
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Mar. Sci. , 19 August 2024
Sec. Deep-Sea Environments and Ecology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1452112
This article is part of the Research Topic From Cold Seeps to Hydrothermal Vents: Geology, Chemistry, Microbiology, and Ecology in Marine and Coastal Environments View all 23 articles
This article is a correction to:
Comparative omics analysis of a deep-sea barnacle species (Cirripedia, Scalpellomorpha) and shallow-water barnacle species provides insights into deep-sea adaptation
By Mao N, Shao W, Cai Y, Kong X, Ji N and Shen X (2024). Front. Mar. Sci. 10:1269411. doi: 10.3389/fmars.2023.1269411
In the published article, there was an error in the article title. Instead of “Comparative omics analysis of a new deep-sea barnacle species (Cirripedia, Scalpellomorpha) and shallow-water barnacle species provides insights into deep-sea adaptation”, it should be “Comparative omics analysis of a deep-sea barnacle species (Cirripedia, Scalpellomorpha) and shallow-water barnacle species provides insights into deep-sea adaptation”.
In the published article, there was an error. We provisionally identified the deep-water species Vulcanolepas fijiensis. As a result of the Chan et al. Commentary Paper (2024), we became aware that our initial identification of Vulcanolepas fijiensis was incorrect. We have confirmed that the barnacle is actually Neolepas marisindica and have revised this paper to include the correct species indentation.
The change to “Neolepas marisindica” or “N. marisindica” applies to all instances of the words “Vulcanolepas fijiensis” and “V. fijiensis” respectively, which appear in the Keywords, Abstract, Introduction, Materials and methods, Results, Discussion, and Conclusions.
A correction has been made to Introduction. This sentence previously stated:
“While studies on other organisms inhabiting extreme deep-sea environments have identified several potential mechanisms, the transcriptome of Vulcanolepas fijiensis (Chan) and Scalpellum stearnsi (Pilsbry) remains unexplored. Moreover, few studies have investigated the molecular adaptive characteristics that assist with barnacles’ survival in deep-sea harsh chemosynthetic habitats. Due to the limited prior investigations concerning the transcriptome of barnacles, particularly with regard to the deep-sea adaptation of V. fijiensis, our study attempted to address this research gap.
In this study, we collected deep-sea (V. fijiensis) and shallow-water (S. stearnsi) individuals at Longqi hydrothermal field on the Southwest Indian Ridge and Guangxi Beibu Gulf, respectively. In order to investigate the adaptability of barnacles to the deep-sea environment, the analysis of positive selection pressure was conducted using the comparative transcriptome of these two individuals, as well as the 13 protein-coding genes (PCGs) of mitogenomes from 11 barnacles, including V. fijiensis. This research endeavor aimed to not only provide valuable genomic resources for deep-sea barnacles but also establish a foundation for comprehending their genetic strategies and adaptation mechanisms in the deep-sea habitat.”
The corrected sentence appears below:
“While studies on other organisms inhabiting extreme deep-sea environments have identified several potential mechanisms, the transcriptome of them remains unexplored. Moreover, few studies have investigated the molecular adaptive characteristics that assist with barnacles’ survival in deep-sea harsh chemosynthetic habitats. Due to the limited prior investigations concerning the transcriptome of barnacles, particularly with regard to the deep-sea adaptation, our study attempted to address this research gap.
In this study, we collected the deep-sea and shallow-water barnacle individuals at Longqi hydrothermal field on the Southwest Indian Ridge and Guangxi Beibu Gulf, respectively. In order to investigate the adaptability of barnacles to the deep-sea environment, the analysis of positive selection pressure was conducted using the comparative transcriptome of these two individuals, as well as the 13 protein-coding genes (PCGs) of mitogenomes from 11 barnacles. This research endeavor aimed to not only provide valuable genomic resources for deep-sea barnacles but also establish a foundation for comprehending their genetic strategies and adaptation mechanisms in the deep-sea habitat.”
A correction has been made to Materials and methods, 2.1 Sample collection, DNA and RNA extraction, and sequencing, paragraph one. This sentence previously stated:
“A variety of manned submarines were used to obtain V. fijiensis from a hydrothermal vent Longqi hydrothermal field on the Southwest Indian Ridge (49°64’E, 37°17’S) at a depth of 2,759 m in the Southwest Indian Ocean. S. stearnsi were obtained from the Guangxi Beibu Gulf (109°12’E, 21°04’N), depth of about 300 m. In contrast to deep-sea species, the depth of 300m can be considered relatively shallow. Therefore, we provisionally classified S. stearnsi as a shallow species. After collection, they were immediately stored at -80°C.”
The corrected sentence appears below:
“Samples of the deep-sea barnacles from a hydrothermal vent Longqi hydrothermal field on the Southwest Indian Ridge (49°64’E, 37°17’S) at a depth of 2,759 m in the Southwest Indian Ocean collected on board of “Explorer 1” scientific research ship of Institute of Deep-sea Science and Engineering (Chinese Academy of Sciences) by using “Deep-sea Warrior” manned submersible during the winter of 2018, we thank the crew for their help during the cruise. Samples of the shallow-water barnacles were obtained from the Guangxi Beibu Gulf (109° 12 ‘ E, 21° 04 ‘ N), depth of about 300 m. In contrast to deep-sea species, the depth of 300m can be considered relatively shallow. Therefore, we provisionally identified the two barnacle species as the deep water species Vulcanolepas fijiensis (Lee et al., 2019) and the shallow water Scalpellum stearnsi (Chan et al., 2010). As a result of the Chen et al. Commentary Paper (Chan et al., 2024) we became aware that our initial identification of V. fijiensis was incorrect. We have confirmed that the barnacle is actually Neolepas marisindica and have revised this paper to include the correct species indentation. After collection, they were immediately stored at -80°C.”
The references for “Lee et al., 2019”, “Chan et al., 2010”, and “Chan et al., 2024” should be:
“Chan, B. K., Prabowo, R. E., Lee, K. S. (2010). North West Pacific deep-sea barnacles (Cirripedia, Thoracica) collected by the Taiwan expeditions, with descriptions of two new species. Zootaxa 2405, 1-47. doi: 10.11646/zootaxa.2405.1.1
Chan, B. K., Watanabe, H. K., Chen, C. (2024) Commentary: Comparative omics analysis of a new deep-sea barnacle species (Cirripedia, Scalpellomorpha) and shallow-water barnacle species provides insights into deep-sea adaptation. Front. Mar. Sci. 11, 1374419. doi: 10.3389/fmars.2024.1374419
Lee, W .K., Mi Kang, H., Chan, B. K., Ju, S. J., Kim, S. J. (2019). Complete mitochondrial genome of the hydrothermal vent stalked barnacle Vulcanolepas fijiensis (Cirripedia, Scalpelliforms, Eolepadidae). Mitochondrial DNA B. 4 (2), 2725-2726. doi: 10.1080/23802359.2019.1644564”
In the published article, there was an error in the Data availability statement. Our initial identification of Vulcanolepas fijiensis was incorrect. We have confirmed that the barnacle mentioned in the name of our deep-sea barnacle species is actually Neolepas marisindica. Therefore, the Neolepas marisindica and Scalpellum stearnsi datasets generated during the current study have been deposited and the statement “The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, BioProject: PRJNA935972.” has been updated. The correct Data Availability statement appears below.
“The Neolepas marisindica and Scalpellum stearnsi datasets generated during the current study are available in the National Center for Biotechnology Information (NCBI) databases in GenBank repository at the following links: https://www.ncbi.nlm.nih.gov/nuccore/MZ772032 and https://www.ncbi.nlm.nih.gov/nuccore/OP345466.”
In the published article, there was no Acknowledgments section. The correct Acknowledgments section appears below.
“We would like to thank the reviewers and Chan et al. for their detailed comments about misidentification. We agree with the commentary paper of Chan et al. and have changed all reference to Vulcanolepas fijiensis to Neolepas marisindica in our corrected paper. We would also like to thank Professor Haibin Zhang from Institute of Deep-sea Science and Engineering (Chinese Academy of Sciences) for providing the samples and pictures of the deep-sea barnacle.”
In the published article, there was an error in Figure 1 as published. The use of photos from the database in Figure 1 has raised copyright issues. We apologize for the error and have corrected them to photos taken at the time of collection to eliminate copyright and plagiarism issues.
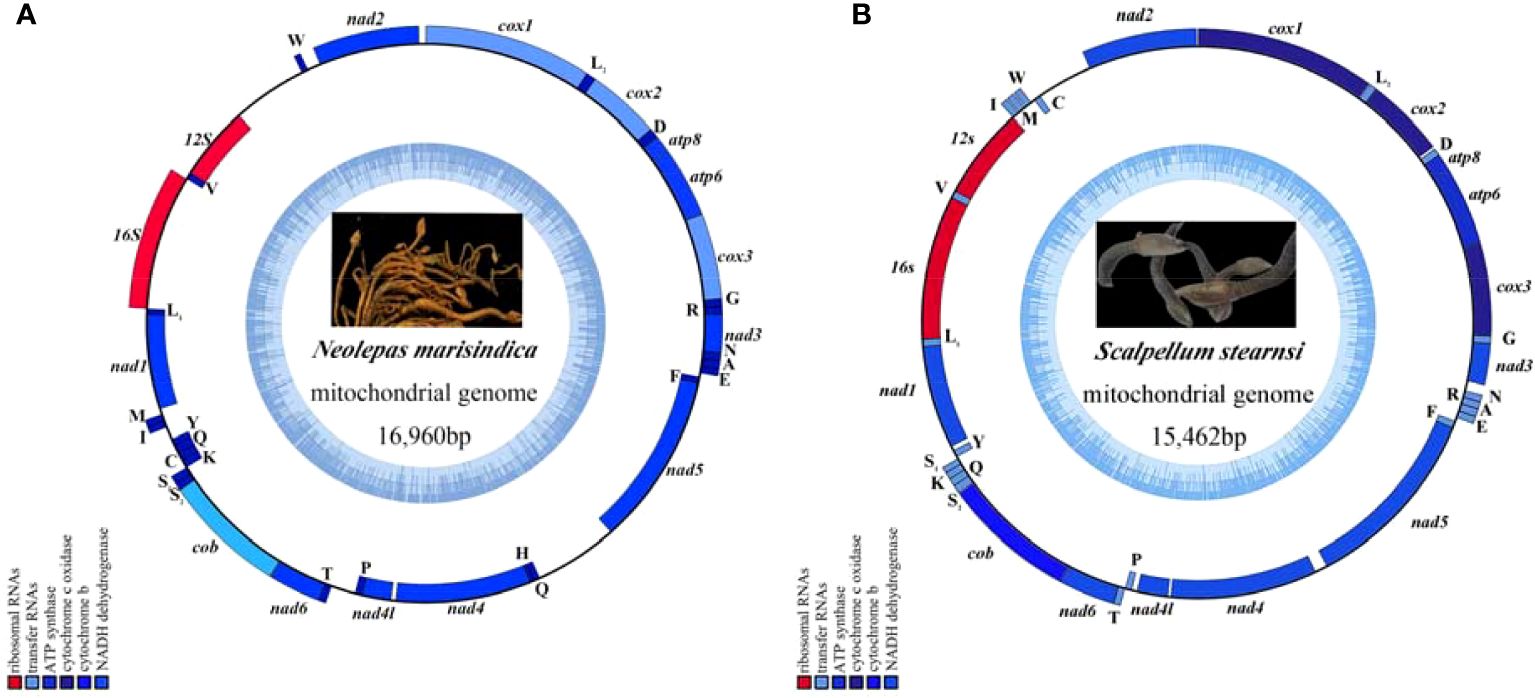
Figure 1. The mitochondrial genome map of deep-sea barnacle Neolepas marisindica (A) and shallow barnacle Scalpellum stearnsi (B) collected for this study, and photographs in the genome map are from preserved samples that were collected for this study, which were collected by Professor Haibin Zhang and kept in Institute of Deep-sea Science and Engineering (Chinese Academy of Sciences). The heavy strand is represented by the outer circle, while the light strand is represented by the inner circle; The tRNA and PCGs are denoted by the blue genes, whereas the 12S rRNA and 16S rRNA are indicated by the red genes.
Furthermore, there was an error in the legend for Figure 1. Our initial identification of Vulcanolepas fijiensis was incorrect. We have confirmed that the barnacle mentioned in the caption of Figure 1 is actually Neolepas marisindica. Therefore, instead of “The mitochondrial genome map of deep-sea barnacle Vulcanolepas fijiensis (A) and shallow barnacle Scalpellum stearnsi (B)”, the legend should read: “The mitochondrial genome map of deep-sea barnacle Neolepas marisindica (A) and shallow barnacle Scalpellum stearnsi (B) collected for this study, and photographs in the genome map are from preserved samples that were collected for this study, which were collected by Professor Haibin Zhang and kept in Institute of Deep-sea Science and Engineering (Chinese Academy of Sciences)”. The corrected Figure 1 and its updated caption appear below.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
In the published article, there was an error in Table 2 as published. Our initial identification of “Vulcanolepas fijiensis” in the third column of Table 2 was incorrect and should be “Neolepas marisindica” instead. Furthermore, the barnacle mentioned in the legends for Table 2 is actually “Neolepas marisindica”, not “Vulcanolepas fijiensis”. The corrected Table 2 and its updated caption appear below.
In the published article, there was an error in Table 3 as published. Our initial identification of “Vulcanolepas fijiensis” in the first column of Table 3 was incorrect and should be “Neolepas marisindica” instead. The corrected Table 3 and its caption appear below.
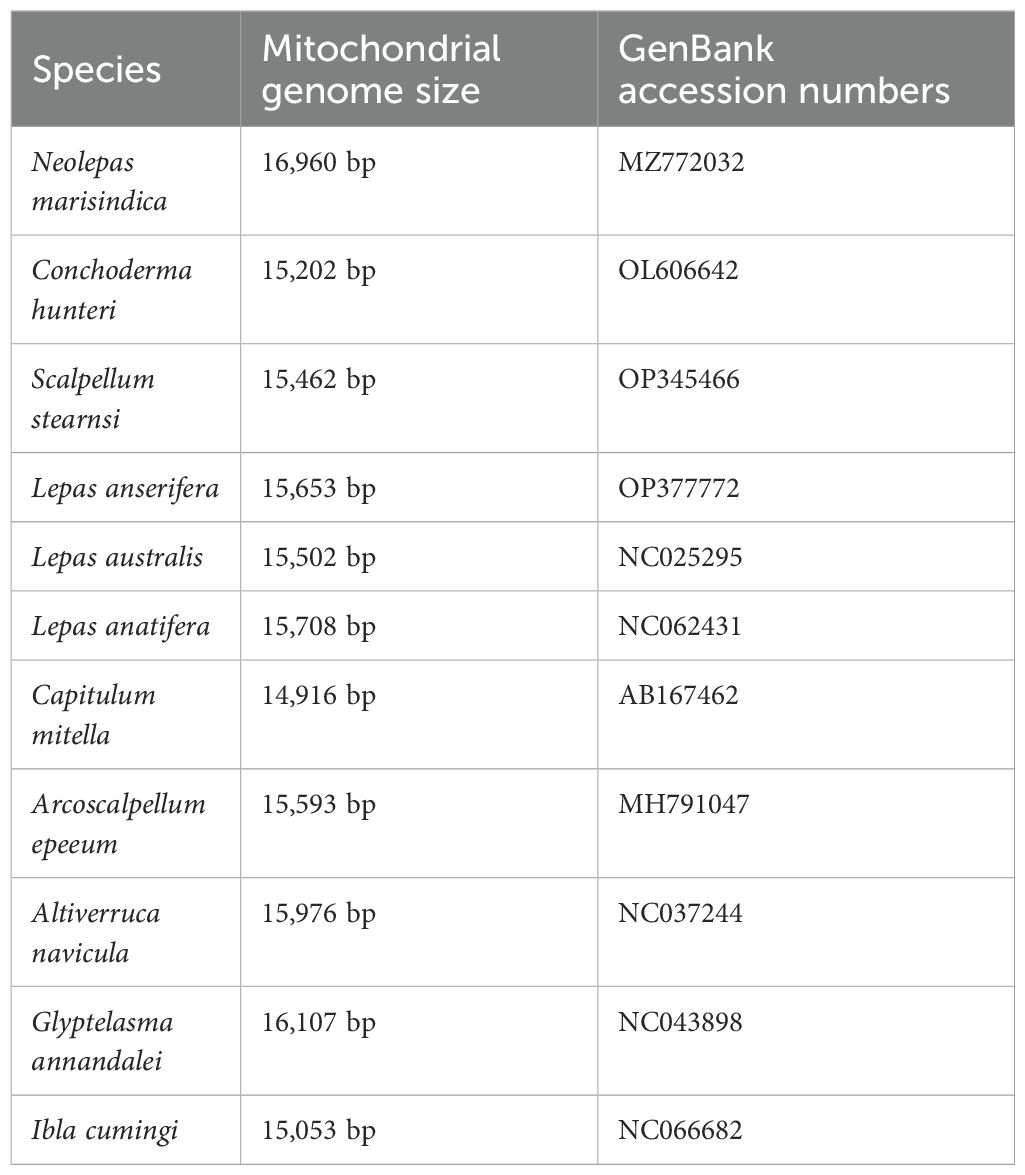
Table 3. Collection and mitogenome information of the specimens that were used for the positive selection analysis.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
In the published article, there was an error in the legends for Figures 2 – 7 and in the legends for Tables 1, 4, and 5 as published. Our initial identification of Vulcanolepas fijiensis was incorrect. We have confirmed that the barnacle mentioned in the legends for Figures 2 – 7 and in the legends for Tables 1, 4, and 5 should be “Neolepas marisindica”. The corrected legends appear below.
[TABLE 1 Summary of the mitogenome features of Neolepas marisindica and Scalpellum stearnsi.]
[FIGURE 2 AT and GC skews in protein-coding genes of the mitochondrial genomes in deep-sea barnacle Neolepas marisindica (A) and shallow barnacle Scalpellum stearnsi (B). The horizontal and vertical coordinates represent gene name and skew value, respectively; The blue line represents the AT skew and the orange line represents the GC skew.]
[FIGURE 3 The codon usage of the protein-coding genes of the mitochondrial genomes of deep- sea barnacle Neolepas marisindica (A) and shallow barnacle Scalpellum stearnsi (B). The different colors represent different kinds of codons, and the different columns represent different amino acids.]
[FIGURE 4 Gene Ontology function annotation of the transcriptomes of deep-sea barnacle Neolepas marisindica (A) and shallow barnacle Scalpellum stearnsi (B). The red terms represent Biological Process, the blue terms represent Cellular Composition, and the green terms represent Molecular Function.]
[FIGURE 5 Classification of the Kyoto Encyclopedia of Genes and Genomes metabolic pathway of the transcriptomes of deep-sea barnacle Neolepas marisindica (A) and shallow barnacle Scalpellum stearnsi (B). Vertical lines represent five main pathways: red terms denote cellular processes, purple terms denote environmental information processing, blue terms denote genetic information processing, green terms denote metabolism, and yellow terms denote organismal systems.]
[TABLE 4 Results of the selective pressure acting on the protein-coding genes of Neolepas marisindica.]
[FIGURE 6 Three-dimensional structural models of the COIII (A) and ND2 (B) genes of deep- sea barnacle Neolepas marisindica. The red represents positive selection site; N represents amino terminus and (C) represents carboxy terminus.]
[TABLE 5 Genes that were subject to positive selection pressure in Neolepas marisindica and Scalpellum stearnsi.]
[FIGURE 7 Enrichment of the Kyoto Encyclopedia of Genes and Genomes metabolic pathway of the positive selection genes in deep-sea barnacle Vulcanolepas fijiensis compared with shallow barnacle Scalpellum stearnsi. The circle size represents the number of genes, and the color from cool to warm represents the smaller q value.]
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: barnacle, Neolepas marisindica, deep-sea adaptation, mitogenome, transcriptome
Citation: Mao N, Shao W, Cai Y, Kong X, Ji N and Shen X (2024) Corrigendum: Comparative omics analysis of a deep-sea barnacle species (Cirripedia, Scalpellomorpha) and shallow-water barnacle species provides insights into deep-sea adaptation. Front. Mar. Sci. 11:1452112. doi: 10.3389/fmars.2024.1452112
Received: 04 July 2024; Accepted: 18 July 2024;
Published: 19 August 2024.
Edited and Reviewed by:
Glen T. Snyder, The University of Tokyo, JapanCopyright © 2024 Mao, Shao, Cai, Kong, Ji and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuefeng Cai, eXVlZmVuZ2NhaUBqb3UuZWR1LmNu; Xin Shen, c2hlbnRoaW5AMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.