
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 14 April 2025
Sec. Aquatic Physiology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1580077
Phaeocystis globosa is a globally distributed harmful algal bloom species that causes severe damage to local environments and economies. The use of the invasive plant Ipomoea cairica as an algaecide for P. globosa is an environmentally friendly management strategy. Although previous studies have found that I. cairica extracts can effectively inhibit P. globosa growth, the molecular mechanism of cellular response to this algaecide remains unclear. Therefore, transcriptome differences between 24 and 48 h after treatment with I. cairica extracts were investigated to explore the P. globosa gene expression changes in response to the algaecide. In this study, control groups were treated with sterile seawater, while experimental groups were exposed to I. cairica extracts (final concentration of 1%). The results of physiological experiments showed no difference in the number of solitary cells and colonies at 24 h, but significant decreases at 48 h. Transcriptomic data also showed significant differences in differentially expressed genes (DEGs) at 24 and 48 h. DEGs were mainly upregulated at 24 h and downregulated at 48 h. Mitogen-activated protein kinase (MAPK) and phosphatidylinositol (PI) signaling pathways that respond stress in the external environment were activated throughout the response process. Genes related to the antioxidant enzyme system and autophagy were upregulated at 24 h, which may contribute to the stress response mechanisms of P. globosa. Downregulation of photosynthesis-related genes leading to inhibition of the photosynthetic system and thus accelerating the accumulation of reactive oxygen species (ROS) may be responsible for the eventual cell death. In addition, 411 known compounds were identified usingultra-high-performance liquid chromatography (UHPLC). Unfortunately, apigenin-7-O-glucoside, a flavonoid, did not have a significant inhibitory effect on P. globosa. This study highlights the mechanism of the toxic effect of I. cairica extracts on P. globosa and provides a theoretical basis for the development of an algaecide targeting P. globosa.
Harmful algal blooms (HABs) have become a serious problem for marine agriculture, tourism, and human health worldwide due to their severe negative impact on aquatic ecosystems (Anderson, 2009). Phaeocystis globosa is one of the most notorious HAB species, secreting hemolytic toxins capable to kill fish and produce foul-smelling foam (Blauw et al., 2010; Tan et al., 2016). In addition, the ‘giant-colony’ ecotype P. globosa may block the water intake of the cold source system of nuclear power plants and endanger their safe operation (Zhang et al., 2022). Therefore, P. globosa algal blooms have drawn increasing attention.
This particular species exhibits a heterotypic life cycle, where solitary cells measure only 3–9 μm in diameter, yet clusters of these cells may create colonies reaching a maximum size of 3 cm in diameter (Chen et al., 2002; Rousseau et al., 2007). Colony formation is one of the ecological characteristics of P. globosa, as the cells essentially exist as a colony during algal blooms (Liang et al., 2020; Wang et al., 2021). The colony consists of a hollow spherical structure with colony cells distributed 15–20 µm below the colony envelope (van Rijssel et al., 1997). There are no cells in the center of the colony, only some seawater and polysaccharide comprise the main component of the colony envelope (Hamm et al., 1999). Colony formation protects cells from predator feeding, as well as viruses and bacteria, thus reducing cell mortality inside the colony (Jakobsen and Tang, 2002; Brussaard et al., 2005). The hollow structure reduces the settling rate and keeps P. globosa above the water column, enhancing light exposure for growth and biomass accumulation (Wang et al., 2010). When resources for growth are limited due to light or nutrient limitations, the colony acts as a reservoir of energy and nutrients that can be used to maintain cell growth and development within the colony (Schoemann et al., 2005). Colony formation enhances the environmental adaptability of P. globosa and facilitates the outbreak of algal blooms. Therefore, finding a method to inhibit colony formation can be pivotal in the control of P. globosa blooms.
The use of physical and chemical techniques to avert of HABs presents numerous drawbacks, such as elevated expenses, significant toxicity, potential for secondary contamination, and challenges in execution (Sun et al., 2018; Zhang et al., 2020). Some biological approaches using living organisms, such as fish, zooplankton, and algae-killing bacteria, as well as viruses, can be used to manage HABs (Kim et al., 1998; Mayali and Azam, 2004; Zohdi and Abbaspour, 2019). However, these approaches lack technical stability and have the potential to alter or destroy existing ecosystems. Recent studies have found that allelopathic substances extracted from plants can be applied against HABs (Mulderij et al., 2005; Patino et al., 2018). This approach has been shown to be effective and environmentally friendly and has great potential for the elimination of harmful algae.
Ipomoea cairica is a semi-woody perennial vine that first emerged in Central America and tropical Africa, and it currently thrives in various subtropical and tropical areas globally (Weber et al., 2008). I. cairica has an allelopathic effect as a major invasive plant in southern China and, therefore, may be a potential algaecide. Previous studies have demonstrated that I. cairica extracts exhibit algicidal activity against P. globosa by disrupting cellular structures and metabolic pathways (Liang et al., 2022). However, the molecular mechanisms underlying this algicidal effect remain poorly understood. This study aims to elucidate the transcriptomic responses of P. globosa to I. cairica extracts, providing insights into the algicidal mechanisms at the molecular level. To investigate these issues, we (i) examined the global gene expression changes of P. globosa at different time points; (ii) identified the effective secondary metabolites using ultra-high-performance liquid chromatography–mass spectrometry (UHPLC-MS); and (iii) verified the algacidal effect of the identified active components.
P. globosa was collected during a bloom period in the Beibu Gulf, located in China at coordinates 20˚55.82′N and 109˚11.43′E. To establish a single-cell isolation strain, solitary cells were carefully selected from the harvested colonies using a micropipette technique, ensuring purity in the cultures obtained. The isolated pure strains were then cultivated in a modified nutrient medium, specifically half-strength Guillard’s “F” solution, as described by Guillard and Ryther in 1962. The cultures were maintained at a stable temperature between 20°C and 21°C, under a controlled environment with a light cycle of 12 h of light followed by 12 h of darkness. The illumination was provided by cool white fluorescent lights, delivering approximately 80 to 100 μmol photos m−2 s−1, creating optimal conditions for the growth of the organism.
The research materials were sourced from the South China Botanical Garden in Guangzhou, China. Fresh and pest-free leaves were selected, ensuring that the samples were representative and consistent. The leaves were harvested from the middle to lower parts of mature plants, with a total weight of 2 kg. After collection, the leaves were quickly placed into sealed bags and transported to the laboratory. Upon arrival, the leaves were initially heated in an oven at 80°C for 30 minutes to remove surface moisture. Subsequently, they were dried at 60°C until they reached a constant weight, ensuring the complete removal of moisture. The dried leaves were then ground into fine powder with a particle size smaller than 20 mesh (0.85 mm) for further extraction. The grinding was performed using a mixer mill (MM400, Retsch, Germany). To prepare the leaf aqueous extract, 8 grams of the powdered leaves were soaked in 150 ml of sterile distilled water for 24 hours. The resulting solution was then filtered through a 0.22 μm filter membrane, and the filtrate was collected as the final leaf aqueous extract.
Algae were cultured in three 5 L conical flasks (n=3), with daily shaking to prevent cell adhesion to the flask walls. P. globosa was cultured until the exponential growth phase was reached, which occurred after 9 days. I. cairica aqueous extracts were then added to achieve a final concentration of 1% (v/v). Control groups were treated with sterile seawater under the same conditions as experimental groups. The cultures were maintained under the conditions described previously. Samples of 10 mL were collected for cell count, while 500 mL samples were collected for transcriptome and metabolite analysis at 0, 24, and 48 hours. The 10 mL samples were fixed with Lugol’s solution, and the abundance of solitary cells was monitored daily by counting cells under an inverted light microscope (LM, Olympus-CKX41, Osaka, Japan). Colony number, colony diameter, and cell abundance in colonies were measured following the methods described by Wang et al. (2010). For the 500 mL samples, centrifugation was performed at 10,000 × g for 15 minutes, and the resulting pellets were flash-frozen in liquid nitrogen and stored at −80°C for future RNA extraction, transcriptome sequencing and metabolite analysis.
The samples were collected at three time points: 0 hours, 24 hours, and 48 hours, with three biological replicates for each time point. Each sample was individually suspended in 2 mL of RNA extraction buffer to ensure the stability and integrity of the RNA. The RNA extraction buffer was composed of equal volumes of water-phenol solution and L-buffer. The L-buffer consisted of 0.5% SDS, 10 mmol L−1 EDTA, 0.2 mol L−1 sodium acetate (pH 5), and β-mercaptoethanol at a 1:100 ratio, which facilitated optimal RNA extraction and stabilization. To remove potential genomic DNA contamination, each sample was treated with DNase I (Takara, Japan) at 37°C for 30 minutes, ensuring the purity of the RNA. After RNA extraction, quality control, cDNA library construction, sequencing, data filtering, and alignment were all performed by Vazyme Biotech Co., Ltd. (Nanjing, China). The sequencing libraries were constructed using the Illumina TruSeq RNA Sample Prep Kit (Cat<ns/> FC-122-1001, Illumina, USA) from 1 µg of total RNA. All sequencing data generated in this study have been stored in the NCBI SRA database and can be accessed under the dataset number PRJNA913698, enabling public sharing and further research analysis.
Differentially expressed genes (DEGs) were identified using the DESeq R package (version 1.10.1), which enables the detection of significant gene expression differences between the 0-hour, 24-hour, and 48-hour time groups. DESeq employs a negative binomial distribution-based statistical method to assess gene expression changes in RNA-seq data. To control the false discovery rate (FDR), the raw P-values obtained from DESeq were adjusted using the Benjamini-Hochberg procedure. Genes were considered significantly differentially expressed if the absolute value of the log2 fold change was greater than 1.5 (|log2(fold-change)| > 1.5) and the adjusted P-value was less than 0.05 (P < 0.05). The identified DEGs were then subjected to functional annotation by comparing them to the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, in order to further explore their biological roles and the pathways they may be involved in.
The compounds were extracted from I. cairica leaf extracts by adding 1000 μL of a methanol/acetonitrile/H2O solvent mixture (2:2:1, v/v/v) to the samples. The mixture was then centrifuged at 14,000 g for 15 minutes at 4°C, and the supernatant was collected and dried using a vacuum concentrator. The dried extract was dissolved in 100 μL of a 1:1 (v/v) acetonitrile/water solvent for subsequent liquid chromatography-mass spectrometry (LC-MS) analysis.
LC-MS analysis was performed using an ultra-high-performance liquid chromatography (UHPLC) system (1290 Infinity LC, Agilent Technologies) coupled with a quadrupole time-of-flight mass spectrometer (AB Sciex TripleTOF 6600) at Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China). Chromatographic separation was conducted on a 2.1 mm × 100 mm ACQUIY UPLC BEH 1.7 μm column (Waters, Ireland), and electrospray ionization (ESI) was applied in both positive and negative ionization modes. The mobile phase consisted of two components: A was 25 mM ammonium acetate and 25 mM ammonium hydroxide in water, and B was acetonitrile. A gradient elution was used, starting with 85% B for 1 minute, followed by a linear reduction to 65% B over 11 minutes; then, B was further reduced to 40% over 0.1 minutes, held for 4 minutes, and finally, B was increased to 85% for a 5-minute re-equilibration. In the LC-MS/MS analysis, compound identification was based on matching with the METLIN database and mzCloud database, with stringent criteria for mass error (≤ 5 ppm) and fragment ion match score (≥ 80%). Additionally, the retention time of each compound was compared with that of standards or reference retention times in the database, allowing a deviation range of ± 0.2 minutes.
Apigenin-7-O-glucuronide (purity ≥ 98%) used in this study was purchased from Chengdu Cuiyirensheng Biotechnology Co., Ltd. (Chengdu, China). Prior to use, the Apigenin-7-O-glucoside solution was filtered through a 0.22 μm filter membrane to remove potential impurities. To evaluate its algal inhibition activity, different concentrations (0 μg/mL, 50 μg/mL, and 100 μg/mL) of Apigenin-7-O-glucoside were used to treat P. globosa samples, with three biological replicates for each concentration. The initial cell density was set at 1 × 10^5 cells/mL. The experiment was conducted under the previously described culture conditions, and measurements were taken after 48 hours to assess the following indicators: colony abundance, colony diameter, and the cell density within the colonies. These measurements were used to evaluate the inhibitory effect of Apigenin-7-O-glucoside on P. globosa.
To assess the significant differences in physiological characteristics (such as solitary cell count, colony abundance, and colony diameter) between the 0-hour, 24-hour, and 48-hour groups, a t-test was performed for statistical analysis. All statistical analyses were conducted using SPSS software (version 22.0, IBM, USA), ensuring the accuracy and reliability of data processing. For determining statistical significance, a P-value less than 0.05 (P < 0.05) was considered significant, meaning that differences between groups were deemed statistically significant when the P-value was below 0.05.
For solitary cells, there was no difference at 24 h, but significant inhibition was observed at 48 h, with cell abundance reduced to 30.2 ± 4.43×103 cells/mL and only half of the control cell abundance (Figure 1a). For colonies, there was a slight increase in the treated group at 24 h, but a significant decrease to 66.6 ± 5.46 colonies/mL at 48 h (Figure 1b). The colony diameter remained essentially unchanged at about 100 mm throughout the experiment (Figure 1c). These results suggested that the cells were not inhibited within 24 h because the tolerance system of P. globosa cells exerted a mitigating effect. When the algacidal effect exceeded the tolerance system, it inhibited cell growth at 48 h.
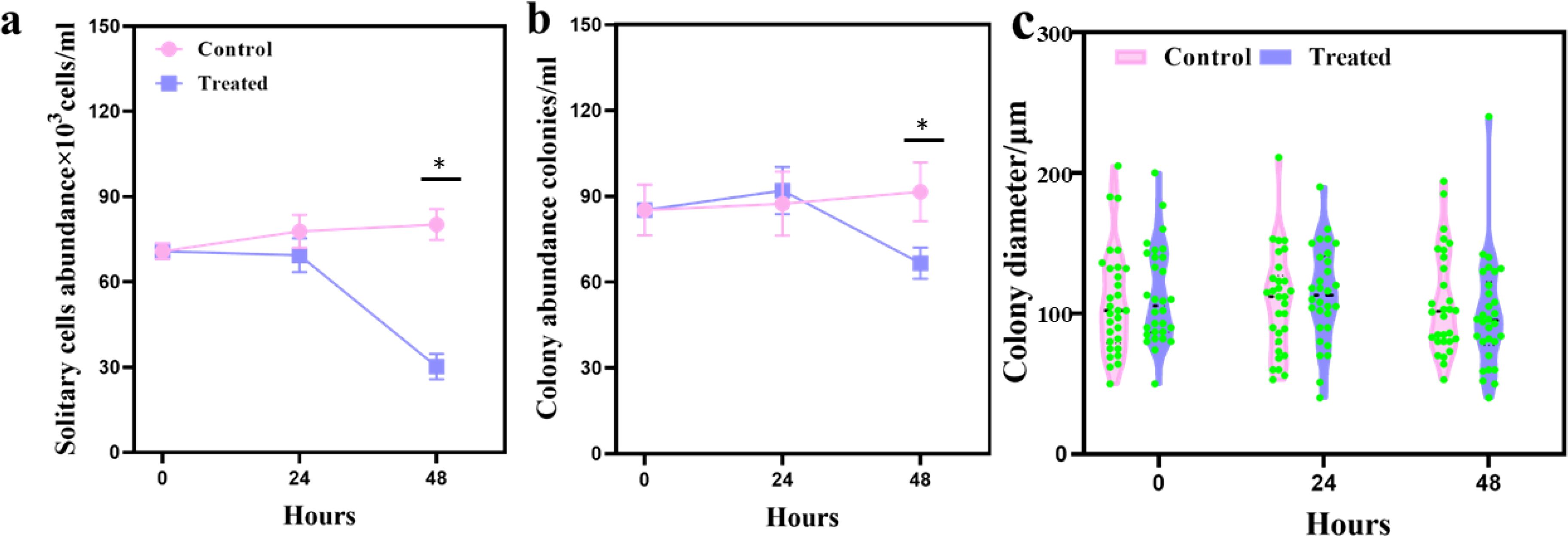
Figure 1. Algacidal effect of the leaf extracts of Ipomoea cairica on Phaeocystis globosa at 24 and 48 h. (a) Abundance of solitary cells; (b) abundance of colonies; (c) mean colony diameter. In all graphs, the control group (untreated) is shown in pink, while the treated group is shown in purple. Values graphed are the means ± the standard deviation of three biological replicates. * represents a statistically significant difference of p < 0.01.
In this study, transcriptome sequencing was performed on samples collected at three time points (0-hour, 24-hour, and 48-hour), with three biological replicates for each time point. Each sample generated between 51,070,404 and 66,254,422 raw reads. After removing low-quality reads and adapter sequences, more than 50 million high-quality clean reads were retained (Supplementary Table S1). After de novo assembly and annotation of the transcriptome data, 68,967 unigenes were obtained, with an average length of 1,118 bp and an N50 of 1,595 bp. Differentially expressed genes (DEGs) were identified, with 23,973 DEGs in the 24-hour group (15,437 upregulated and 8,536 downregulated) and 39,420 DEGs in the 48-hour group (12,488 upregulated and 26,932 downregulated). The distribution of the DEGs is shown in Figure 2a, clearly illustrating the gene expression changes between different time points.
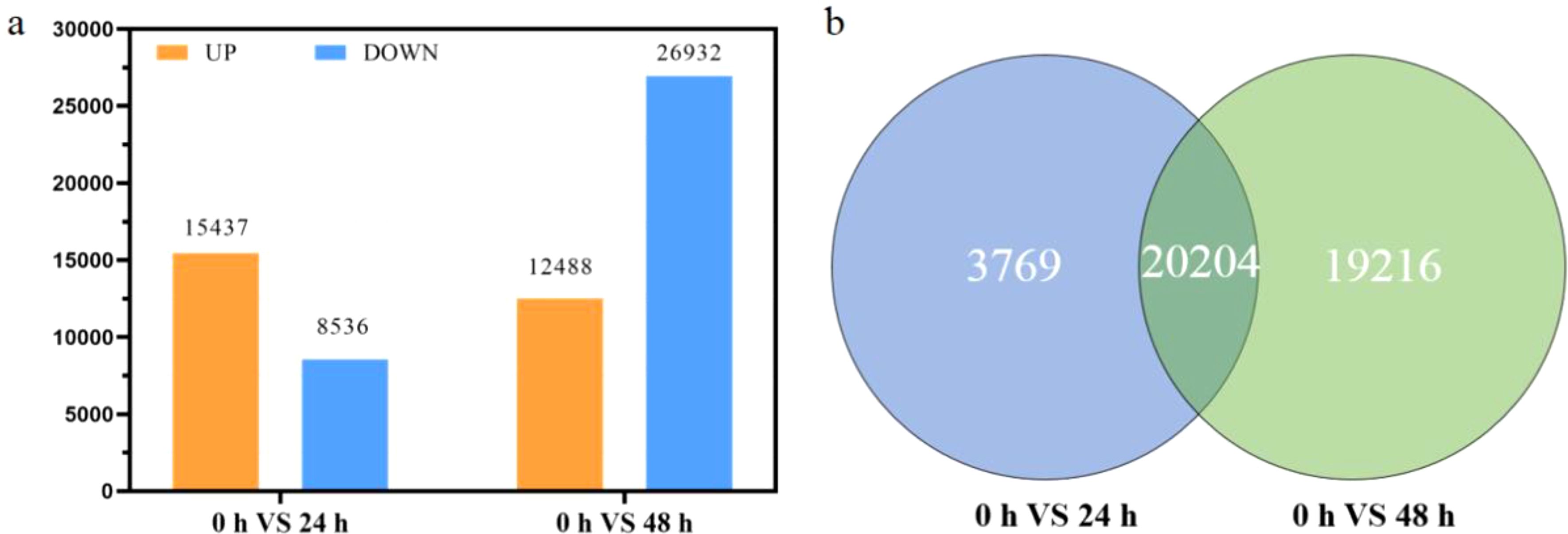
Figure 2. Global changes in gene expression patterns in Phaeocystis globosa. (a) Differentially expressed genes (DEGs) of P. globosa treated with the extract for 24 and 48 h. Upregulated and downregulated genes are shown in orange and blue, respectively; (b) Venn diagram of DEGs at 24 and 48 h.
A comparison of 24 and 48 h DEGs revealed that there were 3,768 genes specific to 24 h, 19,216 genes specific to 48 h, and 20,204 differential genes in common (Figure 2b). Trend analysis of these differential genes revealed that 2,263 of the 24 h-specific genes were first upregulated and then downregulated, and 1,331 genes were upregulated and then remained constant (Figure 3a, profiles 5, 6). Among the 48 h-specific genes 12,269 genes maintained a downregulation trend, while 5,388 genes were unchanged at 24 h and downregulated at 48 h (Figure 3b, profiles 0, 3). Among the DEGs shared at 24 h and 48 h, 7,256 genes were consistently downregulated, and 10,783 genes were first upregulated and then remained stable (Figure 3c, profiles 0, 6). The switch from upregulation to downregulation implied a significant change in the gene regulation pathway in P. globosa cells.

Figure 3. Shaded map of differentially expressed genes (DEGs). (a) DEGs specific to 24 h extract treatment; (b) DEGs specific to 48 h extract treatment; (c) shared DEGs at 24 h and 48 h of extract treatment in Phaeocystis globosa.
The GO enrichment of DEGs at 24 h and 48 h showed a great difference. There were mainly changes in molecular functions at 24 h, such as binding, ion binding, and purine ribonucleotide (Figure 4a). There were also a small number of differences in cellular fractions, such as intracellular anatomical structure, nucleus, and catalytic complex, and practically no differences in biological functions (Figure 4a). However, GO enrichment differences, such as cell protein modification process, glycosylation, protein methylation, and glycoprotein biosynthetic process, were mainly concentrated in biological processes at 48 h (Figure 4b). The above results seem to indicate that the resistance mechanisms (24 h) are dominated by molecular functional regulation, while the algaecide mechanism (48 h) is mainly due to altered biological processes.
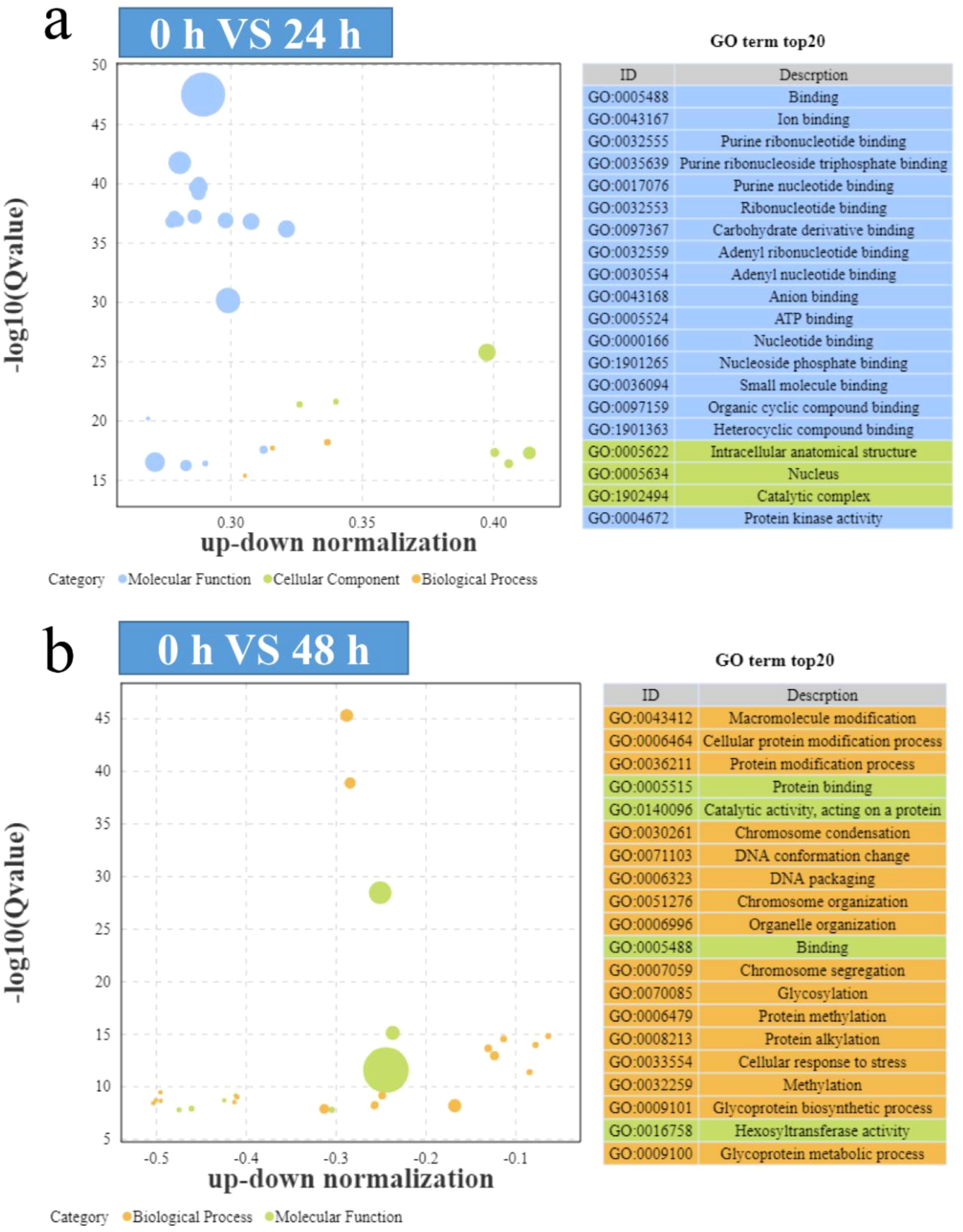
Figure 4. Bubble diagram showing the Gene Ontology (GO) enrichment of the differentially expressed genes (DEGs). (a) GO enrichment of the 24 h-specific DEGs; (b) GO enrichment of the 48 h-specific DEGs.
During the treatment process both the P. globosa signaling pathways were active, for example, pathways such as the mitogen-activated protein kinase (MAPK) signaling pathway and the phosphatidylinositol signaling system were significantly enriched (Figure 4). In addition, the KEGG pathway enrichment analysis revealed enrichment in plant hormone signal transduction, autophagy, and plant–pathogen interaction at 24 h and 48 h (Figure 5). However, some differences in the KEGG pathway enrichment were found, such as ubiquitin-mediated proteolysis and peroxisome at 24 h (Figure 5a), as well as photosynthesis-antenna protein and glycan degradation at 48 h (Figure 5b). Changes in the KEGG pathways indicated changes in molecular metabolic pathways in P. globosa cells at different time points.
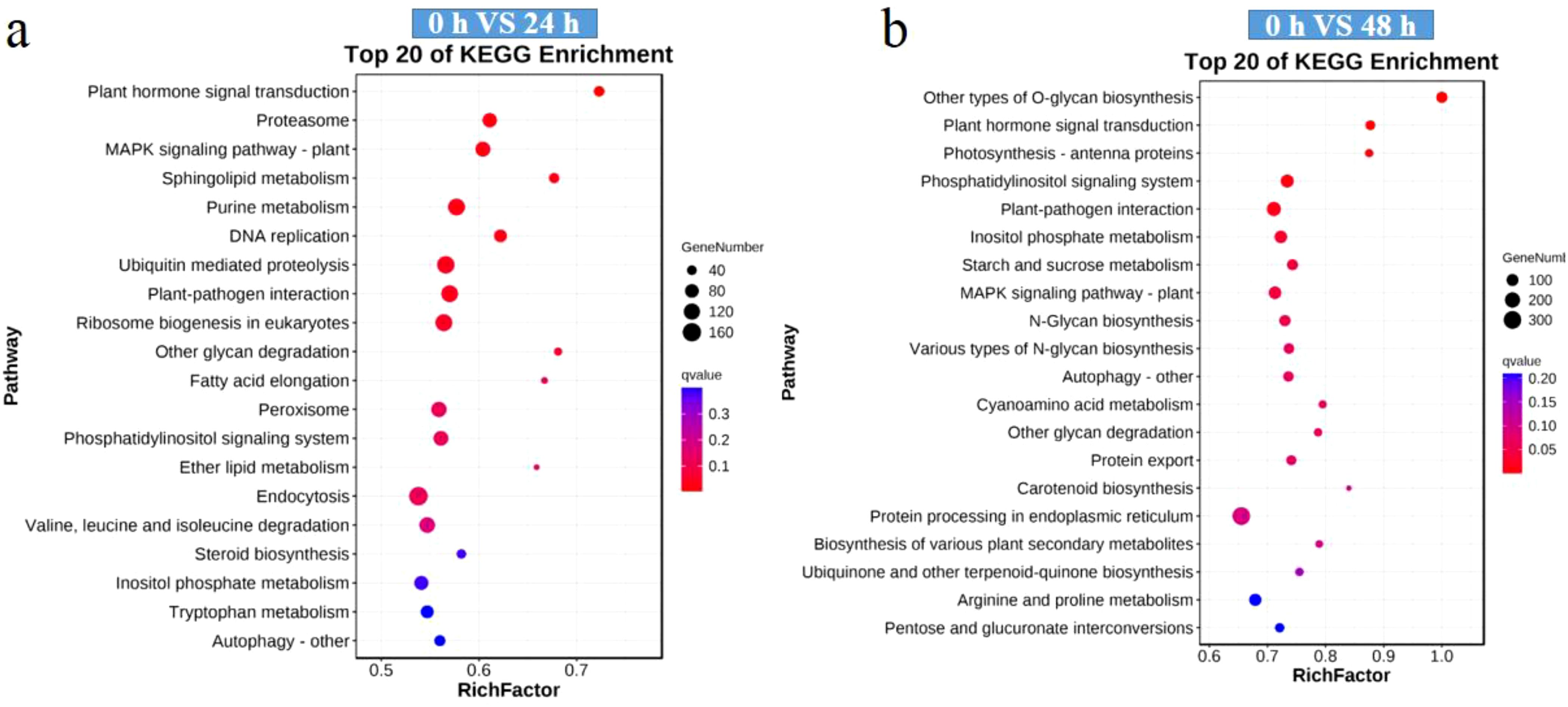
Figure 5. Top 20 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways most enriched in the differentially expressed genes (DEGs). (a) KEGG pathways of the 24 h-specific DEGs; (b) KEGG pathways of the 48 h-specific DEGs. Dot size reflects the number of DAMs associated with the corresponding pathway, while color indicates Q-value (smaller Q-values are redder, larger Q-values are bluer).
At different time points, the response of P. globosa to I. cairica extracts showed significant differences. At 24 h, differentially expressed genes (DEGs) were mainly enriched in molecular function categories such as binding, ion binding, and purine ribonucleotide-related functions, while the activation of the MAPK signaling pathway and phosphatidylinositol signaling system indicated that P. globosa responded to stress by regulating molecular functions and signaling pathways, demonstrating certain resistance. However, these responses might not be sufficient to fully counteract the algicidal effects. By 48 h, DEGs were primarily enriched in biological process categories such as cell protein modification process, glycosylation, and glycoprotein biosynthetic process. Meanwhile, KEGG pathway analysis revealed significant enrichment in pathways related to photosynthesis-antenna proteins and glycan degradation. These changes led to metabolic disorders and cellular dysfunction in P. globosa, ultimately resulting in cell death. In summary, P. globosa exhibited resistance through molecular function regulation in the early stage, while significant changes in biological processes and metabolic pathways in the later stage contributed to the algicidal effects.
DEGs in the different KEGG pathways were evaluated in greater detail. In the MAPK signaling pathway, most of the genes were upregulated at 24 h and then remain unchanged until 48 h (Supplementary Figure S1). For example, the gene encoding mitogen-activated protein kinase 1 (MPK6) was upregulated by 2.82-fold and 3.01-fold at 24 and 48 h, respectively, while the gene encoding mitogen-activated protein kinase (MKK3) was upregulated by 3.69-fold and 3.96-fold at 24 and 48 h, respectively. Interestingly, the gene expression trend of the phosphatidylinositol (PI) signaling pathway was similar to that of the MAPK signaling pathway, with most genes also being upregulated and then remaining unchanged. These upregulated genes all play important roles in the PI signaling pathway, including genes encoding phosphatidylinositol-4, 5-diphosphate 3-kinase, phosphatidylinositol 4-kinase, and phospholipase C-eta2 (Supplementary Figure S2). These results suggest that P. globosa perceived the external environmental stress caused by the extracts and then regulated the cellular metabolic pathways through MAPK and PI signaling pathways.
Metabolic pathways that played a role in the resistance system of P. globosa and algaecide mechanism of I. cairica were identified based on P. globosa DEG trend analysis. Transcriptome analysis showed that the levels of antioxidant gene expression in P. globosa cells were significantly upregulated at 24 h after treatment with the extract, including superoxide dismutase (SOD) genes (SODA, Sod1, and Sod2), catalase (CAT) genes (cat), and PRDX5 genes (PRXIIB) (Supplementary Figure S3). This implies that extract treatment may induce an oxidative stress response in P. globosa cells, which activated the antioxidant system. Some antioxidant enzyme genes (SODCC2, Sod2, and SODCC) were downregulated at 48 h, leading to the accumulation of reactive oxygen species (ROS) and subsequent cell damage. In addition, 13 DEGs in the autophagic pathway were identified, of which 10 genes were upregulated at 24 h and six genes were downregulated at 48 h (Supplementary Figure S3). The gene encoding autophagy-related protein 8 (ATG8) was upregulated by 1.67-fold at 24 h and decreased to 0.55-fold at 48 h. Similarly, the gene encoding ubiquitin-like modifier-activating enzyme (ATG7) was upregulated by 1.67-fold at 24 h and downregulated by 0.58-fold at 48 h. This implies that the upregulation of autophagy-related genes at 24 h could maintain intracellular protein homeostasis. In comparison, the downregulation of genes at 48 h would completely disrupt protein homeostasis, thus causing damage to the cell.
Transcriptome data showed differential photosynthesis-related genes, half of which were downregulated at 24 h and all of which were downregulated at 48 h (Supplementary Figure S4). These genes are involved in the process of photosynthetic light reaction, including antenna proteins (Lhcal, Lhca4, and Lhcb6), photosystem II (PsbA, PsbC, PsbB, PsbE, PsbO, PsbU, PsbZ, and Psb27), photosystem I (PsaA and PsaE), cytochrome b6 (PetA and PetC), photosynthetic electron transport (PetE, PETF, PetH, and PetJ) and F-type ATPase (atpA and atpD). This indicates that I. cairica extracts could significantly inhibit genes related to the photosynthetic system of P. globosa, thus disrupting photosynthesis, which may be the main aspect of its algacidal effect.
A total of 10,837 chemical compounds were identified from I. cairica leaf extracts using UHPLC, of which 441 were known compounds (Supplementary Table S2). Among the known compounds, carboxylic acids and their derivatives were the most diverse, with 155 types. The others major compounds were organooxygen compounds (30), fatty acids (26), benzene and substituted derivatives (30), steroids and steroid derivatives (22), pyridines and their derivatives (26), and flavonoids (10). Because previous studies have shown that flavonoids have good algacidal effects, the focus here was also on flavonoids. The relative content of apigenin-7-O-glucoside was the highest among the flavonoids, so it was inferred that this might be one of the main algacidal active substances in the extracts. Unfortunately, there were no significant differences in the solitary cell abundance, colony number, or colony diameter of P. globosa following the addition of apigenin-7-O-glucoside (50 μg/mL and 100 μg/mL) (Figure 6).
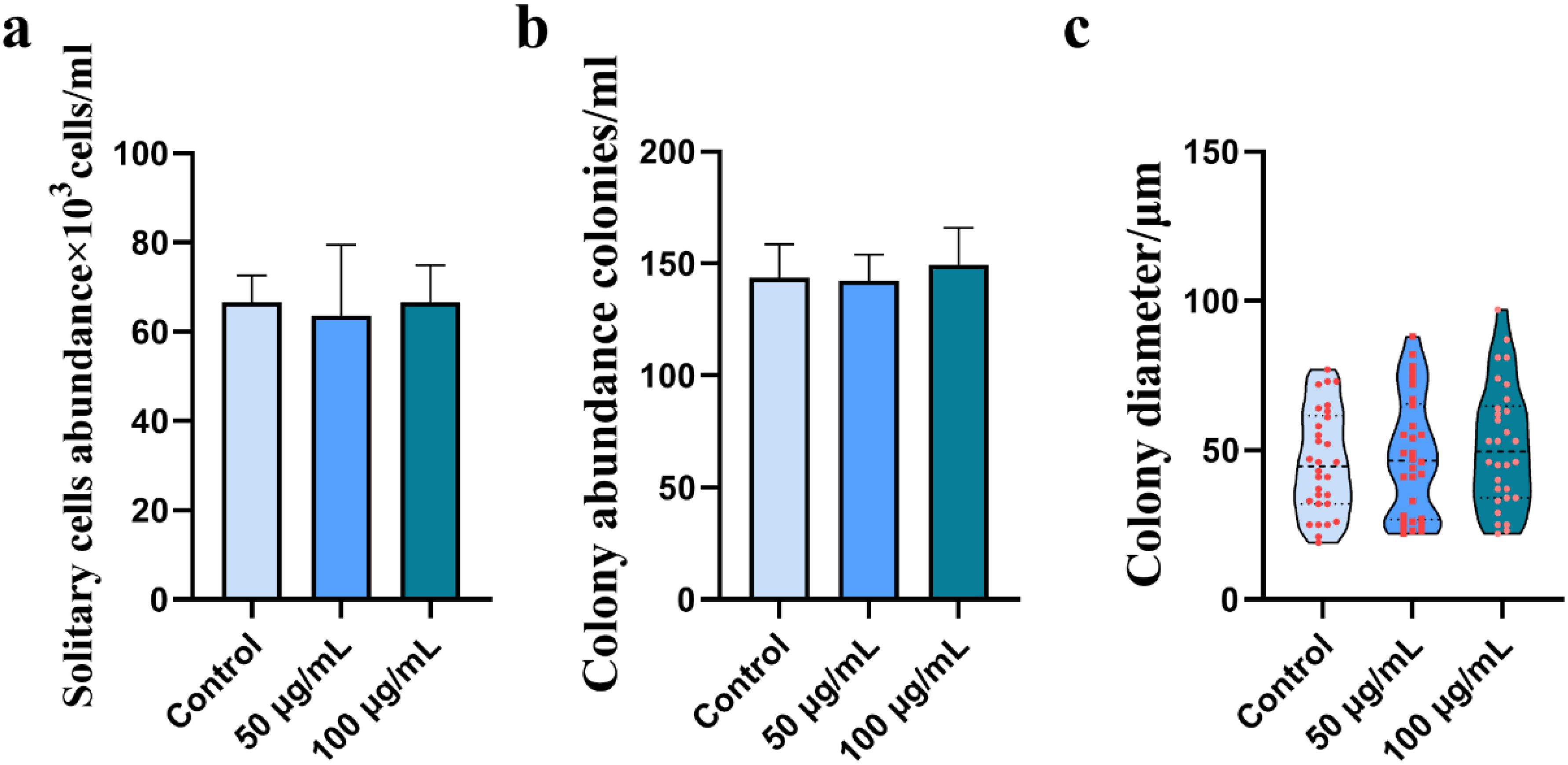
Figure 6. Effects of apigenin-7-O-glucoside (0 μg/mL, 50 μg/mL, and 100 μg/mL) on Phaeocystis globosa. (a) Abundance of solitary cells; (b) abundance of colonies; (c) mean colony diameter.
The use of I. cairica’s allelopathic effect to inhibit the growth of P. globosa is in line with the scientific concept of green environmental protection. In previous studies, it was found that I. cairica extract could rapidly inhibit the growth of solitary cells of P. globosa and could penetrate inside the colonies to kill the cells (Liang et al., 2022). In the present study, although I. cairica extract eventually inhibited the growth of P. globosa cells, there was slight growth at 24 h after treatment, suggesting the existence of some resistance mechanism to protect the cells from damage. The resistance mechanisms of P. globosa and the algaecide mechanism of I. cairica extract remain unclear. Therefore, the present study investigated the transcriptomic response of P. globosa treated with extract (1% final concentration) at different time points to reveal the changes in the gene regulation of cells. This work also identified possible allelopathic compounds in the extracts, but the screened apigenin-7-O-glucoside was not an active algacidal compound.
The processing of external environmental information is a major physiological process by which cells respond to environmental stress. Plants use various signaling pathways to perceive and react to these external pressures. The MAPK signaling pathway plays a crucial role in plants, primarily responsible for transmitting signals from external stress sources. These signals are transmitted to the cell nucleus or cytoplasm, triggering a series of corresponding cellular responses (Taj et al., 2010). In this study, most of DEGs in the MAPK signaling pathway exhibited a pattern of upregulation followed by maintenance, a result that is consistent with findings from other studies, further supporting the key role of the MAPK signaling pathway in plant responses to environmental stress. For example, Ren et al. (2022) found that all genes encoding MAPKs, MAPKKs, and MAPKKKs were upregulated after treatment with modified clay in P. globosa. These overexpressed genes can transduce stimulatory signals to cells and regulate cell growth and antioxidant defense systems (Doczi et al., 2012). The vast majority of genes in the plant MAPK family are involved in response to abiotic stress caused by adverse factors in the environment. In Arabidopsis, MPK6 is gradually activated because MPK6 can alleviate high salt stress by rapidly phosphorylating the Na+/H+ reverse transporter protein, thereby reducing intracellular sodium ions (Kim et al., 2012).
The PI signaling pathway is considered critical for plant responses to many environmental factors. In the PI signaling pathway, extracellular signaling molecules bind to cell-surface G protein-coupled receptors and activate phospholipase on the plasma membrane, producing inositol 1, 4, 5-trisphosphate and diacylglycerol (Munnik et al., 1998). In the present study, most of the PI signaling pathway genes were upregulated and then unchanged, including PLCD1, pikB, and CML3. This is consistent with previous studies, in which P. globosa relied on PI signal transduction during the perception of external stress. Wang et al. (2018) found that gene expression of PI5K and PLC in Pyropia haitanensis was significantly upregulated under high temperature stress. Liang et al. (2023) also found that the PI signaling pathway was upregulated in P. globosa under warming and acidifying environments. Furthermore, phospholipase C has been shown to regulate the transmission of oxidative stress signals from the plasma membrane in response to environmental stress (Cooper et al., 1999). This suggests that the PI signaling pathway mediates the defense system of P. globosa cells.
Many studies have shown that both bacteria and algaecides may lead to the excessive accumulation of ROS in algal cells, implying that treatment with I. cairica extract may induce oxidative stress in P. globosa cells (Zhang et al., 2017).This triggers the intracellular antioxidant system, including superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD). SOD primarily converts superoxide radicals (O2−) into hydrogen peroxide (H2O2), which is subsequently removed by CAT and POD. In this study, all genes encoding SOD, CAT, and PRDX5 were upregulated in P. globosa cells after 24 h treatment. This was also demonstrated in the previous study, where treatment with I. cairica extract increased antioxidant enzyme activity in P. globosa cells (Liang et al., 2022). Similar phenomena have been found in other studies on algaecides. For example, Ren et al. (2022) found that treatment of P. globosa with modified clay resulted in different degrees of enhancement of cellular antioxidant system activity (including SOD, CAT, POD, GPX, GST, and GSH activity), which was associated with the excessive production of ROS. Zhu et al. (2022) discovered Microbulbifer sp. YX04 could induce oxidative damage in P. globosa, leading to enhanced antioxidant enzyme activity and upregulation of related genes. There was no significant change in the number of P. globosa cells at 24 h after I. cairica extract treatment, suggesting that the antioxidant system played a protective role. Most of the antioxidant enzyme genes remained continuously upregulated and a small proportion were downregulated at 48 h, indicating that the cells were under continuous oxidative stress. In addition, a significant decrease in cell number indicated that the accumulation of ROS far exceeded the capabilities of the antioxidant system, eventually leading to cell death.
In eukaryotic cells, organelle transformation is typically achieved through the process of autophagy. Autophagy is a highly regulated process where damaged organelles are encapsulated in a double-membrane vesicle called an autophagosome and transported to vacuoles in yeast and plants, or to lysosomes in animal cells for degradation (Mizushima and Komatsu, 2011). Initially, autophagy was considered primarily a process for the degradation of cytoplasmic components, mainly for nutrient recycling, especially under starvation conditions (Takeshige et al., 1992). In the green algae Chlamydomonas reinhardtii, autophagy-induced degradation of ribosomal proteins has been observed, with the degraded materials subsequently used for triglyceride accumulation, demonstrating the complex roles of autophagy under specific physiological conditions (Jiang et al., 2012). Recent studies have further revealed that autophagy plays an essential role not only in nutrient recycling but also in the selective removal of damaged organelles, particularly in the clearance of dysfunctional mitochondria. Mitochondrial autophagy (mitophagy) is a typical example, which has been widely studied in yeast and mammals (Kanki et al., 2015). The autophagy process relies on a series of key genes known as autophagy-related genes (ATGs). To date, 41 ATGs have been identified in yeast, with 15 core ATGs playing a crucial role in the nucleation and expansion of the autophagosome membrane, and they are essential for all types of autophagy processes (Yao et al., 2015). The significant upregulation of genes encoding autophagy-related genes was also found in the present study, including ATG3, ATG4, ATG7, ATG8, and ATG9. Among them, ATG8 plays an important role in the formation of autophagosomes, target recognition, and vesicle tethering processes. Zhu et al. (2022) found that the treatment of P. globosa with bacteria (Microbulbifer sp. YX04) triggered an increase in the atg8 flux and led to autophagic cell death. However, the present study’s findings suggested that autophagy was not the true cause of cell death because there was no significant change in cell number at 24 h of treatment and autophagy-related genes were significantly expressed. If damaged mitochondria are not cleared through autophagy, ROS can accumulate, leading to oxidative stress. This oxidative stress may result in mutations in mitochondrial DNA, disrupting mitochondrial function and causing toxic effects in the cell (Kurihara et al., 2012). Similarly, chloroplast autophagy plays an essential role in the physiological processes of plant cells. Damaged chloroplasts and chloroplast proteins must be cleared or repaired through autophagy to maintain optimal chloroplast function. This process not only prevents further damage but also ensures the efficiency of photosynthesis in plants, avoiding growth and metabolic issues caused by the loss of chloroplast function (Izumi et al., 2017). Our interpretation is that autophagy can regulate protein homeostasis and play a protective role when cells experience adversity.
ROS are highly reactive molecules generated in cells that can oxidize various biomolecules, including lipids, DNA, and proteins, leading to severe oxidative damage of organelle membranes. The accumulation of ROS has a negative impact on the overall function of the cell, particularly through oxidative damage to key molecules such as membrane lipids and nucleic acids (Han et al., 2001). To counteract this damage, cells have an antioxidant enzyme system that reduces ROS production and protects biomolecules from oxidative stress. However, when the accumulation of ROS exceeds the neutralizing capacity of the antioxidant system, ROS continue to oxidize polyunsaturated fatty acids, amino acids, and nucleic acids, leading to the degradation of cellular components and ultimately triggering cell death (Jacobson, 1996). The primary sources of ROS are the mitochondria and chloroplasts. In mitochondria, electrons are transferred from highly reduced components to molecular oxygen, generating ROS. In chloroplasts, the light reactions of photosynthesis are also a significant source of ROS (Auchère and Rusnak, 2002). In the present study, transcriptome data did not show significant differences in respiration-related genes, while photosynthesis-related genes were significantly downregulated. The results also indicated that the rapid light-response curve of P. globosa cells treated with I. cairica extract (1%) gradually declined over time. In addition, the alpha value, maximum relative electron transport rate, and light saturation point all showed a decreasing trend (Liang et al., 2022). These findings suggest that the I. cairica extract not only inhibited photosynthesis but also impeded the repair of the photosynthetic system through downregulating photosynthetic gene expression. Altogether, these findings suggest that the inhibition of the photosynthetic system caused the accumulation of intracellular ROS.
During the accumulation of chloroplast ROS material, the formation of O2− occurs during the photosynthetic light reaction, and the formation of H2O2 results from the water splitting manganese complex in PS II (Pospisil, 2012). When algal cells are subjected to external environmental stress, electrons in H2O are transferred to Fd via PS II and PS I, and then to O2 to eventually form superoxide (Gururani et al., 2015). The transcriptome data showed that all photosynthesis-related genes were downregulated at 48 h, including antenna proteins, photosystem II, photosystem I, cytochrome b6, photosynthetic electron transport, and F-type ATPase. This further affected the biosynthesis of photosynthetic pigments and the regeneration of electron transport chain complexes, ultimately leading to the complete disruption of photosynthesis and the excessive accumulation of ROS. The accumulation of ROS caused alteration of the properties of the thylakoid membrane, which prevented the transfer of electrons to F-type ATPase and NADP 1, affecting electron transport and eventually causing irreversible damage to the algal cells.
Although the I. cairica leaf extracts effectively inhibited the growth of solitary cells and colonies of P. globosa, the specific algacidal components are not yet known. Tens of thousands of compounds were identified using UHPLC, and this research finally targeted flavonoids. No direct correlation was observed between concentration and peak area, as different compounds exhibit varying ionization efficiencies and ionic stabilities. However, compounds within similar chemical classes often share comparable ionization potentials and chemical properties, which allows for the relative comparison of flavonoid compound concentrations (St-Pierre et al., 2018). The highest flavonoid content was that found in apigenin-7-O-glucoside, which is known to have biological, pharmacological, and medical properties, including anti-inflammatory, antioxidant, and anti-tumor mechanisms that inhibit cancer development and metastasis through affecting cell proliferation, blocking the cell cycle, and promoting apoptosis (Hadrich and Sayadi, 2018; Guo et al., 2019). Zhu et al. (2019) found that the growth of P. globosa was inhibited by luteolin-7-O-glucuronide extracted from the seagrass Enhalus acoroides. Therefore, it was hypothesized that apigenin-7-O-glucoside had possible algacidal effects. However, the growth of P. globosa cells was not affected following the addition of different concentrations of apigenin-7-O-glucoside. This means that algacidal substances may not be limited to flavonoids, and future research may seek to identify algacidal substances in other classes.
Among the identified compounds, Phlorizin, Nobiletin, and Procyanidin stand out as likely contributors due to their known bioactivities and mechanisms of action (Table 1). Phlorizin, a dihydrochalcone glycoside, is known to inhibit glucose transporters, which could disrupt energy metabolism in P. globosa and lead to cell death (Nozu et al., 2023). Nobiletin, a polymethoxyflavone, has demonstrated anti-inflammatory and anticancer properties, and its ability to interfere with cellular signaling pathways in P. globosa may explain its algicidal effects (Moazamiyanfar et al., 2023). Additionally, Procyanidin, a condensed tannin with strong antioxidant activity, may exert its effects by binding to proteins or disrupting cellular membranes, thereby contributing to its algicidal potential (Chen et al., 2022). While these compounds are promising candidates, further experimental validation, such as bioassay-guided fractionation and compound-specific toxicity testing, is necessary to confirm their roles. We have expanded the discussion in the manuscript to address these points and emphasize the need for future studies to identify the specific active compounds responsible for the observed effects.
This study advances the field of algicidal research by identifying novel bioactive compounds from I. cairica and elucidating their mechanisms of action on P. globosa. These findings provide a foundation for developing eco-friendly algaecides as alternatives to traditional chemical treatments. However, we acknowledge several limitations, including the lack of in situ validation and challenges in applying these extracts in natural environments. Factors such as variability in water chemistry, potential ecological impacts on non-target organisms, and scalability issues need to be addressed. Future research should focus on field trials, ecological risk assessments, and optimization of extraction and application methods to bridge the gap between laboratory findings and real-world applications.
This study elucidated the inhibitory mechanism of I. cairica extracts on P. globosa by analyzing transcriptomes at different time points. The results demonstrated that I. cairica extracts (1% final concentration) effectively inhibited P. globosa growth, suppressed free single-cell proliferation and colony formation, and achieved high removal efficiency (Figure 7). At 24 h, P. globosa activated resistance mechanisms through the MAPK and PI signaling pathways to counteract external stress, including ROS scavenging by the antioxidant enzyme system and protein homeostasis maintenance via the autophagy system. By 48 h, the algicidal mechanism dominated, with the extract inhibiting photosynthesis-related gene expression, leading to ROS accumulation in chloroplasts and eventual cell death. Although apigenin-7-O-glucoside was identified as a component of the extract, it did not exhibit algicidal activity. These findings provide valuable insights into the dynamic gene expression changes during P. globosa inhibition and offer a theoretical basis for controlling P. globosa blooms. Future research should focus on testing I. cairica extracts in real-world settings, optimizing their application for large-scale use, and evaluating their ecological impacts to advance sustainable solutions for harmful algal bloom management.
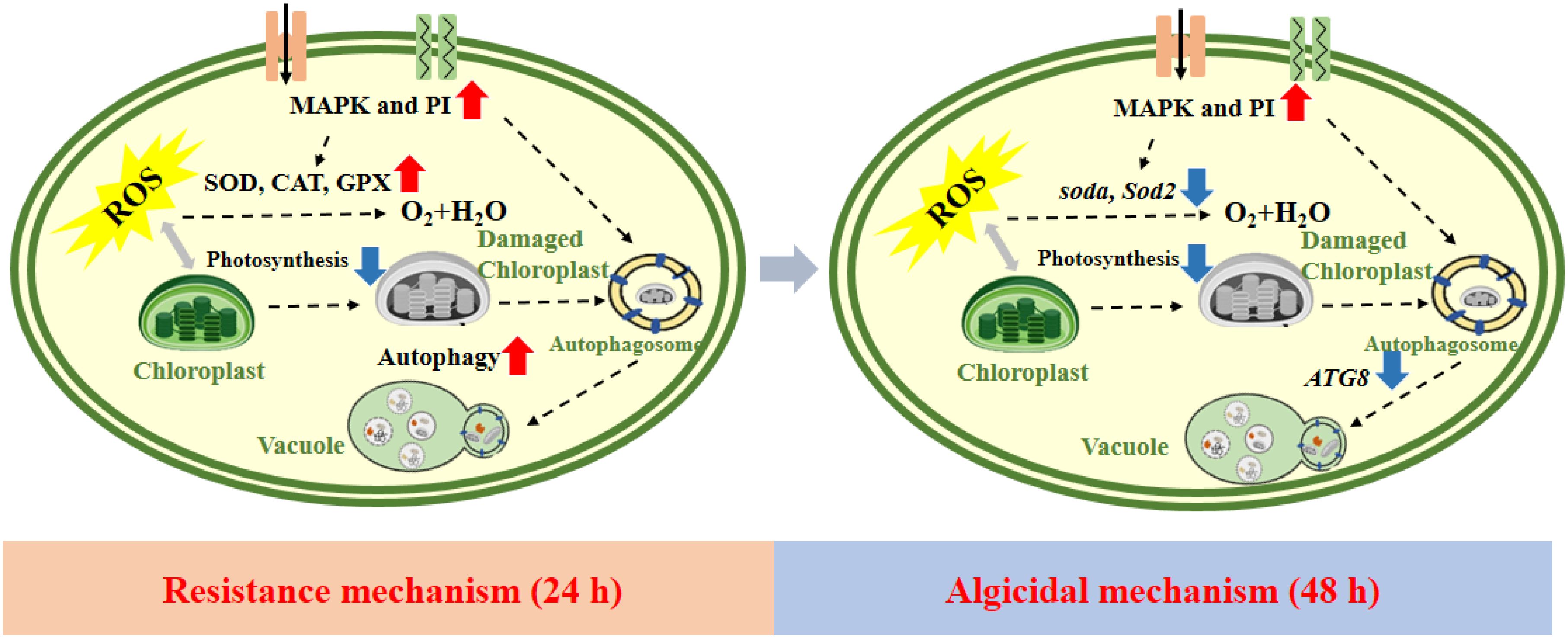
Figure 7. Schematic diagram depicting the algacidal mechanism of Ipomoea cairica extracts against Phaeocystis globosa.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA913698.
DL: Data curation, Project administration, Writing – original draft, Writing – review & editing. HL: Methodology, Supervision, Writing – review & editing. YP: Methodology, Software, Writing – review & editing. ZL: Writing – review & editing. HX: Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by Weifang University Doctoral Start-up Fund (BS44124018, BS44122066) and the Natural Science Youth Fund of Shandong Province (ZR2024QD184).
We thank LetPub (www.letpub.com) for linguistic assistance and pre-submission expert review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1580077/full#supplementary-material
Anderson D. M. (2009). Approaches to monitoring, control and management of harmful algal blooms (HABs). Ocean Coast. Manage. 527, 342–347. doi: 10.1016/j.ocecoaman.2009.04.006
Auchère F., Rusnak F. (2002). What is the ultimate fate of superoxide anion in vivo? J. Biol. Inorganic Chem. 76, 664–667. doi: 10.1007/s00775-002-0362-2
Blauw A. N., Los F. J., Huisman J., Peperzak L. (2010). Nuisance foam events and Phaeocystis globosa blooms in Dutch coastal waters analyzed with fuzzy logic. J. Mar. Syst. 833-4, 115–126. doi: 10.1016/j.jmarsys.2010.05.003
Brussaard C., Kuipers B., Veldhuis M. (2005). A mesocosm study of Phaeocystis globosa population dynamics: I. Regulatory role of viruses in bloom control. Harmful Algae. 45, 859–874. doi: 10.1016/j.hal.2004.12.015
Chen H., Wang W., Yu S., Wang H., Tian Z., Zhu S. (2022). Procyanidins and their therapeutic potential against oral diseases. Molecules 27, 2932. doi: 10.3390/molecules27092932
Chen Y. Q., Wang N., Zhang P., Zhou H., Qu L. H. (2002). Molecular evidence identifies bloom-forming Phaeocystis (Prymnesiophyta) from coastal waters of southeast China as Phaeocystis globosa. Biochem. Syst. Ecol. 301, 15–22. doi: 10.1016/S0305-1978(01)00054-0
Cooper K. F., Mallory M. J., Strich R. (1999). Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol. Cell. Biol. 195, 3338–3348. doi: 10.1128/MCB.19.5.3338
Doczi R., Okresz L., Romero A. E., Paccanaro A., Bogre L. (2012). Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci. 179, 518–525. doi: 10.1016/j.tplants.2012.05.009
Guo H., Li M., Xu L. J. (2019). Apigetrin treatment attenuates LPS- induced acute otitis media though suppressing inflammation and oxidative stress. Biomed. Pharmacother. 109, 1978–1987. doi: 10.1016/j.biopha.2018.07.022
Gururani M. A., Venkatesh J., Tran L. S. P. (2015). Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 89, 1304–1320. doi: 10.1016/j.molp.2015.05.005
Hadrich F., Sayadi S. (2018). Apigetrin inhibits adipogenesis in 3T3-L1 cells by downregulating PPARγ and CEBP-α. Lipids Health Disease 171, 1–8. doi: 10.1186/s12944-018-0738-0
Hamm C. E., Simson D. A., Merkel R., Smetacek V. (1999). Colonies of Phaeocystis globosa are protected by a thin but tough skin. Mar. Ecol. Prog. Ser. 187, 101–111. doi: 10.3354/meps187101
Han D., Williams E., Cadenas E. (2001). Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space. Biochem. J. 3532, 411–416. doi: 10.1042/bj3530411
Izumi M., Ishida H., Nakamura S., Hidema J. (2017). Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell. 292, 377–394. doi: 10.1105/tpc.16.00637
Jacobson M. D. (1996). Reactive oxygen species and programmed cell death. Trends Biochem. Sci. 213, 83–86. doi: 10.1016/S0968-0004(96)20008-8
Jakobsen H. H., Tang K. W. (2002). Effects of protozoan grazing on colony formation in Phaeocystis globosa (Prymnesiophyceae) and the potential costs and benefits. Aquat. Microbial Ecol. 273, 261–273. doi: 10.3354/ame027261
Jiang Q., Zhao L., Dai J. B., Wu Q. Y. (2012). Analysis of autophagy genes in microalgae: chlorella as a potential model to study mechanism of autophagy. PloS One e41826. doi: 10.1371/journal.pone.0041826
Kanki T., Furukawa K., Yamashita S. (2015). Mitophagy in yeast: Molecular mechanisms and physiological role. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 185310, 2756–2765. doi: 10.1016/j.bbamcr.2015.01.005
Kim J. M., Woo D. H., Kim S. H., Lee S. Y., Park H. Y., Seok H. Y., et al. (2012). Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 311, 217–224. doi: 10.1007/s00299-011-1157-0
Kim M.-C., Yoshinaga I., Imai I., Nagasaki K., Itakura S., Ishida Y. (1998). A close relationship between algicidal bacteria and termination of Heterosigma akashiwo (Raphidophyceae) blooms in Hiroshima Bay, Japan. Mar. Ecol. Prog. Series 170, 25–32. doi: 10.3354/meps170025
Kurihara Y., Kanki T., Aoki Y., Hirota Y., Saigusa T., Uchiumi T., et al. (2012). Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J. Biol. Chem. 2875, 3265–3272. doi: 10.1074/jbc.M111.280156
Liang D., Wang X., Huo Y., Wang Y., Li S. (2020). Differences in the formation mechanism of giant colonies in two Phaeocystis globosa strains. Int. J. Mol. Sci. 21 (15), 5393. doi: 10.3390/ijms21155393
Liang D., Xiang H., Jin P., Xia J. (2023). Response mechanism of harmful algae Phaeocystis globosa to ocean warming and acidification. Environ. pollut. 320. doi: 10.1016/j.envpol.2023.121008
Liang D., Xiang H., Xia J. (2022). Inhibitory effects of Ipomoea cairica extracts on the harmful algae Phaeocystis globosa. Mar. pollut. Bulletin 185, 114228. doi: 10.1016/j.marpolbul.2022.114228
Mayali X., Azam F. (2004). Algicidal bacteria in the sea and their impact on algal blooms. J. Eukaryotic Microbiol. 512, 139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x
Mizushima N., Komatsu M. (2011). Autophagy: renovation of cells and tissues. Cell 1474, 728–741. doi: 10.1016/j.cell.2011.10.026
Moazamiyanfar R., Rezaei S., AliAshrafzadeh H., Rastegar-Pouyani N., Jafarzadeh E., Mouludi K., et al. (2023). Nobiletin in cancer therapy; mechanisms and therapy perspectives. Curr. Pharm. Design 29, 1713–1728. doi: 10.2174/1381612829666230426115424
Mulderij G., Mooij W. M., Van Donk E. (2005). Allelopathic growth inhibition and colony formation of the green alga Scenedesmus obliquus by the aquatic macrophyte Stratiotes aloides. Aquat. Ecol. 391, 11–21. doi: 10.1007/s10452-004-1021-1
Munnik T., Irvine R., Musgrave A. (1998). Phospholipid signalling in plants. Biochim. ET Biophys. Acta-lipids Lipid Metab. 13893, 222–272. doi: 10.1016/S0005-2760(97)00158-6
Nozu T., Miyagishi S., Ishioh M., Takakusaki K., Okumura T. (2023). Phlorizin attenuates postoperative gastric ileus in rats. Neurogastroenterol. Motil. 35, e14659. doi: 10.1111/nmo.v35.11
Patino R., Rashel R. H., Rubio A., Longing S. (2018). Growth-suppressing and algicidal properties of an extract from Arundo donax, an invasive riparian plant, against Prymnesium parvum, an invasive harmful alga. Harmful Algae. 71, 1–9. doi: 10.1016/j.hal.2017.11.005
Pospisil P. (2012). Molecular mechanisms of production and scavenging of reactive oxygen species by photosystem II. Biochim. Biophys. Acta (BBA) - Bioenergetics. 18171, 218–231. doi: 10.1016/j.bbabio.2011.05.017
Ren X. Z., Yu Z. M., Song X. X., Zhu J. A., Wang W. T., Cao X. H. (2022). Effects of modified clay on the formation of Phaeocystis globosa colony revealed by physiological and transcriptomic analyses. Sci. Total Environ. 838, 155985. doi: 10.1016/j.scitotenv.2022.155985
Rousseau V., Chretiennot-Dinet M. J., Jacobsen A., Verity P., Whipple S. (2007). The life cycle of Phaeocystis: state of knowledge and presumptive role in ecology. Biogeochemistry 831-3, 29–47. doi: 10.1007/s10533-007-9085-3
Schoemann V., Becquevort S., Stefels J., Rousseau W., Lancelot C. (2005). Phaeocystis blooms in the global ocean and their controlling mechanisms: a review. J. Sea Res. 531-2, 43–66. doi: 10.1016/j.seares.2004.01.008
St-Pierre A., Blondeau D., Lajeunesse A., Bley J., Bourdeau N., Desgagné-Penix I. (2018). Phytochemical screening of quaking aspen (Populus tremuloides) extracts by UPLC-QTOF-MS and evaluation of their antimicrobial activity. Molecules 237, 1739. doi: 10.3390/molecules23071739
Sun R., Sun P. F., Zhang J. H., Esquivel-Elizondo S., Wu Y. H. (2018). Microorganisms-based methods for harmful algal blooms control: A review. Biores. Technol. 248, 12–20. doi: 10.1016/j.biortech.2017.07.175
Taj G., Agarwal P., Grant M., Kumar A. (2010). MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 511, 1370–1378. doi: 10.4161/psb.5.11.13020
Takeshige K., Baba M., Tsuboi S., Noda T. (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1192, 301–311. doi: 10.1083/jcb.119.2.301
Tan S., Hu X., Yin P., Zhao L. (2016). Photosynthetic inhibition and oxidative stress to the toxic Phaeocystis globosa caused by a diketopiperazine isolated from products of algicidal bacterium metabolism. J. Microbiol. 545, 364–375. doi: 10.1007/s12275-016-6012-0
van Rijssel M., Hamm C. E., Gieskes W. W. (1997). Phaeocystis globosa (Prymnesiophyceae) colonies: hollow structures built with small amounts of polysaccharides. Eur. J. Phycol. 322, 185–192. doi: 10.1080/09670269710001737119
Wang W. L., Lin Y. H., Teng F., Ji D. H., Xu Y., Chen C. S., et al. (2018). Comparative transcriptome analysis between heat-tolerant and sensitive Pyropia haitanensis strains in response to high temperature stress. Algal Res.-Biomass Biofuels Bioprod. 29, 104–112. doi: 10.1016/j.algal.2017.11.026
Wang X. D., Song H. Y., Wang Y., Chen N. S. (2021). Research on the biology and ecology of the harmful algal bloom species Phaeocystis globosa in China: Progresses in the last 20 years. Harmful Algae 107, 102057. doi: 10.1016/j.hal.2021.102057
Wang X. D., Tang K. W., Wang Y., Smith W. O. (2010). Temperature effects on growth, colony development and carbon partitioning in three Phaeocystis species. Aquat. Biol. 93, 239–249. doi: 10.3354/ab00256
Weber E., Sun S. G., Li B. (2008). Invasive alien plants in China: diversity and ecological insights. Biol. Invasions 108, 1411–1429. doi: 10.1007/s10530-008-9216-3
Yao Z., Delorme-Axford E., Backues S. K., Klionsky D. J. J. A. (2015). Atg41/Icy2 regulates autophagosome formation. Autophagy. 1112, 2288–2299. doi: 10.1080/15548627.2015.1107692
Zhang Q.-C., Liu C., Wang J.-X., Kong F.-Z., Niu Z., Xiang L., et al. (2022). Intense blooms of Phaeocystis globosa in the South China Sea are caused by a unique “giant-colony” ecotype. Harmful Algae. 114, 102227. doi: 10.1016/j.hal.2022.102227
Zhang H. J., Wang H., Zheng W., Yao Z. Y., Peng Y., Zhang S., et al. (2017). Toxic Effects of Prodigiosin Secreted by Hahella sp KA22 on Harmful Alga Phaeocystis globosa. Front. Microbiol. 8, 999. doi: 10.3389/fmicb.2017.00999
Zhang S., Zheng W., Wang H. (2020). Physiological response and morphological changes of Heterosigma akashiwo to an algicidal compound prodigiosin. J. Hazard. Mater. 385. doi: 10.1016/j.jhazmat.2019.121530
Zhu X. Y., Chen S. S., Luo G. Y., Zheng W., Tian Y., Lei X. Q., et al. (2022). A novel algicidal bacterium, microbulbifer sp. yx04, triggered oxidative damage and autophagic cell death in Phaeocystis globosa, which causes harmful algal blooms. Microbiol. Spectr. 10 (1), e00934-21. doi: 10.1128/spectrum.00934-21
Zhu J., Yang Y., Duan S., Sun D. (2019). The antialgal mechanism of luteolin-7-O-glucuronide on Phaeocystis globosa by metabolomics analysis. Int. J. Environ. Res. Public Health 16 (17), 3222. doi: 10.3390/ijerph16173222
Keywords: algaecide, gene trend analysis, harmful algal blooms, Ipomoea cairica, Phaeocystis globosa
Citation: Liang D, Li H, Pan Y, Liu Z and Xiang H (2025) Transcriptome analysis of Ipomoea cairica algicidal mechanism against Phaeocystis globosa. Front. Mar. Sci. 12:1580077. doi: 10.3389/fmars.2025.1580077
Received: 21 February 2025; Accepted: 25 March 2025;
Published: 14 April 2025.
Edited by:
Vikash Kumar, Central Inland Fisheries Research Institute (ICAR), IndiaReviewed by:
Sofia Priyadarsani Das, National Taiwan Ocean University, TaiwanCopyright © 2025 Liang, Li, Pan, Liu and Xiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua Xiang, eGlhbmdodWEyMEBtYWlscy51Y2FzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.