- 1Department of Respiratory Medicine, The First Hospital of Qinhuangdao, Qinhuangdao, China
- 2Department of Respiratory Medicine, Chinese People's Liberation Army General Hospital, Beijing, China
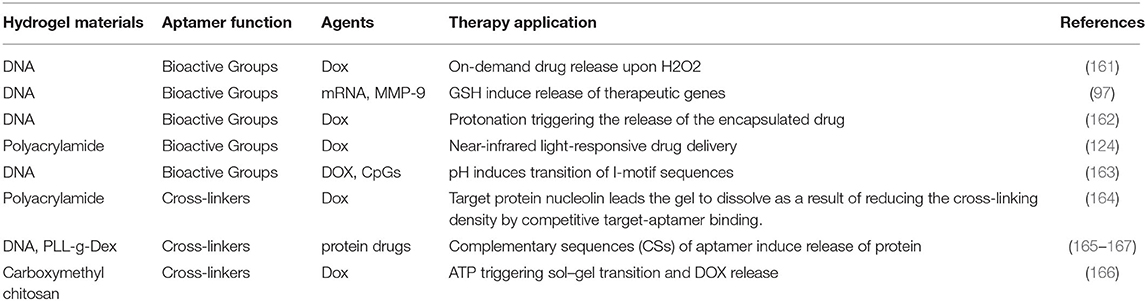
Aptamers are special types of single-stranded DNA generated by a process called systematic evolution of ligands by exponential enrichment (SELEX). Due to significant advances in the chemical synthesis and biotechnological production, aptamers have gained considerable attention as versatile building blocks for the next generation of soft materials. Hydrogels are high water-retainable materials with a three-dimensional (3D) polymeric network. Aptamers, as a vital element, have greatly expanded the applications of hydrogels. Due to their biocompatibility, selective binding, and molecular recognition, aptamer-based hydrogels can be utilized for bioanalytical and biomedical applications. In this review, we focus on the latest strategies of aptamer-based hydrogels in bioanalytical and biomedical applications. We begin this review with an overview of the underlying design principles for the construction of aptamer-based hydrogels. Next, we will discuss some bioanalytical and biomedical applications of aptamer-based hydrogel including biosensing, target capture and release, logic devices, gene and cancer therapy. Finally, the recent progress of aptamer-based hydrogels is discussed, along with challenges and future perspectives.
Introduction
Hydrogels are a kind of high water-retainable material (containing up to 99 wt% water) with a three-dimensional (3D) polymeric network which is similar to natural tissue. Due to the hydrophilic residues in the backbone of polymers, an immense amount of water molecules are retained within their structures (1). In addition, an extremely large surface area with good porosity has abundant interior space for biomolecules to be retained within the system through Coulombic attraction (2), while maintaining their biological activities. Moreover, the polymers or polymer monomers are easily dissolved in water before crosslinking, while after crosslinking, they are in a gel state with a defined shape. Due to their excellent properties, hydrogels have attracted much attention over the past years as the elaborate scaffolds in drug delivery carriers (3), tissue engineering, sensors (4) and cancer therapy (5). Hydrophilic polymer networks of hydrogels are formed through the crosslinking of monomers or polymer chains via covalent bonds and/or non-covalent interactions including hydrogen bonding, electrostatic interactions, host-guest complexation and their combinations (6–11). Plus, hydrogels can be made from a very large range of building blocks including polymers, peptides (12–15), and surfactants (16), with different types, degrees of cross-linking, and properties leading to the nanoscopic structures, size range, physical properties, and functions of hydrogel (17). Great attention has been paid to explore the strategy to control the functionalities of hydrogels. For example, many of the stimuli-responsive hydrogels have been constructed by using polymers modified with specific functional units that can rapidly respond to external stimuli. A variety of physical and chemical changes of the hydrogel, including volume change and sol-gel transition, are particularly sensitive to specific external stimuli due to their component materials (18–21). These stimuli-responsive hydrogels have gained immense consideration because of their potential in drug delivery systems (22–26), sensors (27–32), cancer therapy (33–37), cell culture substrates (38–40), and tissue engineering (38, 41–44). Beyond these stimulants, many specific biomolecules, such as antibodies, nucleic acids (or DNA), and enzymes, that can rapidly respond to target analytes are used for functional materials to modify polymers in order to construct target-responsive hydrogels (45, 46).
Aptamers are used to construct polymer networks as the stimuli-responsive element in aptamer-based hydrogels, and due to their unique characteristics they have gained great attention among the development of hydrogels which are responsive to specific target analytes (45, 47). Nucleic acid aptamers are single-stranded DNA(ssDNA) or RNA molecules, commonly containing 12–80 nucleotides (48, 49), generated by the systematic evolution of ligands by exponential enrichment (SELEX) (50) from a random ssDNA or RNA library (usually 1015~1016 different sequences) by means of three main steps including selection, separation, and amplification. Aptamer DNA hybridization and aptamer-target recognition both have very high specificity. Aptamers also possess high recognition ability toward specific molecular targets including ions (51, 52), small molecules (53, 54), proteins (55, 56), and cells (57, 58), because aptamers fold into a unique secondary or tertiary structure to bind to a target of interest, depending on van der Waals forces, hydrogen bonds, or electrostatic interactions (48, 59–62). Since they were discovered in the 1990s by Tuerk and Gold (63) and Ellington and Szostak (50), aptamers have become smart, specific, and high-affinity probes in bioanalytical, diagnostic, and therapeutic applications. Moreover, compared to other antibodies, aptamers are often called chemical antibodies due to their unique properties: (1) Aptamers are structurally stable with little immunogenicity and are chemically synthesized using standard solid state phosphoramidite reactions, which minimizes the batch-to-batch variation and improves the reproducibility of hydrogel systems. (2) aptamers are highly selective and have an affinity to targets, and low-dissociation constant values (Kds, 1 × 10−12-1 × 10−9 M) (64), so that aptamers can specifically recognize and undergo changes of their substrates even at very low concentrations. (3) The molecular weight of an aptamer is between 5 and 20 kDa, which is smaller than antibodies (ca.150 kDa), leading to better tumor uptake kinetics (6). (4) Aptamers are stable in a wide range of temperature, solvents, and pH. (5) Aptamers can be synthesized by chemical or enzymatic procedures or by a combination of these two methods without any animal-based synthesis. (6) Aptamers are easily modified with other functional moieties and have the capability of directional amplification by polymerase chain reaction (PCR). These excellent characters of aptamers make aptamer-based hydrogels even more versatile, and are excellent components in hydrogel engineering: (1) Aptamers can chemically conjugate with polymers such as acrydite and carboxymethylcellulose to construct the hydrogel (65, 66). (2) Aptamers can be integrated onto the surface of particles by chemical or physical methods and mixed with a pre-gel solution to form particle/hydrogel composites (67, 68). (3) Aptamers can recognize both target molecules and trigger complementary sequences (69–72). Especially to trap or introduce drugs (73–75), nanoparticles (76–78) into aptamer-based hydrogels have greatly expanded the applications in biosensing, target capture and release, cell adhesion and targeted therapy.
In this review, we focus on the latest strategies of aptamer-based hydrogels in bioanalytical and biomedical applications (Scheme 1). We begin this review with an overview of the underlying design principles for the construction of aptamer-based hydrogels. Next, we will discuss some bioanalytical and biomedical applications of aptamer-based hydrogels including biosensing, target capture and release, logic devices, gene and cancer therapy. Finally, recent progress of aptamer-based hydrogels is discussed along with challenges and future perspectives.
Scheme 1. Schematic illustration of aptamer—based hydrogels for bioanalytical and biomedical applications.
Design and Principle of Aptamer-Based Hydrogels
Aptamer as Cross-Linkers
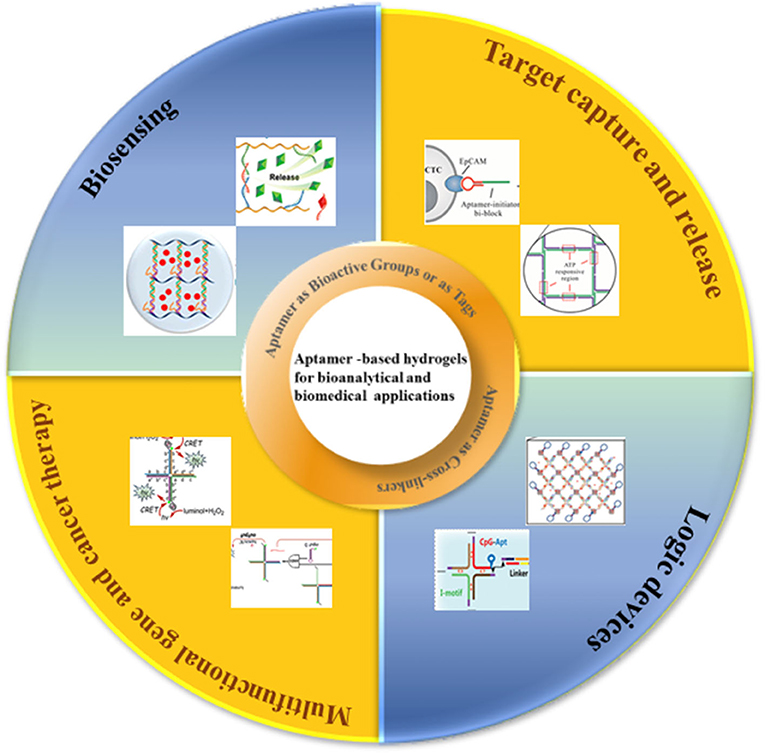
Aptamer-based hydrogels have been prepared based on different design principles. The selected design and preparation methods have a strong impact on the characteristic features of hydrogels and thus determine their respective biomedical applications. Using DNA aptamers as crosslinkers in hydrogels allows the hydrogels to be prepared to recognize the targets. In the absence of the target, the aptamer acts only as a conventional DNA crosslinker, but when the target is present, the aptamer preferentially forms a complex with the target and induces the change of the structure of the hydrogel. Such hydrogels utilize both the smart and programmable features of the DNA components as well as short aptamer sequences acting as supramolecular cross-linking agents (79–81). The first DNA-based polymer hydrogels were reported by Nagahara and Matsuda in which the short DNA sequences were grafted to a poly(acrylamide) polymer chain, and two pathways achieved gelation: (1) two DNA strands grafted to the polymer backbone were hybridized by other DNA sequences to induce the formation of gelation. (2) DNA strands attached to the polymer chain hybridized directly to form gelation without any external cross-linking agents (82). Based on this principle, a series of aptamer-based hydrogels have been prepared responding to target molecules (Figure 1Aa) (85, 86). Aptamers can also be used as crosslinkers in pure DNA hydrogels. A typical example is that a pure DNA hydrogel was constructed using a Y-shaped DNA and a thrombin aptamer linker through DNA self-assembly (Figure 1Ab) (87). The aptamers for ochratoxin A, ATP and adenine were used as DNA linkers to construct pure DNA hydrogels that were sensitive to targets (88–90).
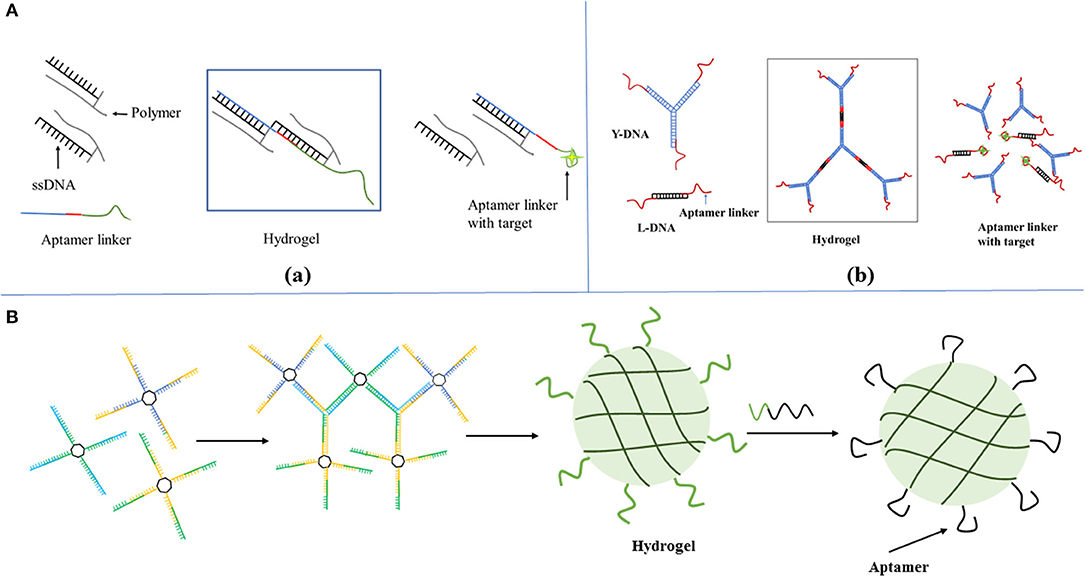
Figure 1. Schematic representation of the design strategy for formation and dissolution of aptamer-based hydrogel based on aptamers as (A) crosslinkers in (a) DNA functional polymer hydrogels [Reprinted with permission from Wang et al. (83). Copyright (2008) American Chemical Society] and (b) pure DNA hydrogels [Reprinted with permission from Previtera et al. (82). Copyright (2013) American Chemical Society]. (B) Bioactive groups or as tags for functionalization [Reprinted with permission from Lai et al. (84). Copyright (2019) American Chemical Society].
Aptamer as Bioactive Groups or as Tags for Functionalization
As we know, DNA segments as functional, bioactive elements rather than structural components were incorporated into hydrogels, and have also been explored for various bioanalytical and biomedical applications (84, 91–93). The presence of aptamer DNA as a bioactive group in these hydrogels typically does not change the mechanical properties and brings their high specificity toward a wide range of biological target molecules. Liu et al. (94) put forward a new strategy for fabricating a protein-scaffolded DNA nanohydrogel. By further incorporating therapeutic agents and tumor-targeting MUC1 aptamer, these SA-scaffolded DNA nanohydrogels can specifically target cancer cells and selectively release the preloaded therapeutic agents via a structure switching. A thrombin-binding aptamer was incorporated into the gel which can bind to adenosine, AMP, and ATP as shown in Figure 1B. DNA-functionalized gold nanoparticles or liposomes to DNA-functionalized hydrogels, when thrombin was added, a stable G-quadruplex structure emerged in the aptamer structure, which looked like a molecular switch between tight and relaxed states (95). X-shaped DNA, a DNA linker, and an aptamer were used to create a DNA hydrogel through the one-pot and the aptamer was only used as a functional unit for the target protein capture (96). A DNA nanohydrogel was developed to efficiently take up cells due to the recognition of an aptamer in the nanohydrogel (97). Moreover, an outstanding advantage of aptamer-functionalized hydrogels, as we know, would overcome the shortcomings of aptamers in bioanalytical and biomedical applications. Compared to antibodies, aptamers with high target-specific binding affinity values, were easily tailored for different targets. However, the drawback is their low cellular uptake for their high negative charge density and the limitation of stability in DNA degrading enzymes which are typically present in cells.
Aptamer-Functionalized hydrogels for bioanalytical and Biomedical applications
Aptamer-Based Hydrogels for Biosensing
Biosensors, as the powerful tools in monitoring biological or biochemical processes, have been applicated in various fields including medicine, disease diagnosis, food safety, and the environment (92). Nucleic acid aptamers are systematically engineered functional nucleic acids that demonstrate a very high affinity and specificity for targets including ions, metabolites, drugs, proteins, and even whole cells (98). Compared with antibodies, aptamers have been thought the ideal candidates as molecular recognition units to develop biosensors. Combined with DNA nanostructures possessing desirable advantages, aptamer-based biosensors hold great promise for the detection of a variety of targets. However, aptamers cannot freely penetrate the cell membrane and some nucleic acid probes are unstable in both intercellular and intracellular environments so that aptamer-based biosensors are usually compromised in intracellular environments (78, 99). Due to the protection of hydrogel nets, aptamer-based hydrogels have gained great attention in biosensing detection for their biocompatibility, chemical stability, and selective binding. Based on the designable conformational changes of aptamers, aptamer-based hydrogels can be combined with a variety of signaling mechanisms including fluorescence, electrochemistry, colorimetry, electroluminescence, and surface plasmon resonance, to construct rapid and sensitive biosensors to detect inorganic ions, organic small molecules, proteins, cells, and tissues. Usually, DNA-functionalized polymers have been simply crosslinked by the hybridization of aptamers with their complementary sequences to construct a hydrogel network structure. Because the binding affinities of aptamers to their target analytes are much stronger than that of simple hybridization, the network structure may deform or disintegrate when the specific target analytes are presented. The deformation of the aptamer-based hydrogel network can be easily detected with naked eyes or various colorimetric or fluorescence agents including, silver, gold nanoparticles, iodine, fluorescent dyes, and quantum dots (83, 100–107). The constructed hydrogel biosensor may achieve visual detection easily.
To detect biological molecules is vital for understanding their physiological and pathological functions. Since aptamers are easily modified and engineered, a large number of aptamer-based sensing hydrogel systems have been developed for the efficient detection of a wide range of biomolecules. Based on the use of DNA aptamers that cross-link with linear polyacrylamide chains, the first reported in this field was an aptamer-based hydrogel based on a gel-sol transition for detecting adenosine (85). In this design, when two oligonucleotide (DNA1, DNA2)-conjugated polyacrylamide chains (P1, P2) were mixed, a transparent and fluid state was obtained. Subsequently, upon the cross-linking of the oligonucleotides DNA3 was added, so that the above fluid system could undergo a sol-gel transition. The linker strand DNA3 contained three functional domains, that is, the complementary domains with DNA1, DNA2, and aptamer sequence domain. When target adenosine molecules were presented in system, aptamers competitively bound to adenosine molecules, leading to the breakdown of the hydrogel and target-responsive payload release. In order to achieve visual detection, gold nanoparticles were used as the indicator to add into the gel to monitor the process of gel-sol transition because of their unique optical properties. In the presence of adenosine, the upper buffer solution turned from colorless to red, indicating that the AuNPs had been released into the solution. The method was generally representative to use a target molecule as a trigger for the dissociation of the aptamer-based hydrogels to develop a biosensing system with high selectivity and visuality. In a similar work, a detection of food toxin, toxin A, was developed by applying this approach. The linear polyacrylamide polymers functionalized with short DNA strands were hybridized by OTA aptamer strands to construct the hydrogel network structure, and gold nanoparticles were still entrapped within the hydrogel using optical agent (108) (Figure 2A). An aptamer-functionalized DNA hydrogel was also prepared though DNA hybridization and incorporated inorganic nanomaterials including gold nanoparticles (AuNPs) and quantum dots (QDs) as signal indicators. The pure DNA hydrogel was directly constructed using Y-shaped DNA, linker DNA, and aptamer sequence with two different recognition sites for thrombin and the complementary sequence. Upon adding thrombin, it competitively bonded with aptamer, leading to the collapse and dissolution of the DNA hydrogel. The released negatively charged AuNPs would meet positively charged polyethyleneimine (PEI)-functionalized QDs and a fluorescence quenching strategy based on the Förster resonance energy transfer (FRET) was developed for the sensitive detection of thrombin in complex matrices (87). It is obvious that great progress has been made in this research area, however, regarding AuNPs, quantum dots etc. as visual indicators, there are several issues that need to be considered. For example, AuNPs exhibited an intense background color in the process of gel dissolution. As a result, this detecting method is not sensitive enough.
Figure 2. (A) Scheme of DNA-induced formation and adenosine-induced dissolution of hydrogel. Reprinted with permission from Wang et al. (83). Copyright (2008) American Chemical Society. (B) The scheme of a target-responsive hydrogel film in capillary tube for visual quantitative detection. [Reprinted with permission from Li et al. (110). Copyright (2019)]. (C) Preparation of aptamer functionalized hydrogels for the sensitive detection of α-fetoprotein using SERS method, Reprinted with permission from Guo et al. (103). Copyright (2020) American Chemical Society.
More recently, a thrombin-binding aptamer was incorporated into hydrogel. When thrombin was presented, a stable G-quadruplex structure in the aptamer structure emerged which changed the form of hydrogel by the molecular switch between tight and relaxed states (109). Li et al. (110) reported a gel film in a capillary tube based on the thermally reversible principle which transformed the analyte-induced small changes inside the DNA hydrogel into visual signals. In the analysis process, the permeability of the DNA hydrogel film will increase because of the small structural changes in the gel induced by the interaction between target molecules and the aptamer linkers, thereby changing the flow velocity of the sample solution in the capillary tube (Figure 2B). The duration time of the target solution flowing through the capillary tube with a specified length was used to characterize the concentration of different solutions. The ultra-trace aptamer DNA hydrogel (0.01 ml) detected cocaine directly with a low detection limitation (1.17 nM) and excellent selectivity as (Figure 1B). A novel SERS biosensing platform was constructed by combining the target-responsive DNA hydrogel for the sensitive detection of α-fetoprotein (AFP) (83). The aptamer as a linker strand in DNA hydrogel specifically recognized AFP and accurately controlled the release of immunoglobulin G (IgG) encapsulated in hydrogel. In the presence of AFP, the hydrogels were disentangled and the IgG was released. Interestingly, the released IgG was captured by SERS probes and bio-functional magnetic beads through the formation of sandwich-like structures to decrease the detecting signals, which significantly improved the detecting sensitivity (Figure 2C).
Furthermore, the exploration and introduction of new functional nanomaterials to produce aptamer-based hydrogel biosensing systems which are highly sensitive are also being developed. Aptamer-incorporated graphene oxide (GO) hydrogel without synthetic polymers was developed for the detection of antibiotics. GO hydrogels were readily prepared by physically mixing GO solution with adenosine. The fast gelation of the GO dispersion in the presence of adenosine would attribute to the strong hydrogen bonding and electrostatic interactions between the adenosine and the GO nanosheets. Aptamer chains flatly lay on the surfaces of GO sheets as a result of the strong π-π stacking interactions between the hexagonal cells of graphene and the ring structure of nucleobases in ssDNA, which had been elucidated as an effective driving force for assembling GO sheets into hydrogels (111, 112). Tan et al. (113) reported a fluorescence biosensor based on GO hydrogel incorporated with aptamers, which could selectively bind to tetracyclines. After GO hydrogels were formed, the fluorescence signal of fluorescence labeled aptamers was quenched for fluorescence resonance energy transfer (FRET). When hydrogel was exposed to the tetracycline, the fluorescence recovered. Using the quenching/recovering of fluorescence, this biosensor of GO hydrogel provided a quantitative analysis of tetracycline with high sensitivity at much lower concentration.
Aptamer-Based hydrogels for Target Capture and Release
Capture and Release of Circulating Tumor Cells (CTCs)
Circulating tumor cells (CTCs) are the collective term for the tumor cells that escape from the primary tumor sites and travel through the circulatory system into the peripheral blood stream, at which point then, the metastases can be ultimately formed in resident organs. Therefore, detection of CTCs at early stages of tumors will increase diagnostic accuracy and therapeutic efficacy. However, CTCs are a very small population. In general, there are <10 CTCs/mL whereas there are approximately 5 × 109 normal cells present in the same volume of blood sample (65, 114, 115). Therefore, a variety of materials have been recently investigated for sensitive catch and release of CTCs. Aptamer-based hydrogels as an emerging biomaterial, have recently attracted great attention in the fields of medical devices for cell catch and separation. For example, aptamer-based hydrogels were reported for in situ identification of live CTCs by cloaking/decloaking of CTCs (93). In this design as shown in Figure 3A, an aptamer DNA strand that specifically recognized epithelial cell adhesion molecule (EpCAM) on the CTCs surface triggered a hybridization chain reaction (HCR) via toehold-initiated branch migration. And an ATP aptamer was incorporated in the clamped HCR to decloak the DNA hydrogel on cell surface in order to achieve the phase transition from hydrogel to solution. The encapsulated AuNPs were exploited as the indicators of hydrogel formation via generating a red color at this state. Moreover, this method allowed to identify a low number of CTCs in whole blood by DNA hydrogel cloaking with high sensitivity and specificity for diagnosis. More significantly, controlled and defined chemical stimuli was used for the decloaking of CTCs without damages for subsequent culture and live cell analysis. Ye et al. (117) proposed an aptamer-trigger-clamped hybridization chain reaction (atcHCR) method for the capture of CTCs by porous 3D DNA hydrogels. The 3D environment of the DNA networks minimizes cell damage, and the CTCs can subsequently be released for live-cell analysis. In their work, initiator DNAs with aptamer-toehold biblocks specifically bind to the epithelial cell adhesion molecule (EpCAM) on the surface of CTCs triggering the atcHCR and the formation of a DNA hydrogel. The DNA hydrogel cloaks the CTCs, which would facilitate quantification with minimal cell damage. 10 MCF-7 cells in a 2-μl blood as sample were used to quantitively identify the decloaking of tumor cells via gentle chemical stimulus (ATP) which is used to release living tumor cells for subsequent cell culture and live-cell analysis. The whole experiment only was about 2.5 d including downstream cell culture and analysis. Aptamer-DNA hydrogels would open new powerful and effective routes for capturing rare live CTCs and their quantification in whole blood so that it can provide a new approach for cancer diagnostics and therapeutics. An aptamer-functionalized hydrogel was also reported that could catch CTCs with a density over 1,000 cells/mm2. When the hydrogel was coated by restriction endonucleases, the bound cells were released from the hydrogel coating because of the endonuclease-mediated sequence-specific hydrolysis of the aptamer sequences. The release efficiency reached 99%. Importantly, 98% of the released cells maintained viability (118). Polyvalent aptamer-functionalized hydrogel could also induce cell attachment on the hydrogel in dynamic flow. The cell density on the hydrogel was increased from 40 cells/mm2 to nearly 700 cells/mm2 when the shear stress was decreased from 0.05 to 0.005 Pa. After the attachment onto the hydrogel surface, approximately 95% of the cells could be triggered to detach within 20 min by using an oligonucleotide complementary sequence that displaced polyvalent aptamer strands from the hydrogel surface (119).
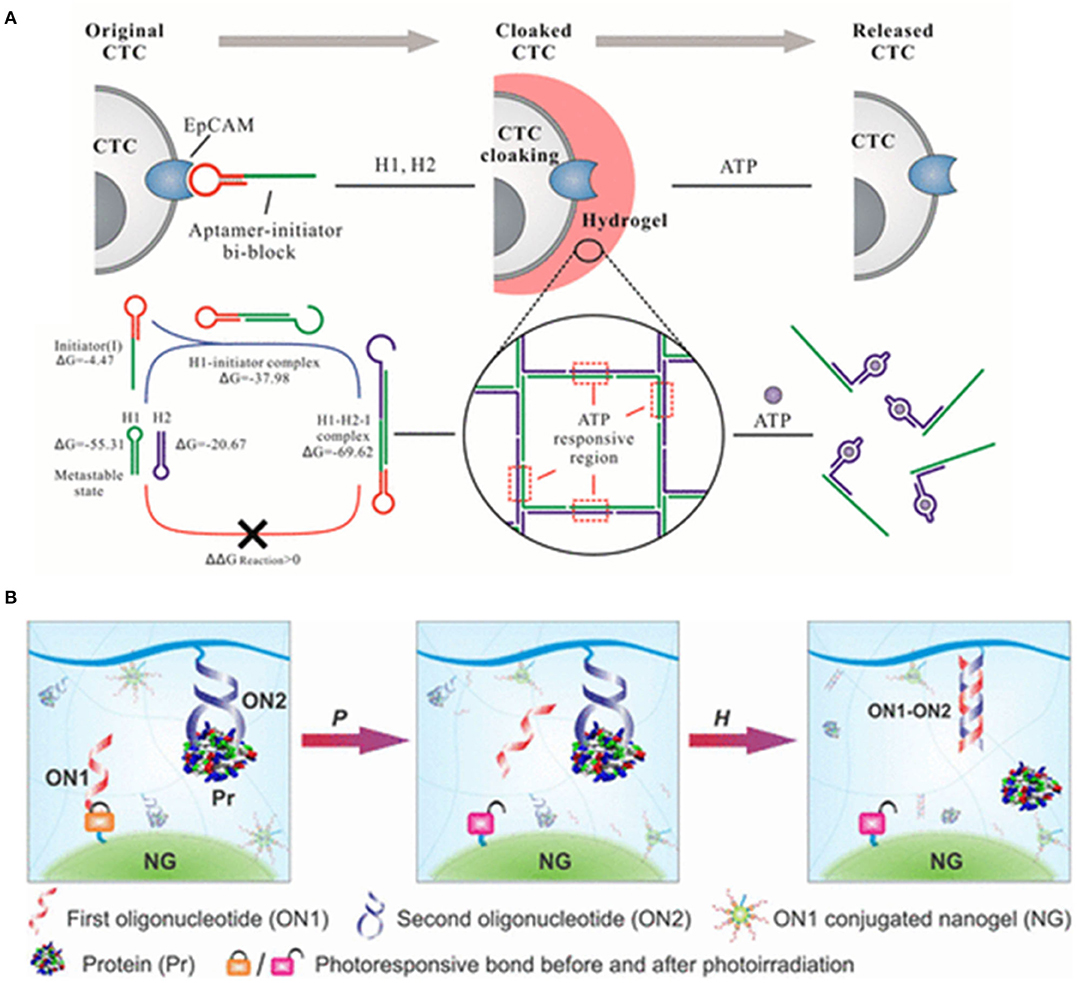
Figure 3. (A) DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis, reprinted with permission from Tan et al. (113). Copyright (2017) American Chemical Society. (B) Programmable self-assembly of protein-scaffolded dna nanohydrogels for tumor-targeted imaging and therapy, reprinted with permission from Pasparakis et al. (116). Copyright (2019) American Chemical Society.
Capture and Release of Protein
To develop the efficient systems for the protection and sustained release of encapsulated molecules would be beneficial in improving how we treat disease and study complex biochemical processes. Exogenous signaling molecules as biochemical cues promoted mesenchymal stem cells (MSCs) survival, presumably because MSCs themselves can release a variety of potent signaling molecules. Zhao et al. (120) examined whether the release of exogenous signaling molecules from hydrogels can promote the survival of MSC spheroids. They thought that aptamer-functionalized fibrin hydrogel (aFn) could release exogenous VEGF and PDGF-BB in a sustained manner. PDGF-BB-loaded aFn could double the survival rate of MSC spheroids in comparison with VEGF-loaded aFn during the 1-week test in vivo. Therefore, aptamer-based hydrogels have been considered as the new and promising materials which could be used for encapsulating a variety of biomacromolecules because they are responsive to environmental changes and multiple stimulus could trigger conformational or chemical changes of elastic network in hydrogels resulting in deswelling or degrading of hydrogels. Aptamer-based hydrogels have been utilized as smart systems with sensitivities toward various non-invasive stimuli. However, several factors, including the pore size of the polymer network, the diffusion rate of the entrapped target molecule, and the affinity between the aptamer and target molecule usually influence the functionality and efficiency of aptamer-based hydrogels as capture-release systems (85, 116, 121–123). Moreover, biochemical signals or biomarkers stimuli are subtle or presented at subnanomolar concentrations. Therefore, a sensitive signal trigger is usually necessary to control the release of preloaded effectors in aptamer-based hydrogels (124–127). Recently, Lai et al. (128) developed new responsive hydrogels for controlled protein release by multistep molecular recognition events. Two oligonucleotides were integrated into the system as pendant motifs. The first oligonucleotide was used to covalently construct a hydrogel nanoparticle via a photolabile linker; and the other aptamer which could form a protein-DNA complex, was covalently conjugated to the bulk hydrogel network. When the hydrogel system was exposed to an external light signal, the nanogel was activated and dissociated. Subsequently, the freed oligonucleotide would hybridize with aptamer strands to induce the dissociation of the protein-DNA complex to release the bound protein (Figure 3B).
Capture and Release of Pollutants
An aptamer-based hydrogel was developed for water remediation with both high selectivity and multiple adsorbing abilities for several pollutants. In water remediation techniques, the contradiction between selectivity and multiple adsorptions limited this approach for environmental crisis previously (129). Aptamers in hydrogel were used to accommodate the molecular structure of pollutants in the scavenger and afforded the perfect selectivity. Meanwhile, Janus nanoparticles with an antibacterial function, in which aptamers were on the anisotropic surfaces to handle different kinds of pollutants. The final hydrogel scavenger was prepared by entrapping aptamer-functionalized Janus nanoparticles into a porous cellulose hydrogel. An aptamer column for the removal of trace pharmaceuticals in drinking water was reported (130). 5′-Aminomodified DNA aptamer bound to CNBr-Sepharose as sorbent was packed into gel as a column to simultaneously test cocaine and diclofenac in drinking water. The removal of pharmaceuticals was as high as 88–95%. The aptamer column was reusable and achieved a high removal efficiency from 4°C to 30°C in normal cation ion concentrations (5–100 mg L−1) for multiple pollutants without cross effects and secondary pollution.
Aptamer-Based Hydrogels for Logic Devices
Nucleic acid molecules can be rationally designed, synthesized, and further integrated into Boolean operations, which provided an unprecedented potential to develop the basic components of molecular computing devices, because nucleic acid have high-capacity and low-maintenance digital information storage due to their predictable structures, high throughput synthesis and sequencing techniques (131, 132). Nucleic acid-based logic devices were first introduced in 1994 by Adleman and Lipton to solve the directed Hamiltonian path problem and the “SAT” question in computer science with single-stranded DNA sequences and enzymes (133, 134). Since then, science has seen the emergence of new logic systems for mimicking mathematical functions, diagnosing disease and even imitating biological systems (135–142). In recent years, logic gate systems based on aptamer-based hydrogels have attracted remarkable attention due to their intelligent responses to the external stimuli and convert input signals into a certain output signal.
Comparing this to silicon-based computation, although many challenges in designing computation devices, aptamer-based hydrogels logic circuits are still developing with great rapidity, due to their stability, biocompatibility, and predictable structure (143–146). Yin et al. (147) exploited a hydrogel structure based on hybridization behavior between crosslinker strands with aptamer sequences of ATP and cocaine molecules onto polymer chains. As detecting signals output, the BSA-modified gold nanoparticles were trapped in the hydrogel. The hydrogel served as an “AND” logic gate, when both cocaine and ATP presented, it was dissolved and led to the release of entrapped AuNPs And, the “OR” logic gate was reached if either cocaine or ATP presented, which led to the collapse of the hydrogel and release of the AuNPs. A novel colorimetric logic system based on an aptamer-crosslinked colloidal crystal hydrogel was also reported (148). When the Hg2+ and Ag+ responsive aptamers was incorporated into hydrogels, the reversible binding between the specific target ion (Hg2+ and Ag+) could induce the conformational change of the aptamers and thus make shrinkage of the hydrogels with different stimuli. The visualization of the logic output signals was realized, the aptamer-crosslinked hydrogel displayed a shrinking response and color change corresponding to a logical “OR” and “AND” gate when the stimuli of Hg2+ and Ag+ at a concentration of 0.1 μM was input.
Bi et al. (149) reported a DNA four-way junction (DNA-4WJ) which is target-catalytically formed through cascade assembly of four DNA hairpins on the basis of DNA TM-SDR. A concatenated logic circuit composed of one YES gate and three AND gates with an automatic reset function by using four DNA hairpins as inputs was fabricated and the formed DNA-4WJs serving as building units to construct DNA nanohydrogels (~120 nm). By incorporating aptamers, bioimaging agents, and drug loading sites into the building unit aptamer-based DNA nanohydrogels were synthesized with high loading capacity, target ability and good biocompatibility (Figure 4).
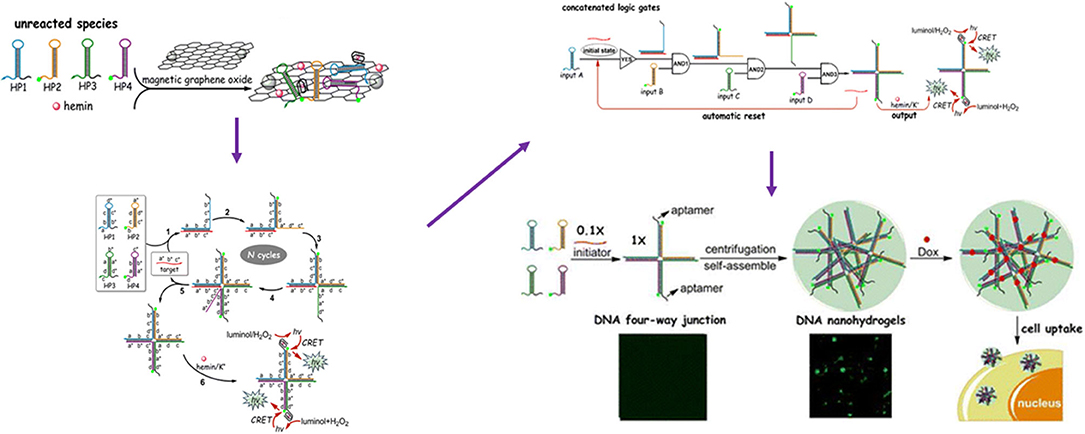
Figure 4. Schematic Illustration of the four-input concatenated logic gates based on target-catalyzed DNA four-way junctions. Reprinted with permission from Ramezani and Dietz (143). Copyright (2015) American Chemical Society.
For traditional silicon circuitry, logic devices of aptamer-based hydrogels showed more powerful functions in medical diagnosis (150, 151), in situ analysis (151), and artificial intelligence. However, this is still in its infancy. Most of the reported works are conceptional, with isolated logic functions and limited applications. The breakthrough and development of novel design and construction might promote the development of logic devices of aptamer-based hydrogels.
Aptamer-Based Hydrogels for Multifunctional Gene and Cancer Therapy
Gene therapy is a promising approach for the treatment of inherited diseases, such as cancers, hemophilia, and viral infections. It depends mainly on the research and development of the delivery vectors for gene. To achieve the safety and efficiency of gene delivery vectors, there remain many technical barriers to explore the potential of gene therapy. To date, gene therapy vectors mainly include viral vectors and non-viral vectors. Viral vectors are widely used for efficient gene transfer, but they are usually high-risk for immunogenicity and mutagenicity. In several clinical cases, their use has resulted in patient death. Compared with viral vectors, non-viral vectors are safer and more desirable. Therefore, the development of safe non-viral vectors is highly desirable (97, 149, 152, 153).
Recently, a variety of non-viral vectors, including liposomes (154), micelles (155), inorganic nanoparticles (156, 157), DNA nanostructures (73), and polymeric nano-hydrogel (158), have been explored as delivery vector for gene therapy. Among these, aptamer-based hydrogels are used as strong delivery vector candidates owing to their high payload capacity, as well as their biocompatibility, flexibility, and mechanical stability (78, 159, 160) as Table 1 shown. Tan (97) created a self-assembly process using three kinds of building units, Y-shaped monomer A with three sticky ends (YMA), Y-shaped monomer B with one sticky end (YMB), and DNA linker (LK) with two sticky ends, to hybridize a DNA nanohydrogel. By incorporating aptamers, disulfide linkages, and therapeutic genes into different building units, the aptamer-based DNA nanohydrogels (Y-gel-Apt) were formatted for targeted and stimuli-responsive gene therapy. And, a new intelligent DNA nano system integrating targeting, immunostimulant, and chemotherapy was also prepared based on unmethylated cytosine-phosphate-guanine oligonucleotides (CpG ODNs) DNA nanohydrogels (CpG-MUC1-hydrogel) (163). The cross-shaped DNAs (C-DNAs) assembled from pH-responsive I-motif sequences and targeted MUC1 aptamer-immunoadjuvant CpG-fused sequences (CpG-MUC1) were integrated into DNA nanohydrogels. DOX was successively intercalated into the base pairs of CpG-MUC1-hydrogel to form the CpG-MUC1-hydrogel/Dox that would controllably release DOX and CpGs at acidic conditions (Figure 5A). Moreover, a new class of physically cross-linked nanogel based on DNA, protein, and biotin as a nanocarrier using for the targeted cancer therapy was reported (162). The specific molecular recognition interaction between biotin and streptavidin was used to explore the cross-linking of a nanogel. The selective uptake of a doxorubicin-loaded nanogel by aptamer-receptor-positive cell lines (CCRF-CEM and HeLa) resulted in the specific interaction between the aptamer DNA decorated on the surface of the nanogel with the PTK7 receptor overexpressed on CCRF-CEM and HeLa cell lines (Figure 5B).
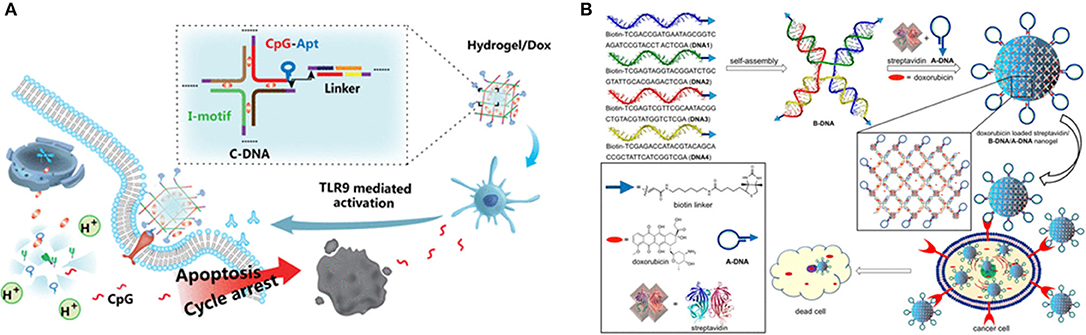
Figure 5. (A) Schematic diagram of the synthesis and action procedure of CpG-MUC1-hydrogel/Dox. Reprinted with permission from Ding et al. (168). Copyright (2019) American Chemical Society. (B) Doxorubicin-loaded nanogels using for delivery of doxorubicin. Reprinted with permission from Mazloumi Tabrizi et al. (154). Copyright (2019) American Chemical Society.
During clinical treatment, the side effects and accuracy of drug molecules in intravenous chemotherapy are the main topic of discussion for treatment. To design multifunctional therapeutic delivery nanoplatforms would overcome these limitations. A protein-scaffolded aptamer DNA nanohydrogel was fabricated by three types of streptavidin (SA)-based DNAtetrad accompanying with the further incorporation of therapeutic agents and tumor-targeting MUC1 aptamer. In an ATP-rich intracellular environment, this aptamer DNA nanohydrogel specifically targeted cancer cells and selectively released the preloaded therapeutic agents via a structure switching to attain the image and treatment of cancer cells (94). Furthermore, a novel class of physically cross-linked nanogels solely made of DNA, protein, and biotin were designed and the biotin–streptavidin molecular recognition interaction was used for the physical cross-linking of DNA nanostructures. Biotin-modified ssDNAs were assembled to form 5′-biotin-tethered X-shaped branched DNA acting as a tetravalent host and then streptavidin-modified aptamer DNAs interacted with them to form the aptamer-based hydrogels which allowed the loading of doxorubicin inside the gel network and delivered in the cancerous environment. The aptamer-functionalized and doxorubicin-loaded nanogels exhibited selective uptake into target cell lines (162). In the clinical treatment of tumors, the delivery of drugs or genes, nanogels nanocarriers need long term circulation in the blood, enhanced permeability and retention effect (EPR effect), enrichment, infiltration, uptake, and release of the drug or gene. Aptamer-based hydrogels as intelligent nanocarriers demonstrated excellent biocompatibility and high selectivity for target cancer cells (156, 169).
Conclusions and Perspectives
In summary, the recent progress in preparing aptamer-based hydrogels has made these kinds of materials accessible for encouraging applications in bioanalytical and biomedical fields. Aptamer DNA as the unique building blocks have prompted the development in sensitive biosensors, drug delivery systems, and cellular scaffolds for regenerative therapies. In this review, we divided aptamer-based hydrogels into two categories according to the gelation mechanism: aptamer as cross-linkers, bioactive groups or as tags for Functionalization. Various synthetic strategies and applications have been detailed. It is worth noting that the aptamer technology enables the design of DNA hydrogels that can detect almost any type of analyte with high selectivity and sensitivity. New aptamers are easy to generate which would lead to continue to growth of addressable targets. Despite the tremendous progress in the development of aptamer-based hydrogels, several challenges remained: (1) Aptamer-based hydrogels, as DNA hydrogels have low storage modulus and consequent thixotropic property, in which the strength is much lower than of most conventional polymers. It is crucial to regulate the mechanical properties for biological applications (78). (2) Mechanism studies on synthesis and responsiveness of aptamer-based hydrogels are needed to promote the development of hydrogels in biosensing, controlled release, and tissue engineering. Although the release profile of some stimuli-responsive hydrogels has been investigated, theoretically kinetic studies are rarely reported to reveal the release characteristics of aptamer-based hydrogels. Deeper kinetic studies will promote the design of aptamer-based hydrogels for biological applications. (3) There are still some problems in large scale applications of aptamer-based hydrogels because of their high cost and difficulty of preparation. To develop more techniques and novel synthetic methods to obtain more aptamer DNA should be mainly considered in order to reduce the cost. Furthermore, to explore new materials to hybrid with aptamer to construct multiple hydrogels is also a solution for cost concern.
Author Contributions
YD: conceptualization and writing-original draft preparation. PW: conceptualization. CL: investigation. QT and TW: investigation and writing-original draft preparation. YT and LG: supervision. SX: writing-reviewing and Editing. All authors read and approved the final manuscript.
Funding
This research was supported by the Key Research and Development Program of Qin Huang Dao (201602A110).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Ganji F, Vasheghani Farahani S, Vasheghani Faraahani E. Theoretical description of hydrogel swelling: a review. Iran Polym J. (2010) 19:375–98.
2. Li P, Poon YF, Li W, Zhu HY, Yeap SH, Cao Y, et al. A polycationic antimicrobial and biocompatible hydrogel with microbe membrane suctioning ability. Nat Mater. (2011) 10:149–56. doi: 10.1038/nmat2915
3. Annabi N, Tamayol A, Uquillas JA, Akbari M, Bertassoni LE, Cha C, et al. 25th anniversary article: rational design and applications of hydrogels in regenerative medicine. Adv Mater. (2014) 26:85–124. doi: 10.1002/adma.201303233
4. Liu S, Li L. Ultrastretchable and self-healing double-network hydrogel for 3d printing and strain sensor. ACS Appl Mater Interfaces. (2017) 9:26429–37. doi: 10.1021/acsami.7b07445
5. Willner I. Stimuli-controlled hydrogels and their applications. Acc Chem Res. (2017) 50:657–8. doi: 10.1021/acs.accounts.7b00142
6. Loebel C, Rodell CB, Chen MH, Burdick JA. Shear-thinning and self-healing hydrogels as injectable therapeutics and for 3D-printing. Nat Protoc. (2017) 12:1521–41. doi: 10.1038/nprot.2017.053
7. Liu X, Chen X, Chua MX, Li Z, Loh XJ, Wu Y-L. Cancer therapy: injectable supramolecular hydrogels as delivery agents of bcl-2 conversion gene for the effective shrinkage of therapeutic resistance tumors. Adv. Healthc Mater. (2017) 6. doi: 10.1002/adhm.201770053
8. Huang F, Scherman OA. Supramolecular polymers. Chem Soc Rev. (2012) 41:5879–80. doi: 10.1039/c2cs90071h
9. Voorhaar L, Diaz MM, Leroux F, Rogers S, Abakumov AM, Van Tendeloo G, et al. Supramolecular thermoplastics and thermoplastic elastomer materials with self-healing ability based on oligomeric charged triblock copolymers. NPG Asia Mate. (2017) 9:e385. doi: 10.1038/am.2017.63
10. Song P, Xu Z, Lu Y, Guo Q. Bio-Inspired hydrogen-bond cross-link strategy toward strong and tough polymeric materials. Macromolecules. (2015) 48:3957–64. doi: 10.1021/acs.macromol.5b00673
11. Zhang X, Qu W, Li D, Shi K, Li R, Han Y, et al. Functional polymer-based nerve guide conduits to promote peripheral nerve regeneration. Adv Mater Interfaces. (2020) 7:2000225. doi: 10.1002/admi.202000225
12. Leach DG, Dharmaraj N, Piotrowski SL, Lopez-Silva TL, Lei YL, Sikora AG, et al. STINGel: controlled release of a cyclic dinucleotide for enhanced cancer immunotherapy. Biomaterials. (2018) 163:67–75. doi: 10.1016/j.biomaterials.2018.01.035
13. Asai D, Kanamoto T, Takenaga M, Nakashima H. In situ depot formation of anti-HIV fusion-inhibitor peptide in recombinant protein polymer hydrogel. Acta Biomater. (2017) 64:116–25. doi: 10.1016/j.actbio.2017.10.024
14. Wu C, Liu J, Liu B, He S, Dai G, Xu B, et al. NIR light-responsive short peptide/2D NbSe2 nanosheets composite hydrogel with controlled-release capacity. J Mater Chem B. (2019) 7:3134–42. doi: 10.1039/C8TB03326A
15. Badeau BA, Comerford MP, Arakawa CK, Shadish JA, DeForest CA. Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery. Nat Chem. (2018) 10:251–8. doi: 10.1038/nchem.2917
16. Chu Z, Dreiss CA, Feng Y. Smart wormlike micelles. Chem Soc Rev. (2013) 42:7174–203. doi: 10.1039/c3cs35490c
17. Qiu H, Guo H, Li D, Hou Y, Kuang T, Ding J. Intravesical hydrogels as drug reservoirs. Trends Biotechnol. (2020) 38:579–83. doi: 10.1016/j.tibtech.2019.12.012
18. Klouda L. Thermoresponsive hydrogels in biomedical applications: a seven-year update. Eur J Pharm Biopharma. (2015) 97:338–49. doi: 10.1016/j.ejpb.2015.05.017
19. Pang X, Xie D, Ge X, Li Y, Zhen X, Yu X. Switchable sol-gel transition controlled by ultrasound and body temperature. Supramol Chem. (2016) 28:335–8. doi: 10.1080/10610278.2015.1102262
20. Xiong X, Wu C, Zhou C, Zhu G, Chen Z, Tan W. Responsive DNA-based hydrogels and their applications. Macromol Rapid Commun. (2013) 34:1271–83. doi: 10.1002/marc.201300411
21. Liu J. Oligonucleotide-functionalized hydrogels as stimuli responsive materials and biosensors. Soft Matter. (2011) 7:6757–67. doi: 10.1039/c1sm05284e
22. Deng G, Wong W-T, Huang M, Wu R, Lai W-F. Chapter 8—self-healing properties of hydrogels based on natural polymers. In: Chen Y, editor. Hydrogels Based on Natural Polymers. Hong Kong: Elsevier (2020). p. 223–45.
23. Cascone S, Lamberti G. Hydrogel-based commercial products for biomedical applications: a review. Int J Pharma. (2020) 573:118803. doi: 10.1016/j.ijpharm.2019.118803
24. Ornell KJ, Lozada D, Phan NV, Coburn JM. Controlling methacryloyl substitution of chondroitin sulfate: injectable hydrogels with tunable long-term drug release profiles. J Mater Chem B. (2019) 7:2151–61. doi: 10.1039/C8TB03020K
25. Pourreza N, Ghomi M. Hydrogel based aptasensor for thrombin sensing by Resonance Rayleigh Scattering. Anal Chim Acta. (2019) 1079:180–91. doi: 10.1016/j.aca.2019.06.049
26. Palmese LL, Thapa RK, Sullivan MO, Kiick KL. Hybrid hydrogels for biomedical applications. Curr Opin Chem Eng. (2019) 24:143–57. doi: 10.1016/j.coche.2019.02.010
27. Guo Z, Gu H, He Y, Zhang Y, Xu W, Zhang J, et al. Dual dynamic bonds enable biocompatible and tough hydrogels with fast self-recoverable, self-healable and injectable properties. Chem Eng J. (2020) 388:124282. doi: 10.1016/j.cej.2020.124282
28. Shao X, Feng Y, Zhu L, Zhang Y, Luo Y, Mei X, et al. Three dimensional DNA nanotracks: a novel method for ultrasensitive and visible mercury (II) detection. Sensors Actuat B: Chem. (2020) 303:126988. doi: 10.1016/j.snb.2019.126988
29. Liu X, Liu J, Lin S, Zhao X. Hydrogel machines. Mater Today. (2020) 36:102–24. doi: 10.1016/j.mattod.2019.12.026
30. Uygun M, Doganci E, Tasdelen MA, Gurek AG. One-pot photoinduced synthesis of dansyl containing acrylamide hydrogels and their chemosensing properties. J Appl Polym Sci. (2019) 136:47096. doi: 10.1002/app.47096
31. Torres L, Daristotle JL, Ayyub OB, Bellato Meinhardt BM, Garimella H, Margaronis A, et al. Structurally colored protease responsive nanoparticle hydrogels with degradation-directed assembly. Nanoscale. (2019) 11:17904–12. doi: 10.1039/C9NR04624K
32. Chen J, Xu L, Yang M, Chen X, Chen X, Hong W. Highly stretchable photonic crystal hydrogels for a sensitive mechanochromic sensor and direct ink writing. Chem Mater. (2019) 31:8918–26. doi: 10.1021/acs.chemmater.9b02961
33. Xu K, Lee F, Gao SJ, Chung JE, Yano H, Kurisawa M. Injectable hyaluronic acid-tyramine hydrogels incorporating interferon-α2a for liver cancer therapy. J Control Release. (2013)166:203–10. doi: 10.1016/j.jconrel.2013.01.008
34. Dosio F, Arpicco S, Stella B, Fattal E. Hyaluronic acid for anticancer drug and nucleic acid delivery. Adv Drug Deliv Rev. (2016) 97:204–36. doi: 10.1016/j.addr.2015.11.011
35. Tripodo G, Trapani A, Torre ML, Giammona G, Trapani G, Mandracchia D. Hyaluronic acid and its derivatives in drug delivery and imaging: recent advances and challenges. Eur J Pharm Biopharm. (2015) 97:400–16. doi: 10.1016/j.ejpb.2015.03.032
36. Weiden J, Tel J, Figdor CG. Synthetic immune niches for cancer immunotherapy. Nat Rev Immunol. (2018) 18:212–9. doi: 10.1038/nri.2017.89
37. Talebian S, Foroughi J, Wade SJ, Vine KL, Dolatshahi-Pirouz A, Mehrali M, et al. Biopolymers for antitumor implantable drug delivery systems: recent advances and future outlook. Adv Mater. (2018) 30:1706665. doi: 10.1002/adma.201706665
38. Schmidt S, Zeiser M, Hellweg T, Duschl C, Fery A, Möhwald H. Adhesion and mechanical properties of pnipam microgel films and their potential use as switchable cell culture substrates. Adv Funct Mater. (2010) 20:3235–43. doi: 10.1002/adfm.201000730
39. Xia Y, Wu H, Tang D, Gao S, Chen B, Zeng Z, et al. Graphene oxide nanosheet-composited poly(n-isopropylacrylamide) hydrogel for cell sheet recovery. Macromol Res. (2019) 27:679–85. doi: 10.1007/s13233-019-7099-z
40. Liu J, Shang J, Chen Y, Tian Y, Yang Q, Chen M, et al. A surface-engineered NIR light-responsive actuator for controllable modulation of collective cell migration. J Mater Chem B. (2019) 7:5528–34. doi: 10.1039/C9TB01038F
41. Afewerki S, Sheikhi A, Kannan S, Ahadian S, Khademhosseini A. Gelatin-polysaccharide composite scaffolds for 3D cell culture and tissue engineering: towards natural therapeutics. Bioeng Transl Med. (2019) 4:96–115. doi: 10.1002/btm2.10124
42. Hosseini V, Maroufi NF, Saghati S, Asadi N, Darabi M, Ahmad SNS, et al. Current progress in hepatic tissue regeneration by tissue engineering. J Transl Med. (2019) 17:383. doi: 10.1186/s12967-019-02137-6
43. Caballero Aguilar LM, Silva SM, Moulton SE. Growth factor delivery: defining the next generation platforms for tissue engineering. J Control Release. (2019) 306:40–58. doi: 10.1016/j.jconrel.2019.05.028
44. Zhu T, Cui Y, Zhang M, Zhao D, Liu G, Ding J. Engineered three-dimensional scaffolds for enhanced bone regeneration in osteonecrosis. Bioact Mater. (2020) 5:584–601. doi: 10.1016/j.bioactmat.2020.04.008
45. Zhu Z, Wu C, Liu H, Zou Y, Zhang X, Kang H, et al. An aptamer cross-linked hydrogel as a colorimetric platform for visual detection. Angew Chem Int Ed. (2010) 49:1052–6. doi: 10.1002/anie.200905570
46. Kim J, Singh N, Lyon LA. Label-free biosensing with hydrogel microlenses. Angew Chem Int Ed. (2006) 45:1446–9. doi: 10.1002/anie.200503102
47. Liu J, Liu H, Kang H, Donovan M, Zhu Z, Tan W. Aptamer-incorporated hydrogels for visual detection, controlled drug release, and targeted cancer therapy. Anal Bioanal Chem. (2012) 402:187–94. doi: 10.1007/s00216-011-5414-4
48. Ostrovidov S, Salehi S, Costantini M, Suthiwanich K, Ebrahimi M, Sadeghian RB, et al. 3D bioprinting in skeletal muscle tissue engineering. Small. (2019). 15:1805530. doi: 10.1002/smll.201805530
49. Liang K, Bae KH, Kurisawa M. Recent advances in the design of injectable hydrogels for stem cell-based therapy. J Mater Chem B. (2019) 7:3775–91. doi: 10.1039/C9TB00485H
50. Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. (1990) 346:818–22. doi: 10.1038/346818a0
51. Li M, Sato Y, Nishizawa S, Seino T, Nakamura K, Teramae N. 2-aminopurine-modified abasic-site-containing duplex dna for highly selective detection of theophylline. J Am Chem Soc. (2009). 131:2448–9. doi: 10.1021/ja8095625
52. Zheng D, Zou R, Lou X. Label-free fluorescent detection of ions, proteins, and small molecules using structure-switching aptamers, sybr gold, and exonuclease I. Anal Chem. (2012) 84:3554–60. doi: 10.1021/ac300690r
53. Shamsipur M, Fsrzin L, Amouzadeh Tabrizi M, Shanesaz M. CdTe amplification nanoplatforms capped with thioglycolic acid for electrochemical aptasensing of ultra-traces of ATP. Mater Sci Eng C. (2016) 69:1354–60. doi: 10.1016/j.msec.2016.08.038
54. Zhang S, Wang L, Liu M, Qiu Y, Wang M, Liu X, et al. A novel, label-free fluorescent aptasensor for cocaine detection based on a G-quadruplex and ruthenium polypyridyl complex molecular light switch. Anal Methods. (2016) 8:3740–6. doi: 10.1039/C6AY00231E
55. Amouzadeh Tabrizi M, Shamsipur M, Farzin L. A high sensitive electrochemical aptasensor for the determination of VEGF165 in serum of lung cancer patient. Biosens Bioelectron. (2015) 74:764–9. doi: 10.1016/j.bios.2015.07.032
56. Shamsipur M, Farzin L, Tabrizi MA. Ultrasensitive aptamer-based on-off assay for lysozyme using a glassy carbon electrode modified with gold nanoparticles and electrochemically reduced graphene oxide. Microchim Acta. (2016) 183:2733–43. doi: 10.1007/s00604-016-1920-6
57. Mir TA, Yoon J-H, Gurudatt NG, Won M-S, Shim Y-B. Ultrasensitive cytosensing based on an aptamer modified nanobiosensor with a bioconjugate: detection of human non-small-cell lung cancer cells. Biosens Bioelectron. (2015) 74:594–600. doi: 10.1016/j.bios.2015.07.012
58. Wang T, Liu J, gu X, Li D, Wang J, Wang E. Label-free electrochemical aptasensor constructed by layer-by-layer technology for sensitive and selective detection of cancer cells. Anal Chim Acta. (2015) 882:32–7. doi: 10.1016/j.aca.2015.05.008
59. Trachman RJ, Ferré-D'Amaré AR. Tracking RNA with light: selection, structure, and design of fluorescence turn-on RNA aptamers. Q Rev Biophy. (2019) 52:e8. doi: 10.1017/S0033583519000064
60. Munzar JD, Ng A, Juncker D. Duplexed aptamers: history, design, theory, and application to biosensing. Chem Soc Rev. (2019) 48:1390–419. doi: 10.1039/C8CS00880A
61. Samanta D, Ebrahimi SB, Mirkin CA. Nucleic-acid structures as intracellular probes for live cells. Adv Mater. (2019) 32:1901743. doi: 10.1002/adma.201901743
62. Agnew HD, Coppock MB, Idso MN, Lai BT, Liang J, McCarthy-Torrens AM, et al. Protein-catalyzed capture agents. Chem Rev. (2019) 119:9950–70. doi: 10.1021/acs.chemrev.8b00660
63. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. (1990) 249:505–10. doi: 10.1126/science.2200121
64. Cho EJ, Lee J-W, Ellington AD. Applications of aptamers as sensors. Annu Rev Anal Chem. (2009) 2:241–64. doi: 10.1146/annurev.anchem.1.031207.112851
65. Gačanin J, Synatschke CV, Weil T. Biomedical applications of DNA-based hydrogels. Adv Funct Mater. (2020) 30:1906253. doi: 10.1002/adfm.201906253
66. Battig MR, Huang Y, Chen N, Wang Y. Aptamer-functionalized superporous hydrogels for sequestration and release of growth factors regulated via molecular recognition. Biomaterials. (2014) 35:8040–8. doi: 10.1016/j.biomaterials.2014.06.001
67. Battig MR, Soontornworajit B, Wang Y. Programmable release of multiple protein drugs from aptamer-functionalized hydrogels via nucleic acid hybridization. J Am Chem Soc. (2012) 134:12410–3. doi: 10.1021/ja305238a
68. Bae KH, Wang L-S, Kurisawa M. Injectable biodegradable hydrogels: progress and challenges. J Mater Chem B. (2013) 1:5371–88. doi: 10.1039/c3tb20940g
69. Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, et al. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. (2002) 419:90–4. doi: 10.1038/nature00963
70. Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, et al. Antidote-mediated control of an anticoagulant aptamer in vivo. Nat Biotechnol. (2004) 22:1423–8. doi: 10.1038/nbt1023
71. Liu J, Lu Y. Non-base pairing dna provides a new dimension for controlling aptamer-linked nanoparticles and sensors. J Am Chem Soc. (2007) 129:8634–43. doi: 10.1021/ja072075+
72. Yang CJ, Jockusch S, Vicens M, Turro NJ, Tan W. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc Natl Acad Sci USA. (2005) 102:17278. doi: 10.1073/pnas.0508821102
73. Wu C, Han D, Chen T, Peng L, Zhu G, You M, et al. Building a multifunctional aptamer-based dna nanoassembly for targeted cancer therapy. J Am Chem Soc. (2013) 135:18644–50. doi: 10.1021/ja4094617
74. Xiang D, Shigdar S, Qiao G, Wang T, Kouzani AZ, Zhou S-F, et al. Nucleic acid aptamer-guided cancer therapeutics and diagnostics: the next generation of cancer medicine. Theranostics. (2015) 5:23–42. doi: 10.7150/thno.10202
75. Hu Q, Li H, Wang L, Gu H, Fan C. DNA nanotechnology-enabled drug delivery systems. Chem Rev. (2019) 119:6459–506. doi: 10.1021/acs.chemrev.7b00663
76. Liang H, Zhang X-B, Lv Y, Gong L, Wang R, Zhu X, et al. Functional DNA-containing nanomaterials: cellular applications in biosensing, imaging, and targeted therapy. Acc Chem Res. (2014) 47:1891–901. doi: 10.1021/ar500078f
77. Li F, Lyu D, Liu S, Guo W. DNA Hydrogels and microgels for biosensing and biomedical applications. Adv Mater. (2020) 32:1806538. doi: 10.1002/adma.201806538
78. Zhou L, Jiao X, Liu S, Hao M, Cheng S, Zhang P, et al. Functional DNA-based hydrogel intelligent materials for biomedical applications. J Mater Chem B. (2020) 8:1991–2009. doi: 10.1039/C9TB02716E
79. Li C, Faulkner-Jones A, Dun AR, Jin J, Chen P, Xing Y, et al. Rapid Formation of a supramolecular polypeptide–dna hydrogel for in situ three-dimensional multilayer bioprinting. Angew Chem Int Ed. (2015) 54:3957–61. doi: 10.1002/anie.201411383
80. Kahn JS, Hu Y, Willner I. Stimuli-responsive dna-based hydrogels: from basic principles to applications. Acc Chem Res. (2017) 50:680–90. doi: 10.1021/acs.accounts.6b00542
81. Lu C-H, Guo W, Hu Y, Qi X-J, Willner I. Multitriggered shape-memory acrylamide–DNA hydrogels. J Am Chem Soc. (2015) 137:15723–31. doi: 10.1021/jacs.5b06510
82. Previtera ML, Chippada U, Schloss RS, Yurke B, Langrana NA. Mechanical properties of DNA-crosslinked polyacrylamide hydrogels with increasing crosslinker density. Biores Open Access. (2012) 1:256–9. doi: 10.1089/biores.2012.9906
83. Wang Q, Hu Y, Jiang N, Wang J, Yu M, Zhuang X. Preparation of aptamer responsive DNA functionalized hydrogels for the sensitive detection of α-fetoprotein using SERS method. Bioconjug Chem. (2020) 31:813–20. doi: 10.1021/acs.bioconjchem.9b00874
84. Lai J, Li S, Shi X, Coyne J, Zhao N, Dong F, et al. Displacement and hybridization reactions in aptamer-functionalized hydrogels for biomimetic protein release and signal transduction. Chem Sci. (2017) 8:7306–11. doi: 10.1039/C7SC03023A
85. Yang H, Liu H, Kang H, Tan W. Engineering target-responsive hydrogels based on aptamer–target interactions. J Am Chem Soc. (2008) 130:6320–1. doi: 10.1021/ja801339w
86. Zhou L, Chen C, Ren J, Qu X. Towards intelligent bioreactor systems: triggering the release and mixing of compounds based on DNA-functionalized hybrid hydrogel. Chem Commun. (2014) 50:10255–7. doi: 10.1039/C4CC04791E
87. Zhang L, Lei J, Liu L, Li C, Ju H. Self-assembled DNA hydrogel as switchable material for aptamer-based fluorescent detection of protein. Anal Chem. (2013) 85:11077–82. doi: 10.1021/ac4027725
88. Zhou L, Sun N, Xu L, Chen X, Cheng H, Wang J, et al. Dual signal amplification by an “on-command” pure DNA hydrogel encapsulating HRP for colorimetric detection of ochratoxin A. RSC Adv. (2016) 6:114500–4. doi: 10.1039/C6RA23462C
89. Muckley ES, Naguib M, Wang H-W, Vlcek L, Osti NC, Sacci RL, et al. Multimodality of structural, electrical, and gravimetric responses of intercalated mxenes to water. ACS Nano. (2017) 11:11118–26. doi: 10.1021/acsnano.7b05264
90. Simon AJ, Walls-Smith LT, Plaxco KW. Exploiting the conformational-selection mechanism to control the response kinetics of a “smart” DNA hydrogel. Analyst. (2018) 143:2531–8. doi: 10.1039/C8AN00337H
91. Wang X, Wang X. Aptamer-functionalized hydrogel diffraction gratings for the human thrombin detection. Chem Commun. (2013) 49:5957–9. doi: 10.1039/c3cc41827h
92. Soontornworajit B, Zhou J, Zhang Z, Wang Y. Aptamer-functionalized in situ injectable hydrogel for controlled protein release. Biomacromolecules. (2010) 11:2724–30. doi: 10.1021/bm100774t
93. Song P, Ye D, Zuo X, Li J, Wang J, Liu H, et al. DNA hydrogel with aptamer-toehold-based recognition, cloaking, and decloaking of circulating tumor cells for live cell analysis. Nano Lett. (2017) 17:5193–8. doi: 10.1021/acs.nanolett.7b01006
94. Li N, Wang X-Y, Xiang M-H, Liu J-W, Yu R-Q, Jiang J-H. Programmable self-assembly of protein-scaffolded dna nanohydrogels for tumor-targeted imaging and therapy. Anal Chem. (2019) 91:2610–4. doi: 10.1021/acs.analchem.8b05706
95. El-Hamed F, Dave N, Liu J. Stimuli-responsive releasing of gold nanoparticles and liposomes from aptamer-functionalized hydrogels. Nanotechnology. (2011) 22:494011. doi: 10.1088/0957-4484/22/49/494011
96. Liu C, Han J, Pei Y, Du J. Aptamer functionalized dna hydrogel for wise-stage controlled protein release. Appl Sci. (2018) 8:1941. doi: 10.3390/app8101941
97. Li J, Zheng C, Cansiz S, Wu C, Xu J, Cui C, et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J Am Chem Soc. (2015) 137:1412–5. doi: 10.1021/ja512293f
98. Srinivas RL, Chapin SC, Doyle PS. Aptamer-functionalized microgel particles for protein detection. Anal Chem. (2011) 83:9138–45. doi: 10.1021/ac202335u
99. Meng HM, Liu H, Kuai H, Peng R, Mo L, Zhang XB. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem Soc Rev. (2016) 45:2583–602. doi: 10.1039/C5CS00645G
100. Li J, Mo L, Lu C-H, Fu T, Yang H-H, Tan W. Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications. Chem Soc Rev. (2016) 45:1410–31. doi: 10.1039/C5CS00586H
101. Baruah U, Chowdhury D. Functionalized graphene oxide quantum dot–PVA hydrogel: a colorimetric sensor for Fe2+, Co2+and Cu2+ions. Nanotechnology. (2016) 27:145501. doi: 10.1088/0957-4484/27/14/145501
102. Tian T, Wei X, Jia S, Zhang R, Li J, Zhu Z, et al. Integration of target responsive hydrogel with cascaded enzymatic reactions and microfluidic paper-based analytic devices (μPADs) for point-of-care testing (POCT). Biosens Bioelectron. (2016) 77:537–42. doi: 10.1016/j.bios.2015.09.049
103. Guo W, Orbach R, Mironi-Harpaz I, Seliktar D, Willner I. Fluorescent DNA hydrogels composed of nucleic acid-stabilized silver nanoclusters. Small. (2013) 9:3748–52. doi: 10.1002/smll.201300055
104. Wang J, Wang X, Tang H, He S, Gao Z, Niu R, et al. A ratiometric magnesium sensor using DNAzyme-templated CdTe quantum dots and Cy5. Sensors Actuat B: Chem. (2018) 272:146–50. doi: 10.1016/j.snb.2018.05.162
105. Wang J, Gao Z, He S, Jin P, Ma D, Gao Y, et al. A universal growth strategy of DNA-programmed quantum dots on graphene oxide surface. Nanotechnology. (2020) 31:24LT02. doi: 10.1088/1361-6528/ab7c42
106. Wang J, Zhai J, Han S. Non-injection one-pot preparation strategy for multiple families of magic-sized CdTe quantum dots with bright bandgap photoemission. Chem Eng J. (2013) 215–216:23–8. doi: 10.1016/j.cej.2012.10.092
107. Zheng Y, Wang X, He S, Gao Z, Di Y, Lu K, et al. Aptamer-DNA concatamer-quantum dots based electrochemical biosensing strategy for green and ultrasensitive detection of tumor cells via mercury-free anodic stripping voltammetry. Biosens Bioelectron. (2019) 126:261–8. doi: 10.1016/j.bios.2018.09.076
108. Liu R, Huang Y, Ma Y, Jia S, Gao M, Li J, et al. Design and synthesis of target-responsive aptamer-cross-linked hydrogel for visual quantitative detection of ochratoxin A. ACS Appl Mater Interfaces. (2015) 7:6982–90. doi: 10.1021/acsami.5b01120
109. Deshpande SR, Hammink R, Nelissen FHT, Rowan AE, Heus HA. Biomimetic stress sensitive hydrogel controlled by DNA nanoswitches. Biomacromolecules. (2017) 18:3310–7. doi: 10.1021/acs.biomac.7b00964
110. Li Y, Ma Y, Jiao X, Li T, Lv Z, Yang CJ, et al. Control of capillary behavior through target-responsive hydrogel permeability alteration for sensitive visual quantitative detection. Nat Commun. (2019) 10:1036. doi: 10.1038/s41467-019-08952-1
111. Xu Y, Wu Q, Sun Y, Bai H, Shi G. Three-dimensional self-assembly of graphene oxide and DNA into multifunctional hydrogels. ACS Nano. (2010) 4:7358–62. doi: 10.1021/nn1027104
112. Zhu Y, Cai Y, Xu L, Zheng L, Wang L, Qi B, et al. Building an aptamer/graphene oxide fret biosensor for one-step detection of bisphenol A. ACS Appl Mater Interfaces. (2015) 7:7492–6. doi: 10.1021/acsami.5b00199
113. Tan B, Zhao H, Du L, Gan X, Quan X. A versatile fluorescent biosensor based on target-responsive graphene oxide hydrogel for antibiotic detection. Biosens Bioelectron. (2016) 83:267–73. doi: 10.1016/j.bios.2016.04.065
114. De Mattos-Arruda L, Tabernero J, Seoane J, Cortes J. Circulating tumour cells in early breast cancer. Lancet Oncol. (2012) 13:e370. doi: 10.1016/S1470-2045(12)70316-9
115. Ji X, Wang J, Niu S, Ding C. Size-controlled DNA-cross-linked hydrogel coated silica nanoparticles served as a ratiometric fluorescent probe for the detection of adenosine triphosphate in living cells. Chem Commun. (2019) 55:5243–6. doi: 10.1039/C9CC01832H
116. Pasparakis G, Manouras T, Vamvakaki M, Argitis P. Harnessing photochemical internalization with dual degradable nanoparticles for combinatorial photo–chemotherapy. Nat Commun. (2014) 5:3623. doi: 10.1038/ncomms4623
117. Ye D, Li M, Zhai T, Song P, Song L, Wang H, et al. Encapsulation and release of living tumor cells using hydrogels with the hybridization chain reaction. Nat Protoc. (2020) 15:2163–85. doi: 10.1038/s41596-020-0326-4
118. Li S, Chen N, Zhang Z, Wang Y. Endonuclease-responsive aptamer-functionalized hydrogel coating for sequential catch and release of cancer cells. Biomaterials. (2013) 34:460–9. doi: 10.1016/j.biomaterials.2012.09.040
119. Gaddes ER, Gydush G, Li S, Chen N, Dong C, Wang Y. Aptamer-based polyvalent ligands for regulated cell attachment on the hydrogel surface. Biomacromolecules. (2015) 16:1382–9. doi: 10.1021/acs.biomac.5b00165
120. Zhao N, Coyne J, Abune L, Shi P, Lian XL, Zhang G, et al. Exogenous signaling molecules released from aptamer-functionalized hydrogels promote the survival of mesenchymal stem cell spheroids. ACS Appl Mater Interfaces. (2020) 12:24599–610. doi: 10.1021/acsami.0c05681
121. Tomatsu I, Peng K, Kros A. Photoresponsive hydrogels for biomedical applications. Adv Drug Deliv Rev. (2011) 63:1257–66. doi: 10.1016/j.addr.2011.06.009
122. Fomina N, Sankaranarayanan J, Almutairi A. Photochemical mechanisms of light-triggered release from nanocarriers. Adv Drug Deliv Rev. (2012) 64:1005–20. doi: 10.1016/j.addr.2012.02.006
123. Yan B, Boyer J-C, Habault D, Branda NR, Zhao Y. Near infrared light triggered release of biomacromolecules from hydrogels loaded with upconversion nanoparticles. J Am Chem Soc. (2012) 134:16558–61. doi: 10.1021/ja308876j
124. Kang H, Trondoli AC, Zhu G, Chen Y, Chang Y-J, Liu H, et al. Near-infrared light-responsive core–shell nanogels for targeted drug delivery. ACS Nano. (2011) 5:5094–9. doi: 10.1021/nn201171r
125. Vermonden T, Censi R, Hennink WE. Hydrogels for protein delivery. Chem Rev. (2012) 112:2853–88. doi: 10.1021/cr200157d
126. Hu Y, Guo W, Kahn JS, Aleman-Garcia MA, Willner I. A shape-memory DNA-based hydrogel exhibiting two internal memories. Angew Chem Int Ed. (2016) 55:4210–4. doi: 10.1002/anie.201511201
127. Huang F, Liao W-C, Sohn YS, Nechushtai R, Lu C-H, Willner I. Light-responsive and pH-responsive DNA microcapsules for controlled release of loads. J Am Chem Soc. (2016) 138:8936–45. doi: 10.1021/jacs.6b04773
128. Lai J, Jiang P, Gaddes ER, Zhao N, Abune L, Wang Y. Aptamer-functionalized hydrogel for self-programmed protein release via sequential photoreaction and hybridization. Chem Mater. (2017) 29:5850–7. doi: 10.1021/acs.chemmater.7b00875
129. Wang J, Shen H, Hu X, Li Y, Li Z, Xu J, et al. A targeted “capture” and “removal” scavenger toward multiple pollutants for water remediation based on molecular recognition. Adv Sci. (2016) 3:1500289. doi: 10.1002/advs.201500289
130. Hu X, Mu L, Zhou Q, Wen J, Pawliszyn J. ssDNA aptamer-based column for simultaneous removal of nanogram per liter level of illicit and analgesic pharmaceuticals in drinking water. Environ Sci Technol. (2011) 45:4890–5. doi: 10.1021/es104241z
131. Church GM, Gao Y, Kosuri S. Next-generation digital information storage in DNA. Science. (2012) 337:1628. doi: 10.1126/science.1226355
132. Goldman N, Bertone P, Chen S, Dessimoz C, LeProust EM, Sipos B, et al. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. (2013) 494:77–80. doi: 10.1038/nature11875
133. Adleman LM. Molecular computation of solutions to combinatorial proteins. Science. (1994) 266:1021. doi: 10.1126/science.7973651
134. Lipton RJ. DNA solution of hard computational problems. Science. (1995) 268:542. doi: 10.1126/science.7725098
135. Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nat Biotechnol. (2003) 21:1069–74. doi: 10.1038/nbt862
136. Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. (2006) 314:1585. doi: 10.1126/science.1132493
137. Zhang DY, Turberfield AJ, Yurke B, Winfree E. Engineering entropy-driven reactions and networks catalyzed by DNA. Science. (2007) 318:1121. doi: 10.1126/science.1148532
138. Qian L, Winfree E. Scaling up digital circuit computation with DNA strand displacement cascades. Science. (2011) 332:1196. doi: 10.1126/science.1200520
139. Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. (2011) 475:368–72. doi: 10.1038/nature10262
140. Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. (2011) 333:1307. doi: 10.1126/science.1205527
141. Douglas SM, Bachelet I, Church GM. A Logic-gated nanorobot for targeted transport of molecular payloads. Science. (2012) 335:831. doi: 10.1126/science.1214081
142. Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. (2013) 31:448–52. doi: 10.1038/nbt.2510
143. Ramezani H, Dietz H. Building machines with DNA molecules. Nat Rev Genet. (2020) 21:5–26. doi: 10.1038/s41576-019-0175-6
144. Harroun SG, Prévost-Tremblay C, Lauzon D, Desrosiers A, Wang X, Pedro L, et al. Programmable DNA switches and their applications. Nanoscale. (2018) 10:4607–41. doi: 10.1039/C7NR07348H
145. Morihiro K, Ankenbruck N, Lukasak B, Deiters A. Small molecule release and activation through dna computing. J Am Chem Soc. (2017) 139:13909–15. doi: 10.1021/jacs.7b07831
146. Yang S, Yang C, Huang D, Song L, Chen J, Yang Q. Recent progress in fluorescence signal design for dna-based logic circuits. Chem Eur J. (2019) 25:5389–405. doi: 10.1002/chem.201804420
147. Yin B-C, Ye B-C, Wang H, Zhu Z, Tan W. Colorimetric logic gates based on aptamer-crosslinked hydrogels. Chem Commun. (2012) 48:1248–50. doi: 10.1039/C1CC15639J
148. Ye B, Wang H, Ding H, Zhao Y, Pu Y, Gu Z. Colorimetric logic response based on aptamer functionalized colloidal crystal hydrogels. Nanoscale. (2015) 7:7565–8. doi: 10.1039/C5NR00586H
149. Bi S, Xiu B, Ye J, Dong Y. Target-catalyzed dna four-way junctions for cret imaging of microrna, concatenated logic operations, and self-assembly of dna nanohydrogels for targeted drug delivery. ACS Appl Mater Interfaces. (2015) 7:23310–9. doi: 10.1021/acsami.5b07827
150. Liu Y-L, Wu H-P, Zhou Q, Song Q-X, Rui J-Z, Guan X-X, et al. Controllable extension of hairpin-structured flaps to allow low-background cascade invasive reaction for a sensitive DNA logic sensor for mutation detection. Chem Sci. (2018) 9:1666–73. doi: 10.1039/C7SC04210H
151. Guo S, Huang H, Deng X, Chen Y, Jiang Z, Xie M, et al. Programmable DNA-responsive microchip for the capture and release of circulating tumor cells by nucleic acid hybridization. Nano Res. (2018) 11:2592–604. doi: 10.1007/s12274-017-1885-8
152. Li Y, Ding J, Xu X, Shi R, Saw PE, Wang J, et al. Dual hypoxia-targeting rnai nanomedicine for precision cancer therapy. Nano Lett. (2020) 20:4857–63. doi: 10.1021/acs.nanolett.0c00757
153. Wang J, Li Z, Wang Z, Yu Y, Li D, Li B, et al. Nanomaterials for combinational radio–immuno oncotherapy. Adv Funct Mater. (2020) 30:10676. doi: 10.1002/adfm.201910676
155. Chen T, Wu CS, Jimenez E, Zhu Z, Dajac JG, You M, et al. DNA micelle flares for intracellular mRNA imaging and gene therapy. Angew Chem Int Ed. (2013) 52:2012–6. doi: 10.1002/anie.201209440
156. Sun Q, Sun X, Ma X, Zhou Z, Jin E, Zhang B, et al. Integration of nanoassembly functions for an effective delivery cascade for cancer drugs. Adv Mater. (2014) 26:7615–21. doi: 10.1002/adma.201401554
157. Antonelli A, Sfara C, Manuali E, Bruce IJ, Magnani M. Encapsulation of superparamagnetic nanoparticles into red blood cells as new carriers of MRI contrast agents. Nanomedicine. (2011) 6:211–23. doi: 10.2217/nnm.10.163
158. Dunn SS, Tian S, Blake S, Wang J, Galloway AL, Murphy A, et al. Reductively responsive siRNA-conjugated hydrogel nanoparticles for gene silencing. J Am Chem Soc. (2012) 134:7423–30. doi: 10.1021/ja300174v
159. Gruber A, Navarro L, Klinger D. Reactive precursor particles as synthetic platform for the generation of functional nanoparticles, nanogels, and microgels. Adv Mater Interfaces. (2020) 7:1901676. doi: 10.1002/admi.201901676
160. Stickdorn J, Nuhn L. Reactive-ester derived polymer nanogels for cancer immunotherapy. Eur Polym J. (2020) 124:109481. doi: 10.1016/j.eurpolymj.2020.109481
161. Li F, Tang J, Geng J, Luo D, Yang D. Polymeric DNA hydrogel: design, synthesis and applications. Prog Polym Sci. (2019) 98:101163. doi: 10.1016/j.progpolymsci.2019.101163
162. Thelu HVP, Atchimnaidu S, Perumal D, Harikrishnan KS, Vijayan S, Varghese R. Self-assembly of an aptamer-decorated, dna–protein hybrid nanogel: a biocompatible nanocarrier for targeted cancer therapy. ACS Appl Bio Mater. (2019) 2:5227–34. doi: 10.1021/acsabm.9b00323
163. Wei H, Zhao Z, Wang Y, Zou J, Lin Q, Duan Y. One-Step self-assembly of multifunctional DNA nanohydrogels: an enhanced and harmless strategy for guiding combined antitumor therapy. ACS Appl Mater Interfaces. (2019) 11:46479–89. doi: 10.1021/acsami.9b15874
164. Zhang L, Abdullah R, Hu X, Bai H, Fan H, He L, et al. Engineering of bioinspired, size-controllable, self-degradable cancer-targeting dna nanoflowers via the incorporation of an artificial sandwich base. J Am Chem Soc. (2019) 141:4282–90. doi: 10.1021/jacs.8b10795
165. Wang Z, Xia J, Cai F, Zhang F, Yang M, Bi S, et al. Aptamer-functionalized hydrogel as effective anti-cancer drugs delivery agents. Colloids Surf B: Biointerfaces. (2015) 134:40–6. doi: 10.1016/j.colsurfb.2015.06.031
166. Zhang Z, Han J, Pei Y, Fan R, Du J. Chaperone copolymer-assisted aptamer-patterned DNA hydrogels for triggering spatiotemporal release of protein. ACS Appl Bio Mater. (2018) 1:1206–14. doi: 10.1021/acsabm.8b00450
167. Zhao M, Zhang Y, Yuan S, Xu X, Wu Z, Wu Z, et al. ATP responsive DNA nanogels grown on biocompatible branches for anticancer drug delivery. Soft Matter. (2019) 15:3655–8. doi: 10.1039/C9SM00480G
168. Ding L, Li J, Wu C, Yan F, Li X, Zhang S. A self-assembled RNA-triple helix hydrogel drug delivery system targeting triple-negative breast cancer. J Mater Chem B. (2020) 8:3527–33. doi: 10.1039/C9TB01610D
Keywords: aptamer-based hydrogels, aptamer, hydrogels, bioanalysis, biomedicine
Citation: Di Y, Wang P, Li C, Xu S, Tian Q, Wu T, Tian Y and Gao L (2020) Design, Bioanalytical, and Biomedical Applications of Aptamer-Based Hydrogels. Front. Med. 7:456. doi: 10.3389/fmed.2020.00456
Received: 12 June 2020; Accepted: 09 July 2020;
Published: 22 October 2020.
Edited by:
Fu Wang, Xidian University, ChinaReviewed by:
Jianxun Ding, Changchun Institute of Applied Chemistry (CAS), ChinaThean Hock Tang, Universiti Sains Malaysia (USM), Malaysia
Copyright © 2020 Di, Wang, Li, Xu, Tian, Wu, Tian and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shufeng Xu, c2h1ZmVuZ3h1MTk2NUBzaW5hLmNvbQ==
 Ya Di1
Ya Di1 Shufeng Xu
Shufeng Xu