
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Mech. Eng. , 13 May 2021
Sec. Tribology
Volume 7 - 2021 | https://doi.org/10.3389/fmech.2021.661370
This article is part of the Research Topic Bioadhesion View all 14 articles
Understanding bacterial adhesion as a first step toward biofilm formation is of fundamental interests in many applications. While adhesion to abiotic surfaces is directly relevant for some applications, it also provides a controlled reference setting to study details of the adhesion process in general. This review describes the traditional approaches from contact mechanics and colloidal science, which treat the bacterium–substratum interaction in a continuous manner. We will discuss its shortcomings and provide an introduction to different approaches, which understand the adhesion process as a result of individual stochastic interactions of many macromolecules with the substratum.
Bacterial biofilms are complex consortia of bacterial cells and extracellular substances that can form on various interfaces (Dunne, 2002). The presence of such biofilms on solid, abiotic surfaces can cause problems in many applications: Formed on ship hulls, they increase hydrodynamic friction and therewith fuel consumption (De Carvalho, 2007), biofilms formed inside pipes reduce the pipes' diameter and therewith flow rates of fluids (Schwermer et al., 2008), biocorrosion caused by biofilms reduces efficiency of cooling water systems in the processing industry (Narenkumar et al., 2019). On medical equipment, such as catheters, implants, protheses, and pacemakers, biofilms are responsible for device-related infections, which can lead to severe diseases and hence are an important health care problem (Magill et al., 2014; Römling et al., 2014; Jamal et al., 2018).
One of the first steps in biofilm formation is the adhesion of single cells to a surface. Therefore, to manage or prevent biofilm formation, a profound understanding of bacterial adhesion to solid surfaces is necessary. In order to gain experimental access to the basic mechanisms of adhesion, the parameters of the system must be kept as controlled as possible. Hence, the presented studies explore bacterial adhesion to abiotic, unconditioned surfaces, i.e., surfaces that are not covered by other biomacromolecules.
First, the approaches of understanding bacterial adhesion on the whole cell level, namely in the framework of colloidal science, i.e., surface thermodynamics and DLVO1 theory, and contact mechanics are briefly presented. We discuss the prospects and limitations of those models and describe the efforts made outside these frameworks in describing bacterial adhesion mediated by cell wall macromolecules.
To understand how experimental results led to the creation of different models for bacterial adhesion, the principle experimental approaches that have been used are briefly explained (see also, e.g., Tandogan et al., 2017). There are predominately two principle types of experimental setups: On the one hand, experiments with a rather high number of planktonic cells that freely adsorb to an interface and eventually desorb again; on the other hand, experiments with single cells that are actively manipulated by external forces to precisely measure their behavior during adhesion and detachment.
For the first setup type, flow chambers are commonly used in which a bacterial solution is flushed over a surface of interest by a laminar flow profile that allows to estimate the forces parallel to the surfaces. Using optical microscopy, quartz crystal micro-balance, or surface plasmon resonance, the number of attached cells in a certain area can be recorded over time (Filion-Côté et al., 2017; Keskin et al., 2018; Alexander et al., 2019). With the help of high-resolution optical techniques, not only the number of cells but also their motion at or above the surface can be quantified (van der Westen et al., 2018; Vissers et al., 2018). These methods can collect data of large numbers of cells simultaneously under controlled (with or without shear flow) conditions tangential to the surface. However, the forces acting during approach of the cells normal to the surface cannot be controlled. In addition, repeating the experiment and the cellular response for one individual cell is hardly possible.
To repeatedly probe single cells and achieve a high force control, optical tweezers (Fällman et al., 2004; Zhang and Liu, 2008) or atomic force microscopy (AFM) (Hinterdorfer and Dufrêne, 2006; Dufrêne and Pelling, 2013; Thewes et al., 2015a; Krieg et al., 2019) are used. While both methods have essentially the same advantages in terms of precise force and position control, the latter places fewer demands on the system itself. Therefore, AFM-based force spectroscopy with individual bacterial probes, termed single cell force spectroscopy (SCFS), is the method of choice for many researchers investigating adhesion properties of bacteria (Berne et al., 2018; Alam et al., 2019). The cells are immobilized at an AFM cantilever and moved toward and then away from a surface. By measuring the deflection of the cantilever as a function of its motion, force-separation curves, such as one schematically shown in Figure 1A can be recorded. SCFS allows to study the adhesion process almost natively by using very small force triggers, i.e., the force threshold at which the cantilever retraction starts2. From these curves, many quantities, such as the adhesion force can be determined. Of note, many experimental force-separation curves recorded with bacterial cells show a very characteristic feature: Before the cells reaches the substratum, a sudden change in the cantilever's deflection and a decrease in the distance between cell and substratum is observed, which is referred to as “snap-in” (Bhushan, 2017). In addition, approach and retraction curves do not necessarily overlap; this is sometimes termed hysteresis. While investigating a significant number of individual cells requires a lot of time, the nature of these experiments allows the repetition of approach and retraction curves with one and the same bacterial cell. This allows to study the role of stochasticity in the adhesion process and to distinguish it from population heterogeneity.
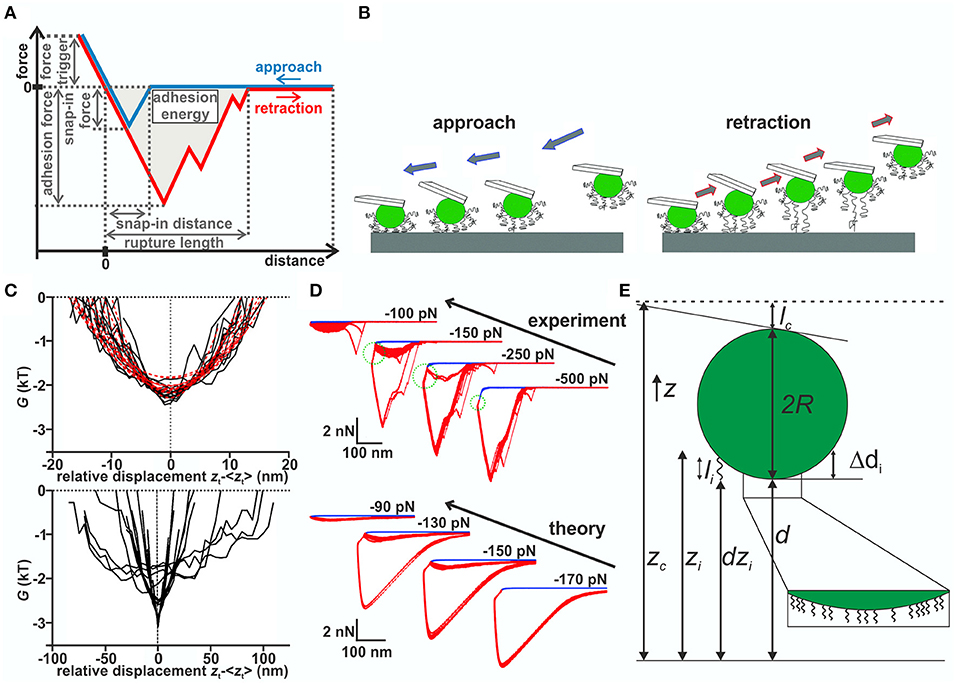
Figure 1. (A) Schematic force–distance curve. (B) Sketch of the model: During approach (upper row), the most extend macromolecules start interacting with the surface and pull the bacterium closer to it, which allows more and more molecules to bind. Upon further approach, some molecules may be compressed. During retraction (lower row), the molecules are decompressed and then stretched before the detaching individually. (C) Gibb's free energy in dependence of the relative displacement of Streptococcus salivarius cells with (top) and without (bottom) fibrillar surface tethers (van der Westen et al., 2018). Image taken from https://pubs.acs.org/doi/10.1021/acs.langmuir.7b04331. For futher information and permissions refer to ACS. (D) Partitioned force–distance curves (from experiment and simulations) with varying force trigger. A smaller negative (larger absolute value) force trigger means that the retraction starts later (Thewes et al., 2015b). Blue curves are individual approach curves, while red curves are individual retraction curves with the same cell. Note the bistable behavior of the retraction curves for intermediate force triggers, where either one or the other type of curve is observed for the same cell. The pulling regime is indicated by the green dotted circle. (E) Model details for Monte Carlo simulations by Thewes et al. (2015b): Sphere of radius R decorated with springs of length li connected at fixed height Δdi from bottom of the sphere. The other end is dzi above the surface and interacts with the potential. In each step, the cantilever's height zc is changed and a new equilibrium position d for the sphere computed.
In contact mechanics exist many models, which extend the Hertz model to include the coupling of adhesion and deformation forces: Very simplified cases that included adhesion are the JKR and DMT model (Johnson et al., 1971; Derjaguin et al., 1975), based on which more accurate models were constructed that account for deformations and longer ranging adhesion forces (Muller et al., 1980; Maugis, 1992; Greenwood, 1997; Ciavarella et al., 2019). The models have also been extended to describe interactions of inhomogeneous objects (Barthel and Perriot, 2007; Stan and Adams, 2016), making them suitable candidates for modeling the adhesion of bacterial cells that have an inhomogeneous surface structure with a lipid bilayer, cross-linked peptidoglycan layer, and eventual cellular appendages (Chen et al., 2014; Loskill et al., 2014). A model including these heterogeneities has been constructed by Chen et al. (2012), who considered a layered structure with different elastic properties along the radial direction. It turns out that this already reduces the extracted Young's modulus to 8–50 kPa, which is about a hundred times smaller than what would be extracted from the Hertz model.
Note that the heterogeneity is limited to the radial direction of the spherical cell. Inhomogeneities within the cell surface, such as clusters of adhesins, and different mechanical properties or lengths of single molecules in the cell wall are not considered. One reason for the fact that Chen et al. (2014) did not experimentally observe effects of these properties can be attributed to their way of preparing bacterial probes: The bacteria, already immobilized on the cantilever, were dried for 2 min, which is likely to alter the proteinaceous cell wall layer and change its original properties, such as heterogeneity (Chen et al., 2012, 2014). This might also explain why no cell-individual adhesion behavior was observed.
Colloidal approaches phrase the problem of bacterial adhesion as minimization of thermodynamic potentials, such as the Gibbs free energy. Thus, the theory does not take into account eventual strengthening of adhesive bonds. In the review article by Perni et al. (2014), it is shown that the simple surface thermodynamics approach of considering only interfacial energies to minimize the Gibbs free energy works only in a few cases and is generally considered too simplistic. A different approach applies the DLVO theory to the bacterium-plane geometry considering electrostatic double layer and van der Waals forces that have shown to influence bacterial adhesion (Van Oss et al., 1990; Boks et al., 2008; Loskill et al., 2012). Various publications use different approximations for these forces that can be quite evolved and in many instances not analytically solvable. However, qualitatively there are only few scenarios possible: If either of the interactions is attractive and the other repulsive, the free energy landscape displays a minimum close to the surface and eventually—depending on the exact relation between attractive and repulsive potentials—also a secondary minimum. Strong adhesion is achieved when the bacterium can overcome the barrier and weak adhesion is achieved inside the secondary minimum. On experimental time scales, weak adhesion manifests itself as reversibility of the adhesion process, not predicted by the surface thermodynamic approach. In DLVO theory, neglected interactions, such as acid–base interactions and steric effects due to the presence of polymers on the bacterium surface, have been incorporated in the so-called xDLVO theory (Van Oss, 1995). These extensions, however, change the interaction potential quantitatively but do not alter the qualitative picture. The failure of these approaches has, according to Perni et al. (2014), been attributed due to neglecting shear forces and the underlying assumption of a homogeneous bacterial surface composition. However, these models do not aim at describing a full approach and retraction cycle. If the derivative of the potential is considered as the force experience by a bacterium, no hysteresis can be observed since the derivative is unique.
To address these limitations, Jasevičius et al. (2015) extended the DMT model of classical adhesion: The snap-in is incorporated by the van der Waals force of sphere-plane geometry acting from the snap-in distance until direct contact of the surfaces. The magnitude, i.e., the snap-in force, as well as the snap-in distance are fitted from experimental data and are not obtained from the constitute equations of DLVO theory. Once the bacterium is in contact, the usual DMT forces in addition to repulsive electrostatic double layer forces and steric repulsion forces of polymer brushes are considered. This is complemented by an energy dissipation mechanism including plastic deformation to produce the adhesion hysteresis.3 Phrased loosely, this model combines xDLVO theory with the Hertzian contact model, while also including an ad hoc snap-in mechanism and energy dissipation. Recently, this model has been extended to mimic flow chamber experiments and determine if a given bacterial strain will adhere to a given surface (Jasevičius and Kruggel-Emden, 2017). Therefore, an initial velocity and viscous drag was included into the model and it was demonstrated that Staphylococcus aureus cells stick to a glass surface.
We point out that all three models assume continuous interactions of the entire bacterium with the surface while neglecting stochasticity and the responses of individual macromolecules in the adhesion process. However, the next section will show that non-continuous interactions are needed to describe certain aspects of bacterial adhesion.
In a different set of studies, the displacement of different bacteria after settling in a flow chamber has been monitored by optical microscopy (Sjollema et al., 2017). These experiments, combined with SCFS, demonstrated that the movement of the cells parallel to the surface decreases with increasing adhesion force. These experimental results combined with an in silico model led to the conclusion that the bacteria adhere via multiple reversibly binding tethers, which repeatedly detach from and attach to the surface without detaching all at the same time (see Figure 1B). An extension of this study has determined if adhering bacteria also exhibit vibrations perpendicular to the surface using internal reflection microscopy (van der Westen et al., 2018). For bacteria without cellular appendages, a comparison of the results with predictions from DLVO theory showed that the surface potential displays two minima with a potential barrier in between that was considered to be too high to be overcome by Brownian motion. The researchers observed on the hydrophobic substrata asymmetric fluctuations inside the secondary minimum with amplitudes fitting to the width of the minimum, independent of ionic strength of the solution.4 In contrast, cells with fibrils showed symmetric fluctuations with five times smaller vibrational amplitudes, regardless of surface hydrophobicity and ionic strength of the solution (see Figure 1C). This lead the authors to distinguish “tether-coupled” and “floating” adhesion where in the latter case adhesion is dominated by the thermal motion inside the secondary minimum predicted by DLVO theory, whereas in the first case the bacterium is bound to the surface by tethers, which penetrate through the potential barrier predicted from DLVO theory.
A different approach toward understanding the adhesion process was taken by analyzing approach curves of S. aureus on hydrophobic surfaces (Thewes et al., 2015b). It has been observed that bacterial contact begins at about 50 nm above the substratum (Thewes et al., 2015b), with the aforementioned snap-in. In buffer solution, attractive forces over such large distances cannot be explained by DLVO forces between the bacterium and the substratum. The snap-in was more detailed by analyzing approach and retraction curves with varying negative force triggers, i.e., retraction starts at a certain distance above the substratum before the cell is in direct contact, in experiment and simulation (see Figure 1D). While for low and high absolute values of the force trigger the same rupture lengths were observed, the adhesion forces were larger for lower absolute values. In between, an unstable behavior with two types of retraction curves was observed. This stochasticity is not caused by difference of individual cells but—since the same cell is repeatedly used—reflects the internal stochasticity of the adhesion process. In particular, the curves with small force triggers displayed an initial attraction to the surface termed “pulling regime” even though the retraction already started.
To explain these observations, Thewes et al. (2015b) built a stochastic model that treats the bacterium as a hard, incompressible sphere decorated with elastic springs representing the cell wall macromolecules (see Figure 1E). One end of the springs is fixed to the bacterium, while the other end fluctuates thermally and interacts with the surface via an interaction potential. In order to mimic SCFS experiments, this sphere is connected to a cantilever, modeled as a spring, which moves toward/away from the surface. After each cantilever step, determined from the step size of the experimental piezo motor, a prescribed number of Monte Carlo (MC) steps is performed in order to incorporate thermal fluctuations. Afterward, the acting force, computed from the length of the connected springs and the deflection of the cantilever, is computed. The separation d of the sphere to the surface is then moved into the mechanical equilibrium position, such that the restoring force of the cantilever FC and the pulling force of the macromolecules FM cancel. The pulling force is generated only by macromolecules, which are in range of the interaction potential. That way the binding of individual macromolecules and the macroscopic movement of the cell are combined in a single model, which reproduces the experimentally observed behavior, namely the adhesion hysteresis, the snap-in event, and the behavior of retraction curves with varying negative force trigger. The model shows that for generating a snap-in, the distribution of spring constants is important, while the form of the interaction potential is not (Thewes et al., 2015b).
The model was extended by replacing the Hookean response of cell wall macromolecules to stretching by the more realistic worm-like chains (WLC) response and by reproducing a high number of experimental force–distance curves from many cells on hydrophilic and hydrophobic surfaces by MC simulations, the adhesion process to abiotic surfaces could be understood in more detail (Maikranz et al., 2020): On hydrophilic surfaces, cell wall macromolecules bind to the substratum (most likely by hydrogen bonds) after overcoming a potential barrier while on hydrophobic surfaces, the molecules tether via hydrophobic interactions without an energy barrier. This leads to rather strong adhesion via many molecules on hydrophobic surfaces and hence rather smooth force–distance curves (where WLC like signatures of single molecules detaching events define the rupture length as shown in the inset in Figure 2A), and to very “spiky,” stochastically varying force–distance curves and rather low adhesion force on hydrophilic surfaces (see Figure 2A) (Thewes et al., 2014; Maikranz et al., 2020).
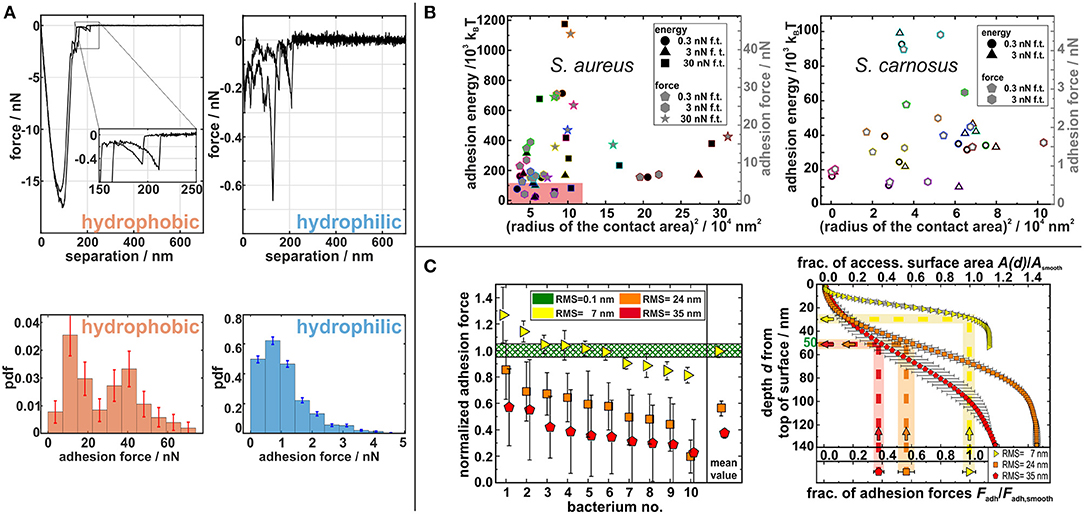
Figure 2. (A) Exemplary force–distance curves (upper row) and probability density function of adhesion forces (lower row) from SCFS with S. aureus cells (Maikranz et al., 2020). (B) Adhesion energy (black symbols) and adhesion force (gray symbols) of S. aureus (left) and S. carnosus (right) in dependence of their contact radius squared. The reddish rectangle displays the size of the complete right graph (Spengler et al., 2017). (C) (Left) Experimentally determined adhesion forces for 10 individual cells each on different rough substrata. (Right) Correlation of adhesion force and accessible surface area in dependence of the depth from the top of the rough surface (Spengler et al., 2019).
Ostvar and Wood (2016) introduced a similar model with individual macromolecules and heterogeneous mechanical responses, but without thermal fluctuations. The flexibility of the cantilever was not considered, and a plane–plane geometry was used: The bacterial cell wall is considered to have a certain roughness (approximately determined by AFM to be about 10–20 nm) that accounts for differing lengths of surface molecules. In the model, the surface molecules are represented by polymers that can either behave like Hookean springs or WLCs. At the end of each polymer, a bead is located that can directly bind to the surface via a Lennard–Jones potential. Upon retraction, every single polymer can either unbind by the bead escaping the potential. Using this model, retraction parts of experimental force–distance curves obtained with Staphylococcus epidermidis cells on glass substrata could be reproduced (Chen et al., 2011; Ostvar and Wood, 2016). In general, the model cannot produce a snap-in event due to the lack of a cantilever that allows the cell to suddenly approach the surface. Both models demonstrate that the adhesion process can be understood as the multi-scale interactions of heterogeneous macromolecules tethering to a surface.
For these models, the number of cell wall macromolecules that are able to bind to the substratum and also the exact knowledge of the cell wall area size that comes in contact with the surface is important. Spengler et al. (2017) investigated the size of this area, i.e., the area of the bacterial cell wall that contributes to the adhesion for S. aureus and S. carnosus cells: both strains have approximately the same (assumed to be circular) interaction area with radii of about 150–300 nm, although S. aureus cells adhere almost one order of magnitude stronger than S. carnosus cells. Even on the single species level, no correlation between the adhesion force and interaction area could be measured (see Figure 2B). In addition, the study demonstrated that the increase of the contact area with the applied force differs for different individual cells proving that the adhesion cannot be described by the Hertzian contact model.
As mentioned before, the knowledge about the interaction area and the thermal fluctuations can be used to describe bacterial adhesion to non-ideal surfaces (Spengler et al., 2019). It has been found that on nano-rough substrata, the adhesion force of S. aureus cells decreases with increasing roughness. The reduced adhesion forces can be directly linked to the decrease in accessible binding area for macromolecules that undergo thermal fluctuations of about 50 nm (see Figure 2C). The study also shows that the thermal fluctuation and hence adhesion can be understood mostly as a passive process: Although cells were killed during SCFS on these spiky surfaces, their adhesion force was not affected (Spengler et al., 2019).
Several approaches to describe and understand bacterial adhesion on unconditioned abiotic surfaces have been reviewed. Many studies demonstrate that traditional approaches to bacterial adhesion from colloid science and contact mechanics have limitations because adhesion, without external load, is primary mediated by the interaction of cell wall macromolecules with the substratum. The force response and stochastic length fluctuations of individual molecules determine the adhesive behavior. This leads to huge differences in adhesion forces of individual cells even within the same population. This mechanical heterogeneity inside a population can be important on the biofilm level, determining the colonization of small cavities, e.g., catheters.
A recent study has also shown that external factors, such as shear stresses, can change the molecules' force response and even “activate” adhesion (Dufrêne and Viljoen, 2020). The complexity caused by these divers mechanical responses is enhanced through the organization of adhesive molecules into patches, which were needed to interpret our own results (Spengler et al., 2021). These patches lead to a strong variation of the adhesion forces, depending on the contact area between patches and the substrate. In SCFS experiments, rotation of the bacteria is excluded, but typically not in the native setting. Reorientation of the bacteria could lead to more adherent areas coming into contact with the surface, which in principle could lead to stronger adhesion, especially on rough surfaces. Incorporating this and more detailed information, such as experimentally determined mechanical properties of cell wall macromolecules, their density and inhomogeneity (for example, in the division plane) are interesting directions for future research.
CS and EM reviewed and selected the content of the paper and wrote the manuscript. LS and KJ supervised the process, discussed the drafts, and helped in writing the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the German Research Foundation (DFG) within the collaborative research center SFB 1027 (Projects B1 and B2). CS and KJ were supported by the DFG-project number JA 905/6. KJ was also supported by the Max Planck School Matter to Life in Heidelberg, Germany.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors gratefully acknowledge funding from the above sources. The authors would also like to thank one of the reviewers for his or her detailed comments on contact mechanics, which helped to improve the manuscript.
1. ^Named after B. Derjaguin, L. D. Landau, E. Verwey, T. Overbeek (Derjaguin and Landau, 1941; Verwey, 1947).
2. ^Even experiments with minimal force triggers do not fully mimic flow chamber experiments since the bacterium is pushed through eventual energy barriers a planktonic bacterium would encounter.
3. ^Different deformation models from contact mechanics display hysteresis even without energy dissipation or plastic deformation (Goryacheva and Makhovskaya, 2001).
4. ^On hydrophilic surfaces, adhesion was too low to determine amplitudes.
Alam, F., Kumar, S., and Varadarajan, K. M. (2019). Quantification of adhesion force of bacteria on the surface of biomaterials: techniques and assays. ACS Biomater. Sci. Eng. 5, 2093–2110. doi: 10.1021/acsbiomaterials.9b00213
Alexander, T. E., Lozeau, L. D., and Camesano, T. A. (2019). QCM-D characterization of time-dependence of bacterial adhesion. Cell Surface 5:100024. doi: 10.1016/j.tcsw.2019.100024
Barthel, E., and Perriot, A. (2007). Adhesive contact to a coated elastic substrate. J. Phys. D Appl. Phys. 40:1059. doi: 10.1088/0022-3727/40/4/021
Berne, C., Ellison, C. K., Ducret, A., and Brun, Y. V. (2018). Bacterial adhesion at the single-cell level. Nat. Rev. Microbiol. 16, 616–627. doi: 10.1038/s41579-018-0057-5
Bhushan, B. (2017). Springer Handbook of Nanotechnology. Berlin; Heidelberg: Springer. doi: 10.1007/978-3-662-54357-3
Boks, N. P., Norde, W., van der Mei, H. C., and Busscher, H. J. (2008). Forces involved in bacterial adhesion to hydrophilic and hydrophobic surfaces. Microbiology 154, 3122–3133. doi: 10.1099/mic.0.2008/018622-0
Chen, Y., Busscher, H. J., van der Mei, H. C., and Norde, W. (2011). Statistical analysis of long- and short-range forces involved in bacterial adhesion to substratum surfaces as measured using atomic force microscopy. Appl. Environ. Microbiol. 77, 5065–5070. doi: 10.1128/AEM.00502-11
Chen, Y., Harapanahalli, A. K., Busscher, H. J., Norde, W., and van der Mei, H. C. (2014). Nanoscale cell wall deformation impacts long-range bacterial adhesion forces on surfaces. Appl. Environ. Microbiol. 80, 637–643. doi: 10.1128/AEM.02745-13
Chen, Y., Norde, W., van der Mei, H. C., and Busscher, H. J. (2012). Bacterial cell surface deformation under external loading. mBio 3:e00378-12. doi: 10.1128/mBio.00378-12
Ciavarella, M., Joe, J., Papangelo, A., and Barber, J. R. (2019). The role of adhesion in contact mechanics. J. R. Soc. Interface 16:20180738. doi: 10.1098/rsif.2018.0738
De Carvalho, C. C. (2007). Biofilms: recent developments on an old battle. Recent Pat. Biotechnol. 1, 49–57. doi: 10.2174/187220807779813965
Derjaguin, B., Muller, V., and Toporov, Y. (1975). Effect of contact deformations on the adhesion of particles. J. Colloid Interface Sci. 53, 314–326. doi: 10.1016/0021-9797(75)90018-1
Derjaguin, B. V., and Landau, L. (1941). Theory of the stability of strongly charged lyophobic sols and of the adhesion of strongly charged particles in solutions of electrolytes. Acta Physicochim. URSS 14, 633–662.
Dufrêne, Y. F., and Pelling, A. E. (2013). Force nanoscopy of cell mechanics and cell adhesion. Nanoscale 5, 4094–4104. doi: 10.1039/c3nr00340j
Dufrêne, Y. F., and Viljoen, A. (2020). Binding strength of gram-positive bacterial adhesins. Front. Microbiol. 11:1457. doi: 10.3389/fmicb.2020.01457
Dunne, W. M. (2002). Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15, 155–166. doi: 10.1128/CMR.15.2.155-166.2002
Fällman, E., Schedin, S., Jass, J., Andersson, M., Uhlin, B. E., and Axner, O. (2004). Optical tweezers based force measurement system for quantitating binding interactions: system design and application for the study of bacterial adhesion. Biosens. Bioelectron. 19, 1429–1437. doi: 10.1016/j.bios.2003.12.029
Filion-Côté, S., Melaine, F., Kirk, A. G., and Tabrizian, M. (2017). Monitoring of bacterial film formation and its breakdown with an angular-based surface plasmon resonance biosensor. Analyst 142, 2386–2394. doi: 10.1039/C7AN00068E
Goryacheva, I., and Makhovskaya, Y. Y. (2001). Adhesive interaction of elastic bodies. J. Appl. Math. Mech. 65, 273–282. doi: 10.1016/S0021-8928(01)00031-4
Greenwood, J. (1997). Adhesion of elastic spheres. Proc. R. Soc. Lond. A Math. Phys. Eng. Sci. 453, 1277–1297. doi: 10.1098/rspa.1997.0070
Hinterdorfer, P., and Dufrêne, Y. F. (2006). Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods 3, 347–355. doi: 10.1038/nmeth871
Jamal, M., Ahmad, W., Andleeb, S., Jalil, F., Imran, M., Nawaz, M. A., et al. (2018). Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 81, 7–11. doi: 10.1016/j.jcma.2017.07.012
Jasevičius, R., Baronas, R., and Kruggel-Emden, H. (2015). Numerical modelling of the normal adhesive elastic-plastic interaction of a bacterium. Adv. Powder Technol. 26, 742–752. doi: 10.1016/j.apt.2015.04.010
Jasevičius, R., and Kruggel-Emden, H. (2017). Numerical modelling of the sticking process of a S. aureus bacterium. Int. J. Adhes. Adhesiv. 77, 15–28. doi: 10.1016/j.ijadhadh.2017.03.015
Johnson, K. L., Kendall, K., and Roberts, A. (1971). Surface energy and the contact of elastic solids. Proc. R. Soc. Lond. A Math. Phys. Sci. 324, 301–313. doi: 10.1098/rspa.1971.0141
Keskin, D., Mergel, O., van der Mei, H. C., Busscher, H. J., and van Rijn, P. (2018). Inhibiting bacterial adhesion by mechanically modulated microgel coatings. Biomacromolecules 20, 243–253. doi: 10.1021/acs.biomac.8b01378
Krieg, M., Fläschner, G., Alsteens, D., Gaub, B. M., Roos, W. H., Wuite, G. J., et al. (2019). Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 1, 41–57. doi: 10.1038/s42254-018-0001-7
Loskill, P., Hähl, H., Thewes, N., Kreis, C. T., Bischoff, M., Herrmann, M., et al. (2012). Influence of the subsurface composition of a material on the adhesion of staphylococci. Langmuir 28, 7242–7248. doi: 10.1021/la3004323
Loskill, P., Pereira, P. M., Jung, P., Bischoff, M., Herrmann, M., Pinho, M. G., et al. (2014). Reduction of the peptidoglycan crosslinking causes a decrease in stiffness of the staphylococcus aureus cell envelope. Biophys. J. 107, 1082–1089. doi: 10.1016/j.bpj.2014.07.029
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370, 1198–1208. doi: 10.1056/NEJMoa1306801
Maikranz, E., Spengler, C., Thewes, N., Thewes, A., Nolle, F., Jung, P., et al. (2020). Different binding mechanisms of staphylococcus aureus to hydrophobic and hydrophilic surfaces. Nanoscale 12, 19267–19275. doi: 10.1039/D0NR03134H
Maugis, D. (1992). Adhesion of spheres: the JKR-DMT transition using a dugdale model. J. Colloid Interface Sci. 150, 243–269. doi: 10.1016/0021-9797(92)90285-T
Muller, V., Yushchenko, V., and Derjaguin, B. (1980). On the influence of molecular forces on the deformation of an elastic sphere and its sticking to a rigid plane. J. Colloid Interface Sci. 77, 91–101. doi: 10.1016/0021-9797(80)90419-1
Narenkumar, J., AlSalhi, M. S., Arul Prakash, A., Abilaji, S., Devanesan, S., Rajasekar, A., et al. (2019). Impact and role of bacterial communities on biocorrosion of metals used in the processing industry. ACS Omega 4, 21353–21360. doi: 10.1021/acsomega.9b02954
Ostvar, S., and Wood, B. D. (2016). Multiscale model describing bacterial adhesion and detachment. Langmuir 32, 5213–5222. doi: 10.1021/acs.langmuir.6b00882
Perni, S., Preedy, E. C., and Prokopovich, P. (2014). Success and failure of colloidal approaches in adhesion of microorganisms to surfaces. Adv. Colloid Interface Sci. 206, 265–274. Manuel G. Velarde. doi: 10.1016/j.cis.2013.11.008
Römling, U., Kjelleberg, S., Normark, S., Nyman, L., Uhlin, B. E., and Åkerlund, B. (2014). Microbial biofilm formation: a need to act. J. Intern. Med. 276, 98–110. doi: 10.1111/joim.12242
Schwermer, C. U., Lavik, G., Abed, R. M., Dunsmore, B., Ferdelman, T. G., Stoodley, P., et al. (2008). Impact of nitrate on the structure and function of bacterial biofilm communities in pipelines used for injection of seawater into oil fields. Appl. Environ. Microbiol. 74, 2841–2851. doi: 10.1128/AEM.02027-07
Sjollema, J., van der Mei, H. C., Hall, C. L., Peterson, B. W., de Vries, J., Song, L., et al. (2017). Detachment and successive re-attachment of multiple, reversibly-binding tethers result in irreversible bacterial adhesion to surfaces. Sci. Rep. 7:4369. doi: 10.1038/s41598-017-04703-8
Spengler, C., Glatz, B. A., Maikranz, E., Bischoff, M., Klatt, M. A., Santen, L., et al. (2021). The adhesion capability of S. aureus cells is heterogeneously distributed over the cell envelope. bioRxiv. doi: 10.1101/2021.01.05.425282
Spengler, C., Nolle, F., Mischo, J., Faidt, T., Grandthyll, S., Thewes, N., et al. (2019). Strength of bacterial adhesion on nanostructured surfaces quantified by substrate morphometry. Nanoscale 11, 19713–19722. doi: 10.1039/C9NR04375F
Spengler, C., Thewes, N., Jung, P., Bischoff, M., and Jacobs, K. (2017). Determination of the nano-scaled contact area of staphylococcal cells. Nanoscale 9, 10084–10093. doi: 10.1039/C7NR02297B
Stan, G., and Adams, G. G. (2016). Adhesive contact between a rigid spherical indenter and an elastic multi-layer coated substrate. Int. J. Solids Struct. 87, 1–10. doi: 10.1016/j.ijsolstr.2016.02.043
Tandogan, N., Abadian, P. N., Huo, B., and Goluch, E. D. (2017). “Characterization of bacterial adhesion and biofilm formation,” in Antimicrobial Coatings and Modifications on Medical Devices, eds Z. Zhang and V. E. Wagner (Cham: Springer), 67–95. doi: 10.1007/978-3-319-57494-3_3
Thewes, N., Loskill, P., Jung, P., Peisker, H., Bischoff, M., Herrmann, M., et al. (2014). Hydrophobic interaction governs unspecific adhesion of staphylococci: a single cell force spectroscopy study. Beilstein J. Nanotechnol. 5, 1501–1512. doi: 10.3762/bjnano.5.163
Thewes, N., Loskill, P., Spengler, C., Hümbert, S., Bischoff, M., and Jacobs, K. (2015a). A detailed guideline for the fabrication of single bacterial probes used for atomic force spectroscopy. Eur. Phys. J. E 38:140. doi: 10.1140/epje/i2015-15140-2
Thewes, N., Thewes, A., Loskill, P., Peisker, H., Bischoff, M., Herrmann, M., et al. (2015b). Stochastic binding of Staphylococcus aureus to hydrophobic surfaces. Soft Matter 11, 8913–9046. doi: 10.1039/C5SM00963D
van der Westen, R., Sjollema, J., Molenaar, R., Sharma, P. K., van der Mei, H. C., and Busscher, H. J. (2018). Floating and tether-coupled adhesion of bacteria to hydrophobic and hydrophilic surfaces. Langmuir 34, 4937–4944. doi: 10.1021/acs.langmuir.7b04331
Van Oss, C. (1995). Hydrophobicity of biosurfaces—origin, quantitative determination and interaction energies. Colloids Surf. B Biointerfaces 5, 91–110. doi: 10.1016/0927-7765(95)01217-7
Van Oss, C. J., Giese, R. F., and Costanzo, P. M. (1990). DLVO and non-DLVO interactions in hectorite. Clays Clay Miner. 38, 151–159. doi: 10.1346/CCMN.1990.0380206
Vissers, T., Brown, A. T., Koumakis, N., Dawson, A., Hermes, M., Schwarz-Linek, J., et al. (2018). Bacteria as living patchy colloids: phenotypic heterogeneity in surface adhesion. Sci. Adv. 4:eaao1170. doi: 10.1126/sciadv.aao1170
Keywords: bacterial adhesion, living colloids, (x)DLVO, tethering cell wall molecules, single-cell force spectroscopy, Monte Carlo simulation, Staphylococcus aureus
Citation: Spengler C, Maikranz E, Santen L and Jacobs K (2021) Modeling Bacterial Adhesion to Unconditioned Abiotic Surfaces. Front. Mech. Eng. 7:661370. doi: 10.3389/fmech.2021.661370
Received: 30 January 2021; Accepted: 10 March 2021;
Published: 13 May 2021.
Edited by:
Stanislav N. Gorb, University of Kiel, GermanyReviewed by:
Yulia Makhovskaya, Institute for Problems in Mechanics (RAS), RussiaCopyright © 2021 Spengler, Maikranz, Santen and Jacobs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ludger Santen, bC5zYW50ZW5AbXgudW5pLXNhYXJsYW5kLmRl; Karin Jacobs, ay5qYWNvYnNAcGh5c2lrLnVuaS1zYWFybGFuZC5kZQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.