- 1Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou, China
- 3Pearl Research Institute, Guangdong Ocean University, Zhanjiang, China
- 4Pearl Breeding and Processing Engineering Technology Research Centre of Guangdong Province, Zhanjiang, China
- 5Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Zhanjiang, China
Titanium dioxide nanoparticles (TiO2-NPs) released into the environment is becoming more prevalent due to their increased usage, marine TiO2-NPs contamination is escalating concerns in coastal areas. To understand the potential impact of TiO2-NPs on transcript changes in pearl oyster (Pinctada fucata martensii), transcriptome analysis on the gill tissues of pearl oysters was conducted after 14-day TiO2-NPs exposure and 7-day brief recovery. A total of 911 differentially expressed genes (DEGs) were identified between the control group (TC) and the experimental group (TE) exposed to 14-day TiO2-NPs. Gene ontology (GO) analyses of the DEGs demonstrated their substantial enrichments in functions related to “hydrolase activity”, “oxidoreductase activity”, and “DNA integration”. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyses of the DEGs indicated enrichment in several pathways, including “ubiquitin-mediated protein hydrolysis”, “ECM-receptor interactions”, “NOD-like receptor signaling pathway”, “Toll-like receptor”, and “FOXO signaling pathway”. This suggests that exposure to TiO2-NPs intensifies oxidative stress and apoptosis in pearls oysters, leading to negative effects such as disrupted protein homeostasis, decreased biomineralization activity, reduced neuronal excitability, weakened immune response, and reduced cellular metabolism. Transcriptome analysis identified 844 DEGs between the TE and recovery group (TR), which underwent a 7-day brief recovery period. GO analyses of the DEGs demonstrated their substantial enrichments in functions related to “DNA integration”, “obsolete oxidation-reduction process”, and “proteolysis”. KEGG pathways analyses of the DEGs indicated enrichment in several pathways, including “lysine degradation”, “glycine, serine, and threonine metabolism”, and “NOD-like receptor signaling pathway”. The findings indicated that although pearl oysters showed only slight relief after 7 days of brief recovery, they continued to experience negative effects from TiO2-NP exposure. Our findings shed light on the complex responses of pearl oysters to TiO2-NPs stress and offer valuable theoretical insights into the toxicological impact of TiO2-NPs on pearl oysters.
1 Introduction
Titanium dioxide nanoparticles (TiO2-NPs) demonstrate remarkable UV-absorbing properties, outstanding chemical stability, high opacity, and excellent resistance to fading. These unique characteristics make TiO2-NPs an essential component in numerous consumer goods, including sunblocks, cosmetics, pharmaceuticals, coatings, fabrics, polymers, paper, and wood treatments (Aitken et al., 2006). TiO2-NPs can enter environments via production, transportation, usage, and disposal stages, thereby posing risks to diverse aquatic organisms and their ecosystem (Shi et al., 2016; Carbuloni et al., 2020). High-concentration TiO2-NPs have been detected in coastal waters, with even greater accumulations observed in sediments (Garner and Keller, 2014). Based on model computations, the TiO2-NP concentration in surface waters and wastewater varies from dozens to several hundred micrograms per liter (Sun et al., 2016). Concentrations have been recorded to be 900 μg/L in the surface water near French Mediterranean beaches (Labille et al., 2020). The titanium dioxide levels were found exceeding the 1.85 ± 0.55 ng/g detection limit by analyzing the bivalve shellfish from northern China coastal cities, indicating notable accumulation (Li and Gao, 2014). Increased levels of TiO2-NPs pose significant threats to various marine organisms, leading to increased concerns about the ecological and physiological impact of TiO2-NPs. A growing body of evidence has demonstrated that TiO2-NPs can lead to various adverse effects, including reduced fertilization success (Gallo et al., 2016), decreased metabolism (Johari et al., 2013; Muller et al., 2014), delayed embryonic development (Libralato et al., 2013), impaired immune response (Ringwood et al., 2009; Rocha et al., 2014), intestinal microbiome disorders (Chen et al., 2022b; Duan et al., 2023; Li et al., 2024a), and DNA damage (Hu et al., 2017).
RNA sequencing (RNA-seq) is used to identify gene expression patterns under environmental stress. This method is crucial for examining the overall functional arrangement of genes and investigating the control mechanisms of biological metabolic pathways. In recent years, transcriptomics technology has been extensively employed in toxicological studies of aquatic organisms exposed to pollutants. By examining changes in gene expression under toxic stress, this technology can reveal the intrinsic toxicity of pollutants in aquatic organisms at the transcriptional level, facilitating the prediction and prevention of pollutant toxicity in aquatic organisms and the environments (Sharon et al., 2013; Liu et al., 2020). Banni et al. (2016) examined the impact of TiO2-NPs and 2,3,7, 8-tetrachlorodibenzo-p-dioxins on genes related to the digestive glands of Mytilus galloprovincialis using a combination of transcriptomics and immunohistochemistry. Zhou et al. (2024) used transcriptomic methods to discover that TiO2-NP exposure triggers lipid metabolism disorders, immune defense disruption, and oxidative stresses in Acipenser schrenckii. These findings underscore the effectiveness of transcriptomics in providing a comprehensive analysis of the impact of TiO2-NPs on aquatic organisms.
Pinctada fucata martensii, native to southern China, Southeast Asia, Australia, India, and Japan (Yang et al., 2019, 2023), are filter-feeding mollusks renowned for their high economic values, contributing near 90% of the world’s marine pearl production (He et al., 2020; Wu et al., 2022). Raft and pile cultures are the primary cultural approaches of offshore pearl oysters (Yang et al., 2017). Nevertheless, escalating nearshore marine pollution has recently placed pearl oysters under significant threat. TiO2-NPs are prone to condensation and deposition on the surface of sediments. The gills of marine bivalves are the primary organs exposed to waterborne pollutants and serve as major detoxification organs in these species, rendering gene expression in this organ highly prone to alterations (Brandts et al., 2018). This study used RNA-Seq analysis of gill tissue to identify the potential genes and pathways affected by TiO2-NPs in pearl oysters after 14-day exposure and 7-day brief recovery, providing a theoretical basis of TiO2-NPs’ effects on pearl oysters.
2 Materials and methods
2.1 Exposure of pearl oysters to TiO2-NPs
The anatase form of TiO2-NPs, approximately 5–10 nm in size, was obtained from Shanghai McLean Biochemical Technology (Shanghai, China). TiO2-NPs were initially added to filtered seawater and homogenized for 20 min using an LC-JY92-TTN ultrasonic homogenizer (65 W; Lichen Bonsi Instrument Technology, China) to prepare 1 g/L nanoparticle suspension. The suspension was subsequently diluted with seawater to a final TiO2-NPs concentration of 5 mg/L.
The pearl oysters with intact and uniform-sized shells (shell width with 26.81 ± 2.53 mm) were selected from a fast-growing, black-selective line of pearl oysters (Deng et al., 2013), sourced from Liusha Bay, China. The pearl oysters were cleaned to remove biofoulings and cultured in cylindrical barrels (300 L) for 7 days. Then the pearl oysters were exposed to 5 mg/L TiO2-NPs for 14 days (treatment phase) and subsequently cultured in TiO2-NP-free seawater for 7 days (recovery phase). Pearl oysters were fed unicellular algae (morning: 20,000 cells/mL Chlorella; evening: 10,000 cells/mL Platymonas subcordiformis) during the study period. The experiment comprised three parallel groups, each consisting of 30 pearl oysters. The water in the barrels was daily refreshed, and TiO2-NPs suspension was added to each barrel maintain a constant TiO2-NPs concentration of 5mg/L. On days 0, 14, and 21, we randomly selected 2 pearl oysters from each parallel group, designated the control (TC), experimental (TE), and recovery (TR). Gill tissues were clipped and frozen in liquid nitrogen and then stored at −80°C for subsequent analyses. No dead pearl oysters were found during the exposure and recovery periods.
2.2 RNA extraction and library construction
RNA extraction and purification from the gill tissue were conducted utilizing TRIzol (Invitrogen, CA, USA) following instructions from the manufacturer. The purity and quantity of total RNA was evaluated utilizing a NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA). Oligo(dT) magnetic beads (Dynabeads Oligo, No. 25-61005, Thermo Fisher, USA) were utilized in two consecutive purification steps to specifically capture mRNA containing PolyA tails. NEBNext® Magnesium RNA Fragmentation technology (product number E6150S, USA) was used to fragment the captured mRNA at a high temperature of 94°C for 5 to 7 min. Subsequently, the DNA-RNA complex double strands into pure DNA double strands. Simultaneously, a dUTP solution (product number R0133, Thermo Fisher, California, USA) was added to smooth the ends of the double-stranded DNA. Subsequently, A bases were added to each end to and used magnetic beads for fragment size screening and purification. The UDG enzyme (NEB, product number m0280, Massachusetts, USA) was then used to digest the double-stranded DNA, followed by PCR amplification. Thus, a library was constructed with a 300 bp ± 50 bp fragment size. In accordance with the standard process provided by LC Bio-Technology Co., Ltd. (Hangzhou, China), Illumina Novaseq™ 6000 was utilized for dual-end PE150 sequencing.
2.3 Bioinformatics analyses of RNA-seq
Cutadapt ver. 1.9 was utilized to discard adapter sequences, polyA sequences, polyG sequences, and other low-quality reads. FastQC ver. 0.11.9 was used to verify the GC content and Q20/30 content of the quality-controlled reads. HISAT2 ver. 2.0.4 was utilized to align the reads to the reference genome (Zheng et al., 2023). StringTie ver. 2.1.6 was employed for gene transcription or assembly using FPKM quantification. The reads were assembled with StringTie ver. 1.3.4d, using default parameters. The gffcompare ver. 0.9.8 was selected for comprehensive transcriptomic reconstruction. The R package EdgeR was then utilized to examine the difference between sample genes, defining significant difference as twice difference ratio > or < 0.5 times and p value < 0.05 (Robinson et al., 2010). Finally, Kyoto Encyclopedia of Genes and Genomes (KEGG) and gene ontology (GO) gene enrichments analyses were performed utilizing DAVID software. Raw transcriptome sequences data have been deposited at Genome Sequence Archive (GSA) under accession CRA017826.
2.4 Gene expression validation
Eight DEGs were randomly selected for qRT-PCR experiments to determine whether the results of RNA-seq data were reliable. β-Actin was used as the internal reference gene. Primers were designed using Primer Premier 5.0 software as shown in Supplementary Table 1. The total mixture of 10 μL of each sample was reacted by the following procedure: denaturation at 95°C for 5 min, followed by 40 amplification cycles (95°C for 10 s, 60°C for 15 s for annealing, and 72°C for 15 s for extension). Expression levels of different genes were analyzed using the 2-ΔΔCT method (Livak and Schmittgen, 2001).
3 Results
3.1 Transcriptome sequencing
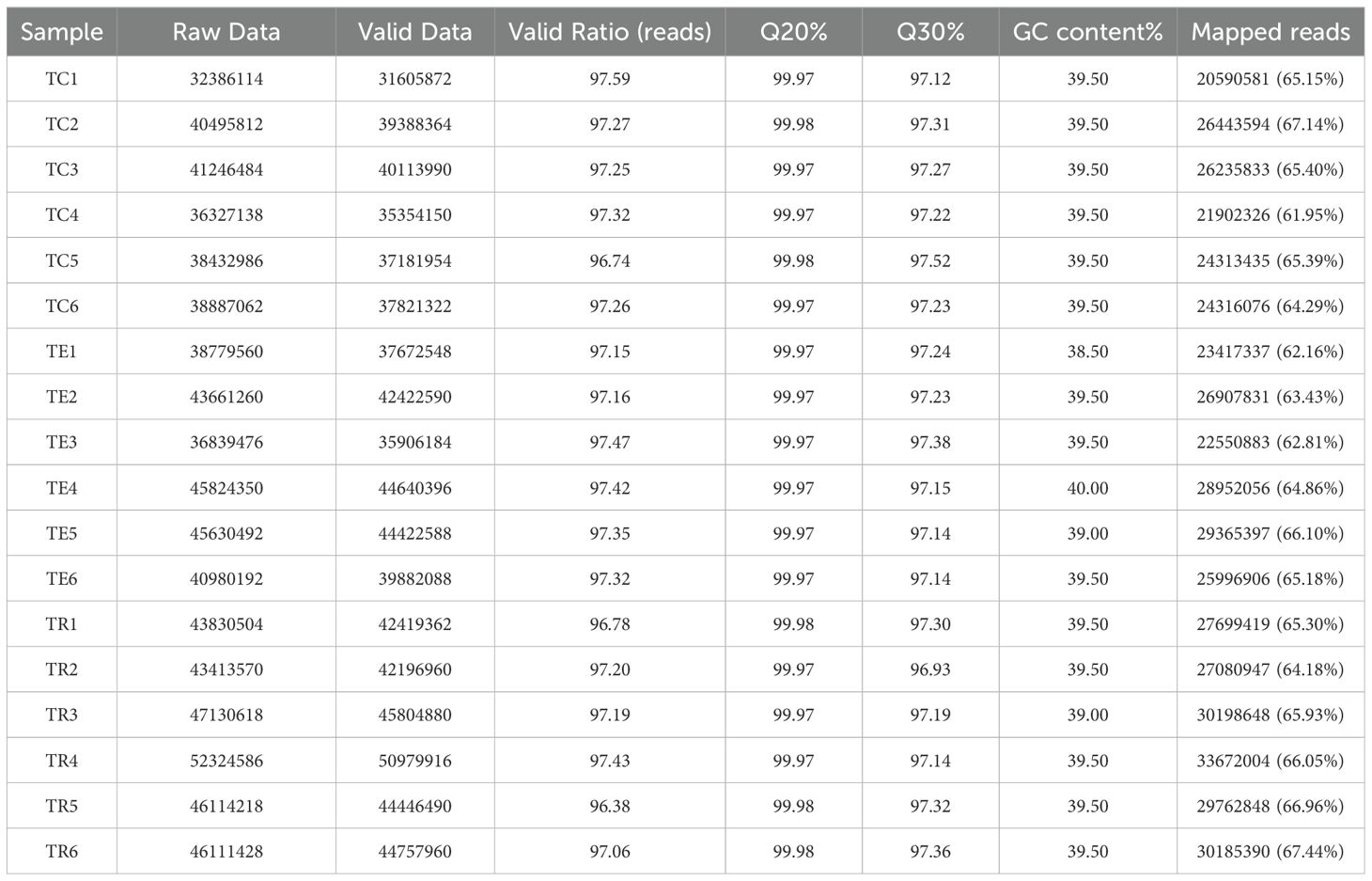
Illumina sequencing generated a significant number of raw reads (758,415,850), which were processed to yield 737,017,614 high-quality clean reads following quality control (QC). The GC contents of these reads ranged from 38.5% to 40%, with Q30 and Q20 quality scores ranging from 96.93% to 97.52% and 99.97% to 99.98%, respectively, as detailed in Table 1. Subsequently, a substantial proportion of the genes (61.95%–67.44%) were aligned successfully with the reference genome. This alignment rate highlights the suitability and reliability of sequencing data for follow-up analytical procedures.
3.2 Detection of differentially expressed genes at different experimental periods
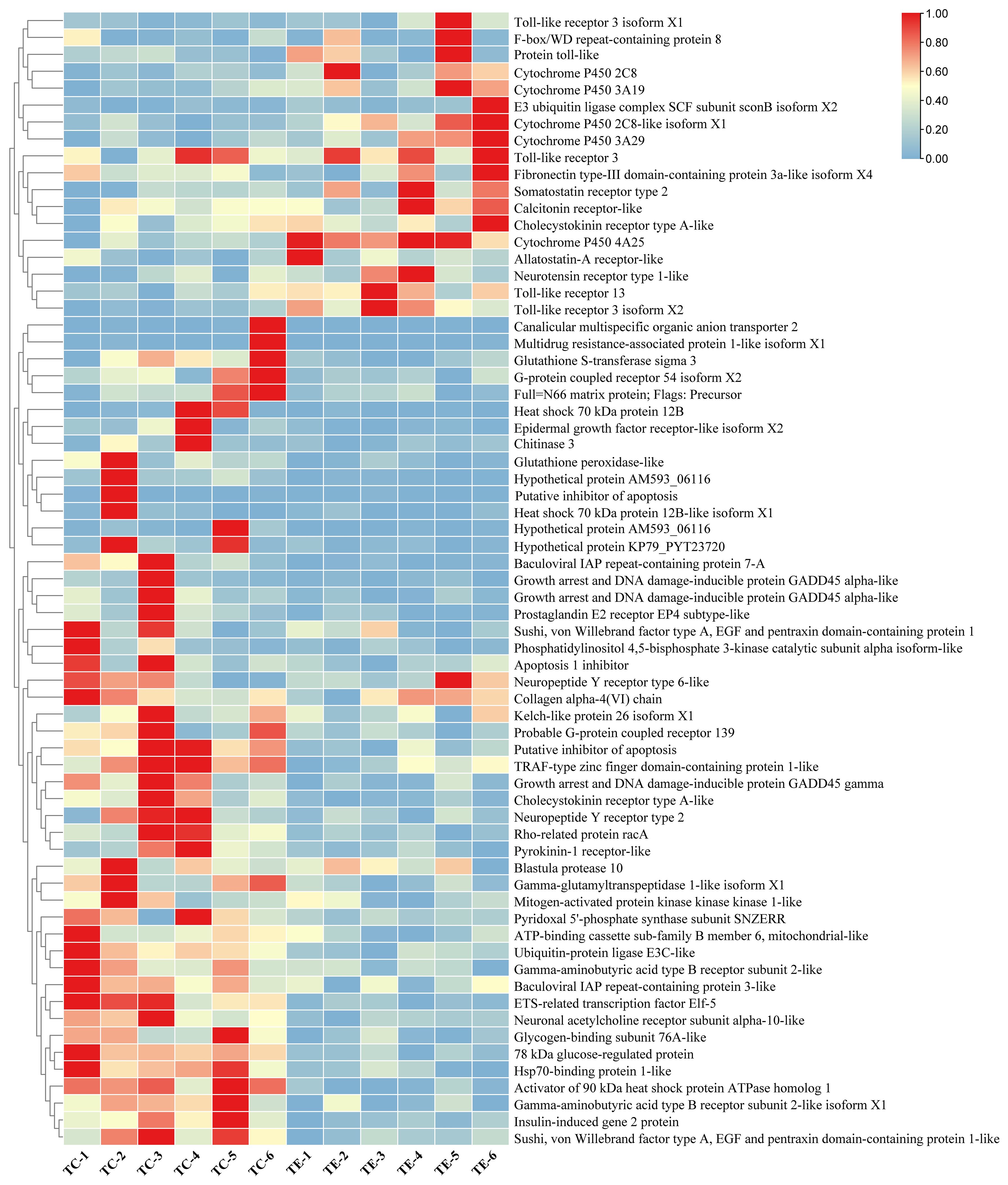
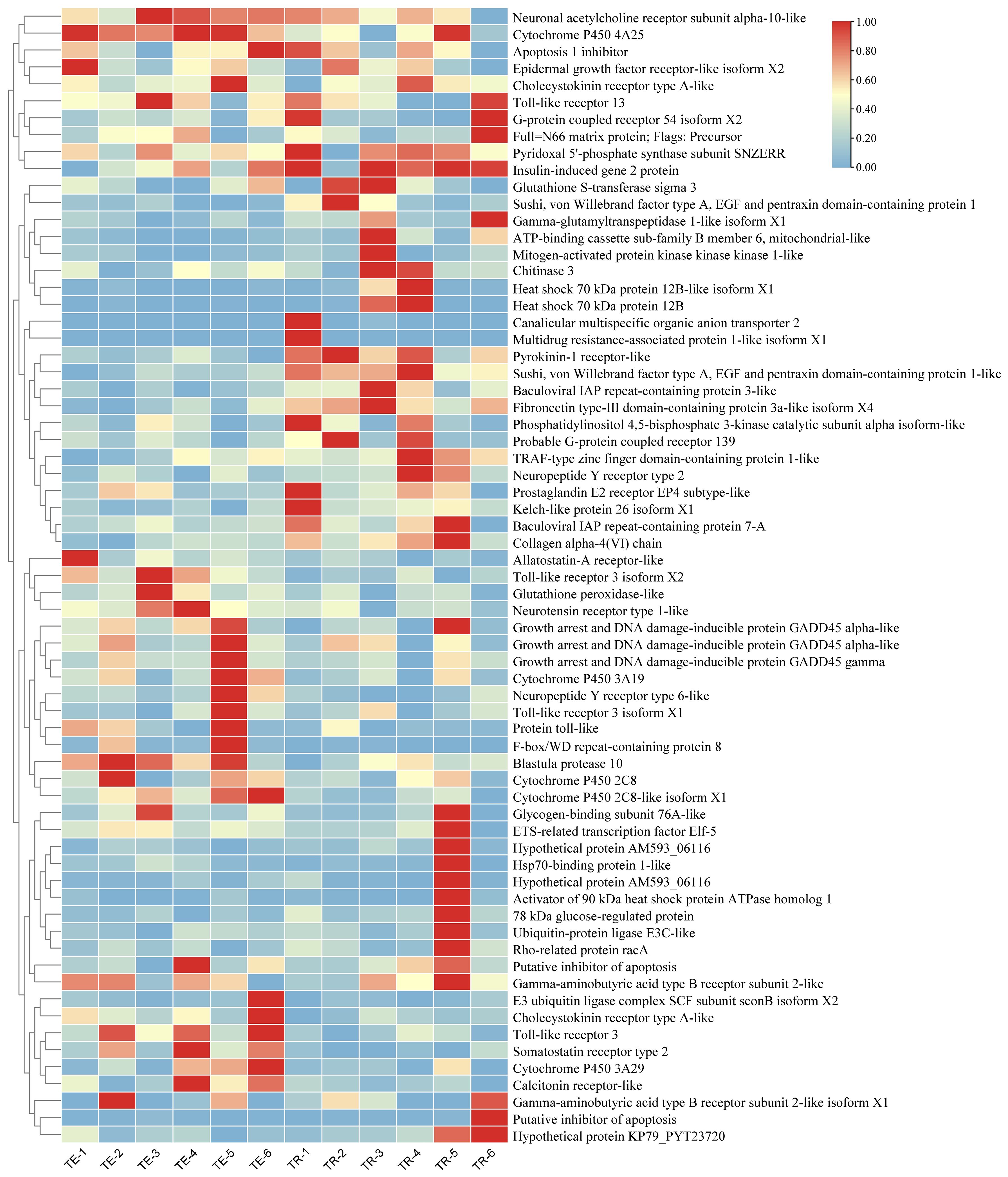
Further differential expression analyses revealed 911 DEGs between the TC and the TE, with 390 downregulated and 521 upregulated in the TC in comparison to the TE (Figure 1). Among these, Cytochrome P450 4A25 (PIN0130055), toll-like receptor 13 (PIN0061698), DNA damage-inducible protein GADD45 gamma (PIN0103192), and several other genes expression exhibited significant upregulation, as illustrated in Figure 2. Conversely, pyridoxal 5’-phosphate synthase subunit SNZERR (PIN0051258), gamma-glutamyltranspeptidase 1-like isoform X1 (PIN0020608), DNA damage-inducible protein GADD45 gamma (PIN0103192), ATP-binding cassette sub-family B member 6 (PIN0010578), and other genes expression exhibited significant downregulation. Furthermore, we identified 844 DEGs between the TE and the TR, including 517 downregulated and 327 upregulated in TE in comparison to the TR (Figure 1). Similarly, gamma-glutamyltranspeptidase 1-like isoform X1 (PIN0020608), ATP-binding cassette (ABC) sub-family B member 6 (PIN0010578), and several other genes expression exhibited significant upregulation after a short recovery. Conversely, genes, such as toll-like receptor 3 (PIN0120448) and heat shock 70 kDa protein 12B (PIN0060012), were substantially downregulated (Figure 3). These significantly altered genes are sensitive to TiO2 NP-induced stress and may play an important role in physiological processes.
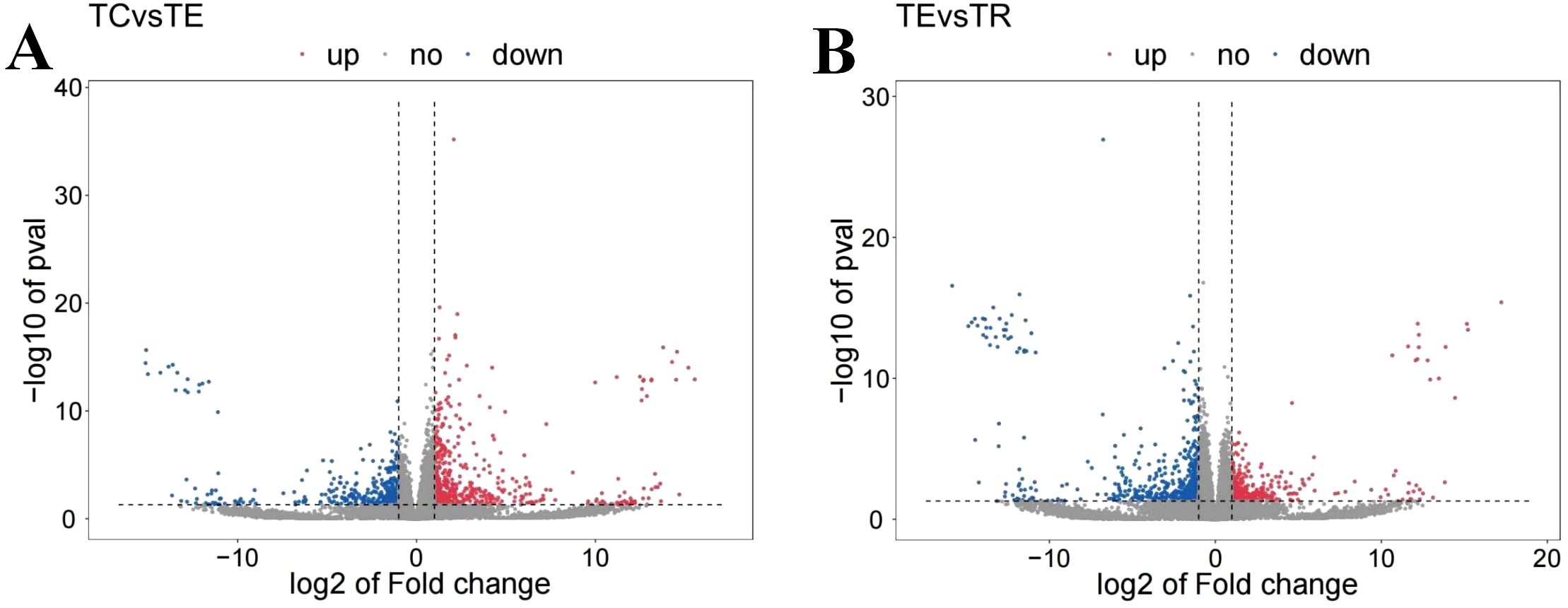
Figure 1. (A) Differential gene expression analyses between TC and TE. (B) Differential gene expression analyses between TE and TR. Red indicates the upregulated gene expression level, blue indicates the downregulated gene expression level, and grey indicated insignificant differential expression.
3.3 GO analyses
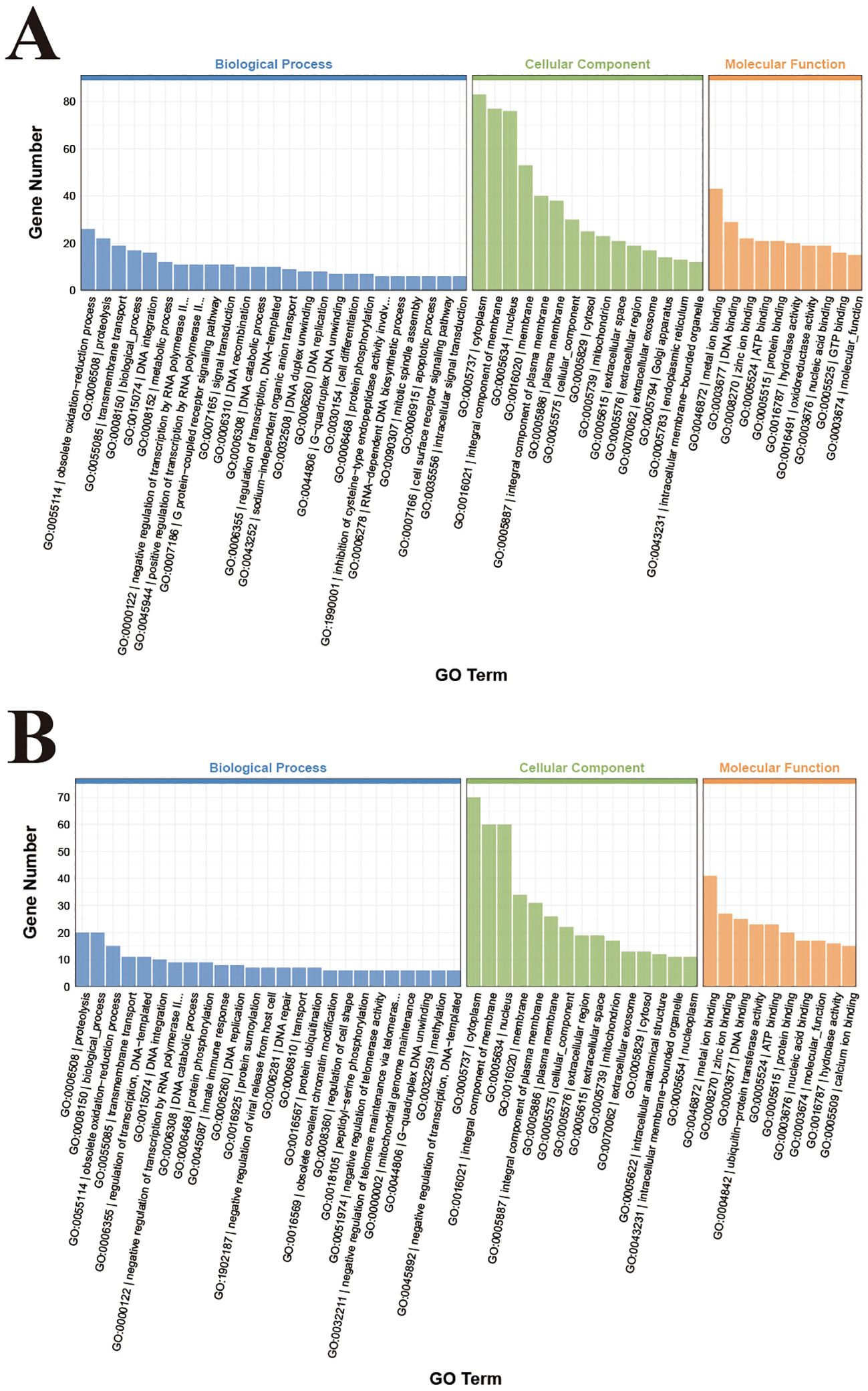
GO enrichments analyses of the DEGs annotated 3373 and 2577 DEGs across molecular functions, cellular components, and biological process in the TC and TE groups and TE and TR groups, respectively. We selected the 30 entries with the highest enrichment levels for bar charts plotting (Figure 4A). Among them, the most annotated DEGs in the TC and TE groups were “metal ion binding”, “DNA binding”, “ATP binding”, “deoxyribonucleotide biosynthetic process”, “DNA recombination”, “mitotic spindle assembly”, “RNA-dependent DNA biosynthetic process”, “DNA integration”, and “nucleus”. In the TE and TR groups, the most annotated DEGs were “biological process”, “proteolysis”, “zinc ion binding”, “metal ion binding”, “obsolete oxidation-reduction process”, “DNA integration”, “DNA binding”, and “integral component of membrane” (Figure 4B).
3.4 KEGG enrichments analyses
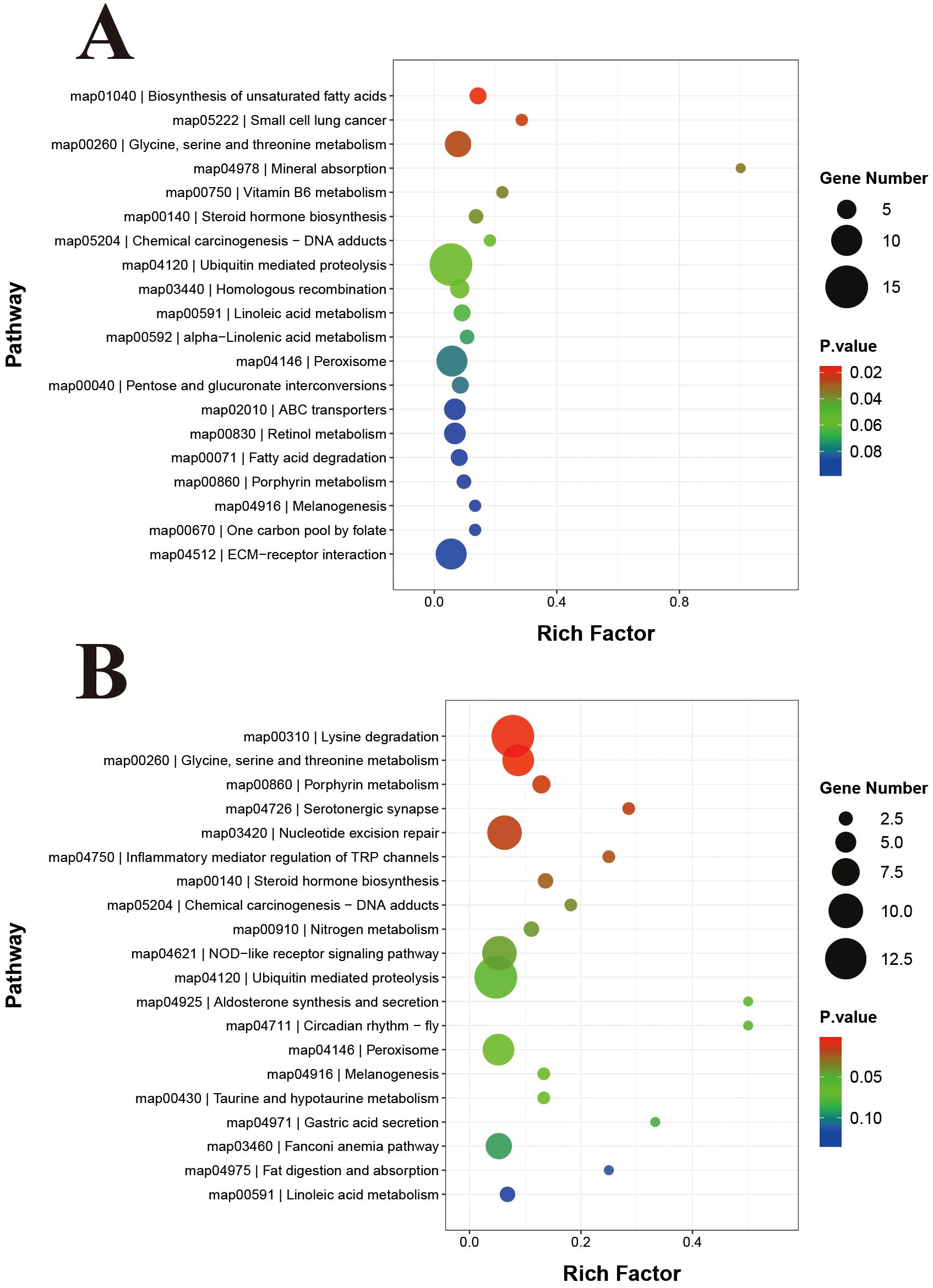
KEGG enrichments analyses demonstrated the enrichments of a total of 369 DEGs in 129 signaling pathways in the TC and TE groups (Figure 5A). Notably, these pathways included significant enrichment in “vitamin B6 metabolism”, “glycine, serine, and threonine metabolism”, “biosynthesis of unsaturated fatty acids”, “ubiquitin-mediated protein hydrolysis”. Furthermore, the “ABC transporter”, and “ECM-receptor interactions”, “NOD-like receptor signaling pathway”, “Toll-like receptor”, “neuroactive ligand-receptor interaction signaling pathway”, “lysosomal pathway”, “phagosomal pathway”, “FOXO signaling pathway”, and “glutathione metabolite” were also identified to be associated with pearl oysters’ responses to TiO2-NPs exposure. Conversely, a total of 284 DEGs in the TE and TR groups were enriched in 102 signaling pathways (Figure 5B). Notably, significant enrichment was observed in “lysine degradation”, “glycine, serine, and threonine metabolism”, “NOD-like receptor signaling pathway”, and other pathways.
3.5 qRT-PCR validation
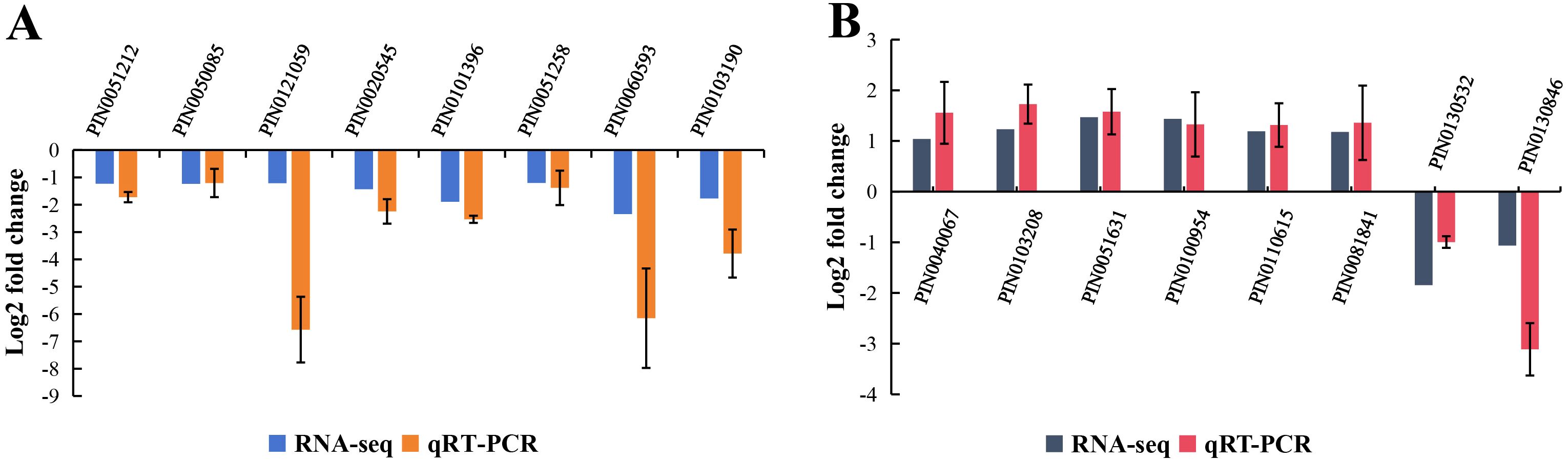
To assess the accuracy of the RNA-seq results, 8 DEGs were randomly selected for qRT-PCR in each of the TC vs TE and TE vs TR groups. qRT-PCR selective gene expression trends were consistent with DEG analyses, suggesting that the transcriptome data were reliable (Figure 6).
4 Discussion
The survival of aquatic organisms is significantly affected by salinity, oxygen content, temperature, and pollutants in the water (Valavanidis et al., 2006). In particular, environmental chemicals pose various toxic threats to aquatic organisms, such as DNA damage, oxidative stress, inflammation, apoptosis, and cell death (Lim et al., 2022; Wang et al., 2023). To withstand complex and variable living environments, intertidal bivalves have developed highly intricate adaptive mechanisms (Falfushynska et al., 2020). Considerable research interest has recently been focused on the potential impacts of TiO2-NPs on marine bivalves (Abdel-Latif et al., 2020; Li et al., 2021). This study revealed the molecular response mechanisms of pearl oysters after exposure to TiO2-NPs, providing crucial insights into evaluating the potential toxic impacts of TiO2-NPs on these organisms.
4.1 TiO2-NPs cause of oxidative stress
TiO2-NPs function as prooxidants that can enter cell membrane, inducing mitochondrial damage (Barmo et al., 2013). It triggers excessive cellular production of reactive oxygen species (ROS), leading to compromised membrane permeability (Girardello et al., 2016). Under such conditions, living beings may experience oxidative stress due to excessive ROS production when the cellular antioxidant defense system becomes overwhelmed (Ray et al., 2012). To mitigate the harmful impacts of ROS, numerous animals can employ antioxidant defensing mechanisms, including non-enzymatic components (such as carotenoids and glutathione) and enzymatic components (such as glutathione peroxidase and superoxide dismutase) (Yang et al., 2017). We detected the substantial enrichment of the DEGs in the glutathione metabolism pathway under TiO2-NP stress. Genes encoding glutathione peroxidase-like (PIN0101396), gamma-glutamyltranspeptidase 1-like isoform X1 (PIN0020608), glutathione S-transferase sigma 3 (PIN0052777), microsomal glutathione S-transferase 2 (PIN0060908), and aminopeptidase N (PIN0020097) were substantially downregulated. According to these results, pearl oysters experience oxidative stress under TiO2-NP stress, being consistent with our prior finding (Li et al., 2024b).
Here, we observed notable enrichment of vitamin B6 metabolism in the KEGG pathway enrichment. Specifically, we found that the pyridoxal 5’-phosphate synthase subunit SNZERR (PIN0051258), which is involved in vitamin B6 metabolism, was downregulated in response to oxidative stress. Pyridoxal-5’-phosphate (PLP) acts as the physiologically active coenzyme of vitamin B6, potentially playing pivotal roles in antioxidation (Schneider et al., 2000). PLP can directly scavenge free radicals for lipid peroxidation inhibition. Alternatively, PLP acts indirectly as a coenzyme of the glutathione antioxidant defense system (Liu et al., 2013). Studies have indicated that deficiencies in vitamin B6 can weaken antioxidant defensing mechanisms and increase oxidative stresses (Danielyan and Chailyan, 2020). This study demonstrated that TiO2-NPs exposure induced downregulation of pyridoxal 5’-phosphate synthase subunit SNZERR (PIN0051258), which is involved in vitamin B6 metabolism, thereby affecting vitamin B6 synthesis and increasing oxidative stress in pearl oysters.
Aquatic species depend on the multi-xenobiotic resistance (MXR) mechanism as a key defensing mechanism against environmental toxins, minimizing intracellular accumulation of toxins and potential damage (Kurelec, 1995), which is mediated by the ABC transporter (Kingtong et al., 2007). The relationship between ABC transporters and nanoparticles has gained increasing attention (Yin et al., 2023). Kingtong et al. (2007) demonstrated the significant roles of ABC transporters in the detoxification metabolic mechanism of Saccostrea forskali after exposure to tributyltin. Research on mussel gill cells has demonstrated the upregulation of Abcb1 by silver NPs, aiding in silver NPs detoxification (Katsumiti et al., 2015). Bhagat et al. (2022) reported that polystyrene and metal oxide nanoparticles disrupt ABC transporters’ functionality in zebrafish embryos, resulting in increased aluminum and cerium ion toxicity. Similarly, Georgantzopoulou et al. (2016) found that the MXR system could be inhibited by silver nanoparticles in Daphnia magna larvae, leading to increased calcineurin accumulation. After TiO2-NPs exposure, significant downregulation was observed in genes linked to the ABC transporters pathway, including ABC sub-family B member 6, mitochondrial-like (PIN0010578), multidrug resistance-associated protein 1-like isoform X1 (PIN0080856), and canalicular multispecific organic anion transporter 2 (PIN0080857). This indicates that TiO2-NPs inhibit the activity of ABC transporter protein activity, thereby worsening oxidative stress and particle accumulation in pearl oysters. After a brief 7-day recovery period, genes associated with ABC transporters and glutathione metabolism pathways exhibited significant upregulation, indicating an increase in oxidative stress in pearl oysters.
Cytochrome P450, a class of monooxygenase, is crucial for the syntheses and metabolisms of various hormones, such as glucocorticoids and mineralocorticoids, which are essential for stress response (Urlacher and Girhard, 2012). Increased P450 production in aquatic organisms typically indicates they are experiencing stress (Ibrahim et al., 2021). For instance, elevated transcription of the cytochrome P450 gene was observed in the kidney, gill, and liver tissues of Nile tilapia following exposure to Aeromonas hydrophila (Hal and El-Barbary, 2020). An upregulation was observed in genes such as cytochrome P450 4A25 (PIN0130055), cytochrome P450 2C8 (PIN0120270), cytochrome P450 3A19 (PIN0100308), cytochrome P450 2C8-like isoform X1 (PIN0140634), and cytochrome P450 3A29 (PIN0100307) following 14-day TiO2-NPs exposure, indicating that TiO2-NPs exposure induces oxidative stress in pearl oysters. After a brief recovery period, the related cytochrome P450 gene expression was downregulated compared with that of the TE group, which also indicates that brief recovery can alleviate stress in pearl oysters.
4.2 TiO2-NPs changes in immune activities
Exposure to TiO2-NPs can adversely affect bivalves’ immune system (Wei et al., 2023). Bivalves primarily depend on innate immunity due to their lack acquired immune system (Jiang et al., 2017). Microbial recognition by innate immunity is mediated by pattern recognition receptors, which bind to pathogen-associated molecular patterns, the conserved microbial molecules. Toll-like receptors detect microorganisms in extracellular cellular lumens and phagosomes, whereas the Nod-like family of receptors recognize intracellular pathogens, triggering transcriptional responses and activating inflammatory vesicles (Chamaillard et al., 2003; Inohara et al., 2005). Innate immune receptors interact with specific microbial ligands to generate signaling pathways triggering host transcriptional response linked to inflammation. Research has demonstrated that bacterial lysis in phagosomes is crucial for acquired and innate immune response development (Herskovits et al., 2007). Phagosome maturation involves regulatory interactions with other membrane organelles, such as regenerating endosomes, late endosomes, and lysosomes. Furthermore, the lysosomes-phagosomes fusion can release toxic compounds that effectively kill and fragment major bacteria (Luzio et al., 2007). Lysosomes play pivotal roles in various physiological functions, such as immune responses, energy metabolism, cell survival, cell membrane repair, and cellular homeostasis, most of which rely on lysosomes’ protease activities (Martínez-Fábregas et al., 2018). We detected substantial upregulation of the toll-like receptor 13 (PIN0061698), toll-like receptor 3 isoform X2 (PIN0061696), and toll-like receptor 3 isoform X1 (PIN0061693) genes after exposure to TiO2-NPs. Furthermore, the expression of some genes in the NOD-like receptor signaling pathway, lysosome pathway, and phagosome pathway was substantially upregulated, indicating that TiO2-NPs exposure activates the immune system of pearl oysters and improves their immune ability to resist the invasion of TiO2-NPs, being consistent with our prior finding (Li et al., 2024a, b). We observed substantial downregulation of the toll-like receptor 3 (PIN0120448) and protein toll-like (PIN0071163) genes after a brief recovery period. In addition, the NOD-like receptor signaling, lysosome, and phagosome pathways remained substantially upregulated compared with the TE group, indicating that brief recovery may alleviate immune stress in pearl oysters.
4.3 TiO2-NPs induces apoptosis
Apoptosis is critical for the immune response of mollusks (Zhang et al., 2022). The FOXO signaling pathway is associated with apoptosis, DNA repair, and several cellular pathways (Chen et al., 2022a). In aquaculture, there is increasing research interest in investigating the changes in the FOXO pathway caused by environmental stress. For instance, studies have demonstrated that acute hypoxia induces changes in the FOXO signaling pathway in Hypophthalmichthys nobilis (Chen et al., 2021) and Oncorhynchus mykiss (Wu et al., 2023). Additionally, Ma et al. (2023) revealed that high-temperature stress mitigates cellular apoptosis by regulating the expressions of FOXO target genes related to apoptosis, thereby mediating liver injury in Brachymystax lenok tsinlingensis. After exposure to TiO2-NPs, the expressions of genes related to the FoxO signaling pathway were substantially downregulated, including epidermal growth factor receptor-like isoform X2 (PIN0140090), growth arrest and DNA damage-inducible protein GADD45 gamma (PIN0103192), growth arrest and DNA damage-inducible protein GADD45 alpha-like (PIN0103191, PIN0103190), growth arrest and DNA-damage-inducible protein GADD45 gamma (PIN0103188), and phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform-like (PIN0101618). This indicates that exposure to TiO2-NPs inhibits the FOXO pathway and induces apoptosis. Gadd45 α is known to be activated in response to DNA damage (Salvador et al., 2013). Martin-Folgar et al. (2022) discovered that exposure of Danio rerio to plastic particles at a concentration of 0.1 ppm resulted in upregulation of GADD450 expression, whereas exposures to 0.5 and 3 ppm led to downregulation of GADD450 gene expression. Consistent with observations of Martin-Folgar et al. (2022), we also observed a significant downregulation of Gadd45 expression in pearl oysters after 14 days of TiO2-NPs exposure, which indicates that TiO2-NPs may have impacted the DNA repair mechanism of pearl oysters by affecting the overall repair capacity of DNA, potentially leading to apoptosis.
Inhibitors of apoptosis proteins (IAP) protect aquatic organisms from the detrimental impacts of atmospheric exposure, thereby extending their survival time (Zhou et al., 2021). A significant upregulation of IAPs in oysters exposed to high temperatures, air, and low salinity has been demonstrated previously (Zhang et al., 2012). Under hypoxia in pearl oysters (Yang et al., 2023) and manila clams (Nie et al., 2020), the expressions of IAPs were substantially upregulated to activate their apoptotic systems in response to survival conditions. Nevertheless, baculovirus IAP repeat sequence protein 3-like (PIN0060593), baculovirus IAP repeat sequence protein 7-A (PIN0060592), and putative apoptosis inhibitory factors (PIN0070159, PIN0070593) were found to be downregulated under the conditions of TiO2-NPs exposure, leading to apoptosis and affecting cell viability and proliferation.
4.4 Energy supply and energy expenditure
Bivalves are particularly vulnerable to nanoparticle toxicity in aquatic environments because of their filter-feeding strategy and sessile lifestyle. Nanoparticles, which are often agglomerated, are trapped in their gills and can enter the digestive system through endocytosis, leading to harmful effects (Gornati et al., 2016; Li et al., 2021). Nanomicroplastics have been demonstrated to disrupt the digestive system of shellfish, impairing nutrient absorption and consequently affecting shellfish health (Cole et al., 2020; Cappello et al., 2021). Furthermore, Mytilus edulis could absorb 100 nm microplastic particles, which affect the filtration activity of these organisms, produce pseudofaeces, and consume energy (Wegner et al., 2012).
Gluconeogenesis and glycolysis are critical metabolic pathways involved in carbohydrate synthesis and catabolism (Jitrapakdee, 2012). Studies have demonstrated that shellfish maintain energy balance under stress conditions by boosting glycolysis (Sun et al., 2021; Hu et al., 2022). During pathogen infections, the host requires increased energy to fight and survive the infection. However, relying solely on sugar metabolism is insufficient to support the higher energy requirements of the host due to its limited carbohydrate content. Proteins and lipids serve as crucial alternative energy sources to meet the increased energy demands of the host. Lipids contribute significantly to animal adaptation to adverse environments, in addition to supporting normal growth and development by providing energy (Chen et al., 2013; Liu et al., 2016). Studies have demonstrated that zebrafish experience increased glucose consumption and energy supply deficits after exposure to NPs (Chen et al., 2020). In Crassostrea gigas larvae, stimulation by OsHV-1 infection resulted in large-scale mortality, likely attributed to reduced oyster feeding activity and significant disruption of lipid metabolism within the organisms (Young et al., 2017). After exposure to TiO2-NPs, KEGG secondary pathways analysis revealed significant enrichment in pathways related to carbohydrate metabolism. Specifically, the expression of most related genes within the glycolytic pathways, pentose and glucuronate interconversion pathway, citrate cycle pathway, and nucleotide and amino sugars metabolism pathway was substantially downregulated. Furthermore, key genes associated with energy metabolisms, such as glycogen-binding subunit 76A-like (PIN0121059), 78 kDa glucose-regulated protein (PIN0111091), and insulin-induced gene 2 protein (PIN0010512) were substantially downregulated. This indicates a reduction in sugar and carbohydrate supply to the organism from pearl oysters exposed to TiO2-NPs. Most genes in the KEGG secondary pathways associated with amino acid metabolism, such as methionine and cysteine metabolism pathways, threonine, serine, and glycine metabolism pathways, lysine degradation pathway, and arginine and proline metabolism pathways, were substantially downregulated. Conversely, most genes involved in pathways associated with lipid metabolism, such as steroid hormone biosynthesis, unsaturated fatty acids biosynthesis, linoleic acid metabolism, and fatty acid degradation pathways, were substantially upregulated. This indicates that under TiO2-NPs stress, lipid metabolism becomes the primary energy supply for pearl oysters. We speculate that sugars, carbohydrates, and proteins may primarily supply energy during the pre-exposure phase of TiO2-NPs, and then switch to lipids during the post-exposure phase of the exposure. However, further studies are required to confirm this hypothesis. After a brief recovery period, secondary pathways related to glycan biosynthesis and metabolism, lipid metabolism, amino acid metabolism, and nucleotide metabolism exhibited significant upregulation, revealing the restored energy supply in pearl oysters after alleviating the stress induced by TiO2-NPs.
Under TiO2-NPs stress, the downregulated DEGs substantially enriched the GO terms associated with “mitotic spindle assembly”, “deoxyribonucleotide biosynthetic process”, “RNA-dependent DNA biosynthetic process”, “DNA recombination”, and “DNA integration”. Extended exposure to TiO2-NPs may cause cellular injury, potentially disrupting metabolic activities within cells. This suppression of cellular processes is hypothesized to be a strategic response to conserve energy under TiO2-NPs-induced stress.
4.5 TiO2-NPs disrupt protein homeostasis
Ensuring protein equilibrium is essential for maintaining physiological function, as it involves a dynamic balance between synthesis and degradation. The accumulation of damaged or misfolded proteins within cells can be toxic and disrupt normal cellular processes (Sherman and Goldberg, 2001). The HSP70 family members are crucial for maintaining protein homeostasis by facilitating newly produced and denatured peptides’ folding into native conformation as well as the refolding of proteins forming aggregates within cells (Fu et al., 2014; Artigaud et al., 2015). Overexpression of HSP70 genes is often pivotal for enhancing resistance to various stresses, a common phenomenon observed among aquatic organisms exposed to pollutants (Ivanina et al., 2009). For instance, the proliferation of the HSP70 gene family and the overexpression of the HSP70 gene are believed to significantly contribute to the capability of oysters’ high-temperature withstanding (Ivanina et al., 2009). Distinct amplification of HSP70 was identified in bivalve shellfish, such as the Crassostrea hongkongensis (Brandts et al., 2018). Conversely, HSP70 downregulation has been reported in Mytilus galloprovincialis after exposure to heat stress and cadmium (Izagirre et al., 2014). Transcript levels of hsp70 in the gill tissues of Mytilus galloprovincialis were observed to decrease after simultaneous exposure to nanoplastics and carbamazepine (Brandts et al., 2018). In our experiments, we observed substantial downregulation of the heat shock 70 kDa protein 12B (PIN0060012), heat shock 70 kDa protein 12B-like isoform X4 (PIN0131632), and HSP70-binding protein 1-like (PIN0050777) genes. However, inactivating these gene transcripts does not necessarily indicate low HSP70 protein levels. High HSP70 level has been demonstrated to cause transcription factors inactivation through a negative feedback mechanism (Pinsino et al., 2017). HSP70 proteins aid in the lysosomal degradation of aggregated proteins, using either molecular chaperone-assisted autophagy or the ubiquitin-proteasome system (Rokutan, 2000). Based on reports, animals can regulate misfolded peptides breakdown through the ubiquitin-proteasome mechanism (Hampton, 2002). Ubiquitin genes activation is stimulated to break down unrepairable proteins in a heat-sensitive abalone lineage (Chen et al., 2019). Whiteleg shrimp (Penaeus vannamei) responds to stress by upregulating ubiquitin gene expression after a short-term microplastic exposure (Han et al., 2021). After exposure to TiO2-NPs, substantial decreased expression of 13 genes, such as ubiquitin-protein ligase E3C-like (PIN0100476) and apoptosis 1 inhibitor (PIN0060591) was observed, indicating that the downregulation of genes involved in the ubiquitin-mediated protein hydrolysis pathway could affect the ability of pearl oyster to remove misfolded proteins, thereby resulting in cytotoxicity. Heat shock 70 kDa protein 12B (PIN0060012) and heat shock 70 kDa protein 12B-like isoform X1 (PIN0081248) remained downregulated following a short-term recovery period of 7 days. In addition, although seven genes in the ubiquitin-mediated proteolysis pathway were upregulated, six genes remained downregulated, indicating that the short-term recovery period was insufficient to restore pearl oysters to normal levels. These findings reveal that TiO2-NPs can significantly affect protein homeostasis in pearl oysters.
4.6 TiO2- NPs disrupt neuronal excitability
In invertebrates, neurons play crucial roles in receiving and processing environmental information. Furthermore, neurons positively regulate synaptic structural plasticity and function, enhancing adaptive capacity under stressful conditions (Konstantinos et al., 2012). In Limulus polyphemus, neurons contribute to osmotic stress tolerance and adaptation through several processes, such as depolarization of membrane potentials, modulation of burst frequencies, and alteration of spiking patterns (David and Sidney, 1981). However, prolonged stress can lead to irreversible neuronal damage (Samokhvalov et al., 2008). For instance, TiO2-NP stress downregulates related genes in pearl oyster neurons, leading to decreased neuronal excitability and inhibited neuronal activity (Yang et al., 2023). Furthermore, Guan et al. (2018) discovered that high-concentration TiO2-NPs induced substantial neurotoxic effects on Tegillarca granosa. In addition, NPs are potentially neurotoxic for mammals and teleosts (Sheng et al., 2016; Ze et al., 2016). Neurofunctions are directly linked to the signaling pathway of neuroactive ligand-receptor interactions (Duan et al., 2016). Neuronal function is affected by neuroactive ligands that bind to intracellular receptors, which in turn can bind with transcription factors and regulate gene expressions (Xu et al., 2012). Disruption of genes associated with neuroactive ligand-receptor interaction results in compromised memory functions (Papassotiropoulos and de Quervain, 2015). Wei et al. (2020) reported that silica nanoparticles cause developmental neurotoxicity in zebrafish embryos by disrupting the signaling pathway of neuroactive ligand-receptor interactions. Similarly, Gainey and Greenberg (2003) observed that neurotransmitters that are both excitatory, such as acetylcholine, and inhibitory, such as gamma-aminobutyric acid, are important for mollusks’ neural signal transduction. Previous studies have highlighted the vital roles of neurotransmitter receptors in responding to changes in neurotransmitters (Mao et al., 2017). Consequently, the downregulation of genes encoding these receptors demonstrates that TiO2-NPs exposure may disrupt various physiological processes regulated by neurotransmitters (Guan et al., 2018). Here, we identified a total of 23 DEGs significantly associated with pathways related to “neuroactive ligand-receptor interaction.” Following 14-day TiO2-NPs exposure, there was an indication of inhibited neuronal activity, marked by substantial downregulation of various neuron-related genes, such as activator of 90 kDa heat shock protein ATPase homolog 1 (PIN0090777), gastrin/cholecystokinin type B receptor-like (PIN2170002), ETS-related transcription factor Elf-5 (PIN0050085), prostaglandin E2 receptor EP4 subtype-like (PIN0100254), cholecystokinin receptor type A-like (PIN0071117), neuronal acetylcholine receptor subunit alpha-10-like (PIN0051212), hypothetical protein KP79_PYT23720 (PIN0110990), pyrokinin-1 receptor-like (PIN0101895), G-protein coupled receptor 54 isoform X2 (PIN0010507), neuropeptide Y receptor type 2 (PIN0030378), probable G-protein coupled receptor 139 (PIN0070411), gamma-aminobutyric acid type B receptor subunit 2-like (PIN0020047), and gamma-aminobutyric acid type B receptor subunit 2-like isoform X1 (PIN0051226). These genes are known to play crucial roles in regulating nervous system processes, such as synaptic transmission, strength, and maturation. After 7 days of recovery, a total of 10 DEGs were significantly associated with pathways related to “neuroactive ligand-receptor interaction,” indicating that the transient recovery period restored neuronal activity. Furthermore, seven neuronal genes, including somatostatin receptor type 2 (PIN0101576), allatostatin-A receptor-like (PIN0111909), calcitonin receptor-like (PIN0010366), blastula protease-10 (PIN0091326), cholecystokinin receptor type A-like (PIN0080212), neuropeptide Y receptor type 6-like (PIN0120125), and neurotensin receptor type 1-like (PIN0090157), remained significantly downregulated. This indicates that even after 7 days of recovery, the neuronal activity of pearl oysters could not return to normal levels.
4.7 Impact of TiO2-NPs on biomineralization activity
Biomineralization is a biological process by which organisms control and promote mineral substances precipitation, such as pearls and shells, through biological mechanisms (Yang et al., 2021). Increased phenanthrene levels, seawater acidification, temperature changes, hypoxia, and pollution can impact the expressions of genes related to shell and pearl formation in economic pearl oysters (Jafari et al., 2023; Yang et al., 2023). Du et al. (2017) discovered that chitin plays a pivotal role as the primary organic matrix in the formation of nacre in the shell of Pinctada fucata martensii. The researchers also hypothesized that genes encoding chitinase could have had substantial effects on the biomineralization process (Du et al., 2017). After exposure to TiO2-NPs, the expression levels of chitinase 1 (PIN0102750), chitinase 3 (PIN0101472), and chitinase-3-like protein 1 (PIN0140499) were substantially downregulated. Studies have demonstrated that proteins containing VWA domains, particularly collagen-related VWA-containing proteins (VWAP), are critical for the organic matrix of nacre and are essential for the formation of nacreous shells in Pinctada fucata martensii (Du et al., 2017). A protein discovered in the shell of Pteria sterna, referred to as N66, has been isolated and investigated, indicating that it exhibits activity similar to carbonic anhydrase and can generate crystalline forms of calcium carbonate under laboratory conditions (Rivera-Perez et al., 2019). The shell matrix serves as a complex system that facilitates interactions between proteins and minerals, proteins and proteins, as well as the epithelium and minerals (Marin et al., 2012), being critical for the biomineralization process (Sedanza et al., 2022). Here, the expressions of von Willebrand factor type A (PIN0110615), Full=N66 matrix protein (PIN0020545), and larval shell matrix protein 3 (PIN0090029) genes associated with pearl oyster biomineralization were substantially downregulated in the TE group compared with the TC group following exposure to TiO2-NPs. This indicates that TiO2-NPs negatively affected the biomineralization capacity of pearl oysters. This finding is consistent with our previous study, which demonstrated that TiO2-NP stress damages pearl oyster microstructure (Li et al., 2024a). Moreover, research has indicated that the kelch-like gene plays a role in the prismatic shell layer formation in pearl oysters (Yang et al., 2022). Shell formation involves a co-expression network of extracellular matrix (ECM)-related proteins (Du et al., 2017). The ECM comprises structural and functional macromolecules, such as fibers (e.g., collagen, fibronectin) (Mariman and Wang, 2010). After 7 days of recovery, the biomineralization-related genes kelch-like protein 26 isoform X1 (PIN0081045), von Willebrand factor type A (PIN0110615, PIN0053245), fibronectin type-III domain-containing protein 3a-like isoform X4 (PIN0081841), and collagen alpha-4(VI) chain (PIN0111490) were observed upregulated, indicating that a short recovery period restored some biomineralization capacity of pearl oysters, being consistent with our prior finding (Li et al., 2024b).
5 Conclusion
In this study, transcriptomic analysis was conducted to assess the effects of TiO2-NPs stress on gene expression in pearl oyster. A total of 911 and 844 DEGs were identified in the TC vs TE groups and the TE vs TR groups, respectively. These findings indicate that exposure to TiO2-NPs conditions disrupts protein homeostasis, reduces biomineralization, decreases neuronal excitability, promotes apoptosis, inhibits immune responses, and induces oxidative stress. These findings help to provided a significant reference to evaluate the threat of TiO2-NPs to pearl oysters in the general ecological environment. Nevertheless, future studies are required for a better assessment of these issues, such as whether a longer recovery period can restore their normal levels, the potential interaction of TiO2-NPs with variable environments (such as water temperature, salinity, and the presence of other pollutants), more comprehensive understanding of the systemic effects of TiO2-NPs, and how TiO2-NPs affect pearl oysters populations and their role in the ecosystem.
Data availability statement
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA017826) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
FL: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. JL: Data curation, Methodology, Writing – original draft. ZG: Data curation, Writing – original draft. CY: Conceptualization, Writing – original draft, Writing – review & editing. LS: Data curation, Writing – original draft. YL: Conceptualization, Writing – original draft. YD: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Science and Technology Program of Guangdong Province (Grant No. 2023A1515011769), National Natural Science Foundation of China (Grant No. 32102817), Department of Education of Guangdong Province (Grant No. 2020ZDZX1045 and 2021KCXTD026), the Earmarked Fund for CARS-49, Special Fund for Guangdong Province’s Science and Technology Innovation Strategy (Grant No. pdjh2024a191), the Program for Scientific Research Start-Up Funds of Guangdong Ocean University (060302022304).
Acknowledgments
We are very grateful to Marine Pearl Science and Technology Backyard in Leizhou of Guangdong for collecting samples. The authors would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript. Transcriptomic analysis was assisted by Biotree Biotech Co.,Ltd. (Shanghai, China).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer [YB] declared a past co-authorship with the author [YD] to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1462589/full#supplementary-material
References
Abdel-Latif H. M. R., Dawood M. A. O., Menanteau-Ledouble S., El-Matbouli M. (2020). Environmental transformation of n-TiO2 in the aquatic systems and their ecotoxicity in bivalve mollusks: A systematic review. Ecotox Environ. Safe 200, 110776. doi: 10.1016/j.ecoenv.2020.110776
Aitken R. J., Chaudhry M. Q., Boxall A. B. A., Hull M. (2006). Manufacture and use of nanomaterials: current status in the UK and global trends. Occup. Med-Oxford 56, 300–306. doi: 10.1093/occmed/kql051
Artigaud S., Richard J., Thorne M. A. S., Lavaud R., Flye-Sainte-Marie J., Jean F., et al. (2015). Deciphering the molecular adaptation of the king scallop (Pecten maximus) to heat stress using transcriptomics and proteomics. Bmc. Genomics 16, 998. doi: 10.1186/s12864-015-2132-x
Banni M., Sforzini S., Balbi T., Corsi I., Viarengo A., Canesi L. (2016). Combined effects of n-TiO2 and 2,3,7,8-TCDD in Mytilus galloprovincialis digestive gland: A transcriptomic and immunohistochemical study. Environ. Res. 145, 135–144. doi: 10.1016/j.envres.2015.12.003
Barmo C., Ciacci C., Canonico B., Fabbri R., Cortese K., Balbi T., et al. (2013). In vivo effects of n-TiO2 on digestive gland and immune function of the marine bivalve Mytilus galloprovincialis. Aquat. Toxicol. 132, 9–18. doi: 10.1016/j.aquatox.2013.01.014
Bhagat J., Zang L. Q., Kaneco S., Nishimura N., Shimada Y. (2022). Combined exposure to nanoplastics and metal oxide nanoparticles inhibits efflux pumps and causes oxidative stress in zebrafish embryos. Sci. Total. Environ. 835, 155436. doi: 10.1016/j.scitotenv.2022.155436
Brandts I., Teles M., Gonçalves A. P., Barreto A., Franco-Martinez L., Tvarijonaviciute A., et al (2018). Effects of nanoplastics on Mytilus galloprovincialis after individual and combined exposure with carbamazepine. Sci. Total. Environ. 643, 775–784. doi: 10.1016/j.scitotenv.2018.06.257
Cappello T., De Marco G., Conti G. O., Giannetto A., Ferrante M., Mauceri A., et al. (2021). Time-dependent metabolic disorders induced by short-term exposure to polystyrene microplastics in the Mediterranean mussel Mytilus galloprovincialis. Ecotoxicol. Environ. Saf. 209, 111780. doi: 10.1016/j.ecoenv.2020.111780
Carbuloni C. F., Savoia J. E., Santos J. S. P., Pereira C. A. A., Marques R. G., Ribeiro V. A. S., et al. (2020). Degradation of metformin in water by TiO2-ZrO2 photocatalysis. J. Environ. 262, 110347. doi: 10.1016/j.jenvman.2020.110347
Chamaillard M., Girardin S. E., Viala J., Philpott D. J. (2003). Nods, Nalps and Naip: intracellular regulators of bacterial-induced inflammation. Cell Microbiol. 5, 581–592. doi: 10.1046/j.1462-5822.2003.00304.x
Chen G., Pang M. X., Yu X. M., Wang J. R., Tong J. O. (2021). Transcriptome sequencing provides insights into the mechanism of hypoxia adaption in bighead carp (Hypophthalmichthys nobilis). Comp. Biochem. Phys. D. 40, 100891. doi: 10.1016/j.cbd.2021.100891
Chen J. Y., Qiu J. Y., Yang C. Y., Liao Y. S., He M. X., Mkuye R., et al. (2022a). Integrated transcriptomic and metabolomic analysis sheds new light on adaptation of Pinctada fucata martensii to short-term hypoxic stress. Mar. pollut. Bull. 187, 114534. doi: 10.1016/j.marpolbul.2022.114534
Chen N., Huang Z. K., Lu C. K., Shen Y. W., Luo X., Ke C. H., et al. (2019). Different transcriptomic responses to thermal stress in heat-tolerant and heat-sensitive pacific abalones indicated by cardiac performance. Front. Physiol. 9. doi: 10.3389/fphys.2018.01895
Chen P. B., Huang J., Rao L. Y., Zhu W. G., Yu Y. H., Xiao F. S., et al. (2022b). Environmental effects of nanoparticles on the ecological succession of gut microbiota across zebrafish development. Sci. Total. Environ. 806, 150963. doi: 10.1016/j.scitotenv.2021.150963
Chen Q. L., Gong Y., Luo Z., Zheng J. L., Zhu Q. L. (2013). Differential effect of waterborne cadmium exposure on lipid metabolism in liver and muscle of yellow catfish Pelteobagrus fulvidraco. Aquat. Toxicol. 142, 380–386. doi: 10.1016/j.aquatox.2013.09.011
Chen Q. Q., Lackmann C., Wang W. Y., Seiler T. B., Hollert H., Shi H. H. (2020). Microplastics lead to hyperactive swimming behaviour in adult zebrafish. Aquat. Toxicol., 224, 105521. doi: 10.1016/j.aquatox.2020.105521
Cole M., Liddle C., Consolandi G., Drago C., Hird C., Lindeque P. K., et al. (2020). Microplastics, microfibres and nanoplastics cause variable sub-lethal responses in mussels (Mytilus spp.). Mar. pollut. Bull. 160, 111552. doi: 10.1016/j.marpolbul.2020.111552
Danielyan K. E., Chailyan S. G. (2020). Chapter 16 - New properties of vitamin B6 or pyridoxine in experimental oxidative stress in the brain. Mol. Nutr. Food. Res. 301, 322. doi: 10.1016/B978-0-12-811907-5.00017-8
David J. P., Sidney K. P. (1981). Adaptation and tolerance of invertebrate nervous systems to osmotic stress. J. Exp. Zool. 215, 237–245. doi: 10.1002/jez.1402150303
Deng Y. W., Fu S., Lu Y. Z., Du X. D., Wang Q. H., Huang H. L., et al. (2013). Fertilization, hatching, survival, and growth of third-generation colored pearl oyster (Pinctada martensii) stocks. J. Appl. Aquac. 25, 113–120. doi: 10.1080/10454438.2013.788311
Du X. D., Fan G. Y., Jiao Y., Zhang H., Guo X. M., Huang R. L., et al. (2017). The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience 6, 100240. doi: 10.5524/100240
Duan Y. F., Yang Y. K., Zhang Z., Xing Y. F., Li H. (2023). Toxicity of titanium dioxide nanoparticles on the histology, liver physiological and metabolism, and intestinal microbiota of grouper. Mar. pollut. Bull. 187, 114600. doi: 10.1016/j.marpolbul.2023.114600
Duan J. C., Yu Y., Li Y., Li Y. B., Liu H. C., Jing L., et al. (2016). Low-dose exposure of silica nanoparticles induces cardiac dysfunction via neutrophil-mediated inflammation and cardiac contraction in zebrafish embryos. Nanotoxicology 10, 575–585. doi: 10.3109/17435390.2015.1102981
Falfushynska H., Piontkivska H., Sokolova I. M. (2020). Effects of intermittent hypoxia on cell survival and inflammatory responses in the intertidal marine bivalves Mytilus edulis and Crassostrea gigas. J. Exp. Biol. 223, 217026. doi: 10.1242/jeb.217026
Fu X., Sun Y., Wang J., Xing Q., Zou J., Li R., et al. (2014). Sequencing-based gene network analysis provides a core set of gene resource for understanding thermal adaptation in Zhikong scallop Chlamys farreri. Mol. Ecol. Resour. 14, 184–189. doi: 10.1111/1755-0998.12169
Gainey L. F., Greenberg M. J. (2003). Nitric oxide mediates seasonal muscle potentiation in clam gills. J. Exp. Biol. 206, 3507–3520. doi: 10.1242/jeb.00573
Gallo A., Boni R., Buttino I., Tosti E. (2016). Spermiotoxicity of nickel nanoparticles in the marine invertebrate Ciona intestinalis (ascidians). Nanotoxicology 10, 1096–1104. doi: 10.1080/17435390.2016.1177743
Garner K. L., Keller A. A. (2014). Emerging patterns for engineered nanomaterials in the environment: a review of fate and toxicity studies. J. Nanopart. Res. 16, 2503. doi: 10.1007/s11051-014-2503-2
Georgantzopoulou A., Cambier S., Serchi T., Kruszewski M., Balachandran Y. L., Grysan P., et al. (2016). Inhibition of multixenobiotic resistance transporters (MXR) by silver nanoparticles and ions in vitro and in Daphnia magna. Sci. Total. Environ. 569, 681–689. doi: 10.1016/j.scitotenv.2016.06.157
Girardello F., Leite C. C., Branco C. S., Roesch-Ely M., Fernandes A. N., Salvador M., et al. (2016). Antioxidant defences and haemocyte internalization in Limnoperna fortunei exposed to TiO2 nanoparticles. Aquat. Toxicol. 176, 190–196. doi: 10.1016/j.aquatox.2016.04.024
Gornati R., Longo A., Rossi F., Maisano M., Sabatino G., Mauceri A., et al. (2016). Effects of titanium dioxide nanoparticle exposure in Mytilus galloprovincialis gills and digestive gland. Nanotoxicology 10, 807–817. doi: 10.3109/17435390.2015.1132348
Guan X. F., Shi W., Zha S. J., Rong J. H., Su W. H., Liu G. X. (2018). Neurotoxic impact of acute TiO2 nanoparticle exposure on a benthic marine bivalve mollusk, Tegillarca granosa. Aquat. Toxicol. 200, 241–246. doi: 10.1016/j.aquatox.2018.05.011
Hal A. M., El-Barbary M. I. (2020). Gene expression and histopathological changes of Nile tilapia (Oreochromis niloticus) infected with Aeromonas hydrophila and Pseudomonas fluorescens. Aquaculture 526, 735392. doi: 10.1016/j.aquaculture.2020.735392
Hampton R. Y. (2002). ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 14, 476–482. doi: 10.1016/S0955-0674(02)00358-7
Han J. E., Choi S. K., Jeon H. J., Park J. K., Han S. H., Jeong J., et al. (2021). Transcriptional response in the whiteleg shrimp (Penaeus vannamei) to short-term microplastic exposure. Aquac 20, 100713. doi: 10.1016/j.aqrep.2021.100713
He C. Z., Hao R. J., Deng Y. W., Yang C. Y., Du X. D. (2020). Response of pearl oyster Pinctada fucata martensii to allograft-induced stress from lipid metabolism. Fish. Shellfish. Immunol. 98, 1001–1007. doi: 10.1016/j.fsi.2019.11.028
Herskovits A. A., Auerbuch V., Portnoy D. A. (2007). Bacterial Ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS. Pathog. 3, e51. doi: 10.1371/journal.ppat.0030051
Hu Z., Feng J., Song H., Zhou C., Yang M. J., Shi P., et al. (2022). Metabolic response of Mercenaria mercenaria under heat and hypoxia stress by widely targeted metabolomic approach. Sci. Total. Environ. 809, 151172. doi: 10.1016/j.scitotenv.2021.151172
Hu M. H., Lin D. H., Shang Y. Y., Hu Y., Lu W. Q., Huang X. Z. (2017). CO2-induced pH reduction increases physiological toxicity of nano-TiO2 in the mussel Mytilus coruscus. Sci. Rep. 2017, 40015. doi: 10.1038/srep40015
Ibrahim D., Neamat-Allah A. N. F., Ibrahim S. M., Eissa H. M., Fawzey M. M., Mostafa D. I. A., et al. (2021). Dual effect of selenium loaded chitosan nanoparticles on growth, antioxidant, immune related genes expression, transcriptomics modulation of caspase 1, cytochrome P450 and heat shock protein and Aeromonas hydrophila resistance of Nile Tilapia (Oreochromis niloticus). Fish. Shellfish. Immunol. 110, 91–99. doi: 10.1016/j.fsi.2021.01.003
Inohara N., Chamaillard M., McDonald C., Nuñez G. (2005). NOD-LRR proteins: Role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 75, 355–383. doi: 10.1146/annurev.biochem.74.082803.133347
Ivanina A. V., Taylor C., Sokolova I. M. (2009). Effects of elevated temperature and cadmium exposure on stress protein response in eastern oysters Crassostrea virginica (Gmelin). Aquat. Toxicol. 91, 245–254. doi: 10.1016/j.aquatox.2008.11.016
Izagirre U., Errasti A., Bilbao E., Múgica M., Marigómez I. (2014). Combined effects of thermal stress and Cd on lysosomal biomarkers and transcription of genes encoding lysosomal enzymes and HSP70 in mussels, Mytilus galloprovincialis. Aquat. Toxicol. 149, 145–156. doi: 10.1016/j.aquatox.2014.01.013
Jafari F., Naeemi A. S., Sohani M. M., Noorinezhad M. (2023). Effect of elevated temperature, sea water acidification, and phenanthrene on the expression of genes involved in the shell and pearl formation of economic pearl oyster (Pinctada radiata). Mar. pollut. Bull. 196, 115603. doi: 10.1016/j.marpolbul.2023.115603
Jiang Y. S., Tang X. X., Zhou B., Sun T. L., Chen H. M., Zhao X. Y., et al. (2017). The ROS-mediated pathway coupled with the MAPK-p38 signalling pathway and antioxidant system plays roles in the responses of Mytilus edulis haemocytes induced by BDE-47. Aquat. Toxicol. 187, 55–63. doi: 10.1016/j.aquatox.2017.03.011
Jitrapakdee S. (2012). Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. Int. J. Biochem. Cell. Biol. 44, 33–45. doi: 10.1016/j.biocel.2011.10.001
Johari S. A., Kalbassi M. R., Soltani M., Yu I. J. (2013). Toxicity comparison of colloidal silver nanoparticles in various life stages of rainbow trout (Oncorhynchus mykiss). Iran. J. Fish. Sci. 12, 76–7V. doi: 10.1071/MF12106
Katsumiti A., Gilliland D., Arostegui I., Cajaraville M. P. (2015). Mechanisms of toxicity of ag nanoparticles in comparison to bulk and ionic ag on mussel hemocytes and gill cells. Plos. One 10, e129039. doi: 10.1371/journal.pone.0129039
Kingtong S., Chitramvong Y., Janvilisri T. (2007). ATP-binding cassette multidrug transporters in Indian-rock oyster Saccostrea forskali and their role in the export of an environmental organic pollutant tributyltin. Aquat. Toxicol. 85, 124–132. doi: 10.1016/j.aquatox.2007.08.006
Konstantinos K., Camilla N., Roger P. (2012). Neuronal responses to physiological stress. Front. Genet. 3. doi: 10.3389/fgene.2012.00222
Kurelec B. (1995). Inhibition of multixenobiotic resistance mechanism in aquatic organisms: ecotoxic consequences. Sci. Total. Environ. 171, 197–204. doi: 10.1016/0048-9697(95)04689-4
Labille J., Slomberg D., Catalano R., Robert S., Apers-Tremelo M. L., Boudenne J. L., et al. (2020). Assessing UV filter inputs into beach waters during recreational activity: A field study of three French Mediterranean beaches from consumer survey to water analysis. Sci. Total. Environ. 706, 136010. doi: 10.1016/j.scitotenv.2019.136010
Li P. M., Gao X. L. (2014). Trace elements in major marketed marine bivalves from six northern coastal cities of China: Concentrations and risk assessment for human health. Ecotox. Environ. Safe. 109, 1–9. doi: 10.1016/j.ecoenv.2014.07.023
Li Z. Q., Hu M. H., Song H. T., Lin D. H., Wang Y. J. (2021). Toxic effects of nano-TiO2 in bivalves-A synthesis of meta-analysis and bibliometric analysis. J. Environ. Sci-China. 104, 188–203. doi: 10.1016/j.jes.2020.11.013
Li F. F., Lin Y. J., Yang C. Y., Yan Y. L., Hao R. J., Mkuye R., et al. (2024a). Effects of titanium dioxide nanoparticle exposure on the gut microbiota of pearl oyster (Pinctada fucata martensii). Comp. Biochem. Phys. C. 280, 109906. doi: 10.1016/J.CBPC.2024.109906
Li F. F., Xie Y. F., Yang C. Y., Ye Q. X., Wang F. Y., Liao Y. S., et al. (2024b). The physiological responses to titanium dioxide nanoparticles exposure in pearl oysters (Pinctada fucata martensii). Mar. Environ. Res. 195, 106345. doi: 10.1016/j.marenvres.2024.106345
Libralato G., Minetto D., Totaro S., Micetic I., Pigozzo A., Sabbioni E., et al. (2013). Embryotoxicity of TiO2 nanoparticles to Mytilus galloprovincialis (Lmk). Mar. Environ. Res. 92, 71–78. doi: 10.1016/j.marenvres.2013.08.015
Lim S., Kang H., Kwon B., Lee J. P., Lee J., Choi K. (2022). Zebrafish (Danio rerio) as a model organism for screening nephrotoxic chemicals and related mechanisms. Ecotoxicol. Environ. Saf. 242, 113842. doi: 10.1016/j.ecoenv.2022.113842
Liu Z. Q., Li Y. M., Perez E., Jiang Q. C., Chen Q., Jiao Y., et al. (2020). Polystyrene nanoplastic induces oxidative stress, immune defense, and glycometabolism change in Daphnia pulex: Application of transcriptome profiling in risk assessment of nanoplastics. J. Hazard. Mater. 402, 123778. doi: 10.1016/j.jhazmat.2020.123778
Liu Z. Q., Lv W. W., Huang Y. H., Fan B., Li Y. M., Zhao Y. L. (2016). Effects of cadmium on lipid metabolism in female estuarine crab, Chiromantes dehaani. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 188, 9–16. doi: 10.1016/j.cbpc.2016.06.002
Liu H. H., Shih T. S., Huang H. R., Huang S. C., Lee L. H., Huang Y. C. (2013). Plasma homocysteine is associated with increased oxidative stress and antioxidant enzyme activity in welders. Sci. World J. 2013, 1–8. doi: 10.1155/2013/370487
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luzio J. P., Pryor P. R., Bright N. A. (2007). Lysosomes: fusion and function. Nat. Rev. Mol. Cell. Biol. 8, 622–632. doi: 10.1038/nrm2217
Ma F., Zhao L., Ma R. L., Wang J., Du L. Q. (2023). FoxO signaling and mitochondria-related apoptosis pathways mediate tsinling lenok trout (Brachymystax lenok tsinlingensis) liver injury under high temperature stress. Int. J. Biol. Macromol. 251, 126404. doi: 10.1016/j.ijbiomac.2023.126404
Mao L. M., Wang H. H., Wang J. Q. (2017). Antagonism of muscarinic acetylcholine receptors alters synaptic ERK phosphorylation in the rat forebrain. Neurochem. Res. 42, 1202–1210. doi: 10.1007/s11064-016-2157-9
Mariman E. C. M., Wang P. (2010). Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life. Sci. 67, 1277–1292. doi: 10.1007/s00018-010-0263-4
Marin F., Le Roy N., Marie B. J. (2012). The formation and mineralization of mollusk shell. Front. Biosci. 4, 1099–1125. doi: 10.2741/s321
Martínez-Fábregas J., Prescott A., van Kasteren S., Pedrioli D. L., McLean I., Moles A., et al. (2018). Lysosomal protease deficiency or substrate overload induces an oxidative-stress mediated STAT3-dependent pathway of lysosomal homeostasis.Nat. Commun 9, 5343. doi: 10.1038/s41467-018-07741-6
Martin-Folgar R., Torres-Ruiz M., de Alba M., Cañas-Portilla A. I., González M. C., Morales M. (2022). Molecular effects of polystyrene nanoplastics toxicity in zebrafish embryos (Danio rerio). Chemosphere 312, 137077. doi: 10.1016/J.chemosphere.2022.137077
Muller E. B., Hanna S. K., Lenihan H. S., Miller R., Nisbet R. M. (2014). Impact of engineered zinc oxide nanoparticles on the energy budgets of Mytilus galloprovincialis. J. Sea. Res. 94, 29–36. doi: 10.1016/j.seares.2013.12.013
Nie H. T., Wang H. M., Jiang K. Y., Yan X. W. (2020). Transcriptome analysis reveals differential immune related genes expression in Ruditapes philippinarum under hypoxia stress: potential HIF and NF-κB crosstalk in immune responses in clam. BMC Genom 21, 318. doi: 10.1186/s12864-020-6734-6
Papassotiropoulos A., de Quervain D. J. F. (2015). Failed drug discovery in psychiatry: time for human genome-guided solutions. Trends. Cogn. Sci. 19, 183–187. doi: 10.1016/j.tics.2015.02.002
Pinsino A., Bergami E., Della Torre C., Vannuccini M. L., Addis P., Secci M., et al. (2017). Amino-modified polystyrene nanoparticles affect signalling pathways of the sea urchin (Paracentrotus lividus) embryos. Nanotoxicology 11, 201–209. doi: 10.1080/17435390.2017.1279360
Ray P. D., Huang B. W., Tsuji Y. (2012). Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal 24, 981–990. doi: 10.1016/j.cellsig.2012.01.008
Ringwood A. H., Levi-Polyachenko N., Carroll D. L. (2009). Fullerene exposures with oysters: embryonic, adult, and cellular responses. Environ. Sci. Technol. 43, 7136–7141. doi: 10.1021/es900621j
Rivera-Perez C., de Areyano J. J. O. R., Hernandez-Saavedra N. Y. (2019). Purification and functional analysis of the shell matrix protein N66 from the shell of the pearl oyster Pteria sterna. Comp. Biochem. Phys. B 235, 19–29. doi: 10.1016/j.cbpb.2019.05.007
Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rocha T. L., Gomes T., Cardoso C., Letendre J., Pinheiro J. P., Sousa V. S., et al. (2014). Immunocytotoxicity, cytogenotoxicity and genotoxicity of cadmium-based quantum dots in the marine mussel Mytilus galloprovincialis. Mar. Environ. Res. 101, 29–37. doi: 10.1016/j.marenvres.2014.07.009
Rokutan K. (2000). Role of heat shock proteins in gastric mucosal protection. J. Gastroenterol. Hepatol. 15, D12–D19. doi: 10.1046/j.1440-1746.2000.02144.x
Salvador J. M., Brown-Clay J. D., Fornace A. J. (2013). Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv. Exp. Med. Biol. 793, 1–19. doi: 10.1007/978-1-4614-8289-5_1
Samokhvalov V., Scott B. A., Crowder C. M. (2008). Autophagy protects against hypoxic injury in C-elegans. Autophagy 4, 1034–1041. doi: 10.4161/auto.6994
Schneider G., Kack H., Lindqvist Y. (2000). The manifold of vitamin B6 dependent enzymes. Structure 8, 1–6. doi: 10.1016/S0969-2126(00)00085-X
Sedanza M. G., Yoshida A., Kim H. J., Yamaguchi K., Osatomi K., Satuito C. G. (2022). Identification and characterization of the larval settlement pheromone protein components in adult shells of crassostrea gigas: A novel function of shell matrix proteins. Int. J. Mol. Sci. 23, 9816. doi: 10.3390/ijms23179816
Sharon D., Tilgner H., Grubert F., Snyder M. (2013). A single-molecule long-read survey of the human transcriptome. Nat. Biotech. 31, 1009. doi: 10.1038/nbt.2705
Sheng L., Wang L., Su M. Y., Zhao X. Y., Hu R. P., Yu X. H., et al. (2016). Mechanism of TiO2 nanoparticle-induced neurotoxicity in zebrafish (Danio rerio). Environ. Toxicol. 31, 163–175. doi: 10.1002/tox.22031
Sherman M. Y., Goldberg A. L. (2001). Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29, 15–32. doi: 10.1016/s0896-6273(01)00177-5
Shi X. ,. M., Li Z. X., Chen W., Qiang L. W., Xia J. C., Chen M., et al. (2016). Fate of TiO2 nanoparticles entering sewage treatment plants and bioaccumulation in fish in the receiving streams. Nanoimpact 3-4, 96–103. doi: 10.1016/j.impact.2016.09.002
Sun T. Y., Bornhöft N. A., Hungerbühler K., Nowack B. (2016). Dynamic probabilistic modeling of environmental emissions of engineered nanomaterials. Environ. Sci. Technol. 50, 4701–4711. doi: 10.1021/acs.est.5b05828
Sun X. J., Tu K., Li L., Biao W., Wu L., Liu Z. H., et al. (2021). Integrated transcriptome and metabolome analysis reveals molecular responses of the clams to acute hypoxia. Mar. Environ. Res. 168, 105317. doi: 10.1016/j.marenvres.2021.105317
Urlacher V. B., Girhard M. (2012). Cytochrome P450 monooxygenases: an update on perspectives for synthetic application. Trends. Biotechnol. 30, 26–36. doi: 10.1016/j.tibtech.2011.06.012
Valavanidis A., Vlahogianni T., Dassenakis M., Scoullos M. (2006). Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol. Environ. Saf. 64, 178–189. doi: 10.1016/j.ecoenv.2005.03.013
Wang K., Che W. A., Duan M. M., Wang C. J., Li X. W., He L. (2023). Effects of broflanilide on oxidative stress and expression of apoptotic genes in zebrafish (Danio rerio) gill. B. Environ. Contam. Tox. 110, 91–91. doi: 10.1007/s00128-023-03733-5
Wegner A., Besseling E., Foekema E. M., Kamermans P., Koelmans A. A. (2012). Effects of nanopolystyrene on the feeding behavior of the blue mussel (Mytilus edulis L.). Environ. Toxicol. Chem. 31, 2490–2497. doi: 10.1002/etc.1984
Wei J. L., Liu J. H., Liang S., Sun M. Q., Duan J. C. (2020). Low-dose exposure of silica nanoparticles induces neurotoxicity via neuroactive ligand-receptor interaction signaling pathway in zebrafish embryos. Int. J. Nanomed. 15, 4407–4415. doi: 10.2147/IJN.S254480
Wei S. S., Sun B. Y., Liu C. H., Sokolova I., Waiho K., Fang J. K. H., et al. (2023). Nano-TiO2 aggravates the adverse effect of pentachlorophenol on antioxidant and immune response in anti-predatory mussels. Sci. Total. Environ. 893, 164836. doi: 10.1016/j.scitotenv.2023.164836
Wu S. J., Huang J. Q., Li Y. J., Pan Y. C. (2023). Dynamic and systemic regulatory mechanisms in rainbow trout (Oncorhynchus mykiss) in response to acute hypoxia and reoxygenation stress. Aquaculture 572, 739540. doi: 10.1016/j.aquaculture.2023.739540
Wu H. L., Yang C. Y., Hao R. J., Liao Y. S., Wang Q. H., Deng Y. W. (2022). Lipidomic insights into the immune response and pearl formation in transplanted pearl oyster Pinctada fucata martensii. Front. Immunol. 13. doi: 10.3389/fimmu.2022.1018423
Xu L. M., Li J. R., Huang Y., Zhao M., Tang X., Wei L. P. (2012). AutismKB: an evidence-based knowledgebase of autism genetics. Nucleic. Acids Res. 40, 1016–1022. doi: 10.1093/nar/gkr1145
Yang C. Y., Hao R. J., Deng Y. W., Liao Y. S., Wang Q. H., Sun R. J., et al. (2017). Effects of protein sources on growth, immunity and antioxidant capacity of juvenile pearl oyster Pinctada fucata martensii. Fish. Shellfish. Immunol. 67, 411–418. doi: 10.1016/j.fsi.2017.06.037
Yang C. Y., Wu H. L., Chen J. Y., Liao Y. S., Mkuye R., Deng Y. W., et al. (2023). Integrated transcriptomic and metabolomic analysis reveals the response of pearl oyster (Pinctada fucata martensii) to long-term hypoxia. Mar. Environ. Res. 191, 106133. doi: 10.1016/j.marenvres.2023.106133
Yang C. Y., Yang J. M., Hao R. J., Du X. D., Deng Y. W. (2019). Molecular characterization of OSR1 in Pinctada fucata martensii and association of allelic variants with growth traits. Aquaculture 516, 734617. doi: 10.1016/j.aquaculture.2019.734617
Yang C. Y., Zeng Y. T., Liao Y. S., Deng Y. W., Du X. D., Wang Q. H. (2021). Integrated GC-MS- and LC-MS-Based Untargeted Metabolomics Studies of the Effect of Vitamin D3 on Pearl Production Traits in Pearl Oyster Pinctada fucata martensii. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.614404
Yang C. Y., Zhang C. S., Huang. J., Hao R. J., Shi S. L., Huang R. L., et al. (2022). Sequence and SNP analysis of Kelch⁃like gene from pearloyster Pinctada fucata martensii. J. Dalian Ocean Univ. 37, 146. doi: 10.16535/j.cnki.dlhyxb.2021-146
Yin J., Hu J., Deng X. D., Zheng Y., Tian J. J. (2023). ABC transporter-mediated MXR mechanism in fish embryos and its potential role in the efflux of nanoparticles. Ecotoxicol. Environ. Saf. 263, 115397. doi: 10.1016/j.ecoenv.2023.115397
Young T., Kesarcodi-Watson A., Alfaro A. C., Merien F., Nguyen T. V., Mae H., et al. (2017). Differential expression of novel metabolic and immunological biomarkers in oysters challenged with a virulent strain of OsHV-1. Dev. Comp. Immunol. 73, 229–245. doi: 10.1016/j.dci.2017.03.025
Ze X., Su M. Y., Zhao X. Y., Jiang H., Hong J., Yu X. H., et al. (2016). TiO2 nanoparticle-induced neurotoxicity may be involved in dysfunction of glutamate metabolism and its receptor expression in mice. Environ. Toxicol. 31, 655–662. doi: 10.1002/tox.22077
Zhang X. K., Fan C., Zhang X. Z., Li Q. Z., Li Y. C., Ma P. Z., et al. (2022). Transcriptome Analysis of Crassostrea sikamea (♀) × Crassostrea gigas (♂) Hybrids Under and After Thermal Stress. J. Ocean Univ. China 21, 213–224. doi: 10.1007/s11802-022-4829-1
Zhang G. F., Fang X. D., Guo X. M., Li L., Luo R. B., Xu F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zheng Z., Hao R. J., Yang C. Y., Jiao Y., Wang Q. H., Huang R. L., et al. (2023). Genome-wide association study analysis to resolve the key regulatory mechanism of biomineralization in Pinctada fucata martensii. Mol. Ecol. Resour. 23, 680–693. doi: 10.1111/1755-0998.13743
Zhou Z., Liu T., Kong J., Zeng S., Zhu J. Q. (2024). Toxicity and transcriptome sequencing analyses of Acipenser schrenckii under titanium dioxide nanoparticles stress. Aquacult. Rep. 36, 102091. doi: 10.1016/J.AQREP.2024.102091
Keywords: TiO2-NPs, Pinctada fucata martensii, transcriptomics, protein homeostasis, oxidative stress
Citation: Li F, Liu J, Gao Z, Yang C, Sun L, Liao Y and Deng Y (2024) Transcriptomic analyses of Pinctada fucata martensii responses under stress of titanium dioxide nanoparticles. Front. Mar. Sci. 11:1462589. doi: 10.3389/fmars.2024.1462589
Received: 10 July 2024; Accepted: 07 August 2024;
Published: 23 August 2024.
Edited by:
Yiming Li, Fishery Machinery and Instrument Research Institute, ChinaCopyright © 2024 Li, Liu, Gao, Yang, Sun, Liao and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chuangye Yang, eWFuZ2N5QGdkb3UuZWR1LmNu
 Fengfeng Li
Fengfeng Li Jiaen Liu
Jiaen Liu Zixin Gao1
Zixin Gao1 Chuangye Yang
Chuangye Yang Liwei Sun
Liwei Sun Yongshan Liao
Yongshan Liao Yuewen Deng
Yuewen Deng