
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 28 March 2025
Sec. Marine Evolutionary Biology, Biogeography and Species Diversity
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1557121
Chloroplast intron infA-62 as a degenerated group II intron family was previously observed to exist specifically in infA genes of chloroplast/plastid genomes (plastomes) in the genus Ulva (Ulvophyceae, Chlorophyta). To understand occurrence frequency, molecular evolution and phylogenetic utility of this intron family in Ulva species, in this study, we conducted more sampling tests based on newly designed specific primers, analyzed evolutionary features of its secondary structures, and employed intron infA-62 for phylogenetic analysis of Ulva species. The 100% occurrence frequency of this intron has been observed in Ulva plastomes, supporting its acquisition by the earliest progenitor of Ulva species. The GC content of this intron family is unprecedentedly low (21.0-25.2%) for group II introns. The intron infA-62 family is classified as an atypical form of ORF-less group IIB-like secondary structures. Some new evolutionary features have been revealed in this intron family, including the extremely low GC content in some domains (e.g. domains IB, ICa, ID2, IDa, II and IV), a very short stem in domain I, a drastically changing domain IC2, and a completely degenerated domain IV. Secondary structures of this intron family showed progressive RNA structural deviations and species-specific variations during the Ulva evolution. Nine mutation hotspots have been detected in loop regions of domains IA, IB, IC1, IC2, ICa, IDa, II, IV and VI. The ML phylogenetic tree constructed based on the nucleotide sequences of intron infA-62 showed that Ulva species were classified into two clades representing two Ulva lineages, Ulva I and II, which was consistent with those based on organelle multigene datasets. Our evidences show that intron infA-62 coevolved with the plastomes during the evolution and speciation of Ulva species. The intron infA-62 that combines primary sequence and secondary structure can be used as an efficient phylogenetic marker for identification and classification of Ulva species.
The green macroalgal genus Ulva Linnaeus is one of the genera with rich species diversity and global distribution in the phylum Chlorophyta. Ulva species have received widespread attention due to the formation of large-scale green tides (Liu et al., 2013; Wang et al., 2019; Zhao et al., 2023) and the usefulness of biomass for edible and biopharmaceutical purposes (Buck and Muki, 2023; Steinhagen et al., 2024). Analyzing the evolution of their organelle genomes has become one of the key fundamental scientific questions for understanding the mechanisms of diversity formation and the evolutionary patterns of global distribution in Ulva species (e.g. Liu and Melton, 2021; Liu et al., 2022a, 2023). So far, a total of 100 species in this genus have been effectively identified in taxonomy, mainly based on phylogenetic analysis as well as morphological features (Guiry and Guiry, 2024). Some common molecular markers (e.g. ITS, rbcL and tufA) are widely used in research on Ulva species identification and cryptic diversity (Hayden and Waaland, 2002; Hofmann et al., 2010; Steinhagen et al., 2019), however, there is still a problem of insufficient differential signals when distinguishing closely related species (e.g. Ulva linza vs U. prolifera). Developing high-resolution molecular markers is of great demand and significance in the identification and classification of Ulva species.
Among the phylum Chlorophyta, chloroplast/plastid genomes (plastomes or cpDNAs) of Ulva species exhibit some unique evolutionary trends compared to those in other lineages of green algae. The Ulva cpDNAs belong to inverted repeat (IR)-lacking circular plastomes with sizes ranging from 86.73 kb in U. linza to 122.72 kb in U. meridionalis. Ulva plastome evolution reflects the strong selection pressure driving the compactness of genome organization and the decrease of overall GC composition (Liu et al., 2023). Their GC content, ranging from 23.89% to 26.25%, is the lowest in sequenced ulvophyceaen plastomes thus far. Phylogenomic analysis based on organelle genome datasets clearly depicts the evolutionary nature of double crown radiation (lineages I and II) in the phylogeny and speciation of Ulva species (Liu et al., 2022b, 2023). The Ulva plastomes contain a considerable number of group I/II introns, and its intron diversity is abundant. Frequent gain or loss of group I/II introns was observed in Ulva plastomes, which leads to sporadic, idiosyncratic distribution patterns in the landscape of chloroplast introns at interspecific and intraspecific levels (Liu et al., 2023). Introns from the same insertion site in Ulva plastomes have high sequence homology, indicating that they have the same origin and belong to the same intron family (Liu and Melton, 2021; Liu et al., 2022a).
Group II introns are a class of autocatalytic ribozymes and retroelements capable of carrying out both self-splicing and retromobility reactions (Robart and Zimmerly, 2005; Peters and Toor, 2015), found in genomes of eubacteria and archaea, and organelle genomes of algae, plants and fungi (Lambowitz and Zimmerly, 2011; Mukhopadhyay and Hausner, 2021). Group II introns exhibit a conserved secondary structure, consisting of six double-helix domains, numbered from domain I to VI, in a central wheel radiating shape (Michel and Dujon, 1983; Michel et al., 1989; Toor et al., 2001). Over the past two decades, the number of known group II introns has seen a dramatic increase, particularly those from organelle genomes (e.g. Liu and Melton, 2021; Kim et al., 2022). Some chloroplast or mitochondrial group II intron sequences yielded well-resolved phylogenies in plants and fungi, and were used as molecular markers for assessment of evolutionary relationships at infrageneric and intergeneric levels (e.g. Vangerow et al., 1999; Clausing and Renner, 2001; Kelchner, 2002; Löhne and Borsch, 2005; Hausner et al., 2006).
It is widely believed that introns are independent evolutionary units in the formation and evolution of biodiversity, and their presence/absence patterns are the results of stochastic processes of gain and loss. In Ulva species, we found that only one chloroplast group II intron, intron infA-62, was shared by all sequenced plastomes. Unlike other group II introns detected in Ulva organelle genomes, the intron infA-62 family lacks an intron-encoding protein (IEP) (Liu and Melton, 2021). To understand occurrence frequency, molecular evolution and phylogenetic utility of this Ulva-specific chloroplast intron infA-62 family, in this study, we conducted more sampling tests based on our newly designed specific primers, performed structural partitioning and differential analysis of secondary structures of this intron family at interspecific and intraspecific (e.g. U. compressa) levels, and employed intron infA-62 as a molecular marker for phylogenetic analysis of Ulva species.
Plastome sequences of Ulva species were selected from the GenBank database for comparative analysis. A total of 42 plastomes from 21 Ulva species, with complete or nearly complete plastome sequences, were retrieved. It was found that there is a specific group II intron in the infA genes in these Ulva plastomes by using the RNAweasel (https://megasun.bch.umontreal.ca/RNAweasel/) (Lang et al., 2007) and by manually aligning nucleotide (nt) sequences of intron-containing infA genes in Ulva species and intronless homologous genes in other ulvophyceaen taxa (e.g. Blidingia minima and Pseudoneochloris marina). Intron insertion site was characterized based on the alignments of nucleotide (nt) sequences of the homologous infA genes with the counterpart in the U. compressa (MW353781) plastome as reference using MEGA7.0 (Kumar et al., 2016). The intron name was defined as host gene (infA) plus intron insertion site in U. compressa.
To further analyze the occurrence frequency of this intron in Ulva plastomes and to test the phylogenetic utility of intron infA-62 sequences, a total of 50 samples of Ulva species collected previously were selected in this study (Supplementary Table 1). The extraction of genomic DNA from fresh or dried tissue of individual algal samples was conducted using a Plant Genome DNA Kit (DP305, Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. These Ulva samples have been identified at the species level based on phylogenetic analyses of two common DNA markers, including the nuclear ITS region including the 5.8S rDNA gene and the chloroplast rbcL gene (Hayden and Waaland, 2002; Liu et al., 2013).
Considering the presence of highly conserved sequences in the flanking exon regions of this intron suitable for designing universal primers for amplification, we developed universal primers to amplify intron infA-62 sequences in Ulva species based on the completely sequenced plastomes from 21 Ulva species (Supplementary Table 2). The intron infA-62 region was amplified in one fragment with the forward primer infA-5exon-F (5’-ATTGAAATGCAAGGTGTCG-3’) that anneals to the 5’ exon of infA and the reverse primer infA-3exon-R (5’-TTTCGTCGTATTTTCCCAG-3’) that anneals to the 3’ exon of infA. PCR amplification was performed in a TC1000-G Thermal Cycler (Scilogex, USA) with initial denaturation (4 min at 94°C), 35 cycles of denaturation (1 min at 94°C), annealing (1 min at 54°C), extension (2 min at 72°C), and a final extension step (10 min at 72°C). PCR reaction was performed in a total volume of 50 μL including 25 μL 2×Mix (Tiangen Biotech, Beijing, China), 1 μL forward primer, 1 μL reverse primer, 2 μl genomic DNA template (1:25), and 21 μL ddH2O. The amplified products were detected by 1.0% agarose gel electrophoresis to check amplicon lengths and then were cut from the gel and purified using a DNA Gel Extraction Kit (BioBasic, Canada). Sequencing reactions were performed from both sides using ABI 3730 XL automated sequencers (Applied Biosystems, USA) by Qingdao Huanuo Biotechnology, Ltd. The sequencing reaction was conducted twice to ensure the accuracy of the DNA sequences.
The most conserved region in group II intron infA-62, known as domain V, was identified using RNAweasel (Lang et al., 2007). The computationally predicted secondary structure of intron infA-62 was constructed using the UNAFold Web Server (Zuker, 2003) based on constraint information which incorporated the principles of electrostatic potential energy, base pair formation energy, and other physical and chemical factors, and the fact that the group II introns shared a consensus structure. The computationally predicted tertiary structure of intron infA-62 was built using the AlphaFold3 and 3dRNA/DNA Web Server (Zhang et al., 2022). Base composition of the intron infA-62 and different domains was determined by using MEGA 7.0 (Kumar et al., 2016). Differences and identity values of DNA sequences were calculated by use of BioEdit v7.1.9 (Hall, 1999).
To our knowledge, the intron infA-62 sequence so far had never been used in phylogenetic studies. The intron infA-62 sequence dataset was compiled from 50 sequenced representatives based on the newly designed primers (Supplementary Table 1) and 42 Ulva plastomes (Supplementary Table 2). Multiple alignments of nucleotide (nt) sequences were conducted by using ClustalX 1.83 with the default settings (Thompson et al., 1997). The RNA secondary structures of intron infA-62 were used to guide the alignment of these intronic sequences. Phylogenetic trees were constructed based on nt sequences of intron infA-62. The phylogenetic relationships were inferred with the Maximum Likelihood (ML) method based on the Tamura-Nei model (Jones et al., 1992). Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach. There were 847 positions in the final intron infA-62 dataset. Phylogenetic analysis was conducted with 1,000 bootstrap replicates by using MEGA 7.0.
Based on our more sampling tests as well as the sequenced Ulva plastomes data, we found that the chloroplast infA genes of all Ulva samples were interrupted by a group II intron infA-62 (Figure 1). The infA gene with initial codon UUG was located between rps8 and rpl36 (Liu et al., 2023). The infA gene is shared by the known chlorophycean plastomes and is highly conserved in size and sequence in Ulva plastomes. This gene encodes the IF1 protein which is one of the factors controlling translation initiation (Kapralou et al., 2009). The insertion site of intron infA-62 is located at the 3’ end of the 62nd nucleotide in the infA gene. The sequences at the intron boundary are highly conserved among this intron family, with GUGUGACUC and AU defining the 5’ and 3’ ends, respectively. Considering the chloroplast infA genes lack this intron in all other chlorophycean taxa whose plastomes have been sequenced thus far (e.g. Turmel et al., 2016; Gao et al., 2022; Liu et al., 2023), the 100% occurrence frequency of this intron observed in Ulva plastomes indicated that this intron should have been acquired by the common ancestor of Ulva species.
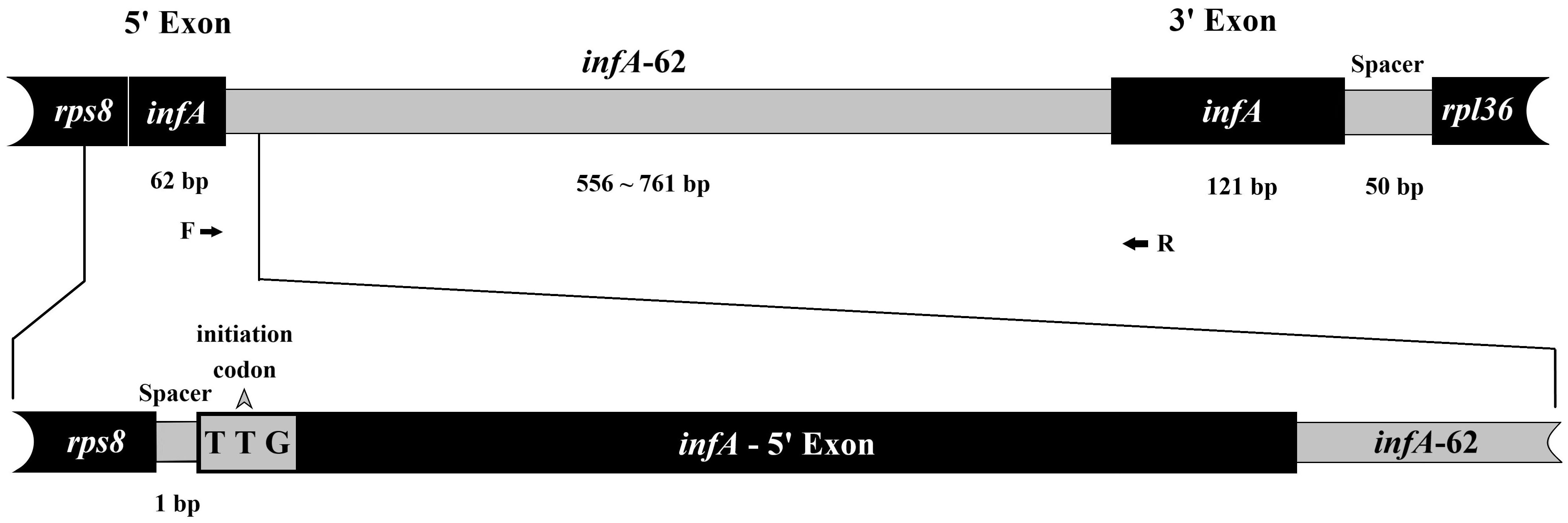
Figure 1. Schematic of the infA gene located between rps8 and rpl36 in Ulva plastomes. Coding stretches and noncoding regions were marked by black and gray boxes, respectively. Size of intergenic spacers and intron infA-62 among Ulva species were indicated below the bar. Arrows mark positions of primers. The intron encoded protein (IEP) has been completely lost from this intron.
The size of the intron infA-62 family ranged from 556 nt in U. fenestrate to 761 nt in U. compressa genotype III (Figure 2), which is much shorter than the other group II introns (1,826 - 2,473 nt) detected in Ulva plastomes (Liu and Melton, 2021; Liu et al., 2023), indicating that it is a severely degenerated intron family in the sequence. Within the genus Ulva, the size of this intron is much longer in U. compressa (728 - 761 nt) than that in the other Ulva species (556 - 650 nt), mainly due to the much larger domain IC2 in U. compressa.
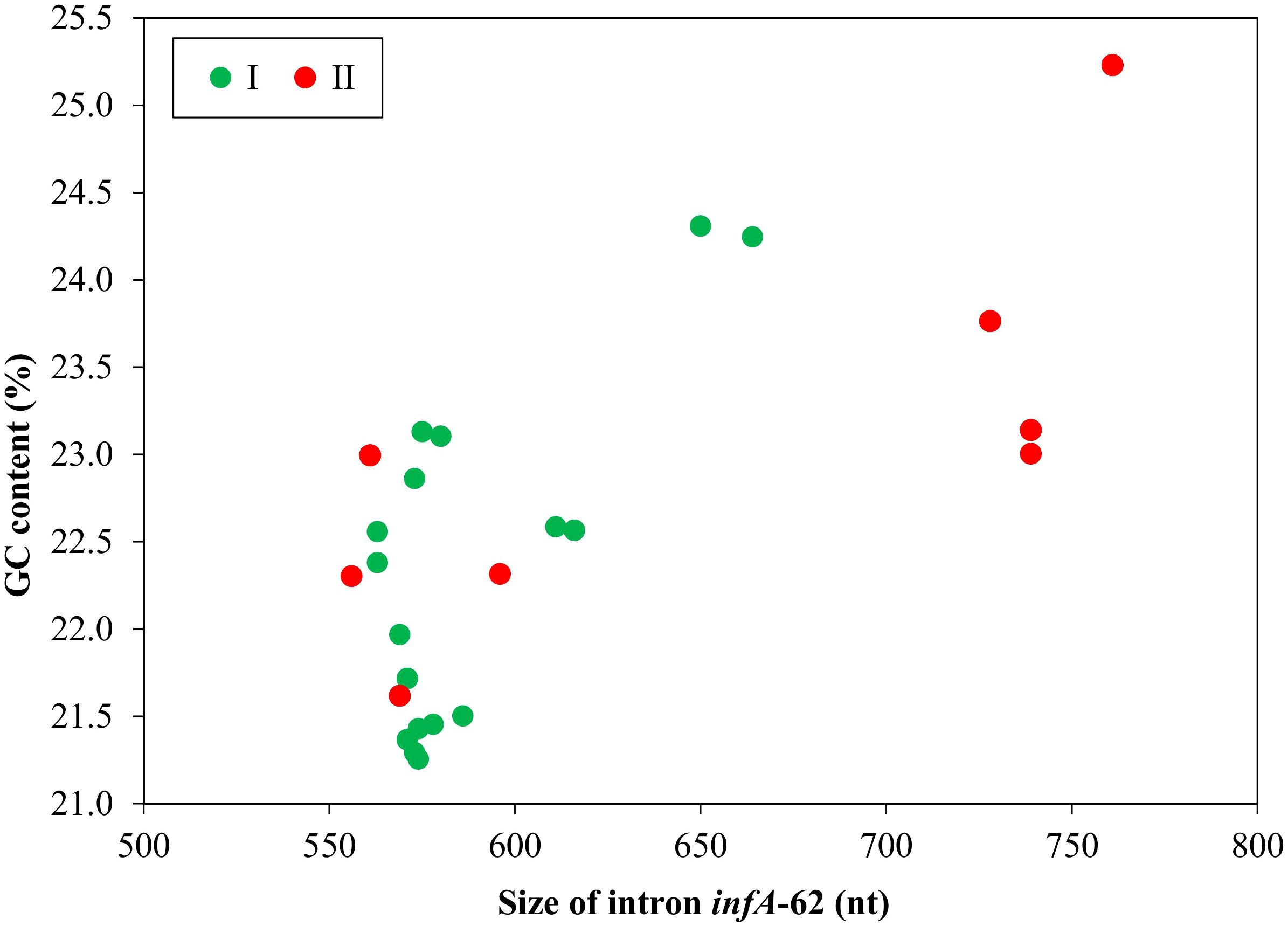
Figure 2. The GC content and size of the group II intron infA-62 in Ulva plastomes. Ulva species in lineages I and II were shown in green and red, respectively (Liu et al., 2023).
The GC content of this intron family is unprecedentedly low for group II introns, ranging from 21.0% in U. paradoxa to 25.2% in U. compressa genotype I (Table 1, Figure 2). It is worth noting that the GC content of this intron family is very close to those of Ulva plastomes (23.9 - 26.2%), but is significantly lower than the other chloroplast group II introns (33.4 - 38.8%) in Ulva (Liu et al., 2023). These facts reveal that this chloroplast intron exhibits coevolution with the plastomes in Ulva species.
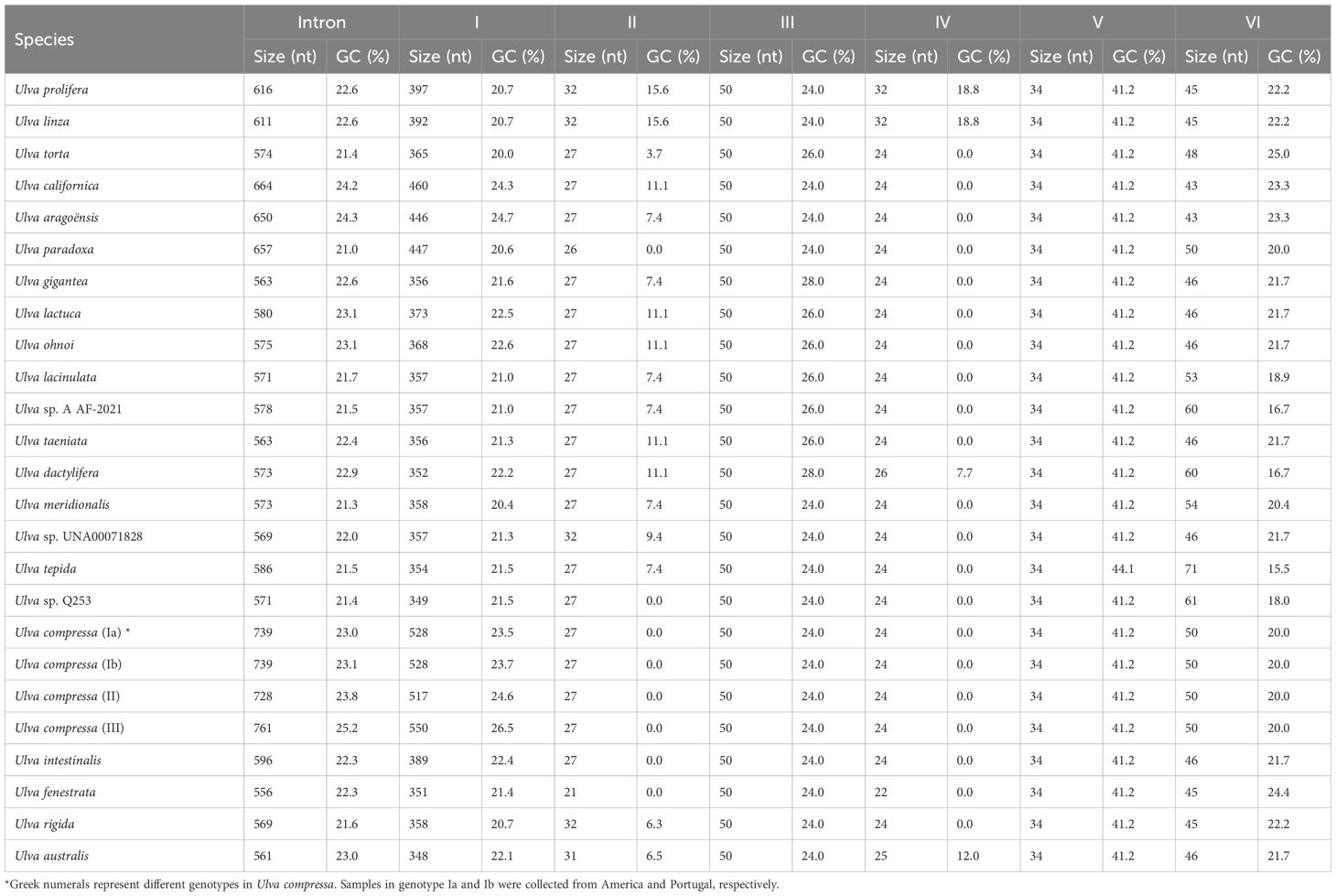
Table 1. Comparison of the size and GC content of six domains in intron infA-62 among different Ulva species.
To supplement the computationally predicted secondary structure of group II intron infA-62 and examine evolution of the intron RNA structures, we selected common conserved sequences to avoid interference from sequences in mutation hotspots and created a structure model using the UNAfold web server (Zuker, 2003). Considering the fact that the group II introns share a consensus structure (Candales et al., 2012; Mukhopadhyay and Hausner, 2021), the intron infA-62 sequences were folded into a conserved secondary structure of group II introns that consists of six double-helix domains (I to VI) in a central wheel radiating shape and a bulging adenosine residue in domain VI, which is located at the eighth nucleotide position upstream of the intron-exon junction (Figure 3).
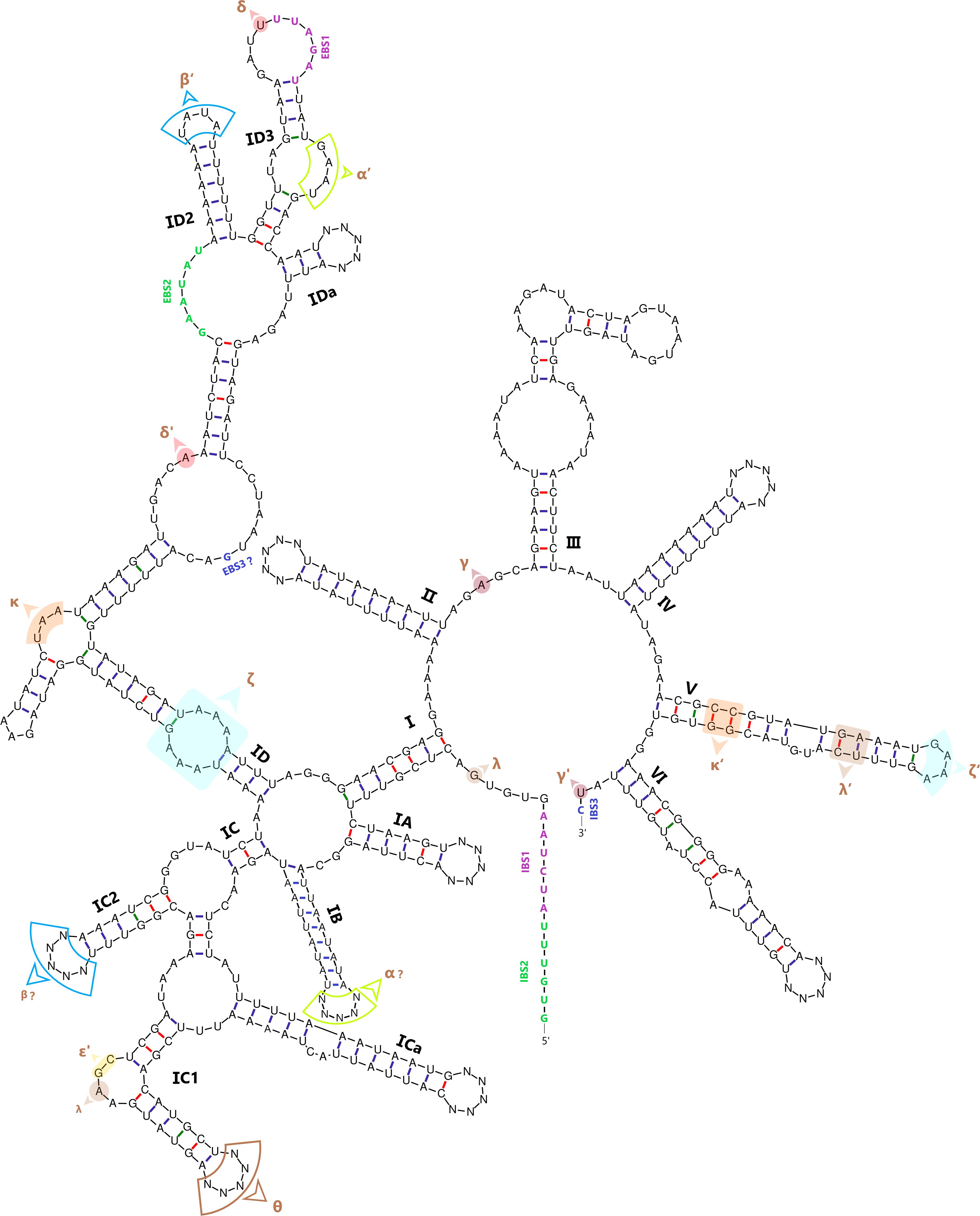
Figure 3. Secondary structural model of group II intron infA-62 in Ulva plastomes. The intron RNA folded into a typical structure of six domains (I to VI), with domain I consisting of four domains (IA to ID). The major structural domains were specified by Roman numbers, and the long-range tertiary interactions were denoted by Greek letters.
The core secondary structure architecture of the intron infA-62 is highly conserved among Ulva species. The shared secondary structure plays an important role in functions such as intron self-splicing (Dai et al., 2003; Pyle, 2016). Most of putative tertiary interaction sites were annotated and characterized in intron infA-62, including the exon binding site 1 (EBS1) and EBS2, intron binding site 1 (IBS1) and IBS2, and regions of tertiary RNA-RNA interactions (e.g. α-α’, β-β’, γ-γ’, δ-δ’, ϵ-ϵ’ and κ-κ’) (Figure 3). The sequences of EBS1 (UUAGAU) and EBS2 (GAAUAU) form base pairs with IBS1 (AUCUAA) and IBS2 (GUGUUU) on the exon, respectively. Intron-exon binding plays an important role in group II intron self-splicing (Bonen and Vogel, 2001). We observed that intron and exon binding sites showed the high complementarity (Figure 3), which facilitates efficient splicing of intron infA-62 from the host gene.
The intron infA-62 is classified as an atypical form of ORF-less group IIB-like secondary structures based on structural characteristics including the 5’-terminal consensus sequence GUGUGACUC, the short basal stem of domain I, the IIB-typical elements (θ, λ and ϵ’) in domain IC1, the stem ID1-less EBS2, the presence of a linker between domains III and IV, and some group IIB specific conserved nucleotides (Toor et al., 2001). The intron-encoding protein (IEP) ORF was completely lost from domain IV in this intron family, which is different from the other group II intron families detected in Ulva plastomes which harbored a reverse transcriptase/maturase (RTM) gene (Liu and Melton, 2021; Mizrahi et al., 2022).
Many examples have demonstrated that the ORF-less group II introns were derived from the ORF-containing introns (e.g. Toor et al., 2001; Toro and Martínez-Abarca, 2013; Zimmerly and Semper, 2015). The absence of IEP is likely the main reason for the loss of mobility competence in this intron family which is significantly different from other intron families that undergo frequent acquisition or loss in Ulva organelle genomes (Jeffares et al., 2006; Liu et al., 2023). However, intron infA-62 must retain splicing competence because they are located in the conserved housekeeping gene infA (Robart and Zimmerly, 2005). In vitro experiments have found that the self-splicing of group II introns only requires two essential components including intron RNA and Mg2+, but they often require recruiting host proteins in vivo to enhance their structure or reactivity (Molina-Sanchez et al., 2011; Pyle, 2016). Given the degeneration of ribozyme structure in intron infA-62, its self-splicing is likely to rely on compensation from host splicing factors.
To gain a deeper understanding of the molecular evolution of intron infA-62, we conducted structural partitioning and differential analysis of secondary structures of this intron family. Some new evolutionary features have been demonstrated in this intron family, including the extremely low GC content in some domains (e.g. domains IB, ICa, ID2, IDa, II and IV), a very short stem in domain I, a drastically changing domain IC2 even at the intraspecific level, and a completely degenerated domain IV (Figure 3). High sequence variations were confined to these mutation hotspots located in apical loop regions in this intron family. The sequences of nine loop regions in domains IA, IB, IC1, IC2, ICa, IDa, II, IV and VI representing mutation hotspots exhibit diverse variations across different Ulva species (Figure 3). The changes of intron size are mainly caused by length variations in these nine loop regions among different Ulva species. The nonpaired regions (e.g. loops and bulges) in mutation hotspots exhibited much greater variations caused by mutations of simple sequence repeats, indels and substitutions than helical regions.
Domain I is the largest and most complex with the size from 348 nt in U. australis to 550 nt in U. compressa genotype III and constitutes more than half of the intron’s size, ranging from 60.4% in U. tepida to 72.3% in U. compressa genotype III (Table 1). This domain consists of four major stem-loop structures, i.e. domain IA, IB, IC and ID (Figure 3). Domains IA and IB are two small stem-loop structures without branch, with the size of 16 - 25 nt and 19 - 26 nt and GC content of 16.0 - 25.0% and 0.0 - 5.3%, respectively (Table 2).
Domain IC contains two common stem-loop regions, IC1 and IC2 (Michel and Dujon, 1983; Toor et al., 2001), as well as an additional stem-loop region ICa. IC1 was the most conserved region in domain IC, with the size of 29 - 37 nt and the GC content of 32.4 - 37.9% (Table 2). The IIB-typical elements (e.g. θ, λ and ϵ’) were located in IC1 (Figure 3). The AT-enriched ICa was inserted into the basal loop of IC1, with the size of 45 - 69 nt. The size of IC2 exhibits great variation across different Ulva species, from 16 - 32 nt in most Ulva species to 44 nt in U. linza-prolifera (LP) (Liu et al., 2013), to 101 - 119 nt in U. californica-aragoënsis-paradoxa to 157 - 190 nt in U. compressa (Table 2), which was caused by different repeat mutations and indels in loop region. It is worth noting that the GC content changed most dramatically in IC2 among the entire intron, ranging from 18.2% in U. rigida to 44.4% in U. ohnoi. The secondary structure of IC2 exhibits significant differences, but the main stem of IC2 is very conserved in Ulva species (Figures 4, 5). In both the Ulva lineage I and II, IC2 exhibits structural complexity and shows convergent evolution characteristics. For example, the IC2 of the vast majority of Ulva species in lineage IB and IC is very similar to that in lineage IIB (Figures 4, 5).
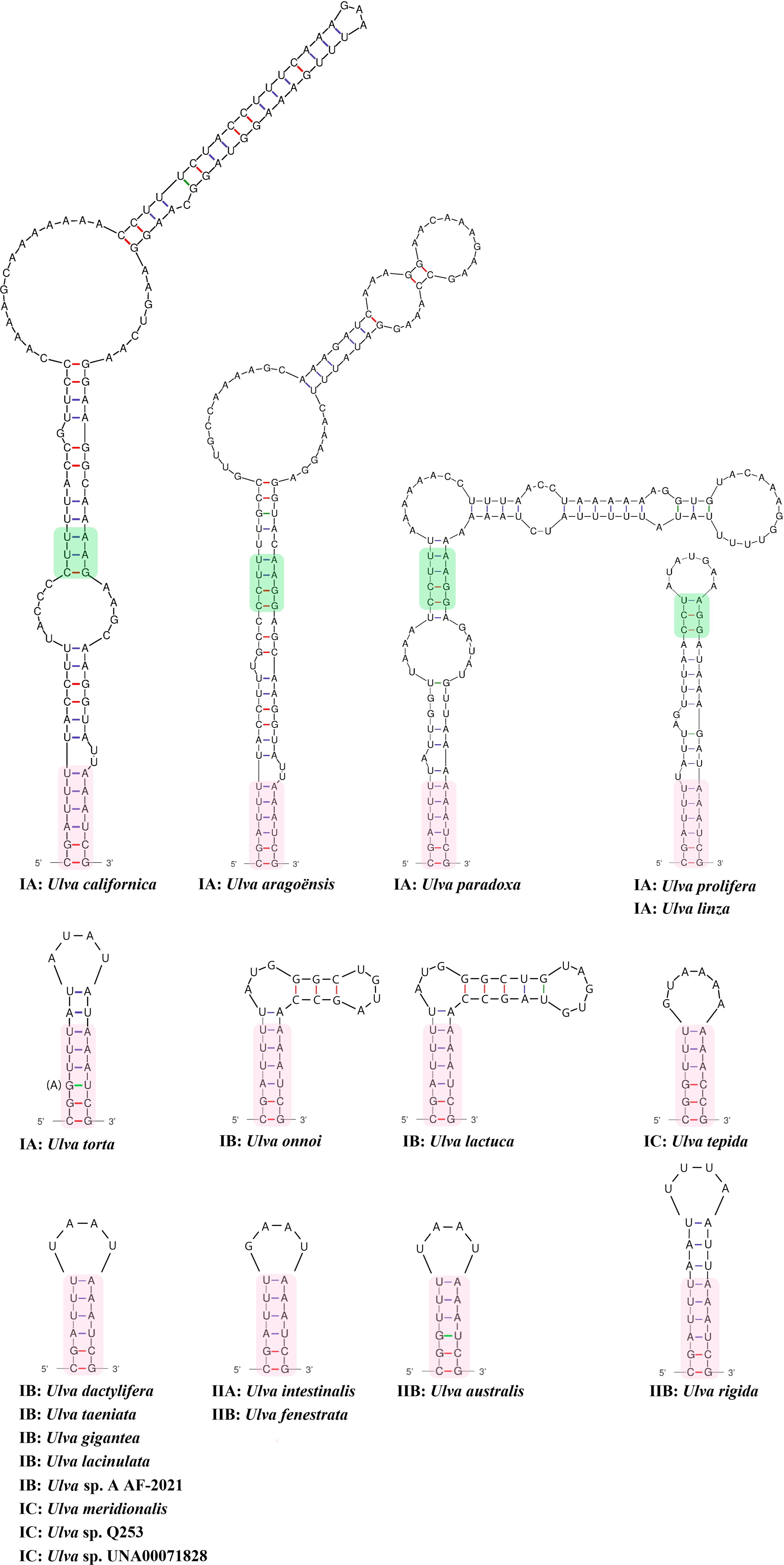
Figure 4. Comparison of secondary structure of domain IC2 in Ulva species. The sequences marked with different colored shadows represent highly conserved sequence regions. IA, IB, IC, IIA and IIB represent different evolutionary lineages in Ulva (Liu et al., 2023).
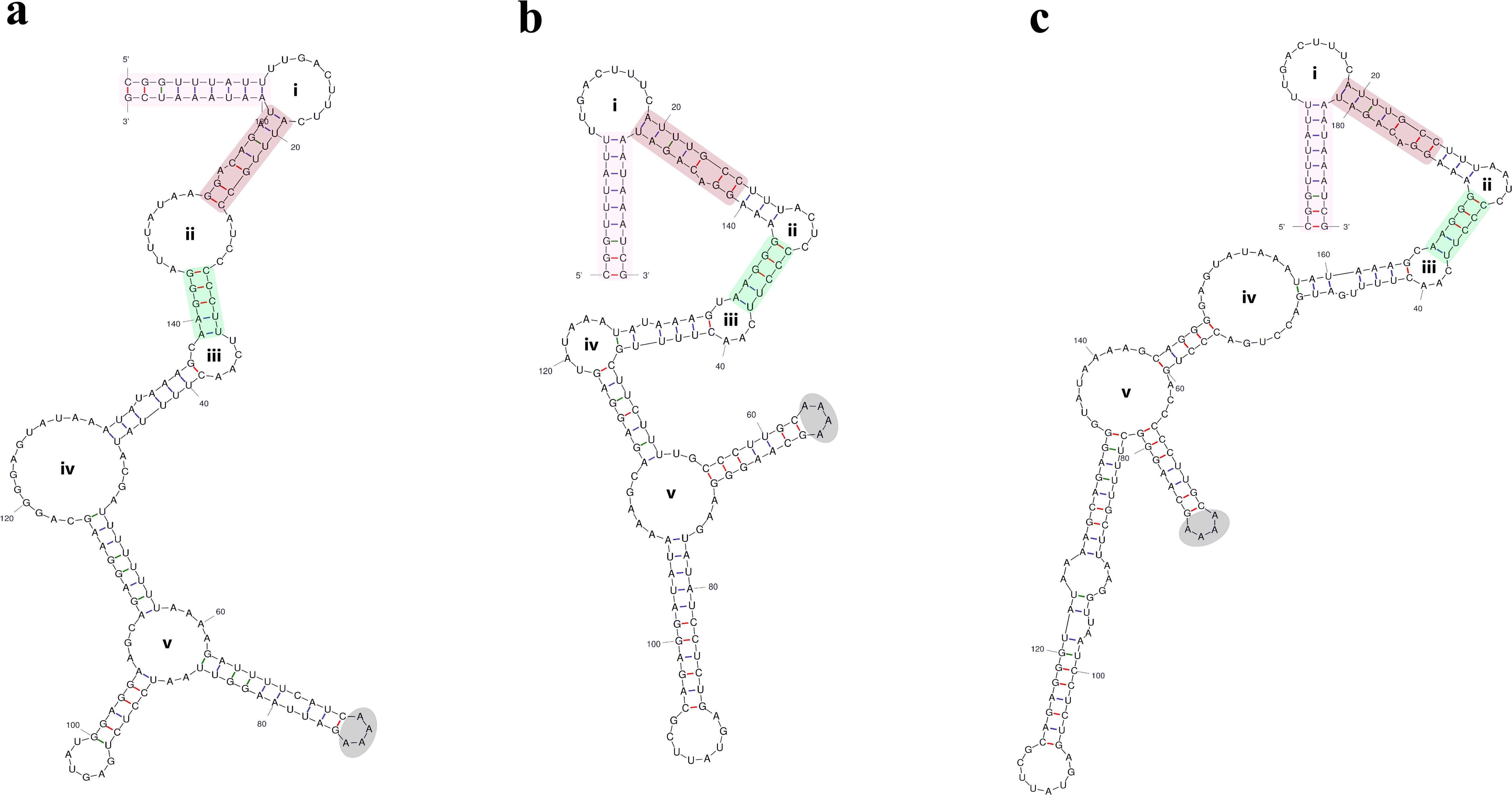
Figure 5. Comparison of secondary structure of domain IC2 among three genotypes of U. compressa. (a) IC2 in genotype I (Uco I). (b) IC2 in genotype II (Uco II). (c) IC2 in genotype III (Uco III). The sequences marked with different colored shadows represent highly conserved sequence regions.
Domain ID provides the recognition sites (EBS1 and EBS2) for sequence-specific exon binding. The stem ID1-less EBS2 is a typical characteristic of group IIB introns (Figure 3). ID2 is a short and AT-enriched stem-loop structure, with the size of 15 - 16 nt and GC content of 0.0 - 6.7% (Table 2). ID3 is one of the most conserved regions with much higher GC content in domain ID and harbored the conserved tertiary interaction sites (EBS1 and δ) (Figure 3). An additional small stem-loop structure IDa occurred in the EBS2 loop, which varied greatly in size from 20 nt in U. tepida to 58 nt in U. intestinalis. The GC content of IDa ranged from 0.0 - 7.7% in the vast majority of species to 22.4% in U. intestinalis caused by a GC-enriched insertion sequence occurring in its loop region (Table 2).
Domains II to VI are five small stem-loop structures in intron infA-62. Domain II displays an atypically stem-loop structure with the size of 21 - 32 nt and GC content of 0.0 - 15.6%. The stem of domain II in Ulva lineage I is 3-bp longer than that in lineage II. The 50-nt domain III is conserved in intron infA-62, with the GC content of 24.0-28.0% (Table 1). The AT-enriched domain IV is the smallest domain with the size of 22-32 nt. The IEP ORF, which usually encoded an enzyme associated with splicing, has been completely degenerated and lost from the domain IV (Figure 3). The loop sequences of domain IV in U. linza-prolifera (LP) are longer than other Ulva species due to the duplication of UGA and ACUCA (Table 1). The 34-nt domain V is the most GC-enriched domain in intron infA-62. Its GC content was 44.1% in U. tepida and 41.2% in all other Ulva species. Domain V was the most structurally conserved domain in group II introns, which is a short hairpin with a conserved catalytic triad at its base. An asymmetric bulge UG is located in the middle of its stem in intron infA-62 (Figure 3). In addition to the triad, it is considered that nucleotides at the junction of domains II and III contribute to the catalytic core that coordinates metal ions and directly participates in catalysis (Toor et al., 2008a; Toor et al., 2008b; Peters and Toor, 2015). Domain VI ranges from 43 nt in U. californica and U. aragoënsis to 71 nt in U. tepida (Table 1). Its stem sequence is highly conserved, but its loop region shows great variations in size and sequence among different Ulva species due to multiple repeated mutations. Domain VI contains the branch-point, a bulged adenosine (Figure 3), the 2’-OH of which is the nucleophile that attacks the phosphate at the 5’ splice site to initiate the first step of splicing (Lambowitz and Zimmerly, 2011; Zhao and Pyle, 2017). This catalytic RNA structure is bound by the IEP for both splicing and retromobility reactions (Cui and Davis, 2007; Qu et al., 2016). Due to the complete loss of IEP in the domain IV of intron infA-62, the retromobility function of this intron family is likely to have been lost (Liu and Melton, 2021).
The tertiary structure of intron infA-62 is achieved through the spatial folding of RNA molecules which involves the interaction of multiple domains, including base pairing, stacking, and spatial repulsion. According to the model diagram, the sequences in mutation hotspots forming the loop regions are distributed in the molecular periphery of tertiary structure of intron infA-62 (Figure 6), and the sequences concentrated in central region are highly conserved. During the self-splicing of intron infA-62, the conformational changes would occur in tertiary structure to facilitate the splicing reaction (Pyle, 2016).

Figure 6. Tertiary structural model of group II intron infA-62 in Ulva plastomes. The hotspot mutation regions were indicated by the red short lines. The infA 5’ exon was shown in cyan line. The 5’ end intron and the junctions of each domain were shown in wheat. Domains I, II, III, IV, V and VI were shown in gray, green, blue, purple, slate and yellow, respectively.
The species-specific variations in unpaired nucleotides from apical loop regions of some domains can serve as useful differential phylogenetic signals for identification of Ulva species. For example, the sequences of domain ICa can effectively distinguish two closely related species, U. linza and U. prolifera, due to a 5-nt indel mutation occurring in the loop region (Figure 7). This interspecific difference has been confirmed through comparative analysis of multiple samples from different regions of China and Korea. Domain IC2 is the most drastically changing domain in intron infA-62. The sequences and secondary structures of intron infA-62 are basically the same in the most regions among the three genotypes in U. compressa, but the only difference among them is the variation in the loop sequences of domain IC2 caused by different repeat mutations and indels (Figure 5). The sequences of domain IC2 in these three genotypes linked to multiple variations in organelle genomes. Although the primary sequences of the IC2 loop region varied greatly in three genotypes, its secondary structure seemed to maintain a conserved form in U. compressa (Figure 5). At the intraspecific level of U. compressa, intron infA-62 can effectively distinguish different genotypes which usually represented different morphotypes (Liu et al., 2020). This region can be used as a molecular probe to distinguish different genotypes in U. compressa. A total of three genotypes (I to III) have been identified in U. compressa thus far. Genotype I was found to be distributed in coasts of America (Ia) and Portugal (Ib). Genotypes II and III were distributed in coasts of Qingdao and Yantai in Shandong Province, and genotype III was also found in the Subei shoal in Jiangsu Province.
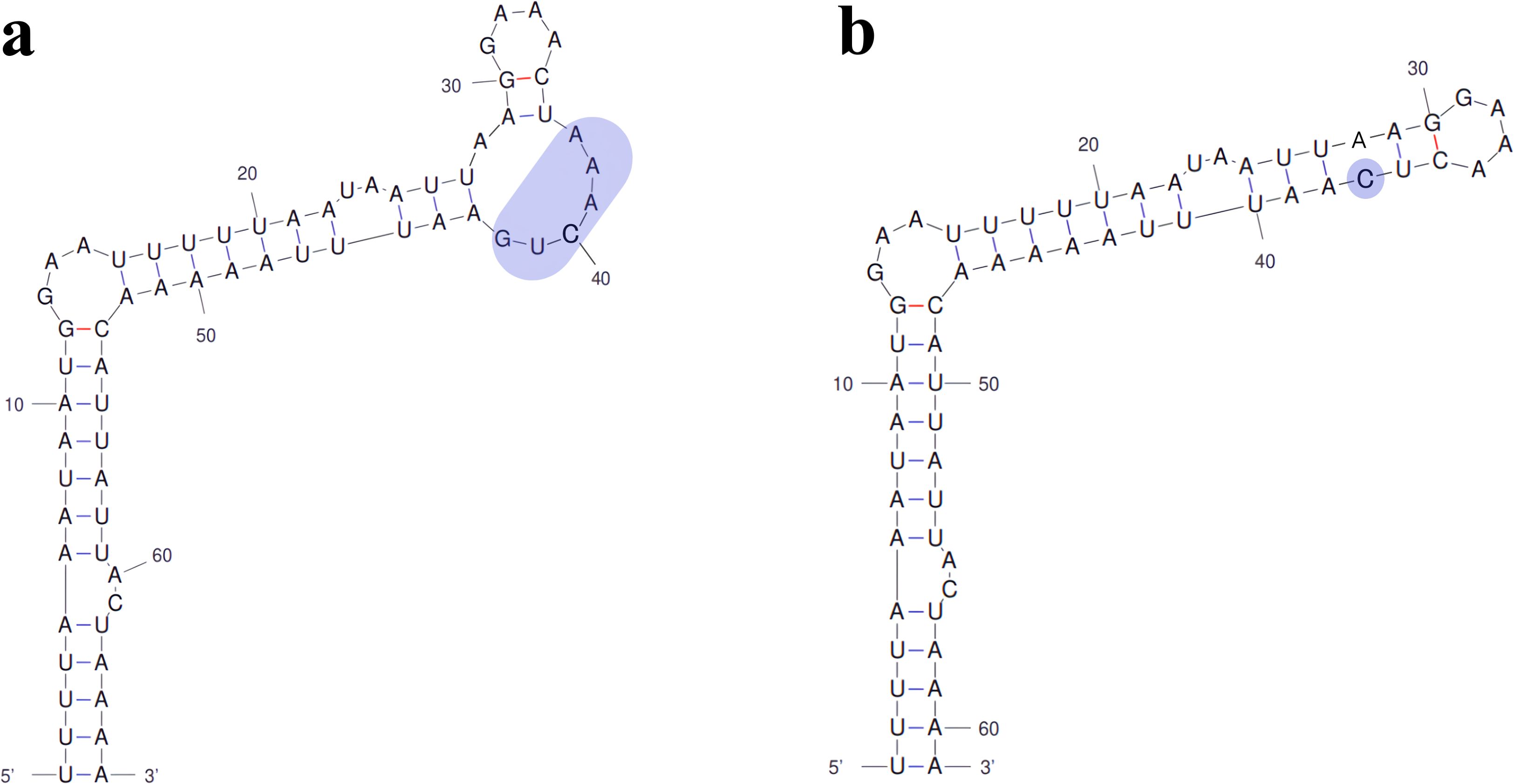
Figure 7. Comparison of secondary structure of domain ICa between U. prolifera and U. linza. (a) ICa in U. prolifera. (b) ICa in U. linza. The shaded sequences represent the regions where the mutation occurred.
Construction of intron infA-62 phylogeny is made feasible because the conserved RNA secondary structures can be used to guide the alignment of these intronic sequences. Sequences of intron infA-62 can be clearly aligned across Ulva species, except for one mutation hotspot located in the loop of domain IC2, and the alignments of the intron infA-62 sequences are used to infer evolutionary trees. The ML phylogenetic tree constructed based on the nucleotide sequences of intron infA-62 from 22 Ulva species yielded well-resolved phylogenies at the species level. These Ulva species were classified into two clades representing two independent Ulva lineages, I and II (Figure 8). Although the topologies of evolutionary trees are affected by different sequence datasets selected, chloroplast intron infA-62 sequences yielded well-resolved phylogenies at the species level. The tree inferred from the intron infA-62 sequences is generally consistent with those obtained from analyses of the nt sequences of 100 common chloroplast genes (Liu et al., 2023) or 61 mitochondrial genes (Liu et al., 2022a; Liu et al., 2022b).
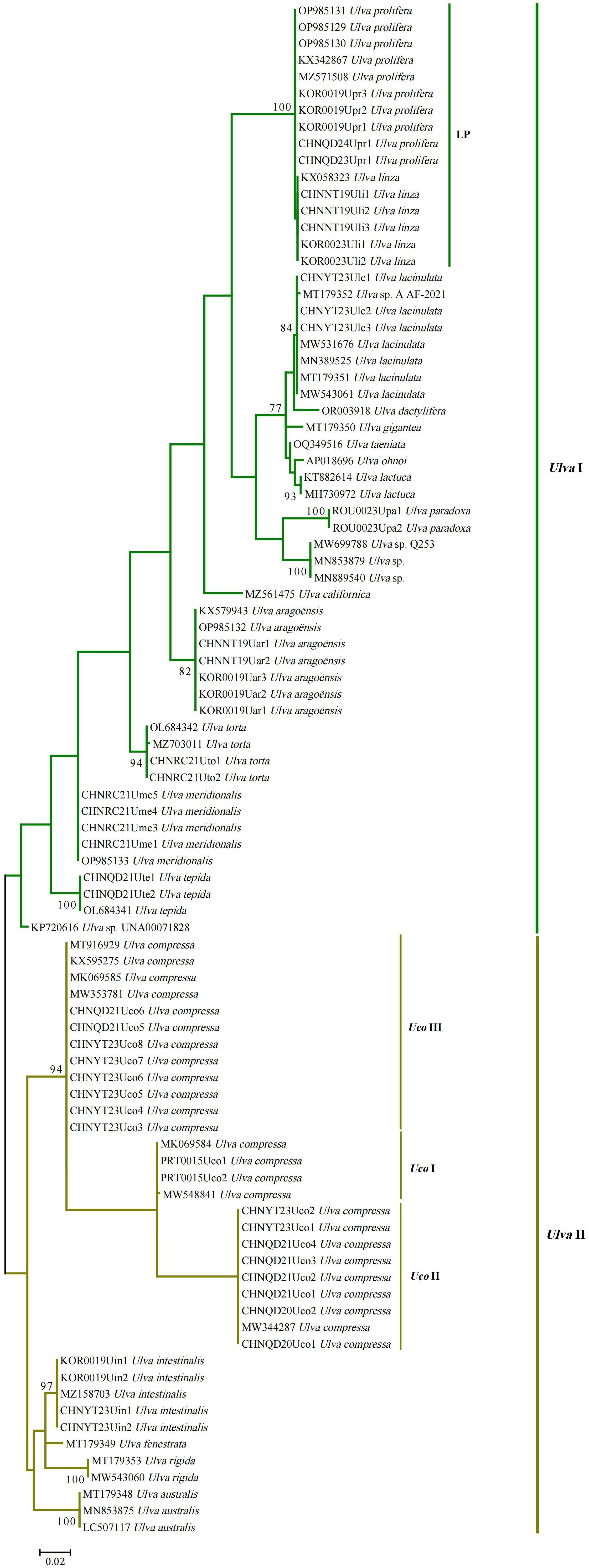
Figure 8. Phylogenetic tree based on Maximum Likelihood (ML) analysis of the nucleotide (nt) sequences of the group II intron infA-62. The ML analysis was conducted with 1,000 bootstrap replicates using MEGA 7.0. The bootstrap support values greater than 70% were displayed at branches. Branch lengths are proportional to the amount of sequence change, which are indicated by the scale bar below the trees.
More evidences support that the intron infA-62 was obtained by the earliest Ulva ancestors, subsequently lost its mobility due to the loss of IEP, then consistently retained in plastomes through vertical transmission and coevolved with the plastomes during the evolution and speciation of Ulva species. First, the 100% occurrence frequency of this intron observed in Ulva plastomes, which was markedly different from that observed in other group II introns. We found that gain and/or loss frequently occurred for most chloroplast group II introns at interspecific and intraspecific levels (Liu and Melton, 2021). Second, we observed that a strong selection pressure drove AT richness in intron infA-62 family, especially domains IB, ICa, ID2, IDa, II and IV, and selection against GC was previously detected in Ulva plastomes not in other ulvophyceaen plastomes. However, the GC content of other group II introns was much higher than that of intron infA-62 family. Third, the intron infA-62 family shows an evolution trend to be more compact in core secondary structure architecture. Marked synchronous degeneration in some domains and completely loss of IEP in domain IV can be detected in secondary structures of intron infA-62 family. A similar strong selection pressure driving the compactness of plastomes was previously found in Ulva species (Liu et al., 2023). Finally, the result of phylogenetic analysis further indicates that this chloroplast intron family exhibits co-evolutionary traits with the plastomes in Ulva species.
The intron infA-62 family shows fast evolution rate in some domains and is short enough in length to be used as a molecular marker. Given that it contains sufficient species-specific mutation accumulation as phylogenetic signals to resolve different Ulva species, this intron family that combines primary sequence and secondary structure can be well suited as an efficient molecular marker to construct Ulva phylogeny for identification and classification of Ulva species.
The chloroplast intron infA-62 with an unprecedentedly low GC content has been found to be present in all tested Ulva samples as well as all of the sequenced Ulva plastomes, indicating its earliest acquisition by the common progenitor of Ulva species. From the perspective of molecular biology, intron infA-62 represents an unusual form of an ORF-less group IIB-like intron. RNA secondary structures of this intron family were folded and analyzed, which revealed progressive RNA structural deviations and species-specific variations throughout evolution and speciation of Ulva species. Some new evolutionary features have been demonstrated in this intron family, including the extremely low GC content in some domains, a very short stem in domain I, a drastically changing domain IC2 even at the intraspecific level, and a completely degenerated domain IV. The loop regions of nine domains (IA, IB, IC1, IC2, ICa, IDa, II, IV and VI) in intron infA-62 exhibit rapid evolutionary rates at both interspecific and intraspecific levels, and microstructural changes occurring in the mutation hotspots were mainly caused by mutations of simple sequence repeats, indels and substitutions. These mutation hotspots are distributed in the molecular periphery of tertiary structure of intron infA-62. Our evidences show that the intron infA-62 was obtained by the earliest Ulva ancestors, subsequently lost its mobility due to the loss of IEP, then consistently retained in plastomes and coevolved with the plastomes during the evolution of Ulva lineages. Based on the rapid evolution rate and sufficient mutation accumulation of this intron, the intron infA-62 can be used as a useful phylogenetic marker for identification and classification of Ulva species. This study provides an important case for further understanding the evolution of group II intron family in organelle genomes of green algae, and establishes a connection between the evolution of introns and the evolution of Ulva species.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
FL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. SJ: Methodology, Software, Writing – review & editing. JK: Data curation, Methodology, Writing – review & editing. XW: Data curation, Methodology, Writing – review & editing. JW: Data curation, Methodology, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (No. 42276133, 41876165), the Strategic Priority Research Program of Chinese Academy of Sciences (No. XDA23050302, XDA23050403), the project from Rongcheng Ocean and Fishery Monitoring and Disaster Mitigation Center and Rongcheng Ocean Development Bureau, the project from Rongcheng Science and Technology Bureau, and the Shandong Provincial Natural Science Foundation (No. ZR2022QD066).
The authors wish to thank Peter Hantz, Wei Luan, Yu Wang, Manman Liu, Hongshu Wang, Jiamin Li, Fan Qu and Hongxiao Yang for their assistance in algal collection, and staffs of marine ecological environment genomics research group in IOCAS for their assistance in data analysis.
Author XW was employed by the company Qingdao Global Marine Safetycare Technology Co., Ltd. GMScare.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1557121/full#supplementary-material
Bonen L., Vogel J. (2001). The ins and outs of group II introns. Trends Genet. 17, 322–331. doi: 10.1016/S0168-9525(01)02324-1
Buck B. H., Muki S. (2023). Ulva: Tomorrow’s “Wheat of the sea”, a model for an innovative mariculture. J. Appl. Phycol. 35, 1967–1970. doi: 10.1007/s10811-023-03003-1
Candales M. A., Duong A., Hood K. S., Li T., Neufeld R. A., Sun R., et al. (2012). Database for bacterial group II introns. Nucleic Acids Res. 40, D187–D190. doi: 10.1093/nar/gkr1043
Clausing G., Renner S. S. (2001). Molecular phylogenetics of Melastomataceae and Memecylaceae: implications for character evolution. Am. J. Bot. 88, 486–498. doi: 10.2307/2657114
Cui X., Davis G. (2007). Mobile group II intron targeting: applications in prokaryotes and perspectives in eukaryotes. Front. Biosci. 12, 4972–4985. doi: 10.2741/2442
Dai L., Toor N., Olson R., Keeping A., Zimmerly S. (2003). Database for mobile group II introns. Nucleic Acids Res. 31, 424–426. doi: 10.1093/nar/gkg049
Gao D., Sun Z., Bi G., Zhang X. (2022). The complete plastome of Blidingia marginata and comparative analysis with the relative species in Ulvales. Aquat. Bot. 183, 103568. doi: 10.1016/j.aquabot.2022.103568
Guiry M. D., Guiry G. M. (2024). AlgaeBase (Galway: National University of Ireland) (Accessed November 3, 2024). World-Wide Electronic Publication.
Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hausner G., Olson R., Simon D., Johnson I., Sanders E. R., Karol K. G., et al. (2006). Origin and evolution of the chloroplast trnK (matK) intron: a model for evolution of group II intron RNA structures. Mol. Biol. Evol. 23, 380–391. doi: 10.1093/molbev/msj047
Hayden H. S., Waaland J. R. (2002). Phylogenetic systematics of the Ulvaceae (Ulvales, ulvophyceae) using chloroplast and nuclear DNA sequences. J. Phycol. 38, 1200–1212. doi: 10.1046/j.1529-8817.2002.01167.x
Hofmann L. C., Nettleton J. C., Neefus C. D., Mathieson A. C. (2010). Cryptic diversity of Ulva (Ulvales, chlorophyta) in the great bay estuarine system (Atlantic USA): introduced and indigenous distromatic species. Eur. J. Phycol. 45, 230–239. doi: 10.1080/09670261003746201
Jeffares D. C., Mourier T., Penny D. (2006). The biology of intron gain and loss. Trends Genet. 22, 16–22. doi: 10.1016/j.tig.2005.10.006
Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kapralou S., Fabbretti A., Garulli C., Gualerzi C. O., Pon C. L., Spurio R. (2009). Characterization of Bacillus stearothermophilus infA and of its product IF1. Gene 428, 31–33. doi: 10.1016/j.gene.2008.09.029
Kelchner S. A. (2002). Group II introns as phylogenetic tools: structure, function, and evolutionary constraints. Am. J. Bot. 89, 1651–1669. doi: 10.3732/ajb.89.10.1651
Kim D., Lee J., Cho C. H., Kim E. J., Bhattacharya D., Yoon H. S. (2022). Group II intron and repeat-rich red algal mitochondrial genomes demonstrate the dynamic recent history of autocatalytic RNAs. BMC Biol. 20, 2. doi: 10.1186/s12915-021-01200-3
Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lambowitz A. M., Zimmerly S. (2011). Group II introns: mobile ribozymes that invade DNA. Cold Spring Harbor Perspect. Biol. 3, a003616. doi: 10.1101/cshperspect.a003616
Lang B. F., Laforest M.-J., Burger G. (2007). Mitochondrial introns: a critical view. Trends Genet. 23, 119–125. doi: 10.1016/j.tig.2007.01.006
Liu F., Chen N., Wang H., Li J., Wang J., Qu F. (2023). Novel insights into chloroplast genome evolution in the green macroalgal genus Ulva (Ulvophyceae, Chlorophyta). Front. Plant Sci. 14, 1126175. doi: 10.3389/fpls.2023.1126175
Liu F., Melton J. T. (2021). Chloroplast genomes of the green-tide forming alga Ulva compressa: comparative chloroplast genomics in the genus Ulva (Ulvophyceae, chlorophyta). Front. Mar. Sci. 8, 668542. doi: 10.3389/fmars.2021.668542
Liu F., Melton J. T., Lopez-Bautista J. M., Chen N. (2020). Multiple intraspecific variations of mitochondrial genomes in the green-tide forming alga, Ulva compressa Linnaeus (Ulvophyceae, Chlorophyta). Front. Mar. Sci. 7, 714. doi: 10.3389/fmars.2020.00714
Liu F., Melton J. T., Wang H., Wang J., Lopez-Bautista J. M. (2022a). Understanding the evolution of mitochondrial genomes in the green macroalgal genus Ulva (Ulvophyceae, chlorophyta). Front. Mar. Sci. 9, 850710. doi: 10.3389/fmars.2022.850710
Liu F., Pang S., Chopin T., Gao S., Shan T., Zhao X., et al. (2013). Understanding the recurrent large-scale green tide in the Yellow Sea: temporal and spatial correlations between multiple geographical, aquacultural and biological factors. Mar. Environ. Res. 83, 38–47. doi: 10.1016/j.marenvres.2012.10.007
Liu F., Wang H., Song W. (2022b). Tandem integration of circular plasmid contributes significantly to the expanded mitochondrial genomes of the green-tide forming alga Ulva meridionalis (Ulvophyceae, chlorophyta). Front. Plant Sci. 13, 937398. doi: 10.3389/fpls.2022.937398
Löhne C., Borsch T. (2005). Molecular evolution and phylogenetic utility of the petD group II intron: a case study in basal angiosperms. Mol. Biol. Evol. 22, 317–332. doi: 10.1093/molbev/msi019
Michel F., Dujon B. (1983). Conservation of RNA secondary structure in two intron families including mitochondrial- chloroplast- and nuclear- encoded members. EMBO J. 2, 33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x
Michel F., Umesono K., Ozeki H. (1989). Comparative and functional anatomy of group II catalytic introns - a review. Gene 82, 5–30. doi: 10.1016/0378-1119(89)90026-7
Mizrahi R., Shevtsov-Tal S., Ostersetzer-Biran O. (2022). Group II intron-encoded proteins (IEPs/maturases) as key regulators of nad1 expression and complex I biogenesis in land plant mitochondria. Genes (Basel) 13, 1137. doi: 10.3390/genes13071137
Molina-Sanchez M. D., Barrientos-Duran A., Toro N. (2011). Relevance of the branch point adenosine, coordination loop, and 3’ exon binding site for in vivo excision of the Sinorhizobium meliloti group II intron RmInt1. J. Biol. Chem. 286, 21154–21163. doi: 10.1074/jbc.m110.210013
Mukhopadhyay J., Hausner G. (2021). Organellar introns in fungi, algae, and plants. Cells 10, 2001. doi: 10.3390/cells10082001
Peters J. K., Toor N. (2015). Group II intron lariat: Structural insights into the spliceosome. RNA Biol. 12, 913–917. doi: 10.1080/15476286.2015.1066956
Pyle A. M. (2016). Group II intron self-splicing. Annu. Rev. Biophys. 45, 183–205. doi: 10.1146/annurev-biophys-062215-011149
Qu G., Kaushal P. S., Wang J., Shigematsu H., Piazza C. L., Agrawal R. K., et al. (2016). Structure of a group II intron complexed with its reverse transcriptase. Nat. Struct. Mol. Biol. 23, 549–557. doi: 10.1038/nsmb.3220
Robart A. R., Zimmerly S. (2005). Group II intron retroelements: function and diversity. Cytogenet. Genome Res. 110, 589–597. doi: 10.1159/000084992
Steinhagen S., Weinberger F., Karez R. (2019). Molecular analysis of Ulva compressa (Chlorophyta, Ulvales) reveals its morphological plasticity, distribution and potential invasiveness on German North Sea and Baltic Sea coasts. Eur. J. Phycol. 54, 102–114. doi: 10.1080/09670262.2018.1513167
Steinhagen S., Wichard T., Blomme J. (2024). Phylogeny and ecology of the green seaweed Ulva. Bot. Mar. 67, 89–92. doi: 10.1515/bot-2024-0005
Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The ClustalX windows interface flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Toor N., Hausner G., Zimmerly S. (2001). Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA Society 7, 1142–1152. doi: 10.1017/S1355838201010251
Toor N., Keating K. S., Taylor S. D., Pyle A. M. (2008a). Crystal structure of a self-spliced group II intron. Science 320, 77–82. doi: 10.1126/science.1153803
Toor N., Rajashankar K., Keating K. S., Pyle A. M. (2008b). Structural basis for exon recognition by a group II intron. Nat. Struct. Mol. Biol. 15, 1221–1222. doi: 10.1038/nsmb.1509
Toro N., Martínez-Abarca F. (2013). Comprehensive phylogenetic analysis of bacterial group II intron-encoded ORFs lacking the DNA endonuclease domain reveals new varieties. PloS One 8, e55102. doi: 10.1371/journal.pone.0055102
Turmel M., Otis C., Lemieux C. (2016). Mitochondrion-to-chloroplast DNA transfers and intragenomic proliferation of chloroplast group II introns in Gloeotilopsis green algae (Ulotrichales, Ulvophyceae). Genome Biol. Evol. 8, 2789–2805. doi: 10.1093/gbe/evw190
Vangerow S., Teerkorn T., Knoop V. (1999). Phylogenetic information in the mitochondrial nad5 gene of pteridophytes: RNA editing and intron sequences. Plant Biol. 1, 235–243. doi: 10.1111/j.1438-8677.1999.tb00249.x
Wang Y., Liu F., Liu X., Shi S., Bi Y., Moejes F. W. (2019). Comparative transcriptome analysis of four co-occurring Ulva species for understanding the dominance of Ulva prolifera in the yellow Sea green tides. J. Appl. Phycol. 31, 3303–3316. doi: 10.1007/s10811-019-01810-z
Zhang Y., Wang J., Xiao Y. (2022). 3dRNA: 3D structure prediction from linear to circular RNAs. J. Mol. Biol. 434, 167452. doi: 10.1016/j.jmb.2022.167452
Zhao C., Pyle A. M. (2017). Structural insights into the mechanism of group II intron splicing. Trends Biochem. Sci. 42, 470–482. doi: 10.1016/j.tibs.2017.03.007
Zhao S., Xia Z., Liu J., Sun J., Zhang J., He P. (2023). Morphology, growth, and photosynthesis of Ulva prolifera O.F. Müller (Chlorophyta, Ulvophyceae) gametophytes, the dominant green tide species in the Southern Yellow Sea. J. Sea Res. 193, 102375. doi: 10.1016/j.seares.2023.102375
Zimmerly S., Semper C. (2015). Evolution of group II introns. Mobile DNA 6, 1–19. doi: 10.1186/s13100-015-0037-5
Keywords: Ulva species, chloroplast genome, molecular evolution, group II intron, secondary structure, molecular marker
Citation: Liu F, Jin S, Kim JK, Wu X and Wang J (2025) Occurrence frequency, molecular evolution and phylogenetic utility of Ulva-specific chloroplast group II intron infA-62 family. Front. Mar. Sci. 12:1557121. doi: 10.3389/fmars.2025.1557121
Received: 08 January 2025; Accepted: 10 March 2025;
Published: 28 March 2025.
Edited by:
Kerstin Johannesson, University of Gothenburg, SwedenReviewed by:
Dahai Gao, Shanghai Ocean University, ChinaCopyright © 2025 Liu, Jin, Kim, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng Liu, bGl1ZmVuZ0BxZGlvLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.