- 1Fisheries College, Guangdong Ocean University, Guangdong Research Center on Reproductive Control and Breeding Technology of Indigenous Valuable Fish Species, Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Zhanjiang, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang, China
- 3Baise Agriculture and Rural Afairs Bureau, Baise, Guangxi, China
- 4Guangdong Evergreen Feed Industry Co., Ltd., Zhanjiang, Guangdong, China
- 5Guangdong Havwii Agriculture Group Co., Ltd, Zhanjiang, China
The redclaw crayfish is sexually dimorphic, with males preferring females in terms of growth rate and size. Currently, the molecular mechanism of sexual dimorphism is not well understood in crustaceans, and studies on the joint analysis of the gonadal metabolome and transcriptome of male and female individuals of redclaw crayfish are limited. In this study, we found that there were differences in relevant metabolites and metabolic pathways between intersex and normal individuals, including metabolites such as testosterone glucuronide, cortisol, pregnenolone sulfate, and estrone glucuronide, as well as steroid hormone synthesis pathways, lipid metabolism pathways, amino acid metabolism, and other metabolic pathways. The joint analysis of metabolome and transcriptome identified differential genes such as cyp3a40, cyp18a1, hsd17b12b, and cyp19a, and co-enriched pathways such as insulin signaling pathway, TGF-β signaling pathway, and FoxO signaling pathway, indicating that they may play a significant role in sex differentiation of redclaw crayfish. Our results provide a reference for the molecular mechanism of gonadal development between male and female individuals of redclaw crayfish.
1 Introduction
The redclaw crayfish (Cherax quadricarinatus), generally known as the Australian red crayfish, is one of the most economically valuable freshwater crustaceans in the world (Ghanawi and Saoud, 2012; Zheng et al., 2019). Originally from the ephemeral catchments of the Gulf of Carpentaria in northern Australia, this species is more easily adapted to freshwater than marine environments (Nguyen et al., 2016; Zheng et al., 2020). With the ability to survive in temperatures ranging 10°C–30°C and salinities of 0.3 ppt–20 ppt, the redclaw crayfish is considered a promising and attractive species for freshwater aquaculture (Garcı́a-Guerrero et al., 2003). The aquaculture of redclaw crayfish is steadily growing into a successful industry in several tropical and subtropical countries worldwide (Rodgers et al., 2006). In China, the species was initially introduced in the early 1990s and has the potential to become a significant agricultural commodity.
In giant freshwater prawn (Macrobrachium rosenbergii) and redclaw crayfish, male individuals are superior to female individuals in terms of size and growth rate. Intersexuality appears to occur frequently in gonochoristic crustaceans and has been reported in several crayfish species (Parnes et al., 2003). It has been observed that intersexual individuals are present in redclaw crayfish, which may have both female and male genitalia and gonads that may contain both testicular and ovarian tissue, or testicular tissue (Bachtrog et al., 2014).
With the continuous development of high-throughput sequencing technology, based on this experiment, the transcriptome analysis of the gonads of intersexual redclaw crayfish has screened some sex-related genes and pathways such as foxl2 and dmrt (Nong et al., 2024). Crustacean omics have been continuously excavated, but there are few reports on intersex individuals. Zhang et al. reported a case of intersex individuals of Procambarus clarkii, which was described and preliminarily discussed (Zhang et al., 2015). A study has analyzed the characteristics of gonad transcriptome of male and female individuals of redclaw crayfish and had a preliminary understanding of the differences between male and female individuals (Li et al., 2023).
With the advancement of detection technology and bioinformatics, metabolomics is widely used in gene function elucidation (Denkert et al., 2006), nutrition (Astarita and Langridge, 2013), stress analysis (Tuffnail et al., 2009), and other fields. Studies on redclaw crayfish have mainly focused on the transcriptome and genomics, with less on the metabolome. These studies have been largely confined to the analysis of Vibrio infection and detection of hepatopancreas, with no reports of gonadal correlation in intersex individuals. However, metabolomic techniques have been employed to study ovarian development in other aquatic organisms, such as Coilia nasus (Japanese grenadier anchovy) (Xu et al., 2016) and Megalobrama amblycephala (blunt snout bream) (Zhou et al., 2017). These studies confirmed the applicability of metabolomics in describing ovarian development in aquatic organisms. Joint metabolomic and transcriptomic analyses have been studied in some fish nutritional and sexual dimorphism studies (Prisingkorn et al., 2017; Qiao et al., 2016).
As the study progressed, the researchers concluded that changes in the concentration of sex hormones were related to ovarian development, as 17β-estradiol and progesterone could only be detected in the mature ovaries of the Homarus americanus (Couch et al., 1987). Meanwhile, hemolymph metabolomic studies of Cherax tenuimanus showed significant differences in metabolites between female and male crayfish due to differences in reproductive activity or life history (Lette et al., 2020). In addition, metabolomic analyses of bait composition and nutritional structure of Trichiurus japonicus at different stages of ovarian development revealed that lipid molecules and amino acids were the primary metabolites that differed between striped bass ovaries at different developmental stages (Feng, 2023), which suggested that these metabolites may play an important role in the development of fish ovaries. However, there is a paucity of studies related to the gonadal metabolome in intersex individuals of the redclaw crayfish.
To explore the differences in phenotype and gonadal anatomy between intersexual redclaw crayfish and normal redclaw crayfish, we combined the gonadal metabolome, and the results of previous gonadal transcriptome (Nong et al., 2024) to screen the differential genes and differential metabolites of sex differentiation and development regulation between intersexual and normal redclaw crayfish, so as to supplement the omics research of intersexual redclaw crayfish.
2 Material methods
2.1 Ethics statement
All experimental protocols in this study were approved by the Animal Research and Ethics Committee of Guangdong Ocean University (NIH Pub. No. 85-23, revised 1996). This study does not involve endangered or protected species.
2.2 Experimental shrimps and tissues
The Cherax quadricarinatus (three female ovaries as M, three male testes as FA, three right and left female, left male gonopores intersexual gonads as TB, and three right and left female, right male gonopores intersexual gonads as TA) were collected from the Hengxing shrimp farm in Zhanjiang city, Guangdong, China. No specific permissions were required for the sampling locations and activities, and the studies did not involve endangered or protected species and locations. The conditions are as follows: freshwater at natural water temperature transient in the workshop pool, water depth 30 cm–35 cm, continuous daily oxygenation, daily feeding bait for 17:00 (Guangdong Hengxing brand ordinary shrimp pellet feed), according to a certain time the shrimp sucked off the feces. Change of 1/3 water was carried out. The samples were collected from October 2022 to April 2023, a total of 200 intersexual individuals of males and females, seven different types and statistical ratios. The normal female and male shrimp gonads were taken as control and stored at −80°C until transcriptomic and metabolomic analyses.
2.3 Analysis of metabolomic data
2.3.1 Metabolite extraction and liquid chromatography-mass spectrometry analysis
The samples were thawed on ice and minced, and 20 mg (± 1 mg) was weighed into the corresponding centrifuge tubes and homogenized for 20 s in a ball mill (30 Hz). After centrifugation at 3,000 r/min for 30 s at 4°C, 400 μL of 70% methanol internal standard extract was added, followed by 5 min of shaking (1,500 r/min) and 15 min in an ice bath. After centrifugation at 12,000 r/min for 10 min at 4°C, 300 μL of supernatant was transferred to the appropriate centrifuge tube and then placed at −20°C for 30 min. Finally, after centrifugation at 12,000 r/min for 3 min at 4°C, 200 μL of supernatant was transferred to the appropriate injection bottle for analysis.
In this paper, an ultra-high-performance liquid chromatography (UHPLC) LC-30A (Shimadzu, Japan) was applied, and each sample was chromatographed on a Waters ACQUITY Premier HSS T3 Column 1.8 µm, 2.1 mm*100 mm liquid chromatography column for the chromatographic separation of the target compounds. The separation was performed on a Waters ACQUITY Premier HSS T3 Column 1.8 µm, 2.1 mm*100 mm. The conditions were as follows: mobile phase A, 0.1% formic acid/water; mobile phase B, 0.1% formic acid/acetonitrile; instrumental column temperature, 40°C; flow rate, 0.4 mL/min; injection volume, 4 μL. Mass spectrometry (MS) analysis was performed on a TripleTOF 6600+ High Resolution Mass Spectrometer (Foster City, CA, USA).
2.3.2 Data processing
Principal component analysis (PCA) and orthogonal projection to latent structure discriminant analysis (OPLS-DA) were performed using multivariate statistical analyses to discriminate the accuracy and validity of the data.
2.3.3 Differential metabolites selected
For two-group analysis, differential metabolites were determined by VIP (VIP > 1) and P-value (P-value < 0.05, Student’s t-test). VIP values were extracted from OPLS-DA results, which also contain score plots and permutation plots, and were generated using the R package MetaboAnalystR. The data were log transformed (Log2FC) and mean centered before OPLS-DA. In order to avoid overfitting, a permutation test (200 permutations) was performed.
2.3.4 Kyoto Encyclopedia of Genes and Genomes annotation and enrichment analysis
The detected metabolites were labeled by the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, and the labeled metabolites were mapped to the KEGG Pathway database, and then the pathways of metabolite enrichment with significant regulatory effects were analyzed. The significance was determined by the P-value of the hypergeometric test.
2.4 Differential transcript and differential metabolite KEGG co-enrichment analysis
Based on the KEGG database, the differential genes and differential metabolites of the same comparative groups were simultaneously mapped onto KEGG pathway maps for significance analysis, and the correlation was analyzed with the significant condition of P-value<0.05. The results were analyzed for correlation. Based on the results of KEGG enrichment analysis of differential metabolites and differential genes, the KEGG pathways jointly enriched by the two histologies were found and analyzed, and the related transcript and metabolite regulatory network maps were constructed.
3 Results
3.1 Gonadal metabolic profiling of normal and intersex redclaw crayfish
In order to analyze the differences in the variation of metabolites in different experimental groups, the scatter plots of PCA scores of the liquid chromatography-mass spectrometry (LC-MS) metabolic profiles of the control normal male spermathecae FA, normal female ovaries M, and the experimental groups of the intersexual gonads TA, intersexual gonads TB, showed that all the samples appeared in ellipses of 95% confidence intervals and that in the positive-negative ion model FA vs. TA, M vs. TA, FA vs. TB, and M vs. TB data points were separated in spatial distribution, indicating that there were differences in metabolic patterns between the two groups and between the two groups (Figures 1A-D). The R2X, R2Y, and Q2 of the OPLS-DA model for FA vs. TA, M vs. TA, FA vs. TB, and M vs. TB were 0.421, 0.99, and 0.827, respectively, 0.469, 0.994, and 0.879, respectively, and 0.469, 0.994, and 0.879, respectively. 0.533, 0.991, 0.822,0.618, 0.993, and 0.943 (Figures 2A-D) are used to discriminate the stability of the models. The R2 of FA vs. TA and M vs. TA is less than 0.5, indicating that the models are less stable; the R2 and Q2 of FA vs. TB and M vs. TB are both greater than 0.5, indicating that the models of these two groups are relatively stable, and the closer the value is to 1, the closer the value is to 1, and the better the model is.
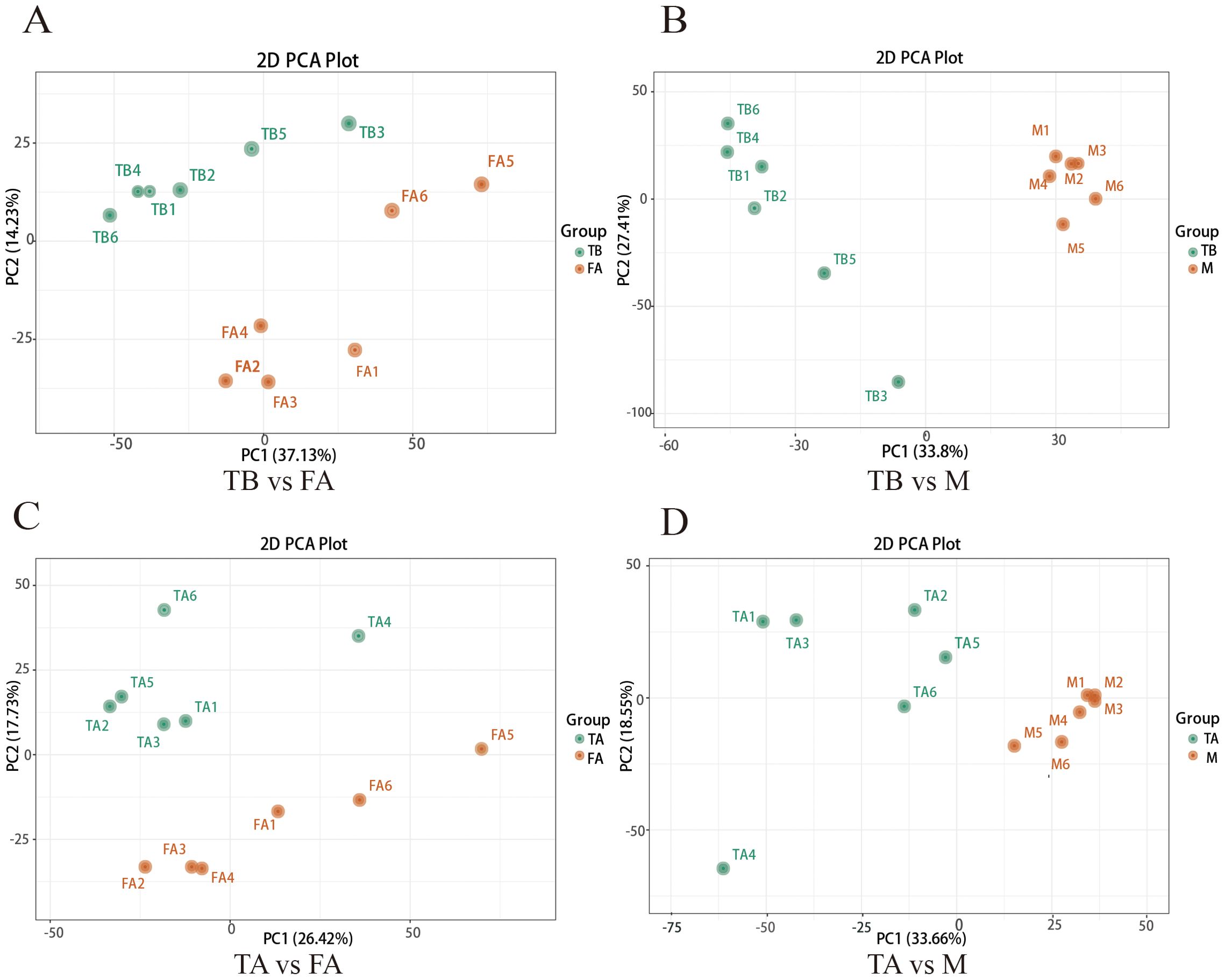
Figure 1. Score plots of PCA between gonads of intersex individuals and normal individuals of the redclaw crayfish. (A, B) represent PCA plots between the TB vs. FA group and with the TB vs. M group, and (C, D) represent PCA plots between the TA vs. FA group and the TA vs. M group. Each PCA plot corresponds to a different grouping. PC1 denotes the first principal component, PC2 denotes the second principal component, and percentage denotes the percentage of variance explained by the principal component for the dataset; each point in the plot denotes a sample, and samples in the same group are denoted by using the same color. Group denotes the grouping.
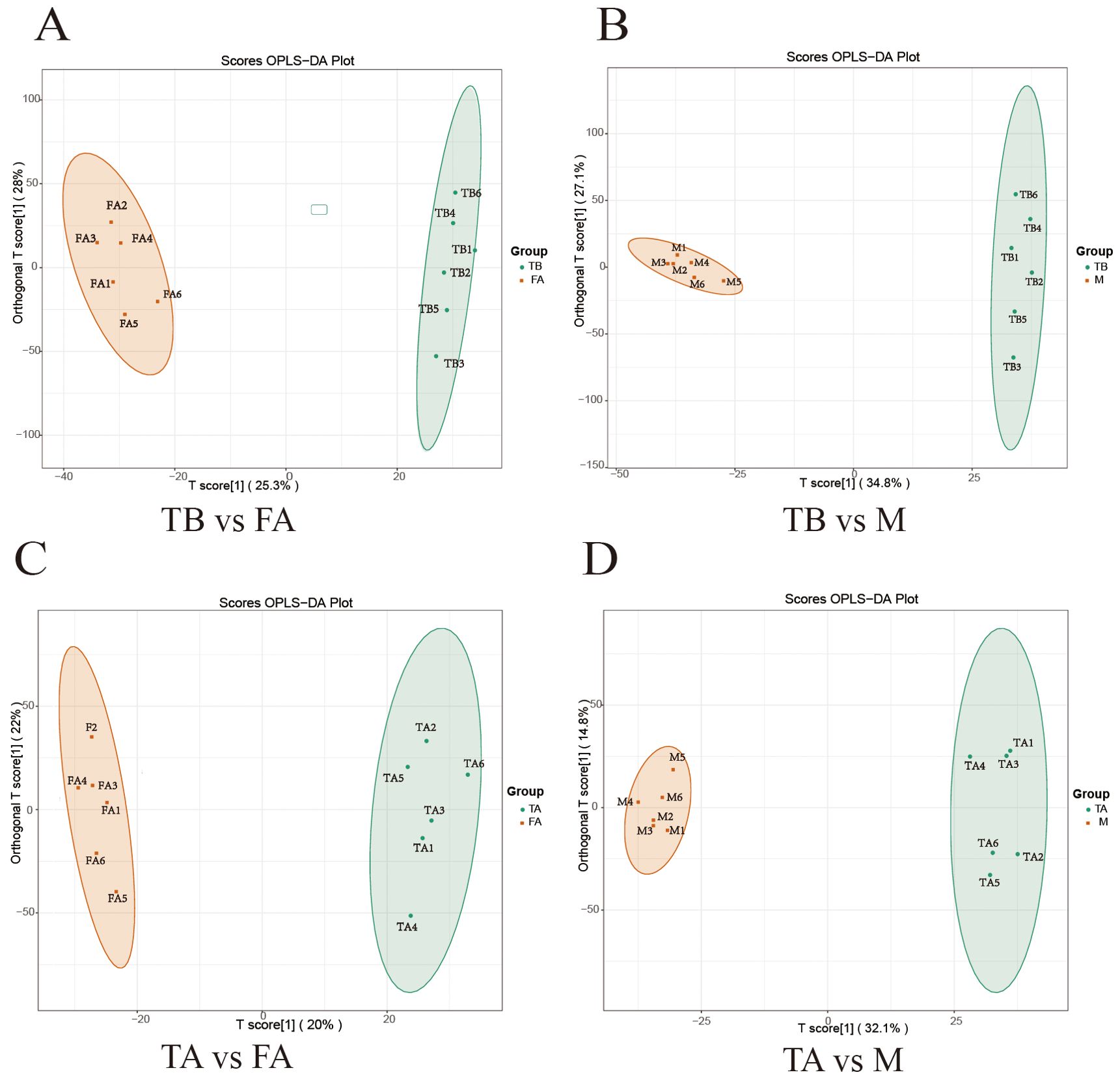
Figure 2. Plot of OPLS-DA scores between gonads of redclaw crayfish intersex individuals and normal individuals. (A, B) represent OPLS-DA plots between the TB vs. FA group and with the TB vs. M group. (C, D) represent OPLS-DA plots between the TA vs. FA group and the TA vs. M group. The horizontal coordinate indicates the predictive component scores, with the horizontal direction showing the gap between groups; the vertical coordinate indicates the orthogonal component scores, with the vertical direction showing the gap within groups; the percentage indicates the rate of explanation of the component for the dataset.
3.2 Identification and screening of differential metabolites
In order to more clearly show the changes of metabolites in different groups, we used non-targeted metabolomics to identify the metabolites of the samples. A total of 3,857 metabolites were screened in the FA, M, TA, and TB groups. Metabolites with VIP > 1 and P < 0.05 were selected from the above metabolites as significantly different metabolites. A total of 673 differential metabolites were screened in TA vs. FA, of which 205 metabolites were upregulated and 467 metabolites were downregulated (Figure 3A). A total of 1,220 differential metabolites were screened in TA vs. M, of which 625 metabolites were upregulated and 595 metabolites were downregulated (Figure 3B). A total of 877 metabolites were screened in TB vs. FA, of which 283 metabolites were upregulated and 594 metabolites were downregulated (Figure 3C). A total of 1,498 differential metabolites were screened in TA vs. M, including 639 upregulated metabolites and 850 downregulated metabolites (Figure 3D). Hierarchical cluster analysis of the differential metabolites showed that the trends of variation among different biological replicates within the groups were relatively consistent, whereas significant differences appeared between the groups. In summary, the results from cluster analysis and principal component analysis showed that there were obvious differences in metabolites among different groups.
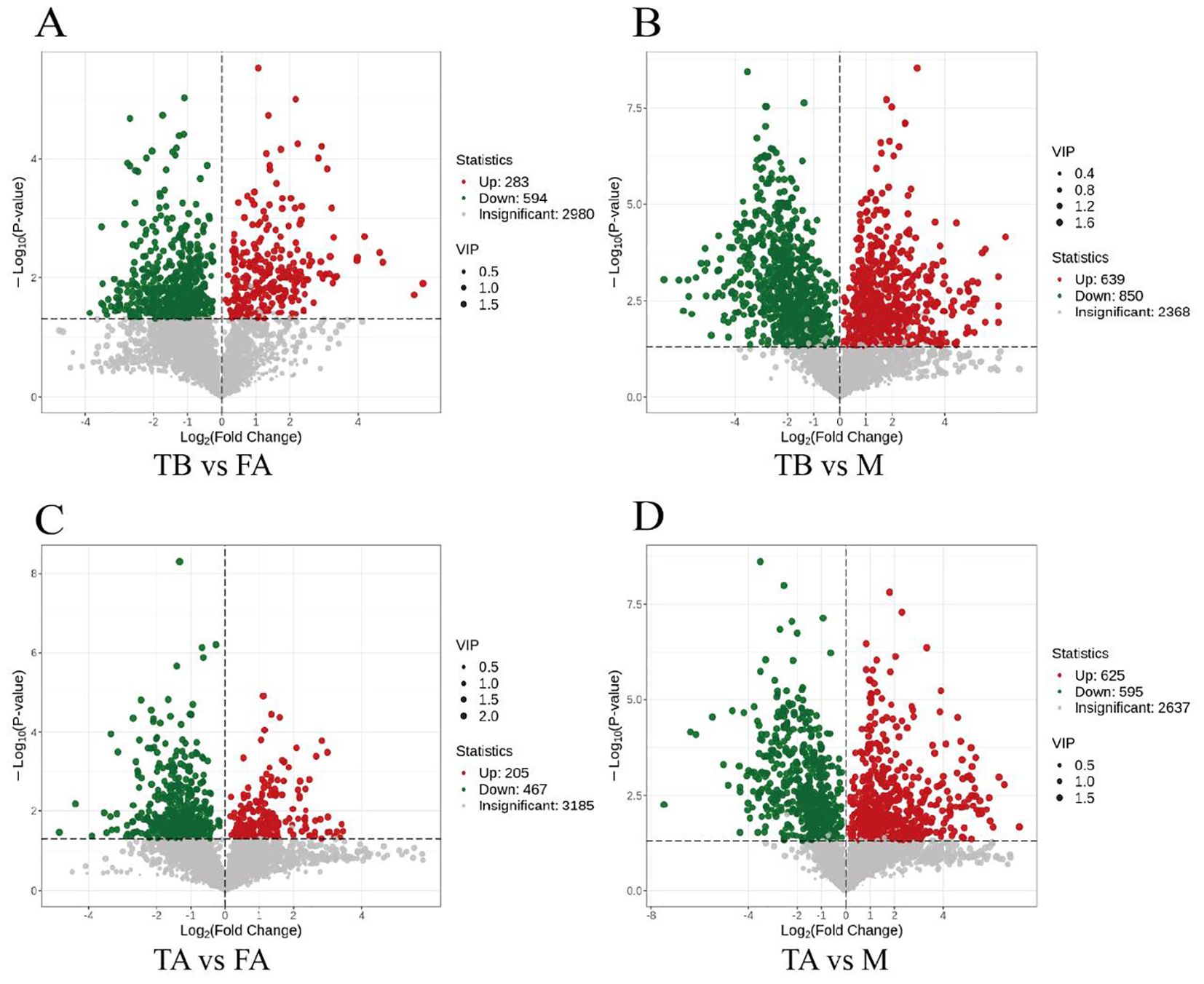
Figure 3. Differential metabolite volcano plots between gonads of intersex individuals and normal individuals of the red crayfish. Figures (A, B) represent the differential metabolite volcano plots between TB and FA groups and between TB and M groups, and (C, D) represent the differential metabolite volcano plots between TA and FA groups and between TA and M groups. Each dot in the volcano diagram represents a metabolite, where green dots represent down-regulated differential metabolites, red dots represent up-regulated differential metabolites, and grey represents metabolites that were detected but not significantly different. The horizontal coordinate indicates the logarithmic value of the multiplicative difference in the relative content of a metabolite in the two sets of samples (log2FC), The greater the absolute value of the horizontal coordinate, the greater the difference in the relative content of the substance between the two groups of samples. VIP+FC+P-value screening condition and the vertical coordinate indicates the level of significance of the difference (-log10P-value), the size of the dot represents the VIP value.
3.3 Pathway analysis of differential gonadal metabolites in normal redclaw crayfish and intersex individuals of both sexes
The different metabolite data were imported into the KEGG database and the MetPA database for analysis and screened for relevant differential metabolic pathways. To investigate the potential differences between metabolic pathways in normal and intersex individuals, we further annotated KEGG pathways for significantly different metabolites in the four groups and generated KEGG difference enrichment bubble maps to demonstrate the enriched metabolic pathways (Figures 4A–D). The study showed that there were 121 differential metabolic pathways in the TA vs. FA group; 152 differential metabolic pathways in the TA vs. M group; 129 differential metabolic pathways in the TB vs. FA group; and 146 differential metabolic pathways in the TA vs. M group.
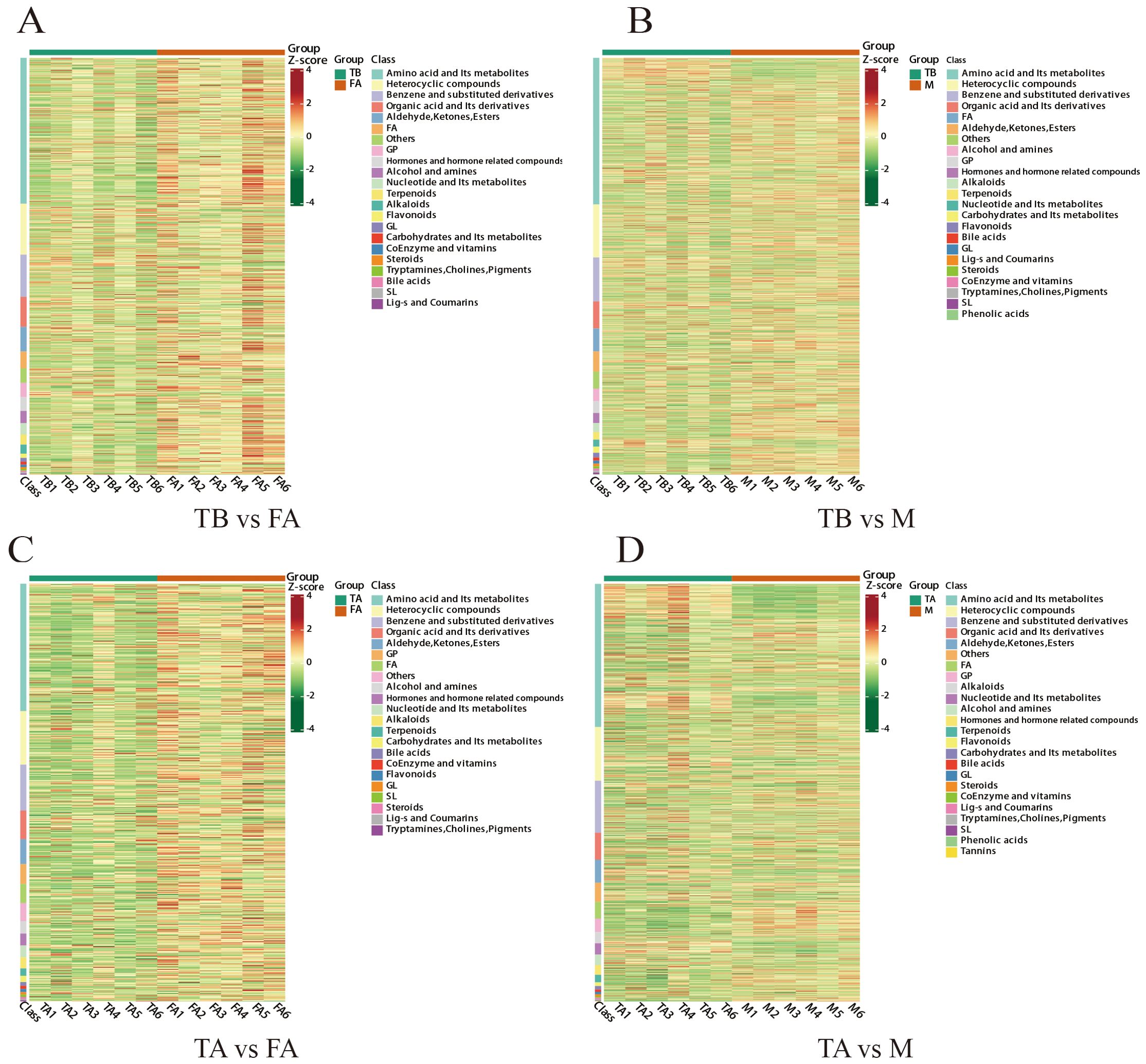
Figure 4. Cluster analysis of differential metabolites between gonads of intersex and normal individuals of the red crayfish. Figures (A, B) represent heat maps of differential metabolite clustering between TB and FA groups and between TB and M groups, and Figures (C, D) represent heat maps of differential metabolite clustering between TA and FA groups and between TA and M groups. Horizontal coordinates represent samples, and vertical coordinates represent metabolites. groups represent different subgroups. Group represents different subgroups, different colours represent different values normalised to relative content, and colour fills represent different values normalised to relative content (red for high content, green for low content). _heatmap_class: heatmap by substance, Class is the first level of substance classification. _heatmap_col-row_cluster: both metabolites and samples were analysed by clustering, the clustering line on the left side of the graph is the metabolite clustering line, the clustering line on the top of the graph is the sample clustering line. _heatmap_row_cluster: only metabolites were analysed for clustering, the clusters on the left side of the figure are metabolite clusters.
All pathways were ranked, and the top 20 pathways in terms of pathway ranking were listed (Figures 5A-D). Pathways were most abundant in TA vs. M and least abundant in TA vs. FA. There were 121 differential enrichment pathways in TA vs. FA, including vitamin B6 metabolism, sulfur metabolism, cysteine and methionine metabolism, purine metabolism, D-amino acid metabolism, and neuroactive ligand–receptor interaction. There were 152 differential enrichment pathways in TA vs. M, including D-amino acid metabolism, butanoate metabolism, protein digestion and absorption, mineral absorption, phenylalanine metabolism, and aminoacyl-tRNA biosynthesis. There were 129 differential enrichment pathways in TB vs. FA, including cysteine and methionine metabolism, D-amino acid metabolism, glutathione metabolism, protein digestion and absorption, antifolate resistance, and purine metabolism. There were 146 differential enrichment pathways in TB vs. M, including protein digestion and absorption, D-amino acid metabolism, ABC transporters, aminoacyl-tRNA biosynthesis, galactose metabolism, and mineral absorption.
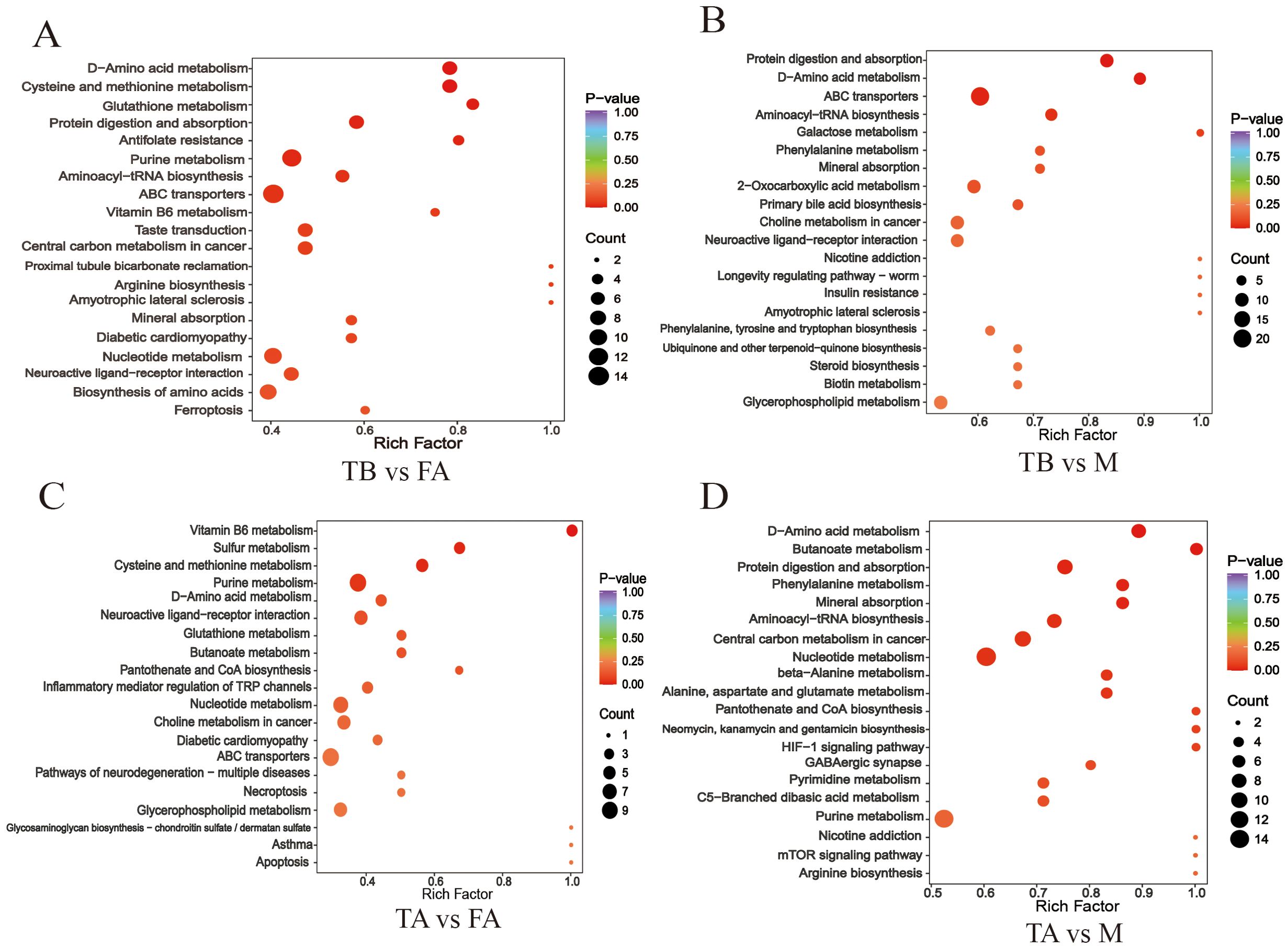
Figure 5. Enrichment analysis of metabolic pathways for significant differences between gonads of intersex individuals and normal individuals of the redclaw crayfish In the figure, (A-D) indicates the top 20 metabolic pathways enriched in TA vs. FA, TA vs. M, TB vs. M, and TB vs. M groups; each dot represents a metabolic pathway, the vertical axis indicates the pathway, the horizontal axis indicates the rate of enrichment, and the size of the dots indicates the amount of the number of different metabolites. The colors indicate the significance of enrichment, i.e., P-value, and the darker red color indicates the significance of enrichment, whereas the color of the right hand side indicates the P-value.
For these four comparisons (TA vs. FA, TA vs. M, TB vs. FA, TB vs. M), there were 17 significantly different pathways (P < 0.05). In the comparison of TA vs. FA, four pathways were significant: vitamin B6 metabolism; sulfur metabolism; cysteine and methionine metabolism; purine metabolism. For TA vs. M, 10 pathways are significant: D-amino acid metabolism; butanoate metabolism; protein digestion and absorption; mineral absorption; phenylalanine metabolism; aminoacyl-tRNA biosynthesis; central carbon metabolism in cancer; nucleotide metabolism alanine; aspartate and glutamate metabolism; beta-alanine metabolism. For TB vs. FA, there were eight significant pathways (P < 0.05): cysteine and methionine metabolism; D-amino acid metabolism; glutathione metabolism; protein digestion and absorption; antifolate resistance; purine metabolism; aminoacyl-tRNA biosynthesis; ABC transporters. There are four significant pathways in TB vs. M: protein digestion and absorption; D-amino acid metabolism; ABC transporters; aminoacyl-tRNA biosynthesis (Table 1). In the comparisons of TA vs. FA, and TA vs. M, the metabolic pathways of significant differences were different, and in the comparisons of TB vs. FA, TB vs. M, the aminoacyl-tRNA biosynthesis, protein digestion and absorption, D-amino acid metabolism, and the ABC transporters pathways were common to them, whereas antifolate resistance, glutathione metabolism, cysteine and methionine metabolism, and purine metabolism, which were significantly different pathways, were TB vs. FA-specific.
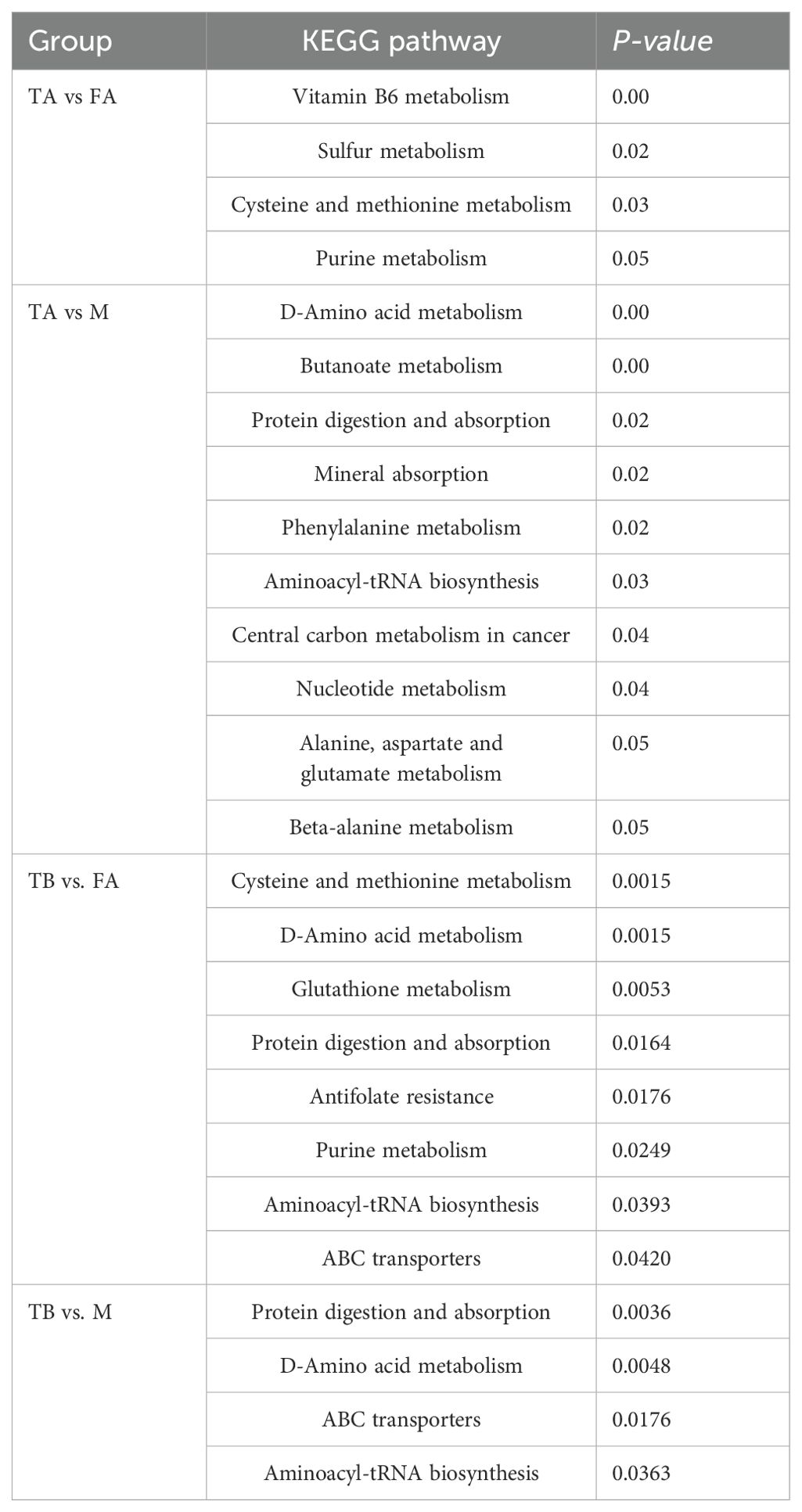
Table 1. Metabolic pathways of significant gonadal differentiators in intersex and normal individuals of the red crayfish.
3.4 Combined transcriptomic and metabolomic analyses
3.4.1 Joint analysis of sample information
In the comparison between the metabolic group and the transcriptome, there were four groups of comparisons (Table 2). Correlation analysis was performed on these four groups of comparisons. The specific samples and grouping information are as follows.
Table 2. Information on the grouping of transcriptome and metabolome samples of red crayfish intersex individuals and normal individuals.
3.4.2 Differential genes and differential metabolites KEGG enrichment bar chart
Combined metabolomics and transcriptomics analyses (Figure 6) showed that the differential genes and metabolites in the TA vs. M group, which were mainly enriched in the front-ranking group, were enriched in pathways related to steroid hormones and lipid metabolism, including lysosome, platelet activation, regulation of lipolysis in adipocytes; cortisol synthesis and secretion, ovarian steroidogenesis, cGMP-PKG signaling pathway, and estrogen signaling pathway. The main enrichment in TA vs. FA was amino acid metabolism, tyrosine metabolism, glutathione metabolism, drug metabolism-cytochrome P450, phenylalanine, tyrosine and tryptophan biosynthesis, folate biosynthesis, glycerophospholipid metabolism, metabolic pathways, PPAR signaling pathway, galactose metabolism, phenylalanine metabolism, and thyroid hormone synthesis. In TB vs. FA, the main enrichments were mainly in amino acid metabolism and lipid metabolism, including tyrosine metabolism; phenylalanine, tyrosine, and tryptophan biosynthesis; FoxO signaling pathway; AMPK signaling pathway; metabolic pathways; sphingolipid signaling pathway; phospholipase D signaling pathway; and D-amino acid metabolism. The major enrichments in TB vs. M were mainly steroid hormones. The major enrichment in TB vs. M was mainly steroid hormones and lipid substitutes, including lysosome, thiamine metabolism, sphingolipid metabolism, ether lipid metabolism, protein digestion and absorption, thyroid hormone signaling pathway, D-amino acid metabolism, hormone signaling pathway, cortisol synthesis and secretion, sphingolipid signaling pathway, pyrimidine metabolism, insulin resistance, and tyrosine metabolism. The pathways that ranked first from the transcriptome and metabolome were mainly steroid metabolism pathway, lipid metabolism, and amino acid metabolism, suggesting that these related pathways may be related to gonadal development.
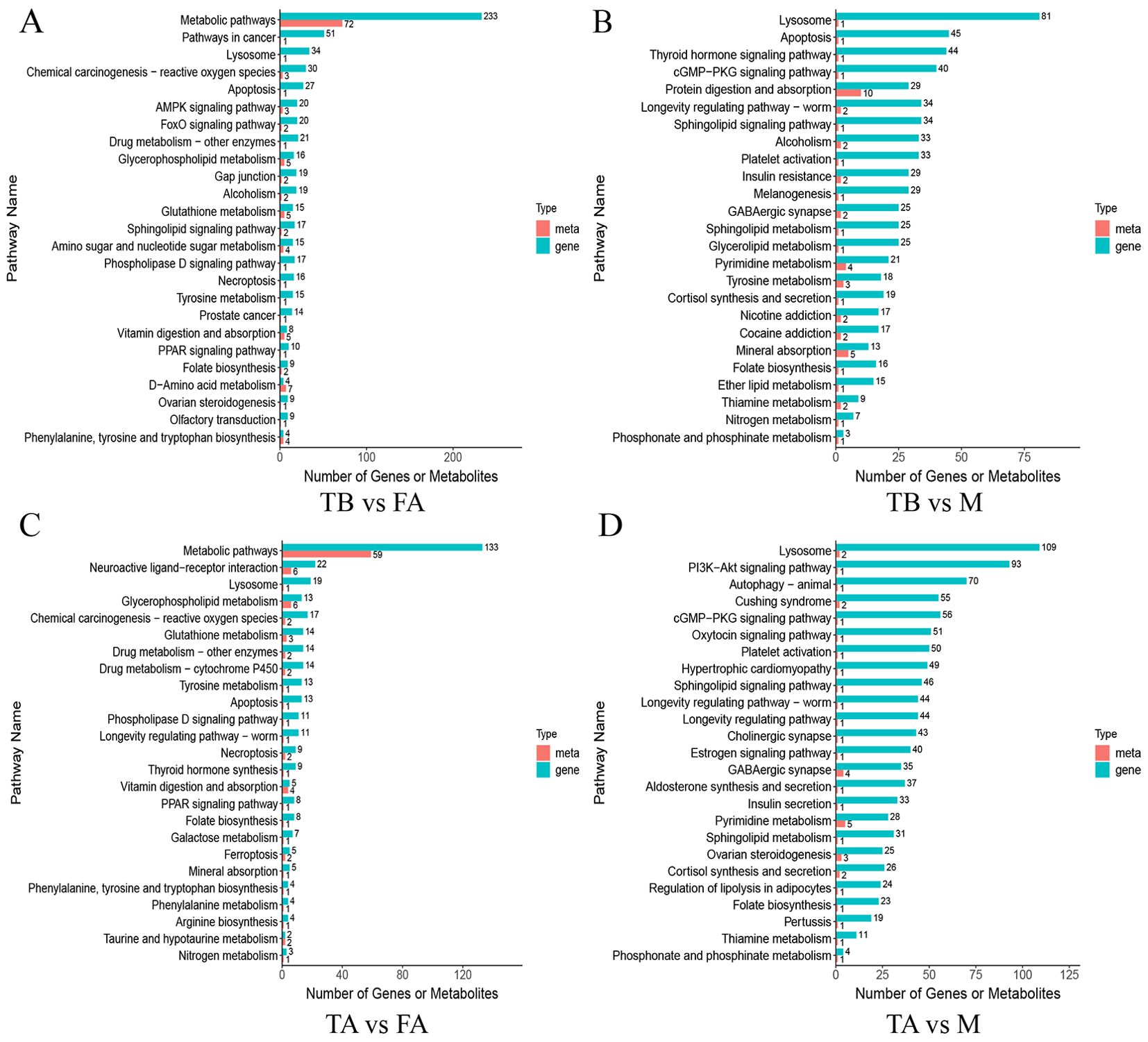
Figure 6. Association analysis of the KEGG pathway for differential genes and differential metabolites in normal and intersex individuals of the red crayfish. Figures (A, B) represent KEGG pathway association analysis plots of differential genes and differential metabolites between TB and FA groups and between TB and M groups, and Figures (C, D) represent KEGG pathway association analysis plots of differential genes and differential metabolites between TA and FA groups and between TA and M groups. The horizontal coordinate is the ratio (Ratio) of the differential metabolites or differential genes enriched in the pathway to the number of metabolites or genes annotated in the pathway, and the vertical coordinate is the KEGG pathway co-enriched by metabolome-transcriptome. count: the number of metabolites or genes enriched in the pathway.
3.4.3 Combined transcript and metabolite analysis of regulatory networks
Transcriptome and metabolome analyses were used to screen KEGG pathways associated with sex from co-enriched pathways, including ovarian steroidogenesis, estrogen signaling pathway, FoxO signaling pathway, glycerolipid metabolism, and fatty acid metabolism pathway. Based on the biological functions of these pathways, the differentially expressed genes and metabolites of these pathways were mapped onto the KEGG pathway map, suggesting that they may play a regulatory role in gonadal development of the redclaw crayfish, and a putative regulatory network was constructed (Figure 7).
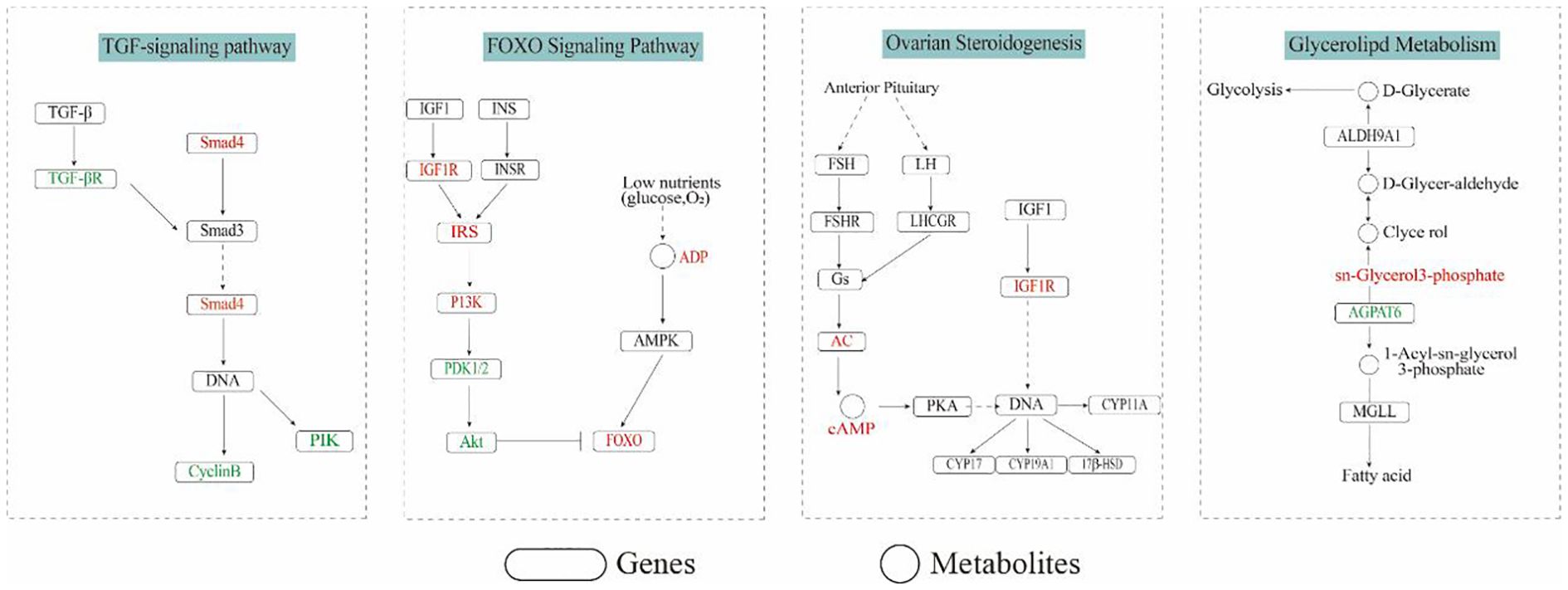
Figure 7. Enrichment maps of gonadal differential genes and metabolites in intersexual and normal individuals of the redclaw crayfish. Green represents downward adjustments and red represents upward adjustments.
4 Discussion
In this study, we analyzed the transcriptome and metabolome of intersex redclaw crayfish and found that intersex individuals differed from normal individuals in relevant transcripts and metabolites, including metabolites such as testosterone glucosinolate, cortisol, pregnenolone sulfate, estrone glucosinolate, steroid hormone synthesis pathway, estrogen signaling pathway, steroid hormone synthesis, and lipid metabolism related to the development of the gonads. At the same time, the expression of cyp18a1, cyp49a1, cyp19a, hsd17b12b, and other genes in the transcriptome may lead to differences in the gonads of intersex individuals and normal individuals.
Recent research has revealed that steroid hormones are not limited to traditional endocrine glands but are also found in tissues like the brain, liver, and adipose tissue. These hormones are significant in sexual development and function in a paracrine or autocrine manner (Beitel et al., 2014; Gennotte et al., 2017; Li et al., 2015; Senthilkumaran, 2015; Tokarz et al., 2015). In sclerotia, gonadotropin-I stimulation prompts the ovaries or spermathecae to synthesize the steroidal precursor pregnenolone from cholesterol, resulting in the production of luteinizing hormone, 17-hydroxyprogesterone, androstenedione, and testosterone in a continuous sequence (Devlin and Nagahama, 2002). At the same time, during vitellogenesis, aromatase converts some of the available testosterone into estradiol-17β (E2). As the ovary gradually matures, the pituitary gland turns to synthesize gonadotropin-II, resulting in an increase in testosterone and estradiol-17β (E2) synthesis, whereas progesterone secretion promotes ovarian maturation (Kime and Scott, 1993; Pankhurst and Thomas, 1998). Our study screened the differential expression of steroid synthesis metabolites such as pregnenolone sulfate, estrone glucuronide, and testosterone glucuronide, indicating that the phenotypic differences between male and female individuals and normal individuals are related, indicating that the steroid hormone pathway may play a role in gonadal differentiation and development.
Steroid hormones are regulated in synthesis and metabolism by two classes of enzymes, cytochrome P45 (CYP) and hydroxysteroid dehydrogenase (HSD) (Hilborn et al., 2017; Uno et al., 2012). A range of CYP enzymes have been shown to play an important role in the synthesis of steroid hormones in adrenal, gonadal, and peripheral tissues (Jefcoate et al., 1992; Waxman et al., 1991). In the combined analysis of metabolome and transcriptome, the key signaling pathways—ovarian steroid and estrogen signaling pathways—were located. Among them, genes such as cyp18a1, cyp40, and hsd17b12b showed significant differences. cyp18a1 and cyp49a1 were significantly different between the normal individual ovary and the intersex individual control group, but there was no significant difference between the normal individual male and the intersex individual. This suggests that they may be involved in the process of ovarian development. Related cyp family genes have been shown to be involved in sex determination. For example, cyp19a is responsible for aromatizing androgens to estrogens in Danio rerio and Oreochromis niloticus (Li et al., 2019; Trant et al., 2001; van Nes and Andersen, 2006; Yin et al., 2017; Zhang et al., 2017). In the gonads of clupeoid fishes, the expression of hsd17b12b was found to be higher in the ovaries compared with the spermathecae and in intersex individuals (Nyuji et al., 2020). Similar results were obtained in the present study, with higher expression of hsd17b12b in the ovary than in hermaphroditic individuals and spermatozoa, although a more in-depth study to validate the experiment was not conducted, which provides preliminary evidence that hsd17b12b expression affects the development of the ovary in the redclaw crayfish.
Cortisol is the main glucocorticoid in scleractinian fish and the hormone most directly associated with stress (Geffroy and Douhard, 2019; Solomon-Lane et al., 2013). Cortisol has been recognized as a potential key mediator between environmental factors and sex reversal, associated with sex determination (Nozu and Nakamura, 2015; Solomon-Lane et al., 2013). Previous research has suggested that 11-deoxycorticosterone in the ovary could act as a regulatory hormone, stimulating oocyte maturation and ovulation (Díaz and Piferrer, 2015). 21-Deoxycortisol is a steroid hormone secreted by the adrenal gland. In this study, it was found that this metabolite showed significant differences between TA vs. M and TB vs. M groups. Its expression was the highest in the ovary of normal individuals, followed by intersex individuals, and the expression of males was almost minimal, indicating that its expression changes were related to ovarian development. Similarly, cortisol plays an important role in a variety of female fish, including Paralichthys olivaceus, Paralichthys lethostigma, Odontesthes bonariensis, and Oryzias latipes (Hayashi et al., 2010). The metabolism of 17α-hydroxypregnenolone is different in TA vs. M and TB vs. M, and the expression of 17α-hydroxypregnenolone in the ovary of normal individuals is higher than that of intersex individuals. In addition, cortisol can promote female sex change in Halichoeres trimaculatus and Parapercis cylindrica cylindrica, and histological sex change can be observed. This indicates that the stagnation of ovarian development in intersex individuals is related to the metabolic levels of these key metabolites (Frisch et al., 2007; Nozu and Nakamura, 2015).
Amino acids are essential building blocks in all living organisms, forming the basis of peptides and proteins. They play crucial roles in various biological functions like growth, reproduction, and immunity (Viant et al., 2003). This study found that there were significant differences in metabolites associated with specific amino acids (such as phenylalanine, lysine, L-glutamic acid, L-proline, and L–D-phenylalanine). Related studies have shown that the metabolism of amino acids such as phenylalanine can significantly affect the growth and survival of marine invertebrates and is also affected by gender (Taboada et al., 2003). The present study showed that L-phenylalanine and D-phenylalanine were significantly upregulated in the TB vs. M group of metabolites and significantly downregulated in the TB vs. FA group of metabolites. Similarly, the metabolic trends of L-glutamate, L-proline, and L–D-phenylalanine were the same as those of L-phenylalanine and D-phenylalanine. The results were consistent with the downregulation of amino acids of lysine differentially expressed proteins such as L-proline, L-glutamine, and L-aspartic acid in the differential metabolites of sea urchin gonads (Cai et al., 2023), indicating that amino acid-related metabolites have an important effect on gonad development. Lysine is a signaling molecule that regulates the body’s endocrine, intermediate metabolism, and reproductive and immune responses (Wu et al., 2014). Recently, many fish studies have also emphasized the effect of lysine on the growth performance of fish (Khan and Abidi, 2011; Lansard et al., 2011). In this study, L-lysine was significantly upregulated in TA vs. M and TB vs. M, but there was no significant difference in TA vs. FA and TB vs. FA. The above studies further indicate that metabolites in the amino acid metabolic pathway play an important role in the male and female differentiation of redclaw claw crayfish.
Lipid digestion and decomposition not only provide energy for gonadal development but also participate in the process of reproductive endocrine regulation (Arukwe, 2008; Jiang et al., 2009). In this study, metabolites related to unsaturated fatty acid metabolic pathways were significantly enriched, namely, linoleic acid, γ-linolenic acid, oleic acid, eicosapentaenoic acid, γ-linolenic acid, and other metabolites. Fatty acids are the main components of oocytes, and their release can be used as lipid energy reserves for oocytes and early embryos (Sturmey et al., 2009). In mammals, oleic acid has a positive effect on oocyte maturation and can overcome the adverse effects of other major saturated fatty acids (McKeegan and Sturmey, 2011). In the study of the ovarian metabolome of Macrobrachium nipponense, oleic acid increased in the first to fifth stages, whereas linoleic acid increased first and then decreased (fourth and fifth decreased) (Zhang et al., 2020). In this study, oleic acid and linoleic acid, as differential metabolites, were expressed more highly in the ovaries of normal individuals than in the intersex individuals and the testis of normal individuals, indicating that fatty acids may promote the gonadal development of redclaw crayfish and the stagnation of intersex individual development is related to fatty acid metabolism. Studies have shown that various fatty acids, such as eicosapentaenoic acid, docosahexaenoic acid, arachidonic acid, and sterols, are closely associated with gonadal development (Nhan et al., 2020; Soudant et al., 1996; Xu et al., 2017). Lipid and eicosanoid lipid metabolites have also been reported to be present at high concentrations in the gonadal tissues of crustaceans (Coffa and Hill, 2000; Lubet et al., 1986). In this study, the metabolites of γ-linolenic acid and eicosapentaenoic acid were significantly different in TA vs. M, but not in TA vs. FA, and their expression levels in the ovaries of normal individuals were higher than those of intersexual individuals. In addition, during the reproduction of mussels (Mytilus galloprovincialis), the total lipid content of the female outer membrane is twice that of the male (Lubet et al., 1986). In crabs, lipids can be transported from the hepatopancreas to the gonads through transporters and key fatty acid transferases (Ghazali et al., 2017; Li et al., 2011; Wen et al., 2001). The related fatty acid metabolites screened in this study were consistent with the changing trend in the existing research results, indicating that the ovarian development of redclaw crayfish was closely related to fatty acid metabolism.
In the transcriptome and metabolome co-enrichment pathway, the insulin signaling pathway and TGF-β signaling pathway in conjunction with the pathways such as FoxO signaling also play a role in the combination (Koutsoulidou et al., 2011; Ogg et al., 1997). Studies have shown that in the Scatophagus argus, the FoxO signaling pathway may be involved in the regulation of ovarian maturation (Wang et al., 2021). Insulin regulates ovarian maturation by interacting with the insulin receptor (IR). Insulin plays various physiological regulatory roles by binding to the insulin receptor (IR). Upon binding to a ligand, the insulin receptor undergoes a conformational change and then activates a variety of cellular substrates for signaling, including insulin receptor substrate (insulin receptor substrate, IRS) (Dupont and Scaramuzzi, 2016). At the same time, the IRS acts on the regulatory subunit of regulatory subunit of PI3K, which activates the catalytic subunit of PI3K (Wijesekara et al., 2005). In this study, we found that the irs1 gene of insulin receptor substrate 1 (Irs1) in the gonads of redclaw crayfish increased in expression after stimulation with Igf1, similar to the results of the mouse study, and the lack of the Irs1 protein may lead to impaired reproductive function (Wijesekara et al., 2005). In addition, the action of igf1 also affects the expression level of pik3ca gene in the gonads of redclaw crayfish, and the differential expressions of ADP metabolites and irs1 and pik3ca genes are co-enriched in the FoxO signaling pathway. The TGF-β signaling pathway controls a variety of cellular processes during animal embryonic development, including cell recognition, proliferation, differentiation, apoptosis, and developmental fate (Shi and Massagué, 2003; Ten Dijke et al., 2002). In Macrobrachium rosenbergii and Pelodiscus sinensis, the insulin signaling pathway and TGF-β signaling pathway are related to ovarian development and maturation, and smad4, as a female-specific gene and female-related gene, has been confirmed in Pelodiscus sinensis (Jiang et al., 2019; Zhou et al., 2023). This is consistent with the upregulated expression of smad4 in the pathway in this study. At the same time, the enrichment of glucolipid metabolism was obtained in KEGG analysis. In Penaeus vannamei, the metabolic level of glucolipid metabolism affects its growth (Wu et al., 2022), indicating that these pathways may play a regulatory role in the gonads of Cherax quadricarinatus.
5 Conclusion
In this study, metabolic pathways and metabolites that may be related to sex differentiation and gonadal development of the redclaw crayfish were screened out through the joint analysis of metabolomic and transcriptomic results. This study supplements the research gap of gonad metabolomics and joint analysis in intersex redclaw crayfish and lays a theoretical foundation for the study of sex development mechanism in intersex redclaw crayfish. Due to the small number of samples and limited basic research, this paper mainly analyzes and demonstrates the discrepancy of the overall data, and the next step is to analyze, identify, and validate the relevant metabolic pathways or specific genes in depth. Meanwhile, the study of the relationship between sex differentiation and related genes of redclaw crayfish can help to improve the efficiency of breeding production and provide a strong theoretical basis for the study of artificially induced sex reversal of redclaw crayfish and the development of unisexual breeding technology.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SUB13985791.
Ethics statement
The animal study was approved by the Animal Research and Ethics Committee of Guangdong Ocean University (NIH Pub. No.85–23, revised 1996). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RH: Data curation, Investigation, Methodology, Writing – original draft. CN: Investigation, Methodology, Software, Writing – review & editing. YC: Conceptualization, Investigation, Methodology, Writing – review & editing. SL: Resources, Software, Supervision, Writing – original draft. HC: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This article was supported by grants from the Natural Science Foundation of China (32273131 and 32172971), the Science and Technology Plan of Zhanjiang City (2022E03002 and 2024E03007), Modern Seed Industry Park for Whiteleg Shrimp of Guangdong Province (K22221), the Youth Science and Technology Innovation Talent of Guangdong Tezhi plan talent (2023TQ07A888).
Acknowledgments
The authors would like to thank all the reviewers who participated in the review.
Conflict of interest
Authors YC and SL were employed by the company Guangdong Evergreen Feed Industry Co., Ltd. Author HC was employed by the company Guangdong Havwii Agriculture Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1506646/full#supplementary-material
References
Arukwe A. (2008). Steroidogenic acute regulatory (StAR) protein and cholesterol side-chain cleavage (P450scc)-regulated steroidogenesis as an organ-specific molecular and cellular target for endocrine disrupting chemicals in fish. Cell Biol. Toxicol. 24, 527–540. doi: 10.1007/s10565-008-9069-7
Astarita G., Langridge J. (2013). An emerging role for metabolomics in nutrition science. J. Nutrigenet. Nutrigenom. 6, 181–200. doi: 10.1159/000354403
Bachtrog D., Mank J. E., Peichel C. L., Kirkpatrick M., Otto S. P., Ashman T.-L., et al. (2014). Sex determination: why so many ways of doing it? PloS Biol. 12, e1001899. doi: 10.1371/journal.pbio.1001899
Beitel S. C., Doering J. A., Patterson S. E., Hecker M. (2014). Assessment of the sensitivity of three North American fish species to disruptors of steroidogenesis using in vitro tissue explants. Aquat Toxicol. 152, 273–283. doi: 10.1016/j.aquatox.2014.04.013
Cai W.-Q., Jiang C.-Y., Shang S., Wang S.-C., Zhu K.-Y., Dong X.-P., et al. (2023). Insight into the relationship between metabolite dynamic changes and microorganisms of sea urchin (S. intermedius) gonads during storage. Food Chem. X 18, 100727. doi: 10.1016/j.fochx.2023.100727
Coffa G., Hill E. M. (2000). Discovery of an 11(R)- and 12(S)-lipoxygenase activity in ovaries of the mussel Mytilus edulis. Lipids 35, 1195–1204. doi: 10.1007/s11745-000-0636-5
Couch E. F., Hagino N., Lee J. W. (1987). Changes in estradiol and progesterone immunoreactivity in tissues of the lobster, Homarus americanus, with developing and immature ovaries. Comp. Biochem. Physiol. Part A: Physiol. 87, 765–770. doi: 10.1016/0300-9629(87)90397-5
Denkert C., Budczies J., Kind T., Weichert W., Tablack P., Sehouli J., et al. (2006). Mass spectrometry-based metabolic profiling reveals different metabolite patterns in invasive ovarian carcinomas and ovarian borderline tumors. Cancer Res. 66, 10795–10804. doi: 10.1158/0008-5472.CAN-06-0755
Devlin R. H., Nagahama Y. (2002). Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquacult. Sex Determination Sex Different. Fish 208, 191–364. doi: 10.1016/S0044-8486(02)00057-1
Díaz N., Piferrer F. (2015). Lasting effects of early exposure to temperature on the gonadal transcriptome at the time of sex differentiation in the European sea bass, a fish with mixed genetic and environmental sex determination. BMC Genomics 16, 679. doi: 10.1186/s12864-015-1862-0
Dupont J., Scaramuzzi R. J. (2016). Insulin signalling and glucose transport in the ovary and ovarian function during the ovarian cycle. Biochem. J. 473, 1483–1501. doi: 10.1042/BCJ20160124
Feng L. (2023). A preliminary study on the feeding habits of striped bass (Trichiurus japonicus) and the physiological changes during its gonadal development in the Wen-Tai fishing ground (Shanghai China: Shanghai Ocean University). doi: 10.27314/d.cnki.gsscu.2023.000373
Frisch A. J., Walker S. P. W., McCormick M. I., Solomon-Lane T. K. (2007). Regulation of protogynous sex change by competition between corticosteroids and androgens: An experimental test using sandperch, Parapercis cylindrica. Hormones Behav. 52, 540–545. doi: 10.1016/j.yhbeh.2007.07.008
Garcı́a-Guerrero M., Villarreal H., Racotta I. S. (2003). Effect of temperature on lipids, proteins, and carbohydrates levels during development from egg extrusion to juvenile stage of Cherax quadricarinatus (Decapoda: Parastacidae). Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 135, 147–154. doi: 10.1016/S1095-6433(02)00354-9
Geffroy B., Douhard M. (2019). The adaptive sex in stressful environments. Trends Ecol. Evol. 34, 628–640. doi: 10.1016/j.tree.2019.02.012
Gennotte V., Akonkwa B., Mélard C., Denoël M., Cornil C. A., Rougeot C. (2017). Do sex reversal procedures differentially affect agonistic behaviors and sex steroid levels depending on the sexual genotype in Nile tilapia? J. Exp. Zool A Ecol. Integr. Physiol. 327, 153–162. doi: 10.1002/jez.2080
Ghanawi J., Saoud I. P. (2012). Molting, reproductive biology, and hatchery management of redclaw crayfish Cherax quadricarinatus (von Martens 1868). Aquaculture 358–359, 183–195. doi: 10.1016/j.aquaculture.2012.06.019
Ghazali A., Azra M. N., Noordin N. M., Abol-Munafi A. B., Ikhwanuddin M. (2017). Ovarian morphological development and fatty acids profile of mud crab (Scylla olivacea) fed with various diets. Aquaculture 468, 45–52. doi: 10.1016/j.aquaculture.2016.09.038
Hayashi Y., Kobira H., Yamaguchi T., Shiraishi E., Yazawa T., Hirai T., et al. (2010). High temperature causes masculinization of genetically female medaka by elevation of cortisol. Mol. Reprod. Dev. 77, 679–686. doi: 10.1002/mrd.21203
Hilborn E., Stål O., Jansson A. (2017). Estrogen and androgen-converting enzymes 17β-hydroxysteroid dehydrogenase and their involvement in cancer: with a special focus on 17β-hydroxysteroid dehydrogenase type 1, 2, and breast cancer. Oncotarget 8, 30552–30562. doi: 10.18632/oncotarget.15547
Jefcoate C. R., McNamara B. C., Artemenko I., Yamazaki T. (1992). Regulation of cholesterol movement to mitochondrial cytochrome P450scc in steroid hormone synthesis. J. Steroid Biochem. Mol. Biol. 43, 751–767. doi: 10.1016/0960-0760(92)90305-3
Jiang W. Z., Min Y., Yang H., Wan W., Zhang X. (2019). De novo transcriptome analysis of eyestalk reveals ovarian maturation related genes in Macrobrachium rosenbergii. Aquaculture 505, 280–288. doi: 10.1016/j.aquaculture.2019.02.073
Jiang H., Yin Y., Zhang X., Hu S., Wang Q. (2009). Chasing relationships between nutrition and reproduction: A comparative transcriptome analysis of hepatopancreas and testis from Eriocheir sinensis. Comp. Biochem. Physiol. Part D: Genomics Proteomics 4, 227–234. doi: 10.1016/j.cbd.2009.05.001
Khan M. A., Abidi S. F. (2011). Effect of dietary l-lysine levels on growth, feed conversion, lysine retention efficiency and haematological indices of Heteropneustes fossilis (Bloch) fry. Aquacut Nutr. 17, e657–e667. doi: 10.1111/j.1365-2095.2010.00815.x
Kime D. E., Scott A. P. (1993). In vitro synthesis of 20α-reduced and of 11- and 21-oxygenated steroids and their sulfates by testes of the goldfish (Carassius auratus): Testicular synthesis of corticosteroids. Fish Physiol. Biochem. 11, 287–292. doi: 10.1007/BF00004577
Koutsoulidou A., Mastroyiannopoulos N. P., Furling D., Uney J. B., Phylactou L. A. (2011). Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev. Biol. 11, 11–34. doi: 10.1186/1471-213X-11-34
Lansard M., Panserat S., Plagnes-Juan E., Dias K., Seiliez I., Skiba-Cassy S. (2011). L-leucine, L-methionine, and L-lysine are involved in the regulation of intermediary metabolism-related gene expression in rainbow trout hepatocytes. J. Nutr. 141, 75–80. doi: 10.3945/jn.110.124511
Lette E., Lawler N., Burnham Q., Boyce C., Duffy R., Koenders A., et al. (2020). Metabolomic profiling of crayfish haemolymph distinguishes sister species and sex: implications for conservation, aquaculture and physiological studies. Freshw. Crayfish 25, 89–101. doi: 10.5869/fc.2020.v25-1.089
Li W., Jin X., He L., Gong Y., Jiang H., Wang Q. (2011). Molecular cloning and tissue expression of the fatty acid-binding protein (Es-FABP9) gene in the reproduction seasons of Chinese mitten crab, Eriocheir sinensis. Mol. Biol. Rep. 38, 5169–5177. doi: 10.1007/s11033-010-0666-1
Li J., Papadopoulos V., Vihma V. (2015). Steroid biosynthesis in adipose tissue. Steroids 103, 89–104. doi: 10.1016/j.steroids.2015.03.016
Li M., Sun L., Wang D. (2019). Roles of estrogens in fish sexual plasticity and sex differentiation. Gen. Comp. Endocrinol. 277, 9–16. doi: 10.1016/j.ygcen.2018.11.015
Li F., Zheng J., Cheng S., Liu S., Liu Y., Jia Y., et al. (2023). Characteristics of the gonadal transcriptiome of intersex red claw crayfish Cherax quadricarinatus. J. Biol. 40, 80-. doi: 10.3969/j.issn.2095-1736.2023.02.080
Lubet P., Brichon G., Besnard J. Y., Zwingelstein G. (1986). Sexual differences in the composition and metabolism of lipids in the mantle of the mussel Mytilus galloprovincialis LMK (Mollusca: Bivalvia). Comp. Biochem. Physiol. Part B: Comp. Biochem. 84, 279–285. doi: 10.1016/0305-0491(86)90077-5
McKeegan P. J., Sturmey R. G. (2011). The role of fatty acids in oocyte and early embryo development. Reprod. Fertil Dev. 24, 59–67. doi: 10.1071/RD11907
Nguyen T. V., Cummins S. F., Elizur A., Ventura T. (2016). Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci. Rep. 6, 38658. doi: 10.1038/srep38658
Nhan H. T., Nhu T. Q., Duc P. M., Liew H. J., Ako H., Jha R. (2020). Effects of dietary arachidonic acid on final maturation, spawning and composition of gonad of black sea urchin Diadema setosum (Leske 1778). Aquacut Nutr. 26, 1771–1779. doi: 10.1111/anu.13127
Nong C., Chen Y., Yang H., Chen N., Tian C., Li S., et al. (2024). Phenotypic sorting of individual male and female intersex Cherax quadricarinatus and analysis of molecular differences in the gonadal transcriptome. Comp. Biochem. Physiol. Part D Genomics Proteomics 49, 101194. doi: 10.1016/j.cbd.2024.101194
Nozu R., Nakamura M. (2015). Cortisol administration induces sex change from ovary to testis in the protogynous Wrasse, Halichoeres trimaculatus. Sex Dev. 9, 118–124. doi: 10.1159/000373902
Nyuji M., Hongo Y., Yoneda M., Nakamura M. (2020). Transcriptome characterization of BPG axis and expression profiles of ovarian steroidogenesis-related genes in the Japanese sardine. BMC Genomics 21, 668. doi: 10.1186/s12864-020-07080-1
Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., Tissenbaum H. A., et al. (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999. doi: 10.1038/40194
Pankhurst N. W., Thomas P. M. (1998). Maintenance at elevated temperature delays the steroidogenic and ovulatory responsiveness of rainbow trout Oncorhynchus mykiss to luteinizing hormone releasing hormone analogue. Aquaculture 166, 163–177. doi: 10.1016/S0044-8486(98)00284-1
Parnes S., Khalaila I., Hulata G., Sagi A. (2003). Sex determination in crayfish: are intersex Cherax quadricarinatus (Decapoda, Parastacidae) genetically females? Genet. Res. 82, 107–116. doi: 10.1017/s0016672303006372
Prisingkorn W., Prathomya P., Jakovlić I., Liu H., Zhao Y.-H., Wang W.-M. (2017). Transcriptomics, metabolomics and histology indicate that high-carbohydrate diet negatively affects the liver health of blunt snout bream (Megalobrama amblycephala). BMC Genomics 18, 856. doi: 10.1186/s12864-017-4246-9
Qiao Q., Le Manach S., Sotton B., Huet H., Duvernois-Berthet E., Paris A., et al. (2016). Deep sexual dimorphism in adult medaka fish liver highlighted by multi-omic approach. Sci. Rep. 6, 32459. doi: 10.1038/srep32459
Rodgers L. J., Saoud P. I., Rouse D. B. (2006). The effects of monosex culture and stocking density on survival, growth and yield of redclaw crayfish (Cherax quadricarinatus) in earthen ponds. Aquaculture 259, 164–168. doi: 10.1016/j.aquaculture.2005.11.056
Senthilkumaran B. (2015). Pesticide- and sex steroid analogue-induced endocrine disruption differentially targets hypothalamo-hypophyseal-gonadal system during gametogenesis in teleosts - A review. Gen. Comp. Endocrinol. 219, 136–142. doi: 10.1016/j.ygcen.2015.01.010
Shi Y., Massagué J. (2003). Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700. doi: 10.1016/s0092-8674(03)00432-x
Solomon-Lane T. K., Crespi E. J., Grober M. S. (2013). Stress and serial adult metamorphosis: multiple roles for the stress axis in socially regulated sex change. Front. Neurosci. 7. doi: 10.3389/fnins.2013.00210
Soudant P., Marty Y., Moal J., Robert R., Quéré C., Le Coz J. R., et al. (1996). Effect of food fatty acid and sterol quality on Pecten maximus gonad composition and reproduction process. Aquaculture 143, 361–378. doi: 10.1016/0044-8486(96)01276-8
Sturmey R. G., Reis A., Leese H. J., McEvoy T. G. (2009). Role of fatty acids in energy provision during oocyte maturation and early embryo development. Reprod. Domest Anim. 44 Suppl 3, 50–58. doi: 10.1111/j.1439-0531.2009.01402.x
Taboada M. C., Gonzalez M., Rodriguez E. (2003). Value and effects on digestive enzymes and serum lipids of the marine invertebrate Holothuria forskali. Nutr. Res. 23, 1661–1670. doi: 10.1016/S0271-5317(03)00175-1
Ten Dijke P., Goumans M.-J., Itoh F., Itoh S. (2002). Regulation of cell proliferation by Smad proteins. J. Cell Physiol. 191, 1–16. doi: 10.1002/jcp.10066
Tokarz J., Möller G., Hrabě de Angelis M., Adamski J. (2015). Steroids in teleost fishes: A functional point of view. Steroids 103, 123–144. doi: 10.1016/j.steroids.2015.06.011
Trant J. M., Gavasso S., Ackers J., Chung B. C., Place A. R. (2001). Developmental expression of cytochrome P450 aromatase genes (CYP19a and CYP19b) in zebrafish fry (Danio rerio). J. Exp. Zool 290, 475–483. doi: 10.1002/jez.1090
Tuffnail W., Mills G. A., Cary P., Greenwood R. (2009). An environmental 1H NMR metabolomic study of the exposure of the marine mussel Mytilus edulis to atrazine, lindane, hypoxia and starvation. Metabolomics 5, 33–43. doi: 10.1007/s11306-008-0143-1
Uno T., Ishizuka M., Itakura T. (2012). Cytochrome P450 (CYP) in fish. Environ. Toxicol. Pharmacol. 34, 1–13. doi: 10.1016/j.etap.2012.02.004
van Nes S., Andersen Ø. (2006). Temperature effects on sex determination and ontogenetic gene expression of the aromatases cyp19a and cyp19b, and the estrogen receptors esr1 and esr2 in atlantic halibut (Hippoglossus hippoglossus). Mol. Reprod. Dev. 73, 1481–1490. doi: 10.1002/mrd.20514
Viant M. R., Rosenblum E. S., Tieerdema R. S. (2003). NMR-based metabolomics: a powerful approach for characterizing the effects of environmental stressors on organism health. Environ. Sci. Technol. 37, 4982–4989. doi: 10.1021/es034281x
Wang T., Jiang D. N., Shi H., Li G. L. (2021). Liver transcriptomic analysis of the effects of dietary fish oil revealed a regulated expression pattern of genes in adult female spotted scat (Scatophagus argus). Front Marine Sci. doi: 10.3389/fmars.2021.784845
Waxman D. J., Lapenson D. P., Aoyama T., Gelboin H. V., Gonzalez F. J., Korzekwa K. (1991). Steroid hormone hydroxylase specificities of eleven cDNA-expressed human cytochrome P450s. Arch. Biochem. Biophys. 290, 160–166. doi: 10.1016/0003-9861(91)90602-f
Wen X., Chen L., Ai C., Zhou Z., Jiang H. (2001). Variation in lipid composition of Chinese mitten-handed crab, Eriocheir sinensis during ovarian maturation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 130, 95–104. doi: 10.1016/s1096-4959(01)00411-0
Wijesekara N., Konrad D., Eweida M., Jefferies C., Liadis N., Giacca A., et al. (2005). Muscle-specific Pten deletion protects against insulin resistance and diabetes. Mol. Cell Biol. 25, 1135–1145. doi: 10.1128/MCB.25.3.1135-1145.2005
Wu G., Bazer F. W., Dai Z., Li D., Wang J., Wu Z. (2014). Amino acid nutrition in animals: protein synthesis and beyond. Annu. Rev. Anim. Biosci. 2, 387–417. doi: 10.1146/annurev-animal-022513-114113
Wu Y., Chen J., Liao G., Hu M., Zhang Q., Meng X., et al. (2022). Down-Regulation of Lipid Metabolism in the Hepatopancreas of Shrimp Litopenaeus vannamei upon Light and Heavy Infection of Enterocytozoon hepatopenaei: A Comparative Proteomic Study. Int. J. Mol. Sci. 23, 11574. doi: 10.3390/ijms231911574
Xu H., Cao L., Wei Y., Zhang Y., Liang M. (2017). Effects of different dietary DHA: EPA ratios on gonadal steroidogenesis in the marine teleost, tongue sole (Cynoglossus semilaevis). Br. J. Nutr. 118, 179–188. doi: 10.1017/S0007114517001891
Xu G., Du F., Li Y., Nie Z., Xu P. (2016). Integrated application of transcriptomics and metabolomics yields insights into population-asynchronous ovary development in Coilia nasus. Sci. Rep. 6, 31835. doi: 10.1038/srep31835
Yin Y., Tang H., Liu Y., Chen Y., Li G., Liu X., et al. (2017). Targeted disruption of aromatase reveals dual functions of cyp19a1a during sex differentiation in zebrafish. Endocrinology 158, 3030–3041. doi: 10.1210/en.2016-1865
Zhang Y., Fu Y., Jiang S., Qiao H., Xiong Y., Fu H., et al. (2020). Comparative metabolomics analysis of ovarian developmental stages in Macrobrachium nipponense. Comp. Biochem. Physiol. Part D Genomics Proteomics 34, 100648. doi: 10.1016/j.cbd.2019.100648
Zhang X., Li M., Ma H., Liu X., Shi H., Li M., et al. (2017). Mutation of foxl2 or cyp19a1a Results in Female to Male Sex Reversal in XX Nile Tilapia. Endocrinology 158, 2634–2647. doi: 10.1210/en.2017-00127
Zhang M., Xu W., Zhu C., Zou J., Wang Y., Hong F., et al. (2015). Description of intersexual Procambarus clarkii (Decapoda: Cambaridae). J. Nanchang Univ. (Science Edition) 39, 502–506. doi: 10.13764/j.cnki.ncdl.2015.05.019
Zheng J., Cai L., Jia Y., Chi M., Cheng S., Liu S., et al. (2020). Identification and functional analysis of the doublesex gene in the redclaw crayfish, Cherax quadricarinatus. Gene Expression Patterns 37, 119129. doi: 10.1016/j.gep.2020.119129
Zheng J., Cheng S., Jia Y., Gu Z., Li F., Chi M., et al. (2019). Molecular identification and expression profiles of four splice variants of Sex-lethal gene in Cherax quadricarinatus. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 234, 26–33. doi: 10.1016/j.cbpb.2019.05.002
Zhou T., Chen G., Cao J., Wang J., Zou G., Liang H. (2023). Single-molecule real-time sequencing for identifying sexual-dimorphism-related transcriptomes and genes in the Chinese soft-shelled turtle (Pelodiscus sinensis). Anim. (Basel) 13, 3704. doi: 10.3390/ani13233704
Keywords: intersexuality, Cherax quadricarinatus, metabolism, transcriptome, pathway
Citation: He R, Nong C, Chen Y, Li S and Chen H (2024) Transcriptome and metabolome analysis of intersex gonads of redclaw crayfish (Cherax quadricarinatus). Front. Mar. Sci. 11:1506646. doi: 10.3389/fmars.2024.1506646
Received: 05 October 2024; Accepted: 19 November 2024;
Published: 12 December 2024.
Edited by:
Ming Li, Ningbo University, ChinaReviewed by:
Bingmiao Gao, Hainan Medical University, ChinaDr. D. K. Meena, Central Inland Fisheries Research Institute (ICAR), India
Copyright © 2024 He, Nong, Chen, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sedong Li, bGlzZWRvbmdAMTYzLmNvbQ==; Huapu Chen, Y2hlbmhwQGdkb3UuZWR1LmNu
†These authors have contributed equally to this work
 Ruiqi He1,2†
Ruiqi He1,2† Huapu Chen
Huapu Chen