- 1Fisheries Research Institute, Sichuan Academy of Agricultural Sciences, Yibin, China
- 2Key Laboratory of Sichuan Province for Fishes Conservation and Utilization in the Upper Reaches of the Yangtze River, Neijiang Normal University, Neijiang, China
- 3College of Life Science, Neijiang Normal University, Neijiang, China
- 4School of Animal Science, Yangtze University, Jingzhou, China
- 5Shenzhen Key Lab of Marine Genomics, Guangdong Provincial Key Lab of Molecular Breeding in Marine Economic Animals, BGI Academy of Marine Sciences, BGI Marine, Shenzhen, China
- 6Department of Animal and Aquatic Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai, Thailand
- 7National Genetic Breeding Center of Channel Catfsh, Freshwater Fisheries Research Institute of Jiangsu Province, Nanjing, China
- 8Yueyang Yumeikang Biotechnology Co., Ltd., Yueyang, China
- 9Laboratory of Aquatic Genomics, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen, China
Fatty acid desaturases (FADs) are rate-limiting enzymes for the biosynthesis of highly unsaturated fatty acids (HUFAs). As a new member of the FAD family, Fad6 and its roles remain unclear in various teleost fishes. In this study, we identified a fad6 gene from yellow catfish (ycfad6) and determined its spatiotemporal expression patterns and responses to different initial diets and nutritional status in yellow catfish. Our results showed that the open reading frame (ORF) of ycfad6 was 1,080 bp in length, encoding a protein of 359 amino acids. Multiple protein sequences alignment proved that fad6 is highly conserved among diverse vertebrates. Meanwhile, phylogenetic analysis revealed that Southern catfish and yellow catfish were clustered into one branch, supporting evolutionary consistence between the fad6 gene and fish morphology. Moreover, comparisons of genomic synteny and gene structure revealed functional and evolutionary conservation of the fad6 gene in various teleost fishes. Tissue distribution analysis by quantitative RT-PCR demonstrated that the ycfad6 gene was extensively expressed in examined tissues, with higher transcription levels in the heart and liver. Meanwhile, ycfad6 gene was widely expressed in various developmental stages, indicating Fad6 may play important roles in HUFA biosynthesis at early developmental stages in yellow catfish. Functional experiments verified that the transcription of ycfad6 decreased significantly with the extension of feeding time (with egg yolk or Artemia nauplii) at the early developmental stages, indicating that a diet rich in HUFA can remarkably inhibit the transcription of ycfad6 in yellow catfish. In addition, ycfad6 transcription was significantly reduced after a short-term (24-h) or long-term food deprivation (1-week) and then continued to decrease during refeeding, suggesting that nutritional states can affect the transcription of ycfad6, which further regulates the metabolism of HUFAs. Anyway, these fundamental findings provide basic references for further investigating evolutionary and physiological functions of the fad6 gene in yellow catfish as well as in other teleost fishes.
Introduction
Highly unsaturated fatty acids (HUFAs) are essential for promotion of normal growth and development in various organisms, and play important roles in maintaining the structure and function of cell membranes, energy metabolism signal transduction, and other physiological activities (Awada et al., 2013; Muhlhausler and Ailhaud, 2013; Nayak et al., 2017). Representative HUFAs, such as docosahexaenoic acid (DHA, 22:6n-3), eicosapentaenoic acid (EPA, 20:5n-3), and arachidonic acid (ARA, 20:4n-6), are critical fatty acids with important roles in fatty acids metabolism, maintaining cell membrane integrity and fluidity, eicosanoid synthesis, inflammation, and cell division signaling and regulation (Kuah et al., 2016; Lopes-Marques et al., 2018). HUFAs are mainly obtained from diets, and fish meat is the primary source of HUFAs consumption for human (Li et al., 2019a, 2020; Xie et al., 2021). Indeed, fishes usually have the capacity to biosynthesize HUFAs through a series of desaturation and elongation reactions from several C18 PUFA precursors, such as α-linolenic acid (ALA, 18:3n-3) and linoleic acid (LA, 18:2n-6) (Monroig et al., 2011; Zou et al., 2019). These reactions are mainly regulated by two rate-limiting enzymes, namely elongases of very long chain fatty acids (Elovls) and fatty acid desaturases (Fads) (Zeng et al., 2023). The former can prolong the carbon chain of fatty acids, while the latter can introduce an unsaturated double bond between two specific carbon atoms (Li et al., 2010; Wen et al., 2020).
Fatty acid desaturases (FADs) have been widely reported in plants and animals, and they play important roles in maintaining the structure and function of biofilms (Xie et al., 2021; Halim et al., 2022). In general, two different Fad-like desaturases have been identified in mammals, including FAD1 (Δ5 desaturase) and FAD2 (Δ6 desaturase) (Guillou et al., 2010). Differently, fad1 gene is only found in cartilaginous fish and early evolutionary fish (such as sea chubs), but fad2 gene is widely identified in almost all teleost fishes (Castro et al., 2016; Dong et al., 2018; Lopes-Marques et al., 2018). Interestingly, teleost usually contain variable number of fad2 gene in various species, such as one copy in zebrafish (Danio rerio) (Blahova et al., 2022), two copies in rabbitfish (Siganus canaliculatus) (Li et al., 2010; Dong et al., 2018; Li et al., 2019b), three copies in Nile tilapia (Oreochromis niloticus) (Castro et al., 2016; Chen et al., 2018; Dong et al., 2018; Jangprai and Boonanuntanasarn, 2018) or even four copies in Atlantic salmon (Salmo salar) (Hastings et al., 2004; Monroig et al., 2010). However, some species such as the pufferfish (Takifugu rubripes) and spotted green pufferfish (Tetraodon nigroviridis) do not have any fad genes (Castro et al., 2016; Dong et al., 2018). The various copies of fad2 gene in different fish species may be caused by the whole-genome duplication events occurred in teleost (Li et al., 2020; Wen et al., 2020). Recently, a new member of the fad gene family (fad6) has been identified and it is shown to be widespread in vertebrates. The teleost fad6 gene was firstly cloned and characterized in golden pompano (Trachinotus ovatus), and functional experiments demonstrated that this Fad6 desaturase owns capacity of Δ8/Δ5/Δ4 enzyme activities and thus participates in endogenous synthesis of HUFAs (Zhu et al., 2018). However, the exact roles of Fad6 are still rarely unknown in teleost, and therefore more studies are required to resolve these issues.
Yellow catfish (Pseudobagrus fulvidraco), belonging to the order Siluriformes and the family Bagridae, is a benthic omnivorous fish that commonly found in freshwater (Wei et al., 2023). Due to its high nutritional value and good taste, yellow catfish has been widely cultured in Asian countries, especially in China (Liu et al., 2019). Thus far, numerous studies have focused on various aspects such as breeding habits, developmental biology, lipid metabolism, and toxicology in yellow catfish (Yang et al., 2010; Liu et al., 2019; Fang et al., 2020). Meanwhile, several live prey species enriched with HUFAs (such as cladoceran) or artificial feeds containing HUFAs (such as egg yolk) are commonly used as initial diets in cultivation of yellow catfish industry (Wei et al., 2014; Zhou et al., 2016). Regarding HUFAs biosynthesis, fad2 and elovl5 genes were cloned and their transcriptional regulation by leptin system was determined (Song et al., 2015). Additionally, a teleost-specific fatty acid elongase (elovl8) gene was identified in yellow catfish, and its potential roles in HUFAs biosynthesis and fatty acids metabolism were investigated (Zeng et al., 2023). However, the structural and functional characteristics of the fad6 gene involved in HUFAs biosynthesis are still unknown in yellow catfish. In the present study, the fad6 gene was identified in yellow catfish (ycfad6), and then its molecular and evolutionary properities were characterized. Meanwhile, the spatio-temporal expression patterns of the ycfad6 gene were also examined. Furthermore, the regulatory effects of different nutritional status on the ycfad6 transcription were also explored. Our findings will not only offer basic data for further investigations on the physiological functions of Fad6 in teleost, but also provide novel insights into the regulatory mechanism for HUFAs synthesis in various vertebrates.
Materials and methods
Fish sampling and feeding trails
The yellow catfish (body weight = 32.6 ± 3.1g) used in this study were obtained from Neijiang Fish Farm (Neijiang, China) and were transported to net cages (1 m × 1 m × 1 m) erected in a 16-m2 micro-flowing water fish pond in the College of Life Sciences at Neijiang Normal University (Neijiang, China). Fishes were kept in the cages under natural light and dark conditions (12 L/12 D) with a constant flow of filtered water at 24-26°C. Fishes were fed with commercial feeds (3-4% of body weight) at 19:00 every day. Fishes were acclimated to these conditions for 2 weeks before the formal experiments, showing a normal feeding pattern during this acclimation period.
After acclimation, five fishes were randomly selected for tissue distribution studies. These fishes were anesthetized on ice before decapitation, and 12 tissue samples including adipose, barbel, brain, gill, gonad, heart, intestine, kidney, liver, muscle, spleen, and stomach, were collected and immediately frozen in liquid nitrogen until use.
For the short-term food deprivation experiment, fishes were divided into five tanks (10 fishes per tank) and these fishes were fed simultaneously at 18:30, then five fishes were separately selected from each tank for sampling livers at 18:30 (0 h), 21:30 (3 h), 00:30 (6 h), 06:30 (12 h), and 18:30 (24 h) after feeding. For the long-term food deprivation experiment, fishes were assigned to three experimental tanks (marked with A, B, and C groups; 10 fishes per tank; A group was fed but B and C groups were fasted) for one week. Subsequently, five fishes were randomly selected from the B group and their livers were sampled at 18:30. Meanwhile, fishes in both A and C groups were fed at 18:30, and they were allowed to feeding for half an hour before sampling their livers at 19:00. All samples were immediately frozen in liquid nitrogen and then stored at -80°C until use.
Meanwhile, fertilized eggs of yellow catfish used in this study were obtained by artificial breeding in our laboratory at Neijiang Normal University. During incubation, the development status of embryos was observed under a microscope, and five embryos or larvae were respectively sampled at each time point including 0, 3, 6, 12, 24, 36, 48, 72, and 96 h after fertilization at the early developmental stages. Similarly, samples were frozen in liquid nitrogen and then stored at -80°C until use.
Diets with different contents of HUFAs, including A. nauplii or egg yolk, were separately used as larval feeds to investigate the potential effects of various diets on transcription of fad6 in yellow catfish. The nutritional content of these diets can be observed in a previous study (Zhou et al., 2016). Meanwhile, approximately 200 larvae were randomly selected and then fed with different diets on day 4 after hatching (three parallel experiments were designed for each group). During the experiment, 25% water changes were carried out daily, the oxygenation pump was kept continuously oxygenated and the dissolved oxygen in the water was maintained above 6 mg/L. Fishes were fed at 8:00, 12:00, 16:00, and 20:00 to ensure sufficient diet supplements in the experimental tanks. The experiment was lasted for three days, and five offsprings from each tank were collected at 24, 48, and 72 h after feeding. Finally, samples were immediately frozen in liquid nitrogen and stored at -80°C.
Total RNA extraction and cDNA template preparation
About 50~100 mg liver tissues were selected for total RNA isolation with a commercial RNA extraction kit (Tiangen, Beijing, China). After validation by agarose gel electrophoresis, the RNA concentration was measured by an Agilent 2100 bioanalyzer (Musen, Shanghai, China), and then the RNA was reversely transcribed to cDNA by using the Revert Aid First Stand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, USA) for subsequent work.
Identification of fad6 gene in yellow catfish
Protein-similarity blast was conducted to identify the fad6 gene from the yellow catfish genome in NCBI database using several known fish Fad6 protein sequences as queries. Then, we designed two pairs of primers (Supplementary Table 1) with the Primer Premier 5.0 software to amplify the ORF regions of ycfad6.
Basic cycling conditions of the PCRs were set as follows: a denaturing stage at 94°C for 30 s, gene-specific annealing at 55°C for 30 s, and an elongation stage at 72°C for 60 s, a total of 34 cycles. Subsequently, target fragments were obtained and purified for Sanger sequencing. Finally, PCR products were cloned into the pMD-19T vector (TaKaRa, Dalian, China) and then sequenced at Sangon Co. Ltd. (Guangzhou, China) to ensure accuracy of the potential fad6 gene in yellow catfish.
Sequence comparison and evolutionary analysis of the fad6 gene
The sequence of ycfad6 was validated by BLAST (http://www.ncbi.nlm.nih.gov/BLAST) in the NCBI database. After validation, the Primer 5.0 software was applied to predict corresponding protein sequence. Multiple sequences alignment was conducted in MEGA11 using the MegAlign model, and the alignment results were annotated using GeneDoc 2.7 (Wei et al., 2023). Meanwhile, comparative analyses of genomic synteny and gene structure were performed to determine the evolution pattern of fad6 genes in teleost.
Additionally, a phylogenetic tree was constructed on the basis of the protein sequences of various teleost to investigate the evolutionary history of the fad6 gene family. All Fad6 protein sequences used for the phylogeny construction were downloaded from the NCBI database. Details of the selected Fad6 family proteins are shown in Supplementary Table 2. The phylogeny was established with the neighbor joining (NJ) method to explore the evolutionary relationship among fad6 gene family in vertebrates.
Quantitative real-time PCR
Total RNA was extracted from fish tissues and first-strand cDNA was synthesized in the same way as described above (in subsection 2.2). Subsequently, the mRNA level of ycfad6 was detected by qRT-PCR on a Light Cycler Real-Time System (Roche Life Science, Indianapolis, IN, USA), and the final volume of this reaction system was maintained at 20 μL. Meanwhile, the relative expression level of mRNA was normalized with β-actin (Supplementary Table 1) after assessing the stability of five reference genes (β-actin, GAPDH, EF1α, Tubα 1 and 18S). Finally, the relative transcription level of fad6 was calculated using the Pfaffl method (Yang et al., 2018; Wen et al., 2019). Sequences of the specific primers used for qRT-PCRs are provided in Supplementary Table 1.
Statistical analysis
All data are expressed as mean ± SEM (standard errors of the mean). Statistical analysis was performed with SPSS 22.0 (IBM, Armonk, NY, USA) and GraphPad Prism 7.0 (GraphPad Prism Software Inc., San Diego, CA, USA). Significant differences were calculated using the one-way analysis of variance (ANOVA) followed by Tukey’s and Duncan’s multiple range tests, after confirming data normality and homogeneity of variances. The difference among groups was considered as significant when P < 0.05.
Results
Molecular characterization of the fad6 gene in yellow catfish
In this study, we identified the fad6 gene from yellow catfish for the first time. The ORF of ycfad6 was 1,080 bp, predicted to encode a protein of 359 amino acids. Multiple sequences alignment showed that protein sequences of Fad6s were unconservative in the N-terminal but were highly conserved in the core FADS-like domains including three highly conserved histidine signature motifs (HXXH, HXXXHH, HXXHH; see more details in Figure 1).

Figure 1 Multiple sequences alignment of Fad6 among yellow catfish and other representative teleost. Three highly conserved histidine regions (HXXH, HXXXH, HXXHH) are highlighted within black boxes. Eight conserved histidine residues are marked in red type. Membrane-FADS-like domains are underlined.
Phylogenetic analysis
In this study, a phylogenetic tree inferred by the neighbor-joining method was constructed with a series of protein sequences of vertebrate fad6 genes, showing high bootstrap values (Figure 2). The tree was divided into two groups, including teleost and the other vertebrates. The topology also displayed that yellow catfish (P. fulvidraco) was clustered with channel catfish (I. Punctatus), striped catfish (P. hypophthalmus), North African catfish (C. gariepinus), and Southern catfish (S. meridionalis), and it showed the closest relationship with the Southern catfish. All clades with high scores and the tetrapods were used as the outgroup (Figure 2).

Figure 2 Phylogeny reveals the evolutionary history of fad6 gene family in vertebrates. Numbers on the branches represent the reliability of this tree. Tetrapod fad6 genes were set as the outgroup. The aimed species investigated in the present study is highlighted in red type.
Genetic synteny and gene structure comparison
Comparative genomics was conducted to investigate the genomic characteristics of the fad6 gene in eight representative fish species. Our results revealed a highly conserved gene cluster mrp138-fdxr-hid1b-ush1gb-fad6-trim16-tvp23b-exoc7-plaub, which was identified in almost all the selected representative fish genomes (Figure 3). Meanwhile, a gene structure comparison of the fad6 gene in eight fishes was also conducted. It appears that fad6 genes in L. rohita, P. tetrazona, C. idella, D. rerio, M. amblycephala, C. gariepinus and P. hypophthalmus had similar gene structures, containing six exons and five introns. However, the P. fulvidraco fad6 contained seven exons and six introns, although it shared a conserved coding sequence (CDS) with the other seven fish fad6 genes (Figure 4).
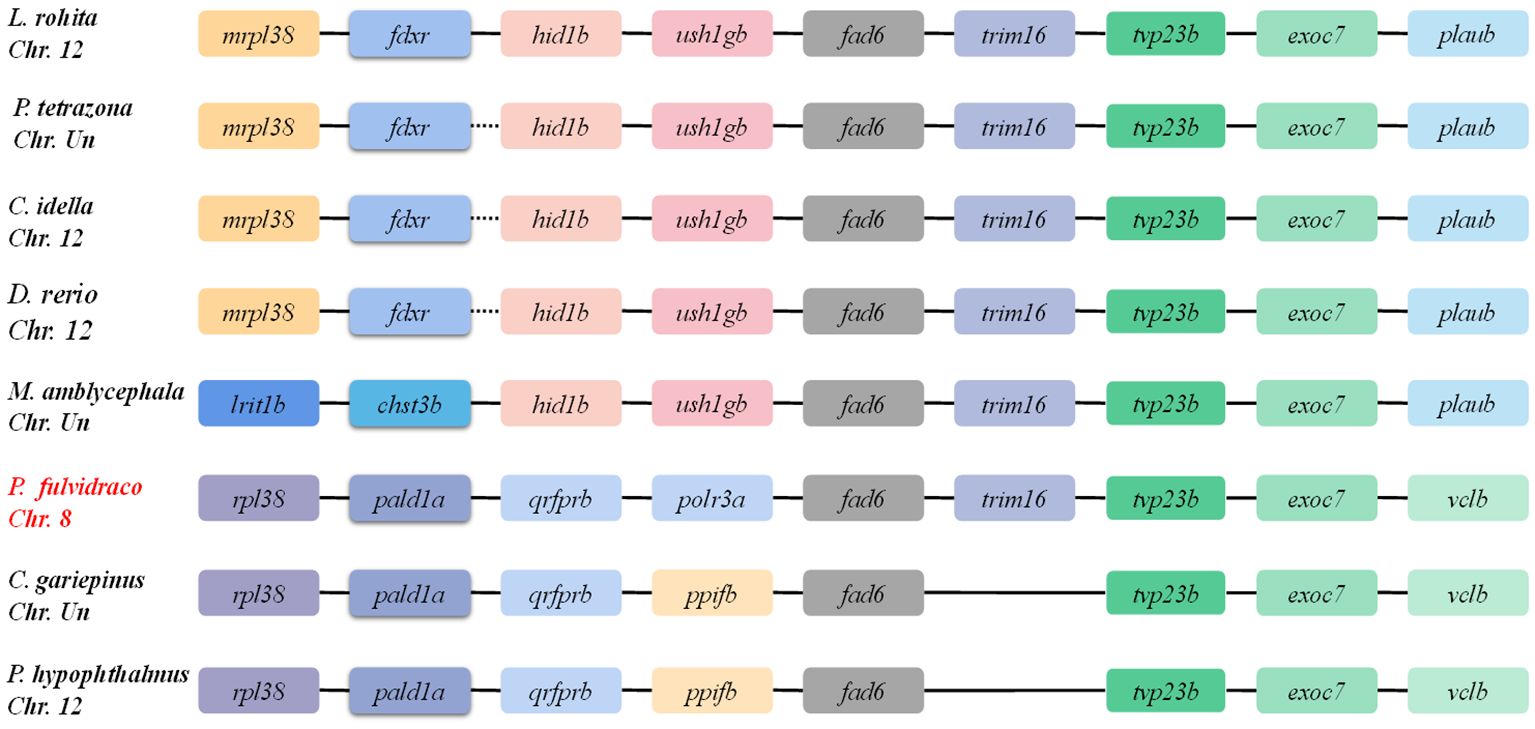
Figure 3 Genetic synteny of fad6 in representative fish genomes. These colorful blocks represent different genes. The solid lines represent intergenic regions. The target yellow catfish is highlighted in red.
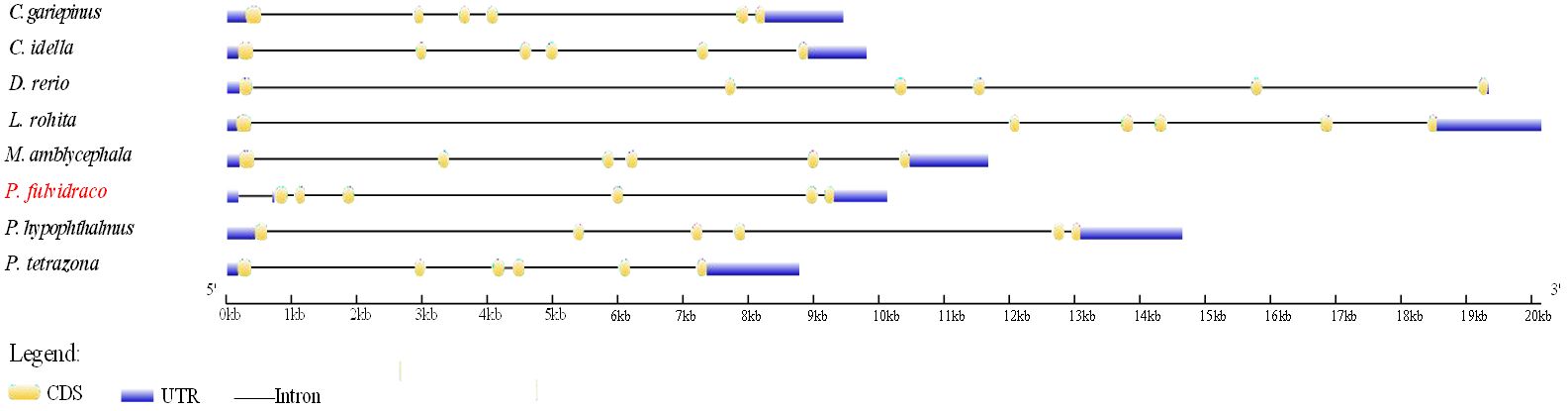
Figure 4 Comparison of the fad6 gene structure in eight representative teleost fishes. The colorful blocks and solid lines represent the exons and introns, respectively. UTR (untranslated region) and CDS (coding sequence) are represented by blue and yellow blocks, respectively. The aimed species investigated in the present study is highlighted in red type.
Tissue distribution pattern of fad6 in yellow catfish
Transcription of the ycfad6 gene in 12 tissues of yellow catfish was measured and analyzed by qRT-PCR. Our results showed that ycfad6 was widely transcribed in various tissues including adipose, barbel, brain, gill, gonad, heart, intestine, kidney, liver, muscle, spleen, and stomach (Figure 5). The highest transcription level was present in the heart, which is followed by the liver, adipose, brain, gill, gonad, intestine, kidney, and muscle, while the least transcription levels were detected in the beard, spleen and stomach (Figure 5).
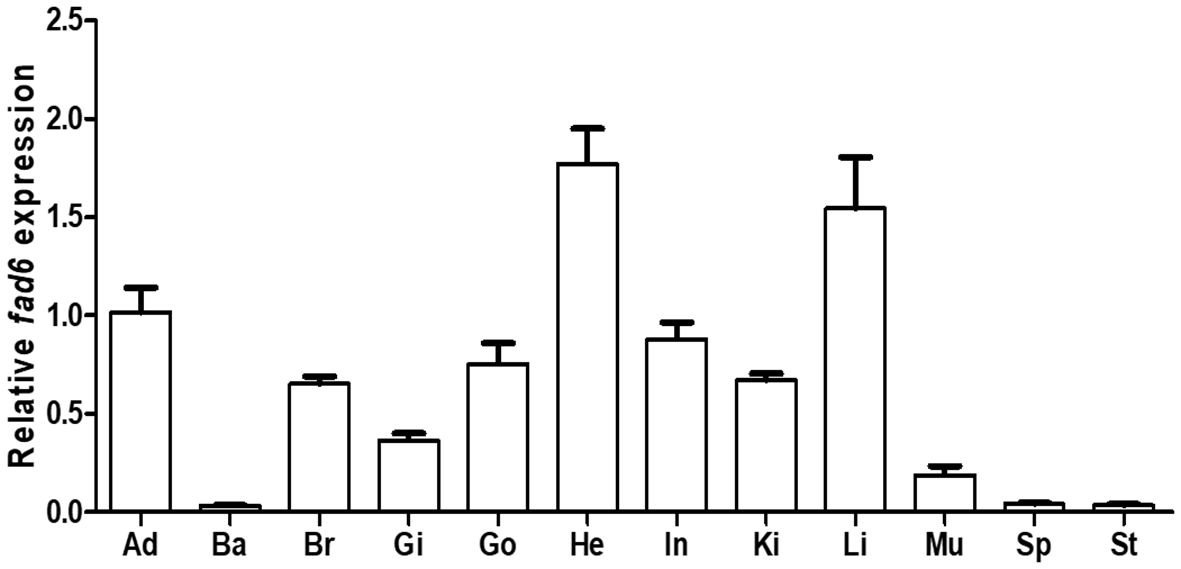
Figure 5 Tissue distribution of the ycfad6 gene in yellow catfish. Examined tissues include adipose (Ad), barbel (Ba), brain (Br), gill (Gi), gonad (Go), heart (He), intestine (In), kidney (Ki), liver (Li), muscle (Mu), spleen (Sp), and stomach (St). β-actin was selected as the internal reference gene. Data are expressed as mean ± standard error (n = 5).
Expression pattern of fad6 gene at early developmental stages in yellow catfish
qRT-PCR was performed to detect the transcription of ycfad6 at different developmental stages, and the results demonstrated that the ycfad6 gene was transcribed at all examined stages (Figure 6). As observed in Figure 6, the transcription level of fad6 gene was significantly decreased at 0, 3 and 6 h after fertilization, and then dramatically increased at 12, 24, 36, 48, and 72 h after fertilization, and finally further elevated to the highest expression level at 96 h after fertilization (Figure 6).
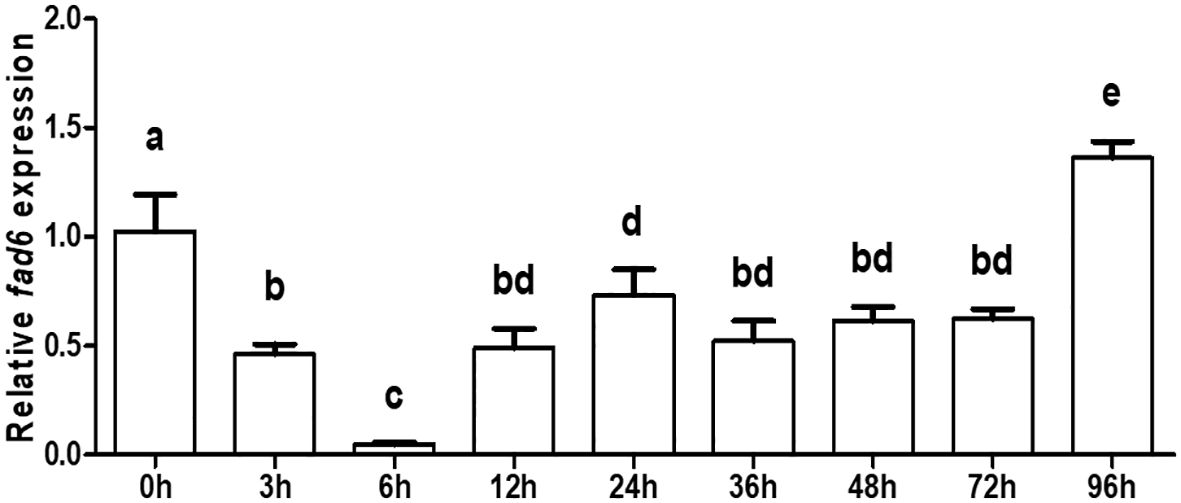
Figure 6 Gene expression pattern of the ycfad6 gene at the early developmental stages. β-actin was selected as the internal reference gene. Data are expressed as mean ± standard error (n = 5). Different lowercase letters above the bars mean significant difference among groups.
Effects of different initial diets on the transcription of fad6 gene in yellow catfish
Transcriptional changes of the yellow catfish larvae fad6 gene in response to different initial diets (with different concentrations of HUFAs) were determined (Figure 7). Our results showed that the transcription level of the ycfad6 gene was significantly decreased at 2 and 3 d after invariably feeding with egg yolk (Figure 7A). Similarly, the transcription level of the ycfad6 gene was also obviously decreased at 2 and 3 d after feeding with A. nauplii as the initial feed (Figure 7B).
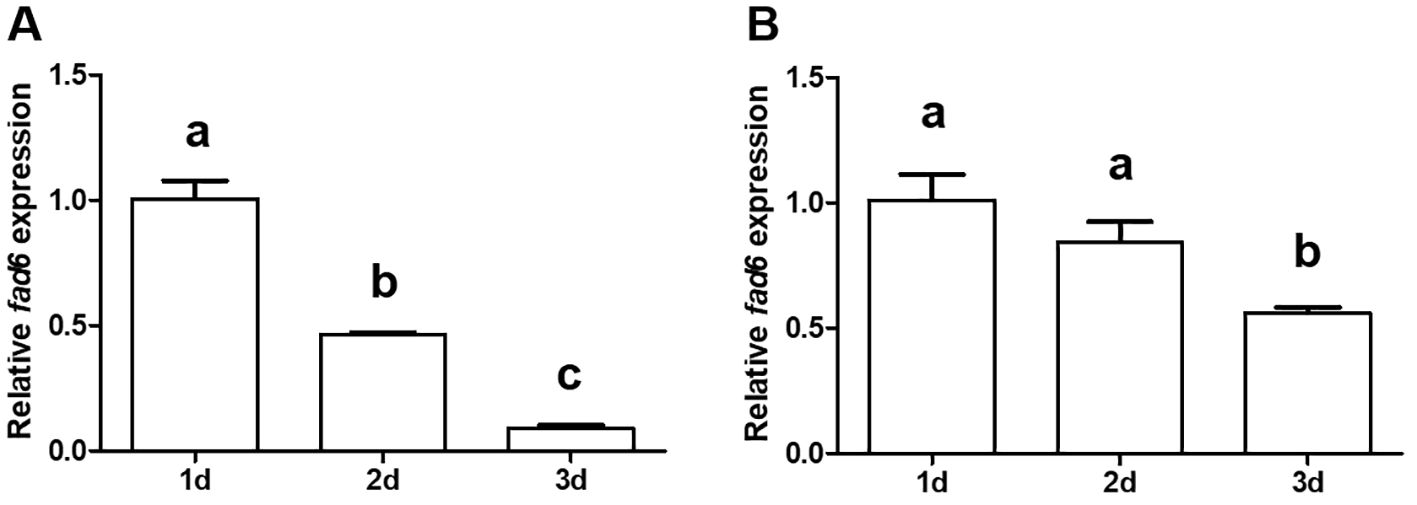
Figure 7 Effect of dietary egg yolk (A) or A. nauplii (B) on the transcription of ycfad6 in yellow catfish larvae. β-actin was selected as the internal reference gene. Data are expressed as mean ± standard error (n = 5). Different lowercase letters above the bars mean significant difference among groups.
Effects of fasting and refeeding on the fad6 transcription in yellow catfish
Hepatic fad6 gene transcription levels were measured after a short-term (24-h) or long-term fasting (1-week) and refeeding to investigate the effects of various nutritional status on the HUFA biosynthesis in yellow catfish. For the short-term fasting experiment, fad6 transcription levels were found to be significantly elevated at 3 h after fasting and then returned to the control levels at 6, 12, and 24 h after food deprivation (Figure 8A). For the long-term experiment, fad6 transcription levels were significantly decreased in fishes from the 1-week fasting group in comparison with those in the control group (Figure 8B). However, refeeding had no dramatical effect on the transcription of hepatic fad6 in yellow catfish (Figure 8B).
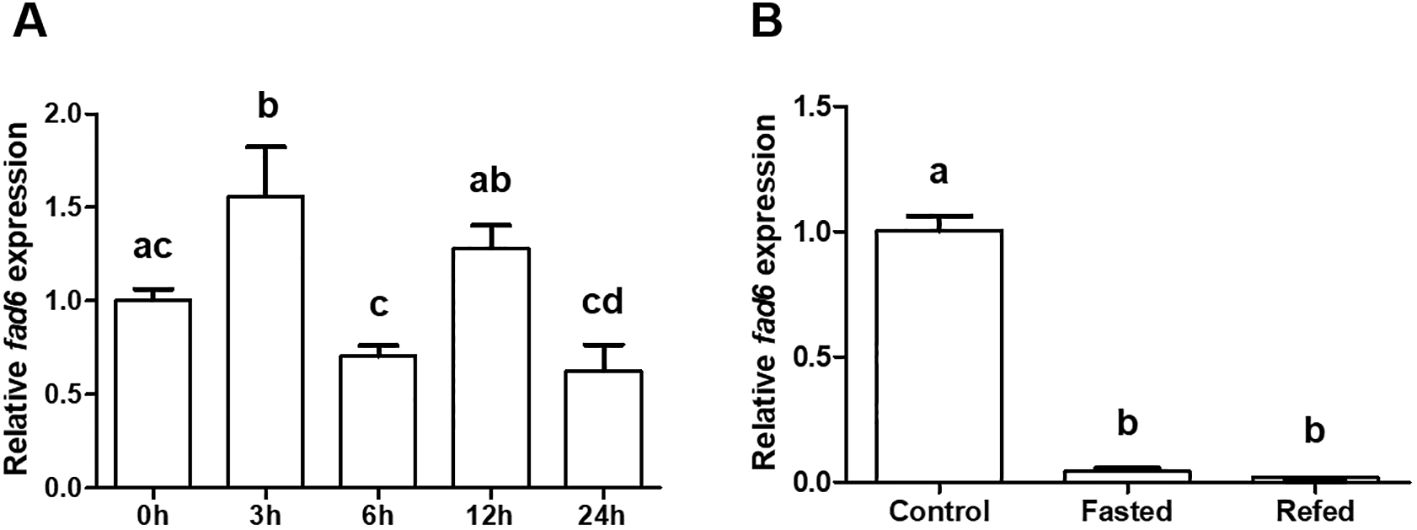
Figure 8 Effects of 24-h fasting (A) or 1-week fasting and refeeding (B) on ycfad6 transcription in yellow catfish. β-actin was selected as the internal reference gene. Data are expressed as mean ± standard error (n = 5). Different lowercase letters above the bars mean significant difference among groups.
Discussion
Fatty acid desaturases play important roles in regulating the synthesis of unsaturated fatty acids in vertebrates (Wu et al., 2018). Thus far, lots of studies have focused on the regulatory roles of Fads in HUFAs biosynthesis both in mammals and teleost. However, as a new member identified recently in the Fads family, Fad6 and its roles of are still largely unknown. In this study, the fad6 gene of yellow catfish was identified and characterized for the first time, and its ORF was validated to be 1,080 bp in length, encoding a protein of 359 amino acids. The length of ycFad6 is shorter than that in golden pompano (T. ovatus) (Zhu et al., 2018), suggesting that the fad6 gene may be species-specific among various fish species. Multiple sequence alignment revealed that the ycFad6 contained three highly conserved histidine motifs (HXXH, HXXXHH, HXXHH) and a FADS-like domain, which were similar to other Fad6 proteins from diverse fish species. Our findings are consistent with the report of golden pompano (Zhu et al., 2018), indicating that these conserved motifs and domains may be essential for iron ligand binding and enzyme activity (Lin et al., 2018; Soo et al., 2020).
Previous studies have shown that gene copy number was varied for fad2 in teleost, such as two copies in rabbitfish (Li et al., 2010; Dong et al., 2018; Li et al., 2019b), three copies in Nile tilapia (Castro et al., 2016; Chen et al., 2018; Dong et al., 2018; Jangprai and Boonanuntanasarn, 2018) and four copies in Atlantic salmon (Hastings et al., 2004; Monroig et al., 2010). Differently, our comparative analyses of genomic synteny and gene structure revealed that the desaturase gene obtained in this study is the fad6 gene and there is only one single copy of this gene in various vertebrate genomes. Meanwhile, a highly conserved gene cluster mrp138-fdxr-hid1b-ush1gb-fad6-trim16-tvp23b-exoc7-plaub was identified in all the selected representative fish genomes, indicating that the physiological functions of Fad6 should be conserved in teleost. Additionally, gene structure comparison revealed that the fad6 gene in teleost commonly contains six exons and five introns, which further supported its conservation and potentially similar functions in various teleost. The phylogenetic analysis showed that the evolutionary history of fad6 was in line with the putative species evolution. Further results showed that fad6 gene in yellow catfish (P. fulvidraco) was clustered with that in Southern catfish (S. meridionalis), suggesting their close relationship; indeed, both catfish species belong to the same order Siluriformes (Wei et al., 2023).
Tissue distribution assays showed that the fad6 gene was transcribed in all the examined tissues, with the highest transcription levels in the heart and liver, which is different from the pattern in golden pompano (Zhu et al., 2018). It seems that the tissue distribution pattern of fad6 is species-specific and the gene may play different roles in diverse teleost. Notably, the ycfad6 was highly transcribed in the liver, which is the primary organ for endogenous HUFAs biosynthesis (Wen et al., 2020; Zeng et al., 2023), implying that ycFad6 may play an important role in the endogenous HUFAs biosynthesis (Xu et al., 2020; Blahova et al., 2022).
The transcription patterns of ycfad6 at different developmental stages were detected by qRT-PCR. These results showed that the transcription of ycfad6 was significantly decreased at 0-6 h, and then dramatically increased at 12-72 h, suggesting that endogenous HUFAs biosynthesis was inhibited at the early stages of embryonic development (0-6 h), and the HUFAs biosynthesis process was initiated to meet the demand of HUFAs requirement from 12 h after fertilization. Similar phenomenon was observed in zebrafish, since its fad mRNA levels were very low until 9 h after fertilization (Leonard et al., 2002; Monroig et al., 2009), suggesting that the HUFAs biosynthesis process may be also restricted during the early embryogenesis of zebrafish. Additionally, the increased tendency of ycfad6 expression was observed at 12 h after fertilization in our current study, which is in line with those findings at the early development stages of zebrafish embryos (Monroig et al., 2009). This implies that HUFAs biosynthesis is required for early embryonic development, although the precise patterns may be different among various species.
Notably, sufficient dietary HUFAs supplement is crucial for healthy growth and development to fishes (Morais et al., 2015). Thus far, several live prey species enriched with HUFAs (such as Limnodrilus hoffmeisteri, rotifers, A. nauplii, and cladoceran) or artificial feeds containing HUFAs (such as egg yolk, artificial compound feeds) are commonly used as initial diets in fish farming (Wei et al., 2014; Zhou et al., 2016). A previous study investigated the effects of different initial diets on the growth performance and survival rate of yellow catfish larvae, and found that adequate palatable diet is an important factor to affect the growth and survive of yellow catfish larvae (Yang et al., 2010). In our present study, we observed that the transcription level of ycfad6 was gradually decreased in 1-3 d after feeding with egg yolk or A. nauplii, respectively. This phenomenon indicates that the initial diets with high concentrations of HUFAs can significantly inhibit the transcription of ycfad6 in yellow catfish. Indeed, HUFAs-rich diets supplement is usually required for the normal growth and development of fish larvae, as well as for quality improvement of the offspring.
Nutritional status can also affect the HUFAs biosynthesis in teleost, and a recent study revealed that food deprivation can induce down-regulation of Δ4fad in the liver of senegalese sole (Solea senegalensis) (Morais et al., 2015). In our present study, we found that fad6 transcription in yellow catfish was significantly increased at 3 h after feeding and then decreased to the control levels, suggesting that food intake may promote the HUFAs biosynthesis but a short-term of fasting may inhibit such process. Consistently, hepatic fad6 was determined to be significantly downregulated after one week of fasting but without significantly changes after refeeding in yellow catfish, indicating that a long-term food deprivation can also restrict the process of HUFAs biosynthesis, although rescue of food supplement cannot return the HUFAs biosynthesis after this long-term food deprivation.
In summary, the fad6 gene was identified and characterized in yellow catfish, and its molecular and biological characteristics were determined. Meanwhile, spatio-temporal expression patterns of this gene were also detected. In addition, transcriptional changes of the ycfad6 gene in response to different initial diets and nutritional status were also investigated. Our findings not only offer basic data for further studies on physiological functions of the fad6 gene in teleost, but also provide novel insights into the regulatory mechanism for HUFAs synthesis in various vertebrates.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Animal Care and Use Committee of Neijiang Normal University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BZ: Formal analysis, Investigation, Writing – original draft. X-YW: Formal analysis, Investigation, Writing – original draft. Z-YW: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. BW: Formal analysis, Investigation, Writing – original draft. Y-YZ: Formal analysis, Investigation, Writing – original draft. W-HZ: Investigation, Software, Writing – original draft. YH: Investigation, Writing – original draft. PP: Investigation, Writing – original draft. Y-PL: Software, Writing – original draft. Y-YL: Investigation, Writing – original draft. JW: Funding acquisition, Investigation, Writing – original draft. RL: Investigation, Writing – original draft. X-GL: Investigation, Writing – original draft. JZ: Investigation, Writing – original draft. S-YZ: Funding acquisition, Investigation, Software, Writing – original draft. J-DF: Investigation, Writing – original draft. QS: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Natural Science Fund of Sichuan Province of China (No. 2023NSFSC1221), the Project of Sichuan Provincial Department of Science and Technology (No. 2022ZYZFSC22004; No. 2023YFN0048), the Research Fund from Key Laboratory of Sichuan Province for Fishes Conservation and Utilization in the Upper Reaches of the Yangtze River (No. NJTCSC23-3), the Important New Varieties Selection Project of Jiangsu Province (No. PZCZ201742), Provincial science and technology innovative program for carbon peak and carbon neutrality of Jiangsu of China (No. BE2022422), and the Cooperation Fund from Sichuan University (No. 2022H013).
Conflict of interest
Author J-DF was employed by the company Yueyang Yumeikang Biotechnology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor YL declared a past co-authorship with the authors JW, ZYW, YL.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1453516/full#supplementary-material
References
Awada M., Meynier A., Soulage C. O., Hadji L., Géloën A., Viau M., et al. (2013). n-3 PUFA added to high-fat diets affect differently adiposity and inflammation when carried by phospholipids or triacylglycerols in mice. Nutr. Metab. 10, 1–14. doi: 10.1186/1743-7075-10-23
Blahova Z., Franek R., Let M., Blaha M., Psenicka M., Mraz J. (2022). Partial fads2 Gene Knockout Diverts LC-PUFA Biosynthesis via an Alternative Delta 8 Pathway with an Impact on the Reproduction of Female Zebrafish (Danio rerio). Genes 13, 700. doi: 10.3390/genes13040700
Castro L. F., Tocher D. R., Monroig O. (2016). Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid. Res. 62, 25–40. doi: 10.1016/j.plipres.2016.01.001
Chen C. Y., Guan W. T., Xie Q. M., Chen G. L., He X. L., Zhang H. L., et al. (2018). n-3 essential fatty acids in Nile tilapia, Oreochromis niloticus: Bioconverting LNA to DHA is relatively efficient and the LC-PUFA biosynthetic pathway is substrate limited in juvenile fish. Aquaculture 495, 513–522. doi: 10.1016/j.aquaculture.2018.06.023
Dong Y. W., Zhao J. H., Chen J. L., Wang S. Q., Liu Y., Zhang Q. H., et al. (2018). Cloning and characterization of Delta 6/Delta 5 fatty acyl desaturase (Fad) gene promoter in the marine teleost Siganus canaliculatus. Gene 647, 174–180. doi: 10.1016/j.gene.2018.01.031
Fang D. A., Yang X. J., Zhou Y. F., Xu D. P., Yang Y., Qin C. (2020). Discovery of the indicator role of period 2 in yellow catfish (Pelteobagrus fulvidraco) food intake during early life development stages. Chronobiol. Int. 37, 629–640. doi: 10.1080/07420528.2020.1752706
Guillou H., Zadravec D., Martin P. G., Jacobsson A. (2010). The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid. Res. 49, 186–199. doi: 10.1016/j.plipres.2009.12.002
Halim N., Ali M. S. M., Leow A. T. C., Rahman R. (2022). Membrane fatty acid desaturase: biosynthesis, mechanism, and architecture. Appl. Microbiol. Biotechnol. 106, 5957–5972. doi: 10.1007/s00253-022-12142-3
Hastings N., Agaba M. K., Tocher D. R., Zheng X., Dickson C. A., Dick J. R., et al. (2004). Molecular cloning and functional characterization of fatty acyl desaturase and elongase cDNAs involved in the production of eicosapentaenoic and docosahexaenoic acids from alpha-linolenic acid in Atlantic salmon (Salmo salar). Mar. Biotechnol. 6, 463–474. doi: 10.1007/s10126-004-3002-8
Jangprai A., Boonanuntanasarn S. (2018). Ubiquitous promoters direct the expression of fatty acid Delta-6 desaturase from Nile tilapia (Oreochromis niloticus) in Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 28, 281–292. doi: 10.1159/000499568
Kuah M. K., Jaya-Ram A., Shu-Chien A. C. (2016). A fatty acyl desaturase (fads2) with dual Δ6 and Δ5 activities from the freshwater carnivorous striped snakehead Channa striata. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 201, 146–155. doi: 10.1016/j.cbpa.2016.07.007
Leonard A. E., Kelder B., Bobik E. G., Chuang L. T., Lewis C. J., Kopchick J. J., et al. (2002). Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 37, 733–740. doi: 10.1007/s11745-002-0955-6
Li Y., Monroig O., Zhang L., Wang S., Zheng X., Dick J. R., et al. (2010). Vertebrate fatty acyl desaturase with Δ4 activity. Proc. Natl. Acad. Sci. U. S. A. 107, 16840–16845. doi: 10.1073/pnas.1008429107
Li Y., Wen Z. Y., You C. H., Xie Z. Y., Tocher D. R., Zhang Y. L., et al. (2020). Genome wide identification and functional characterization of two LC-PUFA biosynthesis elongase (elovl8) genes in rabbitfish (Siganus canaliculatus). Aquaculture 522, 735127. doi: 10.1016/j.aquaculture.2020.735127
Li Y. Y., Yin Z. Y., Dong Y. W., Wang S. Q., Monroig O., Tocher D. R., et al. (2019b). Ppar is involved in the transcriptional regulation of liver LC-PUFA biosynthesis by targeting the 65 fatty acyl desaturase gene in the marine teleost siganus canaliculatus. Mar. Biotechnol. 21, 19–29. doi: 10.1007/s10126-018-9854-0
Li Y., Zhao J., Dong Y., Yin Z., Li Y., Liu Y., et al. (2019a). Sp1 is involved in vertebrate LC-PUFA biosynthesis by upregulating the expression of liver desaturase and elongase genes. Int. J. Mol. Sci. 20, 5066. doi: 10.3390/ijms20205066
Lin Z., Huang Y., Zou W., Rong H., Hao M., Wen X. (2018). Cloning, tissue distribution, functional characterization and nutritional regulation of a fatty acyl Elovl5 elongase in chu’s croaker Nibea coibor. Gene 659, 11–21. doi: 10.1016/j.gene.2018.03.046
Liu Q. N., Yang T. T., Wang C., Jiang S. H., Zhang D. Z., Tang B. P., et al. (2019). A non-mammalian Toll-like receptor 26 (TLR26)gene mediates innate immune responses in yellow catfish Pelteobagrus fulvidraco. Fish. Shellfish. Immunol. 95, 491–497. doi: 10.1016/j.fsi.2019.11.005
Lopes-Marques M., Kabeya N., Qian Y., Ruivo R., Santos M. M., Venkatesh B., et al. (2018). Retention of fatty acyl desaturase 1 (fads1) in Elopomorpha and Cyclostomata provides novel insights into the evolution of long-chain polyunsaturated fatty acid biosynthesis in vertebrates. BMC Evol. Biol. 18, 157. doi: 10.1186/s12862-018-1271-5
Monroig O., Li Y., Tocher D. R. (2011). Delta-8 desaturation activity varies among fatty acyl desaturases of teleost fish: high activity in delta-6 desaturases of marine species. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 159, 206–213. doi: 10.1016/j.cbpb.2011.04.007
Monroig O., Rotllant J., Sánchez E., Cerdá-Reverter J. M., Tocher D. R. (2009). Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim. Biophys. Acta 1791, 1093–1101. doi: 10.1016/j.bbalip.2009.07.002
Monroig O., Zheng X., Morais S., Leaver M. J., Taggart J. B., Tocher D. R. (2010). Multiple genes for functional 6 fatty acyl desaturases (Fad) in Atlantic salmon (Salmo salar L.): gene and cDNA characterization, functional expression, tissue distribution and nutritional regulation. Biochim. Biophys. Acta 1801, 1072–1081. doi: 10.1016/j.bbalip.2010.04.007
Morais S., Mourente G., Martínez A., Gras N., Tocher D. R. (2015). Docosahexaenoic acid biosynthesis via fatty acyl elongase and Δ4-desaturase and its modulation by dietary lipid level and fatty acid composition in a marine vertebrate. Biochim. Biophys. Acta 1851, 588–597. doi: 10.1016/j.bbalip.2015.01.014
Muhlhausler B. S., Ailhaud G. P. (2013). Omega-6 polyunsaturated fatty acids and the early origins of obesity. Curr. Opin. Endocrinol. Diabetes. Obes. 20, 56–61. doi: 10.1097/MED.0b013e32835c1ba7
Nayak M., Saha A., Pradhan A., Samanta M., Giri S. S. (2017). Dietary fish oil replacement by linseed oil: Effect on growth, nutrient utilization, tissue fatty acid composition and desaturase gene expression in silver barb (Puntius gonionotus) fingerlings. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 205, 1–12. doi: 10.1016/j.cbpb.2016.11.009
Song Y. F., Luo Z., Pan Y. X., Zhang L. H., Chen Q. L., Zheng J. L. (2015). Three unsaturated fatty acid biosynthesis-related genes in yellow catfish Pelteobagrus fulvidraco: Molecular characterization, tissue expression and transcriptional regulation by leptin. Gene 563, 1–9. doi: 10.1016/j.gene.2014.12.014
Soo H. J., Sam K. K., Chong J., Lau N. S., Ting S. Y., Kuah M. K., et al. (2020). Functional characterisation of fatty acyl desaturase, Fads2, and elongase, Elovl5, in the Boddart’s goggle-eyed goby Boleophthalmus boddarti (Gobiidae) suggests an incapacity for long-chain polyunsaturated fatty acid biosynthesis. J. Fish. Biol. 97, 83–99. doi: 10.1111/jfb.14328
Wei X. Y., Wang J., Guo S. T., Lv Y. Y., Li Y. P., Qin C. J., et al. (2023). Molecular characterization of a teleost-specific toll-like receptor 22 (tlr22) gene from yellow catfish (Pelteobagrus fulvidraco) and its transcriptional change in response to poly I:C and Aeromonas hydrophila stimuli. Fish. Shellfish. Immunol. 134, 108579. doi: 10.1016/j.fsi.2023.108579
Wei G. L., Xu G. C., Gu R. B., Du F. K., Xu P. (2014). Effects of dietary high unsaturated fatty acids on FAD gene expression in different tissues of Japanese grenadier anchovy (Coilia nasus). Jiangsu. Agric. Sci. 42, 233–236.
Wen Z. Y., Li Y., Bian C., Shi Q., Li Y. Y. (2020). Genome-wide identification of a novel elovl4 gene and its transcription in response to nutritional and osmotic regulations in rabbitfish (Siganus canaliculatus). Aquaculture 529, 735666. doi: 10.1016/j.aquaculture.2020.735666
Wen Z. Y., Wang J., Bian C., Zhang X., Li J., Peng Y., et al. (2019). Molecular cloning of two kcnk3 genes from the Northern snakehead (Channa argus) for quantification of their transcriptions in response to fasting and refeeding. Gen. Comp. Endocrinol. 281, 49–57. doi: 10.1016/j.ygcen.2019.05.016
Wu D. L., Huang Y. H., Liu Z. Q., Yu P., Gu P. H., Fan B., et al. (2018). Molecular cloning, tissue expression and regulation of nutrition and temperature on Δ6 fatty acyl desaturase-like gene in the red claw crayfish (Cherax quadricarinatus). Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 225, 58–66. doi: 10.1016/j.cbpb.2018.07.003
Xie D., Chen C., Dong Y., You C., Wang S., Monroig Ó., et al. (2021). Regulation of long-chain polyunsaturated fatty acid biosynthesis in teleost fish. Prog. Lipid. Res. 82, 101095. doi: 10.1016/j.plipres.2021.101095
Xu H., Ferosekhan S., Turkmen S., Afonso J. M., Zamorano M. J., Izquierdo M. (2020). Influence of Parental Fatty Acid Desaturase 2 (fads2) Expression and Diet on Gilthead Seabream (Sparus aurata) Offspring fads2 Expression during Ontogenesis. Animals 10, 2191. doi: 10.3390/ani10112191
Yang S., Wen Z. Y., Zou Y. C., Qin C. J., Wang J., Yuan D. Y., et al. (2018). Molecular cloning, tissue distribution, and effect of fasting and refeeding on the expression of neuropeptide Y in Channa argus. Gen. Comp. Endocrinol. 259, 147–153. doi: 10.1016/j.ygcen.2017.11.017
Yang R. B., Xie C. X., Fan Q. X., Gao C., Fang L. B. (2010). Effects of feeding frequency and bait type on the growth and survival of Pelteobagrus fulvidraco pups and juveniles. Chin. J. Appl. Environ. 302, 112–123.
Zeng W. H., Wei X. Y., Qin W., Shi Q., Guo S. T., Prathomya P., et al. (2023). Molecular characterization, spatiotemporal expression patterns of fatty acid elongase (elovl8) gene, and its transcription changes in response to different diet stimuli in yellow catfish (Pelteobagrus fulvidraco). Front. Mar. Sci. 10, 1270776. doi: 10.3389/fmars.2023.1270776
Zhou X., Zhan Z. J., Ji Y. P., Yu Q. W., Shang G. L., Xu H. H., et al. (2016). Effects of bait types on growth and survival of Pelteobagrus fulvidraco pups and young fish at different stages of life. Jiangsu. Agric. Sci. 44, 324–327.
Zhu K. C., Song L., Guo H. Y., Guo L., Zhang N., Liu B. S., et al. (2018). Identification of fatty acid desaturase 6 in golden pompano trachinotus ovatus (Linnaeus 1758) and its regulation by the PPARαb transcription factor. Int. J. Mol. Sci. 20, 23. doi: 10.3390/ijms20010023
Keywords: yellow catfish, fad6, HUFA biosynthesis, gene expression, nutritional regulation
Citation: Zhou B, Wei X-Y, Wen Z-Y, Wang B, Zhao Y-Y, Zeng W-H, He Y, Prathomya P, Lv Y-Y, Li Y-P, Wang J, Li R, Li X-G, Zhou J, Zhang S-Y, Fan J-D and Shi Q (2024) Molecular characterization of fad6 gene and its transcriptional changes in response to different initial diets and nutritional status in yellow catfish (Pelteobagrus fulvidraco). Front. Mar. Sci. 11:1453516. doi: 10.3389/fmars.2024.1453516
Received: 23 June 2024; Accepted: 23 July 2024;
Published: 08 August 2024.
Edited by:
Yiming Li, Fishery Machinery and Instrument Research Institute, ChinaReviewed by:
Xiaochen Yuan, Anhui Agricultural University, ChinaShan He, Huazhong Agricultural University, China
Copyright © 2024 Zhou, Wei, Wen, Wang, Zhao, Zeng, He, Prathomya, Lv, Li, Wang, Li, Li, Zhou, Zhang, Fan and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zheng-Yong Wen, emhlbmd5b25nX3dlbkAxMjYuY29t; Qiong Shi, c2hpcWlvbmdAc3p1LmVkdS5jbg==; c2hpcWlvbmdAZ2Vub21pY3MuY24=
†These authors have contributed equally to this work
 Bo Zhou
Bo Zhou Xiu-Ying Wei
Xiu-Ying Wei Zheng-Yong Wen
Zheng-Yong Wen Bin Wang1
Bin Wang1 Wan-Hong Zeng
Wan-Hong Zeng Panita Prathomya
Panita Prathomya Yun-Yun Lv
Yun-Yun Lv Yan-Ping Li
Yan-Ping Li Jun Wang
Jun Wang Shi-Yong Zhang
Shi-Yong Zhang Qiong Shi
Qiong Shi