- 1Aquatic Ecosystem Research, Faculty of Biology, University of Duisburg-Essen, Essen, Germany
- 2HYIG ARJEL, Benthic Ecology, Alfred Wegener Institute Helmholtz Center for Polar and Marine Research, Bremerhaven, Germany
- 3FB2, BreMarE – Bremen Marine Ecology, University of Bremen, Bremen, Germany
- 4British Antarctic Survey, NERC, High Cross, Madingley Road, Cambridge, United Kingdom
Introduction: In the Southern Ocean, myctophids are the most successful pelagic fish group in terms of diversity, biomass, and abundance. They play a crucial role in linking primary consumers and coupling carbon flux between surface and mesopelagic depths. Understanding their trophodynamics is key to assessing pelagic ecosystem resilience under environmental change. Conventional stomach content analyses indicate that myctophids predominantly feed on crustaceans, such as copepods and euphausiids, but are less effective at detecting easily digestible, soft-bodied organisms like gelatinous zooplankton (GZP) and pteropods.
Methods: This study used multimarker (COI Leray-XT and 18S v1-v2) DNA metabarcoding to analyze the diets of two abundant myctophids in the Scotia Sea, Electrona antarctica and Electrona carlsbergi.
Results and discussion: We found a diverse diet dominated by copepods and euphausiids, followed by pteropods and GZP in terms of frequency of occurrence and relative read abundances. Within the GZP, salps and appendicularians were major components of the diet for E. carlsbergi, while hydrozoans were prominent in E. antarctica. With regards seasonal and spatial variability in the dietary contribution of GZP, E. carlsbergi consumed primarily appendicularians, chaetognaths, and hydrozoans during spring (2006) in the northern region, and more salps (predominantly Ilhea racovitzai) and other cnidarians in autumn (2009) in the southern region. Our study reveals the dietary diversity of myctophid fish as well as the importance GZP consumption as a key trophic pathway in the Southern Ocean.
1 Introduction
Myctophids (Family Myctophidae) are among the most diverse and abundant mesopelagic fishes and are globally distributed (Catul et al., 2011). They occupy an intermediate trophic position in most pelagic food webs, feeding on zooplankton, such as copepods and euphausiids, and being preyed upon by sea birds, marine mammals, and large pelagic fish (Cherel et al., 2010). By migrating vertically across the water column, myctophids transport carbon from the sea surface to deeper ocean layers (Collins et al., 2012; Saunders et al., 2014; Belcher et al., 2019; Saba et al., 2021), playing a vital role in energy transfer within the pelagic ecosystem. In the Southern Ocean, myctophids are the most successful pelagic fish group in terms of diversity and abundance, with around 35 species in 12 genera and an estimated biomass that may exceed 200 million tonnes (Hulley, 1981; Dornan et al., 2022). Myctophids can form a significant part of the diet of many higher predators, particularly in regions and seasons when Antarctic krill is scarce (Barrera-Oro, 2002; Davis et al., 2006; Murphy et al., 2007).
The Scotia Sea, located in the Atlantic sector, stands out as one of the most productive regions of the Southern Ocean (Atkinson et al., 2001; Whitehouse et al., 2012). It is also one of the regions where ocean temperatures have risen significantly in recent decades, and winter sea-ice extent has decreased substantially (Curran et al., 2003). In this region, Electrona antarctica and Electrona carlsbergi are two of the most abundant myctophids (Collins et al., 2008; Collins et al., 2012; Saunders et al., 2014). Electrona antarctica is the most numerically abundant species south of the Antarctic Polar Front, with highest abundances in the sea-ice covered sectors. Electrona carlsbergi is more common in more northern regions, either side of the Polar Front (Saunders et al., 2014). The feeding ecology of these two myctophids has been described using optical techniques (Shreeve et al., 2009; Saunders et al., 2014; Saunders et al., 2019; Duan et al., 2024), fatty acids and stable isotopes (Stowasser et al., 2009; Papadimitraki et al., 2023). The overall consensus is that these similar sized species (~90-100 mm standard length - SL) are opportunistic feeders with relatively broad diets, consuming mostly amphipods, large copepods and small euphausiids. However, the available data also suggests dietary differences between the two species. For E. antarctica, amphipods such as Themisto gaudichaudii and small euphausiids (Thysanoessa spp.) are the main prey groups, whilst copepods (e.g., Rhincalanus gigas and Metridia spp.) dominate the diet of E. carlsbergi. Unlike E. antarctica, salps are also an important prey item for E. carlsbergi, whilst Antarctic krill (Euphausia superba) appears rare in its diet (Shreeve et al., 2009; Stowasser et al., 2009; Saunders et al., 2014; Saunders et al., 2019; Papadimitraki et al., 2023). However, the diets of both E. antarctica and E. carlsbergi may vary spatially, seasonally and ontogenetically. For example, E. antarctica appears to consume more copepods in the northern regions of the Scotia Sea and during the spring season, while predation on euphausiids is more predominant in the southern regions during autumn. For E. carlsbergi, regional dietary patterns are less clear due to spatially confined data, but seasonal switches in copepod prey have been reported, with greater proportions of R. gigas consumed in spring than in other seasons (Saunders et al., 2014). Additionally, as the size of the myctophid predators increases, there is a general dietary shift from copepods to larger sized prey such as euphausiids and amphipods (Shreeve et al., 2009; Saunders et al., 2014; Clarke et al., 2020; Riaz et al., 2020).
The contribution of gelatinous zooplankton (GZP), including ctenophores, cnidarian medusae, chaetognaths and pelagic tunicates (e.g. salps – order: Salpida and appendicularians – order: Copelata) is poorly resolved in conventional diet studies of E. antarctica and E. carlsbergi (Shreeve et al., 2009; Saunders et al., 2014; Duan et al., 2024). This is because their watery and soft tissues are rapidly digested, so they are mainly undetected in predators’ stomachs, particularly when using conventional microscopy (Amundsen and Sánchez-Hernández, 2019; Brodeur et al., 2021). In recent years, the application of new genetic-based techniques such as DNA metabarcoding have caused a paradigm shift in our understanding of the role of GZP in marine food webs. Gelatinous zooplankton are no longer considered a trophic dead end, but a common, or even important, part of the diet of various polar fish and arthropods (McInnes et al., 2017; Dischereit et al., 2022; Urban et al., 2022; Ruiz et al., 2024). However, such methods have seldom been applied to Southern Ocean mesopelagic fish and the role of these potentially important soft-bodied organisms in myctophid trophodynamics and the Southern Ocean food web remains virtually unknown. To date, metabarcoding has only been used to study the diet composition of E. antarctica, detecting cnidarians (Coronatae) and salps, among other GZP taxa, based on relatively small sample sizes (Clarke et al., 2020; Vasiliadis et al., 2023). Furthermore, these studies found greater numbers of other previously overlooked prey species, including rare, small, damaged and partially digested organisms such as gastropods. Wider extension of these methods in this group therefore has the potential to cast new light on Southern Ocean food web dynamics.
The requirement for this research focus is becoming increasingly apparent in light of continued evidence of broad-scale environmental change in the Southern Ocean, which may alter local food web structure and ecosystem function (Atkinson et al., 2004; Moline et al., 2004; Bernard et al., 2012; Loeb and Santora, 2012; Fuentes et al., 2016; Atkinson et al., 2019; Yang et al., 2022). At present, there are major uncertainties on how such change may be manifested in the region across all trophic levels. Myctophids have been identified as potentially key for sustaining globally important predator populations and maintaining ecosystem stability in the short to medium term, as local food webs restructure under future scenarios of environmental change (Murphy et al., 2007; Saunders et al., 2015). Yet their resilience to such change and the extent to which they can buffer the local ecosystem in the long term remains uncertain, necessitating the need for continued research on myctophid trophodynamics, particularly regarding understudied trophic pathways. In an ocean warming context, Southern Ocean GZP are emerging as climate change winners, particularly salps (e.g. Salpa thompsoni) which are a major group that may substantially increase in biomass and expand their range poleward under realistic scenarios of future change (Suprenand and Ainsworth, 2017; Rogers et al., 2020; Johnston et al., 2022). Therefore, there is a pressing need to understand the role of such GZP in myctophid trophodynamics and the wider Southern Ocean food web to assess their resilience to future change.
In this paper, we examine the dietary prey composition and the extent of GZP predation in two dominant myctophid species of the Southern Ocean, Electrona antarctica and Electrona carlsbergi, using a multimarker (COI Leray-XT and 18S v1-v2 rRNA) metabarcoding approach. This method allows a novel and comprehensive examination of their potential prey spectrum, complementing conventional identification techniques and improving taxonomic resolution, by providing a semiquantitative assessment of the importance of GZP in the diet of these dominant myctophid species. We discuss these results in the context of how GZP consumption may provide resilience to changes in prey fields resulting from climate change.
In view of the existing evidence outlined above, we test the following hypotheses:
1. Salps and other GZP constitute a crucial component of the diet for E. carlsbergi, whereas they do not significantly contribute to the diet of E. antarctica.
2. Predation on salps occurs more frequently in the Antarctic Polar Front region compared to the southern areas of the Scotia Sea.
2 Methods
2.1 Sampling and specimens’ measurements
Myctophid fish (Family Myctophidae) were collected from the Scotia Sea (southwest Atlantic) during three research cruises onboard RRS James Clark Ross in austral spring (cruise: JR161; October–December 2006), summer (cruise: JR177; January–February 2008), and autumn 2009 (cruise: JR200; March–April 2009) (Figure 1). During all surveys, net hauls were conducted at regular stations along a transect from the sea ice zone to the Antarctic Polar Front. Specimens of Electrona antarctica and E. carlsbergi (among other species) were collected primarily by a depth-stratified 25 m2 rectangular mid-water trawl (RMT25) deployed between 0-200, 200-400, 400-700 and 700-1000 m. On occasion, specimens were also collected from the upper 200 m using an 8 m2 rectangular midwater trawl (RMT8). All nets were towed at ~2.5 knots with deployments being carried out both day and night (see Collins et al. (2012) for further details on net sampling). For each fish specimen, we recorded the standard length to the nearest 0.1 cm and their weight in g. All specimens were frozen at -20°C for subsequent processing in the laboratory.
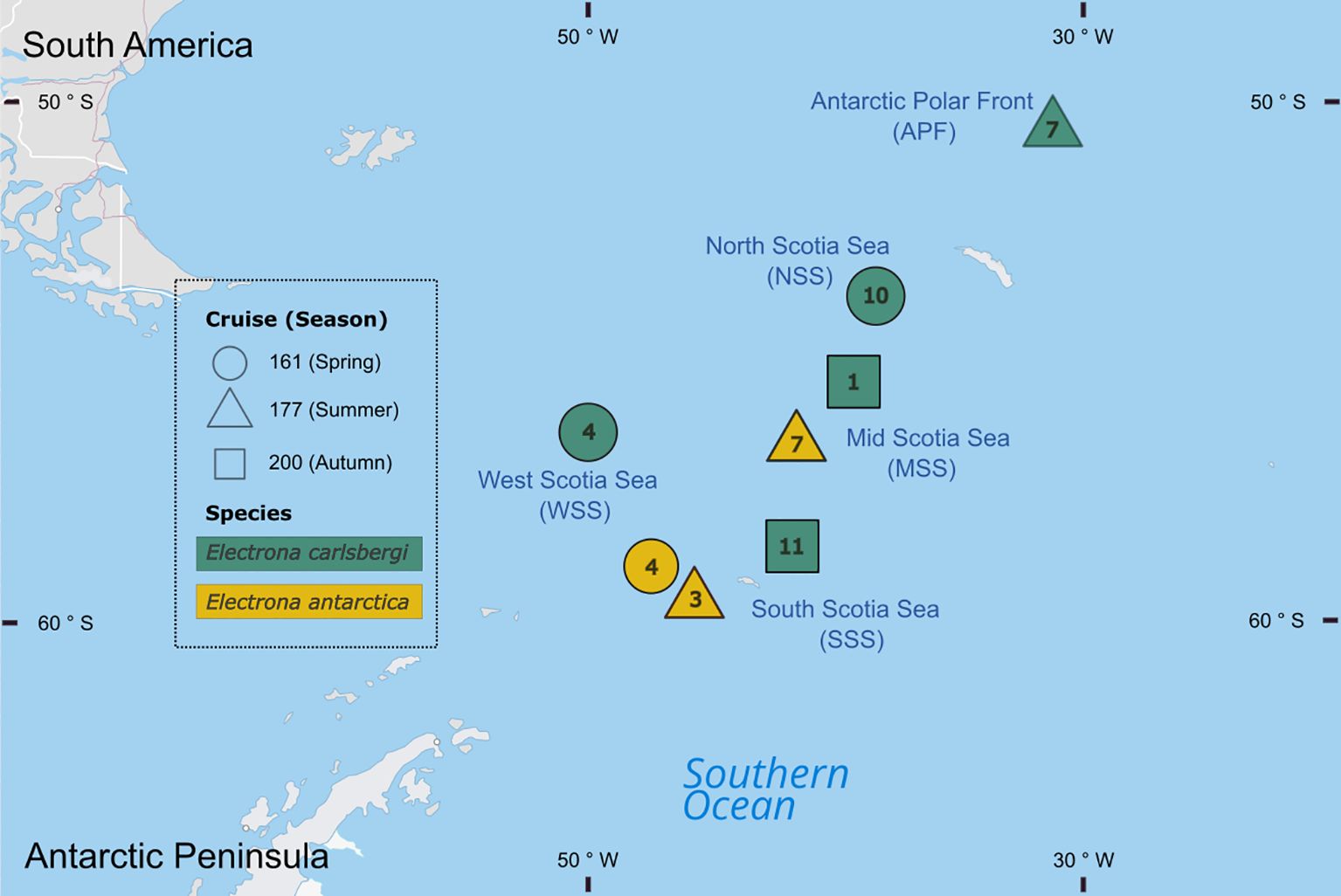
Figure 1. Map of samples location. Shape indicates cruise and season (JR161: spring 2006, JR177: summer 2008, and JR200: autumn 2009), colour indicates species and the numbers inside the symbols indicate the number of samples used in the analysis.
2.2 DNA extraction from stomach contents
All stomachs were dissected, weighed (with 0.01 g precision) and stored at -20°C at the laboratory until further processing. Stomachs were thawed and carefully opened to extract contents whilst avoiding contact with the stomach walls. A blender and grinding tools were then used for the homogenization of contents retrieved from each individual stomach. All lab instruments were cleaned with 10% bleach, water and 70% ethanol, before processing each stomach. After homogenization, DNA extraction was performed using the Qiagen Blood and Tissue extraction kit following the manufacturer’s protocol, using a final elution volume of 100 ul. The complete stomach content was used for the extraction and ranged from 6 to 75 mg. For every round of 24 DNA extractions, a blank extraction control was performed, treating the empty tube as the rest of the samples. DNA quantity and quality were assessed using a Nanodrop ND-1000 (Thermo Fisher Scientific) spectrophotometer. Only extracts with a 260/280 ratio of >1.5 and a concentration >10 ng/ul were used for the following sequencing steps, which resulted in 18 samples of E. antarctica and 33 of E. carlsbergi.
2.3 Library preparation and sequencing
DNA metabarcoding library preparation and sequencing of the “Leray-XT”- fragment of the COI mitochondrial gene (Geller et al., 2013; Wangensteen et al., 2018) and the fragment of the 18S (v1-v2 region) gene (Blaxter et al., 1998; Sinniger et al., 2016) were carried out by AllGenetics & Biology SL (www.allgenetics.eu) following the same protocol detailed in Ruiz et al. (2024). Finished libraries were pooled in equimolar amounts according to the results of a Qubit dsDNA HS Assay (Thermo Fisher Scientific) quantification. Finally, the pool was sequenced in a fraction of an Illumina NovaSeq (2 × 250 bp paired-ends), adding 5% PhiX to the sequencing run, aiming for a total output of 24 Gbp.
2.4 Bioinformatics analysis
Bioinformatic analysis of the COI sequencing data was performed following the MJOLNIR v3.0.0 pipeline (Metabarcoding Joining Obitools and Linkage Networks In R; https://github.com/adriantich/MJOLNIR3), using the package mjolnir v3.0.0 (Dischereit et al., 2024) in R v4.0.4. The pipeline default settings, which were already adjusted for the COI marker gene by the provider, were retained, clustering the sequences into operational taxonomic units (OTUs) with a d value of 13. Taxonomy was assigned against the NJORDR COI v202305 reference set (https://github.com/adriantich/NJORDR-MJOLNIR3). BOLDigger (Buchner and Leese, 2020) and local database blasting and alignment were run to doublecheck taxonomic annotations and define thresholds. Assignments with less than 83% similarity were excluded, 97% similarity threshold was used to define species level, >96% to genus, >93% to family, >90% to order and >85% class level (Wangensteen and Turon, 2016; Macher et al., 2018). Bioinformatic analysis of the 18S v1-v2 sequencing data was performed by applying Cudadapt v.2.8 (Martin, 2011) to remove all primers and leftover adapters. By applying functions of the R package “DADA2” (version 1.18.0) (Callahan et al., 2016), sequence trimming and filtering, sequence denoising was carried out according to the Divisive Amplicon Denoising Algorithm (DADA2). This was followed by paired-end merging, chimera detection-removal and amplicon sequence variant (ASV) annotation against the MetaZooGene (O’Brien et al., 2024) database (https://metazoogene.org/mzgdb/, version MZG_v2023-m07-15). Taxonomic assignments with less than 80% bootstrap support were excluded and a threshold of >98% was used for family-level assignments. Assignments at the genus or species level were made only when COI assignments were consistent with 18S v1-v2 results and when BLASTn searches using the NCBI database returned the same assignment, based on query coverage and percentage identity. After these steps, we carried out a manual correction of taxonomic annotations using the World Register of Marine Species (WoRMS; https://www.marinespecies.org) for both datasets. A final refinement of both datasets was done following the same steps as described in Ruiz et al. (2024), this included a removal of every OTU/ASV for which the abundance in the blank or negative controls was higher than 10% of the total reads of that OTU/ASV; and a removal of non-metazoans and contaminants (predator and human DNA). The final dataset comprised, for E. antarctica, 13 samples for COI and 10 for 18S-v1-2; and for E. carlsbergi 32 samples for COI and 33 for 18S v1-v2.
2.5 Statistical analysis
The data obtained for both markers were analyzed using the R vegan package [R version 4.2.3 (Oksanen et al., 2019)]. Rarefaction curves were run using the function “rarecurve” to check sequencing depth coverage. Graphs and explorative analysis were performed using the TaxonTableTools software (Macher et al., 2021) and ggplot2 package in R (Wickham and Wickham, 2016). We used relative read abundance (number of reads of one prey group divided by the number of total reads in all prey groups, RRA%) and frequency of occurrence (number of samples that contained a given prey item divided by the total number of samples, FOO%) as metrics for analyses and tables.
Multivariate statistical analyses were undertaken using the Phyloseq package (version 1.42.0) and vegan (Oksanen et al., 2019) in R (version 4.2.3). In this process, Principal Coordinate Analysis (PCoA) plots were performed to provide visual checks on dietary patterns in a two-dimensional plane according to their relevant diet similarity. All data were expressed as relative abundances and transformed into a dissimilarity matrix based on the Bray-Curtis distances for each species and marker. Pearson correlations between individual OTUs/ASVs and the PCoA axes were calculated to generate overlay vectors for the PCoA plots, using the “envfit” function. To enhance visualization, OTUs and ASVs were replaced in the plots by their lowest-rank taxonomic assignments. To test the effects of sampling season (spring-2006, summer-2008 and autumn-2009), sampling region (south Scotia Sea-SSS, west Scotia Sea-WSS, north Scotia Sea-NSS, mid Scotia Sea-MSS and the Antarctic polar front-APF; Figure 1) and size on diet composition within species, a PERMANOVA analysis was performed using the “adonis2” vegan function (Oksanen et al., 2019). Each PERMANOVA was run separately with 9999 permutations. Categories for the numerical factor “fish size” were arbitrarily defined as follows: <75 mm = small (S); 75-80 mm = medium (M); 80-99 mm = large (L); for E. antarctica L=8 samples and S=5, for E. carlsbergi L=11, M=14 and S=8. Finally, pairwise multiple comparisons were performed using the “pairwise.adonis” function in the PairwiseAdonis package (Martinez Arbizu, 2020).
3 Results
3.1 Multimarker metabarcoding output
The sequencing runs produced 12,894,808 paired-end raw reads for the COI-library and 6,686,334 paired-end raw reads for the 18S v1-v2 -library. After quality filtering, the final dataset consisted of 4,872,790 reads for COI and 5,466,495 reads for 18S v1-v2. The negative controls, including DNA extraction and PCR controls, showed for COI 50 reads (0.0003% of total reads) clustered into 14 OTUs, assigned to Arthropoda, Chordata, Ctenophora and Platyhelminthes. For 18S v1-v2 negative controls contained 1,120 reads (0.016% of total reads), comprising 30 ASVs assigned to Arthropoda, Chordata, Cnidaria, Platyhelminthes, Annelida, Chaetognatha, Ctenophora and Ochrophyta. Rarefaction curves reached the asymptote for all samples (Supplementary Figure S1), suggesting that sequencing depth was enough to recover the vast majority of biodiversity within each sample. The number of reads per sample, number of OTUs and ASVs and other details before and after curation are shown in Supplementary Table S6.
In general, for both markers and species, the most represented phylum in terms of relative reads abundance (RRA%) and frequency of occurrence (FOO%) was Arthropoda comprising more than 87.54 RRA% and 100 FOO% for COI, for 18S v1-v2 represented 78.72 RRA% and 100 FOO%. The orders Calanoida and Euphausiacea were the most abundant groups at this level. For COI results, the second most abundant and frequently ingested prey items were assigned to the phylum Cnidaria, reaching 4.37 RRA% and 73 FOO%, followed by Mollusca with 1.91 RRA% and 55 FOO%. However, for 18S v1-v2 results, the second most represented group in the 18S v1-v2 results was the phylum Chordata, showing 12.77 RRA% and 81 FOO%, followed by Mollusca with 4.68 RRA% and 56 FOO%. The metazoan prey list detected using the COI and 18S v1-v2 sequences are summarized in Supplementary Tables 1–4, with all the taxa found in the stomach contents of E. antarctica and E. carlsbergi.
3.2 Diet composition of Electrona antarctica
The analysis of prey items in the stomach contents of E. antarctica using COI and 18S v1-v2 metabarcoding sequencing revealed a wide range of taxa, with both gene fragments demonstrating similar patterns of prey importance at the phylum level. For COI, 82.9% of the reads corresponded to the phylum Arthropoda, 13.4% to Mollusca, 2.79% to Cnidaria, and 0.45% to Chordata (comprising mainly Aulopiformes fish). In the 18S v1-v2 analysis, 73.2% of the reads were assigned to Arthropoda, 23.7% to Mollusca, and 2.5% to Cnidaria, with the remaining phyla each representing less than 0.3% of the reads.
Based on the relative read abundances (RRA%s) and frequency of occurrence (FOO%) of prey items, both COI and 18S v1-v2 markers indicated that the most represented prey group was Euphausiacea at the order level, with Antarctic krill being the most abundant species (Table 1). This was followed by Copepoda (primarily calanoids) and Pteropoda. Notably, even at this resolution, both markers yielded similar patterns of prey importance (Figure 2). Table 1 lists the top prey items for COI and 18S v1-v2, demonstrating that, whilst the taxonomic resolution of the COI marker is superior to that of 18S v1-v2, both markers showed similar patterns of prey composition. In terms of GZP, the class Hydrozoa was ranked highly for COI, whilst scyphozoans (order Coronatae) were among the main prey items for 18S v1-v2 (Table 1; Supplementary Table S7).
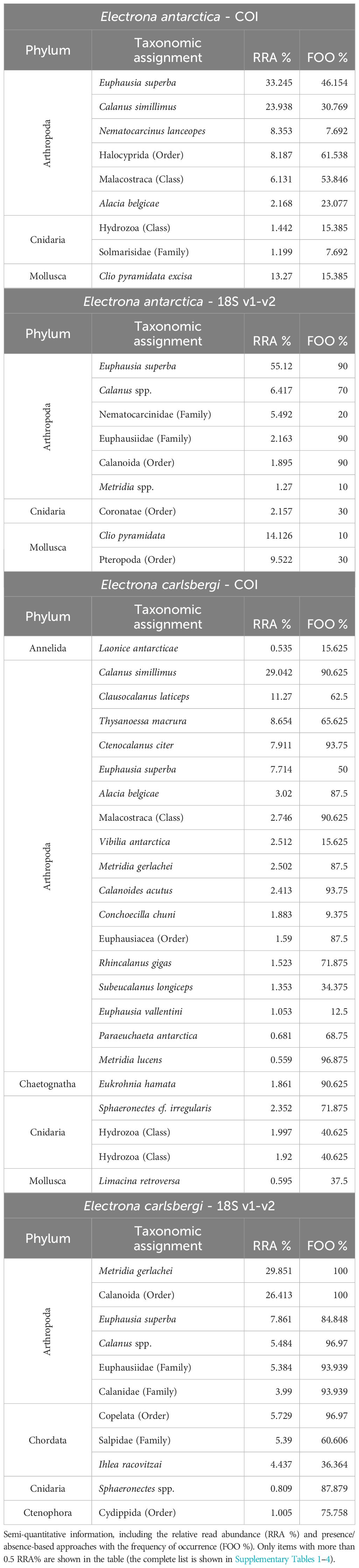
Table 1. Top prey items for E. antarctica and E. carlsbergi that showed the higher number of reads and its taxonomic assignments.
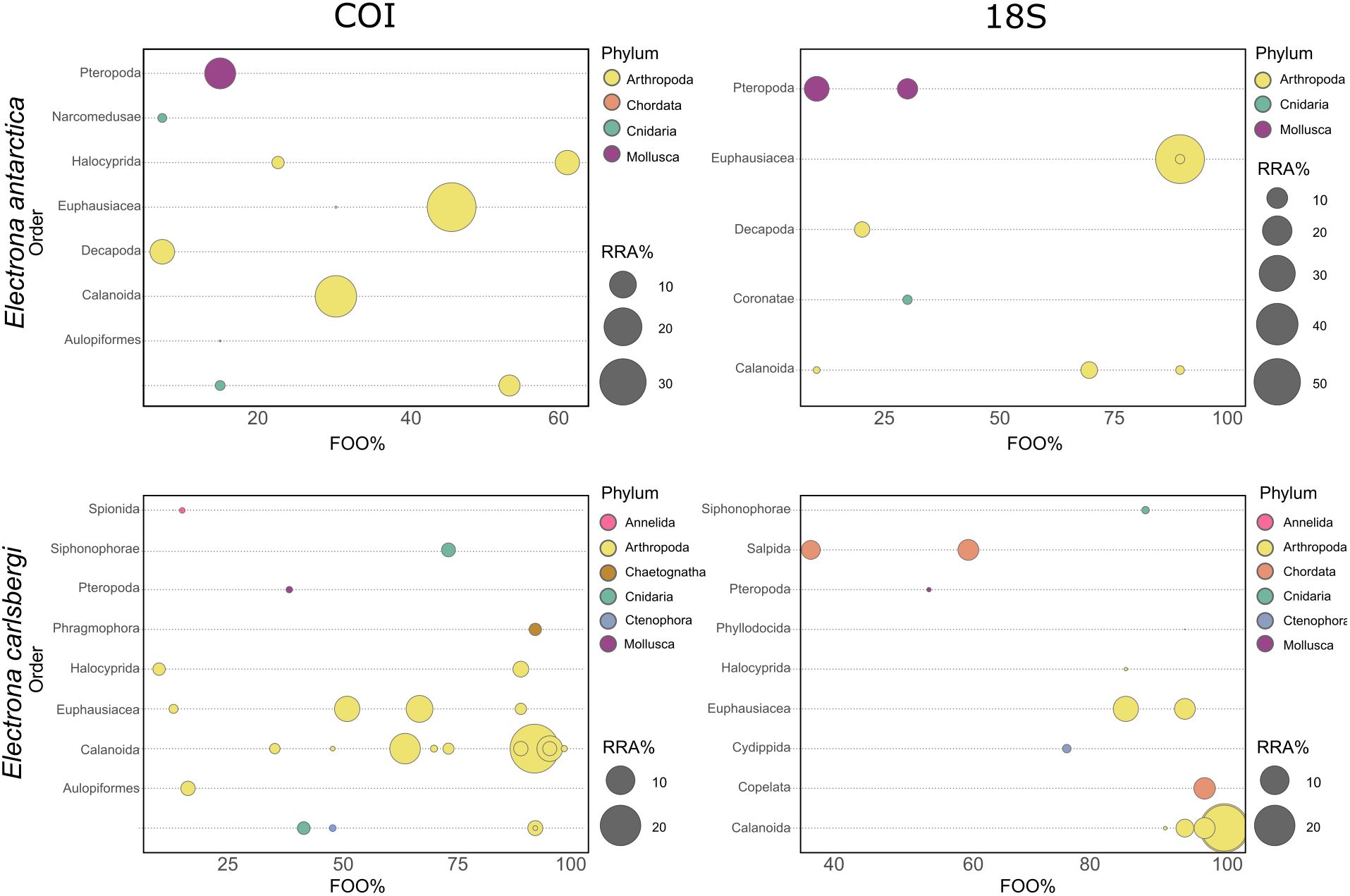
Figure 2. Bubble plot showing the dominant prey orders by E. antarctica (upper panels) and E. carlsbergi (lower panels). Values corresponding to COI results are shown on the left and 18S v1-v2 results on the right. FOO% is the frequency of occurrence of the prey orders. Orders on the y-axis are those most represented OTUs and ASVs (>0.2 RRA%). The size of the bubble corresponds to the relative read abundances (RRA%), with bubbles colored according to phylum (see legend).
3.3 Multivariate analysis of 18S v1-v2 and COI metabarcoding of Electrona antarctica stomach contents
Our multivariate analyses of COI dietary data revealed a significant effect for cruise-season (PERMANOVA; F = 2.6731, p = 0.0027) and body size (PERMANOVA; F = 2.7618, p = 0.0027). Analyses of 18S v1-v2 data found no significant dietary effects for the available covariate factors (Supplementary Table S8; Figure 3).
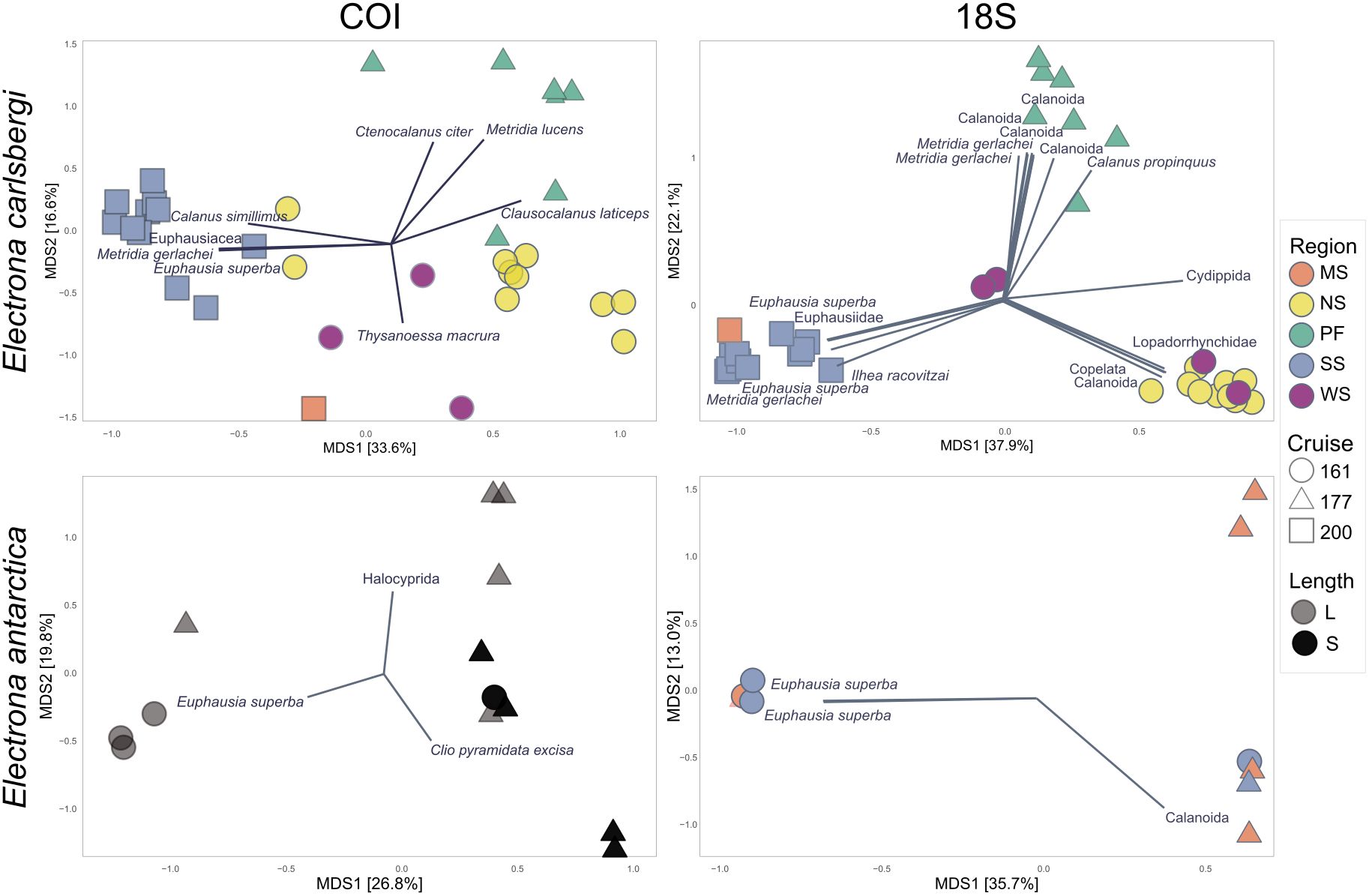
Figure 3. Principal Coordinate Analysis (PCoA) for Electrona carlsbergi and Elecrona antarctica diet composition; Left: COI ordination, right: 18S v1-v2 ordination. Analysis for E. carlsbergi was run with 32 samples and 146 OTUs for COI, and 33 samples and 1077 ASVs for 18S v1-v2. Analysis for E. antarctica was run with 13 samples and 47 OTUs for COI, and 10 samples and 528 ASVs for 18S v1-v2. Data were transformed into proportions and the distance matrix was calculated using Bray-Curtis. Shapes indicate season (JR161: spring 2006, JR177: summer 2008, and JR200: autumn 2009 and colours indicate sampling region. Vectors were only drawn when Pearson correlations for E. carlsbergi showed p<0.001 and for E. antarctica showed p<0.05.
Ordination of COI data showed a cluster of samples collected in spring-2006 in the SSS region and comprising only large myctophids that preyed mainly on Antarctic krill (Supplementary Figure S10). The second cluster, including small myctophids collected in summer-2008, consisted almost exclusively of those preying on the mollusc Clio pyramidata excise (Supplementary Figure S11). A third group comprised large myctophids collected in summer-2008 (mid water temperature 0.5-1°C) in at the MSS, with a diet dominated by the order Halocyprida (small crustaceans) (Supplementary Figure S12).
3.4 Diet composition of Electrona carlsbergi
The range of prey items detected in the stomachs of E. carlsbergi using both COI and 18S v1-v2 metabarcoding sequencing was diverse, with similar patterns of dietary prey rankings between the two markers, except for salps and appendicularians, which were important only in the 18S v1-v2 analysis (Table 1). For COI, 88% of the reads corresponded to the phylum Arthropoda, 6.46% to Cnidaria, 1.98% to Chaetognatha, 1.29% to Annelida, 0.7% to Mollusca, and 0.66% to Ctenophora. In the 18S v1-v2 analysis, 80% of the reads corresponded to Arthropoda, 15.6% to Chordata (mainly salps and appendicularians), 1.2% to Ctenophora, 0.86% to Cnidaria, 0.43% to Mollusca, and 0.32% to Annelida. The remaining phyla each represented less than 0.3% of the reads.
At the order level, both markers indicated that the most represented prey group was Copepoda (based on RRA% and the FOO%), primarily calanoid species. This was followed by Euphausiacea for both markers (Figure 2). While the taxonomic resolution of the COI marker was found to be superior to that of 18S v1-v2, there were still congruent taxonomic assignments between both datasets (Table 1). In terms of GZP, the siphonophore Sphaeronectes cf. irregularis was among the top ten most represented OTUs for COI (Supplementary Table S7), whilst 18S v1-v2, appendicularians and salps (at family level and at species level for Ihlea racovitzai) were among the top ten ASVs for 18S v1-v2 (Supplementary Table S7). However, while the phylum Chaetognatha was well represented in the COI data, it was less so in the 18S v1-v2 data. Conversely, the salps and appendicularians were prominent in the 18S v1-v2 results but nearly absent in the COI output. Additionally, Amphipoda appeared as one of the most represented arthropod orders in the COI data, whereas this was not the case in the 18S v1-v2 data. Finally, within the phylum Annelida, the order Spionida was well-represented in the COI results, while the order Phyllodocida was the most abundant in the 18S v1-v2 results.
3.5 Multivariate analysis of 18S v1-v2 and COI metabarcoding of Electrona carlsbergi stomach contents
Multivariate analyses on E. carlsbergi diet composition (Figure 3) showed a significant effect of season-cruise (PERMANOVA; F = 12.3833, p = 0.0001) and region (PERMANOVA; F = 4.258, p = 0.0001) for COI (Supplementary Table S9). Further pairwise comparisons indicated that all seasons-cruises were significantly different from each other (“pairwise.adonis”, p < 0.05; Supplementary Table S9). Regarding the regional pairwise comparison, the SSS was significantly different to APF, NSS and WSS. The Polar Front was also different to the NSS, as was the NSS to the WSS (“pairwise.adonis”, p < 0.05; Supplementary Table S9). Likewise, a significant effect of season-cruise (PERMANOVA; F = 12.3833, p = 0.0001) and region (PERMANOVA; F = 4.258, p = 0.0001) was found for the 18S v1-v2 data (Supplementary Table S9). Again, further pairwise comparisons showed that all season-cruises were significantly different from each other (“pairwise.adonis”, p < 0.05; Supplementary Table S9). Regarding region, pairwise comparison showed that SS was significantly different to APF, NSS and WSS; the APF was different to the NSS and the WSS (“pairwise.adonis”, p < 0.05; Supplementary Table S9). There was no significant difference in diet between myctophid sizes in our analyses.
Visual clustering of samples is displayed on the PCoA plots (Figure 3). Ordination of COI data showed a cluster of samples (blue squares) collected in autumn-2009 (low water temperature <0.5°C) and in SSS region, with these animals preying mainly upon euphausiids (Euphausia superba and Thysanoessa macrura) and copepods (Calanus simillimus and Metridia gerlachei). A second group (green triangles) comprised samples collected in summer-2008 (relatively high-water temperature 1-3°C) at the Antarctic Polar Front and only in the epipelagic zone (0-200m), which consumed mainly copepods (Clausocalanus laticeps, Ctenocalanus citer and C. simillimus). Finally, the third group (yellow circles) consisted of specimens collected in spring-2006 (mid temperatures 0.5-1°C) and in the epipelagic zone (0-200 m) of the NSS, which also preyed upon copepods, of which C. laticeps and C. simillimus were the most represented prey items, followed by the euphausiid T. macrura. Ordination of 18S v1-v2 output clustered the samples in a similar pattern. The first group (blue squares) comprised samples collected in autumn-2009 and in the SSS region, where fish consumed mainly euphausiids (E. superba), copepods (M. gerlachei), but also salps (Ihlea racovitzai). The second group (green triangles) comprised samples collected in summer-2008 at the APF, which consumed mainly copepods (Calanus sp. and M. gerlachei). The third group (yellow circles), collected in spring-2006 and the NSS except for two collected in the WSS, showed a very high number of reads assigned to copepods, Lopadorrhynchidae (Polichaeta) and appendicularians (see Supplementary Figures S13–S15).
3.6 Focus on GZP
Among the prey items found in the stomachs of E. antarctica and E. carlsbergi, GZP were detected in both species using both markers (COI and 18S v1-v2). These gelatinous prey items belonged to the phylum Chordata (salps and appendicularians), Cnidaria, Ctenophora and Chaetognatha, which were present in different read proportions depending on the marker and species. Electrona carlsbergi consumed more GZP (Figure 4; Supplementary Table S16) compared to its congener. Discrepancies between the markers in identifying the most important prey items are notable: whereas Hydrozoans were more represented in the COI output, reads assigned to salps were more abundant in the 18S v1-v2 results, while appendicularians were only assigned in 18S v1-v2 output. The complete list of taxonomic resolutions for each marker and species is available in Supplementary Tables 1, 2 for E. antarctica and Supplementary Tables 3, 4 for E. carlsbergi.
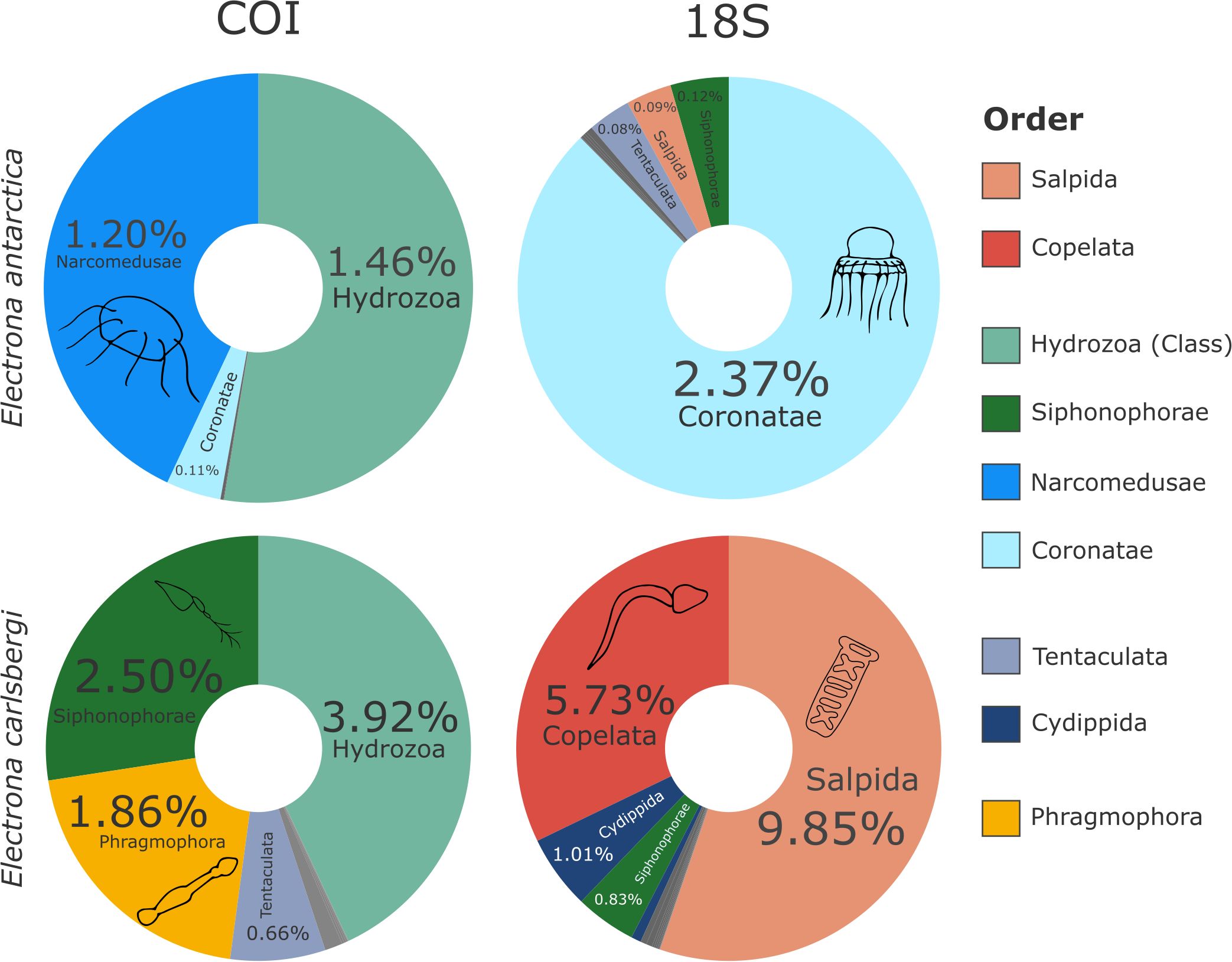
Figure 4. Relative read proportions (relative to all prey items) of GZP present in the diet of E. antarctica and E. carlsbergi for COI and 18S v1-v2 datasets. Categories are at order level, else class level where order level was not attained. Colour assignments as for Figure 2 except grey which represent other GZP showing low number of reads (for the complete taxonomic prey list see Supplementary Tables 1–4). Schematic drawings were added for the most represented orders.
4 Discussion
We present a high-resolution taxonomic analysis of the diets of two abundant mesopelagic fish species in the Scotia Sea, Electrona antarctica and Electrona carlsbergi. Utilizing a multimarker DNA metabarcoding approach, our study specifically examined the role of easily digestible prey that may have been overlooked in previous research. Our findings reveal a diverse array of prey taxa in the diets of these species, predominantly copepods and euphausiids, followed by pteropods and GZP (salps, appendicularians, and other cnidarians). This comprehensive approach revealed that GZP are common in the diets of both species. Specifically, salps and appendicularians are crucial components of the diet for E. carlsbergi, while hydrozoans play a secondary role in the diet of E. antarctica. Consumption of GZP species was higher for the animals collected during the spring (2006) and autumn (2009) compared to the ones collected in summer (2008). In particular, E. carlsbergi, showed higher read abundances of appendicularians, chaetognaths, and hydrozoans during spring (2006), and more salps and other cnidarians in autumn (2009). Additionally, our results indicate a geographical variation in predation patterns: salps (predominantly Ilhea racovitzai) are more frequently consumed in the SSS whereas appendicularians are more commonly consumed in the NSS. Although our study is limited in time and space, sampling the remote Southern Ocean across seasons in consecutive years and cruises is challenging and our data are the best available to date. We therefore maintain that our results contribute to a more comprehensive understanding of the dietary habits of these mesopelagic fish species and the ecological dynamics of the Scotia Sea.
4.1 Prey spectrum breadth
The diet composition of E. antarctica recovered using both conventional and metabarcoding methods was broadly similar, with Arthropoda (euphausiiids and copepods) being the most abundant prey groups across all studies (Shreeve et al., 2009; Stowasser et al., 2009; Saunders et al., 2014; Clarke et al., 2020; Papadimitraki et al., 2023; Vasiliadis et al., 2023; Duan et al., 2024). The differences between methods were clear, with metabarcoding revealing the importance of gastropods in the diet of E. antarctica, which also represented the second most abundant prey group in the study of Vasiliadis et al. (2023). We recovered a high number of reads corresponding to the order Pteropoda, with the species Clio pyramidata and Limacina helicina being well represented in the COI dataset. In contrast, Saunders et al. (2014). showed that pteropods were less prevalent in terms of relative mass and frequency and this prey group was unresolved at genus or species level. Additionally, we detected the GZP taxa Chaetognatha, Chordata (salps and appendicularians), Ctenophora, and Cnidaria (mostly Hydrozoa) in this species diet.
Using conventional stomach analysis methods, the diet of E. carlsbergi was previously reported to be dominated by copepods (Saunders et al., 2014), followed by euphausiids and amphipods, with salps recorded as an important prey item (Shreeve et al., 2009; Stowasser et al., 2009; Saunders et al., 2014; Saunders et al., 2019; Papadimitraki et al., 2023). Here, both metabarcoding markers indicated that the most represented group were copepods, primarily calanoid species, followed by euphausiids. Within the order Amphipoda, Vibilia antarctica ranked among the most represented OTUs (Table 1; Supplementary Table S7), along with Themisto spp., aligning with the findings of Saunders et al. (2014). and Shreeve et al. (2009). Our results also identified salps, particularly Ihlea racovitzai, as an important prey group, which is consistent with previous studies. Similarly, the order Copelata (appendicularians) was well represented across studies.
GZP were better represented in our study than previous molecular studies (Clarke et al., 2020; Vasiliadis et al., 2023), particularly the classes Tentaculata, Hydrozoa and the order Siphonophorae, which were abundant in both the COI and 18S v1-v2 outputs. The genus Sphaeronectes in particular, was preyed upon by most analyzed specimens. Also, other groups, such as Polychaeta and Ostracoda, have not been detected as a major dietary component in conventional stomach analysis studies (Shreeve et al., 2009; Saunders et al., 2014; Duan et al., 2024), but were often present in the diet of E. carlsbergi in this study. Specifically, in terms of COI results, seven species of the ostracod Order Halocyprida, were found in E. carlsbergi, while three species were found in E. antarctica.
In accordance with previous studies, large copepods such as Rhincalanus gigas and Metridia spp. were the predominant copepods consumed by the two Electrona species (Shreeve et al., 2009; Saunders et al., 2014). However, our results further revealed the presence of many other copepod species not previously described in Electrona diets in the region, particularly the small copepods Clausocalanus laticeps and Ctenocalanus citer, which showed particularly high RRA% in E. carlsbergi stomachs. It has been postulated that the absence of such small copepods in conventional stomach analyses was because they were not consumed, either because they were too small to be retained by gill rakers, or they were selected against because their energy content was too low (Shreeve et al., 2009). However, our findings suggest these smaller copepod species are consumed by these myctophid species.
The sensitivity of metabarcoding raises the possibility of secondary predation influencing the results, namely detecting taxa consumed by the actual myctophid prey species (Sakaguchi et al., 2017). Clarke et al. (2020). applied high-throughput sequencing to individual diet items from Southern Ocean mesopelagic fish stomachs (copepods, amphipods and euphausiids) to explore whether secondary predation was a likely influence on the results. They found that sequences indicative of secondary predation was generally uncommon and suggested that, although secondary predation could potentially be detected, the relative abundance of indirectly ingested prey items would be < 1% and an unlikely source of error. Given that we used strict criteria in our processing steps whereby very low read abundance items are discarded from the analysis (Ruiz et al., 2024), we consider that secondary predation effects were negligible in the interpretation of our results. Nevertheless, it is worthy to mention that diatoms (Phylum Bacillariophyta) and dinoflagellates were found in the stomach of E. carlsbergi using the COI marker (Supplementary Table 3), a likely case of accidental consumption and secondary ingestion (Tercel et al., 2021).
4.2 Metabarcoding resolution of GZP
The importance of GZP in the diet of Antarctic fishes is becoming increasingly recognized, particularly the consumption of Salpa thompsoni (Shreeve et al., 2009; Kruse et al., 2015; Diaz Briz et al., 2017; Clarke et al., 2020; Ruiz et al., 2024). We show that GZP taxa are common in the diets of both Southern Ocean Electrona species. Specifically, salps (principally Ilhea racovitzai) and appendicularians were primary (high RRA% and FOO%) components of the diet of E. carlsbergi, while hydrozoans played a secondary role in the diet of E. antarctica.
Both Clarke et al. (2020) and Vasiliadis et al. (2023) previously used metabarcoding to examine the diet of E. antarctica using the 18S v7-v9 region. In both studies, sequences were taxonomically assigned to Cnidaria, Siphonophorae, Ctenophora, Coronatae, Trachymedusae, and Salpida; although none were identified as major dietary components. Our results indicate that E. antarctica prey on Hydrozoa, Siphonophorae, Narcomedusae and Coronatae, as well as Phragmophora and Tentaculata. The importance of GZP predation for this species was also secondary in our results, except for pelagic tunicates since nearly all individuals (90%) consumed salps and appendicularians, according to the 18S-v1-v2 analysis.
Congruent with Ruiz et al. (2024), we found COI and 18S v1-v2 markers to be relatively consistent in the detection of GZP. This can be explained either by differential amplification and sequencing success of each marker for specific groups, or taxonomic coverage of their associated reference databases. Although the mitochondrial COI gene is the most commonly used marker in metazoan (meta)barcoding studies, providing high resolution taxon discrimination, the nuclear marker 18S is frequently used to target a broad spectrum of eukaryotic phyla because it has highly conserved regions. However, 18S often has a lower discriminatory power compared to mitochondrial markers (Clarke et al., 2017; Wangensteen et al., 2018). In our results, COI Leray-XT reads allowed for a better taxonomic resolution than 18S-v1-v2, but in terms of GZP, both markers showed similar results with the only major discrepancies being for pelagic tunicates (salps and appendicularians). Such a discrepancy has been described in previously, with differences attributed to less successful amplification of GZP DNA using the COI compared to other markers (van der Reis et al., 2018; Pappalardo et al., 2021). This is related to the high degeneracy of the COI gene (Deagle et al., 2014) which is particularly true for tunicates, where the high mutation rate in the primer binding sites can affect the amplification success (Goodall-Copestake, 2014; Goodall-Copestake, 2017; Ruiz et al., 2020).
4.3 Congeneric differences in diet
Evidence of dietary segregation between the two myctophid congeners was reported previously, based predominantly on the hard-bodied crustacean component of their diets. In general, E. antarctica fed principally on amphipods (Shreeve et al., 2009; Stowasser et al., 2009), euphausiids, including Antarctic krill, and copepods (Saunders et al., 2014; Saunders et al., 2019), whereas E. carlsbergi consumed mostly copepods. Interestingly, E. carlsbergi was the only species among the wider myctophid community that appeared to consume salps based on the limited conventional studies (Shreeve et al., 2009; Stowasser et al., 2009; Saunders et al., 2014; Saunders et al., 2019). Our multi-marker metabarcoding approach revealed a similar pattern, albeit with a higher detection of soft-bodied prey that also varied between congeners. Notably, we found that pteropods comprised a substantial part of the diet of E. antarctica, while pelagic tunicates (salps and appendicularians) were important in the diet of E. carlsbergi.
The dietary differentiation of these species may be linked to their horizontal distribution patterns (Collins et al., 2008; Shreeve et al., 2009; Saunders et al., 2014; Saunders et al., 2018). While E. antarctica is widespread throughout the Scotia Sea, it was shown to be more abundant in the SSS (Saunders et al., 2014; Saunders et al., 2018). We found euphausiids to be a primary dietary item of E. antarctica, with Antarctic krill being the most consumed species. These findings align with the horizontal distribution of small Antarctic krill stages, which are abundant in sea ice areas (Murphy et al., 2007) primarily found in the SSS. The distribution of E. carlsbergi is generally associated with water masses around the Antarctic Polar Front, being caught predominantly in the NSS during spring, and around the MSS and WSS during summer and autumn with the species being rare in southern regions (Collins et al., 2008; Saunders et al., 2014; Saunders et al., 2018). We found that specimens collected in autumn and in the SSS (<0.5°C net mean temperatures) predominantly consumed salps (Ihlea racovitzai), while those collected in spring and the NSS and WSS (0 – 1°C net mean temperatures) showed a very high number of reads assigned to copepods and appendicularians. This is in line with evidence showing that appendicularians are abundant in 0°C and subzero surface temperature waters around the Antarctic continent (Hunt and Hosie, 2005); and I. racovitzai being abundant in the ice-covered regions closer to the Antarctic continent in <0°C water masses (Ono and Moteki, 2013).
4.4 Spatio-temporal variation in diet
We found three distinct spatio-temporal feeding types within the sampled E. carlsbergi population, while there was no such segregation in E. antarctica. The three E. carlsbergi feeding types consisted of (1) specimens collected in spring-2006 and in the NSS (net mean temperature: 0.5-1°C), which predominantly preyed upon copepods, followed by appendicularians, and then euphausiids, (2) samples collected in summer-2008 around the APF (net mean temperature: 1-3°C), which consumed mainly copepods and (3) specimens collected in autumn-2009 in the SSS (net mean temperature <0.5°C), which preyed mainly upon euphausiids, copepods, and Ilhea racovitzai. This feeding segregation in E. carsbergi partially correlates with it known feeding patterns. Although appendicularians are understudied in the Southern Ocean, it is known that they are generally abundant in waters around 0°C (Hunt and Hosie, 2005; Atkinson et al., 2012). These organisms discard houses, producing marine snow; both the discarded houses and the appendicularians themselves have been documented as food sources for carnivorous zooplankton and fish (Gorsky and Fenaux, 1998). Therefore, this study adds to the evidence that when appendicularians are abundant (e.g., during spring in the NSS), they form an important part of E. carlsbergi’s diet. Euphausiids were consumed more often and in more quantities during spring-2006 in the NSS (represented by Thysanoessa macrura) and autumn-2009 in the SSS (represented mostly by Antarctic krill); where mean net water temperatures are lower than 1°C. This can be explained by the distributional patterns and thermal window of the Antarctic krill, of which highest biomass is reported mainly from the shelf areas (Atkinson et al., 2008; Siegel and Watkins, 2016), whereas it decreases towards the Polar Front (Atkinson et al., 2008; Hill et al., 2009; Silk et al., 2016), which is outside its optimal thermal range (Atkinson et al., 2006; Tarling et al., 2006). The small euphausiid species T. macrura was preyed upon in all three sampled seasons-cruises, but in terms of RRA%, it was a more abundant prey in spring-2006. This species is the most numerically abundant euphausiid species in the Southern Ocean (Nordhausen, 1992), where it is found in pelagic and coastal waters across a wide thermal gradient, from near freezing water to warm waters (10°C).
The biomass of copepods in the Southern Ocean is dominated by large species from the genera Calanoides, Calanus, Rhincalanus, and Metridia (Atkinson et al., 2012; Johnston et al., 2022). Our results indicate that these copepods were consumed throughout all three sampled seasons-cruises, with their highest consumption by E. carlsbergi occurring in summer-2008 in the APF. This pattern aligns with their seasonal vertical migration and the higher total copepod abundance near the APF recorded in the Scotia Sea (Atkinson and Sinclair, 2000). Additionally, Metridia gerlachei was consumed more frequently and its DNA reads were present in higher abundances during autumn-2009 in the SSS, consistent with observations of high numbers of these specimens in shallow waters (0-100 meters) during autumn in the Scotia Sea (Atkinson and Sinclair, 2000).
Interestingly, our results revealed that feeding on salps was greatest during autumn-2009 and in the SSS, where the in-situ water temperatures were below 0.5°C. We initially hypothesized the consumption of GZP to be higher during summer and autumn, and salps to be more frequently preyed upon in the APF compared to the southern areas of the Scotia Sea. This hypothesis was based on the assumption that Salpa thompsoni achieves high densities in the warmer waters of the Antarctic Polar Front (Pakhomov et al., 2002) and is one of the most commonly consumed GZP species in myctophids according to previous metabarcoding studies (Queirós et al., 2024; Ruiz et al., 2024). However, Ihlea racovitzai was the most represented salp species in our specimens. This species shows higher nutritional value in comparison to other salps in terms of carbon and protein content (Dubischar et al., 2012). This lesser-known salp species is typically found in waters below 0°C (Ono and Moteki, 2013) and reproduces sexually during autumn, with young solitaries dominating during winter and growing through summer. It rarely forms large blooms, generally being present at low densities throughout its range (Atkinson et al., 2004; Dubischar et al., 2012; Ono and Moteki, 2013). According to our findings, E. carlsbergi preyed on the aggregates formed in autumn-2009 in the SSS, where water temperatures are suitable for I. racovitzai.
4.5 Implications of GZP consumption by Southern Ocean myctophids
The Antarctic Peninsula region has experienced rapid climate-related change over recent decades including increases in sea surface temperature (Meredith and King, 2005) and reductions in sea ice extent (Stammerjohn et al., 2008; Turner et al., 2016). Associated with these changes, many of the species previously identified as dominant myctophid prey items are predicted to shift distributions, including euphausiids such as Euphausia superba, Thysanoessa macrura, and copepods such as Calanus simillimus, Clausocalanus laticeps and Ctenocalanus citer (Atkinson et al., 2008; Daufresne et al., 2009; Hill et al., 2013; Driscoll et al., 2015; Richerson et al., 2015; Horne et al., 2016; Murphy et al., 2017; Atkinson et al., 2019). Particularly, salp populations (mainly S. thompsoni) have shifted significantly southward, expanding into areas generally dominated by Antarctic krill (Pakhomov et al., 2002; Suprenand and Ainsworth, 2017; Atkinson et al., 2019; Henschke and Pakhomov, 2019). The continued success of myctophids in this region will depend on their ability to either follow shifting prey distributions or adapt to alternative food sources, such as GZP, as well as their ability to adapt physiologically to warming temperatures (Deutsch et al., 2022).
In terms of the initial hypotheses of this study, although there were differences in the GZP taxa consumed by E. carlsbergi and E. antarctica, GZP were a prominent part of the diet of both species if secondary to the consumption of copepods and euphausiids. Therefore, both Electrona species are likely to have a similar capacity to adapt to a prey field that may increase in GZP biomass as a result of climate change. Furthermore, we did not find evidence that salp consumption by these species in particular was greatest in the northern warmer parts of the Southern Ocean, towards the Polar Front. Indeed, some of the highest levels of salp consumption was further south and in the colder autumn months through consumption of the polar salp species, Ihlea racovitzai. This indicates dietary diversity and a capacity to adapt consumption in response to GZP availability in the prey field.
The use of multimarker metabarcoding by the present study has shown that the myctophid diet is much broader than has previously been recognized with many different taxa of GZP being either primary (salps and appendicularians) or secondary dietary items (Hydrozoa, Siphonophorae, Narcomedusae and Coronatae). This new knowledge provides a different perspective on the resilience of Electrona species to shifts in its trophic environment given that it is now apparent that it has a large diversity of prey types with which to fulfil their dietary requirements. Nevertheless, greater information is required on the nutritional value of GZP to myctophids to provide a more complete understanding of their role as alternate dietary items.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Sequence Read Archive (SRA) under accession numbers BioProject ID PRJNA1131325, and BioSamples ID SAMN42277734 - SAMN42277847. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1131325.
Ethics statement
The animal study was approved by As required by the Antarctic Treaty (www.ats.aq), all research was conducted after a preliminary environmental assessment and under permits issued by the UK Foreign and Commonwealth Office and the Government of South Georgia and South Sandwich Islands. Fishes were collected as part of faunal surveys and no animal experiments were conducted. Therefore, no ethical approval was required. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. RS: Conceptualization, Investigation, Methodology, Validation, Writing – review & editing. GT: Investigation, Project administration, Resources, Supervision, Writing – review & editing. AM: Writing – review & editing, Validation, Software, Methodology. FL: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. CH: Writing – review & editing, Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) in the framework of the priority program SPP 1158 “Antarctic Research with comparative investigations in Arctic ice areas” by the following grant HA 7627/3-1, LE 2323/11-1. CH is supported by the Helmholtz Young Investigator Group “ARJEL – Arctic Jellies” with the project number VH-NG-1400, funded by the Helmholtz Society and the Alfred Wegener Institute Helmholtz Centre for Polar and Marine Research. Open Access funding enabled and organized by Projekt DEAL. RAS and GAT were funded under the British Antarctic Survey’s National Capability Antarctic Logistics and Infrastructure programme CONSEC, supported by the Natural Environment Research Council, a part of UK Research and Innovation.
Acknowledgments
We are grateful to the ARJEL team members for their help during laboratory work. We also thank Stefan Neuhaus for his support and help in the bioinformatics analysis. Finally, we would like to thank the reviewers, whose efforts have greatly improved the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1474424/full#supplementary-material
References
Amundsen P., Sánchez-Hernández J. (2019). Feeding studies take guts–critical review and recommendations of methods for stomach contents analysis in fish. J. Fish. Biol. 95, 1364–1373. doi: 10.1111/jfb.14151
Atkinson A., Sinclair J. (2000). Zonal distribution and seasonal vertical migration of copepod assemblages in the Scotia Sea. Polar. Biol. 23, 46–58. doi: 10.1007/s003000050007
Atkinson A., Whitehouse M. J., Priddle J., Cripps G. C., Ward P., Brandon M. A. (2001). South Georgia, Antarctica: a productive, cold water, pelagic ecosystem. Mar. Ecol. Prog. Ser. 216, 279–308. doi: 10.3354/meps216279
Atkinson A., Siegel V., Pakhomov E., Rothery P. (2004). Long-term decline in krill stock and increase in salps within the Southern Ocean. Nature 432, 100–103. doi: 10.1038/nature02996
Atkinson A., Shreeve R. S., Hirst A. G., Rothery P., Tarling G. A., Pond D. W., et al. (2006). Natural growth rates in Antarctic krill (Euphausia superba): II. Predictive models based on food, temperature, body length, sex, and maturity stage. Limnol. Oceanogr. 51, 973–987. doi: 10.4319/lo.2006.51.2.0973
Atkinson A., Siegel V., Pakhomov E. A., Rothery P., Loeb V., Ross R. M., et al. (2008). Oceanic circumpolar habitats of Antarctic krill. Mar. Ecol. Prog. Ser. 362, 1–23. doi: 10.3354/meps07498
Atkinson A., Ward P., Hunt B., Pakhomov E., Hosie G. (2012). An overview of Southern Ocean zooplankton data: abundance, biomass, feeding and functional relationships. Ccamlr. Sci. 19, 171–218.
Atkinson A., Hill S. L., Pakhomov E. A., Siegel V., Reiss C. S., Loeb V. J., et al. (2019). Krill (Euphausia superba) distribution contracts southward during rapid regional warming. Nat. Climate Change 9, 142–147. doi: 10.1038/s41558-018-0370-z
Barrera-Oro E. (2002). The role of fish in the Antarctic marine food web: differences between inshore and offshore waters in the southern Scotia Arc and west Antarctic Peninsula. Antarctic. Sci. 14, 293–309. doi: 10.1017/S0954102002000111
Belcher A., Saunders R., Tarling G. (2019). Length, weight and abundance data of fish species captured in RMT-25 net surveys in the Scotia Sea, Southern Ocean in 2006, 2008, and 2009 (Version 1.0)[Data set. UK. Polar. Data Centre. Br. Antarctic. Survey. Natural Environ. Res. Council. UK. Res. Innovation.
Bernard K. S., Steinberg D. K., Schofield O. M. E. (2012). Summertime grazing impact of the dominant macrozooplankton off the Western Antarctic Peninsula. Deep. Sea. Res. Part I.: Oceanographic. Res. Papers. 62, 111–122. doi: 10.1016/j.dsr.2011.12.015
Blaxter M. L., De Ley P., Garey J. R., Liu L. X., Scheldeman P., Vierstraete A., et al. (1998). A molecular evolutionary framework for the phylum Nematoda. Nature 392, 71–75. doi: 10.1038/32160
Brodeur R. D., Buckley T. W., Lang G. M., Draper D. L., Buchanan J. C., Hibpshman R. E. (2021). Demersal fish predators of gelatinous zooplankton in the Northeast Pacific Ocean. Mar. Ecol. Prog. Ser. 658, 89–104. doi: 10.3354/meps13489
Buchner D., Leese F. (2020). BOLDigger–a Python package to identify and organise sequences with the Barcode of Life Data systems. Metabarcoding. Metagenom. 4, e53535. doi: 10.3897/mbmg.4.53535
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Catul V., Gauns M., Karuppasamy P. K. (2011). A review on mesopelagic fishes belonging to family Myctophidae. Rev. Fish. Biol. Fisheries. 21, 339–354. doi: 10.1007/s11160-010-9176-4
Cherel Y., Fontaine C., Richard P., Labatc J.-P. (2010). Isotopic niches and trophic levels of myctophid fishes and their predators in the Southern Ocean. Limnol. Oceanogr. 55, 324–332. doi: 10.4319/lo.2010.55.1.0324
Clarke L. J., Beard J. M., Swadling K. M., Deagle B. E. (2017). Effect of marker choice and thermal cycling protocol on zooplankton DNA metabarcoding studies. Ecol. Evol. 7, 873–883. doi: 10.1002/ece3.2017.7.issue-3
Clarke L. J., Trebilco R., Walters A., Polanowski A. M., Deagle B. E. (2020). DNA-based diet analysis of mesopelagic fish from the southern Kerguelen Axis. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 174, S0967064517304046. doi: 10.1016/j.dsr2.2018.09.001
Collins A. G., Bentlage B., Lindner A., Lindsay D., Haddock S. H. D., Jarms G., et al. (2008). Phylogenetics of Trachylina (Cnidaria: Hydrozoa) with new insights on the evolution of some problematical taxa. J. Mar. Biol. Ass. 88, 1673–1685. doi: 10.1017/S0025315408001732
Collins M. A., Stowasser G., Fielding S., Shreeve R., Xavier J. C., Venables H. J., et al. (2012). Latitudinal and bathymetric patterns in the distribution and abundance of mesopelagic fish in the Scotia Sea. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 59–60, 189–198. doi: 10.1016/j.dsr2.2011.07.003
Curran M. A., van Ommen T. D., Morgan V. I., Phillips K. L., Palmer A. S. (2003). Ice core evidence for Antarctic sea ice decline since the 1950s. Science 302, 1203–1206. doi: 10.1126/science.1087888
Daufresne M., Lengfellner K., Sommer U. (2009). Global warming benefits the small in aquatic ecosystems. Proc. Natl. Acad. Sci. 106, 12788–12793. doi: 10.1073/pnas.0902080106
Davis D., Staniland I. J., Reid K. (2006). Spatial and temporal variability in the fish diet of Antarctic fur seal (Arctocephalus gazella) in the Atlantic sector of the Southern Ocean. Can. J. Zool. 84, 1025–1037. doi: 10.1139/z06-071
Deagle B. E., Jarman S. N., Coissac E., Pompanon F., Taberlet P. (2014). DNA metabarcoding and the cytochrome c oxidase subunit I marker: not a perfect match. Biol. Lett. 10, 20140562. doi: 10.1098/rsbl.2014.0562
Deutsch C., Penn J. L., Verberk W. C. E. P., Inomura K., Endress M.-G., Payne J. L. (2022). Impact of warming on aquatic body sizes explained by metabolic scaling from microbes to macrofauna. Proc. Natl. Acad. Sci. 119, e2201345119. doi: 10.1073/pnas.2201345119
Diaz Briz L., Sánchez F., Marí N., Mianzan H., Genzano G. (2017). Gelatinous zooplankton (ctenophores, salps and medusae): an important food resource of fishes in the temperate SW Atlantic Ocean. Mar. Biol. Res. 13, 630–644. doi: 10.1080/17451000.2016.1274403
Dischereit A., Beermann J., Lebreton B., Wangensteen O. S., Neuhaus S., Havermans C. (2024). DNA metabarcoding reveals a diverse, omnivorous diet of Arctic amphipods during the polar night, with jellyfish and fish as major prey. Front. Mar. Sci. 11, 1327650. doi: 10.3389/fmars.2024.1327650
Dischereit A., Wangensteen O. S., Præbel K., Auel H., Havermans C. (2022). Using DNA metabarcoding to characterize the prey spectrum of two co-occurring themisto amphipods in the rapidly changing atlantic-arctic gateway fram strait. Genes 13, 2035. doi: 10.3390/genes13112035
Dornan T., Fielding S., Saunders R. A., Genner M. J. (2022). Large mesopelagic fish biomass in the Southern Ocean resolved by acoustic properties. Proc. R. Soc. B. 289, 20211781. doi: 10.1098/rspb.2021.1781
Driscoll R. M., Reiss C. S., Hentschel B. T. (2015). Temperature-dependent growth of Thysanoessa macrura: inter-annual and spatial variability around Elephant Island, Antarctica. Mar. Ecol. Prog. Ser. 529, 49–61. doi: 10.3354/meps11291
Duan M., Zhang C., Luo Y., Gao L., Liu G., Liu C., et al. (2024). The feeding strategies of the antarctic lanternfish electrona Antarctica (pisces: myctophidae) in the amundsen and cosmonaut seas (southern ocean), assessed with a classification tree analysis. Polar. Biol. 47, 515–532. doi: 10.1007/s00300-024-03249-6
Dubischar C. D., Pakhomov E. A., Von Harbou L., Hunt B. P. V., Bathmann U. V. (2012). Salps in the Lazarev Sea, Southern Ocean: II. Biochemical composition and potential prey value. Mar. Biol. 159, 15–24. doi: 10.1007/s00227-011-1785-5
Fuentes V., Alurralde G., Meyer B., Aguirre G. E., Canepa A., Wölfl A.-C., et al. (2016). Glacial melting: an overlooked threat to Antarctic krill. Sci. Rep. 6, 27234. doi: 10.1038/srep27234
Geller J., Meyer C., Parker M., Hawk H. (2013). Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol. Ecol. Resour. 13, 851–861. doi: 10.1111/men.2013.13.issue-5
Goodall-Copestake W. P. (2014). Morphological and molecular characterization of salps (Thalia spp.) from the Tristan da Cunha archipelago. J. Plankton. Res. 36, 883–888. doi: 10.1093/plankt/fbu013
Goodall-Copestake W. P. (2017). One tunic but more than one barcode: evolutionary insights from dynamic mitochondrial DNA in Salpa thompsoni (Tunicata: Salpida). Biol. J. Linn. Soc. 120, 637–648. doi: 10.1111/bij.12915
Gorsky G., Fenaux R. (1998). The role of Appendicularia in marine food webs. Biol. Pelagic. Tunicates., 161–169.
Henschke N., Pakhomov E. A. (2019). Latitudinal variations in Salpa thompsoni reproductive fitness. Limnol. Oceanogr. 64, 575–584. doi: 10.1002/lno.11061
Hill S. L., Phillips T., Atkinson A. (2013). Potential climate change effects on the habitat of Antarctic krill in the Weddell quadrant of the Southern Ocean. PloS One 8, e72246. doi: 10.1371/journal.pone.0072246
Hill S. L., Trathan P. N., Agnew D. J. (2009). The risk to fishery performance associated with spatially resolved management of Antarctic krill (Euphausia superba) harvesting. ICES. J. Mar. Sci. 66, 2148–2154. doi: 10.1093/icesjms/fsp172
Horne C. R., Hirst A. G., Atkinson D., Neves A., Kiørboe T. (2016). A global synthesis of seasonal temperature–size responses in copepods. Global Ecol. Biogeogr. 25, 988–999. doi: 10.1111/geb.2016.25.issue-8
Hulley P. A. (1981). Results of the research cruises of FRV ‘Walther Herwig’to South America. 58. Family Myctophidae (Osteichthyes, Myctophiformes). Arch. Fischereiwiss. 31, 1.
Hunt B. P., Hosie G. W. (2005). Zonal structure of zooplankton communities in the Southern Ocean South of Australia: results from a 2150 km continuous plankton recorder transect. Deep. Sea. Res. Part I.: Oceanographic. Res. Papers. 52, 1241–1271. doi: 10.1016/j.dsr.2004.11.019
Johnston N. M., Murphy E. J., Atkinson A., Constable A. J., Cotté C., Cox M., et al. (2022). Status, change, and futures of zooplankton in the southern ocean. Front. Ecol. Evol. 9, 624692. doi: 10.3389/fevo.2021.624692
Kruse S., Pakhomov E. A., Hunt B. P. V., Chikaraishi Y., Ogawa N. O., Bathmann U. (2015). Uncovering the trophic relationship between Themisto gaudichaudii and Salpa thompsoni in the Antarctic Polar Frontal Zone. Mar. Ecol. Prog. Ser. 529, 63–74. doi: 10.3354/meps11288
Loeb V., Santora J. (2012). Population dynamics of Salpa thompsoni near the Antarctic Peninsula: growth rates and interannual variations in reproductive activity (1993–2009). Prog. Oceanogr. 96, 93–107. doi: 10.1016/j.pocean.2011.11.001
Macher T., Beermann A. J., Leese F. (2021). TaxonTableTools: A comprehensive, platform-independent graphical user interface software to explore and visualise DNA metabarcoding data. Mol. Ecol. Resour. 21, 1705–1714. doi: 10.1111/1755-0998.13358
Macher J., Vivancos A., Piggott J. J., Centeno F. C., Matthaei C. D., Leese F. (2018). Comparison of environmental DNA and bulk-sample metabarcoding using highly degenerate cytochrome. c Oxidase. I. Primers. Mol. Ecol. Resour. 18, 1456–1468. doi: 10.1111/men.2018.18.issue-6
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
McInnes J. C., Alderman R., Deagle B. E., Lea M.-A., Raymond B., Jarman S. N. (2017). Optimised scat collection protocols for dietary DNA metabarcoding in vertebrates. Methods Ecol. Evol. 8, 192–202. doi: 10.1111/mee3.2017.8.issue-2
Meredith M. P., King J. C. (2005). Rapid climate change in the ocean west of the Antarctic Peninsula during the second half of the 20th century. Geophys. Res. Lett. 32 (19). doi: 10.1029/2005GL024042
Moline M. A., Claustre H., Frazer T. K., Schofield O., Vernet M. (2004). Alteration of the food web along the Antarctic Peninsula in response to a regional warming trend. Global Change Biol. 10, 1973–1980. doi: 10.1111/j.1365-2486.2004.00825.x
Murphy E. J., Watkins J. L., Trathan P. N., Reid K., Meredith M. P., Thorpe S. E., et al. (2007). Spatial and temporal operation of the Scotia Sea ecosystem: a review of large-scale links in a krill centred food web. Philos. Trans. R. Soc. B.: Biol. Sci. 362, 113–148. doi: 10.1098/rstb.2006.1957
Murphy E. J., Thorpe S. E., Tarling G. A., Watkins J. L., Fielding S., Underwood P. (2017). Restricted regions of enhanced growth of Antarctic krill in the circumpolar Southern Ocean. Sci. Rep. 7, 6963. doi: 10.1038/s41598-017-07205-9
Nordhausen W. (1992). Distribution and growth of larval and adult Thysanoessa macrura(Euphausiacea) in the Bransfield Strait Region, Antarctica. Mar. Ecol. Prog. Series. Oldendorf. 83, 185–196. doi: 10.3354/meps083185
O’Brien T. D., Blanco-Bercial L., Questel J. M., Batta-Lona P. G., Bucklin A. (2024). “MetaZooGene atlas and database: reference sequences for marine ecosystems,” in DNA Barcoding: Methods and Protocols (New York, NY: Springer), 475–489.
Oksanen J. , Blanchet G., Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2019). Vegan: Community ecology package (version 2.5-6). The Comprehensive R Archive Network.
Ono A., Moteki M. (2013). Spatial distributions and population dynamics of two salp species, Ihlea racovitzai and Salpa thompsoni, in the waters north of Lützow-Holm Bay (East Antarctica) during austral summers of 2005 and 2006. Polar. Biol. 36, 807–817. doi: 10.1007/s00300-013-1305-9
Pakhomov E. A., Froneman P. W., Perissinotto R. (2002). Salp/krill interactions in the Southern Ocean: spatial segregation and implications for the carbon flux. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 49, 1881–1907. doi: 10.1016/S0967-0645(02)00017-6
Papadimitraki M., Maar K., Jónasdóttir S. H. (2023). Meso- and bathypelagic fish feeding ecology: A meta-analysis on fatty acids and stable isotope trophic studies. Deep. Sea. Res. Part I.: Oceanographic. Res. Papers. 198, 104083. doi: 10.1016/j.dsr.2023.104083
Pappalardo P., Collins A. G., Pagenkopp Lohan K. M., Hanson K. M., Truskey S. B., Jaeckle W., et al. (2021). The role of taxonomic expertise in interpretation of metabarcoding studies. ICES. J. Mar. Sci. 78, 3397–3410. doi: 10.1093/icesjms/fsab082
Queirós J. P., Borras-Chavez R., Friscourt N., Groß J., Lewis C. B., Mergard G., et al. (2024). Southern Ocean food-webs and climate change: A short review and future directions. PloS Clim. 3, e0000358. doi: 10.1371/journal.pclm.0000358
Riaz J., Walters A., Trebilco R., Bestley S., Lea M.-A. (2020). Stomach content analysis of mesopelagic fish from the southern Kerguelen Axis. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 174, 104659. doi: 10.1016/j.dsr2.2019.104659
Richerson K., Watters G. M., Santora J. A., Schroeder I. D., Mangel M. (2015). More than passive drifters: a stochastic dynamic model for the movement of Antarctic krill. Mar. Ecol. Prog. Ser. 529, 35–48. doi: 10.3354/meps11324
Rogers A. D., Frinault B. A. V., Barnes D. K. A., Bindoff N. L., Downie R., Ducklow H. W., et al. (2020). Antarctic futures: an assessment of climate-driven changes in ecosystem structure, function, and service provisioning in the Southern Ocean. Annu. Rev. Mar. Sci. 12, 87–120. doi: 10.1146/annurev-marine-010419-011028
Ruiz M. B., Moreira E., Novillo M., Neuhaus S., Leese F., Havermans C. (2024). Detecting the invisible through DNA metabarcoding: The role of gelatinous taxa in the diet of two demersal Antarctic key stone fish species (Notothenioidei). Environ. DNA 6, e561. doi: 10.1002/edn3.v6.3
Ruiz M. B., Taverna A., Servetto N., Sahade R., Held C. (2020). Hidden diversity in Antarctica: Molecular and morphological evidence of two different species within one of the most conspicuous ascidian species. Ecol. Evol. 10, 8127–8143. doi: 10.1002/ece3.v10.15
Saba G. K., Burd A. B., Dunne J. P., Hernández-León S., Martin A. H., Rose K. A., et al. (2021). Toward a better understanding of fish-based contribution to ocean carbon flux. Limnol. Oceanogr. 66, 1639–1664. doi: 10.1002/lno.11709
Sakaguchi S. O., Shimamura S., Shimizu Y., Ogawa G., Yamada Y., Shimizu K., et al. (2017). Comparison of morphological and DNA-based techniques for stomach content analyses in juvenile chum salmon Oncorhynchus keta: a case study on diet richness of juvenile fishes. Fisheries. Sci. 83, 47–56. doi: 10.1007/s12562-016-1040-6
Saunders R. A., Collins M. A., Foster E., Shreeve R., Stowasser G., Ward P., et al (2014). The trophodynamics of southern ocean electrona (Myctophidae) in the scotia sea. Polar. Biol. 37, 789–807. doi: 10.1007/s00300-014-1480-3
Saunders R. A., Collins M. A., Ward P., Stowasser G., Shreeve R., Tarling G. A. (2015). Trophodynamics of protomyctophum (Myctophidae) in the scotia sea (Southern ocean). J. Fish. Biol. 87, 1031–1058. doi: 10.1111/jfb.2015.87.issue-4
Saunders R. A., Collins M. A., Shreeve R., Ward P., Stowasser G., Hill S. L., et al. (2018). Seasonal variation in the predatory impact of myctophids on zooplankton in the Scotia Sea (Southern Ocean). Prog. Oceanogr. 168, 123–144. doi: 10.1016/j.pocean.2018.09.017
Saunders R. A., Hill S. L., Tarling G. A., Murphy E. J. (2019). Myctophid fish (Family myctophidae) are central consumers in the food web of the scotia sea (Southern ocean). Front. Mar. Sci. 6, 530. doi: 10.3389/fmars.2019.00530
Shreeve R. S., Collins M. A., Tarling G. A., Main C. E., Ward P., Johnston N. M. (2009). Feeding ecology of myctophid fishes in the northern Scotia Sea. Mar. Ecol. Prog. Ser. 386, 221–236. doi: 10.3354/meps08064
Siegel V., Watkins J. L. (2016). Distribution, biomass and demography of Antarctic krill, Euphausia superba. Biol. Ecol. Antarctic. krill., 21–100. doi: 10.1007/978-3-319-29279-3
Silk J. R. D., Thorpe S. E., Fielding S., Murphy E. J., Trathan P. N., Watkins J. L., et al. (2016). Environmental correlates of Antarctic krill distribution in the Scotia Sea and southern Drake Passage. ICES. J. Mar. Sci. 73, 2288–2301. doi: 10.1093/icesjms/fsw097
Sinniger F., Pawlowski J., Harii S., Gooday A. J., Yamamoto H., Chevaldonné P., et al. (2016). Worldwide analysis of sedimentary DNA reveals major gaps in taxonomic knowledge of deep-sea benthos. Front. Mar. Sci. 3. doi: 10.3389/fmars.2016.00092
Stammerjohn S. E., Martinson D. G., Smith R. C., Iannuzzi R. A. (2008). Sea ice in the western Antarctic Peninsula region: Spatio-temporal variability from ecological and climate change perspectives. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 55, 2041–2058. doi: 10.1016/j.dsr2.2008.04.026
Stowasser G., Pond D. W., Collins M. A. (2009). Using fatty acid analysis to elucidate the feeding habits of Southern Ocean mesopelagic fish. Mar. Biol. 156, 2289–2302. doi: 10.1007/s00227-009-1256-4
Suprenand P. M., Ainsworth C. H. (2017). Trophodynamic effects of climate change-induced alterations to primary production along the western Antarctic Peninsula. Mar. Ecol. Prog. Ser. 569, 37–54. doi: 10.3354/meps12100
Tarling G. A., Shreeve R. S., Hirst A. G., Atkinson A., Pond D. W., Murphy E. J., et al. (2006). Natural growth rates in Antarctic krill (Euphausia superba): I. Improving methodology and predicting intermolt period. Limnol. Oceanogr. 51, 959–972. doi: 10.4319/lo.2006.51.2.0959
Tercel M. P. T. G., Symondson W. O. C., Cuff J. P. (2021). The problem of omnivory: A synthesis on omnivory and DNA metabarcoding. Mol. Ecol. 30, 2199–2206. doi: 10.1111/mec.v30.10
Turner J., Lu H., White I., King J. C., Phillips T., Hosking J. S., et al. (2016). Absence of 21st century warming on Antarctic Peninsula consistent with natural variability. Nature 535, 411–415. doi: 10.1038/nature18645
Urban P., Præbel K., Bhat S., Dierking J., Wangensteen O. S. (2022). DNA metabarcoding reveals the importance of gelatinous zooplankton in the diet of Pandalus borealis, a keystone species in the Arctic. Mol. Ecol. 31, 1562–1576. doi: 10.1111/mec.16332
van der Reis A. L., Laroche O., Jeffs A. G., Lavery S. D. (2018). Preliminary analysis of New scampi (Metanephrops challengeri) diet using metabarcoding. PeerJ 6, e5641. doi: 10.7717/peerj.5641
Vasiliadis M. M., Freer D. J. J., Collins D. M. A., Cleary D. A. C. (2023). Assessing the trophic ecology of Southern Ocean Myctophidae: the added value of DNA metabarcoding. doi: 10.1139/cjfas-2023-0079
Wangensteen O. S., Palacín C., Guardiola M., Turon X. (2018). DNA metabarcoding of littoral hard-bottom communities: high diversity and database gaps revealed by two molecular markers. PeerJ 6, e4705. doi: 10.7717/peerj.4705
Wangensteen O. S., Turon X. (2016). “Metabarcoding techniques for assessing biodiversity of marine animal forests,” in Marine Animal Forests. Eds. Rossi S., Bramanti L., Gori A., Orejas Saco Del Valle C. (Springer International Publishing, Cham), 1–29. doi: 10.1007/978-3-319-17001-5_53-1
Whitehouse M. J., Atkinson A., Korb R. E., Venables H. J., Pond D. W., Gordon M. (2012). Substantial primary production in the land-remote region of the central and northern Scotia Sea. Deep. Sea. Res. Part II.: Topical. Stud. Oceanogr. 59, 47–56. doi: 10.1016/j.dsr2.2011.05.010
Keywords: eDNA, salps, soft-bodied prey, Southern Ocean, stomach content, trophodynamics
Citation: Ruiz MB, Saunders RA, Tarling GA, Murray A, Leese F and Havermans C (2024) The secret meal of Antarctic mesopelagic fish (Myctophidae: Electrona) revealed by multi-marker metabarcoding. Front. Mar. Sci. 11:1474424. doi: 10.3389/fmars.2024.1474424
Received: 01 August 2024; Accepted: 24 October 2024;
Published: 19 November 2024.
Edited by:
Lorenzo Zane, University of Padua, ItalyReviewed by:
Álvaro Roura, Independent researcher, Vigo, SpainAnn Bucklin, University of Connecticut Avery Point, United States
Filip Volckaert, KU Leuven, Belgium
Copyright © 2024 Ruiz, Saunders, Tarling, Murray, Leese and Havermans. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Micaela Belen Ruiz, bWljYWVsYS5ydWl6QGF3aS5kZQ==; bWljYXJ1aXo5MEBnbWFpbC5jb20=
 Micaela Belen Ruiz
Micaela Belen Ruiz Ryan Alexander Saunders
Ryan Alexander Saunders Geraint A. Tarling
Geraint A. Tarling Ayla Murray
Ayla Murray Florian Leese
Florian Leese Charlotte Havermans
Charlotte Havermans