
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Med. , 30 November 2020
Sec. Precision Medicine
Volume 7 - 2020 | https://doi.org/10.3389/fmed.2020.612393
This article is part of the Research Topic Nucleic Acids-Based Cancer Theranostics View all 29 articles
Long non-coding RNAs (lncRNAs) are major components of cellular transcripts that are arising as important players in various biological pathways. They have received extensive attention in recent years, regarded to be involved in both developmental processes and various diseases. Due to their specific expression and functional diversity in a variety of cancers, lncRNAs have promising applications in cancer diagnosis, prognosis and therapy. Studies have shown that lncRNAs with high specificity and accuracy have the potential to become biomarkers in cancers. LncRNAs can be noninvasively extracted from body fluids, tissues and cells, and can be used as independent or auxiliary biomarkers to improve the accuracy of diagnosis or prognosis. Currently, the most well-recognized lncRNA is PCA3, which has been approved for use in the diagnosis of prostate cancer. Moreover, the underlying mechanisms of lncRNAs were explored as therapeutic targets, which have been investigated in clinical trials of several cancers. In this review, we presented a compilation of recent publications, clinical trials and patents, addressing the potential of lncRNAs that could be considered as biomarkers or therapeutic targets, with the hopes of providing promised implications for future cancer therapy.
Cancer is a life-threating disease with rising morbidity and mortality (1). Despite tremendous progress made in recent years, there are still a number of issues in cancer treatment that need improvements, such as delayed diagnosis and poor prognosis (2). Most tumor biomarkers or therapeutic targets currently in clinical use are proteins. However, only 2% of human genome is translated into proteins. Therefore, we may need to focus more on non-coding regions, where more cancer mutations occur than in coding regions (2, 3). In recent years, long non-coding RNAs (lncRNAs), which occupy the majority of non-coding RNAs (ncRNAs), are hotspots in cancer research. Due to the large number of lncRNAs, with an estimate of 102,000, lncRNA-based research holds great promise in cancer treatment (3, 4).
LncRNAs are non-coding transcripts with more than 200 nucleotides in length, and most of them remain in the nucleus after transcription (5, 6). Due to their low expression levels, lncRNAs were initially considered to be transcription noise. With better understanding, lncRNAs are found to be involved in transcriptional and post-transcriptional regulation, through interactions with DNA, RNA or proteins (6). LncRNAs promote or inhibit the formation of transcription loops, and recruit or block regulators, regulating gene transcription (7–9). Besides, lncRNAs also regulate mRNA splicing and act as precursors to other ncRNAs, such as microRNAs (miRNAs) (10). LncRNAs function as oncogenes or tumor suppressors, taking part in various signaling pathways (11). Of note, by analyzing the expression of lncRNAs in peripheral blood, urine sediments or tissue samples, a series of lncRNAs were identified with great promise as auxiliary or independent biomarkers in cancer diagnosis and prognosis (12).
There are currently few biomarkers or therapeutic agents targeting lncRNAs. Prostate cancer antigen 3 (PCA3), an early diagnostic biomarker for prostate cancer (PCa), is the first and only approved lncRNA for clinical use at this time (13). There are also some lncRNAs undergoing clinical trials or having been patented, which we will discuss in more details below. Moreover, other research of lncRNA-based drug discoveries, including UBE3A-ATS in Angelman syndrome, SCN1ANAT in Dravet syndrome and SMN-AS1 in spinal muscular atrophy, also illustrate the potential of lncRNAs (14–17).
In this review, we discuss the mechanisms by which lncRNAs function. A thorough understanding of these mechanisms is critical for the development of anti-tumor drugs. We next summarize a collection of recent publications, clinical trials, and patents and also discuss the potential of lncRNAs that could be considered as biomarkers or therapeutic targets in cancer diagnosis, prognosis and treatment.
Understanding how lncRNAs work is critical to know how they cause diseases such as cancers, and therefore to their potential applications in cancer treatment. Based on current studies, lncRNAs are implicated in many intracellular molecular interaction networks. The levels of their expression are regulated by many factors, and they are also involved in complex networks as regulatory factors. The myriad mechanisms behind these complex regulations can be summarized in four ways, including signal, scaffold, decoy, and guide (Figure 1) (5, 18, 19).
(i) Some lncRNAs are expressed at different levels in various cell states. Thus, they can be turned into signals, serving as indicators to reflect development or disease status (18, 20). For example, Xist, which is typically transcribed by the inactive X chromosome, can be used to indicate X chromosome inactivation (21, 22).
(ii) LncRNAs can bind proteins and act as scaffolds to assist in the assembly of regulatory complexes (23). In this way, HOTAIR interacts with polycomb repressive complex 2 (PRC2) to recruit EZH2 to promote H3K27 trimethylation or LSD1 to demethylate H3K4me2 (23, 24).
(iii) As decoys, lncRNAs regulate gene expression by preventing the binding of transcription regulators (19, 25). For example, p53-dependent PANDA inhibits proptosis by directly sequestering of NF-YA (26). In addition, as competing endogenous RNAs (ceRNAs), lncRNAs also bind miRNAs and prevent RNA degradation (27). This is common in cancers. H19 acts as ceRNAs both for miR-17-5P in thyroid cancer and for miR-152 in breast cancer (27, 28).
(iv) LncRNAs can also guide the transcription factors to specific sites (29). In this way, MEG3 guides PRC2 and forms a complex with DNA (30). It is noteworthy that each type is not mutually exclusive, and an individual lncRNA may have one or more of these functions (20).
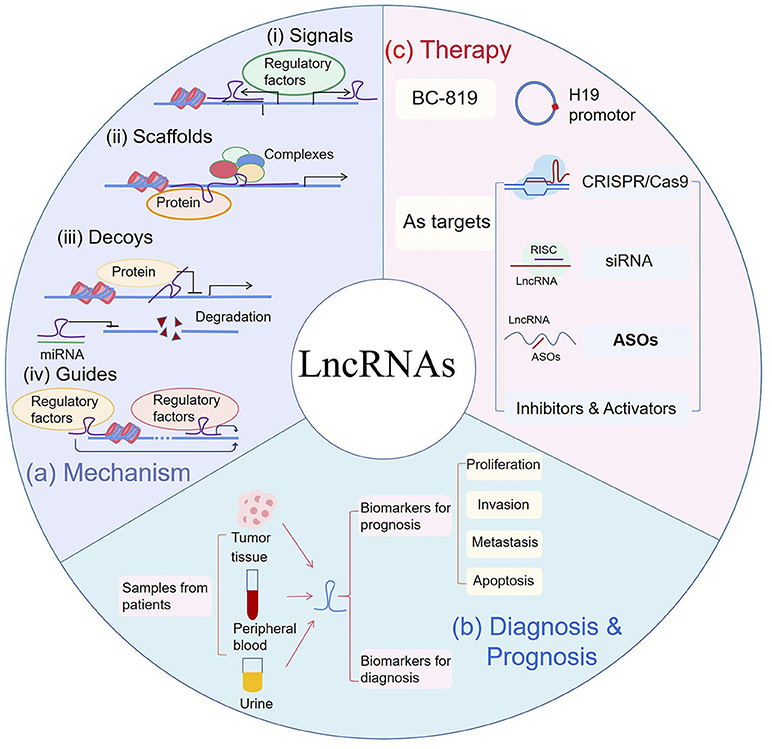
Figure 1. (a) Mechanism: (i) LncRNAs can serve as signals to reflect the activity of pathways or developmental status. (ii) LncRNAs act as scaffolds by recruiting proteins to regulate gene expression. (iii) LncRNAs can be used as decoys to block activities of proteins and can also bind to microRNA (miRNA) to inhibit miRNA-induced degradation. (iv) As guides, lncRNAs recruit transcription regulators to specific sites. (b) Diagnosis & Prognosis: As biomarkers for cancer diagnosis and prognosis, lncRNAs can be extracted from tumor tissues, peripheral blood and urine samples of patients. In prognosis, they are correlated to patient's proliferation, metastasis, invasion or survival. (c) Therapy: BC-819, fused with H19 promotor and Diphtheria toxin gene, was used in phase 2 clinical trial. Gene editing such as CRISPR/Cas9, small interfering RNA (siRNA) and antisense oligonucleotides (ASOs), were used to silence targeted lncRNAs. RISC RNA-induced silencing complex.
Some lncRNAs are highly tissue specific and abnormally expressed in cancer, which can be extracted noninvasively from the circulation (31, 32). These features render them potential candidates for cancer diagnosis (Table 1). The one most well-recognized is PCA3, a biomarker for early diagnosis of prostate cancer (PCa) (67). As mentioned above, it has been approved in clinical use (13). The diagnosis of PCa currently relies on the elevation of serum prostate-specific antigen (PSA) (12). However, its specificity in discriminating benign and malignant tumors is low, which may lead to over-diagnosis in low-risk patients (68). PSA is a good predictor when its level is above 10 ng/ml. However, in the gray area of PSA, some auxiliary indicators are still needed to improve diagnostic accuracy (12).
Urine PCA3 diagnosis is not only highly sensitive (58–82%) but also has excellent specificity (59–76%) (33). PCA3 is up-regulated 60 to 100-folds in more than 95% of PCa specimens (18, 33). It is noteworthy that PCA3-based assays are still effective when cancer cells make up <10% of the examined sample (33). PCA3 silence leads to an increase in the expression of E-cadherin, Claudin-3, and Keratin-18, while a decrease of Vimentin. The association between PCA3 and these traditional protein biomarkers provides more support for its application as diagnostic marker (34).
MALAT1 can be used as an auxiliary biomarker to improve the accuracy of early diagnosis, especially in the gray area of PSA. Its diagnostic accuracy is higher than the previous index, free/total PSA ratio (12, 69). In fact, one of the MALAT1 assay has been patented in PCa diagnosis (CN104498495).
H19 is another lncRNA with high diagnostic sensitivity and specificity. In breast cancer, patient plasma H19 levels were elevated, with an sensitivity of 0.81 (AUC, area under the curve), higher than traditional diagnostic biomarkers (42). H19 is related to many important miRNAs in the network of cancer-related functions (70). It is a precursor of miR-675 which has downstream targets like c-CbI, CbI-b and Igf1r (71, 72). H19/miR-675 also causes the activation of EGRF and c-Met, which leads to the sustained activation of Akt and Erk (43). Besides, H19 acts as a ceRNA for Let-7 to maintain the activation of breast cancer stem cells (73). The potential of miRNAs like miR-675 and Let-7 as biomarkers has been reported in cancers, which implicate the diagnostic potential of H19 (74). In gastric cancer (GC), H19 had a high diagnostic ability with an AUC of 0.838. A patent has been filed for gastric cancer diagnosis with HOTAIR and MALAT1 (CN105586399A), showing possible applications. Notably, single nucleotide polymorphisms (SNPs) of H19 are used to predict the risk of cancer, such as rs2839698 and rs2107425 genotypes are found to be related to decreased risk of bladder cancer (75).
LncRNAs are usually aberrantly expressed and can be extracted noninvasively from the circulation (32). Although some lncRNAs overlap in various cancer, ~60% of these abnormally expressed lncRNAs are cancer type-specific (31). Recently, the diagnostic potential of some lncRNAs has been implicated by clinical trials (76). One report have shown that UCA1 was sensitive for bladder cancer, especially in patients with superficial G2-G3 (77). Another clinical trial is underway to explore the possible application of CCAT1 in CRC (NCT04269746). Moreover, lncRNAs can not only be used as an independent biomarkers, but also can be combined with other lncRNAs or proteins to improve the sensitivity and accuracy of diagnosis (76).
The expression of lncRNAs in cancer correlates with overall survival (OS), metastasis, tumor stage or grade, thus can potentially serve as markers for prognosis (Table 1).
HOTAIR is transcribed from HOXC locus located at 12q13.13 (78). It has been proved to be poor prognostic indicators of various cancers. Bladder transitional cell carcinoma (TCC) patients with high HOTAIR have lower overall survival, and positively associated with histological grade (41).
An analysis of a large cohort containing 300 samples showed that the increased expression level of HOTAIR in GC tissues was correlated with peritoneal diffusion (55). In addition, in diffuse GC, tissues with a high HOTAIR level showed more venous infiltration and poorer overall survival (56). Besides, HOTAIR is involved in tumorigenicity in pancreatic cancer and can also cause CRC proliferation and metastasis mediated by PCR2 complex (51, 52).
MALAT1, located on chromosome 11q13.1, is dysregulated in many cancers (49). A cohort analysis of 169 patients showed that patients with high MALAT1 expression levels had a worse prognosis than the normal group (79). Moreover, upregulated MALAT1 is closely related to hepatocellular carcinoma (HCC) progression, and can be an independent biomarker for recurrence after liver transplantation (80). However, the roles of MALAT1 in breast cancer are still controversial. In previous reports, MALAT1 acts as a ceRNA for miR-1/CDC42 axis to enhance cell migration and invasion (44). On the contrary, Kim et al. reported that MALAT1 acted as a metastasis suppressor by preventing the binding of transcription factor TEAD and its co-activator YAP (45). In addition, MALAT1 knockout leads to different phenotypes in various cell lines and models, and there is no clear explanation for this variation. Therefore, further investigation is required before MALAT1 could be use as a potential prognositic biomarker (81, 82).
CCAT2 showed extensive effects during proliferation and metastasis in a variety of cancers (47). In fact, patients with high CCAT2 had a lower overall survival and almost twice the risk of death (48).
The aberrant expression of lncRNAs and their involvement in diverse cellular processes make them possible targets for cancer therapy. Clinical studies have demonstrated the importance of studying the mechanisms of lncRNA. BC-819, a plasmid containing the promoter of H19 and coding sequence of diphtheria toxin, has been applied in clinical trials of bladder, pancreatic and ovarian cancer (83, 84). The H19 promoter allows diphtheria toxin to be specifically expressed in tumor tissues. Thus, BC-819 can effectively ablate tumors, reduce tumor growth, prolong recurrence time, and has low local toxicity (83–85). We can also learn about the prospect of lncRNAs from patent applications (86). For example, an inhibitor of LINC01212 is used to treat melanoma (US2016271163). What's more, lncMyoD, acts directly as functional element on IMP1 and IMP2 for sarcoma therapy (WO2015020960). Furthermore, although lncRNAs are considered with no protein-coding ability, some special lncRNAs can be translated into micropeptides (87). In fact, some patents utilized these polypeptides for antibodie design in cancer diagnosis and treatment, such as lncRNA-6585 and its antibody in cervical cancers (CN109337903A).
LncRNAs and their loci can be targeted for the design and synthesis of specific nucleic acid sequences in therapy, such as CRISPR/Cas9 design, small interfering RNA (siRNA) and antisense oligonucleotides (ASOs). However, unlike mRNA, most lncRNAs are located in the nucleus and have high-level structure (88, 89). Oligonucleotide drugs must enter cells and bind to their target RNA to function, which raises challenges for drug delivery and intrinsic affinity (90). To address these issues, a common approach is to modify the sequence of oligonucleotides, and the development of nanomedicine to improve drug delivery.
(i) CRISPR/Cas9 silencing of NEAT1 or MALAT1 was reported to inhibit metastasis of cancer cells (91). A patent used CRISPR/Cas9 to silence UCA1 inhibited the growth of cancer cells (CN106399306B). However, due to the overlap of loci, CRISPR/Cas9 cannot be applied to the silencing of all lncRNAs. In a genome-wide study including 15,929 lncRNA loci, only 38% were successfully silenced as expected, while the remaining had a severe negative impact on the expression of neighboring genes (91).
(ii) Both of siRNA and ASOs can effectively and specifically silence the expression of target genes, making them essential tools for research and clinical uses. Despite some challenges, the progress of siRNAs as therapy drugs has evolved from pre-design to clinical trials (92). Recently, a siRNA targeting DDX11-AS1 has been patented in liver cancer (CN108546702A). Compare to siRNA, ASOs enter the nucleus more efficiently and bind to precursor RNAs near the intron and exon junction, affecting the alternative splicing process (93).
(iii) Methods for affinity and delivery improvement: Affinity of oligonucleotides can be improved by constructing aptamers (94). They can also be modified to reduce nuclease degradation and increase their internal affinity (95, 96). For delivery, one effective approach is to construct drug carriers, such as in the form of gold nanoparticles (97).
In recent years, many studies have been devoted to lncRNAs in cancer progression and treatment. Due to their highly specific expression and diverse functions, lncRNAs hold great promise for cancer diagnosis, prognosis and therapy. To the best of our knowledge, however, the sole lncRNA that has been approved in clinical use is PCA3 in the diagnosis of PCa. Although multiple lncRNAs have been extensively investigated in clinical trials or have been patented, their applications still have a long way to go. Here we review potential lncRNAs that could be considered as biomarkers or therapeutic targets and discuss some of the issues that deserve special consideration.
First, the actual mechanisms by which lncRNA act are not fully understood. Indeed, the developing of oligonucleotide drug Genasense can be served as an informative case. Due to the lack of in-depth understanding of the mechanism, the development of Genasense failed, revealing the importance of understanding the mechanism in drug development (93). Second, the low conservation of lncRNAs, some of which are expressed only in primates, makes it difficult to establish universal experimental models (98). For the majority of lncRNAs, we have yet to establish a suitable animal model, which is essential for understanding the functions better. Third, although some experiments have been conducted on the applications of lncRNAs, they are not very reliable due to the small sample size. Moreover, for therapeutic targets, it is important to study whether the dysregulated expression of lncRNAs is the cause or a result of cancer.
Although there are many challenges, the prospects and clinical significance of lncRNAs cannot be overlooked in the long run. A distinctive feature of lncRNAs is their high specificity in tumor tissues and cells, making it possible for them to be specific and accurate biomarkers (99). In addition, abnormally expressed lncRNAs can be extracted non-invasively, showing great potential to be more economical and less harmful. Compared to protein-based anti-tumor drugs, lncRNA are more refined and less toxic, and the low expression of lncRNA means that only a small amount of inhibitors are needed to make a difference (16). Besides, bioinformatics and computational tools provide new opportunities for lncRNA biomarker development (100). However, due to the lack of experimental evidence and no further clinical validation, we do not discuss it here. Although there are currently no lncRNA-based oncology drugs, drugs targeting lncRNAs in other diseases will provide useful clinical insights.
In conclusion, the intensive study of lncRNAs has brought new hope for the diagnosis and treatment of cancer. Although limitations exist, such as mechanisms, conservative, and animal models, the successful application of PCA3 is a great source of inspiration and impetus for clinical research on lncRNAs. A comprehensive understanding of lncRNA's expression, structure, and mechanisms will help to open up a new intervention, identifying novel and sensitive biomarkers and therapeutic targets.
YQ and LS participated in the design of the study, prepared table, figure, and wrote the manuscript. LS and ZL conceived the study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Fundamental Research Funds for the Central Universities (2019CDYGYB024), the National Natural Science Foundation of China (31300726), and Chongqing Primary and Middle School Innovation Talent Training Project (CY200118).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
LncRNA, Long Non-coding RNA; ncRNA, Non-coding RNA; miRNA, MicroRNA; PCA3, Prostate Cancer Antigen 3; PCa, Prostate Cancer; ceRNA, Competing Endogenous RNA; PSA, Prostate-specific Antigen; AUC, Area Under the Curve; EGRF, Epidermal Growth Factor Receptor; GC, Gastric Cancer; SNPs, Single Nucleotide Polymorphisms; CRC, Colorectal Cancer; OS, Overall Survival; TCC, Bladder Transitional Cell Carcinoma; EMT, Epithelial to Mesenchymal Transition; HCC, Hepatocellular Carcinoma.
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients, from biomarkers to therapeutic targets. Int J Cancer. (2017) 140:1955–67. doi: 10.1002/ijc.30546
3. Cuykendall TN, Rubin MA, Khurana E. Non-coding genetic variation in cancer. Curr Opin Syst Biol. (2017) 1:9–15. doi: 10.1016/j.coisb.2016.12.017
4. Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, et al. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. (2016) 44:D203–8. doi: 10.1093/nar/gkv1252
5. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08.018
6. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
7. Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. (2013) 498:516–20. doi: 10.1038/nature12210
8. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature. (2013) 494:497–501. doi: 10.1038/nature11884
9. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. (2009) 106:11667–72. doi: 10.1073/pnas.0904715106
10. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. (2017) 1008:1–46. doi: 10.1007/978-981-10-5203-3_1
11. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. (2015) 21:1253–61. doi: 10.1038/nm.3981
12. Smolle MA, Bauernhofer T, Pummer K, Calin GA, Pichler M. Current insights into long non-coding RNAs (LncRNAs) in prostate cancer. Int J Mol Sci. (2017) 18:473. doi: 10.3390/ijms18020473
13. Groskopf J, Aubin SM, Deras IL, Blase A, Bodrug S, Clark C, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. (2006) 52:1089–95. doi: 10.1373/clinchem.2005.063289
14. Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. (2015) 518:409–12. doi: 10.1038/nature13975
15. Hsiao J, Yuan TY, Tsai MS, Lu CY, Lin YC, Lee ML, et al. Upregulation of haploinsufficient gene expression in the brain by targeting a long non-coding RNA improves seizure phenotype in a model of dravet syndrome. EBioMedicine. (2016) 9:257–77. doi: 10.1016/j.ebiom.2016.05.011
16. d'Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, et al. The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron. (2017) 93:66–79. doi: 10.1016/j.neuron.2016.11.033
17. Woo CJ, Maier VK, Davey R, Brennan J, Li G, Brothers J, et al. Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. Proc Natl Acad Sci USA. (2017) 114:E1509–18. doi: 10.1073/pnas.1616521114
18. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. (2016) 73:2491–509. doi: 10.1007/s00018-016-2174-5
19. Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. (2018) 3:108–17. doi: 10.1016/j.ncrna.2018.03.001
20. Kazemzadeh M, Safaralizadeh R, Orang AV. LncRNAs: emerging players in gene regulation and disease pathogenesis. J Genet. (2015) 94:771–84. doi: 10.1007/s12041-015-0561-6
21. Pontier DB, Gribnau J. Xist regulation and function explored. Hum Genet. (2011) 130:223–36. doi: 10.1007/s00439-011-1008-7
22. Zylicz JJ, Bousard A, Zumer K, Dossin F, Mohammad E, da Rocha ST, et al. The implication of early chromatin changes in X chromosome inactivation. Cell. (2019) 176:182–97.e23. doi: 10.1016/j.cell.2018.11.041
23. Zappulla DC, Cech TR. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc Natl Acad Sci USA. (2004) 101:10024–9. doi: 10.1073/pnas.0403641101
24. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
25. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
26. Puvvula PK, Desetty RD, Pineau P, Marchio A, Moon A, Dejean A, et al. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nat Commun. (2014) 5:5323. doi: 10.1038/ncomms6323
27. Li Z, Li Y, Li Y, Ren K, Li X, Han X, et al. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol. (2017) 31:e21933. doi: 10.1002/jbt.21933
28. Liu L, Yang J, Zhu X, Li D, Lv Z, Zhang X. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. (2016) 283:2326–39. doi: 10.1111/febs.13741
29. Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. (2010) 7:582–5. doi: 10.4161/rna.7.5.13216
30. Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun. (2015) 6:7743. doi: 10.1038/ncomms8743
31. Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. (2015) 28:529–40. doi: 10.1016/j.ccell.2015.09.006
32. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
33. Hessels D, Klein Gunnewiek JM, van Oort I, Karthaus HF, van Leenders GJ, van Balken B, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. (2003) 44:8–15. discussion: 15–6. doi: 10.1016/S0302-2838(03)00201-X
34. Lemos AE, Ferreira LB, Batoreu NM, de Freitas PP, Bonamino MH, Gimba ER. PCA3 long noncoding RNA modulates the expression of key cancer-related genes in LNCaP prostate cancer cells. Tumour Biol. (2016) 37:11339–48. doi: 10.1007/s13277-016-5012-3
35. Salameh A, Lee AK, Cardo-Vila M, Nunes DN, Efstathiou E, Staquicini FI, et al. PRUNE2 is a human prostate cancer suppressor regulated by the intronic long noncoding RNA PCA3. Proc Natl Acad Sci USA. (2015) 112:8403–8. doi: 10.1073/pnas.1507882112
36. Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. (2013) 49:2949–59. doi: 10.1016/j.ejca.2013.04.026
37. Luo J, Wang K, Yeh S, Sun Y, Liang L, Xiao Y, et al. LncRNA-p21 alters the antiandrogen enzalutamide-induced prostate cancer neuroendocrine differentiation via modulating the EZH2/STAT3 signaling. Nat Commun. (2019) 10:2571. doi: 10.1038/s41467-019-09784-9
38. Zheng R, Du M, Wang X, Xu W, Liang J, Wang W, et al. Exosome-transmitted long non-coding RNA PTENP1 suppresses bladder cancer progression. Mol Cancer. (2018) 17:143. doi: 10.1186/s12943-018-0880-3
39. Xue M, Pang H, Li X, Li H, Pan J, Chen W. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. (2016) 107:18–27. doi: 10.1111/cas.12844
40. Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. (2017) 388:281–91. doi: 10.1016/j.canlet.2016.12.005
41. Sun X, Du P, Yuan W, Du Z, Yu M, Yu X, et al. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. (2015) 6:e1907. doi: 10.1038/cddis.2015.269
42. Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. (2016) 17:187–94. doi: 10.3233/CBM-160630
43. Zhan H, Tu S, Zhang F, Shao A, Lin J. MicroRNAs and long non-coding RNAs in c-Met-regulated cancers. Front Cell Dev Biol. (2020) 8:145. doi: 10.3389/fcell.2020.00145
44. Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, et al. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. (2016) 472:262–9. doi: 10.1016/j.bbrc.2016.02.102
45. Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. (2018) 50:1705–15. doi: 10.1038/s41588-018-0252-3
46. Younger ST, L J. Rinn: 'Lnc'-ing enhancers to MYC regulation. Cell Res. (2014) 24:643–4. doi: 10.1038/cr.2014.54
47. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. (2013) 23:1446–61. doi: 10.1101/gr.152942.112
48. Zhang X, Xu Y, He C, Guo X, Zhang J, He C, et al. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. (2015) 111:834–9. doi: 10.1002/jso.23888
49. Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3' end functional motif in colorectal cancer metastasis. Int J Oncol. (2011) 39:169–75. doi: 10.3892/ijo.2011.1007
50. Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. (2003) 88:5119–26. doi: 10.1210/jc.2003-030222
51. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. (2011) 71:6320–6. doi: 10.1158/0008-5472.CAN-11-1021
52. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. (2013) 32:1616–25. doi: 10.1038/onc.2012.193
53. Zhang J, Qiu WQ, Zhu H, Liu H, Sun JH, Chen Y, et al. HOTAIR contributes to the carcinogenesis of gastric cancer via modulating cellular and exosomal miRNAs level. Cell Death Dis. (2020) 11:780. doi: 10.1038/s41419-020-02946-4
54. Elsayed ET, Salem PE, Darwish AM, Fayed HM. Plasma long non-coding RNA HOTAIR as a potential biomarker for gastric cancer. Int J Biol Markers. (2018) 33:1724600818760244. doi: 10.1177/1724600818760244
55. Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. (2014) 35:2731–9. doi: 10.1093/carcin/bgu200
56. Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS ONE. (2013) 8:e77070. doi: 10.1371/journal.pone.0077070
57. Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. (2016) 142:2231–7. doi: 10.1007/s00432-016-2183-7
58. Li L, Geng Y, Feng R, Zhu Q, Miao B, Cao J, et al. The human RNA surveillance factor UPF1 modulates gastric cancer progression by targeting long non-coding RNA MALAT1. Cell Physiol Biochem. (2017) 42:2194–206. doi: 10.1159/000479994
59. Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. (2016) 7:56209–18. doi: 10.18632/oncotarget.10941
60. Abbastabar M, Sarfi M, Golestani A, Khalili E. lncRNA involvement in hepatocellular carcinoma metastasis and prognosis. EXCLI J. (2018) 17:900–13. doi: 10.17179/excli2018-1541
61. Liu D, Zhu Y, Pang J, Weng X, Feng X, Guo Y. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem. (2018) 119:1368–80. doi: 10.1002/jcb.26297
62. Li C, Miao R, Liu S, Wan Y, Zhang S, Deng Y, et al. Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis. Oncotarget. (2017) 8:28683–95. doi: 10.18632/oncotarget.15640
63. Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS, et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour Biol. (2015) 36:2501–7. doi: 10.1007/s13277-014-2863-3
64. Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell Signal. (2015) 27:275–82. doi: 10.1016/j.cellsig.2014.11.011
65. Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. (2015) 367:122–8. doi: 10.1016/j.canlet.2015.03.027
66. Di W, Li Q, Shen W, Guo H, Zhao S. The long non-coding RNA HOTAIR promotes thyroid cancer cell growth, invasion and migration through the miR-1-CCND2 axis. Am J Cancer Res. (2017) 7:1298–1309.
67. Xu YH, Deng JL, Wang G, Zhu YS. Long non-coding RNAs in prostate cancer: functional roles and clinical implications. Cancer Lett. (2019) 464:37–55. doi: 10.1016/j.canlet.2019.08.010
68. Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. (2014) 65:534–42. doi: 10.1016/j.eururo.2012.11.014
69. Wang F, Ren S, Chen R, Lu J, Shi X, Zhu Y, et al. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. (2014) 5:11091–02. doi: 10.18632/oncotarget.2691
70. Ghafouri-Fard S, Esmaeili M, Taheri M. H19 lncRNA: roles in tumorigenesis. Biomed Pharmacother. (2020) 123:109774. doi: 10.1016/j.biopha.2019.109774
71. Vennin C, Spruyt N, Dahmani F, Julien S, Bertucci F, Finetti P, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. (2015) 6:29209–23. doi: 10.18632/oncotarget.4976
72. Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. (2012) 14:659–65. doi: 10.1038/ncb2521
73. Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. (2017) 8:e2569. doi: 10.1038/cddis.2016.438
74. Chirshev E, Oberg KC, Ioffe YJ, Unternaehrer JJ. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin Transl Med. (2019) 8:24. doi: 10.1186/s40169-019-0240-y
75. Verhaegh GW, Verkleij L, Vermeulen SH, den Heijer M, Witjes JA, Kiemeney LA. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur Urol. (2008) 54:1118–26. doi: 10.1016/j.eururo.2008.01.060
76. Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif . (2017) 50:e12329. doi: 10.1111/cpr.12329
77. Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clin Cancer Res. (2006) 12:4851–8. doi: 10.1158/1078-0432.CCR-06-0134
78. Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. (2015) 12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006
79. Zheng HT, Shi DB, Wang YW, Li XX, Xu Y, Tripathi P, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. (2014) 7:3174–81.
80. Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. (2012) 29:1810–6. doi: 10.1007/s12032-011-0004-z
81. Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol. (2019) 16:860–3. doi: 10.1080/15476286.2019.1592072
82. Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. (2013) 91:791–801. doi: 10.1007/s00109-013-1028-y
83. Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. (2010) 12:607–16.
84. Hanna N, Ohana P, Konikoff FM, Leichtmann G, Hubert A, Appelbaum L, et al. Phase 1/2a, dose-escalation, safety, pharmacokinetic and preliminary efficacy study of intratumoral administration of BC-819 in patients with unresectable pancreatic cancer. Cancer Gene Ther. (2012) 19:374–81. doi: 10.1038/cgt.2012.10
85. Gofrit ON, Benjamin S, Halachmi S, Leibovitch I, Dotan Z, Lamm DL, et al. DNA based therapy with diphtheria toxin-A BC-819: a phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J Urol. (2014) 191:1697–702. doi: 10.1016/j.juro.2013.12.011
86. Zhang Y, Tang L. The application of lncRNAs in cancer treatment and diagnosis. Recent Pat Anticancer Drug Discov. (2018) 13:292–301. doi: 10.2174/1574892813666180226121819
87. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. (2015) 160:595–606. doi: 10.1016/j.cell.2015.01.009
88. Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. (2016) 44:863–77. doi: 10.1093/nar/gkv1206
89. Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. (2015) 16:20. doi: 10.1186/s13059-015-0586-4
90. Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu Rev Pharmacol Toxicol. (2010) 50:259–93. doi: 10.1146/annurev.pharmtox.010909.105654
91. Goyal A, Myacheva K, Gross M, Klingenberg M, Duran Arque B, Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. (2017) 45:e12. doi: 10.1093/nar/gkw883
92. Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv Drug Deliv Rev. (2015) 87:108–9. doi: 10.1016/j.addr.2015.01.007
93. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. (2017) 16:167–79. doi: 10.1038/nrd.2016.117
94. Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. (2017) 1008:199–222. doi: 10.1007/978-981-10-5203-3_7
95. Nulf CJ, Corey D. Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs). Nucleic Acids Res. (2004) 32:3792–8. doi: 10.1093/nar/gkh706
96. Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. (1991) 254:1497–500. doi: 10.1126/science.1962210
97. Gong N, Teng X, Li J, Liang XJ. Antisense oligonucleotide-conjugated nanostructure-targeting lncRNA MALAT1 inhibits cancer metastasis. ACS Appl Mater Interfaces. (2019) 11:37–42. doi: 10.1021/acsami.8b18288
98. Boon RA, Jae N, Holdt L, Dimmeler S. Long noncoding RNAs: from clinical genetics to therapeutic targets? J Am Coll Cardiol. (2016) 67:1214–26. doi: 10.1016/j.jacc.2015.12.051
99. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. (2017) 77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634
Keywords: long non-coding RNAs, cancer, biomarker, diagnosis, prognosis, therapy
Citation: Qian Y, Shi L and Luo Z (2020) Long Non-coding RNAs in Cancer: Implications for Diagnosis, Prognosis, and Therapy. Front. Med. 7:612393. doi: 10.3389/fmed.2020.612393
Received: 30 September 2020; Accepted: 09 November 2020;
Published: 30 November 2020.
Edited by:
Guo Chen, Jinan University, ChinaReviewed by:
Zhaojun Qiu, The Ohio State University, United StatesCopyright © 2020 Qian, Shi and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Shi, c2hpbEBjcXUuZWR1LmNu; Zhong Luo, bHVvemhvbmc5MThAY3F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.