- 1Marine Science Center, Northeastern University, Nahant, MA, United States
- 2Department of Marine and Environmental Sciences, Northeastern University, Boston, MA, United States
Diseases have caused unprecedent mortality in Caribbean coral communities. White band disease (WBD) has killed up to 95% of all endangered Caribbean Acroporids since it was first observed in 1979. Despite the devastating impacts of WBD, its etiology is currently unknown although recent research identified two bacterial strains – ASVs classified as a Cysteiniphilum litorale and a Vibrio sp., as the most likely pathogens. To better understand the disease etiology of WBD, we pretreated corals with antibiotics to determine how prophylactic use of antibiotics impacts the transmission of WBD in a replicated tank-based experiment. We found the prophylactic use of antibiotics led to significantly reduced infection rates in disease exposed corals with a 30-percentage point decrease in the infection rate. Analyses of 16S rRNA amplicon gene sequencing data in the disease exposed corals demonstrated that antibiotic pretreatment resulted in coral microbiomes which were less speciose and contained relatively fewer Vibrio spp. than untreated corals, indicating that the benefit of the antibiotic pretreatment was its ability to reduce the relative abundance of intrinsic secondary opportunists and/or opportunistic pathogens suggesting their likely importance to the etiology of WBD. We propose two distinct etiologies involving either an extrinsic keystone pathogen (Cysteiniphilum litorale) or overgrowth of intrinsic opportunistic pathogens (Vibrio spp.). Future research should isolate these strains to confirm the etiology of white band disease.
Introduction
Diseases have caused unprecedent mortality in Caribbean coral communities with white band disease (WBD) killing up to 95% of the formerly dominant Caribbean Acropora spp. since 1979 (Gladfelter, 1982; Aronson and Precht, 2001) and now stony coral tissue loss disease (SCTLD) causing high mortality in over 20 coral species (Precht et al., 2016; Alvarez-Filip et al., 2022). White band disease, sometimes referred to generally as rapid tissue loss (RTL; Williams and Miller, 2005; Miller et al., 2014), develops as an advancing disease interface of dead or dying tissue that progresses distally toward the apical branch tip (Gladfelter, 1982; Ritchie and Smith, 1998). Two forms of the disease have been described based on the presence (Type 2; Ritchie and Smith, 1998) or absence (Type 1; Gladfelter, 1982) of a band of bleached tissue at the margin of the disease lesion. WBD is transmitted via direct contact, snail vectors, and through the water column (Gignoux-Wolfsohn et al., 2012) with infection rates increasing in warmer temperatures, exacerbating the effects of both WBD and increased temperature (Randall and van Woesik, 2015; Gignoux-Wolfsohn et al., 2020; Selwyn et al., 2024).
The prophylactic and therapeutic use of broad-spectrum antibiotics has emerged as a promising method to combat the in situ spread of coral diseases (Sheridan et al., 2013; Neely et al., 2021) and as an experimental tool to manipulate the transmission of coral bacterial pathogens (Kline and Vollmer, 2011; Sweet et al., 2014). Experimental application of antibiotics was used to demonstrate that white band disease is caused by a bacterial pathogen where antibiotics arrest disease transmission (Kline and Vollmer, 2011) and disease progression (Sweet et al., 2014). The efficacy of antibiotics in treating coral diseases has also been demonstrated for white syndrome in Indo-Pacific Acroporid corals and for SCTLD in multiple Caribbean corals (Sweet and Bythell, 2015; Aeby et al., 2019; Neely et al., 2021).
While the specific causal agent and disease etiology are unknown, previous work has implicated several potential pathogens including several Vibrio spp (Ritchie and Smith, 1998; Gil-Agudelo et al., 2006; Rosales et al., 2019; Selwyn et al., 2024), Sphingobium yanoikuyae (Rosales et al., 2019), and Cysteiniphilum litorale (Gignoux-Wolfsohn et al., 2017; Selwyn et al., 2024). Early bacterial culturing identified a strong association of Vibrio charcharia (now synonymized with V. harveyi) on WBD infected Acropora cervicornis (Ritchie and Smith, 1998) and in situ grafting of uncharacterized Vibrio cultures elicited WBD disease signs (Gil-Agudelo et al., 2006). Vibrio spp. are well known opportunistic pathogens in corals (Bourne et al., 2009; Munn, 2015). Rosales et al. (2019) concluded that Sphingobium yanoikuyae (family Sphingomonadacea) was the most likely WBD pathogen using in situ transmission assays to A. cervicornis and A. palmata in Florida, and yet Sphingobium spp. are rarely pathogenic (Glaeser and Kämpfer, 2014; however see: Miyamatsu et al., 2024). Most recently, our multiyear analysis of 269 healthy and 143 WBD infected A. cervicornis from Panama identified a Vibrio sp. strain and Cysteiniphilum litorale strain as the two top candidate pathogens (Selwyn et al., 2024). Cysteiniphilum is a new genus that was previously described as a Francisella (Liu et al., 2017; Qian et al., 2023) and can cause skin infections in humans (Xu et al., 2021). Cysteiniphilum spp. possess a partial copy of the Francisella pathogenicity island (Qian et al., 2023) which makes Francisella spp. particularly virulent (Nano and Schmerk, 2007; Cowley and Elkins, 2011). Parasitic infection by the alpha-proteobacterium “Candidatus Aquarickettsia rohweri” has also been associated with increased WBD susceptibility in A. cervicornis (Casas et al., 2004; Klinges et al., 2020).
In this study, we conducted a replicated tank-based disease transmission experiment where we first pre-treated Acropora cervicornis fragments with a broad-spectrum antibiotic cocktail and then exposed the coral fragments to diseased or healthy tissue slurries. Survivorship analyses were used to examine the effect of antibiotic pretreatment on disease transmission. 16S rRNA amplicon gene sequencing was used to profile the change in bacterial communities after antibiotic treatment and after exposure to disease. Antibiotic pretreatment allowed us to test whether intrinsic bacterial pathogens and/or opportunists generally living commensally on healthy corals contributed to WBD transmission. The comparison of corals exposed to disease which develop symptoms or remain healthy allowed the identification of top bacterial ASVs associated with WBD which could be classified as primary pathogens or secondary opportunistic pathogens based on their response to the antibiotic pretreatment.
Methods
Sampling and experimental design
We conducted a tank-based transmission experiment to compare the effects of antibiotic pre-treatment (yes or no) and the subsequent exposure to white band disease slurries (diseased vs. healthy doses) in a two-factor experiment (antibiotic x disease exposure) with five tank replicates per level and 20 replicate fragments from each of ten healthy A. cervicornis genotypes (Figure 1). We examined the effects of antibiotic pretreatment and disease exposure on the infection rate of the coral fragments and changes in their associated microbiomes using 16S rRNA amplicon gene sequencing. Twenty replicate fragments from ten healthy coral genotypes were collected from Sebastian’s reef (9° 15’ 16.4” N, 82° 7’ 37.8” W), Bocas del Toro in July 2017 and one fragment of each genotype was randomly assigned to each of the 20 18-liter recirculating tanks filled with UV sterilized seawater, which were held at ambient seawater temperatures in the flow-through seawater system. After six hours of acclimation, ten tanks were treated twice with 100mg/l each of Kanamycin, Ampicillin, Chloramphenicol, and Tetracycline, with 24 hours between treatments. Both Ampicillin and Tetracycline have previously been shown to inhibit WBD transmission when used separately through the inhibition of cell wall synthesis and protein synthesis respectively (Kline and Vollmer, 2011). Due to the light sensitivity of Tetracycline it was supplemented with two additional antibiotics which act to inhibit protein synthesis, Kanamycin and Chloramphenicol. Two antibiotic doses were used to ensure the treatment was effective and not affected by the light sensitivity of Tetracycline. In the morning after the second antibiotic dose, the seawater was replaced with new UV sterilized seawater, a post-antibiotic, pre-exposure sample of each fragment was taken (day 0, see below for tissue sampling methods), and a Waterpik with 0.2 um filtered seawater (FSW) was used to create small (ca. 0.25 cm2) experimental lesions in the coral tissue to facilitate transmission (Gignoux-Wolfsohn et al., 2012). Five of the antibiotic treated tanks and five untreated tanks were designated as disease exposure with the remaining tanks designated as healthy exposure. The disease exposure tanks were dosed with 50ml of disease slurry produced from 22 WBD infected coral fragments while healthy exposed tanks were dosed with 50ml of healthy slurry created from 22 healthy fragments. Healthy and diseased coral fragments were collected from Sebastian’s reef 1 hour prior to dosing the tanks. Tissue slurries were produced by liberating diseased or healthy coral tissue from the skeleton of sampled corals using a Waterpik containing filtered seawater (FSW), normalizing the slurry doses to a standard ocular density of 0.4 at 600nm with FSW, and then dosing each tank with 50ml of slurry using sterile centrifuge tubes.

Figure 1. Schematic showing the experimental design and timing of the transmission experiment used to assess the efficacy of prophylactic antibiotic treatment on disease transmission and determine differences in the microbial communities of disease exposed coral fragments. Half the samples were pretreated with antibiotics prior to half of each antibiotic pretreatment group being exposed to disease or healthy slurries. Samples were observed for eight days following slurry exposure with six genotypes from each of three disease exposed experimental tanks sampled for 16S analysis before exposure and after exposure (days two and eight, combined into a post-exposure treatment).
Effects of antibiotic pretreatment and disease exposures on coral infection rate
During the experiment, corals were monitored every 12 hours and new disease signs recorded for a total of eight days after slurry exposure. Kaplan-Meier survival curves (Kaplan and Meier, 1958) comparing the four treatment groups - antibiotic pretreated and untreated corals crossed with disease versus healthy exposure – were used to analyze the rate of infection over time across coral fragments and test for differences in infection rates using a log-rank test (Harrington and Fleming, 1982). To determine which combinations of treatment groups differed significantly from each other, we performed post-hoc pairwise log-rank tests, adjusting the p-values to account for the familywise error rate using sequential Bonferroni correction (Holm, 1979).
16S rRNA amplicon gene sequencing
16S rRNA amplicon gene sequencing was obtained by haphazardly sampling two polyps near the lesion site from six genotypes, so as to not sacrifice the whole fragment, in three disease exposed tanks for both antibiotic treated and untreated tanks sets across three time points: post-antibiotic treatment, pre-exposure (day 0, see above), two days post-exposure (day 2), and eight days post-exposure or when WBD symptoms developed, whichever occurred first (day 8). Fragments of the same six genotypes in the same three antibiotic treated and three untreated tanks were repeatedly sampled with repeated measurements statistically accounted for by including fragment nested within genotype and tank as random effects (see below). In all subsequent analyses the two post-exposure samples (day 2 and day 8) are analyzed as a single post-exposure treatment. Post-exposure samples at day 2 and day 8 were combined to improve statistical power while accounting for repeated sampling through the use of random effects to accommodate the sampling design (Hurlbert, 1984; Millar and Anderson, 2004). At each timepoint, polyps were sampled adjacent to the tissue lesion or disease interface using flame sterilized tweezers, the sampled polyps were placed into 150 ul of DNA/RNA shield (Zymo Research) and stored at -20C until extraction. Diseased corals were removed from the tank to prevent disease amplification. Genomic DNA was extracted from each sample using CHAOS extraction buffer (Fukami et al., 2004) and GenElute DNA extraction kits. 16S rRNA amplicon gene sequencing of the V3-V4 region was produced using Klindworth et al.’s (2013) protocol, V3-V4 (341F/785R) primer sets, and four lanes of Illumina MiSeq 2x300 bp sequencing. Reads were quality trimmed, overlapped and assembled into amplicon sequencing variants (ASVs) using the dada2 denoising algorithm and pipeline in R v4.2.1 (Callahan et al., 2016; R Core Team, 2022). Chimeras were removed and taxonomy was assigned to each ASV, first by using a Bayesian taxonomic classifier based on the NCBI 16S microbial database (downloaded on 3 Feb 2024) and classified to the lowest taxonomic level possible with greater than 80% classification confidence (Gao et al., 2017) and then using the Silva SSU r138 database modified for decipher for ASVs not classified by the Bayesian classifier (Quast et al., 2013). ASV sequences were aligned using decipher (Wright, 2016) and a neighbor-joining tree of the aligned ASV data was constructed using phangorn (Schliep, 2011). The resulting ASV table, taxa table and 16S rRNA tree was imported into phyloseq (McMurdie and Holmes, 2013) and merged with the sample metadata for downstream analyses. Samples were pruned to keep only samples with more than 1,000 16S rRNA gene reads and ASVs identified as cyanobacteria, mitochondria, and/or chloroplast sequences were removed as potential host or algal contaminants (Hanshew et al., 2013; Thomas et al., 2020). This removed 150 putative cyanobacteria ASVs, 133 of which would have been filtered due to low abundance with 17 passing the 10% prevalence filter. All 17 of these ASVs had significant BLAST hits against Symbiodiniaceae genomes suggesting they are host symbiont contamination (Supplementary Table S1). Read counts were normalized for variable sequencing depth accounting for the compositional nature of 16S sequencing data using the robust centered log-ratio (rclr) of the number of reads (Gloor et al., 2017; Martino et al., 2019). This normalization method improves upon the additive log-ratio method used in the popular analysis software ANCOM in using the geometric mean of all taxa as the reference rather than one designated reference taxa and also improves the incorporation of 0 data to avoid the use of pseudo-counts which can bias results (Mandal et al., 2015; Kaul et al., 2017; Lin and Peddada, 2020b). Furthermore, the use of this normalization method, allows for planned post-hoc analyses (see below) using the same linear mixed-effects model framework which are not yet possible in the ANCOM or ANCOM-BC analysis software (Mandal et al., 2015; Lin and Peddada, 2020a, 2024).
Microbial community composition
To fully document the alpha diversity, we used a suite of common metrics assessing various aspects of the communities; these included observed richness, Camargo evenness (Camargo, 1992), Shannon Diversity (Shannon, 1948), Gini inequality (Gini, 1921) to assess community dominance, and Faith’s phylogenetic diversity (Faith, 1992). Prior to calculation of the alpha diversity metrics we rarified samples to an equal sequencing depth, with samples below that sequencing depth being removed, 1,000 times and calculated each alpha diversity metric. To determine the sampling depth which balances removing additional samples and having adequate sequencing depth to characterize the community we calculated rarefaction curves and Good’s coverage (Good, 1953; Sanders, 1968). The average of each diversity metric across all bootstrap rarefaction subsamples was modeled using linear mixed effects models using the lme4 package in R v4.2.1 (Bates et al., 2015; R Core Team, 2022).
Each alpha diversity metric was modeled using the same model as the individual ASV models with a fixed treatment effect combining the sampling time (before/after disease exposure), antibiotic treatment, and disease state, resulting in five unique combinations (Table 1). All metrics were also modeled with random effects for tank, genotype, and fragment nested within genotype to represent the experimental design. We used a priori contrasts to distinguish the effects of time, antibiotics, and disease. To test for an effect of time, we compared the average alpha diversity metric of healthy fragments, regardless of antibiotic pretreatment, before and after disease slurry exposure. To test for an effect of antibiotic treatment, we compared the average alpha diversity metric of the healthy fragments before and after disease exposure between antibiotic treated and untreated fragments. Finally, we examined the effect of disease exposure by comparing diseased fragments to the average alpha diversity metric of untreated healthy fragments before and after disease exposure.
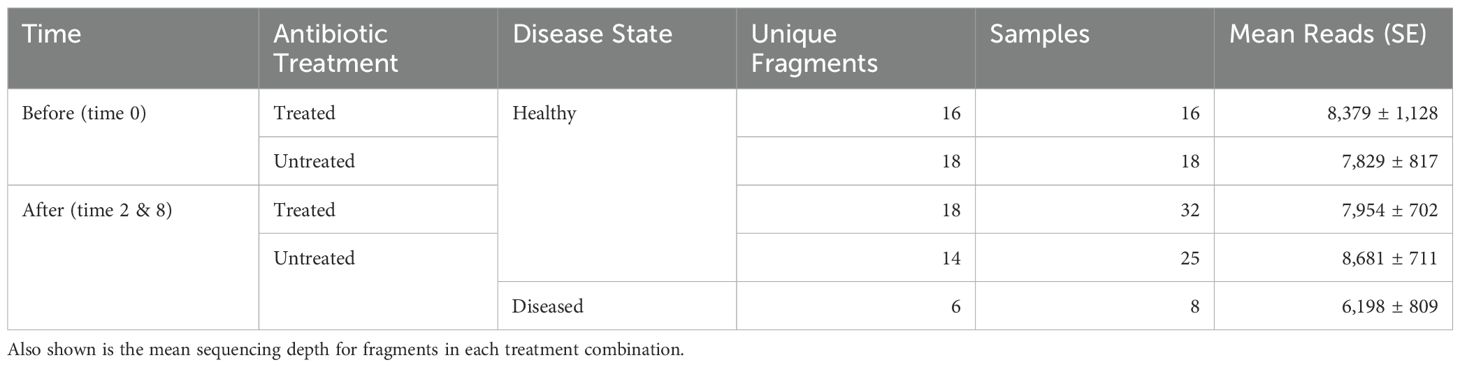
Table 1. Experimental design and sampling summary showing the number of unique coral fragments and total samples which passed quality filtering in each of the five treatment combinations of time, antibiotic treatment, and disease state.
ASVs rarely detected in coral microbiomes, less than 10% of samples, were removed prior to the analysis of beta diversity. Community differences in coral microbiomes were analyzed using distance-based redundancy analysis using the robust Aitchison distance metric, which is scale invariant, obviating the need for rarefaction (Aitchison, 1982; McArdle and Anderson, 2001; Martino et al., 2019). Specifically, we investigated differences in microbial community composition between coral fragments prior to disease homogenate exposure and after exposure, as well as among corals treated with antibiotics and those left untreated and those which develop the disease. Significance was assessed using permutational MANOVAs with 10,000 permutations (McArdle and Anderson, 2001). Homogeneity of dispersions was tested for to distinguish differences in microbial community composition from differences in microbiome variability within treatments (O’Neill and Mathews, 2000; Anderson, 2006). As post-hoc analyses for both the community composition change and homogeneity of dispersion analyses, we tested for differences between healthy corals before and after dosing with the disease homogenate and between antibiotic treated and untreated fragments, excluding in both cases corals which develop disease symptoms. Finally, we looked at community differences between diseased and healthy corals, excluding corals treated with antibiotics. All beta diversity analyses were performed using the R package vegan (Oksanen et al., 2013).
Individual ASV analysis
To analyze differences in individual ASVs that were retained after the low prevalence filter and which drove changes in alpha and beta diversity, abundances were modeled using linear mixed effects models using lme4 (Bates et al., 2015). We modeled ASV abundance as an independent effect, combining the effects of time, antibiotic treatment, and disease outcome for a total of five treatment combinations, as no antibiotic treated samples developed disease signs after disease exposure (Table 1). To control for repeated measurements and potential tank effects, we included random effects of tank and fragment nested within genotype. Main effect p-values were calculated using Satterthwaite’s method of calculating denominator degrees of freedom and adjusted to account for multiple testing (Satterthwaite, 1946; Benjamini and Hochberg, 1995), with planned post-hoc contrasts applied only to those found to have a significant main effect. Given a significant treatment effect (i.e. at least one of the five treatment combinations had significantly different ASV abundance than the others), we used the following a priori contrasts to test for different effects. To test for an effect of time, we compared the average robust centered log ratio (rclr) of the healthy fragments, regardless of antibiotic pretreatment, before and after disease slurry exposure. To test for an effect of antibiotic treatment, we compared the average rclr of the healthy fragments before and after disease exposure between antibiotic treated and untreated fragments. Finally, we examined the effect of disease exposure by comparing diseased fragments to the average rclr of untreated healthy fragments before and after disease exposure. These p-values were adjusted to account for multiple comparisons with the significance and direction (positive vs. negative association) of these three effects being used to categorize ASVs based on how they responded to the experimental procedures.
Results
Effects of antibiotic pretreatment and disease exposures on coral infection rate
Infection rates in our experimental transmission were impacted by the combination of antibiotic pretreatments and disease exposure (χ2(3) = 236.7, p < 0.0001, Figure 2). Corals that were exposed to disease without first being treated with antibiotics had significantly higher infection rates than any other treatment combinations (all p < 0.0001), with 37.2% (± 3.3% SE) of coral fragments becoming infected with WBD by day seven. In contrast, corals that received antibiotics prior to disease exposure had significantly higher non-infection rates with 93.1% (± 2.3 SE %) of the antibiotic treated, disease exposed corals remaining uninfected until the end of the experiment. These high non-infection rates were comparable to untreated fragments exposed to the healthy slurry (92.4% ± 2.5% SE, χ2(1) = 0.61, p = 0.44). Antibiotic pretreatment even conferred an advantage to corals dosed with healthy slurries, as no corals in the antibiotic treated, healthy exposed treatment developed WBD infections after seven days.
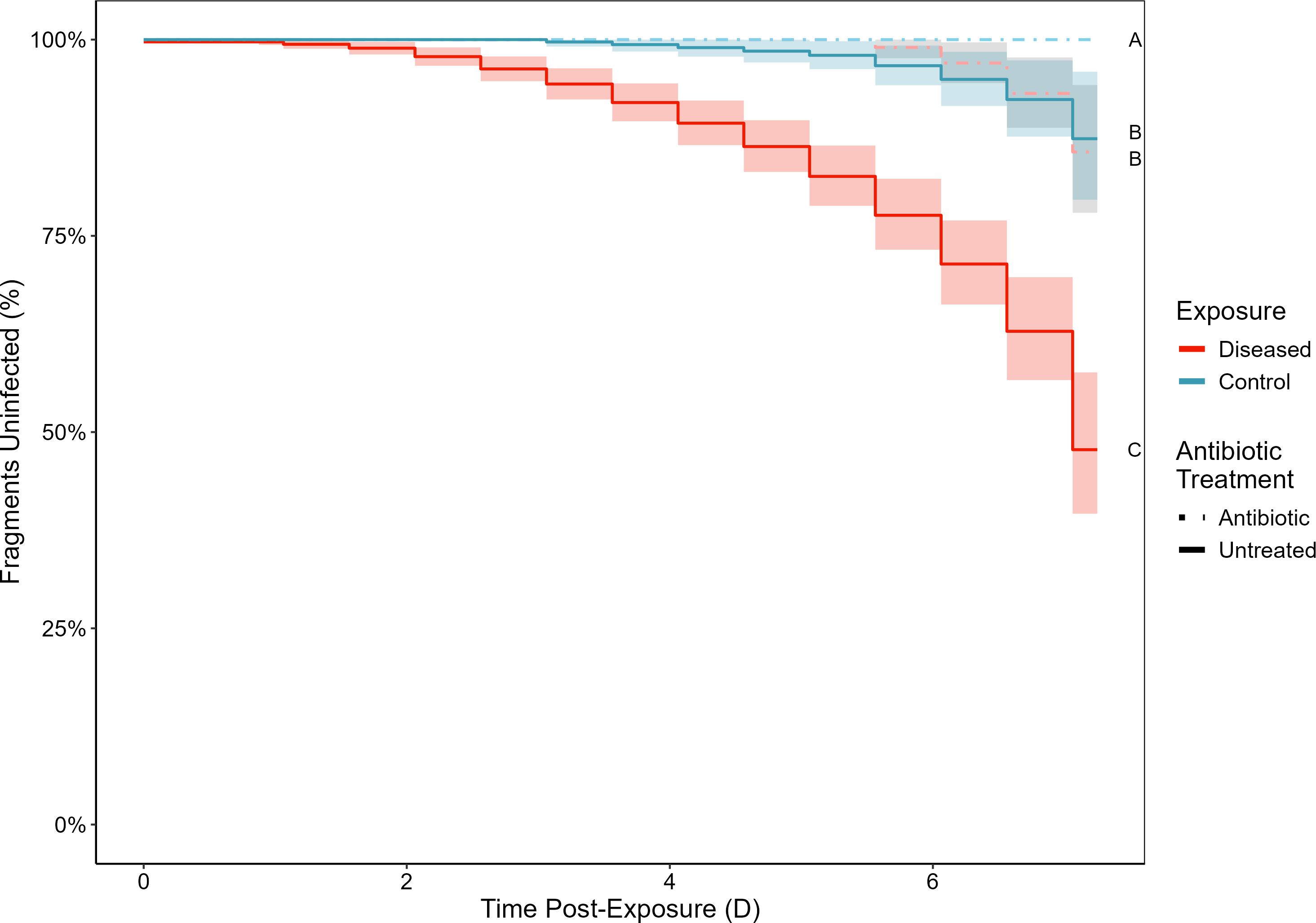
Figure 2. Kaplan-Meier survival curves showing the percent of corals without signs of white band disease after being treated with antibiotics (solid lines) or left untreated (dashed lines) which were subsequently either exposed to an experimental disease dose (red) or a healthy control dose (blue). Letters indicate groupings with non-significantly different infection rates at the end of the experiment. Bands indicate 95% confidence intervals.
Microbial community composition
16S rRNA gene sequencing allowed us to examine how antibiotic pretreatment and subsequent exposure to disease changed coral microbiomes over time. 16S rRNA gene sequencing data was obtained for 36 replicate coral fragments from six coral genotypes from which 18 samples were treated with antibiotics and 18 left untreated all of which were exposed to disease slurries. Over the course of the experiment six of the untreated fragments developed WBD. Two polyps from each fragment were sampled prior to disease exposure, on day two post-exposure, and either on day eight post-exposure or when WBD symptoms first developed, whichever came first. A total of 106 samples were taken, less than the planned 108 samples as two fragments developed WBD symptoms and were removed following the day 2 sampling. In an additional two samples the PCR failed to amplify leaving 104 samples which were sequenced. After quality control filtering to remove samples with fewer than 1,000 reads, we were left with 99 samples for the analysis. The 16S rRNA gene dataset contained 4,705 unique bacterial ASVs across 427 genera from 42 classes and 206 families (Figure 3). Removal of low prevalence ASVs (< 10%) left 1,182 ASVs from 22 classes and 94 families. The average number of reads of the removed ASVs was 0.125 ± 0.009 SE), 54 times less than the average number of reads of the retained ASVs (6.8 ± 0.25 SE, t(117,333) = 26.9, p < 0.001). Samples were collected from and analyzed as five unique treatment combinations of before/after disease dosage either with or without antibiotic pretreatment, with the fifth treatment combination being those corals after disease exposure that were not pretreated with antibiotics which became infected with WBD.
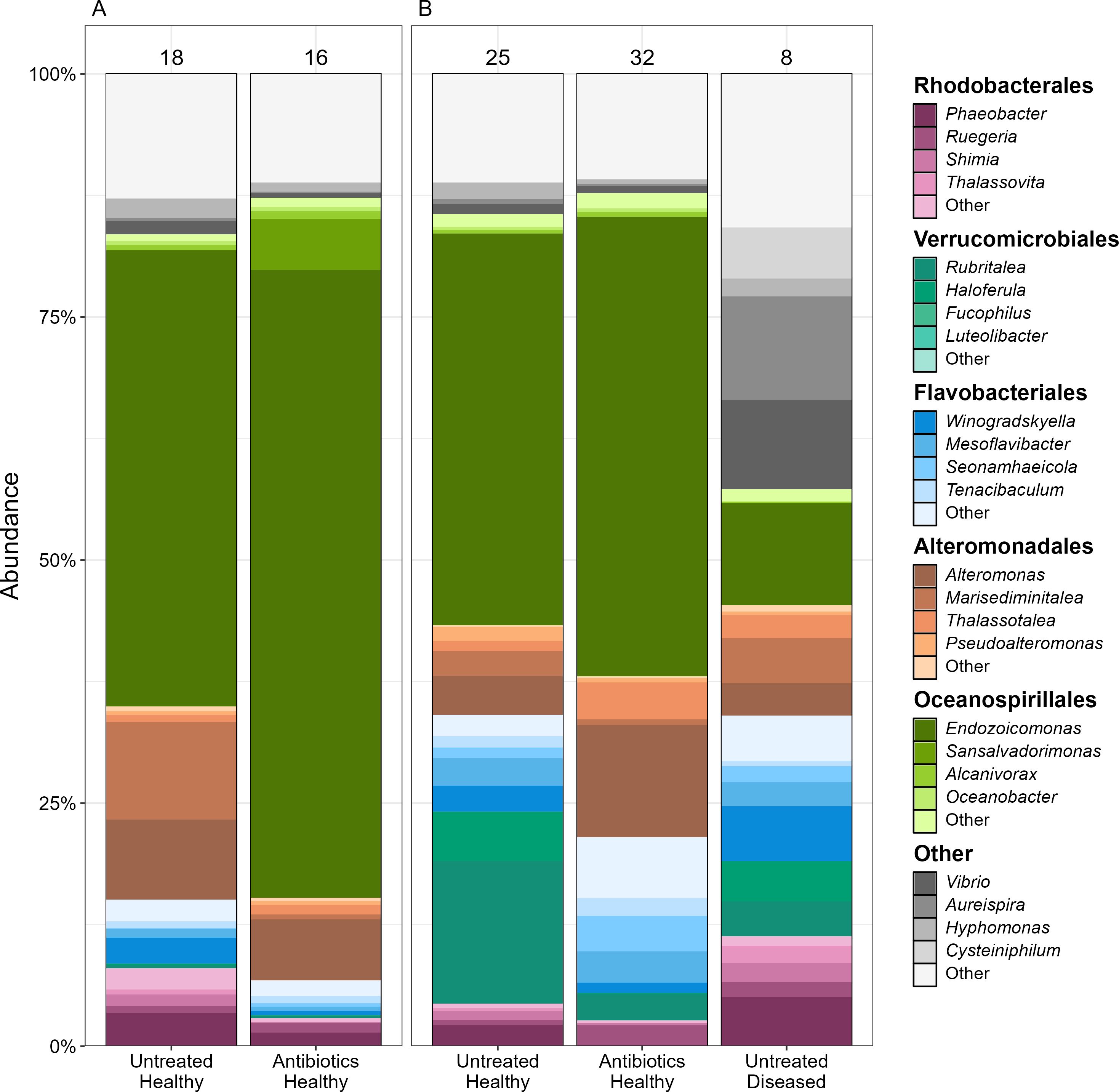
Figure 3. Microbial community compositions of coral fragments (A) before, and (B) after exposure to a disease slurry. Corals were either treated with antibiotics prior to exposure and then separated into those which developed signs of white band disease and those which remained apparently healthy. Colors indicate the major microbial families present in the coral microbiomes with different shades showing the dominant genera in each family. Numbers above each bar show the total number of samples represented by the bar.
The lowest Good’s coverage value for all 99 samples was 95.9%, indicating that in all samples at most 4.1% of the reads are ASVs which appear only once in the sample (Supplementary Figure S1). This along with the sample rarefaction curves shows that there are diminishing returns of increased sequencing depth leading to the observation of new ASVs (Supplementary Figure S2). Given that these ASVs are definitionally rare and that filtering to improve the minimum Good’s coverage requires the removal of 20 additional samples with lower sequencing depth we decided to filter to the sequencing depth of the least sequenced sample (1,056 reads) to prioritize the breadth of the samples at the slight expense of sampling depth.
Antibiotic treatment led to changes in the coral microbiome ASV richness (F(4, 91.1) = 2.74, p = 0.03), and dominance (F(4, 22.6) = 2.53, p = 0.069), but not diversity (F(4, 12.2) = 1.74, p = 0.21), evenness (F(4, 9.75) = 1.45, p = 0.29) or phylogenetic diversity (F(4, 25.5) = 1.60, p = 0.20, Figure 4, Table 2). Microbiomes of untreated corals had 30.5 more ASVs (± 9.5 SE; z = 3.21, p = 0.023) than antibiotic treated corals. As a result, antibiotic treated samples were more dominated by a few ASVs (z = 2.91, p = 0.034).
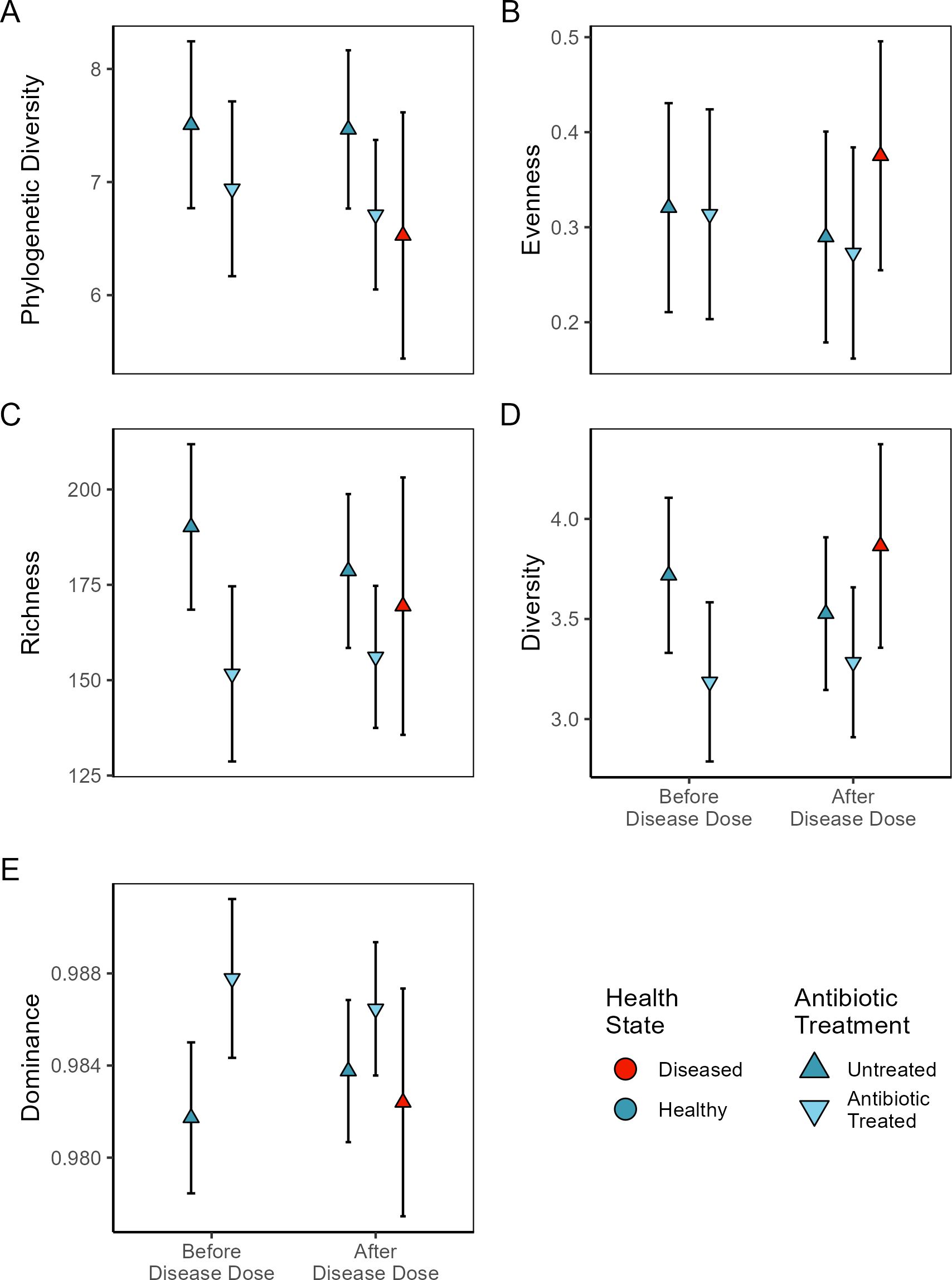
Figure 4. Plots of ASV level alpha diversity metrics (with 95% confidence intervals) before and after exposure to disease homogenate slurries. Some coral fragments were previously treated with antibiotics (inverted triangle) while others were left untreated (triangle). Those corals which develop disease symptoms are marked in red while healthy corals are blue. (A) Faith’s phylogenetic diversity (Faith, 1992), (B) Camargo Evenness (Camargo, 1992), (C) Observed ASV richness, (D) Shannon Diversity (Shannon, 1948), (E) GINI inequality (Gini, 1921).
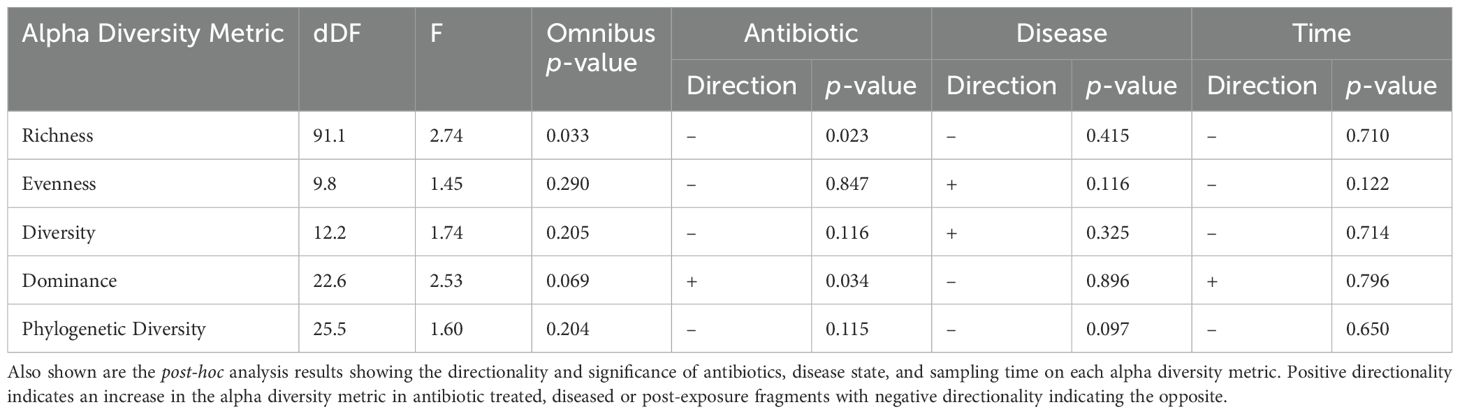
Table 2. Alpha diversity test summary showing the Satterthwaite estimated denominator degrees of freedom (dDF) and F-statistic given four numerator degrees of freedom for each alpha diversity metric including the omnibus p-value.
Corals in different treatment combinations had significantly different microbial community compositions (r2 = 0.10, F(4, 94) = 2.67, p < 0.0001, Figure 5) with some treatment combinations having significant within treatment variation among coral fragments (F(4, 94) = 2.54, p = 0.045). Antibiotic treatment resulted in coral fragments with different microbial communities than untreated fragments (r2 = 0.039, F(1, 89) = 3.65, p < 0.0001), likely attributable to significant within treatment variation in microbial communities among antibiotic treated coral fragments (F(1, 89) = 5.47, p = 0.022), suggesting that the effect of antibiotic treatment on ASV richness is primarily through the removal of low abundance/rare ASVs. Diseased coral fragments were associated with different microbial communities than healthy fragments (r2 = 0.037, F(1, 49) = 1.91, p = 0.006) without significant within treatment variation in community composition (F(1, 49) = 0.007, p = 0.93) indicating a shift from a healthy to diseased microbial composition (Figure 3). Finally, there is evidence that microbial communities change in composition (r2 = 0.026, F(1, 89) = 2.37, p = 0.0005) without significant variation within treatments (F(1, 89) = 2.54, p = 0.11) after exposure to coral slurries regardless of the slurry type.
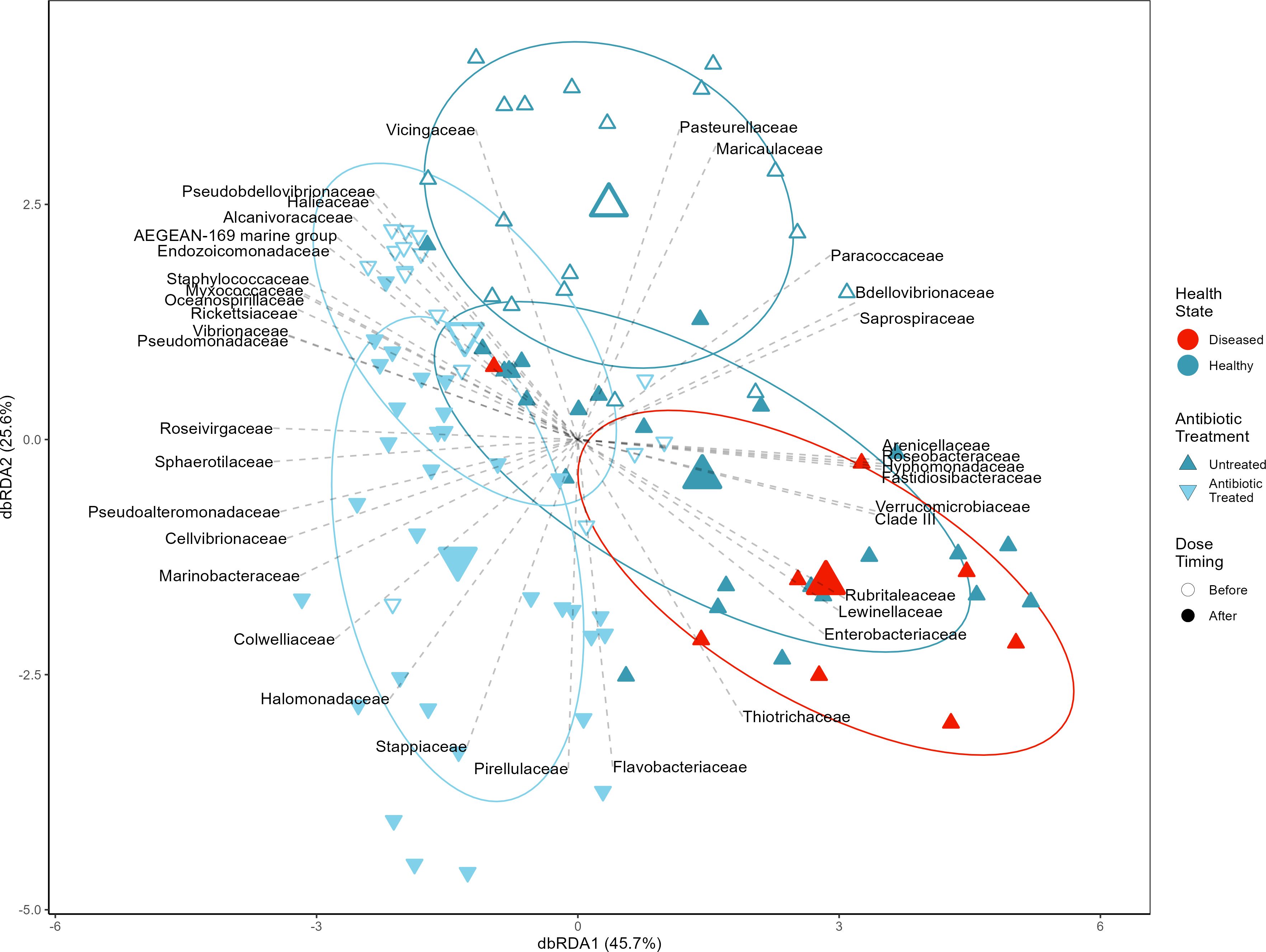
Figure 5. Robust Aitchison distance-based redundancy analysis shows microbial community compositional changes between antibiotic (upside-down triangles) treated and untreated (triangles) coral fragments before (open) and after (closed) exposure to disease homogenate slurries, including distinguishing those which developed disease symptoms (red). Ellipses represent one standard deviation from the group centroid. Lines show the direction of maximum correlation between microbial families and ordinated coral fragments filtered to only show families significantly correlated with the ordination (pFDR < 0.05 and r2 > 0.09).
We used linear mixed effect models to identify individual ASVs that differ significantly due to antibiotic treatment, disease outcome, and/or across time. These differential abundance analyses identified 14 out of 1,182 ASVs that differ significantly in our main effect model combining antibiotic pretreatment, disease outcome, and time (i.e. before/after exposure to the disease dose, Table 3, Supplementary Table S2). Out of the 14 ASVs, 9 ASVs differed due to antibiotic pretreatment, 6 ASVs differed due to coral disease outcome, and 11 ASVs differed depending on if the coral fragment was sampled before or after disease exposure (Table 3).
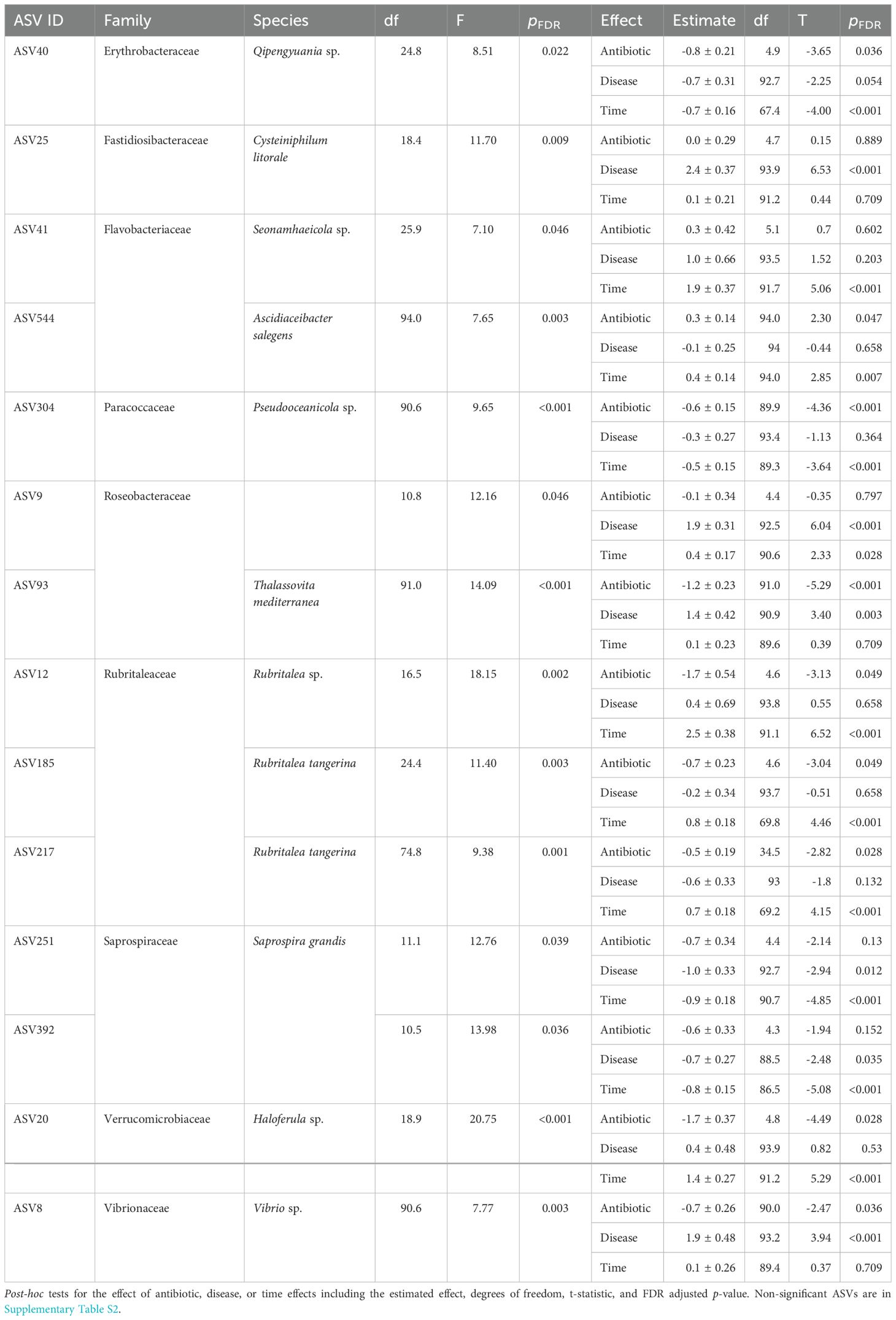
Table 3. The denominator degrees of freedom (df), F statistic and FDR adjusted p-value are shown for the omnibus significance tests of the 14 ASVs found to significantly differ in at least one treatment combination given four numerator degrees of freedom.
The ASVs which were affected by antibiotic pretreatment included one Flavobacteriaceae (ASV 544, Ascidiaceibacter salegens), which was more abundant in antibiotic treated corals, and eight ASVs which were negatively affected by antibiotic treatment (Table 3), including three strains of Rubritaleaceae (ASV 12, 185, 217) and one strain each of Erthrobacteraceae (ASV 40), Paracoccaceae (ASV 304), Roseobacteraceae (ASV 93), Verrucomicrobiaceae (ASV 20), and Vibrionaceae (ASV8, Figure 6, Table 3). The Vibrionaceae (ASV 8, Vibrio sp.) and Roseobacteraceae (ASV 93, Thalassovita mediterranea) were both also more abundant in diseased corals, indicating that they were impacted by antibiotic treatment and were also strongly associated with WBD outcomes. Specifically, ASV 8 and 93 had low initial abundances before the antibiotic pretreatment (0.66 ± 0.26, 1.08 ± 0.17, respectively), and were knocked-down by the antibiotics (0.01 ± 0.26, -0.16 ± 0.17, respectively). In untreated corals, these ASVs increased 5-fold in abundance on diseased corals after disease exposure (2.56 ± 0.47, 2.49 ± 0.38, respectively). Our differential abundance analyses detected one ASV that was only associated with disease outcome, a Fastidiosibacteraceae (ASV 25, Cysteiniphilum litorale), and another which was associated with disease but increased in healthy corals after disease exposure, an unclassified Roseobacteraceae (ASV 9, Figure 6, Table 3). In contrast to ASV 8 and 93 these ASVs were not detected initially (ASV 25: 0.14 ± 0.2, ASV 9: 0.15 ± 0.24) on healthy corals and thus were unaffected by the antibiotic treatment. Both ASVs were highly abundant on diseased corals (2.58 ± 0.37, 2.04 ± 0.36, respectively, Figure 6) and strongly associated with disease outcome, with ASV 9 also inhabiting healthy corals after receiving the disease dose (0.57 ± 0.26), unlike ASV 25 (0.35 ± 0.24).
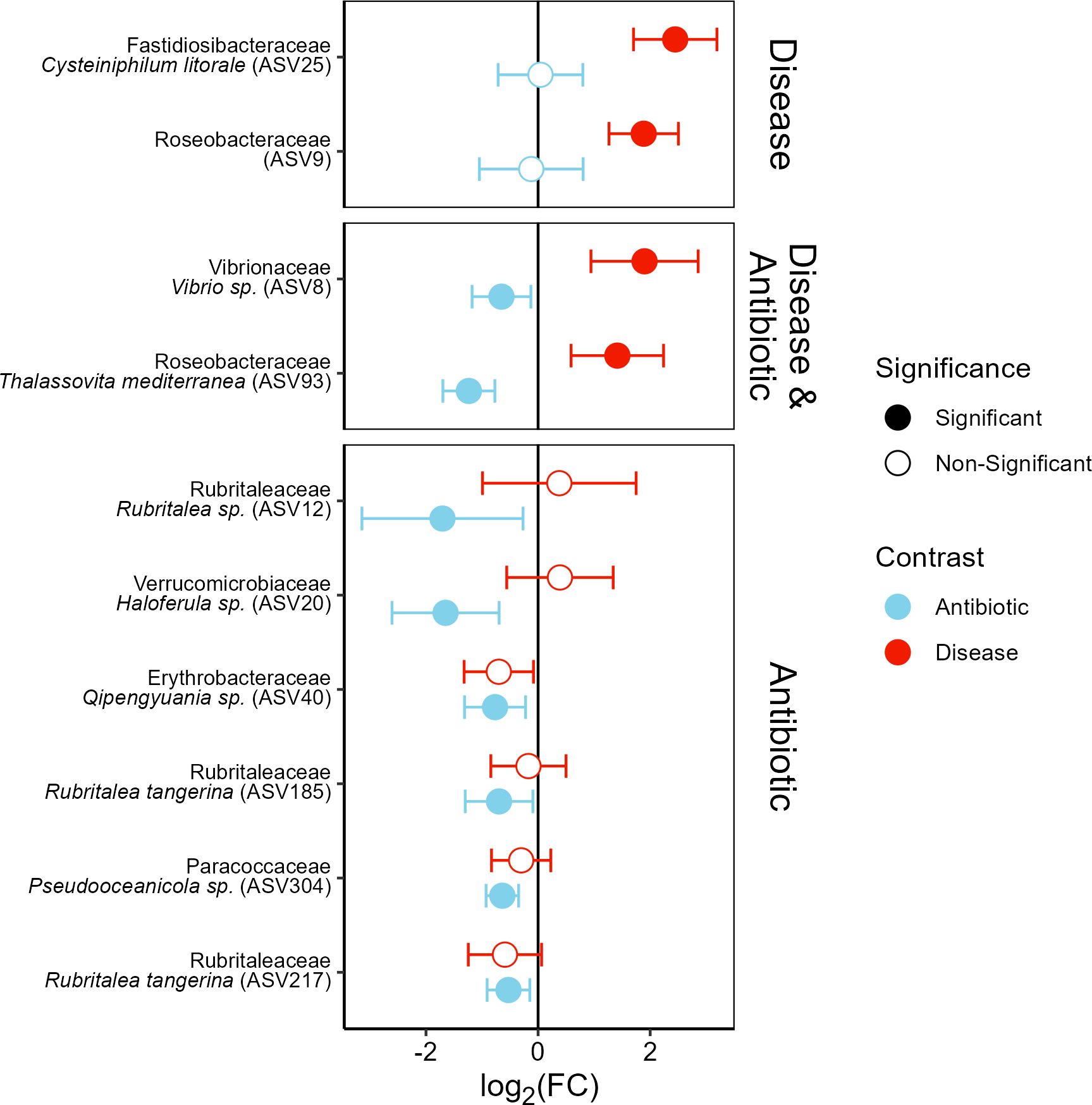
Figure 6. Fold-change differences in the abundance of ASVs significantly differentially abundant depending on antibiotic treatment (blue) and/or disease state (red). Error bars show 95% confidence intervals with filled circles indicating significance. Facets group ASVs based on the significance of the effect of coral disease state (Disease), antibiotic treatment (Antibiotic), or both (Disease & Antibiotic).
Discussion
Prior research has shown that antibiotic treatment suppressed the transmission (Kline and Vollmer, 2011) and arrests the progression of white band disease in Acropora cervicornis (Sweet et al., 2014). Prophylactically, pretreating healthy A. cervicornis prior to experimental disease exposure increased the non-infection rate by 30-percentage points to 93%. We even documented improved non-infection in antibiotic treated corals within our control healthy exposed tank treatments. These results demonstrate the efficacy of the prophylactic use of broad-spectrum antibiotics to lower disease transmission risks in land-based and in situ nursery settings, including as a mitigation strategy in quarantine settings, and as a tool to reduce disease transmission risks to wild coral populations during coral out planting efforts. The prophylactic benefit of antibiotics suggests that either intrinsic secondary opportunistic bacteria, which are constituents of healthy microbiomes, contribute to WBD pathogenicity or that the recent treatment with antibiotics prevents the transmission of WBD causing pathogens upon exposure.
16S rRNA amplicon gene sequencing indicates that antibiotic pretreatment shifted the staghorn coral microbiomes to a less diverse bacterial community. Differential abundance analyses identified eight ASVs that were significantly suppressed by the antibiotic pretreatment, including one of the candidate pathogens identified in a multi-year analysis, Vibrio sp. (ASV 8; Selwyn et al., 2024), and Thalassovita mediterranea (ASV 93). Both of which had initially low abundances on healthy corals that were knocked down by antibiotics (Figure 6). Untreated corals which developed disease symptoms after exposure had a 5-fold increase in ASV 8 and 93 through growth on the fragment and/or by being introduced in the disease dose. Similarly, the most likely WBD pathogen identified by Selwyn et al. (2024), Cysteiniphilum litorale (ASV 25) and an unidentified Roseobacteraceae (ASV 9) were both highly abundant on diseased corals (Figure 6). However, in contrast neither of these ASVs were detected on healthy coral fragments prior to disease exposure and as such were not affected by the antibiotic treatment. Unlike ASV 25, ASV 9 was also abundant on healthy fragments which were exposed to the disease dose (Figure 6) suggesting an opportunistic relationship.
Both Cysteiniphilum litorale (ASV 25) and Vibrio sp. (ASV 8) have been identified previously as the top two potential pathogens causing WBD in a multi-year analysis of diseased and healthy A. cervicornis (Selwyn et al., 2024), whereas Thalassovita mediterranea (ASV 93), previously Thalassobius mediterranea, and ASV 9 are members of the Roseobacter group (Arahal et al., 2005; Deshmukh and Oren, 2023) and were either not consistently associated with disease (ASV 93) or were highly correlated with more explanatory ASVs (ASV 9) in our multiyear analysis (Selwyn et al., 2024). Clear cases can be made for both Cysteiniphilum litorale and Vibrio sp. as being likely WBD pathogens (see below). While Roseobacters are a diverse bacterial lineage that have sometimes been associated with diseased corals (Cooney et al., 2002; Pantos et al., 2003; Buchan et al., 2005; MacKnight et al., 2021), they are more typically observed as mutualists with eukaryotes (Simon et al., 2017). The temporal variability in T. mediterranea abundance and ASV 9’s high degree of correlation with more explanatory ASVs (Selwyn et al., 2024) and their ability to participate in quorum sensing (Zan et al., 2014) suggests an opportunistic relationship with the observed disease association being primarily a result of opportunistic growth and the fact that the experiment took place during a peak of T. mediterranea abundance in Bocas del Toro, Panama (Selwyn et al., 2024).
Hypothesized etiology
The two top pathogen ASVs identified by the multi-year analysis in Selwyn et al. (2024) – Cysteiniphilum litorale (ASV 25) and Vibrio sp. (ASV 8) – both were strongly associated with disease outcomes in our tank-based experiment. Furthermore, Vibrio sp. was significantly negatively affected by antibiotic pretreatment while C. litorale was not detected on coral fragments prior to being dosed with the disease slurry. While both ASVs are clearly important to the WBD pathogenicity, it is unclear what roles they play in the WBD etiology.
Cysteiniphilum litorale is a recently described Fastidiosibacteraceae that has been linked to shrimp farm derived skin infections in humans (Liu et al., 2017; Xu et al., 2021). Prior to its description, it was described as Francisella-like (Liu et al., 2017; Qian et al., 2023), a genus which has been associated with WBD (Gignoux-Wolfsohn et al., 2017; Walton, 2017). In Florida a recent transmission experiment found Cysteiniphilum on A. cervicornis which developed disease symptoms following the experimental grafting of disease tissue (ASV 5b79cf6d5a5a9bf0bb866aed449eff44; Rosales et al., 2019) with additional studies in Florida finding Cysteiniphilum was present on both WBD resistant (Klinges et al., 2023) and WBD susceptible (Klinges et al., 2022) nursery reared Acropora cervicornis genotypes. The Cysteiniphilum genome, isolated in Wenzhou, China, contains a partial copy of the Francisella pathogenicity island (Qian et al., 2023) which facilitates Francisella being pathogens across a broad taxonomic range, including multiple marine species (Nano and Schmerk, 2007; Birkbeck et al., 2011; Colquhoun and Duodu, 2011).
Vibrio are well known opportunistic pathogens (Munn, 2015), which have been implicated in numerous coral diseases (Bourne et al., 2009) and are frequently commensally associated with corals under homoeostatic environmental conditions (Munn, 2015) where they can exist on ~20% of healthy corals (Ben-Haim et al., 2003; Gibbin et al., 2019; Selwyn et al., 2024). However, these vibrios become pathogenic under various environmental conditions, including increased temperature (e.g. V. coralliilytics causing tissue lysis in Pocillopora damicornis; Ben-Haim et al., 2003). One key trigger for the conversion of commensalist vibrios into pathogens is by quorum sensing initiated by the introduction of autoinducers (Liu et al., 2013). When autoinducers are introduced to healthy coral microbiomes, WBD symptoms develop (Certner and Vollmer, 2015) and the spread of WBD can be arrested by introducing quorum sensing inhibitors (Certner and Vollmer, 2018). Our results suggest that pretreating corals with antibiotics may prevent the initiation of quorum sensing, as quorum sensing is a density dependent behavior and pretreatment significantly reduced the relative abundance of Vibrio in the microbiome, which we assume to result from an absolute reduction in Vibrio abundance rather than an increase in other taxa following antibiotic treatment; this prevents the Vibrio sp. from becoming pathogenic and arrests any further cascade of opportunistic bacterial growth, preventing the shift to a diseased microbial community.
Quorum sensing is the process of intercellular communication among bacteria via the release and detection of small signaling molecules called autoinducers. This allows bacteria to alter behaviors based on bacterial density, such as activating virulence factors in situations of high cell density (Abisado et al., 2018). Bacterial quorum sensing is important in the transmission of WBD (Certner and Vollmer, 2015, 2018). Generally, autoinducers fall into two classes, species-specific (AI-1) and universal (AI-2), with AI-1 type autoinducers being variations of the class of molecules known as acylated homoserine lactones (AHLs) (Fuqua et al., 2001; Miller and Bassler, 2001; Schauder and Bassler, 2001; Xavier and Bassler, 2003). The enzymatic pathways required for both AI-1 (AHL) and AI-2 production and detection are well characterized across many Vibrio spp (Schauder et al., 2001; Winzer et al., 2002; Henke and Bassler, 2004), including those known to cause other coral diseases (Tait et al., 2010), and have been shown to regulate pathogenicity in many marine Vibrio spp (Henke and Bassler, 2004; Natrah et al., 2011).
In WBD, it is unclear what causes the initial production of autoinducers which initiates quorum sensing and triggers pathogenesis. We propose two potential initiators: either an overabundance of Vibrio sp., or the introduction of C. litorale. If vibrios are the initiators, then the densities of Vibrio spp. likely pass a threshold, initiating pathogenesis; this is analogous to infection by Clostridium difficile in humans, which are commensal bacteria in the digestive system and become pathogenic only after reaching high densities, often induced through the removal of competitors with antibiotics (Ng et al., 2010; Kamada et al., 2013). In our study, the disease dose likely introduced sufficient Vibrio sp. to pass this threshold in the untreated corals, which still contained indigenous Vibro sp., but not in the antibiotic treated corals where Vibrio ASV 8 was knocked down to be either absent or in very low abundance. In nature, changes in Vibrio sp. density could be caused by environmental changes such as increased temperature, which has been associated with increased WBD prevalences (Randall and van Woesik, 2015; Selwyn et al., 2024). The second mechanism is that C. litorale is a keystone pathogen which promotes pathogenicity in the indigenous bacteria (Hajishengallis and Lamont, 2016; Vega Thurber et al., 2020) through the production of autoinducers and initiation of quorum sensing, analogous to the role of enterotoxigenic Bacteroides fragilis which produces a biofilm that causes inflammation and changes in the gut microbiome resulting in colon cancer (Sears and Pardoll, 2011; Cheng et al., 2020).
Conclusions
Prophylactic antibiotic treatment of A. cervicornis colonies reduces the transmission of WBD through the alteration of the coral microbiome and the removal of potential pathogenic strains like Vibrio sp. ASV 8. In general, our results 1) advance our mechanistic understanding of WBD including the roles of Vibrio sp. and C. litorale and 2) demonstrate antibiotic pretreatment can be an effective strategy to lower disease transmission risks in land-based and in situ nursery settings, including as a mitigation strategy in quarantine settings, and as a tool to reduce disease transmission risks to wild coral populations. This may be especially useful in the future as temperatures continue to increase leading to not only increased thermal stress but increased disease transmission (Gignoux-Wolfsohn et al., 2020; Reimer et al., 2024; Selwyn et al., 2024). Our results confirm the association of C. litorale (ASV 25) and Vibrio sp. (ASV 8) with WBD (Selwyn et al., 2024). Furthermore, understanding the etiology of WBD can allow for the creation of more targeted effective treatments. Future research is needed to delineate the two hypothesized etiologies proposed here through the cultivation of both C. litorale (ASV 25) and Vibrio sp. (ASV 8) in pure cultures.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1106053. All code used in this analysis can be found here: https://github.com/VollmerLab/Panama_Antibiotics, raw 16S rRNA amplicon sequencing data is available at NCBI BioProject PRJNA1106053, sample accession numbers: SAMN41116617 – SAMN41116658, SAMN41116665 – SAMN41116676, SAMN41116683 – SAMN41116688, SAMN41116701 – SAMN41116706, SAMN41116713 – SAMN41116718, SAMN41116725 – SAMN41116730, SAMN41116737 – SAMN41116747, SAMN41116754 – SAMN41116759, SAMN41116772 – SAMN41116777 and SAMN41116784 – SAMN41116787.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements. Sample collections were permitted with approval of Autoridad Nacional del Ambiente, Panama CITES permits (SEX/A-116-16 and SEX/A-98-19).
Author contributions
JS: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. BD: Investigation, Methodology, Project administration, Visualization, Writing – review & editing. KG-D: Writing – review & editing. ET: Writing – review & editing. SV: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Grant funding was provided to SVV by National Science Foundation Division of Ocean Sciences grants (NSF OCE-1458158 and OCE-1924145).
Acknowledgments
We would like to thank the staff of the Smithsonian Tropical Research Institute staff in Bocas del Toro, Panama for their help with this project. Sample collections were permitted with approval of Autoridad Nacional del Ambiente, Panama CITES permits (SEX/A-116-16 and SEX/A-98-19).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1491476/full#supplementary-material
References
Abisado R. G., Benomar S., Klaus J. R., Dandekar A. A., Chandler J. R. (2018). Bacterial quorum sensing and microbial community interactions. mBio 9, 1–13. doi: 10.1128/mbio.02331-17
Aeby G. S., Ushijima B., Campbell J. E., Jones S., Williams G. J., Meyer J. L., et al. (2019). Pathogenesis of a tissue loss disease affecting multiple species of corals along the florida reef tract. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00678
Aitchison J. (1982). The statistical analysis of compositional data. J. R. Stat. Soc.: Ser. B Method. 44, 139–160. doi: 10.1111/j.2517-6161.1982.tb01195.x
Alvarez-Filip L., González-Barrios F. J., Pérez-Cervantes E., Molina-Hernández A., Estrada-Saldívar N. (2022). Stony coral tissue loss disease decimated Caribbean coral populations and reshaped reef functionality. Commun. Biol. 5, 1–10. doi: 10.1038/s42003-022-03398-6
Anderson M. J. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. doi: 10.1111/j.1541-0420.2005.00440.x
Arahal D. R., Macián M. C., Garay E., Pujalte M. J. (2005). Thalassobius mediterraneus gen. nov., sp. nov., and reclassification of Ruegeria gelatinovorans as Thalassobius gelatinovorus comb. nov. Int. J. Syst. Evol. Microbiol. 55, 2371–2376. doi: 10.1099/ijs.0.63842-0
Aronson R. B., Precht W. F. (2001). “White-band disease and the changing face of Caribbean coral reefs,” in The ecology and etiology of newly emerging marine diseases. Ed. Porter J. W. (Springer Netherlands, Dordrecht), 25–38. doi: 10.1007/978-94-017-3284-0_2
Bates D., Mächler M., Bolker B., Walker S. (2015).Fitting Linear Mixed-Effects Models Using lme4. Available online at: https://ideas.repec.org//a/jss/jstsof/v067i01.html (Accessed April 28, 2023).
Ben-Haim Y., Thompson F. L., Thompson C. C., Cnockaert M. C., Hoste B., Swings J., et al. (2003). Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53, 309–315. doi: 10.1099/ijs.0.02402-0
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc.: Ser. B Method. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Birkbeck T. H., Feist S. W., Verner – Jeffreys D. W. (2011). Francisella infections in fish and shellfish. J. Fish Dis. 34, 173–187. doi: 10.1111/j.1365-2761.2010.01226.x
Bourne D. G., Garren M., Work T. M., Rosenberg E., Smith G. W., Harvell C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. doi: 10.1016/j.tim.2009.09.004
Buchan A., González J. M., Moran M. A. (2005). Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Camargo J. A. (1992). New diversity index for assessing structural alterations in aquatic communities. Bull. Environ. Contam. Toxicol. 48, 428–434. doi: 10.1007/BF00195643
Casas V., Kline D. I., Wegley L., Yu Y., Breitbart M., Rohwer F. (2004). Widespread association of a Rickettsiales-like bacterium with reef-building corals. Environ. Microbiol. 6, 1137–1148. doi: 10.1111/j.1462-2920.2004.00647.x
Certner R. H., Vollmer S. V. (2015). Evidence for autoinduction and quorum sensing in white band disease-causing microbes on acropora cervicornis. Sci. Rep. 5, 11134. doi: 10.1038/srep11134
Certner R. H., Vollmer S. V. (2018). Inhibiting bacterial quorum sensing arrests coral disease development and disease-associated microbes. Environ. Microbiol. 20, 645–657. doi: 10.1111/1462-2920.13991
Cheng W. T., Kantilal H. K., Davamani F. (2020). The mechanism of bacteroides fragilis toxin contributes to colon cancer formation. Malays J. Med. Sci. 27, 9–21. doi: 10.21315/mjms2020.27.4.2
Colquhoun D. J., Duodu S. (2011). Francisella infections in farmed and wild aquatic organisms. Vet. Res. 42, 47. doi: 10.1186/1297-9716-42-47
Cooney R. P., Pantos O., Le Tissier M. D. A., Barer M. R., O’Donnell A. G., Bythell J. C. (2002). Characterization of the bacterial consortium associated with black band disease in coral using molecular microbiological techniques. Environ. Microbiol. 4, 401–413. doi: 10.1046/j.1462-2920.2002.00308.x
Cowley S., Elkins K. (2011). Immunity to francisella. Front. Microbiol. 2, 1–21. doi: 10.3389/fmicb.2011.00026
Deshmukh U. B., Oren A. (2023). Proposal of Thalassovita gen. nov. and Alloyangia gen. nov. as replacement names for the illegitimate prokaryotic generic names Thalassobius and Yangia, respectively. Int. J. Syst. Evol. Microbiol. 73, 1–5. doi: 10.1099/ijsem.0.006025
Faith D. P. (1992). Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. doi: 10.1016/0006-3207(92)91201-3
Fukami H., Budd A. F., Levitan D. R., Jara J., Kersanach R., Knowlton N. (2004). Geographic differences in species boundaries among members of the montastraea annularis complex based on molecular and morphological markers. Evolution 58, 324–337. doi: 10.1111/j.0014-3820.2004.tb01648.x
Fuqua C., Parsek M. R., Greenberg E. P. (2001). Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35, 439–468. doi: 10.1146/annurev.genet.35.102401.090913
Gao X., Lin H., Revanna K., Dong Q. (2017). A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinf. 18, 247. doi: 10.1186/s12859-017-1670-4
Gibbin E., Gavish A., Krueger T., Kramarsky-Winter E., Shapiro O., Guiet R., et al. (2019). Vibrio coralliilyticus infection triggers a behavioural response and perturbs nutritional exchange and tissue integrity in a symbiotic coral. ISME J. 13, 989–1003. doi: 10.1038/s41396-018-0327-2
Gignoux-Wolfsohn S. A., Aronson F. M., Vollmer S. V. (2017). Complex interactions between potentially pathogenic, opportunistic, and resident bacteria emerge during infection on a reef-building coral. FEMS Microbiol. Ecol. 93, 1–10. doi: 10.1093/femsec/fix080
Gignoux-Wolfsohn S. A., Marks C. J., Vollmer S. V. (2012). White Band Disease transmission in the threatened coral, Acropora cervicornis. Sci. Rep. 2, 804. doi: 10.1038/srep00804
Gignoux-Wolfsohn S. A., Precht W. F., Peters E. C., Gintert B. E., Kaufman L. S. (2020). Ecology, histopathology, and microbial ecology of a white-band disease outbreak in the threatened staghorn coral Acropora cervicornis. Dis. Aquat Organ 137, 217–237. doi: 10.3354/dao03441
Gil-Agudelo D. L., Smith G. W., Weil E. (2006). The white band disease type II pathogen in Puerto Rico. Rev. Biología Trop. 54, 59–67.
Gladfelter W. B. (1982). White-band disease in acropora palmata: implications for the structure and growth of shallow reefs. Bull. Mar. Sci. 32, 639–643.
Glaeser S. P., Kämpfer P. (2014). “The family sphingomonadaceae,” in The prokaryotes: alphaproteobacteria and betaproteobacteria. Eds. Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., Thompson F. (Springer, Berlin, Heidelberg), 641–707. doi: 10.1007/978-3-642-30197-1_302
Gloor G. B., Macklaim J. M., Pawlowsky-Glahn V., Egozcue J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02224
Good I. J. (1953). The population frequencies of species and the estimation of population parameters. Biometrika 40, 237–264. doi: 10.1093/biomet/40.3-4.237
Hajishengallis G., Lamont R. J. (2016). Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 24, 477–489. doi: 10.1016/j.tim.2016.02.010
Hanshew A. S., Mason C. J., Raffa K. F., Currie C. R. (2013). Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Methods 95, 149–155. doi: 10.1016/j.mimet.2013.08.007
Harrington D. P., Fleming T. R. (1982). A class of rank test procedures for censored survival data. Biometrika 69, 553–566. doi: 10.1093/biomet/69.3.553
Henke J. M., Bassler B. L. (2004). Three parallel quorum-sensing systems regulate gene expression in vibrio harveyi. J. Bacteriol. 186, 6902–6914. doi: 10.1128/jb.186.20.6902-6914.2004
Hurlbert S. H. (1984). Pseudoreplication and the design of ecological field experiments. Ecol. Monogr. 54, 187–211. doi: 10.2307/1942661
Kamada N., Chen G. Y., Inohara N., Núñez G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Kaplan E. L., Meier P. (1958).Nonparametric estimation from incomplete observations. Available online at: https://www.tandfonline.com/doi/abs/10.1080/01621459.1958.10501452 (Accessed May 13, 2024).
Kaul A., Mandal S., Davidov O., Peddada S. D. (2017). Analysis of microbiome data in the presence of excess zeros. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02114
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1. doi: 10.1093/nar/gks808
Kline D. I., Vollmer S. V. (2011). White band disease (type I) of endangered caribbean acroporid corals is caused by pathogenic bacteria. Sci. Rep. 1, 7. doi: 10.1038/srep00007
Klinges G., Maher R. L., Vega Thurber R. L., Muller E. M. (2020). Parasitic “Candidatus Aquarickettsia rohweri” is a marker of disease susceptibility in Acropora cervicornis but is lost during thermal stress. Environ. Microbiol. 22, 5341–5355. doi: 10.1111/1462-2920.15245
Klinges J. G., Patel S. H., Duke W. C., Muller E. M., Vega Thurber R. L. (2022). Phosphate enrichment induces increased dominance of the parasite Aquarickettsia in the coral Acropora cervicornis. FEMS Microbiol. Ecol. 98, fiac013. doi: 10.1093/femsec/fiac013
Klinges J. G., Patel S. H., Duke W. C., Muller E. M., vega thurber R. L. (2023). Microbiomes of a disease-resistant genotype of Acropora cervicornis are resistant to acute, but not chronic, nutrient enrichment. Sci. Rep. 13, 3617. doi: 10.1038/s41598-023-30615-x
Lin H., Peddada S. D. (2020a). Analysis of compositions of microbiomes with bias correction. Nat. Commun. 11, 3514. doi: 10.1038/s41467-020-17041-7
Lin H., Peddada S. D. (2020b). Analysis of microbial compositions: a review of normalization and differential abundance analysis. NPJ Biofilms Microbiomes 6, 1–13. doi: 10.1038/s41522-020-00160-w
Lin H., Peddada S. D. (2024). Multigroup analysis of compositions of microbiomes with covariate adjustments and repeated measures. Nat. Methods 21, 83–91. doi: 10.1038/s41592-023-02092-7
Liu H., Srinivas S., He H., Gong G., Dai C., Feng Y., et al. (2013). Quorum sensing in vibrio and its relevance to bacterial virulence. J. Bacteriol. Parasitol. 4, 3. doi: 10.4172/2155-9597.1000172
Liu L., Salam N., Jiao J.-Y., E S.-M., Chen C., Fang B.-Z., et al. (2017). Cysteiniphilum litorale gen. nov., sp. nov., isolated from coastal seawater. Int. J. Syst. Evol. Microbiol. 67, 2178–2183. doi: 10.1099/ijsem.0.001917
MacKnight N. J., Cobleigh K., Lasseigne D., Chaves-Fonnegra A., Gutting A., Dimos B., et al. (2021). Microbial dysbiosis reflects disease resistance in diverse coral species. Commun. Biol. 4, 679. doi: 10.1038/s42003-021-02163-5
Mandal S., Van Treuren W., White R. A., Eggesbø M., Knight R., Peddada S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microbial Ecol. Health Dis. 26, 27663. doi: 10.3402/mehd.v26.27663
Martino C., Morton J. T., Marotz C. A., Thompson L. R., Tripathi A., Knight R., et al. (2019). A novel sparse compositional technique reveals microbial perturbations. mSystems 4, e00016-19. doi: 10.1128/mSystems.00016-19
McArdle B. H., Anderson M. J. (2001). Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Millar R. B., Anderson M. J. (2004). Remedies for pseudoreplication. Fish. Res. 70, 397–407. doi: 10.1016/j.fishres.2004.08.016
Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Miller M. W., Lohr K. E., Cameron C. M., Williams D. E., Peters E. C. (2014). Disease dynamics and potential mitigation among restored and wild staghorn coral, Acropora cervicornis. PeerJ 2, e541. doi: 10.7717/peerj.541
Miyamatsu Y., Tanizaki R., Yamada S. (2024). Sphingobium yanoikuyae bacteremia, Japan. Emerging Infect. Dis. 30, 1060–1062. doi: 10.3201/eid3005.231514
Munn C. B. (2015). The role of vibrios in diseases of corals. Microbiol. Spectr. 3, 1–12. doi: 10.1128/microbiolspec.ve-0006-2014
Nano F. E., Schmerk C. (2007). The francisella pathogenicity island. Ann. New York Acad. Sci. 1105, 122–137. doi: 10.1196/annals.1409.000
Natrah F. M. I., Ruwandeepika H. A. D., Pawar S., Karunasagar I., Sorgeloos P., Bossier P., et al. (2011). Regulation of virulence factors by quorum sensing in Vibrio harveyi. Vet. Microbiol. 154, 124–129. doi: 10.1016/j.vetmic.2011.06.024
Neely K. L., Shea C. P., Macaulay K. A., Hower E. K., Dobler M. A. (2021). Short- and long-term effectiveness of coral disease treatments. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.675349
Ng J., Hirota S. A., Gross O., Li Y., Ulke-Lemee A., Potentier M. S., et al. (2010). Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology 139, 542–552. doi: 10.1053/j.gastro.2010.04.005
Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., et al. (2013). vegan: community ecology package. Available online at: http://CRAN.R-project.org/package=vegan (Accessed November 14, 2013).
O’Neill M. E., Mathews K. (2000). A weighted least squares approach to levene’s test of homogeneity of variance. Aust. New Z. J. Stat 42, 81–100. doi: 10.1111/1467-842X.00109
Pantos O., Cooney R. P., Le Tissier M. D. A., Barer M. R., O’Donnell A. G., Bythell J. C. (2003). The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 5, 370–382. doi: 10.1046/j.1462-2920.2003.00427.x
Precht W. F., Gintert B. E., Robbart M. L., Fura R., van Woesik R. (2016). Unprecedented disease-related coral mortality in southeastern florida. Sci. Rep. 6, 31374. doi: 10.1038/srep31374
Qian C., Xu M., Huang Z., Tan M., Fu C., Zhou T., et al. (2023). Complete genome sequence of the emerging pathogen Cysteiniphilum spp. and comparative genomic analysis with genus Francisella: Insights into its genetic diversity and potential virulence traits. Virulence 14, 2214416. doi: 10.1080/21505594.2023.2214416
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Randall C. J., van Woesik R. (2015). Contemporary white-band disease in Caribbean corals driven by climate change. Nat. Clim Change 5, 375–379. doi: 10.1038/nclimate2530
Reimer J. D., Peixoto R. S., Davies S. W., Traylor-Knowles N., Short M. L., Cabral-Tena R. A., et al. (2024). The Fourth Global Coral Bleaching Event: Where do we go from here? Coral Reefs 43, 1121–1125. doi: 10.1007/s00338-024-02504-w
R Core Team (2022). R: A language and environment for statistical computing. Available online at: https://www.R-project.org/ (Accessed September 25, 2023).
Rosales S. M., Miller M. W., Williams D. E., Traylor-Knowles N., Young B., Serrano X. M. (2019). Microbiome differences in disease-resistant vs. susceptible Acropora corals subjected to disease challenge assays. Sci. Rep. 9, 18279. doi: 10.1038/s41598-019-54855-y
Sanders H. L. (1968). Marine benthic diversity: A comparative study. Am. Nat. 102, 243–282. doi: 10.1086/282541
Satterthwaite F. E. (1946). An approximate distribution of estimates of variance components. Biom. Bull. 2, 110–114. doi: 10.2307/3002019
Schauder S., Bassler B. L. (2001). The languages of bacteria. Genes Dev. 15, 1468–1480. doi: 10.1101/gad.899601
Schauder S., Shokat K., Surette M. G., Bassler B. L. (2001). The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41, 463–476. doi: 10.1046/j.1365-2958.2001.02532.x
Schliep K. P. (2011). phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593. doi: 10.1093/bioinformatics/btq706
Sears C. L., Pardoll D. M. (2011). Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 203, 306–311. doi: 10.1093/jinfdis/jiq061
Selwyn J. D., Despard B. A., Vollmer M. V., Trytten E. C., Vollmer S. V. (2024). Identification of putative coral pathogens in endangered Caribbean staghorn coral using machine learning. Environ. Microbiol. 26, e16700. doi: 10.1111/1462-2920.16700
Shannon C. (1948).A mathematical theory of communication. Available online at: http://www.agent.ai/doc/upload/200302/shan48.pdf (Accessed March 12, 2014).
Sheridan C., Kramarsky-Winter E., Sweet M., Kushmaro A., Leal M. C. (2013). Diseases in coral aquaculture: causes, implications and preventions. Aquaculture 396–399, 124–135. doi: 10.1016/j.aquaculture.2013.02.037
Simon M., Scheuner C., Meier-Kolthoff J. P., Brinkhoff T., Wagner-Döbler I., Ulbrich M., et al. (2017). Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499. doi: 10.1038/ismej.2016.198
Sweet M., Bythell J. (2015). White Syndrome in Acropora muricata: Nonspecific bacterial infection and ciliate histophagy. Mol. Ecol. 24, 1150–1159. doi: 10.1111/mec.13097
Sweet M. J., Croquer A., Bythell J. C. (2014). Experimental antibiotic treatment identifies potential pathogens of white band disease in the endangered Caribbean coral Acropora cervicornis. Proc. R. Soc. B: Biol. Sci. 281, 20140094. doi: 10.1098/rspb.2014.0094
Tait K., Hutchison Z., Thompson F. L., Munn C. B. (2010). Quorum sensing signal production and inhibition by coral-associated vibrios. Environ. Microbiol. Rep. 2, 145–150. doi: 10.1111/j.1758-2229.2009.00122.x
Thomas F., Dittami S. M., Brunet M., Le Duff N., Tanguy G., Leblanc C., et al. (2020). Evaluation of a new primer combination to minimize plastid contamination in 16S rDNA metabarcoding analyses of alga-associated bacterial communities. Environ. Microbiol. Rep. 12, 30–37. doi: 10.1111/1758-2229.12806
Vega Thurber R., Mydlarz L. D., Brandt M., Harvell D., Weil E., Raymundo L., et al. (2020). Deciphering coral disease dynamics: integrating host, microbiome, and the changing environment. Front. Ecol. Evol. 8, 575927. doi: 10.3389/fevo.2020.575927
Walton C. (2017). Bacterial Communities Associated with Healthy and Diseased Acropora cervicornis (Staghorn Coral) Using High-Throughput Sequencing (United States – Florida: Nova Southeastern University). Available at: https://nsuworks.nova.edu/occ_stuetd/449 (Accessed March 19, 2024).
Williams D. E., Miller M. W. (2005). Coral disease outbreak: pattern, prevalence and transmission in Acropora cervicornis. Mar. Ecol. Prog. Ser. 301, 119–128. doi: 10.3354/meps301119
Winzer K., Hardie K. R., Burgess N., Doherty N., Kirke D., Holden M. T. G., et al. (2002). LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148, 909–922. doi: 10.1099/00221287-148-4-909
Wright E. S. (2016). Using DECIPHER v2.0 to analyze big biological sequence data in R. R J. 8, 352. doi: 10.32614/RJ-2016-025
Xavier K. B., Bassler B. L. (2003). LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6, 191–197. doi: 10.1016/S1369-5274(03)00028-6
Xu C., Zhang X., Wu Q., Chen L., Qu P., Zhang Y., et al. (2021). Skin and soft tissue infection caused by Cysteiniphilum litorale in an immunocompetent patient: A case report. Indian J. Med. Microbiol. 39, 545–547. doi: 10.1016/j.ijmmb.2021.08.002
Keywords: white band disease, Acropora, keystone pathogen, opportunistic pathogen, Vibrio, Cysteiniphilum litorale
Citation: Selwyn JD, Despard BA, Galvan-Dubois KA, Trytten EC and Vollmer SV (2025) Antibiotic pretreatment inhibits white band disease infection by suppressing the bacterial pathobiome. Front. Mar. Sci. 12:1491476. doi: 10.3389/fmars.2025.1491476
Received: 04 September 2024; Accepted: 03 February 2025;
Published: 20 February 2025.
Edited by:
Neus Garcias-Bonet, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Julie L. Meyer, University of Florida, United StatesAlejandra Prieto-Davó, National Autonomous University of Mexico, Mexico
Amanda Shore, Farmingdale State College, United States
Copyright © 2025 Selwyn, Despard, Galvan-Dubois, Trytten and Vollmer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jason D. Selwyn, amFzb24uc2Vsd3luQHRhbXVjYy5lZHU=
†Present address: Jason D. Selwyn, Genomics Core Laboratory, Texas A&M University - Corpus Christi, Corpus Christi, TX, United States
 Jason D. Selwyn
Jason D. Selwyn Brecia A. Despard
Brecia A. Despard Kai A. Galvan-Dubois
Kai A. Galvan-Dubois Emily C. Trytten
Emily C. Trytten Steven V. Vollmer
Steven V. Vollmer