- 1Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, China
- 2South China Institute of Environmental Sciences, Ministry of Ecology and Environment, Guangzhou, China
- 3University of Chinese Academy of Sciences, Beijing, China
Introduction: Coral reef ecosystems are undergoing significant restructuring due to climate-driven marine heatwaves. Understanding how calcareous macroalgae, the important primary producers and reef-builders, respond to temperature fluctuations is crucial.
Methods: In our study, we investigated the physiological and molecular responses of the green macroalga Halimeda macroloba to different temperatures (25, 28, 31°C) through quantitative proteomics and untargeted metabolomics analyses.
Results and discussion: H. macroloba showed an optimal growth at 28°C, with the highest levels of Chl a, calcium content, and photosynthetic efficiency. At the sub-optimal temperature of 25°C, the alga modulated its fatty acid composition to maintain membrane fluidity and accumulated selenium-containing metabolites to mitigate oxidative stress. At the supra-optimal temperature of 31°C, despite reduced photosynthetic performance, the alga maintained high metabolic activity to facilitate growth and calcification by increasing the abundance of proteins involved in light-harvesting, photosystems, and carbon fixation, while redistributing substantial carbon into sugars and fatty acids. Enhanced nitrogen reserves, shown by increased levels of nitrogen uptake and assimilation proteins and amino acids such as pyroglutamic and N-Methyl-D-aspartic acids, likely contributed to its high-temperature tolerance. These findings highlight the resilience of H. macroloba to moderate thermal stress and suggest its important implications for reef conservation and potential applications in aquaculture as ocean temperatures rise.
1 Introduction
Contemporary biogenic reef ecosystems are primarily composed of coral and calcareous algae, and include a wave-resistant framework constructed by large non-coral skeletal organisms. These ecosystems face various stressors from climate change and human impacts, such as ocean acidification (OA), global warming, overfishing, and eutrophication (Hughes et al., 2003; Hoegh-Guldberg et al., 2007; Ponti et al., 2021). Repeated mass coral bleaching events have profoundly impacted marine biodiversity and the stability of reef ecosystems (Sully et al., 2019). The decline of coral reefs often coincides with the proliferation of benthic algal turfs and crustose coralline algae, which competes with corals for space and dominates the benthic landscape in many tropical reefs (Enochs et al., 2015; Tebbett et al., 2023). Moreover, crustose coralline algae are reported to contribute as much or even more to carbonate production as corals in some biogenic reefs (Cornwall et al., 2023). As climate change continues to diminish coral cover (Kayanne et al., 2005), understanding the role of calcareous algae in primary production, net calcium carbonate production and the structural stability of coral reefs is becoming increasing critical.
An investigation of the biogenic reef column in core NK-1 from the South China Sea reveals that calcareous macroalgae and corals were principal reef builders in ancient oceans, with peak biomass occurring at distinct intervals (Yi et al., 2021). Calcification in algae has evolved independently through different mechanisms over a long period. The earliest sheath-calcified cyanobacteria are known to arise 750–700 million years ago (Mya) (Knoll et al., 1991), and subsequently calcification phenotype spans across green (Chlorophyta), red (Rhodophyta), brown (Phaeophyta), and golden (Haptophyta) algae (Pentecost, 1991). Calcareous algae can decompose distinct crystalline forms (aragonite or calcite) in specialized regions, such as intracellular organelle, sheath, cell wall, and thallus surface (Borowitzka and Larkum, 1987). Continuous Ca2+ supply and efficient regional regulation of pH in the calcifying fluid that directly affect ΩCaCO3 are crucial for algal calcification (Lowenstam, 1981). Environmental changes, particularly temperature variations, significantly influence calcification traits by affecting the algal photosystem and enzymatic activity, as well as the seawater carbonate system, pH, and Ca2+ concentration.
With global warming and the increasing frequency of local marine heatwaves (Sun et al., 2023), fluctuations in seawater temperature are becoming more pronounced on both seasonal and diurnal scales. The distinct responses of calcifying organisms, such as corals and algae, to temperature changes are anticipated to exert a pivotal influence on their biomass dynamics within reef ecosystems. Certain calcareous algal physiological processes, including photosynthesis, respiration, and calcification, are more adversely affected by elevated temperatures than OA (Vásquez-Elizondo and Enríquez, 2016). A meta-analysis of published studies reveals that significant negative impacts on algal calcification emerge at 5.2°C above and 2.0°C below ambient temperature (Cornwall et al., 2019). Contemporary studies on the molecular responses of marine algae to temperature changes increasingly rely on multi-omics approaches, revealing adaptive strategies like nutrient acquisition regulation, element flux reallocation, membrane composition modification, and biosynthesis of antioxidant metabolites and protective solutes (Toseland et al., 2013; Liang et al., 2019; Zhao et al., 2020; Dougan et al., 2023). However, molecular responses to temperature changes in calcareous macroalgae remain relatively understudied compared to microalgae.
Recognizing the inevitability of future coral reef transformations underscores the need for continued investigation into reef-building organism dynamics. The genus Halimeda is a worldwide distributed reef-building organism characterized by fast growth and a rapid turnover rate in tropical and sub-tropical oceans (Xu et al., 2015). Halimeda sp. thalli exhibit diverse morphological forms, such as erect, pendant, and sprawling structures, and are secured by rhizoids originating from both inner and outer cortical cells (Hillis-Colinvaux, 1980). It is furthermore an excellent candidate for calcareous algae research because its calcification occurs extracellularly, which deposits aragonite in the semi-enclosed inter-utricle space (IUS) (Wizemann et al., 2014). In this study, we sampled the thalli of Halimeda macroloba in Sanya Bay of the South China Sea, where the mean winter and summer water temperatures in Sanya Bay were 25.05°C and 28.72°C, respectively (Geng et al., 2023). Marine heatwaves are typically defined as periods when sea surface temperatures exceed the 90th percentile of 30-year historic values for a specific location for at least five consecutive days, with anomalies ranging from 2°C to over 4°C above the long-term average (Oliver et al., 2018). In order to cover the natural temperature range and marine heatwave events, a factorial experimental design with three treatments (25°C, 28°C, and 31°C) was set. Our goal was to identify molecular and metabolic traits that can enable H. macroloba to maintain high productivity and active physiology under different temperatures. Using quantitative proteomics and untargeted metabolomics, we investigated the protein and metabolite profiles of H. macroloba. Our results showed that H. macroloba exhibited temperature-dependent physiological and molecular adjustments, with resource reallocation across pigments, sugars, fatty acids, and amino acids to optimize key metabolic processes and enhance thermal acclimation.
2 Materials and methods
2.1 Sample collection and experimental design
We collected thalli of Halimeda from a coastal rocky reef in Sanya Bay, China (18°12′ N, 109°28′ E) at depth of 2 m in July 2022, where the seawater temperature was approximately 28°C. The sampling area is located in the subtidal zone that consists of sand, stones, macroalgae, and branched corals. All specimens of Halimeda were carefully uprooted with the holdfast intact from the substrate. After collection, the samples were placed in a polyethene-free, continuous-flow, seawater system for transport to the laboratory. Subsequent to seawater incubation and the removal of living and attached eukaryotes, the algae were cultured in laboratory at 25°C under a light intensity of 150 µmol m-2 s-1 with a 14:10 light/dark cycle in a 20 L tank, using a continuous circulation system of autoclaved natural seawater with a salinity of 32 PSU. An air pump is employed to continuously infuse air to sustain the vitality of algae cells, while the temperature of the culture water is regulated through a digital temperature controller and a titanium alloy heating rod. A filter is installed along the edges of the tank to ensure water quality.
Three temperature treatments (25°C, 28°C, and 31°C) were set to simulate the seasonal temperature variations and marine heatwave events of Sanya Bay. Ten individuals were placed in each temperature-controlled aquarium containing 20 L of seawater. To maintain stable seawater chemistry conditions, filtered seawater were changed twice weekly. The seawater temperature was gradually increased by 1°C every 12 hours using water thermostats (TC10, Teco Italy) until the desired treatment temperature was reached. Once each temperature stabilized, the treatment was continued for 21 days before sampling.
2.2 Molecular phylogenetic analyses
For genetic study, approximately 0.6 g of the algae were ground with silica beads using a pestle, followed by DNA extraction using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Here, we investigated the molecular phylogenetic relationship of the collected Halimeda using the plastid elongation factor Tu (tufA) and ribulose-1, 5-bisphosphate carboxylase/oxygenase large subunit (rbcL) DNA sequences (Famà et al., 2002; Kojima et al., 2015). Polymerase chain reaction (PCR) amplification of the large subunit of rbcL gene sequences were carried out in a total volume of 10 μL composed of 5 μL 2 × PCR buffer for KOD FX, 2 mM dNTPs, 20 μM of each Primer, 0.2 units KOD FX and 0.5 μL DNA solution with a PCR Thermal Cycler Dice (TaKaRa Bio, Shiga, Japan). The primers used for PCR amplification of the tufA region were as follows: tufA-A1 (Forward), ATGRCWCGHGMAAAATTTSAACG; tufA-H2 (Forward), ATTTTAGTVGTNTCNGGTGC; and tufA-R (Reverse), CCTTCNCGAATMGCRAAWCGC. The touch down PCR protocol consisted of the following steps: an initial denaturation at 94°C for 2 min; three cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and extension at 68°C for 30 s; three cycles of denaturation at 98°C for 10 s, annealing at 48°C for 30 s, and extension at 68°C for 30 s; followed by 28 cycles of denaturation at 98°C for 10 s, annealing at 46°C for 30 s, extension at 68°C for 30 s, and a final extension at 72°C for 10 min. The primers used for PCR amplification of the rbcL region were as follows: GAACATGGACAACRGTTTGGAC (Forward) and GACTCCATTTAGCAGCATCACG (Reverse). The profile of PCR conditions was as follows: an initial denaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 50 s, annealing at 55°C for 40 s, extension at 72°C for 70 s, and a final extension at 72°C for 10 min.
PCR products were examined on ethidium bromide-stained 1.0% agarose gels. After PEG purification, PCR products were directly sequenced using the GenomeLab Dye Terminator Cycle sequencing with Quick Start Kit (Beckman Coulter, Fullerton, CA, USA) and the CEQ8000 DNA Analysis System (Beckman Coulter) according to the manufacturer’s instructions. Sequences from the other Halimeda species and the outgroup species of Caulerpa racemosa and C. cuneate were downloaded from the NCBI database for the molecular phylogenetic analyses. All the sequences were preliminarily aligned together with reported sequence data using the MEGA11 (Tamura et al., 2021) and then manually aligned for the phylogenetic analyses. Phylogenetic trees were obtained using maximum likelihood (ML). Evolutionary distance was computed using the Kimura two-parameter method (Kimura, 1980). The ML phylogenetic tree was inferred in MEGA using the GTR + G + I model. The reliability of each branch was checked using 1 000 bootstrap replications.
2.3 Measurements of Fv/Fm, Chl a and tissue calcium contents
The photosynthetic maximum quantum yield of photosystem II (Fv/Fm) was measured at the end of the experiment using a DIVING-PAM fluorometer (Heinz Walz GmbH, Germany) with an 8.0 mm diameter fiberoptic cable. The mature Halimeda segments (~3.0 cm) were randomly selected, removed, and dark-adapted for approximately 10 minutes before measurement. To minimize photochemical quenching caused by the measuring light, measurements were performed with a constant light intensity (setting “8”) in the “Burst mode”. Initial measurements of Fv and Fm were recorded to test for bias and ensure accurate PS II photophysiology activity. Chlorophyll a (Chl a) concentration was determined spectrophotometrically following the published method (Wellburn, 1994). The fresh weight (FW) of 50.0 mg of mature Halimeda segments was ground in 10 mL of 90% acetone, and stored in the dark at 4°C for 24 hours for extraction. Samples were then centrifuged at 5,000 g for 10 min at 4°C (Eppendorf centrifuge 5810 R, Germany). The supernatant was transferred to a quartz cuvette and its absorbance was measured at 664, 647 and 470 nm using a spectrophotometer (UV 530, Beckman counlter, USA).
After the completion of the experiment, algae samples at each temperature were collected and rinsed with sterile seawater for 3-5 cycles. The surface moisture of the algae was then dried using absorbent paper. Subsequently, the samples were placed in an oven at 60°C and obtained its dry weight (Wt0). Following this, 5% of 12 nM/L HCl (W/V) was added to the dried algae to dissolve the tissue CaCO3. After no effervescence was generated by the addition of acid, the algae were then rinsed with distilled water for 3-5 cycles and dried again at 60°C until a constant weight (Wt1) was reached. The calcium content was calculated using the formula (Neustupa and Nemcova, 2022): PCaCO3 = 100 * (Wt0 – Wt1)/Wt0, where PCaCO3 represents the percentage of CaCO3 within the dry weight of the algae at each treatment.
All the physiological parameter results were presented as the mean ± standard deviation (SD) of triplicate measurements. A one-way analysis of variance (ANOVA) was used to assess the effect of temperature on the response variables. Significant differences were defined as P<0.05.
2.4 Protein extraction, digestion, peptide labeling and LC-MS/MS analysis
After three weeks of treatment at temperatures of 25°C, 28°C, and 31°C, approximately 1.2 g of thalli from each triplicate at each temperature were harvested, purified to remove contaminants, rinsed with autoclaved seawater, and immediately frozen in liquid nitrogen. For protein extraction, thalli powders, ground in liquid nitrogen, were suspended in 1 mL of Trizol reagent (Invitrogen, Carlsbad, USA), following the established protocol (Wang et al., 2013). Proteins (100 μg per sample) were digested by adjusting the pH to 8.5 with 1 M ammonium bicarbonate, then reduced with dithiothreitol at 60°C for 1 hour, and alkylated with iodoacetamide in the dark for 45 minutes at room temperature. Proteins were double-digested using Trypsin Gold (Promega, Madison, WI, USA) at a 30:1 protein/trypsin ratio for 14 hours at 37°C. Post-digested samples were desalted with a Strata X C18 solid-phase extraction column (Phenomenex, Torrance, CA, USA), evaporated, and reconstituted in 0.2 M triethylammonium bicarbonate. Desalted peptides from nine samples (three biological replicates for each temperature) were labeled with TMT reagents from the 12-plex Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions: Tag113, Tag114 and Tag115, 25°C; Tag116, Tag117 and Tag118, 28°C; Tag119, Tag120 and Tag121, 31°C. After a 2 h incubation, the labeled samples were combined, desalted with a Strata X C18 column (Phenomenex) and subsequently vacuum-dried.
The reconstituted peptides, resuspended in UPLC loading buffer [2% acetonitrile (ammonia to pH 10)], were separated using a high pH liquid phase using a reversed-phase C18 column ACQUITY UPLC BEH C18 Column1.7 µm, 2.1 mm × 150 mm (Waters, USA). Peptides were separated with a gradient of elution (Phase A: 2% acetonitrile, pH 10; Phase B: 80% acetonitrile, pH 10) over 48 min at a flow rate of 200 μL/min, following the gradient: 0-1.9 min, 0-0% B; 1.9-2 min, 0-5% B; 2-17 min, 5-5% B; 17-18 min, 5-10% B; 18-35.5 min, 10-30% B; 35.5-38 min, 30-36% B; 38-39 min, 36-42% B; 39-40 min, 42-100%B; 40-44 min, 100% B; 44-45 min, 100-0% B; 45-48 min, 0% B. A total of 28 fractions were collected based on peak shape and time, then combined into 14 fractions, and concentrated by vacuum centrifugation.
Two-dimensional analysis was performed by liquid chromatography tandem mass spectrometry (Evosep One combined with Obitrap Exploris 480 mass spectrometer) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Each peptide fraction, resuspended in buffer C (2% acetonitrile and 0.1% formic acid), underwent centrifugation at 16,000 g for 10 min. The supernatant was loaded onto a C18 column (150 μm × 15 cm, Evosep, Denmark) for liquid phase separation in solvent A (water with 0.1% formic acid) and a linear gradient of solvent B (100% acetonitrile with 0.1% formic acid) at a flow rate of 300 nL/min. Peptides were eluted using the following gradient: 0-2 min, 5-5% B; 2-30 min, 5-38% B; 30-40 min, 38-90% B; 40-44 min; 90-90% B. The Orbitrap Exploris 480 was operated in the data-dependent acquisition mode (DDA) to automatically switch between full scan MS and MS/MS acquisition. Full scan MS spectra (m/z 350–1500) were acquired in the Orbitrap with a resolution of 60K. Precursor ions were then selected for fragmentation by higher-energy collision dissociation (HCD) in the collision cell. The MS/MS resolution was set at 15K, and dynamic exclusion was set to 30 s.
2.5 Protein identification, quantification and bioinformatics analysis
The raw MS/MS data were analyzed using ProteomeDiscoverer 2.4 (Thermo Scientific, Waltham, MA) against a database encompassing translated protein sequences from the genome of H. opuntia (Zhang et al., 2024). The parameters were set as follows: trypsin was designated as the specific enzyme with a maximum allowance of two missed cleavages per peptide; fixed modifications included carbamidomethyl (C), iTRAQ8-plex (N-term), and iTRAQ8-plex (K); variable modifications comprised oxidation (M); peptide charge states of 2+, 3+, and 4+ were considered; 20 ppm was set for peptide mass tolerance, and 0.02 Da for fragment mass tolerance. The false discovery rate (FDR) for peptide identification was set at FDR ≤ 0.01. High-confidence proteins, characterized by at least one unique peptide and two unique spectra, were selected, and differentially expressed proteins (DEPs) were filtered with fold ratios ≥ 1.2 or ≤ 0.83 and P < 0.05 (calculated by t-test). Functional annotations were conducted against the NCBI non-redundant protein database (NCBInr) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). Subcellular location of identified proteins was predicted using Cello (http://cello.life.nctu.edu.tw/).
2.6 Metabolite extraction and LC-ESI/MS analysis
Thalli were collected concurrently with proteomic sampling, purified to remove contaminants, rinsed with autoclaved seawater, and ground in liquid nitrogen. Thalli powders were suspended in 2 mL centrifuge tubes containing 1 mL extraction buffer (acetonitrile: methanol: water; 2:2:1, v/v/v) and 100 mg glass beads. Following a freeze-thaw cycle involving liquid nitrogen freezing for 5 min and subsequent room temperature thawing, the samples underwent grinding at 55 Hz for 2 min. This freeze-thaw-grind cycle was repeated three times. After centrifugation (12,000 g, 10 min, 4°C), the supernatant was transferred to a new tube, vacuum-dried, and re-dissolved in 300 μL of 2-amino-3-(2-chloro-phenyl)-propionic acid (4 ppm). The solution was filtered through a 0.2 μm membrane (PALL Life Sciences, USA) before LC-MS/MS analysis.
LC-MS/MS analysis was conducted using a Vanquish UHPLC System (Thermo Fisher Scientific, USA) equipped with an ACQUITY UPLC ® HSS T3 column (150×2.1 mm, 1.8 μm) (Waters, Milford, MA, USA), maintained at 40°C. The flow rate and injection volume were set at 250 µL/min and 2 μL, respectively. For LC-ESI (+)/MS analysis, mobile phases included buffer E (0.1% formic acid in CAN) and buffer F (0.1% formic acid in water), and the separation gradient was as follows: 2% E, 0-1 min; 2-50% E, 1-9 min; 50-98% E, 9-12 min; 98% E, 12-13.5 min; 98-2% E, 13.5-14 min, 2% E, 14-20 min. LC-ESI (-)-MS analysis employed ACN and ammonium formate (5mM) as mobile phases, with a separation gradient of 2% ACN, 0-1 min; 2-50% ACN, 1-9 min; 50-98% ACN, 9-12 min; 98% ACN, 12-13.5 min; 98%-2% ACN, 13.5-14 min; 2% ACN, 14-17 min.
Metabolite mass spectrometric detection was carried out on a Q Exactive instrument (Thermo Fisher Scientific, USA) with an ESI ion source, using simultaneous MS1 and MS/MS (Full MS-ddMS2 mode, data-dependent MS/MS) acquisition. The parameters were set as follows: sheath gas pressure, 30 arb; auxiliary gas flow, 10 arb; spray voltage, 3.50 kV and -2.50 kV for ESI(+) and ESI(-), respectively; capillary temperature, 325°C; MS1 range, m/z 81-1000; MS1 resolving power, 70,000 FWHM; number of data-dependent scans per cycle, 10; MS/MS resolving power, 17,500 FWHM; normalized collision energy, 30%; dynamic exclusion time, automatic.
2.7 Metabolite identification, quantification and bioinformatics analysis
The raw data underwent conversion to mzXML format through MSConvert in the ProteoWizard software package (v3.0.8789) (Smith et al., 2006) and subsequent processing employing XCMS for feature detection, retention time correction, and alignment. Metabolites were identified based on mass accuracy (<30 ppm) and matched with MS/MS data from HMDB, Massbank, LipidMaps, mzcloud, and KEGG. Robust LOESS signal correction (QC-RLSC) was applied for data normalization to rectify any systematic bias. Following normalization, only ion peaks with relative standard deviations (RSDs) less than 30% in QC were retained, ensuring accurate metabolite identification.
Multivariate data analyses were conducted using Ropls software (Thévenot et al., 2015). After data scaling, models were constructed using principal component analysis, orthogonal partial least-square discriminant analysis (PLS-DA), and partial least-square discriminant analysis (OPLS-DA). Metabolite profiles were visualized as score plots, with each point representing a sample. Corresponding loading plots and S-plots were generated to provide insights into the metabolites influencing sample clustering. All models were assessed for overfitting through permutation tests. Descriptive performance was determined by R2X (cumulative) and R2Y (cumulative) values, while prediction performance was measured by Q2 (cumulative) and a permutation test. The permuted model was employed for predicting classes, with R2 and Q2 values at the Y-axis intercept lower than those of Q2 and the R2 of the non-permuted model. OPLS-DA facilitated the identification of discriminating metabolites using variable importance on projection (VIP). The p-value, VIP, and fold change (FC) were applied to ascertain contributable variables for classification. Finally, metabolites with a p-value < 0.05 and VIP values > 1 were considered statistically differentially expressed metabolites (DEMs). The identified metabolites were functionally annotated using the KEGG database. Subsequently, the correlation between metabolites and associated proteins was validated through additional KEGG-based functional analysis. Consistent expression patterns of metabolites and proteins further strengthen the reliability of the findings, suggesting a temperature-regulated pathway. Moreover, functional analysis linked these molecular alterations to corresponding physiological responses, providing a more comprehensive understanding of the underlying biological processes.
3 Results
3.1 Phylogenetic position and physiological parameters
Based on ML analysis of tufA and rbcL genes, molecular phylogenetic tree showed that the species used in this study is H. macroloba (Figure 1). Photosynthetic performance, Chl a and tissue calcium content showed similar responses to temperature changes, with no significant variation observed across three temperatures (Figure 2). The average Fv/Fm value was highest at 28°C (0.75), followed by at 25°C (0.72) and 31°C (0.727). Chl a content averaged 223.6 μg/(g FW) at 25°C, 237.5 μg/(g FW) at 28°C, and 234.8 μg/(g FW) at 31°C. The average tissue calcium content was 80.6% at 25°C, 82.2% at 28°C, and 80.4% at 31°C.
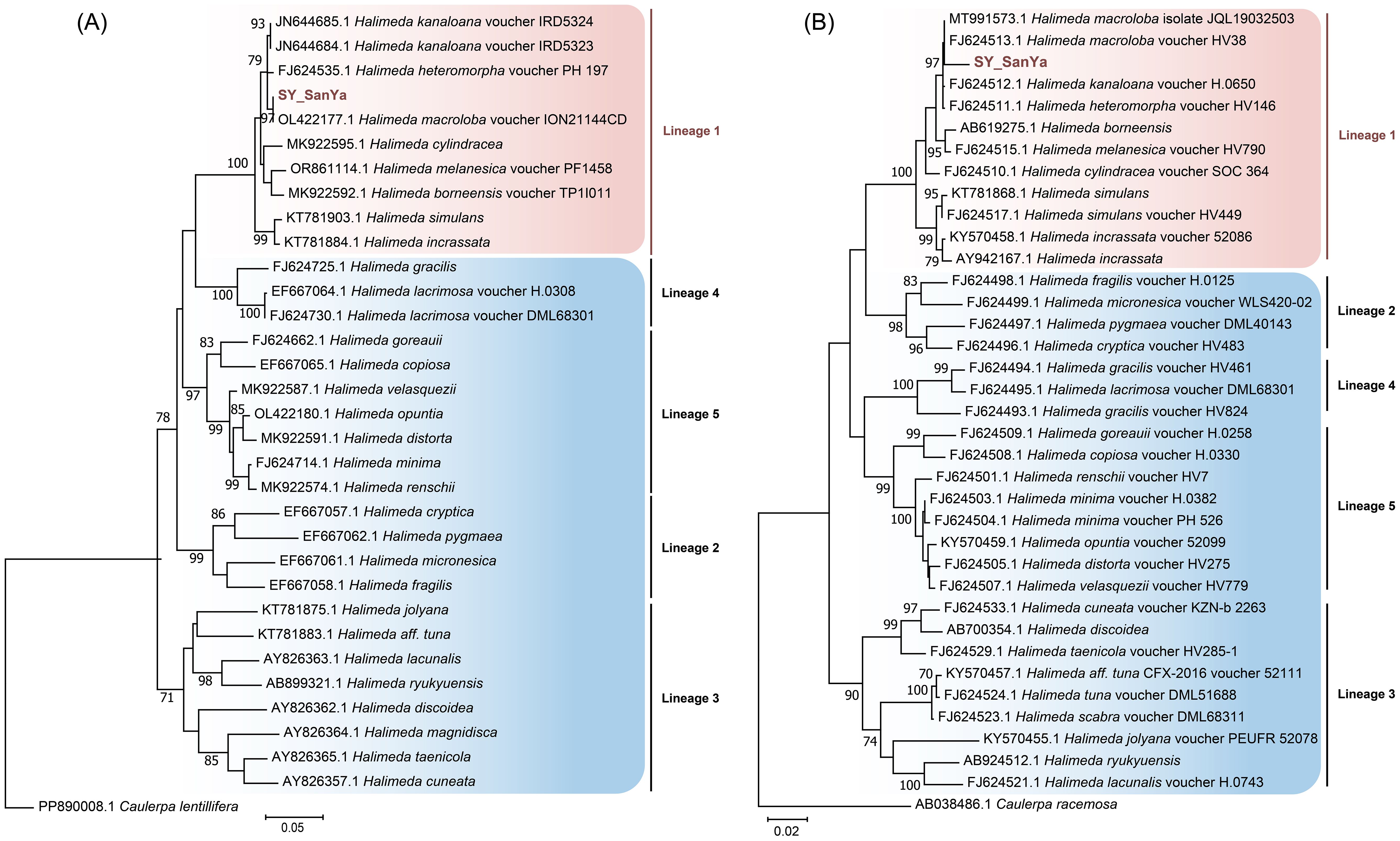
Figure 1. Maximum likelihood (ML) molecular phylogenetic tree of the genus Halimeda based on tufA (A) and rbcL (B) genes. Numbers on branches indicatebootstrap values from ML analysis and SY_SanYa represents the alga used in this study. The vertical line indicates that these algae belong to the same lineage.
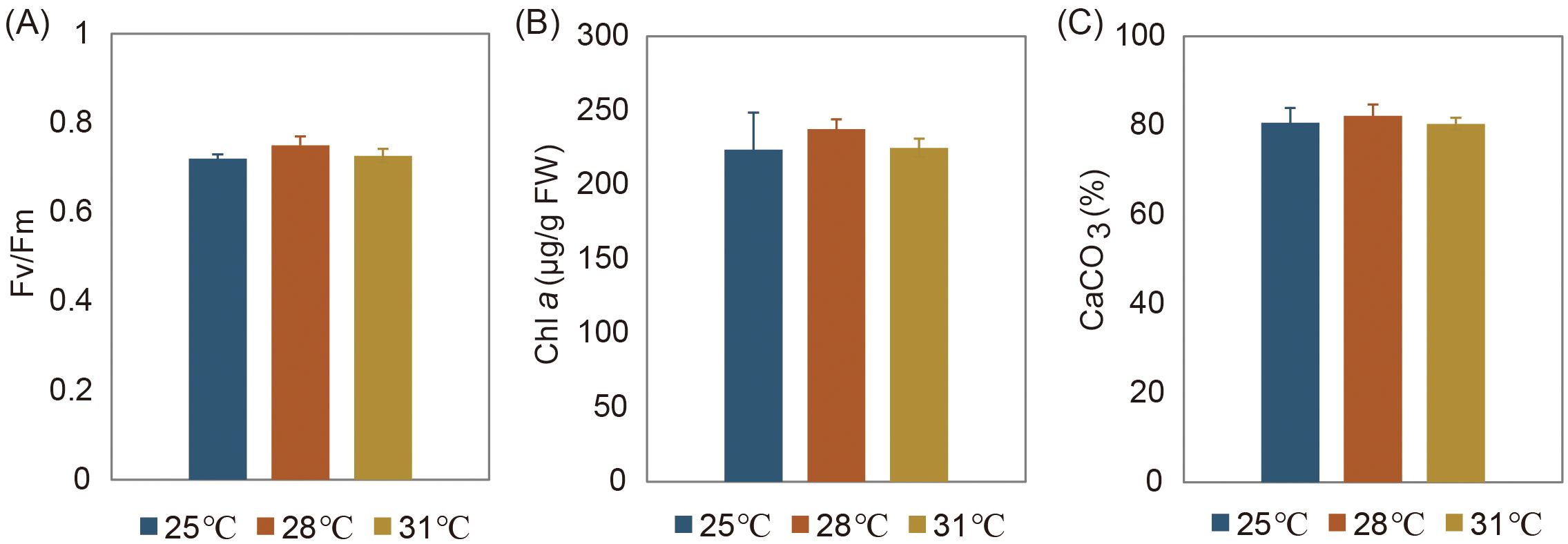
Figure 2. Physiological responses of H macroloba to temperature changes. (A) Photosynthetic maximum quantum yield (Fv/Fm), (B) Chl a content, and (C) tissue calcium content. No significant variation in the three variables was observed across three temperatures (P>0.05).
3.2 Overview of quantitative iTRAQ proteomics and untargeted metabolomics
We detected a total of 1,058 high-confidence proteins at three temperatures (Supplementary Table S1). Hierarchical clustering heatmap showed that the protein expression profiles at 25°C and 28°C exhibited a high similarity, while they largely differed from those at 31°C (Supplementary Figure S1A). Compared with 28°C, 166 and 454 DEPs were identified at 25°C and 31°C, respectively (Supplementary Figure S1B). The enrichment analysis of DEPs revealed distinct temperature-dependent patterns in KEGG pathways (Figure 3). Specifically, we observed a significant upregulation of photosynthesis-related processes, encompassing both antenna proteins and core photosynthetic activities, between 28°C and 25°C. Shifts in metabolic pathways associated with terpenoid backbone biosynthesis, fatty acid biosynthesis, and nitrogen metabolism were prominently evident between 31°C and 25°C. Furthermore, plant-pathogen interactions, ascorbate and aldarate metabolism, and oxidative phosphorylation, were notably enriched between 31°C and 28°C.
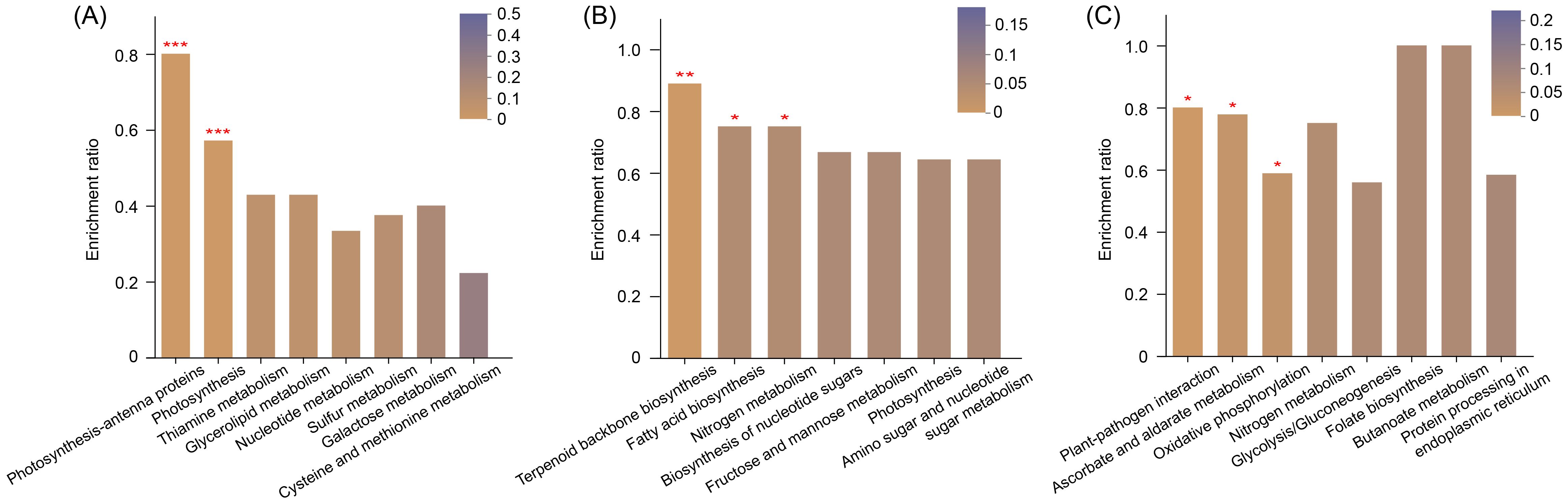
Figure 3. The top KEGG terms of differentially expressed proteins (DEPs) enriched at 28°C vs 25°C (A), 31°C vs 25°C (B), and 31°C vs 28°C (C). The enrichment ratio is calculated by dividing the number of DEPs by the total number of detected proteins in a given KEGG pathway. *P < 0.05, **P < 0.01, and ***P < 0.001.
We identified a total of 204 metabolites from cells growing at three temperatures (Supplementary Table S2). Among them, 28, 47, and 22 DEMs were detected at 28°C vs 25°C, 31°C vs 25°C, and 31°C vs 28°C, respectively (Supplementary Figure S1C). Most of DEMs were categorized into fatty acids and conjugates, amino acids, peptides and their analogues, followed by carbohydrates and carbohydrate conjugates, steroids and steroid derivatives, and steroids and steroid derivatives (Figure 4). These metabolites are involved in lipid and amino acid metabolism, membrane transport, carbohydrate metabolism, and other pathways (Supplementary Table S2).
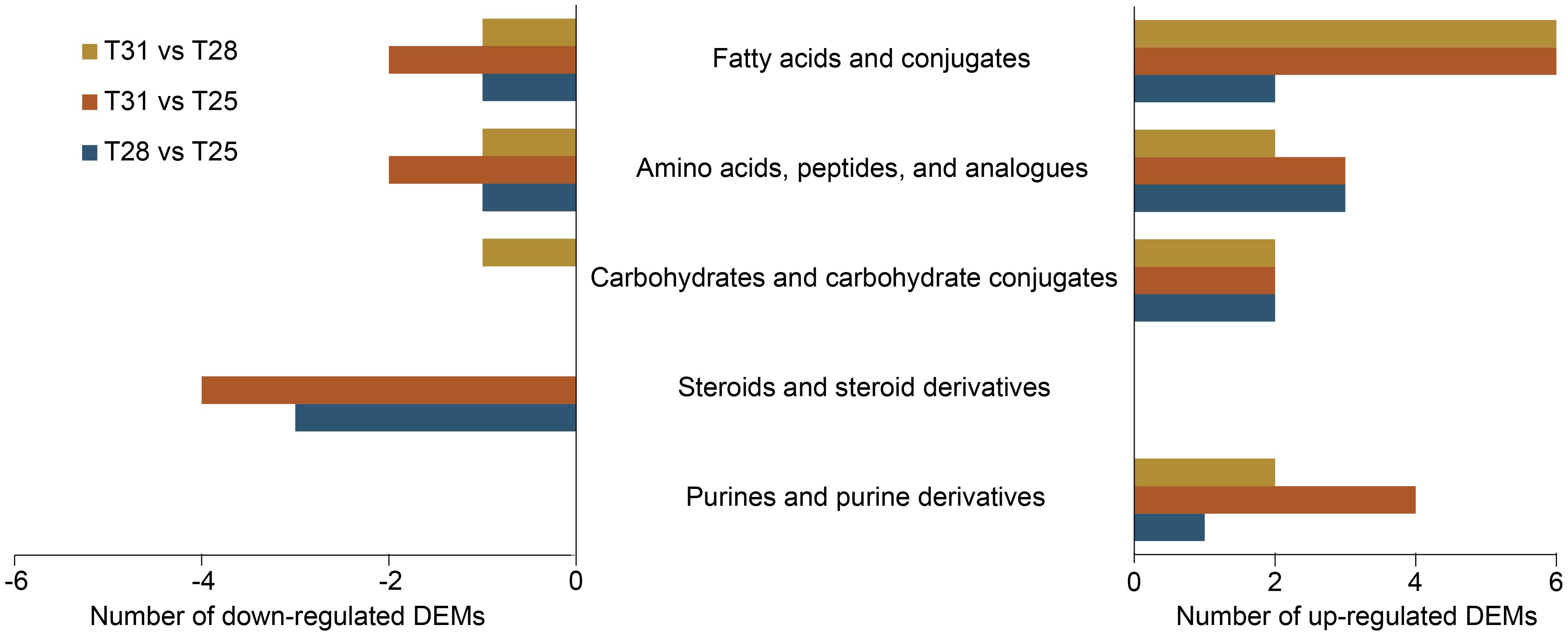
Figure 4. Classification of differentially expressed metabolites (DEMs) between temperature comparisons. Up-regulated DEMs at T31 vs T28 represents higher levels at 31°C compared to 28°C.
3.3 Classification of key DEPs and DEMs
We generated two heatmaps to represent the expression patterns of DEPs and DEMs associated with some crucial biological processes (Figure 5). The abundance of certain proteins belong to light-harvesting complexes I and II (LHCA1, LHCA3, LHCB1, LHCB2, LHCB4, and LHCB5), photosystem I subunits (psaD, psaF, psaH, psaK, psaL, and psaN), ferredoxin (petF), and cytochrome b6-f complex iron-sulfur subunit (petC) increased significantly from 25°C to 28°C, while the abundance of LHCA1, LHCB4, LHCB5, psaD, psaF, psaK, and petF increased significantly from 28°C to 31°C. The essential carbon fixation protein rbcL reached its highest abundance at 31°C. Regarding sugars and carbohydrate conjugates, the abundance of glyceric acid peaked at 28°C, while mannitol and D-mannose showed maximum levels at 31°C. Among fatty acids and conjugates, only one polyunsaturated type of oleic acid exhibited significant accumulation at 25°C, whereas eicosadienoic acid, 9,10-epoxyoctadecenoic acid, 12,13-DHOME, adrenic acid, nonadecanoic acid, and myristic acid exhibited increased abundances from 25°C to 31°C.
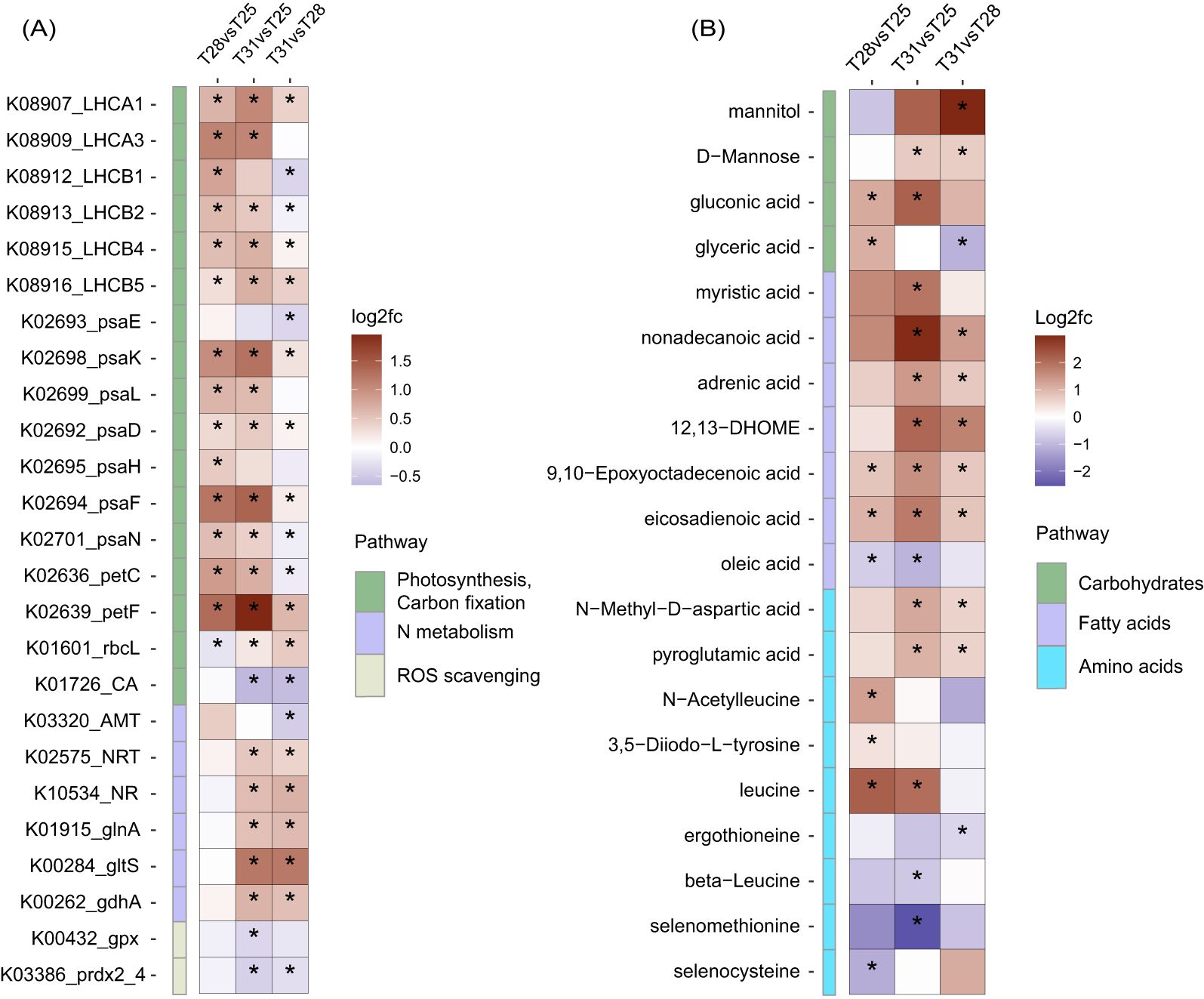
Figure 5. Heatmap of key DEPs (A) and DEMs (B) of H macroloba under temperature changes. T31 vs T28 means 31°C compared to 28°C, with an asterisk denoting a statistically significant difference.
Proteins associated with nitrogen metabolism, such as nitrate/nitrite transporter (NRT2), nitrate reductase (NR), glutamate dehydrogenase (NADP+) (gdhA), glutamine synthetase (glnA), and glutamate synthase (ferredoxin) (gltS) exhibited minor variations between 25°C and 28°C, followed by significant increases from 28°C to 31°C. Since nitrogen is an essential component of amino acid, we observed that the abundances of beta-leucine, and ergothioneine peaked at 25°C, the abundances of leucine, 3,5-Diiodo-L-tyrosine, and N-Acetylleucine peaked at 28°C, and the abundances of pyroglutamic acid and N-Methyl-D-aspartic acid peaked at 31°C.
Consistently, Se-containing oxidoreductases of glutathione peroxidase (gpx) and peroxiredoxin 2/4 (prdx2_4) gradually decreased in abundance from 25°C to 31°C, and the amounts of selenocysteine and selenomethionine peaked at 25°C. Additionally, one calcification gene carbonic anhydrase (CA) showed a decreasing trend from 25°C to 31°C.
4 Discussion
Halimeda (Bryopsidales), a genus of calcified, segmented green macroalgae, is widely distributed in lagoons and reefs across the subtropical and tropical ecosystems. Its thallus consists of a singular tubular cell that branches to form an organized network of siphons, with distinct medullary and cortical regions facilitating mineral deposition in IUS (Hillis-Colinvaux, 1980; Wizemann et al., 2014). Environmental conditions strongly influence the shape and size of Halimeda segments, complicating species identification based on morphology alone. Phylogenetic studies using nuclear ribosomal and plastid DNA sequences have identified five distinct lineages within the genus, suggesting geographic isolation among sibling taxa during Pleistocene or Pliocene climatic cooling periods (Kooistra et al., 2002; Verbruggen et al., 2005). Our phylogenetic analysis based on tufA and rbcL sequences places our sample close to H. macroloba, which falls within lineage 1 (Figure 1). Lineage 1 is characterized by medullary siphons connecting via pores and enlarged middle and lower segment structures (Kooistra et al., 2002). These algae typically exhibit adaptive responses to water motion, with certain species exhibiting flexible upper thalli branches that stiffen in shallow, wave-exposed environments. Due to the significant impact of ocean warming on calcareous organisms, we conducted a multi-omics analysis of H. macroloba to assess its temperature response. Recognizing the constraints of a small sample size and a single omics approach, we increased the reliability of our findings by integrating physiological data with consistent metabolomic and transcriptomic results.
The relationship between algal growth and temperature typically follows a bell-shaped curve, with gradual increases in growth rate and physiological performance up to an optimal temperature, followed by steep declines at supra-optimal temperatures (Ras et al., 2013). In this study, we observed lower Fv/Fm and Chl a values at 25°C compared to 28°C, indicating sub-optimal conditions for H. macroloba (Figure 2). These physiological changes align with modifications in the algal lipid profile, particularly in the fatty acid composition. Algae adjust their plasma membrane composition to optimize the liquid/crystalline physical structure and membrane fluidity necessary for proper function (Uemura et al., 2006). The balance between saturated and polyunsaturated fatty acids determine membrane fluidity, and this composition shifts in response to temperature stress (Leblond et al., 2006; Hyun et al., 2016; Willette et al., 2018). Our findings of peak oleic acid levels in H. macroloba at 25°C suggest its role in enhancing membrane fluidity under sub-optimal temperatures (Figure 5B). This increase in oleic acid correlates with the observed decrease in Fv/Fm and Chl a at 25°C, suggesting that although the alga is adjusting its lipid profile to maintain membrane functionality, these changes are not sufficient to sustain optimal photosynthetic performance. The shift in fatty acid composition may therefore be a compensatory mechanism to maintain cellular function under stress, but it does not fully prevent the physiological decline at sub-optimal temperatures. Additionally, we observed a significant accumulation of selenocysteine and selenomethionine at 25°C from the metabolomics data (Figure 5B). Selenium (Se), primarily exists as selenocysteine, is largely incorporated into selenoproteins that are known to scavenge reactive oxygen species (ROS) in various organisms (Gobler et al., 2013; Jiang et al., 2020; Patel et al., 2022). Under low temperatures, plants can generate excess ROS due to decreased photosynthetic rates and excessive electron transfer to O2 (Mittler, 2002). In line with this, the consistent accumulations of selenocysteine and selenomethionine, as well as Se-containing proteins like glutathione peroxidase and peroxiredoxin 2/4 in H. macroloba at 25°C (Figure 5) underscore their role in ROS scavenging. This finding provides new insights into how H. macroloba utilizes selenium to counteract oxidative stress under sub-optimal temperatures.
Conversely, at 31°C, we observed lower Fv/Fm and Chl a values compared to 28°C, indicating supra-optimal conditions for H. macroloba (Figure 2). Despite this, the increased abundance of key proteins involved in photosynthesis, such as LHCs, psaD, psaF, psaK, petF, and rbcL at 31°C (Figure 5A) indicated that the algal cells sustained high metabolic activity under moderate thermal stress. This suggests that while photosynthetic efficiency (as reflected in Fv/Fm and Chl a) was compromised, the cells were compensated by enhancing metabolic processes to support cellular function in response to heat stress. In parallel, we observed significant shifts in the carbohydrate profile, particularly the accumulation of mannitol, D-mannose, and gluconic acid at 31°C (Figure 5B). Mannitol, a major photosynthetic product, serve as crucial carbon storage and antioxidant against environmental stress in algae (Tonon et al., 2017). Previous studies show significant mannitol accumulation during summer and at supra-optimal temperatures in some brown algae and dinoflagellates (Adams et al., 2011; Zhang et al., 2022). The higher heat capacities of D-mannose, attributed to its larger molecular weight and the solid–liquid transition observed in heat capacity curve (Wang et al., 2018), suggest its powerful heat tolerance and dissipation abilities at higher temperatures. It is plausible that H. macroloba cells likely accumulate specific sugars such as mannitol and D-mannose to acclimate to moderate heat stress. Additionally, fatty acids represent another important carbon pool. We observed increased levels of eicosadienoic acid, 9,10-epoxyoctadecenoic acid, adrenic acid, 12,13-DHOME, adrenic acid, nonadecanoic acid, and myristic acid in H. macroloba from 28°C to 31°C (Figure 5B). These increases suggest a substantial redistribution of carbon into these fatty acids under higher temperatures, likely as part of the algal strategy to regulate membrane stability and fluidity, which are critical for maintaining cellular integrity under heat stress. Despite these significant metabolic changes, the Fv/Fm ratio remained relatively stable between 28°C and 31°C. This phenomenon suggests that H. macroloba possesses a robust photosynthetic system capable of maintaining high efficiency within this moderate temperature range. Indeed, in siphonous algae like Halimeda sp., millions of chloroplasts can move freely through cytoplasmic streaming, a mechanism that helps cope with stress and maximize photosynthetic efficiency (Grant and Borowitzka, 1984). Even though metabolic pathways and carbon storage patterns are altered, the movement of chloroplasts may play a key role in protecting the photosynthetic apparatus.
Our study showed a significant increase in proteins NRT2, NR, gdhA, glnA, and gltS from 28°C to 31°C (Figure 5A). NRT2 and NR facilitate the uptake and initial reduction of nitrate from the environment, while gdhA, glnA, and gltS involve in the subsequent assimilation of ammonium and the synthesis of key amino acids. Upregulation of these proteins at 31°C suggests enhanced nitrogen uptake and active nitrogen metabolism, which is further supported by our metabolomic data. Notably, we observed significant accumulations of pyroglutamic acid and N-Methyl-D-aspartic acid at 31°C (Figure 5B), metabolites associated with amino acid metabolism that could play a role in mitigating the moderate thermal stress. This metabolic shift is consistent with findings in heat-induced brown macroalgae kelp, where nitrogen accumulation, rather than an enhancement of photosynthetic apparatus, is central to thermal tolerance (Gerard, 1997; Endo et al., 2020). Nitrogen in algae is primarily assimilated into chlorophylls, amino acids, and proteins. In our study, the decrease in Chl a content from 28°C to 31°C (Figure 2B) suggests that nitrogen in H. macroloba may be rerouted from chlorophyll synthesis toward the accumulation of amino acids and proteins. Therefore, H. macroloba likely manages the trade-off between reduced photosynthetic capacity and enhanced nitrogen reserves mediated through both protein and metabolite regulation to cope with thermal stress.
As an energy-consuming process heavily reliant on the ΩCaCO3 (Millero et al., 2006), any factors influencing ΩCaCO3 will affect the algal calcification process. To cope with the environmental constraints, calcareous algae have evolved various physio-chemical mechanisms to modulate calcification. One such mechanism is to build a favorable environment for mineral deposition through altering gene expressions associated with the regional redistribution of calcifying ions (Zhang et al., 2024). Notably, we detected no plasma-membrane proteins such as Ca2+-ATPases, potassium-dependent sodium calcium exchanger, Na+/H+ exchanger, and bicarbonate transporter, which are essential for transporting H+, HCO3- and Ca2+ in and out of the cells (Brini and Carafoli, 2011; Read et al., 2013; Hu et al., 2018). These proteins, crucial for regulation of pH value and calcifying ion concentration at calcification sites, serves as vital indicators of calcification in Halimeda sp (Zhang et al., 2024). However, due to the strong hydrophobicity, embeddability, and aggregation propensity of plasma-membrane proteins, conventional methods, including the one utilized in this study, often encounter challenges in their extraction, separation, and identification (Tao et al., 2023). Additionally, CAs catalyze the reversible conversion of CO2 and water to HCO3- and H+ ions, playing vital roles in both carbon-concentrating mechanism (CCM) for photosynthesis (Reinfelder, 2010), and calcification in calcareous algae (Rahman et al., 2008; Read et al., 2013; Zhang et al., 2024). Genome analysis of H. opuntia reveals an expansion of 17 CA copies and different isoforms are expected to exhibit varying expression patterns to adapt to environment changes (Zhang et al., 2024). In this study, we identified three CAs, with only one showing a significant decrease from 25°C to 31°C (Figure 5A). Physiochemically, the solubility of DIC tends to increase with rising temperature, consequently reducing the ΩCaCO3. Considering the energy consumption of DIC catalysis, the ample supply of HCO3- and CO2 at higher temperatures may alleviate the need for up-regulated CA expression. Furthermore, subcellular localization models predict that the one CA with significant variations localizes within the chloroplast (Supplementary Table S3). Given that H. macroloba calcifies extracellularly, this CA is more likely to play a crucial role in CCM rather than calcification under rising temperatures.
Photosynthesis in calcareous algae plays a vital role in regulating the regional calcifying fluid chemistry. The relationships between photosynthesis and calcification have long been explored owing to their close yet delicate linkage. Typically, photosynthesis elevates calcifying fluid pH by CO2 uptake to stimulate CaCO3 deposition, and calcification consumes HCO3- and ATP while releasing CO2 for photosynthesis (De Beer and Larkum, 2001; Buapet and Sinutok, 2021). The calcifying fluid pH and DIC can be measured through techniques such as microelectrodes, pH-sensitive dyes, and boron isotopic systematics. For instance, two coralline algae exhibited higher pH values at the site of calcification compared to seawater, as indicated by δ11B as a proxy (Cornwall et al., 2017). Using microsensors, the thallus surface pH of H. discoidea gradually increased with photosynthesis activity, with calcium dynamics closely following pH dynamics (De Beer and Larkum, 2001). These studies highlight the integral relationship between photosynthesis and calcification in calcareous algae. While previous studies have documented the negative effects of rising temperatures on calcification (Comeau et al., 2016; Cornwall et al., 2019), our findings suggest that H. macroloba employs a mechanism to mitigates temperature stress on calcification by maintaining efficient photosynthetic processes. This is reflected in the stable Fv/Fm values and the elevated expression of proteins related to LHC, photosystem I, and cytochrome b6-f complex from 25°C to 31°C (Figures 2A, 5A).
Based on chemical catalytic reactions, calcareous algae contribute to the calcification process by fixing HCO3- and releasing CO2, acting as a “carbon source”. However, over time, they convert inorganic carbon into CaCO3, resulting in the formation of calcite that is preserved on the seafloor. Conducting quantitative analysis of mineral density and properties formed by the calcifying organisms under varying temperature gradients is highly significant. Our study found that the tissue calcium content of H. macroloba peaked at 28°C and decreased at 31°C (Figure 2C). The tissue calcium content of H. cylindracea, H. opuntia, and H. lacunalis, three other species found in the SCS, also peaked at 28°C and largely decreased above 32°C (Wei et al., 2020). Given the mean summer water temperature in Sanya Bay is recorded as 28.72°C (Geng et al., 2023), these findings suggest a reduced carbonate deposition in Halimeda during certain summer months in the SCS, with potential exacerbation in a warming ocean. Additionally, most coral reefs inhabit in tropical regions between 22 °S and 22 °N, experiencing average maximum temperatures of approximately 30°C (Kleypas et al., 1999). For instance, the calcification and growth of some corals decline above 28°C, and the optimal temperature for Symbiodiniaceae algae is around 26°C (Castillo et al., 2014; Dilernia et al., 2023). Whereas reef-associated algae like Halimeda sp. and Turbinaria ornata have higher thermal limits than certain coral species (Campbell et al., 2016; Anton et al., 2020; Wei et al., 2020). This higher thermal tolerance of macroalgae like genus Halimeda implies that they may achieve a dominant status in some reefs as the ocean temperatures continue to rise.
5 Conclusion
This study provides valuable insights into the physiological mechanisms employed by the calcareous macroalga H. macroloba to sustain growth across a temperature gradient. We observed that H. macroloba exhibited increasing Chl a and tissue calcium contents, as well as higher Fv/Fm values from 25°C to 28°C. Along this gradient, key pathways related to photosynthesis-antenna proteins and photosynthesis were significantly enriched. Notably, at the lower temperature of 25°C, H. macroloba accumulated substantial levels of selenocysteine and selenomethionine, with corresponding upregulation of Se-containing proteins such as glutathione peroxidase and peroxiredoxin. This suggests that selenium plays a crucial role in supporting growth and acclimating to cooler temperatures. In contrast, thalli exposed to moderate heat stress at 31°C exhibited a decrease in both chlorophyll a content and Fv/Fm values, reflecting the onset of thermal stress. However, H. macroloba increased the expression of proteins involved in light-harvesting, photosystems, and carbon fixation, while redirecting carbon into sugars and fatty acids to maintain metabolic activity. Accumulation of sugars, such as mannitol and D-mannose, and fatty acids, including eicosadienoic acid and adrenic acid, at 31°C suggests active mechanisms to mitigate heat stress. Additionally, elevated nitrogen reserves, indicated by increased nitrogen assimilation proteins and amino acid levels, further support the acclimation of cells to higher temperatures. Together, these findings highlight the integrated role of selenium, carbon, nitrogen, and other metabolic pathways in the thermal acclimation of H. macroloba. The combination of proteomics and metabolomics provides a comprehensive understanding of the algal response to temperature stress, offering insights into strategies that may enhance the resilience of reef ecosystems in the face of ocean warming.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
HZ: Writing – original draft, Formal analysis, Investigation. YW: Writing – review & editing. LZ: Methodology, Writing – review & editing. ZW: Data curation, Writing – review & editing. ZZ: Data curation, Writing – review & editing. LL: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2022YFC3102002), the Science and Technology Planning Project of Guangdong Province (2023B1212060047), and the National Natural Science Foundation of China (42076155).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1543591/full#supplementary-material
References
Adams J. M. M., Ross A. B., Anastasakis K., Hodgson E. M., Gallagher J. A., Jones J. M., et al. (2011). Seasonal variation in the chemical composition of the bioenergy feedstock Laminaria digitata for thermochemical conversion. Biores. Technol. 102, 226–234. doi: 10.1016/j.biortech.2010.06.152
Anton A., Randle J. L., Garcia F. C., Rossbach S., Ellis J. I., Weinzierl M., et al. (2020). Differential thermal tolerance between algae and corals may trigger the proliferation of algae in coral reefs. Global Change Biol. 26, 4316–4327. doi: 10.1111/gcb.15141
Borowitzka M. A., Larkum A. W. D. (1987). Calcification in algae: Mechanisms and the role of metabolism. Crit. Rev. Plant Sci. 6, 1–45. doi: 10.1080/07352688709382246
Brini M., Carafoli E. (2011). The plasma membrane Ca²+ ATPase and the plasma membrane sodium calcium exchanger cooperate in the regulation of cell calcium. LID - 10.1101/cshperspect.a004168 [doi] LID - a004168. Cold Spring Harbor Perspect. Biol. 3, a004168. doi: 10.1101/cshperspect.a004168
Buapet P., Sinutok S. (2021). Calcification in three common calcified algae from Phuket, Thailand: Potential relevance on seawater carbonate chemistry and link to photosynthetic process. Plants 10, 2537. doi: 10.3390/plants10112537
Campbell J. E., Fisch J., Langdon C., Paul V. J. (2016). Increased temperature mitigates the effects of ocean acidification in calcified green algae (Halimeda spp.). Coral Reefs 35, 357–368. doi: 10.1007/s00338-015-1377-9
Castillo K., Ries J., Bruno J., Westfield I. (2014). The reef-building coral Siderastrea siderea exhibits parabolic responses to ocean acidification and warming. Proc. Biol. sciences/The R. Soc. 281, 20141856. doi: 10.1098/rspb.2014.1856
Comeau S., Carpenter R. C., Lantz C. A., Edmunds P. J. (2016). Parameterization of the response of calcification to temperature and pCO2 in the coral Acropora pulchra and the alga Lithophyllum kotschyanum. Coral Reefs 35, 929–939. doi: 10.1007/s00338-016-1425-0
Cornwall C. E., Carlot J., Branson O., Courtney T. A., Harvey B. P., Perry C. T., et al. (2023). Crustose coralline algae can contribute more than corals to coral reef carbonate production. Commun. Earth Environ. 4, 105. doi: 10.1038/s43247-023-00766-w
Cornwall C. E., Comeau S., McCulloch M. T. (2017). Coralline algae elevate pH at the site of calcification under ocean acidification. Global Change Biol. 23, 4245–4256. doi: 10.1111/gcb.2017.23.issue-10
Cornwall C. E., Diaz-Pulido G., Comeau S. (2019). Impacts of ocean warming on coralline algal calcification: Meta-analysis, knowledge gaps, and key recommendations for future research. Front. Mar. Sci. 6, 186. doi: 10.3389/fmars.2019.00186
De Beer D., Larkum A. W. D. (2001). Photosynthesis and calcification in the calcifying algae Halimeda discoidea studied with microsensors. Plant Cell Environ. 24, 1209–1217. doi: 10.1046/j.1365-3040.2001.00772.x
Dilernia N. J., Camp E. F., Bartels N., Suggett D. J. (2023). Contrasting the thermal performance of cultured coral endosymbiont photo-physiology. J. Exp. Mar. Biol. Ecol. 561, 151865. doi: 10.1016/j.jembe.2022.151865
Dougan K. E., Deng Z.-L., Wöhlbrand L., Reuse C., Bunk B., Chen Y., et al. (2023). Multi-omics analysis reveals the molecular response to heat stress in a “red tide. dinoflagellate. Genome Biol. 24, 265. doi: 10.1186/s13059-023-03107-4
Endo H., Inomata E., Gao X., Kinoshita J., Sato Y., Agatsuma Y. (2020). Heat stress promotes nitrogen accumulation in meristems via apical blade erosion in a brown macroalga with intercalary growth. Front. Mar. Sci. 7, 575721. doi: 10.3389/fmars.2020.575721
Enochs I. C., Manzello D. P., Donham E. M., Kolodziej G., Okano R., Johnston L., et al. (2015). Shift from coral to macroalgae dominance on a volcanically acidified reef. Nat. Climate Change 5, 1083–1088. doi: 10.1038/nclimate2758
Famà P., Wysor B., Kooistra W. H. C. F., Zuccarello G. C. (2002). Molecular phylogeny of the genus Caulerpa (Caulerpales, Chlorophyta) inferred from chloroplast tufA gene. J. Phycol. 38, 1040–1050. doi: 10.1046/j.1529-8817.2002.t01-1-01237.x
Geng X., Wu C., Yang Z., Zhu J., Tang K., Lin J., et al. (2023). Spatiotemporal variations and impact factors of nutrients in the Sanya Bay, northern South China Sea. Environ. Sci. pollut. Res. 30, 76784–76797. doi: 10.1007/s11356-023-27527-8
Gerard V. A. (1997). The role of nitrogen nutrition in high-temperature tolerance of the kelp, Laminaria saccharina (Chromophyta). J. Phycol. 33, 800–810. doi: 10.1111/j.0022-3646.1997.00800.x
Gobler C. J., Lobanov A. V., Tang Y.-Z., Turanov A. A., Zhang Y., Doblin M., et al. (2013). The central role of selenium in the biochemistry and ecology of the harmful pelagophyte, Aureococcus anophagefferens. ISME J. 7, 1333–1343. doi: 10.1038/ismej.2013.25
Grant B. R., Borowitzka M. A. (1984). The Chloroplasts of giant-celled and coenocytic algae: Biochemistry and structure. Bot. Rev. 50, 267–307. doi: 10.1007/BF02862634
Hillis-Colinvaux L. (1980). “Ecology and taxonomy of Halimeda: primary producer of coral reefs,” in Advances in Marine Biology. Eds. Blaxter J. H. S., Russell F. S., Yonge M. (Academic Press), 1–327. doi: 10.1016/S0065-2881(08)60303-X
Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. doi: 10.1126/science.1152509
Hu M. Y., Yan J. J., Petersen I., Himmerkus N., Bleich M., Stumpp M. (2018). A SLC4 family bicarbonate transporter is critical for intracellular pH regulation and biomineralization in sea urchin embryos. Elife 7, e36600. doi: 10.7554/eLife.36600.019
Hughes T. P., Baird A. H., Bellwood D. R., Card M., Connolly S. R., Folke C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Hyun B., Ju S.-J., Ko A.-R., Choi K.-H., Jung S. W., Jang P.-G., et al. (2016). Thermal effects on the growth and fatty acid composition of four harmful algal bloom species: Possible implications for ichthyotoxicity. Ocean Sci. J. 51, 333–342. doi: 10.1007/s12601-016-0029-5
Jiang L., Lu Y., Zheng L., Li G., Chen L., Zhang M., et al. (2020). The algal selenoproteomes. BMC Genomics 21, 699. doi: 10.1186/s12864-020-07101-z
Kayanne H., Hata H., Kudo S., Yamano H., Watanabe A., Ikeda Y., et al. (2005). Seasonal and bleaching-induced changes in coral reef metabolism and CO2 flux. Global Biogeochem. Cycles 19, GB3015. doi: 10.1029/2004GB002400
Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kleypas J. A., McManus J. W., MeÑEz L. A. B. (1999). Environmental limits to coral reef development: Where do we draw the line? Am. Zool. 39, 146–159. doi: 10.1093/icb/39.1.146
Knoll A. H., Swett K., Mark J. (1991). Paleobiology of a Neoproterozoic tidal flat/lagoonal complex: the Draken Conglomerate Formation, Spitsbergen. J. Paleontol. 65, 531–570. doi: 10.1017/S0022336000030663
Kojima R., Hanyuda T., Kawai H. (2015). Taxonomic re-examination of Japanese alimeda species using genetic markers, and proposal of a new species alimeda ryukyuensis (Bryopsidales, Chlorophyta). Phycol. Res. 63, 178–188. doi: 10.1111/pre.2015.63.issue-3
Kooistra W. H. C. F., Coppejans E. G. G., Payri C. (2002). Molecular systematics, historical ecology, and phylogeography of Halimeda (Bryopsidales). Mol. Phylogenet. Evol. 24, 121–138. doi: 10.1016/S1055-7903(02)00221-X
Leblond J., Anderson B., Kofink D., Logares R., Rengefors K., Kremp A. (2006). Fatty acid and sterol composition of two evolutionary closely related dinoflagellate morphospecies from cold Scandinavian brackish and freshwaters. Eur. J. Phycol. 41, 303–311. doi: 10.1080/09670260600804843
Liang Y., Koester J. A., Liefer J. D., Irwin A. J., Finkel Z. V. (2019). Molecular mechanisms of temperature acclimation and adaptation in marine diatoms. ISME J. 13, 2415–2425. doi: 10.1038/s41396-019-0441-9
Lowenstam H. A. (1981). Minerals formed by organisms. Science 211, 1126. doi: 10.1126/science.7008198
Millero F. J., Graham T. B., Huang F., Bustos-Serrano H., Pierrot D. (2006). Dissociation constants of carbonic acid in seawater as a function of salinity and temperature. Mar. Chem. 100, 80–94. doi: 10.1016/j.marchem.2005.12.001
Mittler R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Neustupa J., Nemcova Y. (2022). Geometric morphometrics shows a close relationship between the shape features, position on thalli, and CaCO3 content of segments in Halimeda tuna (Bryopsidales, Ulvophyceae). Hydrobiologia 849, 2581–2594. doi: 10.1007/s10750-022-04876-y
Oliver E. C. J., Donat M. G., Burrows M. T., Moore P. J., Smale D. A., Alexander L. V., et al. (2018). Longer and more frequent marine heatwaves over the past century. Nat. Commun. 9, 1324. doi: 10.1038/s41467-018-03732-9
Patel A., Mulder D. W., Söll D., Krahn N. (2022). Harnessing selenocysteine to enhance microbial cell factories for hydrogen production. Front. Catal. 2, 1089176. doi: 10.3389/fctls.2022.1089176
Pentecost A. (1991). “Calcification processes in algae and cyanobacteria,” in Calcareous Algae and Stromatolites. Ed. Riding R. (Springer Berlin Heidelberg, Berlin, Heidelberg), 3–20.
Ponti M., Linares C., Cerrano C., Rodolfo-Metalpa R., Hoeksema B. W. (2021). Editorial: Biogenic reefs at risk: facing globally widespread local threats and their interaction with climate change. Front. Mar. Sci. 8, 793038. doi: 10.3389/fmars.2021.793038
Rahman M. A., Oomori T., Uehara T. (2008). Carbonic anhydrase in calcified endoskeleton: novel activity in biocalcification in alcyonarian. Mar. Biotechnol. 10, 31–38. doi: 10.1007/s10126-007-9030-4
Ras M., Steyer J. P., Bernard O. (2013). Temperature effect on microalgae: A crucial factor for outdoor production. Rev. Environ. Sci. Bio/Technol. 12, 153–164. doi: 10.1007/s11157-013-9310-6
Read B. A., Kegel J., Klute M. J., Kuo A., Lefebvre S. C., Maumus F., et al. (2013). Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213. doi: 10.1038/nature12221
Reinfelder J. R. (2010). Carbon concentrating mechanisms in eukaryotic marine phytoplankton. Annu. Rev. Mar. Sci. 3, 291–315. doi: 10.1146/annurev-marine-120709-142720
Smith C. A., Want E. J., O’Maille G., Abagyan R., Siuzdak G. (2006). XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787. doi: 10.1021/ac051437y
Sully S., Burkepile D. E., Donovan M. K., Hodgson G., van Woesik R. (2019). A global analysis of coral bleaching over the past two decades. Nat. Commun. 10, 1264. doi: 10.1038/s41467-019-09238-2
Sun D., Li F., Jing Z., Hu S., Zhang B. (2023). Frequent marine heatwaves hidden below the surface of the global ocean. Nat. Geosci. 16, 1099–1104. doi: 10.1038/s41561-023-01325-w
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tao X., Zhao C., MacKinnon R. (2023). Membrane protein isolation and structure determination in cell-derived membrane vesicles. Proc. Natl. Acad. Sci. 120, e2302325120. doi: 10.1073/pnas.2302325120
Tebbett S. B., Connolly S. R., Bellwood D. R. (2023). Benthic composition changes on coral reefs at global scales. Nat. Ecol. Evol. 7, 71–81. doi: 10.1038/s41559-022-01937-2
Thévenot E. A., Roux A., Xu Y., Ezan E., Junot C. (2015). Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 14, 3322–3335. doi: 10.1021/acs.jproteome.5b00354
Tonon T., Li Y., McQueen-Mason S. (2017). Mannitol biosynthesis in algae: more widespread and diverse than previously thought. New Phytol. 213, 1573–1579. doi: 10.1111/nph.2017.213.issue-4
Toseland A., Daines S. J., Clark J. R., Kirkham A., Strauss J., Uhlig C., et al. (2013). The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Climate Change 3, 979–984. doi: 10.1038/nclimate1989
Uemura M., Tominaga Y., Nakagawara C., Shigematsu S., Minami A., Kawamura Y. (2006). Responses of the plasma membrane to low temperatures. Physiol. Plant. 126, 81–89. doi: 10.1111/j.1399-3054.2005.00594.x
Vásquez-Elizondo R. M., Enríquez S. (2016). Coralline algal physiology is more adversely affected by elevated temperature than reduced pH. Sci. Rep. 6, 19030. doi: 10.1038/srep19030
Verbruggen H., Clerck O. D., Schils T., Kooistra W. H. C. F., Coppejans E. (2005). Evolution and phylogeography of Halimeda section Halimeda (Bryopsidales, Chlorophyta). Mol. Phylogenet. Evol. 37, 789–803. doi: 10.1016/j.ympev.2005.06.015
Wang S., Zhang Y., Zhang J., Wang S., Tan Z.-C., Shi Q. (2018). Heat capacities and thermodynamic functions of d-ribose and d-mannose. J. Thermal Anal. Calorim. 133, 1049–1059. doi: 10.1007/s10973-018-7128-8
Wang D. Z., Zhang Y. J., Zhang S. F., Zhang S. F., Lin L., Hong H. S. (2013). Quantitative proteomic analysis of cell cycle of the dinoflagellate Prorocentrum donghaiense (Dinophyceae). PloS One 8, e63659. doi: 10.1371/journal.pone.0063659
Wei Z., Mo J., Huang R., Hu Q., Long C., Ding D., et al. (2020). Physiological performance of three calcifying green macroalgae Halimeda species in response to altered seawater temperatures. Acta Oceanologica Sinica, 39 (2), 89–100. doi: 10.1007/s13131-019-1471-3
Wellburn A. R. (1994). The spectral determination of Chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Willette S., Gill S. S., Dungan B., Schaub T. M., Jarvis J. M., St. Hilaire R., et al. (2018). Alterations in lipidome and metabolome profiles of Nannochloropsis salina in response to reduced culture temperature during sinusoidal temperature and light. Algal Res. 32, 79–92. doi: 10.1016/j.algal.2018.03.001
Wizemann A., Meyer F. W., Westphal H. (2014). A new model for the calcification of the green macro-alga Halimeda opuntia (Lamouroux). Coral Reefs 33, 951–964. doi: 10.1007/s00338-014-1183-9
Xu H., Zhu Y., Eberli G. P., Luo W., Zhao X., Cai Y., et al. (2015). Characteristics of porosity and permeability layer of fossil Halimeda reef mineral rock of Miocene in the Xisha Islands and its genetic model. Acta Oceanol. Sin. 34, 74–83. doi: 10.1007/s13131-015-0638-3
Yi L., Deng C., Yan W., Wu H., Zhang C., Xu W., et al. (2021). Neogene–quaternary magnetostratigraphy of the biogenic reef sequence of core NK–1 in Nansha Qundao, South China Sea. Sci. Bull. 66, 20–203. doi: 10.1016/j.scib.2020.08.014
Zhang H., Gu B., Zhou Y., Ma X., Liu T., Xu H., et al. (2022). Multi-Omics profiling reveals resource allocation and acclimation strategies to temperature changes in a marine dinoflagellate. Appl. Environ. Microbiol. 88, e01213–e01222. doi: 10.1128/aem.01213-22
Zhang H., Wang X., Qu M., Yu H., Yin J., Liu X., et al. (2024). Genome of Halimeda opuntia reveals differentiation of subgenomes and molecular bases of multinucleation and calcification in algae. Proc. Natl. Acad. Sci. United States America 24, e2403222121. doi: 10.1073/pnas.2403222121
Keywords: temperature, macroalga, Halimeda, proteomics, metabolomics, calcification
Citation: Zhang H, Wang Y, Zhang L, Wang Z, Zhu Z and Long L (2025) Protein and metabolite acclimations to temperature variability in a calcareous green macroalga Halimeda macroloba. Front. Mar. Sci. 12:1543591. doi: 10.3389/fmars.2025.1543591
Received: 11 December 2024; Accepted: 13 January 2025;
Published: 12 February 2025.
Edited by:
Yongyu Zhang, Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Yida Gao, Florida Fish and Wildlife Research Institute, United StatesLingjie Zhou, University of California, San Diego, United States
Copyright © 2025 Zhang, Wang, Zhang, Wang, Zhu and Long. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijuan Long, bGlqdWFubG9uZ0BzY3Npby5hYy5jbg==
 Hao Zhang
Hao Zhang Yuyu Wang2
Yuyu Wang2