
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Med. , 30 October 2020
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 7 - 2020 | https://doi.org/10.3389/fmed.2020.582793
This article is part of the Research Topic Coronavirus Disease (COVID-19): Pathophysiology, Epidemiology, Clinical Management and Public Health Response, Volume II View all 165 articles
Diverse populations worldwide are differentially affected by coronavirus disease 2019 (COVID-19). While socioeconomic background has been studied extensively, little is known about the genetic variation underlying this phenomenon. This study is aimed at examining the genetic basis behind the great discrepancies among diverse ethnic groups in terms of COVID-19 susceptibility for viral infection, disease prognosis, and mortality. To this end, in silico analysis of single-nucleotide polymorphisms (SNPs) within regulatory sequences of the human angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2)—the virus's gateway to host cells—and their plausible implications on expression levels was conducted. We provide indication that the variation in the human ACE2 and TMPRSS2 regulatory sequences is likely to be involved in and contribute to this phenomenon. SNPs that are abundant in the more susceptible populations introduce binding sites (BSs) for transcription factors or they may invalidate BSs for transcription repressor—both may enhance target gene (ACE2 or TMPRSS2) expression in the relevant target tissues. SNPs that are abundant in the more resistant populations may invalidate BSs for a transcriptional repressor or they may introduce BSs for a transcriptional repressor or initiator of mRNA degradation, which may reduce target gene expression levels. This aspect, when added to the socioeconomic factors, can be a cause for the divergent prevalence of the disease and the different mortality rates within diverse populations. This demonstration may call for a shift in the paradigm of searching for COVID-19 biomarkers, such that SNPs within regulatory sequences should be of high importance.
The coronavirus disease 2019 (COVID-19) pandemic, which began in late 2019 in Wuhan, Hubei province, China (1, 2), has spread throughout the world and affected every aspect of human life. The most common clinical signs and symptoms of the disease are fever, fatigue, dry cough, and breathlessness, while expectoration, headache, myalgia, diarrhea, nausea, vomiting, loss of taste or smell, cutaneous eruptions, and renal failure have also been reported (3). Countries throughout the world, and even subpopulations within countries, present great variation in death rate as well as in case fatality ratios (4). The important role of demography, particularly age structure of a population, was demonstrated and may help explain differences in fatality rates across countries (5). These differences can also be caused by variations between countries in the number of people tested, characteristics of the local healthcare system, the tactics and actions taken to fight against COVID-19, the presence of possible subtypes of the virus, as well as inequalities in socioeconomic, ethnic, geographical, and social determinants of health (6, 7). The following risk factors have been associated with COVID-19: advanced age, obesity, male gender, heart diseases (8), diabetes and immunodeficiency, ethnicity/race (9, 10), and minorities. For example, in the USA (11) and the UK, COVID-19 death rates among African descent populations were higher than among Asian descent or white populations. Noteworthy, ethnicity is a complex entity composed of genetic makeup, social and economic constructs, cultural identity, lifestyle habits, and behavioral patterns (12). Thus far, disparities in COVID-19 disease burden and outcomes among racial and ethnic minorities were mostly associated with socioeconomic conditions, baseline health states, as well as social and health behaviors/behavioral risk factors (13–16), and a call for proper representation and race reporting in clinical trials has emerged (16, 17). Yet, data on COVID-19 by ethnicity/race are scant, and the genetic component has been largely overlooked in most studies.
COVID-19 infection depends on a specific interaction between host angiotensin-converting enzyme 2 (ACE2) as the entry receptor and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus receptor binding domain of the surface spike glycoprotein (18–20). The cellular serine protease transmembrane protease serine 2 (TMPRSS2) is employed for the Spike protein priming, a cleavage that allows the fusion of viral and cellular membranes (21) and viral spread in the infected host (22). This process potentially involves other proteins, such as the human exopeptidase CD26 (23), also known as DPP4—a key immunoregulatory factor for hijacking and virulence, which are out of the scope of this paper.
ACE2 expression is highly abundant in the lungs and the epithelial cells of the gastrointestinal tract (GIT) and to a lesser extent in the kidney, liver, and male reproductive tissues (24, 25). Expression of TMPRSS2 is high in the GIT and proximal digestive tract and moderate in adult lungs—mainly in bronchial epithelial cells—and also abundant in the prostate gland, kidney, and urinary bladder (26). Both TMPRSS2 and ACE2 are expressed in human corneal epithelium, suggesting that ocular surface cells could serve as a potential entry point and as a reservoir for person-to-person transmission of this virus (27). Recently, the expression and function of coding regions and other variants in ACE2 and TMPRSS2 among different populations were systematically analyzed, implying different susceptibilities or responses to COVID-19 in different populations (28–34). In addition, variants located at regulatory regions of TMPRSS2 were found to influence its expression (35). For example, delC allele (rs35074065, located in the shared 3′ regulatory region of TMPRSS2) leads to overexpression of TMPRSS2 [probably by disrupting a binding site (BS) for the repressor IRF2], thus facilitating entry of the D614G COV-19 subtype into host cells and accelerating its spread in Europe and North America where the allele is common (36). Moreover, a single-nucleotide polymorphism (SNP) within the androgen response element in an enhancer located 13 kb upstream of TMPRSS2 transcription start site reduces binding and transactivation by the androgen receptor (37)—a signaling pathway that also modulates both TMPRSS2 and ACE2 expression and is associated with severe COVID-19 symptoms in men (38, 39).
Little attention has hitherto been given to polymorphism in the ACE2 and TMPRSS2 promoters and the possible association with COVID-19 infection, prognosis, and mortality in different ethnicities. Of note, while no association was observed between genetic variants located in or near ACE2 and TMPRSS2 genes and human quantitative phenotypes (40), some polymorphisms with relatively high frequencies in different human populations have possible functional effects of COVID-19 infection as they generate BSs for transcription factors (TFs) (41).
This study aims to propose possible variants in the regulatory regions of ACE2 and TMPRSS2 that may underlie the marked geographic and race variations in COVID-19 prevalence and mortality. These may further serve in genetic association studies in patients with SARS-CoV-2 infection.
In order to gain insights on SNPs that might be relevant to the marked COVID-19 geographic and race variations, the following inclusion criteria were applied: (1) SNPs with a relatively high allelic frequency in specific populations; (2) SNPs for which there is a marked difference in their frequencies among Asian and Africans descents. The frequencies of each allele among diverse ethnic groups were obtained from the following studies: 1000 Genomes, gnomAD–Genomes, ExAc, and TopMed, and when the sample size was big enough, other studies of more specific populations were utilized. These SNPs and the relevant findings are described in detail in Table 1.
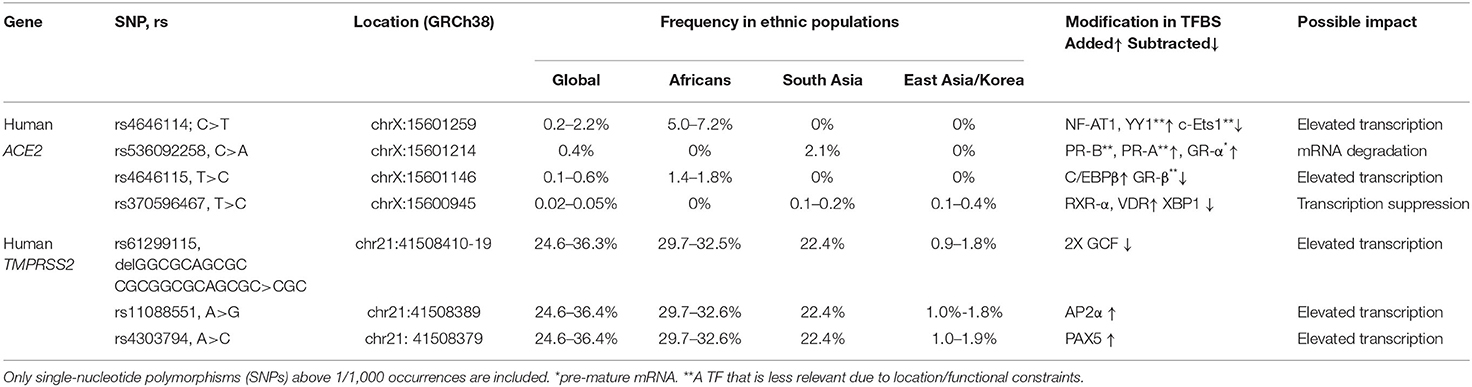
Table 1. REFSEQ ID (rs), chromosomal location, occurrence, ethnicity, added/subtracted transcription factor (TF) prediction, and estimated expression outcome in the human angiotensin-converting enzyme 2 (ACE2) and transmembrane protease, serine 2 (TMPRSS2) proximal, and core promoter sequence.
To examine the potential impact of the more abundant SNPs in ACE2 and TMPRSS2 regulatory sequences on their transcriptional regulation, expression, and mRNA stability, PROMO (42) was used to predict transcription factor binding sites (TFBSs) and their modifications in the presence of a given SNP. The diverse possible mechanisms through which these SNPs modulate ACE2 and TMPRSS2 levels are schematically described in Figures 1A,B, respectively. A summary of the expression pattern of ACE2, TMPRSS2, and the related key TFs, based on the Human Protein Atlas (43), is provided in Table 2.
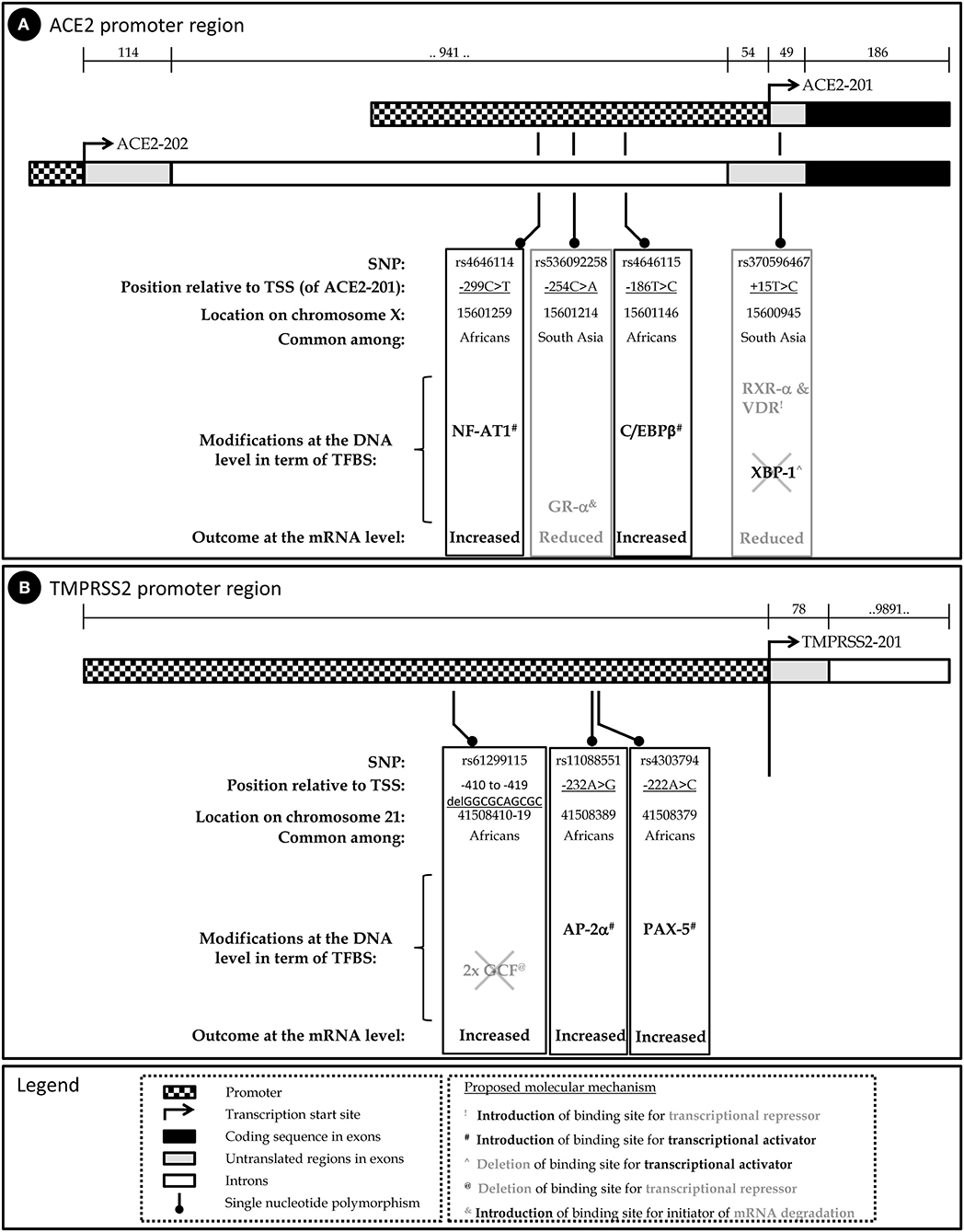
Figure 1. Schematic representation of the human ACE2 (A) and TMPRSS2 (B) genes, the relevant SNPs and their predicted implication on mRNA level.
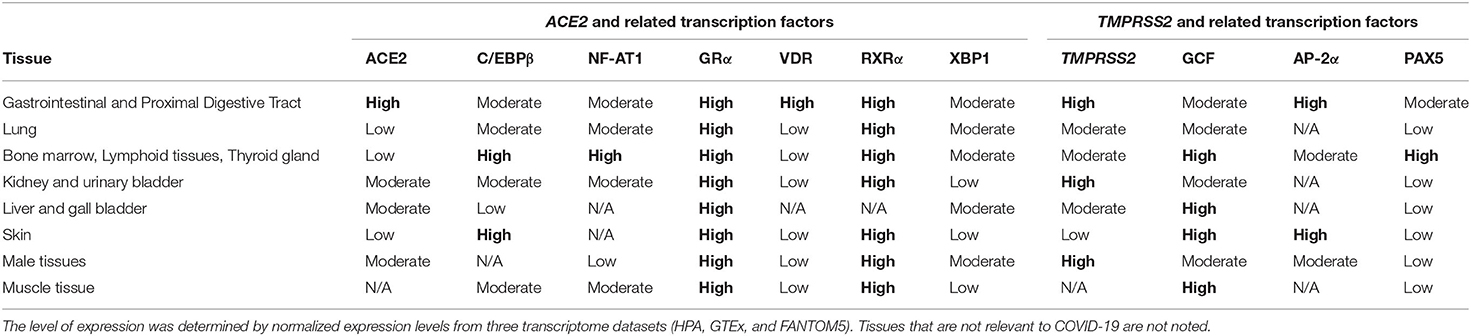
Table 2. Expression levels of angiotensin-converting enzyme 2 (ACE2), transmembrane protease, serine 2 (TMPRSS2), and related transcription factors in different tissues.
Expression of the human ACE2 gene is derived by alternative promoters; the former generates an alternative 5′-untranslated exon (44) and both encoding a similar 805 amino acid precursor: the predominant transcript ACE2-201 (ENST00000252519.8) and ACE2-202 (ENST00000427411.1). Additionally, alternative splicing may generate a shorter isoform due to termination after coding exon 12. The expression of the TMPRSS2 gene is derived from a single promoter, yet two main transcripts are generated (45), of which the major one is TMPRSS2-201 (ENST00000332149.10).
rs4646114 is the most abundant SNP (5–7.2%), mainly among African descent populations. It forms an additional TFBS to nuclear factor of activated T cells (NF-AT1). Viral infection activates T cells that induce NF-AT1 dephosphorylation, nuclear translocation, and transcriptional activation of target genes primarily involved in cell–cell interactions (46). NF-AT1 is expressed throughout the body, but especially in the lymphoid tissues, muscles, urinary bladder, kidneys, and lungs, all reported to be infected in many cases of COVID-19. Thus, following initial infection in certain cells expressing a high level of ACE2, NF-AT1 is proposed to further induce ACE2 transcription during the immune response, which in turn enables substantial penetration and spread of COVID-19 to the other host cells during infection. This forms a positive feedback loop that accelerates penetration and spread of the virus in host cells.
rs536092258 is highly abundant in Asian populations (>2%). It forms a TFBS to the steroid nuclear receptor GR-α, which functions as an expression regulator of glucocorticoid-responsive genes. GR-α has a posttranscriptional role, acting as an RNA-binding protein and initiating mRNA degradation (47) and thus reducing protein levels. This potential effect is limited to the ACE2-202 variant, but not to the ACE2-201 variant, as only the former harbors this variation in the primary transcript.
rs4646115 is prevalent in African descent populations (1.4–1.8%). The SNP multiplies the TFBS of CCAAT/enhancer binding protein beta (C/EBPβ)—a leucine zipper-type TF that is involved in inflammation and acute-phase response and it is highly expressed in the lungs and liver. The multiplication of TFBSs has been shown to increase the expression of a given gene, and thus rs4646115 is likely to enhance ACE2 expression in the lungs and liver and facilitate COVID-19 infection that spreads through the lungs. Interestingly, C/EBPβ is also highly abundant in the adipose tissue, and a high-fat diet or saturated fatty acid exposure has been shown to directly activate C/EBPβ protein expression in the liver, adipocytes, and macrophages. It also influences the development of abdominal obesity and phenotypes related to the development of type 2 diabetes mellitus and cardiovascular disease, all reported as COVID-19 risk factors (8).
rs370596467 is quite rare though an interesting SNP. It is frequent in South and East Asian populations (0.1–0.4%). TFBSs to both retinoid X receptor alpha (RXR-α) (that is expressed in the lungs, skin, and GIT) and vitamin D receptor (VDR) (which is most abundant in the GIT) are introduced by this variation. VDR is a zinc finger protein containing a DNA-binding domain and two protein interaction surfaces. One of those surfaces is a site for the formation of a heterodimer with the partner protein, RXR-α. Together, this heterodimer suppresses gene activity, although the exact mechanism is currently unclear (48). The SNP also subtracts TFBSs for X-box binding protein 1 (XBP-1), a transcription activator that can increase ACE2 activation. Together, the subtraction of activator (XBP-1) TFBS and the introduction of BSs to repressors can lead to ACE2 gene repression and, consequently, lower ACE2 expression.
Altogether, this analysis implies that carriers of SNPs rs4646114 and rs4646115, which are relatively more abundant among Africans, may present higher susceptibility to COVID-19. On the other hand, SNPs rs536092258 and rs370596467, which are relatively more abundant among individuals of South and East Asian origin, may provide tolerance, at least to some extent, against COVID-19 (Figure 1A).
rs61299115, rs11088551, and rs4303794 are all highly frequent in the global population (25–36%); however, they appear in East Asian and Korean populations at a much lower extent (<2%).
rs61299115 introduces a deletion of 10 bp. Due to this deletion, an overlapping double BS for the transcriptional repressor GC factor [GC-Rich Sequence DNA-Binding Factor (GCF)] (49) is deleted, potentially enhancing TMPRSS2 transcription. Therefore, among East Asian populations, where the minor allele is much less frequent compared to the rest of the world population, TMPRSS2 expression is expected to be relatively lower among the higher share of the population, conferring lower COVID-19 infection.
rs11088551 introduces a BS for activating enhancer binding protein 2 alpha (AP-2α), which belongs to a family of transcriptional regulators and involved in diverse developmental processes, apoptosis, and cell cycle (50, 51). AP-2α also interacts with inducible viral and cellular enhancer elements to regulate the transcription of selected genes. This suggests—similarly to rs61299115—that among East Asian populations, where the minor allele is much less frequent compared to the rest of the world population, TMPRSS2 expression is expected to be relatively lower among a higher share of the population, conferring lower COVID-19 infection.
rs4303794 introduces a BS for paired box 5 (PAX5), a pluripotent transcriptional activator of B-cell development and cancerous processes (52). This suggests that lack of rs4303794 is consistent with lower expression levels of TMPRSS2, and this scenario is prevalent among the East Asian populations.
Together, the three SNPs that are highly prevalent in the general population (25–36%) and are quite rare in the East Asian and Korean populations, hint toward a lower expression of TMPRSS2. This, in addition to the variations found in the promoter of ACE2, can suggest a different COVID-19 etiology and prognosis in different populations (Figure 1B).
This study presents a novel approach and intriguing initial findings possibly underlying the relationship between genetic variations and ethnic susceptibility to COVID-19, which are of high and immediate interest, particularly to the biomedical community and more generally to civil societies worldwide. It brings to light five possible mechanisms by which the modification of TFBS (either production or subtraction) might impact mRNA levels of genes related to COVID-19 entry into host cells. Yet, the potential effects of the SNPs on ACE2 and TMPRSS2 expression levels should be further validated first by expression studies in diverse ethnic populations as well as in healthy and infected individuals, and also by mechanistic studies, to infer differential SNP-derived TF binding and activity in target host cells of the virus.
Noteworthy, as ACE2 is located on chromosome X, allele distribution and impact are expected to be different among males and females. For instance, all males carrying a given SNP are considered hemizygous and would be affected, whereas only homozygote females carrying this SNP, but not heterozygote ones, would be affected. This should be further evaluated epidemiologically, while taking into account variations in the coding region of ACE2.
This study represents a proof of concept for a possible relationship of genetic variations within the ACE2 and TMPRSS2 regulatory sequences and COVID-19 etiologies, which, in addition to socioeconomic gaps, may explain discrepancies among diverse ethnic groups. It broadens the biological outlook on the COVID-19 pandemic to gene regulatory regions, rather than the more obvious and frequently investigated coding sequences. The variation presented in the human ACE2 and TMPRSS2 regulatory sequences is assumed, at least partially, to contribute to the different disease etiologies—including susceptibility to viral infection, disease prognosis, severity, and mortality—among, for example, African/African descent and Asian populations. Genetic evidence from human samples of infected and healthy individuals of diverse ethnicities around the world could further confirm and validate the proposed relationship. This approach should also be applied to other COVID-19-related human genes in the pursuit of COVID-19 biomarkers. Such information on variations in regulatory and coding sequences may pave the way for designing a diagnostic tool and perhaps also for formulating future population-sensitive government policies, i.e., setting priorities for preventive programs, quarantine, and (in the future) vaccination.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Prof. Ran Friedman from the Department of Chemistry and Biomedical Sciences, Linnaeus University, Kalmar, Sweden, for his insightful comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2020.582793/full#supplementary-material
ACE2, angiotensin-converting enzyme 2; AP-2α, activating enhancer binding protein 2 alpha; C/EBPβ, CCAAT/enhancer binding protein beta; COVID-19, coronavirus disease 2019; GCF, GC-Rich Sequence DNA-Binding Factor; GIT, gastrointestinal tract; GR-α, glucocorticoid receptor alpha; NF-AT1, nuclear factor of activated T cells; PAX5, paired box 5; RXR-α, retinoid X receptor alpha; SNP, single-nucleotide polymorphism; TFBS, transcription factor binding site; TMPRSS2, transmembrane protease, serine 2; VDR, vitamin D receptor; XBP-1, X-box binding protein 1.
1. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
2. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
3. Balla M, Merugu GP, Patel M, Koduri NM, Gayam V, Adapa S, et al. COVID-19, modern pandemic: a systematic review from front-line health care providers' perspective. J Clin Med Res. (2020) 12:215–29. doi: 10.14740/jocmr4142
4. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. (2020) 20:533–4. doi: 10.1016/S1473-3099(20)30120-1
5. Dowd JB, Andriano L, Brazel DM, Rotondi V, Block P, Ding X, et al. Demographic science aids in understanding the spread and fatality rates of COVID-19. Proc Natl Acad Sci USA. (2020) 117:9696–8. doi: 10.1073/pnas.2004911117
6. Bambra C, Riordan R, Ford J, Matthews F. The COVID-19 pandemic and health inequalities. J Epdemiol Commun Health. (2020) 74:964–8. doi: 10.1136/jech-2020-214401
7. Biswas NK, Majumder PP. Analysis of RNA sequences of 3636 SARS-CoV-2 collected from 55 countries reveals selective sweep of one virus type. Indian J Med Res. (2020) 151:450–8. doi: 10.4103/ijmr.IJMR_1125_20
8. Mehra MR, Desai SS, Kuy S, Henry TD, Patel AN. Cardiovascular disease, drug therapy, and mortality in Covid-19. N Engl J Med. (2020) 382:e102. doi: 10.1056/NEJMoa2007621
9. Bhala N, Curry G, Martineau AR, Agyemang C, Bhopal R. Sharpening the global focus on ethnicity and race in the time of COVID-19. Lancet. (2020) 395:1673–6. doi: 10.1016/S0140-6736(20)31102-8
10. Webb Hooper M, Nápoles AM, Pérez-Stable EJ. COVID-19 and racial/ethnic disparities. JAMA. (2020) 323:2466–7. doi: 10.1001/jama.2020.8598
12. Lee C. “Race” and “ethnicity” in biomedical research: how do scientists construct and explain differences in health? Soc Sci Med 1982. (2009). 68:1183–90. doi: 10.1016/j.socscimed.2008.12.036
13. Niedzwiedz CL, O'Donnell CA, Jani BD, Demou E, Ho FK, Celis-Morales C, et al. Ethnic and socioeconomic differences in SARS-CoV-2 infection: prospective cohort study using UK Biobank. BMC Med. (2020) 18:160. doi: 10.1186/s12916-020-01640-8
14. Verhagen MD, Brazel DM, Dowd JB, Kashnitsky I, Mills MC. Forecasting spatial, socioeconomic and demographic variation in COVID-19 health care demand in England and Wales. BMC Med. (2020) 18:203. doi: 10.1186/s12916-020-01646-2
15. Pareek M, Bangash MN, Pareek N, Pan D, Sze S, Minhas JS, et al. Ethnicity and COVID-19: an urgent public health research priority. Lancet. (2020) 395:1421–2. doi: 10.1016/S0140-6736(20)30922-3
16. Chastain DB, Osae SP, Henao-Martínez AF, Franco-Paredes C, Chastain JS, Young HN. Racial disproportionality in covid clinical trials. N Engl J Med. (2020) 383:e59. doi: 10.1056/NEJMp2021971
17. Borno HT, Zhang S, Gomez S. COVID-19 disparities: an urgent call for race reporting and representation in clinical research. Contemp Clin Trials Commun. (2020) 19:100630. doi: 10.1016/j.conctc.2020.100630
18. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. (2020) 581:221–4. doi: 10.1038/s41586-020-2179-y
19. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. (2020) 581:215–20. doi: 10.1038/s41586-020-2180-5
20. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
21. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271–80.e8. doi: 10.1016/j.cell.2020.02.052
22. Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. (2019) 93:e01815-18. doi: 10.1128/JVI.01815-18
23. Vankadari N, Wilce JA. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect. (2020) 9:601–4. doi: 10.1080/22221751.2020.1739565
24. Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. (2020) 9:45. doi: 10.1186/s40249-020-00662-x
25. Albini A, Di Guardo G, Noonan DM, Lombardo M. The SARS-CoV-2 receptor, ACE-2, is expressed on many different cell types: implications for ACE-inhibitor- and angiotensin II receptor blocker-based cardiovascular therapies. Int Emerg Med. (2020) 15:1–8. doi: 10.1007/s11739-020-02436-7
26. Thunders M, Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J Clin Pathol. (2020) doi: 10.1136/jclinpath-2020-206987
27. Zhou L, Xu Z, Castiglione GM, Soiberman US, Eberhart CG, Duh EJ. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul Surf. (2020) 18:537–44. doi: 10.1016/j.jtos.2020.06.007
28. Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. (2020) 6:11. doi: 10.1038/s41421-020-0147-1
29. Devaux CA, Rolain J-M, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. (2020) 53:425–35. doi: 10.1016/j.jmii.2020.04.015
30. Lee IH, Lee JW, Kong SW. A survey of genetic variants in SARS-CoV-2 interacting domains of ACE2, TMPRSS2 and TLR3/7/8 across populations. Infect Genet Evol. (2020) 2020:104507. doi: 10.1016/j.meegid.2020.104507
31. Fujikura K, Uesaka K. Genetic variations in the human severe acute respiratory syndrome coronavirus receptor ACE2 and serine protease TMPRSS2. J Clin Pathol. (2020). doi: 10.1136/jclinpath-2020-206867
32. Stawiski EW, Diwanji D, Suryamohan K, Gupta R, Fellouse FA, Sathirapongsasuti JF, et al. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.04.07.024752
33. Paniri A, Hosseini MM, Akhavan-Niaki H. First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. J Biomol Struct Dynam.(2020) 2020:1–18. doi: 10.1080/07391102.2020.1767690
34. Asselta R, Paraboschi EM, Mantovani A, Duga S. ACE2 and TMPRSS2 variants and expression as candidates to sex and country differences in COVID-19 severity in Italy. Aging. (2020) 12:10087–98. doi: 10.18632/aging.103415
35. Senapati S, Kumar S, Singh AK, Banerjee P, Bhagavatula S. Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J Genet. (2020) 99:53. doi: 10.1007/s12041-020-01217-7
36. Bhattacharyya C, Das C, Ghosh A, Singh AK, Mukherjee S, Majumder PP, et al. Global spread of SARS-CoV-2 subtype with spike protein mutation D614G is shaped by human genomic variations that regulate expression of TMPRSS2 and MX1 genes. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.05.04.075911
37. Clinckemalie L, Spans L, Dubois V, Laurent M, Helsen C, Joniau S, et al. Androgen regulation of the TMPRSS2 gene and the effect of a SNP in an androgen response element. Mol Endocrinol. (2013) 27:2028–40. doi: 10.1210/me.2013-1098
38. Ghazizadeh Z, Majd H, Richter M, Samuel R, Zekavat SM, Asgharian H, et al. Androgen regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. bioRxiv [Preprint]. (2020) doi: 10.1101/2020.05.12.091082
39. Mjaess G, Karam A, Aoun F, Albisinni S, Roumeguère T. COVID-19 and the male susceptibility: the role of ACE2, TMPRSS2 and the androgen receptor. Prog Eurol. (2020) 30:484–7. doi: 10.1016/j.purol.2020.05.007
40. Lopera Maya EA, van der Graaf A, Lanting P, van der Geest M, Fu J, Swertz M, et al. Lack of association between genetic variants at ACE2 and TMPRSS2 genes involved in SARS-CoV-2 infection and human quantitative phenotypes. Front Genet. (2020) 11:613. doi: 10.3389/fgene.2020.00613
41. Vargas-Alarcón G, Posadas-Sánchez R, Ramírez-Bello J. Variability in genes related to SARS-CoV-2 entry into host cells (ACE2, TMPRSS2, TMPRSS11A, ELANE, and CTSL) and its potential use in association studies. Life Sci. (2020) 260:118313. doi: 10.1016/j.lfs.2020.118313
42. Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. (2002) 18:333–4. doi: 10.1093/bioinformatics/18.2.333
43. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. tissue-based map of the human proteome. Science. (2015) 347:1260419. doi: 10.1126/science.1260419
44. Itoyama S, Keicho N, Hijikata M, Quy T, Phi NC, Long HT, et al. Identification of an alternative 5′-untranslated exon and new polymorphisms of angiotensin-converting enzyme 2 gene: lack of association with SARS in the Vietnamese population. Am J Med Genet A. (2005) 136:52–7. doi: 10.1002/ajmg.a.30779
45. Jacquinet E, Rao NV, Rao GV, Zhengming W, Albertine KH, Hoidal JR. Cloning and characterization of the cDNA and gene for human epitheliasin. Eur J Biochem. (2001) 268:2687–99. doi: 10.1046/j.1432-1327.2001.02165.x
46. Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. (2002) 109(2 Suppl. 1):S67–79. doi: 10.1016/S0092-8674(02)00699-2
47. Cho H, Park OH, Park J, Ryu I, Kim J, Ko J, et al. Glucocorticoid receptor interacts with PNRC2 in a ligand-dependent manner to recruit UPF1 for rapid mRNA degradation. Proc Natl Acad Sci USA. (2015) 112:E1540–9. doi: 10.1073/pnas.1409612112
48. Pike JW, Meyer MB, Bishop KA. Regulation of target gene expression by the vitamin D receptor - an update on mechanisms. Rev Endocr Metab Disord. (2012) 13:45–55. doi: 10.1007/s11154-011-9198-9
49. Johnson AC, Kageyama R, Popescu NC, Pastan I. Expression and chromosomal localization of the gene for the human transcriptional repressor GCF. J Biol Chem. (1992) 267:1689–94.
50. Hilger-Eversheim K, Moser M, Schorle H, Buettner R. Regulatory roles of AP-2 transcription factors in vertebrate development, apoptosis and cell-cycle control. Gene. (2000) 260:1–12. doi: 10.1016/S0378-1119(00)00454-6
51. Eckert D, Buhl S, Weber S, Jäger R, Schorle H. The AP-2 family of transcription factors. Genome Biol. (2005) 6:246. doi: 10.1186/gb-2005-6-13-246
Keywords: SNP, COVID-19, biomarkers, regulatory sequence elements, ACE2, TMPRSS2
Citation: Barash A, Machluf Y, Ariel I and Dekel Y (2020) The Pursuit of COVID-19 Biomarkers: Putting the Spotlight on ACE2 and TMPRSS2 Regulatory Sequences. Front. Med. 7:582793. doi: 10.3389/fmed.2020.582793
Received: 13 July 2020; Accepted: 23 September 2020;
Published: 30 October 2020.
Edited by:
Eugenie Ruth Lumbers, The University of Newcastle, AustraliaReviewed by:
Jeffrey Wilusz, Colorado State University, United StatesCopyright © 2020 Barash, Machluf, Ariel and Dekel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaron Dekel, eWFyb25kQGdyaS5vcmcuaWw=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.