- Division of Bacteriology, Indian Council of Medical Research (ICMR)-National Institute of Cholera and Enteric Diseases, Kolkata, India
The convergence of a vulnerable population and a notorious pathogen is devastating, as seen in the case of sepsis occurring during the first 28 days of life (neonatal period). Sepsis leads to mortality, particularly in low-income countries (LICs) and lower-middle-income countries (LMICs). Klebsiella pneumoniae, an opportunistic pathogen is a leading cause of neonatal sepsis. The success of K. pneumoniae as a pathogen can be attributed to its multidrug-resistance and hypervirulent-pathotype. Though the WHO still recommends ampicillin and gentamicin for the treatment of neonatal sepsis, K. pneumoniae is rapidly becoming untreatable in this susceptible population. With escalating rates of cephalosporin use in health-care settings, the increasing dependency on carbapenems, a “last resort antibiotic,” has led to the emergence of carbapenem-resistant K. pneumoniae (CRKP). CRKP is reported from around the world causing outbreaks of neonatal infections. Carbapenem resistance in CRKP is largely mediated by highly transmissible plasmid-encoded carbapenemase enzymes, including KPC, NDM, and OXA-48-like enzymes. Further, the emergence of a more invasive and highly pathogenic hypervirulent K. pneumoniae (hvKP) pathotype in the clinical context poses an additional challenge to the clinicians. The deadly package of resistance and virulence has already limited therapeutic options in neonates with a compromised defense system. Although there are reports of CRKP infections, a review on neonatal sepsis due to CRKP/ hvKP is scarce. Here, we discuss the current understanding of neonatal sepsis with a focus on the global impact of the CRKP, provide a perspective regarding the possible acquisition and transmission of the CRKP and/or hvKP in neonates, and present strategies to effectively identify and combat these organisms.
Introduction
The infiltration of sterile regions of the body with microorganisms and the manifestation of a reaction to it thereof, is known as sepsis. When this occurs within the first 28 days of life consequences can be dire, as the newborns first encounter a world of pathogens. Their immune system has not yet developed to equip them in this battle. Thus, the total number of neonatal deaths due to sepsis is a staggering half-million per year (1, 2).
The pathogens that a neonate encounters are essentially present everywhere, starting with the birth canal, the crib, the hands of the nurse, or even the nasogastric tube. For the ones who are premature or low-birth-weight and require prolonged hospitalization or life support systems, the chances of infection are very high (3). In low-income countries (LICs) and lower-middle-income countries (LMICs), where nearly all resources are inadequate, breaches in care or infection control can lead to sepsis.
If one had to name an organism that readily becomes resistant to antibiotics, can harbor numerous plasmids, can survive in the environment & within the human gut, and is a dread in the neonatal intensive care units (NICU) in LMICs, it would be none other than Klebsiella pneumoniae (4, 5). This organism has increasingly shown various facets of a successful pathogen. Resistance to several antibiotics at a low fitness cost makes it capable of causing outbreaks in neonatal units. K. pneumoniae is resistant to a repertoire of antibiotics. Resistance to carbapenems, considered as the “last resort” against serious infections caused by Gram-negative bacilli, has limited therapeutic options immensely. As options of treatment were slowly failing, resistance to carbapenems was a cul-de-sac, because carbapenem-resistant K. pneumoniae (CRKP) are also resistant to several other antibiotics. Carbapenem-resistant genes are frequently harbored on plasmids that can spread from one species to the other. These resistance genes code for enzymes that efficiently hydrolyze carbapenems and all other β-lactam antibiotics. Further, most are also resilient against inhibition by the commercially viable β-lactamase inhibitors. In addition to carbapenem resistance, K. pneumoniae also had another ace up its sleeve, hypervirulence. Hypervirulent K. pneumoniae (hvKP) possess features that arm them to evade the host immune system to cause infections in immunocompetent hosts. hvKP are invasive and can disseminate to multiple sites.
With the World Health Organization (WHO) guidelines of treatment inadequately poised to tackle CRKP and hvKP in the neonatal population, this review presents different aspects of CRKP and hvKP and their impact on the newborn.
Neonatal Sepsis: A Disease That Can't Be Ignored
According to the “Levels and Trends in Child Mortality report, 2019” the estimated global rate of neonatal deaths was 17 per 1,000 live births, and worldwide ~6,700 neonates died each day in 2019 (6). A notable disparity exists between high-income countries (HICs) and LMICs in terms of the rate of neonatal death (1, 6). Eighty percent of the entire global burden was from two regions: Sub-Saharan Africa (42%) and Central and Southern Asia (37%). In both these regions, the neonatal mortality rate is around 24 deaths per 1,000 live births whereas in North America and Europe the rate is three deaths per 1,000 live births (6). Conditions related to infections such as sepsis and pneumonia as well as conditions not related to infections such as preterm birth complications, intrapartum-related events (e.g., birth asphyxia), and congenital anomalies are the predominant causes of neonatal death (1).
Sepsis is a dysregulated host response to systemic infections leading to shock and multi-organ dysfunction (7, 8) and may or may not be associated with a positive blood culture (9). When a pathogen can be isolated from the blood or cerebrospinal fluid of a neonate (a child within 28 days from birth) with noticeable hemodynamic changes, it is defined as neonatal septicemia (3). Overall, the rate of neonatal sepsis varies between 6.5 and 38 per 1,000 live births (hospital born only) in LMICs with bloodstream infections (BSIs) ranging between 1.7 and 33 per 1,000 live births (10). These rates are 3–20 times higher than the rates of the industrialized countries which ranges between 1 and 5 per 1,000 live births (4). The neonatal sepsis rates from underdeveloped countries are not exactly represented in the above figures because in these countries many children are born at home. In the present COVID-19 situation, hospitalization for delivery has decreased further (11). Conversion of health management facilities to COVID-19 hospitals or shutting down of medical facilities due to COVID-19 spread has caused non-availability of proper treatment to all other critical life-threatening conditions (12, 13), including neonatal care (11, 14, 15).
In the most widely accepted notion, if sepsis is manifested within 72 h of life, it is defined as Early-onset sepsis (EOS), in which case, infections are conventionally thought to be transmitted from the mother. Any sepsis presenting after 72 h of life is defined as Late-onset sepsis (LOS) and the infection in such cases is thought to be hospital or community-acquired (3, 8). Premature and low-birth-weight neonates are more susceptible to infections caused by microorganisms and thus, for low-birth-weight neonates, every infection should be considered as hospital-acquired (3). In a recent study, a significant difference observed between EOS and LOS was associated with gestational age, as premature neonates showed higher rates of LOS and it is well-known that they are at higher risk of exposure to nosocomial infections as these neonates require longer hospital stays, central venous access, and often mechanical ventilation (16). In the same study, it was also noted that birth by cesarean section was more associated with LOS than birth by normal vaginal delivery (16). In LICs and LMICs, EOS and LOS cannot be properly distinguished. Due to poor hygienic practices in labor rooms and nurseries, every infection regardless of the time of onset can be hospital-acquired (10).
The disparity between LMICs and HICs is also reflected in the etiological agents causing the infections. In LMICs like India or Jordan, the causative agents of EOS are similar to causative agents of LOS (17–19). Studies show that Gram-negative bacteria, mainly K. pneumoniae and Acinetobacter baumannii play a pivotal role in causing neonatal septicemia along with Escherichia coli and Gram-positive Staphylococcus aureus in resource-poor settings (17–21). Whereas, in developed countries, group B Streptococcus, E. coli, and S. aureus are the major pathogens causing neonatal septicemia (3, 8).
The Bug
Klebsiella pneumoniae is a Gram-negative, non-motile, and usually encapsulated bacillus of the Enterobacteriaceae family. This organism is omnipresent in the environment; K. pneumoniae is found in the soil, water, plants, and sewage. K. pneumoniae is also a part of the microbiome of the nasopharynx and gastrointestinal (GI) tract of healthy human beings (22, 23). It is an opportunistic pathogen and causes both hospital-acquired and community-acquired infections (24). In hospitals, K. pneumoniae causes both endemic infections and outbreaks of epidemic strains; chances of acquisition of K. pneumoniae in nasopharynx, GI tract, and skin increases with longer hospital stays and use of invasive devices (22). K. pneumoniae is one of the most predominant pathogens isolated from the intensive care units (ICUs) (23) and causes infections such as bacteremia, respiratory tract infections, urinary tract infection (UTI), invasive liver abscesses, endophthalmitis, and endocarditis (22, 25). Its capability of biofilm formation in the catheter enables it to cause catheter-associated UTI (26).
K. pneumoniae is also the predominant causative pathogen of neonatal sepsis (19, 27–30). Often a localized infection or colonization of the urinary tract, GI tract, or respiratory tract disseminates into the blood and leads to sepsis (26). The role of K. pneumoniae in causing neonatal sepsis is discussed later in detail.
Typing of strains is an integral part of epidemiological studies and presently multi-locus sequence typing (MLST), a method of distinguishing strains based on DNA sequences of internal fragments of multiple house-keeping genes, is prevalent. In K. pneumoniae, MLST is based on seven conserved housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) (31). The extensive drug-resistant (XDR) epidemic clones of K. pneumoniae are ST11, ST14, ST15, ST17, ST37, ST101, ST147, ST258, ST512, and these are reviewed elsewhere (32). K. pneumoniae can also be serotyped based on its capsular antigens (78 K antigens) (33).
Virulence of K. pneumoniae is essentially linked with its capsule which serves a dual purpose in the cell: it protects the cell from phagocytosis mediated by polymorpho-nuclear granulocytes and serum resistance by inactivation of one of the complement components (C3B). The pili or fimbriae is another component of the bacterial cell that helps in pathogenicity by mediating the adhesion of the pathogen to the mucosal layer and/or epithelial cells of the lower urinary tract, respiratory tract, and GI tract. Type 1 pili mediate adherence and then colonization of urinary and respiratory tract. Mannose-resistant Klebsiella-like hemagglutinin (MR/K-HA), a Type 3 pili, helps in the adhesion to Bowman's capsule, renal vessels, and tubular basal membranes of the human kidney (22). K. pneumoniae often possess large virulence plasmids (pLVPK) which harbor rmpA, rmpA2, and aerobactin biosysnthesis genes (34). With the possession of such virulence traits such as hypercapsule production, aerobactin and yersiniabactin synthesis, the bacterial cell becomes hypervirulent (34, 35).
For more than two decades, the increase in septicemia and meningitis in newborns caused by Klebsiella has been a matter of concern (22). Acquired resistance to critical antibiotics and hypervirulence in this fast-evolving pathogen is bound to make the existing scenario even more unmanageable.
The Resistance of the Bug to the Drugs
The indiscriminate use of antimicrobials has inadvertently lead to the emergence of resistance to different drugs (32, 36). The stark difference between antibiotic resistance rates of K. pneumoniae between HICs and LMICs is evident in Figure 1.
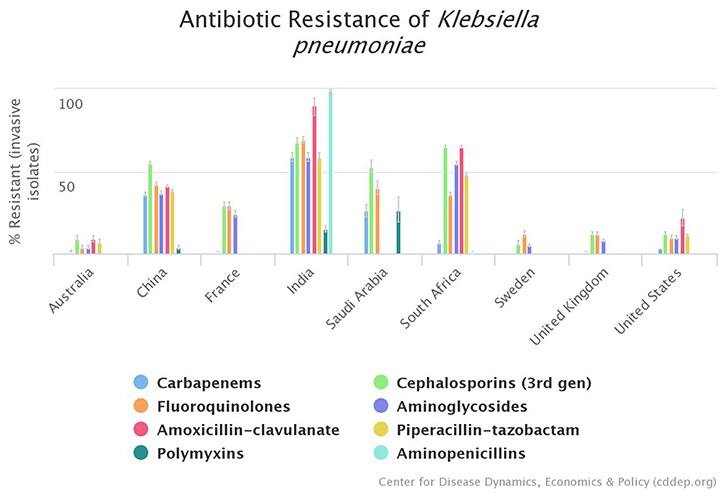
Figure 1. Graphical representation of existing disparity between high-income countries (HICs) and lower-middle-income countries (LMICs) regarding antimicrobial resistance profile of K. pneumoniae; according to Center for Disease Dynamics, Economics, & Policy (CDDEP) data (37).
Klebsiella pneumoniae is notorious for its ability to acquire antibiotic resistance determinants and it belongs to the ‘critical’ category in the WHO global priority pathogen list (38). It is one of the ESKAPE pathogens [Enterococcus faecium, Staphylococcus aureus, K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp.] which are mostly responsible for the spread of antibiotic resistance in hospital-acquired infections (36). The presence of Klebsiella in both the environment and the human body allows them to acquire a large variety of antibiotic resistance determinants. K. pneumoniae has a repertoire of around 400 antibiotic resistance genes which is almost double that of other pathogens (24). As the soil and the gut are both hot-spots for the inter-genus transfer of antibiotic resistance, K. pneumoniae has a selective advantage as it dwells in both these niches (24). Most of the antibiotic resistance determinants either appear first in K. pneumoniae or they are quickly acquired by this organism. K. pneumoniae also show higher variability in the G+C contents of its genomes than its other counterparts, indicating that it acquires external DNA from varied sources (24).
As with other Enterobacteriaceae, the majority of antimicrobial resistance (AMR) determinants are plasmid-mediated in K. pneumoniae (24, 36). Most pathogenic K. pneumoniae carry three or more AMR plasmids and the stability of the plasmids are relatively more in this organism compared to E. coli (24). The segment of the plasmid responsible for replication control to maintain a specific plasmid copy number is called a replicon (32). PCR-based replicon typing method (39) and advances in genomics have helped in recognizing plasmid types of K. pneumoniae. Various replicons have been found in K. pneumoniae, either alone or in combinations, which are IncFIIK, IncN1, IncX3, IncA/C, IncR, IncHI1-FIA, IncHIB-FIB, IncHI2, IncL (32). One of the replicons found frequently in K. pneumoniae, IncFIIK replicon, is present in multi-replicon plasmids which also possess IncFIB replicons (32). IncFIIK plasmids have a narrow host range and are rarely found outside this genus. Whereas, other replicons, IncR, IncA/C, IncX3, IncHI1 are of broad host range and thus act as a shuttle for inter-genus horizontal gene transfer (32). Apart from the transfer of genes via plasmids, mobile genetic elements such as insertion elements (e.g., IS26), transposons (e.g., Tn4401a), and integrons (e.g., Integron1), present in the plasmids, mediate mobilization of the resistance genes (often gene cassettes) between different plasmids or between chromosome and plasmids (32).
The Crkp Menace
In the 1980s, extended-spectrum β-lactamases (ESBLs) producing K. pneumoniae emerged and spread throughout the world (22, 40). This led to the use of the carbapenems (meropenem, imipenem, ertapenem), a β-lactam antibiotic, which became the antibiotic of choice to treat infections caused by ESBL-positive K. pneumoniae (40, 41). Eventually, with use of carbapenems, a new group of enzymes emerged- the carbapenemases, which could hydrolyze most of the β-lactam antibiotics including the carbapenems. The first plasmid-mediated carbapenemase IMP-1 was identified in K. pneumoniae in 1991 (42). KPC-1-producing K. pneumoniae was reported from the USA in 1996 (43). This carbapenemase was named KPC-1, for K. pneumoniae carbapenemase. Since then other carbapenemases have also emerged and CRKP has rapidly spread worldwide (23, 44).
Carbapenem resistance in K. pneumoniae or other bacteria occurs by two main mechanisms: (i) production of carbapenemases (45), and (ii) porin loss (OmpK35 and OmpK36) combined with the presence of AmpC cephalosporinases or ESBLs such as CTX-M-15 and/or overexpression of efflux pumps (21, 46, 47).
As carbapenemases are primarily mediated via mobile genetic elements contributing to the spread of carbapenem resistance, a more detailed discussion of these enzymes are done here. Carbapenemases belong to the molecular class A (e.g., KPC, GES, IMI), class B (e.g., IMP, VIM, NDM), and class D (e.g., OXA-48-like) of β-lactamases according to the Ambler classification. Molecular class A and D enzymes are called serine carbapenemases as they contain serine molecule in their active site and molecular class B enzymes are called metallo-β-lactamase (MBLs) as they contain two Zn2+ ions in their active site. MBLs cannot hydrolyze the monobactam aztreonam. Class A serine carbapenemases are predominantly inhibited by tazobactam. Class B MBLs are inhibited by EDTA, dipicholinic acid, or 1,10-o-phenanthroline in vitro. Class D carbapenemases (e.g., OXA-48-like) which hydrolyze oxacillin, cloxacillin, and carbenicillin, are inhibited in vitro by NaCl (46). As different carbapenemases have emerged over time, many have several enzymatic variants (such as KPC-2, KPC-3, NDM-5, NDM-7, OXA-48, OXA-232, IMP-4, IMP-8, etc.) with higher catalytic efficiency, stability, or better metal ion binding capacity (47). All such carbapenemases are harbored on plasmids which have shown both intra- and inter-species transmission.
KPC is a plasmid-mediated molecular class A serine carbapenemase. blaKPC gene is found within a unique transposon Tn4401 variant which has led to the mobilization of the gene to different types of plasmids and through the plasmids to other organisms (32). This carbapenemase has spread vastly in Italy, Greece (48), and the USA (49). Currently, there are 75 alleles reported in the NCBI pathogens database (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#blaKPC), of which KPC-2 and KPC-3 are prevalent. Association of the KPC-2 and KPC-3 with epidemic clone ST258 is the pivotal factor for the spread of these genes.
New Delhi metallo-β-lactamase (NDM-1) is the most widely disseminated class B metallo-β-lactamase. It was first reported in a K. pneumoniae and E. coli in 2009 and was recovered from a Swedish patient returning from India (50). Although NDM-1 was the most prevalent variant to date, variants such as NDM-4, NDM-5, NDM-7 which are more stable in the zinc-deprived condition due to an M154L mutation have rapidly emerged (51, 52). Currently, there are 31 alleles of blaNDM reported in the NCBI pathogens database (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#blaNDM). blaNDM is found across various sequence types of K. pneumoniae, no association with any particular ST was reported (53). Co-existence of many other antibiotic resistance determinants like armA, rmtB (aminoglycoside resistance), qnrB, qnrS, aac(6′)-Ib-cr (plasmid-mediated fluoroquinolone resistance), and blaCTX−M−15 (ESBL) (21, 28, 54) is often noticed in these strains. blaNDM−1 is present in varied broad host plasmids (e.g., IncX3, IncA/C, IncHIB-M/FIB-M) (32). The blaNDM gene is almost always found bracketed by a truncated or entire ISAba125 element (upstream) and a bleMBL gene (downstream) (55).
Molecular class D carbapenemases such as OXA-48 is also a very potent transferable carbapenemase emerging in Enterobacteriaceae (45). It was first reported in 2001 from Turkey in a multidrug-resistant (MDR) K. pneumoniae isolate which possessed MBLs and lacked outer membrane proteins (53). OXA-48 has now spread to all continents except Antarctica (56). OXA-48 and OXA-48-like carbapenemases (e.g., OXA-181 and OXA-232) cannot hydrolyze extended-spectrum cephalosporins and can selectively hydrolyze carbapenems (imipenem and ertapenem). Although, blaOXA−48 is associated with diverse STs, epidemic STs such as ST101, ST147, ST15, and ST395 are more common than others. blaOXA−48 is generally bracketed by IS1999 in transposon Tn1999 and blaOXA−181 is associated with ISEcp1 (57). OXA-181 differs from OXA-232 by a single amino acid substitution and both have a similar genetic environment, suggesting that blaOXA−181 is the probable progenitor of blaOXA−232. OXA-181 was first reported from Indian hospitals and is endemic to Indian subcontinent. It is now reported from Asia, Africa, Middle East, Europe, North America, and Oceania (56). OXA-232 was first isolated in France from three patients who just returned from India. The blaOXA−232 gene was carried in a ColE2 plasmid, situated within a Tn2013 transposon, downstream a ISEcp1 element (58). OXA-232 has been majorly associated with ST14, ST15, and ST16. OXA-232 is endemic in India and has now been reported from other parts of Asia, USA, Africa, and Europe (56).
IMP and VIM are two other plasmid-mediated MBLs. IMP (imipenemase) carbapenemase was first reported in the year 1991 from Japan from an Serratia marcescens strain (59). Followed by its identification in K. pneumoniae strains from Japan and Singapore, IMP-4-positive K. pneumoniae was reported from Australia in 2002 and IMP-8-positive K. pneumoniae was reported from Taiwan 2001–2002 and later from various other countries. VIM (Verona integron-encoded metallo-β-lactamases) are spread in Southern Europe and also in other countries (23). VIM-1 and VIM-2 were discovered in P. aeruginosa and later found in Enterobacteriaceae. IncN plasmid carrying blaVIM−1 was later reported from K. pneumoniae in Greece (60). Both these enzymes are associated with class 1 integrons and various insertion sequences such as IS26, IS6100 which are associated with specific plasmid types (61).
Drug-Resistant Bug and the Newborn
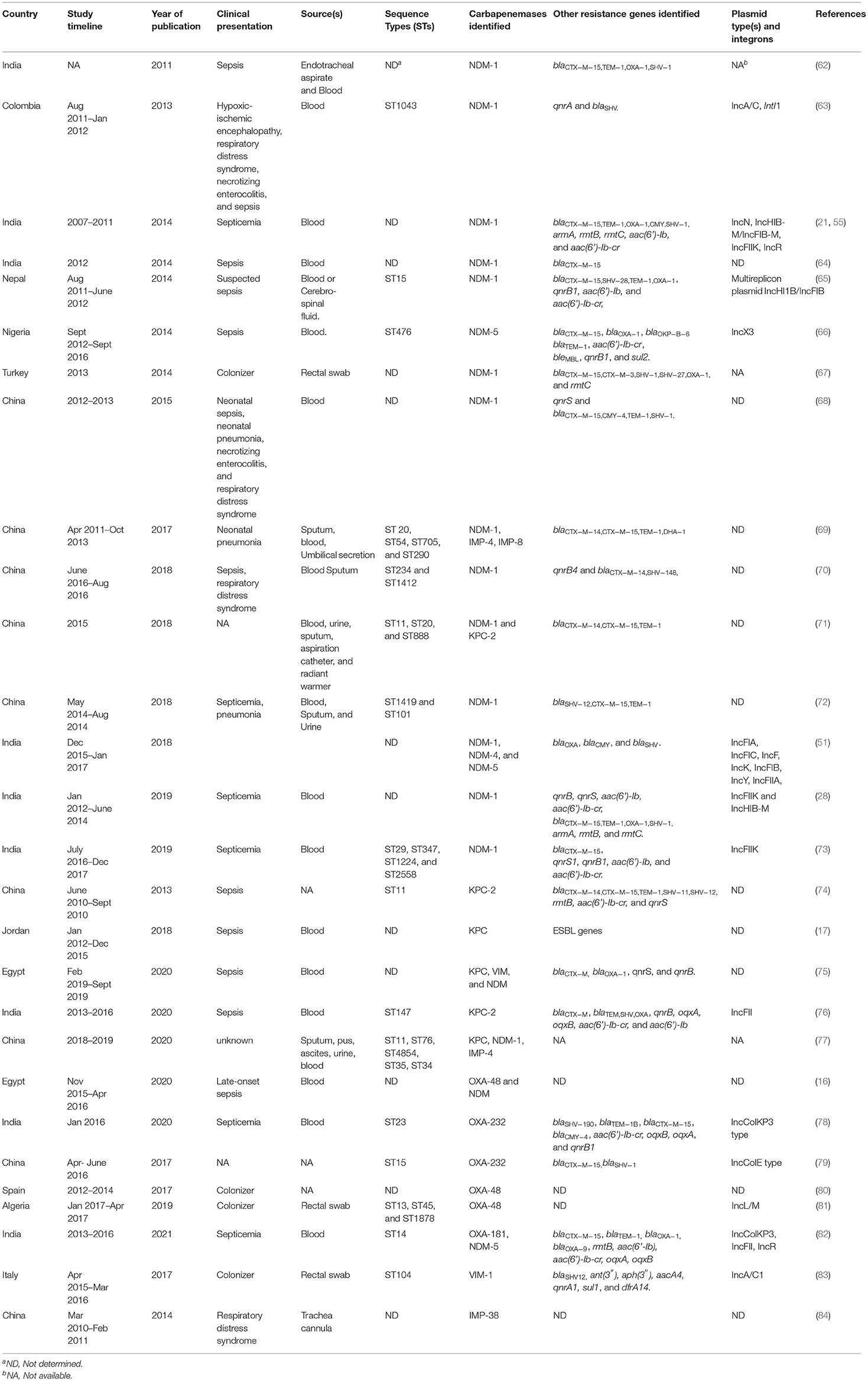
Various studies have been published regarding the spread of carbapenem-resistant Enterobacteriaceae but the data on neonatal sepsis is infrequent. Here we present the studies related to neonatal sepsis caused by CRKP (Table 1). We have focused on the major carbapenemases such as KPC, NDM, and to some extent OXA-48, IMP, and VIM. The genetic aspects of these carbapenemases are already discussed in the above section.
Over the last few years, KPC-producing K. pneumoniae have been reported in neonates from various countries such as Egypt (75), Jordan (17), China (71, 74, 77), and India (76). blaKPC−2-positive K. pneumoniae caused infections in China and India. The sequence types of the corresponding strains from China and India were however different: strains from China belonged to ST11 whereas, strains from India belonged to ST147 (76). blaKPC was harbored in large plasmids along with other resistance determinants such as blaCTX−M−15, TEM, SHV, OXA−1, rmtB, aac(6′)-Ib-cr, qnrB, and qnrS. The genetic environment of blaKPC−2 in ST147 strains from India corroborated with the genetic environment of the other blaKPC−2-possessing strains recovered from adults, where blaKPC−2 was associated with IS elements ISKpn6 and ISKpn7, plasmid type IncFII, and transposon Tn 4401. All neonates in this study had an overlapping stay in the hospital so a chance of transmission from one neonate to another was predicted (76). In one of the above studies from China, three of the four infants died due to the infection caused by KPC-producing K. pneumoniae. Only one neonate responded to the therapy of amikacin in combination with imipenem (74). The other study from China mentioned the isolation of KPC-producing CRKP not only from blood but also from sputum, urine, aspiration catheter, and hospital environment, indicating that the hospital environment can harbor CRKP strains which may cause disease later (71). The study from Jordan interestingly showed that infection due to KPC-producing K. pneumoniae and Acinetobacter spp. led to higher mortality and previous exposure to carbapenems and vancomycin significantly increase this risk (17). Further, the study from Egypt showed that neonatal mortality was inversely related to gestational age and birth-weight. The same study also showed that neonates who eventually succumbed to the infection had a significant reduction in platelet count and hemoglobin levels (75).
Shortly after the report of blaNDM−1 in an adult patient in 2009, blaNDM−1−possessing K. pneumoniae causing neonatal septicemia was identified in 2011 from India (62). Three retrospective studies from the same unit showed that carbapenem-resistant K. pneumoniae not only persisted in the unit but gradually became the most predominant carbapenem-resistant organism causing septicemia (21, 28, 55). One of these studies also reported the in vivo interspecies plasmid transfer event of blaNDM−1 in a neonate from whom Enterobacter cloacae was isolated initially and E. coli later. The study showed that the blaNDM−1 plasmid in both the species was identical indicating its possible transmission between Enterobacter cloacae and E. coli (55). The study of Datta et al. reported that male sex, low birth weight, birth at extramural centers were significantly associated with sepsis caused by NDM-1-positive isolates. However, sepsis caused due to these isolates did not result in a higher mortality rate (21). NDM-positive K. pneumoniae-mediated neonatal infections are now reported from India (51, 85), Nepal (65), China (68–70, 72, 77), Nigeria (66), Colombia (63), and Turkey (67). Although blaNDM−1 was the most prevalent allele to date, alleles such as blaNDM−4 and blaNDM−5 are slowly emerging in K. pneumoniae causing neonatal infections (51, 52). It was observed that isolates harboring blaNDM−1 are not associated with any particular sequence type and these strains belonged to varied STs (ST15, ST17, ST20, ST29, ST76, ST101, ST234, ST347, ST433, ST476, ST888, ST1043, ST1412, ST1419, ST1224, ST2558, ST4854) (Table 1). These isolates also possessed several antibiotic resistance determinants such as armA, rmtB (aminoglycoside resistance), qnrB, qnrS, aac(6′)-Ib-cr (plasmid-mediated fluoroquinolone resistance), blaCTX−M−15 (ESBL) (21, 28, 86) along with blaNDM−1 gene in different plasmid types such as IncFIIK, IncHIB-M, IncFII, IncFIA, IncFIB, IncF, IncA/C, IncL/M, IncA/C, IncX3, etc. (28, 51).
Isolates harboring blaNDM−1 have also caused outbreaks in several healthcare settings. During August 2011–Jan 2012, an outbreak occurred due to K. pneumoniae ST1043 in a neonatal unit in Colombia infecting six neonates. As the neonates had no contact with people from countries that reported NDM-1-producing bacteria, the authors proposed that autochthonous clones were acquiring the blaNDM gene (63). Another outbreak around the same time was reported from Nepal by a blaNDM−1-positive K. pneumoniae ST15. This outbreak caused high mortality among the neonates. Apart from the outbreak cluster, three smaller genetically close clusters were also identified in this study (65). An outbreak of NDM-5-producing K. quasipneumoniae was reported from Nigeria in April 2016. The outbreak occurred when the neonatal ward was overcrowded and less critical neonates often shared cots. blaNDM−5 gene was carried on an IncX3 plasmid (66). Five separate studies from different parts of China (Nanjing, Wuhan, Hunan, Jiangsu, Shandong) reported outbreaks of blaNDM−1-possessing K. pneumoniae in neonatal units. Strains were isolated from blood or sputum or umbilical secretions of neonates and belonged to different sequence types (68–70, 72, 77). In another study from China, five CRKP isolated from neonates (blood, urine, and catheter tips) possessed blaNDM−1 and belonged to ST20 (n = 4) and ST888 (n = 1), which were susceptible to gentamicin, amikacin, aztreonam, ciprofloxacin, and levofloxacin. NDM-1-producing ST20 strains (n = 2) were also isolated from the hospital environment (71). A systematic review from China on carbapenem-resistant Enterobacteriaceae reviewed seventeen studies of neonatal infections among which seven studies were of NDM-1-producing K. pneumoniae (87).
OXA-48-positive CRKP causing neonatal infection or intestinal colonization was reported from Algeria, Spain, and Egypt (75, 80, 81). In the study from Algeria, blaOXA−48-carrying K. pneumoniae of two different STs (ST13 and ST1878) were found colonizing the gut of the neonates in two maternity wards. Carriage of carbapenem-resistant strains was significantly related to the low-birth-weight of the neonates (81). In the study from Egypt, K. pneumoniae was the predominant organism causing LOS but not EOS. Eventually, the mortality was significantly higher in neonates suffering from LOS (16). OXA-232, another OXA-48-like enzyme, was reported from a hypermucoviscous K. pneumoniae causing septicemia from India. The gene was carried in a ColKP3 type plasmid (73). Another clonal outbreak of OXA-232-producing K. pneumoniae ST15 was reported from a NICU in Shanghai, China (79).
The other carbapenemases such as IMP or VIM have not been reported frequently in neonatal infections. IMP-38-positive CRKP causing respiratory distress syndrome in neonates was reported from China. The nine IMP-38-positive strains were clonal and were isolated from the trachea cannula of neonates. blaIMP−38 was a novel allele and differed by a single mutation from IMP-4 which was found in the other wards of the same hospital (84). In another recent study from China, fourteen blaIMP−38-possessing K. pneumoniae ST307 were recovered from neonates suffering from sepsis (88). Another variant, IMP-4 has also been reported from China causing neonatal infections (69, 77, 87). VIM-positive CRKP was recovered from the neonates in the USA, Italy, and Egypt (75, 83, 89). In the study from Italy, VIM-1, associated with an IncA/C plasmid, was primarily found in K. pneumoniae ST104 recovered from the rectal swabs of neonates admitted in a NICU from 2015 to 2016. The strains were susceptible to fluoroquinolones, amikacin, and colistin (83).
It is noteworthy that earlier studies reported the presence of a single carbapenemase in K. pneumoniae but in recent years reports of co-occurrence of multiple carbapenemases are emerging (16, 75, 82). In a recent study from India OXA-181/OXA-232 was concomitantly present with NDM-5 in K. pneumoniae causing neonatal septicemia (82). A study from Egypt also reported (previously mentioned) presence of blaNDM and blaVIM in K. pneumoniae causing LOS. blaKPC was also present in 96% of these strains (75). Another (previously mentioned) study from Egypt reported the presence of blaOXA−48 in 61% strains and co-occurrence of blaNDM and blaOXA−48 in 52% strains (16). The presence of multiple carbapenemases and other resistance genes pose additional limitations to the treatment protocols.
Hypervirulent K. Pneumoniae (hvKP)—Another Dimension to the Problem
hvKP and cKP: We Beg to Differ
Recently, K. pneumoniae has gained a revised and serious global attention due to the emergence of the hypervirulent pathotype. Over the last few decades, the majority of hospital-acquired infections reported globally were due to the classical K. pneumoniae (cKP). However, since the mid-1980s, the emergence of hypervirulent K. pneumoniae (hvKP) in the clinical context poses a far greater challenge to the clinicians (90). Although, both cKP and hvKP pathotypes have their own global importance, the incidence of infections due to hvKP has been reported at an escalating rate over the last three decades (91). Unlike cKP, hvKP pathotypes are more virulent and have the potential to cause several community-acquired invasive, life-threatening, and unusual infections, such as pyogenic liver abscess, lung abscess, meningitis, endophthalmitis, brain abscess, and necrotizing fasciitis in otherwise healthy adults (92). Initially, hvKP infections were reported primarily from Taiwan and South-East Asia, but several sporadic reports of hvKP have now been observed in other Asian, European, and American countries (25, 91, 93–98). Although hvKP cause community-acquired diseases, some current reports argued that the infiltration of these notorious strains is increasing in the healthcare settings also (99–104). Infections due to hvKP are found to be more complicated due to their ability to metastatically disseminate to other organs or systems and subsequently cause multiple sites of infection (90). This type of dissemination is common for some selected Gram-positive pathogens, such as S. aureus, but it is unusual for an enteric Gram-negative bacillus to involve multiple sites of infection (105). In addition, unlike cKP, hvKP pathotypes commonly possess a hypermucoviscous phenotype, produce a robust capsule, synthesize several iron-scavenging siderophore molecules, especially salmochelin and aerobactin, and harbor several chromosomal and large virulence plasmid-encoded factors (91, 106). Due to their enhanced virulence, these K. pneumoniae are considered hypervirulent. An overview of the differences between cKP and hvKP strains is depicted in Table 2.
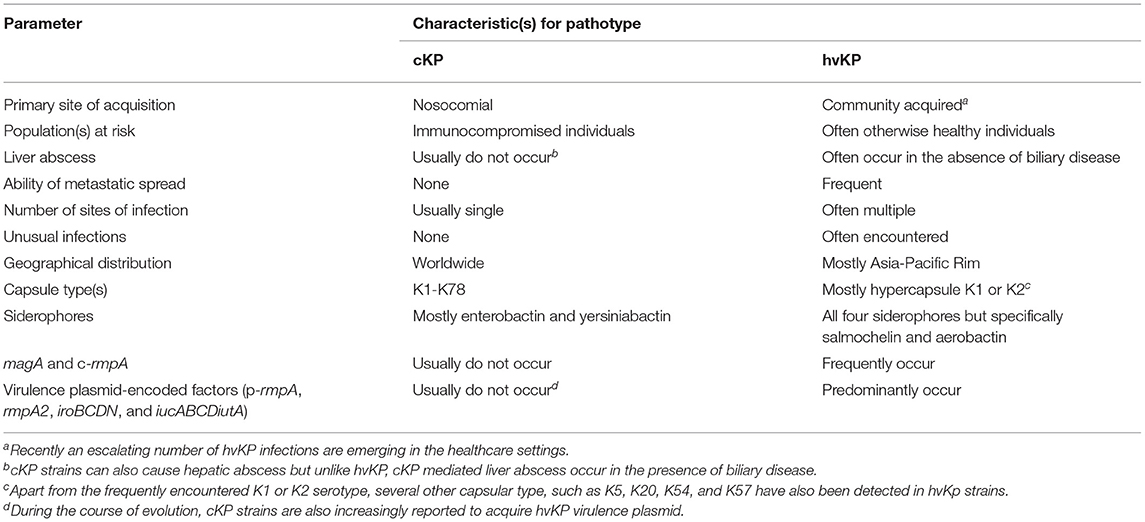
Table 2. Characteristic features of classical K. pneumoniae (cKP) and hypervirulent K. pneumoniae (hvKP) strains.
hvKP and cKP: Molecular Markers
Recently, the advancement of whole-genome sequencing (WGS) in the clinical context has identified a set of important and unique hypervirulent biomarkers which can accurately differentiate hvKP from cKP, including several capsular serotypes (K1, K2, K5, K20, K54, and K57), mucoviscosity-associated gene A (magA), regulator of mucoid phenotype A (rmpA) genes, biosynthetic genes responsible for the production of siderophore salmochelin (iro cluster), aerobactin producing biosynthetic genes (iuc cluster), virulence plasmid-encoded gene with unknown function (peg-344), and the virulence plasmid-encoded heavy metal resistance genes for tellurite (ter cluster) and silver (silS) (78, 91, 106). Due to presence of these factors, hvKP strains are more resistant to macrophage- or neutrophil-mediated phagocytosis & the complement-mediated bactericidal activity of human serum, they exhibit enormous biofilm-forming capability, and also display enhanced lethality in the in vivo mouse model compared to cKP (107). In the following section, we discuss briefly the several hvKP-specific biomarkers detected till date which are responsible for the increased virulence and severe clinical expression.
(a) Capsular Serotypes
Capsule is the most established virulence factor of K. pneumoniae. Both cKP and hvKP possess chromosomally-encoded capsular polysaccharide (K antigens) genes, known as the cps cluster (108). Till date, around 78 capsular serotypes (K1–K78) have been reported in K. pneumoniae (33). However, majority of reports have shown the strong association of K1 and/or K2 serotypes with hvKP strains (109, 110). Recent studies, especially from China, reported the occurrence of K2 serotypes in at least 70% and 42% of hvKP strains (93, 111) and K1 in 24% (93). Why are the increased incidences of virulence associated typically with K1 and K2 serotypes? There are several explanations for this. One study suggested that strains with K1 and/or K2 serotypes have better survival probably because of the induction of slightly greater amount of reactive oxygen species released by neutrophils than other serotypes (112). In addition, several studies have argued that the strains of the K1 and K2 serotypes are significantly more resistant to phagocytosis and subsequently intracellular killing by macrophages and neutrophils than other serotypes (113, 114). Moreover, others have suggested reduced uptake of K1 and K2 serotypes by the innate immune cells probably due to the presence of a significant amount of sialic acid on their surfaces, which may mimic the host cell and allow them to easily evade the immune response (115). Apart from the K1 and K2 serotypes, recent studies revealed the occurrence of other serotypes, such as K5, K20, K54, and K57 in hvKP strains causing various invasive infections (91). However, significant reports of immune evasion are still scarce for these capsular types.
(b) Major Players in Hypercapsule Production: rmpA and magA
The capsule is found in both cKP and hvKP, but the hvKP strains produce an increased amount of capsular polysaccharide compared to that of cKP. This robust capsule synthesis in hvKP strains is chiefly mediated by the rmpA and magA genes. In hvKP strains, a total of three rmpA genes are generally found, of which two are large virulence plasmid-encoded (p-rmpA and p-rmpA2) and one is chromosomally-encoded (c-rmpA) (116). Several reports suggested that the rmpA genes along with the regulation of capsule synthesis A and B genes (rcsAB) can induce the transcription of entire cps operon, resulting in hypercapsule production (106). In separate studies, Hsu et al. and Li et al. revealed that about 55–100% hvKP strains express at least one copy of rmpA or rmpA2 (116, 117). In the absence of rmpA or rmpA2, hypercapsule biosynthesis can be triggered alone by the chromosomally-encoded magA gene which was isolated from hypermucoviscous liver abscess-causing K. pneumoniae (118, 119). Subsequent bioinformatics and genetic experiments determined that magA is a K1-specific factor and encodes a polymerase gene termed wzy in the cps operon (118, 120, 121).
(c) The Iron Scavengers: Aerobactin and Salmochelin
Iron, a crucial and limiting metal, essential for bacterial growth and plays a pivotal role in the progression of bacterial infection, especially in the case of K. pneumoniae. However, this essential metal is not readily available in the host during the infection because of the non-specific immune response exhibited by the host where the host efficiently sequesters this metal ion (Fe3+) with several iron-binding molecules, such as transferrin and lactoferrin, eventually restricting the growth of many opportunistic pathogens (122). Therefore, to acquire iron from such an iron-poor environment, K. pneumoniae secrete several small proteins with high iron-scavenging ability, called siderophores (123). Molecular epidemiological studies have shown that hvKP strains produce all the siderophores (enterobactin, yersiniabactin, salmochelin, and aerobactin) compared to cKP which produce only enterobactin and yersiniabactin (124). Apart from producing all siderophores, hvKP strains are also capable of synthesizing quantitatively more siderophores than cKP (125, 126). Among the four siderophores, the activity of enterobactin and yersiniabactin is greatly hindered by host molecule lipocalin-2 and transferrin, respectively (127, 128). Therefore, K. pneumoniae with only enterobactin and/or yersiniabactin are unlikely to cause systemic infection in immunocompetent individuals (128). In contrast, the functionality of both salmochelin and aerobactin cannot be inhibited by these host proteins. Several studies have shown that salmochelin is more prevalent in hvKP strains, sometimes more than 90% of pyogenic liver abscess-causing hvKP strains possess this scavenger (128, 129). Aerobactin, a citrate-hydroxamate siderophore, is also rarely detected in cKP (124). Studies revealed its presence in 93–100% of hvKP strains (130). Immense genome-based analysis in the molecular epidemiologic studies confirmed that in most cases iro gene cluster for salmochelin and iuc gene cluster for aerobactin specifically reside on the large virulence plasmid of hvKP (131).
(d) Plasmids That Matter: pLVPK
WGS analysis of initially identified hvKP strains revealed the presence of the unique ~220 kb large virulence plasmids pK2044 (224,152bp) and pLVPK (219,385bp) (34, 35). All the best described genetic markers which confer the hypervirulent phenotype are located on these plasmids, including complete biosynthetic gene cluster of aerobactin (iucABCDiutA) and salmochelin (iroBCDN), regulators of hypercapsule production (rmpA, rmpA2, and rcsA), resistance genes for tellurite (terZABCDE and terWXY) and silver (silS) (Figure 2). Recently, bioinformatics studies revealed that all the hvKP strains possessed either pK2044-like or pLVPK-like plasmid. Struve et al. showed that all 30 studied community-acquired liver abscess-causing hvKP strains harbored pLVPK-like plasmids (131). Similarly, another study revealed the presence of pK2044-like plasmids in 94 hvKP strains (132). The lateral gene transfer in bacteria is largely mediated by plasmids and as the hypervirulent biomarkers mostly reside in the non-conjugative virulence plasmids, it is quite likely that hvKP strains acquire drug-resistant plasmids from the cKP strains due to their conjugative nature and this molecular incidence is now increasingly evident in recent studies.
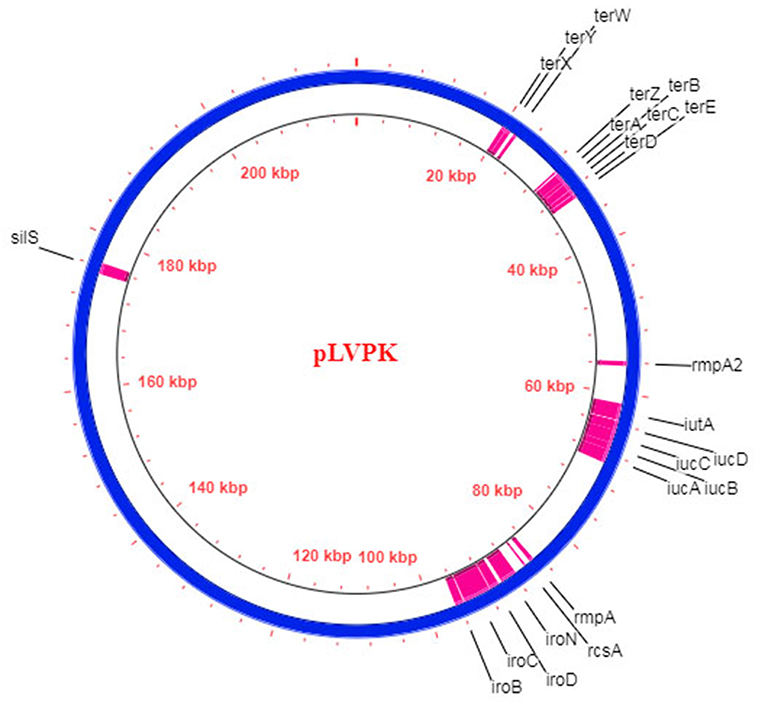
Figure 2. Schematic diagram of the hvKP virulence plasmid pLVPK (Blue circle; 219,385 bp) (35). The respective CDS of the hypervirulent biomarkers are demarcated in pink.
(e) hvKP and Sequence Type 23 (ST23): In Search of an Association
Several studies showed that the genes conferring hypervirulence are widely distributed across diverse STs (133) but some selected STs, such as ST23, ST65, and ST86 are found to be predominantly associated with hvKP (134). Recently, core genome multi-locus sequence typing (CG-MLST) and/or WGS revealed that strains of clonal group 23 (CG23) are strongly associated with K1 capsular type causing severe and invasive disease which occur in typical hvKP infection (131). ST23 was frequently associated with hvKP strains especially in the Asia-Pacific Rim. Studies from China and South Korea show that majority of the hvKP strains belonging to ST23 possessed K1 capsular type (134, 135). Recently, a study from India in 2020 also described a case of community-acquired neonatal septicemia caused by a carbapenem-resistant ST23 hvKP strain (78). Although the reasons behind the association of the hvKP and/or K1 capsular type with ST23 are still uncertain, it is hypothesized that the CG23 probably has a discrete genetic infrastructure that confers the hypervirulence. However, in-depth research will be required to fulfill the existing knowledge gaps.
The Next Generation Super-Bug: Carbapenem-Resistant hvKP (CR-hvKP)
It is debatable whether the association of virulence and antibiotic resistance is deleterious or helpful for the microbe (90, 124). However, recent reports on K. pneumoniae that are both antimicrobial-resistant and virulent have surely put an end to this debate. K. pneumoniae are extremely capable of receiving and incorporating DNA segments from other bacteria, mostly via large plasmids (26). Carbapenem resistance genes, as discussed earlier can easily spread across species via these mobile genetic elements. Their presence in hvKP has challenged the health system and the emerging CR-hvKP pathotypes are now being considered as “the next generation super-bug.” The epidemiological data of some CR-hvKP strains is given in Table 3 and the worldwide distribution of CR-hvKP strains is depicted in Figure 3.
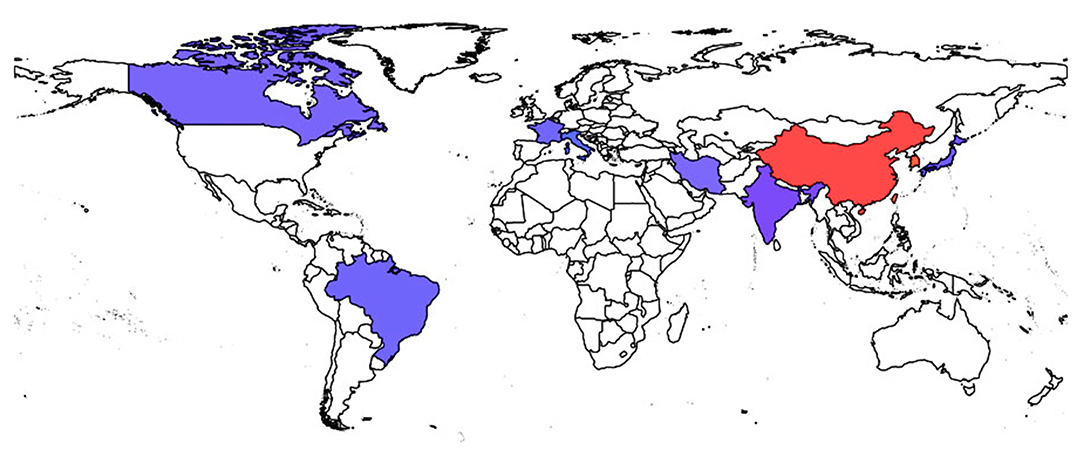
Figure 3. Worldwide spread of CR-hvKP strains. Endemic spread of CR-hvKP strains were reported from the countries indicated in red while the blue colored regions represent the sporadic occurrence of CR-hvKP strains.
From the standpoint of the microbe, the convergence of resistance and virulence is beneficial particularly under antibiotic pressure. Exchange of genes can happen in two ways, hvKP strains can acquire the antibiotic-resistant plasmids from the XDR K. pneumoniae strains (155) or the XDR cKP strains can acquire hvKP-specific virulence plasmids (145). The first option seems more plausible given the fact that majority of pLVPK-like plasmids are non-conjugative, lacking the transfer (tra) genes (131, 134). On the other hand, carbapenem resistance genes are primarily found on conjugative plasmids. However, some recent in-depth investigations on hvKP revealed that due to the immense antibiotic selection pressure and an extraordinary ability of K. pneumoniae to acquire highly antibiotic-resistance- and hypervirulence-encoding genetic determinants, new hybrid and/or conjugative plasmids with both carbapenem-resistant and hypervirulent markers have emerged in the K. pneumoniae strains as a result of convergent evolution. This molecular phenomenon has eventually led to the emergence of new and perilous variants of CR-hvKP strains (156, 157).
Since 2015, multiple epidemiological studies from China showed the prevalence of KPC-2-producing CR-hvKP strains in the clinical settings causing several infections in otherwise healthy individuals, including UTI, pneumonia, septicemia, bacteremia, abdominal infections, septic arthritis, catheter-associated bacteremia, and ventilator-associated pneumonia (140–142, 144). Majority of the reported blaKPC−2-harboring CR-hvKP strains from China belonged to ST11. Although ST11 clone of K. pneumoniae is an internationally categorized high-risk clone known to harbor blaKPC−2 as the major carbapenemase gene, these recent studies have confirmed its association with the hypervirulence attributes.
Typical to hvKP, pLVPK-like virulence plasmid pVir-CR-hvKP4 was identified. However, compared to the pLVPK, pVir-CR-hvKP4 had a 41.231 bp deletion. The subsequent plasmid-curing experiment suggested that this modified pLVPK-like plasmid was responsible for enhanced virulence phenotype both in vitro and in vivo (145). Although majority of the CR-hvKP reports were initially detected with blaKPC−2, recently the occurrence of other classes of carbapenemases (Class B -MBL-type and Class D—OXA-48-like) have also been detected in hvKP strains. For example, a recent study from China showed the emergence of an ST86 CR-hvKP K2 strain that co-harbored two different classes of clinically important carbapenemase-producing genes blaNDM−1 and blaKPC−2. This strain was also responsible for significant mouse lethality (148), refuting the fact that association of resistance and virulence is deleterious for the microbe.
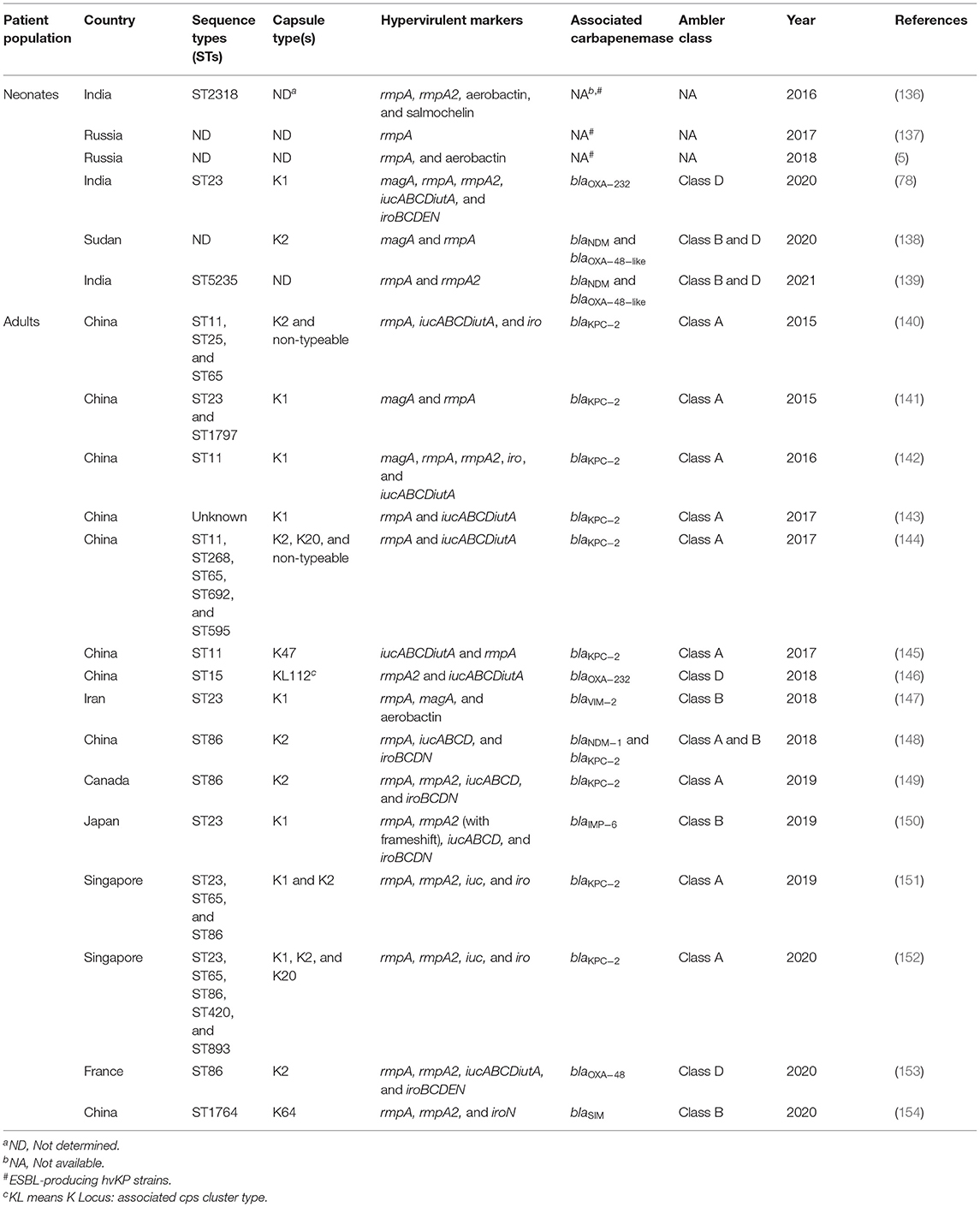
hvKP/CR-hvKP and the Newborn
Till date, majority of the cases of hvKP and/or CR-hvKP infections have been reported in adults and reports regarding hvKP infection in cases of neonatal sepsis are just a handful (Table 3). When resistance and virulence meet to cause infection in a vulnerable population, the outcome is fatal, as was evident from most neonatal studies (5, 137, 139). Previous studies have concluded that the risk of development of severe forms of neonatal infection can be associated with virulence factors (5). In 2016, a study from India reported the occurrence of an ESBL-producing hvKP ST2318 strain causing neonatal sepsis. The strain was isolated from the blood of a very low-birth-weight neonate suffering from hepatosplenomegaly with severe thrombocytopenia, coagulopathy, and metabolic acidosis. Cefoperazone-sulbactam and amikacin were administered as an empirical treatment which was later changed to meropenem with amikacin after receiving the clinical microbiology report. The neonate was discharged after proper medical supervision. Genome analysis revealed that the strain harbored rmpA, rmpA2, and several siderophores, including aerobactin, enterobactin, yersiniabactin, and salmochelin (136). Khaertynov et al. in 2017 reported another fatal case of pyogenic neonatal meningitis caused by an ESBL-producing hvKP. The strain was isolated from the blood and cerebrospinal fluid of a 12-day-old neonate exhibiting several clinical symptoms, such as pallor, loss of appetite, fever, seizures, and a bulging anterior fontanelle. In addition, low Pediatric Glasgow Coma Scale (PGCS) score indicated that the neonate suffered from severe brain injury. Though the newborn was initially treated with ampicillin and amikacin it was changed to meropenem and later cefoperazone due to unsatisfactory clinical outcome and isolation of ESBL-producing K. pneumoniae from the infection sites. Apart from antibiotic treatment, IgM-enriched intravenous immunoglobulin and infusion therapy were also given to the neonate but the neonate succumbed to the infection. Laboratory-based characterization revealed that the strain exhibited hypermucoviscous phenotype and also harbored rmpA gene (137). The following year another report from Kazan, Russia revealed the occurrence of ESBL-producing neonatal septicemic hvKP strains in the clinical settings with fatal outcome, harboring rmpA and aerobactin biosynthetic genes. In this study, two groups of neonates were registered, infected by K. pneumoniae. The first group comprised of 10 neonates with culture-positive sepsis and the second group consisted of neonates with UTI. All the 10 septicemic neonates received a comprehensive treatment, including antibiotics, intravenous immunoglobulins, and infusion therapy. However, three neonates died despite the therapy, of which two suffered from purulent meningitis and one from necrotizing enterocolitis. The most grievous forms of sepsis in neonates, were correlated with meningitis (5). The first report of CR-hvKP was in 2020 from our laboratory showing the occurrence of a unique community-acquired neonatal septicemic case caused by a hypermucoviscous CR-hvKP ST23 K1 strain. The neonate was low-birth-weight, delivered at home in a rural area and was given cow milk since birth. During admission, the baby displayed several clinical signs and symptoms of sepsis. As an empirical treatment, piperacillin/tazobactam and netilmicin were given to the neonate. The baby was hospitalized for 22 days before taking a high-risk discharge. The strain displayed all hvKP-associated features, including hypermucoviscous phenotype, magA, rmpA, rmpA2, rcsAB, iroBCDEN, and iucABCDiutA. In addition, the strain also harbored blaOXA−232 as the candidate carbapenemase gene. Molecular typing and WGS analysis revealed that the studied strain belonged to an internationally high-risk clone, ST23 and possessed a pLVPK-like virulence plasmid sequence in the studied genome. Moreover, comparative genomic analysis with the other hvKP ST23 reported genomes showed >99.5% of inter-genomic resemblance (78). The virulence determinants such as magA, rmpA, rmpA2, iroN, iucA, and iutA were not transmitted along with the carbapenem resistance gene, blaOXA−232, in conjugation experiments. However, the possibility of such transmission is worrisome. The same year another study from Khartoum, Sudan revealed that about 16.7% CR-hvKP strains were recovered from the blood of septicemic neonates and adults. Molecular characterization suggested that among the recovered CR-hvKP strains, eight were positive for blaOXA−48−like gene and two for blaNDM. Moreover, the strains belonged to K2 capsular serotype and also harbored rmpA and magA (138). In 2021, a study from India showed the occurrence of nine XDR hvKP ST5235 strains causing sepsis in neonates. All the neonates in this study were empirically treated with piperacillin/tazobactam and amikacin followed by imipenem/meropenem and vancomycin. However, 100% neonatal mortality was recorded in this study even after the treatment with polymyxin B. Molecular characterization showed that the strains harbored rmpA/rmpA2 in various combinations. Although all strains showed resistance against carbapenems in phenotypic tests, blaOXA−48−like gene and blaNDM were detected only in three strains (139).
Clearly, more and more reports of hvKP and CR-hvKP in cases of neonatal sepsis are being published. Clinical laboratories do not evaluate K. pneumoniae on the basis of their hypervirulent features and thus such strains are underreported. In neonates, the combined effect of an immature immune system, use of several invasive devices, the involvement of extensively drug-resistant cKP, and the emergence of hvKP/CR-hvKP is worrisome. Comprehensive analysis of cKP and hvKP, their differences, and also the distinguishing biomarkers for hvKP will probably help the clinicians in future to take a more strain-specific therapeutic approach.
The Possible Acquisition of cKP and hvKP in Neonates
Just like the other members of the Enterobacteriaceae family, K. pneumoniae is also found in the normal human microbiota (158). Although the acquisition of K. pneumoniae appears to be obligatory for the infection, this scenario is not obvious always (159). The time of pathogenic exposure, bacterial inoculum size, immune status of the individual, and the virulence potential of the causative microorganism collectively manifest the clinical expression of an infection (3). At birth, the neonates do not possess their endogenous microbiota which is naturally acquired through the perinatal transfer of the maternal flora and from environmental sources. Due to the lack of an established microbiota, immaturely developed gut barrier, and the high permeability of the mucosa in the GI tract, neonates get infected easily, particularly low-birth-weight and premature neonates requiring prolonged hospitalization (160). The probable modes of infiltration of K. pneumoniae in the neonates are largely accomplished by in-utero acquisition, acquisition from maternal flora, and postnatal acquisition from the hospital or community (3).
(a) K. pneumoniae Acquisition From Maternal Flora
In-utero infection in neonatal septicemic cases is largely reported to be a result of chorioamnionitis (161). This is an acute inflammation of fetal membranes, probably due to the bacterial infiltration and this clinical condition is often found to be caused by two specific genital mycoplasmas, such as Ureaplasma parvum and Ureaplasma urealyticum (3). The vaginal microenvironment also provides a suitable ground for the colonization of pathogenic bacteria. Ascending movement of these microorganisms followed by the premature rupture of amniotic membrane results in the contamination of amniotic fluid. The inhalation or swallowing of the infected amniotic fluid by the neonate can subsequently lead to the colonization of pathogenic K. pneumoniae in their gut which can lead to intrapartum sepsis (162–164). However, reports regarding in-utero infection due to K. pneumoniae are extremely scarce. Two exclusive studies confirmed the occurrence of K. pneumoniae in intrauterine infection. In the first study in 2005, Sheikh et al. reported a unique case of acute chorioamnionitis and acute villitis due to K. pneumoniae infection in a 40-year-old woman at 18 weeks of gestation (165). Torabi et al. also reported a case of severe chorioamnionitis with umbilical cord and chorionic plate vasculitis due to K. pneumoniae in a woman who had suffered from fetal expiration at 20 weeks of gestation (166).
Acquisition can also frequently happen during the process of vaginal delivery, the neonates might get infected by the pathogens residing in the birth canal (167). Besides, the neonates can also acquire maternal flora through the contaminated hands, from the skin during the kangaroo care, and/or from the infected breast milk. Recent studies have revealed the incidence of acquisition of ESBL- or carbapenemase-producing K. pneumoniae from maternal flora to the neonates. For example, a study from Italy in 2017, reported a case of mother to child transmission of KPC-3-producing CRKP at birth (168). In 2018, Herindrainy et al. reported that the acquisition of about 24% (n = 20) of ESBL-producing K. pneumoniae detected in the neonates were from the maternal gut flora (169).
(b) Nosocomial Acquisition of K. pneumoniae
Till date, the majority of neonatal septicemic cases from LMICs are reported to be caused by hospital-associated drug-resistant classical K. pneumoniae strains which we have already discussed earlier. In several studies, we also have noticed the prevalence of hospital-acquired K. pneumoniae and/or CRKP infections in the neonates. The MDR or XDR nature of the K. pneumoniae strains offer them an immanent selective advantage by which they can easily persist in the flora of hospitalized patients as well as in the nosocomial environment (170). In a study from India, Das et al. revealed that among the studied bacterial strains, K. pneumoniae was detected as the predominant microorganism colonizing the neonatal gut. Neonates with longer stay in the NICU and those with prolonged feeding through an enteral tube were predisposed toward such colonization. Moreover, molecular typing showed that about 50% of the K. pneumoniae strains isolated from the blood were genotypically identical with their gut counterparts, reflecting the possible association between gut colonization and neonatal sepsis (162).
(c) Acquisition of K. pneumoniae From the Community
Till date, the majority of community-acquired K. pneumoniae have been reported to cause infections in healthy adult individuals. But the reports regarding community-acquired K. pneumoniae infections in neonates are very limited. In a study, Waters et al. showed that about 11.6% of K. pneumoniae strains caused community-acquired neonatal sepsis in the LMICs (171). In another study, Khatri et al. reported a case of community-acquired pyelonephritis caused by a KPC-2-producing K. pneumoniae (172). Recently, in a study we reported a unique case of CR-hvKP, causing sepsis in a neonate who was delivered at home and was given cow milk after birth (also discussed earlier). In that study, we hypothesized that the consumption of cow milk after birth was probably a cause of community-acquired hvKP infection (78). Due to various factors like low female literacy rate, unavailability of clinicians and other healthcare workers, the inadequate number of hospitals and/or well-equipped healthcare facilities, improper transport system, and most importantly inequality in the society, some of the neonates in the LMICs are still delivered either at home or at resource-poor primary care facilities where they might acquire infections from the environmental sources.
Detection of CRKP and hvKP: Seek and You Shall Find
The initial detection of carbapenemase-producing organisms in a lab is carried out by the trusted disk diffusion method which uses a carbapenem disk placed on a lawn of sample organism (173, 174). If there is a zone of inhibition, the diameter is measured and according to the cut-off mark delineated by CLSI (175) or EUCAST (176), the organism is declared resistant, or intermediate, or susceptible. Automated machines such as VITEK® 2 from bioMérieux and Phoenix™ from BD are also frequently used in setups that can afford them. Phenotypic detection methods (though some are still not cost-effective) may be considered in resource-poor settings. A suitable test should be chosen based upon the sensitivity, specificity, turn-around time, and most importantly, cost of the test. Some phenotypic methods suitable for detection of carbapenemase-producing organisms are presented here in brief, exhaustive reading on these methods are available elsewhere (177). For easy detection of KPC and MBLs, combined disk test was developed. This test uses a disk of meropenem and another disk of a meropenem supplemented with an inhibitor (EDTA/ aminophenyl boronic acid/ dipicholinic acid). If the difference between the zone of inhibition of the two disks is >5 mm, a positive result is indicated (178). These tests are now commercially available (179, 180). mCIM uses a simple method and has high sensitivity and specificity. In this test, a meropenem disk is incubated with 1 μl loopful bacterial culture in Tryptic soy broth for 4 h. After that the disk is placed on a plate spread with a susceptible isolate. If the initial sample is carbapenem-producer then the disk is inactivated and zone of inhibition will be absent on the plate. Both these tests yield results after an overnight incubation (181). On the other hand, CarbaNP test uses the property of a carbapenemase-producer to hydrolyze imipenem within 2 h (182). This hydrolysis produces an acid which lowers the pH and triggers phenol red to change its color from red to yellow. There are several variants of this test. These tests detect all carbapenemases but has low sensitivity for OXA-48-like carbapenemases. The modified CarbaNP test uses a different buffer (0.02% cetyltrimethylammonium bromide lysis buffer) of higher (pH 7.8 instead of pH 7.5) and has higher sensitivity than CarbaNP test. For K. pneumoniae the modified test is beneficial as mucoid cells tend to give false negative results in the original test. To differentiate between classes of carbapenemase, CarbaNP test II is used which discriminates based on the use of inhibitors tazobactam (for KPC) or EDTA (for MBLs) (177).
Colorimetric methods to detect carbapenemases directly from blood without blood culture are cost-effective and less time consuming (3–4 h). A study evaluated two variations of CarbaNP, CarbaNPT-direct which uses Triton X-100 as enzymatic extractor and Blue-carba which uses bromothymol blue instead of phenol red, for detection of carbapenemases directly from blood samples (183). The study reported high sensitivity for MBLs and KPC by colorimetric assays but less sensitivity for detection of GES and OXA-48 carbapenemases. A new method with increased sensitivity is the CarbaLux test which uses a fluorescent carbapenem substrate and substrate with cloxacillin to detect not only all carbapenems but also carbapenem-hydrolyzing AmpC enzymes from a bacterial culture (184). Another highly sensitive and specific method is lateral flow immunoassay which detects widely spread carbapenemases NDM, KPC, OXA-48-like, IMP, VIM on a single strip within 15 min from a bacterial culture (185, 186). These require minimal infrastructure.
Several phenotypic tests and commercial kits are available for the detection of CRKP strains (179, 180), such tests and/or kits are yet unavailable for the hvKP strains. The clinical microbiology laboratories are thus, not equipped to distinguish cKP from hvKP during routine diagnosis. Since the hvKP strains are often found to harbor hypercapsule, the hypermucoviscosity appearance of the colonies on the agar plate can be determined by a string test in which a viscous string is generated (>5 mm) when a colony is stretched by an inoculation loop (90). However, the strain discrimination based on only the hypermucoviscous appearance of the colonies and/or the positive string test can be misleading because studies have argued that hypermucoviscosity and hypervirulence are two different phenomena (91, 187). It is noteworthy that several rmpA and/or rmpA2-positive non-hypermucoviscous strains and also rmpA and/or rmpA2-negative hypermucoviscous strains were detected in causing invasive syndrome (103, 188, 189). hvKP strains produce quantitatively more siderophores than cKP, and the siderophore assay can be an option for detection. However, as of date, the conclusive discrimination of hvKP is carried out by genotypic methods, such as via specifically amplifying several hvKP-associated genes (magA, rmpA, rmpA2, iroB, iroN, iucA, and iutA) using polymerase chain reaction (PCR). These hypervirulent biomarkers hold the promise of a sensitive and specific diagnostic test in future. Since, the methodology is largely genotype-based, it is restricted to the research laboratories and may not be readily available in the healthcare settings of LMICs. Research is on-going for the development of a cost-effective commercial test which will not only help the clinical laboratories but can also be used in surveillance and research studies (189). In some resource-poor settings where the basic blood culture facilities are still unavailable or not properly standardized, the detection of either CRKP or hvKP is far from sight. Moreover, the major hindrance in detecting hvKP is probably the lack of awareness of such strains and the need to detect them.
Treatment: Is There a Silver Lining?
Treatment of neonatal sepsis is a great challenge, the signs of the disease are non-specific, the pathogens are numerous and drug-resistant, the diagnosis has limitations and the patient is vulnerable. Antibiotics remain the primary treatment with supportive respiratory and circulatory treatments along with treatment for metabolic derangements. In contrast to the well-established supportive treatments, the protocols for antimicrobial treatments are often found to be inadequate and region-specific. The choice of antibiotics largely depends on the etiology of the prevailing pathogens, their antimicrobial susceptibility profiles, the age of onset of sepsis, and the microbiology laboratory support available (3). The protocol is often specific for a given unit and greatly varies between and within countries. In this review, we discuss the limitations of the currently available antimicrobial therapy particularly in the LMICs in the current situation of drug resistance.
Empirical Antibiotic Therapy: Are the Guidelines Still Adequate for Neonates?
In general, the treatment of neonatal sepsis can be broadly divided into suspected or empirical therapy and known or definitive therapy. In clinical microbiology, it is always recommended to obtain the blood cultures for ascertaining bacterial sepsis before the initiation of antimicrobial therapy. However, culture results often require at least 48–72 h to be reported. Considering the non-specific clinical manifestations of neonatal sepsis, the initial antimicrobial therapy should not be unnecessarily delayed for the culture report in case of severely ill and high-risk neonates. In most cases the treatment should start within 1 h of decision to treat (190). This initial antibiotic therapy which is implemented before obtaining the blood culture report is defined as the empirical therapy. Empirical antibiotic therapy should be guided by the prevalent spectrum of pathogenic bacterial strains and their resistance profiles commonly detected in the given NICU or in the community settings. Microbiology support is varied and insufficient in resource-poor settings and treatment is largely empirical in such situations.
Antibiotic resistance has limited the therapeutic options in neonates and this problem is compounded in resource-limited heath infrastructure in LMICs. CRKP strains in neonatal units have challenged the healthcare settings, as these strains not only produce broad-spectrum carbapenemases but also harbor a repertoire of other plasmid-mediated resistance determinants that confer resistance to almost all clinically important antimicrobial classes, including third- and/or fourth-generation cephalosporins, cephamycins, aztreonam, aminoglycosides, and fluoroquinolones (67). Under these circumstances, the WHO treatment guidelines for neonates were found to be ineffective for the LMICs.
In response to the antibiotic resistance crisis, recently, the WHO has revised the treatment guidelines and launched a global action plan on antimicrobial resistance. The aim of this global campaign is to reduce the spread of AMR through optimizing the use of antibiotics, and also to reduce the adverse events and overall costs. The WHO Essential Medicine List (EML) Working Group adopted a tool, AWaRe. This tool classifies the antibiotics into three groups: Access, Watch, and Reserve (191). The Access group generally includes narrow-spectrum antimicrobials against a wide range of commonly encountered susceptible pathogens. The WHO EML enlisted 19 antibiotics recommended as first or second choice of empiric treatment options for clinical infections. These antibiotics are affordable, greatly assured, and are generally available at all times. The Watch group contains broader spectrum antimicrobial classes that have higher resistance potential and includes most of the highest priority agents among critically important antimicrobials. Among the 110 Watch group antibiotics, WHO EML enlisted 11 antibiotics as first or second choice of empiric treatment options. However, these antibiotics are recommended only for specific and limited indications. In addition, the Reserve group consists of antibiotics and antimicrobial classes for the treatment of MDR infections. Antibiotics in this group should be considered as “last resort” and also should be highly specific for patients and settings when all other antibiotics have failed. Till date, 22 antimicrobials have been classified as reserve group. In the recent global action plan, WHO enlisted seven reserve group antibiotics in the EML (191).
For both early and late onset neonatal sepsis, the most commonly recommended and used antibiotics, as per the WHO guidelines, is a β-lactam antibiotic (most commonly ampicillin) combined with an aminoglycoside (most commonly gentamicin) (191). Recently, in 2019, the use of AwaRe group of antibiotics was assessed in a pediatric survey across 56 countries (192). The study revealed that among the Access group of antibiotics, gentamicin and ampicillin were commonly given to the septicemic neonates in most of the countries, including Africa, America, Eastern Mediterranean, Europe, and South-East Asia. However, in the Western Pacific region, amoxicillin and β-lactamase inhibitor were used as empiric treatment for neonates. In addition, the study also showed that among the Watch group of antibiotics, globally meropenem and/or cefotaxime were prescribed to the neonates suffering from bacterial sepsis. Moreover, in South-East Asia, among the Reserve group of antibiotics, colistin was given to the critically ill neonates (192).
Due to the emergence and spread of CRKP strains in the healthcare settings, the treatment options for neonates are narrowing. Added to this, are the challenges in sepsis diagnosis. This alarming scenario demands new antimicrobials or additional alternatives for the treatment of CRKP-infected septicemic neonates. Although currently there is a scarcity of new antimicrobials globally, combination therapy is an alternative. Such therapies can expand the spectrum of antibiotic coverage and synergism between the antibiotics with enhanced killing effect. In addition, new alternatives have also been suggested for CRKP infections though all may not be suitable for neonates. For example, avibactam (a non-β-lactam β-lactamase inhibitor) showed high effectivity against KPC- and OXA-48-producing CRKP strains when combined with ceftazidime (193). Similarly, other studies revealed that vaborbactam (a boronic acid derivative) when combined with meropenem can exert bactericidal activity against the KPC-2-producing CRKP strains (194). Current studies have also argued that plazomicin (a novel semisynthetic aminoglycoside) showed impressive activity against most of the CRKP strains (195). The use of polymyxins (including both colistin and polymyxin B) and fosfomycin have also been documented in several case studies for the treatment of CRKP infections in the neonates (196, 197). In the current AwaRe classification, WHO has enlisted these antibiotics in the reserve group for which significant clinical data are still missing (191).
In response to the critical priority pathogens, recently WHO has published a third review of the clinical antibacterial pipeline where they enlisted a total of 8 new antibiotics that gained market authorization since July 2017 (198). Of them, two β-lactam and β-lactamase inhibitor combinations (vaborbactam + meropenem and relebactam + imipenem), one aminoglycoside (plazomicin), and one tetracycline (eravacycline) were found to be active against the CRE. Although all of these newly launched antibiotics are active against the Class A and Class D carbapenemases but found ineffective against the MBLs. To combat the MBL-producing CRE, WHO has also enlisted several other antimicrobial agents that are now in clinical trials (Phase 1–3), such as cefiderocol (Phase 3), taniborbactam + cefepime (Phase 3), BOS-288 (Phase 2), zidebactam + cefepime (Phase 1), and nacubactam + meropenem (Phase 1) (198). All of these currently developed antibiotics and/or antibiotic combinations are active against the CRE but the clinical data regarding their effectiveness against neonatal septicemia are yet to come. Although several clinical trials are ongoing for neonatal sepsis, additional large-scale clinical trials, strict infection control measures, and antimicrobial stewardship programmes should be undertaken in future to treat infections caused by drug-resistant and virulent K. pneumoniae to arrest their further dissemination.
Alternative Combat Approaches
The gut of a term neonate having normal-body-weight is mainly comprised of Bifidobacterium spp., Bacteroides, Clostridium spp. and Lactobacillus spp. (in minor proportion) (160, 199, 200) and serves key role in providing nutrients, providing defense against gut colonization by pathogens, and development of immunity of the neonate (160). Whereas, in preterm and/or low-birth-weight neonates, intestinal flora lacks colonization by the above mentioned favorable microorganisms (199). Preterm and/or low-birth-weight neonates are often colonized by Gram-negative bacilli, some of which can be opportunistic pathogens and may trigger an inflammatory response which plays the key role in the initiation of the necrotizing enterocolitis and sepsis (160, 200). If the neonates are given supplements of probiotics (consisting of bacteria which forms the healthy gut flora of term neonates) with human breast milk, it helps in reduction of inflammation (200) as well as translocation of pathogenic bacteria (160). The use of Bifidobacterium spp. and Lactobacillus spp, in different probiotic doses has shown significant decrease in necrotizing enterocolitis and neonatal death (199–201). Despite pre-existing skepticism that probiotics may increase sepsis instead of decreasing it (200, 202), systemic reviews and meta analyses of randomized control trials of large sample sizes have shown that probiotics indeed play a beneficial role in decreasing the rate of LOS in preterm low-birth-weight neonates (203–205). The bacterial strains used in the randomized control trials were mainly Bifidobacterium bifidum, B. lactis, B. breve, B. infantis, Lactobacillus acidophilus, L. rhamnosus, L. reuteri. Streptococcus thermophilus in varied doses (203). As probiotics not only provide a low cost, non-invasive, safer way to replenish the preterm neonatal gut with natural gut flora of a healthy term neonate, but also provides defense against LOS, it can be considered for routine use in LMICs where the load of LOS is overwhelming.
Preterm neonates of <32 weeks of gestational age lack maternal transplacental immunoglobulin; transfer of which, from mother to fetus, occurs only after 32 weeks of gestation. In the search for suitable alternative therapy for sepsis in preterm neonates, studies were conducted by administering intravenous immunoglobulin, granulocytes, granulocyte colony stimulating factor and granulocyte-macrophage colony stimulating factor, and pentoxifylline to neonates. The results were not promising except for pentoxifylline which caused decrease in all-cause mortality (206, 207). However, an monoclonal antibody (mAb)—Pagibaximab has shown some positive result (208). Recently, broadly reactive mAbs were raised against the capsular antigens of CRKP and those mAbs showed positive result in efficient killing of K. pneumoniae in mice model (209). Although, the efficacy of mAbs can be properly assessed only after clinical trials on neonates, but it shows some ray of hope in an otherwise grim situation.
Conclusion
Antibiotic resistance in Gram-negative bacteria has clearly exposed that the World Health Organization (WHO) guidelines for the management of neonatal sepsis, which is currently ampicillin plus gentamicin, is in dire need of modification. The major burden of neonatal sepsis is borne by the LMICs where antibiotic resistance is high and microbiology laboratory support is inadequate. The number of documented neonatal infections caused by CRKP and hvKP are just the tip of the iceberg. Most cases are not recorded as the microbiology laboratories in resource-limited countries lack infrastructure and capability. This is probably likelier for hvKP, as these strains are not specifically detected in clinical microbiology laboratories. This lack of information is thus translated into treatment protocols. Studies have shown that the most severe forms of neonatal sepsis with an unfavorable outcome were due to virulent strains of K. pneumoniae (5). The clinician is however unaware of the hvKP phenotype. Some carbapenem resistance genes such as blaKPC and blaOXA−48 are also difficult to recognize by routine disc diffusion tests. The presence of these genes may not be detectable by such tests as their capability to hydrolyze carbapenems are variable.
A recurring premise about carbapenemases is their ability to hydrolyze most β-lactam antibiotics, emergence of variants and their promiscuous nature. Plasmids are numerous and have facilitated intra and interspecies transmission of these genes particularly under antibiotic pressure. Globalization has encouraged their spread and most genes have within a few years of identification crossed boundaries and invaded new terrains. Clonal spread is uncommon but not unknown. The evolution of variants of enzymes that are more efficient or have better stability have also made them difficult to contain. Plasmids carrying carbapenem-resistant genes also carry other resistant genes in a bid to make a panel of antibiotics ineffective. With new carbapenem resistance genes being identified and new variants evolving, the problem of CRKP and hvKP that we recognize presently is incomplete.
Another overlooked aspect of K. pneumoniae is that it commonly resides in the human gut. The rate of colonization of K. pneumoniae increases drastically in hospitalized patients with invasive devices, antibiotic exposure, and prolonged stay. Needless to say, that this would also happen with premature, low-birth-weight neonates with a pristine gut. K. pneumoniae that colonize the gut in the hospital are resistant to antibiotics and the gut provides an environment apt for the exchange of resistance genes. Studies have shown that such exchange of resistance genes frequently happen in the gut (210). Further, bacteria from the gut can translocate and cause sepsis in neonates. This can happen in neonates who have an immature immune system, lower levels of mucus and gastric acid production. The bacteria evade the gut barrier and cause sepsis. Our study on the neonatal flora showed that K. pneumoniae was not only the predominant organism that colonized the neonatal gut, but also the most common organism isolated from the neonatal blood specimens (162). The exchange of carbapenem resistance genes in the gut and subsequent translocation can further complicate the situation.
It is difficult to change things overnight and the economic implications of these changes may also be a constraint on health systems that are poorly funded. The COVID-19 situation has further exposed the fragility of health systems around the world and newborns have also been affected. Measures that can reduce infection rates can also reduce CRKP and hvKP. These include surveillance systems to recognize changes in etiology and drug resistance profile, improved laboratories for better and timely detection of pathogenic strains, setting empirical treatment guidelines based on the profiles and proper education of mothers and healthcare workers regarding sanitation. We have come a long way from the days when we understood microbes through the lens of a microscope. We now seem to have an unprecedented power over them by being able to understand their genetic make-up to the last nucleotide. Whole genome sequencing has opened up newer avenues to understand the bacterial genomes. This enormous amount of information generated can be honed to create new cures and products. The challenge is big but there is light at the end of the tunnel.
Author Contributions
SB: conception, design, wrote the introduction, and discussion. SMu and SMi: performed literature search, wrote the initial draft, and prepared all the tables and figures. SB and SD: critically revised the manuscript. All authors read and approved the final manuscript.
Funding
SMi was supported by a fellowship from Indian Council of Medical Research (ICMR).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We extend our thanks to the curators of the Center for Disease Dynamics, Economics & Policy (CDDEP) and CGView server.
References
1. Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. (2010) 375:1969–87. doi: 10.1016/S0140-6736(10)60549-1
3. Shane AL, Sánchez PJ, Stoll BJ. Neonatal sepsis. Lancet. (2017) 390:1770–80. doi: 10.1016/S0140-6736(17)31002-4
4. Thaver D, Zaidi AKM. Burden of neonatal infections in developing countries. Pediatr Infect Dis J. (2009) 28:S3–S9. doi: 10.1097/INF.0b013e3181958755
5. Khaertynov KS, Anokhin VA, Rizvanov AA, Davidyuk YN, Semyenova DR, Lubin SA, et al. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Front Med. (2018) 5:225. doi: 10.3389/fmed.2018.00225
7. Singer M, Bellomo R, Bernard GR, Chiche J, Craig M, Hotchkiss RS, et al. The third international concesus definitions for Sepsis and Septic shock (Sepsis 3). Jama. (2016) 315:801–10. doi: 10.1001/jama.2016.0287.The
8. Ershad M, Mostafa A, Dela Cruz M, Vearrier D. Neonatal Sepsis. Curr Emerg Hosp Med Rep. (2019) 7:83–90. doi: 10.1007/s40138-019-00188-z
9. Wynn JL. Defining neonatal sepsis. Curr Opin Pediatr. (2016) 28:135–40. doi: 10.1097/MOP.0000000000000315
10. Zaidi AKMM, Huskins WC, Thaver D, Bhutta ZA, Abbas Z, Goldmann DA. Hospital-acquired neonatal infections in developing countries. Lancet. (2005) 365:1175–88. doi: 10.1016/S0140-6736(05)71881-X
11. KC A, Gurung R, Kinney MV, Sunny AK, Moinuddin M, Basnet O, Paudel P, et al. Effect of the COVID-19 pandemic response on intrapartum care, stillbirth, and neonatal mortality outcomes in Nepal: a prospective observational study. Lancet Glob Heal. (2020) 8:e1273–e81. doi: 10.1016/S2214-109X(20)30345-4
12. Masroor S. Collateral damage of COVID-19 pandemic: delayed medical care. J Card Surg. (2020) 35:1345–7. doi: 10.1111/jocs.14638
13. Sud A, Jones ME, Broggio J, Loveday C, Torr B, Garrett A, et al. Collateral damage: the impact on cancer outcomes of the COVID-19 Pandemic. SSRN Electron J. (2020) 31:1065–74. doi: 10.2139/ssrn.3582775
14. Jacob CM, Briana DD, Carlo G, Renzo D, Modi N, Bustreo F, et al. Viewpoint Building resilient societies after COVID-19 : the case for investing in maternal, neonatal, and child health. Lancet Public Heal. (2020) 2667:19–22. doi: 10.1016/S2468-2667(20)30200-0
15. Juan J, Gil MM, Rong Z, Zhang Y, Yang H, Poon LC. Effect of coronavirus disease 2019 (COVID-19) on maternal, perinatal and neonatal outcome: systematic review. Ultrasound Obstet Gynecol. (2020) 56:15–27. doi: 10.1002/uog.22088
16. Ghaith DM, Zafer MM, Said HM, Elanwary S, Elsaban S, Al-Agamy MH, et al. Genetic diversity of carbapenem-resistant Klebsiella pneumoniae causing neonatal sepsis in intensive care unit, Cairo, Egypt. Eur J Clin Microbiol Infect Dis. (2020) 39:583–91. doi: 10.1007/s10096-019-03761-2
17. Yusef D, Shalakhti T, Awad S, Algharaibeh H, Khasawneh W. Clinical characteristics and epidemiology of sepsis in the neonatal intensive care unit in the era of multi-drug resistant organisms: a retrospective review. Pediatr Neonatol. (2018) 59:35–41. doi: 10.1016/j.pedneo.2017.06.001
18. Viswanathan R, Singh AK, Mukherjee S, Mukherjee R, Das P, Basu S. Aetiology and antimicrobial resistance of neonatal sepsis at a tertiary care centre in eastern india: a 3 year study. Indian J Pediatr. (2011) 78:409–12. doi: 10.1007/s12098-010-0272-1
19. Viswanathan R, Singh AK, Basu S, Chatterjee S, Sardar S, Isaacs D. Multi-drug resistant gram negative bacilli causing early neonatal sepsis in India. Arch Dis Child - Fetal Neonatal Ed. (2012) 97:F182–F7. doi: 10.1136/archdischild-2011-300097
20. Chatterjee S, Datta S, Roy S, Ramanan L, Saha A, Viswanathan R, et al. Carbapenem Resistance in Acinetobacter baumannii and Other Acinetobacter spp. Causing neonatal sepsis: focus on NDM-1 and its linkage to ISAba125. Front Microbiol. (2016) 7:1126. doi: 10.3389/fmicb.2016.01126
21. Datta S, Roy S, Chatterjee S, Saha A, Sen B, Pal T, et al. A five-year experience of carbapenem resistance in enterobacteriaceae causing neonatal septicaemia: predominance of NDM-1. PLoS ONE. (2014) 9:e112101. doi: 10.1371/journal.pone.0112101
22. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. (1998) 11:589–603. doi: 10.1128/CMR.11.4.589
23. Spagnolo AM, Orlando P, Panatto D, Perdelli F, Cristina ML. An overview of carbapenem-resistant Klebsiella pneumoniae. Rev Med Microbiol. (2014) 25:7–14. doi: 10.1097/MRM.0b013e328365c51e
24. Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. (2018) 45:131–9. doi: 10.1016/j.mib.2018.04.004
25. Chew KL, Lin RTP, Teo JWP. Klebsiella pneumoniae in singapore: hypervirulent infections and the carbapenemase threat. Front Cell Infect Microbiol. (2017) 7:515. doi: 10.3389/fcimb.2017.00515
26. Martin RM, Bachman MA. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. (2018) 8:1–15. doi: 10.3389/fcimb.2018.00004
27. Malaki M. Klebsiella pneumoniae bacteremia presenting on a neonatal intensive care unit during the first week of life. J Pediatr Infect Dis. (2015) 1:12–5. doi: 10.1055/s-0035-1554971
28. Mitra S, Mukherjee S, Naha S, Chattopadhyay P, Dutta S, Basu S. Evaluation of co-transfer of plasmid-mediated fluoroquinolone resistance genes and blaNDM gene in Enterobacteriaceae causing neonatal septicaemia. Antimicrob Resist Infect Control. (2019) 8:46. doi: 10.1186/s13756-019-0477-7
29. Viswanathan R, Singh AK, Ghosh C, Dasgupta S, Mukherjee S, Basu S. Profile of neonatal septicaemia at a district-level sick newborn care unit. J Heal Popul Nutr. (2012) 30:41–8. doi: 10.3329/jhpn.v30i1.11274
30. Sands K, Carvalho MJ, Portal E, Thomson K, Dyer C, Akpulu C, et al. Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat Microbiol. (2021) 6:512–23. doi: 10.1038/s41564-021-00870-7
31. Diancourt L, Passet V, Verhoef J, Grimont PAD, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. (2005) 43:4178–82. doi: 10.1128/JCM.43.8.4178-4182.2005
32. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. (2017) 41:252–75. doi: 10.1093/femsre/fux013
33. Pan Y-J, Fang H-C, Yang H-C, Lin T-L, Hsieh P-F, Tsai F-C, et al. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae Serotype K57 and a new capsular serotype. J Clin Microbiol. (2008) 46:2231–40. doi: 10.1128/JCM.01716-07
34. Chen Y-T, Chang H-Y, Lai Y-C, Pan C-C, Tsai S-F, Peng H-L. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. (2004) 337:189–98. doi: 10.1016/j.gene.2004.05.008
35. Wu K-M, Li L-H, Yan J-J, Tsao N, Liao T-L, Tsai H-C, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. (2009) 191:4492–501. doi: 10.1128/JB.00315-09
36. Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. (2013) 11:297–308. doi: 10.1586/eri.13.12
37. The Center for Disease DE& P. ResistanceMap: Antibiotic Resistance. (2020). Available online at: https://resistancemap.cddep.org/AntibioticResistance.php. (accessed October 3, 2020).
38. Asokan GV, Ramadhan T, Ahmed E, Sanad H. WHO global priority pathogens list: a bibliometric analysis of medline-pubmed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Med J. (2019) 34:184–93. doi: 10.5001/omj.2019.37
39. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. (2005) 63:219–28. doi: 10.1016/j.mimet.2005.03.018
40. Gupta A, Ampofo K, Rubenstein D, Saiman L. Extended spectrum β lactamase-producing Klebsiella pneumoniae infections: a review of the literature. J Perinatol. (2003) 23:439–43. doi: 10.1038/sj.jp.7210973
41. Lee C-H, Su L-H, Tang Y-F, Liu J-W. Treatment of ESBL-producing Klebsiella pneumoniae bacteraemia with carbapenems or flomoxef: a retrospective study and laboratory analysis of the isolates. J Antimicrob Chemother. (2006) 58:1074–7. doi: 10.1093/jac/dkl381
42. Haruta S, Yamaguchi H, Yamamoto ET, Eriguchi Y, Nukaga M, O'Hara K, et al. Functional analysis of the active site of a metallo-β-lactamase proliferating in Japan. Antimicrob Agents Chemother. (2000) 44:2304–9. doi: 10.1128/AAC.44.9.2304-2309.2000
43. Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing -lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. (2001) 45:1151–61. doi: 10.1128/AAC.45.4.1151-1161.2001
44. Reyes J, Aguilar AC, Caicedo A. Carbapenem-Resistant Klebsiella pneumoniae: microbiology key points for clinical practice. Int J Gen Med. (2019) 12:437–46. doi: 10.2147/IJGM.S214305
45. Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. (2016) 7:1–30. doi: 10.3389/fmicb.2016.00895
46. Klundert JAM, Gestel MH, Meerdink G, Marie S. Emergence of bacterial resistance to cefamandole in vivo due to outer membrane protein deficiency. Eur J Clin Microbiol Infect Dis. (1988) 7:776–8. doi: 10.1007/BF01975046
47. Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother. (2010) 54:177–83. doi: 10.1128/AAC.00715-09
48. van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. (2017) 8:460–9. doi: 10.1080/21505594.2016.1222343
49. Arnold RS, Thom KA, Sharma S, Phillips M, Kristie Johnson J, Morgan DJ. Emergence of Klebsiella pneumoniae carbapenemase-producing bacteria. South Med J. (2011) 104:40–5. doi: 10.1097/SMJ.0b013e3181fd7d5a
50. Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-β-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. (2009) 53:5046–54. doi: 10.1128/AAC.00774-09
51. Ahmad N, Khalid S, Ali SM, Khan AU. Occurrence of blaNDM variants among enterobacteriaceae from a neonatal intensive care unit in a northern india hospital. Front Microbiol. (2018) 9:407. doi: 10.3389/fmicb.2018.00407
52. Basu S, Basu S. Variants of the New Delhi metallo-β-lactamase: new kids on the block. Future Microbiol. (2020) 15:465–67. doi: 10.2217/fmb-2020-0035
53. Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. (2019) 32:e00115–18. doi: 10.1128/CMR.00115-18
54. Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK : a molecular, biological, and epidemiological study. Lancet Infect Dis. (2010) 3099:1–6. doi: 10.1016/S1473-3099(10)70143-2
55. Datta S, Mitra S, Chattopadhyay P, Som T, Mukherjee S, Basu S. Spread and exchange of bla NDM-1 in hospitalized neonates: role of mobilizable genetic elements. Eur J Clin Microbiol Infect Dis. (2017) 36:255–65. doi: 10.1007/s10096-016-2794-6
56. Pitout JDD, Peirano G, Kock MM, Strydom K, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. (2019) 33:1–48. doi: 10.1128/CMR.00102-19
57. Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. (2012) 67:1597–606. doi: 10.1093/jac/dks121
58. Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, et al. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D β-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. (2013) 41:325–9. doi: 10.1016/j.ijantimicag.2012.11.007
59. Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-beta-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. (1995) 39:824–9. doi: 10.1128/AAC.39.4.824
60. Miriagou V, Papagiannitsis CC, Kotsakis SD, Loli A, Tzelepi E, Legakis NJ, et al. Sequence of pNL194, a 79.3-Kilobase IncN Plasmid Carrying the blaVIM-1 Metallo-β-Lactamase Gene in Klebsiella pneumoniae. Antimicrob Agents Chemother. (2010) 54:4497–502. doi: 10.1128/AAC.00665-10
61. Matsumura Y, Peirano G, Bradford PA, Motyl MR, DeVinney R, Pitout JDD. Genomic characterization of IMP and VIM carbapenemase-encoding transferable plasmids of Enterobacteriaceae. J Antimicrob Chemother. (2018) 73:3034–8. doi: 10.1093/jac/dky303
62. Roy S, Viswanathan R, Singh AK, Das P, Basu S. Sepsis in neonates due to imipenem resistant Klebsiella pneumoniae producing NDM-1 in India. J Antimicrob Chemother. (2011) 66:1411–13. doi: 10.1093/jac/dkr068
63. Escobar Pérez JA, Olarte Escobar NM, Castro-Cardozo B, Valderrama Márquez IA, Garzón Aguilar MI, Martinez de la Barrera L, et al. Outbreak of NDM-1-Producing Klebsiella pneumoniae in a neonatal unit in Colombia. Antimicrob Agents Chemother. (2013) 57:1957–60. doi: 10.1128/AAC.01447-12
64. Khajuria A, Praharaj A, Kumar M, Grover N, Aggarwal A. Multidrug resistant NDM-1 metallo-beta-lactamase producing Klebsiella pneumoniae sepsis outbreak in a neonatal intensive care unit in a tertiary care center at central India. Indian J Pathol Microbiol. (2014) 57:65. doi: 10.4103/0377-4929.130900
65. Stoesser N, Giess A, Batty EM, Sheppard AE, Walker AS, Wilson DJ, et al. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob Agents Chemother. (2014) 58:7347–57. doi: 10.1128/AAC.03900-14
66. Brinkac LM, White R, D'Souza R, Nguyen K, Obaro SK, Fouts DE. Emergence of New Delhi Metallo-β-Lactamase (NDM-5) in Klebsiella quasipneumoniae from neonates in a nigerian hospital. mSphere. (2019) 4:e00685–18. doi: 10.1128/mSphere.00685-18
67. Poirel L, Yilmaz M, Istanbullu A, Arslan F, Mert A, Bernabeu S, et al. Spread of NDM-1-producing Enterobacteriaceae in a neonatal intensive care unit in Istanbul, Turkey. Antimicrob Agents Chemother. (2014) 58:2929–33. doi: 10.1128/AAC.02047-13
68. Zhang X, Li X, Wang M, Yue H, Li P, Liu Y, et al. Outbreak of NDM-1-Producing Klebsiella pneumoniae causing neonatal infection in a teaching hospital in mainland China. Antimicrob Agents Chemother. (2015) 59:4349–51. doi: 10.1128/AAC.03868-14
69. Jin Y, Song X, Liu Y, Wang Y, Zhang B, Fan H, et al. Characteristics of carbapenemase-producing Klebsiella pneumoniae as a cause of neonatal infection in Shandong, China. Exp Ther Med. (2017) 13:1117–26. doi: 10.3892/etm.2017.4070
70. Huang X, Cheng X, Sun P, Tang C, Ni F, Liu G. Characteristics of NDM-1-producing Klebsiella pneumoniae ST234 and ST1412 isolates spread in a neonatal unit. BMC Microbiol. (2018) 18:186. doi: 10.1186/s12866-018-1334-1
71. Yu J, Tan K, Rong Z, Wang Y, Chen Z, Zhu X, et al. Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect Dis. (2016) 16:563. doi: 10.1186/s12879-016-1870-y
72. Yu J, Wang Y, Chen Z, Zhu X, Tian L, Li L, et al. Outbreak of nosocomial NDM-1-producing Klebsiella pneumoniae ST1419 in a neonatal unit. J Glob Antimicrob Resist. (2017) 8:135–9. doi: 10.1016/j.jgar.2016.10.014
73. Mukherjee S, Bhattacharjee A, Naha S, Majumdar T, Debbarma SK, Kaur H, et al. Molecular characterization of NDM-1-producing Klebsiella pneumoniae ST29, ST347, ST1224, and ST2558 causing sepsis in neonates in a tertiary care hospital of North-East India. Infect Genet Evol. (2019) 69:166–75. doi: 10.1016/j.meegid.2019.01.024
74. Liu Y, Li X-Y, Wan L-G, Jiang W-Y, Yang J-H, Li F-Q. Acquisition of carbapenem resistance in multiresistant Klebsiella pneumoniae isolates of sequence type 11 at a university hospital in China. Diagn Microbiol Infect Dis. (2013) 76:241–3. doi: 10.1016/j.diagmicrobio.2013.02.002
75. Hassuna NA, AbdelAziz RA, Zakaria A, Abdelhakeem M. Extensively-Drug Resistant Klebsiella pneumoniae recovered from neonatal sepsis cases from a major NICU in Egypt. Front Microbiol. (2020) 11:1375. doi: 10.3389/fmicb.2020.01375
76. Naha S, Sands K, Mukherjee S, Roy C, Rameez MJ, Saha B, et al. KPC-2-producing Klebsiella pneumoniae ST147 in a neonatal unit: clonal isolates with differences in colistin susceptibility attributed to AcrAB-TolC pump. Int J Antimicrob Agents. (2020) 55:105903. doi: 10.1016/j.ijantimicag.2020.105903
77. Kong Z, Liu X, Li C, Cheng S, Xu F, Gu B. Clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Jiangsu Province, China. Infect Drug Resist. (2020) 13:4627–35. doi: 10.2147/IDR.S293206
78. Mukherjee S, Naha S, Bhadury P, Saha B, Dutta M, Dutta S, et al. Emergence of OXA-232-producing hypervirulent Klebsiella pneumoniae ST23 causing neonatal sepsis. J Antimicrob Chemother. (2020) 75:2004–6. doi: 10.1093/jac/dkaa080
79. Yin D, Dong D, Li K, Zhang L, Liang J, Yang Y, et al. Clonal dissemination of OXA-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob Agents Chemother. (2017) 61:e00385–17. doi: 10.1128/AAC.00385-17
80. Herruzo R, Ruiz G, Perez-Blanco V, Gallego S, Mora E, Vizcaino MJ, et al. Bla-OXA48 gene microorganisms outbreak, in a tertiary Children's Hospital, Over 3 years (2012–2014). Medicine. (2017) 96:e7665. doi: 10.1097/MD.0000000000007665
81. Mairi A, Touati A, Ait Bessai S, Boutabtoub Y, Khelifi F, Sotto A, et al. Carbapenemase-producing Enterobacteriaceae among pregnant women and newborns in Algeria: Prevalence, molecular characterization, maternal-neonatal transmission, and risk factors for carriage. Am J Infect Control. (2019) 47:105–108. doi: 10.1016/j.ajic.2018.07.009
82. Naha S, Sands K, Mukherjee S, Saha B, Dutta S, Basu S. OXA-181-like carbapenemases in Klebsiella pneumoniae ST14, ST15, ST23, ST48, and ST231 from septicemic neonates: coexistence with ndm-5, resistome, transmissibility, and genome diversity. mSphere. (2021) 6:1–0. doi: 10.1128/mSphere.01156-20
83. Esposito EP, Gaiarsa S, Del Franco M, Crivaro V, Bernardo M, Cuccurullo S, et al. A Novel IncA/C1 group conjugative plasmid, encoding VIM-1 metallo-beta-lactamase, mediates the acquisition of carbapenem resistance in ST104 Klebsiella pneumoniae isolates from neonates in the intensive care unit of V. Monaldi hospital in Naples. Front Microbiol. (2017) 8:2135. doi: 10.3389/fmicb.2017.02135
84. Jian Z, Li Y, Liu W, Li H, Zhang Y, Li Y, et al. Detection of the novel IMP-38 among carbapenemase-producing Enterobacteriaceae in a university hospital, China. J Infect Dev Ctries. (2014) 8:1044–48. doi: 10.3855/jidc.4179
85. Datta S, Mitra S, Viswanathan R, Saha A, Basu S. Characterization of novel plasmid-mediated b-lactamases (SHV-167 and ACT-16) associated with New Delhi metallo-b-lactamase-1 harbouring isolates from neonates in India. J Med Microbiol. (2014) 63:480–2. doi: 10.1099/jmm.0.067223-0
86. Poirel L, Hombrouck-Alet C, Freneaux C, Bernabeu S, Nordmann P. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect Dis. (2010) 10:832. doi: 10.1016/s1473-3099(10)70279-6
87. Ding Y, Wang Y, Hsia Y, Sharland M, Heath PT. Systematic review of carbapenem-resistant Enterobacteriaceae causing neonatal sepsis in China. Ann Clin Microbiol Antimicrob. (2019) 18:36. doi: 10.1186/s12941-019-0334-9
88. Wang S, Zhao J, Liu N, Yang F, Zhong Y, Gu X, et al. IMP-38-producing high-risk sequence type 307 Klebsiella pneumoniae strains from a neonatal unit in China. mSphere. (2020) 5:e00407–20. doi: 10.1128/mSphere.00407-20
89. Yaffee AQ, Roser L, Daniels K, Humbaugh K, Brawley R, Thoroughman D, et al. Notes from the field: verona integron-encoded metallo-beta-lactamase-producing carbapenem-resistant enterobacteriaceae in a neonatal and adult intensive care unit–kentucky, 2015. MMWR Morb Mortal Wkly Rep. (2016) 65:190. doi: 10.15585/mmwr.mm6507a5
90. Shon AS, Bajwa RPS, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Virulence. (2013) 4:107–18. doi: 10.4161/viru.22718
91. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. (2019) 32: doi: 10.1128/CMR.00001-19
92. Prokesch BC, TeKippe M, Kim J, Raj P, TeKippe EM, Greenberg DE. Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. (2016) 16:e190–e5. doi: 10.1016/S1473-3099(16)30021-4
93. Guo Y, Wang S, Zhan L, Jin Y, Duan J, Hao Z, et al. Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front Cell Infect Microbiol. (2017) 7:24. doi: 10.3389/fcimb.2017.00024
94. Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, et al. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol. (2016) 6:165. doi: 10.3389/fcimb.2016.00165
95. Hao Z, Duan J, Liu L, Shen X, Yu J, Guo Y, et al. Prevalence of Community-Acquired, Hypervirulent Klebsiella pneumoniae Isolates in Wenzhou, China. Microb Drug Resist. (2020) 26:21–7. doi: 10.1089/mdr.2019.0096
96. Liu C, Guo J. Hypervirulent Klebsiella pneumoniae (hypermucoviscous and aerobactin positive) infection over 6 years in the elderly in China: antimicrobial resistance patterns, molecular epidemiology and risk factor. Ann Clin Microbiol Antimicrob. (2019) 18:4. doi: 10.1186/s12941-018-0302-9
97. Harada S, Aoki K, Yamamoto S, Ishii Y, Sekiya N, Kurai H, et al. Clinical and molecular characteristics of Klebsiella pneumoniae isolates causing bloodstream infections in japan: occurrence of hypervirulent infections in health care. J Clin Microbiol. (2019) 57:e01206–19. doi: 10.1128/JCM.01206-19
98. Hentzien M, Rosman J, Decré D, Brenkle K, Mendes-Martins L, Mateu P. Seven hypervirulent ST380 Klebsiella pneumoniae septic localizations. Médecine Mal Infect. (2017) 47:171–3. doi: 10.1016/j.medmal.2016.10.002
99. Li J, Ren J, Wang W, Wang G, Gu G, Wu X, et al. Risk factors and clinical outcomes of hypervirulent Klebsiella pneumoniae induced bloodstream infections. Eur J Clin Microbiol Infect Dis. (2018) 37:679–89. doi: 10.1007/s10096-017-3160-z
100. Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, et al. High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob Agents Chemother. (2016) 60:6115–20. doi: 10.1128/AAC.01127-16
101. Lan Y, Zhou M, Jian Z, Yan Q, Wang S, Liu W. Prevalence of pks gene cluster and characteristics of Klebsiella pneumoniae-induced bloodstream infections. J Clin Lab Anal. (2019) 33:e22838. doi: 10.1002/jcla.22838
102. Wang X, Xie Y, Li G, Liu J, Li X, Tian L, et al. Whole-Genome-Sequencing characterization of bloodstream infection-causing hypervirulent Klebsiella pneumoniae of capsular serotype K2 and ST374. Virulence. (2018) 9:510–21. doi: 10.1080/21505594.2017.1421894
103. Cubero M, Grau I, Tubau F, Pallarés R, Dominguez MA, Liñares J, et al. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin Microbiol Infect. (2016) 22:154–60. doi: 10.1016/j.cmi.2015.09.025
104. Shankar C, Veeraraghavan B, Nabarro LEB, Ravi R, Ragupathi NKD, Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiol. (2018) 18:6. doi: 10.1186/s12866-017-1148-6
105. Cheng D-L. Septic metastatic lesions of pyogenic liver abscess. Arch Intern Med. (1991) 151:1557. doi: 10.1001/archinte.1991.00400080059010
106. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. (2016) 80:629–61. doi: 10.1128/MMBR.00078-15
107. Pomakova DK, Hsiao C-B, Beanan JM, Olson R, MacDonald U, Keynan Y, et al. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis. (2012) 31:981–9. doi: 10.1007/s10096-011-1396-6
108. Shu H-Y, Fung C-P, Liu Y-M, Wu K-M, Chen Y-T, Li L-H, et al. Genetic diversity of capsular polysaccharide biosynthesis in Klebsiella pneumoniae clinical isolates. Microbiology. (2009) 155:4170–83. doi: 10.1099/mic.0.029017-0
109. Fang C-T, Lai S-Y, Yi W-C, Hsueh P-R, Liu K-L, Chang S-C. Klebsiella pneumoniae Genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. (2007) 45:284–93. doi: 10.1086/519262
110. Yu W-L, Chan K-S, Ko W-C, Lee C-C, Chuang Y-C. Lower prevalence of diabetes mellitus in patients with Klebsiella pneumoniae primary liver abscess caused by isolates of k1/k2 than with non-k1/k2 capsular serotypes. Clin Infect Dis. (2007) 45:1529–30. doi: 10.1086/523006
111. Zhao J, Chen J, Zhao M, Qiu X, Chen X, Zhang W, et al. Multilocus sequence types and virulence determinants of hypermucoviscosity-positive Klebsiella pneumoniae isolated from community-acquired infection cases in harbin, North China. Jpn J Infect Dis. (2016) 69:357–60. doi: 10.7883/yoken.JJID.2015.321
112. Sahly H, Aucken H, Benedi VJ, Forestier C, Fussing V, Hansen DS, et al. Impairment of respiratory burst in polymorphonuclear leukocytes by extended-spectrum beta-lactamase-producing strains of Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. (2004) 23:20–6. doi: 10.1007/s10096-003-1047-7
113. Kabha K, Nissimov L, Athamna A, Keisari Y, Parolis H, Parolis LAS, et al. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect Immun. (1995) 63:847–52. doi: 10.1128/IAI.63.3.847-852.1995
114. Lin J-C, Chang F-Y, Fung C-P, Xu J-Z, Cheng H-P, Wang J-J, et al. High prevalence of phagocytic-resistant capsular serotypes of Klebsiella pneumoniae in liver abscess. Microbes Infect. (2004) 6:1191–8. doi: 10.1016/j.micinf.2004.06.003
115. Lee C-H, Chang C-C, Liu J-W, Chen R-F, Yang KD. Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence. (2014) 5:673–79. doi: 10.4161/viru.32076
116. Hsu C-R, Lin T-L, Chen Y-C, Chou H-C, Wang J-T. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology. (2011) 157:3446–57. doi: 10.1099/mic.0.050336-0
117. Li W, Sun G, Yu Y, Li N, Chen M, Jin R, et al. Increasing occurrence of antimicrobial-resistant hypervirulent (Hypermucoviscous) Klebsiella pneumoniae Isolates in China. Clin Infect Dis. (2014) 58:225–32. doi: 10.1093/cid/cit675
118. Fang C, Lai S, Yi W, Hsueh P, Liu K. The function of wzy_ K1 (magA), the serotype K1 polymerase gene in Klebsiella pneumoniae cps gene cluster. J Infect Dis. (2010) 201:1268–9. doi: 10.1086/652183
119. Chuang Y, Fang C, Lai S, Chang S, Wang J. Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. (2006) 193:645–54. doi: 10.1086/499968
120. Struve C, Bojer M, Nielsen EM, Hansen DS, Krogfelt KA. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. J Med Microbiol. (2005) 54:1111–3. doi: 10.1099/jmm.0.46165-0
121. Yeh K-M, Chang F-Y, Fung C-P, Lin J-C, Siu LK. magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of K. pneumoniae serotype K1. J Med Microbiol. (2006) 55:803–4. doi: 10.1099/jmm.0.46368-0
122. Bullen JJ, Rogers HJ, Griffiths E. Iron binding proteins and infection. Br J Haematol. (1972) 23:389–92. doi: 10.1111/j.1365-2141.1972.tb07073.x
123. Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. (2007) 71:413–51. doi: 10.1128/MMBR.00012-07
124. Podschun R, Sievers D, Fischer A, Ullmann U. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of klebsiella isolates causing human urinary tract infections. J Infect Dis. (1993) 168:1415–21. doi: 10.1093/infdis/168.6.1415
125. Russo TA, Olson R, MacDonald U, Metzger D, Maltese LM, Drake EJ, et al. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (Hypermucoviscous) Klebsiella pneumoniae. Infect Immun. (2014) 82:2356–67. doi: 10.1128/IAI.01667-13
126. Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. (2018) 56:e00776–18. doi: 10.1128/JCM.00776-18
127. Lawlor MS, O'Connor C, Miller VL. Yersiniabactin Is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect Immun. (2007) 75:1463–72. doi: 10.1128/IAI.00372-06
128. Bachman MA, Oyler JE, Burns SH, Caza M, Lépine F, Dozois CM, et al. Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect Immun. (2011) 79:3309–16. doi: 10.1128/IAI.05114-11
129. Hsieh P, Lin T, Lee C, Tsai S, Wang J. Serum-induced iron-acquisition systems and tonb contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J Infect Dis. (2008) 197:1717–27. doi: 10.1086/588383
130. El Fertas-Aissani R, Messai Y, Alouache S, Bakour R. Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol Biol. (2013) 61:209–16. doi: 10.1016/j.patbio.2012.10.004
131. Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, et al. Mapping the Evolution of Hypervirulent Klebsiella pneumoniae. MBio. (2015) 6:e00630–15. doi: 10.1128/mBio.00630-15
132. Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, et al. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb genomics. (2018) 4:196. doi: 10.1099/mgen.0.000196
133. Lin J-C, Koh T, Lee N, Fung C-P, Chang F-Y, Tsai Y-K, et al. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. (2014) 6:21. doi: 10.1186/1757-4749-6-21
134. Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. (2015) 112:E3574–81. doi: 10.1073/pnas.1501049112
135. Chung DR, Lee HR, Lee SS, Kim SW, Chang H-H, Jung S-I, Ko KS, Kang C-I, Peck KR, et al. Evidence for clonal dissemination of the serotype K1 Klebsiella pneumoniae strain causing invasive liver abscesses in Korea. J Clin Microbiol. (2008) 46:4061–3. doi: 10.1128/JCM.01577-08
136. Shankar C, Santhanam S, Kumar M, Gupta V, Devanga Ragupathi NK, Veeraraghavan B. Draft genome sequence of an extended-spectrum-β-lactamase-positive hypervirulent Klebsiella pneumoniae strain with novel sequence type 2318 isolated from a neonate. Genome Announc. (2016) 4:1–2. doi: 10.1128/genomeA.01273-16
137. Khaertynov KS, Anokhin VA, Davidyuk YN, Nicolaeva IV, Khalioullina SV, Semyenova DR, et al. Case of meningitis in a neonate caused by an extended-spectrum-beta-lactamase-producing strain of hypervirulent Klebsiella pneumoniae. Front Microbiol. (2017) 8:1–6. doi: 10.3389/fmicb.2017.01576
138. Albasha AM, Osman EH, Abd-Alhalim S, Alshaib EF, Al-Hassan L, Altayb HN. Detection of several carbapenems resistant and virulence genes in classical and hyper-virulent strains of Klebsiella pneumoniae isolated from hospitalized neonates and adults in Khartoum. BMC Res Notes. (2020) 13:1–7. doi: 10.1186/s13104-020-05157-4
139. Banerjee T, Wangkheimayum J, Sharma S, Kumar A, Bhattacharjee A. Extensively drug-resistant hypervirulent Klebsiella pneumoniae from a series of neonatal sepsis in a tertiary care hospital, India. Front Med. (2021) 8:645955. doi: 10.3389/fmed.2021.645955
140. Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis. (2015) 37:107–12. doi: 10.1016/j.ijid.2015.06.023
141. Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. (2015) 71:553–60. doi: 10.1016/j.jinf.2015.07.010
142. Wei D, Wan L-G, Deng Q, Liu Y. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn Microbiol Infect Dis. (2016) 85:192–4. doi: 10.1016/j.diagmicrobio.2015.03.012
143. Wu H, Li D, Zhou H, Sun Y, Guo L, Shen D. Bacteremia and other body site infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb Pathog. (2017) 104:254–62. doi: 10.1016/j.micpath.2017.01.049
144. Zhan L, Wang S, Guo Y, Jin Y, Duan J, Hao Z, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol. (2017) 7:182. doi: 10.3389/fcimb.2017.00182
145. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. (2018) 18:37–46. doi: 10.1016/S1473-3099(17)30489-9
146. Shu L, Dong N, Lu J, Zheng Z, Hu J, Zeng W, et al. Emergence of OXA-232 carbapenemase-producing Klebsiella pneumoniae that carries a pLVPK-like virulence plasmid among elderly patients in China. Antimicrob Agents Chemother. (2018) 63:e02246–18. doi: 10.1128/AAC.02246-18
147. Mohammad Ali Tabrizi A, Badmasti F, Shahcheraghi F, Azizi O. Outbreak of hypervirulent Klebsiella pneumoniae harbouring blaVIM-2 among mechanically-ventilated drug-poisoning patients with high mortality rate in Iran. J Glob Antimicrob Resist. (2018) 15:93–8. doi: 10.1016/j.jgar.2018.06.020
148. Liu Y, Long D, Xiang T-X, Du F-L, Wei DD, Wan L-G, et al. Whole genome assembly and functional portrait of hypervirulent extensively drug-resistant NDM-1 and KPC-2 co-producing Klebsiella pneumoniae of capsular serotype K2 and ST86. J Antimicrob Chemother. (2019) 74:1233–240. doi: 10.1093/jac/dkz023
149. Mataseje LF, Boyd DA, Mulvey MR, Longtin Y. Two Hypervirulent Klebsiella pneumoniae isolates producing a blaKPC-2 carbapenemase from a canadian patient. Antimicrob Agents Chemother. (2019) 63:1–4. doi: 10.1128/AAC.00517-19
150. Harada S, Aoki K, Ishii Y, Ohno Y, Nakamura A, Komatsu M, et al. Emergence of IMP-producing hypervirulent Klebsiella pneumoniae carrying a pLVPK-like virulence plasmid. Int J Antimicrob Agents. (2019) 53:873–5. doi: 10.1016/j.ijantimicag.2019.05.007
151. Octavia S, Kalisvar M, Venkatachalam I, Ng OT, Xu W, Sridatta PSR, et al. Klebsiella pneumoniae and Klebsiella quasipneumoniae define the population structure of blaKPC-2Klebsiella: a 5 year retrospective genomic study in Singapore. J Antimicrob Chemother. (2019) 74:3205–10. doi: 10.1093/jac/dkz332
152. Chen Y, Marimuthu K, Teo J, Venkatachalam I, Cherng BPZ, De Wang L, et al. Acquisition of plasmid with carbapenem-resistance gene bla KPC2 in hypervirulent Klebsiella pneumoniae, Singapore. Emerg Infect Dis. (2020) 26:549–59. doi: 10.3201/eid2603.191230
153. Beyrouthy R, Dalmasso G, Birer A, Robin F, Bonnet R. Carbapenem resistance conferred by OXA-48 in K2-ST86 hypervirulent Klebsiella pneumoniae, France. Emerg Infect Dis. (2020) 26:1529–33. doi: 10.3201/eid2607.191490
154. Li Q, Zhu J, Kang J, Song Y, Yin D, Guo Q, et al. Emergence of NDM-5-Producing carbapenem-resistant Klebsiella pneumoniae and SIM-producing hypervirulent Klebsiella pneumoniae isolated from aseptic body fluid in a large tertiary hospital, 2017–2018: genetic traits of blaNDM-Like and blaSIM-like genes as. Infect Drug Resist. (2020) 13:3075–89. doi: 10.2147/IDR.S261117
155. Feng Y, Lu Y, Yao Z, Zong Z. Carbapenem-resistant hypervirulent Klebsiella pneumoniae of sequence type 36. Antimicrob Agents Chemother. (2018) 62:e02644–17. doi: 10.1128/AAC.02644-17
156. Yang X, Dong N, Chan EWC, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. (2021) 29:65–83. doi: 10.1016/j.tim.2020.04.012
157. Tang M, Kong X, Hao J, Liu J. Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. (2020) 11:1–10. doi: 10.3389/fmicb.2020.581543
158. Gupta A. Hospital-acquired infections in the neonatal intensive care unit-Klebsiella pneumoniae. Semin Perinatol. (2002) 26:340–45. doi: 10.1053/sper.2002.36267
159. Martin RM, Cao J, Brisse S, Passet V, Wu W, Zhao L, et al. Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere. (2016) 1:e00261–16. doi: 10.1128/msphere.00261-16
160. Basu S. Neonatal sepsis: the gut connection. Eur J Clin Microbiol Infect Dis. (2014) 34:215–22. doi: 10.1007/s10096-014-2232-6
161. Wortham JM, Hansen NI, Schrag SJ, Hale E, Van Meurs K, Sánchez PJ, et al. Chorioamnionitis and culture-confirmed, early-onset neonatal infections. Pediatrics. (2016) 137:e20152323. doi: 10.1542/peds.2015-2323
162. Das P, Singh AK, Pal T, Dasgupta S, Ramamurthy T, Basu S. Colonization of the gut with Gram-negative bacilli, its association with neonatal sepsis and its clinical relevance in a developing country. J Med Microbiol. (2011) 60:1651–60. doi: 10.1099/jmm.0.033803-0
163. Simonsen KA, Anderson-Berry AL, Delair SF, Davies HD. Early-onset neonatal sepsis. Clin Microbiol Rev. (2014) 27:21–47. doi: 10.1128/CMR.00031-13
164. Chereau F, Herindrainy P, Garin B, Huynh B-T, Randrianirina F, Padget M, et al. Colonization of extended-spectrum-beta-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob Agents Chemother. (2015) 59:3652–5. doi: 10.1128/AAC.00029-15
165. Sheikh SS, Amr SS, Lage JM. Acute placental infection due to Klebsiella pneumoniae: report of a unique case. Infect Dis Obstet Gynecol. (2005) 13:49–52. doi: 10.1080/10647440400028177
166. Torabi R, Charnova S, Abellar RG, Pinar H, De Paepe ME. Intrauterine infection with Klebsiella pneumoniae: report of a case and literature review. Pediatr Dev Pathol. (2008) 11:152–5. doi: 10.2350/07-09-0337.1
167. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. (2006) 118:511–21. doi: 10.1542/peds.2005-2824
168. Bonfanti P, Bellù R, Principe L, Caramma I, Condò M, Giani T, et al. Mother-to-child transmission of KPC carbapenemase-producing Klebsiella pneumoniae at birth. Pediatr Infect Dis J. (2017) 36:228–9. doi: 10.1097/INF.0000000000001403
169. Herindrainy P, Rabenandrasana MAN, Andrianirina ZZ, Rakotoarimanana FMJ, Padget M, de Lauzanne A, et al. Acquisition of extended spectrum beta-lactamase-producing enterobacteriaceae in neonates: a community based cohort in Madagascar. PLoS ONE. (2018) 13:e0193325. doi: 10.1371/journal.pone.0193325
170. Asensio A, Oliver A, González-Diego P, Baquero F, Pérez-Díaz JC, Ros P, et al. Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin Infect Dis. (2000) 30:55–60. doi: 10.1086/313590
171. Waters D, Jawad I, Ahmad A, Lukšić I, Nair H, Zgaga L, et al. Aetiology of community-acquired neonatal sepsis in low- and middle-income countries. J Glob Health. (2011) 1:154–70.
172. Khatri A, Naeger Murphy N, Wiest P, Osborn M, Garber K, Hecker M, et al. Community-acquired pyelonephritis in pregnancy caused by KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother. (2015) 59:4375–8. doi: 10.1128/AAC.00553-15
173. Gooch JW. “Disk diffusion method”. In: Encyclopedic Dictionary of Polymers. New York, NY: Springer New York. (2016). p. 888–888. doi: 10.1007/978-1-4419-6247-8_13582
174. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests: Approved standard - Eleventh edition. M02-A11. Wayne: Scientific Research Publishing (SCIRP) (2012).
175. Clinical and Laboratory Standards Institute. M100 Performance Standards for Antimicrobial Susceptibility Testing. Wayne: Scientific Research Publishing (SCIRP) (2020).
176. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 11.0. Växjö: European society of Clinical Microbiology and Infectious Diseases (ESCMID) (2021).
177. Tamma PD, Simner PJ. Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J Clin Microbiol. (2018) 56:1–3. doi: 10.1128/JCM.01140-18
178. Giske CG, Gezelius L, Samuelsen Ø, Warner M, Sundsfjord A, Woodford N. A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin Microbiol Infect. (2011) 17:552–6. doi: 10.1111/j.1469-0691.2010.03294.x
179. Polsfussa S, Bloemberga GV, Gigera J, Meyera V, Böttgera EC, Hombach M. Evaluation of a rapid extended spectrum beta-lacta mases/ carbapenemases (ESBL/CRO) detection kit in a diagnostic microbiology laboratory. Can J Infect Dis Med Microbiol. (2013) 18:1194–204.
180. Hojabri Z, Arab M, Darabi N, Kia NS, Lopes BS, Pajand O. Evaluation of the commercial combined disk test and minimum inhibitory concentration (MIC) determination for detection of carbapenemase producers among Gram-negative bacilli isolated in a region with high prevalence of bla OXA-48 and bla NDM. Int Microbiol. (2019) 22:81–9. doi: 10.1007/s10123-018-0030-1
181. Pierce VM, Simner PJ, Lonsway DR, Roe-Carpenter DE, Johnson JK, Brasso WB, et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among enterobacteriaceae. J Clin Microbiol. (2017) 55:2321–33. doi: 10.1128/JCM.00193-17
182. Dortet L, Bréchard L, Poirel L, Nordmann P. Rapid detection of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. (2014) 20:340–4. doi: 10.3201/eid1809.120355
183. Lima-Morales D de, Ávila H, Soldi T, Dalmolin TV, Lutz L, Aquino V, Zavascki AP, et al. Rapid detection of carbapenemase production directly from blood culture by colorimetric methods: evaluation in a routine microbiology laboratory. J Clin Microbiol. (2018) 56:1–5. doi: 10.1128/JCM.00325-18
184. Pfaendler HR, Schmidt H-U, Freidank H. The novel carbalux test for carbapenemases and carbapenem deactivating ampc beta-lactamases. Front Microbiol. (2020) 11:588887. doi: 10.3389/fmicb.2020.588887
185. Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, Cotellon G, et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. (2018) 73:909–15. doi: 10.1093/jac/dkx521
186. Jenkins S, Ledeboer NA, Westblade LF, Burnham C-AD, Faron ML, Bergman Y, et al. Evaluation of NG-test carba 5 for rapid phenotypic detection and differentiation of five common carbapenemase families: results of a multicenter clinical evaluation. J Clin Microbiol. (2020) 58:1–1. doi: 10.1128/JCM.00344-20
187. Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence. (2017) 8:1111–23. doi: 10.1080/21505594.2017.1317412
188. Lee CH, Liu JW, Su LH, Chien CC, Li CC, Yang KD. Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int J Infect Dis. (2010) 14:e688–92. doi: 10.1016/j.ijid.2010.01.007
189. Harada S, Doia Y. Hypervirulent Klebsiella pneumoniae: A call for consensus definition and international collaboration. J Clin Microbiol. (2018) 56:e00959–18. doi: 10.1128/JCM.00959-18
190. Korang SK, Safi S, Gluud C, Lausten-Thomsen U, Jakobsen JC. Antibiotic regimens for neonatal sepsis - a protocol for a systematic review with meta-analysis. Syst Rev. (2019) 8:306. doi: 10.1186/s13643-019-1207-1
191. WHO. Executive Summary: Report of the 22nd WHO Expert Committee on Selection and Use of Essential Medicines. Geneva: WHO Med web (2019).
192. Hsia Y, Lee BR, Versporten A, Yang Y, Bielicki J, Jackson C, et al. Use of the WHO Access, Watch, and Reserve classification to define patterns of hospital antibiotic use (AWaRe): an analysis of paediatric survey data from 56 countries. Lancet Glob Heal. (2019) 7:e861–71. doi: 10.1016/S2214-109X(19)30071-3
193. Zasowski EJ, Rybak JM, Rybak MJ. The β-lactams strike back: ceftazidime-avibactam. Pharmacother J Hum Pharmacol Drug Ther. (2015) 35:755–70. doi: 10.1002/phar.1622
194. Hecker SJ, Reddy KR, Totrov M, Hirst GC, Lomovskaya O, Griffith DC, et al. Discovery of a cyclic boronic acid β-lactamase inhibitor (RPX7009) with utility vs class a serine carbapenemases. J Med Chem. (2015) 58:3682–92. doi: 10.1021/acs.jmedchem.5b00127
195. Livermore DM, Mushtaq S, Warner M, Zhang JC, Maharjan S, Doumith M, et al. Activity of aminoglycosides, including ACHN-490, against carbapenem-resistant Enterobacteriaceae isolates. J Antimicrob Chemother. (2011) 66:48–53. doi: 10.1093/jac/dkq408
196. Thomas R, Velaphi S, Ellis S, Walker AS, Standing JF, Heath P, et al. The use of polymyxins to treat carbapenem resistant infections in neonates and children. Expert Opin Pharmacother. (2019) 20:415–22. doi: 10.1080/14656566.2018.1559817
197. Chiotos K, Han JH, Tamma PD. Carbapenem-resistant enterobacteriaceae infections in children. Curr Infect Dis Rep. (2016) 18:1–1. doi: 10.1007/s11908-015-0510-9
198. World Health Organization. Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline. Geneva (2019).
199. Bin-Nun A, Bromiker R, Wilschanski M, Kaplan M, Rudensky B, Caplan M, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight neonates. J Pediatr. (2005) 147:192–6. doi: 10.1016/j.jpeds.2005.03.054
200. Lin H-C, Hsu C-H, Chen H-L, Chung M-Y, Hsu J-F, Lien R, et al. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. (2008) 122:693–700. doi: 10.1542/peds.2007-3007
201. Braga TD, da Silva GAP, de Lira PIC, de Carvalho Lima M. Efficacy of Bifidobacterium breve and Lactobacillus casei oral supplementation on necrotizing enterocolitis in very-low-birth-weight preterm infants: a double-blind, randomized, controlled trial. Am J Clin Nutr. (2011) 93:81–6. doi: 10.3945/ajcn.2010.29799
202. Sohn K, Underwood MA. Prenatal and postnatal administration of prebiotics and probiotics. Semin Fetal Neonatal Med. (2017) 22:284–9. doi: 10.1016/j.siny.2017.07.002
203. Rao SC, Athalye-Jape GK, Deshpande GC, Simmer KN, Patole SK. Probiotic supplementation and late-onset sepsis in preterm infants: a meta-analysis. Pediatrics. (2016) 137:e20153684. doi: 10.1542/peds.2015-3684
204. Aceti A, Maggio L, Beghetti I, Gori D, Barone G, Callegari M, et al. Probiotics prevent late-onset sepsis in human milk-fed, very low birth weight preterm infants: systematic review and meta-analysis. Nutrients. (2017) 9:904. doi: 10.3390/nu9080904
205. Zhang G-Q, Hu H-J, Liu C-Y, Shakya S, Li Z-Y. Probiotics for preventing late-onset sepsis in preterm neonates. Medicine (Baltimore). (2016) 95:e2581. doi: 10.1097/MD.0000000000002581
206. Cohen-Wolkowiez M, Benjamin DK, Capparelli E. Immunotherapy in neonatal sepsis: advances in treatment and prophylaxis. Curr Opin Pediatr. (2009) 21:177–181. doi: 10.1097/MOP.0b013e32832925e5
207. Pammi M, Weisman LE. Late-onset sepsis in preterm infants: update on strategies for therapy and prevention. Expert Rev Anti Infect Ther. (2015) 1–18. doi: 10.1586/14787210.2015.1008450
208. Weisman LE, Thackray HM, Steinhorn RH, Walsh WF, Lassiter HA, Dhanireddy R, et al. A randomized study of a monoclonal antibody (pagibaximab) to prevent staphylococcal sepsis. Pediatrics. (2011) 128:271–9. doi: 10.1542/peds.2010-3081
209. Diago-Navarro E, Motley MP, Ruiz-Peréz G, Yu W, Austin J, Seco BMS, et al. Novel, broadly reactive anticapsular antibodies against carbapenem-resistant Klebsiella pneumoniae protect from infection. MBio. (2018) 9:1–1. doi: 10.1128/mBio.00091-18
Keywords: neonatal sepsis, Klebsiella pneumoniae, carbapenem resistance, hypervirulence, CR-hvKP, antibiotics, treatment, lower-middle-income countries
Citation: Mukherjee S, Mitra S, Dutta S and Basu S (2021) Neonatal Sepsis: The Impact of Carbapenem-Resistant and Hypervirulent Klebsiella pneumoniae. Front. Med. 8:634349. doi: 10.3389/fmed.2021.634349
Received: 27 November 2020; Accepted: 26 April 2021;
Published: 11 June 2021.
Edited by:
Alessandro Russo, University of Magna Graecia, ItalyReviewed by:
Terri J. Harford, Cleveland Clinic, United StatesChaitra Shankar, Christian Medical College and Hospital, India
Copyright © 2021 Mukherjee, Mitra, Dutta and Basu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sulagna Basu, YmFzdXMubmljZWRAZ292Lmlu; c3VwYWJhc3VAeWFob28uY28uaW4=
†Present Address: Subhankar Mukherjee, Department of Zoology, Government General Degree College, Singur, India
‡These authors share first authorship
 Subhankar Mukherjee
Subhankar Mukherjee Shravani Mitra
Shravani Mitra Shanta Dutta
Shanta Dutta Sulagna Basu
Sulagna Basu