- Pulmonary, Critical Care and Sleep Medicine Section, Department of Internal Medicine, Yale University School of Medicine, New Haven, CT, United States
Cellular senescence is a cell fate implicated in the pathogenesis of idiopathic pulmonary fibrosis (IPF) and chronic obstructive pulmonary disease (COPD). Cellular senescence occurs in response to cellular stressors such as oxidative stress, DNA damage, telomere shortening, and mitochondrial dysfunction. Whether these stresses induce cellular senescence or an alternative cell fate depends on the type and magnitude of cellular stress, but also on intrinsic factors regulating the cellular stress response. Non-coding RNAs, including both microRNAs and long non-coding RNAs, are key regulators of cellular stress responses and susceptibility to cellular senescence. In this review, we will discuss cellular mechanisms that contribute to senescence in IPF and COPD and highlight recent advances in our understanding of how these processes are influenced by non-coding RNAs. We will also discuss the potential therapeutic role for targeting non-coding RNAs to treat these chronic lung diseases.
Introduction
Chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF) are chronic lung diseases that disproportionately affect the elderly and impose a significant global health burden. IPF is characterized by chronic and progressive lung scarring while COPD is characterized by heterogenous manifestations of emphysema, small airway disease, and chronic bronchitis. Despite their distinct pathologic features, both diseases are epidemiologically and biologically associated with aging (1–3). The prevalence of COPD amongst individuals aged ≥75 years is approximately 10% compared with 3–4% in those 25–44 years (1, 4) and the prevalence of IPF amongst individuals aged ≥75 years is 0.2–0.3% compared to 0.004–0.012% cases in those aged 35–44 years (5). Additionally, the pathogenesis of IPF and COPD involve biologic “hallmarks of aging” (1, 2, 6–10). These “hallmarks of aging,” first described by Lopez-Otín et al., are cellular processes that occur more frequently with age, contribute to aging-related functional decline, and can be experimentally manipulated to accelerate or slow aging in model organisms (11). One biologic “hallmark of aging” that has emerged as a therapeutic target for ILD, COPD, and other age-related disorders is cellular senescence.
Cellular senescence is a cell fate that occurs in response to diverse causes of cellular stress, such as DNA damage, oxidative stress, telomere shortening, and oncogene activation (12, 13). Cellular senescence is characterized by permanent cell cycle arrest due to persistent activation of p16INK4a-RB (retinoblastoma) and p53-p21CIP1/WAF1 pathways (14, 15). However, senescent cells frequently have altered cellular metabolisms, reorganized chromatin, and activated damage sensing pathways (e.g., p38 MAPK and NF-κB), and are apoptosis resistant. They also adopt a senescence associated secretory phenotype (SASP) and secrete high levels of cytokines, chemokines, and matrix metallopeptidases (MMPs). The biologic consequences of cellular senescence are complex because senescence has both beneficial and detrimental effects. Cellular senescence is critical for embryogenesis, promotes wound healing, and mitigates malignant transformation. However, the accumulation of senescent cells with age also causes chronic inflammation, extracellular matrix alterations, a decline in tissue regeneration, and an increased risk for many aging-related disorders (16).
Cellular senescence is just one of many potential cell fates. Cells maintain diverse stress responses that can resolve cellular stress or activate alternative cell fate pathways such as programmed cell death (e.g., apoptosis/necroptosis), quiescence, or differentiation. While cell fate is influenced by the type, magnitude, and duration of cellular stress, microenvironmental and intracellular factors also influence cell fate “decisions” through modulation of intracellular signaling networks. Consequently, susceptibility to cellular senescence varies across cell/tissue types and with age and in disease (17). There is increasing recognition for the important role of non-coding RNAs in the regulation of signaling networks that influence susceptibility to cellular senescence (18, 19). In this review, we will highlight non-coding RNAs that regulate senescence-associated molecular pathways in the context of IPF and COPD pathogenesis and discuss current approaches and challenges for therapeutically targeting non-coding RNAs for these diseases.
Cellular Senescence and Its Causes in the Pathogenesis of IPF and COPD
Cellular senescence is now considered an important mechanism of IPF and COPD pathogenesis (20, 21). In IPF, cellular senescence markers are increased in epithelial and mesenchymal cells within remodeled areas of fibrotic lung, and eliminating senescent cells using genetically modified mice or pharmacologic agents decreases disease severity in animal models of pulmonary fibrosis (22–26). Singe-cell RNA sequencing studies suggest epithelial senescence in IPF occurs in a unique subpopulation of cells that reside adjacent to myofibroblasts and may arise from the persistence of a transitional alveolar epithelial cell state (27, 28). These cells express p16, p21, certain basal cell markers, developmental and epithelial-mesenchymal transition markers, and may be a source of pro-fibrotic SASP signaling (29, 30). Additionally, while fibroblast senescence is important for normal wound healing, IPF pathogenesis may involve the persistence of senescent fibroblasts that secrete pro-fibrotic mediators and senescent myofibroblasts that are apoptosis resistant (23, 31). Lungs of patients with COPD demonstrate increased markers of cellular senescence in epithelial cells, fibroblasts, and endothelial cells and many (albeit not all) studies have demonstrated genetic or pharmacologic inhibition of cellular senescence can mitigate disease severity in animal models of COPD (32–37). It has also been postulated that chronic inflammation and airway remodeling in COPD may arise from the production of pro-inflammatory SASP factors (38). Additionally, impaired tissue repair may be the result of reduced replicative capacity in senescent progenitor cells.
Cellular senescence in IPF and COPD is commonly caused by oxidative stress and DNA damage (39). Oxidative stress refers to an imbalance between reactive oxygen/nitrogen species (ROS/RNS) and cellular antioxidants. While ROS can arise from many exogenous sources (e.g., cigarette smoke) and chronic inflammation, one of the most abundant sources of ROS are mitochondria. There is increased mitochondrial ROS production and mitochondrial dysfunction with age and in IPF and COPD (40–42). In addition, IPF and COPD are associated with increased oxidative biomarkers, and consequences of oxidative stress including macromolecular damage (e.g., protein, DNA, and organelle damage), inflammation, cellular senescence and cell death (26, 32, 43). DNA damage is another important cause of cellular senescence, through activation of p53, increased p21 and p16 transcription, and stabilization of GATA4 (12, 15, 44). DNA damage is increased in IPF and COPD, and inadequate DNA repair capacity may contribute to disease progression in COPD (45–47). Telomere shortening can also activate DNA damage responses (17, 48). Normally, a shelterin complex protects telomeric strands from being recognized by DNA damage responses. However, telomere shortening can cause loss of the shelterin complex, telomere “uncapping,” DNA damage response activation, and cellular senescence. In IPF, shortened telomeres and mutations in telomere maintenance genes are well-described risk factors for disease (49–51). Similarly, telomerase mutations are risk factors for early onset emphysema and telomere length is associated with severity of airflow limitation, increased risk for acute exacerbations, and increased mortality (34, 52–55).
To combat cellular stress, cells maintain a repertoire of cellular stress responses, but many of these adaptive responses wane with age or are decreased in IPF and COPD. For example, NRF2 is a transcription factor that promotes the production of cellular antioxidants and detoxifying enzymes. However, NRF2 activity decreases with age and impaired NRF2 activity is implicated in the pathogenesis of IPF and COPD (31, 56–59). Another adaptive stress response is autophagy, a process in which cells degrade and “recycle” damaged proteins and organelles through lysosome-dependent pathways. With age, there is reduced autophagy and mitophagy, a selective type of autophagy for the specific degradation of mitochondria. Consequently, there is a reduced capacity to alleviate consequences of oxidative stress and increased susceptibility to cellular senescence (60, 61). In IPF, reduced autophagy in epithelial cells and fibroblasts increase susceptibility to cellular senescence and disease pathogenesis (62–66). Similarly, deficient mitophagy and its mediators, including PINK1 and SIRT3, impair mitochondrial function, increase mitochondrial ROS production, and contribute to progressive fibrosis in IPF (9, 10, 67). While autophagy is an adaptive response, persistent autophagy can cause activation of cell death and cell senescence pathways as well (68, 69). Autophagy and mitophagy are increased in severe COPD, and both insufficient and excess autophagy and mitophagy are implicated in COPD pathogenesis (70–74).
Collectively, these findings underscore the increasing evidence that the pathogeneses of IPF and COPD involve cellular senescence and dysregulation of stress responses that mitigate cellular senescence. Therefore, regulatory factors that increase or reduce susceptibility to cellular senescence may represent novel therapeutic targets for these diseases.
Non-Coding RNAs in Aging, IPF, AND COPD
Non-coding RNAs lack protein coding capacity but still regulate diverse cellular processes including those implicated in aging biology, cellular senescence, and the pathogeneses of IPF and COPD. Non-coding RNAs are mainly classified into two groups, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). miRNAs are small 18–25 base single stranded RNA molecules (75). They are initially transcribed as primary-miRNA (pri-miRNA) molecules that fold into a stem loop structure. Subsequently these pri-miRNAs undergo sequential processing by enzymes Drosha and Dicer to generate miRNA strand duplexes. The mature miRNA strand of the duplex is then loaded into a miRNA-induced silencing complex (miRISC) where it binds complementary mRNA sequence to inhibit mRNA translation or promote mRNA degradation. Typically, miRNAs bind the 3′ untranslated region (UTR) of mRNA but can bind other regions as well. Because one single miRNA can target hundreds of mRNAs, miRNAs can modulate complex biologic processes including those related to lifespan and aging (76, 77). In humans, age-related changes in miRNA expression have been identified in lung, peripheral blood mononuclear cells (PBMCs) and serum (18, 78–81).
LncRNAs are a diverse group of non-coding RNAs longer than 200 nucleotides (82, 83). Certain lncRNAs are transcribed from intergenic regions (long intergenic non-coding RNAs or lincRNAs), while others are derived from excised introns. Sense lncRNAs are located in proximity to a coding gene on the sense strand while antisense lncRNAs are transcribed from the opposite strand of a coding gene. Certain lncRNAs undergo capping, splicing, and polyadenylation much like mRNAs, while others undergo alternative post-transcriptional processing, such as forming circular molecules or processing by RNase P to form stabilizing triple helix structures at their 3′ ends (84). LncRNAs are also functionally diverse. They can act as cis- or trans- regulatory elements to enhance or inhibit mRNA transcription and/or mRNA translation. LncRNAs can mediate their regulator effects by affecting chromosomal architecture, modulating the recruitment of chromatin modifiers, binding DNA directly to form complex structures that interfere with transcriptional machinery, or binding complementary mRNA transcripts to regulate their stability, splicing, or post-transcriptional modification (84–86). Similar to microRNAs, lncRNAs have also been implicated in aging biology (87, 88).
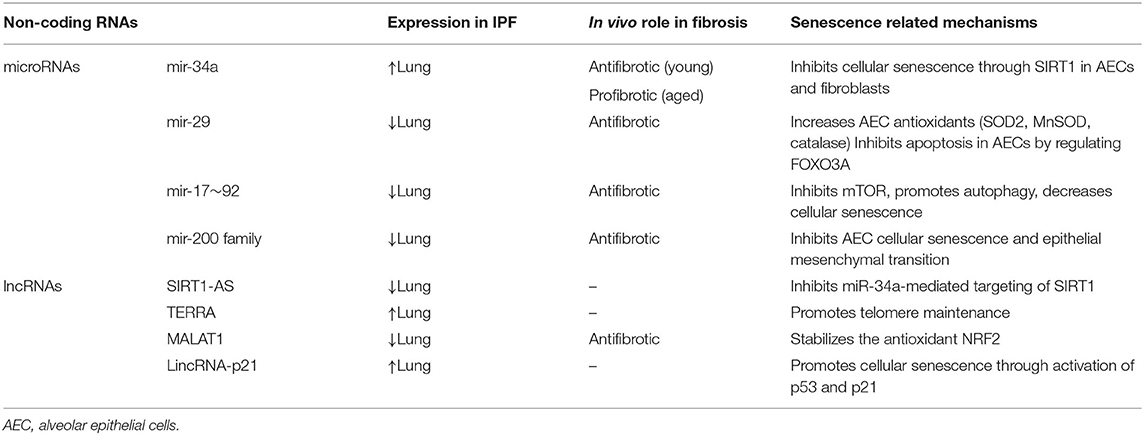
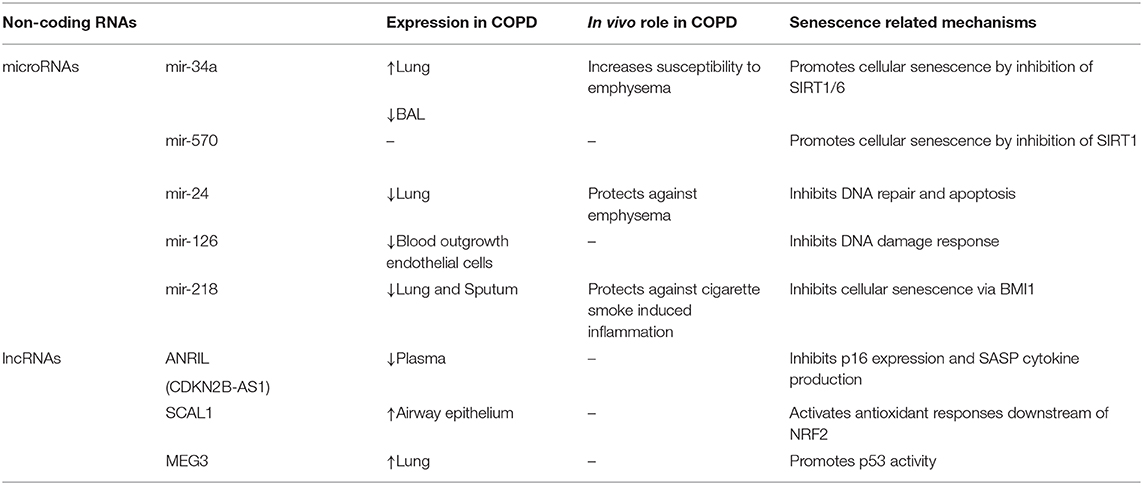
Non-coding RNAs are implicated in the pathogeneses of IPF and COPD and are emerging targets for therapeutic intervention. Approximately 10% of miRNAs are significantly changed in IPF lungs, including a decrease in miR-29, miR-30, let-7, miR-96, and miR-17–92 family members and an increase in miR-154, miR-155, miR-34, miR-26, miR-200 and miR-21 family members (89–91). miRNA expression can be altered by cigarette smoke or by the presence of COPD (92, 93). Studies profiling miRNAs in COPD lung tissue samples have demonstrated increased expression of miR-34a, miR-146a, miR-144, miR-15b, miR-570, and decreased expression of miR-24 and mir-218 (94–99). Other studies evaluating the expression of miRNAs in serum and sputum samples have found differential expression of miR-21, let-7c, miR-610, miR-34 a/b/c, let-7c, miR-146a, miR-125b, and miR-199a with COPD (93, 95, 97, 100–102). The differentially expressed lncRNAs in IPF or animal models of lung fibrosis include MEG3, TERRA, SIRT1-AS, MALAT1, FENDRR, and DNM3OS (103–108). Studies of lncRNAs in COPD lung tissue have identified differential expression of MEG3, ANRIL, SAL-RNA, and SCAL1 with COPD (97, 109, 110). Many of these differentially expressed non-coding RNAs in IPF and COPD have been shown to regulate various aspects of aging biology and cellular senescence. Below, we provide examples of such non-coding RNAs and discuss how their regulation of aging biology and cellular senescence may contribute to disease pathogenesis (Tables 1, 2).
Non-coding RNAs in COPD and IPF
miR-34 and miR-570 Regulation of Sirtuins in IPF and COPD
The miR-34 family consists of three members: miR-34a, miR-34b, and miR-34c. They are direct transcriptional target of p53 and therefore can be induced by oxidative and genotoxic stress (111). Members of the miR-34 family are encoded by two different genes; miR-34a is encoded within chromosome 1 whereas miR-34b and miR-34c are encoded within chromosome 11. Studies show miR-34a expression increases with age and can promote cellular senescence in part through negative regulation sirtuins, particularly SIRT1 and SIRT6 (111–113). Sirtuins are nicotinamide adenine dinucleotide (NAD)-dependent molecules that promote longevity by regulating diverse cellular processes including: cellular senescence, inflammation, DNA repair, autophagy, mitochondrial generation and mitochondrial ROS production (114, 115). For example, SIRT1 can function as a histone deacetylase to negatively regulate NF-κB, mitochondrial biogenesis, p53, p21, and p16 (116, 117). In the lung, miR-34a is expressed in type II alveolar epithelial cells (AECs) and fibroblasts, and increased miR-34a expression coupled with reduced SIRT1 and SIRT6 expression are associated with IPF and COPD (118–123).
Previous studies demonstrate miR-34a is increased in type II AECs from patients with IPF and in murine models of lung fibrosis (124, 125). Both in vitro and in vivo experiments demonstrate miR-34a promotes cellular senescence in part through inactivation of SIRT1 and increased mitochondrial dysfunction (118, 119, 124). Interestingly, the consequences of miR-34a genetic deletion in mice are age-dependent. miR-34a protects against lung fibrosis by increasing fibroblast susceptibility to cellular senescence in young mice, while miR-34a promotes lung fibrosis by increasing alveolar epithelial susceptibility to cellular senescence and apoptosis in old mice (119, 124). The divergent roles for miR-34a in young and old mice underscore the complex temporal- and cell type-specific roles for cellular senescence in disease pathogenesis.
miR-34a is also increased COPD lungs. In airway epithelial cells and lung tissue, miR-34a expression is induced by oxidative stress and inversely correlates with SIRT1 and SIRT6 expression (99, 126). In a murine model of COPD, miR-34a inhibitors increase SIRT1 and SIRT6 expression and reduce NF-κB signaling, matrix metalloproteinase expression, cellular senescence, and emphysema severity. (121–123, 126). Another sirtuin regulator in COPD is miR-570, which is located at chromosome 3 and targets the 3′-UTR of SIRT1 mRNA for degradation. miR-570 expression is induced by oxidative stress and increased in lung tissue and airway epithelial cells from patients with COPD (98). Inhibition of miR-570 reduces cellular senescence and the secretion of SASP factors such as IL-6, IL-1, and CXCL8. Together, these data demonstrate the important roles of miR-34a and miR-570 in regulation of cellular senescence and susceptibility to IPF and COPD through modulation of sirtuins.
miR-29 and IPF
The roles of miR-29 family members in IPF are context dependent and underscore the complex interactions of microRNAs, aging biology, and disease pathogenesis. There are three mature members of the miR-29 family, miR-29a, miR-29b, and miR-29c, which are encoded within two bicistronic clusters (miR-29a/miR-29b-1 located on chromosome 7 and miR-29b-2/mir-29c located on chromosomes 1) (127, 128). In the lung, miR-29 is largely expressed in mesenchymal and epithelial cells where its expression is associated with oxidative stress, DNA damage, and cellular senescence (129–131). However, miR-29c is decreased in IPF lung tissue samples and experimentally induced fibrosis in mouse lungs (124, 132). miR-29c deficiency in type II AECs increases susceptibility to apoptosis and reduces their capacity for epithelial renewal while miR-29c mimics protect type II AECs from apoptosis by regulating FOXO3A and increasing expression of ROS-neutralizing enzymes such as SOD2, MnSOD and catalase (133). miR-29b mimics can inhibit bleomycin-induced lung fibrosis, fibroblast production of extracellular matrix, expression of IGF-1 and production of inflammatory cytokines such as IL-4 and IL-12 (128, 134). Therefore, an increase in miR-29 with oxidative stress, cellular senescence, or with age may be an endogenous response that protects against fibrosis, and a loss of this adaptive response may contribute to the pathogenesis of IPF.
miR-17~92 Cluster and miR-200 Family in IPF
Both the miR-17~92 cluster and miR-200 family regulate susceptibility to cellular senescence in IPF. The miR-17~92 cluster encodes 6 miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a) on chromosome 13 and is frequently decreased in multiple tissue types with age and in senescent cells (135). miR-17~92 decreases susceptibility to cellular senescence through diverse mechanisms including targeting cell cycle proteins, inhibition of the mechanistic target of rapamycin (mTOR), and activation of autophagy (136). Members of the miR-17~92 cluster are hypermethylated in lung tissue samples and fibroblasts from IPF patients, and the use of epigenetic methylation inhibitors to promote expression of the miR-17~92 cluster attenuates fibrosis in bleomycin-murine models (90). Similarly, mice overexpressing miR-17 have highly proliferative, albeit poorly differentiated, epithelial cells and decreased number of senescent cells in their lung (137, 138).
The miR-200 family consists of five members within two clusters, miRs-200a/b/429 on chromosome 1 and miRs-200c/141 on chromosome 12. These microRNAs can regulate oxidative stress, DNA repair, and cellular senescence, although the direction of effect can be context dependent (139, 140). Levels of miR-200a and miR-200c are significantly decreased in IPF lungs and in the lungs of mice with experimental lung fibrosis (141). Transfection of AECs with miR-200a and miR-141 reduces epithelial mesenchymal transition (EMT) and the expression of cellular senescence markers including p16 and p21, but does not improve AEC proliferation capacity. In contrast, transfection with miR-200b/c increases differentiation of senescent type II AECs into type I AECs, decreases EMT, and reduces disease severity in animal models of pulmonary fibrosis (142–145).
miR-24 and miR-126 Regulate DNA Damage Responses in COPD
miR-24 is a member of a poly-cistronic miR-23~27~24 miRNA clusters that occur in two genomic loci in humans. The miR-23b-27b-24-2 cluster is located in an intronic region of chromosome 9 while the miR-23a~27a~24-1 cluster is located in an intergenic region of chromosome 19 (146, 147). Dysregulation of miR-23~27~24 signaling has been identified in multiple age-related disorders including diabetes and Alzheimer's disease, and both oxidative and genotoxic stress have been shown to modulate expression of these miRNAs, although the direction of effect is context dependent (148, 149). In COPD, miR-24 expression inversely correlates with COPD disease severity as measured by FEV1 percent predicted and radiographic emphysema (150). miR-27a and miR-23a expression also inversely correlates with disease severity, albeit it to a lesser degree that miR-24. In a mouse model, inhibition of miR-24 increases susceptibility to cigarette smoke-induced emphysema. Others have found that inhibition miR-24-27-23 cluster in T-cells increases allergic airway inflammation and goblet metaplasia (151). miR-24 can inhibit the expression of p16 by targeting its 3′ UTR to inhibit cellular senescence (152). However, miR-24 can also inhibit DNA repair and the translation of DNA repair genes including H2AX, TOP1, and BRCA1, which can promote cellular senescence in certain contexts (148). Interestingly, another miRNA that inhibits DNA damage responses and is decreased in COPD is miR-126 (153–155). These collective findings suggest that microRNA inhibition of DNA damage responses may protect against COPD pathogenesis, although whether this occurs by changing susceptibility to cellular senescence remains to be determined.
miR-218 and COPD
The mature form of miR-218 can be transcribed from intronic regions of SLIT2 and SLIT3 located on chromosomes 4 and 5, respectively (156). miR-218 is decreased in bronchial epithelial cells of smokers and in lungs and sputum from COPD patients (93, 96). In a murine model of COPD, inhibition of miR-218 increases susceptibility to emphysema and airway inflammation with increased production of IL-8 and CCL2 (96). Notably, one of the downstream targets of miR-218 is BMI-1, a polycomb repressive group protein which inhibits p16 expression and cellular senescence (157). This raises the possibility that decreased miR-218 expression promotes cellular senescence and disease progression in COPD, although further studies are warranted.
LncRNA
Lnc ANRIL (CDKN2B-AS1) and Lnc SIRT1-AS
ANRIL is transcribed from the antisense strand of CDKN2A/2B, the genes that encode cyclin-dependent kinase inhibitors p15 and p16, on chromosome 9 (158). ANRIL mediates transcriptional repression of these antisense genes through RNA-RNA interactions, as well as histone methylation and chromatin remodeling of polycomb repressive complexes (PRC) (158, 159). ANRIL activity is highly variable and dependent on tissue type. There are 21 ANRIL splice variants, including linear and circular isoforms, and ANRIL activity is highly influenced by methylation activity in its promoter region (158). In addition to its role in regulating p15 and p16, ANRIL suppresses NF-κB and can inhibit chronic inflammation (160). In one study, ANRIL expression in plasma was decreased during acute exacerbations of COPD and ANRIL expression negatively correlated with SASP related cytokines such as TNF-α, IL-1β, IL-8 and LTB-4 in stable COPD patients (161). LncRNA SIRT1 antisense (SIRT1-AS), is transcribed from the antisense strand of SIRT1 and can form RNA hybrid double strands with SIRT1 mRNA to increase its stability (162). SIRT-AS protects SIRT1 mRNA degradation by inhibiting miR-34a binding to the 3′UTR of SIRT1 (162). In one study of bleomycin-induced lung fibrosis, SIRT1-AS overexpression inhibited TGF-β-mediated EMT (163). Despite these data, more studies will be necessary to confirm the roles of ANRIL and SIRT1-AS in COPD and pulmonary fibrosis.
TERRA (Telomere Repeat-Containing RNA)
TERRAs are important for telomere maintenance and characterized by 5′-(UUAGGG)-3′ repeats (164, 165). These lncRNAs are commonly transcribed from the subtelomeric 20q locus in humans in response to cellular stress and telomere shortening, the later as a consequence of reduced methylation marks and loss of telomeric heterochromatin (166). TERRAs are recruited to telomeres where they form DNA-RNA hybrid R-loops. This R-loop formation regulates telomere maintenance through interactions with chromatin modifiers, telomerase, and promoting DNA repair (166, 167). TERRAs also facilitate telomere replication and promote the assembly of shelterin proteins (168, 169). However, TERRA expression is increased in the PBMCs from IPF patients and inversely correlated with the percentage of predicted force vital capacity (106). While not well-defined, TERRAs may have an important role in IPF pathogenesis.
MALAT1 (Metastasis Associated in Lung Adenocarcinoma Transcript-1) and SCAL1 (Cancer-Associated lncRNA-1)
Both MALAT1 and SCAL1 are lncRNAs that regulate cellular responses to oxidative stress and cellular senescence. MALAT1 is an 8.7kbp lncRNA transcribed from human chromosome 11 and is ubiquitously express in almost all human tissue (170). MALAT1 is frequently found in nuclear “speckles” and can interact with pre-mRNA splicing factors to modulate alternative mRNA splicing (171, 172). Consequently, MALAT1 can regulate the expression of cell cycle genes and can also stabilize NRF2 to attenuate oxidative stress and DNA damage (173). MALAT1 is decreased in senescent cells and in bleomycin-induced murine fibrosis where myeloid deletion of MALAT1 increases susceptibility to fibrosis and the number of profibrotic M2 macrophages (19, 108). SCAL1, a lncRNA located on chromosome 5, can be induced by oxidative stress through NRF2-mediated transcriptional activity and is increased in the airway epithelium of smokers compared to nonsmokers (97, 174). Inhibition of SCAL1 in airway epithelial cells augments cytotoxicity induced by cigarette smoke extract in vitro, suggesting SCAL1 may act downstream of NRF2 to mediate protective antioxidant responses.
LincRNA-p21 (Long Intergenic Non-coding RNA p21) and MEG3 (Maternally Expressed Gene 3)
Both lincRNA-21 and MEG3 are downstream targets of p53 and mediate many p53-dependent transcriptional responses. LincRNA-p21 is a transcriptional target of p53 located approximately 15 kb upstream from CDKN1A (175). LincRNA-p21 functions as a repressor of p53-dependent transcription by binding to hnRNP-K (heterogeneous nuclear ribonucleoprotein K) and interacting with PRC1 and PRC2, although these same interactions also promote p53 activity at the p21 promoter to increase p21 transcription (176, 177). In one study, lincRNA-p21 inhibited fibroblast collagen expression through downregulation of THY1 expression (178). Maternally expressed gene 3 (MEG3) is a maternally imprinted gene located on chromosome 14, and increases with age in human lung tissue and PBMCs due to changes in promoter methylation (87, 179). Like lincRNA-21, MEG3 also promotes p53 activity. MEG3 interactions with p53 inhibit p53 ubiquitination and MDM2-mediated degradation. MEG3 can also selectively upregulate certain p53 target genes, such as GDF15, and interact with PRC1/2 to mediate p53-dependent gene silencing (180–182). Intriguingly, there are 27 known splice variants of MEG3, and changes in the relative abundance of these slice variants in response to cellular stress can modulate p53 activity (183). MEG3 is increased in the lungs of patients with COPD (184, 185). Additionally, epithelial MEG3 expression has been shown to be induced by cigarette smoke, correlate with disease severity, and promote inflammation and apoptosis through a mechanism involving miR-218 (186). MEG3 expression is also increased in atypical IPF epithelial cells and can impair basal cell differentiation, which may contribute to abnormal tissue remodeling (105). Notably, p53 can induce both cellular senescence and apoptosis in a context-dependent manner, but the role of lincRNA-p21 and MEG3 in regulating p53-mediated cell fate responses in the lung remain unknown. Additionally, while p53 is implicated in the pathogenesis of IPF and COPD, more studies are necessary to determine the roles of lincRNA-p21 and MEG3 in these diseases (187).
Therapeutic Targeting of Non-coding RNAs
There is a growing interest in targeting non-coding RNAs to treat chronic lung diseases due to their regulatory functions and roles in disease pathogenesis (85, 188). Therapeutic approaches for RNA targeting utilize nucleotides with complementary sequences to prevent RNA transcription, promote RNA degradation, or interfere with post-transcriptional processing of target RNAs. Catalytically dead RNA-guided CAS9 endonucleases that target specific DNA sequences can be used to hinder RNA transcription. Single-stranded antisense oligonucleotides (ASOs) that bind RNA molecules through complementary sequences promote RNA degradation through RNAase-H dependent cleavage, although newer ASOs inhibit mRNA translation through steric hindrance or interfering with normal mRNA splicing. Similarly, double-stranded RNA molecules, including small interfering RNA (siRNA) or miRNA mimics, utilize the RISC complex to inhibit transcription or promote RNA degradation.
Nucleotide-based approaches for targeting non-coding RNAs are attractive for a variety of reasons. Many non-coding RNAs, particularly lncRNAs, are expressed in a tissue- or cell- specific manner (189). Therefore, augmenting or inhibiting their expression in a cell- or tissue- specific manner can reduce off-target effects and increase the therapeutic window. Additionally, generating oligonucleotide sequences complementary to their target sequence is a much easier task using currently available technologies than identifying small-molecule inhibitors or antibodies that can specifically target proteins of interest. Even if targeted antibodies or small molecules are identified, they commonly reduce rather than augment target molecule activity. In contrast, oligonucleotide therapies can increase the concentration of target molecule production through inhibition of negative regulators such as miRNAs. Finally, many therapeutic targets, while pathologic in certain contexts, also have important homeostatic functions. For example, oxidative stress is deleterious, but ROS are critical intracellular signaling molecules. Similarly, cellular senescence promotes aging related disorders but also prevents malignant transformation. Rather than inhibiting such integral pathways completely, a more effective therapeutic strategy may be to focus on modulating these pathways by targeting regulatory non-coding RNAs.
However, nucleic acid-based therapies are not without challenges (190). First, oligonucleotides are susceptible to degradation by extracellular and intracellular nucleases. To overcome this challenge and increase oligonucleotide stability, researchers have used chemically modified phosphate backbones. For example, antagomirs are ASOs that commonly contain 2′-O-methyl or phosphonothioate modifications to improve stability. Locked nucleic acids are another commonly used ASO that utilizes a modified RNA-DNA-RNA backbone to increase binding affinity and improve stability. Certain oligoribonucleotides possess targeting moieties that can deliver nucleic acid-based therapies to specific tissue. Another challenge is that nucleotides are large negatively charged molecules and therefore do not easily cross the cell membrane. Therefore, lipid-, peptide-, and polymer-based nanoparticles have been used to deliver oligonucleotides to the cytosol. Some of these nanoparticles promote the specific uptake of oligonucleotides into the lung or increase retention within the lung following inhalation. (191, 192). Nucleic acid-based therapies are capable of promoting inflammation through toll-like receptors and other innate immune receptors for foreign DNA and RNA, although this problem can be mitigated through assays to test for immune activation and reducing CpG elements (85). Finally, non-coding RNAs can target hundreds of genes and/or function through diverse mechanisms, and therefore targeting non-coding RNAs may cause unwanted effects.
Several oligonucleotide therapies that target mRNAs have already been approved by the U.S. Food and Drug Administration for treating disease, and there are currently multiple clinical trials targeting non-coding RNAs. For example, Remlarsen, a first-generation miR-29 mimic, is currently being evaluated in a Phase 2 clinical trial assessing its safety and efficacy in skin fibrosis (193). MRX-34 is a liposomal nanoparticle formulation of a miR-34 mimic that was under investigation in a Phase I trial for cancer, but the study was stopped short because of serious adverse events (194). While lncRNAs have not been tested in clinical trials, therapeutic manipulation of MALAT1 and MEG3 have shown benefit in preclinical transgenic and xenograft models of cancer (195–197). Lin et al. reported that the knockdown of lncRNA MALAT1 via tail injection of RNAi can improve septic lung injury in mice (198). Additionally, intraperitoneal administration of ASO targeting lncRNA DNM3OS, a regulator of the TGF-β pathway, attenuated bleomycin-induced lung fibrosis in mice (104). These pre-clinical and clinical studies suggest the possibility that non-coding RNAs may have a potential therapeutic role for treating lung diseases such as IPF and COPD.
Conclusion
Diverse cellular processes implicated in aging biology, including cellular senescence, contribute to the pathogenesis of IPF and COPD. In these diseases, cellular senescence can occur from oxidative stress, DNA damage, telomere shortening, or mitochondrial dysfunction. While these processes occur commonly with age, their impact on cell fate and disease susceptibility are influenced by diverse regulatory factors. Additionally, many of the cellular responses to these stressors, including senescence, have homeostatic functions and are not universally pathologic. Therefore, nuanced therapeutic approaches will be required to target these processes. Such approaches may need to be cell- or tissue- specific or have modulatory rather than inhibitor effects on key pathways. Because of the fundamental regulatory role of non-coding RNAs, and the growing capacity for cell-specific targeting, non-coding RNAs may emerge as ideal therapies to target chronic lung disease and other age-related disorders.
Author Contributions
NO and MS wrote the manuscript and built the tables, which were original. All authors have read and approved the submitted manuscript version.
Funding
This work was in part supported by Yale Claude D. Pepper Older Americans Independence Center Center P30 AG021342. Flight Attendant Medical Research Institute (grant YCSA 142017) and the NIH (National Heart, Lung, and Blood Institute grant K08-HL-135402) (MS) and The Japanese Cell Science Research Foundation (NO).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Thannickal VJ, Murthy M, Balch WE, Chandel NS, Meiners S, Eickelberg O, et al. Blue journal conference. Aging and susceptibility to lung disease. Am J Respir Crit Care Med. (2015) 191:261–9. doi: 10.1164/rccm201410-1876PP
2. Cho SJ, Stout-Delgado HW. Aging and lung disease. Annu Rev Physiol. (2020) 82:433–59. doi: 10.1146/annurev-physiol-021119-034610
3. Meiners S, Eickelberg O, Konigshoff M. Hallmarks of the ageing lung. Eur Respir J. (2015) 45:807–27. doi: 10.1183/0903193600186914
4. Ford ES, Croft JB, Mannino DM, Wheaton AG, Zhang X, Giles WH. COPD surveillance–United States, 1999–2011. Chest. (2013) 144:284–305. doi: 10.1378/chest13-0809
5. Raghu G, Weycker D, Edelsberg J, Bradford WZ, Oster G. Incidence and prevalence of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2006) 174:810–6. doi: 10.1164/rccm200602-163OC
6. Chilosi M, Carloni A, Rossi A, Poletti V. Premature lung aging and cellular senescence in the pathogenesis of idiopathic pulmonary fibrosis and COPD/emphysema. Transl Res. (2013) 162:156–73. doi: 10.1016/j.trsl.2013.06004
7. Faner R, Rojas M, Macnee W, Agusti A. Abnormal lung aging in chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2012) 186:306–13. doi: 10.1164/rccm201202-0282PP
8. Mercado N, Ito K, Barnes PJ. Accelerated ageing of the lung in COPD: new concepts. Thorax. (2015) 70:482–9. doi: 10.1136/thoraxjnl-2014-206084
9. Zank DC, Bueno M, Mora AL, Rojas M. Idiopathic pulmonary fibrosis: aging, mitochondrial dysfunction, and cellular bioenergetics. Front Med (Lausanne). (2018) 5:10. doi: 10.3389/fmed.201800010
10. Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, et al. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest. (2015) 125:521–38. doi: 10.1172/JCI74942
11. Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. (2013) 153:1194–217. doi: 10.1016/j.cell.2013.05039
12. Gorgoulis V, Adams PD, Alimonti A, Bennett DC, Bischof O, Bishop C, et al. Cellular senescence: defining a path forward. Cell. (2019) 179:813–27. doi: 10.1016/j.cell.2019.10005
13. Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest. (2018) 128:1238–46. doi: 10.1172/JCI95148
14. Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. (1961) 25:585–621. doi: 10.1016/0014-4827(61)90192-6
15. Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. (2013) 75:685–705. doi: 10.1146/annurev-physiol-030212-183653
16. Karin O, Agrawal A, Porat Z, Krizhanovsky V, Alon U. Senescent cell turnover slows with age providing an explanation for the Gompertz law. Nat Commun. (2019) 10:5495. doi: 10.1038/s41467-019-13192-4
17. Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun. (2012) 3:708. doi: 10.1038/ncomms1708
18. ElSharawy A, Keller A, Flachsbart F, Wendschlag A, Jacobs G, Kefer N, et al. Genome-wide miRNA signatures of human longevity. Aging Cell. (2012) 11:607–16. doi: 10.1111/j.1474-9726.2012.00824x
19. Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH, et al. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. (2013) 12:890–900. doi: 10.1111/acel12115
20. Barnes PJ, Baker J, Donnelly LE. Cellular senescence as a mechanism and target in chronic lung diseases. Am J Respir Crit Care Med. (2019) 200:556–64. doi: 10.1164/rccm201810-1975TR
21. Parikh P, Wicher S, Khandalavala K, Pabelick CM, Britt RD Jr, Prakash YS. Cellular senescence in the lung across the age spectrum. Am J Physiol Lung Cell Mol Physiol. (2019) 316:L826–42. doi: 10.1152/ajplung.004242018
22. Zhang Y, Jiang M, Nouraie M, Roth MG, Tabib T, Winters S, et al. GDF15 is an epithelial-derived biomarker of idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. (2019) 317:L510–21. doi: 10.1152/ajplung.000622019
23. Schafer MJ, White TA, Iijima K, Haak AJ, Ligresti G, Atkinson EJ, et al. Cellular senescence mediates fibrotic pulmonary disease. Nat Commun. (2017) 8:14532. doi: 10.1038/ncomms14532
24. Lehmann M, Korfei M, Mutze K, Klee S, Skronska-Wasek W, Alsafadi HN, et al. Senolytic drugs target alveolar epithelial cell function and attenuate experimental lung fibrosis ex vivo. Eur Respir J. (2017) 50:1602367. doi: 10.1183/1399300302367-2016
25. Wiley CD, Brumwell AN, Davis SS, Jackson JR, Valdovinos A, Calhoun C, et al. Secretion of leukotrienes by senescent lung fibroblasts promotes pulmonary fibrosis. JCI Insight. (2019) 4:e130056. doi: 10.1172/jci.insight130056
26. Alvarez D, Cardenes N, Sellares J, Bueno M, Corey C, Hanumanthu VS, et al. IPF lung fibroblasts have a senescent phenotype. Am J Physiol Lung Cell Mol Physiol. (2017) 313:L1164–73. doi: 10.1152/ajplung.002202017
27. Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv. (2020) 6:eaba1983. doi: 10.1126/sciadvaba1983
28. Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv. (2020) 6:eaba1972. doi: 10.1126/sciadvaba1972
29. Strunz M, Simon LM, Ansari M, Kathiriya JJ, Angelidis I, Mayr CH, et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat Commun. (2020) 11:3559. doi: 10.1038/s41467-020-17358-3
30. Kobayashi Y, Tata A, Konkimalla A, Katsura H, Lee RF, Ou J, et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat Cell Biol. (2020) 22:934–46. doi: 10.1038/s41556-020-0542-8
31. Hecker L, Logsdon NJ, Kurundkar D, Kurundkar A, Bernard K, Hock T, et al. Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance. Sci Transl Med. (2014) 6:231ra47. doi: 10.1126/scitranslmed3008182
32. Tsuji T, Aoshiba K, Nagai A. Alveolar cell senescence in patients with pulmonary emphysema. Am J Respir Crit Care Med. (2006) 174:886–93. doi: 10.1164/rccm200509-1374OC
33. Cottage CT, Peterson N, Kearley J, Berlin A, Xiong X, Huntley A, et al. Targeting p16-induced senescence prevents cigarette smoke-induced emphysema by promoting IGF1/Akt1 signaling in mice. Commun Biol. (2019) 2:307. doi: 10.1038/s42003-019-0532-1
34. Amsellem V, Gary-Bobo G, Marcos E, Maitre B, Chaar V, Validire P, et al. Telomere dysfunction causes sustained inflammation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2011) 184:1358–66. doi: 10.1164/rccm201105-0802OC
35. Kim SJ, Shan P, Hwangbo C, Zhang Y, Min JN, Zhang X, et al. Endothelial toll-like receptor 4 maintains lung integrity via epigenetic suppression of p16(INK4a). Aging Cell. (2019) 18:e12914. doi: 10.1111/acel12914
36. Sundar IK, Rashid K, Gerloff J, Li D, Rahman I. Genetic ablation of p16(INK4a) does not protect against cellular senescence in mouse models of chronic obstructive pulmonary disease/emphysema. Am J Respir Cell Mol Biol. (2018) 59:189–99. doi: 10.1165/rcmb2017-0390OC
37. Yao H, Yang SR, Edirisinghe I, Rajendrasozhan S, Caito S, Adenuga D, et al. Disruption of p21 attenuates lung inflammation induced by cigarette smoke, LPS, and fMLP in mice. Am J Respir Cell Mol Biol. (2008) 39:7–18. doi: 10.1165/rcmb2007-0342OC
38. Kumar M, Seeger W, Voswinckel R. Senescence-associated secretory phenotype and its possible role in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. (2014) 51:323–33. doi: 10.1165/rcmb2013-0382PS
39. Ryter SW, Rosas IO, Owen CA, Martinez FJ, Choi ME, Lee CG, et al. Mitochondrial dysfunction as a pathogenic mediator of chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Ann Am Thorac Soc. (2018) 15(Suppl. 4):S266–72. doi: 10.1513/AnnalsATS201808-585MG
40. Mora AL, Bueno M, Rojas M. Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. J Clin Invest. (2017) 127:405–14. doi: 10.1172/JCI87440
41. Schmidlin CJ, Dodson MB, Madhavan L, Zhang DD. Redox regulation by NRF2 in aging and disease. Free Radic Biol Med. (2019) 134:702–7. doi: 10.1016/j.freeradbiomed.2019.01016
42. Wiegman CH, Michaeloudes C, Haji G, Narang P, Clarke CJ, Russell KE, et al. Oxidative stress-induced mitochondrial dysfunction drives inflammation and airway smooth muscle remodeling in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. (2015) 136:769–80. doi: 10.1016/j.jaci.2015.01046
43. Hecker L. Mechanisms and consequences of oxidative stress in lung disease: therapeutic implications for an aging populace. Am J Physiol Lung Cell Mol Physiol. (2018) 314:L642–53. doi: 10.1152/ajplung.002752017
44. Kang C, Xu Q, Martin TD, MZ Li, Demaria M, Aron L, et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. (2015) 349:aaa5612. doi: 10.1126/scienceaaa5612
45. Sauler M, Lamontagne M, Finnemore E, Herazo-Maya JD, Tedrow J, Zhang X, et al. The DNA repair transcriptome in severe COPD. Eur Respir J. (2018) 52:1701994. doi: 10.1183/1399300301994-2017
46. Kuwano K, Kunitake R, Kawasaki M, Nomoto Y, Hagimoto N, Nakanishi Y, et al. P21Waf1/Cip1/Sdi1 and p53 expression in association with DNA strand breaks in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (1996) 154:477–83. doi: 10.1164/ajrccm.154.28756825
47. Sears CR, Zhou H, Justice MJ, Fisher AJ, Saliba J, Lamb I, et al. Xeroderma pigmentosum group c deficiency alters cigarette smoke DNA damage cell fate and accelerates emphysema development. Am J Respir Cell Mol Biol. (2018) 58:402–11. doi: 10.1165/rcmb2017-0251OC
48. Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. (2007) 8:299–309. doi: 10.1038/nrg2047
49. Cronkhite JT, Xing C, Raghu G, Chin KM, Torres F, Rosenblatt RL, et al. Telomere shortening in familial and sporadic pulmonary fibrosis. Am J Respir Crit Care Med. (2008) 178:729–37. doi: 10.1164/rccm200804-550OC
50. McDonough JE, Martens DS, Tanabe N, Ahangari F, Verleden SE, Maes K, et al. A role for telomere length and chromosomal damage in idiopathic pulmonary fibrosis. Respir Res. (2018) 19:132. doi: 10.1186/s12931-018-0838-4
51. Alder JK, Chen JJ, Lancaster L, Danoff S, Su SC, Cogan JD, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci U S A. (2008) 105:13051–6. doi: 10.1073/pnas0804280105
52. Wan ES, Goldstein RL, Fan VS, Nguyen HQ, Hart JE, Garshick E, et al. Telomere length in COPD: Relationships with physical activity, exercise capacity, and acute exacerbations. PLoS ONE. (2019) 14:e0223891. doi: 10.1371/journal.pone0223891
53. Stanley SE, Chen JJ, Podlevsky JD, Alder JK, Hansel NN, Mathias RA, et al. Telomerase mutations in smokers with severe emphysema. J Clin Invest. (2015) 125:563–70. doi: 10.1172/JCI78554
54. Albrecht E, Sillanpaa E, Karrasch S, Alves AC, Codd V, Hovatta I, et al. Telomere length in circulating leukocytes is associated with lung function and disease. Eur Respir J. (2014) 43:983–92. doi: 10.1183/0903193600046213
55. Rode L, Bojesen SE, Weischer M, Vestbo J, Nordestgaard BG. Short telomere length, lung function and chronic obstructive pulmonary disease in 46,396 individuals. Thorax. (2013) 68:429–35. doi: 10.1136/thoraxjnl-2012-202544
56. Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke–induced emphysema in mice. J Clin Invest. (2004) 114:1248–59. doi: 10.1172/JCI200421146
57. Milara J, Peiro T, Serrano A, Cortijo J. Epithelial to mesenchymal transition is increased in patients with COPD and induced by cigarette smoke. Thorax. (2013) 68:410–20. doi: 10.1136/thoraxjnl-2012-201761
58. Suzuki M, Betsuyaku T, Ito Y, Nagai K, Nasuhara Y, Kaga K, et al. Down-regulated NF-E2-related factor 2 in pulmonary macrophages of aged smokers and patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. (2008) 39:673–82. doi: 10.1165/rcmb2007-0424OC
59. Goven D, Boutten A, Lecon-Malas V, Marchal-Somme J, Amara N, Crestani B, et al. Altered Nrf2/Keap1-Bach1 equilibrium in pulmonary emphysema. Thorax. (2008) 63:916–24. doi: 10.1136/thx.2007091181
60. Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. (2011) 146:682–95. doi: 10.1016/j.cell.2011.07030
61. Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, et al. Measuring in vivo mitophagy. Mol Cell. (2015) 60:685–96. doi: 10.1016/j.molcel.2015.10009
62. Araya J, Kojima J, Takasaka N, Ito S, Fujii S, Hara H, et al. Insufficient autophagy in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. (2013) 304:L56–69. doi: 10.1152/ajplung.002132012
63. Hill C, Li J, Liu D, Conforti F, Brereton CJ, Yao L, et al. Autophagy inhibition-mediated epithelial-mesenchymal transition augments local myofibroblast differentiation in pulmonary fibrosis. Cell Death Dis. (2019) 10:591. doi: 10.1038/s41419-019-1820-x
64. Ricci A, Cherubini E, Scozzi D, Pietrangeli V, Tabbi L, Raffa S, et al. Decreased expression of autophagic beclin 1 protein in idiopathic pulmonary fibrosis fibroblasts. J Cell Physiol. (2013) 228:1516–24. doi: 10.1002/jcp24307
65. Kuwano K, Araya J, Hara H, Minagawa S, Takasaka N, Ito S, et al. Cellular senescence and autophagy in the pathogenesis of chronic obstructive pulmonary disease (COPD) and idiopathic pulmonary fibrosis (IPF). Respir Investig. (2016) 54:397–406. doi: 10.1016/j.resinv.2016.03010
66. Patel AS, Lin L, Geyer A, Haspel JA, An CH, Cao J, et al. Autophagy in idiopathic pulmonary fibrosis. PLoS ONE. (2012) 7:e41394. doi: 10.1371/journal.pone0041394
67. Jablonski RP, Kim SJ, Cheresh P, Williams DB, Morales-Nebreda L, Cheng Y, et al. SIRT3 deficiency promotes lung fibrosis by augmenting alveolar epithelial cell mitochondrial DNA damage and apoptosis. FASEB J. (2017) 31:2520–32. doi: 10.1096/fj201601077R
68. Horikawa I, Fujita K, Jenkins LM, Hiyoshi Y, Mondal AM, Vojtesek B, et al. Autophagic degradation of the inhibitory p53 isoform Delta133p53alpha as a regulatory mechanism for p53-mediated senescence. Nat Commun. (2014) 5:4706. doi: 10.1038/ncomms5706
69. Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, et al. Autophagy mediates degradation of nuclear lamina. Nature. (2015) 527:105–9. doi: 10.1038/nature15548
70. Fujii S, Hara H, Araya J, Takasaka N, Kojima J, Ito S, et al. Insufficient autophagy promotes bronchial epithelial cell senescence in chronic obstructive pulmonary disease. Oncoimmunology. (2012) 1:630–41. doi: 10.4161/onci20297
71. Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS ONE. (2008) 3:e3316. doi: 10.1371/journal.pone0003316
72. Ahmad T, Sundar IK, Lerner CA, Gerloff J, Tormos AM, Yao H, et al. Impaired mitophagy leads to cigarette smoke stress-induced cellular senescence: implications for chronic obstructive pulmonary disease. FASEB J. (2015) 29:2912–29. doi: 10.1096/fj14-268276
73. Mizumura K, Cloonan SM, Nakahira K, Bhashyam AR, Cervo M, Kitada T, et al. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J Clin Invest. (2014) 124:3987–4003. doi: 10.1172/JCI74985
74. Ryter SW, Nakahira K, Haspel JA, Choi AM. Autophagy in pulmonary diseases. Annu Rev Physiol. (2012) 74:377–401. doi: 10.1146/annurev-physiol-020911-153348
75. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. (2008) 9:102–14. doi: 10.1038/nrg2290
76. Smith-Vikos T, Slack FJ. MicroRNAs and their roles in aging. J Cell Sci. (2012) 125:7–17. doi: 10.1242/jcs099200
77. Zhou Y, Wang X, Song M, He Z, Cui G, Peng G, et al. A secreted microRNA disrupts autophagy in distinct tissues of Caenorhabditis elegans upon ageing. Nat Commun. (2019) 10:4827. doi: 10.1038/s41467-019-12821-2
78. Huan T, Chen G, Liu C, Bhattacharya A, Rong J, Chen BH, et al. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell. (2018) 17:e12687. doi: 10.1111/acel12687
79. Hooten NN, Fitzpatrick M, Wood WH III, De S, Ejiogu N, Zhang Y, et al. Age-related changes in microRNA levels in serum. Aging (Albany NY). (2013) 5:725–40. doi: 10.18632/aging100603
80. Hooten NN, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS ONE. (2010) 5:e10724. doi: 10.1371/journal.pone0010724
81. Ong J, Woldhuis RR, Boudewijn IM, van den Berg A, Kluiver J, Kok K, et al. Age-related gene and miRNA expression changes in airways of healthy individuals. Sci Rep. (2019) 9:3765. doi: 10.1038/s41598-019-39873-0
82. Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. (2014) 48:433–55. doi: 10.1146/annurev-genet-120213-092323
83. Lee JT. Epigenetic regulation by long noncoding RNAs. Science. (2012) 338:1435–9. doi: 10.1126/science1231776
84. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. (2019) 21:542–51. doi: 10.1038/s41556-019-0311-8
85. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in cancer. Trends Mol Med. (2018) 24:257–77. doi: 10.1016/j.molmed.2018.01001
86. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. (2011) 43:904–14. doi: 10.1016/j.molcel.2011.08018
87. Marttila S, Chatsirisupachai K, Palmer D, de Magalhaes JP. Ageing-associated changes in the expression of lncRNAs in human tissues reflect a transcriptional modulation in ageing pathways. Mech Ageing Dev. (2020) 185:111177. doi: 10.1016/j.mad.2019111177
88. Grammatikakis I, Panda AC, Abdelmohsen K, Gorospe M. Long noncoding RNAs(lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY). (2014) 6:992–1009. doi: 10.18632/aging100710
89. Pandit KV, Milosevic J, Kaminski N. MicroRNAs in idiopathic pulmonary fibrosis. Transl Res. (2011) 157:191–9. doi: 10.1016/j.trsl.2011.01012
90. Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, et al. Epigenetic regulation of miR-17~92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. (2013) 187:397–405. doi: 10.1164/rccm201205-0888OC
91. Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2010) 182:220–9. doi: 10.1164/rccm200911-1698OC
92. Savarimuthu Francis SM, Davidson MR, Tan ME, Wright CM, Clarke BE, Duhig EE, et al. MicroRNA-34c is associated with emphysema severity and modulates SERPINE1 expression. BMC Genom. (2014) 15:88. doi: 10.1186/1471-2164-15-88
93. Van Pottelberge GR, Mestdagh P, Bracke KR, Thas O, van Durme YM, Joos GF, et al. MicroRNA expression in induced sputum of smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2011) 183:898–906. doi: 10.1164/rccm201002-0304OC
94. Dang X, Qu X, Wang W, Liao C, Li Y, Zhang X, et al. Bioinformatic analysis of microRNA and mRNA Regulation in peripheral blood mononuclear cells of patients with chronic obstructive pulmonary disease. Respir Res. (2017) 18:4. doi: 10.1186/s12931-016-0486-5
95. Molina-Pinelo S, Pastor MD, Suarez R, Romero-Romero B, Gonzalez De la Pena M, Salinas A, et al. MicroRNA clusters: dysregulation in lung adenocarcinoma and COPD. Eur Respir J. (2014) 43:1740–9. doi: 10.1183/0903193600091513
96. Conickx G, Mestdagh P, Avila Cobos F, Verhamme FM, Maes T, Vanaudenaerde BM, et al. MicroRNA profiling reveals a role for MicroRNA-218-5p in the pathogenesis of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2017) 195:43–56. doi: 10.1164/rccm201506-1182OC
97. De Smet EG, Mestdagh P, Vandesompele J, Brusselle GG, Bracke KR. Non-coding RNAs in the pathogenesis of COPD. Thorax. (2015) 70:782–91. doi: 10.1136/thoraxjnl-2014-206560
98. Baker JR, Vuppusetty C, Colley T, Hassibi S, Fenwick PS, Donnelly LE, et al. MicroRNA-570 is a novel regulator of cellular senescence and inflammaging. FASEB J. (2019) 33:1605–16. doi: 10.1096/fj201800965R
99. Mizuno S, Bogaard HJ, Gomez-Arroyo J, Alhussaini A, Kraskauskas D, Cool CD, et al. MicroRNA-199a-5p is associated with hypoxia-inducible factor-1alpha expression in lungs from patients with COPD. Chest. (2012) 142:663–72. doi: 10.1378/chest11-2746
100. Ezzie ME, Crawford M, Cho JH, Orellana R, Zhang S, Gelinas R, et al. Gene expression networks in COPD: microRNA and mRNA regulation. Thorax. (2012) 67:122–31. doi: 10.1136/thoraxjnl-2011-200089
101. Conickx G, Avila Cobos F, van den Berge M, Faiz A, Timens W, Hiemstra PS, et al. microRNA profiling in lung tissue and bronchoalveolar lavage of cigarette smoke-exposed mice and in COPD patients: a translational approach. Sci Rep. (2017) 7:12871. doi: 10.1038/s41598-017-13265-8
102. Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A. (2009) 106:2319–24. doi: 10.1073/pnas0806383106
103. Huang C, Liang Y, Zeng X, Yang X, Xu D, Gou X, et al. lncRNA FENDRR exhibits anti-fibrotic activity in pulmonary fibrosis. Am J Respir Cell Mol Biol. (2020) 62:440–53. doi: 10.1165/rcmb2018-0293OC
104. Savary G, Dewaeles E, Diazzi S, Buscot M, Nottet N, Fassy J, et al. The long noncoding RNA DNM3OS is a reservoir of FibromiRs with major functions in lung fibroblast response to TGF-β and pulmonary fibrosis. Am J Respir Crit Care Med. (2019) 200:184–98. doi: 10.1164/rccm201807-1237OC
105. Gokey JJ, Snowball J, Sridharan A, Speth JP, Black KE, Hariri LP, et al. MEG3 is increased in idiopathic pulmonary fibrosis and regulates epithelial cell differentiation. JCI Insight. (2018) 3:e122490. doi: 10.1172/jci.insight122490
106. Gao Y, Zhang J, Liu Y, Zhang S, Wang Y, Liu B, et al. Regulation of TERRA on telomeric and mitochondrial functions in IPF pathogenesis. BMC Pulm Med. (2017) 17:163. doi: 10.1186/s12890-017-0516-1
107. Qian W, Cai X, Qian Q, Peng W, Yu J, Zhang X, et al. lncRNA ZEB1-AS1 promotes pulmonary fibrosis through ZEB1-mediated epithelial-mesenchymal transition by competitively binding miR-141-3p. Cell Death Dis. (2019) 10:129. doi: 10.1038/s41419-019-1339-1
108. Cui H, Banerjee S, Guo S, Xie N, Ge J, Jiang D, et al. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight. (2019) 4:e124522. doi: 10.1172/jci.insight124522
109. Gu C, Li Y, Liu J, Ying X, Liu Y, Yan J, et al. LncRNAmediated SIRT1/FoxO3a and SIRT1/p53 signaling pathways regulate type II alveolar epithelial cell senescence in patients with chronic obstructive pulmonary disease. Mol Med Rep. (2017) 15:3129–34. doi: 10.3892/mmr.20176367
110. Devadoss D, Long C, Langley RJ, Manevski M, Nair M, Campos MA, et al. Long noncoding transcriptome in chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. (2019) 61:678–88. doi: 10.1165/rcmb2019-0184TR
111. Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. (2012) 12:613–26. doi: 10.1038/nrc3318
112. Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, et al. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. (2012) 482:519–23. doi: 10.1038/nature10810
113. Li N, Muthusamy S, Liang R, Sarojini H, Wang E. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech Ageing Dev. (2011) 132:75–85. doi: 10.1016/j.mad.2010.12004
114. Nakagawa T, Guarente L. SnapShot: sirtuins, NAD, and aging. Cell Metab. (2014) 20:192. doi: 10.1016/j.cmet.2014.06001
115. Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. (2009) 136:62–74. doi: 10.1016/j.cell.2008.10052
116. Yao H, Sundar IK, Ahmad T, Lerner C, Gerloff J, Friedman AE, et al. SIRT1 protects against cigarette smoke-induced lung oxidative stress via a FOXO3-dependent mechanism. Am J Physiol Lung Cell Mol Physiol. (2014) 306:L816–28. doi: 10.1152/ajplung.003232013
117. Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-kappaB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. (2007) 292:L567–76. doi: 10.1152/ajplung.003082006
118. Disayabutr S, Kim EK, Cha SI, Green G, Naikawadi RP, Jones KD, et al. miR-34 miRNAs regulate cellular senescence in type II alveolar epithelial cells of patients with idiopathic pulmonary fibrosis. PLoS ONE. (2016) 11:e0158367. doi: 10.1371/journal.pone0158367
119. Cui H, Ge J, Xie N, Banerjee S, Zhou Y, Antony VB, et al. miR-34a inhibits lung fibrosis by inducing lung fibroblast senescence. Am J Respir Cell Mol Biol. (2017) 56:168–78. doi: 10.1165/rcmb2016-0163OC
120. Rajendrasozhan S, Yang SR, Kinnula VL, Rahman I. SIRT1, an antiinflammatory and antiaging protein, is decreased in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2008) 177:861–70. doi: 10.1164/rccm200708-1269OC
121. Takasaka N, Araya J, Hara H, Ito S, Kobayashi K, Kurita Y, et al. Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J Immunol. (2014) 192:958–68. doi: 10.4049/jimmunol1302341
122. Yao H, Chung S, Hwang JW, Rajendrasozhan S, Sundar IK, Dean DA, et al. SIRT1 protects against emphysema via FOXO3-mediated reduction of premature senescence in mice. J Clin Invest. (2012) 122:2032–45. doi: 10.1172/JCI60132
123. Minagawa S, Araya J, Numata T, Nojiri S, Hara H, Yumino Y, et al. Accelerated epithelial cell senescence in IPF and the inhibitory role of SIRT6 in TGF-beta-induced senescence of human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. (2011) 300:L391–401. doi: 10.1152/ajplung.000972010
124. Cui H, Ge J, Xie N, Banerjee S, Zhou Y, Liu RM, et al. miR-34a promotes fibrosis in aged lungs by inducing alveolarepithelial dysfunctions. Am J Physiol Lung Cell Mol Physiol. (2017) 312:L415–24. doi: 10.1152/ajplung.003352016
125. Shetty SK, Tiwari N, Marudamuthu AS, Puthusseri B, Bhandary YP, Fu J, et al. p53 and miR-34a Feedback Promotes Lung Epithelial Injury and Pulmonary Fibrosis. Am J Pathol. (2017) 187:1016–34. doi: 10.1016/j.ajpath.2016.12020
126. Baker JR, Vuppusetty C, Colley T, Papaioannou AI, Fenwick P, Donnelly L, et al. Oxidative stress dependent microRNA-34a activation via PI3Kalpha reduces the expression of sirtuin-1 and sirtuin-6 in epithelial cells. Sci Rep. (2016) 6:35871. doi: 10.1038/srep35871
127. van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. (2008) 105:13027–32. doi: 10.1073/pnas0805038105
128. Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, et al. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med. (2014) 6:1347–56. doi: 10.15252/emmm201303604
129. Heid J, Cencioni C, Ripa R, Baumgart M, Atlante S, Milano G, et al. Age-dependent increase of oxidative stress regulates microRNA-29 family preserving cardiac health. Sci Rep. (2017) 7:16839. doi: 10.1038/s41598-017-16829-w
130. Hu Z, Klein JD, Mitch WE, Zhang L, Martinez I, Wang XH. MicroRNA-29 induces cellular senescence in aging muscle through multiple signaling pathways. Aging (Albany NY). (2014) 6:160–75. doi: 10.18632/aging100643
131. Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. (2011) 45:287–94. doi: 10.1165/rcmb2010-0323OC
132. Matsushima S, Ishiyama J. MicroRNA-29c regulates apoptosis sensitivity via modulation of the cell-surface death receptor, Fas, in lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. (2016) 311:L1050–61. doi: 10.1152/ajplung.002522016
133. Xie T, Liang J, Geng Y, Liu N, Kurkciyan A, Kulur V, et al. MicroRNA-29c prevents pulmonary fibrosis by regulating epithelial cell renewal and apoptosis. Am J Respir Cell Mol Biol. (2017) 57:721–32. doi: 10.1165/rcmb2017-0133OC
134. Lian X, Chen X, Sun J, An G, Li X, Wang Y, et al. MicroRNA-29b inhibits supernatants from silica-treated macrophages from inducing extracellular matrix synthesis in lung fibroblasts. Toxicol Res (Camb). (2017) 6:878–88. doi: 10.1039/C7TX00126F
135. Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Mück C, et al. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell. (2010) 9:291–6. doi: 10.1111/j.1474-9726.2010.00549x
136. Dellago H, Bobbili MR, Grillari J. MicroRNA-17-5p: at the crossroads of cancer and aging - a mini-review. Gerontology. (2017) 63:20–8. doi: 10.1159/000447773
137. Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, et al. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. (2009) 11:1031–8. doi: 10.1038/ncb1917
138. Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. (2007) 310:442–53. doi: 10.1016/j.ydbio.2007.08007
139. Cortez MA, Valdecanas D, Zhang X, Zhan Y, Bhardwaj V, Calin GA, et al. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol Ther. (2014) 22:1494–503. doi: 10.1038/mt.201479
140. Magenta A, Cencioni C, Fasanaro P, Zaccagnini G, Greco S, Sarra-Ferraris G, et al. miR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. (2011) 18:1628–39. doi: 10.1038/cdd.201142
141. Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, et al. Participation of miR-200 in pulmonary fibrosis. Am J Pathol. (2012) 180:484–93. doi: 10.1016/j.ajpath.2011.10005
142. Schliekelman MJ, Gibbons DL, Faca VM, Creighton CJ, Rizvi ZH, Zhang Q, et al. Targets of the tumor suppressor miR-200 in regulation of the epithelial-mesenchymal transition in cancer. Cancer Res. (2011) 71:7670–82. doi: 10.1158/0008-5472CAN-11-0964
143. Cao Y, Liu Y, Ping F, Yi L, Zeng Z, Li Y. miR-200b/c attenuates lipopolysaccharide-induced early pulmonary fibrosis by targeting ZEB1/2 via p38 MAPK and TGF-beta/smad3 signaling pathways. Lab Invest. (2018) 98:339–59. doi: 10.1038/labinvest.2017123
144. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. (2008) 10:593–601. doi: 10.1038/ncb1722
145. Moimas S, Salton F, Kosmider B, Ring N, Volpe MC, Bahmed K, et al. miR-200 family members reduce senescence and restore idiopathic pulmonary fibrosis type II alveolar epithelial cell transdifferentiation. ERJ Open Res. (2019) 5:00138–2019. doi: 10.1183/2312054100138-2019
146. Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3'UTR microRNA recognition elements. Mol Cell. (2009) 35:610–25. doi: 10.1016/j.molcel.2009.08020
147. Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a~27a~24-2 cluster and its implication in human diseases. Mol Cancer. (2010) 9:232. doi: 10.1186/1476-4598-9-232
148. Bu H, Baraldo G, Lepperdinger G, Jansen-Durr P. mir-24 activity propagates stress-induced senescence by down regulating DNA topoisomerase 1. Exp Gerontol. (2016) 75:48–52. doi: 10.1016/j.exger.2015.12012
149. Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, et al. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal. (2009) 2:ra69. doi: 10.1126/scisignal2000442
150. Nouws J, Wan F, Finnemore E, Roque W, Kim S, Bazan I, et al. Decreased miR-24-3p potentiates DNA damage responses and increases susceptibility to COPD. bioRxiv. doi: 10.1101/2020.05.22.108688
151. Pua HH, Steiner DF, Patel S, Gonzalez JR, Ortiz-Carpena JF, Kageyama R, et al. MicroRNAs 24 and 27 suppress allergic inflammation and target a network of regulators of T helper 2 cell-associated cytokine production. Immunity. (2016) 44:821–32. doi: 10.1016/j.immuni.2016.01003
152. Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E, et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol. (2009) 16:492–8. doi: 10.1038/nsmb1589
153. Paschalaki KE, Zampetaki A, Baker JR, Birrell MA, Starke RD, Belvisi MG, et al. Downregulation of MicroRNA-126 augments DNA damage response in cigarette smokers and patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2018) 197:665–8. doi: 10.1164/rccm201706-1304LE
154. Cao DW, Jiang CM, Wan C, Zhang M, Zhang QY, Zhao M, et al. Upregulation of MiR-126 delays the senescence of human glomerular mesangial cells induced by high glucose via telomere-p53-p21-Rb signaling pathway. Curr Med Sci. (2018) 38:758–64. doi: 10.1007/s11596-018-1942-x
155. Olivieri F, Bonafè M, Spazzafumo L, Gobbi M, Prattichizzo F, Recchioni R, et al. Age- and glycemia-related miR-126-3p levels in plasma and endothelial cells. Aging (Albany NY). (2014) 6:771–87. doi: 10.18632/aging100693
156. Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. (2010) 107:1336–44. doi: 10.1161/CIRCRESAHA.110227926
157. Cheng Y, Yang X, Deng X, Zhang X, Li P, Tao J, et al. MicroRNA-218 inhibits bladder cancer cell proliferation, migration, and invasion by targeting BMI-1. Tumour Biol. (2015) 36:8015–23. doi: 10.1007/s13277-015-3532-x
158. Kong Y, Hsieh CH, Alonso LC. ANRIL: a lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front Endocrinol (Lausanne). (2018) 9:405. doi: 10.3389/fendo.201800405
159. He J, Tu C, Liu Y. Role of lncRNAs in aging and age-related diseases. Aging Med (Milton). (2018) 1:158–75. doi: 10.1002/agm212030
160. Mirza AH, Berthelsen CH, Seemann SE, Pan X, Frederiksen KS, Vilien M, et al. Transcriptomic landscape of lncRNAs in inflammatory bowel disease. Genome Med. (2015) 7:39. doi: 10.1186/s13073-015-0162-2
161. Ge J, Geng S, Jiang H. Long noncoding RNAs antisense noncoding RNA in the INK4 locus (ANRIL) correlates with lower acute exacerbation risk, decreased inflammatory cytokines, and mild GOLD stage in patients with chronic obstructive pulmonary disease. J Clin Lab Anal. (2019) 33:e22678. doi: 10.1002/jcla22678
162. Wang Y, Pang WJ, Wei N, Xiong Y, Wu WJ, Zhao CZ, et al. Identification, stability and expression of Sirt1 antisense long non-coding RNA. Gene. (2014) 539:117–24. doi: 10.1016/j.gene.2014.01037
163. Qian W, Cai X, Qian Q. Sirt1 antisense long non-coding RNA attenuates pulmonary fibrosis through sirt1-mediated epithelial-mesenchymal transition. Aging. (2020) 12:4322–36. doi: 10.18632/aging102882
164. Porro A, Feuerhahn S, Delafontaine J, Riethman H, Rougemont J, Lingner J. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat Commun. (2014) 5:5379. doi: 10.1038/ncomms6379
165. Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet. (2015) 6:143. doi: 10.3389/fgene.201500143
166. Montero JJ, Lopez-Silanes I, Megias D, Fraga MF, Castells-Garcia A, Blasco MA. TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat Commun. (2018) 9:1548. doi: 10.1038/s41467-018-03916-3
167. Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L, Lingner J. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature. (2020) 587:303–8. doi: 10.1038/s41586-020-2815-6
168. Bettin N, Oss Pegorar C, Cusanelli E. The emerging roles of TERRA in telomere maintenance and genome stability. Cells. (2019) 8:246. doi: 10.3390/cells8030246
169. Koch L. Non-coding RNA: a protective role for TERRA at telomeres. Nat Rev Genet. (2017) 18:453. doi: 10.1038/nrg.201758
170. Arun G, Aggarwal D, Spector DL. MALAT1 long non-coding RNA: functional implications. Noncoding RNA. (2020) 6:22. doi: 10.3390/ncrna6020022
171. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. (2010) 39:925–38. doi: 10.1016/j.molcel.2010.08011
172. Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. (2014) 159:188–99. doi: 10.1016/j.cell.2014.08018
173. Zeng R, Zhang R, Song X, Ni L, Lai Z, Liu C, et al. The long non-coding RNA MALAT1 activates Nrf2 signaling to protect human umbilical vein endothelial cells from hydrogen peroxide. Biochem Biophys Res Commun. (2018) 495:2532–8. doi: 10.1016/j.bbrc.2017.12105
174. Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. (2013) 49:204–11. doi: 10.1165/rcmb2013-0159RC
175. He C, Ding JW, Li S, Wu H, Jiang YR, Yang W, et al. The role of long intergenic noncoding RNA p21 in vascular endothelial cells. DNA Cell Biol. (2015) 34:677–83. doi: 10.1089/dna.20152966
176. Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. (2014) 54:777–90. doi: 10.1016/j.molcel.2014.04025
177. Groff AF, Sanchez-Gomez DB, Soruco MML, Gerhardinger C, Barutcu AR, Li E, et al. In vivo characterization of linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. (2016) 16:2178–86. doi: 10.1016/j.celrep.2016.07050
178. Zhou WQ, Wang P, Shao QP, Wang J. Lipopolysaccharide promotes pulmonary fibrosis in acute respiratory distress syndrome (ARDS) via lincRNA-p21 induced inhibition of Thy-1 expression. Mol Cell Biochem. (2016) 419:19–28. doi: 10.1007/s11010-016-2745-7
179. Hooten NN, Evans MK. Age and poverty status alter the coding and noncoding transcriptome. Aging (Albany NY). (2019) 11:1189–203. doi: 10.18632/aging101823
180. Uroda T, Anastasakou E, Rossi A, Teulon JM, Pellequer JL, Annibale P, et al. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol Cell. (2019) 75:982–95 e9. doi: 10.1016/j.molcel.2019.07025
181. Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. (2014) 53:290–300. doi: 10.1016/j.molcel.2013.11012
182. Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. (2007) 282:24731–42. doi: 10.1074/jbcM702029200
183. Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, et al. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: isoform structure, expression, and functions. Endocrinology. (2010) 151:939–47. doi: 10.1210/en2009-0657
184. Tang W, Shen Z, Guo J, Sun S. Screening of long non-coding RNA and TUG1 inhibits proliferation with TGF-beta induction in patients with COPD. Int J Chron Obstruct Pulmon Dis. (2016) 11:2951–64. doi: 10.2147/COPDS109570
185. Li X, Zheng M, Pu J, Zhou Y, Hong W, Fu X, et al. Identification of abnormally expressed lncRNAs induced by PM2.5 in human bronchial epithelial cells. Biosci Rep. (2018) 38:BSR20171577. doi: 10.1042/BSR20171577
186. Song B, Ye L, Wu S, Jing Z. Long non-coding RNA MEG3 regulates CSE-induced apoptosis and inflammation via regulating miR-218 in 16HBE cells. Biochem Biophys Res Commun. (2020) 521:368–74. doi: 10.1016/j.bbrc.2019.10135
187. Kusko RL, Brothers JF II, Tedrow J, Pandit K, Huleihel L, Perdomo C, et al. Integrated genomics reveals convergent transcriptomic networks underlying chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2016) 194:948–60. doi: 10.1164/rccm201510-2026OC
188. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–22. doi: 10.1038/nrd.2016246
189. Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genet. (2017) 33:665–76. doi: 10.1016/j.tig.2017.08002
190. Sanchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic Acid Ther. (2013) 23:15–20. doi: 10.1089/nat.20120414
191. Bolte C, Ustiyan V, Ren X, Dunn AW, Pradhan A, Wang G, et al. Nanoparticle delivery of proangiogenic transcription factors into the neonatal circulation inhibits alveolar simplification caused by hyperoxia. Am J Respir Crit Care Med. (2020) 202:100–11. doi: 10.1164/rccm201906-1232OC
192. Hoesel LM, Flierl MA, Niederbichler AD, Rittirsch D, McClintock SD, Reuben JS, et al. Ability of antioxidant liposomes to prevent acute and progressive pulmonary injury. Antioxid Redox Signal. (2008) 10:973–81. doi: 10.1089/ars.20071878
193. Gallant-Behm CL, Piper J, Lynch JM, Seto AG, Hong SJ, Mustoe TA, et al. A MicroRNA-29 mimic (remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Invest Dermatol. (2019) 139:1073–81. doi: 10.1016/j.jid.2018.11007
194. Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. (2017) 35:180–8. doi: 10.1007/s10637-016-0407-y
195. Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. (2016) 30:34–51. doi: 10.1101/gad.270959115
196. Fu WM, Lu YF, Hu BG, Liang WC, Zhu X, Yang HD, et al. Long noncoding RNA Hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget. (2016) 7:4712–23. doi: 10.18632/oncotarget6731
197. Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH, Guo RH. Decreased expression of long noncoding RNA MEG3 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer. Tumour Biol. (2015) 36:4851–9. doi: 10.1007/s13277-015-3139-2
Keywords: COPD–Chronic Obstructive Pulmonary Disease, IPF–Idiopathic Pulmonary Fibrosis, cellular senescence, non-coding RNA, micro-RNA (miRNA), long-non coding RNA
Citation: Omote N and Sauler M (2020) Non-coding RNAs as Regulators of Cellular Senescence in Idiopathic Pulmonary Fibrosis and Chronic Obstructive Pulmonary Disease. Front. Med. 7:603047. doi: 10.3389/fmed.2020.603047
Received: 04 September 2020; Accepted: 09 November 2020;
Published: 23 December 2020.
Edited by:
Claudia A. Staab-Weijnitz, Helmholtz Zentrum München, GermanyReviewed by:
Marta Bueno, University of Pittsburgh, United StatesJonathan Baker, Imperial College London, United Kingdom
Copyright © 2020 Omote and Sauler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maor Sauler, bWFvci5zYXVsZXJAeWFsZS5lZHU=
 Norihito Omote
Norihito Omote Maor Sauler
Maor Sauler