
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 31 January 2025
Sec. Aquatic Microbiology
Volume 12 - 2025 | https://doi.org/10.3389/fmars.2025.1442210
 Peihao Liu1
Peihao Liu1 Qijun Luo2
Qijun Luo2 Shanshan Zhu1
Shanshan Zhu1 Juanjuan Chen1
Juanjuan Chen1 Rui Yang1
Rui Yang1 Tingting Niu1
Tingting Niu1 Tiegan Wang3
Tiegan Wang3 Peng Zhang3
Peng Zhang3 Haimin Chen1*
Haimin Chen1*Introduction: The aquaculture in intertidal mudflat ponds on the coast of the East China Sea typically coexists with oceanic seaweed cultivation in the nearby open water area, forming an interconnected aquatic ecosystem. This study aimed to understand the impact of the seaweed cultivation on the water quality and microbial communities in their neighboring intertidal mudflat ponds.
Methods: A unique recirculating culture system was established, integrating seaweed farming in marine areas with aquaculture ponds on mudflats, and incorporating an effluent treatment process for added efficiency. Water samples were collected from three sites, including open water areas with or without seaweed (Pyropia haitanensis) cultivation, the aquaculture mudflat ponds, and effluent water ponds, with control sites for comparison. The dynamic diversity patterns and structure of microbial communities were determined by 16S and 18S rRNA sequencing, and the environmental factors were measured.
Results: In the offshore aquaculture area, the cultivation of P. haitanensis increased the dissolved oxygen and pH, decreased inorganic nutrients, reduced the alpha diversity of bacteria, established the dominance of Proteobacteria, but did not influence the composition of microeukaryotes. These changes in the seawater subsequently affected the dynamics in the corresponding aquaculture ponds, where the water quality was improved, the microbial diversity was enhanced, and the abundance of functional bacteria such as Microbacteriaceae was increased. According to the co-occurrence network analysis, the cultivation of P. haitanensis potentially improved the stability of the microbial communities in both the offshore aquaculture area and the onshore aquaculture ponds and allowed them to resist stress better. In addition, after the effluent from the aquaculture ponds was purified in the treatment ponds with mangroves and oysters, the water quality improved and the abundances of Dinophyceae and Bacillariophyta decreased.
Discussion: This study highlights the positive effects of P. haitanensis cultivation on water quality and microbial stability in adjacent aquaculture ponds.
In China, coastal mudflat areas are an important land resource whose utilization has accelerated in recent years. Regions with extensive tidal mudflats, most prominently in the provinces of Jiangsu, Zhejiang and Guangdong, now have many aquaculture dyke ponds, which are excavated in saline-alkali land and used to produce fish, shrimp, and crab (Long et al., 2016). The intensive farming of animals such as shrimp, crab, and fish in the aquaculture ponds poses environmental challenges, as the nutrient in the effluent discharged from the ponds can exacerbate eutrophication. In addition, the ponds typically host microbial species at high densities, including pathogenic bacteria. These ponds typically draw water from the neighboring open water area, and their effluent is discharged back to the sea, thus forming a cycle. As a result, the discharged effluent water may impose a burden on the marine environment (Hou et al., 2023). Meanwhile, seawater, as a key water source for aquaculture, directly determines the quality of aquaculture water, which in turn significantly affects the growth of farmed animals. In some regions of China, intertidal seaweed is cultivated in nearby coastal waters, which could potentially improve water quality in pond systems by reducing nutrient pollution and promoting ecosystem health.
Macroalgae play pivotal and defining roles in coastal ecosystems. Macroalgae can remove nitrogen and phosphorus from coastal waters and achieve carbon storage (Buschmann et al., 2017; Duarte et al., 2022; Seghetta et al., 2016; Xiao et al., 2017; Yang et al., 2015). The cultivation of seaweed offers substantial environmental benefits as it can help to mitigate ocean eutrophication (Alleway, 2023), and it may also enhance the seawater oxygen concentration and reduce seawater acidification (Xiao et al., 2021). Seaweed is also employed in aquaculture to improve water quality. Efforts have been made to develop integrated multi-trophic aquaculture systems with the capacity for highly efficient bioremediation, aiming to mitigate eutrophication in aquaculture. In these systems, seaweed is cultured on the surface of the cultivation area in the coastal water and animals (e.g., fish and bivalves) are farmed in the middle or lower layers, thus creating co-culture environments where the seaweed and animals share the same space. These systems have been operated in open-water environments (Chopin et al., 2012; Reid et al., 2020) as well as in land-based aquaculture ponds (Ashkenazi et al., 2019; Kang et al., 2021) and have proven effective in reducing nutrients and pollutants (Xiao et al., 2017; Yang et al., 2015). Nevertheless, this pattern diverges from the intertidal seaweed farming integrated with aquaculture ponds on nearby coastal mudflats.
China is a leading country in seaweed cultivation, and its total output of cultivated seaweed in 2022 is 2,713,900 tons (China Fishery Bureau, 2023). Porphyra sensu lato make up one of the largest groups of red algae in the world, extensively cultivated in the provinces of Jiangsu, Zhejiang, and Fujian, and the harvest of Porphyra in 2022 alone is approximately 218,658 tons (China Fishery Bureau, 2023). In the context of eutrophication abatement via large-scale seaweed cultivation, Porphyra is an excellent choice due to its high productivity and its higher capability (63%–170% higher than other seaweeds) to rapidly assimilate nitrogen and phosphorus (Chopin and Yarish, 1999; Neori et al., 2004). Porphyra cultivation is predominantly carried out in the intertidal zone, and in certain regions, this seaweed farming in offshore aquaculture areas is integrated with the aquaculture of fish, shrimp, and crab in adjacent mudflat ponds (Liu et al., 2023). However, the environmental impacts of this practice have not been thoroughly investigated to date, raising an intriguing question: Can water from seaweed cultivation indirectly enhance the quality of aquaculture water in adjacent mudflat ponds? If so, through what mechanisms can this positive effect be achieved?
Microorganisms participate in the decomposition of organic matters and the recycling of dissolved nutrients, and thus play crucial ecological roles in coastal and freshwater ecosystems. Recent studies on the epiphytic and endophytic microbial communities associated with various seaweeds, including green algae, red algae, and brown algae, have revealed that seaweeds can influence or regulate microbial dynamics within their cultivation areas (Xie et al., 2017). For instance, Wang et al. (2020) observed differences in the microbial communities of seawater in the presence of P. haitanensis cultivation compared to its absence. The cultivation of P. haitanensis led to an increase in the abundance of Rhodobacteraceae while decreasing the presence of Cyanobacteria. The taxonomically diverse microbial communities associated with seaweed are vital to their hosts and contribute to the overall health of the marine environment. Therefore, it is likely that seaweed cultivation may indirectly affect microbial communities in aquaculture ponds when these ponds draw water from areas utilized for seaweed farming.
The current study, carried out at the Zhejiang province in China, examined aquaculture ponds at coastal mudflats situated in the vicinity of intertidal seaweed farming, where the open water area provided the aquaculture water for the ponds and received their discharge. High-throughput sequencing based on 16S and 18S rRNA genes was employed to assess the diversity and structure of the microbial communities. The analysis focused on the seawater with or without P. haitanensis cultivation, the water from ponds with different sources of aquaculture water, and the water in the treatment ponds before discharge. The environmental factors of these water bodies were closely monitored. This study deepens our understanding of the differences in microbial communities between marine and farmed areas with and without algal cultivation and reveals the complex linkages between seaweed farming and microbial communities and environmental factors, providing valuable scientific evidence for optimizing farming models and enhancing ecosystem management.
The study was conducted on December 15, 2022, at two sites in Xiangshan County, Ningbo City, Zhejiang Province, China. There was extensive aquaculture of P. haitanensis throughout the Hongweitang Bay (29.095°N, 121.910°E). The Heping Bay (29.168°N, 121.958°E) served as the control (Figure 1).
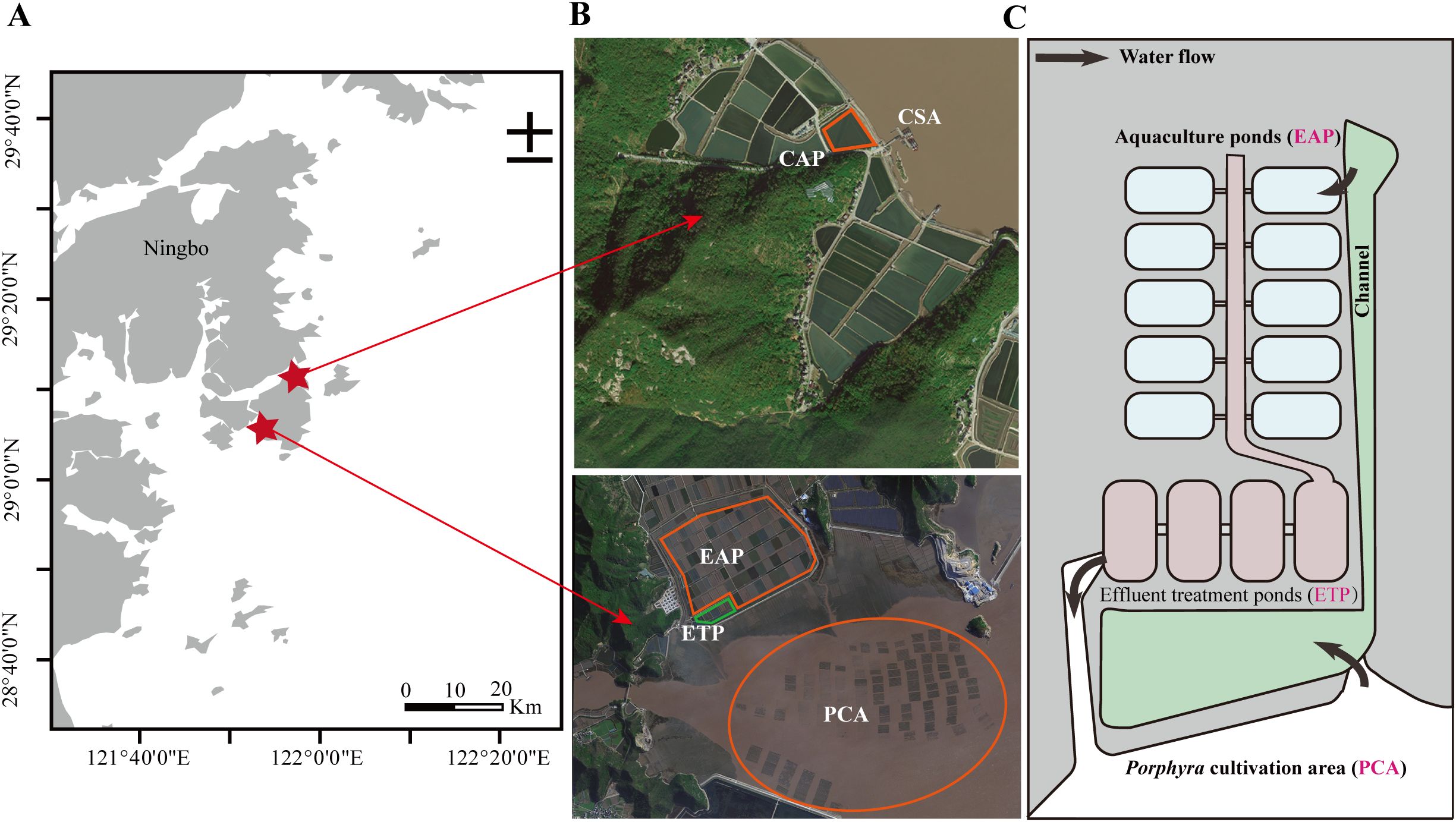
Figure 1. Distribution map of sampling sites and a schematic representation of the aquaculture system. (A) The locations of Hongweitang Bay and Heping Bay. (B) The satellite images of the survey areas consisting of the five groups. Here, PCA represents the Pyropia haitanensis cultivation area, EAP represents the experimental aquaculture ponds, and ETP represents the effluent treatment ponds, while CSA and CAP represent the control offshore aquaculture area and the control aquaculture ponds, respectively. (C) Schematic representation of the water circulation at Hongweitang Bay (The seawater flows from PCA to EAP, then to ETP, and back to PCA).
At Hongweitang Bay, samples were collected from three groups, i.e., the P. haitanensis cultivation area (PCA), the experimental aquaculture ponds (EAP), and the effluent treatment ponds (ETP). The PCA was representative of the Hongweitang Bay and featured extensive cultivation of P. haitanensis. The EAP consisted of open aquaculture ponds situated along the coastline of the PCA and received natural seawater pumped from a sedimentation pond that was connected to the PCA through a tidal gate. The ponds of EAP were utilized for the polyculture of crabs (mainly Portunus trituberculatus), shrimps (primarily Exopalaemon carinicauda), and bivalves (mainly Sinonovacula constrzcta and Meretrix meretrix) in a semi-intensive system. The effluents from these ponds were channeled to the ETP for storage and treatment. The ponds of ETP were cultured with Ostrea gigas and featured extensive plantings of mangroves (primarily Rhizophora mangle).
At Heping Bay, samples were collected from two groups, i.e., the control sea area (CSA), which was a natural open water area without seaweed cultivation, and the control aquaculture ponds (CAP), which consisted of the aquaculture ponds in the neighboring mudflat. The CAP received the supply of aquaculture water from the CSA (Figure 1). Both CAP and EAP had identical aquaculture species and breeding density, and their daily management practices were the same.
The water in the ponds of EAP and CAP was refreshed every 15 days. The water in the ponds of ETP was discharged into the sea after treatment for 15 days.
For each of the five groups (PCA, EAP, ETP, CSA, and CAP), water samples were collected at eight randomly chosen sampling spots. For EAP and CAP, water was collected on the seventh day after the introduction of fresh seawater. For ETP, all eight sampling spots were near the gate through which the treated water was discharged. At each spot, surface water samples were harvested at 0.5 m depth using a 5 L water collector. About 1 L of the collected water was promptly passed through a 100 μm nylon filter and then a 0.2 μm polycarbonate membrane (Millipore, USA). The membrane was frozen using liquid nitrogen and stored at −80°C until DNA extraction. The remaining water was used for the analysis of water chemistry in the lab, including total nitrogen (TN), phosphate (PO3-4-P), nitrite (NO-2-N), nitrate (NO-3-N), and ammonium (NH4+-N), according to the standard methods stipulated by GAQSIQ (2007) (AQSIQ, 2007). Total carbon (TC) and total organic carbon (TOC) were quantified using a vario TOC select analyzer (Elementar, Germany). Water temperature, conductivity, dissolved oxygen (DO), and pH were measured on site using a HACH HQ4300 portable multimeter (HACH, USA).
The total DNA was extracted from the membranes using the OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, USA) following the manufacturer’s instructions. The universal primers 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) were used for the amplification of the V3-V4 regions of the bacterial 16S rRNA, and the primers TAReuk454F (5′-CCAGCASCYGCGGTAATTCC-3′) and TAReukR (5′-ACTTTCGTTCTTGATYRA-3′) were used for the amplification of the V4 region of the eukaryotic 18S rRNA genes.
The reaction mixture of the PCR amplification included the buffer (5×) containing 1.25 U Fast pfu DNA Polymerase and 250 μM dNTPs (5 μL), the primer (10 μM, 1 μL), the DNA Template (1 μL), and ddH2O (14.75 μL). A standard protocol was applied as follows: initial denaturation, 98°C for 5 min; 25 cycles of denaturation (98°C for 30 s), annealing (53°C for 30 s), and extension (72°C for 45 s); final extension, 72°C for 5 min. All amplifications were run in triplicate. The PCR amplicons were purified using the Vazyme VAHTS DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Sequencing was executed by Shanghai Personal Biotechnology Co., Ltd (Shanghai, China) on the Illumina MiSeq platform using MiSeq Reagent Kit v3.
The analyses of microbiome bioinformatics were conducted using QIIME2 2019.4 (Bolyen et al., 2019) with minor modifications. The demux plugin was used for the demultiplexing of the raw sequencing data, and the primers were removed using the cutadapt plugin (Martin, 2011). The amplicon sequence variants (ASVs) were generated from paired-end sequences using DADA2 in QIIME2, which sequentially filtered out low-quality reads, denoised and spliced the data, and eliminated chimeric sequences. The ‘qiime dada2 denoise paired’ option was employed (Callahan et al., 2016). The taxonomic annotation of each ASV in the bacteria and the microeukaryotes was assigned according to the SILVA database (http://www.arb-silva.de). The raw sequencing data were deposited in the NCBI Sequence Read Archive under the accession number PRJNA1045075 and PRJNA1045086. The ASV abundance table after draw levelling was used for all subsequent analyses.
The physical and chemical parameters of the water samples were analyzed by one-way ANOVA using SPSS 26.0 (n = 8). The Duncan test was used for multiple comparisons. The sequencing data were primarily analyzed using QIIME2 and the relevant packages in R (v. 4.3.2). The alpha diversity indexes (Chao1, Shannon, and Simpson) of the ASVs were computed using the ASV table in QIIME2 and presented as boxplots. Venn diagrams were drawn to visualize the unique and shared microorganic taxa at the family level. The structural variations of the microbial communities across samples were examined by the beta diversity analysis based on the Bray–Curtis dissimilarity (Bray and Curtis, 1957) and visualized by principal coordinate analysis (PCoA). The relative abundance of microbial communities was the average of 8 parallel samples. The ten most abundant phyla were selected to examine the shifts in the community composition. A t-test was employed to analyze the differences in the abundance of the top ten families.
The Pearson correlation analysis was further utilized to assess the relationship between the dominant (with the relative abundance in the top 15) bacteria/microeukaryotes and the environmental parameters. Canonical correlation analysis (CCA) was employed to determine the association between the microbial communities and the environmental factors. The function envfit in the R package vegan was used to evaluate the association of microbial community variation and each environmental variable in CCA. The heatmap package in R (v. 4.3.1) was used to create the Pearson correlation heatmaps.
To explore the associations among multiple species, the co-occurrence network analysis was conducted based on the Spearman correlation, and the result was visualized using the Gephi software (Barberán et al., 2014). The genera with a relative abundance of less than 0.05% in at least five samples were filtered out, which streamlined the network analysis and facilitated the identification of the key taxa. A valid co-occurrence event was defined as a robust correlation when the absolute value of the Spearman’s correlation coefficient (|r|) exceeded 0.8 and the statistical significance satisfied P < 0.01. To minimize the risk of false positive results, the P values underwent adjustment through multiple testing correction based on the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). All correlation analyses were conducted in R v. 4.0.3 using the vegan and igraph packages (Csardi and Nepusz, 2006). The topology of the constructed networks was characterized by calculating the average degree, density, average clustering coefficient, average path distance, and modularity (Newman, 2006).
After the removal of low-quality sequences, 1,771,106 bacterial sequences and 3,051,651 microeukaryotic sequences were obtained, which gave 47,277 ASVs for bacteria and 15,419 ASVs for microeukaryotes. According to the alpha diversity analysis of the 16S rRNA (Figure 2), the bacteria from CAP had the lowest Chao1, Shannon, and Simpson indexes. The Shannon and Simpson indexes of the bacteria were higher at EAP than at CAP (P < 0.001), but lower at PCA than at CSA (P < 0.001). The Simpson indices of the bacteria was significantly higher at EAP than at PCA (P < 0.01).
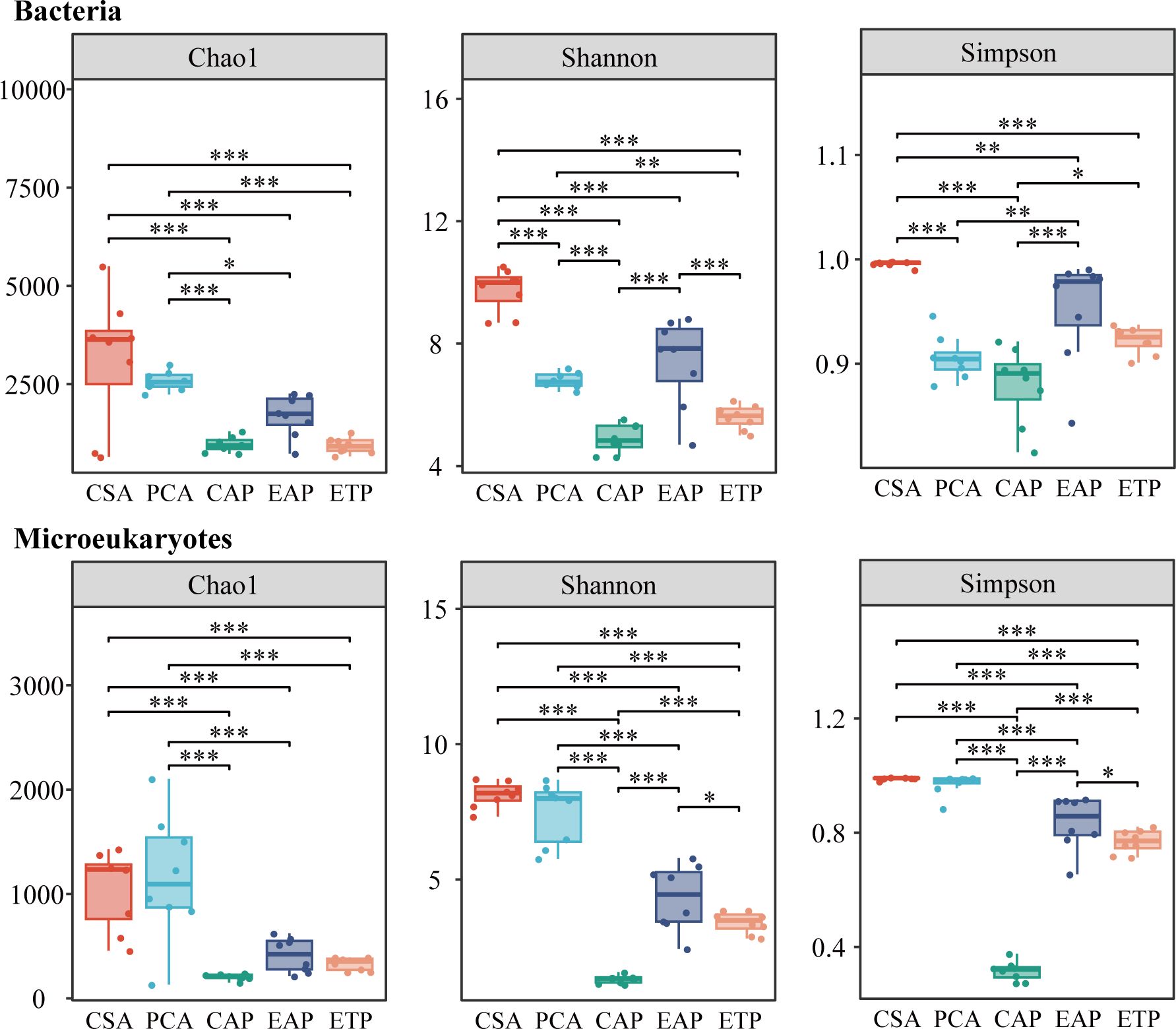
Figure 2. Alpha diversity of bacterial and microeukaryotic communities in the water samples. *P < 0.05, **P < 0.01, ***P < 0.001.
According to the alpha diversity analysis of the 18S rRNA (Figure 2), no significant difference was identified in the Chao1, Shannon, and Simpson indexes of the microeukaryotes between the two open water areas (PCA and CSA, P > 0.05). The indexes of the microeukaryotes were always higher at the open water area than at the respective aquaculture ponds (i.e., PCA vs EAP, CSA vs CAP). The effluent treatment reduced the diversity and abundance for both bacteria and microeukaryotes (i.e., ETP vs EAP).
According to PCoA, there were notable differences in the bacterial community composition among PCA, EAP, ETP, CSA, and CAP (Figure 3A). A total of 63 bacterial phyla were identified across the five groups, and Figure 3B gives the histogram of the relative abundances of the top 10 phyla. The predominant bacteria were Proteobacteria (53.81%), Actinobacteria (31.86%), Cyanobacteria (3.01%), Firmicutes (2.58%), and Acidobacteria (1.95%). The relative abundance of Proteobacteria was the highest in PCA, which was 17.30% higher than in CSA (P < 0.01). Proteobacteria had lower relative abundances in EAP and ETP than in PCA, CSA, and CAP. Actinobacteria had significantly higher relative abundances in CAP, EAP, and ETP than in PCA and CSA (P < 0.001). Firmicutes had higher relative abundance in PCA than in CSA. Cyanobacteria had the highest average relative abundance in EAP (8.85%). At the family level (Supplementary Figure 1), the dominant bacteria were Moraxellaceae (24.09%), Microbacteriaceae (19.44%), Rhodobacteraceae (17.11%), PeM15 (4.74%), and Ilumatobacteraceae (2.29%).
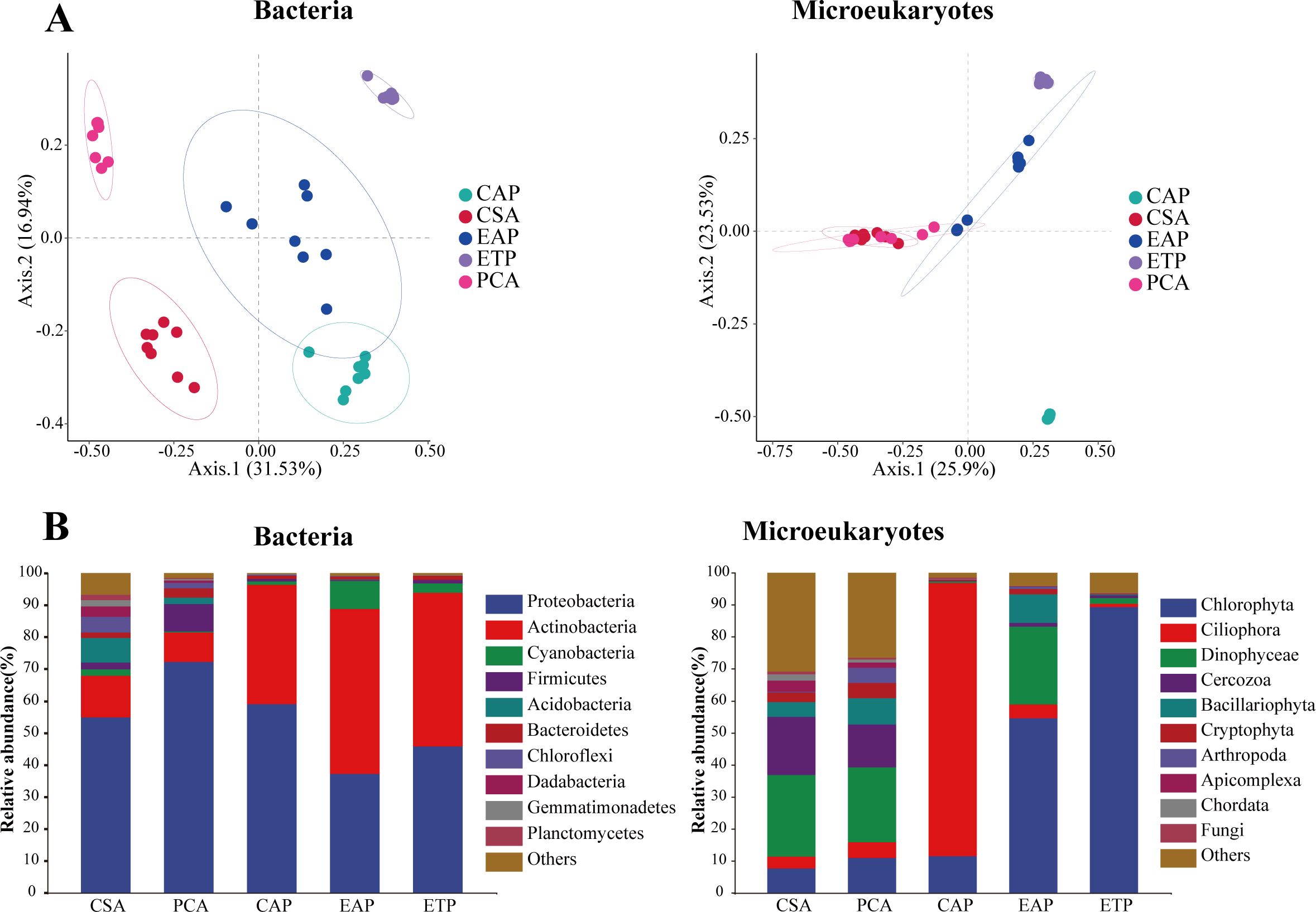
Figure 3. The structure and composition of microbial communities across different regions. (A) Distribution patterns of the bacterial and microeukaryotic communities in the water samples from PCoA based on the Bray–Curtis distance matrix. (B) Relative abundances of the dominant bacterial and microeukaryotic phyla.
In PCoA, the microeukaryotic communities of PCA and CSA clustered together, and both were close to EAP. Across the five groups, a total of 61 eukaryotic phyla were identified from the water samples. The dominant phyla included Chlorophyta (34.78%), Ciliophora (19.91%), Dinophyceae (15.07%), Cercozoa (6.75%), and Bacillariophyta (4.43%). The relative abundance of Chlorophyta was highest in ETP (89.22%), followed by EAP (54.51%), and lower in CSA, CAP, and PCA. Dinophyceae and Bacillariophyta had higher relative abundance in CSA (25.39%, 4.73%), PCA (23.45%, 8.15%), and EAP (24.30%, 8.97%) than in CAP (0.37%, 0.01%) and ETP (1.83%, 0.29%). Interestingly, Ciliophora had an exceedingly high relative abundance in CAP (85.38%) but lower relative abundance in all other groups. At the family level (Supplementary Figure 1), Chlorellaceae (19.85%) and Strombidiidae (16.90%) were the dominant taxa.
Figure 4A presents the contrast in the microbial community compositions between different groups, where the top 10 families of the bacteria and the microeukaryotes were examined for each pair (PCA/CSA, EAP/CAP, and EAP/ETP). Regarding bacteria, PCA had significantly higher relative abundances of Moraxellaceae, Rhodobacteraceae, Microbacteriaceae, and Planococcaceae compared to CSA (P < 0.01), while exhibiting significantly lower relative abundances of Dadabacteriales, Microtrichaceae, and Woeseiaceae (P < 0.01). Compared to CAP, EAP had significantly higher relative abundance of Microbacteriaceae (P < 0.001) and significantly lower relative abundance of Moraxellaceae (P < 0.001). Compared to EAP, ETP had significantly higher relative abundances of Rhodobacteraceae, PeM15, and Chloroplast (P < 0.001). Regarding microeukaryotes, PCA had significantly higher relative abundance of Pyramimonas (P < 0.05) and is otherwise similar to CSA. Compared to CAP, EAP had significantly higher relative abundances of Chlorellaceae and Dinophyceae (P < 0.05), but significantly lower relative abundance of Strombidiidae (P < 0.001). Compared to EAP, ETP had significantly higher relative abundance of Chlorellaceae (P < 0.001), but significantly lower relative abundance of Dinophyceae (P < 0.05).
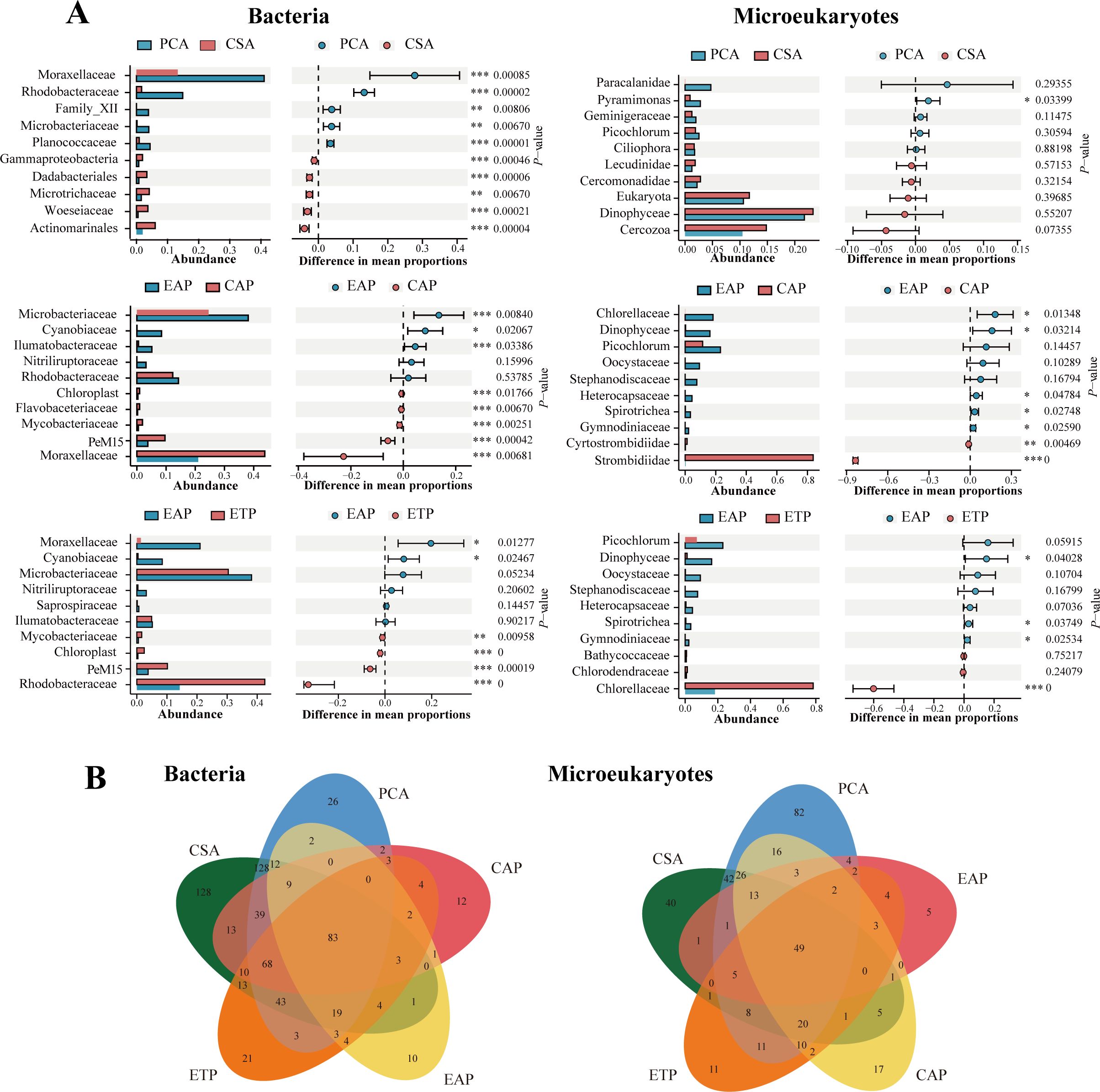
Figure 4. The microbial community differences at the level of families. (A) Differences in the dominant microbial families between groups. *P < 0.05, **P < 0.01, ***P < 0.001. (B) Shared microbial families among water samples from different groups.
Figure 4B illustrates the shared and unique families of the bacteria and the microeukaryotes of the five groups. A total of 39 bacterial families and 130 eukaryotic families were identified in PCA but not in CSA (Supplementary Table 1). Five bacterial families, i.e., Sphingobacteriales, Enterobacteriaceae, Corynebacteriales, Euzebyaceae, and Rhodospirillales, were present in both PCA and EAP, but not in CSA or CAP.
For each group, the co-occurrence network was constructed based on Spearman’s rank correlation coefficients to illustrate the relationships among the microbial communities (Figure 5; Supplementary Table 2). The edges and nodes in the network reflect the complexity of microbial interactions. Compared to the PCA network, the CSA network had more nodes and edges but lower clustering coefficient, graph density, average path length, and modularity. The PCA network also had a higher proportion of negative correlations (27.84%) than the CSA network (22.27%). Compared to the CAP and ETP networks, the EAP network had more nodes and edges, as well as higher clustering coefficient, graph density, average path length, and network diameter. However, the EAP network had a lower modularity than the ETP network. The proportion of negative correlations ranked as ETP (37.07%) > EAP (33.76%) > CAP (27.75%).
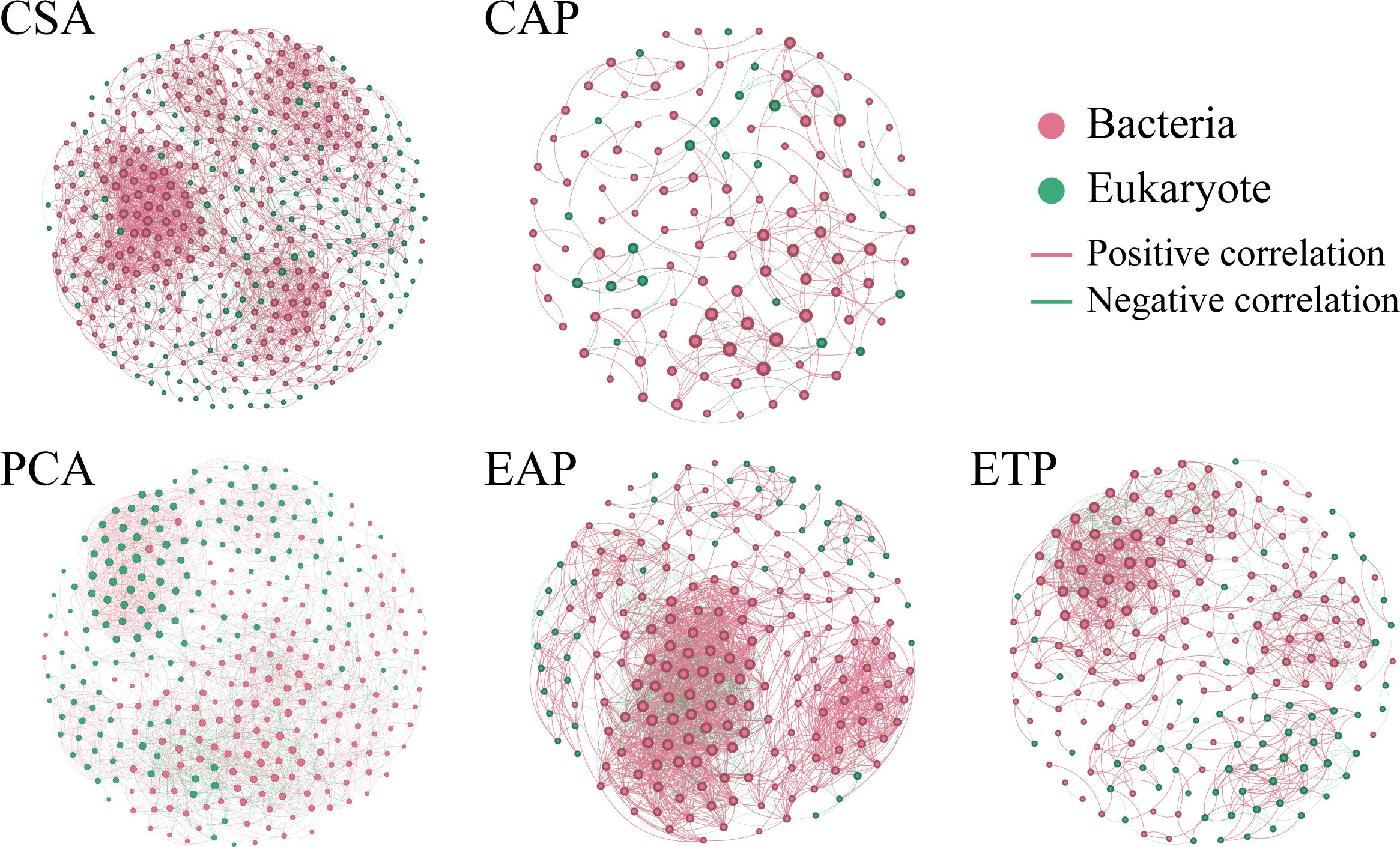
Figure 5. Co-occurrence networks of microbial communities constructed based on Spearman’s rank correlation coefficients. The edges and nodes in the network reflect the complexity of microbial interactions. Each connection signifies a robust (Spearman’s |q| > 0.8) and statistically significant (FDR-adjusted P < 0.01) correlation. The size of each node is proportional to the number of degrees. Networks containing both bacterial and microeukaryotic taxa, with red dots representing bacteria and green dots representing microeukaryotes.
Figure 6 summarizes the water quality indicators of the five groups. The temperature ranged from 9.2 to 13.7°C across the five groups, and the offshore aquaculture areas (CSA and PCA) were warmer than the ponds (CAP, EAP, ETP) (P < 0.01). The pH ranged from 7.99 to 8.31, and the highest pH was at PCA (8.29 ± 0.01). The open water areas had higher pH than the corresponding ponds (CSA vs CAP, PCA vs EAP and ETP) (P < 0.05), and EAP had higher pH than CAP (P < 0.05). The DO concentration varied significantly among the groups, down to a minimum of 10.29 ± 0.05 mg·L-1 at CAP and up to a maximum of 11.43 ± 0.12 mg·L-1 at ETP. EAP had a significantly higher DO concentration (11.38 ± 0.11 mg·L-1) than CAP (10.29 ± 0.05 mg·L-1) (P < 0.05), and PCA had a significantly higher DO concentration (10.83 ± 0.06 mg·L-1) than CSA (10.37 ± 0.07 mg·L-1) (P < 0.05). The open water areas (CSA and PCA) had significantly lower conductivity than the ponds (CAP, EAP, and ETP) (P < 0.05), but there was no significant difference in the conductivity between the offshore aquaculture areas or between the ponds (P > 0.05).
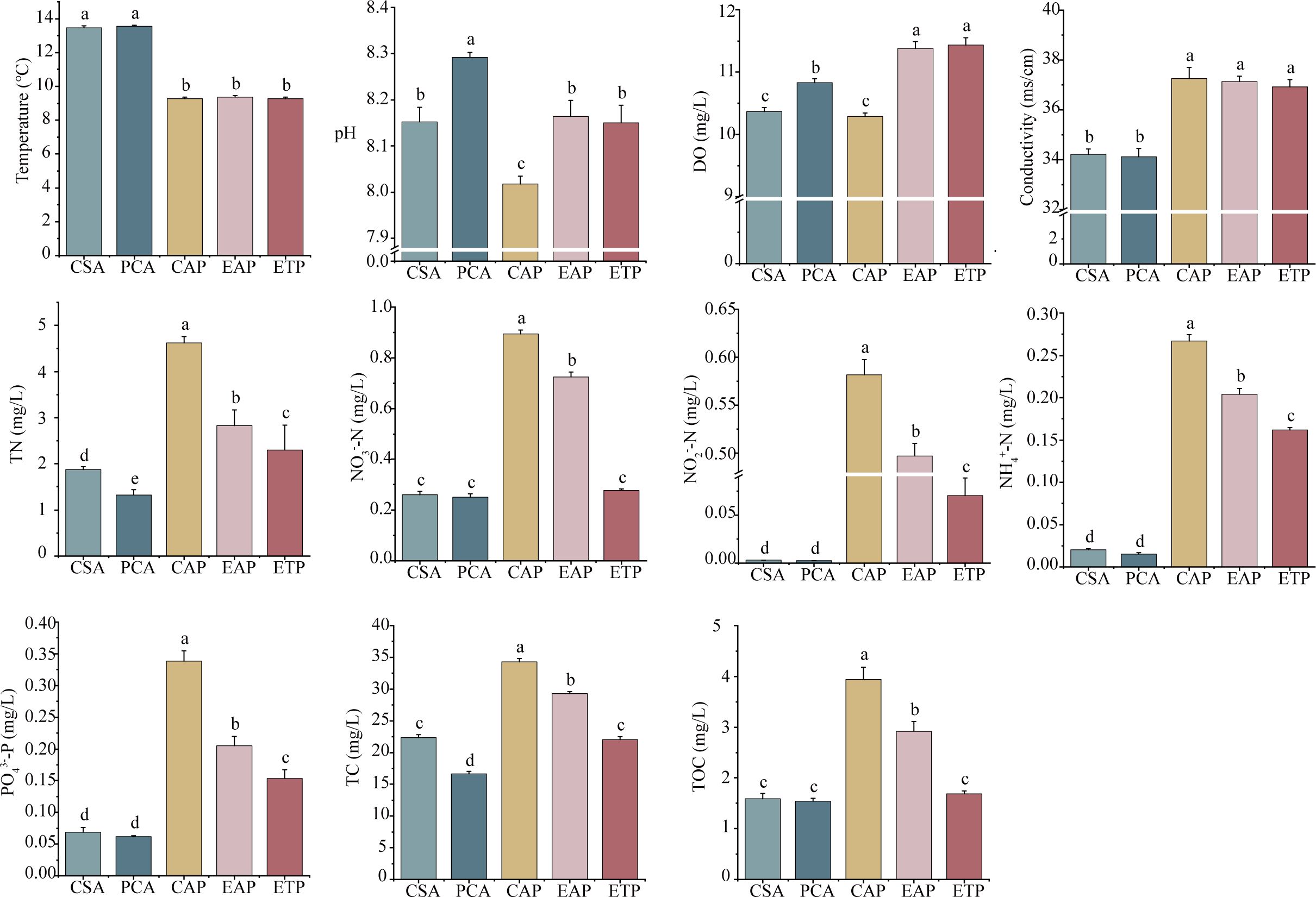
Figure 6. The water chemistry of different groups. The letters on the bars denote statistically significant differences (P < 0.05). DO, dissolved oxygen; TN, total nitrogen; NO-3-N, nitrate; NO-2-N, nitrite; NH4+-N, ammonium; PO3-4-P, phosphate; TC, total carbon; TOC, total organic carbon.
The concentrations of nutrients, including NH+ 4-N, NO-3-N, NO-2-N, PO3-4-P, TN, TC, and TOC, varied significantly among the five groups, but all followed the general pattern of CSA > PCA and CAP > EAP > ETP. The ponds typically had higher nutrient concentrations than the open water areas. For instance, the NO-2-N levels of PCA and CSA were 214.5 and 204.8 times lower than those of EAP and CAP (P < 0.01), and the NH+ 4-N levels of PCA and CSA were 13.7 and 13.4 times lower than those of EAP and CAP (P < 0.01), respectively. Compared to CSA, PCA had significantly lower TN and TC (P < 0.05). For the ponds, the contents of all nutrients fell in the order of CAP > EAP > ETP (P < 0.05). The ETP removed 61.60% and 85.90% of the NO-3-N and NO-2-N from EAP, respectively.
The environmental variables associated with the changes in the microbial communities were identified by CCA and envfit test (Figure 7; Supplementary Table 3). It was found that the bacterial communities were correlated strongly with temperature, conductivity, pH, PO3-4-P and NH+ 4-N (r > 0.5, P < 0.001), while the microeukaryotic communities were correlated strongly with all examined parameters (r > 0.6, P < 0.001). In comparison to the bacterial communities, the association between the microeukaryotic communities and environmental factors was stronger. In the Pearson correlation analysis, it was found that, for samples sourced from marine environments, variations in bacterial phyla exhibited positive correlations with TC, TN, NH+ 4-N, and NO-2-N, except for Bacteroidetes, Proteobacteria and Firmicutes, which were positively correlated with temperature, DO, and pH, but negatively with other factors. While the correlations between the microeukaryotic communities and environmental factors were relatively weak. In the case of pond samples, environmental factors, excluding temperature and conductivity, demonstrated significant correlations with certain bacterial phyla. In particular, the phyla Chloroflexi, Dadabacteria, Acidobacteria, Gemmatimonadetes, and Proteobacteria were positively correlated with TOC, TC, TN, NH+ 4-N, NO-3-N and NO-2-N (P < 0.01). On the other hand, the microeukaryotic phyla Ascomycota, Ciliophora, and Fungi displayed positive correlations with environmental factors excluding DO, pH, conductivity, and temperature (P < 0.001). In contrast, Chlorophyta and Dinophyceae showed an opposing trend.
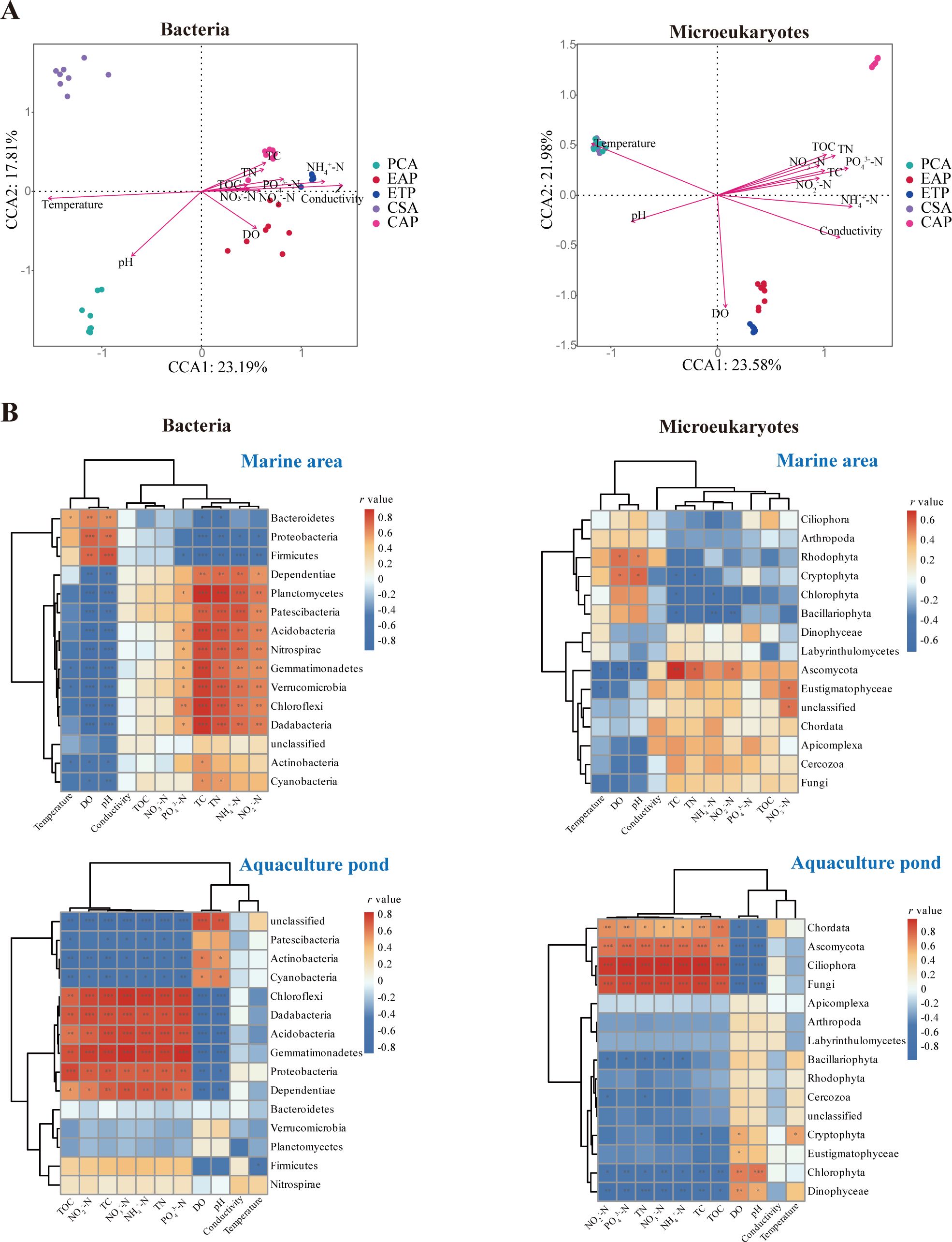
Figure 7. The relationship between environmental factors and microbial communities. (A) Canonical correlation analysis of the distribution of microbial communities in relation to environmental variables. (B) Heatmap of the Spearman correlation between environmental factors and the dominant microbial phyla (r > 0 indicates a positive correlation and r < 0 indicates a negative correlation). *P < 0.05, **P < 0.01, ***P < 0.001.
The pH is a key indicator of water quality, and it is governed by the amount of carbon dioxide (Howland et al., 2000). The seaweed can consume carbon dioxide via photosynthesis, which promotes the decomposition of HCO3− in the water and increases the pH value. Indeed, the cultivation of P. haitanensis significantly increased the pH of the water in the offshore aquaculture area (PCA vs CSA). The water of EAP came from PCA, and the pH was significantly higher at EAP than at CAP, although EAP had lower pH than PCA due to the respiration of aquatic animals and the decomposition of organic matters.
The DO level is an indicator of aquatic growth conditions and pollution status (Zang et al., 2011). The DO at PCA was higher than those at CSA, largely due to the photosynthetic oxygen production by P. haitanensis, which served as a major oxygen input. These results align with previous studies that reported higher DO levels in regions with seaweed cultivation compared to regions without (Huang et al., 2017; Liu et al., 2019; Xie et al., 2017; Yang et al., 2015). Similarly, EAP exhibited higher DO levels than CAP, as the water source significantly impacted water quality in the aquaculture ponds, even when farming the same animals at the same density. The higher DO levels in ETP compared to EAP could be attributed to the photosynthetic oxygen production by mangroves in ETP.
In terms of nutrients, the various N and P indices were consistently lower in PCA compared to CSA, with PCA showing reductions of 25.4% in TC and 29.6% in TN relative to CSA. Recent studies have indicated that mariculture of seaweeds (e.g., P. haitanensis, Gracilaria lemaneiformis, and Saccharina japonica) can improve water quality by removing NO-3-N and PO3-4-P and increasing DO (Xiao et al., 2021; Xu et al., 2022). Both EAP and CAP had higher nutrient concentrations (NH+ 4-N, NO-3-N, NO-2-N, PO3-4-P) than their corresponding seawater sources (PCA and CSA), possibly due to the aquaculture processes in the ponds, which produce substantial amounts of nutrients that contribute to eutrophication. However, nutrient enrichment was significantly lower in EAP compared to CAP, suggesting that the source of aquaculture water affected the quality of pond water. Various factors may facilitate the removal of nutrients generated in aquaculture ponds, thereby indirectly enhancing water quality.
Microorganisms are mediators that link organisms with ecosystems (Bahram and Netherway, 2022), and microbial communities are shaped by strong selective forces arising from their environments (Reis et al., 2009; Yakimov et al., 2006). Many research works have examined the responses of microbial communities to environmental alterations and the potential bioremediation of contaminated environments in mariculture areas. In this work, we explored the impact of oceanic seaweed cultivation on the microbial communities in land-based aquaculture ponds and their potential roles in water quality improvement.
The bacterial communities in PCA and CSA were discerned unambiguously by PCoA, and their Shannon and Simpson indexes were lower at PCA than at CSA. The lower diversity of the bacterial communities at PCA may be due to the impacts of the seaweed cultivation on the environmental factors. Large algae release substantial amounts of organic matters during their growth (Croft et al., 2005), which can be absorbed and utilized by the surrounding epiphytic bacteria, potentially inhibiting and shaping specific microbial communities. The dominant bacterial phylum in the five groups was Proteobacteria, followed by Actinobacteria, Cyanobacteria, and Firmicutes. Research has shown that Proteobacteria, Cyanobacteria, and Firmicutes are generally the most abundant bacterial communities associated with seaweeds (Ahmed et al., 2021; Selvarajan et al., 2019), while Actinobacteria are dominant on the surface of the brown alga Laminaria digitata (Singh and Reddy, 2014). The carbon-rich constituents in the cell wall of macroalgae (e.g., agar in P. haitanensis) may facilitate bacterial proliferation, as the algal polysaccharides are potential sources of carbon and energy for marine bacteria (Goecke et al., 2010; Hehemann et al., 2012). Specific enzymatic activities have been detected in marine bacteria capable of degrading the cell wall of macroalgae (Egan et al., 2013). For example, Proteobacteria are known to digest galactan sulfates in the cell wall of red algae (Miranda et al., 2013), and marine Rhodobacteraceae, which were also highly abundant in PCA, generally function in the desulfonation of fucoidan (Dogs et al., 2017). These factors may explain why PCA exhibited decreased bacterial diversity compared to CSA, despite showing no significant changes in bacterial relative abundance.
In this context, we observed a robust correlation between the bacterial community and factors such as temperature, pH, PO3-4-P, as well as NH+ 4-N (Figure 7). Previous research has indicated that the variability of the bacterial communities related to Neopyropia yezoensis and seawater is predominantly shaped by nitrate (NO3−), ammonium (NH4+), and temperature (Ahmed et al., 2021). In this study, Proteobacteria in seawater showed a positive correlation with pH and a negative correlation with TN, NH+ 4-N, and NO-2-N. Proteobacteria typically play a role in processes such as sulfate reduction, sulfur oxidation, and nitrate assimilation. The oxidation of sulfides and the conversion of organic carbon in the seawater can foster anaerobic conditions to trigger denitrification processes that convert NO3− to N2, NO, NO2, and NH4+. Moreover, PCA had more Moraxellaceae than CSA. Moraxellaceae, which belong to Pseudomonadales and are used as a health indicator, have been suggested by Deng et al. (2021) to play a role in removing nitrogen and phosphorus by denitrification and phosphate solubilization. This may explain why PCA had lower TN concentration and the highest pH, suggesting that the seaweed not only absorb nitrogen directly but also reduce nitrogen concentration by regulating microbial communities.
Contrary to the findings of Wang et al. (2020) in Fujian, our study revealed that the cultivation of P. haitanensis had no significant impact on the relative abundance and diversity of microeukaryotes in seawater, regardless of the presence or absence of P. haitanensis. However, the relative abundance of Dinophyceae and Bacillariophyta appeared to increase in the open water areas (PCA and CSA) and at EAP. One possible explanation for this observation is that Typhoon Muifa, which occurred in September 2022, may have triggered a microalgal bloom in the region where this study was conducted, as typhoons that occur in summer are known to sometimes cause red tides in coastal bays (Zhang et al., 2024).
The microbial communities in the aquaculture ponds were influenced by the introduction of seawater sourced from the seaweed cultivation area. Following the introduction of seawater into the ponds for seven days, notable changes were observed in various water quality parameters, including nutrients and pH. These alterations indicated a decline in water quality during the aquaculture process, which subsequently affected the composition of microorganisms. The Chao1 and Simpson indices for both bacteria and microeukaryotes showed significant differences between EAP and PCA, with distinct clustering evident in PCoA. At the phylum level, the water from the ponds had significantly higher relative abundance of Actinobacteria and Cyanobacteria compared to the samples from the sea. These phyla, along with Cyanobacteria, are commonly dominant in shrimp, crab, and fish aquaculture ponds (Shen et al., 2020). Both Actinobacteria and Cyanobacteria are known to have positive correlations with nutrient levels and negative correlations with pH (Zhang et al., 2022). The high relative abundance of Cyanobacteria may serve as an indicator of increased nutrient availability (Thacker and Paul, 2001). Aquaculture ponds typically exhibit low pH and high nutrient concentrations due to the metabolic waste produced during aquaculture (Dauda et al., 2019). Consequently, both Actinobacteria and Cyanobacteria thrived in the ponds more effectively than in the open sea, leading to their higher relative abundance in this environment.
CAP had a strikingly high relative abundance of Ciliophora. This group showed a significant positive correlation with nutrient levels and a significant negative correlation with pH and DO. Notably, CAP had the highest nutrient levels and the lowest pH and DO among the five groups. While there was no significant difference in the Chao1 index for bacteria or microeukaryotes between EAP and CAP, the Shannon and Simpson indices were consistently higher in EAP (P < 0.001). At the phylum level, Actinobacteria and Cyanobacteria exhibited relatively high relative abundance in the EAP. Correlation analysis of pond samples indicated a negative correlation between these phyla and nutrients in the samples of pond. Wang et al. (2021) also found that Actinobacteria have significant negative correlations with TN, and both Cyanobacteria and Actinobacteria are negatively correlated with TOC. At the family level, EAP had a relatively high relative abundance of Microbacteriaceae, the bacteria known for their ability to degrade waste in soil (Jacques et al., 2007, 2008). The bacterial families identified in both EAP and PCA but not in CAP or CSA included Sphingobacteriales, Enterobacteriaceae, Corynebacteriales, Euzebyaceae, and Rhodospirillales. These bacteria in EAP originated from the seawater exposed to seaweed cultivation and remained present in the ponds after aquaculture for seven days. Sphingobacteriales are predominantly marine-associated and can hydrolyze and utilize complex carbon sources. The members of Sphingobacteriales play a crucial role in breaking down complex organic compounds in various environments, a function that has been demonstrated in the activated sludge process in wastewater treatment (Dennis et al., 2013; Shen et al., 2017). Certain members of Corynebacteriales (Boada et al., 2021; Nahar, 2020) and Rhodospirillales (Grevesse et al., 2022; Sosa et al., 2019) share similar functions, potentially participating in the decomposition of organic matters and contributing to nutrient cycling in aquatic environments, and they are promising bacteria in wastewater treatment processes. In an aquaculture setting, some members of Rhodospirillales are photosynthetic bacteria capable of converting carbon dioxide into organic compounds (Prabu and Santhiya, 2016), and this ability may contribute to the primary production and influence the overall nutrient dynamics in aquaculture systems. In addition, certain members of Rhodospirillales may establish symbiotic relationships with specific aquatic species, which affects the overall health and balance of the aquaculture environment (Degli Esposti et al., 2019).
The co-occurrence networks reflect the potential interactions within the microbiome (Jiao et al., 2021). The average degree of edges and nodes indicates the extent of connection and the closeness between microbial interactions, and the modularity shows the resistance of the system to environmental coercion (Trivedi et al., 2020). The cultivation of P. haitanensis had a positive effect on the complexity of the microbial network, as PCA and EAP had a more complex network than CSA and CAP, respectively. A short average path length of a network is related to the rapid response of the ecosystem to disturbance (Watts and Strogatz, 1998), and negative correlations are essential to increasing the stability of the microbial communities (Deng et al., 2022). Indeed, PCA and EAP had shorter average path length, higher modularity, and a greater number of negative links than CSA and CAP, respectively. Thus, our results reflected the relative stability of the PCA microbial system and suggested that the cultivation of P. haitanensis improved the resilience of the ecosystem. The introduction of seawater from PCA into EAP as aquaculture water also helped to stabilize the microbial ecosystem. In addition, the network parameters of ETP were better than those of EAP. Hence, the oysters and mangrove improved the ecosystem of microbial communities in ETP.
Large-scale oceanic seaweed cultivation often coexists with intertidal mudflat pond aquaculture along the coast of the East China Sea, and they form an interconnected system. This work assessed the impact of intertidal P. haitanensis cultivation on the adjacent mudflat aquaculture ponds. Seaweed cultivation directly improved the water quality and the microbial communities in the open water area, and indirectly improved the ecosystem in the aquaculture ponds that received the water supply from the sea. The ponds next to seaweed farming had higher microbial diversity and preserved functional bacteria. With mangroves and oysters, the treatment ponds could effectively purify the effluent from the aquaculture ponds. The findings provide empirical evidence of the positive effects of P. haitanensis cultivation on the microbial community in seawater, and they contribute to the knowledge of the indirect impacts of seaweed cultivation on the aquaculture in mudflat ponds.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
PL: Data curation, Investigation, Visualization, Writing – original draft. QL: Funding acquisition, Writing – review & editing. SZ: Writing – review & editing, Methodology. JC: Writing – review & editing, Data curation. RY: Writing – review & editing, Resources. TN: Writing – review & editing, Data curation. TW: Resources, Writing – review & editing. PZ: Resources, Writing – review & editing. HC: Funding acquisition, Conceptualization, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Major Scientific and Technological Project of Ningbo (No. 2021Z004, 2021Z103), NSFC (32373099), Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural (Aquaculture) Varieties (No. 2021C02069-9), China Agriculture Research System of MOF and MARA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2025.1442210/full#supplementary-material
Ahmed A., Khurshid A., Tang X., Wang J., Khan T. U., Mao Y. (2021). Structural and functional impacts of microbiota on Pyropia yezoensis and surrounding seawater in cultivation farms along coastal areas of the Yellow Sea. Microorganisms 9, 1291. doi: 10.3390/microorganisms9061291
Alleway H. K. (2023). Climate benefits of seaweed farming. Nat. Sustain. 6, 356–357. doi: 10.1038/s41893-022-01044-x
Aqsiq, P (2007). The specification for marine monitoring of China-Part 4: seawater analysis (GB 17378.4-2007) (Beijing, China: General Administration of Quality Supervision, Inspection Quarantine of People’s Republic of China).
Ashkenazi D. Y., Israel A., Abelson A. (2019). A novel two-stage seaweed integrated multi-trophic aquaculture. Rev. Aquac. 11, 246–262. doi: 10.1111/raq.2019.11.issue-1
Bahram M., Netherway T. (2022). Fungi as mediators linking organisms and ecosystems. FEMS Microbiol. Rev. 46, fuab058. doi: 10.1093/femsre/fuab058
Barberán A., Bates S. T., Casamayor E. O., Fierer N. (2014). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 8, 952. doi: 10.1038/ismej.2013.236
Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc Ser. B-Stat. Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Boada E., Santos-Clotas E., Cabrera-Codony A., Martín M. J., Bañeras L., Gich F. (2021). The core microbiome is responsible for volatile silicon and organic compounds degradation during anoxic lab scale biotrickling filter performance. Sci. Total Environ. 798, 149162. doi: 10.1016/j.scitotenv.2021.149162
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bray J. R., Curtis J. T. (1957). An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27, 326–349. doi: 10.2307/1942268
Buschmann A. H., Camus C., Infante J., Neori A., Israel Á., Hernández-González M. C., et al. (2017). Seaweed production: overview of the global state of exploitation, farming and emerging research activity. Eur. J. Phycol. 52, 391–406. doi: 10.1080/09670262.2017.1365175
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
China Fishery Bureau (2023). China fishery statistical yearbook. Fishery production (Chinese Agriculture Express).
Chopin T., Cooper J. A., Reid G., Cross S., Moore C. (2012). Open-water integrated multi-trophic aquaculture: environmental biomitigation and economic diversification of fed aquaculture by extractive aquaculture. Rev. Aquac. 4, 209–220. doi: 10.1111/j.1753-5131.2012.01074.x
Chopin T., Yarish C. (1999). Aquaculture does not only mean finfish monoculture: seaweeds must be a significant component for an integrated ecosystem approach. Bull. Aquac. Assoc. Canada 98, 15–21.
Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438, 90–93. doi: 10.1038/nature04056
Csardi G., Nepusz T. (2006). The igraph software. Complex Sys. 1695, 1–9. Available at: https://igraph.org.
Dauda A. B., Ajadi A., Tola-Fabunmi A. S., Akinwole A. O. (2019). Waste production in aquaculture: Sources, components and managements in different culture systems. Aquac. Fish. 4, 81–88. doi: 10.1016/j.aaf.2018.10.002
Degli Esposti M., Lozano L., Martínez-Romero E. (2019). Current phylogeny of Rhodospirillaceae: A multi-approach study. Mol. Phylogenet. Evol. 139, 106546. doi: 10.1016/j.ympev.2019.106546
Deng Y., Debognies A., Zhang Q., Zhang Z., Zhou Z., Zhang J., et al. (2022). Effects of ofloxacin on the structure and function of freshwater microbial communities. Aquat. Toxicol. 244, 106084. doi: 10.1016/j.aquatox.2022.106084
Deng Y., Mao C., Chen H., Wang B., Cheng C., Ma H., et al. (2021). Shifts in pond water bacterial communities are associated with the health status of sea bass (Lateolabrax maculatus). Ecol. Indic. 127, 107775. doi: 10.1016/j.ecolind.2021.107775
Dennis P. G., Seymour J., Kumbun K., Tyson G. W. (2013). Diverse populations of lake water bacteria exhibit chemotaxis towards inorganic nutrients. ISME J. 7, 1661–1664. doi: 10.1038/ismej.2013.47
Dogs M., Wemheuer B., Wolter L., Bergen N., Daniel R., Simon M., et al. (2017). Rhodobacteraceae on the marine brown alga Fucus spiralis are abundant and show physiological adaptation to an epiphytic lifestyle. Syst. Appl. Microbiol. 40, 370–382. doi: 10.1016/j.syapm.2017.05.006
Duarte C. M., Bruhn A., Krause-Jensen D. (2022). A seaweed aquaculture imperative to meet global sustainability targets. Nat. Sustain. 5, 185–193. doi: 10.1038/s41893-021-00773-9
Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. (2013). The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiol. Rev. 37, 462–476. doi: 10.1111/1574-6976.12011
Goecke F., Labes A., Wiese J., Imhoff J. F. (2010). Chemical interactions between marine macroalgae and bacteria. Mar. Ecol.-Prog. Ser. 409, 267–299. doi: 10.3354/meps08607
Grevesse T., Guéguen C., Onana V. E., Walsh D. A. (2022). Degradation pathways for organic matter of terrestrial origin are widespread and expressed in Arctic Ocean microbiomes. Microbiome 10, 237. doi: 10.1186/s40168-022-01417-6
Hehemann J. H., Correc G., Thomas F., Bernard T., Barbeyron T., Jam M., et al. (2012). Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J. Biol. Chem. 287, 30571–30584. doi: 10.1074/jbc.M112.377184
Hou Y., Liu Y., Zhang J., Yu X. (2023). Temporal dynamics of lateral carbon export from an onshore aquaculture farm. Sci. Total Environ. 859, 160258. doi: 10.1016/j.scitotenv.2022.160258
Howland R., Tappin A., Uncles R., Plummer D., Bloomer N. (2000). Distributions and seasonal variability of pH and alkalinity in the Tweed Estuary, UK. Sci. Total Environ. 251, 125–138. doi: 10.1016/S0048-9697(00)00406-X
Huang Y., Ou L., Yang Y. (2017). Nutrient competition between macroalgae Gracilaria lemaneiformis and phytoplankton in coastal waters of Nan’Ao Island, Guangdong. Oceanol. Limnol. Sin. 48, 806–813. doi: 10.11693/hyhz20170200031
Jacques R. J., Okeke B. C., Bento F. M., Peralba M. C., Camargo F. A. (2007). Characterization of a polycyclic aromatic hydrocarbon–degrading microbial consortium from a petrochemical sludge landfarming site. Bioremediat. J. 11, 1–11. doi: 10.1080/10889860601185822
Jacques R. J., Okeke B. C., Bento F. M., Teixeira A. S., Peralba M. C., Camargo F. A. (2008). Microbial consortium bioaugmentation of a polycyclic aromatic hydrocarbons contaminated soil. Bioresour. Technol. 99, 2637–2643. doi: 10.1016/j.biortech.2007.04.047
Jiao Y., Yuan Q., Wang W., Yan L., Mu X., Li H., et al. (2021). Vallisnerian natans tolerance and response of microbial community in wetlands to excess nutrients loading. Ecol. Indic. 131, 108179. doi: 10.1016/j.ecolind.2021.108179
Kang Y. H., Kim S., Choi S. K., Lee H. J., Chung I. K., Park S. R. (2021). A comparison of the bioremediation potential of five seaweed species in an integrated fish-seaweed aquaculture system: implication for a multi-species seaweed culture. Rev. Aquac. 13, 353–364. doi: 10.1111/raq.12478
Liu Z., Luo H., Wu Y., Ren H., Yang Y. (2019). Large-scale cultivation of Gracilaria lemaneiformis in Nan’ao Island of Shantou and its effects on the aquatic environment and phytoplankton. J. Fish. Sci. China 26, 99–107. doi: 10.3724/SP.J.1118.2019.18373
Liu Y., Wang Z., Yang X., Wang S. Q., Liu X. L., Liu B., et al. (2023). Changes in mariculture and offshore seawater quality in China during the past 20 years. Ecol. Indic. 157, 111220. doi: 10.1016/j.ecolind.2023.111220
Long X. H., Liu L. P., Shao T. Y., Shao H. B., Liu Z. P. (2016). Developing and sustainably utilize the coastal mudflat areas in China. Sci. Total Environ. 569, 1077–1086. doi: 10.1016/j.scitotenv.2016.06.170
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMB J. 17, 10–12. doi: 10.14806/ej.17.1.200
Miranda L. N., Hutchison K., Grossman A. R., Brawley S. H. (2013). Diversity and abundance of the bacterial community of the red macroalga Porphyra umbilicalis: did bacterial farmers produce macroalgae? PloS One 8, e58269. doi: 10.1371/journal.pone.0058269
Nahar A. (2020). Characterisation and insight into the metabolism of oleaginous sub-Antarctic Corynebacteriales through genomic, lipidomic and proteomic analysis. [dissertation/doctor’s thesis]. University of Tasmania, Australia.
Neori A., Chopin T., Troell M., Buschmann A. H., Kraemer G. P., Halling C., et al. (2004). Integrated aquaculture: rationale, evolution and state of the art emphasizing seaweed biofiltration in modern mariculture. Aquaculture 231, 361–391. doi: 10.1016/j.aquaculture.2003.11.015
Newman M. E. (2006). Modularity and community structure in networks. Proc. Natl. Acad. Sci. U. S. A. 103, 8577–8582. doi: 10.1073/pnas.0601602103
Prabu E., Santhiya A. A. V. (2016). An overview of bioremediation towards aquaculture. J. Aquac. Trop. 31, 155.
Reid G. K., Lefebvre S., Filgueira R., Robinson S. M., Broch O. J., Dumas A., et al. (2020). Performance measures and models for open-water integrated multi-trophic aquaculture. Rev. Aquac. 12, 47–75. doi: 10.1111/raq.12304
Reis A., Araújo S. Jr., Moura R., Francini-Filho R., Pappas G. Jr., Coelho A., et al. (2009). Bacterial diversity associated with the Brazilian endemic reef coral Mussismilia Braziliensis. J. Appl. Microbiol. 106, 1378–1387. doi: 10.1111/j.1365-2672.2008.04106.x
Seghetta M., Tørring D., Bruhn A., Thomsen M. (2016). Bioextraction potential of seaweed in Denmark—An instrument for circular nutrient management. Sci. Total Environ. 563, 513–529. doi: 10.1016/j.scitotenv.2016.04.010
Selvarajan R., Sibanda T., Venkatachalam S., Ogola H. J., Christopher Obieze C., Msagati T. A. (2019). Distribution, interaction and functional profiles of epiphytic bacterial communities from the rocky intertidal seaweeds, South Africa. Sci. Rep. 9, 19835. doi: 10.1038/s41598-019-56269-2
Shen L., Liu Y., Xu B., Wang N., Zhao H., Liu X., et al. (2017). Comparative genomic analysis reveals the environmental impacts on two Arcticibacter strains including sixteen Sphingobacteriaceae species. Sci. Rep. 7, 2055. doi: 10.1038/s41598-017-02191-4
Shen X., Xu M., Li M., Zhao Y., Shao X. (2020). Response of sediment bacterial communities to the drainage of wastewater from aquaculture ponds in different seasons. Sci. Total Environ. 717, 137180. doi: 10.1016/j.scitotenv.2020.137180
Singh R. P., Reddy C. (2014). Seaweed–microbial interactions: key functions of seaweed-associated bacteria. FEMS Microbiol. Ecol. 88, 213–230. doi: 10.1111/fem.2014.88.issue-2
Sosa O. A., Repeta D. J., DeLong E. F., Ashkezari M. D., Karl D. M. (2019). Phosphate-limited ocean regions select for bacterial populations enriched in the carbon–phosphorus lyase pathway for phosphonate degradation. Environ. Microbiol. 21, 2402–2414. doi: 10.1111/emi.2019.21.issue-7
Thacker R. W., Paul V. J. (2001). Are benthic cyanobacteria indicators of nutrient enrichment? Relationships between cyanobacterial abundance and environmental factors on the reef flats of Guam. Bull. Mar. Sci. 69, 497–508.
Trivedi P., Leach J. E., Tringe S. G., Sa T., Singh B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Wang C., Wang Y., Liu P., Sun Y., Song Z., Hu X. (2021). Characteristics of bacterial community structure and function associated with nutrients and heavy metals in coastal aquaculture area. Environ. pollut. 275, 116639. doi: 10.1016/j.envpol.2021.116639
Wang W., Wu L., Xu K., Xu Y., Ji D., Chen C., et al. (2020). The cultivation of Pyropia haitanensis has important impacts on the seawater microbial community. J. Appl. Phycol. 32, 2561–2573. doi: 10.1007/s10811-020-02068-6
Watts D. J., Strogatz S. H. (1998). Collective dynamics of ‘small-world’networks. Nature 393, 440–442. doi: 10.1038/30918
Xiao X., Agusti S., Lin F., Li K., Pan Y., Yu Y., et al. (2017). Nutrient removal from Chinese coastal waters by large-scale seaweed aquaculture. Sci. Rep. 7, 46613. doi: 10.1038/srep46613
Xiao X., Agustí S., Yu Y., Huang Y., Chen W., Hu J., et al. (2021). Seaweed farms provide refugia from ocean acidification. Sci. Total Environ. 776, 145192. doi: 10.1016/j.scitotenv.2021.145192
Xie X., He Z., Hu X., Yin H., Liu X., Yang Y. (2017). Large-scale seaweed cultivation diverges water and sediment microbial communities in the coast of Nan’ao Island, South China Sea. Sci. Total Environ. 598, 97–108. doi: 10.1016/j.scitotenv.2017.03.233
Xu N., Wang W., Xu K., Xu Y., Ji D., Chen C., et al. (2022). Cultivation of different seaweed species and seasonal changes cause divergence of the microbial community in coastal seawaters. Front. Microbiol. 13, 988743. doi: 10.3389/fmicb.2022.988743
Yakimov M. M., Cappello S., Crisafi E., Tursi A., Savini A., Corselli C., et al. (2006). Phylogenetic survey of metabolically active microbial communities associated with the deep-sea coral Lophelia pertusa from the Apulian plateau, Central Mediterranean Sea. Deep-Sea Res. Part I-Oceanogr. Res. Pap. 53, 62–75. doi: 10.1016/j.dsr.2005.07.005
Yang Y., Chai Z., Wang Q., Chen W., He Z., Jiang S. (2015). Cultivation of seaweed Gracilaria in Chinese coastal waters and its contribution to environmental improvements. Algal. Res. 9, 236–244. doi: 10.1016/j.algal.2015.03.017
Zang C., Huang S., Wu M., Du S., Scholz M., Gao F., et al. (2011). Comparison of relationships between pH, dissolved oxygen and chlorophyll a for aquaculture and non-aquaculture waters. Water Air Soil pollut. 219, 157–174. doi: 10.1007/s11270-010-0695-3
Zhang P., Long H., Li Z., Chen R., Peng D., Zhang J. (2024). Effects of typhoon events on coastal hydrology, nutrients, and algal bloom dynamics: Insights from continuous observation and machine learning in semi-enclosed Zhanjiang Bay, China. Sci. Total Environ. 924, 171676. doi: 10.1016/j.scitotenv.2024.171676
Keywords: intertidal mudflat aquaculture, Pyropia haitanensis, seaweed cultivation, microbial communities, water quality
Citation: Liu P, Luo Q, Zhu S, Chen J, Yang R, Niu T, Wang T, Zhang P and Chen H (2025) The extensive cultivation of Pyropia haitanensis along the coastal areas influences the ecological dynamics of nearby intertidal mudflat pond water. Front. Mar. Sci. 12:1442210. doi: 10.3389/fmars.2025.1442210
Received: 01 June 2024; Accepted: 15 January 2025;
Published: 31 January 2025.
Edited by:
Jesus L. Romalde, University of Santiago de Compostela, SpainReviewed by:
Ajaya Kumar Rout, Rani Lakshmi Bai Central Agricultural University, IndiaCopyright © 2025 Liu, Luo, Zhu, Chen, Yang, Niu, Wang, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haimin Chen, Y2hlbmhhaW1pbkBuYnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.