
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mech. Eng., 18 June 2020
Sec. Micro- and Nanoelectromechanical Systems
Volume 6 - 2020 | https://doi.org/10.3389/fmech.2020.00040
Collecting samples for diagnosis of respiratory infections can be invasive and highly uncomfortable for patients; with sampling techniques ranging from nasal or throat swabs to bronchoalveolar lavage. In this work, we explore the electrostatic capture of exhaled pathogens as a non-invasive sampling method for rapid diagnosis in a primary care setting. A pilot study at primary care centers in Belgium enrolled 20 patients presenting with flu-like symptoms whose diagnosis was determined by nasopharyngeal (NP) swabs. We collected exhaled aerosol particles from infected patients using a breath electrostatic sampler (BESS) and using filters, following an extremely light procedure consisting of five normal exhales. All samples were analyzed using a commercial multiplex q-RT-PCR panel assay. Staphylococcus aureus (S. aureus) was detected from BESS samples of seven patients; three of whom were in agreement with their corresponding diagnoses. The BESS method was negative for exhaled viruses. We detected viruses, but no bacteria, with the filters, but the results showed no correlation with the corresponding diagnosis. Our results indicate that electrostatic sampling of exhaled breath is a technique and approach potentially suited for the primary care setting, where it might constitute a helpful diagnostic device.
Exhaled breath analysis is a developing field with the possibility to provide individualized and non-invasive disease diagnostics. Traditionally, diagnosis of exhaled breath only considered the analysis of volatile organic compounds (VOCs) and gases. During the last 25 years, breath research has expanded beyond gas-phase analysis to include expelled aerosols containing non-volatile compounds, such as lipids, proteins, DNA and bacteria (Pleil et al., 2018). Much effort has been put into analyzing these non-volatile biomarkers, mostly using condensation capture methods (Davis et al., 2012), but often relying on detecting a set of various volatile biomarkers to determine the disease state of a patient. With the emergence of cheap, highly sensitive and miniaturized molecular diagnostic technologies, an interesting alternative is the capture and direct detection of specific disease-causing pathogens in exhaled aerosol (Brankston et al., 2007; Fabian et al., 2011; Johnson et al., 2011). Such tests could prove extremely helpful in the case of epidemics, such as previous Influenza (Yan et al., 2018), Severe Acute Respiratory Syndromes (SARS) (Booth et al., 2005; Chowell et al., 2015) or Corona virus outbreaks (Santarpia et al., 2020).
To capture aerosolized pathogens from exhaled breath, several techniques have emerged (Grinshpun et al., 1996; Fung and Mykhaylova, 2013): impactors (Fennelly et al., 2004; Wainwright et al., 2009; Gralton et al., 2013), filters (Fabian et al., 2008; McDevitt et al., 2013; Hatagishi et al., 2014), liquid impingers (Lindsley et al., 2010) and condensators (Xu et al., 2012; Zheng et al., 2018). An alternative method for sampling aerosols has yet to be considered more thourgouhly: electrostatic sampling (Parker, 2003). Electrostatic sampling has the advantages of handling high-volume air flows, compatible with human exhaled stream, and has demonstrated high capture efficiencies (>90% for particles larger than 1 μm; Parker, 2003). Compared to impaction devices, electrostatic samplers have a lower impaction velocity (Mainelis, 1999; Mainelis et al., 2002) and pressure drop, which could play a role in preserving the integrity of pathogens. Furthermore, compared to liquid impingers, electrostatic samples have demonstrated 5–10 times higher concentration output (Yao et al., 2009; Wei et al., 2014; Rufino de Sousa et al., 2020). There is potential to develop this technology for clinical use as an alternative sampling technique for exhaled breath aerosols (Han and Mainelis, 2008; Kettleson et al., 2009; Miller et al., 2010; Christensen et al., 2011; Tan et al., 2011; Lancereau et al., 2013); however, we found no reports yet of direct exhaled breath sampling using electrostatic methods.
In this work, we performed a clinical pilot study in which we electrostatically captured breath aerosols from infected patients at a point-of-care setting (Figure 1). Additionally, we aimed to validate the electrostatic sampling device for the capture of pathogens from exhaled breath. We adapted a previously developed electrostatic sampler for in-vitro capture of aerosolized influenza virus (Pardon et al., 2015; Ladhani et al., 2017) into a Breath ElectroStatic Sampler (BESS) for patient use. We report, to our knowledge, the first investigation of breath aerosol sampling using an electrostatic sampler.
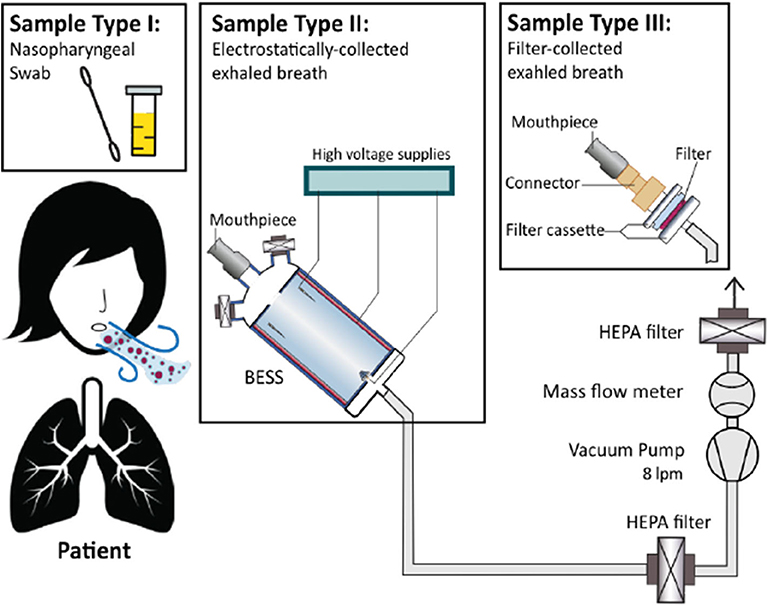
Figure 1. Experimental setup and study design. Three sample types were collected from each patient: a nasopharyngeal (NP) swab, breath aerosol sample using the BESS device, and a breath aerosol sample using a gelatin filter.
We conducted the study at primary care centers in Antwerp, Belgium. We obtained ethical approval from the committee of the University Hospital Antwerp (UZA) and the University of Antwerp, Eudract number 2015-005724-26. Patients presenting with flu-like symptoms were referred to the study team by their family physician. After an explanation of the study, we asked the patients for their written consent, and three types of samples were collected: (1) a nasopharyngeal (NP) swab sample, (2) a breath aerosol sample using the BESS, and (3) a breath aerosol sample using a gelatin filter. Diagnosis was established from analysis of NP swabs. Filter collection was employed as a control for breath sampling, due to its widely accepted use and its excellent collection efficiency.
The BESS is a multi-point-to-plane electrostatic precipitator contained within a hollow 3D-printed cylindrical housing (Figure 2). The multi-point discharge electrode is composed of five tungsten needles placed cylindrically around the aerosol inlet. The collector electrode is composed of an electrically-connected 200 μL liquid droplet housed within a disposable 3D-printed cage collector, placed 5 cm downstream of the discharge needles. A third shielding electrode lines the internal wall of the cylindrical housing and is composed of a sheet of stainless steel. We applied a DC electrostatic potential of −7, +2.8, and −4 kV, respectively, to the discharge, collector, and wall electrodes. The high electrical potential applied at the sharp needle induces a corona discharge which ionizes the surrounding air. These ions are attracted by electrostatic force toward the electrode of opposite electrical charge, here the liquid collector, creating an electrohydrodynamic wind. Airborne particles traveling through this ion stream become electrically charged by collision with the charged ions, thereby also experiencing the effect of electrical force toward the liquid collector electrode. The third shielding wall electrode serves the purpose of concentrating the electrical field line toward the collector electrode, while simultaneously avoiding uncontrolled electrostatic charging of the wall surface, which may otherwise attenuate the discharge and electrostatic precipitation onto the collector electrode. We fitted two disposable HEPA filters and a disposable mouthpiece at the inlet and connected the outlet to further HEPA filters and a vacuum pump. These extra inlets allow for creating a sheath flow, which help creating a more controlled airflow stream in the device, concentrating the exhaled breath in the center, and preventing direct contact between the discharge electrode and the exhaled breath.
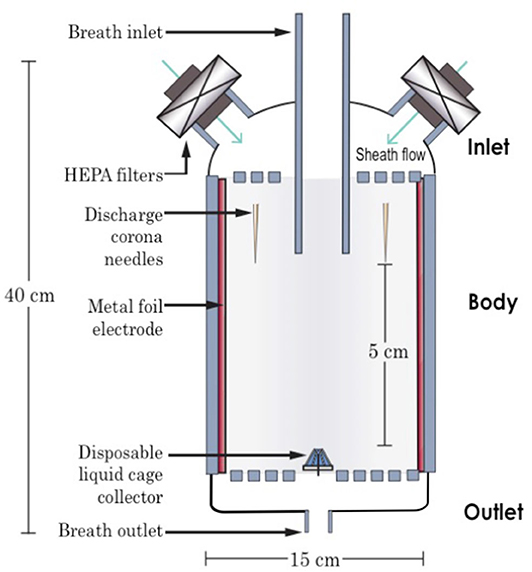
Figure 2. Cross-sectional schematic of the breath electrostatic sampler (BESS). The device consists of a breath inlet, breath outlet, and a body housing a multi-point-to-plane electrostatic precipitator, along with a disposable liquid collector.
To limit the experimental burden on the sick patients and minimize sample collection time, we restricted the breathing maneuver to five slow exhales. Breath aerosols were sampled first with the BESS, within a liquid collector loaded with a 200 μL droplet of universal transport medium (UTM) (COPAN, U.S.A.); and next with a 25 mm gelatine filter (SKC Inc.), placed inside its cassette holder. Please refer to Supplementary Material for details of the experimental setup and methods. We stored all samples at 4°C until the end of each day, after which we transferred them to the UZA hospital lab where we stored them at −80°C until analysis was performed.
We prescreened the diagnoses of all 20 patients by analyzing their NP swabs using the FTD Respiratory pathogens 21 plus panel (Fast Track diagnostics, Luxembourg), see Supplementary Table 1 for a list of the pathogens and their limits of detection. Breath samples from BESS and filters were then analyzed using the FTD panel according the manufacturer's instructions.
Prescreening of 20 patients via their NP swabs revealed the following diagnoses: 1 with InfA, 5 with Inf B, 2 with Inf B and S. aur, 1 with CoOC43, 2 with CoOC43 and S. aur, 1 with CoHKU1 and adenovirus, 2 with rhinovirus, and 4 with no pathogens detected. The remaining two patients had positive samples from breath but not swab. Four patients had multiple diagnoses, i.e., multiple pathogens were detected from their NP swabs. Please refer to Supplementary Table 1 for detailed Ct values. Unfortunately, after this point, breath samples from 12 patients were discarded due to a handling error. We tested the remaining eight patient breath samples using the FTD panel, see Table 1.
Analysis of BESS samples indicated seven patients who showed positive for S. aureus: 2 who had no diagnoses (BE110 and BE115); 3 diagnosed with S. aureus (BE116, BE117, and BE043), and; 2 diagnosed with HRV (BE047 and BE058). No viruses were detected.
Analysis of BESS samples indicated seven patients who showed positive for S. aureus: 2 who had no diagnoses (BE110 and BE115); 3 diagnosed with S. aureus (BE116, BE117, and BE043), and; 2 diagnosed with HRV (BE047 and BE058). No viruses were detected.
This pilot study provides a proof-of-concept for electrostatic sampling at the point-of-care and investigates this type of device for capturing exhaled aerosols, a previously unattempted sampling method for breath. Despite the limited study size (n = 8) of patient samples tested, we discern a few potential trends. Combining the BESS with downstream qPCR analysis, we were able to non-invasively detect Staphylococcus aureus bacteria from the exhaled breath of patients. Additionally, filter-collected exhaled breath samples indicate capture of coronavirus and rhinovirus. From the ten positive breath samples detected, three were in agreement with their corresponding patient diagnosis, all captured using the BESS. These results indicate the BESS as a potentially non-invasive sampling device for bacterial detection. The 30% agreement between breath and swab samples is corroborated by Zheng et al. (2018), who find a 35% agreement between their breath samples and throat swabs [where analysis is made using the loop mediated isothermal amplification (LAMP) method].
The capture of S. aureus from exhaled breath with the BESS device but not filters could be attributed either to low recovery efficiencies for S. aureus from the filters, as reported elsewhere (Chang and Wang, 2015); or to sources of error such as contamination during sample handling or collection. S. aureus is a known commensal organism, thus prone to spread through contact easily.
The size-dependent collection efficiency of electrostatic samplers, specifically their reduced efficiency for sub-micron particles (Parker, 2003), may explain the lack of virus capture by the BESS. S. aureus with size ~1 μm (Monteiro et al., 2015) are thus more likely to be captured, rather than viruses whose sizes range from 20 to 500 nm. Future improvements could involve increased collection efficiency of the BESS for submicron particles by integrating soft x-rays (Kulkarni et al., 2002) or a non-thermal plasma pre-charger (Thonglek and Kiatsiriroat, 2014). This will require further technical development and testing.
The differences in virus presence between the filter and the swab samples may indicate a more significant presence of viruses in the nasopharynx vs. airways. Unfortunately, we could not repeat the analysis due to the limited amount of sample available.
The differences in the presence of S. aureus between breath and swab samples may be attributed to either the previously mentioned contamination, poor swabbing techniques, or low bacterial loads present in the nasal passages vs. airways.
The presence of pathogens in breath aerosols were expected to occur in minimal amounts, based on previous investigations. Fabian et al. (2008) captured influenza from breath, detecting as few as 3–20 RNA cp/min of breathing; while Milton et al. (2013) demonstrated minimum 21 cp/30 min of breathing and 10× coughing. Thus, it is crucial to consider the limit of detection (LoD) of not only the analysis technique (qPCR in this work) but also the breath sampling methods (filters or the BESS in this work) to ensure detection of these low viral loads. The FTD panel assay used for sample analysis features an at-best LoD of 102–105 cp/mL, refer to Supplementary Table 3 for LoDs of each pathogen. Minimizing the collection volume of the BESS would reduce sample dilution and deliver a potentially lower LoD. The BESS device, with a sample volume of 230 μL and assuming 100% capture efficiency, is potentially 6.5 times more sensitive than filters, which are diluted into 1.5 mL of buffer. Together, the BESS + FTD sampling+detection scheme offers a theoretical LoD of 23–104 copies while the filter + FTD scheme offers a theoretical LoD of 150–105 copies. Considering a viral load of Influenza in breath of 20 cp/min (Fabian et al., 2008), these schemes would require minimum 1 min breathing time into the BESS device, but 7 min of breathing time into filters, providing further motivation for considering the BESS device for sampling breath aerosols.
PCR-based assays, although relatively sensitive, might not form the best detection strategy. Alternative molecular methods could be considered, such as targetting nucleoproteins of virus capsids, which have a higher copy-number as compared with nucleic acids (Leirs et al., 2016) to push the limits of the technique and potentially yield an even lower LoD (Decrop et al., 2017).
Table 2 summarizes results from previous studies, which emphasize the minute amounts of pathogens present in exhaled breath (Fennelly et al., 2004; Huynh et al., 2007; Fabian et al., 2008; Stelzer-Braid et al., 2009; Lindsley et al., 2010; St George et al., 2010). Moreover, results can vary significantly depending on the day of illness onset, and from patient to patient. The substantial variation of breathing/coughing maneuvers and analysis methods utilized in these studies creates difficulties in comparing results and establishing conclusive outcomes.
The sparse datasets available on pathogen loads in breath, differences in breathing maneuvers, and the small population sizes in the studies emphasize the need for further in-depth and large-scale clinical studies to understand the pathophysiology of respiratory pathogens.
The aim of our study was not to demonstrate the potency of direct pathogen sampling for exhaled breath, but rather to explore the clinical potential of our approach. Our results and that of others indicate that exhaled breath as a sampling medium is yet to be better explored. With the emergence of novel technologies enable sampling with higher efficiencies, a better understanding of the benefits of breathing maneuver, and more sensitive detection technologies, we may discover areas in which exhaled breath may represent a useful sample source for non-invasive diagnostic testing. However, more work is yet needed before to be able to draw such conclusions.
We demonstrate, for the first time, the use of electrostatic-based methods for the capture of breath aerosols from patients at primary care centers. Our BESS device, particularly adapted for point-of-care settings, demonstrated capture of S. aureus from the breath aerosols of patients with a 30% agreement to patient diagnosis, whereas filters captured exhaled viruses. Discrepancies were observed between the two breath sampling methods and between breath and nasopharyngeal swab samples, indicating the need for further investigations. A review of previous studies indicates the exceptionally low pathogen loads available in exhaled breath, indicating the need for further investigations and large-scale studies. To address this low pathogen load, we suggest optimizing patient breathing maneuvers and targeting viral proteins, rather than nucleic acids, to achieve a sensitive sampling-detection scheme.
Breath sampling presents a promising non-invasive solution for studying transmission or pathophysiology of lower respiratory tract infections; and further research is required to potentially allow reliable detection of respiratory pathogens using electrostatic breath samplers.
All datasets generated for this study are included in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Commissie voor Medische Ethiek UZA/UAntwerpen. The patients/participants provided their written informed consent to participate in this study.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by the Innovative Medicines Initiative (IMI), a public-private partnership between the European Union, and the European Federation of Pharmaceutical Industries and Associations as part of the RAPP-ID project (grant agreement number 115153).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the patients who participated in this study, their physicians, as well as the primary care centers around Antwerp for hosting the study team. We would also like to acknowledge the laboratory technicians at the University of Antwerp for their help in analyzing patient samples.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmech.2020.00040/full#supplementary-material
Booth, T. F., Kournikakis, B., Bastien, N., Ho, J., Kobasa, D., Stadnyk, L., et al. (2005). Detection of airborne severe acute respiratory syndrome (SARS) coronavirus and environmental contamination in SARS outbreak units. J. Infect. Dis. 191, 1472–1477. doi: 10.1086/429634
Brankston, G., Gitterman, L., Hirji, Z., Lemieux, C., and Gardam, M. (2007). Transmission of influenza A in human beings. Lancet Infect. Dis. 7, 257–265. doi: 10.1016/S1473-3099(07)70029-4
Chang, C. W., and Wang, L.-J. (2015). Impact of culture media and sampling methods on Staphylococcus aureus aerosols. Indoor Air. 25, 488–498. doi: 10.1111/ina.12162
Chowell, G., Abdirizak, F., Lee, S., Lee, J., Jung, E., Nishiura, H., et al. (2015). Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 13, 1–12. doi: 10.1186/s12916-015-0450-0
Christensen, L. S., Brehm, K. E., Skov, J., Harlow, K. W., Christensen, J., and Haas, B. (2011). Detection of foot-and-mouth disease virus in the breath of infected cattle using a hand-held device to collect aerosols. J. Viral Methods 177, 44–48. doi: 10.1016/j.jviromet.2011.06.011
Davis, M. D., Montpetit, A., and Hunt, J. (2012). Exhaled breath condensate: an overview. Immunol. Allergy Clin. North Am. 32, 363–375. doi: 10.1016/j.iac.2012.06.014
Decrop, D., Pardon, G., Brancato, L., Kil, D., Zandi Shafagh, R., Kokalj, T., et al. (2017). Single-step imprinting of femtoliter microwell arrays allows digital bioassays with attomolar limit of detection. ACS Appl. Mater. Interfaces 9, 10418–10426. doi: 10.1021/acsami.6b15415
Fabian, P., Brain, J., Houseman, E. A., Gern, J., and Milton, D. K. (2011). Origin of exhaled breath particles from healthy and human rhinovirus-infected subjects. J. Aerosol Med Pulm Drug Deliv. 24, 137–147. doi: 10.1089/jamp.2010.0815
Fabian, P., McDevitt, J. J., DeHaan, W. H., Fung, R. O. P., Cowling, B. J., Chan, K. H., et al. (2008). Influenza virus in human exhaled breath: an observational study. PLoS ONE 3:e2691. doi: 10.1371/journal.pone.0002691
Fennelly, K. P., Martyny, J. W., Fulton, K. E., Orme, I. M., Cave, D. M., and Heifets, L. B. (2004). Cough-generated aerosols of Mycobacterium tuberculosis: a new method to study infectiousness. Am. J. Resp. Crit. Care 169, 604–609. doi: 10.1164/rccm.200308-1101OC
Fung, A. O., and Mykhaylova, N. (2013). Analysis of airborne biomarkers for point-of-care diagnostics. J. Lab Autom. 19, 225–247. doi: 10.1177/2211068213517119
Gralton, J., Tovey, E. R., McLaws, M.-L., and Rawlinson, W. D. (2013). Respiratory virus RNA is detectable in airborne and droplet particles. J. Med. Virol. 85, 2151–2159. doi: 10.1002/jmv.23698
Grinshpun, S. A., Willeke, K., Ulevicius, V., Donnelly, J., Lin, X., and Mainelis, G. (1996). Collection of airborne microorganisms: advantages and disadvantages of different methods. J. Aerosol. Sci. 27, S247–S248. doi: 10.1016/0021-8502(96)00196-6
Han, T., and Mainelis, G. (2008). Design and development of an electrostatic sampler for bioaerosols with high concentration rate. J. Aerosol. Sci. 39, 1066–1078. doi: 10.1016/j.jaerosci.2008.07.009
Hatagishi, E., Okamoto, M., Ohmiya, S., Yano, H., Hori, T., Saito, W., et al. (2014). Establishment and clinical applications of a portable system for capturing influenza viruses released through coughing. PLoS ONE 9:e103560. doi: 10.1371/journal.pone.0103560
Huynh, K. N., Oliver, B. G., Stelzer, S., Rawlinson, W. D., and Tovey, E. R. (2007). A new method for sampling and detection of exhaled respiratory virus aerosols. Clin. Infect. Dis 46, 93–95. doi: 10.1086/523000
Johnson, G. R., Morawska, L., Ristovski, Z. D., Hargreaves, M., Mengersen, K., Chao, C. Y. H., et al. (2011). Modality of human expired aerosol size distributions. J. Aerosol Sci. 42, 839–851. doi: 10.1016/j.jaerosci.2011.07.009
Kettleson, E. M., Ramaswami, B., Hogan, C. J. Jr., Lee, M.-H., Statyukha, G. A., Biswas, P., et al. (2009). Airborne virus capture and inactivation by an electrostatic particle collector. Environ. Sci. Technol. 43, 5940–5946. doi: 10.1021/es803289w
Kulkarni, P., Namiki, N., Otani, Y., and Biswas, P. (2002). Charging of particles in unipolar coronas irradiated by in-situ soft X-rays: enhancement of capture efficiency of ultrafine particles. J. Aerosol. Sci. 33, 1279–1296. doi: 10.1016/S0021-8502(02)00067-8
Ladhani, L., Pardon, G., Meeuws, H., Van Wesenbeeck, L., Schmidt, K., Stuyver, L., et al. (2017). Sampling and detection of airborne influenza virus towards point-of-care applications. PLoS ONE 12:e0174314. doi: 10.1371/journal.pone.0174314
Lancereau, Q., Roux, J.-M., and Achard, J.-L. (2013). Influence of secondary flows on the collection efficiency of a cylindrical electrostatic precipitator. J. Aerosol. Sci. 63, 146–160. doi: 10.1016/j.jaerosci.2013.05.002
Leirs, K., Tewari Kumar, P., Decrop, D., Pérez-Ruiz, E., Leblebici, P., Van Kelst, B., et al. (2016). Bioassay development for ultrasensitive detection of influenza a nucleoprotein using digital ELISA. Anal. Chem. 88, 8450–8458. doi: 10.1021/acs.analchem.6b00502
Lindsley, W. G., Blachere, F. M., Beezhold, D. H., Thewlis, R. E., Noorbakhsh, B., Othumpangat, S., et al. (2016). Viable influenza a virus in airborne particles expelled during coughs versus exhalations. Influenza Other Respir. Viruses 10, 404–413. doi: 10.1111/irv.12390
Lindsley, W. G., Blachere, F. M., Thewlis, R. E., Vishnu, A., Davis, K. A., Cao, G., et al. (2010). Measurements of airborne influenza virus in aerosol particles from human coughs. PLoS ONE 5:e15100. doi: 10.1371/journal.pone.0015100
Mainelis, G. (1999). Collection of airborne microorganisms by electrostatic precipitation. Aerosol. Sci. Technol. 30, 127–144. doi: 10.1080/027868299304732
Mainelis, G., Adhikari, A., Willeke, K., Lee, S. A., Reponen, T., and Grinshpun, S. A. (2002). Collection of airborne microorganisms by a new electrostatic precipitator. J. Aerosol. Sci. 33, 1417–1432. doi: 10.1016/S0021-8502(02)00091-5
McDevitt, J. J., Koutrakis, P., Ferguson, S. T., Wolfson, J. M., Fabian, M. P., Martins, M., et al. (2013). Development and performance evaluation of an exhaled-breath bioaerosol collector for influenza virus. Aerosol. Sci. Technol. 47, 444–451. doi: 10.1080/02786826.2012.762973
Miller, A., Frey, G., King, G., and Sunderman, C. (2010). A handheld electrostatic precipitator for sampling airborne particles and nanoparticles. Aerosol. Sci. Technol. 44, 417–427. doi: 10.1080/02786821003692063
Milton, D. K., Fabian, M. P., Cowling, B. J., Grantham, M. L., and McDevitt, J. J. (2013). Influenza virus aerosols in human exhaled breath: particle size, culturability, and effect of surgical masks. PLoS Pathog. 9:e1003205. doi: 10.1371/journal.ppat.1003205
Monteiro, J. M., Fernandes, P. B., Vaz, F., Pereira, A. R., Tavares, A. C., Ferreira, M. T., et al. (2015). Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 6, 8055–8012. doi: 10.1038/ncomms9055
Pardon, G., Ladhani, L., Sandstrom, N., Ettori, Maxime, L. G., and van der Wijngaart, W. (2015). Aerosol sampling using an electrostatic precipitator integrated with a microfluidic interface. Sensor Actuat. B Chem. 212, 344–352. doi: 10.1016/j.snb.2015.02.008
Parker, K. (2003). Electrical Operation of Electrostatic Precipitators, 1st Edn. London, UK: Institution of Engineering and Technology.
Pleil, J. D., Wallace, M. A. G., and Madden, M. C. (2018). Exhaled breath aerosol (EBA): the simplest non-invasive medium for public health and occupational exposure biomonitoring. J. Breath Res. 12:027110. doi: 10.1088/1752-7163/aa9855
Rufino de Sousa, N., Sandstrom, N., Shen, L., Håkansson, K., Vezozzo, R., Udekwu, K. I., et al. (2020). A fieldable electrostatic air sampler enabling tuberculosis detection in bioaerosols. Tuberculosis 120:101896. doi: 10.1016/j.tube.2019.101896
Santarpia, J. L., Rivera, D. N., Herrera, V., Morwitzer, M. J., Creager, H., Santarpia, G. W., et al. (2020). Transmission potential of SARS-CoV-2 in viral shedding observed at the University of Nebraska Medical Center. medRxiv. doi: 10.1101/2020.03.23.20039446
St George, K., Fuschino, M. E., Mokhiber, K., Triner, W., and Spivack, S. D. (2010). Exhaled breath condensate appears to be an unsuitable specimen type for the detection of influenza viruses with nucleic acid-based Methods. J. Virol Methods 163, 144–146. doi: 10.1016/j.jviromet.2009.08.019
Stelzer-Braid, S., Oliver, B. G., Blazey, A. J., Argent, E., Newsome, T. P., Rawlinson, W. D., et al. (2009). Exhalation of respiratory viruses by breathing, coughing, and talking. J. Med. Virol. 81, 1674–1679. doi: 10.1002/jmv.21556
Tan, M., Shen, F., Yao, M., and Zhu, T. (2011). Development of an automated electrostatic sampler (AES) for bioaerosol detection. Aerosol. Sci. Technol. 45, 1154–1160. doi: 10.1080/02786826.2011.582193
Thonglek, V., and Kiatsiriroat, T. (2014). Agglomeration of sub-micron particles by a non-thermal plasma electrostatic precipitator. J. Electrostat. 72, 33–38. doi: 10.1016/j.elstat.2013.11.006
Wainwright, C. E., France, M. W., O'Rourke, P., Anuj, S., Kidd, T. J., Nissen, M. D., et al. (2009). Cough-generated aerosols of Pseudomonas aeruginosa and other Gram-negative bacteria from patients with cystic fibrosis. Thorax 64, 926–931. doi: 10.1136/thx.2008.112466
Wei, K., Zou, Z., and Yao, M. (2014). Charge levels and Gram (±) fractions of environmental bacterial aerosols. J. Aerosol. Sci. 74, 52–62. doi: 10.1016/j.jaerosci.2014.04.002
Xu, Z., Shen, F., Li, X., Wu, Y., Chen, Q., Jie, X., et al. (2012). Molecular and microscopic analysis of bacteria and viruses in exhaled breath collected using a simple impaction and condensing method. PLoS ONE 7:e41137. doi: 10.1371/journal.pone.0041137
Yan, J., Grantham, M., Pantelic, J., Bueno de Mesquita, P. J., Albert, B., Liu, F., et al. (2018). Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc. Nat. Acad. Sci. U. S. A. 115, 1081–1086. doi: 10.1073/pnas.1716561115
Yao, M., Zhang, H., Dong, S., Zhen, S., and Chen, X. (2009). Comparison of electrostatic collection and liquid impinging methods when collecting airborne house dust allergens, endotoxin and (1,3)-beta-d-glucans. J. Aerosol. Sci. 40, 492–502. doi: 10.1016/j.jaerosci.2009.02.002
Keywords: exhaled breath aerosol, electrostatic sampling, point-of-care test, non-invasive diagnostics, respiratory infectious diseases
Citation: Ladhani L, Pardon G, Moons P, Goossens H and van der Wijngaart W (2020) Electrostatic Sampling of Patient Breath for Pathogen Detection: A Pilot Study. Front. Mech. Eng. 6:40. doi: 10.3389/fmech.2020.00040
Received: 10 January 2020; Accepted: 18 May 2020;
Published: 18 June 2020.
Edited by:
Ellis Meng, University of Southern California, Los Angeles, United StatesReviewed by:
Marwan Nafea, University of Nottingham Malaysia Campus, MalaysiaCopyright © 2020 Ladhani, Pardon, Moons, Goossens and van der Wijngaart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wouter van der Wijngaart, d291dGVyQGt0aC5zZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.