- 1ESI Group Chair @ PIMM Lab, ENSAM Institute of Technology, Paris, France
- 2ESI Group Chair @ LAMPA Lab, ENSAM Institute of Technology, Paris, France
- 3CNRS@CREATE Ltd, Singapore, Singapore
- 4ESI Group, Paris, France
In the context of parametric surrogates, several nontrivial issues arise when a whole curve shall be predicted from given input features. For instance, different sampling or ending points lead to non-aligned curves. This also happens when the curves exhibit a common pattern characterized by critical points at shifted locations (e.g., in mechanics, the elastic-plastic transition or the rupture point for a material). In such cases, classical interpolation methods fail in giving physics-consistent results and appropriate pre-processing steps are required. Moreover, when bifurcations occur into the parametric space, to enhance the accuracy of the surrogate, a coupling with clustering and classification algorithms is needed. In this work we present several methodologies to overcome these issues. We also exploit such surrogates to quantify and propagate uncertainty, furnishing parametric stastistical bounds for the predicted curves. The procedures are exemplified over two problems in Computational Mechanics.
1 Introduction
In a large variety of engineering applications, parametric surrogates are thoroughly powerful tools (Simpson et al., 2001; Prud’homme et al., 2002; Audouze et al., 2013; Mainini and Willcox, 2015; Hesthaven et al., 2016; Benner et al., 2020a). They allow a real-time monitoring and control of the most relevant physical quantities describing a given phenomenon. Moreover, they empower smart decision making, optimizing time and manufacturing costs. The uncertainty propagation in such models is also fundamental to operate efficiently in diagnosis and prognosis. In a non-intrusive framework, given an engineering problem, a Design of Experiments–DoE–based on the problem parameters is established and the corresponding responses of the system are collected into databases, which are used as training data to build the surrogate model via Machine Learning–ML–and Model Order Reduction–MOR–algorithms (Wang and Shan, 2007; Benner et al., 2015; Hesthaven and Ubbiali, 2018; Rajaram et al., 2020; Franchini et al., 2022; Khatouri et al., 2022). Such responses are usually the ensemble of several Quantities of Interest–QoI–observed, for instance, over time (i.e., time series) and can come both from experiments and numerical simulations. Therefore, each QoI is usually a curve, discretized according to the number of sampling points. This is the case when, for example, a material is tested and the force-displacement curve is extracted for different parameters p defining the material itself. It is also the case when a sensor placed on a mould records the pressure evolution during the mould filling from a resin injected into a mould. In this paper, we propose several strategies to build parametric curves, illustrating the procedure over two applications in computational solid mechanics.
The target quantities representing the system response are univariate functions, depending on d features (parameters), that is
where
Our procedure mainly consists in the application of non-intrusive nonlinear regressions based on the sparse Proper Generalized Decomposition–sPGD– (Chinesta et al., 2011; Borzacchiello et al., 2017; Ibáñez et al., 2018; Sancarlos et al., 2021), these being efficient under the scarce data availability constraint. Indeed, in real engineering applications, when dealing with simulation-based metamodels, data availability is largely limited by the complexity of the Finite Element–FE–computations. From the High-Fidelity–HiFi–offline simulations, it is often possible to define a Reduced Order Model–ROM–, for instance, by extracting the most relevant Proper Orthogonal Decomposition–POD–modes from the training data (Raghavan et al., 2013; Fareed and Singler, 2019). Consequently, since the curve can be expressed into the extracted POD reduced basis through a set of weighting coefficients, the nonlinear regressions can be applied to predict such coefficients. A similar workflow is applied by Gooijer et al. (2021), where the POD-based surrogate models employ Radial Basis Function–RBF–interpolations. For the sake of completeness, it shall be noticed that the use of POD-based interpolations–PODI–has several limitations and drawbacks, particularly when dealing with non-linear solution manifolds. To alleviate such issues, several works have been conducted in the framework of interpolations on Grassmann manifolds and its tangent space, improving the model robustness over the parametric space (Amsallem and Farhat, 2008; Mosquera et al., 2018, 2021; Friderikos et al., 2020, 2022).
However, ad-hoc physics-based data pre-processing is a fundamental step to be embedded in the procedure. Indeed, when different choices of the parameters carry radically different physical behaviours, the interpolation in the parametric space can lead to nonphysical solutions. In such cases, separate regression sub-models are built, requiring the coupling with some clustering and classification algorithms, leading to the so-called multi-regression strategy.
Another non-trivial issue comes when the curves exhibit a common pattern characterized by some critical points resulting from a change in the physical behaviour. Indeed, a shift among the locations of such critical points in the different curves would cause nonphysical results when employing a classical interpolation. To overcome this matter, we propose a parametrization of the curves accounting for the locations of such critical points and allowing a curve alignment prior to the interpolation.
The main points addressed in this work are:
1. the parametric modeling of a quantity of interest using advanced sparse nonlinear regressions;
2. the parametric modeling of a curve where a data alignement is needed;
3. the statistical parametric modeling based on a parametrized physical model;
4. the statistical parametric model learned from scarce data (measurements);
5. and, finally, the concept of data clustering to overcome bifurcations in the parametric space.
The paper is structured as follows. Section 2 is mainly a review of some well-known techniques, excepting SubSection 2.3 which illustrates the implementation of the sPGD algorithm for the prediction of functions defined over an interval (i.e., curves). Elements of novelty are introduced in Section 3 and 4, where 1) we propose a curve alignment prior to regression; 2) we define a statistical model for uncertainty propagation, furnishing confidence bounds for a parametric curve; 3) we employ a multi-regression, based on clustering and classification, to tackle bifurcations in the parametric space, enhancing the model accuracy. We exemplify the methodologies over two engineering applications in computational solid mechanics. The first application concerns a reduced order model for virtual materials characterized by a parametric Krupkowski hardening law; the second application is related to crack propagation analysis in parametric notched specimens under tensile loading. Section 5 is a short conclusion, in which possible further developments and approaches are discussed.
2 Methods
In this section we briefly summarize the main tools in MOR employed in this work. For a complete description of the most recent advances in the MOR community, we refer to the handbooks by Benner et al. (2020c,b,a) and the plentiful bibliography therein.
2.1 POD
We assume that a numerical approximation of the unknown field of interest u (x, t) is known at the nodes xi of a spatial mesh for discrete times tj = (j − 1)Δt, with i ∈ [1, … , nx] and j ∈ [1, … , nt]. We use the notation
which leads to the following eigenvalue problem Cϕ = λϕ, where
is the two-point correlation matrix (symmetric and positive definite). Defining the matrix
we have C = QQT.
In order to obtain a reduced-order model, we first solve the eigenvalue problem and select the r eigenvectors ϕi associated with the highest eigenvalues (truncated SVD at rank r), with in practice r ≪ nx. Thus r eigenvectors are placed in the columns of a matrix B that allows reducing U into its reduced counterpart γ, according to U = Bγ. Then, considering the full-size system KU = F, we have KBγ = F. Premultiplying by BT one gets BTKBγ = BTF and, with new definitions, the reduced counterpart becomes kγ = f.
The main drawback related to such a procedure is the size of the eigenproblem to be solved, the size of the correlation matrix C, nx × nx, with nx scaling with the number of nodes considered in the problem discretization that can reach in some applications millions and much more. The so-called Snapshot-POD allows alleviating the just referred issue (Hilberg et al., 1994). The basic concept is that, when nt ≪ nx, it is much more convenient solving the eigenvalue problem for
2.2 PODI
The origin of the non-intrusive POD, comes from the so-called POD with interpolation. PODI consider different snapshots related with different values of the model parameter p, U (pi), i = 1, … , ns, without loss of generality assumed scalar and ordered, i.e.
Then, as usual in POD-based MOR, the reduced basis is extracted, ϕ1, … , ϕr. Now, for a given parameter p, with
• Sampling: U (pi) ≡Ui, i = 1, … , ns;
• Reduced basis extraction: POD is applied to extract the reduced basis ϕ1, … , ϕr;
• Reproduction: calculation of γi. For that, we look to express
Repeating for all i ∈ {1, … , ns} and k ∈ {1, … , r}, we obtain γi (the reduced counterpart of
• Interpolation: with the reduced solution representations γi ≡γ(p = pi), one is tempted for any other p to proceed by interpolation, i.e.
with
• Reconstruction: with γ(p) obtained, the solution can be reconstructed everywhere from the nodal values U(p) = Bγ(p).
2.2.1 Extension to Multi-Parametric Settings
The just discussed procedure seems very appealing, however, its extension to highly-multidimensional settings remains difficult because of usual approximation bases suffer from the so-called curse of dimensionality.
In the case of moderate dimensionality, the PODI algorithm is easily generalizable. For that purpose we first reformulate the PODI described above as follows: the reconstruction U(p) = Bγ(p) can be expressed in the equivalent form:
with
that is directly generalizable to the multi-parametric setting where the scalar p is replaced by the parameters vector p, with the interpolation expressed now as
As previously mentioned the main difficulty associated with the technique just described is the difficulty of interpolating when the number of parameters (the size of vector p) increases too much. Separated representations in sparse settings, addressed in Subsection 2.3, succeed in circumventing the just referred difficulty.
2.3 Advanced Sparse PGD-Based Nonlinear Regressions
Here we discuss the PGD-based regression methods to build metamodels depending on d features. In particular, we focus on the case where, for a given choice of the parameters.
1. a single-valued output is measured;
2. a vector-valued output is measured;
3. a single-valued output is measured over a certain interval.
2.3.1 Single-Valued Output
In the case of a scalar output, the general problems consists of constructing the function
that depends on d features (parameters) pk, k = 1, … , d, taking values in the parametric space Ω, where a sparse sample of ns points and the corresponding outputs are known.
The so-called sparse PGD (sPGD) expresses the function f from a low-rank separated representation
constructed from rank-one updates within a greedy constructor. In the previous expression
Functions
where D represents the number of degrees of freedom (nodes) of the chosen approximation and
In the context of usual regression the approximation
where
The approximation coefficients of each one-dimensional function are computed by employing a greedy algorithm, such that, once the approximation up to order M − 1 is known, the Mth order term reads
The computed function is expected to approximate f not only in the training set but in any point p ∈ Ω.
The main issue is how to ally rich approximations and scarce available data, while avoiding overfitting. For that purpose a modal adaptivity strategy–MAS–was associated to the sPGD, however, it has been observed that the desired accuracy is not achieved before reaching overfitting or the algorithm stops too early when using MAS in some cases. This last issue implies a PGD solution composed of low order approximation functions, thus not getting an as rich as desired function. Some papers describing the just referred techniques are (Borzacchiello et al., 2017; Ibáñez et al., 2018).
In addition, in problems where just a few terms of the interpolation basis are present (that is, there are just some sparse non-zero elements in the interpolation basis to be determined), the strategy fails in recognizing the true model and therefore lacks accuracy.
To solve these difficulties, different regularizations were proposed (Sancarlos et al., 2021), combining L2 and L1 norms affecting the coefficients
2.3.2 Vector-Valued Output
In the case of a multidimensional output, we seek the function
This is a trivial extension of the single-valued function, where each component fi (p1, … , pd), for i = 1, … , n, is fitted independently using the procedures explained in Subsection 2.3.1.
2.3.3 Single-Valued Output Over an Interval
Let us now consider the case when, d features (parameters), the system output is a univariate function of the variable x, that is
where
Usually, the target function g(x) is evaluated (known) in a finite number nx of sampling points, that is the discrete ensemble
In this case, the coordinate x can be considered as an additional parameter, and the approximation problem can be reformulated as seeking the function
We have dropped the subscript X related to the variable x since the approximation problem has been recast into a new parametric framework defined by Ξ. The newly defined parametric coordinate pd+1 accounts for the location in which g(x) shall be approximated, that is:
Such coordinate is thus much richer than the others, given the very fine discretization in nx points available along this direction, compared to the sparse knowledge concerning the first d parametric coordinates belonging to Ω.
Equation 1 now reads:
where the univariate functions of the first d parameters
where nx is the number of discretization and
with h denoting an uniform discretization step. In particular,
The minimization problem (Eq. 3) can also be recast as
With these definitions made, the algorithm runs as previously explained.
2.3.3.1 POD Modes Extraction
Here we reformulate the approximation problem of curves within a POD-based MOR builder, which can be seen as a data pre-compression and dimensionality reduction approach. Indeed, considering the training data
where
A reduced factorization of the snapshots matrix is then obtained via a standard truncated POD of rank r:
where
In particular, the matrix Φ contains, by columns, the functions of the reduced POD basis
and, in particular, its discrete form reads
where Λk,• denotes the kth row of the matrix Λ.
Let us consider now a parametric curve depending on d features
The above equation suggests that a reduced-order parametric metamodel for the curves can be built considering only the set of coefficients
from the available training dataset
2.4 Multi-Regression
Creating a unique regression in large physical and parametric domains is a tricky issue. From one side, constructing a regression of a quantity of interest is much more accurate than creating the parametric curve (e.g., the parametric time evolution of the solution at a certain point), that in turn, becomes much more accurate than creating a regression of a field. The reason is that in general regressions are constructed by using the L2-norm, and consequently, if a given field exhibits strong localizations, these local behaviors are sacrificed in benefit of a quite good solution everywhere (on average).
Thus, a valuable route for enhancing accuracy consists in partitioning the physical space, in order to perform a regression in each of the resulting patches. Local quasi-linear regressions perform in general better than rich nonlinear regressions in the whole space domain.
The main issue in using multiple regressions, one per patch, is that the continuity can be lost on the patch boundaries. One could try to enforce the continuity, for example within a Partition of Unity–PU–framework, however, continuity is not compulsory, and then, on the patch borders (or in its neighborhood) one could compute the regressions from both sides and average them. Another possibility is taking profit of those discontinuities for refinement purposes, as usually considered within the finite element method framework.
In the case of parametric models the issue that we just discussed not only affects the spatial domain, but also the parametric one. In that case, making a partition of the multi-parametric space is not simple. One possibility consists in clustering the solutions related to the considered sampling, for example by invoking the k-means. Then, a nonlinear regression is created from the solutions in each cluster. Finally, the trickiest issue becomes the way of associating a cluster to any parameters choice, that is, performing an accurate classification. The procedure can be summarized in the following steps:
1. clustering high-fidelity solutions related to a design of experiments;
2. creating a regression model in each cluster (for instance, via the algorithms presented in Subsection 2.3);
3. constructing a classifier able to associate a cluster to any parameters choice and to select the most suitable regression model.
2.5 k-Means
k-means is one of the earliest methods for non-supervised vector quantization in artificial intelligence (MacQueen, 1967). In essence, as the Support Vector Machines–SVMs– (Cristianini and Shawe-Taylor, 2000) would do in the context of supervised learning models, k-means performs cluster analysis. In other words, this technique groups a set of objects such that every member of the group or cluster is more similar (closer) to the other members of the cluster than to any member of the rest of clusters.
In the case of k-means, this partition is made on the basis that each experimental data pertains to the cluster with the nearest mean. As can be readily noticed, this is equivalent to computing Voronoi cells in the data. Formally, if we have a set of observations in the form of high-dimensional vectors (x1, x2, … , xM), we aim at partitioning these M observations into k sets (k ≤ M),
where μi is the mean of each cluster.
3 Data Alignment and Uncertainty Propagation
In this Section we will present the curve parameterization based on data alignment to obtain an accurate physics-informed interpolation. We will exemplify the procedure to study the mechanical response of parametric materials loaded in tension.
In this Section we consider a parametric study over dog bone tensile test samples, as sketched in Figure 1. We are interested in the influence of the 3 parameters (n, K, ɛ0) characterizing the Krupkowski hardening law (also known as Swift hardening law), widely used in FEM software
linking the True Strength and the True Strain. ɛ denotes the effective plastic strain, ɛ0 the offset strain, n the strain hardening exponent and K the material constant.
The image in Figure 2 top shows two patterns of the Force-Displacement curve, obtained for two different choices of the Krupkowski parameters (blue and orange lines). A classical interpolation of these two patterns would result in the non-physical black dashed pattern.
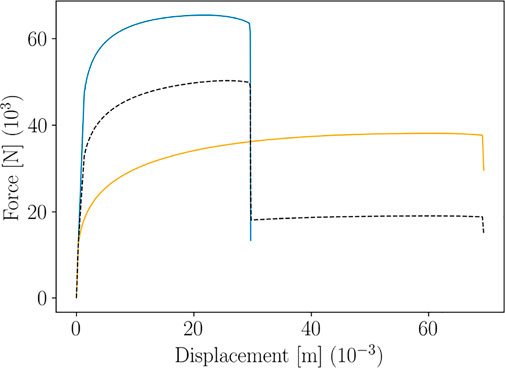
FIGURE 2. Main issue encountered when using standard interpolations on non-aligned curves (the black dashed line represents the interpolation between the two colored lines).
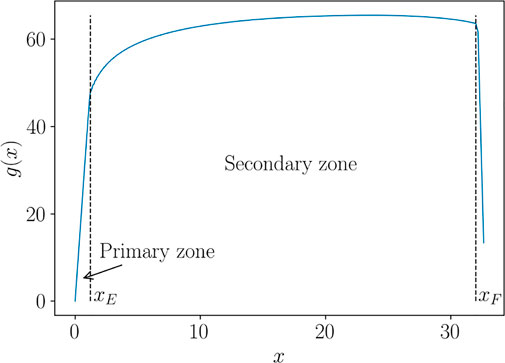
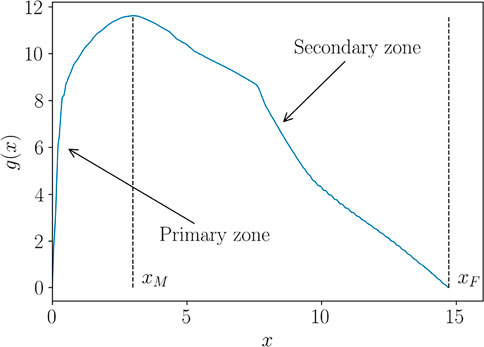
In what follows, we propose a procedure to overcome such spurious effects, based on a curve alignment prior to interpolate. The method is illustrated over the Force-Displacement curves. However, for the sake of generality, we refer to such curves as generic functions g(x), presenting two characteristic behaviors in the so-called primary and secondary zones. In the specific case of Force-Displacement, the primary zone is the elastic response of the material, up to the yield point xE. The secondary zone is the post yield behaviour up to the failure point xF, as illustrated in Figure 3. We will also refer to xE as the “transition point” and to xF as the “end point”, related to the specimen fracture.
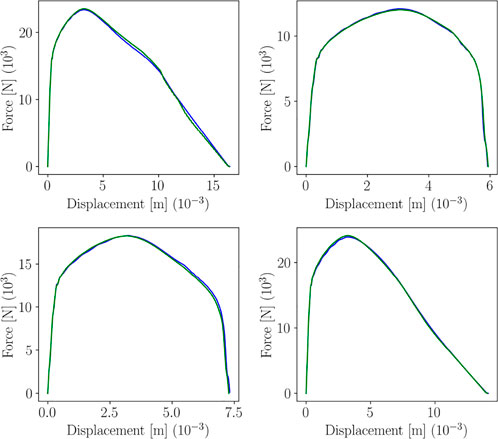
We assume that the behaviors in the primary and secondary zone, g1(x) and g2(x) respectively, and the transition and end points, xE and xF respectively, depend on a series of parameters grouped in vector p, i.e. g1 (x; p) ≡ g (x ∈ [0, xE]; p), g2 (x; p) ≡ g (x ∈ [xE, xF]; p), xE(p) and xF(p). Indeed, when considering different choices of the model parameter pi = (Ki, ni, ɛ0,i), i = 1, … , ns, one obtains a set of curves, as the ones shown in Figure 4, for instance. Such curves correspond to a sparse DoE (Latin Hypercube) of 20 points in the 3-dimensional parametric space

FIGURE 4. Curves g (x; pi) related to different choices of the model features pi = (Ki, ni, ɛ0,i), i = 1, … , ns.
Once the transition and end points of each curve have been determined, the curves can be rediscretized over the same number of points (through a standard piecewise linear interpolation, for instance). To align them, we define a dimensionless coordinate in each zone, y in the primary zone, x ∈ [0, xE], and z in the secondary zone, x ∈ [xE, xF], both defined through the change of variable
and
expressions that hold for each curve g (x; pi), i = 1, … , ns, with
and
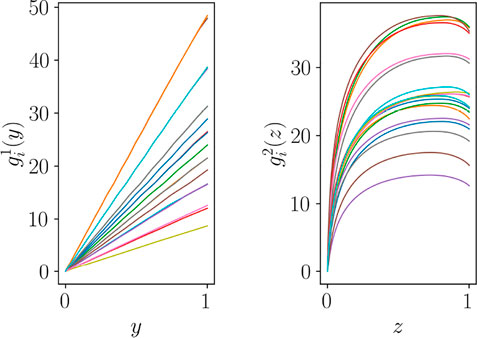
Figure 5 depicts functions
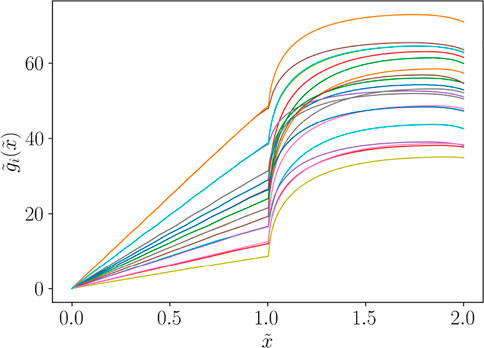
Actually, this procedure amounts at performing an alignment based on a dilatation of the curves in the first and secondary zone, as shown in Figure 6. In such case, we can express the aligned curves as functions of
Once the curves have been aligned, the nonlinear regressor presented in Subsection 2.3.3 can be invoked to build the parametric metamodel of the curve. This can be done separately in each zone or over the whole newly defined coordinate
3.1 POD Modes Extraction
In order to extract the most significant modes able to describe these functions, the POD can be applied in each group of curves in Figure 5. This amounts to build the snapshot matrix within each group and perform a truncated SVD. In the case that serves here to illustrate the procedure, a single mode suffices for describing the almost linear functions in the primary zone, that will be noted by ξ1(y), whereas in the secondary zone two functions are needed, ϕ1(z) and ϕ2(z).
Thus, any function
whereas functions
The α and β coefficients can be easily computed by simple projection, i.e.
where the normality of ξ1(y) was used. In the same way, and taking into account the orthonormality of functions ϕ1(z) and ϕ2(z),
and
Thus, for each curve gi(x) we succeeded to extract its five main descriptors:
Now, each of these descriptors can be expressed parametrically, xE(p), xF(p), α1(p), β1(p) and β2(p), by using the regression techniques described in Subsection 2.3.1 for scalar quantities.
3.2 Curves Reconstruction
When considering a choice of the parameters p, the curves descriptors are extracted from the regressions xE(p), xF(p), α1(p), β1(p) and β2(p), the dimensionless coordinates defining both zones calculated from
and
and, finally, the curve in each zone reconstructed according to
and
from which the curve g (x; p) can be straightforward obtained via
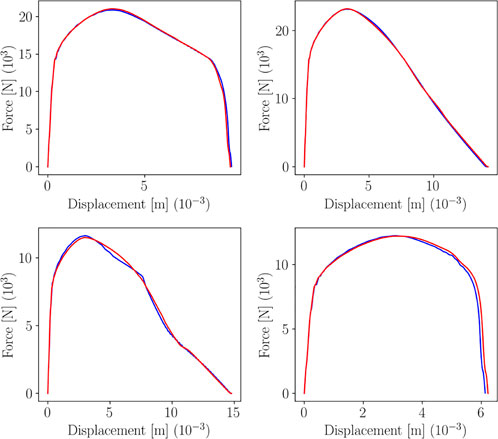
To build the parametric metamodel, 17 curves have been used to train the sPGD regressor, while the remaining 3 for testing. Figure 7 shows the resulting predictions over 3 training points and test points.
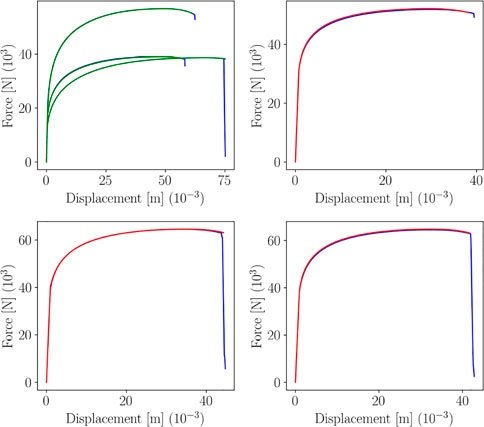
FIGURE 7. sPGD predictions (green line for training, red for testing) versus true curve (blue line).
3.3 Real-Time Calibration
Now, given an experimental curve g(x), its parameters are extracted according to.
• xE from the point at which the change of behavior occurs (for instance, computing the function derivatives by means of finite differences);
• xF is the terminal point;
• α1 follows from
• β1 follows from
• β2 follows from
Then, from the regression models xE(p), xF(p), x1(p), β1(p) and β2(p), the inverse problem is solved for extracting the associated parameters, p.
3.4 Statistical Model Derived by Parametric Curves
With the previously built surrogate model, the curve related to any possible value of p can be computed in real-time, i.e. g (x; p). In this section, this surrogate will be employed for uncertainty quantification.
We assume that each feature pk in vector p is assumed characterized by a Gaussian distribution defined its mean value μk and its variance
where diag (•) is the diagonal matrix of diagonal •.
The aim is linking the sensitivity over the input features with the one over the output curve. This means computing some estimators of the average M and the variance Σ of the curve descriptors for different choices of μ and σ, and from them, by using the regressions presented in Subsection 2.3, build the set of statistical surrogates:
where
To build the surrogate (5), for instance for the curve descriptor
This can be achieved by means of a Monte Carlo sampling, which gives the estimators of mean and variance for the curves g (x; pj (μj, σj)), and of any descriptor
The whole procedure is summarized in Algorithm 1.
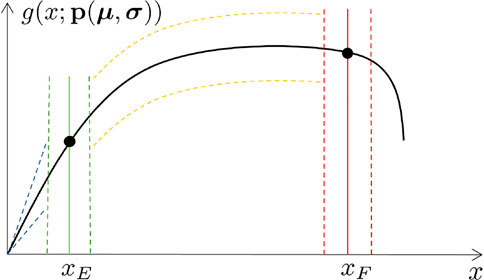
Figure 9 shows the parametric curve and its statistical sensing, for a given choice of the input features distribution parameters. Confidence Intervals have been computed using Algorithm 1, for the curve and the rupture point.
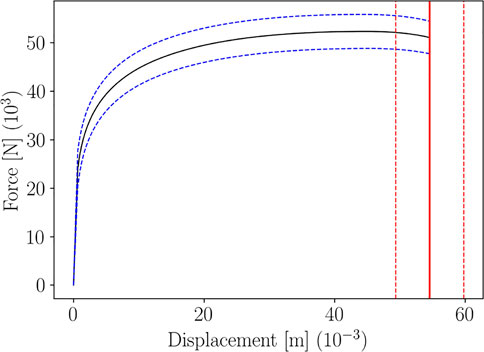
FIGURE 9. Confidence Interval of level 0.95 for the parametric Force-Displacement curve and for the rupture point, for a given choice of μ and σ.
3.5 Statistical Model Derived From Measures
In this Section we consider that for different choices of the problem features pi, the measure gm (x; pi) is collected. We assume that measures contain a significant uncertainty, modeled again, without loss of generality, by a Gaussian distribution of null average and variance σ, that is,
In these circumstances applying a regression to fit those values gm (x; pi), that is fX,m (pi) = gm (x; pi), according to the techniques described in Subsection 2.3 is not a valuable route. The most valuable solution consists of looking for the baseline regression fX(p) such that the deviation
The just described procedure is very close to standard Bayesian inference.
3.6 Model Enrichment
When two regression models are known, for the sake of simplicity assumed scalar, one related to a physics based model fX,model(p) and the second one to the measures fX,measure(p), both associated with the average values in case of uncertainty in the model and the measures, one could define the gap model ΔfX(p) from fX,measure(p) − fX,model(p) ≡ΔfX(p).
Thus, the enriched model reads
As in general the nonlinear character of fX,measure(p) is expected being much higher than the one of the gap, ΔfX(p), a more valuable route consists in calculating the discrete gap
4 Data Alignment and Data Clustering
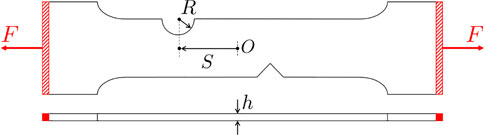
Here we focus on the study of crack propagation in notched specimens loaded in tension, whose geometry is sketched in Figure 10. The test piece has a V-shaped notch defect which is always at the same location (almost bottom-middle). On the other side of the test piece there is a half-circle groove. The goal is to predict the crack propagation from the defect based off of different locations (S) and radii (R) of the groove, as well as different test piece thicknesses (h). Depending on the location of the groove, the crack will propagate differently from the defect to the groove.
We have considered a sparse DoE (Latin Hypercube) of 50 points in the 3-dimensional parametric space Ω = IR × IS × Ih, with the parameters bounds specified in Table 2. Numerical simulations (carried out in VPS software from ESI Group) employ an Explicit Analysis and the EWK rupture model (Kamoulakos, 2005), using a mesh of 1096218 solid elements.
We focus on the prediction of the Force-Displacement curves plotted in Figure 11, which are considered as the generic functions g(x), following the same notation of Section 3.
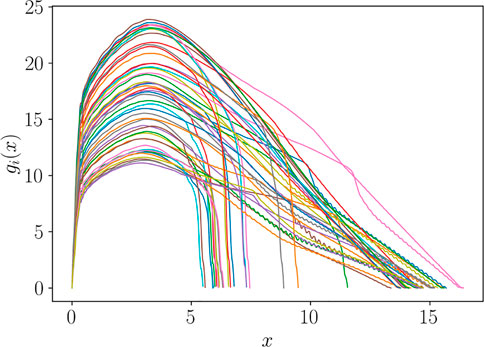
FIGURE 11. Curves gi(x) = g (x; pi) related to different choices of the model features pi = (Ri, Si, hi), i = 1, … , ns.
It can be observed that all the curves present a similar pattern in the first zone, monotonically increasing, while the response appears much different in the secondary zone. A first pre-processing step consists in splitting the zones as illustrated in Figure 12, where xM denotes the point where the curve reaches its maximum value, while xF its endpoint.
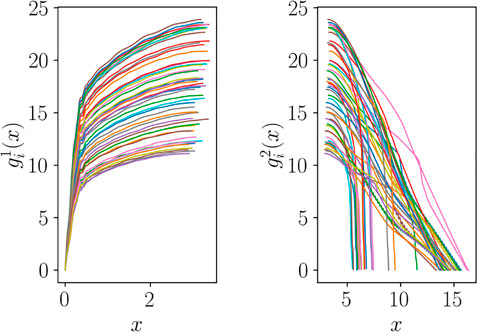
Cutting the curves, we obtain the two groups of functions plotted in Figure 13, which are of course not aligned. However, they can be expressed as functions of normalized coordinates y and z, respectively, and aligned following the dilatation procedure discussed in Section 3.
Once the alignment has been performed, using the usual nonlinear regression techniques of Subsection 2.3 and same notations of Section 3, two regression models, one for each group, can be established:
In Eq. 6, for the sake of clarity, we have specified xM and xF since these points are involved into the parametrization of the functions g1(x) and g2(x), respectively, and thus expressed parametrically.
As we have previously pointed out, the second group of functions
4.1 Clustering
To exemplify the bifurcation problem in the parametric space, we consider two different configurations of the model parameters, resulting into the specimens shown in Figure 14.

FIGURE 14. Two different parameters configurations. Top: R = 7.59, S = 18.23, h = 0.84; bottom: R = 3.75, S = 5.58, h = 1.51 (all dimensions are provided in mm). The red zone is the part subject to rigid body constraints.
Figure 15 shows four snapshots of the displacement field related to the specimens in Figure 14, under axial tensile loading. The crack propagation follows two completely different patterns, drastically influencing the Force-Displacement curve, as shown in Figure 16.
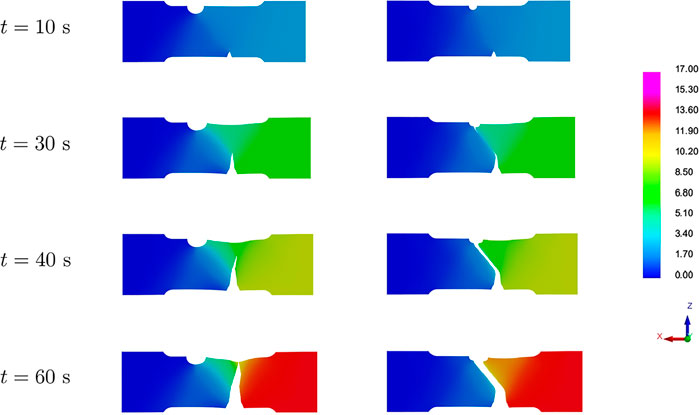
FIGURE 15. Bifurcation in the parametric space causing completely different crack propagation dynamics.
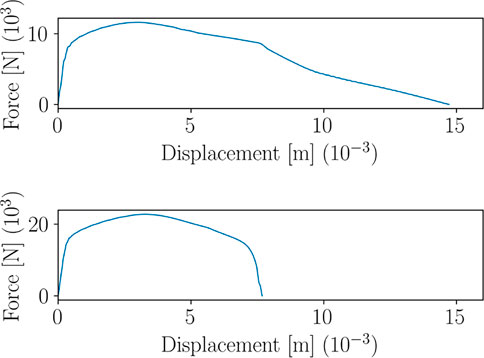
FIGURE 16. Force-Displacement curves corresponding to the two parameters configurations in Figure 14.
The clustering step can be performed automatically by using a hierarchical clustering based on the curves shape or on the location of damaged elements into the finite element mesh. Once the clusters
Figure 17 shows the functions in the secondary zone after the clustering.
In particular, one can remark that fracture occurs early on for tests belonging to cluster
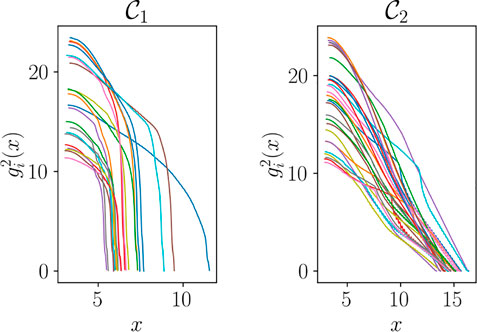
FIGURE 17. Functions
4.2 Curves Reconstruction and Classification
For a newly defined choice of model features p*, the curve g (x; p*) is obtained via
where g1 and g2 are obtained through Eq. 7.
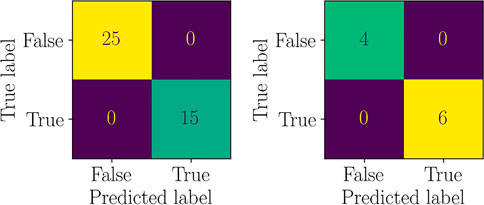
The training of the regression models has been performed using 40 points of the DoE, remaining 10 have been used for testing. Moreover, a Support Vector Machine classifier (a Random Forest classifier could also be used, for instance) has been trained to select the best regression submodel to predict g2 (x; p*). Such classifier has shown perfect accuracy, as shown by the Confusion Matrices in Figure 18. Moreover, Figure 19 shows the separating surface and classified points in the 3-dimensional parametric space.
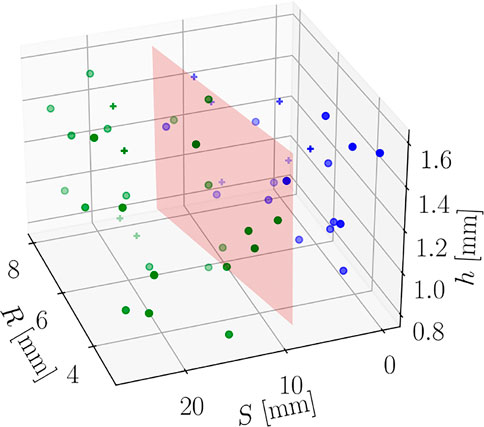
FIGURE 19. Parametric space and classified points (marker + is used for test points). The red plane is the separation surface.
Figures 20, 21 represent the plots of predictions for train and test, respectively, for 4 data points.
5 Conclusion
In this paper we have focused on several nontrivial issues encountered when a whole curve shall be predicted from a given number of features. A major argument is the data alignment to achieve physics-consistent interpolations among curves and the data clustering to detect bifurcations in the parametric space. The proposed methodologies rely on adopting specific parametrizations of the curve and a physics-based pre-processing prior to the application of any regression technique. We have also suggested a reduced order parametrization of the curve via POD coefficients, requiring the prediction of a few scalar quantities (i.e., the POD coefficients) instead of the whole curve. Here, without loss of generality, we have preferred sPGD-based nonlinear regressions, these being efficient in high-dimensional parametric spaces under the scarce data limit constraint. Indeed, since our data come from numerical simulations of complex engineering problems, due to the high computational complexity of the offline simulations, not much data are usually available. Moreover, one important achievement of the work is the definition of a statistical sensing for uncertainty propagation based on the parametric model.
We have focused on two applications in computational mechanics: 1) plastic materials with parametric hardening law, 2) crack propagation in parametric notched specimens. However, these methodologies can be applied to any time series or generic curve stem from any context. For instance, in our current research, we are successfully applying these techniques to solve many other problems (to cite some, the study of a two-phase flow dynamics in a heated channel, the composite forming processes involving a reactive resin injection molding). Moreover, we are focusing on other physics-based curves interpolation strategies based on Optimal Transport–OT–(Torregrosa et al., 2022) and other mappings.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
All the authors participated in the definition of techniques and algorithms. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors acknowledge the support of ESI Group through its research chair at ENSAM ParisTech. This research is part of the programme DesCartes and is supported by the National Research Foundation, Prime Minister’s Office, Singapore under its Campus for Research Excellence and Technological Enterprise (CREATE) programme.
References
Amsallem, D., and Farhat, C. (2008). Interpolation Method for Adapting Reduced-Order Models and Application to Aeroelasticity. AIAA J. 46, 1803–1813. doi:10.2514/1.35374
Audouze, C., De Vuyst, F., and Nair, P. B. (2013). Nonintrusive Reduced-Order Modeling of Parametrized Time-dependent Partial Differential Equations. Numer. Methods Partial Differ. Eq. 29, 1587–1628. doi:10.1002/num.21768
Benner, P., Schilders, W., Grivet-Talocia, S., Quarteroni, A., Rozza, G., and Silveira, L. M. (2020a). Model Order Reduction: Applications. Berlin: De Gruyter.
Benner, P., Schilders, W., Grivet-Talocia, S., Quarteroni, A., Rozza, G., and Silveira, L. M. (2020b). Model Order Reduction: Snapshot-Based Methods and Algorithms. Berlin: De Gruyter.
Benner, P., Schilders, W., Grivet-Talocia, S., Quarteroni, A., Rozza, G., and Silveira, L. M. (2020c). Model Order Reduction: System- and Data-Driven Methods and Algorithms. Berlin: De Gruyter.
Benner, P., Gugercin, S., and Willcox, K. (2015). A Survey of Projection-Based Model Reduction Methods for Parametric Dynamical Systems. SIAM Rev. 57, 483–531. doi:10.1137/130932715
Borzacchiello, D., Aguado, J. V., and Chinesta, F. (2017). Non-intrusive Sparse Subspace Learning for Parametrized Problems. Arch. Comput. Methods Eng. 26, 303–326. doi:10.1007/s11831-017-9241-4
Chinesta, F., Ladeveze, P., and Cueto, E. (2011). A Short Review on Model Order Reduction Based on Proper Generalized Decomposition. Arch. Comput. Methods Eng. 18, 395–404. doi:10.1007/s11831-011-9064-7
Cristianini, N., and Shawe-Taylor, J. (2000). An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. Cambridge: Cambridge University Press.
de Gooijer, B. M., Havinga, J., Geijselaers, H. J. M., and Van den Boogaard, A. H. (2021). Evaluation of Pod Based Surrogate Models of Fields Resulting from Nonlinear Fem Simulations. Adv. Model. Simul. Eng. Sci. 8. doi:10.1186/s40323-021-00210-8
Fareed, H., and Singler, J. R. (2019). A Note on Incremental Pod Algorithms for Continuous Time Data. Appl. Numer. Math. 144, 223–233. doi:10.1016/j.apnum.2019.04.020
Franchini, A., Sebastian, W., and D'Ayala, D. (2022). Surrogate-based Fragility Analysis and Probabilistic Optimisation of Cable-Stayed Bridges Subject to Seismic Loads. Eng. Struct. 256, 113949. doi:10.1016/j.engstruct.2022.113949
Friderikos, O., Baranger, E., Olive, M., and Néron, D. (2022). On the Stability of Pod Basis Interpolation on Grassmann Manifolds for Parametric Model Order Reduction. Comput Mech. Cham: Springer. doi:10.1007/s00466-022-02163-0
Friderikos, O., Olive, M., Baranger, E., Sagris, D., and David, C. N. (2020). A Space-Time Pod Basis Interpolation on Grassmann Manifolds for Parametric Simulations of Rigid-Viscoplastic Fem. MATEC Web Conf. 318, 01043. doi:10.1051/matecconf/202031801043
Hesthaven, J. S., Rozza, G., and Stamm, B. (2016). Certified Reduced Basis Methods for Parametrized Partial Differential Equations. Cham: Springer. doi:10.1007/978-3-319-22470-1
Hesthaven, J. S., and Ubbiali, S. (2018). Non-intrusive Reduced Order Modeling of Nonlinear Problems Using Neural Networks. J. Comput. Phys. 363, 55–78. doi:10.1016/j.jcp.2018.02.037
Hilberg, D., Lazik, W., and Fiedler, H. E. (1994). The Application of Classical Pod and Snapshot Pod in a Turbulent Shear Layer with Periodic Structures. Appl. Sci. Res. 53, 283–290. doi:10.1007/bf00849105
Ibáñez, R., Abisset-Chavanne, E., Ammar, A., González, D., Cueto, E., Huerta, A., et al. (2018). A Multidimensional Data-Driven Sparse Identification Technique: The Sparse Proper Generalized Decomposition. Complexity 2018, 1–11. doi:10.1155/2018/5608286
Kamoulakos, A. (2005). “The ESI-Wilkins-Kamoulakos (EWK) Rupture Model,” in Continuum Scale Simulation of Engineering Materials: Fundamentals - Microstructures - Process Applications (Hoboken: John Wiley & Sons), 795–804. doi:10.1002/3527603786.ch43
Khatouri, H., Benamara, T., Breitkopf, P., and Demange, J. (2022). Metamodeling Techniques for Cpu-Intensive Simulation-Based Design Optimization: a Survey. Adv. Model. Simul. Eng. Sci. 9. doi:10.1186/s40323-022-00214-y
MacQueen, J. B. (1967). “Some Methods for Classification and Analysis of Multivariate Observations,” in Proc. Of the Fifth Berkeley Symposium on Mathematical Statistics and Probability. Editors L. M. L. Cam., and J. Neyman (California: University of California Press), 281–297.
Mainini, L., and Willcox, K. (2015). Surrogate Modeling Approach to Support Real-Time Structural Assessment and Decision Making. AIAA J. 53, 1612–1626. doi:10.2514/1.J053464
Mosquera, R., El Hamidi, A., Hamdouni, A., and Falaize, A. (2021). Generalization of the Neville-Aitken Interpolation Algorithm on Grassmann Manifolds: Applications to Reduced Order Model. Int. J. Numer. Meth Fluids 93, 2421–2442. doi:10.1002/fld.4981
Mosquera, R., Hamdouni, A., Hamdouni, A., El Hamidi, A., and Allery, C. (2018). Pod Basis Interpolation via Inverse Distance Weighting on Grassmann Manifolds. Discrete Continuous Dyn. Syst. - S 12, 1743–1759. doi:10.3934/dcdss.2019115
Prud’homme, C., Rovas, D. V., Veroy, K., Machiels, L., Maday, Y., Patera, A. T., et al. (2002). Reliable Real-Time Solution of Parametrized Partial Differential Equations: Reduced-Basis Output Bound Methods. J. Fluids Eng. 124, 70–80. doi:10.1115/1.1448332
Raghavan, B., Hamdaoui, M., Xiao, M., Breitkopf, P., and Villon, P. (2013). A Bi-level Meta-Modeling Approach for Structural Optimization Using Modified Pod Bases and Diffuse Approximation. Comput. Struct. 127, 19–28. doi:10.1016/j.compstruc.2012.06.008
Rajaram, D., Perron, C., Puranik, T. G., and Mavris, D. N. (2020). Randomized Algorithms for Non-intrusive Parametric Reduced Order Modeling. AIAA J. 58, 5389–5407. doi:10.2514/1.J059616
Sancarlos, A., Champaney, V., Duval, J., Cueto, E., and Chinesta, F. (2021). Pgd-based Advanced Nonlinear Multiparametric Regressions for Constructing Metamodels at the Scarce-Data Limit. CoRR abs/2103.05358, ArXiv.
Simpson, T. W., Poplinski, J. D., Koch, P. N., and Allen, J. K. (2001). Metamodels for Computer-Based Engineering Design: Survey and Recommendations. Eng. Comput. 17, 129–150. doi:10.1007/PL00007198
Torregrosa, S., Champaney, V., Ammar, A., Herbert, V., and Chinesta, F. (2022). Surrogate Parametric Metamodel Based on Optimal Transport. Math. Comput. Simul. 194, 36–63. doi:10.1016/j.matcom.2021.11.010
Keywords: parametric curves, data-driven modeling, uncertainty quantification and propagation, POD, PGD
Citation: Champaney V, Pasquale A, Ammar A and Chinesta F (2022) Parametric Curves Metamodelling Based on Data Clustering, Data Alignment, POD-Based Modes Extraction and PGD-Based Nonlinear Regressions. Front. Mater. 9:904707. doi: 10.3389/fmats.2022.904707
Received: 25 March 2022; Accepted: 11 May 2022;
Published: 24 June 2022.
Edited by:
Chady Ghnatios, Notre Dame University, LebanonReviewed by:
Attilio Frangi, Politecnico di Milano, ItalyYongxing Shen, Shanghai Jiao Tong University, China
Nicolas Montes, Universidad CEU Cardenal Herrera, Spain
Copyright © 2022 Champaney, Pasquale, Ammar and Chinesta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Victor Champaney, dmljdG9yLmNoYW1wYW5leUBlbnNhbS5ldQ==
 Victor Champaney
Victor Champaney Angelo Pasquale1,2
Angelo Pasquale1,2 Francisco Chinesta
Francisco Chinesta