
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 15 December 2015
Sec. Pathology
Volume 2 - 2015 | https://doi.org/10.3389/fmed.2015.00085
This article is part of the Research Topic Non-Coding RNA and Cancer View all 10 articles
Human aging is characterized by chronic low-grade inflammation known as “inflammaging.” Persistent low-level inflammation also plays a key role in all stages of breast cancer since “inflammaging” is the potential link between cancer and aging through NF-kB pathways highly influenced by specific miRs. Micro-RNAs (miRNAs) are small non-coding RNAs that negatively regulate gene expression at a posttranscriptional level. Inflamma-miRs have been implicated in the regulation of immune and inflammatory responses. Their abnormal expression contributes to the chronic pro-inflammatory status documented in normal aging and major age-related diseases (ARDs), inflammaging being a significant mortality risk factor in both cases. Nevertheless, the correct diagnosis of inflammaging is difficult to make and its hidden contribution to negative health outcomes remains unknown. This methodological work flow was aimed at defining crucial unanswered questions about inflammaging that can be used to clarify aging-related miRNAs in serum and cell lines as well as their targets, thus confirming their role in aging and breast cancer tumorigenesis. Moreover, we aim to highlight the links between the pro-inflammatory mechanism underlying the cancer and aging processes and the precise function of certain miRNAs in cellular senescence (CS). In addition, miRNAs and cancer genes represent the basis for new therapeutic findings indicating that both cancer and ARDs genes are possible candidates involved in CS and vice versa. Our goal is to obtain a focused review that could facilitate future approaches in the investigation of the mechanisms by which miRNAs control the aging process by acting as efficient ARDs inflammatory biomarkers. An understanding of the sources and modulation of inflamma-miRs along with the identification of their specific target genes could enhance their therapeutic potential.
Aging is an inherently multifactorial process that is manifested within an organism at the genetic, molecular, cellular, organ, and system levels. Gene expression changes in aging at both protein and mRNA levels. An omnipresent feature of the aging process and most all age-related diseases (ARDs) is persistent inflammation. Research has recently shown the potential of Micro-RNAs (miRNAs) to modulate the development period, thus playing a major role in lifespan and the aging process. The spectrum of miRNAs is highly specific for different pathologies contributing to distinct patterns of gene expression (1–3).
The fact that a single miRNA has multiple targets is crucial for understanding the role of miRNAs in normal and pathological processes. The target mRNAs of a given miRNA could be predicted using homologies between the miRNA “seed” region and the complementary site on the target mRNA 3′-untranslated region (UTR) (1, 2). Furthermore, miRNAs are critical regulators of cellular processes such as cell division, differentiation, apoptosis, and senescence. The discovery of extracellular miRNAs enabled such small non-coding molecules to be used for monitoring both cancer and the biomarkers of aging. However, patients need additional investigations. In breast cancer, miRNAs have the potential to facilitate diagnosis and prognosis, predict the response to therapy, and act as therapeutic targets for miRNA-based replacement treatment (4–6). Age is known to be a major risk factor for several pathological conditions, including cancer, cardiovascular diseases, diabetes, and neurodegeneration (7). Breast cancer is the most frequent malignancy and the second cause of carcinoma-related death in women (5, 7).
Recent research demonstrated that miRNAs function in a multiple-to-multiple relationship with their specific target genes. Therefore, a certain miRNA can modulate expression of up to thousand mRNAs, and a specific mRNA can be coordinated by multiple miRNAs, suggesting that the interference of miRNAs in aging, inflammation, and cancer processes is complicated (1). Since miRNAs and their target mRNAs take effect cooperatively, it will be needed of a miRNAs network-based systems biology approach. Recently constructed, the human protein interaction network that consists of approximately 10,000 proteins has over 200,000 documented specific interactions (3). Moreover, the network of the aging process was proved to be a very efficient tool for integrating multiple strong links between the Human protein–protein interaction (PPI) network, the Human longevity network (HLN), and ARD transcriptional regulatory network nodes (proteins) (8).
microRNAs directly and indirectly connect senescence and tumorigenesis through HLN (1) (Table 1). Furthermore, from yeast to mouse, the pattern of evolutionary conservation of CS genes is almost identical to that of cancer-associated genes, this similarity being the result of the coevolution between these two processes. In addition, cellular senescence (CS) could be considered the tumor-suppresor mechanism of a molecular program that inexorably arrests cells at risk for malignant transformation. However, recent research reveals that senescent cells can also have detrimental effects on the tissue microenvironment. The most convincing of these effects is the acquisition of a senescence-associated secretory phenotype (SASP) that transforms senescent fibroblasts into pro-inflammatory cells able to promote tumor growth. CS genes, LAGs (longevity-associated) and cancer genes could be linked through miRNAs. Forty CS genes were validated as targets of 39 miRNAs. From these miRNAs many of them (including miR-21, miR-17, miR-29b) have targets involved in cancer, associated with other ARDs and longevity as well (1, 7, 9–11).
The canonical miRNAs biogenesis pathway starts in the nucleus (7). MiRNAs are firstly transcribed by RNA polymerase II (RNA Pol II) as an approximately 70-nucleotide (nt) long stem-loop primary structure named primary-miRNA transcripts, pri-miRNAs (long miRNA precursors), which are processed by DROSHA RNase III enzyme into a precursor to generate premiRNAs structure (10). Finally, the two strands of the duplex are separated from each other by the Dicer–TRBP complex. Next, the RNA-induced silencing complex (RISC), which also consists of the Argonaute protein and the target mRNA, is complementary bound by specific miRNAs. Consequently, the target mRNAs translation is repressed resulting in translational silencing (10, 11, 36) [Biogenesis of miRNAs is presented in Figure 1 adapted from Ref. (7, 10, 11)].
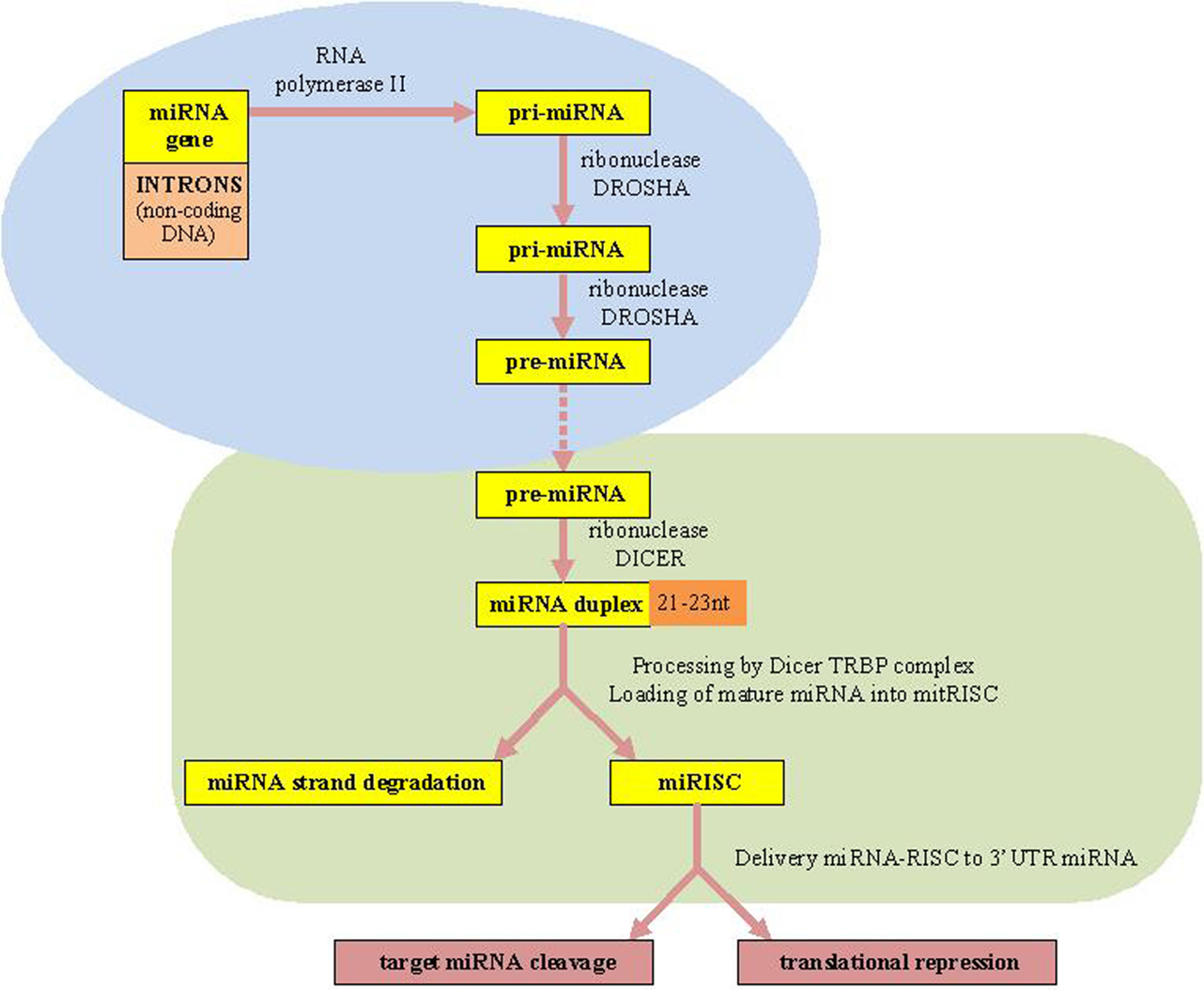
Figure 1. microRNAs biogenesis. miRNAs, microRNAs; RISC, RNA-induced silencing complex; TRBP, transactivating response (TAR) RNA-binding protein as a protein partner of human Dicer.
microRNAs are unique due to their small size, which is approximately 22-nt-long. miRNAs control gene activity by interacting with the RNA that has been copied from DNA. There was no agreement on whether miRNAs were the “key” players in aging or not until 2008 (37). First, researchers were particularly interested in the inflammatory response and found an important role for a few miRNAs. They also showed that these small non-coding RNAs modulate the immune cell development as well as inflammation. In that research work, genetically engineered mice, blood stem cells, and in vitro techniques were used. Work was extended to examine the importance of miRNAs in the aging process, using microarrays in young and old mice (38). Subsequent research also revealed for the first time a signature of genotype-by-age changes in the circulating levels of miRNAs in the long-lived Ames mice (39).
The first evidence for the involvement of endogenous miRNAs in the control of lifespan was reported in 2005 (40, 41). The overexpression of miRNAs lin-4 led to an extended lifespan in Caenorhabditis elegans whereas its loss of it had the opposite effect (42). We also wish to focus on the important role of miRNAs in the immune function. Immunosenescence is “the key” to human aging, all aging-associated diseases (cancer, Alzheimer’s disease, metabolic diseases, and atherosclerosis) being caused by the chronic inflammation coordinated by oxidative stress and manifested by the increase in the level of proinflammatory cytokines, IL (interleukin)-1, IL-6, IL-17 coded by genes activated by the kappa B transcription factor (TF), NF-kB (nuclear factor kappa B) (43).
The comparative analysis between patients with ARDs (neurological or cardiovascular pathology) and controls revealed statistically significant differences in the anti (IL-10) and pro (IL-6, IL-17)-inflammatory investigated cytokines (44) (Figure 2).
Another reason to focus on miRNAs is that they are involved in chronic inflammation, a characteristic of aging and tumorigenesis. NF-κB is the master modulator of the pro-inflammatory status in these processes. A recent paper has shown that in mice, inhibiting one DNA-binding protein can revert aging skin to a pattern of gene activity characteristic to young skin. For example, inducible genetic blockade of NF-κB for 2 weeks in the epidermis of chronologically aged mice reverted the tissue characteristics and global gene expression programs to those of young mice. The chronic activation of this protein is indicative of an inflammatory condition, which strengthens the association between aging and inflammation and suggests that the examination of inflammatory miRNAs may be useful (45). Besides its major role in chronic inflammation, the downregulation of the NF-κB-signaling pathway could inhibit tumor cell growth in breast cancer (46).
The cellular participants involved in the pro-inflammatory status known as “inflammaging” cause senescence by inducing genotoxic stress and the SASP (1, 44, 47). These findings may be relevant for tumor growth, aging, and the senescence-activated inflammation responsible for the increased cancer incidence associated with aging. The inflammatory loop is fueled by SASP. Multiple doses of rapamycin, a promising therapeutic agent with both anti-tumor and immunosuppressive properties selectively splits the cascade of pro-inflammatory events associated with CS (48). Aging is plastic, as it can be shaped by adequate interventions such as specific miRNAs, which could be active components of SASP (49). This fact might contribute to the disease-free phenotype known as “Healthy Aging” (50, 51).
Researchers identified miRNAs that can be associated with cancer (oncomiRs), inflammation (inflamma-miRs), and aging/senescence (SA-miRs). Three of them, namely, miR-21, miR-126, and miR-146a and their target mRNAs belong to the NF-kB pathway, which is the master modulator of the pro-inflammatory status in ARDs (49, 52). MiR-126 and miR-126⋆, a miRNA pair derived from a single precursor, repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis, which demonstrates a correlation between miR-126/126⋆ downregulation and poor metastasis-free survival in breast cancer patients (26).
A senescent cell phenotype reduced the expression of proliferation-stimulating/apoptosis-suppressing miR-21, miR-214, and miR-92 and increased the expression of tumor suppressors and apoptotic markers; inflammation-repressing miR-126 was reduced, whereas inflammatory proteins had a higher level in senescent human aortic endothelial cells (53). For example, high levels of miR-21 occur in invasive breast tumors. There are two tumor suppressors among the miR-21 targets: tropomyosin I (TPMI) and phosphatase and tensin homolog (PTEN), plus the proteins involved in suppression, invasion, and metastasis: programed cell death 4 protein (PDCD4) and maspin (54–56).
In cancer cells, all these proteins are inhibited by high levels of miR-21, while inhibition of this miRNA has the opposite effect of decreasing tumor cell growth, migration, and invasion (57). It has recently been demonstrated that miR-146a plays an important role in the modulation of the innate immune response (58). Several studies have shown the relevance of the upregulation of NF-kB/miR-146a in breast cancer. In addition, by counteracting the pro-inflammatory effects of CS, miR-146 provides anti-inflammatory effects and general suppressive action (52).
Normal cells must acquire several characteristics in order to become a tumor. Furthermore, cancerous cells thrive if they maintain a proliferative status, survive despite strict environmental conditions, induce local angiogenesis, invade other tissues, metastasize and avoid being recognized by the immune system (59). Non-coding small miRNAs play an important role in cancer (60–62). The association between miRNAs and breast cancer has been recognized since 2005, including multiple functions such as suppression of tumorigenesis, promotion or inhibition of metastasis, and sensitivity or resistance to chemotherapy. According to the role of miRNAs in cancer cell phenotypes, some are oncogenic (oncomiRs) and others are tumor-suppressive (TS-miRNAs), depending on the cellular context and on their specific targets in each cellular event (62, 63).
Underexpressed miRNAs in cancers, such as the let-7 family members (TS-miRs) function as tumor suppressor genes. They also induce p53 mediated apoptosis, inhibit the cell cycle and cause breast cancer resistance to tamoxifen. Moreover, experiments carried out between 2008 and 2013 suggest that let-7 based therapy in breast cancer could be available in the near future, especially for drug resistant and estrogen-positive metastatic variants (13). In addition, let-7 is a well-known enhancer of CS (14).
Conversely, overexpressed miRNAs in cancer such as miR-17-92 may function as oncogenes and promote cancer development by negatively regulating tumor suppressor genes that control apoptosis or cell differentiation. In breast cancer, excellent information coverage on certain miRNAs named oncomiRs was also provided (64, 65) [Table 2; (5, 7, 18–22, 66–71)].
Tumor suppressor miRNAs are extremely effective in breast cancer progression because they suppress cell growth, proliferation, angiogenesis, invasion, and metastasis. They also enhance cell death and immune recognition, as well as cancer therapy. For most of the above processes, TS-miRNAs are downregulated in cancer tissues. Upon re-expression they suppress tumorigenesis, including proliferation, apoptosis, and migration (72) [Table 3; (5, 7, 73–98)].
microRNAs such as TS-miRNAs (let-7, miR-15a/16, and miR 143/145) act as tumor suppressors while miR-21, miR-17-92, and miR-155 act as oncogenes by suppressing and promoting tumorigenesis. Highly specific miRNAs also control the development of the metastatic phenotype and are present in different types of cancer (72).
In a certain stage of the same cancer, they are used as a marker for diagnosis, prognosis, and therapy response. In invasive breast tumor, two miR-21 targets mentioned before (TPMI, PTEN) are inhibited by high levels of miR-21, while inhibition of this miRNA has the opposite effect of decreasing tumor cell growth, migration and invasion (19, 20, 55, 57).
Interestingly, all ARDs, including cancer share inflammation as a common denominator, CS being at the basis of the chronic inflammatory state in aging (99). Many individual miRNAs identified in humans target “key” proteins involved in aging and cancer, thus demonstrating their crucial role in aging and breast cancer tumorigenesis. For example, miR-146b increase with age and repress the senescence-associated proinflammatory cytokines IL-6 and IL-8 Activator of transcription 3 (STAT3) and interleukin-6 (IL-6)-mediated signal transducer could be mechanisms by which chronic inflammation contributes to breast cancer such as a common oncogenic event. The gene encoding the TS miRNA, miR-146b is a direct STAT3 target gene, its expression being decreased in tumor cells but increased in normal breast epithelial cells. Moreover, miR-146b inhibits NF-κB-dependent production of IL-6, subsequent STAT3 activation, and IL-6/STAT3-driven invasion and migration in breast cancer cells. Therefore, higher expression of miR-146b was positively correlated with patient survival in breast cancer subtypes with increased IL6 expression and STAT3 phosphorylation. The use of antagomirs directed against breast tumor cells represents a new and highly important therapeutic method (98, 100).
MiR-21 promoter contains binding sites for the TF STAT-3 activated by IL-6 signaling pathway (37). In the elderly, an increased production of proinflammatory cytokines such as IL-6 was observed, which proves the link between tumorigenesis and the aging process; miR-21 was also found elevated in other ARDs such as hypertrophic heart, neointimal formation and Alzheimer’s disease. High levels of pro-inflammatory molecules found in centenarians were offset by large amounts of anti-inflammatory molecules such as TGF-β and IL-10. Besides CDK (cyclin-dependent kinase) regulators driving cell cycle progression in all eukaryotes, new strategies for controlling the NF-kB related inflammation pathway in pathological aging should be used (101, 102).
MiR-519 represses the production of HuR, an RNA-binding protein very abundantly found in tumor cells and less expressed in untransformed cells, while the overexpression of HuR delays the senescent phenotype (83).
MiR-155, which is only one example, out of the many miRNAs could be considered a bridge between inflammation and breast cancer. The overexpression of miR-155 in breast cancer cells leads to constitutive activation of signal transducer and activator of transcription 3 (STAT3) through the Janus-activated kinase (JAK) pathway, and stimulation of breast cancer cells by the inflammatory cytokines IFN-γ and interleukin-6 (IL-6), lipopolysaccharide (LPS), and polyriboinosinic: polyribocytidylic acid [poly(I:C)] significantly upregulates mir-155 expression. In addition, the suppressor of cytokine signaling 1 (socs1) is a new target of miR-155 in breast cancer cells, thus contributing as a consequence to the constitutive STAT3 activation. The cross talk between miR-155, SOCS1, and STAT3 signaling may provide a new mechanism for inflammation-associated tumorigenesis, which suggests that miR-155 and SOCS1 could potentially be used in cancer therapy (30, 31).
Furthermore, serum miR-155 could be a potential biomarker for differentiating between cancer patients and healthy subjects, as well as an indicator of treatment response. Low levels of miR-155 were also observed after surgery and chemotherapy (103, 104). In addition, serum miR-155 is particularly upregulated in more advanced cancer stages than in low-grade breast tumor (105).
The overexpression of miR-34a, a TS-miRNA that represses the production of Bcl-2 and SirT1 (two proteins displaying high levels of expression in breast tumors, which are involved in cell growth and proliferation) induces the senescence (the phenomenon by which normal diploid cells cease to divide) of cancer cells. This miR impairs angiogenesis by induction of senescence via SirT. Higher miR-34c was observed in senescent cells and also in the blood of breast cancer women, the reduced expression of miR-34c being particularly important for progression to the most advanced stages. It also controls the NF NF-kB and could be considered an inflamma-miR (105). Let-7 miRNAs are also upregulated in senescent cells, thus suppressing tumor growth and contributing to the aging process (52).
MiR-195, which is involved in the activation of NF-kB, is highly increased in the blood of breast cancer patients and in CS. The abrogation of the miR-195 expression is a promising therapy in elderly patients. Therefore, miR-195 could serve as an inflammatory biomarker in ARDs (105, 106). Based on the finding that certain miRNAs decreased tumorigenesis, and it was proposed that the coordinated action of upregulated-senescence inflamma-miRs could block cancerous cell growth by reducing oncogenes and tumor promoters’ levels. Their regulation chelp treat ARDs (83, 107).
IL-6 leads to the activation of Stat3, which then dimerizes and binds to its cognate sites located in the regulatory regions of genes. The Stat3-mediated transcription of several antiapoptotic genes contributes to the survival of cancer cells. In a separate mechanism, Stat3 directs the expression of miR-21, resulting in the suppression of apoptosis possibly through the inhibition of TPM1 and/or other proteins [Figure 3 adapted from Ref. (108)].
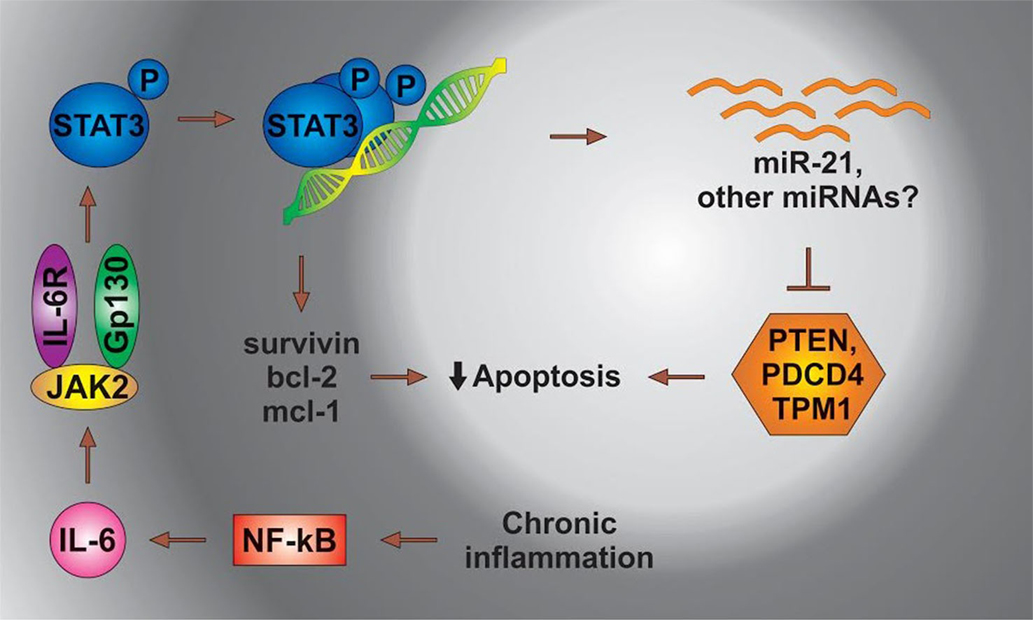
Figure 3. The status of miRs in chronic inflammation. STAT3, signal transducer and activator of transcription 3; NFkB, nuclear factor kappa; PDCD4, programed cell death 4 (neoplastic transformation inhibitor); TPM1, tropomyosin 1 (alpha); PTEN, phosphatase and tensin homolog B; Bcl-2, B cell lymphoma 2; mcl-1, myeloid cell leukemia-1; JAK 2, Janus kinase-2 (the Janus family of tyrosine kinase plays an essential role in coupling cytokine receptors to downstream intracellular signaling pathways).
The inflammaging phenotype results from age-related cell and tissue adaptation/remodeling interacting with the genetic/epigenetic background. This is a complex phenotype involving not only innate but also adaptive immunity and affecting a range of tissues and organs such as the gut, liver, muscle, and brain. Importantly, inflammaging appears to be accelerated in a variety of age-associated diseases. Tissue and circulating inflamma-miRs could restrain the activity of the senescent cell secretome and check the destruction induced by the activation of the inflammatory response (109).
Inflamma-miRs have been involved in the regulation of the immune and inflammatory response, and their abnormal expression may contribute to the low-level chronic inflammation that has been documented both in normal aging and in the major ARDs (109).
Circulating inflamma-miRs could thus have diagnostic/prognostic relevance in human diseases with a common inflammatory background, such as cancer. Recent studies have shown upregulation of inflamma-miRs in the circulation of healthy elderly and chronically ill old individuals: the increase is less pronounced in centenarians and greater in patients with ARDs. It is generally accepted that the main sources of circulating inflamma-miRs in aging and ARDs are immunity circulating/tissue cells and endothelial circulating/resident cells. Inflammatory stimulation and cell senescence can induce and perpetuate systemic inflammation over time, by inducing the upregulation of inflamma-miRs through the excessive activation of inflammatory pathways (109, 112) [Table 4; (105, 106, 109–112)].
Circulating miR-21, miR-34, miR-126, miR-195, let-7a, miR-146a, and miR-155 could be considered as inflamma-miRs that modulate the NF-kB signaling pathway in aging, inflammation, and breast cancer. The similar features of miRNAs in cancer and aging elucidate the functions and therapeutic implications of these common small RNAs. Interestingly, a number of such circulating miRNAs seem to be promising biomarkers for major ARDs that share a common chronic, low-level pro-inflammatory status, such as breast cancer and other ARDs.
A better understanding of the sources and modulation of inflamma-miRs, along with the identification of their specific target genes may enhance their therapeutic potential. New drug development and biological understanding of the inflammatory processes need to be improved. The seven miRs are promising targets as they can be an extremely selective and effective therapeutic strategy against aging and breast cancer.
IB-N and CC initiated this review, searched the literature, and contributed largely to the writing of the manuscript. GC contributed with his own opinions to this review, surveyed, and advised the literature search. IB-N revised all the versions of the manuscript and gave the final approval of the version to be published. IB-N, GC, and CC drafted the article and revised it critically for important intellectual content.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Dr. GC is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), AIM at Melanoma Foundation and the Miriam and Jim Mulva research funds, the Brain SPORE (2P50CA127001), the Center for Radiation Oncology Research Project, the Center for Cancer Epigenetics Pilot project, a 2014 Knowledge GAP MDACC grant, a CLL Moonshot pilot project, the UT MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment, a SINF grant in colon cancer, the Laura and John Arnold Foundation, the RGK Foundation, and the Estate of C. G. Johnson, Jr. POSCCE 709/2010 financed Dr. IB-N work grant with title: “Clinical and economical impact of proteome and transcriptome molecular profiling in neoadjuvant therapy of triple negative breast cancer (BREASTIMPACT).”
1. Huang JT, Wang J, Srivastava V, Sen S, Liu SM. MicroRNA machinery genes as novel biomarkers for cancer. Front Oncol (2014) 4:113. doi: 10.3389/fonc.2014.00113
2. Wei YN, Hu HY, Xie GC, Fu N, Ning ZB, Zeng R, et al. Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging. Genome Biol (2015) 16:41. doi:10.1186/s13059-015-0608-2
3. Franceschi C, Campisi J. Chronic Inflammation (Inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci (2014) 69(Suppl 1):S4–9. doi:10.1093/gerona/glu057
4. Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin Oncol (2006) 33(2):167–73; Modulation of cancer traits by tumor suppressor microRNAs. Int J Mol Sci 14:1822–42. doi:10.1053/j.seminoncol.2006.01.010
5. Wang W, Luo YP. MicroRNAs in breast cancer: oncogene and tumor suppressors with clinical potential. J Zhejiang Univ Sci B (2015) 16(1):18–31. doi:10.1631/jzus.B1400184
6. Voutetakis K, Chatziioannou A, Gonos ES, Trougakos IP. Comparative meta-analysis of transcriptomics data during cellular senescence and in vivo tissue ageing. Oxid Med Cell Longev (2015) 2015:732914. doi:10.1155/2015/732914
7. Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res (2015) 17:21. doi:10.1186/s13058-015-0526-y
8. Lee SK, Calin G. Non-coding RNAs and cancer: new paradigms in oncology. Discov Med (2011) 11(58):245–54. doi:10.1007/s13148-011-0040-8
9. Piano A, Titorenko VI. The intricate interplay between mechanisms underlying aging and cancer. Aging Dis (2014) 6(1):56–75. doi:10.14336/AD.2014.0209
10. Smith-Vikos T, Slack JF. MicroRNAs and their roles in aging. J Cell Sci (2012) 125:7–17. doi:10.1242/jcs.099200
11. Bullrich F, Fujii H, Calin G, Mabuchi H, Negrini M, Pekarski Z, et al. Characterization of the 13q14 tumor suppressor locus in CLL. Identification of ALT1, an alternative splice variant of the LEU2 gene 1. Cancer Res (2001) 61(18):6640–8. doi:10.261/rna.2146903
12. Barh D, Parida S, Parida BP, Viswanathan G. Let-7, miR-125, miR-205 and miR-296 are prospective therapeutic agents in breast cancer molecular medicine. Gene Ther Mol Biol (2008) 12:189–206.
13. Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Research (2015) 17:40. doi:10.1186/s13058-015-0542-y
14. Guo R, Abdelmohsen K, Morin PJ, Gorospe M. Novel microRNA reporter uncovers repression of let-7 by GSK-3β. PLoS One (2013) 8(6):e66330. doi:10.1371/journal.pone.0066330
15. Pichler M, Winter E, Ress AL, Bauernhofer T, Gerger A, Kiesslich T, et al. miR-181a is associated with poor clinical outcome in patients with colorectal cancer treated with EGFR inhibitor. J Clin Pathol (2013). doi:10.1136/jclinpath-2013-201904
16. Luo Q, Li X, Li J, Kong X, Zhang J, Chen L, et al. MiR-15a is underexpressed and inhibits the cell cycle by targeting CCNE1 in breast cancer. Int J Oncol (2013) 43(4):1212–8. doi:10.3892/ijo.2013.2034
17. Ouchida M, Kanzaki H, Ito S, Hanafusa H, Jitsumori Y, Tamaru S, et al. Novel direct targets of miR-19a identified in breast cancer cells by a quantitative proteomic approach. PLoS One (2012) 7(8):e44095. doi:10.1371/journal.pone.0044095
18. Rebbeck TR, Mitra N, Wan F, Sinilnihova OM, Healey S, McGuffog L, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA (2015) 313. doi:10.1001/jama.2014.5985
19. Frankel L, Christoffersen N, Jacobsen A, Lindow M, Krogh A, Lund A. Programmed cell death 4 (pdcd4) is an important functional target of the microRNA mir-21 in breast cancer cells. J Biol Chem (2008) 283:1026–33. doi:10.1074/jbc.M707224200
20. Kieffer-Kwon P, Happel C, Uldrick TS, Ramalingam D, Ziegelbauer JM. KSHV microRNAs repress tropomyosin 1 and increase anchorage-independent growth and endothelial tube formation. PLoS One (2015) 10(8):e0135560. doi:10.1371/journal.pone.0135560
21. Cowley D, Pandya K, Khan I, Kerwin J, Owen K, Griner E. Registered report: a coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Elife (2015) 4. doi:10.7554/eLife.08245
22. Song B, Wang C, Liu J, Wang X, Lv L, Wei L, et al. MicroRNA-21 regulates breast cancer invasion partly by targeting the tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res (2010) 29:29. doi:10.1186/1756-9966-29-29
23. Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat Rev Cardiol (2015) 12:135–42. doi:10.1038/nrcardio.2014.207
24. Srivastava N, Manvati S, Srivastava A, Pal R, Kalaiarasan P, Chattopadhyay S, et al. miR-24-2 controls H2AFX expression regardless of gene copy number alteration and induces apoptosis by targeting antiapoptotic gene BCL-2: a potential for therapeutic intervention. Breast Cancer Res (2011) 13:R39. doi:10.1186/bcr2861
25. Zhang H, Wang Q, Zhao Q, Di W. MiR-124 inhibits the migration and invasion of ovarian cancer cells by targeting SphK1. J Ovarian Res (2013) 6:84. doi:10.1186/1757-2215-6-84
26. Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y, et al. miR-126 and miR-126⋆ repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nat Cell Biol (2013) 15:284–94. doi:10.1038/ncb2690
27. Hu J, Guo H, Li H, Liu Y, Liu J, Chen L, et al. MiR-145 regulates epithelial to mesenchymal transition of breast cancer cells by targeting Oct4. PLoS One (2012) 7:e45965. doi:10.1371/journal.pone.0045965
28. Wu H, Xiao Z, Wang K, Liu W, Hao Q. MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys Res Commun (2013) 441(4):693–700. doi:10.1016/j.bbrc.2013.10.053
29. Zhou L, Zhao X, Han Y, Lu Y, Shang Y, Liu C. Regulation of UHRF1 by miR-146a/b modulates gastric cancer invasion and metastasis. FASEB J (2013) 27(12):4929–39. doi:10.1096/fj.13-233387
30. Van Schooneveld E, Wildiers H, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res (2015) 17(1):21. doi:10.1186/s13058-015-0526-y
31. Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X, et al. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS One (2012) 7(10):e46082. doi:10.1371/journal.pone.0046082
32. Schwarzenbach H, Milde-Langosch K, Steinbach B, Müller V, Pantel K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res Treat (2012) 134(3):933–41. doi:10.1007/s10549-012-1988-6
33. Lambertini E, Lolli A, Vezzali F, Penolazzi L, Gambari R, Piva R. Correlation between Slug transcription factor and miR-221 in MDA-MB-231 breast cancer cells. BMC Cancer (2012) 12:445. doi:10.1186/1471-2407-12-445
34. Goldberger N, Walker RC, Kim CH, Winter S, Hunter KW. Inherited variation in miR-290 expression suppresses breast cancer progression by targeting the metastasis susceptibility gene Arid4b. Cancer Res (2013) 73(8):2671–81. doi:10.1158/0008-5472.CAN-12-3513
35. Yan GR, Xu SH, Tan ZL, Liu L, He QY. Global identification of miR-373-regulated genes in breast cancer by quantitative proteomics. Proteomics (2011) 11(5):912–20. doi:10.1002/pmic.201000539
36. Jung HJ, Suh Y. Regulation of IGF-1 signaling by microRNAs. Front Genet (2015) 5:472. doi:10.3389/fgene.2014.00472
37. Wolfson M, Tacutu R, Budovsky A, Aizenberg N, Fraifeld VE. MicroRNAs: relevance to aging and age-related diseases. Open Longev Sci (2008) 2:66–75. doi:10.2174/1876326X00802010066
38. Linton PJ, Thoman M. Immunosenescence in monocytes, macrophages, and dendritic cells: lessons learned from the lung and heart. Immunol Let (2014) 162. doi:10.1016/j.imlet.2014.06.017
39. Victoria B, Dhahbi JM, Nunez Lopez YO, Spinel L, Atamna H, Spindler SR, et al. Circulating microRNA signature of genotype-by-age interactions in the long-lived Ames dwarf mouse. Aging Cell (2015). doi:10.1111/acel.12373
40. Nehammer C, Podolska A, Mackowiak SD, Kagias K. Specific microRNAs regulate heat stress responses in Caenorhabditis elegans. Sci Rep (2015) 5:8866. doi:10.1038/srep08866
41. Boehm M, Slack FJ. MicroRNAs control lifespan and metabolism. Cell Cycle (2006) 5(8):837–40. doi:10.4161/cc.5.8.2688
42. Antebi A. Physiology. The tick-tock of aging? Science (2005) 310(5756):1911–3. doi:10.1126/science.1122816
43. El Assar M, Anguolo J, Rodriguez-Manas L. Oxidative stress and vascular inflammation in aging. Free Radic Biol Med (2013) 65:380–401. doi:10.1016/j.freeradbiomed.2013.07.003
44. Cătană CS, Berindan-Neagoe I. Aging and Immunity. Age-Related Changes in Immunological, Biochemical and Genetic Markers. Saarbrücken: Lambert Academic Publising (2012).
45. Ljubuncic P, Gochman E, Reznick AZ. Nitrosative stress in aging. Its importance and biological implications in NF-kB signaling. In: Bondy S, Maiese K, editors. Oxidative Stress in Applied Basic Research and Clinical Practice. Aging and Age-Related Disorders. Armstrong: Springer (2010). p. 27–54.
46. Zhai Z, Qu X, Li H, Ouyang Z, Yan W, Liu G, et al. Inhibition of MDA-MB-231 breast cancer cell migration and invasion activity by andrographolide via suppression of nuclear factor-κB-dependent matrix metalloproteinase-9 expression. Mol Med Rep (2015) 11(2):1139–45. doi:10.3892/mmr.2014.2872
47. Ostan R, Lanzarini C, Pini E, Scurti M, Vianello D, Bertarelli C. Inflammaging and cancer: a challenge for the Mediterranean diet. Nutrients (2015) 7(4):2589–621. doi:10.3390/nu7042589
48. Laberge RM, Sun Y, Orjalo AV, Patil CK, Freund A, Zhou L, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol (2015) 17:1049–61. doi:10.1038/ncb3195
49. Bonafe M, Olivieri F. Circulating microRNAs in aging. Oncotarget (2015) 6(3):1340–1. doi:10.18632/oncotarget.3175
50. Sikora E, Scapagnini G, Barbagallo M. Curcumin, inflammation, aging and age- related diseases. Immun Ageing (2010) 7(1):1–4. doi:10.1186/1742-4933-7-1
51. Sood S, Gallagher IJ, Lunnon K, Rullman E, Keohane A, Crossland H, et al. A novel multi-tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biol (2015) 16:185. doi:10.1186/s13059-015-0750-x
52. Olivieri F, Rippo MR, Monsurro V, Salvioli S, Capri M, Procopio AD, et al. Micro-RNAs linking inflamm-aging, cellular senescence and cancer. Ageing Res Rev (2013) 12(4):1056–68. doi:10.1016/j.arr.2013.05.001
53. Rippe C, Blimline M, Magerko KA, Lawson BR, LaRocca TJ, Donato AJ, et al. MicroRNA changes in human arterial endothelial cells with senescence: relation to apoptosis, eNOS and inflammation. Exp Gerontol (2012) 47(1):45–51. doi:10.1016/j.exger.2011.10.004
54. LeBlanc VC, Morin P. Exploring miRNA-Associated Signatures with Diagnostic Relevance in Glioblastoma Multiforme and Breast Cancer Patients. J Clin Med (2015) 4(8):1612–30. doi:10.3390/jcm4081612
55. Qi L, Bart J, Ping-Tan L, Platteel I, Van der Sluis T, Huitema S, et al. Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer (2009) 9:163. doi:10.1186/1471-2407-9-163
56. Chen J, Xu T, Chen C. The critical roles of miR-21 in anti-cancer effects of curcumin. Ann Transl Med (2015).
57. Fatima R, Akhade VS, Pal D, MR Rao S. Long non-coding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther (2015) 3. doi:10.1186/s40591-015-0042-6
58. Labbaye C, Testa U. The emerging role of miR-146a in the control of immune function and cancer. J Hematol Oncol (2012) 5:13. doi:10.1186/1756-8722-5-13
59. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144:646–74. doi:10.1016/j.cell.2011.02.013
60. Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med (2012) 12(12):580–7. doi:10.1016/j.molmed.2006.10.006
61. Calin GA, Croce CM. MicroRNAs signatures in human cancers. Nat Rev Cancer (2006) 6(11):857–66. doi:10.1038/nrc1997
62. Cojocneanu Petric R, Braicu C, Raduly L, Zanoaga O, Dragos N, Monroig P, et al. Phytochemicals modulate carcinogenic signaling pathways in breast and hormone-related cancers. Onco Targets Ther (2015) 8:2053–66. doi:10.2147/OTT.S83597
63. Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics (2015) 5(10):1122–43. doi:10.7150/thno.11543
64. Blenkiron C, Goldstein LD, Thome NP. MicroRNAs expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol (2007) 8:R214. doi:10.1186/bcr1867
65. Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res (2008) 10:203. doi:10.1186/bcr1867
66. Jang S, Zhang H, Lu M, He X, Li Y, Gu H, et al. MicroRNA-155 functions as an oncomir in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res (2010) 70:3119–27. doi:10.1158/0008-5472.CAN-09-4250
67. Ren YQ, Fu F, Han J. MiR-27a modulates radiosensitivity of triple-negative breast cancer (TNBC) cells by targeting CDC27. Med Sci Monit (2015) 21:1297–303. doi:10.12659/MSM.893974
68. Guttilla I, White B. Coordinate regulation of foxo1 by mir-27a, mir-96 and mir-192 in breast cancer cells. J Biol Chem (2009) 284:23204–16. doi:10.1074/jbc.M109.031427
69. Masri S, Liu Z, Phung S, Wang E, Yuan Y, Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res Treat (2010) 124:89–99. doi:10.1007/s10549-009-0716-3
70. Liu Y, Zhao J, Zhang PY, Zhang Y, Sun SY, Yu SY, et al. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit (2012) 18(8):Br299–308. doi:10.12659/MSM.883262
71. Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol (2010) 12(3):247–56. doi:10.1038/ncb2024
72. Costa PM, Pedroso de Lima MC. MicroRNAs as molecular targets for cancer therapy: on the modulation of microRNA expression. Pharmaceuticals (Basel) (2013) 6(10):1195–220. doi:10.3390/ph6101195
73. Li L, Yuang L, Luo J, Gao J, Guo J, Xie X. MiR-34° inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med (2013) 13(2):109–17. doi:10.1007/s10238-012-0186-5
74. Muluhngwi P, Klinge CM. Roles for miRNAs in endocrine resistance in breast cancer. Endocr Relat Cancer (2015) 22(5):R279–300. doi:10.1530/ERC-15-0355
75. Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ, et al. MiR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res (2011) 71:3552–62. doi:10.1158/0008-5472.CAN-10-2435
76. Zhao AH, Zeng Q, Xie XY, Zhou JN, Yue W, Li YL, et al. MicroRNAs-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. J Genet Genomics (2012) 39:29–35. doi:10.1016/j.jgg.2011.12.003
77. Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JK, Gill RM, Huillard E, et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene (2012) 31:1884–95. doi:10.1038/onc.2011.380
78. Garimella SV, Gehlhaus K, Dine JL, Pitt JJ, Grandin M, Chakka S, et al. Identification of novel molecular regulators of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in breast cancer cells by RNAi screening. Breast Cancer Res (2014) 16(2):R41. doi:10.1186/bcr3645
79. Chen F, Hu SJ. Effect of microRNA-34a in cell cycle, differentiation and apoptosis: a review. J Biochem Mol Toxicol (2012) 26:79–86. doi:10.1002/jbt.20412
80. Zou C, Xu Q, Mao F, Li D, Bian CX, Liu LZ, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle (2012) 11:2137–45. doi:10.4161/cc.20598
81. Xu Q, Liu LZ, Qian X, Chen Q, Jiang Y, Li D, et al. MiR-145 directly targets p70S6K1 in cancer cells to inhibit tumor growth and angiogenesis. Nucleic Acids Res (2012) 40:761–74. doi:10.1093/nar/gkr730
82. Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA (2010) 1:214–29. doi:10.1002/wrna.4
83. Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. MiR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci U S A (2008) 105:20297–302. doi:10.1073/pnas.0809376106
84. Abdelmohsen K, Kim MM, Srikantan S, Mercken EM, Brennan SE, Wilson GM, et al. MiR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle (2010) 9:1354–9. doi:10.4161/cc.9.7.11164
85. Abdelmohsen K, Srikantan S, Tominaga K, Kang MJ, Yaniv Y, Martindale JL, et al. Growth inhibition by miR-519 via multiple p21- inducing pathways. Mol Cell Biol (2012) 32:2530–48. doi:10.1128/MCB.00510-12
86. Abdelmohsen K, Gorospe M. Noncoding RNA control of cellular senescence. Wiley Interdiscip Rev RNA (2015) 6(6):615–29. doi:10.1002/wrna.1297
87. Ristimaki A. Tumor suppressor effect of the microRNA miR-519 is mediated via the mRNA-binding protein HuR. Cell Cycle (2010) 9(7):1234. doi:10.4161/cc.9.7.11322
88. Wu ZS, Wu Q, Wang CQ, Wang XN, Huang XZ, Zhang GH, et al. MiR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer (2011) 117:2842–52. doi:10.1002/cncr.25860
89. Ramasamy S, Duraisamy S, Barbashov S, Kawano T, Kharbanda S, Kufe D. The MUCI and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol Cell (2007) 27:992–1004. doi:10.1016/j.molcel.2007.07.031
90. Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Treviro R, Bai S, et al. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet (2012) 8(6):e1002751. doi:10.1371/journal.pgen.1002751
91. Hu XW, Guo JY, Zheng L, Li C, Zheng TM, Tanyi JL, et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Mol Cancer Res (2013) 11(3):240–50. doi:10.1158/1541-7786.MCR-12-0432
92. Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC, et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest (2014) 124(7):3093–106. doi:10.1172/JCI73351
93. Yan DS, Zhou XT, Chen XY, Hu DN, Da Dong X, Wang J, et al. MicroRNA-34a inhibits uveal melanoma cell proliferation and migration through downregulation of c-Met. Invest Ophthalmol Vis Sci (2009) 50:1559–65. doi:10.1167/iovs.08-2681
94. Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer (2010) 10:502. doi:10.1186/1471-2407-10-502
95. Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang TH, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through down-regulated expression of Fra-1 proto-oncogene. Oncogene (2014) 33(23):3014–23. doi:10.1038/onc.2013.258
96. Jiang H, Wang P, Li X, Wang Q, Deng ZB, Zhuang X, et al. Restoration of miR17/20a in solid tumor cells enhances the natural killer cell antitumor activity by targeting Mekk2. Cancer Immunol Res (2014) 2(8):789–99. doi:10.1158/2326-6066.CIR-13-0162
97. Fan M, Sethuraman A, Brown M, Sun W, Pfeffer LM. Systematic analysis of metastasis-associated genes identifies miR-17-5p as a metastatic suppressor of basal-like breast cancer. Breast Cancer Res Treat (2014) 146(3):487–502. doi:10.1007/s10549-014-3040-5
98. Xiang M, Birbak NJ, Vafaizadeh V, Walker SR, Yeh JE, Liu S, et al. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci Signal (2014) 7(310):ra11. doi:10.1126/scisignal.2004497
99. Youm YH, Grant RW, McCabe LR, Albarado DC, Nguyen KY, Ravussin A, et al. Canonical Nlrp3 inflammasome links systemic low-grade inflammation to functional decline in aging. Cell Metab (2013) 18(4):519–32. doi:10.1016/j.cmet.2013.09.010
100. Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther (2013) 21:185–91. doi:10.1038/mt.2012.180
101. Ling H, Samarasinghe S, Kulasiri D. Computational experiments reveal the efficacy of targeting CDK2 and CKIs for significantly lowering cellular senescence bar for potential cancer treatment. Biosystems (2013) 111(2):71–82. doi:10.1016/j.biosystems.2012.12.001
102. Soria-Valles C, Osorio FG, Gutiérrez-Fernández A, De Los Angeles A, Bueno C, Menéndez P, et al. NF-κB activation impairs somatic cell reprogramming in ageing. Nat Cell Biol (2015) 17. doi:10.1038/ncb3207
103. Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, et al. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One (2012) 7(10):e47003. doi:10.1371/journal.pone.0047003
104. Schroen B, Heyman S. Small but smart – microRNAs in the centre of inflammatory processes during cardiovascular diseases, the metabolic syndrome, and ageing. Cardiovasc Res (2012) 93(4):605–13. doi:10.1093/cvr/cvr268
105. Taylor MA. Circulating microRNAs as biomarkers and mediators of cell–cell communication in cancer. Biomedicines (2015) 3. doi:10.3390/biomedicines3040270
106. Kondol H, Kim HW, Millard RW, Wangl YG. Abrogation of senescence-associated microrna-195 in aged skeletal myoblasts facilitates reprogramming to produce induced pluripotent stem cells. Circulation (2014) 130:Abstract15481.
107. Sinkam N, Croce SP. Clinical applications for microRNAs in Cancer. Clin Pharmacol Ther (2013) 93:98–104. doi:10.1038/clpt.2012.192
108. Chan JK, Blansit K, Kiet T, Sherman A, Wong G, Earle C, et al. The inhibition of miR-21 promotes apoptosis and chemosensitivity in ovarian cancer. Gynecol Oncol (2014) 132(3):739–44. doi:10.1016/j.ygyno.2014.01.034
109. Olivieri F, Rippo MR, Procopio AD, Fazioli F. Circulating inflamma-miRs in aging and age-related diseases. Front Genet (2013) 4:121. doi:10.3389/fgene.2013.00121
110. Wang K, Li L, Wu J, Qiu Q, Zhou F, Wu H. The different expression profiles of microRNAs in elderly and young human dental pulp and the role of miR-433 in human dental pulp cells. Mech Ageing Dev (2015) 146-148:1–11. doi:10.1016/j.mad.2015.03.001
111. Mar-Aguilar F, Mendoza-Ramírez JA, Malagón-Santiago I, Espino-Silva PK, Santuario-Facio SK, Ruiz-Flores P, et al. Serum circulating microRNA profiling for identification of potential breast cancer biomarkers. Dis Markers (2013) 34:163–9. doi:10.3233/DMA-120957
Keywords: inflammaging, inflamma-miRs, microRNAs, aging, breast cancer
Citation: Cătană CS, Calin GA and Berindan-Neagoe I (2015) Inflamma-miRs in Aging and Breast Cancer: Are They Reliable Players? Front. Med. 2:85. doi: 10.3389/fmed.2015.00085
Received: 28 September 2015; Accepted: 13 November 2015;
Published: 15 December 2015
Edited by:
Alfredo Fusco, IEOS – Consiglio Nazionale delle Ricerche, ItalyCopyright: © 2015 Cătană, Calin and Berindan-Neagoe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ioana Berindan Neagoe, aW9hbmFuZWFnb2UyOUBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.