
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med., 08 March 2021
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 8 - 2021 | https://doi.org/10.3389/fmed.2021.645955
This article is part of the Research TopicSpotlight on the Relationship between Sepsis and Infection: from Mechanisms to TherapyView all 26 articles
The recent emergence of multidrug-resistant (MDR) Klebsiella pneumoniae with hypervirulent traits causing severe infections and considerable mortality is a global cause for concern. The challenges posed by these hypermucoviscous strains of K. pneumoniae with regard to their optimal treatment, management, and control policies are yet to be answered. We studied a series of extensively drug-resistant (XDR) and hypervirulent K. pneumoniae ST5235 isolates with resistance to carbapenems and polymyxins causing neonatal sepsis in a tertiary care hospital in India. A total of 9 K. pneumoniae isolates from 9 cases of neonatal sepsis were studied with respect to their clinical relevance, antimicrobial susceptibility profile, presence of extended spectrum β lactamase (ESBL) production, and responsible genes, carbapenemases (classes A, B, and D), and aminoglycoside-resistant genes. Hypervirulence genes encoding hypermucoid nature, iron uptake, and siderophores were detected by multiplex PCR. The plasmid profile was studied by replicon typing. Isolates were typed by multilocus sequence typing (MLST) and enterobacterial repetitive intergenic consensus (ERIC) PCR to study the sequence types (STs) and clonal relation, respectively. The neonates in the studied cases had history of pre-maturity or low birth weight with maternal complications. All the cases were empirically treated with piperacillin–tazobactam and amikacin followed by imipenem/meropenem and vancomycin and polymyxin B as a last resort. However, all the neonates finally succumbed to the condition (100%). The studied isolates were XDR including resistance to polymyxins harboring multiple ESBL genes and carbapenemase genes (blaNDM and blaOXA−48). Hypervirulence genes were present in various combinations with rmpA/A2 genes present in all the isolates. IncFI plasmids were detected in these isolates. All belonged to ST5235. In ERIC PCR, 6 different clusters were seen. The study highlighted the emergence and burden of XDR hypervirulent isolates of K. pneumoniae causing neonatal sepsis in a tertiary care hospital.
Drug resistance in Klebsiella pneumoniae is ever increasing with adoption of several evolutionary routes by this “World Health Organization (WHO) critical priority pathogen” (1). With tremendous ability to acquire resistant determinants and hypervirulent elements, K. pneumoniae is now a global threat causing considerable mortality in the affected patients. Evolving gradually from multidrug-resistant (MDR) to extensively drug-resistant (XDR), K. pneumoniae isolates had not exhibited simultaneous drug resistance and hypervirulence for long until recently with the emergence of carbapenem-resistant hypervirulent K. pneumoniae (CR-hvKP) (2). The primary reason for this delayed emergence has been the successive convergence of two distinct traits of drug resistance and virulence through recombination of several plasmids, resulting in MDR-virulent strains (3). While hvKP has been associated with community-acquired infections, increasing reports of healthcare-associated infections are also being reported (4).
Over the years, hvKP has spread like an epidemic in Asian countries like China, South Korea, and Japan, with the first case being reported from Taiwan in 1986 (5). Widely prevalent and diverse convergent MDR-virulent strains of K. pneumoniae have already been reported from the South Asian region including India (6). There have been sporadic reports from India on CR-hvKP in invasive infections revealing the seriousness of the situation. The first report on CR-hvKP in sepsis from the subcontinent came in 2018 followed by a single case of neonatal sepsis recently (7, 8). We report a series of neonatal sepsis due to CR-hvKP along with simultaneous resistance to polymyxins causing high mortality from a tertiary care center.
The study was conducted at the Institute of Medical Sciences, Banaras Hindu University, in Varanasi, and Assam University, Silchar, Assam, India. During a retrospective audit on mortality in the neonatal intensive care unit (NICU), a cluster of cases of neonatal sepsis due to MDR K. pneumonia with significant mortality was found during the months of April to June 2017. The isolates were revived from stock cultures and studied as detailed below. The study was approved by the Institute's ethical committee.
A total of 9 isolates that were biochemically identified as K. pneumoniae were revived by subculture on MacConkey agar and were re-identified by a VITEK® 2 compact system (bioMerieux, USA).
Susceptibility testing was done using both disk diffusion method and VITEK® 2 compact system on the following antibiotics: amoxicillin/clavulanic acid, piperacillin/tazobactam, cefuroxime, ceftriaxone, cefoperazone, cefepime, ertapenem, imipenem, meropenem, amikacin, gentamicin, ciprofloxacin, levofloxacin, and trimethoprim/sulfamethoxazole. For susceptibility against polymyxin B and colistin, broth microdilution was performed. Results were interpreted based on CLSI 2020 guidelines (9). For quality control, Escherichia coli ATCC 25922 was used.
All the isolates were tested for the presence of ESBL genes, as well as carbapenemase and aminoglycoside resistance-encoding genes. Briefly, multiplex PCR was performed for the amplification of ESBL genes (blaTEM, blaSHV, blaCTXM1,2,9) along with class A (blaSME, blaNMC, blaGES, blaKPC), class B (blaIMP, blaVIM, blaNDM), and class D (blaOXA-48), carbapenemase genes using primers and reaction conditions as described elsewhere (10, 11). The isolates were further screened for the presence of aminoglycoside resistance genes by three multiplex PCR assays targeting different aminoglycoside resistance genes, viz., ant(2″)-Ia, ant(3″)-I, ant(4′)-Ia, aac(3)-I, aac(3)-IIc, aac(6′)-Ib, aac(6′)-II, aph(2″)-Ib, aph(2″)-Ic, aph(2″)-Id, aph(3′)-I, aph(3′)-IIb, aph(3′)-IIIa, aph(3′)-VI a, and aph(4)-Ia (12).
All the isolates were selected for the characterization of different virulence genes by five different multiplex PCR assays. For amplification and characterization of hypervirulence genes, a set of 29 primers was designed in-house using the NCBI primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/); from the nucleotide sequence of virulence genes, the amplified products were further sequenced to confirm the presence of virulence genes. The primer sequences and running conditions are summarized in Supplementary Table 1.
For performing PCR-based replicon typing, plasmids from the K. pneumoniae isolates were extracted (QIAprep Spin Miniprep Kit, QIAGEN, Germany) and were transformed into E. coli JM107. Transformants were selected in the media containing 100 μg/ml of ampicillin. Plasmids isolated from transformants were subjected to PCR assay. Eighteen pairs of primers were used to perform five multiplex and three simplex PCR assays recognizing FIA, FIB, FIC, HI1, HI2, I1-Ig, L/M, N, P, W, T, A/C, K, B/O, X, Y, F, and FIIA (13).
Multilocus sequence typing (MLST) was carried out for all K. pneumoniae isolates. Genomic DNA was extracted using QIAamp® DNA mini kit as per manufacturer instruction. Amplification of the seven housekeeping genes rpoB, gapA, mdh, pgi, PhoE, infB, and tonB was done using the primer pairs and conditions as described earlier (14). The amplified product from all nine isolates was sequenced by Sanger sequencing (EzeDiagnon Healthcare PVT., LTD, India). The obtained sequences were assembled by BioEdit v7.2.5 software. The sequence types were determined after analyzing with the help of the Institut Pasteur K. pneumoniae MLST database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
The heterogeneity of the isolates was determined by enterobacterial repetitive intergenic consensus (ERIC) PCR using the universal primer ERIC-F (5′-ATGTAAGCTCCTGGGGATTCAC-3′) and ERIC-R (5′-AAGTAAGTGACTGGGGTGAGCG-3′). Amplification was done under previously described reaction conditions, and the bands patterns were analyzed by agarose gel electrophoresis (15). Based on the band patterns, the dendrogram was constructed by using computer program NTSYS-pc version 2.0.
A total of 9 isolates of K. pneumoniae subspecies pneumoniae were characterized. The characteristics of the patients who were the source of these isolates are shown in Table 1. Majority of the neonates were of very low birth weight (<1,500 g, 66.6%), were pre-term/pre-mature (55.5%), and presented with respiratory distress (6, 66.6%). All but 1 (88.8%) mother of these cases of neonatal sepsis had complications during or prior to delivery in form of preeclampsia, heart disease, and abnormal vaginal discharges. In majority of the cases (6, 66.6%), the mode of delivery was by lower-segment Cesarean section (LSCS). All the cases were empirically treated with piperacillin–tazobactam and amikacin followed by imipenem/meropenem and vancomycin. As a last resort, treatment with polymyxin B was also given. However, all the neonates finally succumbed to the condition (100%).
All the isolates were extensively drug resistant (XDR) including resistance to polymyxins as per the resistance profile shown in Table 2. The MIC range noted for polymyxin B was 4–32 μg/mL whereas that for colistin was 4–16 μg/mL. Presence of ESBL genes and carbapenemase- and aminoglycoside-encoding genes is shown in Table 3. All the isolates harbored blaSHV, blaCTXM, and aac(6′)-Ib. Among the class B carbapenemases, blaNDM and/or blaOXA−48 were detected in 4 isolates while 5 isolates did not harbor these genes. None of the isolates had class A carbapenemase genes.
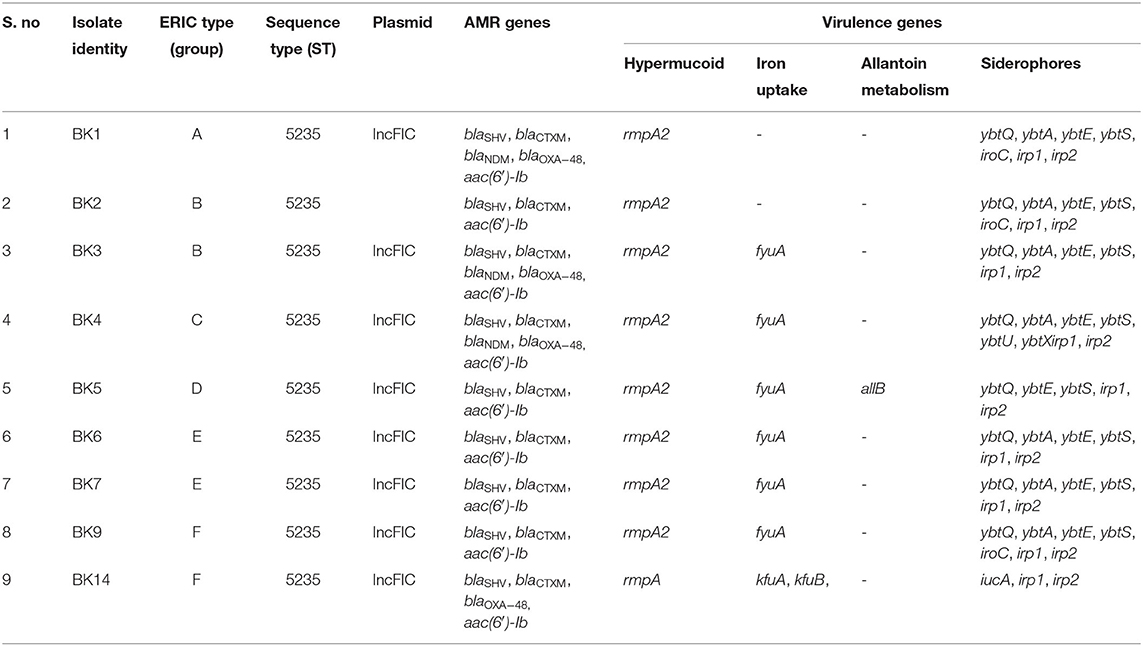
Table 3. Summary of K. pneumoniae isolates showing resistance genes, ERIC, MLST and plasmid profiles, and virulence genes.
Among the hypervirulence genes, rmpA2 was present in 8 isolates while 1 isolate harbored the rmpA gene. Iron uptake-encoding genes (fyuA/kfuA, kfuB) were present in 7 isolates while the gene for allantoin metabolism (allB) was seen in a single isolate. Siderophore-encoding genes (iroC, ybt, irp, iucA) were present in all the isolates. The distribution of these hypervirulence genes is shown in Table 3.
All the isolates harbored the IncFIc plasmid. Strain typing by ERIC-PCR revealed 100% similarity in 2 isolates each in groups B, E, and F (Figure 1). All the isolates belonged to ST5235.
Carbapenem resistance in K. pneumoniae is of critical importance owing to its widespread dissemination. Concurrently, hypervirulence in K. pneumoniae is also a serious public health threat. In this study, we report a series of neonatal sepsis due to carbapenem and polymyxin (polymyxin B and colistin)-resistant hvKP causing 100% mortality. To the best of our knowledge, this is the first extensive series of neonatal sepsis caused by XDR-hvKP from the subcontinent.
Hypervirulence has been reported to be associated with very high mortality with rates ranging from 3 to 31%, often increasing up to 35% in case of sepsis (4). This rate further increases when hvKP additionally presents with drug resistance, where the rate as high as 100% has been noted in cases of sepsis due to CR-hvKP (16). Extremely high mortality (84.2%) associated with meropenem-resistant phenotypic and genotypic hvKP-causing sepsis has also been reported from India (6). There has been another single case study on neonatal sepsis from Kolkata, India, with CR-hvKP with inconclusive data on survival as the neonate was lost to follow-up (7). Unfortunately, 100% mortality was seen in the neonates in this study where isolates were also resistant to polymyxins. There have been sporadic reports of colistin-resistant hvKP where isolates retained susceptibility to aminoglycosides, ciprofloxacin, piperacillin–tazobactam, imipenem, and tigecycline despite colistin resistance (17). Consequently, mortality rates were low due to available treatment options.
For long, it was thought and observed that hvKP isolates remained susceptible to antimicrobials until the recent introduction of “convergent” K. pneumoniae strains that are both drug resistant and hypervirulent (5). Few of the previous studies have mentioned low drug resistance potential of the hvKP isolates based on either their susceptibilities to the first line of antimicrobials or due to the sporadic presence of ESBL genes not limiting therapeutic options to a greater extent (18). On the other hand, there has been a noticeable upsurge in reports on MDR hvKP especially from developing countries like China, India, Iran, and Brazil where tackling of these isolates has been a big challenge from the treatment point of view (6, 16, 17, 19, 20). The situation is exemplified in the current study where XDR isolates with resistance to last-resort drugs like carbapenems and polymyxins from cases of neonatal sepsis displayed hypervirulence, posing a huge challenge for control. Management of such infections encompasses both adequate source control and active antimicrobial treatment (4). However, it should be mentioned that the neonatal unit in the study followed strict disinfection policy and appropriate infection control practices and even after thorough routine environmental surveillance, no such isolates have been found in the environment or in any of the surveillance cultures from the neonates.
There has been dearth of data and absence of any trial assessing the antimicrobials best suited for treatment of infections caused by hvKP. In contrast, prevalence of MDR in hvKP has been increasing in parallel with the increase in healthcare-associated infections. Suggestions on the use of newer antimicrobial combinations like ceftazidime–avibactam, meropenem–vaborbactam, and imipenem–relebactam for effective treatment of infections by CR-KP have been made (4). However, these antimicrobials have a limited role in infections due to class B carbapenemases, which is one of the major mechanisms of carbapenem resistance in these isolates as revealed in this study and those from developing countries. The blaNDM and blaOXA−48 genes have been often reported to be associated with the hvKP isolates in most of the studies from Asian countries (6, 19). Against antibiotic selection pressure, studies have shown that hvKP isolates can harbor resistance gene plasmids including ESBL, carbapenemases, and those for colistin resistance (5).
It is a well-known fact that several virulence factors of K. pneumoniae have facilitated its spread and emergence as a global threat. Among the several genetic markers to define the hypervirulence in K. pneumoniae, presence of regulators of the mucoid phenotype, rmpA and rmpA2 genes, produced siderophores like aerobactin, enterobactin, yersiniabactin, and salmochelin, and genes involved in iron uptake and allantoin metabolism are the major ones (18, 21). There has been evidence that rmpA/rmpA2 genes along with siderophores are closely associated with invasive infections (22). All the XDR hvKP isolates in the present study were phenotypically and genotypically hypermucoviscous with multiple types of siderophore-encoding genes. The highly invasive profile of these isolates was reflected in their ability to cause 100% mortality in the neonatal unit.
Emergence of new sequence types among hvKP strains has been noted in few of the recently conducted studies. Though ST23 has been proposed as the major dominant clone of hvKP in Asia (5); most of the infections caused by this clone are community acquired (18). All the XDR hvKP isolates in this study belonged to ST5235, the significance of which could not be discussed owing to paucity of literature on this sequence type.
Despite being an addition to the scarce data on XDR hvKP in neonatal sepsis, this study was not without limitations. While the sources of these isolates could not be traced, classification of the nature of the infections as community or hospital acquired could not be made. Whole-genome sequences of the isolates could have provided better insight into the genome of these isolates. Nevertheless, this study puts forward several yet-to-be-answered issues related to the emergence of XDR hvKP in hospitals. While the origin of these isolates still remains debated, their exact control measures are yet to be determined. More importantly, consensus decisions on optimal therapy against these XDR K. pneumoniae isolates with hypervirulence could be the best armament, thus changing the epidemiology of these infections.
The study analyzed a fatal case series of neonatal sepsis caused by hvKP with extensive drug resistance to carbapenems and polymyxins in a tertiary-care hospital in India, thus revealing the challenges posed by these emerging pathogens in developing countries.
The datasets generated in this article are not readily available because it is a collaborative study and requires administrative approval for availability. Requests to access the datasets should be directed to YWIwNDA0QGdtYWlsLmNvbQ==.
TB designed the concept and prepared the manuscript. JW performed the experimental work, data collection, and analysis and preparation of the manuscript. SS collected samples and carried out susceptibility profiling. AK helped in clinical analysis of the study. AB supervised the whole work. All authors read and approved the final manuscript.
ICMR SRF was awarded to JW Vide Letter No.; AMR/Fellowship/19/2019-ECD-II/ID No. 2019-3677 dated 21.08.2019.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank BHU for providing basic infrastructure for the study and Institute Merieux, France, for laboratory grant (IAMM-Merieux Award 2018 to TB, R/Dev/M-14-60).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.645955/full#supplementary-material
1. World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: WHO Press (2017). p. 1-7.
2. Liu B, Su W. Whole genome sequencing of NDM-1-producing serotype K1 ST23 hypervirulent Klebsiella pneumoniae in China. J Med Microbiol. (2019) 68:866–73. doi: 10.1099/jmm.0.000996
3. Yang X, Dong N, Chan EW, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. (2021) 29:65–83. doi: 10.1016/j.tim.2020.04.012
4. Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae clinical and molecular perspectives. J Intern Med. (2020) 287:283–300. doi: 10.1111/joim.13007
5. Marr CM, Russo TA. Expert review of anti-infective therapy hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther. (2019) 17:71–3. doi: 10.1080/14787210.2019.1555470
6. Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, Dance DA, et al. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med. (2020) 12:1–6. doi: 10.1186/s13073-019-0706-y
7. Shankar C, Nabarro LE, Anandan S, Ravi R, Babu P, Munusamy E, et al. Extremely high mortality rates in patients with carbapenem-resistant, hypermucoviscous Klebsiella pneumoniae blood stream infections. J Assoc Phys India. (2018) 66:13–6.
8. Mukherjee S, Naha S, Bhadury P, Saha B, Dutta M, Dutta S, et al. Emergence of OXA-232-producing hypervirulent Klebsiella pneumoniae ST23 causing neonatal sepsis. J Antimicrob Chemother. (2020) 75:2004–6. doi: 10.1093/jac/dkaa080
9. CLSI M100-ED30:2020. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed. Wayne, PA: CLSI (2019).
10. Hong SS, Kim K, Huh JY, Jung B, Kang MS, Hong SG. Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann Lab Med. (2012) 32:359–61. doi: 10.3343/alm.2012.32.5.359
11. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. (2011) 70:119–23. doi: 10.1016/j.diagmicrobio.2010.12.002
12. Nie L, Lv Y, Yuan M, Hu X, Nie T, Yang X, et al. Genetic basis of highlevel aminoglycoside resistance in Acinetobacter baumannii from Beijing, China. Acta Pharm Sin B. (2014) 4:295–300. doi: 10.1016/j.apsb.2014.06.004
13. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. (2005) 63:219–28. doi: 10.1016/j.mimet.2005.03.018
14. Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. (2005) 43:4178–82. doi: 10.1128/JCM.43.8.4178-4182.2005
15. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. (1991) 19:6823–31. doi: 10.1093/nar/19.24.6823
16. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. (2018) 18:37–46. doi: 10.1016/S1473-3099(17)30489-9
17. Lu Y, Feng Y, McNally A, Zong Z. The occurence of colistin-resistant hypervirulent Klebsiella pneumoniae in China. Front Microbiol. (2018) 9:2568. doi: 10.3389/fmicb.2018.02568
18. Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. (2018) 18:6. doi: 10.1186/s12866-017-1148-6
19. Pajand O, Darabi N, Arab M, Ghorbani R, Bameri Z, Ebrahimi A, et al. The emergence of the hypervirulent Klebsiella pneumoniae (hvKp) strains among circulating clonal complex 147 (CC147) harbouring blaNDM/OXA-48 carbapenemases in a tertiary care center of Iran. Ann Clin Microbiol Antimicrob. (2020) 19:12. doi: 10.1186/s12941-020-00349-z
20. Ferreira RL, da Silva BCM, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Front Microbiol. (2019) 9:3198. doi: 10.3389/fmicb.2018.03198
21. Effah CY, Sun T, Liu S, Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. (2020) 19:1. doi: 10.1186/s12941-019-0343-8
22. Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. (2015) 112:E3574–81. doi: 10.1073/pnas.1501049112
Keywords: polymyxins, hypermucoviscous, MDR, carbapenems, fatal, ST5325
Citation: Banerjee T, Wangkheimayum J, Sharma S, Kumar A and Bhattacharjee A (2021) Extensively Drug-Resistant Hypervirulent Klebsiella pneumoniae From a Series of Neonatal Sepsis in a Tertiary Care Hospital, India. Front. Med. 8:645955. doi: 10.3389/fmed.2021.645955
Received: 24 December 2020; Accepted: 04 February 2021;
Published: 08 March 2021.
Edited by:
David Paterson, The University of Queensland, AustraliaReviewed by:
Nitya Singh, University of Florida, United StatesCopyright © 2021 Banerjee, Wangkheimayum, Sharma, Kumar and Bhattacharjee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amitabha Bhattacharjee, YWIwNDA0QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.