- 1Qingjiang Station, Zhejiang Mariculture Research Institute, Wenzhou, China
- 2Zhejiang Key Laboratory of Coastal Biological Germplasm Resources Conservation and Utilization, Wenzhou, China
- 3Wenzhou key Laboratory of Marine Biological Genetics and Breeding, Wenzhou, China
- 4Division of Culture and Food Processing Technology, Zhejiang Mariculture Research Institute, Wenzhou, China
Introduction: In the context of Litopenaeus vannamei aquaculture, the incorporation of oyster shells has proven beneficial for enhancing water quality and the growth conditions of the shrimp. Nonetheless, the specific effects of in-situ water treatment using oyster shells on water quality and shrimp growth, along with the composition and succession dynamics of the microbial community within oyster shell biofilms, have yet to be thoroughly investigated.
Methods: This study established control, low-concentration, and high-concentration oyster shell addition groups to emulate the in-situ water treatment environment with oyster shells, with the objective of elucidating the impacts of oyster shell addition on the aquaculture setting.
Results: The results showed that the addition of oyster shells could significantly improve the length (F = 12.248, P = 0.005), weight(F = 138.234, P < 0.001), and survival rate (F = 15.248, P < 0.001) of shrimp, while there were no significant differences in the length (F = -1.233, P = 0.267) and survival rate (F = -2.143, P = 0.076) between the high and low concentration groups. Additionally, oyster shell addition resulted in elevated phosphate levels (F = 74.92, P < 0.001 in Day 70), diminished nitrite levels (F = 5.276, P = 0.031 in Day 56), and increased nitrate concentrations (F = 9.421, P = 0.006 in Day 70). Within the biofilms, the relative abundances of Ruegeria, Tenacibaculum, BD2- 11_terrestrial_group, and Kapabacteriales exhibited significant declines over time, whereas the relative abundance of Nitrospira demonstrated a marked increase, ultimately emerging as the predominant bacterium (Relative abundance 31.8%) in the biofilms during the latter stages of the experiment. Nitrospira also exhibited a notably higher relative abundance in the microbial community of the experimental water group relative to the control group (F = 2.265, P = 0.001).
Discussion: The biofilm provided conditions for the proliferation of Nitrospira, thereby accelerating the transformation of nitrite into nitrate in the aquaculture system, which subsequently improved the shrimp farming conditions. This research offers valuable insights for the application of oyster shells in shrimp farming and contributes to the theoretical underpinnings necessary for advancing our understanding of the mechanisms through which oyster shell biofilms enhance water quality and foster shrimp health.
Introduction
The Pacific White Shrimp (Litopenaeus vannamei), celebrated for its rapid growth, robust survival rate, and formidable adaptability, has emerged as a pivotal industry in global marine aquaculture, commanding substantial economic value (Liao and Chien, 2011). The aquaculture of the Litopenaeus vannamei is significantly impacted by a range of factors, encompassing water quality conditions, environmental variables, aquaculture infrastructure, seed stock quality, and dietary composition (Monsalve and Quiroga, 2022; Huang et al., 2022). As a tropical shrimp species highly sensitive to water quality, the growth trajectory, health status, and survival rates of the Litopenaeus vannamei are intricately tied to key water parameters such as dissolved oxygen, pH levels, ammonia nitrogen, nitrite, and salinity (Villarreal and Juarez, 2022; Supriatna et al., 2017; Han et al., 2017). Dissolved oxygen is an essential element for the shrimp’s survival, insufficient levels can retard growth and may even lead to asphyxiation (Simbeye and Yang, 2014). Fluctuations in pH beyond normal ranges can disturb the shrimp’s osmotic equilibrium, compromising their immune function and thereby weakening their disease resistance (Mustapha and Scientific, 2020). Maintaining an optimal salinity range can mitigate the osmotic pressure regulation demands on the shrimp, thereby enhancing their environmental adaptability (Li et al., 2024). Ammonia nitrogen and nitrite are prevalent water contaminants; at elevated concentrations, they pose a toxic threat to the shrimp (Valencia-Castañeda et al., 2018). Prolonged exposure to these substances can hinder shrimp growth, impair immune function, and potentially culminate in widespread mortality (Huang et al., 2020a).
Oyster shells play a significant role in enhancing aquaculture water quality during shrimp cultivation (Jones et al., 2002; Huang et al., 2020b). Abundant in calcium carbonate, these shells are instrumental in maintaining optimal pH levels within the water, thereby supporting the molting and new shell formation processes in shrimp (Shen Yuchun et al., 2007). Moreover, oyster shells exhibit a high adsorption capacity, effectively binding and sequestering harmful substances such as ammonia nitrogen, nitrite, and organic pollutants (Patil et al., 2024; An et al., 2024). This capability significantly diminishes the concentration of these detrimental elements in the water, thereby reducing the associated risks to shrimp health. Additionally, oyster shells provide an ideal substrate for microbial colonization, promoting the establishment of beneficial microbial films within the aquatic environment (Lu et al., 2010; Ivanov et al., 2006).
Microbial films are characterized by their diverse composition, encompassing bacteria, algae, protozoa, and fungi, which together create a dynamic ecosystem on the surfaces of oyster shells (Ivanov et al., 2006). These microorganisms play a crucial role in the cycling of organic matter and nutrients within the aquatic environment (Geng et al., 2024). Specifically, Nitrosomonas bacteria can take advantage of the special microenvironment formed in the biofilm on the surface of oyster shells to promote their growth, which may be related to the surface roughness and chemical composition provided by the oyster shells (Sedlacek et al., 2019). These bacteria catalyze the oxidation of ammonia nitrogen in the water to nitrite, a process followed by Nitrobacter bacteria which convert nitrite into nitrate (Daims et al., 2015; Van Kessel et al., 2015). Algae then utilize these nitrate and phosphate compounds as essential nutrients for their growth and reproduction. Additionally, the microorganisms present in the microbial film can be integrated into the food chain, ultimately affecting the microecological balance within the guts of the shrimp (Zeng et al., 2024).
Although the positive role of oyster shells in shrimp aquaculture has gained increasing recognition, the regulatory efficacy of oyster shells in shrimp farming still warrants further validation (Huang et al., 2020b). Moreover, the mechanisms through which the microbial film on oyster shells influences water quality and the intestinal environment of shrimp remain to be fully understood. This study is designed to explore these aspects by setting up groups with different concentrations of oyster shell additions and monitoring the shrimp cultivation outcomes. Specifically, the research aims to: 1) assess whether there are differences in shrimp cultivation performance among groups with varying concentrations of oyster shells; 2) evaluate the differential effects of different oyster shell concentrations on water quality; and 3) determine whether the microbial film on oyster shells at different concentrations has varying impacts on the shrimp.
Materials and methods
Biofilm preparation, water environment monitoring, and sample collection
This experiment was conducted at Zhejiang Mariculture Research Institute in Wenzhou, Zhejiang Province, China. Twelve culture tanks, each with an approximate volume of 800L (diameter 1 m, height 1 m), were selected for the aquaculture experiment system. Each experimental tank had a water volume of about 700L and was stocked with 140 shrimp (body length: 4.054 ± 0.326 cm, body weight: 0.367 ± 0.086 g), resulting in a stocking density of 200 shrimp/m³. The 12 culture systems were divided into three groups, each consisting of four replicates: the control group, the high oyster shell concentration group, and the low oyster shell concentration group. The experimental groups were supplemented with oyster shells at concentrations of 4‰ (approximately 2.8L) and 16‰ (approximately 11.2L) of the culture water volume, respectively (shell length: 4.34 ± 0.89 cm; width: 2.73 ± 0.71 cm). The bottom of the oyster shells was aerated to form an upward flow through the shells, while no oyster shells were added to the control group. During the experiment, each tank was continuously aerated to increase oxygen levels, and an equal amount of feed was provided three times daily (8:00, 12:00, and 16:00). Starting from the 15th day, water was exchanged daily, with the control group having a 10% water change and the oyster shell addition groups having a 2% water change.
Every 7 days, 1L water samples were collected from each experimental tank for the determination of physicochemical parameters of the water. Every 14 days, biofilm samples from the oyster shells were collected by randomly selecting three shells of consistent size from each experimental tank. The oyster shell samples were rinsed with sterile physiological saline and the biofilm was separated using an SB-5200DT ultrasonic cleaner (40kHz, 15min). The suspension was then filtered through a 0.2-μm polycarbonate membrane (Millipore Type GTTP, USA), and the samples collected on the filter membrane represented the biofilm (Figure 1A).
At each sampling time point, the concentrations of ammonia nitrogen, nitrite, nitrate, and active phosphate in the water were measured. The measurement of the physicochemical parameters of the water was conducted in accordance with the “Specifications for Oceanographic Survey” (GB 17378.4-2007): ammonia nitrogen was determined using the indophenol blue spectrophotometric method; nitrite was measured using the sulfanilamide and hydrochloric acid naphthylethylenediamine diazo method; active phosphate was determined using the ascorbic acid reduction phosphomolybdenum blue method.
At the beginning of the experiment (D0) and at the end (D70), the body length and body weight of 10 individual shrimp were measured. At the end of the experiment (D70), shrimp yield was recorded, and survival rate was calculated.
DNA extraction and bacterial 16S rRNA gene sequencing
Genomic DNA was extracted using the Power Soil® DNA Isolation Kit (MOBIO, USA) and the QIAamp® DNA Stool Mini Kit (Qiagen, Germany) according to the manufacturers’ instructions. DNA concentration and quality levels were measured using an ND-2000 spectrophotometer (NanoDrop Technologies, Delaware, USA). The V4 region of the 16S rRNA gene was amplified using primers 515F-Y (5’-GTGYCAGCMGCCGCGGTAA-3’) and 806R-B (5’-GGACTACNVGGGTWTCTAAT-3’). The PCR products were purified using a PCR Fragment Purification Kit (TaKaRa, Japan), quantified with an Agilent 2100 Bioanalyzer (Agilent, USA), and sequenced on the MiSeq platform (Illumina, USA).
Processing of Illumina sequencing data
For the 16S rRNA data, we used the QIIME2 command line platform version 2024.2 with default parameters for quality control, filtering, chimera detection, de-noising, detection of sequence variants and taxonomic classification. Taxonomy was assigned to representative sequences using the feature-classifier plugin with the Naive Bayes SILVA132 515F/806R classifier available on the QIIME2 website (https://docs.qiime2.org/2024.2/data-resources/) based on 97% sequence identity (Hall and Beiko, 2018; Quast et al., 2012). After assigning taxonomic information, OTUs related to Archaea, Chloroplast, Unclassified (not affiliated with Bacteria), and singleton OTUs were removed from the dataset. To correct for unequal sequencing depth, the OTU table was diluted at the level of 10,000 sequences per sample to calculate alpha- and beta-diversity estimates, as well as Bray-Curtis dissimilarity between samples. The research data from the article has been submitted to National Center for Biotechnology Information (SUB14648373).
Statistical analyses
Pearson correlation coefficient was used to calculate the correlation between parameter variables. One-way ANOVA and T-test were used to determine whether there were significant differences between groups. PCA clustering based on Bray-Curtis (BC) distance was used to analyze microbial community clustering, and analysis of similarity (ANOSIM) was used to calculate whether there were significant differences between communities (Granato et al., 2018). Bacteria with a relative abundance greater than 1% were used to plot the bacterial community composition abundance chart. The intersection of bacteria with high contributions predicted by the random forest and bacteria with significant differences identified by one-way ANOVA was taken to obtain the differential bacteria between groups (Rigatti, 2017).
Results
Shrimp growth and water quality outcomes
Throughout the cultivation process, there was a general upward trend in the levels of chemical oxygen demand (COD), suspended substance, phosphorus, nitrite, ammonium, and nitrate (Figure 1B; Supplementary Tables S1, S2). Significant inter-group differences in COD emerged starting from the 7th day (F = 12.717, P = 0.002), escalating to extremely significant variations among groups from the 49th day onwards (F = 151.26, P < 0.001). However, a consistent trend of continuous inter-group differences over time was not observed. Similarly, no consistent inter-group differences were noted in suspended substance. Ammonium levels rose significantly and rapidly from the 7th day, with the control group exhibiting consistently higher concentrations than the experimental groups (F = 7.586, P = 0.012). By the 28th day, the ammonium content in the control group experienced a sharp decline, dropping below that of the experimental groups (F = 5.038, P = 0.034). Nitrite concentrations in the control group were initially lower than those in the experimental groups until the 14th day, after which they remained consistently higher from the 35th day onwards (F = 244.227, P < 0.001). Starting from the 7th day, nitrate levels in the control group were persistently lower than those in the experimental groups (F = 6.068, P = 0.021), while no consistent significant differences were discerned over time between the low-concentration and high-concentration groups. Phosphorus concentrations were consistently lower in the control group compared to the experimental groups, significant differences were observed starting from the 7th day between the control group and the high-concentration group (F = -7.664, P < 0.001), and significant differences were noted from the 21st day between the control group and the low-concentration group (F = -7.367, P < 0.001).
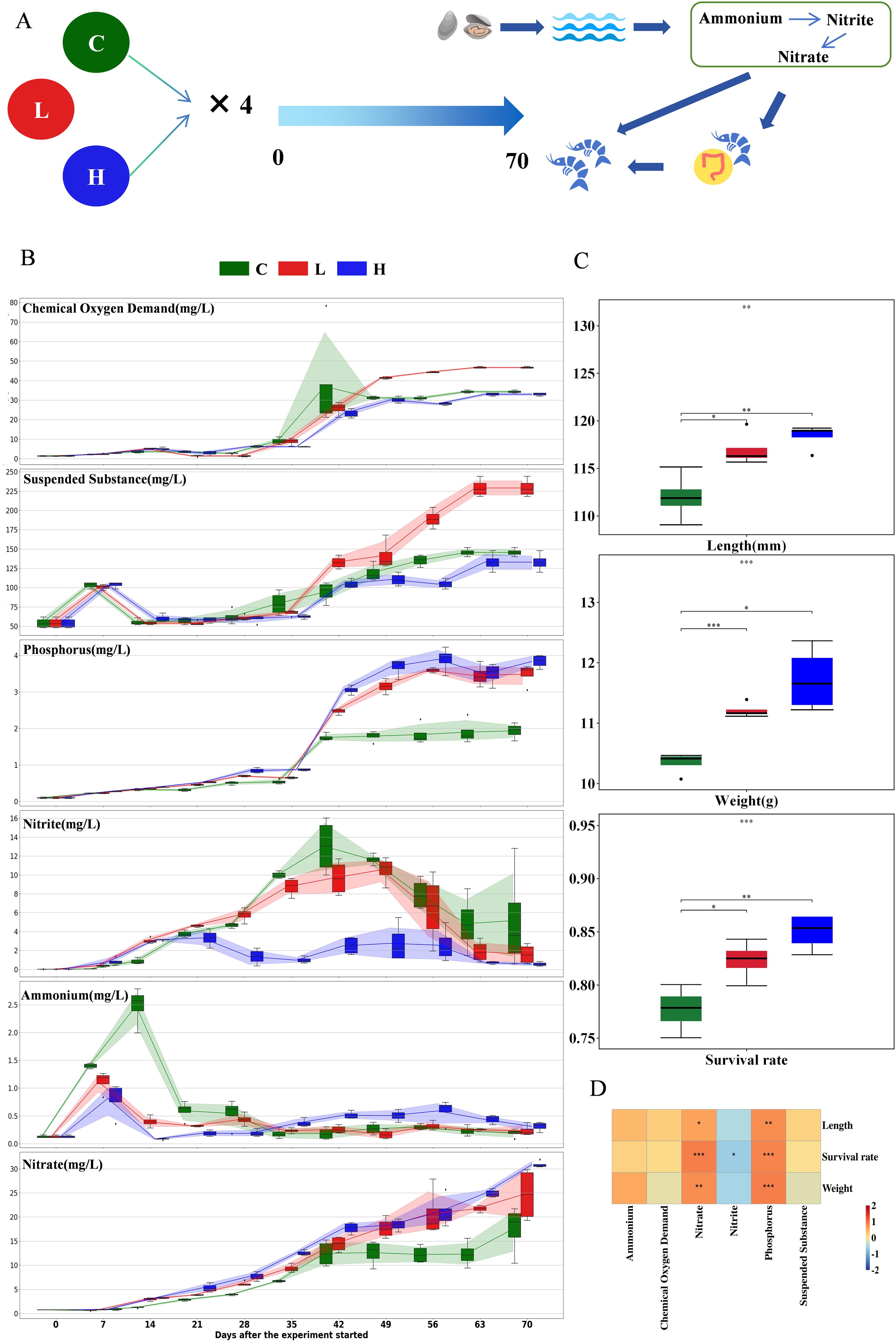
Figure 1. (A) Schematic of the experimental design: The study is divided into three groups: the control group, the low-concentration oyster shell addition group, and the high-concentration oyster shell addition group, with four replicates for each group. The entire aquaculture cycle spans 70 days, with monitoring of water quality indicators (COD, suspended solids, phosphates, nitrites, ammonia, and nitrates) at days 0, 7, 14, 21, 28, 35, 42, 49, 56, 63, and 70. Routine parameter tests (OD, temperature, pH, and salinity) are conducted at days 0, 14, 28, 42, 56, and 70, and microbial film sampling is performed for the two experimental groups. At day 70, the length, weight, and survival rate of the shrimp in each culture bucket are calculated, followed by sampling of the culture water and shrimp guts. (B) Box plot showing the temporal variation of aquaculture water quality indicators. (C) Bar chart illustrating the differences in shrimp length, weight, and survival rate among the groups (T-test). (D) Heatmap of the Pearson correlation analysis results between aquaculture water quality indicators and shrimp growth, with asterisks indicating significant correlations. * p-value < 0.05, ** p-value < 0.01, and *** p-value < 0.001.
On the 70th day of the cultivation period, a statistical analysis was conducted on the length (F = 12.248, P = 0.005), weight(F = 138.234, P < 0.001), and survival rate (F = 15.248, P < 0.001) of the shrimp in each group. The findings revealed that the control group had significantly lower values in terms of body length, weight, and survival rate when compared to the experimental groups (Figure 1C; Supplementary Table S3). Within the experimental groups, a significant difference was observed in weight, with the high-concentration group exhibiting a higher average weight than the low-concentration group (F = -9.559, P < 0.001). However, no significant differences were detected in body length (F = -1.223, P = 0.267) and survival rate (F = -2.143, P = 0.067) between the low and high concentration groups. The increased concentration of oyster shell addition did not result in superior biological enhancement, indicating that an addition rate of 4‰ may have already reached the functional limit for oyster shell enhancement within the aquaculture system, or other factors may be constraining the biological enhancement potential of oyster shells. The excessive addition of oyster shells could potentially disturb the water’s acid-base equilibrium and elevate pH levels. However, an addition of 16‰ oyster shells did not constitute an excessive level, suggesting that the threshold for detrimental effects has not yet been reached.
The correlation analysis between the growth indices of shrimp and the hydro chemical factors revealed that there was a significant negative correlation between the concentration of nitrite and the body length of the shrimp (rho = -1, p = 0.006), while a significant positive correlation was observed between phosphate and body length (rho = 0.999, p = 0.023, Figure 1D; Supplementary Table S4).
Bacterial communities in water, shrimp guts, and biofilms
On the 70th day, there were significant differences in the beta diversity of bacterial communities in the water as a whole (R = 0.544, P = 0.002), but no significant differences between the control and experimental groups. In the shrimp guts, there were significant differences in the beta diversity of bacterial communities as a whole (R = 0.610, P = 0.001), with significant differences between the control and experimental groups, but no significant differences between the high and low concentration groups (R = 0.313, P = 0.057, Figure 2A; Supplementary Table S5). Multiple indicators of alpha diversity in the low concentration group showed significant differences (Figure 2B; Supplementary Tables S6, S7).
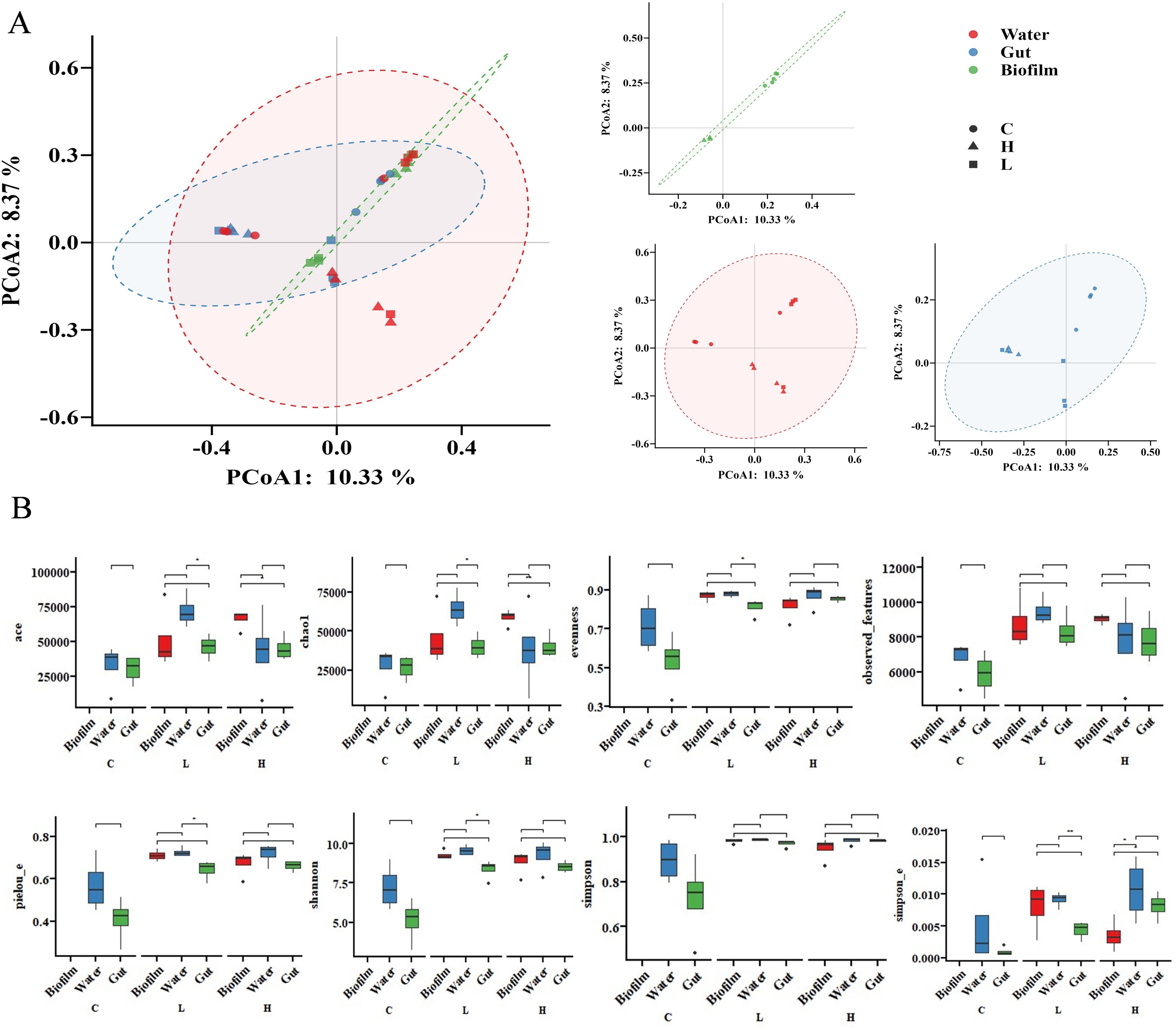
Figure 2. (A) PCA cluster plot of BC distances for the bacterial communities in the biofilms, water, and guts on the 70th day. (B), Box plot of alpha diversity indices for the bacterial communities in the biofilms, water, and guts on the 70th day. * Indicates significant differences. * p-value < 0.05, ** p-value < 0.01, and *** p-value < 0.001.
In the biofilms, Ruegeria, Tenacibaculum, Nitrospira, BD2-11_terrestrial_group, and Kapabacteriales were the dominant bacteria on day 0 (Figure 3A; Supplementary Table S8). Among these, Ruegeria (F = 6.103, P < 0.001), Tenacibaculum (F = 5.110, P < 0.001), BD2-11_terrestrial_group (F = 8.170, P = 0.001), Kapabacteriales (F = 2.444, P = 0.050) demonstrated a significant decrease in relative abundance over time. In contrast, Nitrospira showed a significant increase in relative abundance over time (F = 5.951, P < 0.001). Beginning on day 56, Nitrospira became the most abundant dominant bacterium in the biofilms, reaching relative abundances of 19.6% and 44% in the low and high concentration groups, respectively, by day 70. Additionally, Nitrospira was also the dominant bacterium in the water, with concentrations as high as 12.3% and 7.1% in the experimental groups, and only 0.1% in the treatment groups (Figure 3B; Supplementary Table S9). Nitrospira was not the dominant bacterium in the shrimp gut. Burkholderia-Caballeronia-Paraburkholderia, Ruegeria, PeM15, and Mycobacterium were dominant in both water and the shrimp gut, with significant inter-group differences observed for Burkholderia-Caballeronia-Paraburkholderia (F = 17.548, P = 0.001), Ruegeria (F = 4.985, P = 0.035), and PeM15 (F = 5.947, P = 0.022) in the gut.
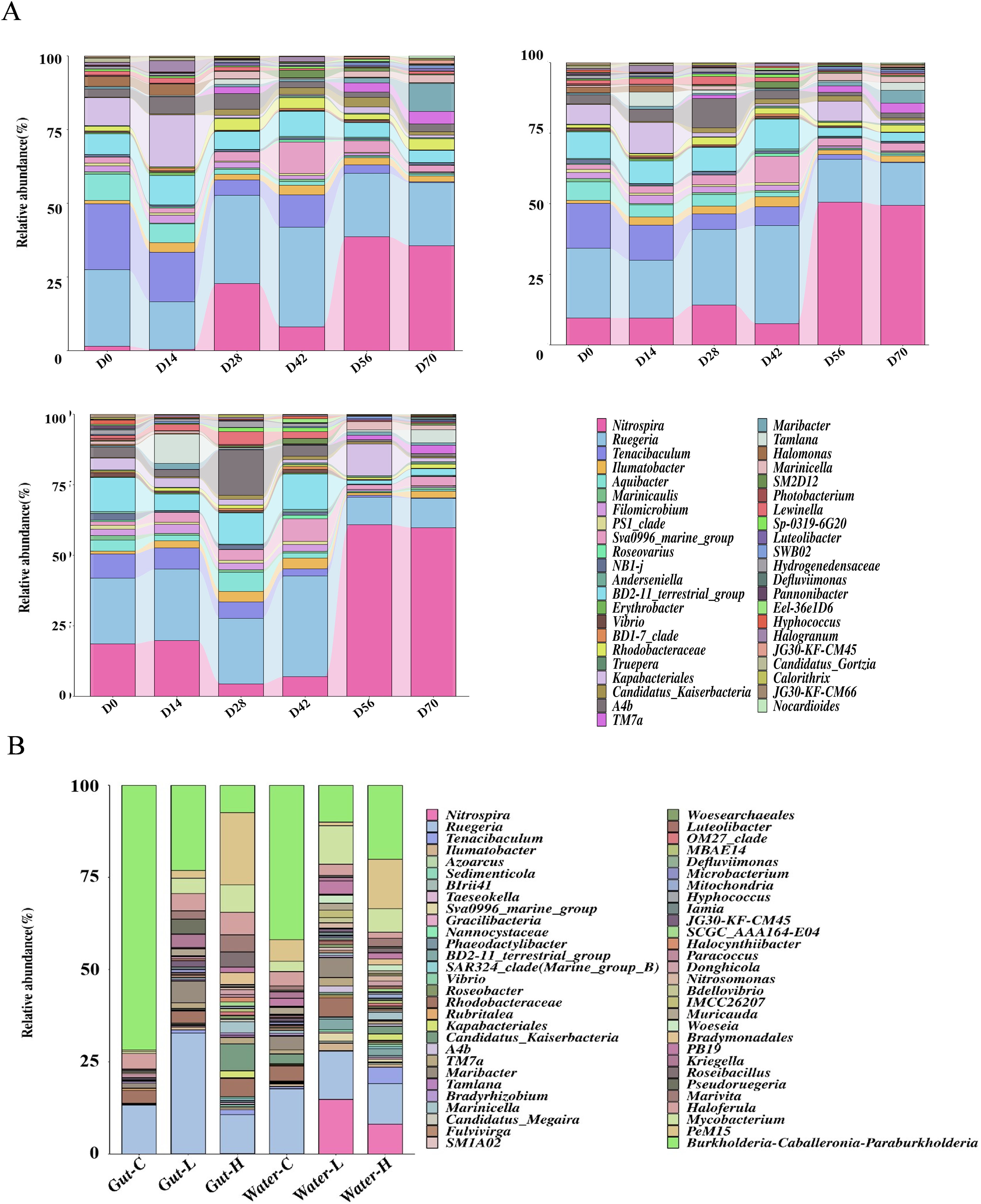
Figure 3. (A) Bar stacked chart of the relative abundance of dominant bacteria at various time points in the biofilms. (B), Bar stacked chart of the relative abundance of dominant bacteria in water and gut.
Through one-way ANOVA and random forest analysis, we identified nine key bacterial genera, WPS-2, Bauldia, SWB02, KI89A_clade, Nitrosomonas, Woeseia, PS1_clade, BD2-11_terrestrial_group, and Truepera, in water that exhibited significant inter-group differences (Figures 4A, B, Supplementary Tables S9, S10). WPS-2, Bauldia, and KI89A_clade were significantly positively correlated with shrimp weight, and KI89A_clade was also significantly positively correlated with survival rate (Figure 4C; Supplementary Table S13). Among these, WPS-2 and Bauldia were significantly positively correlated with Chemical Oxygen Demand and Suspended Substance, SWB02 and KI89A_clade were significantly positively correlated with phosphate, and Nitrosomonas was significantly positively correlated with nitrate and phosphorus (Figure 4D; Supplementary Table S12).
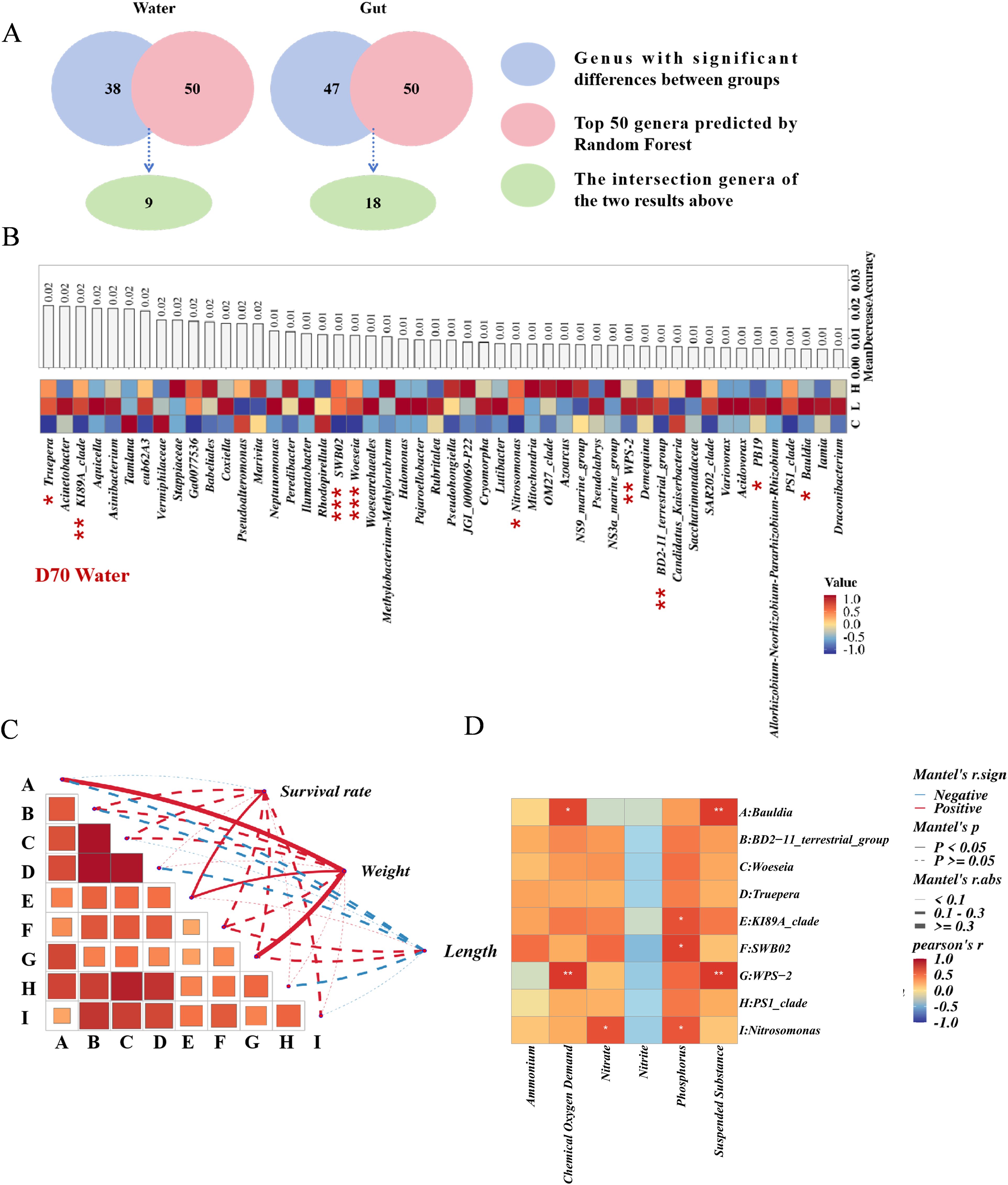
Figure 4. (A) Schematic diagram for the selection of differential bacteria between water and gut. (B) Schematic diagram for the selection of differential bacteria in water, with heat maps representing bacterial abundance and bar charts representing the contribution of random forests, asterisks indicate significant differences in single-factor variance analysis. (C) Correlation analysis chart of differential bacteria with the growth length, weight, and survival rate of shrimp. (D), Correlation analysis heat map of differential bacteria with environmental factors in water. * p-value < 0.05, ** p-value < 0.01, and *** p-value < 0.001.
Tenacibaculum, Kriegella, KI89A_clade, Iamia, JG30-KF-CM45, Fulvivirga, Maribacter, Ruegeria, Mycobacterium, BIrii41, Aureimarina, Ilumatobacter, Halocynthiibacter, SWB02, Nitrosomonas, A4b, Thioclava, and Gracilibacteria were identified as bacteria with significant inter-group differences in the gut (Figures 4A, 5A; Supplementary Tables S9, S11). Among these, Tenacibaculum was significantly positively correlated with nitrate and phosphorus. Kriegella, KI89A_clade, Iamia, JG30-KF-CM45, Fulvivirga, Maribacter, and Ruegeria were all significantly positively correlated with Chemical Oxygen Demand and Suspended Substance, while BIrii41 and Mycobacterium were significantly positively correlated with nitrate (Figure 5B; Supplementary Table S14). Tenacibaculum was significantly positively correlated with shrimp length and survival rate, while Ilumatobacter was significantly positively correlated with shrimp weight and survival rate (Figure 5C; Supplementary Table S15).
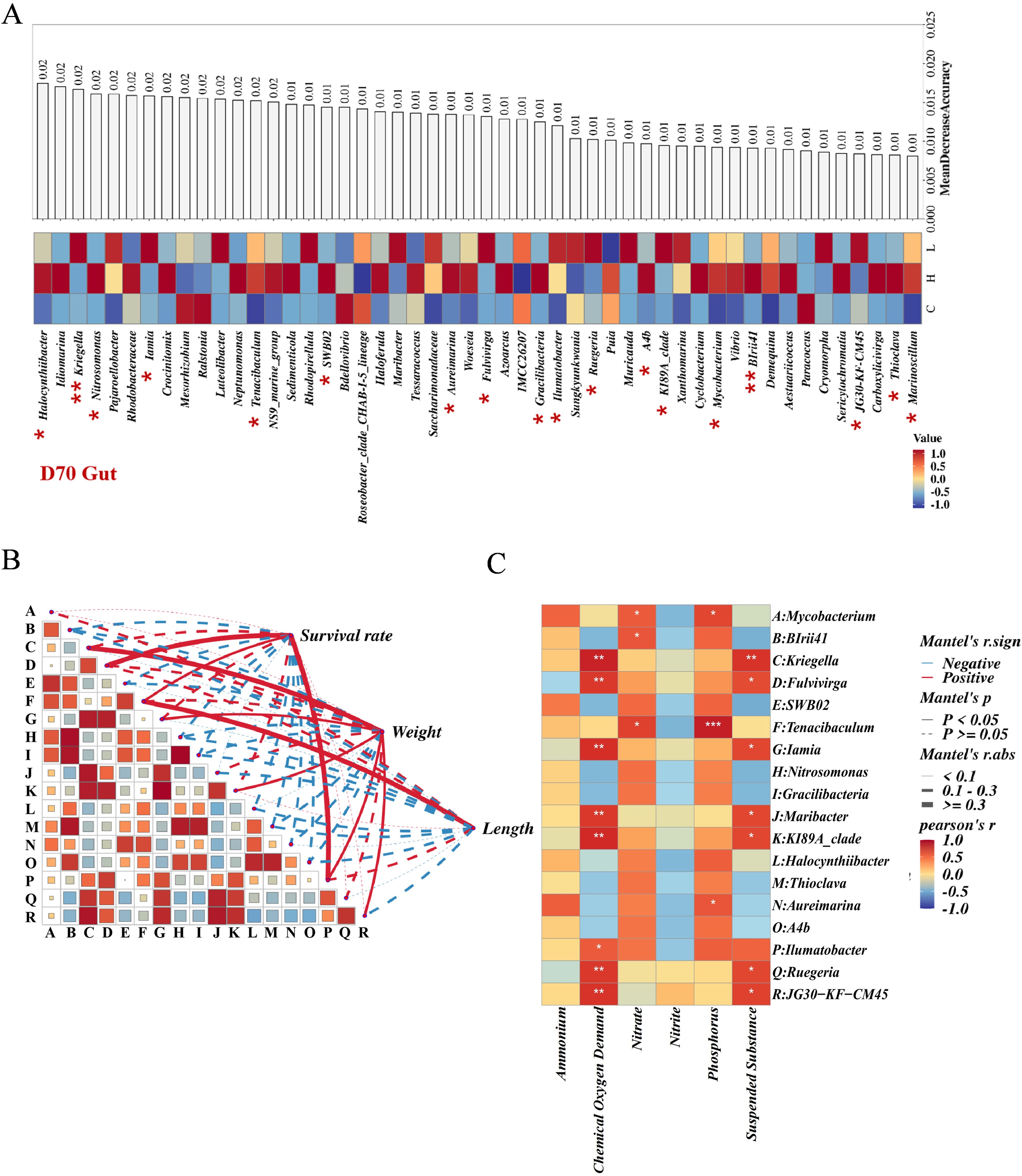
Figure 5. (A) Schematic diagram for the selection of differential bacteria in gut, with heat maps representing bacterial abundance and bar charts representing the contribution of random forests, asterisks indicate significant differences in single-factor variance analysis. (B), Correlation analysis chart of differential bacteria with the growth length, weight, and survival rate of shrimp. (C), Correlation analysis heat map of differential bacteria with environmental factors in water. * p-value < 0.05, ** p-value < 0.01, and *** p-value < 0.001.
Discussion
The addition of oyster shells could improve the water quality in aquaculture
The monitoring of hydrochemical indicators throughout the aquaculture process demonstrated that oyster shells had a marked impact on enhancing water quality. Phosphate, among the hydrochemical factors, correlated positively with shrimp body length, weight, and survival rate, with concentrations notably lower in the control group than in the experimental group. As a vital water buffer, phosphate stabilizes pH levels and bolsters the shrimp’s immune system and resilience, thereby fostering robust growth (Shimoda et al., 2005; Vijuksungsith et al., 2023; Nathanailides et al., 2023).
The biofilm accelerated the conversion of nitrite to nitrate in the nitrogen cycle. Shrimp feed is rich in protein and nitrogen-containing organic matter (Eggink et al., 2024). During the farming process, especially in the later stages, the substantial increase in feed input led to a significant influx of nitrogen sources into the farming system. Decomposed residual feed, shrimp excretion, and plankton debris largely existed in the form of ammonia nitrogen (Chen et al., 2024). However, the control group’s daily 10% water exchange rate effectively decelerated ammonia nitrogen accumulation, thereby mitigating nitrite buildup. Conversely, the experimental group, with a daily 2% water exchange rate, witnessed more rapid nitrite accumulation relative to the control group. Nitrosomonas could convert ammonia nitrogen into hydroxylamine through ammonia monooxygenase, which was then oxidized to nitrite by hydroxylamine oxidase (Mctavish et al., 1993). The adverse effects of nitrite on shrimp aquaculture are widely recognized (Gross et al., 2004; Tomasso, 2012). The substantial influx of ammonia nitrogen allowed Nitrosomonas to occupy a dominant ecological niche in the water, accelerating the conversion process from ammonia nitrogen to nitrite. Nitrospira, a group of nitrite-oxidizing bacteria, was dominant in biofilms and could oxidize nitrite to nitrate through nitrite oxidoreductase (Daims and Wagner, 2018; Daims et al., 2015). Oyster shell biofilms provided an attachment matrix for Nitrospira, creating favorable conditions for their proliferation and accelerating the nitrogen cycle in the farming system from nitrite to nitrate.
Nitrospira, Ruegferia, and Tenacibaculum were the dominant bacteria in the biofilms
In the bacterial community of the biofilm, prior to the emergence of Nitrospira as the dominant bacteria, Ruegeria and Tenacibaculum were the predominant species within the biofilms. Ruegeria, an aerobic chemoheterotrophic bacterium, was noted for its high capacity for polysaccharide metabolism, which enabled it to contribute to the degradation of algal cell walls (Yin et al., 2023; Huo et al., 2011). Research had demonstrated that Tenacibaculum could rapidly establish biofilms on polystyrene surfaces, exhibiting hydrophobic characteristics that were likely associated with biofilm formation. Additionally, Tenacibaculum maritimum, recognized as a fish pathogen, was known to possess the ability to form biofilms (Levipan et al., 2019; Avendaño-Herrera et al., 2006).
Regardless of whether in the control or experimental groups, the concentrations of ammonia and nitrite in the water were notably high. However, Nitrospira was not the dominant bacterial genus within the bacterial communities of the control sample waters. It is hypothesized that Nitrospira in both high and low concentration groups originated from biofilms. Microorganisms on the surfaces of oyster shells interconnected through an extracellular polymeric substance matrix, forming a stable community structure. This structure not only provided physical protection for the microorganisms but also conferred advantages in resource acquisition and substance exchange (Flemming and Wingender, 2010). The favorable microenvironment within the biofilms promoted the growth and activity of Nitrospira (Zhou et al., 2023; Vijayan et al., 2021). The significant enrichment of Nitrospira within the biofilms led to increased accumulation of this genus in water, which, through water and feed, influenced the distribution of Nitrospira within the gut (F = 6.253, P = 0.020); however, it did not dominate the gut microbiota.
Nitrospira faced more intense competition in the complex aquatic environment, and it possibly aggregated back onto the biofilm due to sedimentation (Shao et al., 2024; Wan et al., 1995). In the high concentration group, the contact area between the oyster shells and the water was greater, resulting in a larger surface area of the biofilm that could support more Nitrospira. This led to lower abundance of Nitrospira in the water of the high concentration group compared to the low concentration group. The processes of Nitrospira migrating from the biofilm into the water and the sedimentation of Nitrospira from the water onto the biofilm likely occurred simultaneously. We will design more complex concentration gradients and denser sampling intervals in subsequent studies to explore the evolution of this dynamic process.
In both water and gut environments, besides Nitrospira, Ruegeria, and Tenacibaculum, Burkholderia-Caballeronia-Paraburkholderia were identified as important differential dominant bacteria. Burkholderia was a diverse and environmentally adaptable plant-associated bacterium (Compant et al., 2008). Burkholderia cepacia was reported as a common opportunistic pathogen in plants and was capable of degrading both natural and artificial pollutants (Mahenthiralingam et al., 2005). The ecological niche changes brought about by the addition of oyster shells made bacteria associated with biofilm formation or nitrogen cycling more competitive, which led to a reduction in the abundance of Burkholderia-Caballeronia-Paraburkholderia.
Tenacibaculum, KI89A_clade, SWB02, and Nitrosomonas were the inter-group differential bacteria in water and gut
Through the application of random forest analysis and one-way ANOVA, we identified significant inter-group differences in the relative abundance of Tenacibaculum within the shrimp gut. This abundance was notably correlated with shrimp growth indicators, floating particulate matter, and chemical oxygen demand. The relative abundance of Tenacibaculum in the biofilms exhibited a gradual decline. We hypothesized that this phenomenon is attributable to the critical role of Tenacibaculum in the formation and structural stability of biofilms (Levipan et al., 2019). During the initial stages of biofilm formation, Tenacibaculum, which aids in establishing a stable biofilm structure, enjoys a competitive advantage in the ecological niche. However, as the biofilm structure matures and the concentration of nitrite increases, Nitrospira, with its nitrogen-fixing capabilities, gains a competitive edge, leading to a decrease in the relative abundance of Tenacibaculum (Palomo et al., 2022). Throughout the aquaculture process, Tenacibaculum continually permeated from the biofilm into the water and the shrimp gut, resulting in a positive correlation between Tenacibaculum abundance and shrimp growth.
KI89A_clade, SWB02, and Nitrosomonas were significantly positively correlated with phosphate, while Nitrosomonas was also significantly positively correlated with nitrate in water. KI89A_clade is a bacterial group whose relationship with shrimp growth remains unclear, previous study found that after infection with Vibrio parahaemolyticus, the relative abundance of KI89A in the microbial community significantly decreased in Apocyclops royi(Amparyup, 2024). SWB02 was found to enhance ammonia-oxidizing capacity and produce large quantities of extracellular polymeric substances (EPS). These substances can form granules in ammonia-oxidizing systems, aiding in the biofilms formation and potentially influencing the structure and function of microbial communities in biotreatment processes (Han et al., 2024). Nitrosomonas, an important group of nitrifying bacteria, plays a key role in the first stage of nitrification by oxidizing ammonia (NH4+) to nitrite (NO2-) (Arp et al., 2002). However, due to the rapid conversion of nitrite to nitrate by Nitrospira, Nitrosomonas showed a significant correlation with nitrate concentration. These processes may have indirect interactions with phosphate, which is a limiting factor for algae growth, and algae are a crucial component of primary producers in the aquatic environment (Lobus, 2022).
Conclusion
The addition of oyster shells improved the survival rate, body length, and weight of shrimp. The surface of the oyster shells formed a biofilm, in which the relative abundance of Nitrospira gradually increased over time, becoming the dominant bacteria in the biofilm. Nitrospira infected the water from the biofilm, leading to an increase in the abundance of Nitrospira in the water. This accelerated the nitrogen cycle in the aquaculture system, transforming nitrite into nitrate, which in turn improved the survival rate, body length, and weight of the shrimp. Nitrospira did not directly colonize the gut microbiota of the shrimp. The addition of oyster shells at different concentrations also altered the abundance of bacteria such as KI89A_clad, SWB02, Nitrosomonas, and Tenacibaculum in the water and gut. Compared to the low concentration group, the high concentration group increased the weight of the shrimp. This study elucidated the dynamic changes in the bacterial composition of the biofilm on the oyster shell surface and its role in the nitrogen cycle of the aquaculture system, helping us better understand the positive significance of adding oyster shells to shrimp farming.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1176290.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
XKH: Writing – original draft. GX: Formal analysis, Writing – original draft. XZ: Data curation, Writing – original draft. ST: Methodology, Writing – original draft. ML: Formal analysis, Writing – original draft. YC: Conceptualization, Investigation, Writing – original draft. RC: Investigation, Writing – original draft. XLH: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by Zhejiang Provincial Science and Technology Project of China (LGN21C190008), and Special Program for Research Institutes of Zhejiang Province(2024YS002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1495938/full#supplementary-material
References
Amparyup P. (2024). Transcriptomic and Microbiome Analyses of copepod Apocyclops royi in Response to an AHPND-causing strain of Vibrio parahaemolyticus. Dev Comp Immunol. 105277.
An W., Liu Y., Chen H., Sun X., Wang Q., Hu X., et al. (2024). Adsorption properties of Pb (II) and Cd (II) in acid mine drainage by oyster shell loaded lignite composite in different morphologies. Sci. Rep. 14, 11627. doi: 10.1038/s41598-024-62506-0
Arp D. J., Sayavedra-Soto L. A., Hommes N. G. (2002). Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 178, 250–255. doi: 10.1007/s00203-002-0452-0
Avendaño-Herrera R., Toranzo A. E., Magariños B. (2006). Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis. Aquat. organisms 71, 255–266. doi: 10.3354/dao071255
Chen F., Qiu T., Xu J., Zhang J., Du Y., Duan Y., et al. (2024). Rapid real-time prediction techniques for ammonia and nitrite in high-density shrimp farming in recirculating aquaculture systems. Fishes 9, 386. doi: 10.3390/fishes9100386
Compant S., Nowak J., Coenye T., Clement C., Ait Barka E. (2008). Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol. Rev. 32, 607–626. doi: 10.1111/j.1574-6976.2008.00113.x
Daims H., Lebedeva E. V., Pjevac P., Han P., Herbold C., Albertsen M., et al. (2015). Complete nitrification by Nitrospira bacteria. Nature 528, 504–509. doi: 10.1038/nature16461
Daims H., Wagner M. (2018). Nitrospira. Trends In Microbiol. 26, 462–463. doi: 10.1016/j.tim.2018.02.001
Eggink K. M., Gonçalves R., Skov P. V. (2024). Shrimp processing waste in aquaculture feed: nutritional value, applications, challenges, and prospects. Rev. Aquaculture. doi: 10.1111/raq.12975
Flemming H.-C., Wingender J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Geng Y., Yang L., Lian C.-A., Pavlostathis S. G., Qiu Z., Xiong Z., et al. (2024). Resourceful application and mechanism of oyster shell-microalgae synergistic system: Sustainable treatment of harsh low carbon nitrogen ratio actual wastewater. Environ. Res. 252, 118775. doi: 10.1016/j.envres.2024.118775
Granato D., Santos J. S., Escher G. B., Ferreira B. L., Maggio R. M. (2018). Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 72, 83–90. doi: 10.1016/j.tifs.2017.12.006
Gross A., Abutbul S., Zilberg D. (2004). Acute and Chronic Effects of Nitrite on white shrimp, Litopenaeus vannamei, cultured in low-salinity brackish water. J. World aquaculture Soc. 35, 315–321. doi: 10.1111/j.1749-7345.2004.tb00095.x
Hall M., Beiko R. G. (2018). 16S rRNA gene analysis with QIIME2. Microbiome Analysis: Methods Protoc. 1849, 113–129. doi: 10.1007/978-1-4939-8728-3_8
Han N.-N., Yang J.-H., Fan N.-S., Jin R.-C. (2024). Mechanistic insight into microbial interaction and metabolic pattern of anammox consortia on surface-modified biofilm carrier with extracellular polymeric substances. Bioresource Technol. 407, 131092. doi: 10.1016/j.biortech.2024.131092
Han S., Wang B., Wang M., Liu Q., Zhao W., Wang L. (2017). Effects of ammonia and nitrite accumulation on the survival and growth performance of white shrimp Litopenaeus vannamei. Invertebrate Survival J. 14, 221–232. doi: 10.25431/1824-307X/isj.v14i1.221-232
Huang X., Du S., Zhang H., Chen C., Xiao G., Huang L., et al. (2020b). Temporal patterns of bacterial communities in shrimp (Litopenaeus vannamei) culture systems loaded with oyster shells. Aquaculture 526, 735424. doi: 10.1016/j.aquaculture.2020.735424
Huang C., Luo Y., Zeng G., Zhang P., Peng R., Jiang X., et al. (2022). Effect of adding microalgae to whiteleg shrimp culture on water quality, shrimp development and yield. Aquaculture Rep. 22, 100916. doi: 10.1016/j.aqrep.2021.100916
Huang M., Xie J., Yu Q., Xu C., Zhou L., Qin J. G., et al. (2020a). Toxic effect of chronic nitrite exposure on growth and health in Pacific white shrimp Litopenaeus vannamei. Aquaculture 529, 735664. doi: 10.1016/j.aquaculture.2020.735664
Huo Y.-Y., Xu X.-W., Li X., Liu C., Cui H.-L., Wang C.-S., et al. (2011). Ruegeria marina sp. nov., isolated from marine sediment. Int. J. Systematic EvolutionaryMicrobiology 61, 347–350. doi: 10.1099/ijs.0.022400-0
Ivanov V., Stabnikova O., Sihanonth P., Menasveta P. (2006). Aggregation of ammonia-oxidizing bacteria in microbial biofilm on oyster shell surface. World J. Microbiol. Biotechnol. 22, 807–812. doi: 10.1007/s11274-005-9107-z
Jones A., Preston N., Dennison W. (2002). The efficiency and condition of oysters and macroalgae used as biological filters of shrimp pond effluent. Aquaculture Res. 33, 1–19. doi: 10.1046/j.1355-557X.2001.00637.x
Levipan H. A., Tapia-Cammas D., Molina V., Irgang R., Toranzo A. E., Magariños B., et al. (2019). Biofilm development and cell viability: an undervalued mechanism in the persistence of the fish pathogen Tenacibaculum maritimum. Aquaculture 511, 734267. doi: 10.1016/j.aquaculture.2019.734267
Li Z., Chang T., Han F., Fan X., Liu W., Wu P., et al. (2024). Effects of myo-inositol on growth and biomarkers of environmental stress and metabolic regulation in Pacific white shrimp (Litopenaeus vannamei) reared at low salinity. Comp. Biochem. Physiol. Part D: Genomics Proteomics 50, 101216. doi: 10.1016/j.cbd.2024.101216
Liao I. C., Chien Y.-H. (2011). The pacific white shrimp, Litopenaeus vannamei, in Asia: The world’s most widely cultured alien crustacean. Alien Mar. Crustaceans: Distribution Biology Impacts. 6, 489–819.
Lobus N. V. (2022). Biogeochemical role of algae in aquatic ecosystems: Basic research and applied biotechnology (MDPI).
Lu M., Ouyang T., Ye Z., Liu Y. (2010). “Raw oyster shells as carrier for nitrifying biofilm in aerated biofilter,” in 2010 4th International Conference on Bioinformatics and Biomedical Engineering. (IEEE), 1–4.
Mahenthiralingam E., Urban T. A., Goldberg J. B. (2005). The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 3, 144–156. doi: 10.1038/nrmicro1085
Mctavish H., Fuchs J. A., Hooper A. B. (1993). Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J. bacteriology 175, 2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993
Monsalve E. R., Quiroga E. (2022). Farmed shrimp aquaculture in coastal wetlands of Latin America—A review of environmental issues. Mar. pollut. Bull. 183, 113956. doi: 10.1016/j.marpolbul.2022.113956
Nathanailides C., Kolygas M., Tsoumani M., Gouva E., Mavraganis T., Karayanni H. (2023). Addressing phosphorus waste in open flow freshwater fish farms: Challenges and solutions. Fishes 8, 442. doi: 10.3390/fishes8090442
Palomo A., Dechesne A., Pedersen A. G., Smets B. F. (2022). Genomic profiling of Nitrospira species reveals ecological success of comammox Nitrospira. Microbiome 10, 204. doi: 10.1186/s40168-022-01411-y
Patil M. P., Lee D. I., Hwang U.-G., Joo Y. S., Kim K. (2024). Application of oyster shells in the remediation of marine sediment. J. Soils Sediments 24, 1030–1038. doi: 10.1007/s11368-023-03674-w
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Sedlacek C. J., Mcgowan B., Suwa Y., Sayavedra-Soto L., Laanbroek H. J., Stein L. Y., et al. (2019). A physiological and genomic comparison of Nitrosomonas cluster 6a and 7 ammonia-oxidizing bacteria. Microbial Ecol. 78, 985–994. doi: 10.1007/s00248-019-01378-8
Shao Y.-H., Wu J.-H., Chen H.-W. (2024). Comammox Nitrospira cooperate with anammox bacteria in a partial nitritation–anammox membrane bioreactor treating low-strength ammonium wastewater at high loadings. Water Res. 257, 121698. doi: 10.1016/j.watres.2024.121698
Shen Yuchun S. Y., Xiong Bangxi X. B., Wang Hui W. H., Ye Fuliang Y. F. (2007). A case study on optimal culture structure of prawn-fish-shellfish-algae. Acta Hydrobiologica Sinica. 31, 38.
Shimoda T., Fujioka Y., Srithong C., Aryuthaka C. (2005). Phosphorus budget in shrimp aquaculture pond with mangrove enclosure and aquaculture performance. Fisheries Sci. 71, 1249–1255. doi: 10.1111/j.1444-2906.2005.01090.x
Simbeye D. S., Yang S. F. (2014). Water quality monitoring and control for aquaculture based on wireless sensor networks. J. Networks 9, 840. doi: 10.4304/jnw.9.4.840-849
Supriatna S., Marsoedi M., Hariati A., Mahmudi M. (2017). Dissolved oxygen models in intensive culture of whiteleg shrimp, Litopenaeus vannamei, in East Java, Indonesia.
Tomasso J. (2012). Environmental nitrite and aquaculture: a perspective. Aquaculture Int. 20, 1107–1116. doi: 10.1007/s10499-012-9532-6
Valencia-Castañeda G., Frías-Espericueta M. G., Vanegas-Pérez R. C., Pérez-Ramírez J. A., Chávez-Sánchez M. C., Páez-Osuna F. (2018). Acute toxicity of ammonia, nitrite and nitrate to shrimp Litopenaeus vannamei postlarvae in low-salinity water. Bull. Environ. contamination Toxicol. 101, 229–234. doi: 10.1007/s00128-018-2355-z
Van Kessel M. A., Speth D. R., Albertsen M., Nielsen P. H., Op Den Camp H. J., Kartal B., et al. (2015). Complete nitrification by a single microorganism. Nature 528, 555–559. doi: 10.1038/nature16459
Vijayan A., Vattiringal Jayadradhan R. K., Pillai D., Prasannan Geetha P., Joseph V., Isaac Sarojini B. S. (2021). Nitrospira as versatile nitrifiers: Taxonomy, ecophysiology, genome characteristics, growth, and metabolic diversity. J. Basic Microbiol. 61, 88–109. doi: 10.1002/jobm.202000485
Vijuksungsith P., Satapanajaru T., Chokejaroenrat C., Jarusutthirak C., Sakulthaew C., Kambhu A., et al. (2023). Removal and reuse of phosphorus from aquaculture water using activated carbon-based CaO2 nanoparticles. Environ. Technol. Innovation 29, 102990. doi: 10.1016/j.eti.2022.102990
Villarreal H., Juarez L. (2022). Super-intensive shrimp culture: Analysis and future challenges. J. World Aquaculture Soc. 53. doi: 10.1111/jwas.12929
Wan J., Tokunaga T. K., Tsang C. F. (1995). Bacterial sedimentation through a porous medium. Water Resour. Res. 31, 1627–1636. doi: 10.1029/95WR01255
Yin Q.-J., Zhu F.-C., Tang H.-Z., Chen X.-Y., Liu X., Tang L.-C., et al. (2023). Complete genome sequence of marine Roseobacter lineage member Ruegeria sp. YS9 with five plasmids isolated from red algae. Mar. Genomics 67, 100997. doi: 10.1016/j.margen.2022.100997
Zeng S., He J., Huang Z. (2024). The intestine microbiota of shrimp and its impact on cultivation. Appl. Microbiol. Biotechnol. 108, 1–8. doi: 10.1007/s00253-024-13213-3
Keywords: Litopenaeus vannamei, oyster shells, biofilm, microbial community, water quality
Citation: Huang X, Xiao G, Zhang X, Teng S, Li M, Cai Y, Chen R and Huang X (2024) Effects of oyster shell addition on shrimp aquaculture and the dynamic succession of surface biofilm microbial communities. Front. Mar. Sci. 11:1495938. doi: 10.3389/fmars.2024.1495938
Received: 13 September 2024; Accepted: 08 October 2024;
Published: 20 November 2024.
Edited by:
Antonio Luna-González, National Polytechnic Institute (IPN), MexicoReviewed by:
Fawen Hu, Marine Biology Institute of Shandong Province, ChinaCarlos Angulo, Northwestern Center of Biological Research, S.C. (CIBNOR), Mexico
Copyright © 2024 Huang, Xiao, Zhang, Teng, Li, Cai, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaolin Huang, eGlhb2xpbm5saEBob3RtYWlsLmNvbQ==
 Xianke Huang
Xianke Huang Guoqiang Xiao2,3,4
Guoqiang Xiao2,3,4