- 1Department of Clinical Laboratory, Affiliated Provincial Hospital of Anhui Medical University, Hefei, China
- 2Department of Clinical Laboratory, First Affiliated Hospital of University of Science and Technology of China, Hefei, China
- 3Department of Clinical Laboratory, Peking Union Medical College Hospital, Beijing, China
- 4Nanjing Medical laboratory, Beijing Genomics Institute, Nanjing, China
We report a case of hospital-acquired Legionella pneumonia that was detected by metagenomic next-generation sequencing (mNGS) of blood from a 7-year-old girl after umbilical cord blood stem cell transplantation (UCBT) with myelodysplastic syndrome. UCBT is traditionally associated with an increased risk of infection, particularly during the first 3 months after transplantation. Controlling interstitial pneumonia and severe infection is the key to reducing patient mortality from infection. Legionella pneumophila can cause a mild cough to rapidly fatal pneumonia. After mNGS confirmed that the pathogen was L. pneumophila, azithromycin, cefoperazone sulbactam, and posaconazole were used for treatment, and the patient's temperature decreased and remained normal. The details of this case highlight the benefits of the timely use of metagenomic NGS to identify pathogens for the survival of immunocompromised patients.
Introduction
Legionella, which is widely found in the natural environment, including in water sources and soil, includes 58 species and 3 subspecies (1). More than 90% of legionellosis is caused by L. pneumophila, of which Lp1 is the most common strain (1–4). Mortality from Legionella depends on the severity of the disease, the suitability of the initial antimicrobial treatment, the location where the Legionella infection was contracted, and host factors (2, 5). Immunocompromised patients are the most susceptible hosts. Legionella is a commonly misdiagnosed pathogen that causes severe hospital-acquired pneumonia, and multiple studies have shown it to be both underdiagnosed and undertreated (2). But mNGS technology based on high-throughput sequencing has been widely studied for use as a non-targeted and broad-spectrum pathogen screening technology for identifying clinically moderate and severe infections (6, 7). Here, we report the first case of an umbilical-cord blood stem cell patient with pulmonary L. pneumophila infection who was diagnosed by mNGS before the blood culture results were obtained.
Case Description
A 7-year-old girl with refractory anemia with excess blasts and type 2 myelodysplastic syndrome was admitted to the hematology department in March 2019, and blood analysis showed that the number of primordial blood cells had reached 19%. The patient was treated with allogeneic cord blood stem cell transplantation under electrocardiographic monitoring; 15 days after transplantation, the patient's neutrophils exceeded 0.5 × 109/L, and thus she was transferred to the general ward for further treatment.
Thirty-nine days after transplantation, the patient developed a high fever, and her blood neutrophil count and inflammation indicators increased (Figure 1). Therefore, hematologists treated the patient with imipenem, cilastatin, amikacin, linezolid, and liposomal amphotericin B. After 1 week of treatment, the patient's body temperature had not returned to normal, and CT showed double pneumonia, in which the upper lobe of the left lung was consolidated (Figure 2A). We sent the patient's blood samples for mNGS, and 8,112 raw reads belonged to L. pneumophila (Figure 3A). The anti-infection regimen was changed to azithromycin combined with cefoperazone, sulbactam, and posaconazole on May 10, 2019 (Figure 1). We then tested the urine for Legionella antigen and sent the bronchoalveolar lavage fluid (BALF) for Gram staining, mNGS, and culture on Legionella MWY selective agar. The phagocytosis of gram-negative bacilli by leukocytes in BALF was observed under a microscope (Figures 4A,B). The number of raw reads of L. pneumophila in BALF was 1,960 (Figure 3B). Six days later, several off-white colonies were visible on the MWY medium (Figure 4C). After Gram staining of the colonies, a large number of gram-negative bacilli were observed under microscopy (Figure 4D). The off-white colonies grown on the MWY medium were subsequently identified as L. pneumophila using time-of-flight mass spectrometry. The patient's body temperature returned to normal after 1 day of treatment with azithromycin and remained normal for 1 week (Figure 1), but 10 days after initiating azithromycin treatment, a CT scan still showed inflammation in both lungs (Figure 2B). A CT scan revealed that multiple lung lesions were absorbed 18 days later (Figure 2C). Finally, the patient's general condition improved after follow-up treatment, and she was discharged from hospital on June 3.
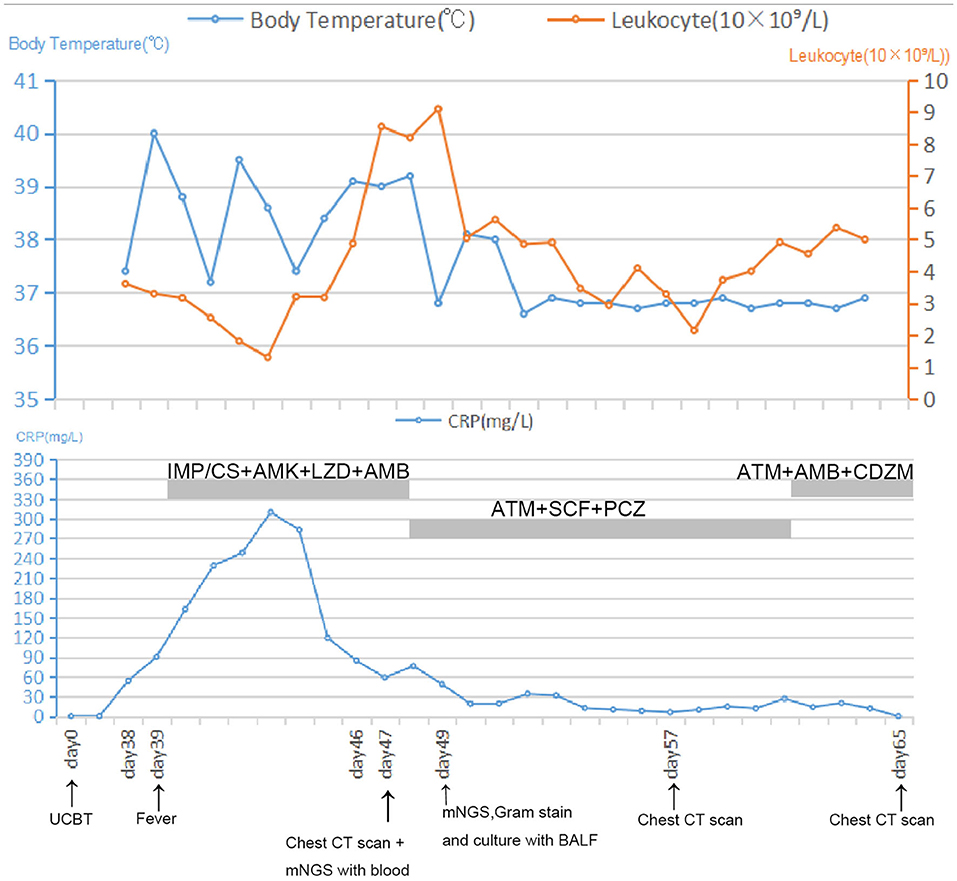
Figure 1. Timeline with relevant data from the episode of care; body temperature curves, leukocyte counts, and CRP. Major events are indicated with arrows. Yellow line shows the leukocyte counts in peripheral blood. Blue line on the top shows body-temperature values. Blue line on the bottom shows CRP values. Horizontal thick gray lines show the medications administered: IMP/CS, Imipenem and cilastatin sodium; AMK, Amikacin; LZD, Linezolid; AMB, Amphotericin B; ATM, Azithromycin; SCF, Sulbactam and cefoperazone sodium; PCZ, Posaconazole; CDZM, Cefodizime.
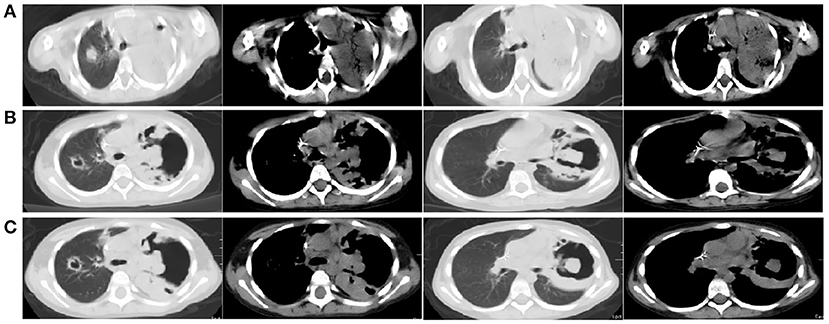
Figure 2. (A) Chest CT scan showing double pneumonia: the upper lobe of the left lung was consolidated; (B) Chest CT scan showing inflammation in both lungs after 10 days of treatment with azithromycin; (C) Chest CT scan revealed that multiple lung lesions were absorbed 18 days later.
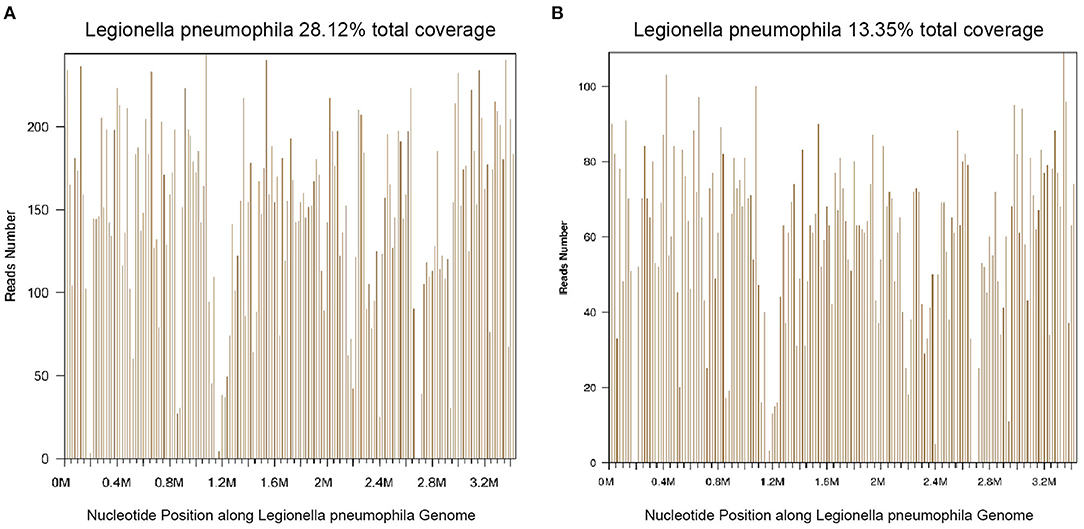
Figure 3. Diagnosis of Legionella pneumophila infection using mNGS. (A) The majority of reads mapped to the L. pneumophila genome with coverage of 28.12%. (B) The majority of reads mapped to the L. pneumophila genome with coverage of 13.35%.

Figure 4. (A,B) Gram stain of bronchoalveolar lavage fluid (BALF) with arrows indicating Legionella pneumophila, magnification ×1,000. (C) L. pneumophila colonies on MWY selective agar (red arrows) from BALF. (D) Gram stain of L. pneumophila colonies (magnification ×1,000).
Discussion
The most susceptible hosts of hospital-acquired Legionella pneumonia are immunocompromised patients, including organ transplant recipients (8) and cancer patients (9), as well as those receiving glucocorticoid therapy. Multiple studies have shown Legionella infections to be underdiagnosed and undertreated (1, 10, 11). In this case report, the patient was fortunate to be treated in time and was discharged from the hospital in good health.
There are many kinds of Legionella detection techniques. The immunological methods are limited by the patient's immune status and cross-reactions with other pathogens (12), and at present, this technology can only detect L. pneumophila serogroups 1, 3, and 6 (13). For immunocompromised patients, it is important to identify the infectious agent as early as possible in the evolution of the disease. The culture of Legionella species from respiratory specimens is the gold standard for the diagnosis of Legionella pneumonia (14); however, a Legionella culture requires more than 3 days and a medium containing L-cysteine (15, 16). When the patient is in the acute infection phase, samples should be quickly transported to the laboratory for culture, preferably before initiating antimicrobial therapy. The diagnosis of Legionnaires' disease is confirmed by means of culture in only 5% of cases (1). The widely used time-of-flight mass spectrometry detection method can only be used to identify bacterial species after the colonies are cultivated by traditional methods. Thus, the turn-around time is too long for critically ill patients. Obviously, the culture results in this report were obtained very much later than the mNGS results. Other techniques for detecting Legionella nucleic acids include PCR (1), isothermal amplification (17), probe hybridization (18), and second-generation sequencing.
This is the first report describing the use of mNGS to detect hospital-acquired Legionella pneumonia in a patient after umbilical cord blood transplantation. NGS has unique advantages for the detection of pathogens that are difficult to cultivate (19), especially for cases without target pathogens (20, 21). In addition, mNGS is extremely suitable for detecting unknown pathogens (22, 23), rare pathogens (24, 25), and between-species transmitted pathogens (26, 27). Of course, there are also shortcomings, such as high testing costs, analytical sensitivity, a complex laboratory workflow, and susceptibility to contamination (28). In this case, the mNGS provided directions for choosing traditional diagnosis methods, and the cultured colonies and immunological results were consistent with the mNGS. Although mNGS is relatively expensive, the patient's family considered it worthwhile to identify the pathogen as early as possible to reduce the patient's symptoms. We believe that, with the reductions in sequencing costs and the continuous improvements in medical standards in China, mNGS-based pathogenic diagnosis can be increasingly used to greatly improve the pertinence and timeliness of clinical pathogenic treatment.
Data Availability Statement
The datasets for this study can be found in the NCBI https://www.ncbi.nlm.nih.gov/bioproject/PRJNA722154 under accession number PRJNA722154.
Ethics Statement
Written informed consent was obtained from the minor(s)' legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
Author Contributions
YW and YD designed the study. HL, WC, and ZW helped collect data. ZL performed the statistical analysis. FM and XM revised the manuscript, which was written by YW. All authors read and approved the final manuscript.
Funding
This paper was supported by National Natural Science Foundation of China (Grant Number: 81972001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Suzanne Leech, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
References
1. Cunha BA, Burillo A, Bouza E. Legionnaires' disease. Lancet. (2016) 387:376–85. doi: 10.1016/S0140-6736(15)60078-2
2. Chahin A, Opal SM. Severe pneumonia caused by legionella pneumophila: differential diagnosis and therapeutic considerations. Infect Dis Clin North Am. (2017) 31:111–21. doi: 10.1016/j.idc.2016.10.009
3. Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, Widmer A, et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis. (2002) 186:127–8. doi: 10.1086/341087
4. Beaute J, Plachouras D, Sandin S, Giesecke J, Sparen P. Healthcare-associated legionnaires' disease, Europe, 2008–2017. Emerg Infect Dis. (2020) 26:2309–18. doi: 10.3201/eid2610.181889
5. Heath CH, Grove DI, Looke DF. Delay in appropriate therapy of Legionella pneumonia associated with increased mortality. Eur J Clin Microbiol Infect Dis. (1996) 15:286–90. doi: 10.1007/BF01695659
6. Grumaz S, Stevens P, Grumaz C, Decker SO, Weigand MA, Hofer S, et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. (2016) 8:73. doi: 10.1186/s13073-016-0326-8
7. Xie Y, Du J, Jin W, Teng X, Cheng R, Huang P, et al. Next generation sequencing for diagnosis of severe pneumonia: China, 2010–2018. J Infect. (2019) 78:158–69. doi: 10.1016/j.jinf.2018.09.004
8. Servais S, Hannon M, Peffault De, Latour R, Socie G, Beguin Y, Reconstitution of adaptive immunity after umbilical cord blood transplantation: impact on infectious complications. Stem Cell Investig. (2017) 4:40. doi: 10.21037/sci.2017.05.03
9. Ehret KM, Chamberlain AT, Berkelman RL, Fridkin SK, Preventing hospital-acquired Legionnaires' disease: A snapshot of clinical practices and water management approaches in US acute-care hospitals. Infect Control Hosp Epidemiol. (2018) 39:1470–2. doi: 10.1017/ice.2018.240
10. Spiegelman J, Pedutem T, Francisco MJ. Legionnaires' disease cases at a large community hospital-common and underdiagnosed. Int J Environ Res Public Health. (2020) 17:332. doi: 10.3390/ijerph17010332
11. Sreenath K, Dey AB, Kabra SK, Thakur B, Guleria R, Chaudhry R. Legionella pneumophila in Patients with Pneumonia at a Referral Hospital, New Delhi, India, 2015–2020. Am J Trop Med Hyg. (2020) 104:854–60. doi: 10.4269/ajtmh.20-0653
12. Edelstein PH, Meyer RD, Finegold SM. Laboratory diagnosis of Legionnaires' disease. Am Rev Respir Dis. (1980) 121:317–27.
13. Elverdal PL, Svarrer CW, Jorgensen CS, Skovsted IC, Uldum SA. Development and validation of ELISA for detection of antibodies to Legionella pneumophila serogroup 1, 3 and 6 in human sera. J Microbiol Methods. (2011) 86:298–303. doi: 10.1016/j.mimet.2011.05.022
14. Fields BS, Benson RF, Besser RE. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev. (2002) 15:506–26. doi: 10.1128/CMR.15.3.506-526.2002
15. Fard SY, Nomanpour B, Fatolahzadeh B, Mobarez AM, Darban-Sarokhalil D, Fooladi AA, et al. Hospital acquired pneumonia: comparison of culture and real-time PCR assays for detection of Legionella pneumophila from respiratory specimens at Tehran hospitals. Acta Microbiol Immunol Hung. (2012) 59:355–65. doi: 10.1556/amicr.59.2012.3.6
16. Mayock R, Skale B, Kohler RB. Legionella pneumophila pericarditis proved by culture of pericardial fluid. Am J Med. (1983) 75:534–6. doi: 10.1016/0002-9343(83)90362-5
17. Olabarria G, Eletxigerra U, Rodriguez I, Bilbao A, Berganza J, Merino S. Highly sensitive and fast Legionella spp. in situ detection based on a loop mediated isothermal amplification technique combined to an electrochemical transduction system. Talanta. (2020) 217:121061. doi: 10.1016/j.talanta.2020.121061
18. Nuthong B, Wilailuckana C, Tavichakorntrakool R, Boonsiri P, Daduang S, Bunyaraksyotin G, et al. One step for Legionella pneumophila detection in environmental samples by DNA-gold nanoparticle probe. J Appl Microbiol. (2018) 125:1534–40. doi: 10.1111/jam.14047
19. Kolb M, Lazarevic V, Emonet S, Calmy A, Girard M, Gaia N, et al. Next-generation sequencing for the diagnosis of challenging culture-negative endocarditis. Front Med. (2019) 6:203. doi: 10.3389/fmed.2019.00203
20. Lin Y, Wang BX, Zhang NN, Zhang L, Gao ZB, Tian J, et al. Metagenomic analysis identified stenotrophomonas maltophilia pneumonia in an infant suffering from unexplained very severe pneumonia. Front Pediatr. (2019) 7:380. doi: 10.3389/fped.2019.00380
21. Xiao N, Gai W, Hu WG, Li JX, Zhang Y, Zhao XY. Next-generation-sequencing technology used for the detection of Mycoplasma hominis in renal cyst fluid: a case report. Infect Drug Resist. (2019) 12:1073–9. doi: 10.2147/IDR.S198678
22. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. (2020) 579:265–9. doi: 10.1038/s41586-020-2008-3
23. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
24. Wilson MR, Shanbhag NM, Reid MJ, Singhal NS, Gelfand JM, Sample HA, et al. Diagnosing balamuthia mandrillaris encephalitis with metagenomic deep sequencing. Ann Neurol. (2015) 78:722–30. doi: 10.1002/ana.24499
25. Du B, Tao Y, Ma J, Weng X, Gong Y, Lin Y, et al. Identification of sparganosis based on next-generation sequencing. Infect Genet Evol. (2018) 66:256–61. doi: 10.1016/j.meegid.2018.10.005
26. Harrison EM, Paterson GK, Holden MT, Larsen J, Stegger M, Larsen AR, et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med. (2013) 5:509–15. doi: 10.1002/emmm.201202413
27. Ai JW, Zhang HC, Cui P, Xu B, Gao Y, Cheng Q, et al. Dynamic and direct pathogen load surveillance to monitor disease progression and therapeutic efficacy in central nervous system infection using a novel semi-quantitive sequencing platform. J Infect. (2018) 76:307–10. doi: 10.1016/j.jinf.2017.11.002
Keywords: metagenomic next-generation sequencing, Legionella pneumophila, myelodysplastic syndrome, umbilical cord blood stem cell transplantation, hospital acquired
Citation: Wang Y, Dai Y, Lu H, Chang W, Ma F, Wang Z, Liu Z and Ma X (2021) Case Report: Metagenomic Next-Generation Sequencing in Diagnosis of Legionella pneumophila Pneumonia in a Patient After Umbilical Cord Blood Stem Cell Transplantation. Front. Med. 8:643473. doi: 10.3389/fmed.2021.643473
Received: 21 January 2021; Accepted: 30 April 2021;
Published: 11 June 2021.
Edited by:
George Weinstock, Jackson Laboratory for Genomic Medicine, United StatesReviewed by:
Alberto Cagigi, Karolinska Institutet (KI), SwedenBeiwen Zheng, Zhejiang University, China
Copyright © 2021 Wang, Dai, Lu, Chang, Ma, Wang, Liu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoling Ma, bWF4aWFvbGluZyYjeDAwMDQwO1VTVEMuZWR1LmNu
 Yangyan Wang1
Yangyan Wang1 Yuanyuan Dai
Yuanyuan Dai Ziran Wang
Ziran Wang Xiaoling Ma
Xiaoling Ma