-presenting cells is encapsulation or conjugation of antigens with nanoparticles in order to preserve their structure and protect them from degradation (155). Nanoparticles designed to either deliver antigens or act as adjuvants can be administered intranasally to induce immunity against lower respiratory tract virus infections, such as influenza, RSV and adenovirus (155). Bacteriophage-derived nanoparticles from Escherichia virus Q-beta were incorporated into a H1N1 vaccine of high immunogenicity and low safety concerns in a phase 1 clinical trial (156).
Adenovirus (class I-dsDNA virus), adeno-associated virus (AAV, class II-ssDNA virus), human papilloma virus (HPV) or even human immunodeficiency virus (HIV) can be modified into carriers for targeted gene/protein delivery (153, 157–161). Bacteria can be engineered for nanoparticle biosynthesis such as Bacillus cereus and Bacillus subtilis for silver nanoparticles (162), Pseudomonas aeruginosa and Pseudomonas fluorescens for gold nanoparticles (163), Shewanella algae for platinum nanoparticles (164), and Pseudomonas aeruginosa for Lanthanum nanoparticles (165).
Nanotechnology is a fast growing industry. The current $60 billion market is expected to double to $120 billion in 5 years. The main market prospects involve the utilization of nanoparticles for medicine, food, agriculture, conductors and computers. In medicine, nanoparticles are used and developed for applications inside of the body (e.g., drug delivery, repair of tissues) and for external purposes.
To decrease the spread of SARS-CoV-2, nanoparticles with a potential to inactivate the virus can enhance physical barriers, sterilize commonly contacted surfaces or air filters, and be incorporated into hand sanitizers and disinfectants (166–169) (Figure 4). Personal Protective Equipment (PPE) such as masks and gloves could be upgraded with nanoparticles that have antimicrobial or antiviral capabilities. Iron-oxide nanoparticles (IO-NPs) and Silver nanoparticles (Ag-NPs) have been shown to neutralize various strains of Influenza and Coronaviruses by physically binding to the SARS-CoV-2 virion and preventing internalization into host cells (166, 167, 170, 171). Moreover, Copper Oxide nanoparticles (CO-NPs) possess antimicrobial capabilities against a plethora of respiratory tract pathogens such as Staphylococcus aureus and Pseudomonas aeruginosa (172). Antimicrobial nanoparticles use chemical and biological mechanisms to eliminate microbes such as cell membrane disruption, DNA and protein damage, gene silencing, heavy metal ion toxicity, Reactive Oxygen Species (ROS) formation, and prevention of biofilm formation (173).
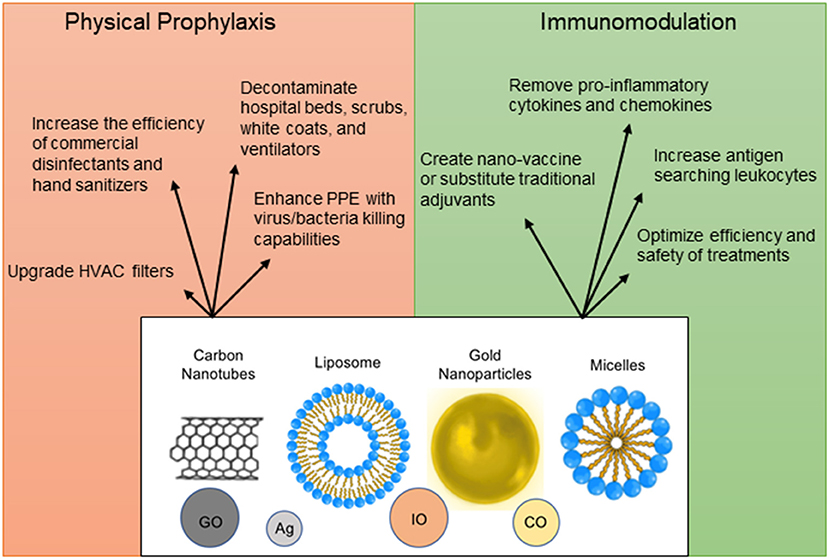
Figure 4. Nanoparticle applications as prophylactic and therapeutic measures. Nanoparticles can be used in a plethora of ways for protection against infection, for immunomodulation, vaccine design, and optimization of detection methods. The structural design of nanomaterials is diverse and includes engineering of carbon nanotubes [Graphene Oxide (GO)], liposomes, micelles, precious metals [Silver (Ag), Copper Oxide (CO), and Iron Oxide (IO)].
One of the strongest arguments for the use of nanoparticles in drug enhancement is that modern drug delivery can lack target specificity due to a poor cellular uptake, insufficient stability under physiological conditions, non-target effects, and excessive immunogenicity (174). A novel approach to avoid such problems uses short interactive RNA molecules bound to nanoparticles, which can interact with a biomarker on the desired cell population, thus localizing the drug's effects to avoid unnecessary contact with other cells and reduce overall toxicity (175, 176). Nanoparticles coated with specific antibodies against a cellular receptor such as human ACE2 or against SARS-CoV-2 spike protein comprise an elegant delivery system for any drug that requires cell specificity and may help reduce the dose of medication and off-target side effects (174). Many nanoparticle types can be used, such as polymers, dendrimers and quantum dots. Nanobots, microscopic robots that can carry out localized drug delivery, could be controlled by a user and might advance drug delivery even further in future (176).
The properties of the molecules mentioned above could also be engineered to reduce the chances of secondary infections that are associated with COVID-19 pathophysiology. PPE, patient gowns, scrubs, white coats and commonly contacted surfaces could be coated with a mixture of nanoparticles to protect healthy and infected individuals. Cotton fabrics can be enriched with zinc oxide nanoflowers to trap and denature SARS-CoV-2 spike protein (177). Additionally, enhancing conventional hand sanitizers and upgrading air filter systems with antimicrobial nanoparticles could be useful for disinfection and containment of SARS-CoV-2 spread. FDA-approved iron-oxide nanoparticles (IO-NPs) were recently found to bind to the envelope and spike protein subunits of SARS-CoV-2 and alter their conformation, thus inactivating the virus (178). Nanoformulations can help to reduce the needed quantities of precious elements such as gold, silver and copper.
The SpyCatcher/SpyTag technology allows irreversible conjugation of a recombinant protein by adding a sequence of the SpyTag peptide (13 amino acids) to its DNA sequence. The SpyTag spontaneously reacts with the SpyCatcher protein and allows for oligomerization (179). This system was employed to generate mosaic nanoparticles that display multivalent antigens of SARS-CoV-2 spike RBD along with RBDs from different animal betacoronaviruses to enhance B cell responses and elicit high-titers of cross-reactive, neutralizing antibodies (180).
Early and rapid detection is key for lowering the basic reproduction number of infected individuals. Nanoparticles can be engineered as biosensors for the detection of biomarkers, including nucleic acids (DNA, RNA), specific antigens (proteins, enzymes), or antibodies in order to rapidly and accurately detect SARS-CoV-2 (143, 149, 181, 182). Recent advancements in nanotechnology have allowed for the release of a SARS-CoV-2 detection platform that uses graphene conjugated to an anti-spike antibody. This novel kit requires no sample pretreatment or labeling and is impressively effective in detecting SARS-CoV-2 at very low concentrations (183). Alternative detection methods have been designed such as dual-functioning plasmonic biosensors, which tap into the energetics of DNA-RNA hybridization, as well as Graphene Oxide particles coated with fluorophore-bound DNA target strands that can detect viral helicase (184, 185).
Nanoparticles can be engineered to directly target SARS-CoV-2 or as immunomodulatory factors to prime and alarm the immune system and reduce the inflammatory response during COVID-19.
Small-interfering RNAs against conserved regions of SARS-CoV-2 were incorporated into lipid nanoparticle formulations and upon delivery into lungs suppressed viral replication and improved survival of infected mice (186).
Graphene Oxide Nanoparticles (GO-NPs) have been shown to increase leukocyte numbers such as macrophages and T cells. This effect boosts adaptive immunity, thus allowing for a better immune response and viral clearance, or a possible use as vaccine adjuvants. In the scenario of uncontrolled hyperinflammation, nanodiamonds elicit an anti-inflammatory state in macrophages, while carbon and graphene sheets can be repurposed to remove pro-inflammatory cytokines and interleukins from the blood of patients (149).
Most importantly, nanotechnology may offer solutions to some of the major problems of traditional vaccines and medications such as sensitivity to acidity, water insolubility, or absorption. Nanoparticles can increase drug delivery efficiency by binding or encapsulating hydrophobic or pH-sensitive drugs and creating a targeted release. For example, certain nanoparticles bound to drugs can be modified using organic molecules that provide better release characteristics, such as Cholesterol-modified-Hydroxychloroquine. Other nanoparticles can facilitate the transport of two or more drugs, thus decreasing each dose as well as the side effects, while augmenting the combined outcome (187).
Another proposal claims that a simple and unconventional vaccine design could combine layered double hydroxide (LDH-NPs) nanoparticles and a plasmid encoding short hairpin RNA to silence the expression of targeted genes, such as essential SARS-CoV-2 proteins to stop infection early. The LDH-nanoparticles are compatible with mammalian cell lines and can insulate the shRNA against degradation, thus providing a promising delivery mechanism (188).
The current COVID-19 mRNA vaccines (Moderna, Pfizer/BioNtech) contain mRNA wrapped in lipid nanoparticles. More nanoparticle vaccines are under development. For example, NVX-CoV2373 (Novavax) is a recombinant nanoparticle-based vaccine, which incorporates the full trimeric spike glycoprotein with a saponin-based adjuvant (Matrix-M1) (189). Testing on macaques and later in phase 1–2 human clinical trials revealed that this vaccine could elicit neutralizing antibodies such as anti-spike IgG antibodies, as well as a specific T-cell response (189).
Sinovac Biotec Company also designed a nanovaccine against SARS-CoV-2 and successfully tested it in mice. This NP based vaccine incorporates the RBD subunit of the spike protein combined with two adjuvants: Monophosphoryl Lipid A (MPLA) and CpG-ODN, which stimulate TLR4 and TLR9, respectively. The vaccination of mice was achieved in three stages (original shot and two boosters) and resulted in a potent and protective T cell response accompanied by neutralizing IgA antibodies (190). This nanovaccine is currently in clinical phase 3 testing and in light of the promising results, a large-scale manufacturing plant is under construction (191).
Of note, another nanotechnology vaccine was recently found to induce a persistent antibody production and long-lasting memory response for at least 7 months in mice (192). In this vaccine design, the RBD of spike protein was conjugated via the SpyTag/SpyCatcher technology to ferritin nanoparticles. Hence, the unique capabilities of nanoparticles could revolutionize the processes of vaccine design, manufacture, and delivery.
While the widespread use of nanoparticles in medicine is an exciting idea, a few drawbacks may delay the realization of these endeavors. An overall examination of literature surrounding the design and application of nanoparticles in pharmacology has shown that there is a lot of variability between the results of independent research studies, and translating the efficacy of these particles from an in vitro to an in vivo situation is difficult (2). Additionally, critics have emphasized that the large-scale production of nanoparticles will be a high hurdle to overcome, especially when trying to keep these treatments affordable (193). The required sophistication of the manufacturing processes of nanoparticles and intellectual property rights can drive up their prices, although overall health care expenditures could be saved if nanomaterials and nano-vaccines accomplish to prevent COVID-19. Another limitation of nanoparticles are risks of unwanted tissue interactions and toxicity, unwanted spread and deposition in the body including unwanted crossing of the blood-brain barrier (194, 195). Accidental inhalation into the lungs is feared to cause epithelial injury, pulmonary inflammation and contribute to fibrosis depending on the size and chemical composition of the nanoparticles (196). Moreover, nanoparticles have been shown to interfere with biological processes like inflammation, oxidative stress, mitochondrial function, macrophage phagocytosis and platelet function (2). Acute or chronic toxicity of nanoparticles may be caused via ROS generation, cell membrane binding, DNA damage, altered cell cycle regulation and protein denaturation (197). Another important issue is the incomplete understanding of long-term effects of nanoparticles in humans and the environment. For example, a study on the effect of chronic administration of nanoparticles to rats resulted in structural damage in their testis, including disorganization of spermatogenic cells, misoriented testis and reduction of germ cells (198, 199). Allergic reactions and anaphylaxis to the mRNA lipid nanoparticle vaccines (Moderna, Pfizer/BioNtech) for COVID-19 have been blamed on the nanoparticle design and composition (200).
These limitations, and other unknown risks, should be taken into consideration when evaluating the actual potential of nanoparticles to form a reasonable approach toward nanomedicine.
Nanotechnology is an emerging field that can alter the way we approach the diagnosis, treatment, and prevention of human diseases. Nanomedicine offers unique potentials to address future epidemiological challenges with other emerging viruses. The ongoing COVID-19 pandemic has shown that health care systems were underprepared for such a large-scale event. Nanotechnology seems very promising, but one must not forget that it is a young and unexplored field. The current state of the field leans in favor of nanoparticles supporting modern medicine, but risks and long-term side effects remain hard to assess.
Effective therapies of COVID-19 remain elusive, but fortunately, the widespread public distribution of vaccines has begun. The promising potential of nanoparticles is not limited to diagnostic and therapeutic approaches but can also be applied to global prophylactic measures that aim toward limiting the spread and symptoms of SARS-CoV-2 infection. Theranostics, a new discipline of medical science, focuses on detecting and eliminating new viral or bacterial threats using nanomedicine and nanodrugs for diagnostics and therapy. This field has demonstrated futuristic applications of nanotechnology, such as spike protein-specific nanoparticles and neutralizing nanomaterials. It may even become a pharmacological standard of care once the side effects are well-understood and mitigated. While true benefit of nanomedicine in the fight against COVID-19 remains to be seen, it is worthy of in-depth considerations and efforts.
AS and KK wrote the manuscript and prepared the figures. MB wrote and edited the manuscript and supervised and funded the work. All the authors are responsible for the contents of this publication.
This work was supported (to MB) by the National Institutes of Health (1R01HL141513, 1R01HL139641, 1R01AI153613, 1UL1TR001430) and the Deutsche Forschungsgemeinschaft (BO3482/3-3, BO3482/4-1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Deepthi Sree Vamaraju and Ana N. Kimber for assisting with the figure preparation and manuscript editing. We thank Catherine O'Neal for reading and editing the final draft of the manuscript. We thank the Evans Center for Interdisciplinary Biomedical Research at Boston University School of Medicine for their support of the Affinity Research Collaborative on Respiratory Viruses: A Focus on COVID-19.
1. Kirtane AR, Verma M, Karandikar P, Furin J, Langer R, Traverso G. Nanotechnology approaches for global infectious diseases. Nat Nanotechnol. (2021) 16:369–84. doi: 10.1038/s41565-021-00866-8
2. De Jong WH, Borm PJ. Drug delivery and nanoparticles: applications and hazards. Int J Nanomed. (2008) 3:133–49. doi: 10.2147/IJN.S596
3. Abd El-Aziz TM, Stockand JD. Recent progress and challenges in drug development against COVID-19 coronavirus (SARS-CoV-2) - an update on the status. Infect Genet Evol. (2020) 83:104327. doi: 10.1016/j.meegid.2020.104327
4. Wu P, Hao X, Lau EHY, Wong JY, Leung KSM, Wu JT, et al. Real-time tentative assessment of the epidemiological characteristics of novel coronavirus infections in Wuhan, China, as at 22 January, 2020. Euro Surveill. (2020) 25:2000044. doi: 10.2807/1560-7917.ES.2020.25.3.2000044
5. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
6. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. (2020) 323:1239–42. doi: 10.1001/jama.2020.2648
7. Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. (2020) 55:105924. doi: 10.1016/j.ijantimicag.2020.105924
8. Khafaie MA, Rahim F. Cross-country comparison of case fatality rates of COVID-19/SARS-COV-2. Osong Public Health Res Perspect. (2020) 11:74–80. doi: 10.24171/j.phrp.2020.11.2.03
9. Sorci G, Faivre B, Morand S. Explaining among-country variation in COVID-19 case fatality rate. Sci Rep. (2020) 10:18909. doi: 10.1038/s41598-020-75848-2
10. Ke R, Romero-Severson E, Sanche S, Hengartner N. Estimating the reproductive number R0 of SARS-CoV-2 in the United States and eight European countries and implications for vaccination. J Theor Biol. (2021) 517:110621. doi: 10.1016/j.jtbi.2021.110621
11. Chen TM, Rui J, Wang QP, Zhao ZY, Cui JA, Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infect Dis Poverty. (2020) 9:24. doi: 10.1186/s40249-020-00640-3
12. Petersen E, Koopmans M, Go U, Hamer DH, Petrosillo N, Castelli F, et al. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect Dis. (2020) 20:e238–e44. doi: 10.1016/S1473-3099(20)30484-9
13. Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J Clin Med. (2020) 9:1225. doi: 10.3390/jcm9041225
14. Fani M, Teimoori A, Ghafari S. Comparison of the COVID-2019 (SARS-CoV-2) pathogenesis with SARS-CoV and MERS-CoV infections. Future Virol. (2020) 15:317–23. doi: 10.2217/fvl-2020-0050
15. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
16. Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. (2020) 16:1686–97. doi: 10.7150/ijbs.45472
17. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. (2020) 181:1016–35 e19. doi: 10.1016/j.cell.2020.04.035
18. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. (2020) 581:215–20. doi: 10.1038/s41586-020-2180-5
19. Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. (2020) 10:102–8. doi: 10.1016/j.jpha.2020.03.001
20. McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA. (1967) 57:933–40. doi: 10.1073/pnas.57.4.933
21. Abdul-Rasool S, Fielding BC. Understanding human coronavirus HCoV-NL63. Open Virol J. (2010) 4:76–84. doi: 10.2174/1874357901004010076
22. Bradburne AF, Bynoe ML, Tyrrell DA. Effects of a “new” human respiratory virus in volunteers. Br Med J. (1967) 3:767–9. doi: 10.1136/bmj.3.5568.767
23. Poutanen SM. Human coronaviruses. Princip Pract Pediatr Infect Dis. (2012) 2012:1117–20.e4. doi: 10.1016/B978-1-4377-2702-9.00224-5
24. Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. (2006) 44:2063–71. doi: 10.1128/JCM.02614-05
25. Fraser C, Donnelly CA, Cauchemez S, Hanage WP, Van Kerkhove MD, Hollingsworth TD, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. (2009) 324:1557–61. doi: 10.1126/science.1176062
26. Hammad MA, Syed Sulaiman SA, Aziz NA, Mohamed Noor DA. Prescribing statins among patients with type 2 diabetes: the clinical gap between the guidelines and practice. J Res Med Sci. (2019) 24:15. doi: 10.4103/jrms.JRMS_100_18
27. Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect. (2020) 54:159–63. doi: 10.1016/j.jmii.2020.03.022
28. Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K. COVID-19 (Novel Coronavirus 2019) - recent trends. Eur Rev Med Pharmacol Sci. (2020) 24:2006–11. doi: 10.26355/eurrev_202002_20378
29. Bianchi M, Benvenuto D, Giovanetti M, Angeletti S, Ciccozzi M, Pascarella S. Sars-CoV-2 envelope and membrane proteins: structural differences linked to virus characteristics? Biomed Res Int. (2020) 2020:4389089. doi: 10.1155/2020/4389089
30. Hu Y, Wen J, Tang L, Zhang H, Zhang X, Li Y, et al. The M protein of SARS-CoV: basic structural and immunological properties. Genomics Proteomics Bioinformatics. (2003) 1:118–30. doi: 10.1016/S1672-0229(03)01016-7
31. Schoeman D, Fielding BC. Coronavirus envelope protein: current knowledge. Virol J. (2019) 16:69. doi: 10.1186/s12985-019-1182-0
32. Robson B. COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed Achilles' heel conserved region to minimize probability of escape mutations and drug resistance. Comput Biol Med. (2020) 121:103749. doi: 10.1016/j.compbiomed.2020.103749
33. Arya R, Kumari S, Pandey B, Mistry H, Bihani SC, Das A, et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. (2021) 433:166725. doi: 10.1016/j.jmb.2020.11.024
34. Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. (2020) 24:91–8. doi: 10.1016/j.jare.2020.03.005
35. Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. (2020) 5:eabc3582. doi: 10.1126/sciimmunol.abc3582
36. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271–80 e8. doi: 10.1016/j.cell.2020.02.052
37. Zeng F, Hon CC, Yip CW, Law KM, Yeung YS, Chan KH, et al. Quantitative comparison of the efficiency of antibodies against S1 and S2 subunit of SARS coronavirus spike protein in virus neutralization and blocking of receptor binding: implications for the functional roles of S2 subunit. FEBS Lett. (2006) 580:5612–20. doi: 10.1016/j.febslet.2006.08.085
38. Hoffmann M, Kleine-Weber H, Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. (2020) 78:779–84 e5. doi: 10.1016/j.molcel.2020.04.022
39. Huang J, Hume AJ, Abo KM, Werder RB, Villacorta-Martin C, Alysandratos KD, et al. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell. (2020) 27:962–73 e7. doi: 10.1016/j.stem.2020.09.013
40. Gao H, Yao H, Yang S, Li L. From SARS to MERS: evidence and speculation. Front Med. (2016) 10:377–82. doi: 10.1007/s11684-016-0466-7
41. Huang Y, Yang C, Xu XF, Xu W, Liu SW. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. (2020) 41:1141–9. doi: 10.1038/s41401-020-0485-4
42. Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. (2007) 20:660–94. doi: 10.1128/CMR.00023-07
43. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
44. Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. (2020) 11:1800. doi: 10.3389/fmicb.2020.01800
45. Jary A, Leducq V, Malet I, Marot S, Klement-Frutos E, Teyssou E, et al. Evolution of viral quasispecies during SARS-CoV-2 infection. Clin Microbiol Infect. (2020) 26:1560 e1–4. doi: 10.1016/j.cmi.2020.07.032
46. Laha S, Chakraborty J, Das S, Manna SK, Biswas S, Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect Genet Evol. (2020) 85:104445. doi: 10.1016/j.meegid.2020.104445
47. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. (2021) 592:116–21. doi: 10.1038/s41586-020-2895-3
48. Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature. (2021) 592:122–7. doi: 10.1038/s41586-021-03361-1
49. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. (2020) 182:812–27 e19. doi: 10.1016/j.cell.2020.06.043
50. Zhang L, Jackson CB, Mou H, Ojha A, Rangarajan ES, Izard T, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. (2020) doi: 10.1101/2020.06.12.148726
51. Abdool Karim SS, de Oliveira T, Loots G. Appropriate names for COVID-19 variants. Science. (2021) 371:1215. doi: 10.1126/science.abh0836
52. Mosselhy DA, Virtanen J, Kant R, He W, Elbahri M, Sironen T. COVID-19 pandemic: what about the safety of anti-coronavirus nanoparticles? Nanomaterials. (2021) 11:796. doi: 10.3390/nano11030796
53. Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants - clinical, public health, and vaccine implications. N Engl J Med. (2021) 384:1866–68. doi: 10.1056/NEJMc2100362
54. Nile SH, Nile A, Qiu J, Li L, Jia X, Kai G. COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. (2020) 53:66–70. doi: 10.1016/j.cytogfr.2020.05.002
55. Polak SB, Van Gool IC, Cohen D, von der Thusen JH, van Paassen J. A systematic review of pathological findings in COVID-19: a pathophysiological timeline and possible mechanisms of disease progression. Mod Pathol. (2020) 33:2128–38. doi: 10.1038/s41379-020-0603-3
56. Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: a clinical-therapeutic staging proposal. J Heart Lung Transplant. (2020) 39:405–7. doi: 10.1016/j.healun.2020.03.012
57. Jordan RE, Adab P, Cheng KK. Covid-19: risk factors for severe disease and death. BMJ. (2020) 368:m1198. doi: 10.1136/bmj.m1198
58. Bohn MK, Lippi G, Horvath A, Sethi S, Koch D, Ferrari M, et al. Molecular, serological, and biochemical diagnosis and monitoring of COVID-19: IFCC taskforce evaluation of the latest evidence. Clin Chem Lab Med. (2020) 58:1037–52. doi: 10.1515/cclm-2020-0722
59. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. (2020) 395:507–13. doi: 10.1016/S0140-6736(20)30211-7
60. Sun X, Wang T, Cai D, Hu Z, Chen J, Liao H, et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. (2020) 53:38–42. doi: 10.1016/j.cytogfr.2020.04.002
61. Gao Z, Xu Y, Sun C, Wang X, Guo Y, Qiu S, et al. A systematic review of asymptomatic infections with COVID-19. J Microbiol Immunol Infect. (2021) 54:12–6. doi: 10.1016/j.jmii.2020.05.001
62. Grant A, Hunter PR. Immunisation, asymptomatic infection, herd immunity and the new variants of COVID 19. medRxiv. (2021). doi: 10.1101/2021.01.16.21249946
63. George PM, Wells AU, Jenkins RG. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med. (2020) 8:807–15. doi: 10.1016/S2213-2600(20)30225-3
64. Sagar M, Reifler K, Rossi M, Miller NS, Sinha P, White LF, et al. Recent endemic coronavirus infection is associated with less-severe COVID-19. J Clin Invest. (2021) 131:e143380. doi: 10.1172/JCI143380
65. Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. (2020) 583:290–5. doi: 10.1038/s41586-020-2349-y
66. Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. (2020) 369:731–6. doi: 10.1126/science.abc7424
67. Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol. (2020) 20:709–13. doi: 10.1038/s41577-020-00460-4
68. Kreye J, Reincke SM, Pruss H. Do cross-reactive antibodies cause neuropathology in COVID-19? Nat Rev Immunol. (2020) 20:645–6. doi: 10.1038/s41577-020-00458-y
69. Caress JB, Castoro RJ, Simmons Z, Scelsa SN, Lewis RA, Ahlawat A, et al. COVID-19-associated Guillain-Barre syndrome: the early pandemic experience. Muscle Nerve. (2020) 62:485–91. doi: 10.1002/mus.27024
70. Kabeerdoss J, Pilania RK, Karkhele R, Kumar TS, Danda D, Singh S. Severe COVID-19, multisystem inflammatory syndrome in children, and Kawasaki disease: immunological mechanisms, clinical manifestations and management. Rheumatol Int. (2021) 41:19–32. doi: 10.1007/s00296-020-04749-4
71. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
72. O'Driscoll M, Ribeiro Dos Santos G, Wang L, Cummings DAT, Azman AS, Paireau J, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. (2021) 590:140–5. doi: 10.1038/s41586-020-2918-0
73. Pastor-Barriuso R, Perez-Gomez B, Hernan MA, Perez-Olmeda M, Yotti R, Oteo-Iglesias J, et al. Infection fatality risk for SARS-CoV-2 in community dwelling population of Spain: nationwide seroepidemiological study. BMJ. (2020) 371:m4509. doi: 10.1136/bmj.m4509
74. Carlotti A, Carvalho WB, Johnston C, Rodriguez IS, Delgado AF. COVID-19 diagnostic and management protocol for pediatric patients. Clinics. (2020) 75:e1894. doi: 10.6061/clinics/2020/e1894
75. Shenoy AT, Brissac T, Gilley RP, Kumar N, Wang Y, Gonzalez-Juarbe N, et al. Streptococcus pneumoniae in the heart subvert the host response through biofilm-mediated resident macrophage killing. PLoS Pathog. (2017) 13:e1006582. doi: 10.1371/journal.ppat.1006582
76. Debnath M, Banerjee M, Berk M. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. (2020) 34:8787–95. doi: 10.1096/fj.202001115R
77. Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. (2020) 27:1451–4. doi: 10.1038/s41418-020-0530-3
78. Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, et al. Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. bioRxiv. (2020) doi: 10.1101/2020.03.02.972935
79. Takayama K. In vitro and animal models for SARS-CoV-2 research. Trends Pharmacol Sci. (2020) 41:513–7. doi: 10.1016/j.tips.2020.05.005
80. Kumar S, Sarma P, Kaur H, Prajapat M, Bhattacharyya A, Avti P, et al. Clinically relevant cell culture models and their significance in isolation, pathogenesis, vaccine development, repurposing and screening of new drugs for SARS-CoV-2: a systematic review. Tissue Cell. (2021) 70:101497. doi: 10.1016/j.tice.2021.101497
81. Alflen A, Aranda Lopez P, Hartmann AK, Maxeiner J, Bosmann M, Sharma A, et al. Neutrophil extracellular traps impair fungal clearance in a mouse model of invasive pulmonary aspergillosis. Immunobiology. (2020) 225:151867. doi: 10.1016/j.imbio.2019.11.002
82. Lakdawala SS, Menachery VD. The search for a COVID-19 animal model. Science. (2020) 368:942–3. doi: 10.1126/science.abc6141
83. Johansen MD, Irving A, Montagutelli X, Tate MD, Rudloff I, Nold MF, et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. (2020) 13:877–91. doi: 10.1038/s41385-020-00340-z
84. Kumar S, Yadav PK, Srinivasan R, Perumal N. Selection of animal models for COVID-19 research. Virusdisease. (2020) 31:1–6. doi: 10.1007/s13337-020-00637-4
85. Munoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al. Animal models for COVID-19. Nature. (2020) 586:509–15.
86. Lin DC, Xu L, Ding LW, Sharma A, Liu LZ, Yang H, et al. Genomic and functional characterizations of phosphodiesterase subtype 4D in human cancers. Proc Natl Acad Sci USA. (2013) 110:6109–14. doi: 10.1158/1538-7445.AM2013-586
87. Huang WE, Lim B, Hsu CC, Xiong D, Wu W, Yu Y, et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. (2020) 13:950–61. doi: 10.1111/1751-7915.13586
88. Dao Thi VL, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. (2020) 12:eabc7075. doi: 10.1126/scitranslmed.abc7075
89. Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. (2020) 38:870–4. doi: 10.1038/s41587-020-0513-4
90. Kruttgen A, Cornelissen CG, Dreher M, Hornef M, Imohl M, Kleines M. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J Clin Virol. (2020) 128:104394. doi: 10.1016/j.jcv.2020.104394
91. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. (2020) 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045
92. Eurosurveillance Editorial Team. Erratum for Euro Surveill. (2020). 25(3). Euro Surveill. (2021) 26:210204e. doi: 10.2807/1560-7917.ES.2021.26.5.210204e
93. Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, et al. Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci. (2020) 6:591–605. doi: 10.1021/acscentsci.0c00501
94. Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. (2020) 14:3822–35. doi: 10.1021/acsnano.0c02624
95. Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. (2020) 9:386–9. doi: 10.1080/22221751.2020.1729071
96. Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. (2020) 4:1140–9. doi: 10.1038/s41551-020-00603-x
97. Dabbous HM, El-Sayed MH, El Assal G, Elghazaly H, Ebeid FFS, Sherief AF, et al. Safety and efficacy of favipiravir versus hydroxychloroquine in management of COVID-19: a randomised controlled trial. Sci Rep. (2021) 11:7282. doi: 10.1038/s41598-021-85227-0
98. Group RC, Horby P, Mafham M, Linsell L, Bell JL, Staplin N, et al. Effect of hydroxychloroquine in hospitalized patients with Covid-19. N Engl J Med. (2020) 383:2030–40. doi: 10.1056/NEJMoa2022926
99. Al-Bari MA. Chloroquine analogues in drug discovery: new directions of uses, mechanisms of actions and toxic manifestations from malaria to multifarious diseases. J Antimicrob Chemother. (2015) 70:1608–21. doi: 10.1093/jac/dkv018
100. Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. (2020) 56:105949. doi: 10.1016/j.ijantimicag.2020.105949
101. Beigel JH, Tomashek KM, Dodd LE. Remdesivir for the treatment of Covid-19 - preliminary report. Reply. N Engl J Med. (2020) 383:994. doi: 10.1056/NEJMc2022236
102. Rubin D, Chan-Tack K, Farley J, Sherwat A. FDA approval of remdesivir - a step in the right direction. N Engl J Med. (2020) 383:2598–600. doi: 10.1056/NEJMp2032369
103. Barnes PJ. How corticosteroids control inflammation: Quintiles Prize Lecture 2005. Br J Pharmacol. (2006) 148:245–54. doi: 10.1038/sj.bjp.0706736
104. Group RC, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, et al. Dexamethasone in hospitalized patients with Covid-19. N Engl J Med. (2021) 384:693–704. doi: 10.1056/NEJMoa2021436
105. Guaraldi G, Meschiari M, Cozzi-Lepri A, Milic J, Tonelli R, Menozzi M, et al. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. (2020) 2:e474–84. doi: 10.1016/S2665-9913(20)30285-X
106. Xu X, Han M, Li T, Sun W, Wang D, Fu B, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. (2020) 117:10970–5. doi: 10.1073/pnas.2005615117
107. Takla M, Jeevaratnam K. Chloroquine, hydroxychloroquine, and COVID-19: systematic review and narrative synthesis of efficacy and safety. Saudi Pharm J. (2020) 28:1760–76. doi: 10.1016/j.jsps.2020.11.003
108. Schogler A, Kopf BS, Edwards MR, Johnston SL, Casaulta C, Kieninger E, et al. Novel antiviral properties of azithromycin in cystic fibrosis airway epithelial cells. Eur Respir J. (2015) 45:428–39. doi: 10.1183/09031936.00102014
109. Molina JM, Delaugerre C, Le Goff J, Mela-Lima B, Ponscarme D, Goldwirt L, et al. No evidence of rapid antiviral clearance or clinical benefit with the combination of hydroxychloroquine and azithromycin in patients with severe COVID-19 infection. Med Mal Infect. (2020) 50:384. doi: 10.1016/j.medmal.2020.03.006
110. Lo MK, Jordan R, Arvey A, Sudhamsu J, Shrivastava-Ranjan P, Hotard AL, et al. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci Rep. (2017) 7:43395. doi: 10.1038/srep43395
111. WHO Rapid Evidence Appraisal for COVID-19 Therapies (REACT) Working Group Sterne JAC, Murthy S, Diaz JV, Slutsky AS, Villar J, et al. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. JAMA. (2020) 324:1330–41. doi: 10.1001/jama.2020.17023
112. Mihara M, Ohsugi Y, Kishimoto T. Tocilizumab, a humanized anti-interleukin-6 receptor antibody, for treatment of rheumatoid arthritis. Open Access Rheumatol. (2011) 3:19–29. doi: 10.2147/OARRR.S17118
113. Wang L. C-reactive protein levels in the early stage of COVID-19. Med Mal Infect. (2020) 50:332–4. doi: 10.1016/j.medmal.2020.03.007
114. Roumier M, Paule R, Vallee A, Rohmer J, Ballester M, Brun AL, et al. Tocilizumab for severe worsening COVID-19 pneumonia: a propensity score analysis. J Clin Immunol. (2021) 41:303–14. doi: 10.1007/s10875-020-00911-6
115. Sarfraz A, Sarfraz Z, Sarfraz M, Aftab H, Pervaiz Z. Tocilizumab and COVID-19: a meta-analysis of 2120. Patients with severe disease and implications for clinical trial methodologies. Turk J Med Sci. (2020) doi: 10.3906/sag-2010-131. [Epub ahead of print].
116. Horby PW, Pessoa-Amorim G, Peto L, Brightling CE, Sarkar R, Thomas K, et al. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): preliminary results of a randomised, controlled, open-label, platform trial. medRxiv. (2021) doi: 10.1101/2021.02.11.21249258
117. Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. (2020) 369:1010–4. doi: 10.1126/science.abd0827
118. Fischer JC, Zanker K, van Griensven M, Schneider M, Kindgen-Milles D, Knoefel WT, et al. The role of passive immunization in the age of SARS-CoV-2: an update. Eur J Med Res. (2020) 25:16. doi: 10.1186/s40001-020-00414-5
119. Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA. (2020) 323:1582–9. doi: 10.1001/jama.2020.4783
120. Young MK. The indications and safety of polyvalent immunoglobulin for post-exposure prophylaxis of hepatitis A, rubella and measles. Hum Vaccin Immunother. (2019) 15:2060–5. doi: 10.1080/21645515.2019.1621148
121. Corti D, Passini N, Lanzavecchia A, Zambon M. Rapid generation of a human monoclonal antibody to combat Middle East respiratory syndrome. J Infect Public Health. (2016) 9:231–5. doi: 10.1016/j.jiph.2016.04.003
122. Zheng Z, Monteil VM, Maurer-Stroh S, Yew CW, Leong C, Mohd-Ismail NK, et al. Monoclonal antibodies for the S2 subunit of spike of SARS-CoV-1 cross-react with the newly-emerged SARS-CoV-2. Euro Surveill. (2020) 25:2000291. doi: 10.2807/1560-7917.ES.2020.25.28.2000291
123. Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. (2020) 369:1014–8. doi: 10.1126/science.abd0831
124. Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. (2021) 384:229–37. doi: 10.1056/NEJMoa2029849
125. Dyer O. Covid-19: Eli Lilly pauses antibody trial for safety reasons. BMJ. (2020) 371:m3985. doi: 10.1136/bmj.m3985
126. Group A-TL-CS, Lundgren JD, Grund B, Barkauskas CE, Holland TL, Gottlieb RL, et al. A neutralizing monoclonal antibody for hospitalized patients with Covid-19. N Engl J Med. (2021) 384:905–14. doi: 10.1056/NEJMoa2033130
127. Gottlieb RL, Nirula A, Chen P, Boscia J, Heller B, Morris J, et al. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA. (2021) 325:632–44. doi: 10.1001/jama.2021.0202
128. Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. (2021) 384:238–51. doi: 10.1056/NEJMoa2035002
129. Osama El-Gendy A, Saeed H, Ali AMA, Zawbaa HM, Gomaa D, Harb HS, et al. Bacillus Calmette-Guerin vaccine, antimalarial, age and gender relation to COVID-19 spread and mortality. Vaccine. (2020) 38:5564–8. doi: 10.1016/j.vaccine.2020.06.083
130. Hotez PJ, Corry DB, Bottazzi ME. COVID-19 vaccine design: the Janus face of immune enhancement. Nat Rev Immunol. (2020) 20:347–8. doi: 10.1038/s41577-020-0323-4
131. Le TT, Cramer JP, Chen R, Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat Rev Drug Discov. (2020) 19:667–8. doi: 10.1038/d41573-020-00151-8
132. Koirala A, Joo YJ, Khatami A, Chiu C, Britton PN. Vaccines for COVID-19: the current state of play. Paediatr Respir Rev. (2020) 35:43–9. doi: 10.1016/j.prrv.2020.06.010
133. Parker EPK, Shrotri M, Kampmann B. Keeping track of the SARS-CoV-2 vaccine pipeline. Nat Rev Immunol. (2020) 20:650. doi: 10.1038/s41577-020-00455-1
134. Team CC-R, Food, Drug A. Allergic reactions including anaphylaxis after receipt of the first dose of moderna COVID-19 vaccine - United States, December 21, 2020-January 10, 2021. MMWR Morb Mortal Wkly Rep. (2021) 70:125–9. doi: 10.15585/mmwr.mm7004e1
135. Tregoning JS, Brown ES, Cheeseman HM, Flight KE, Higham SL, Lemm NM, et al. Vaccines for COVID-19. Clin Exp Immunol. (2020) 202:162–92. doi: 10.1111/cei.13517
136. Soleimanpour S, Yaghoubi A. COVID-19 vaccine: where are we now and where should we go? Expert Rev Vaccines. (2021) 20:23–44. doi: 10.1080/14760584.2021.1875824
137. Badiani AA, Patel JA, Ziolkowski K, Nielsen FBH. Pfizer: the miracle vaccine for COVID-19?. Public Health in Practice. (2020) 1:100061. doi: 10.1016/j.puhip.2020.100061
138. Chagla Z. The BNT162b2 (BioNTech/Pfizer) vaccine had 95% efficacy against COVID-19 >/=7 days after the 2nd dose. Ann Intern Med. (2021) 174:JC15. doi: 10.7326/ACPJ202102160-015
139. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
140. Livingston EH, Malani PN, Creech CB. The Johnson & Johnson vaccine for COVID-19. JAMA. (2021) 325:1575. doi: 10.1001/jama.2021.2927
141. Krammer F, Srivastava K, Alshammary H, Amoako AA, Awawda MH, Beach KF, et al. Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. (2021) 384:1372–4. doi: 10.1056/NEJMc2101667
142. Lavine JS, Bjornstad ON, Antia R. Immunological characteristics govern the transition of COVID-19 to endemicity. Science. (2021) 371:741–5. doi: 10.1126/science.abe6522
143. Palestino G, Garcia-Silva I, Gonzalez-Ortega O, Rosales-Mendoza S. Can nanotechnology help in the fight against COVID-19? Expert Rev Anti Infect Ther. (2020) 18:849–64. doi: 10.1080/14787210.2020.1776115
144. Rivera-Gil P, Jimenez de Aberasturi D, Wulf V, Pelaz B, del Pino P, Zhao Y, et al. The challenge to relate the physicochemical properties of colloidal nanoparticles to their cytotoxicity. Acc Chem Res. (2013) 46:743–9. doi: 10.1021/ar300039j
145. Austin LA, Mackey MA, Dreaden EC, El-Sayed MA. The optical, photothermal, and facile surface chemical properties of gold and silver nanoparticles in biodiagnostics, therapy, and drug delivery. Arch Toxicol. (2014) 88:1391–417. doi: 10.1007/s00204-014-1245-3
146. Wahajuddin, Arora S. Superparamagnetic iron oxide nanoparticles: magnetic nanoplatforms as drug carriers. Int J Nanomed. (2012) 7:3445–71. doi: 10.2147/IJN.S30320
147. Leister H, Luu M, Staudenraus D, Lopez Krol A, Mollenkopf HJ, Sharma A, et al. Pro- and anti-tumorigenic capacity of immunoproteasomes in shaping the tumor microenvironment. Cancer Immunol Res. (2021) doi: 10.1158/2326-6066.CIR-20-0492. [Epub ahead of print].
148. Douziech-Eyrolles L, Marchais H, Herve K, Munnier E, Souce M, Linassier C, et al. Nanovectors for anticancer agents based on superparamagnetic iron oxide nanoparticles. Int J Nanomed. (2007) 2:541–50.
149. Weiss C, Carriere M, Fusco L, Capua I, Regla-Nava JA, Pasquali M, et al. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano. (2020) 14:6383–406. doi: 10.1021/acsnano.0c03697
150. Wolfram J, Zhu M, Yang Y, Shen J, Gentile E, Paolino D, et al. Safety of nanoparticles in medicine. Curr Drug Targets. (2015) 16:1671–81. doi: 10.2174/1389450115666140804124808
151. Wu Z, Li T. Nanoparticle-mediated cytoplasmic delivery of messenger RNA vaccines: challenges and future perspectives. Pharm Res. (2021) 38:473–8. doi: 10.1007/s11095-021-03015-x
152. Iravani S. Bacteria in nanoparticle synthesis: current status and future prospects. Int Sch Res Notices. (2014) 2014:359316. doi: 10.1155/2014/359316
153. Wen AM, Steinmetz NF. Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem Soc Rev. (2016) 45:4074–126. doi: 10.1039/C5CS00287G
154. Fujita Y, Taguchi H. Nanoparticle-based peptide vaccines. Micro Nanotechnol Vaccine Dev. (2017) 149–70. doi: 10.1016/B978-0-323-39981-4.00008-7
155. Al-Halifa S, Gauthier L, Arpin D, Bourgault S, Archambault D. Nanoparticle-based vaccines against respiratory viruses. Front Immunol. (2019) 10:22. doi: 10.3389/fimmu.2019.00022
156. Butkovich N, Li E, Ramirez A, Burkhardt AM, Wang SW. Advancements in protein nanoparticle vaccine platforms to combat infectious disease. Wiley Interdiscip Rev Nanomed Nanobiotechnol. (2020) 13:e1681. doi: 10.1002/wnan.1681
157. Li M, Cripe TP, Estes PA, Lyon MK, Rose RC, Garcea RL. Expression of the human papillomavirus type 11 L1 capsid protein in Escherichia coli: characterization of protein domains involved in DNA binding and capsid assembly. J Virol. (1997) 71:2988–95. doi: 10.1128/JVI.71.4.2988-2995.1997
158. Morikawa Y, Goto T, Momose F. Human immunodeficiency virus type 1 Gag assembly through assembly intermediates. J Biol Chem. (2004) 279:31964–72. doi: 10.1074/jbc.M313432200
159. Kaiser CR, Flenniken ML, Gillitzer E, Harmsen AL, Harmsen AG, Jutila MA, et al. Biodistribution studies of protein cage nanoparticles demonstrate broad tissue distribution and rapid clearance in vivo. Int J Nanomed. (2007) 2:715–33.
160. Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, et al. Biodistribution, pharmacokinetics, and blood compatibility of native and PEGylated tobacco mosaic virus nano-rods and -spheres in mice. Virology. (2014) 449:163–73. doi: 10.1016/j.virol.2013.10.035
161. Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, et al. Bio-distribution, toxicity and pathology of cowpea mosaic virus nanoparticles in vivo. J Control Release. (2007) 120:41–50. doi: 10.1016/j.jconrel.2007.04.003
162. Sunkar S, Nachiyar CV. Biogenesis of antibacterial silver nanoparticles using the endophytic bacterium Bacillus cereus isolated from Garcinia xanthochymus. Asian Pac J Trop Biomed. (2012) 2:953–9. doi: 10.1016/S2221-1691(13)60006-4
163. Husseiny MI, El-Aziz MA, Badr Y, Mahmoud MA. Biosynthesis of gold nanoparticles using Pseudomonas aeruginosa. Spectrochim Acta A Mol Biomol Spectrosc. (2007) 67:1003–6. doi: 10.1016/j.saa.2006.09.028
164. Konishi Y, Ohno K, Saitoh N, Nomura T, Nagamine S, Hishida H, et al. Bioreductive deposition of platinum nanoparticles on the bacterium Shewanella algae. J Biotechnol. (2007) 128:648–53. doi: 10.1016/j.jbiotec.2006.11.014
165. Mullen MD, Wolf DC, Ferris FG, Beveridge TJ, Flemming CA, Bailey GW. Bacterial sorption of heavy metals. Appl Environ Microbiol. (1989) 55:3143–9. doi: 10.1128/AEM.55.12.3143-3149.1989
166. Yang D. Application of nanotechnology in the COVID-19 pandemic. Int J Nanomed. (2021) 16:623–49. doi: 10.2147/IJN.S296383
167. Vahedifard F, Chakravarthy K. Nanomedicine for COVID-19: the role of nanotechnology in the treatment and diagnosis of COVID-19. Emergent Mater. (2021) 4:75–99. doi: 10.1007/s42247-021-00168-8
168. Mukherjee S, Mazumder P, Joshi M, Joshi C, Dalvi SV, Kumar M. Biomedical application, drug delivery and metabolic pathway of antiviral nanotherapeutics for combating viral pandemic: a review. Environ Res. (2020) 191:110119. doi: 10.1016/j.envres.2020.110119
169. Upadhyay SK, Dan S, Girdhar M, Rastogi K. Recent advancement in SARS-CoV-2 diagnosis, treatment, and vaccine formulation: a new paradigm of nanotechnology in strategic combating of COVID-19 pandemic. Curr Pharmacol Rep. (2021) 7:1–14. doi: 10.1007/s40495-021-00250-z
170. Kumar R, Nayak M, Sahoo GC, Pandey K, Sarkar MC, Ansari Y, et al. Iron oxide nanoparticles based antiviral activity of H1N1 influenza A virus. J Infect Chemother. (2019) 25:325–9. doi: 10.1016/j.jiac.2018.12.006
171. Bar-On YM, Flamholz A, Phillips R, Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. (2020) 9:e57309. doi: 10.7554/eLife.57309
172. Ren G, Hu D, Cheng EW, Vargas-Reus MA, Reip P, Allaker RP. Characterisation of copper oxide nanoparticles for antimicrobial applications. Int J Antimicrob Agents. (2009) 33:587–90. doi: 10.1016/j.ijantimicag.2008.12.004
173. Shaikh S, Nazam N, Rizvi SMD, Ahmad K, Baig MH, Lee EJ, et al. Mechanistic insights into the antimicrobial actions of metallic nanoparticles and their implications for multidrug resistance. Int J Mol Sci. (2019) 20:2468. doi: 10.3390/ijms20102468
174. Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. (2010) 464:1067–70. doi: 10.1038/nature08956
175. Sharma A, Steven S, Bosmann M. The pituitary gland prevents shock-associated death by controlling multiple inflammatory mediators. Biochem Biophys Res Commun. (2019) 509:188–93. doi: 10.1016/j.bbrc.2018.12.101
176. Sharma A, Kumar P, Ambasta RK. Cancer fighting SiRNA-RRM2 loaded nanorobots. Pharm Nanotechnol. (2020) 8:79–90. doi: 10.2174/2211738508666200128120142
177. Adhikari A, Pal U, Bayan S, Mondal S, Ghosh R, Darbar S, et al. Nanoceutical fabric prevents COVID-19 spread through expelled respiratory droplets: a combined computational, spectroscopic and anti-microbial study. bioRxiv. (2021) doi: 10.1101/2021.02.20.432081
178. Abo-Zeid Y, Ismail NSM, McLean GR, Hamdy NM. A molecular docking study repurposes FDA approved iron oxide nanoparticles to treat and control COVID-19 infection. Eur J Pharm Sci. (2020) 153:105465. doi: 10.1016/j.ejps.2020.105465
179. Khairil Anuar INA, Banerjee A, Keeble AH, Carella A, Nikov GI, Howarth M. Spy&Go purification of SpyTag-proteins using pseudo-SpyCatcher to access an oligomerization toolbox. Nat Commun. (2019) 10:1734. doi: 10.1038/s41467-019-09678-w
180. Cohen AA, Gnanapragasam PNP, Lee YE, Hoffman PR, Ou S, Kakutani LM, et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. (2021) 371:735–41. doi: 10.1126/science.abf6840
181. Roewe J, Stavrides G, Strueve M, Sharma A, Marini F, Mann A, et al. Bacterial polyphosphates interfere with the innate host defense to infection. Nat Commun. (2020) 11:4035. doi: 10.1038/s41467-020-17639-x
182. Puri N, Niazi A, Srivastava AK, Rajesh. Synthesis and characterization of reduced graphene oxide supported gold nanoparticles-poly(pyrrole-co-pyrrolepropylic acid) nanocomposite-based electrochemical biosensor. Appl Biochem Biotechnol. (2014) 174:911–25. doi: 10.1007/s12010-014-0997-9
183. Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, et al. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. (2020) 14:5135–42. doi: 10.1021/acsnano.0c02823
184. Jang H, Ryoo SR, Kim YK, Yoon S, Kim H, Han SW, et al. Discovery of hepatitis C virus NS3 helicase inhibitors by a multiplexed, high-throughput helicase activity assay based on graphene oxide. Angew Chem Int Ed Engl. (2013) 52:2340–4. doi: 10.1002/anie.201209222
185. Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. (2020) 14:5268–77. doi: 10.1021/acsnano.0c02439
186. Idris A, Davis A, Supramaniam A, Acharya D, Kelly G, Tayyar Y, et al. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. bioRxiv. (2021). doi: 10.1101/2021.04.19.440531
187. Chauhan G, Madou MJ, Kalra S, Chopra V, Ghosh D, Martinez-Chapa SO. Nanotechnology for COVID-19: therapeutics and vaccine research. ACS Nano. (2020) 14:7760–82. doi: 10.1021/acsnano.0c04006
188. Acharya R. Prospective vaccination of COVID-19 using shRNA-plasmid-LDH nanoconjugate. Med Hypotheses. (2020) 143:110084. doi: 10.1016/j.mehy.2020.110084
189. Keech C, Albert G, Cho I, Robertson A, Reed P, Neal S, et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N Engl J Med. (2020) 383:2320–32. doi: 10.1056/NEJMoa2026920
190. Liu L, Liu Z, Chen H, Liu H, Gao Q, Cong F, et al. Subunit nanovaccine with potent cellular and mucosal immunity for COVID-19. ACS Appl Bio Mater. (2020) 3:5633–8. doi: 10.1021/acsabm.0c00668
191. Ye T, Zhong Z, Garcia-Sastre A, Schotsaert M, De Geest BG. Current status of COVID-19 (Pre)clinical vaccine development. Angew Chem Int Ed Engl. (2020) 59:18885–97. doi: 10.1002/anie.202008319
192. Wang W, Huang B, Zhu Y, Tan W, Zhu M. Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell Mol Immunol. (2021) 18:749–51. doi: 10.1038/s41423-021-00643-6
193. Abd Ellah NH, Gad SF, Muhammad K, G EB, Hetta HF. Nanomedicine as a promising approach for diagnosis, treatment and prophylaxis against COVID-19. Nanomedicine. (2020) 15:2085–102. doi: 10.2217/nnm-2020-0247
194. Hoet PH, Bruske-Hohlfeld I, Salata OV. Nanoparticles - known and unknown health risks. J Nanobiotechnology. (2004) 2:12. doi: 10.1186/1477-3155-2-12
195. Ray PC, Yu H, Fu PP. Toxicity and environmental risks of nanomaterials: challenges and future needs. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. (2009) 27:1–35. doi: 10.1080/10590500802708267
196. Byrne JD, Baugh JA. The significance of nanoparticles in particle-induced pulmonary fibrosis. Mcgill J Med. (2008) 11:43–50. doi: 10.26443/mjm.v11i1.455
197. Gupta R, Xie H. Nanoparticles in daily life: applications, toxicity and regulations. J Environ Pathol Toxicol Oncol. (2018) 37:209–30. doi: 10.1615/JEnvironPatholToxicolOncol.2018026009
198. Thakur M, Gupta H, Singh D, Mohanty IR, Maheswari U, Vanage G, et al. Histopathological and ultra structural effects of nanoparticles on rat testis following 90 days (Chronic study) of repeated oral administration. J Nanobiotechnology. (2014) 12:42. doi: 10.1186/s12951-014-0042-8
199. Noon JB, Sharma A, Platten J, Quinton LJ, Reinhardt C, Bosmann M. IL-27 enhances the lymphocyte mediated innate resistance to primary hookworm infection in the lungs. bioRxiv. (2020). doi: 10.1101/2020.08.12.248021
Keywords: SARS-CoV-2 virus, vaccine, nanotechnology, drug delivery systems, sepsis, acute respiratory distress syndrome
Citation: Sharma A, Kontodimas K and Bosmann M (2021) Nanomedicine: A Diagnostic and Therapeutic Approach to COVID-19. Front. Med. 8:648005. doi: 10.3389/fmed.2021.648005
Received: 05 January 2021; Accepted: 04 May 2021;
Published: 04 June 2021.
Edited by:
Zisis Kozlakidis, International Agency For Research On Cancer (IARC), FranceReviewed by:
Hejun Liu, The Scripps Research Institute, United StatesCopyright © 2021 Sharma, Kontodimas and Bosmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Markus Bosmann, bWJvc21hbm5AYnUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.