- 1School of Natural Sciences, Manchester Institute of Biotechnology, University of Manchester, Manchester, United Kingdom
- 2Florida Museum of Natural History, University of Florida, Gainesville, FL, United States
- 3Institute of Archaeology, University College London, London, United Kingdom
- 4UMR 7194 Histoire Naturelle de l’Homme Préhistorique, Département Homme et Environnement, Muséum National d’Histoire Naturelle, Paris, France
- 5Department of Natural Sciences, National Museums Scotland, Edinburgh, United Kingdom
- 6School of Geosciences, University of Edinburgh, Edinburgh, United Kingdom
- 7Department of Anthropology, University of Miami, Miami, FL, United States
- 8Georgia Museum of Natural History and Department of Anthropology, University of Georgia, Athens, GA, United States
- 9Department of Anthropology, University of Oregon, Eugene, OR, United States
Introduction: Elasmobranchs, such as sharks and rays, are among the world’s most endangered vertebrates, with over 70% loss in abundance over the past 50 years due to human impacts. Zooarchaeological baselines of elasmobranch diversity, distribution, and exploitation hold great promise for contributing essential historical contexts in the assessment of contemporary patterns in their taxonomic diversity and vulnerability to human-caused extinction. Yet, the historical ecology of elasmobranchs receives relatively less archaeological attention compared to that of ray-finned fishes or marine mammals, largely due to issues of taxonomic resolution across zooarchaeological identifications.
Methods: We explore the use of Zooarchaeology by Mass Spectrometry (ZooMS) for species identification in this unstudied group, using an archaeological case study from the marine environments of the Florida Keys, a marine biodiversity hotspot that is home to an array of elasmobranch species and conservation efforts. By comparison with 39 modern reference species, we could distinguish 12 taxa within the zooarchaeological assemblage from the Clupper archaeological site (Upper Matecumbe Key) that included nine sharks, two rays and a sawfish.
Results and discussion: The results indicate that, through additional complexity of the collagen peptide mass fingerprint, obtained due to the presence of the cartilaginous type II collagen, ZooMS collagen peptide mass fingerprinting provides exceptionally high taxonomic resolution in this group, yielding species-level identifications in all cases where sufficient reference material was used. This case study also highlights the added value of ZooMS for taxa that are more difficult to distinguish in zooarchaeological analyses, such as vertebrae of the Atlantic sharpnose shark (Rhizoprionodon terraenovae) and the hammerhead sharks (Sphyrna spp.) in the Florida Keys. Therefore, the application of collagen peptide mass fingerprinting to elasmobranchs offers great potential to improve our understanding of their archaeological past and historical ecology.
Introduction
Elasmobranchs (sharks, skates, and rays) are essential to the health of marine ecosystems, with many (particularly sharks) often playing vital roles as keystone taxa and apex predators, helping to maintain species diversity and ecosystem health (Heithaus et al., 2007; Motivarash Yagnesh et al., 2020). Today, despite having existed for over 400 million years, elasmobranchs are among the world’s most endangered vertebrate groups due to human impacts, with over 70% loss in abundance over the past 50 years (Pacoureau et al., 2021). This has been particularly devastating to shark populations, whose life histories are characterized by low fecundity, slow growth, late maturity, and relatively long-life spans, all of which makes them particularly vulnerable to population decline via anthropogenic threats such as over-fishing, habitat degradation, and accelerated climate change (Dulvy et al., 2021; Field et al., 2009; Sherman et al., 2023). Moreover, increases in shark exploitation are also linked to declines in popular bony fishes available for harvest (Camhi et al., 1998), as well as rising demand within the international shark fin trade network (Worm et al., 2024).
The historical ecology of elasmobranchs, and especially sharks, demonstrates that humans have engaged with and harvested taxa across the world for millennia (e.g., the Mediterranean [Giovos et al., 2021; Mojetta et al., 2018]; the North Sea [Bom et al., 2022]; the Gulf of Mexico [Martínez-Candelas et al., 2020]; Southeast Asia [Boulanger, 2023; Boulanger et al., 2021]). Utilizing historical perspectives of shark diversity and harvest patterns, garnered through sources such as oral histories (including traditional ecological knowledge, and indigenous traditional ecological knowledge), ethnohistoric texts, art, photography, and more recent (e.g., decade-scale) survey data, make it clear that the worldwide decline in taxa has been driven by human overfishing, with particular emphasis on loss over the past century (e.g., Bradshaw et al., 2008; Juan-Jordá et al., 2022; Roff et al., 2018). This relatively recent timescale for diversity loss presents challenges for creating long-term baselines needed in conservation decision-making and human-wildlife conflict management, whereby historical patterns of species diversity, distribution (including habitat use), and community composition for many taxa are either understudied or unknown (e.g., Ferretti et al., 2008, 2018; Heithaus et al., 2007; McClenachan et al., 2016). This is particularly the case for century-to-millennia-scales of comparison (e.g., Burg Mayer & de Freitas, 2023).
Archaeology has much to contribute to deep-time baselines and elucidating the antiquity of human engagements with elasmobranch taxa through the study of zooarchaeological specimens. Robust zooarchaeological shark, and other elasmobranch, assemblages have been recovered from many archaeological sites across the world (the Americas [e.g., Prieto, 2023; Betts et al., 2012; Colvin, 2014; Rick et al., 2002; Gilson and Lessa, 2021; Lopes et al., 2016], the African continent and Arabian peninsula [e.g., Charpentier et al., 2020; Roberts et al., 2019], the Mediterranean [van Neer et al., 2005] across Oceania [Wright et al., 2016; Weisler and McNiven, 2016] and Asia [e.g., Boulanger, 2023; Langley et al., 2023; Boulanger et al., 2023, 2022, 2021, 2019; Kealy et al., 2020; O’Connor et al., 2019]). However, relative to bony fish specimens and other types of marine vertebrate taxa, zooarchaeological elasmobranch specimens are often difficult to identify beyond order, family, or genus based on morphology alone (Ono and Intoh, 2011), making it challenging to construct deep-time baselines with enough taxonomic resolution to support conservation needs. There are several reasons for this, but primarily it is because chondrichthyan skeletons are composed of hyaline cartilage, with some elements only partly calcified. As a result, it is the vertebral centra, teeth, spines, and dermal denticles that are mostly preserved in archaeological deposits, and although these are the most readily distinguishable elasmobranch parts, they are particularly difficult to identify to the species level due to uniformity and ubiquity among elements within and between many species (e.g., Rick et al., 2002). For example, centrum size, shape and number of vertebrae can be variable among individuals of the same species as well as between species (Springer & Garrick, 1964). Thus, ease of identification to species and quantifications of relative abundance (e.g., % number of identified specimens [NISP], % minimum number of individuals [MNI]) may not be straightforward between regions of the same vertebral column. Shark tooth morphology also differs interspecifically and depending on tooth position along the upper and lower jaws within species. Furthermore, it is important to note that challenges of specimen identification may also be compounded by lack of access to adequate (e.g., taxonomically and elementally representative) modern comparative skeletal collections for many shark species (Burg Mayer and de Freitas, 2023; Shepherd and Campbell, 2021). All these factors mean that it can be difficult to identify elasmobranch zooarchaeological specimens, and even more so within assemblages from world regions characterized by high elasmobranch taxonomic diversity within orders, families, and genera (e.g., Carcharhinus spp., Sphyrna spp.).
Motivated by a desire to better leverage zooarchaeological data within elasmobranch archaeology and historical ecological fisheries baselines, we present the results of an initial Zooarchaeology by Mass Spectrometry (ZooMS) analysis of elasmobranch specimens from a zooarchaeological assemblage at the Clupper site (8MO17). Clupper is an Ancestral Period1 indigenous archaeological site located in the Florida Keys (Keys), USA (Figure 1); the Keys comprise a small-island archipelago renowned for its breadth of marine biodiversity, including several elasmobranch species (Table 1). The marine and estuary habitats of the Keys are essentially made up of “Bay” and “Ocean” waters, including shallower waters (e.g., < 2 m deep) protected by mangrove islands and deeper waters (e.g., 10-30 m deep) beyond embayments, respectively (Tinari and Hammerschlag, 2021).
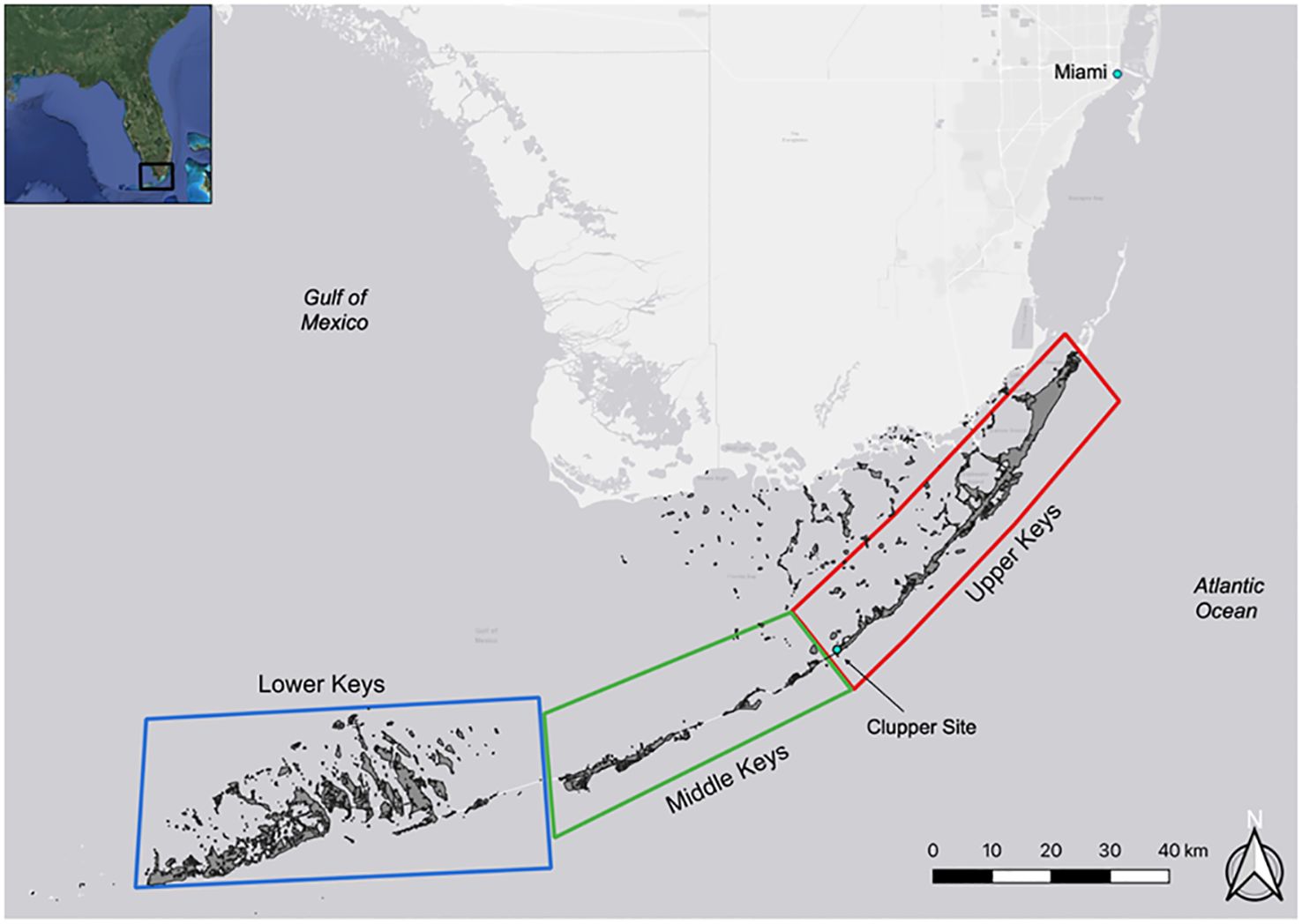
Figure 1. Location of the Clupper archaeological site (8MO17) within the Upper Keys, and distinctions between Upper, Middle, and Lower Keys.
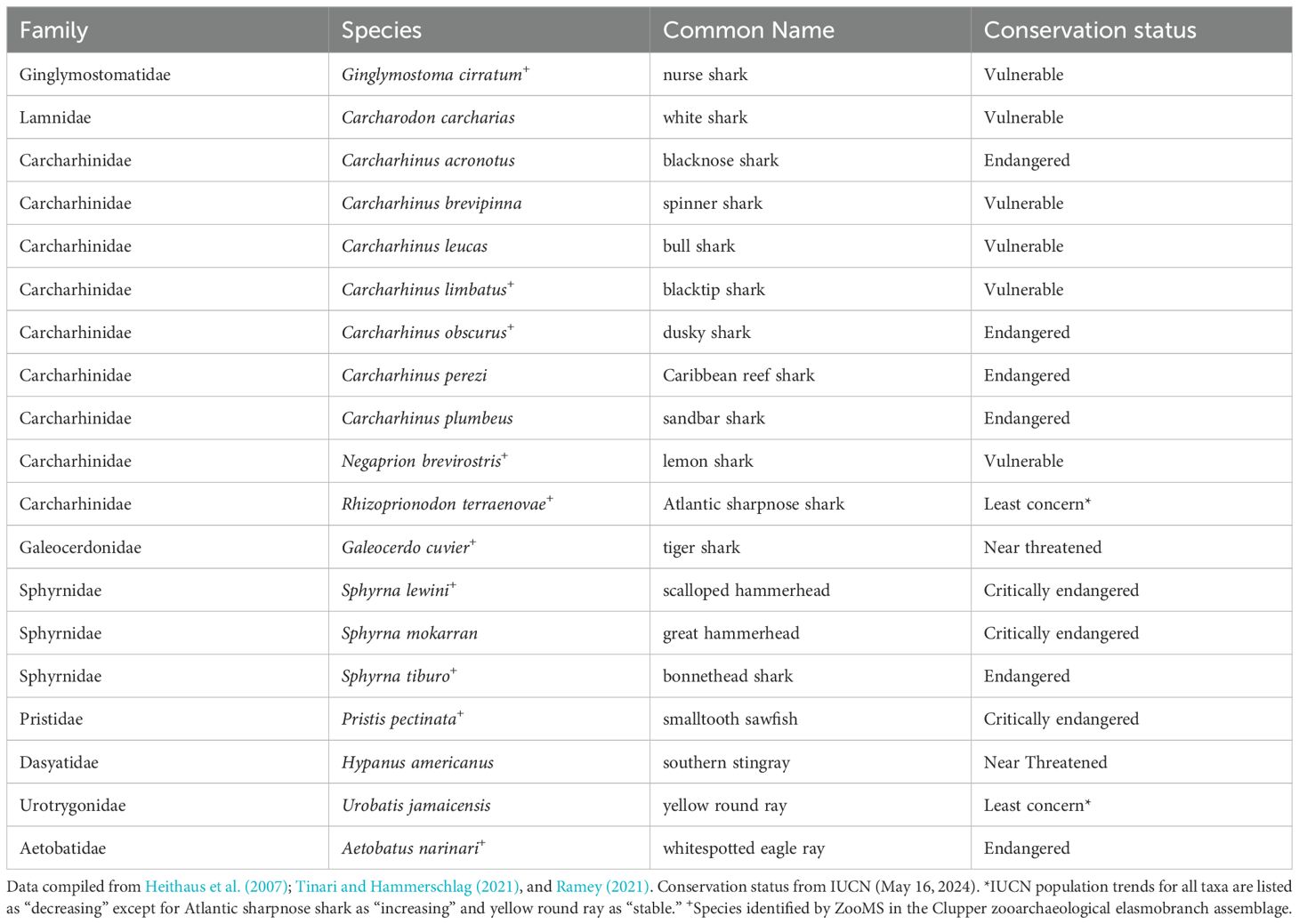
Table 1. List of shark and ray species observed through contemporary survey efforts in south Florida (e.g., Miami and Florida Keys).
The Keys can be characterized as a historical fishery with shifting baselines of fisheries health and persistent decline across a diversity of species throughout the 20th century (e.g., Ault et al., 2005; McClenachan, 2009), including sharks, rays, and sawfishes (Heithaus et al., 2007; Powers et al., 2013). Zooarchaeological assemblages from the Keys provide a record of deep-time fisheries, including centennial-to millennial-scale perspectives of taxonomic diversity, distribution, and human engagement (e.g., LeFebvre et al., 2022), but given the abovementioned limitations in the zooarchaeology of elasmobranch remains, the goals of this study were to: 1) investigate the efficacy of the collagen-fingerprint-based approach known as ZooMS (Zooarchaeology by Mass Spectrometry) in identifying zooarchaeological elasmobranch specimens to species; and 2) begin to build a species-level baseline of elasmobranch diversity present in the Keys approximately 700-1,000 years ago (Ardren et al., 2018) for the advancement of archaeological and historical ecological research.
We begin with background summaries of biomolecular methods in species identifications as well as elasmobranch skeletal structure to contextualize the ZooMS approach, followed by an overview of the Clupper site and current zooarchaeological analysis of elasmobranch specimens. Then we review the materials and methods of analysis with an emphasis on building a modern comparative baseline for collagen peptide mass fingerprints of this previously untried taxonomic group. The results of the analysis demonstrate the ability of ZooMS to achieve species-level identifications of zooarchaeological specimens and are presented within the context of contemporary elasmobranch diversity and habitats. Using the Clupper data as an exemplar, we highlight the implications of the results from archaeological perspectives of Ancestral Indigenous harvest, elasmobranch historical ecology, and future zooarchaeological research.
Background
Biomolecular methods of species identification
Although biomolecular methods offer an objective solution to the issue of species identification, they are largely underutilized for several reasons, including costs per sample, damage to specimens, and speed of interpretation in some cases. The analysis of ancient DNA is becoming increasingly more cost-effective (e.g., Seersholm et al., 2018), and will undoubtedly increase in use in the future, yet its greatest limitation is that of molecular preservation, whereby the few studies that have attempted to do so for shark taxa have focused on very recent material less than a few hundred years old (Ahonen and Stow, 2008; Nielsen et al., 2017) and even these can yield <50% success rates (e.g., Shepherd and Campbell, 2021).
Proteins on the other hand, particularly bone collagen, are thought to survive much longer, or at least better in warmer environments, such as those inhabited by most tropical shark species. Harnessing this greater longevity, a method of species identification by ‘fingerprinting’ collagen using mass spectrometry, referred to as ‘ZooMS’ (Zooarchaeology by Mass Spectrometry), was created over a decade and a half ago (Buckley et al., 2009; 2010), based initially on mammals for investigating animal husbandry practices (e.g., Buckley and Kansa, 2011; Price et al., 2013), with wider taxonomic use in studies on reptiles (Harvey et al., 2019; Guiry et al., 2024), birds (Eda et al., 2020), amphibians (Buckley and Cheylan, 2020), and fishes (Richter et al., 2011; Harvey et al., 2018; 2022; Rick et al., 2019; Guiry et al., 2020; Buckley et al., 2021; 2022) in comparison to the other ‘lower vertebrate’ groups.
Although there are more than 28 types of collagen known to exist within humans at some point in life (Ricard-Blum, 2011), 80-90% of the collagen in the body consists of types I, II and III, of which type I collagen is by far the most abundant. The triple helical structure common to all collagens is maintained through the presence of repeating amino acids, proline (Pro) and its modified form hydroxyproline (Hyp), which induce the twisting structure in each chain. Some collagen types are formed from three identical trains, with the cartilage-dominant type II collagen being a prime example. In contrast, the dominant type in bone, type 1 collagen, is made of genetically distinct chains, which in most vertebrates are made up of two identical chains (alpha 1 chains) and one genetically distinct (alpha 2) chain. In bony ray-finned fishes, type 1 collagen is composed of three distinct chains, where the third (α3(I)) is a duplicate of the (α1(I)) gene (Morvan-Dubois et al., 2003). However, this distinct third chain does not appear to exist in sharks and lampreys (Kimura and Ohno, 1987), given the timing of the duplication event that gave rise to this (Harvey et al., 2021).
Elasmobranchs and their skeletal structure
The vertebrates are a subphylum (Vertebrata) within the phylum Chordata. The jawed vertebrates (gnathostomes) are a group comprising all tetrapods as well as the bony fishes (Osteichthyes), and the cartilaginous fishes (Chondrichthyes). During ontogeny, most vertebrate skeletons are initially composed mostly of hyaline cartilage that is largely replaced by bone via endochondral ossification (Hall, 1975); the biomineralization of skeletal tissues occurs through the deposition of biological apatite into collagen-rich (or amelogenin-rich, in the case of enamel and enameloid) matrices (Donoghue et al., 2006). However, as their name suggests, the skeletal structure of cartilaginous fishes remains primarily composed of cartilage, as they do not develop osseous skeletons, having secondarily lost this ability to produce endoskeletal bone (Coates et al., 2008). Elasmobranchs develop a relatively thin outer layer of cortical mineralization over most of their skeleton (Dean et al., 2015; Seidel et al., 2016), which is typically characterized by type II collagen, a triple helical molecule made up of three identical alpha chains (COL2A1).
The living chondrichthyans are split into two main groups, the Holocephali and the Elasmobranchii. The former are divided into three families, the Callorhincidae, Chimaeridae and the Rhinochimaeridae, whereas the latter are a much more diverse group, including the sharks (selachians; nine recognized orders) and the rays, skates, sawfishes and guitarfishes (batoids; four recognized orders). The cartilaginous elements (jaws, fins, and vertebrae) are covered by mineralized polygonal tiles called tesserae, more abundant in elasmobranchs, but also increasingly recognized in holocephalans (Pears et al., 2020; Seidel et al., 2020). It is the internal part of these tesserae, the round cells enclosed in lacunae, that contain the cartilaginous type II collagen (Seidel et al., 2017), whereas the external parts located on the perichondrial side are characterized by flatter cells that are engulfed in a type I collagen matrix (Orvig, 1951; Kemp and Westrin, 1979), known as fibrous perichondrium (Dean et al., 2015). In addition to this, as reviewed by Dean and Summers (2006), there are other types of mineralized tissues in the elasmobranch endoskeleton. Areolar mineralization characterizes the vertebral centra, and lamellar mineralization, comparable to bone tissue, has been recorded in the neural arches. The latter was first termed osseous tissue due to the presence of elongated cells like the osteoblasts of bone (Peignoux-Deville et al., 1982), expressing type I collagen genes (Enault et al., 2016), and because they are enclosed within a type I collagen-rich extracellular matrix that is able to mineralize (Eames et al., 2007); see Berio et al. (2021) for review. Therefore, although dominated by type II collagen, type I collagen typical of bone is estimated to account for about one third of the total collagen content of shark cartilage (Rama & Chandrakasan, 1984).
The Clupper site and elasmobranch specimens
Elasmobranch specimens and artifacts, especially from sharks (e.g., shark tooth drills), have been documented from coastal, island, and inland Ancestral Period Indigenous archaeological sites across greater Florida, attesting to the cultural significance of such taxa in the past, including their use as food, tools, and items of personal adornment, ritual significance, and/or trade for several millennia (e.g., Early Archaic, ca. 6,000 BC [Farrell, 2021]; see also Keller and Thompson, 2013). The zooarchaeological specimens used in this study were selected from the Clupper site, an Indigenous village inhabited ca. AD 650-1,250 (Ardren et al., 2018) (Figure 1). The site is located on Upper Matecumbe Key, surrounded by or within proximity to a variety of maritime habitats, including sandy bottom flats, coral reefs, sea grass beds, rocky substrate, mangroves, marshes, and deeper waters. The site comprises a black earth midden. The midden is characterized by an abundance of well-preserved vertebrate fauna, invertebrate fauna, pottery and shell artifacts (Ardren et al., 2018; LeFebvre et al., 2022). Current zooarchaeological results from midden samples show that while bony fishes are the most abundant vertebrate taxon as represented per number of individual specimens (NISP = 9,350), followed by marine and freshwater turtles (NISP = 7,683), elasmobrachs are the third most relatively well-represented taxon with 578 NISP (Oliveira, 2024; see Supplementary Table S1). However, it is possible that elasmobranchs are underrepresented within the assemblage due to their cartilaginous structure, with representation being limited to vertebrae and teeth (LeFebvre et al., 2022; Rick et al., 2002).
The Clupper elasmobranch specimens were identified based on morphology using modern comparative skeletal specimens from the Environmental Archaeology Program (EAP) and South Florida Archaeology collections at the Florida Museum of Natural History, USA (LeFebvre et al., 2022). The taxonomic resolution achieved (Table 2) included specimens identified at the level of order (n = 2), family (n = 1), genus (n=3), species (n=4), and cf. (compares with) species (n=3). Order-level identifications to Carcharhiniformes (ground sharks; n = 229) and Rajiformes (flattened cartilaginous fishes; n = 101) accounted for the majority of individual specimen identifications. Carcharhinidae was the only family-level identification. The most abundantly identified shark taxon beyond order or family was the hammerhead genus (Sphyrna spp.). Spotted eagle ray (Aetobatus narinari) specimens were the most common Rajiformes identified beyond order. Other levels of genus, species, and cf. species identifications included nurse shark (Ginglymostoma cirratum), tiger shark (Galeocerdo cuvier), sandbar shark (cf. Carcharhinus plumbeus), lemon shark (cf. Negaprion brevirostris), Atlantic sharpnose shark (Rhizoprionodon terraenovae), sawfish (Pristis spp.), stingray (Hypanus spp.), and Atlantic stingray (Hypanus cf. sabinus).
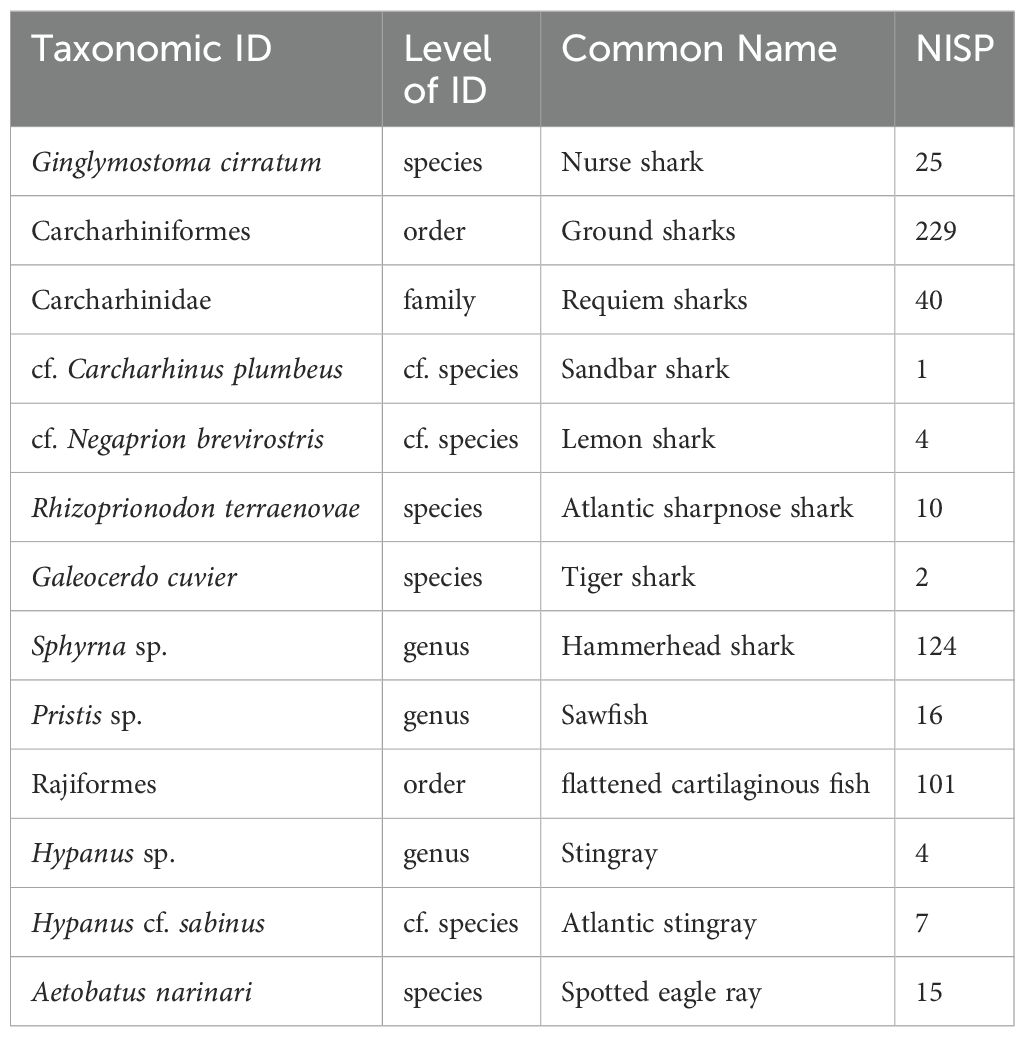
Table 2. Elasmobranch taxonomic identifications based on morphology from the Clupper site, Upper Matecumbe Key, Florida, U.S.A. Levels of taxonomic identification span order to species.
Interpretations of Indigenous fishing practices at Clupper indicate emphases on inshore habitats and a generalist, or multi-species approach, to fishing versus targeted or selective (LeFebvre et al., 2022; Oliveira, 2024). However, it is important to note that a generalist (i.e., multi-species) fishing strategy does not preclude targeted or preferential consumption of species caught. Rather, it refers to fisheries strategies that usually employ gear suited to capturing a diversity of species versus single or a selected few (Bieg et al., 2018). It is also the case that generalist fishing strategies are often not exclusively practiced and may include selective fishing approaches depending on species behaviors and availability (e.g., migrations, aggregations), as well as environmental conditions throughout a year (e.g., Ziegler et al., 2018).
The suggestion of a generalist strategy to fishing at Clupper is based on the identifications of bony fish and sea turtle taxa at genus or species levels and the overall high taxonomic diversity present in the zooarchaeological assemblage (LeFebvre et al., 2022). Moreover, genus- and species-level identifications indicate a fishery primarily focused on inshore habitats (Oliveira, 2024). Patterns of harvest over time have also been noted, including a decrease in almost exclusively marine taxa (e.g., sea turtles, grunts [Haemulon spp.]) concomitant with an increase in taxa that also frequent brackish waters, such as catfishes (e.g., Ariopsis felis and Bagre marinus) (Oliveira, 2024). Based on the schooling habits and daily migration patterns of the majority of bony fishes identified, fishing technology likely employed a mix of mass capture with nets and smaller catches via traps (Oliveira, 2024). Interpreting elasmobranch harvest has been less nuanced due to the lack of taxonomic resolution beyond order or family. Thus, shark, ray, and sawfish species harvest has been generally described as possible across essentially any habitat a shark or ray may inhabit (LeFebvre et al., 2022).
Materials and methods
Both modern and zooarchaeological elasmobranch specimens were sampled for this study. The selection and analysis of modern specimens was based on establishing a modern baseline of species identification, using ZooMS. Zooarchaeological specimens were selected to test the ability of ZooMS to confirm, refine, or refute previous identifications based on morphology.
Modern reference materials
Modern skeletal tissues (jaw and/or vertebrae; see Supplementary Table S2) were obtained for 39 species of 17 taxonomic families of elasmobranch, including all of those known to inhabit the oceanic and estuarine environments surrounding peninsular Florida and the Keys (Table 3). The specimens were selected from the modern skeletal comparative collection in the Environmental Archaeology Program at the Florida Museum of Natural History supplemented by specimens of two Pristis species sampled in triplicate from National Museums Scotland due to uncertainty in the taxonomic identification of the Florida Museum of Natural History (FLMNH) reference specimen. With a focus on sampling as much taxonomic diversity as possible relevant to the study region represented in the comparative collection, specimens included vertebrae, teeth, as well as dry-preserved cartilaginous materials.
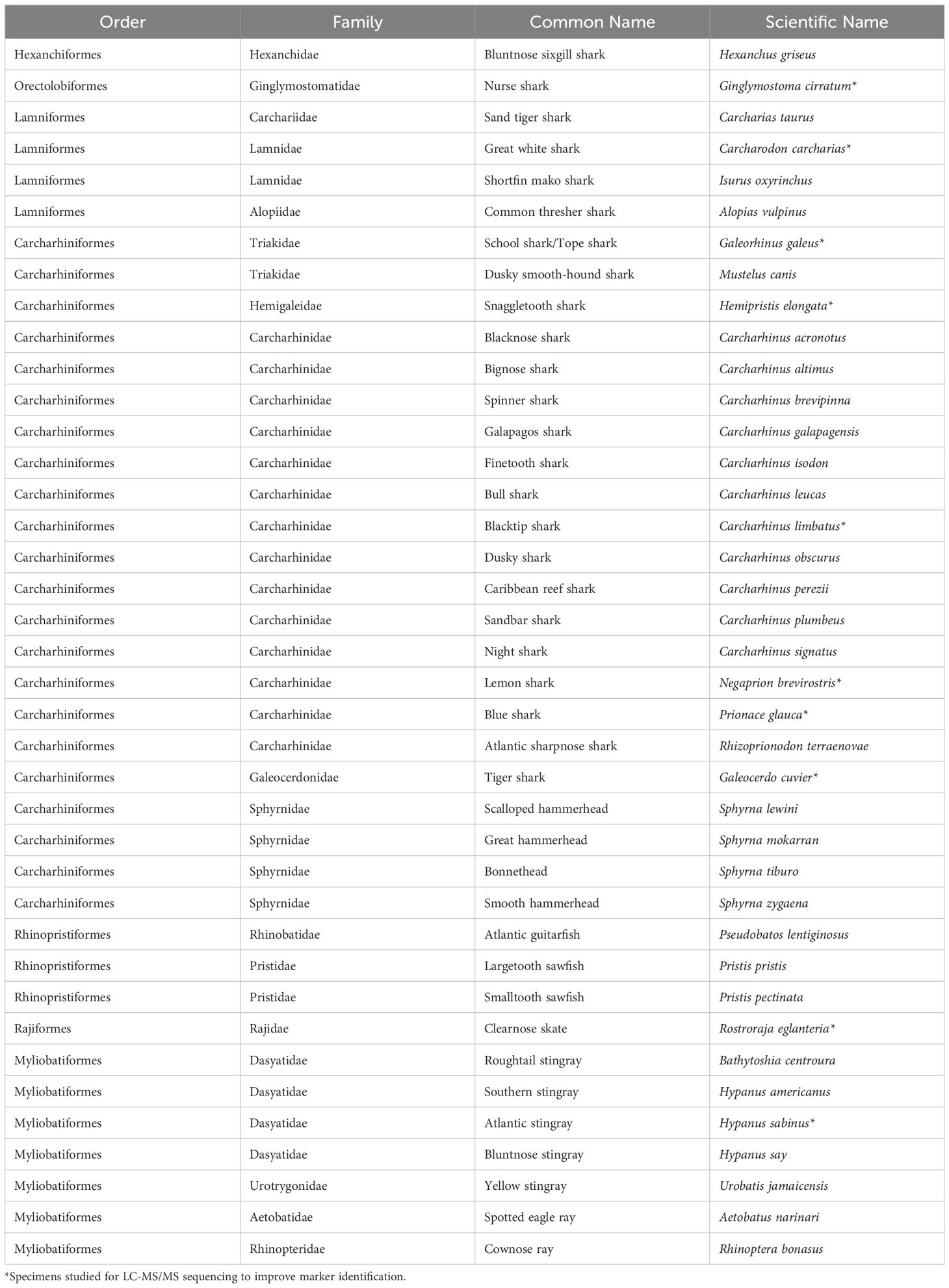
Table 3. List of species included in this study (specimen details given in Supplementary Table S2).
Archaeological materials
Selection of zooarchaeological specimens from currently analyzed Clupper samples was aimed at either refining or testing the taxonomic breadth of specimen identifications made based on morphology (Supplementary Table S3). Specimens were analyzed from the following levels of identification: Order (Carcharhiniformes, Rajiformes), Family (Carcharhinidae), Genus (Sphyrna sp., Hypanus sp., Pristis sp.), species (Aetobatus narinari, Rhizoprionodon terraenovae, Ginglymostoma cirratum), and likely species (cf. Hypanus sabinus, cf. Negaprion brevirostris, cf. Galeocerdo cuvier). Analyzed specimens were from two 50 × 50 cm test pits excavated in 10 cm arbitrary levels. It is important to note that the focus of this initial effort was not quantitative in terms of identifying relative abundances of species represented at the site or trends in elasmobranch taxonomic diversity through time (i.e., across levels of excavation), but rather to gain as refined a taxonomic baseline as possible of species represented at the Clupper site during its Ancestral history from which to (re)consider continuing and future approaches to elasmobranch identification, quantification, and interpretation at Clupper and across the Keys more broadly.
ZooMS collagen peptide mass fingerprinting
Approximately 25-50 mg of modern and archaeological tissues were processed for collagen peptide mass fingerprinting following Buckley (2013), with modern tissues degreased beforehand by fully submerging twice in 83%/17% chloroform/methanol (first for 20 minutes, then for 3 hours). For collagen extraction, 1 mL 0.6 M hydrochloric acid (Sigma-Aldrich, UK) was added to each intact sample to enable decalcification. Half of the acid-soluble fraction was then ultrafiltered into 50 mM ammonium bicarbonate (Sigma-Aldrich, UK), with two centrifugation steps (20 min; 12,400 rpm) and recovered in 0.1 mL solution. This was then diluted 1/20 and 1 µL co-crystalized with 1 µL 10 mg/mL alpha-cyano hydroxycinnamic acid (Sigma-Aldrich, UK) in 50% acetonitrile (ACN)/0.1% trifluoroacetic acid (TFA) onto a stainless-steel Matrix Assisted Laser Desorption Ionization Time-of-Flight (MALDI-ToF) mass spectrometric target plate. Using a Bruker Rapiflex MALDI-ToF instrument, up to 20,000 laser shots were acquired over the mass/charge (m/z) range 700-3,700 and resultant spectra of archaeological samples were manually categorized into ‘groups’ that each were composed of their own set of peptide markers. Peaks that appeared to differ between these groups were then compared with those present or absent in the associated reference material within this study, taking into consideration morphological identification for the majority in each archaeological group. Tandem mass spectra were also acquired via LC-MS/MS sequencing of selected modern specimens for improved results from database searching to assist confirmation of homologous markers between taxa.
LC-MS/MS sequencing
LC-MS/MS was carried out to improve understanding of the peaks observed in the ZooMS spectra (see Supplementary Table S3 for taxa selected to span range of biomarkers). Digested samples were analyzed using an UltiMate® 3000 Rapid Separation LC (RSLC, Dionex Corporation, Sunnyvale, CA) coupled to a QE HF (Thermo Fisher Scientific, Waltham, MA) mass spectrometer. Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile, and the column used was a 75 mm × 250 μm internal diameter 1.7 μM CSH C18, analytical column (Waters, UK). A 1 μl aliquot of the sample was transferred to a 5 μl loop and loaded on to the column at a flow of 300 nl/min for 5 minutes at 5% B. The loop was then taken out of line and the flow was reduced from 300 nl/min to 200nl/min in 0.5 minute. Peptides were separated using a gradient that went from 5% to 18% B in 34.5 minutes, then from 18% to 27% B in 8 minutes and finally from 27% B to 60% B in 1 minute. The column is washed at 60% B for 3 minutes before re-equilibration to 5% B in 1 minute. At 55 minutes the flow is increased to 300 nl/min until the end of the run at 60 minutes. Mass spectrometry data were acquired in a data-directed manner for 60 minutes in positive mode. Peptides were selected for fragmentation automatically by data-dependent analysis on a basis of the top 12 peptides with m/z between 300 to 1750 Th and a charge state of 2, 3 or 4 with a dynamic exclusion set at 15 seconds. The MS Resolution was set at 120,000 with an AGC target of 3 x106 and a maximum fill time set at 20 ms. The MS2 Resolution was set to 30,000, with an AGC target of 2 ×105, a maximum fill time of 45 ms, isolation window of 1.3 Th and a collision energy of 28. All data were collected in centroid mode. Raw files were then converted to mascot generic format (.MGF) files, which were searched against a locally curated database (Supplementary Table S4) of elasmobranch collagen type 1 (COL1A1 and COL1A2) and type 2 (COL2A1) sequences obtained from the protein BLAST search of the elephant shark (Callorhinchus milii). To assist evaluation of the manually selected peptide biomarkers, the resultant.MGF files for each Error Tolerant search were combined, filtered by removing peptides of <10 ion score, and sorted via m/z value.
Results
Peptide composition of the mass spectrometric fingerprints
The results indicate that, despite additional complexity of the collagen peptide mass fingerprint obtained due to the presence of the cartilaginous type II collagen, ZooMS provides exceptionally high taxonomic resolution in this group, yielding species-level identifications. Although only three of the species included in this study had known collagen type I (COL1A1 and COL1A2) and type II (COL2A1) sequences, Hypanus sabinus, Pristis pectinata and Carcharodon carcharias (see Supplementary Tables S5–S7 respectively for LC-MS/MS sequencing results), it was clear that a large proportion of peptides in the fingerprints derived from COL2A1, both in the archaeological (Figure 2) and the modern samples (Supplementary Figures S1, S2). Although differences in relative abundance of particular peaks could be observed in some of our peptide mass fingerprint comparisons between each sampled modern tissue type (e.g., tooth vs. vertebrae; Supplementary Figures S3–S5), they were not unexpected, reflecting the difference in dominant protein, whether COL1 or COL2.
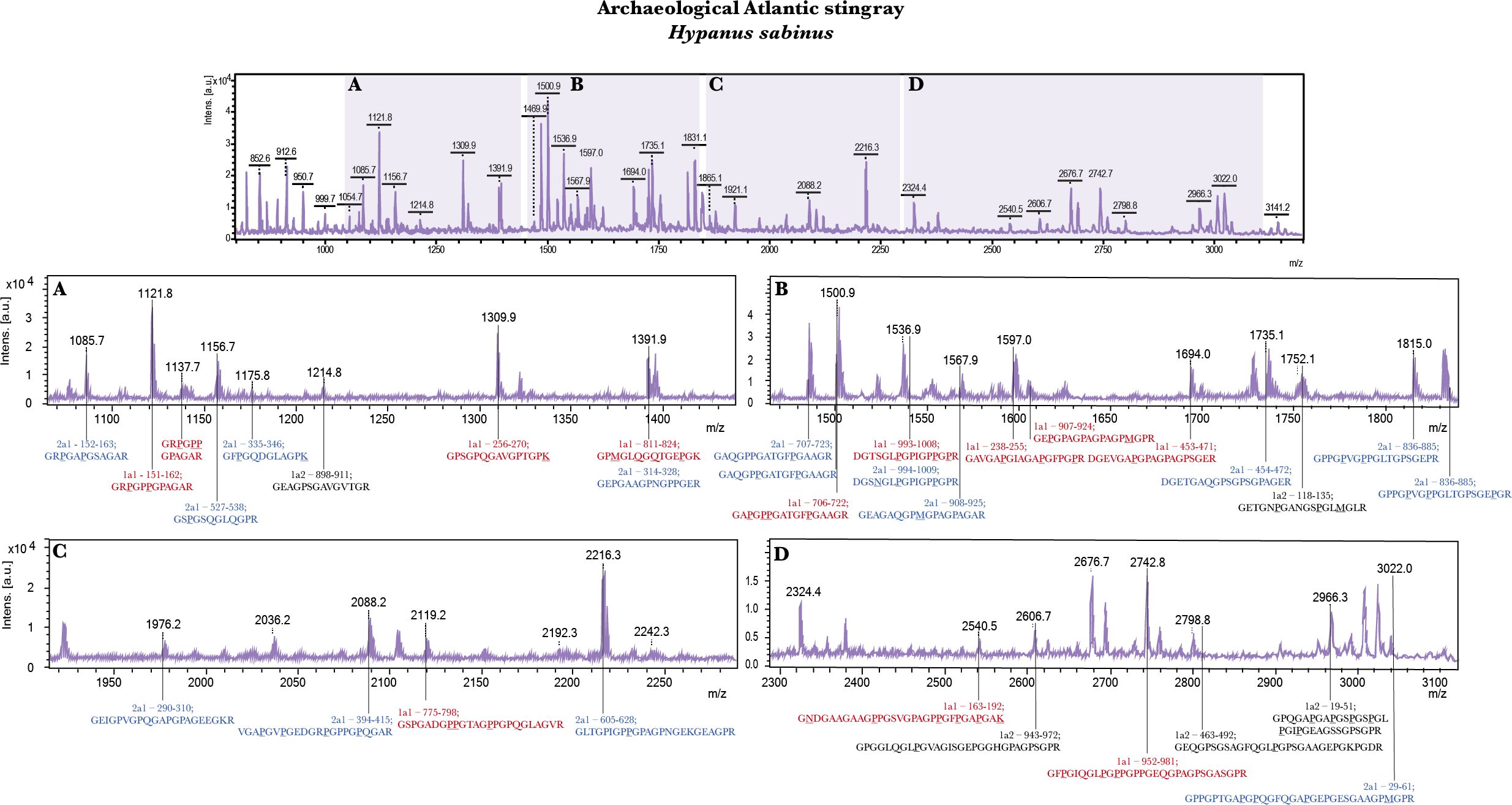
Figure 2. Sequence-annotated MALDI peptide mass fingerprint (top spectrum) of dominant proteins extracted from a confidently identified archaeological Hypanus sabinus vertebra (black text = COL1A1, red text = COL1A2, blue text = COL2A1; matched to peptides listed in Supplementary Table S5) with zoomed in sections (A–D) in order of increasing m/z.
Taxonomic resolution: modern comparative baseline
Species-level differences between the collagen peptide mass fingerprints for modern comparative specimens could readily be observed (Supplementary Figures S6–S16), even for each one of the 12 species of Carcharhinus and the four species of Sphyrna analyzed here (Table 3). Sampling of the two Pristis species of relevance to this study showed that the initially supplied reference material from P. pristis was incorrect, deriving from P. pectinata, more in keeping with geographical origins of the reference material. These differences, proposed as species biomarkers (Supplementary Table S8; Supplementary Figures S17–S51), were clearly visible in the peptide mass fingerprints of the archaeological specimens also (Figure 3).
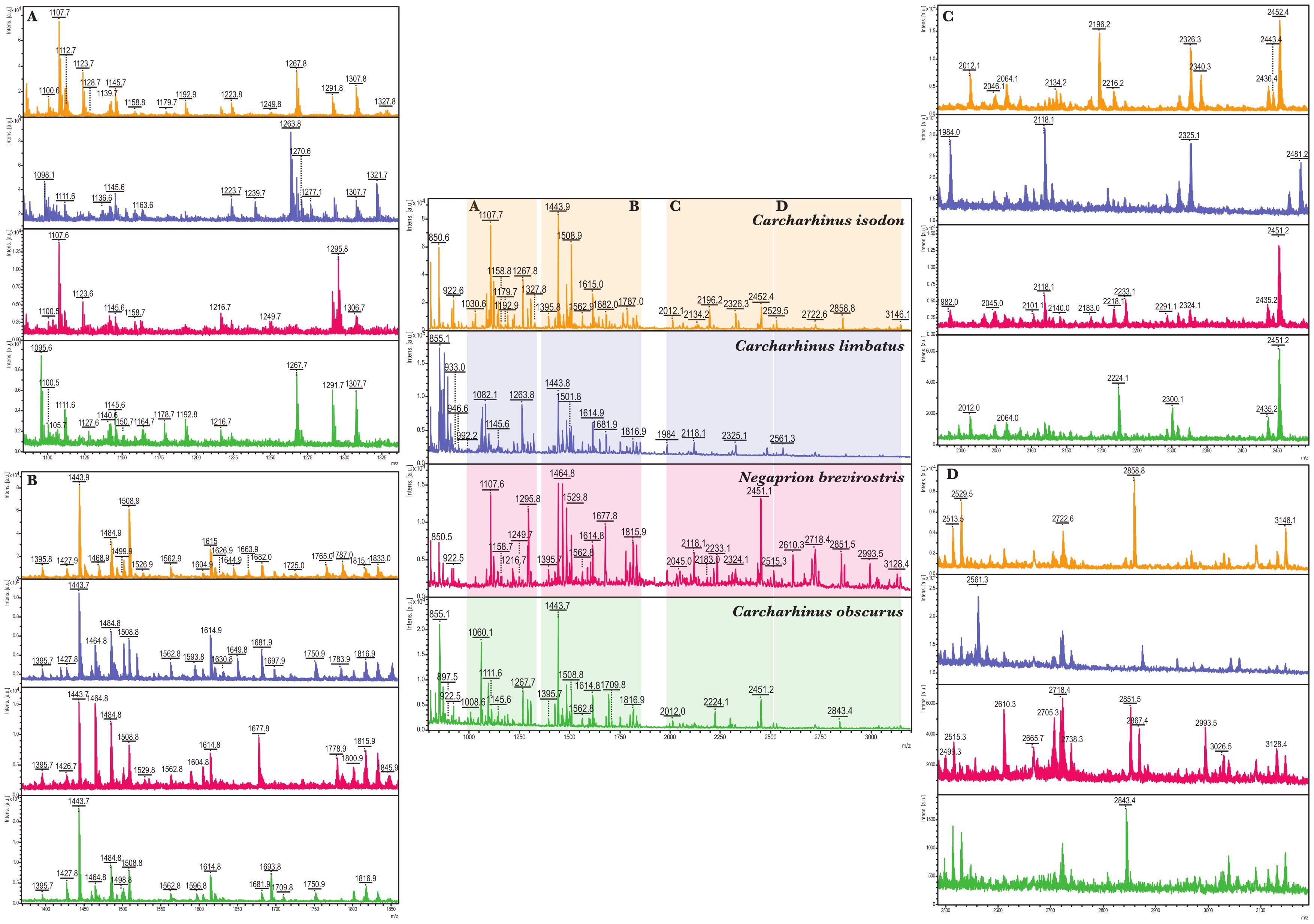
Figure 3. Example peptide mass fingerprints showing taxonomic resolution amongst archaeological samples of carcharhinids; central portion shows the most diagnostic regions (A–D) for species determination of the mass spectra for each of the four elasmobranches Carcharinus isodon (orange; top), C. limbatus (blue; second to top), Negaprion brevirostris (red; second to bottom) and C. obscurus (green; bottom) surrounded by zoomed in spectra (A–D) of increasing m/z.
Taxonomic resolution: zooarchaeological baseline
Although just over 30% (n=41) of the 137 analyzed archaeological samples did not yield suitable quality collagen fingerprints, the 98 specimens that did covered a greater than expected range of taxonomic diversity based on morphological identification, with 12 distinct groups identified (Figure 4). While the majority of these results derived from specimens that could not be morphologically identified below the family level (i.e., Carcharhinidae) and often order level, 48 of our samples did derive from a genus-level or lower suggested identification, though mostly (n=33) from the hammerheads Sphyrna spp. (Supplementary Table S3); of the 40 of these that were successful (7 of the 8 poor collagen samples were of Sphyrna, with one from Galeocerdo), 10 were initially incorrect on the original morphological identification, and one of these (#78) cautioned/corrected prior to ZooMS analysis (initially considered Rhizoprionodon terraenovae and later suggested on morphological grounds as potentially deriving from hammerhead [Sphryna]), an identification confirmed by ZooMS and improved to the level of the bonnethead (S. tiburo). All other misidentifications were from specimens morphologically identified as hammerhead, corrected to Atlantic sharpnose shark (R. terraenovae) in six instances (#70, 87, 99, 101, 103, 110), the lemon shark (Negaprion brevirostris) in two instances (#66, 95), and the blacktip shark (C. limbatus) in one instance (#61). These results are suggestive of misleading morphological criteria used between the two taxonomic groups during initial identification. However, we also note the value in morphological identification, which remains important given the substantial number of specimens that do not yield collagen/biomolecular information, which, in this case comprised as much as approximately one third of the samples analyzed in this study, and is likely to have taphonomic biases that relate to species and element.
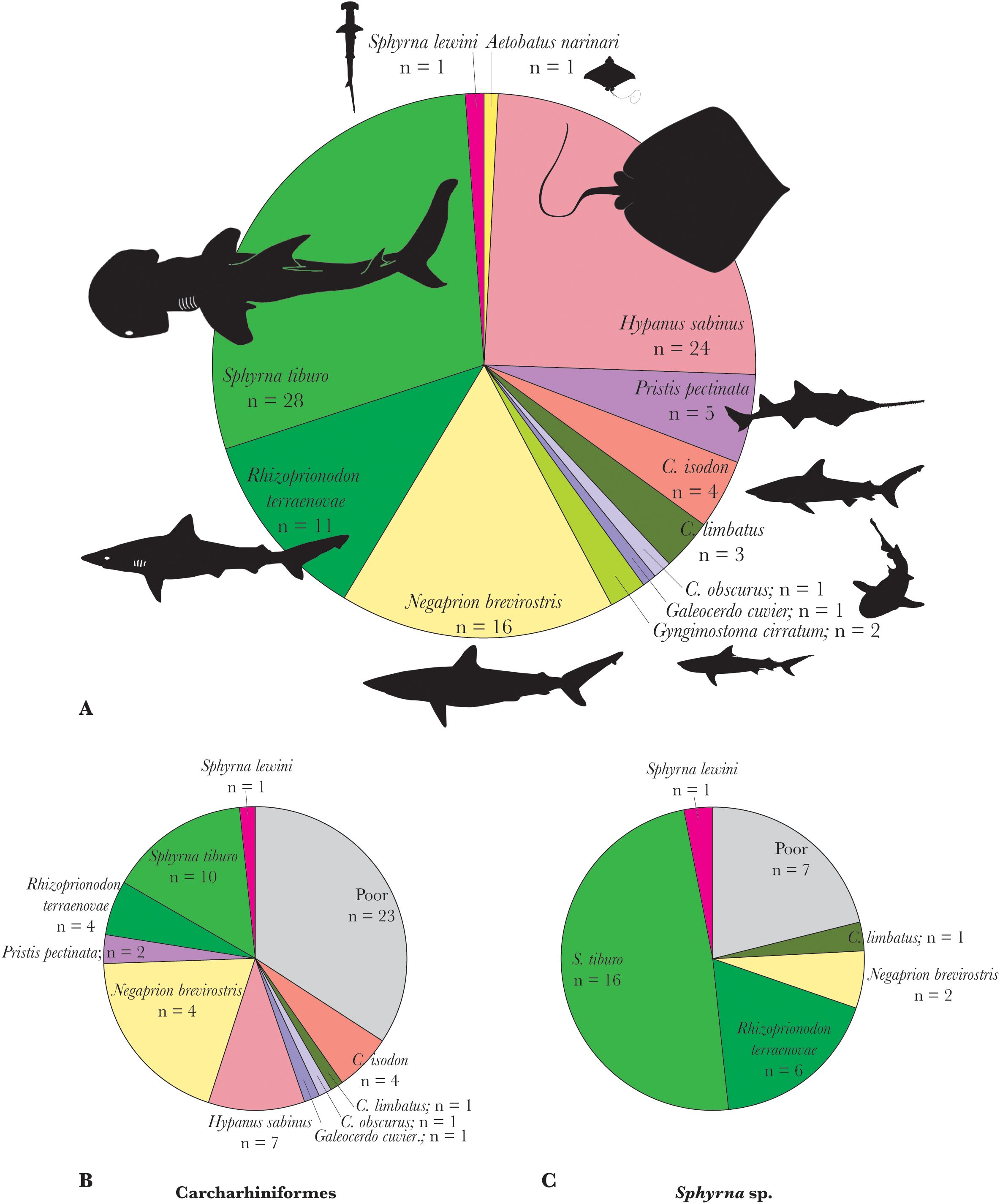
Figure 4. Pie charts showing (A) species composition of archaeological remains analyzed by ZooMS, (B) the ZooMS results of species identified morphologically as Carchariniformes, and (C) the ZooMS results of species identified morphologically as Sphyrna (bottom right).
Consistent with the morphological identifications, the most abundant shark taxon observed via ZooMS among the Clupper specimens was Carcharhinidae (i.e., migratory live-bearing sharks of warm seas). Within this family, the lemon shark was the most frequently identified among the sampled specimens. There are only two species of lemon shark within the genus Negaprion. The lemon shark (N. brevirostris) inhabits oceanic waters of the Americas, and the sicklefin lemon shark (N. acutidens) inhabits the Indo-Pacific. Lemon sharks prefer shallow coastal waters, often feeding at night (Wetherbee et al., 1990) and growing to >3 m in length (Dibattista et al., 2007). Lemon sharks are abundant in the Keys and are known to inhabit inshore marine habitats, such as coral reefs or flats, as well as estuaries (Compagno, 1984; Jennings et al., 2008; Tinari and Hammerschlag, 2021; Wiley and Simpfendorfer, 2007).
Carcharhinus is the largest genus of the Carcharhinidae family, containing at least 35 different species. Of those identified at Clupper, the finetooth shark (C. isodon) is commonly found in the western Atlantic from North Carolina to Brazil. Relatively small (1.6-1.7 m) and fast swimming, it forms large schools in shallow inshore coastal waters and habitats (Compagno, 1984). It is known to migrate seasonally to follow warm waters (Castro, 1993), including those surrounding Florida during the winter. Also identified within the Clupper assemblage, the blacktip shark (C. limbatus) has a worldwide distribution in tropical waters and is usually found in groups of varying size in waters less than 30 m deep, including mangrove and estuary habitats, as well as favoring island lagoons and drop-offs near coral reefs (Castro, 1996). The blacktip is described as a stocky species, typically reaching between 1.2 to 1.9 m, and only rarely reaching 2 m in length (Castro, 1996; Compagno, 1984). While this species is present in waters of the Gulf of Mexico, southern Florida and the Keys year-round (Tinari and Hammerschlag, 2021; Wiley and Simpfendorfer, 2007), it tends to be most abundant during winter months after migrating southward from nursery grounds along the northern Gulf coast, Carolinas, and Georgia (Castro, 1996; Heithaus et al., 2007). It occurs in inshore habitats with shallower waters. The dusky shark (C. obscurus) is one of the largest members of this genus, at 3-4 m in adult length; it spends most of its time at 10-80 m depths and within the western Atlantic migrates south during the winter (Natanson and Kohler, 1996).
The Atlantic sharpnose shark (R. terraenovae), the third most identified shark in this study, is also a relatively small shark (~1 m adult length) (Compagno, 1984). It is one of the most frequently encountered sharks in subtropical waters off the north-western Atlantic Ocean today, including Florida, and the Gulf of Mexico (Branstetter, 1990; Marquez-Farias and Castillo-Geniz, 1998; Parsons and Hoffmayer, 2005). It is considered a coastal species that often engages in regular inshore and offshore migration to depths of up to 280 m (Compagno, 1984). It is found in a variety of habitats such as shallow seagrass beds to deep non-vegetated sand or mud and utilizes a series of coastal bays and estuaries as nurseries for juvenile development (Carlson et al., 2008). Atlantic sharpnose sharks are year-round residents of the Gulf of Mexico, Florida and the Keys, but are most common during the winter (Tinari and Hammerschlag, 2021); they are similar in length to bonnetheads, with the morphology of the vertebrae being especially difficult to distinguish between the two species.
The tiger shark (Galeocerdo cuvier) is the only extant species in the family Galeocerdonidae. It is a large (>5 m), solitary, mostly nocturnal hunter, moving closer to shore to feed (Compagno, 1984). It inhabits coastal and open ocean environments, and is highly mobile, engaging in ontogenetic and seasonal migrations (Ajemian et al., 2020; Lea et al., 2015). It prefers tropical waters during the winter (e.g., Caribbean and Florida) and moves to high-latitude oceanic areas in the summer (Lea et al., 2015). Although it is known to inhabit a variety of inshore habitats, such as estuaries, reefs, and lagoons (Compagno, 1984), there is also an increased use of continental-slope and deep-water habitats with increasing size (Ajemian et al., 2020). Off the coast of Florida, it is most often found in deeper oceanic waters (Tinari and Hammerschlag, 2021). The nurse shark (Ginglymostoma cirratum) is the only species of shark from a different order than ground sharks, the Orectolobiformes. It is a very common inshore, bottom-dwelling shark, often inhabiting waters of one meter or less deep and up to 12 m. It is typically ~3 m in size and is found on rocky reefs, channels between mangrove keys, and sand flats (Compagno, 1984). It is the most common shark found year-round in the shallow waters of the Gulf of Mexico, Florida, and the Keys (Heithaus et al., 2007; Tinari and Hammerschlag, 2021; Wiley and Simpfendorfer, 2007). Nurse sharks are largely nocturnal, often described as sluggish during the daytime, and frequently found aggregating in large sedentary resting groups (Compagno, 1984).
After the requiem sharks mentioned above, the second most frequently identified shark taxonomic group was the hammerheads, of the genus Sphyrna. Hammerheads are common, coastal inshore sharks among the Keys, and like most sharks, they are solitary hunters in the evening. The hammerhead species identified from Clupper were the bonnethead (S. tiburo) and scalloped hammerhead (S. lewini). The bonnethead is the smallest of hammerhead species, typically <1 m in length, but can reach up to 1.3 m in size (Carlson and Parsons, 1997; Compagno, 1984). This species prefers inshore, coastal habitats such as seagrass beds, shallow bays, and estuaries primarily ranging from 10-25 m in depth (Compagno, 1984; Heupel et al., 2006). In fact, this species tends to be a long-term resident of these environments (e.g., Pine Island Sound, Florida Bay, and Lower Florida Keys) with some residents not undertaking long coastal migrations (Heithaus et al., 2007; Heupel et al., 2006; Wiley and Simpfendorfer, 2007). Although bonnetheads do not engage in true schooling, they are known to aggregate together frequently (Compagno, 1984). The scalloped hammerhead is a large hammerhead species, reaching a maximum size of about 3.7 to 4 m in length (Compagno, 1984). It frequents coastal warm temperate and tropical seas, alternating between coastal and pelagic phases. It can be found over continental and insular shelves (<200 m depth) and in adjacent deep waters (Gallagher et al., 2014; Wells et al., 2018). The scalloped hammerhead also frequents shallow bays and estuaries, with nurseries typically located inshore (Corgos & Rosende-Pereiro, 2022). It is considered partly migratory and highly mobile with natural reefs and hard-bottom outcroppings being important foraging areas (Compagno, 1984; Wells et al., 2018). Scalloped hammerheads are the only species of large-bodied shark that engages in highly organized complex social schooling behavior (Gallagher et al., 2014; Klimley, 1985; Klimley and Nelson, 1981).
With regards to the batoids, three species were identified in this study, with a possible unconfirmed fourth (#18). There were at least two rays of the order Myliobatiformes, including the Atlantic stingray (Hypanus sabinus) and the spotted eagle ray (Aetobatus narinari), which is the only species of its genus found in the Atlantic (Richards et al., 2009). They can both be found regularly in the western Atlantic Ocean, including the Keys, but with different habitat preferences. The Atlantic stingray can reside in low salinity and has a high tolerance for shallow estuarine and freshwater habitats; spotted eagle rays are known to occur in inshore marine waters, including reef and mangrove habitats, but the species largely prefers open waters (DeGroot et al., 2021).
The smalltooth sawfish (Pristis pectinata) was the third identified batoid of the order Rhinopristiformes from among the analyzed specimens. While once found on both sides of the Atlantic Ocean (e.g., Bigelow and Schroeder, 1953), this species is now restricted to waters surrounding Florida, particularly within the Everglades National Park, and the Bahamas (Carlson et al., 2013; Simpfendorfer and Wiley, 2005; Wiley and Simpfendorfer, 2007). The species is present all year in south Florida coastal waters, and individual animal size tends to correlate with habitat preferences; with small individuals or juveniles observed in shallow coastal habitats, such as mangroves and estuaries, while larger juveniles or adults are observed among deeper or open waters (Waters et al., 2014).
Discussion
Mitigating limitations in identification
Given the challenges of elasmobranch identification in zooarchaeology (e.g., Gilson and Lessa, 2021; Kozuch and Fitzgerald, 1989; Prieto, 2023), and especially within tropical or subtropical environments with high biodiversity such as the Keys, the results of our analysis with the Clupper specimens are an exemplar of the methodological promise that ZooMS holds for improving elasmobranch species identifications across several world regions and taxa. From the perspective of taxonomic diversity and overcoming limitations in zooarchaeological identification via ZooMS, there are now at least nine different confirmed (including corrected identifications) shark species, two stingray species, and one sawfish species documented for the site. The high species-level taxonomic resolution achieved, for the first time, has implications for advancing archaeological and historical ecology research in the Keys and elsewhere.
Implications for Ancestral Indigenous harvest and elasmobranch historical ecology in the Keys
When considered within the context of contemporary elasmobranch diversity and distribution, the results provide a new opportunity to think beyond generalities gleaned from order- or family-level identifications and information (e.g., LeFebvre et al., 2022; Oliveira, 2024). Here we focus on contemporary species occurrence and co-occurrence observations from the Keys as a foundation for deeper interrogation of elasmobranch harvest and historical ecology represented by the Clupper data.
As part of a recent study investigating shark assemblages spanning state and federal management areas from Miami into the middle Keys (including Upper Matecumbe Key), Tinari and Hammerschlag (2021) (see also Table 1) assessed relationships between species occurrences and abundances by habitat type (i.e., Bay or Ocean), depth, and season (i.e., wet season from May-October and dry season from November-April). They also considered correlations between species size and occurrence. The authors found that while shark assemblages within the study region are characterized by year-round occurrences for the majority of observed species [see Table 1 in Tinari and Hammerschlag (2021)], there are also species-level nuances to occurrences. For example, nurse, tiger, and Atlantic sharpnose sharks have higher probabilities of occurrence in offshore habitats (i.e., ocean), compared to blacktip and lemon sharks found in inshore habitats (i.e., bay). Also, while temperature is not a significant driver of shark occurrence, salinity is for two of the most common species in the Keys, i.e., nurse sharks (i.e., decrease in occurrences with increases in salinity) and lemon sharks (i.e., increase in occurrences with increases in salinity). Depth is also a factor; tiger sharks occur in deeper waters, and nurse, blacktip, and lemon are found in shallower waters. Finally, there are variable correlations between some species sizes (i.e., total length) and season and habitat; whereby, occurrences of larger nurse and bull sharks are higher in the Keys, usually in offshore waters, during the dry season compared to Miami, larger lemon sharks have relatively higher occurrence in offshore habitats overall, and larger Atlantic sharpnose sharks occur more frequently in the Keys compared to waters near Miami.
Like species occurrences, fishery species co-occurrences and community composition are usually related to overlapping habitat preferences and dietary habits, both of which may vary throughout a day (e.g., nocturnal versus diurnal feeding) and/or season, as well as across sex, age, and reproductive cycles of different species (Heithaus et al., 2007). For the Keys and most germane to this study, nurse and bonnethead sharks are among the more frequently observed sharks in the Keys today and they have high rates of co-occurrence with several other elasmobranch species, including an inshore habitat overlap with sharpnose and lemon sharks, as well as with the southern and white-spotted eagle rays (Ramey, 2021).
Assuming contemporary patterns in elasmobranch species occurrence and co-occurrence, as well as the current environmental conditions of the Keys and surrounding waters, extend into the deep-past, our species-level results from the Clupper site suggest that several of the sharks and rays represented would have been opportunistically, and/or predictably, available for harvest from the same habitats as part of a generalist, primarily inshore, fishery at Clupper (e.g., LeFebvre et al., 2022; Oliveira, 2024). Common among inshore, shallow water (e.g., Bay) habitats, we can hypothesize that species, such as lemon, nurse, blacktip, and bonnethead sharks, were the most commonly harvested species based on their likely proximity to the shorelines of Upper Matecumbe Key and that their smaller overall body size was amenable to net and trap capture. We can also hypothesize that co-occurrences between elasmobranch and bony fishes also likely shaped the fishery given that the most frequently identified bony fish taxa from Clupper include catfishes (Ariopsis felis and Bagre marinus), grunts (Haemulon spp.), jacks (Caranx spp.), snappers (Lutjanus spp.), and groupers (Epinephelus spp. and Mycteroperca spp.) - all of which are commonly encountered among the same inshore habitats as the identified elasmobranch species. Species dietary patterns also have implications for hypotheses regarding elasmobranch harvest. For example, scalloped hammerhead, tiger, and lemon sharks are known to feed on bony fishes (e.g., jacks, catfishes), sea turtles, rays, and/or relatively smaller sharks (e.g., blacktip shark, nurse shark) within inshore habitats (e.g., Compagno, 1984), all of which are represented within the Clupper zooarchaeological record.
However, these hypotheses, largely based on co-occurrence inferences, do not necessarily preclude possible offshore or deeper-water harvest practices at Clupper that aimed to target more pelagic species. Three of the largest-growing elasmobranch species identified among the study specimens, the dusky shark, scalloped hammerhead, and white-spotted eagle ray, are also known to inhabit deeper, offshore waters as adults, depending on the time of day, seasonal migrations, and reproduction habits. This is similar to the largest-growing bony fish identified from Clupper, the sailfish (Istiophorus platypterus) (Oliveira, 2024). These taxa may have been harvested while crossing between offshore (i.e., ocean) and inshore (i.e., bay) habitats of the Keys. The Clupper site is located adjacent to a natural deep-water pass that would have made such crossings possible.
We can also begin to think more critically about technological approaches to elasmobranch harvest at Clupper, all of which would have been informed by traditional ecological knowledge developed across generations of cultural practices at the village and more broadly across the Keys (Oliveira, 2024). Approaches likely included several types of gear, including multi-species harvest techniques, such as nets and/or traps intended to capture a diversity of species at once (e.g., elasmobranchs and bony fishes), or those intended for one individual at a time, such as single hook and line and/or spear (Walker 2000). For example, contemporary commercial and recreational fisheries data show that finetooth sharks may be efficiently harvested using gillnets (Portnoy et al., 2016), while blacktip, dusky, and bonnethead sharks are captured using baited hook and line (e.g., individual angling or longline) (e.g., Ulrich et al., 2007). Species such as nurse sharks are also known to take bait intended for other fishes and raid fish nets or traps (Castro, 2000). Testing these ideas and hypotheses will require larger sample sizes identified to species for the calculation of relative abundances, as well as measurements of individual sizes and approximate ages represented across taxa.
From a geographic perspective of contemporary species occurrences, all the elasmobranch species identified from Clupper, except for finetooth shark, are known to inhabit the Keys today. The finetooth shark is not regarded as common in the Keys based on contemporary survey data of species distribution. While the finetooth shark has long been known to inhabit the waters off the peninsular coast of Florida during the fall and into winter (Castro, 1993), there was in 2007 the report of a new southern extent of the species occurrence to include Florida Bay waters along the southern terminus of peninsular Florida (Wiley and Simpfendorfer, 2007), well beyond the previously documented southern extent of the species around Lemon Bay on the southwest coast and Port Salerno on southeast coast of the peninsula; the authors reported that the presence of finetooth sharks in Florida Bay was likely rare overall, but significant from the perspective of possible exchange between Gulf and Atlantic stocks and learning more about the species’ seasonal migration movements and relationship to water temperature. The species identification of finetooth shark via ZooMS at Clupper may have several implications from historical ecological and archaeological perspectives, including: 1) finetooth sharks were more common in the island region and/or greater Florida Bay in the past; 2) there is a potentially deeper history for stock exchanges through time; and 3) a finetooth shark, or portion of an individual, was transported to the Clupper site post mortem from a more northern location along the Florida peninsula. It could also indicate sea temperatures in the past implying environmental/climatic change. To test these possibilities, it will be imperative to identify more finetooth shark specimens within the Clupper zooarchaeological assemblage, from additional zooarchaeological assemblages from the greater Keys region, and ideally from across southern portions of peninsular Florida that are dated to the Ancestral Period.
In summary, we now have a more taxonomically refined and accurate understanding of the elasmobranch diversity represented at Clupper than we did prior to the ZooMS analysis, presenting an opportunity to think beyond generalities gleaned from primarily order- or family-level identifications and information. The data indicate that inshore, and likely to a lesser extent offshore, habitats local to or near the site supported a taxonomically diverse elasmobranch fishery ca. AD 1,000-1,300. It is reasonable to assert that elasmobranch harvest may have been linked to the harvest of other taxa (e.g., bony fishes and sea turtles) and facilitated using multi-species capture nets and/or traps. This may have been most prevalent among smaller species, such as finetooth sharks, blacktip sharks, and bonnetheads in shallower-water habitats. Multi-species capture methods may have also provided opportunities for opportunistic spear fishing of elasmobranchs attracted to fishes caught in nets or traps, such as nurse or bull sharks. Concomitant with inshore harvest and variable uses of fishery technology, some of the species represented may have also been pursued through more targeted approaches, such as hook and line, following seasonal availability depending on aggregation (e.g., S. lewini and S. tiburo) and/or migration habits across both inshore and offshore waters, including larger, migrating species more abundant within the Keys during the winter months (e.g., hammerhead and tiger sharks).
Implications for future zooarchaeological research and supporting conservation
The application of ZooMS to the study of elasmobranchs has potential for improved historical ecological baselines derived from zooarchaeological assemblages, and particularly in support of conservation. Because marine resource managers, conservation practitioners, and fisheries scientists often work with species-level data, the results offer an opportunity to consider how to better leverage zooarchaeological analyses and data to support species-specific as well as community-level historical baselines of elasmobranch diversity and distribution in the Keys (e.g., Tinari and Hammerschlag, 2021), albeit through a cultural lens of human selection and post-depositional taphonomic processes (e.g., preservation). Nonetheless, the species identified among the Clupper specimens are now known to have a deep-time history of human engagement and harvest, pre-dating 20th and 21st centuries scales of impact. Approaching the Clupper zooarchaeological species list as a historical survey, similar to more recent survey lists (e.g., Ramey, 2021; Tinari and Hammerschlag, 2021; see also Table 2), but providing a baseline of species occurrences well before the precipitous losses of recent history, we suggest three lines of possible future research, aimed at elucidating trends (e.g., stasis or change) in species diversity through time, across space, and among groups of people (i.e., cultural practices).
1. Given that almost all identified species from Clupper are extant, we can target specific species for genetic analyses leveraging aDNA (e.g., Shepherd and Campbell, 2021), and ultimately having the potential to contribute to critically needed long-term phylogenetic perspectives of elasmobranch species diversity and loss through time. The one as yet unidentified ray leaves open the potential for new species discovery, at least likely locally extinct.
2. With the use of high-throughput digital scanning techniques linked with ZooMS (e.g., Buckley et al., 2021), we can compare fragmented, species-identified elasmobranch zooarchaeological specimens with modern comparative specimens for the possible identification of species-specific morphological hallmarks not previously recognized.
3. Because elasmobranch species occurrences can be highly variable in terms of individual age and size within a given habitat, depending on many factors (e.g., nursery locations, water depth, salinity, adult migration patterns), and particularly within habitat-diverse tropical locations such as the Keys, we can begin to leverage ZooMS-identified specimens, including bulk-assemblage approaches (e.g.,Buckley et al., 2016; Oldfield et al., 2024), to link specimen identification with allometric size data across species (e.g., species length and age correlations). These data would allow us to measure species sizes (and approximate ages), represented within the zooarchaeological record, and quantify relative species abundances accordingly (e.g., MNI, NISP), ultimately contributing to deep-time perspectives of species sizes beyond more recent time scales (e.g., decadal) as well as more specific interpretations of elasmobranch harvest habitats and practices in the past.
Not surprisingly, difficulties surrounding species-level elasmobranch identification of zooarchaeological specimens can significantly limit the information that can be garnered from preserved bone, teeth, and denticles, including insights into possible habitats of harvest and capture methods used in the past. Furthermore, identifications only to order or family level seriously diminish the historical ecological implications that can be drawn, which often necessitate species-level resolution and inference to support conservation and understanding the impacts of human predation through time (e.g., harvest for subsistence, shark fin markets). Archaeological elasmobranch assemblages, such as Clupper, are composed of specimens that hold critical potential to constructing as taxonomically, geographically, and temporally as possible baselines of diversity through time, and particularly within the context of long-term human engagement. The species identified from Clupper include several taxa considered to be vulnerable, endangered, or critically endangered (Table 1), particularly those characterized by K-selected traits, making them incredibly vulnerable to loss as noted earlier. While the use of ZooMS in establishing robust species-level identifications of elasmobranchs has been successful, its future use will allow us to answer many more questions and develop methodological advances that are not possible based on morphological identifications alone, perhaps most suitably to applications involving wildlife forensics, including the population-decimating shark fin trade (Cardeñosa et al., 2022). However, we assert that both ZooMS and morphology are integral to realizing the full potential of zooarchaeological data to contribute to conservation baselines and deep-time historical perspectives in the Keys and beyond.
Data availability statement
The original contributions presented in the study are available on FigShare at https://figshare.com/articles/dataset/Clupper_elasmobranch_species_identification_using_ZooMS/27087190?file=49358374 and presented as additional figures in the Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because deceased animal tissues from museum collections were used.
Author contributions
MB: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. E-MO: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. CO: Data curation, Formal analysis, Methodology, Validation, Visualization, Writing – original draft. CB: Data curation, Formal analysis, Visualization, Writing – original draft. AK: Resources, Writing – original draft. NF: Resources, Writing – review & editing. TA: Resources, Writing – review & editing. VT: Writing – review & editing. SF: Writing – review & editing. ML: Resources, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding for excavation was provided by the University of Miami, University of Oregon, and University of Georgia. Zooarchaeological analysis was funded by the Florida Museum of Natural History John S. and James L. Knight Professorship in Archaeology; the PhD studentship for Oldfield was funded for by the BBSRC. The BBSRC funded the PhD studentship for Ellie-May Oldfield, grant number BB/T008725/1.
Acknowledgments
From the Florida Museum of Natural History, we thank Kitty Emery for her support in the use of the Environmental Archaeology Program modern comparative skeletal collection, Rob Robins and Gavin Naylor for their guidance on taxonomy, and Coleman Sheehy for leading compliance with CITES regulations for international studies. We also thank Laura Kozuch and Simon-Pierre Gilson for the invitation to share some of this work at the 2024 Society for American Archaeology Annual Meeting. The Clupper archaeological research was conducted under permit 1415.015 to Thompson from the Division of Historical Resources, Florida Department of State (2014) permit 11051415 to Thompson from the Florida Park Service, Florida Department of Environmental Protection (2015). The authors thank avocational archaeologist Jim Clupper, after whom the site is named, for invaluable assistance with this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1500595/full#supplementary-material
Supplementary Figure 1 | Sequence-annotated MALDI peptide mass fingerprint (top) of dominant proteins extracted from modern Hypanus sabinus tissue (black text = COL1A1, red text = COL1A2, blue text = COL2A1; matched to peptides listed in Supplementary Table S5) with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 2 | Sequence-annotated MALDI peptide mass fingerprint (top) of dominant proteins extracted from modern Carcharodon carcharias tissue (black text = COL1A1, red text = COL1A2, blue text = COL2A1; matched to peptides listed in Supplementary Table S5) with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 3 | MALDI spectra showing peptide mass fingerprints (top) of different modern tissues (vertebral process cartilage and tooth) from modern Carcharhinus limbatus, with zoomed in sections (A-C) in order of increasing m/z.
Supplementary Figure 4 | MALDI spectra showing peptide mass fingerprints (center) of different modern tissues (cranial cartilage and vertebra) from Ginglymostoma cirratum, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 5 | MALDI spectra showing peptide mass fingerprints (centre) of different modern tissues (jaw, tooth and ‘cranial cartilage’) from Galeocerdo cuvier, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 6 | MALDI spectra showing peptide mass fingerprints (top) of modern Carcharhinus isodon, Carcharhinus limbatus, Carcharhinus signatus and Carcharhinus leucas, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 7 | MALDI spectra showing peptide mass fingerprints (top) of modern Carcharhinus galapagensis, Carcharhinus brevipinna, Carcharhinus obscurus and Carcharhinus altimus, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 8 | MALDI spectra showing peptide mass fingerprints (top) of modern Carcharhinus perezi, Carcharhinus plumbeus, and Carcharhinus acronotus, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 9 | MALDI spectra showing peptide mass fingerprints (top) of modern Sphryna lewini, Sphryna mokarran, Sphryna tiburo and Sphryna zygaena, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 10 | MALDI spectra showing peptide mass fingerprints (centre) of modern Galeorhinus galeus, Carcharias taurus, Pristis pristis and Pristis pectinata, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 11 | MALDI spectra showing peptide mass fingerprints (top) of modern Rhizoprionodon terraenovae, Mustelus canis, Alopias vulpinus and Isurus oxyrinchus, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 12 | MALDI spectra showing peptide mass fingerprints (centre) of Hexanchus griseus, Negaprion brevirostris, Hemipristis elongata and Prionace glauca, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 13 | MALDI spectra showing peptide mass fingerprints (top) of modern Bathytoshia centroura, Raja eglanteria and Aetobatus narinari, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 14 | MALDI spectra showing peptide mass fingerprints (top) of modern Rhinoptera bonasus, Hypanus say, Urobatis jamaicensis and Hypanus americanus, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 15 | MALDI spectra showing peptide mass fingerprints (top) of remaining archaeological vertebra examples; Aetobatus narinari, Hypanus sabinus, Pristis pristis and one from a confidently identified Hypanus sabinus barb, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figure 16 | MALDI spectra showing peptide mass fingerprints (top) of remaining archaeological examples 2 – Galeocerdo cuvier, Ginglymostoma. cirratum, Rhizoprionodon, Sphyrna tiburo and Sphyrna lewini, with zoomed in sections (A-D) in order of increasing m/z.
Supplementary Figures 17–S51 | LC-MS/MS spectra of peptides of potential interest as biomarkers.
Supplementary Table 1 | Current zooarchaeological results from the Clupper site midden across vertebrate classes.
Supplementary Table 2 | Modern reference specimens with details.
Supplementary Table 3 | Archaeological specimens with details.
Supplementary Table 4 | Local protein sequence database accession numbers.
Supplementary Table 5 | COL1A1, COL1A2 and COL2A1 matches to Hypanus sabinus.
Supplementary Table 6 | COL1A1, COL1A2 and COL2A1 matches to Carcharodon carcharias.
Supplementary Table 7 | COL1A1, COL1A2 and COL2A1 matches to Pristis pectinata.
Supplementary Table 8 | Selected peptide biomarkers.
Footnotes
- ^ Here, within Florida archaeology, the Ancestral Period refers to time spans prior to European colonization. The use of this term as a chronological descriptor of Indigenous history is supported and preferred by some U.S. federally recognized contemporary Indigenous Peoples, including the Seminole Tribe of Florida in south Florida (see https://stofthpo.com/seminole-history/).
References
Ahonen H., Stow A. J. (2008). Shark jaws and teeth: an unexploited resource for population genetic studies. J. Fish Biol. 73, 450–455. doi: 10.1111/j.1095-8649.2008.01896.x
Ajemian M. J., Drymon J. M., Hammerschlag N., Wells R. D., Street G., Falterman B., et al. (2020). Movement patterns and habitat use of tiger sharks (Galeocerdo cuvier) across ontogeny in the Gulf of Mexico. PloS One 15, e0234868. doi: 10.1371/journal.pone.0234868
Ardren T., Thompson V. D., Fitzpatrick S. M., Stevenson J., Sierra R. L. (2018). “When foragers are managers: Social complexity and persistent foraging in the Florida Keys,” in The Archaeology of Caribbean and Circum-Caribbean Farmers (6000 BC-AD 1500) (Oxford: Routledge), 311–326.
Ault J. S., Bohnsack J. A., Smith S. G., Luo J. (2005). Towards sustainable multispecies fisheries in the Florida, USA, coral reef ecosystem. Bull. Mar. Sci. 76, 595–622.
Berio F., Broyon M., Enault S., Pirot N., López-Romero F. A., Debiais-Thibaud M. (2021). Diversity and evolution of mineralized skeletal tissues in chondrichthyans. Front. Ecol. Evol. 9, 660767. doi: 10.3389/fevo.2021.660767
Betts M. W., Blair S. E., Black D. W. (2012). Perspectivism, mortuary symbolism, and human-shark relationships on the Maritime Peninsula. Am. Antiquity 77, 621–645. doi: 10.7183/0002-7316.77.4.621
Bieg C., McCann K. S., McMeans B. C., Rooney N., Holtgrieve G. W., Lek S., et al. (2018). Linking humans to food webs: a framework for the classification of global fisheries. Front. Ecol. Environ. 16, 412–420. doi: 10.1002/fee.2018.16.issue-7
Bigelow h. B., Schroeder W. C. (1953). Sawfishes, guitarfishes, skates and rays. Fishes of the western north Atlantic. Mem. Sears Found. Mar. Res. (New Haven: Yale University Press), Vol. 1. 1–514.
Bom R. A., Brader A., Batsleer J., Poos J. J., van der Veer H. W., Van Leeuwen A. (2022). A long-term view on recent changes in abundance of common skate complex in the North Sea. Mar. Biol. 169, 146. doi: 10.1007/s00227-022-04132-w
Boulanger C. (2023). The exploitation of fishing resources and the maritime skills of early modern humans in island Southeast Asia (No. S3153) (Oxford: BAR Publishing). doi: 10.1016/j.jasrep.2023.104222
Boulanger C., Hawkins S., Samper Carro S. C., Ono R., O’connor S. (2022). Continuity and variability in prehistoric fishing practices by Homo sapiens in Island Southeast Asia: new ichthyofaunal data from Asitau Kuru, Timor-Leste. World Archaeol. 54, 288–316. doi: 10.1080/00438243.2023.2192518
Boulanger C., Ingicco T., Piper P. J., Amano N., Grouard S., Ono R., et al. (2019). Coastal subsistence strategies and mangrove swamp evolution at Bubog I Rockshelter (Ilin Island, Mindoro, Philippines) from the Late Pleistocene to the mid-Holocene. J. Island Coast. Archaeol. 14, 584–604. doi: 10.1080/15564894.2018.1531957
Boulanger C., Ingicco T., Sémah A. M., Hawkins S., Ono R., Reyes M. C., et al. (2023). 30,000 years of fishing in the Philippines: New ichthyoarchaeological investigations in Occidental Mindoro. J. Archaeological Sci.: Rep. 52, 104222.
Boulanger C., Puaud S., Ly V., Glémarec L., Heng S., Forestier H. (2021). Fishbone artefacts from the Samrong Sen site, Cambodia, cast new light on Bronze Age networking between inland and coastal communities. Int. J. Osteoarchaeol. 31, 29–37. doi: 10.1002/oa.v31.1
Bradshaw C. J., Fitzpatrick B. M., Steinberg C. C., Brook B. W., Meekan M. G. (2008). Decline in whale shark size and abundance at Ningaloo Reef over the past decade: the world’s largest fish is getting smaller. Biol. Conserv. 141, 1894–1905. doi: 10.1016/j.biocon.2008.05.007
Branstetter S. (1990). Early life-history implications of selected carcharhinoid and lamnoid sharks of the northwest Atlantic. (U.S. Department of Commerce, National Oceanic and Atmospheric Administration, National Marine Fisheries Service). Available online at: https://scholarworks.wm.edu/vimsbooks/40.
Buckley M. (2013). A molecular phylogeny of Plesiorycteropus reassigns the extinct mammalian order ‘Bibymalagasia’. PloS One 8, e59614. doi: 10.1371/journal.pone.0059614
Buckley M., Cheylan M. (2020). Collagen fingerprinting for the species identification of archaeological amphibian remains. Boreas 49, 709–717. doi: 10.1111/bor.12443
Buckley M., Collins M., Thomas-Oates J., Wilson J. C. (2009). Species identification by analysis of bone collagen using matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun. Mass Spectrometry 23, 3843–3854. doi: 10.1002/rcm.v23:23
Buckley M., Harvey V. L., Petiffer D., Russ H., Wouters W., Van Neer W. (2022). Medieval fish remains on the Newport ship identified by ZooMS collagen peptide mass fingerprinting. Archaeological Anthropological Sci. 14, 41. doi: 10.1007/s12520-021-01478-y
Buckley M., Kansa S. W. (2011). Collagen fingerprinting of archaeological bone and teeth remains from Domuztepe, South Eastern Turkey. Archaeological Anthropological Sci. 3, 271–280. doi: 10.1007/s12520-011-0066-z
Buckley M., Kansa S. W., Howard S., Campbell S., Thomas-Oates J., Collins M. (2010). Distinguishing between archaeological sheep and goat bones using a single collagen peptide. J. Archaeological Sci. 37, 13–20. doi: 10.1016/j.jas.2009.08.020
Buckley M., Pinsonneault M., Brassey C., Rolett B. (2021). High-throughput microCT and ZooMS collagen fingerprinting of Scombrid bone from the Marquesas Islands. J. Archaeological Sci. 136, 105475. doi: 10.1016/j.jas.2021.105475
Buckley M., Gu M., Shameer S., Patel S., Chamberlain A. T. (2016). High‐throughput collagen fingerprinting of intact microfaunal remains; a low‐cost method for distinguishing between murine rodent bones. Rapid Communications in Mass Spectrometry 30(7), 805–812.
Burg Mayer G., de Freitas R. H. A. (2023). Archaeological sharks: changes in the trophic ecology between late Holocene and modern shark communities in South Brazil. Mar. Biol. 170(8), 102. doi: 10.1007/s00227-023-04252-x
Camhi M., Fowler S. L., Musick J. A., Bräutigam A., Fordham S. V. (1998). Sharks and their relatives - Ecology and conservation. IUCN/SSC Shark Specialist Group: IUCN, Gland, Switzerland and Cambridge, UK.
Cardeñosa D., Shea S. K., Zhang H., Fischer G. A., Simpfendorfer C. A., Chapman D. D. (2022). Two thirds of species in a global shark fin trade hub are threatened with extinction: Conservation potential of international trade regulations for coastal sharks. Conserv. Lett. 15, e12910. doi: 10.1111/conl.12910
Carlson J. K., Heupel M. R., Bethea D. M., Hollensead L. D. (2008). Coastal habitat use and residency of juvenile Atlantic sharpnose sharks (Rhizoprionodon terraenovae). Estuaries Coasts 31, 931–940. doi: 10.1007/s12237-008-9075-2
Carlson J. K., Parsons G. R. (1997). Age and growth of the bonnethead shark, Sphyrna tiburo, from northwest Florida, with comments on clinal variation. Environ. Biol. Fishes 50, 331–341. doi: 10.1023/A:1007342203214
Carlson J., Wiley T., Smith K. (2013). Pristis pectinata Vol. 2013 (The IUCN Red List of Threatened Species), e.T18175A43398238. doi: 10.2305/IUCN.UK.2022-2.RLTS.T18175A58298676.en (accessed November 4, 2024).
Castro J. I. (1993). The shark nursery of Bulls Bay, South Carolina, with a review of the shark nurseries of the southeastern coast of the United States. Environ. Biol. fishes 38, 37–48. doi: 10.1007/BF00842902
Castro J. I. (1996). Biology of the blacktip shark, Carcharhinus limbatus, off the southeastern United States. Bull. Mar. Sci. 59, 508–522.
Castro J. I. (2000). The biology of the nurse shark, Ginglymostoma cirratum, off the Florida east coast and the Bahama Islands. Environ. Biol. Fishes 58, 1–22. doi: 10.1023/A:1007698017645
Charpentier V., Adnet S., Cappetta H. (2020). The tooth of a giant sea creature Otodus (Megaselachus) in the material culture of Neolithic maritime hunter-gatherers at Sharbithat (Sultanate of Oman). Int. J. Osteoarchaeol. 30, 835–842. doi: 10.1002/oa.v30.6
Coates M. I., Ruta M., Friedman M. (2008). Ever since Owen: changing perspectives on the early evolution of tetrapods. Annu. Rev. Ecol. Evolution Systematics 39, 571–592. doi: 10.1146/annurev.ecolsys.38.091206.095546
Colvin G. H. (2014). Shark teeth from Ohio archaeological sites: an update based on newly discovered teeth. Ohio Archaeologist 64, 55–60.
Compagno L. V.J. (1984). Sharks of the world. An annotated and illustrated catalogue of shark species known to date. Part 2: carcharhiniformes. FAO fish synop Vol. 4 (Rome: FAO), 251–655.
Corgos A., Rosende-Pereiro A. (2022). Nursery habitat use patterns of the scalloped hammerhead shark, Sphyrna lewini, in coastal areas of the central Mexican Pacific. J. Fish Biol. 100, 117–133. doi: 10.1111/jfb.v100.1
Dean M. N., Ekstrom L., Monsonego-Ornan E., Ballantyne J., Witten P. E., Riley C., et al. (2015). “Mineral homeostasis and regulation of mineralization processes in the skeletons of sharks, rays and relatives (Elasmobranchii),” in Seminars in Cell & Developmental Biology, vol. 46. (Academic Press), 51–67.
Dean M. N., Summers A. P. (2006). Mineralized cartilage in the skeleton of chondrichthyan fishes. Zoology 109, 164–168. doi: 10.1016/j.zool.2006.03.002
DeGroot B. C., Bassos-Hull K., Wilkinson K. A., Lowerre-Barbieri S., Poulakis G. R., Ajemian M. J. (2021). Variable migration patterns of whitespotted eagle rays Aetobatus narinari along Florida’s coastlines. Mar. Biol. 168, 1–21. doi: 10.1007/s00227-021-03821-2
Dibattista J. D., Feldheim K. A., Gruber S. H., Hendry A. P. (2007). When bigger is not better: selection against large size, high condition and fast growth in juvenile lemon sharks. Journal of Evolutionary Biology 20 (1), 201–212.
Donoghue P. C. J., Sansom I. J., Downs J. P. (2006). Early evolution of vertebrate skeletal tissues and cellular interactions, and the canalization of skeletal development. J. Exp. Zoology Part B: Mol. Dev. Evol. 306, 278–294. doi: 10.1002/jez.b.v306b:3
Dulvy N. K., Pacoureau N., Rigby C. L., Pollom R. A., Jabado R. W., Ebert D. A., et al. (2021). Overfishing drives over one-third of all sharks and rays toward a global extinction crisis. Curr. Biol. 31, 4773–4787. doi: 10.1016/j.cub.2021.08.062
Eames B. F., Allen N., Young J., Kaplan A., Helms J. A., Schneider R. A. (2007). Skeletogenesis in the swell shark Cephaloscyllium ventriosum. J. Anat. 210, 542–554. doi: 10.1111/j.1469-7580.2007.00723.x
Eda M., Morimoto M., Mizuta T., Inoué T. (2020). ZooMS for birds: Discrimination of Japanese archaeological chickens and indigenous pheasants using collagen peptide fingerprinting. J. Archaeological Sci.: Rep. 34, 102635. doi: 10.1016/j.jasrep.2020.102635
Enault S., Adnet S., Debiais-Thibaud M. (2016). Skeletogenesis during the late embryonic development of the catshark Scyliorhinus canicula (Chondrichthyes; Neoselachii). MorphoMuseuM 1, e2. doi: 10.18563/m3.1.4.e2
Farrell A. D. (2021). A Use-Wear and Functional Analysis of Precontact Shark Teeth Assemblages from Florida (The Florida State University). Available online at: https://purl.lib.fsu.edu/diginole/2020_Summer_Fall_Farrell_fsu_0071N_16488.
Ferretti F., Curnick D., Liu K., Romanov E. V., Block B. A. (2018). Shark baselines and the conservation role of remote coral reef ecosystems. Sci. Adv. 4, eaaq0333. doi: 10.1126/sciadv.aaq0333
Ferretti F., Myers R. A., Serena F., Lotze H. K. (2008). Loss of large predatory sharks from the Mediterranean Sea. Conserv. Biol. 22, 952–964. doi: 10.1111/j.1523-1739.2008.00938.x
Field I. C., Meekan M. G., Buckworth R. C., Bradshaw C. J. (2009). Susceptibility of sharks, rays and chimaeras to global extinction. Adv. Mar. Biol. 56, 275–363. doi: 10.1016/S0065-2881(09)56004-X
Gallagher A. J., Hammerschlag N., Shiffman D. S., Giery S. T. (2014). Evolved for extinction: the cost and conservation implications of specialization in hammerhead sharks. Bioscience 64, 619–624. doi: 10.1093/biosci/biu071
Gilson S. P., Lessa A. (2021). Capture, processing and utilization of sharks in archaeological context: Its importance among fisher-hunter-gatherers from southern Brazil. J. Archaeological Sci.: Rep. 35, 102693. doi: 10.1016/j.jasrep.2020.102693
Giovos I., Spyridopoulou R. A., Doumpas N., Glaus K., Kleitou P., Kazlari Z., et al. (2021). Approaching the “real” state of elasmobranch fisheries and trade: A case study from the Mediterranean. Ocean Coast. Manage. 211, 105743. doi: 10.1016/j.ocecoaman.2021.105743
Guiry E. J., Buckley M., Orchard T. J., Hawkins A. L., Needs-Howarth S., Holm E., et al. (2020). Deforestation caused abrupt shift in Great Lakes nitrogen cycle. Limnol. Oceanogr. 65, 1921–1935. doi: 10.1002/lno.11428
Guiry E., Kennedy J. R., Malcom C., Miller M., Hall O., Buckley M., et al. (2024). Archaeological evidence for long-term human impacts on sea turtle foraging behaviour. R. Soc. Open Sci. 11, 240120. doi: 10.1098/rsos.240120
Hall B. K. (1975). Evolutionary consequences of skeletal differentiation. Am. Zoologist 15, 329–350. doi: 10.1093/icb/15.2.329
Harvey V. L., Daugnora L., Buckley M. (2018). Species identification of ancient Lithuanian fish remains using collagen fingerprinting. J. Archaeological Sci. 98, 102–111. doi: 10.1016/j.jas.2018.07.006
Harvey V. L., Keating J. N., Buckley M. (2021). Phylogenetic analyses of ray-finned fishes (Actinopterygii) using collagen type I protein sequences. R. Soc. Open Sci. 8, 201955. doi: 10.1098/rsos.201955
Harvey V. L., LeFebvre M. J., DeFrance S. D., Toftgaard C., Drosou K., Kitchener A. C., et al. (2019). Preserved collagen reveals species identity in archaeological marine turtle bones from Caribbean and Florida sites. R. Soc. Open Sci. 6, 191137. doi: 10.1098/rsos.191137
Harvey V. L., LeFebvre M. J., Sharpe A. E., Toftgaard C., DeFrance S. D., Giovas C. M., et al. (2022). Collagen fingerprinting of Caribbean archaeological fish bones: Methodological implications for historical fisheries baselines and anthropogenic change. J. Archaeological Sci. 145, 105642. doi: 10.1016/j.jas.2022.105642
Heithaus M. R., Burkholder D., Hueter R. E., Heithaus L. I., Pratt H. L., Carrier J. C. (2007). Spatial and temporal variation in shark communities of the lower Florida Keys and evidence for historical population declines. Can. J. Fisheries Aquat. Sci. 64, 1302–1313. doi: 10.1139/f07-098
Heupel M. R., Simpfendorfer C. A., Collins A. B., Tyminski J. P. (2006). Residency and movement patterns of bonnethead sharks, Sphyrna tiburo, in a large Florida estuary. Environ. Biol. Fishes 76, 47–67. doi: 10.1007/s10641-006-9007-6
Jennings D. E., Gruber S. H., Franks B. R., Kessel S. T., Robertson A. L. (2008). Effects of large-scale anthropogenic development on juvenile lemon shark (Negaprion brevirostris) populations of Bimini, Bahamas. Environ. Biol. Fishes 83, 369–377. doi: 10.1007/s10641-008-9357-3
Juan-Jordá M. J., Murua H., Arrizabalaga H., Merino G., Pacoureau N., Dulvy N. K. (2022). Seventy years of tunas, billfishes, and sharks as sentinels of global ocean health. Science 378, eabj0211. doi: 10.1126/science.abj0211
Kealy S., O’Connor S., Sari D. M., Shipton C., Langley M. C., Boulanger C., et al. (2020). Forty-thousand years of maritime subsistence near a changing shoreline on Alor Island (Indonesia). Quaternary Sci. Rev. 249, 106599. doi: 10.1016/j.quascirev.2020.106599
Keller B., Thompson V. D. (2013). A preliminary report on the role of shark teeth at fort center. Florida Anthropologist 66, 23–30.
Kemp N. E., Westrin S. K. (1979). Ultrastructure of calcified cartilage in the endoskeletal tesserae of sharks. J. Morphol. 160, 75–101. doi: 10.1002/jmor.1051600106
Kimura S., Ohno Y. (1987). Fish type I collagen: tissue-specific existence of two molecular forms (α1) 2α2 and α1α2α3, in Alaska pollack. Comp. Biochem. Physiol. Part B: Comp. Biochem. 88, 409–413.
Klimley A. P. (1985). Schooling in the large predator, Sphyrna lewini, a species with low risk of predation: A non-egalitarian state. Z. für Tierpsychologie 70, 297–319. doi: 10.1111/j.1439-0310.1985.tb00520.x
Klimley A. P., Nelson D. R. (1981). Schooling of the scalloped hammerhead shark, Sphyrna lewini, in the Gulf of California. Fisheries Bull. 79, 356–360.
Kozuch L., Fitzgerald C. (1989). A guide to identifying shark centra from southeastern archaeological sites. Southeastern Archaeol. 8(2), 146–157.
Langley M. C., Duli A., Stephenson B., Nur M., Matherson C., Burhan B., et al. (2023). Shark-tooth artefacts from middle Holocene Sulawesi. Antiquity 97, pp.1420–1435. doi: 10.15184/aqy.2023.144
Lea J. S., Wetherbee B. M., Queiroz N., Burnie N., Aming C., Sousa L. L., et al. (2015). Repeated, long-distance migrations by a philopatric predator targeting highly contrasting ecosystems. Sci. Rep. 5, 11202. doi: 10.1038/srep11202
LeFebvre M. J., Ardren T., Thompson V. D., Fitzpatrick S. M., Ayers-Rigsby S. (2022). In support of sustainability: The historical ecology of vertebrate biodiversity and Native American harvest practices in the Florida Keys, USA. Sustainability 14, 6552. doi: 10.3390/su14116552
Lopes M. S., Bertucci T. C. P., Rapagnã L., Tubino R. D. A., Monteiro-Neto C., Tomas A. R. G., et al. (2016). The path towards endangered species: prehistoric fisheries in southeastern Brazil. PloS One 11, e0154476. doi: 10.1371/journal.pone.0154476
Márquez-Farias J. F., Castillo-Geniz J. L. (1998). Fishery biology and demography of the Atlantic sharpnose shark, Rhizoprionodon terraenovae, in the southern Gulf of Mexico. Fisheries Res. 39, 183–198. doi: 10.1016/S0165-7836(98)00182-9
Martínez-Candelas I. A., Pérez-Jiménez J. C., Espinoza-Tenorio A., McClenachan L., Méndez-Loeza I. (2020). Use of historical data to assess changes in the vulnerability of sharks. Fisheries Res. 226, 105526. doi: 10.1016/j.fishres.2020.105526
McClenachan L. (2009). Documenting loss of large trophy fish from the Florida Keys with historical photographs. Conserv. Biol. 23, 636–643. doi: 10.1111/j.1523-1739.2008.01152.x
McClenachan L., Cooper A. B., Dulvy N. K. (2016). Rethinking trade-driven extinction risk in marine and terrestrial megafauna. Curr. Biol. 26, 1640–1646. doi: 10.1016/j.cub.2016.05.026
Mojetta A. R., Travaglini A., Scacco U., Bottaro M. (2018). Where sharks met humans: The Mediterranean Sea, history and myth of an ancient interaction between two dominant predators. Regional Stud. Mar. Sci. 21, 30–38. doi: 10.1016/j.rsma.2017.10.001
Morvan-Dubois G., Le Guellec D., Garrone R., Zylberberg L., Bonnaud L. (2003). Phylogenetic analysis of vertebrate fibrillar collagen locates the position of zebrafish α3 (I) and suggests an evolutionary link between collagen α chains and Hox clusters. J. Mol. Evol. 57, 501–514.
Motivarash Yagnesh B., Fofandi Durga C., Dabhi Raj M., Makrani Rehanavaz A., Tanna Poojaben D. (2020). Importance of sharks in ocean ecosystem. J. Entomol. Zoology Stud. 8, 611–613.
Natanson L. J., Kohler N. E. (1996). A preliminary estimate of age and growth of the dusky shark Carcharhinus obscurus from the south-west Indian Ocean, with comparisons to the western North Atlantic population. South Afr. J. Mar. Sci. 17, 217–224. doi: 10.2989/025776196784158572
Nielsen E. E., Morgan J. A. T., Maher S. L., Edson J., Gauthier M., Pepperell J., et al. (2017). Extracting DNA from ‘jaws’: high yield and quality from archived tiger shark (Galeocerdo cuvier) skeletal material. Mol. Ecol. Resour. 17, 431–442. doi: 10.1111/men.2017.17.issue-3
O’Connor S., Mahirta K. S., Boulanger C., Maloney T., Hawkins S., Langley M. C., et al. (2019). Kisar and the archaeology of small islands in the Wallacean Archipelago. J. Island Coast. Archaeol. 14, 198–225. doi: 10.1080/15564894.2018.1443171
Oldfield E. M., Dunstan M., Chowdhury M. P., Slimak L., Buckley M. (2024). AutoZooMS: Integrating robotics into high-throughput ZooMS for the species identification of archaeofaunal remains at Grotte Mandrin, France. Archaeological Anthropological Sci. doi: 10.1007/s12520-024-02073-7
Oliveira C. I. R. (2024). Zooarchaeological Insights into Traditional Ecological Knowledge and Historical Ecology of the Florida Keys [Master’s thesis]. (Gainesville (FL): University of Florida).
Ono R., Intoh M. (2011). Island of pelagic fishermen: Temporal changes in prehistoric fishing on Fais, Micronesia. J. Island Coast. Archaeol. 6, 255–286. doi: 10.1080/15564894.2010.540531
Ørvig T. (1951). Histologic studies of ostracoderms, placoderms and fossil elasmobranchs. 1. The endoskeleton, with remarks on the hard tissues of lower vertebrates in general. Arkiv för Zoologi 2, 321–454.
Pacoureau N., Rigby C. L., Kyne P. M., Sherley R. B., Winker H., Carlson J. K., et al. (2021). Half a century of global decline in oceanic sharks and rays. Nature 589, 567–571. doi: 10.1038/s41586-020-03173-9
Parsons G. R., Hoffmayer E. R. (2005). Seasonal changes in the distribution and relative abundance of the Atlantic sharpnose shark Rhizoprionodon terraenovae in the north central Gulf of Mexico. Copeia 2005, 914–920. doi: 10.1643/0045-8511(2005)005[0914:SCITDA]2.0.CO;2
Pears J. B., Johanson Z., Trinajstic K., Dean M. N., Boisvert C. A. (2020). Mineralization of the Callorhinchus vertebral column (Holocephali; Chondrichthyes). Front. Genet. 11, 571694. doi: 10.3389/fgene.2020.571694
Peignoux-Deville J., Lallier F., Vidal B. (1982). Evidence for the presence of osseous tissue in dogfish vertebrae. Cell Tissue Res. 222, 605–614. doi: 10.1007/BF00213858
Portnoy D. S., Hollenbeck C. M., Bethea D. M., Frazier B. S., Gelsleichter J., Gold J. R. (2016). Population structure, gene flow, and historical demography of a small coastal shark (Carcharhinus isodon) in US waters of the Western Atlantic Ocean. ICES J. Mar. Sci. 73, 2322–2332. doi: 10.1093/icesjms/fsw098
Powers S. P., Fodrie F. J., Scyphers S. B., Drymon J. M., Shipp R. L., Stunz G. W. (2013). Gulf-wide decreases in the size of large coastal sharks documented by generations of fishermen. Mar. Coast. Fisheries 5, 93–102. doi: 10.1080/19425120.2013.786001
Price M. D., Buckley M., Kersel M. M., Rowan Y. M. (2013). Animal management strategies during the chalcolithic in the Lower Galilee: New data from Marj Rabba (Israel). Paléorient 39(2), 183–200. doi: 10.3406/paleo.2013.5527
Prieto G. (2023). Shark fisheries during the second millennium BC in Gramalote, north coast of Peru. J. Island Coast. Archaeol. 18, 165–195. doi: 10.1080/15564894.2021.1910386
Rama S., Chandrakasan G. (1984). Distribution of different molecular species of collagen in the vertebral cartilage of shark (Carcharius acutus). Connective Tissue Res. 12, 111–118.
Ramey E. (2021). Sharks in the shallows: an assessment of coastal shark distribution patterns in the Florida keys archipelago (UC San Diego: Center for Marine Biodiversity and Conservation). Available at: https://escholarship.org/uc/item/7k57r99z.
Ricard-Blum S. (2011). The collagen family. Cold Spring Harbor Perspect. Biol. 3, a004978. doi: 10.1101/cshperspect.a004978
Richards V. P., Henning M., Witzell W., Shivji M. S. (2009). Species delineation and evolutionary history of the globally distributed spotted eagle ray (Aetobatus narinari). J. Heredity 100, 273–283. doi: 10.1093/jhered/esp005
Richter K. K., Wilson J., Jones A. K., Buckley M., van Doorn N., Collins M. J. (2011). Fish’n chips: ZooMS peptide mass fingerprinting in a 96 well plate format to identify fish bone fragments. J. Archaeological Sci. 38, 1502–1510. doi: 10.1016/j.jas.2011.02.014
Rick T. C., Erlandson J. M., Glassow M. A., Moss M. L. (2002). Evaluating the economic significance of sharks, skates, and rays (Elasmobranchs) in prehistoric economies. J. Archaeological Sci. 29, 111–122. doi: 10.1006/jasc.2000.0637
Rick T., Harvey V. L., Buckley M. (2019). Collagen fingerprinting and the chumash billfish fishery, Santa Barbara Channel, California, USA. Archaeological Anthropological Sci. 11, 6639–6648. doi: 10.1007/s12520-019-00930-4
Roberts J., Weeks L., Fillios M., Cable C., Carter M., al Aali Y. Y., et al. (2019). The exploitation of marine resources at Saruq al-Hadid: Insights into the movement of people and resources in Bronze and Iron Age south-eastern Arabia. Arabian Archaeol. Epigraphy 30, 179–198. doi: 10.1111/aae.12137
Roff G., Brown C. J., Priest M. A., Mumby P. J. (2018). Decline of coastal apex shark populations over the past half century. Commun. Biol. 1, 223. doi: 10.1038/s42003-018-0233-1
Seersholm F. V., Cole T. L., Grealy A., Rawlence N. J., Greig K., Knapp M., et al. (2018). Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey. Proc. Natl. Acad. Sci. 115, 7771–7776. doi: 10.1073/pnas.1803573115
Seidel R., Blumer M., Chaumel J., Amini S., Dean M. N. (2020). Endoskeletal mineralization in chimaera and a comparative guide to tessellated cartilage in chondrichthyan fishes (sharks, rays and chimaera). J. R. Soc. Interface 17, 20200474. doi: 10.1098/rsif.2020.0474
Seidel R., Blumer M., Pechriggl E. J., Lyons K., Hall B. K., Fratzl P., et al. (2017). Calcified cartilage or bone? Collagens in the tessellated endoskeletons of cartilaginous fish (sharks and rays). J. Struct. Biol. 200, 54–71. doi: 10.1016/j.jsb.2017.09.005
Seidel R., Lyons K., Blumer M., Zaslansky P., Fratzl P., Weaver J. C., et al. (2016). Ultrastructural and developmental features of the tessellated endoskeleton of elasmobranchs (sharks and rays). J. Anat. 229, 681–702. doi: 10.1111/joa.2016.229.issue-5
Shepherd L. D., Campbell M. (2021). Ancient DNA analysis of an archaeological assemblage of Chondrichthyes vertebrae from South Auckland, New Zealand. J. Archaeological Sci.: Rep. 36, 102830. doi: 10.1016/j.jasrep.2021.102830
Sherman C. S., Simpfendorfer C. A., Pacoureau N., Matsushiba J. H., Yan H. F., Walls R. H., et al. (2023). Half a century of rising extinction risk of coral reef sharks and rays. Nat. Commun. 14, 15. doi: 10.1038/s41467-022-35091-x
Simpfendorfer C. A., Wiley T. R. (2005). determination of the distribution of Florida’s remnant sawfish population and identification of areas critical to their conservation. Final Report (Tallahassee, Florida: Florida Fish and Wildlife Conservation Commission), 40.
Springer V. G., Garrick J. A. F. (1964). A survey of vertebral numbers in sharks. Proc. United States Natl. Museum 116, 73–96. doi: 10.5479/si.00963801.116-3496.73
Tinari A. M., Hammerschlag N. (2021). An ecological assessment of large coastal shark communities in South Florida. Ocean Coast. Management 211, 105772. doi: 10.1016/j.ocecoaman.2021.105772
Ulrich G. F., Jones C. M., Driggers W. B., Drymon J. M., Oakley D., Riley C. (2007). “Habitat utilization, relative abundance, and seasonality of sharks in the estuarine and nearshore waters of South Carolina,” in American Fisheries Society Symposium (Bethesda, Maryland: American Fisheries Society), Vol. 50, 125.
Van Neer W., Zohar I., Lernau O. (2005). The emergence of fishing communities in the eastern Mediterranean region: A survey of evidence from pre-and protohistoric periods. Paléorient 31(1), 131–157. doi: 10.3406/paleo.2005.4793
Waters J. D., Coelho R., Fernandez-Carvalho J., Timmers A. A., Wiley T., Seitz J. C., et al. (2014). Use of encounter data to model spatio-temporal distribution patterns of endangered smalltooth sawfish, Pristis pectinata, in the western Atlantic. Aquat. Conservation: Mar. Freshw. Ecosyst. 24, 760–776. doi: 10.1002/aqc.2461
Weisler M. I., McNiven I. J. (2016). Four thousand years of western Torres Strait fishing in the Pacific-wide context. J. Archaeological Sci.: Rep. 7, 764–774. doi: 10.1016/j.jasrep.2015.05.016
Wells R. D., TinHan T. C., Dance M. A., Drymon J. M., Falterman B., Ajemian M. J., et al. (2018). Movement, behavior, and habitat use of a marine apex predator, the scalloped hammerhead. Front. Mar. Sci. 5, 321. doi: 10.3389/fmars.2018.00321
Wetherbee B. M., Gruber S. H., Cortés E. (1990). Diet, feeding habits, digestion and consumption in sharks, with special reference to the lemon shark. Negrapion brevirostris. NOAA Technical Report, NM FS, Vol. 90. 29–47.
Wiley T. R., Simpfendorfer C. A. (2007). The ecology of elasmobranchs occurring in the Everglades National Park, Florida: implications for conservation and management. Bull. Mar. Sci. 80, 171–189.
Worm B., Orofino S., Burns E. S., D’Costa N. G., Manir Feitosa L., Palomares M. L., et al. (2024). Global shark fishing mortality still rising despite widespread regulatory change. Science 383, 225–230. doi: 10.1126/science.adf8984
Wright D., Langley M. C., May S. K., Johnston I. G., Allen L. (2016). Painted shark vertebrae beads from the Djawumbu–Madjawarrnja complex, western Arnhem Land. Aust. Archaeol. 82, 43–54. doi: 10.1080/03122417.2016.1164356
Ziegler F., Groen E. A., Hornborg S., Bokkers E. A., Karlsen K. M., de Boer I. J. (2018). Assessing broad life cycle impacts of daily onboard decision-making, annual strategic planning, and fisheries management in a northeast Atlantic trawl fishery. Int. J. Life Cycle Assess. 23, 1357–1367. doi: 10.1007/s11367-015-0898-3
Keywords: collagens, cartilage, ZooMS, species identification, Florida Keys, proteomics
Citation: Buckley M, Oldfield E-M, Oliveira C, Boulanger C, Kitchener AC, Fuller NR, Ardren T, Thompson VD, Fitzpatrick SM and LeFebvre MJ (2024) Species identification of modern and archaeological shark and ray skeletal tissues using collagen peptide mass fingerprinting. Front. Mar. Sci. 11:1500595. doi: 10.3389/fmars.2024.1500595
Received: 23 September 2024; Accepted: 24 October 2024;
Published: 26 November 2024.
Edited by:
Anna Rita Rossi, Sapienza University of Rome, ItalyReviewed by:
Alberto Sánchez-González, National Polytechnic Institute (IPN), MexicoThomas J. Kean, University of Central Florida, United States
Copyright © 2024 Buckley, Oldfield, Oliveira, Boulanger, Kitchener, Fuller, Ardren, Thompson, Fitzpatrick and LeFebvre. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Buckley, bS5idWNrbGV5QG1hbmNoZXN0ZXIuYWMudWs=
 Michael Buckley
Michael Buckley Ellie-May Oldfield1
Ellie-May Oldfield1 Clara Boulanger
Clara Boulanger Andrew C. Kitchener
Andrew C. Kitchener Victor D. Thompson
Victor D. Thompson Scott M. Fitzpatrick
Scott M. Fitzpatrick Michelle J. LeFebvre
Michelle J. LeFebvre