
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 10 January 2025
Sec. Microbial Symbioses
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1463968
 Katrina M. Pagenkopp Lohan1*
Katrina M. Pagenkopp Lohan1* Ruth DiMaria1
Ruth DiMaria1 Daniel L. Martin2
Daniel L. Martin2 A. Randall Hughes3
A. Randall Hughes3 Bradley J. Peterson4
Bradley J. Peterson4 Katharyn E. Boyer5
Katharyn E. Boyer5 John J. Stachowicz6
John J. Stachowicz6 Pablo Jorgensen7
Pablo Jorgensen7 Gregory M. Ruiz8
Gregory M. Ruiz8 Cliff Ross2
Cliff Ross2Seagrass wasting disease, caused by parasitic slime nets in the genus Labyrinthula, affects seagrasses globally with outbreaks occurring at local to regional scales. Though prior research showed variation in pathogenicity across strains, little is known about the broad-scale diversity and biogeography of Labyrinthula species. We used targeted amplicon-based high throughput sequencing (HTS) to examine the phylogeography of Labyrinthula species associated with two seagrass hosts (Zostera marina, Thalassia testudinum) collected from across North America, including locations in the USA and Mexico. After testing 189 seagrass blades from 16 locations across North America, Labyrinthula spp. were detected at 14 sites with PCR prevalence ranging from 20 - 100%. We generated 10,936,628 sequences for analyses, which yielded 25 unique Labyrinthula amplicon sequence variants (ASVs), which were phylogenetically grouped into three species of Labyrinthula. All three species detected were previously identified as pathogenic, indicating that Labyrinthula species capable of causing wasting disease are present across these regions, including in locations where disease outbreaks have not been reported. The highest strain level variation was detected in Florida, which is likely due to larger sample sizes from Florida compared to other sampling locations. Assessing the phylogeography of strains was limited by sample size in each location, but indicates that the genetic connectivity of strains appears high, with location and seagrass host being the most important factors explaining the variation in Labyrinthula strains detected. The high connectivity, but low overall species diversity of Labyrinthula, is consistent with a parasite that is a host generalist. Given the high connectivity of strains and broad geographic range of pathogenic Labyrinthula reported here, this study provides valuable insight regarding how parasite diversity impacts seagrass disease.
Seagrasses are a critical foundation species in coastal habitats that create a globally significant carbon stock (Fourqurean et al., 2012), habitat and nurseries for fish and invertebrates, coastal protection, sediment stabilization, and improved water clarity (Mtwana Nordlund et al., 2016). Unfortunately, global seagrass declines threaten to decrease the abundance of associated and dependent species (Hughes et al., 2009; FFWCC, 2021). Infectious disease is a major cause of seagrass decline, namely mortality and pathology from seagrass wasting disease (SWD). In the 1930s, SWD was the primary cause of mass mortality of the seagrass Zostera marina along both coasts of the Atlantic Ocean, resulting in total loss of seagrass beds and their ecosystem functions in some areas (Short et al., 1987). This infectious disease continues to affect seagrasses globally at local to regional scales (Sullivan et al., 2013). Though outbreaks are observed globally, we know relatively little about the broad-scale diversity and biogeography of Labyrinthula species, the etiological agent of SWD.
The Labyrinthulomycetes are classified under the Super-group Chromalveolata, first rank Stramenopiles, and second rank Labyrinthulomycetes (Adl et al., 2012), clearly distinguishing them from “true fungi”; however, they do have characteristics similar to fungi, particularly in their use of a saprotrophic mode of nutrition (Tsui et al., 2009). Though many slime nets are decomposers in marine environments (Raghukumar and Damare, 2011), some taxa have adopted a different strategy as parasites. Phylogenetic work of the Labyrinthulomycetes showed that this group consists of three distinct lineages including the thraustochytrids, labyrinthulids, and aplanochytrids (Tsui et al., 2009; Leander and Porter, 2011), all of which contain organisms associated with diseases of various hosts (Burge et al., 2013). Culture independent examinations of diversity across the Labyrinthulomycetes show these taxa are incredibly diverse and ubiquitous within aquatic habitats globally (Pan et al., 2017; Xie et al., 2018; Menning et al., 2021).
Parasites in the genus Labyrinthula comprise the monotypic family Labyrinthulaceae (Stramenopiles, Labyrinthulomycetes) (Handley et al., 2007). Currently, there are only three described species of Labyrinthula: L. zosterae (Muehlstein et al., 1991), L. terrestris (Bigelow et al., 2005), and L. diatomea (Popova et al., 2020). However, further culture independent surveys indicate many additional presumed terrestrial and marine species (Chitrampalam et al., 2015; Martin et al., 2016; Sullivan et al., 2017; Trevathan-Tackett et al., 2018; Popova et al., 2020), and genetic methods are increasingly being used to delineate species and strain-level taxonomy of Labyrinthula across hosts and geographic regions. For example, Chitrampalam et al. (2015) reported four phylogenetically distinct and thus potentially separate, yet unidentified species of Labyrinthula from turfgrass collected in Arizona, USA and New Mexico, USA. Douhan et al. (2009) found geographically separated isolates from turfgrass consisted of likely additional unidentified species of Labyrinthula. Surveys of seagrass species have also yielded evidence of new species of Labyrinthula. A global survey to assess and delineate species of Labyrinthula revealed between 15–20 phylogenetically distinct and likely different species of Labyrinthula (Martin et al., 2016). One major clade in this study consisted of at least 5–7 species of Labyrinthula that appeared to be pathogenic and specific to seagrasses (Martin et al., 2016). In a survey of five seagrass species from Australian waters, Trevathan-Tackett et al. (2018) used phylogenetic analyses to differentiate five presumed species of Labyrinthula. Additionally, Pagenkopp Lohan et al. (2020) previously reported eight presumptive species of Labyrinthula associated with multiple species of seagrass in the Indian River Lagoon in Florida. Clearly, the species-level diversity within these taxa remains mostly unexplored.
Much less is known about the sub-species or strain-level genetic variation within Labyrinthula species. Douhan et al. (2009) reported multiple genotypes of Labyrinthula terrestris from turf grass, indicating high strain-level variation likely exists within this species. From seagrass hosts, Trevathan-Tackett et al. (2018) reported 2–3 strains for multiple presumed species of Labyrinthula from Australia waters. Similarly, Pagenkopp Lohan et al. (2020) recovered multiple presumed strains (2–5 zero-radius operational taxonomic units or ZOTUs) from two of the presumed Labyrinthula species detected in Florida waters. Additional strains of L. zosterae associated with Z. marina are also suspected in the North Pacific, but no phylogenetic evidence exists to confirm strain delineation (Dawkins et al., 2018). Examining the genetic diversity of this parasite at the species and strain levels is important for linking this sub-species genetic diversity with traits, particularly host specificity and pathogenicity, which experimental data has shown can vary across strains and species of Labyrinthula (Martin et al., 2016; Dawkins et al., 2018; Trevathan-Tackett et al., 2018).
We used targeted amplicon-based high throughput sequencing (HTS) of the first internal transcribed spacer region (ITS1) to examine the phylogeography of Labyrinthula species and strains associated with two seagrass species collected from both coasts of North America, including the USA and Mexico. Our objectives were to (1) identify species of Labyrinthula in North American waters, (2) explore the diversity of Labyrinthula species and strains within and across locations and hosts, and (3) examine the genetic connectivity of these species across locations and seagrass hosts. We expected to find high diversity of Labyrinthula species within and across locations. We also expected to find high connectivity across these parasites at the species and strain levels across broad geographic locations and hosts.
Turtlegrass (Thalassia testudinum) samples from Florida were collected as described in Pagenkopp Lohan et al. (2020) and Trevathan-Tackett et al. (2013). Eelgrass (Zostera marina) samples were collected from 11 sites within the Zostera Experimental Network (ZEN; zenscience.org) across Virginia, New York, Massachusetts, California and Mexico (Table 1). Within the seagrass beds, 10 shoots were collected at least 5 m apart from each other. As the goal was to assess the diversity of Labyrinthula spp. present, shoots that appeared diseased were preferentially selected when present. The shoot was then cut as close to the rhizome as possible and placed into a plastic bag, which was placed on ice until lab processing. If no lesions were observed, then blades were collected at random and processed in the same manner as those with lesions. In the lab, the blade (often the 3rd rank leaf) was separated from the rest of the shoot. The full length of the blade was measured (mm) and epibionts were removed. The disease index score (%) was recorded based on the wasting key index from Burdick et al. (1993). Blades were stored in 95% ethanol and shipped to the Smithsonian Environmental Research Center (Edgewater, Maryland) for genetic processing.
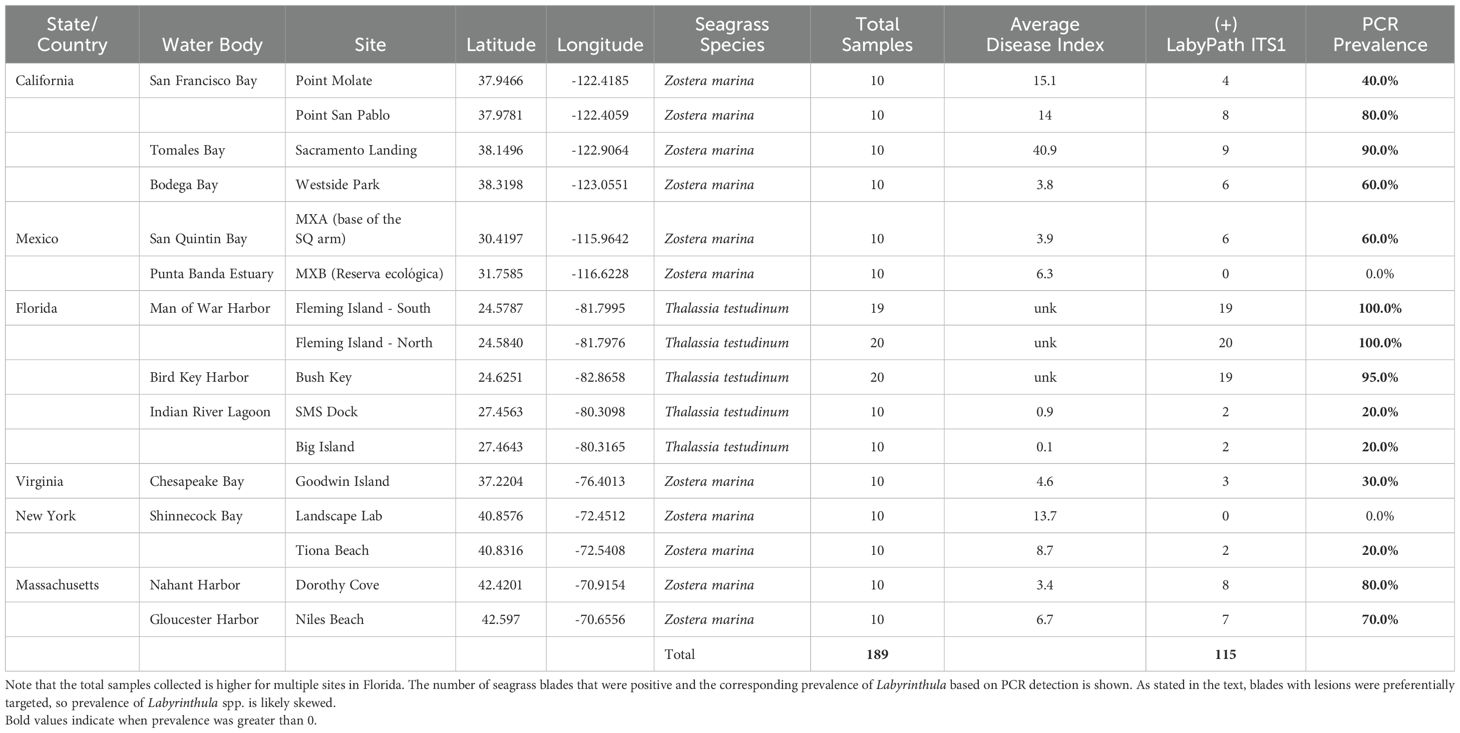
Table 1. The US state or country, water body and site for the two seagrass species that were collected and sampled in this study.
Genetic methods followed those described in Pagenkopp Lohan et al. (2020). Briefly, sections of seagrass blades (with lesions when present) were combined with one stainless steel bead and beaten for 2 min at 30 Hz using a TissueLyser (Qiagen, Germantown, Maryland). This process was repeated 1–2X until the plant tissue was pulverized. Genomic DNA was extracted from seagrass blades using the Qiagen DNeasy Plant Mini DNA Isolation kit (Qiagen, Germantown, Maryland) following the manufacturer’s protocols. All extractions within the same day included a blank extraction, which served as a negative extraction control for PCR. Extraction blanks were treated exactly like samples to identify contaminants that may have been introduced during genetic processing.
A general plant PCR primer set, ITS1-P (T. D. Bruns, https://nature.berkeley.edu/brunslab/tour/primers.html) and ITS1-4 (White et al., 1990), was used to determine if amplifiable DNA was present in the extracted DNAs. PCR reagents consisted of 1X PCR Gold Buffer (150 mM Tris-HCl, pH 8.0; 500 mM KCl; Applied Biosystems, Carlsbad, California), 1.5 mM MgCl2, 0.2 mM dNTP, 0.5 μM each primer, 0.2 mg mL−1 bovine serum albumin (BSA), 0.03 units AmpliTaq Gold polymerase, 1 µL of DNA template (DNA concentration ranged from 2.5 – 37 ng/μL, with average 8.5 ng/μL and median of 6.5 ng/μL) and water to a final volume of 19 μL. Thermocycling was carried out using a Bio-Rad C1000 or S1000 ThermoCycler with an initial denaturation of 94 °C for 10 min; followed by 35 cycles of 94 °C for 30 s, 50 °C for 1 min, and 72 °C for 1 min; and a final extension of 72 °C for 5 min. An aliquot of the PCR product (5 μL) was electrophoresed on agarose gel (2% w/v) and visualized under ultraviolet (UV) light after GelRed staining. Only seagrass samples positive for this PCR reaction were subsequently screened for the presence of Labyrinthula.
The primers Ill-LabPathITS1-2F and Ill-LabPathITS1-2R (Pagenkopp Lohan et al., 2020) were used to amplify and sequence ~316 bp fragment of the first internal transcribed spacer region (ITS1) of the ribosomal gene complex. PCR reactions consisted of the following final concentrations: 1X PCR Gold Buffer (150 mM Tris-HCl, pH 8.0; 500 mM KCl; Applied Biosystems, Carlsbad, California), 1.5 mM MgCl2, 0.2 mM each nucleotide, 0.3 µM each primer, 0.4 mg mL-1 bovine serum albumin (BSA; New England Biolabs), 0.025 units µL-1 of AmpliTaq Gold polymerase, 3 µL of DNA template (DNA concentration ranged from 2.5 – 37 ng/μL, with average 8.5 ng/μL and median of 6.5 ng/μL), and water to a final volume of 25 µL. Thermocycling was carried out using a C1000 Thermal Cycler (Bio-Rad, Hercules, California). The ITS1 PCR protocol followed an initial denaturation of 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 53 °C for 45 s, 72 °C for 45 s and a final elongation of 72 °C for 5 min. Assay screening used an aliquot of PCR product (5 µL) that was electrophoresed on agarose gel (2% w/v) stained with GelRed (Phenix Research) and visualized under UV light. All PCRs were conducted in triplicate, then triplicate PCR amplicons were pooled for each sample based on gel band intensity.
We used dual-indexing with Nextera adapters using a unique combination for each sample. Indexing PCR reagents consisted of 12.5 µL KAPA Ready Mix, 1 µL each index (i7 or i5), 1 µL amplicon (pooled product), and 9.5 µL water for a final reaction volume of 25 µL. Thermocycling was carried out with an initial denaturation of 95 °C for 5 min, followed by 12 cycles of 98 °C for 20 s, 60 °C for 45 s, 72 °C for 45 s, and a final extension of 72 °C for 5 min. To verify that indexing was successful, an aliquot of indexed product and unindexed product were both electrophoresed on agarose gel (2% w/v) stained with GelRed and visualized under UV light. The indexed product was purified with AMPure XP Beads (Beckman-Coulter) following the manufacturer’s instructions for 10 µL sample reaction volume and 1.5X ratio. The bead-cleaned samples were then quantified using Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific) with a Qubit 2.0 Fluorometer following the manufacturer’s instructions. Samples were pooled based on equimolar concentrations into a single library. The final pooled libraries were sequenced using a MiSeq v3 600 Reagent Kit (Illumina) on an Illumina MiSeq platform at the Laboratories of Analytical Biology at the Smithsonian National Museum of Natural History. Raw sequence data were deposited in the Sequence Read Archive (SRA) under BioProject PRJNA1180629.
Primer sequences were removed using cutadapt (Martin, 2011). Sequences were quality trimmed, and merged using the DADA2 package (Callahan et al., 2016) in R (Team, 2020). Chimeras were identified and removed using the consensus method in DADA2. Amplicon sequence variants (ASVs) were generated in DADA2. Only sequences between 247–261 bp were kept, based on the expected sizes of the fragments as well as the size distribution of sequences. The LULU package (Froslev et al., 2017) was used with default parameters to further denoise the ITS1 sequences (n = 1,283 ASVs before LULU curation), given that the ITS1 region is known to have high rates of evolution across Labyrinthula species (Martin et al., 2016). The LULU curated ASVs were identified using the Basic Local Alignment Search Tool (BLAST) (Madden, 2002) within Geneious Primer (San Diego, California) against the NCBI nucleotide (nt/nr) database. One ASV was not identified as Labyrinthula, so it was removed from the dataset.
To refine the taxonomy further, the LULU curated ASVs were aligned with sequences from Martin et al. (2016) using the Clustal Omega plugin (Sievers et al., 2011) with default parameters in Geneious Prime. We used the online Gblocks server v0.91.1 (https://ngphylogeny.fr) with the default parameters and allowing half the gaps. For both alignments, we used the online server for the IQTree Model Finder (Nguyen et al., 2015) to determine the best substitution model for the alignment based on Akaike Information Criterion corrected (AICc) values using the appropriate number of available substitution models for Bayesian analyses. We used MrBayes 3.2.6 plug-in (Huelsenbeck and Ronquist, 2001) with the following parameters GTR+I+G. Labyrinthula species-level designation was assigned based on the clade the sequences were recovered from in the Bayesian tree (Figure 1). With taxonomy assigned to the ASVs, the prevalence method in the decontam package (Davis et al., 2018) was used to explore potential contaminant ASVs from the negative controls. All ASVs were kept as they were at least one, and often additional, orders of magnitude more abundant in samples compared to controls.
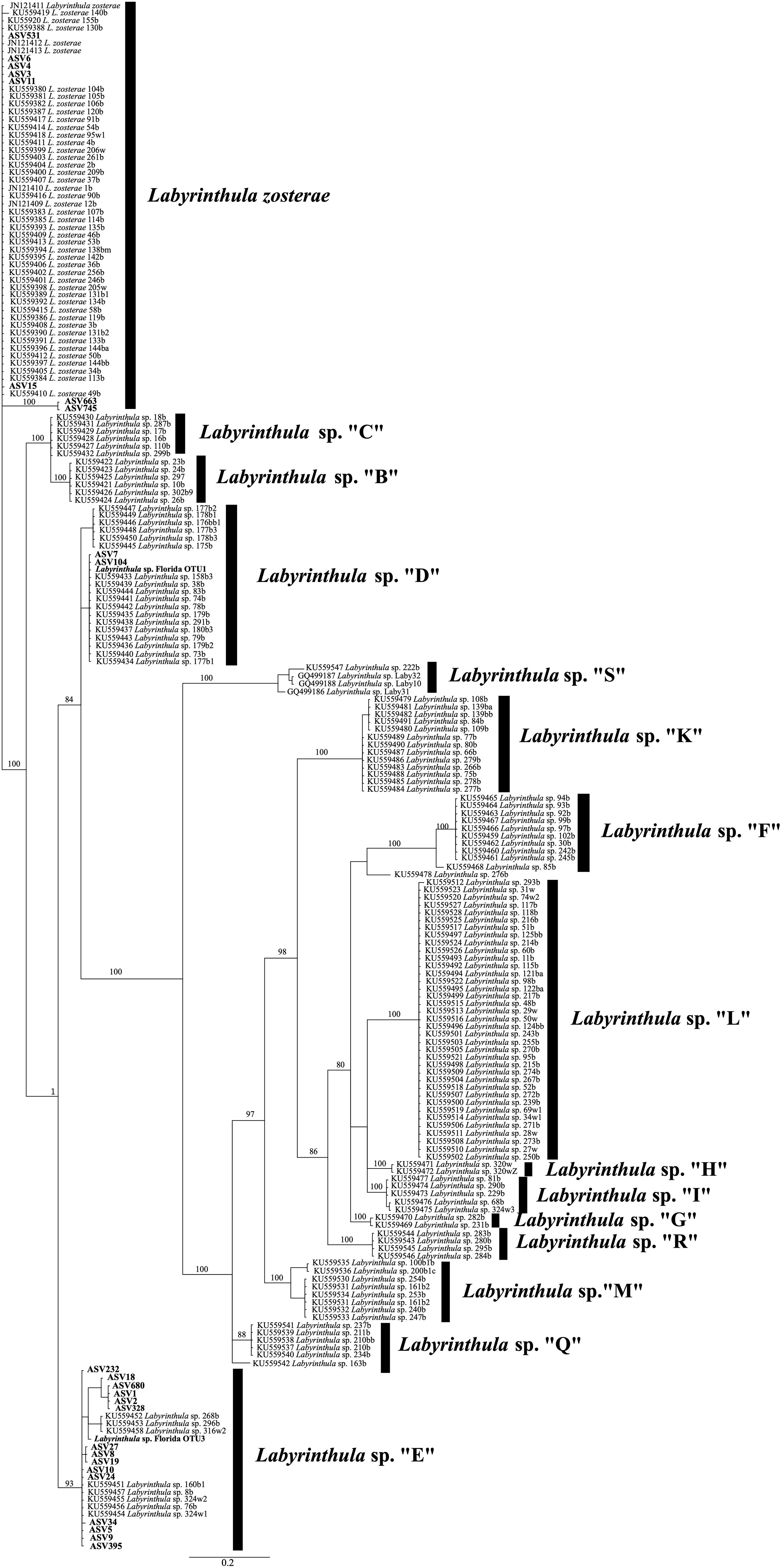
Figure 1. Phylogram for Labyrinthula spp. constructed using ASVs generated from ITS1 sequences from in this study (in bold) and GenBank sequences. The GTR+I+G substitution model for the Bayesian analysis was used to compute this topology, and posterior probabilities (>90) are included at the nodes. See text for additional details.
Alpha diversity metrics (Chao 1 and Shannon) and taxa barplots were calculated in the phyloseq package (McMurdie and Holmes, 2013) in R. Principal Coordinate Analysis (PCoA) was conducted in the phyloseq package, using the Jaccard distance method. To assess if Labyrinthula communities were significantly different across seagrass host species or geographic regions, we used the vegan package (Okasanen et al., 2014) in R to create a distance matrix and conduct a permutational analysis of variance (PERMANOVA) using the Jaccard similarity index and 1,000 permutations. We examined the beta dispersion significance to determine the influence of the spread of objects from the centroid. Also within the vegan package, we calculated which ASVs were significantly contributing to the ordination analysis using the envfit function and 1,000 permutations. For all statistical analyses, we use an alpha of 0.05 to determine significance. Rarefaction and completeness curves for ASVs were generated using the iNEXT package with 1,000 bootstraps and an endpoint of 100 (Hsieh et al., 2016). To create the ASV network, the ASVs generated in this study were aligned using the Clustal Omega plugin (Sievers et al., 2011) with default parameters in Geneious Prime and terminal gaps were filled in with N bases for those sequences that were shorter than the longest sequence. ASV networks were generated using the TCS algorithm (Clement et al., 2000) in PopArt (Leigh et al., 2015).
In total, we analyzed 189 seagrass blades from 16 locations across five US and one Mexican state (Table 1). Labyrinthula spp. were not detected at two locations: Landscape Lab in Shinnecock Bay, New York and Estero Punta Banda in the Punta Banda Estuary, Mexico. Where Labyrinthula spp. were detected, PCR prevalence from targeted collection of potentially diseased leaves ranged from 20% (SMS Dock & Big Island, Florida; Tiona Beach, New York) to 100% (Fleming Key, Florida). In general, PCR prevalence was lowest in sites in Virginia and New York, while highest in sites in Florida (Florida Keys and Dry Tortugas), Massachusetts, and central California.
We successfully amplified and sequenced the ITS1 region from 115 blades. In total, 14,206,358 raw reads were generated, which was reduced to 10,936,646 reads after initial filtering, merging, and chimera removal. With the removal of 1) negative control samples, and 2) sequences not identified as Labyrinthula, 10,936,628 sequences remained, which after curation through LULU yielded 25 unique Labyrinthula ASVs.
Our phylogenetic analyses indicate that these 25 unique ASVs fell into three clades (Figure 1). Eight ASVs were assigned to a clade indicating these are strains of Labyrinthula zosterae. Two ASVs were identified as Labyrinthula sp. “D”, while 15 ASVs were identified as Labyrinthula sp. “E”. All three of these species of Labyrinthula were previously identified as pathogenic (Martin et al., 2016). Labyrinthula zosterae had the highest relative abundance in every location except in Florida, with 97.8% of sequences assigned to this species detected from Z. marina from locations outside of Florida (Figure 2A; Table 1). Both Labyrinthula sp. “E” (99.8%, n=211,545 of 211,919 reads) and “D” (99.8%, n= 6,597,883 of 6,605,370 reads) had the most reads recovered in Florida (Figure 2A). This coincides with locations where Thalassia testudinum was sampled (Table 1).
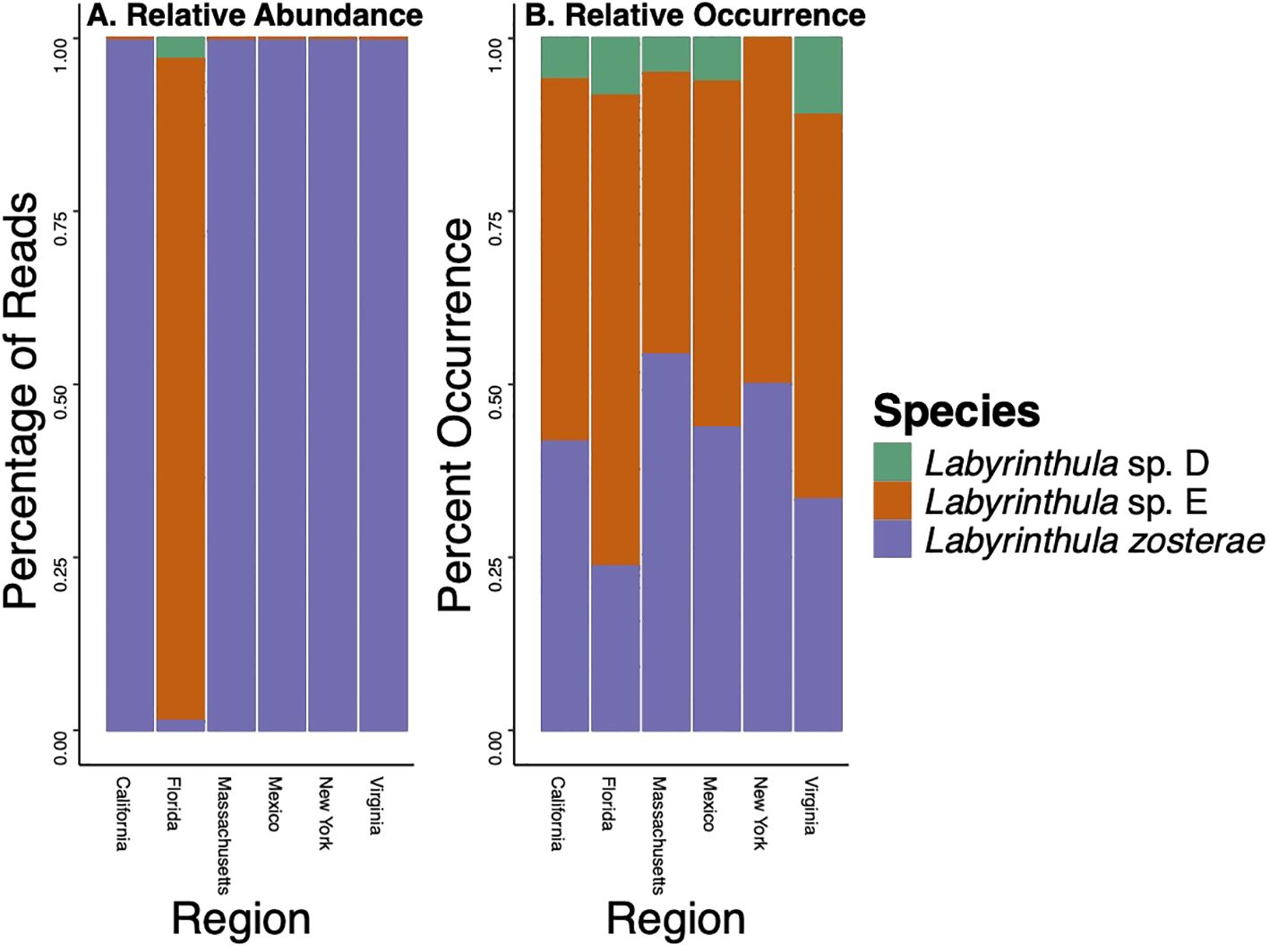
Figure 2. Relative abundance (A, defined as the percentage of sequences obtained) and relative occurrence (B, defined as the percentage of blades each species was detected from) of each Labyrinthula species detected across the US states or country from which the seagrass host was sampled. Note that only Thalassia testudinum was collected in Florida and only Zostera marina was collected in all other locations.
Narrowing down to the strain level, of the 25 ASVs detected, 13 ASVs were detected in only a single location. Of the 12 ASVs detected in multiple locations (Figure 3), for 7 of those ASVs (i.e., 15, 7, 1, 19, 27, 34, 9), 99% of the sequences were detected in a single location. While Labyrinthula sp. “E” had the highest genetic diversity detected (Figure 1; Figure 3), 87% of the ASVs (n = 13) were primarily detected in Florida waters (with >99% of sequences of these ASVs detected only in Florida). For the other two ASVs identified as Labyrinthula sp. “E”, most of the sequences (61-81%) were detected from Z. marina in California. Labyrinthula zosterae had the second highest genetic diversity detected (Figures 1, 3), with the most variability in the distribution of ASVs. Two ASVs were detected only from T. testudinum in Florida (ASV531 and ASV11 in Man of War Harbor), while two others were detected only from Z. marina in California (ASV663 in San Francisco Bay, ASV745 in Tomales Bay). For the other four ASVs, two were most abundant in Massachusetts, one in California, and one in New York (Figure 3). The ASVs most frequently detected were ASV1 (n = 79 detections), ASV4 (n = 49), ASV3 (n = 40), and ASV9 (n = 30). The Chao 1 and Shannon alpha diversity estimates show similar overall patterns, with the highest ASV richness estimates found in Florida and Massachusetts (Supplementary Figure 1). While the relative abundance of these ASVs shows much higher abundance in few locations, the ASV network (Figure 4) shows how these ASVs occur across geographic areas. The ASV network shows 12 haplotypes, as those ASVs with minor differences (e.g., sequence length, gaps) were collapsed for this analysis (Figure 4). Thus, though one Labyrinthula species and some strains appear to dominate within each state sampled based on relative abundance, all species occurred in all states sampled with a single exception, Labyrinthula sp. “D” was not detected on Z. marina from NY (Figures 2B, 4). Additionally, the haplotype network shows two ASVs (one from Labyrinthula sp. “E” and one from Labyrinthula zosterae) that occurred in all six regions, indicating high genetic connectivity, though their relative abundance differs across these areas (Figure 3).
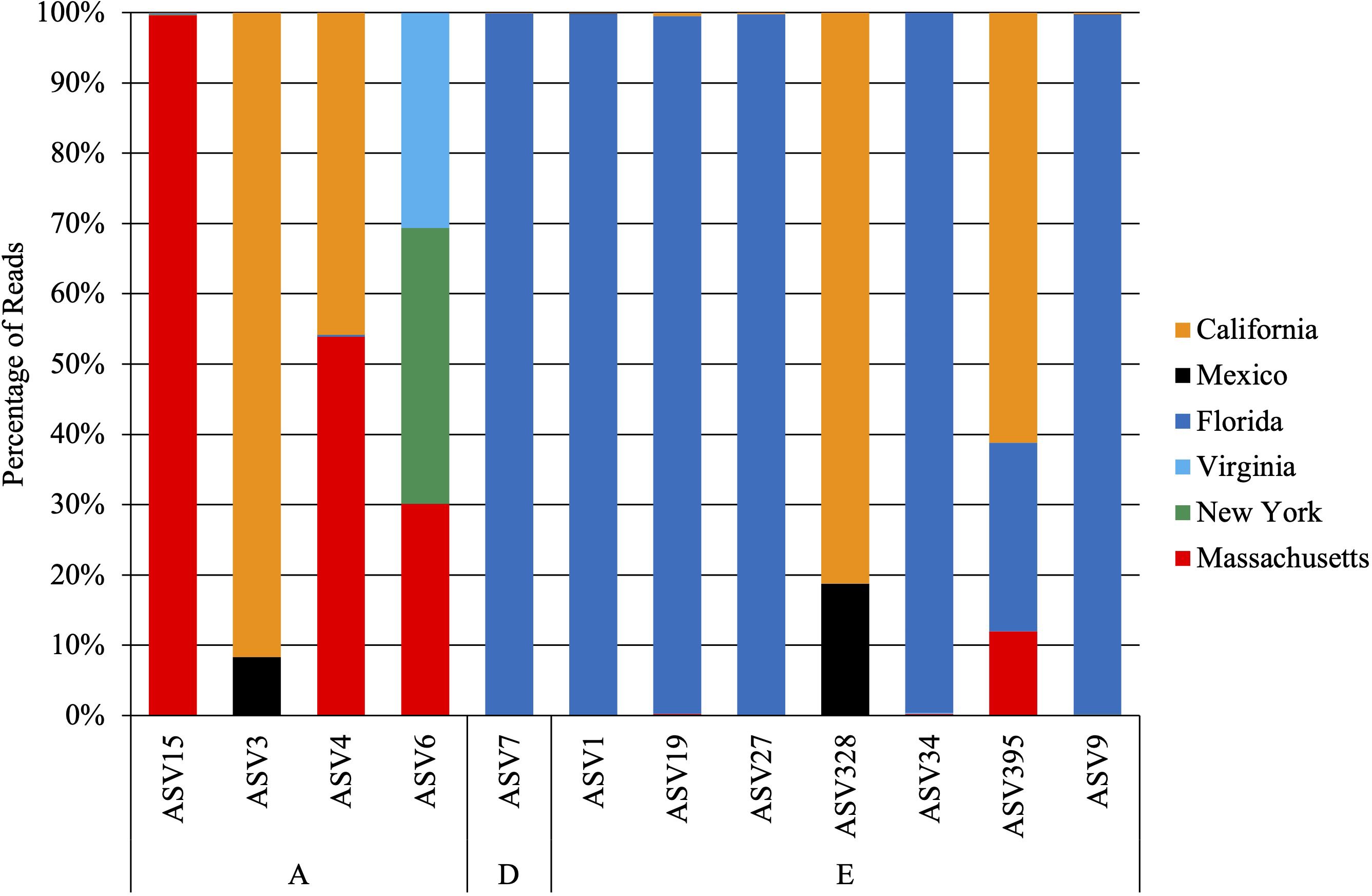
Figure 3. Percentage of reads detected across geographic locations from the 12 ASVs that were detected in more than one location. Note that 11 ASVs were detected only in Florida and 2 were detected only in California.
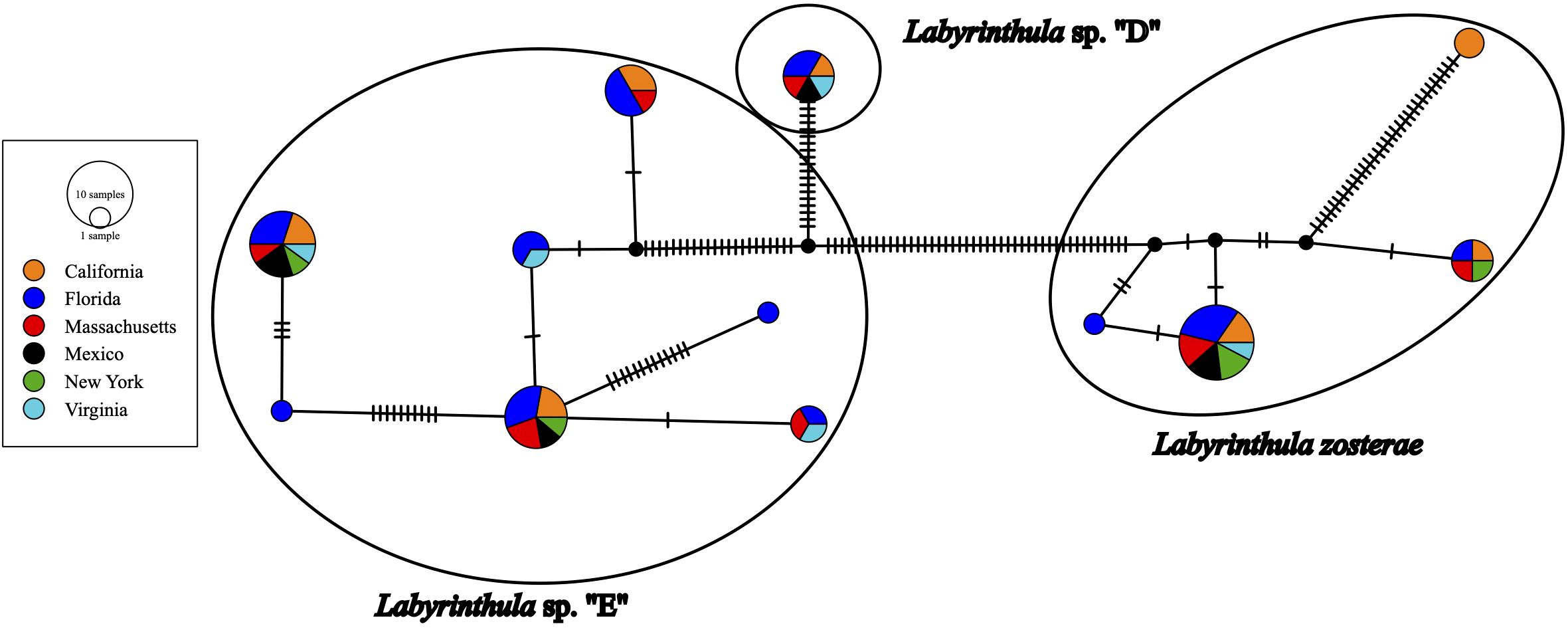
Figure 4. Network analysis for Labyrinthula ASVs generated with the PopArt software using sequences generated in this study. Colors indicate locations. Each cross-bar indicates a single nucleotide polymorphism or gap. The proportion of color in pie charts with >1 sequence indicates the proportion of unique sequences.
Community composition of Labyrinthula ASVs appears driven by a combination of host species and sampling region (Figure 5). Communities of Labyrinthula from some blades of Florida T. testudinum appear to separate from all other blades along Axis 1, while communities from other T. testudinum blades group with communities on Z. marina blades collected from all other North American locations. Upon examining the ASV scores, four ASVs were identified as significantly contributing to the ordination results (ASV1, ASV4, ASV3, ASV2; p<0.01). Despite the apparent delineation of these communities from the Florida blades along Axis 1, further analyses exploring the Labyrinthula communities on the blades collected in Florida show that these communities do not consistently group by sampling location in Florida, as blades collected from Key West and the Indian River Lagoon spread across Axis 1, while those from Bush Key are more similar and only spread across Axis 2 (Supplementary Figure 2). Communities of Labyrinthula from Z. marina blades vary across region and coast primarily along Axis 2, with communities from some locations being highly similar (e.g., Virginia, Massachusetts) while others are less similar (e.g., Mexico, California) (Figure 4, Supplementary Figure 2). Though three different Bays were sampled in California, the Labyrinthula communities on the blades show lots of overlap, particularly communities on blades from Tomales and Bodega Bays, with there being more differentiation between blades from these Bays and those collected in San Francisco Bay (Supplementary Figure 2). The variation in Labyrinthula ASVs across seagrass host species (PERMANOVA: df = 1, sum of squares = 5.3, Fmodel = 18.98, R2 = 0.14, p = 0.001) and across geographic regions (PERMANOVA: df = 5, sum of squares = 9.8, Fmodel = 7.8, R2 = 0.26, p = 0.001) were significantly different, with these two parameters explaining ~58% of the variation. Dispersion was significant for host species (Fspecies = 24.48, pspecies <0.0001) and geographic regions (Fregion = 18.4, pregion < 0.0001).
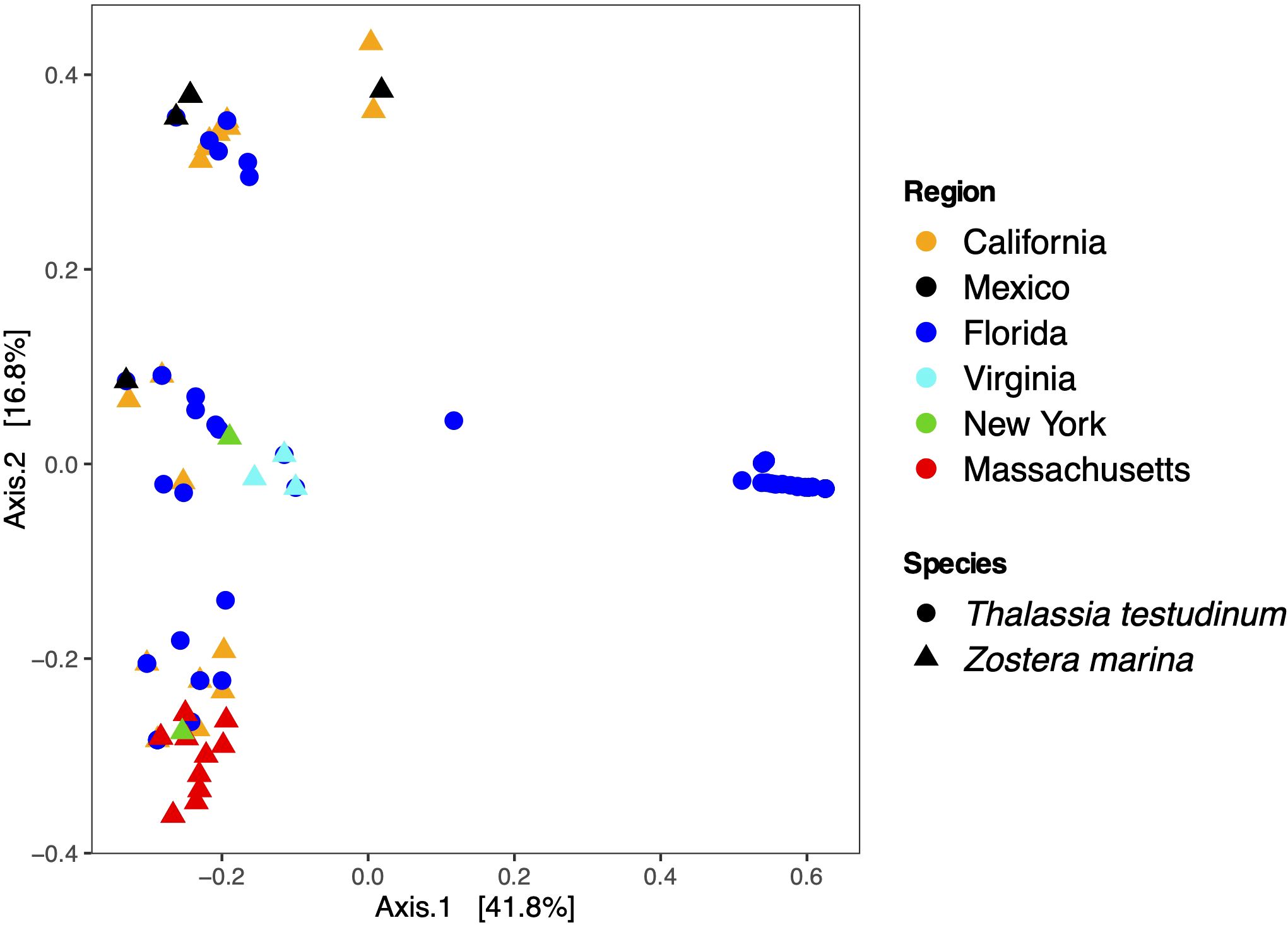
Figure 5. Principal Coordinate Analysis (PCoA) of Labyrinthula communities recovered from seagrass blades. The regions from which the ASVs were detected are noted by color while the seagrass hosts collected are distinguished by shape.
Rarefaction curves generated to assess the limits of the current sampling indicate that none of the sampling in any region was sufficient to detect all the ASVs likely present (Figure 6A). While some of the curves appear close to an asymptote (e.g., New York), the small sample sizes collected in each region require interpreting these curves with caution. Upon extrapolating these curves, most regions require collection of >90 blades to fully assess the ASV diversity within the region (Figure 6B).
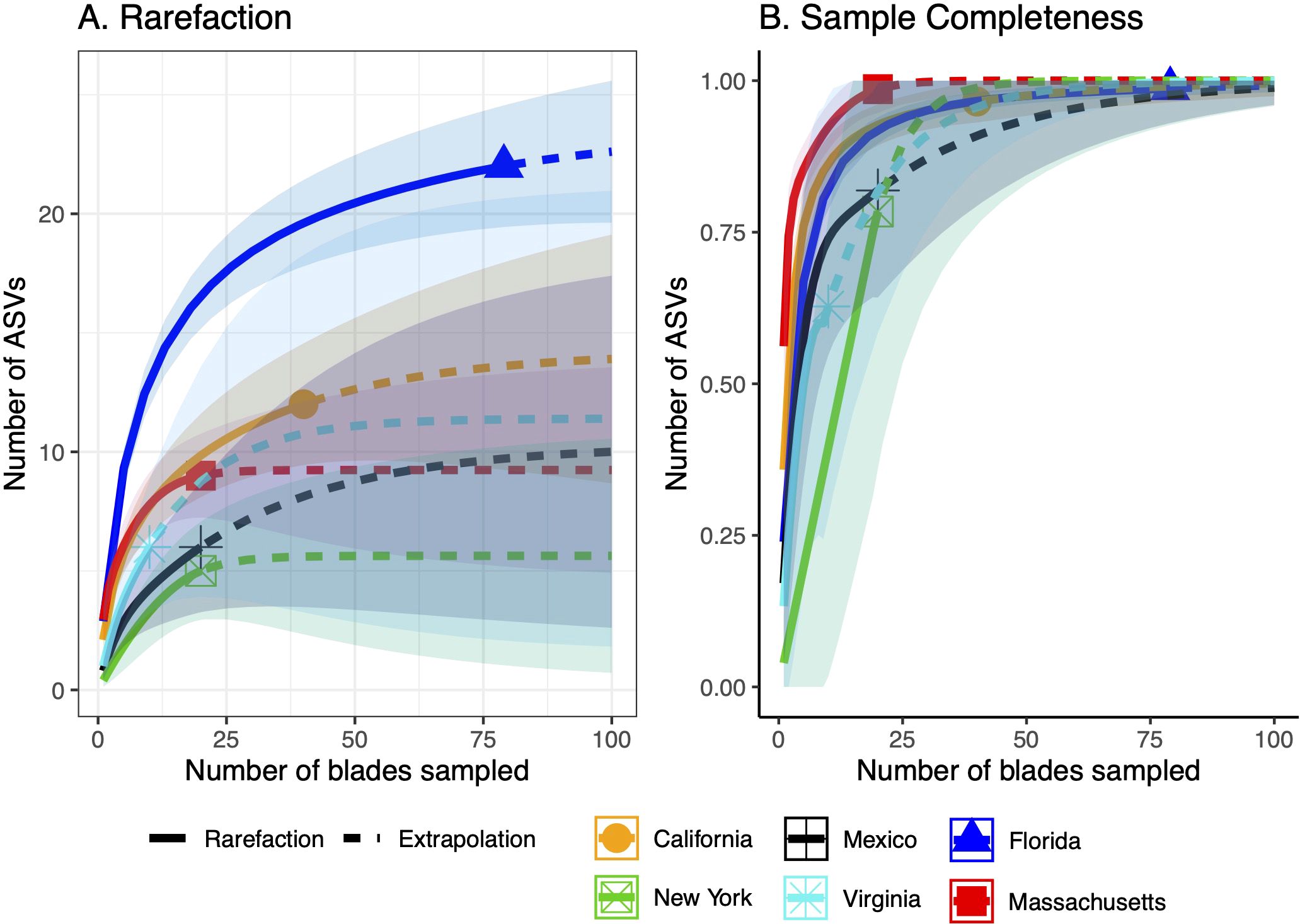
Figure 6. Rarefaction (A) and sample completeness (B) curves with extrapolation (dashed lines) to assess the number of ASVs detected and predicted based on the number of seagrass blades sampled. The color surrounding the lines is the 95% standard error.
This is the first North American wide assessment of the phylogeography of Labyrinthula species and strains. We detected three species of Labyrinthula, represented by 25 ASVs (or strains) at 14 of 16 sites sampled across North America from two seagrass hosts. We detected the highest strain level variation in Florida, which is likely due to larger sample sizes from Florida compared to other sampling locations, but could be a result of higher parasite richness in tropical locations. All three species detected were previously identified as pathogenic, indicating that Labyrinthula species capable of causing wasting disease exist across the regions where seagrasses were sampled for this study. While assessing the genetic connectivity among regions was limited by sample sizes, some strains and species appeared more geographically isolated than others, likely due to limited connectivity or rarity of certain strains across sites.
Since the massive epizootic in the Atlantic Ocean during the 1930s (Renn, 1936), smaller-scale epizootics of SWD have continued to occur around the globe, impacting many seagrass species (Sullivan et al., 2013). Critical knowledge gaps remain regarding the underlying mechanisms that resulted in such a widespread and devastating epizootic and whether something similar could occur in the future (Sullivan et al., 2018). Much research has explored the various factors influencing the timing and severity of SWD epizootics. Currently, host demography (Groner et al., 2014), dosage and strain of parasite (Dawkins et al., 2018), higher temperature (Bishop et al., 2017; Groner et al., 2021; Aoki et al., 2022), salinity (Trevathan et al., 2011; Bishop et al., 2017), hypoxia and sulfide (Bishop et al., 2017), host immunity (Duffin et al., 2020; Schenck et al., 2023), and parasite diversity (Pagenkopp Lohan et al., 2020; Schenck et al., 2023) all appear to be important factors to consider for this disease, though some of these factors are more thoroughly studied than others, particularly across geographic locations and host species. Most of these studies used either a single strain (Trevathan et al., 2011; Bishop et al., 2017) or did not assess if the disease examined was caused by one or more species or strains of the parasite (Groner et al., 2014; Duffin et al., 2020; Groner et al., 2021). Thus, critical knowledge gaps for understanding this pathosystem include the lack of information about the genetic diversity, phylogeography, and connectivity of these parasites so that the potential variation in disease severity within and across hosts, as well as how these vary across different locations, can be examined.
We detected pathogenic species of Labyrinthula in every region and in most sites where seagrasses were sampled, indicating that these parasites are geographically widespread. Our analyses demonstrate that the richness of ASVs detected was significantly correlated with the number of blades tested (R2 = 0.62, F = 19.8, p = 0.0007; Supplementary Figure 3). Additionally, our rarefaction and network analyses indicate that our data likely underestimate the richness of pathogenic Labyrinthula strains infecting Z. marina and T. testudinum across their ranges. Though these parasites are present, major epizootics have not occurred in many of these locations in decades, or are not known to ever have occurred. Rather, some seagrass beds have experienced localized, smaller scale disease events (Trevathan-Tackett et al., 2018; Duffin et al., 2021; Groner et al., 2021), or lesions are present without evidence of damage to the beds. While many environmental factors are known to impact the severity of seagrass wasting disease, experimental work has shown that many environmental conditions appear to favor the seagrass host while negatively impacting the parasite. For example, Trevathan et al. (2011) demonstrated that hypersaline conditions had little impact on T. testudinum, but negatively impacted the growth of the parasite. While multi-stressor experiments by Bishop et al. (2017) showed that environmental stressors can more negatively impact the parasite than the seagrass host particularly under ambient conditions, Schenck et al. (2023) and DuBois et al. (2020) found that temperature enhanced disease severity. Thus, how biotic and abiotic factors combine to create disease and potentially lead to future epizootics appear context dependent, making understanding how these factors vary across the diversity of species and strains of Labyrinthula even more critical to understanding this pathosystem.
The host specificity of Labyrinthula spp. has been explored previously and, like the data presented here, prior studies indicated that these parasites are host generalists. For example, Martin et al. (2016) was able to experimentally inoculate and cause disease in seagrasses of the same species from different geographic locations as well as different seagrass species from different geographic locations from where Labyrinthula isolates were obtained, indicating that at least some strains can infect multiple genera of seagrass hosts. Trevathan-Tackett et al. (2018) similarly experimentally inoculated and caused disease in two different seagrass hosts (Zostera muelleri and Heterozostera nigricaulis). Additionally, Garcias-Bonet et al. (2011) experimentally demonstrated that different strains and species of the parasite can infect seagrass hosts across different genera. Similar to Trevizan Segovia et al. (2021), we report high connectivity within strains and species, but low overall species diversity of Labyrinthula, which is consistent with a parasite generalist. Additionally, though Z. marina and T. testudinum are ecologically and evolutionarily distinct seagrass species (Larkum et al., 2006), many Labyrinthula ASVs were detected from both seagrass hosts. Despite this high connectivity, one Labyrinthula species dominated in relative abundance within each region sampled, further indicating that abiotic factors may be differentially favorable to Labyrinthula strains.
In this study, we used a targeted metabarcode approach to assess the intra- and inter-specific diversity of pathogenic Labyrinthula species infecting seagrass blades. Labyrinthula spp. lack defining morphological characteristics, so previous attempts to define species turned to a phylogenetic approach to tease apart suitable species level delineations (Martin et al., 2016). We chose to use the same approach here, which has limitations given the lack of a suitable outgroup resulting in an odd tree topology creating a polytomy for L. zosterae sequences. More detailed analyses are warranted to truly delineate species across this genus, perhaps even using genome-scale analyses (Tan et al., 2021). At the intra-specific level, there was a relatively high potential for cross contamination, both in the processing of the blades in the lab and in the library preparation steps. Thus, to mitigate any impacts of this potential contamination, we took several approaches. First, seagrass blades were collected and initially processed (e.g., lesions were removed from blades) primarily by the labs who collected them, making cross-contamination from blades collected from disparate regions extremely unlikely. Next, we used strict laboratory methods to reduce contamination, such as using spin-column extractions and strip tubes with lids, only opening one sample at a time, and working in separate pre- and post-PCR spaces. We also utilized dual-indexing of our amplicons, which can also decrease the likelihood of cross-contamination (MacConaill et al., 2018). Then, after our initial processing with DADA2, we ended up with 1,283 ASVs. As it was virtually impossible for us to determine if these ASVs resulted from sequencing error or from the hypervariable ITS1 region that was amplified, we included a second denoising step, utilizing the LULU package to compare sequence similarity and occurrence patterns to remove likely erroneous sequences. Given these precautions, we have confidence that our results showing high connectivity and strain and species dominance across geographic locations are unlikely to be solely attributable to sequencing error or cross-contamination.
We present a broad-scale phylogeographic examination of Labyrinthula strain and species diversity across North America, providing additional evidence regarding the host generality of this parasite. Yet many questions remain, and future research is needed to examine how the pathogenicity of the Labyrinthula species found here vary across these two seagrass hosts. Additionally, future research will need to explore if different strains or species of Labyrinthula respond differently in terms of pathogenicity or prevalence under different environmental conditions. Finally, it is unclear how Labyrinthula strains and species could be spread across these vast geographic distances, though it is possible that maritime shipping, and specifically ballast water, could play a role in the long-distance dispersal of this parasite. Previous studies have detected labyrinthulids in ships’ ballast water (Pagenkopp Lohan et al., 2016, 2022), yet it is unknown whether the parasite survives and is infective following discharge.
The raw sequence data presented in the study are publicly available. This data can be found here: Sequence Read Archive under Bioproject PRJNA1180629.
KP: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. RD: Data curation, Investigation, Writing – original draft, Writing – review & editing. DM: Methodology, Visualization, Writing – original draft, Writing – review & editing. AH: Investigation, Methodology, Writing – original draft, Writing – review & editing. BP: Investigation, Writing – original draft, Writing – review & editing. KB: Investigation, Writing – original draft, Writing – review & editing. JS: Investigation, Writing – original draft, Writing – review & editing. PJ: Investigation, Writing – original draft, Writing – review & editing. GR: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. CR: Conceptualization, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded through a Grand Challenges grant from the Smithsonian Institution to KP and GR and a National Science Foundation OCE 1652320 to AH. This research was made possible in part by support from the Florida Institute of Oceanography to CR.
We would like to thank the following individuals who contributed to the collection and processing of specimens for this study including Diana Chin and Tim Palmer from SUNY Stony Brook University, Forest Schenck from Northeastern University, Stephanie Kiriakopolos from San Francisco State University, Joshua Chow from the University of California at Davis, Clara M. Hereu from the Universidad Autónoma de Baja California, and Shelby Ziegler from the Virginia Institute of Marine Science.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1463968/full#supplementary-material
Adl S. M., Simpson A. G., Lane C. E., Lukes J., Bass D., Bowser S. S., et al. (2012). The revised classification of eukaryotes. J. Eukaryot Microbiol. 59, 429–493. doi: 10.1111/j.1550-7408.2012.00644.x
Aoki L. R., Rappazzo B., Beatty D. S., Domke L. K., Eckert G. L., Eisenlord M. E., et al. (2022). Disease surveillance by artificial intelligence links eelgrass wasting disease to ocean warming across latitudes. Limnology Oceanography 67, 1577–1589. doi: 10.1002/lno.12152
Bigelow D., Olsen M., Gilbertson R. (2005). Labyrinthula terrestris sp. nov., a new pathogen of turf grass. Mycologia 97, 185–190. doi: 10.1080/15572536.2006.11832852
Bishop N., Martin D. L., Ross C. (2017). Effects of multi-stress exposure on the infection dynamics of a Labyrinthula sp.-turtle grass pathosystem. Mar. Ecol. Prog. Ser. 581, 119–133. doi: 10.3354/meps12318
Burdick D. M., Short F., Wolf J. (1993). An index to assess and monitor the progression of wasting disease in eelgrass Zostera marina. Mar. Ecol. Prog. Ser. 94, 83–90. doi: 10.3354/meps094083
Burge C. A., Kim C. J., Lyles J. M., Harvell C. D. (2013). Special issue Oceans and Humans Health: the ecology of marine opportunists. Microb. Ecol. 65, 869–879. doi: 10.1007/s00248-013-0190-7
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chitrampalam P., Goldberg N., Olsen M. W. (2015). Labyrinthula species associated with turfgrasses in Arizona and New Mexico. Eur. J. Plant Pathol. 143, 485–493. doi: 10.1007/s10658-015-0701-0
Clement M., Posada D., Crandall K. A. (2000). TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x
Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226. doi: 10.1186/s40168-018-0605-2
Dawkins P. D., Eisenlord M. E., Yoshioka R. M., Fiorenza E., Fruchter S., Giammona F., et al. (2018). Environment, dosage, and pathogen isolate moderate virulence in eelgrass wasting disease. Dis. Aquat Organ 130, 51–63. doi: 10.3354/dao03263
Douhan G. W., Olsen M. W., Herrell A., Winder C., Wong F., Entwistle K. (2009). Genetic diversity of Labyrinthula terrestris, a newly emergent plant pathogen, and the discovery of new Labyrinthulid organisms. Mycol Res. 113, 1192–1199. doi: 10.1016/j.mycres.2009.08.002
DuBois K., Williams S. L., Stachowicz J. J. (2020). Experimental warming enhances effects of eelgrass genetic diversity via temperature-induced niche differentiation. Estuaries Coasts 44, 545–557. doi: 10.1007/s12237-020-00827-9
Duffin P., Martin D. L., Furman B. T., Ross C. (2021). Spatial Patterns of Thalassia testudinum Immune Status and Labyrinthula spp. Load Implicate Environmental Quality and History as Modulators of Defense Strategies and Wasting Disease in Florida Bay, United States. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.612947
Duffin P., Martin D. L., Pagenkopp Lohan K. M., Ross C. (2020). Integrating host immune status, Labyrinthula spp. load and environmental stress in a seagrass pathosystem: Assessing immune markers and scope of a new qPCR primer set. PloS One 15, e0230108. doi: 10.1371/journal.pone.0230108
FFWCC, F.F.a.W.C.C (2021). Manatee mortality event along the east coast: 2020-2021 (Tallahassee, FL: Florida Fish and Wildlife Conservation Commission). Available online at: https://myfwc.com/research/manatee/rescue-mortality-response/ume/ (Accessed July 9 2021).
Fourqurean J. W., Duarte C. M., Kennedy H., Marbà N., Holmer M., Mateo M. A., et al. (2012). Seagrass ecosystems as a globally significant carbon stock. Nat. Geosci. 5, 505–509. doi: 10.1038/ngeo1477
Froslev T. G., Kjoller R., Bruun H. H., Ejrnaes R., Brunbjerg A. K., Pietroni C., et al. (2017). Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1188. doi: 10.1038/s41467-017-01312-x
Garcias-Bonet N., Sherman T. D., Duarte C. M., Marbà N. (2011). Distribution and Pathogenicity of the Protist Labyrinthula sp. in western Mediterranean Seagrass Meadows. Estuaries Coasts 34, 1161–1168. doi: 10.1007/s12237-011-9416-4
Groner M. L., Burge C. A., Couch C. S., Kim C. J., Siegmund G. F., Singhal S., et al. (2014). Host demography influences the prevalence and severity of eelgrass wasting disease. Dis. Aquat Organ 108, 165–175. doi: 10.3354/dao02709
Groner M. L., Eisenlord M. E., Yoshioka R. M., Fiorenza E. A., Dawkins P. D., Graham O. J., et al. (2021). Warming sea surface temperatures fuel summer epidemics of eelgrass wasting disease. Mar. Ecol. Prog. Ser. 679, 47–58. doi: 10.3354/meps13902
Handley L., Altsman D., DeMay R. (2007). “Seagrass status and trends in the Northern Gulf of Mexico: 1940-2002,” in U.S. Geological survey scientific investigations report 2006-5287. Reston, VA: U.S. Geological Survey.
Hsieh T. C., Ma K. H., Chao A., McInerny G. (2016). iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456. doi: 10.1111/2041-210x.12613
Huelsenbeck J. P., Ronquist F. (2001). MRBAYES : Bayesian inference of phylogenetic trees. Bioinf. Appl. Notes 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Hughes A. R., Williams S. L., Duarte C. M., Heck K. L., Waycott M. (2009). Associations of concern: declining seagrasses and threatened dependent species. Front. Ecol. Environ. 7, 242–246. doi: 10.1890/080041
Larkum A. W. D., Orth R. J., Duarte C. M. (2006). Seagrasses: biology, ecology, and conservation. (The Netherlands: Springer).
Leander C. A., Porter D. (2011). The Labyrinthulomycota is compared is three distinct lineages. Mycologia 93, 459–464. doi: 10.1080/00275514.2001.12063179
Leigh J. W., Bryant D., Nakagawa S. (2015). popart: full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210x.12410
MacConaill L. E., Burns R. T., Nag A., Coleman H. A., Slevin M. K., Giorda K., et al. (2018). Unique, dual-indexed sequencing adapters with UMIs effectively eliminate index cross-talk and significantly improve sensitivity of massively parallel sequencing. BMC Genomics 19, 30. doi: 10.1186/s12864-017-4428-5
Madden T. (2002). “"Chapter 16: the BLAST sequence analysis tool,",” in The NCBI handbook, vol. 13 . Eds. McEntyre J., Ostell J. (National Center for Biotechnology Information, Bethesda, MD).
Martin D. L., Chiari Y., Boone E., Sherman T. D., Ross C., Wyllie-Echeverria S., et al. (2016). Functional, phylogenetic and host-geographic signatures of labyrinthula spp. Provide for putative species delimitation and a global-scale view of seagrass wasting disease. Estuaries Coasts 39, 1403–1421. doi: 10.1007/s12237-016-0087-z
Martin M. (2011). Cutadapt removes adaptor sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
McMurdie P. J., Holmes S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Menning D. M., Gravley H. A., Cady M. N., Pepin D., Wyllie-Echeverria S., Ward D. H., et al. (2021). Metabarcoding of environmental samples suggest wide distribution of eelgrass (Zostera marina) pathogens in the north Pacific. Metabarcoding Metagenomics 5, 35–42. doi: 10.3897/mbmg.5.62823
Mtwana Nordlund L., Koch E. W., Barbier E. B., Creed J. C. (2016). Seagrass ecosystem services and their variability across genera and geographical regions. PloS One 11, e0163091. doi: 10.1371/journal.pone.0163091
Muehlstein L. K., Porter D., Short F. (1991). Labyrinthula zosterae sp. nov., the causative agent of wasting disease of eelgrass, Zostera marina. Mycologia 83, 180–191. doi: 10.1080/00275514.1991.12025994
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Oksanen J., Kindt R., Legendre P., O’Hara B., Stevens M. H. H., Oksanen M. J., et al. (2007). The vegan package. Community ecology package. 10, 719. Available online at: https://www.researchgate.net/profile/Gavin-Simpson-2/publication/228339454_The_vegan_Package/links/0912f50be86bc29a7f000000/The-vegan-Package.pdf.
Pagenkopp Lohan K. M., Darling J. A., Ruiz G. M. (2022). International shipping as a potent vector for spreading marine parasites. Diversity Distributions 28, 1922–1933. doi: 10.1111/ddi.13592
Pagenkopp Lohan K. M., DiMaria R., Martin D. L., Ross C., Ruiz G. M. (2020). Diversity and microhabitat associations of Labyrinthula spp. in the Indian River Lagoon System. Dis. Aquat Organ 137, 145–157. doi: 10.3354/dao03431
Pagenkopp Lohan K. M., Fleischer R. C., Carney K. J., Holzer K. K., Ruiz G. M. (2016). Amplicon-based pyrosequencing reveals high diversity of protistan parasites in ships' Ballast water: implications for biogeography and infectious diseases. Microb. Ecol. 71, 530–542. doi: 10.1007/s00248-015-0684-6
Pan J., Del Campo J., Keeling P. J. (2017). Reference tree and environmental sequence diversity of labyrinthulomycetes. J. Eukaryot Microbiol. 64, 88–96. doi: 10.1111/jeu.12342
Popova O. V., Belevich T. A., Golyshev S. A., Kireev I. I., Aleoshin V. V. (2020). Labyrinthula diatomea n. sp.-A Labyrinthulid Associated with Marine Diatoms. J. Eukaryot Microbiol. 67, 393–402. doi: 10.1111/jeu.12789
Raghukumar S., Damare V. S. (2011). Increasing evidence for the important role of Labyrinthulomycetes in marine ecosystems. Botanica Marina 54, 3–11. doi: 10.1515/bot.2011.008
Renn C. E. (1936). Persistence of the eel-grass disease and parasite on the american atlantic coast. Nature 138, 507–508. doi: 10.1038/138507a0
Schenck F. R., DuBois K., Kardish M. R., Stachowicz J. J., Hughes A. R. (2023). The effect of warming on seagrass wasting disease depends on host genotypic identity and diversity. Ecology 104, e3959. doi: 10.1002/ecy.3959
Short F., Muehlstein L. K., Porter D. (1987). Eelgrass wasting disease: Cause and Recurrence of a Marine Epidemic. Biol. Bull. 173, 557–562. doi: 10.2307/1541701
Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Sullivan B. K., Robinson K. L., Trevathan-Tackett S. M., Lilje E. S., Gleason F. H., Lilje O. (2017). The first isolation and characterisation of the protist labyrinthula sp. in southeastern Australia. J. Eukaryot Microbiol. 64, 504–513. doi: 10.1111/jeu.12387
Sullivan B. K., Sherman T. D., Damare V. S., Lilje O., Gleason F. H. (2013). Potential roles of Labyrinthula spp. in global seagrass population declines. Fungal Ecol. 6, 328–338. doi: 10.1016/j.funeco.2013.06.004
Sullivan B. K., Trevathan-Tackett S. M., Neuhauser S., Govers L. L. (2018). Review: Host-pathogen dynamics of seagrass diseases under future global change. Mar. pollut. Bull. 134, 75–88. doi: 10.1016/j.marpolbul.2017.09.030
Tan M. H., Loke S., Croft L. J., Gleason F. H., Lange L., Pilgaard B., et al. (2021). First Genome of Labyrinthula sp., an Opportunistic Seagrass Pathogen, Reveals Novel Insight into Marine Protist Phylogeny, Ecology and CAZyme Cell-Wall Degradation. Microb. Ecol. 82, 498–511. doi: 10.1007/s00248-020-01647-x
Team R. C. (2020). R: Foundation for statistical computing (Vienna, Austria: R Foundation for Statistical Computing).
Trevathan S. M., Kahn A., Ross C. (2011). Effects of short-term hypersalinity exposure on the susceptibility to wasting disease in the subtropical seagrass Thalassia testudinum. Plant Physiol. Biochem. 49, 1051–1058. doi: 10.1016/j.plaphy.2011.06.006
Trevathan-Tackett S. M., Lauer N., Loucks K., Rossi A. M., Ross C. (2013). Assessing the relationship between seagrass health and habitat quality with wasting disease prevalence in the Florida Keys. J. Exp. Mar. Biol. Ecol. 449, 221–229. doi: 10.1016/j.jembe.2013.10.004
Trevathan-Tackett S. M., Sullivan B. K., Robinson K., Lilje O., Macreadie P. I., Gleason F. H. (2018). Pathogenic Labyrinthula associated with Australian seagrasses: Considerations for seagrass wasting disease in the southern hemisphere. Microbiol. Res. 206, 74–81. doi: 10.1016/j.micres.2017.10.003
Trevizan Segovia B., Sanders-Smith R., Adamczyk E. M., Forbes C., Hessing-Lewis M., O'Connor M. I., et al. (2021). Microeukaryotic communities associated with the seagrass zostera marina are spatially structured. J. Eukaryot Microbiol. 68, e12827. doi: 10.1111/jeu.12827
Tsui C. K., Marshall W., Yokoyama R., Honda D., Lippmeier J. C., Craven K. D., et al. (2009). Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol. Phylogenet Evol. 50, 129–140. doi: 10.1016/j.ympev.2008.09.027
White T. J., Bruns T, Lee S., Taylor J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ eds. PCR protocols: a guide to methods and applications. (New York, NY: Academic Press), p 315–322.
Keywords: slime net, parasite, seagrass wasting disease, Labyrinthula zosterae, Zostera marina, Thalassia testudinium, eelgrass wasting disease, metabarcode
Citation: Pagenkopp Lohan KM, DiMaria R, Martin DL, Hughes AR, Peterson BJ, Boyer KE, Stachowicz JJ, Jorgensen P, Ruiz GM and Ross C (2025) Phylogeography of Labyrinthula species and strains shows high connectivity and low genetic variation across seagrass hosts and geographic locations in North America. Front. Mar. Sci. 11:1463968. doi: 10.3389/fmars.2024.1463968
Received: 12 July 2024; Accepted: 11 December 2024;
Published: 10 January 2025.
Edited by:
David William Waite, Ministry for Primary Industries, New ZealandReviewed by:
Kelly Ugarelli, University of Florida, United StatesCopyright © 2025 Pagenkopp Lohan, DiMaria, Martin, Hughes, Peterson, Boyer, Stachowicz, Jorgensen, Ruiz and Ross. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katrina M. Pagenkopp Lohan, bG9oYW5rQHNpLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.