- School of Marine Sciences and Biotechnology, Guangxi Key Laboratory for Polysaccharide Materials and Modifications, Guangxi Marine Microbial Resources Industrialization Engineering Technology Research Center, Guangxi Minzu University, Nanning, China
Cadmium could induce various degrees of harm to aquatic organisms. A 30-day feeding trial was conducted to investigate the effects of cadmium on growth, muscle composition, digestive enzyme activity, gene expression of antioxidants and lipid metabolism in juvenile genetic improvement of farmed tilapia (GIFT, Oreochromis niloticus, Initial weight: 21.36 ± 0.24 g). Four cadmium concentrations of aquaculture water were designed: 0, 0.2, 0.4, and 0.6 mg/L Cd2+. The main results are as follows: Compared with the control group (0 mg/L Cd2+), the weight gain rate (WGR), specific growth rate (SGR), daily growth index (DGI), and spleen index (SI) of juvenile GIFT under cadmium stress were significantly decreased (p< 0.05). The contents of crude protein and crude lipid in muscle were significantly decreased (p< 0.05), and the ash was significantly increased (p< 0.05). The activities of trypsin, lipase, and α-amylase in the intestinal were significantly decreased (p< 0.05). The relative expression levels of carnitine palmityl transferase 1 (cpt-1), peroxisome proliferator-activated receptor α (pparα), pparγ, hormone-sensitive lipase (hsl), lipoprotein lipase (lpl), malate dehydrogenase (mdh), leptin (lep), fatty acid synthetase (fas), cholesterol response element binding protein 1 (srebp1), squalene cyclooxygenase (sqle), and stearoyl-CoA desaturase (scd) genes in liver were significantly decreased (p< 0.05). The relative expression levels of catalase (cat), superoxide dismutase (sod), glutathione S-transferase (gst), and glutathione peroxidase (gsh-px) genes in the liver were significantly decreased (p< 0.05). In conclusion, exposure to cadmium stress could impact growth, muscle composition, digestive enzyme activity, gene expression of antioxidant and lipid metabolism in juvenile GIFT.
1 Introduction
The heavy metal cadmium (Cd) is a prevalent environmental pollutant and a potential toxin with detrimental health effects on aquatic organisms (Johri et al., 2010). Cadmium has been identified as a prominent environmental contaminant and non-essential metal in China, exerting harmful impacts on organisms even at low concentrations (Chellaiah, 2018). According to the national standards of China, the concentration of cadmium in aquaculture water should not exceed 100 μg/L (Jiang et al., 2014). However, certain fishponds in specific regions of China have been found to have cadmium levels that surpass the regulations set by the China Food Administration (Cheung et al., 2008; Leung et al., 2014). Previous studies have demonstrated that cadmium could induce various degrees of harm to aquatic organisms, including hepatic impairment, reproductive dysfunction, lymphocyte proliferation, cardiovascular system lesions, and oxidative stress-induced damage to the antioxidant defense system (Lee et al., 2023).
The antioxidant system plays a crucial role in mitigating oxidative stress induced by environmental factors. Therefore, ecotoxicological studies commonly employ antioxidant parameters such as catalase (CAT), superoxide dismutase (SOD), glutathione-S-transferase (GST), and glutathione peroxidase (GSH-Px) (Atli et al., 2016). This system aids in detoxifying fish, establishing cellular REDOX potential, and regulating key cellular processes (Da Rocha et al., 2009; Ji and Yeo, 2021). The lipid metabolism of fish is commonly employed to investigate energy metabolism, providing direct insights into lipid deposition and energy balance in fish (Wang et al., 2018). Additionally, it serves as a vital source of essential fatty acids for growth, health, reproduction, and overall physiological functions (Turchini et al., 2009). Research has demonstrated that lipid metabolism plays a pivotal role in nutrient uptake, lipid transportation, and the β-oxidation pathway in fish (Lu et al., 2013; Saito et al., 2021). However, there is a paucity of relevant studies on the impact of cadmium on fish lipid metabolism, necessitating further research to ascertain whether cadmium can induce fish lipid metabolism dysfunction.
Genetic improvement of farmed tilapia (GIFT, Oreochromis niloticus) has emerged as a highly cultivated economic fish globally and is recognized as one of the dominant fish species endorsed by the Food and Agriculture Organization of the United Nations. These improvements include large size, rapid growth, high protein content, and strong stress resistance (Munguti et al., 2022; Zhang et al., 2024). With the continuous advancement of tilapia aquaculture, there has been a concurrent rise in the prevalence of heavy metal contamination among tilapia (Younis et al., 2020; Hamouda and Abd Alkareem, 2021). However, there is a scarcity of research investigating the effects of heavy metal pollution on the antioxidant and lipid metabolism of tilapia. Therefore, this study focused on GIFT as the research subject and utilized a laboratory simulation static method to introduce varying doses of cadmium into aquaculture water. The objective was to investigate the effects of cadmium stress on growth performance, muscle composition, digestive enzyme activity, gene expression of antioxidant and lipid metabolism in juvenile GIFT. This study aims to provide scientific references regarding the effects of heavy metal pollution during aquaculture processes.
2 Materials and methods
2.1 Experimental feed and fish
The experimental feed was procured from Nanning Haida Biotechnology Co., LTD., Nanning, China. The crude lipid content of the feed was 3.61 ± 0.15%. The crude protein content of the feed was 33.86 ± 0.68%. The ash content of the feed was 6.93 ± 0.17%, and the moisture content of the feed was 9.34 ± 0.26%.
The juvenile GIFT was purchased from Guangxi ANGUI Aquaculture Co., LTD, Nanning, China. The use of juvenile GIFT for this study was approved by the Ethics Committee of Guangxi Minzu University (Approval number: GXMZU-2022-001).
2.2 Acclimatization and culture
After a 15-minute disinfection period using a 10 mg/L potassium permanganate solution, the juvenile GIFT was acclimated in a laboratory aquaculture system for 14 days. The acclimatization conditions were water temperature with 26 ± 1 °C, pH 7-8, dissolved oxygen content ≥ 6 mg/L, and light-dark cycle of 12 hours each. The juvenile GIFT in each group were fed three times daily at 9:00, 14:00, and 19:00, ensuring that the daily feeding quantity was sufficient until satiation was observed.
After a 14-day acclimatization period, a total of 360 juvenile GIFT with an initial body weight of 21.21 ± 0.24 g were randomly allocated into four experimental groups in triplicate, making a total of 12 tanks with 30 fish in each tank. Each tank had a volume of 720 L (150 cm × 80 cm × 60 cm). The juvenile GIFT was reared under the same conditions as used during the 14-day acclimatization period.
According to the previous experimental findings, the cadmium levels (Cd2+) in the water environment cannot exceed 5 μg/L (Joseph, 2009). The concentration of cadmium in the experimental water was determined to be 40-fold, 80-fold, and 120-fold. Then the concentrations of Cd2+ in the four groups were 0 mg/L (control group), 0.2 mg/L, 0.4 mg/L, and 0.6 mg/L, respectively. The cadmium chloride (CdCl2) utilized in this study was of analytical purity and branded as Aldrich, which was procured from Sigma-Aldrich (Shanghai) Trading Co. Ltd., Shanghai, China. During the entire duration of the experiment, all tanks were still water-farmed at the aquaculture systems. Daily feces removal and one-third water exchange were conducted once a day in each tank while maintaining a constant concentration of Cd2+ venom by adding the same volume and concentration to each tank. The experimental period lasted for 30 days.
2.3 Sampling
After being starved for 24 h, 9 fish were randomly selected from each experimental group (3 fish per tank). The sampled fish were individually anesthetized using 0.2 g/L ethyl 3-aminobenzoate mesylate (MS-222, Adamas, Shanghai Adamas Reagent Co., Ltd., Shanghai, China). Then their body weight and length were measured individually. The sample fish was placed on ice and a pair of scissors was utilized to carefully remove one side of the fish’s muscle, exposing its internal organs completely. The samples of the intestinal, dorsal muscle, and liver were subsequently extracted with meticulous precision using ophthalmic forceps placed in labeled sample bags and stored in liquid nitrogen. After all the samples were collected, they were meticulously preserved at a bone-chilling temperature of -80°C in an ultra-low temperature refrigerator for subsequent analysis.
2.4 Calculation of growth performance
The survival rate (SR), weight gain rate (WGR), specific growth rate (SGR), daily growth index (DGI), body length gain rate (BLG), feed conversion ratio (FCR), condition factor (CF), hepatosomatic index (HSI), and viscera index (VSI) of juvenile GIFT were calculated using the following formula:
2.5 Determination of muscle composition
The crude protein, crude lipid, ash, and moisture content of dorsal muscle were determined according to the methods of the Association of Official Analytic Chemists (AOAC, 2005). Briefly, crude protein was determined by the Kjeldahl nitrogen determination method. Crude lipid was determined by the Soxhlet extraction method. Ash was determined by the burning method in a muffle furnace at 550 °C. Moisture was determined by the oven drying constant weight method at 105°C.
2.6 Determination of digestive enzyme activity
The method of Liu Y. et al. (2023) was applied to determine the α-amylase, trypsin, and lipase activity in the intestinal tract of juvenile GIFT. The α-amylase, trypsin, and lipase activity was determined using an ELISA analyzer (RT-6100, Rayto, Shenzhen, China) and assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). All the instruction manuals can be found and downloaded at http://www.njjcbio.com/ (accessed on 10 May 2024).
In simple terms: α-amylase activity was determined by starch-iodine colorimetry method. The unit of α-amylase was defined as the interaction between each milligram of tissue protein and the substrate starch at 37°C for 30 minutes, and every ten milligrams of hydrolyzed starch corresponded to one unit of enzyme activity (U/mgprot). Trypsin activity was determined by the ultraviolet colorimetry method. The unit of trypsin was defined as a change of 0.003 per minute in the absorbance value of the reaction substrate per milligram of tissue protein at pH 8.0 and 37°C, representing one unit of enzyme activity (U/mgprot). Lipase activity was determined by methylation-specific oligonucleotide ligation assay and microplate method. The unit of lipase was defined as per gram of tissue protein reacting with the substrate in the reaction system for 1 minute at 37°C, and every 1 μmol substrate consumed was one unit of enzyme activity (U/gprot).
2.7 Determination of gene expression
The gene expression of carnitine palmityl transferase 1 (cpt-1), peroxisome proliferator-activated receptor α (pparα), peroxisome proliferator-activated receptor γ (pparγ), hormone-sensitive lipase (hsl), lipoprotein lipase (lpl), malate dehydrogenase (mdh), leptin (lep), fatty acid synthetase (fas), cholesterol response element binding protein 1 (srebp1), squalene cyclooxygenase (sqle), stearoyl-CoA desaturase (scd), catalase (cat), superoxide dismutase (sod), glutathione S-transferase (gst), and glutathione peroxidase (gsh-px) in the liver of juvenile GIFT were determined by the real-time quantitative polymerase chain reaction (RT-qPCR) method. The RT-qPCR methods of Zhang et al. were applied (Zhang et al., 2023), and the brief steps were as follows:
First, total RNA was extracted from the liver of juvenile GIFT using the Steady Pure Universal RNA Extraction Kit (Accurate Biology Biotechnology Engineering Ltd., Changsha, China) and assessed for concentration and purity by measuring absorbance ratios at 260:280 nm with an ND-2000 spectrophotometer (Thermo, Waltham, MA, USA).
Second, the reverse transcription of 1000 ng total RNA into cDNA was performed using the Evo M-MLV reverse transcription kit (Accurate Biology Biotechnology Engineering Ltd., Changsha, China). The process involved incubation at 30 °C for 10 min, followed by incubation at 42 °C for 15 min, a final step at 95 °C for 5 min, and cooling to 5 °C for an additional 5 min.
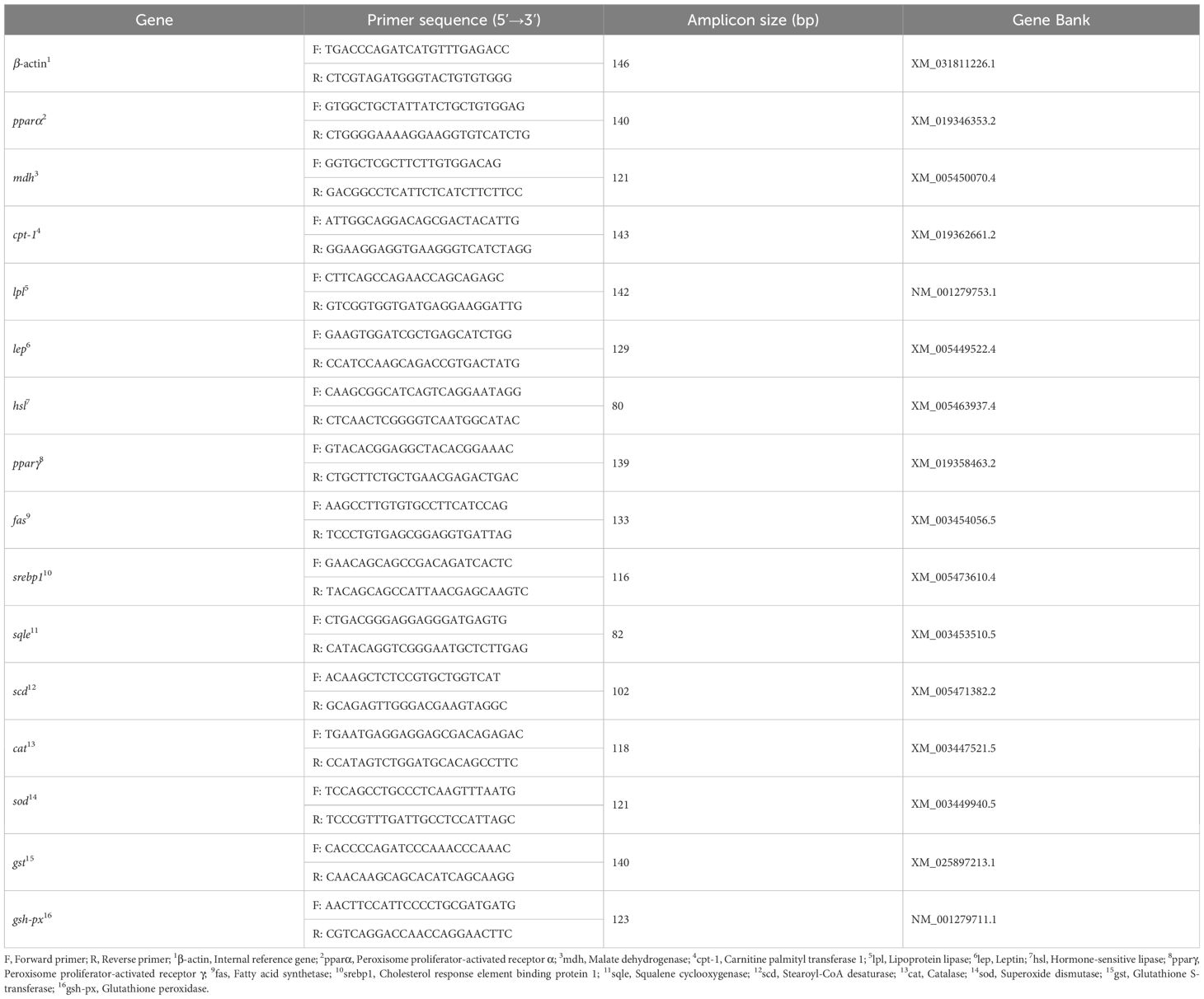
Third, the forward and reverse primers of cpt-1, pparα, pparγ, hsl, lpl, mdh, lep, fas, srebp1, sqle, scd, cat, sod, gst, and gsh-px genes in the liver of juvenile GIFT were designed based on the mRNA sequences of tilapia available in the National Center for Biotechnology Information (NCBI) database, and synthesized by Shanghai Sangon Bioengineering Technology Service Co., LTD, Shanghai, China. β-actin was employed as the internal reference gene. Detailed primer information is provided in Table 1.
Fourth, the RT-qPCR assays were conducted using a LightCycler 96 RT-qPCR Detection System (Roche, Basel, Switzerland) and the TB Green Premix Ex TaqTM II (Tli RNaseH Plus) RT-qPCR kit produced by Takara Biomedical Technology (Beijing) Co., Ltd., Beijing, China. The thermal cycling conditions for the RT-qPCR reactions included an initial denaturation step at 95 °C for 10 s followed by amplification with 40 cycles consisting of denaturation at 95 °C for 60 s; annealing at 60 °C for 30 s; and extension at 72 °C for 90 s. The specificity of the reactions was confirmed by analyzing the melting curve obtained during heating to 95 °C.
The 2−ΔΔCT method (Schmittgen and Livak, 2008) was applied to calculate the relative expression levels of cpt-1, pparα, pparγ, hsl, lpl, mdh, lep, fas, srebp1, sqle, scd, cat, sod, gst, and gsh-px genes in the liver of juvenile GIFT.
2.8 Data analyses
The data underwent initial processing in Microsoft Excel 2023 (Version number: 16.78 [23100802], Microsoft Corporation, Washington, WA, USA). Subsequently, a one-way analysis of variance (ANOVA) was performed using the IBM SPSS 26 software package (International Business Machines Corporation, Armonk, NY, USA). Normality and homogeneity of variances among the groups were evaluated before conducting the statistical tests. For comparisons between groups, Duncan’s multiple-range test was utilized. Statistical significance was set at p< 0.05 level and results are reported as mean ± standard error (mean ± SE).
3 Results
3.1 Effects of cadmium on the growth performance of juvenile GIFT
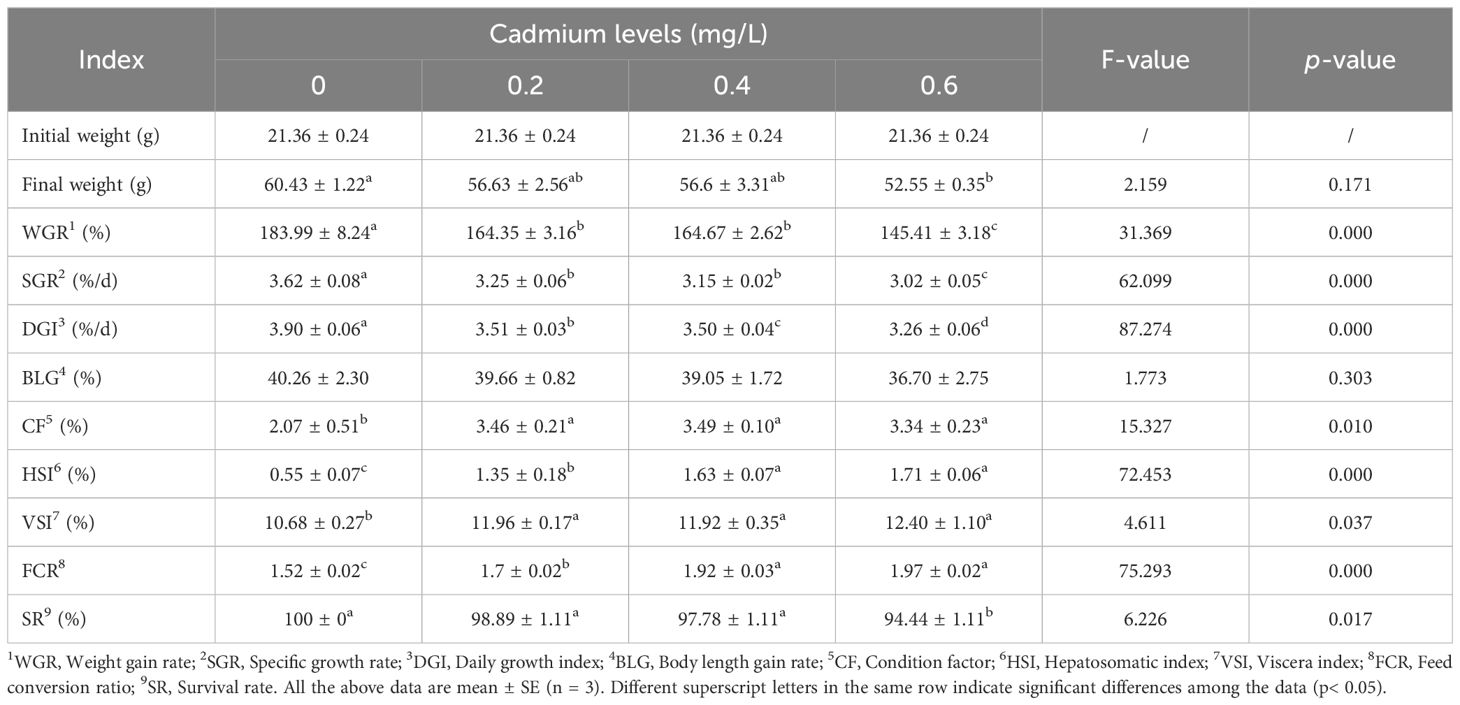
Compared to the control group (0 mg/L Cd2+), the weight gain rate (WGR), specific growth rate (SGR), and daily growth index (DGI) of juvenile GIFT significantly decreased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the lowest WGR, SGR, and DGI, which were significantly lower (p< 0.05) than those in the 0.2 and 0.4 mg/L Cd2+ groups, as shown in Table 2.
Compared to the control group (0 mg/L Cd2+), the feed conversion ratio (FCR), condition factor (CF), hepatosomatic index (HSI), and viscera index (VSI) of juvenile GIFT significantly increased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.4 mg/L Cd2+ group exhibited the highest CF, but no significant difference (p > 0.05) was observed compared to the 0.2 and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the highest HSI and FCR, which were significantly higher (p< 0.05) than those in the 0.2 mg/L Cd2+ group, but no significant difference (p > 0.05) was observed compared to the 0.4 mg/L Cd2+ group. The 0.6 mg/L Cd2+ group exhibited the highest VSI, but no significant difference (p > 0.05) was observed compared to the 0.2 and 0.4 mg/L Cd2+ groups, as shown in Table 2.
Compared to the control group (0 mg/L Cd2+), the final weight and survival rate (SR) of juvenile GIFT significantly decreased (p< 0.05) in the 0.6 mg/L Cd2+ group. However, there was no significant difference (p > 0.05) between 0, 0.2, and 0.4 mg/L Cd2+ groups, as shown in Table 2.
There was no significant difference (p > 0.05) between 0, 0.2, 0.4, and 0.6 mg/L Cd2+ groups on the body length gain rate (BLG), as shown in Table 2.
The relationship between cadmium levels and final weight, WGR, SGR, DGI, BLG, CF, HSI, VSI, FCR, and SR of juvenile GIFT was shown in Figures 1-10 based on a linear regression analysis, in which the order of coefficient of determination (R2) from largest to smallest was: R2 (FCR) > R2 (SR) > R2 (SGR) > R2 (final weight) > R2 (WGR) > R2 (DGI) > R2 (BLG) > R2 (HSI) > R2 (VSI) > R2 (CF).
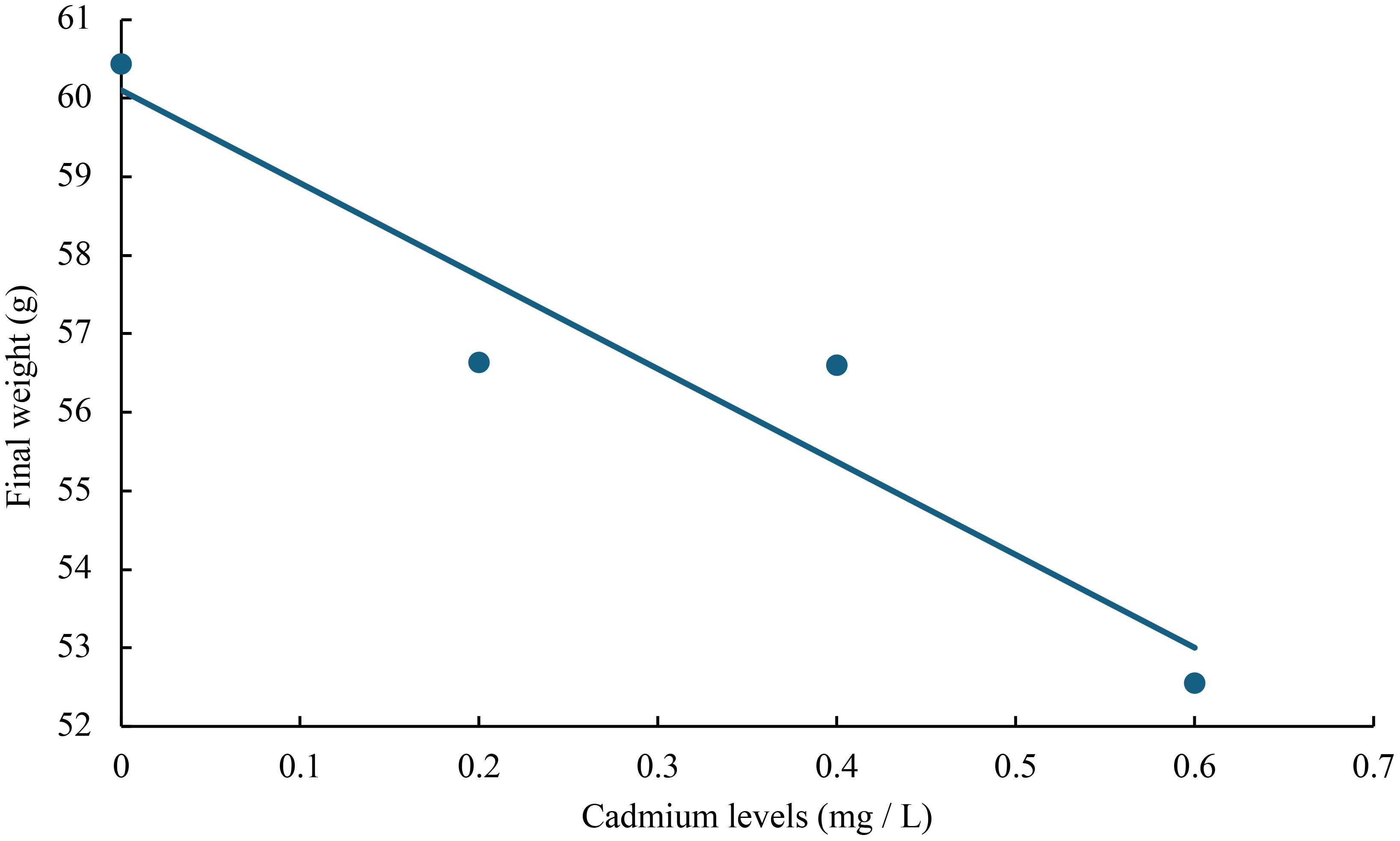
Figure 1 Relationship between cadmium levels and final weight of juvenile GIFT based on a linear regression analysis, in which Y = -11.835X + 60.103, R2 = 0.9018.
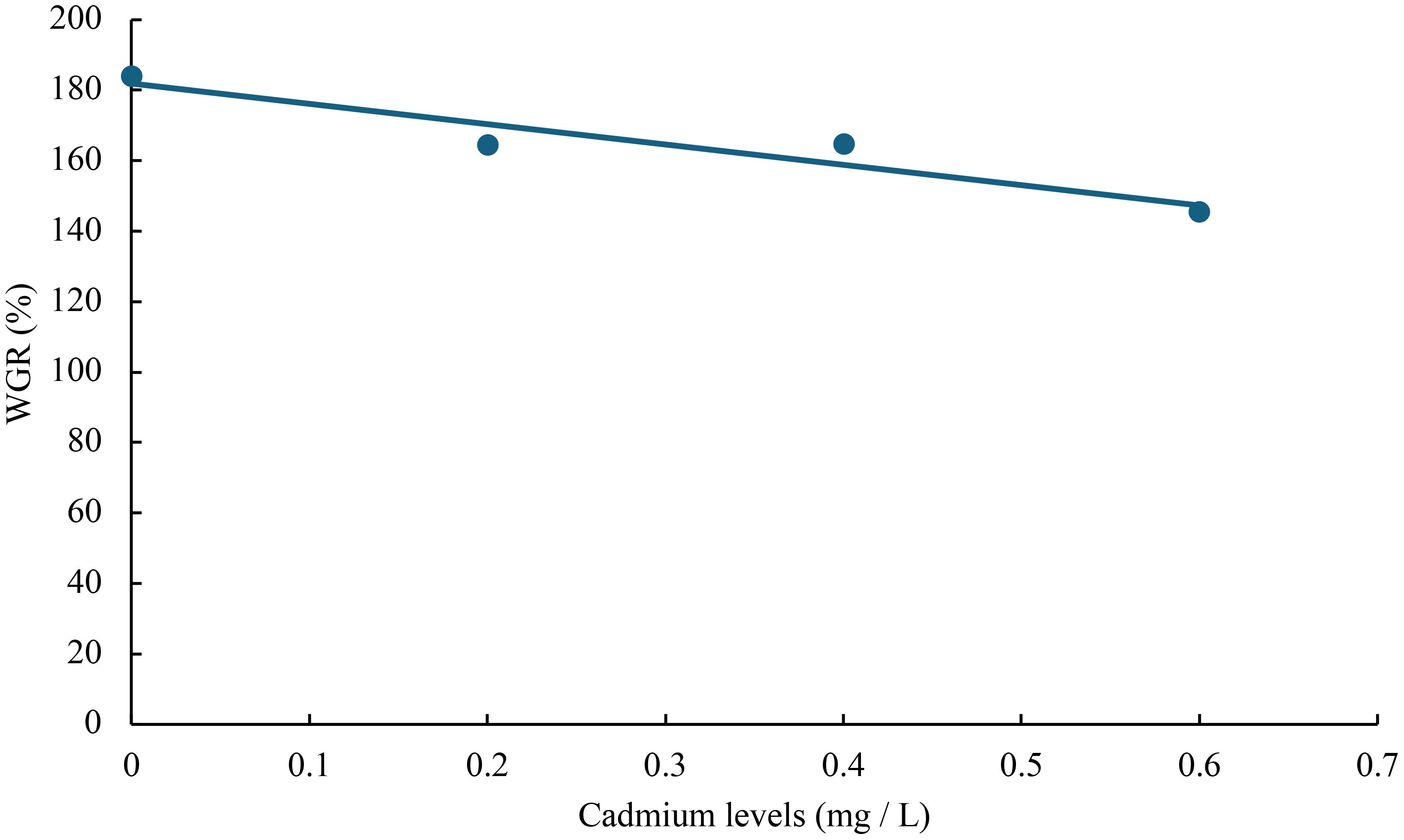
Figure 2 Relationship between cadmium levels and WGR of juvenile GIFT based on a linear regression analysis, in which Y = -57.71X + 181.92, R2 = 0.8949.
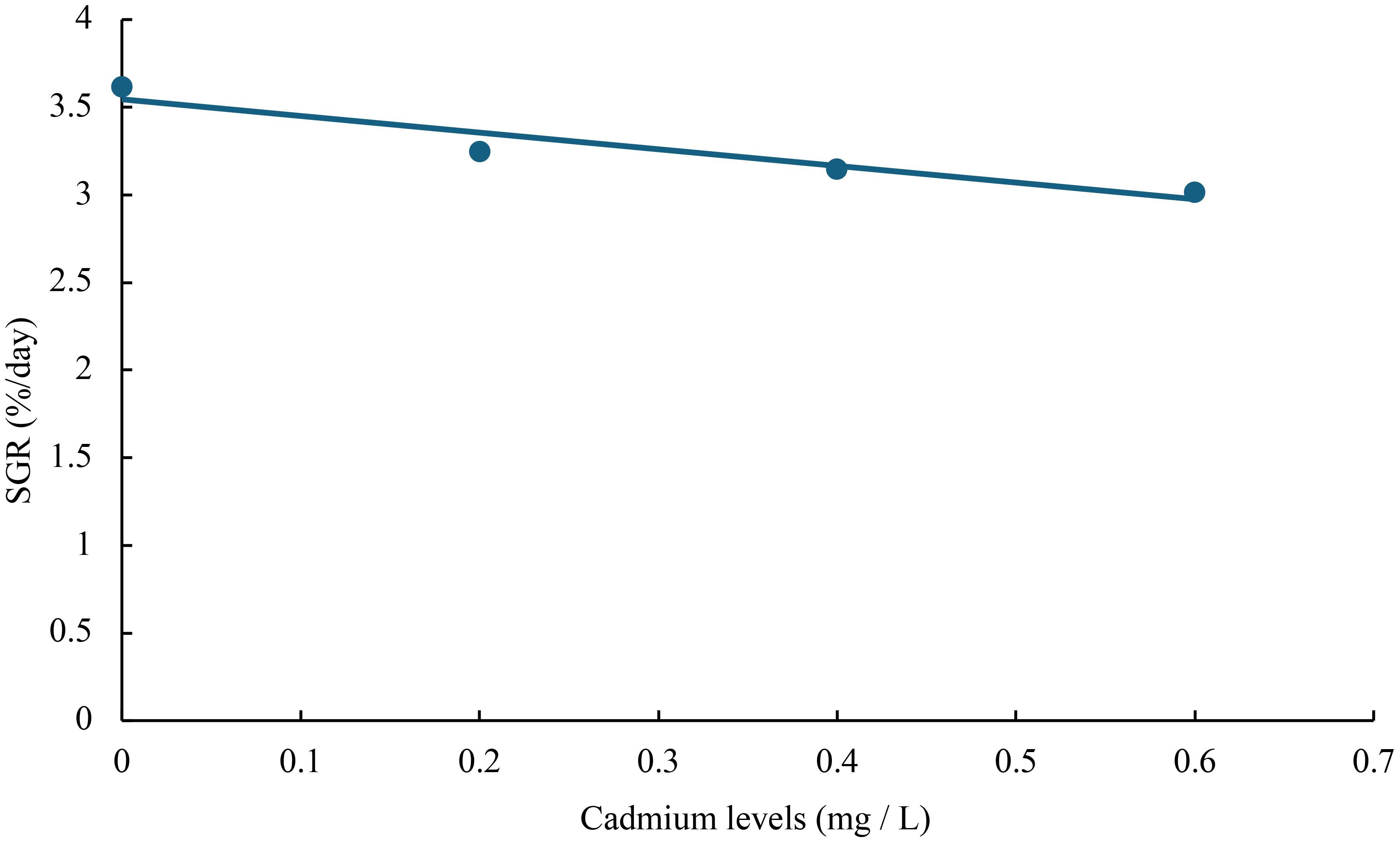
Figure 3 Relationship between cadmium levels and SGR of juvenile GIFT based on a linear regression analysis, in which Y = -0.95X + 3.545, R2 = 0.9052.
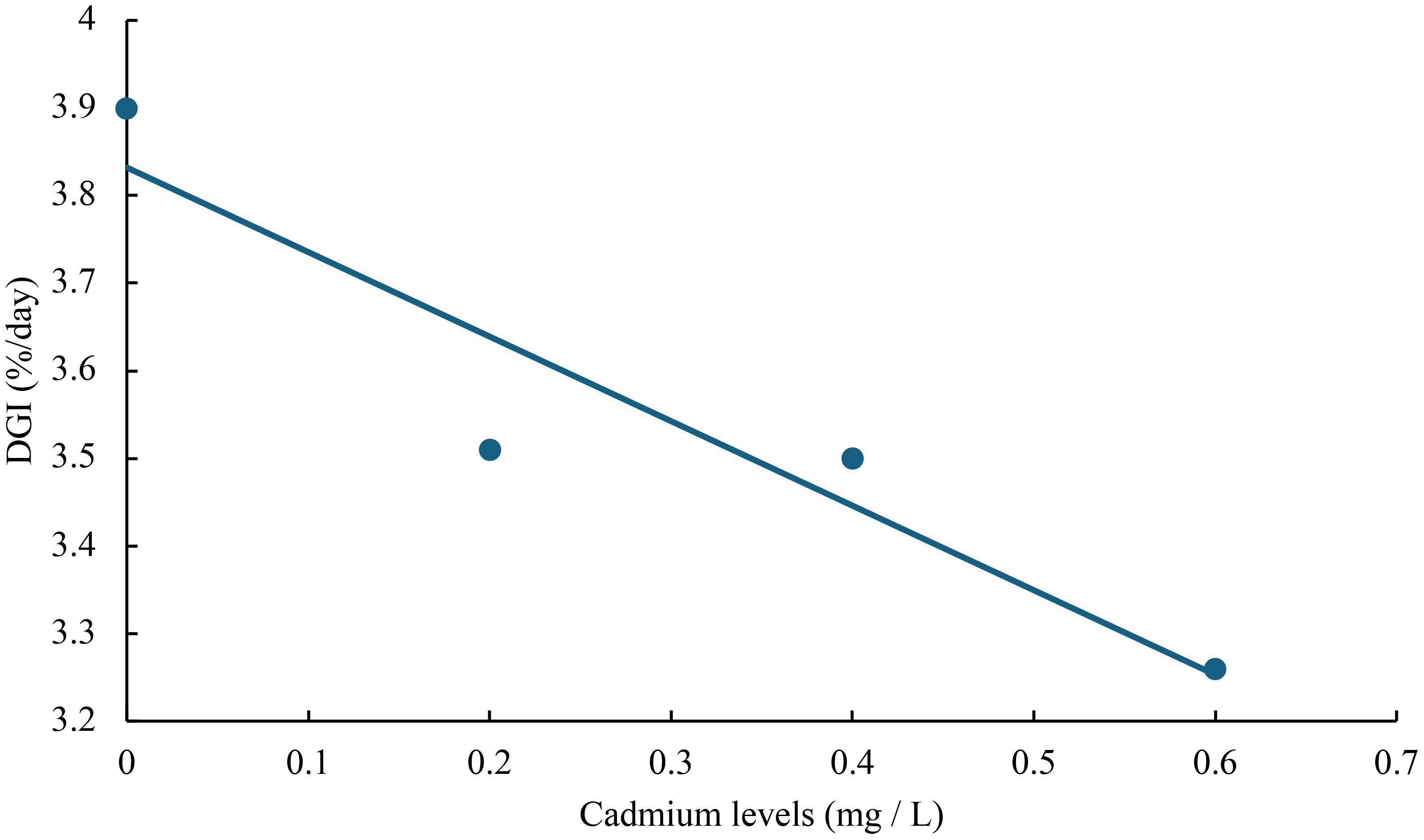
Figure 4 Relationship between cadmium levels and DGI of juvenile GIFT based on a linear regression analysis, in which Y = -0.965X + 3.832, R2 = 0.8849.
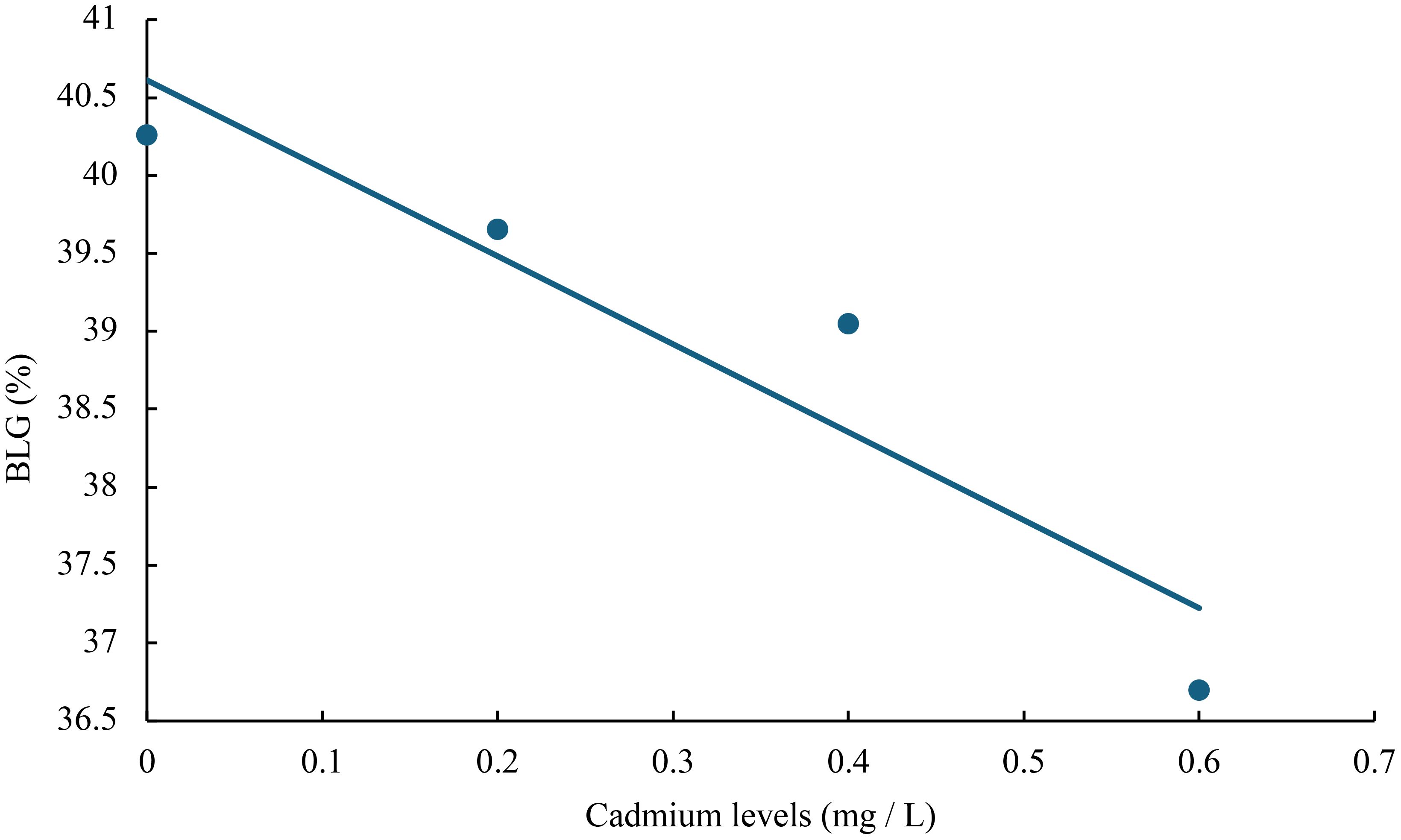
Figure 5 Relationship between cadmium levels and BLG of juvenile GIFT based on a linear regression analysis, in which Y = -5.645X + 40.611, R2 = 0.8744.
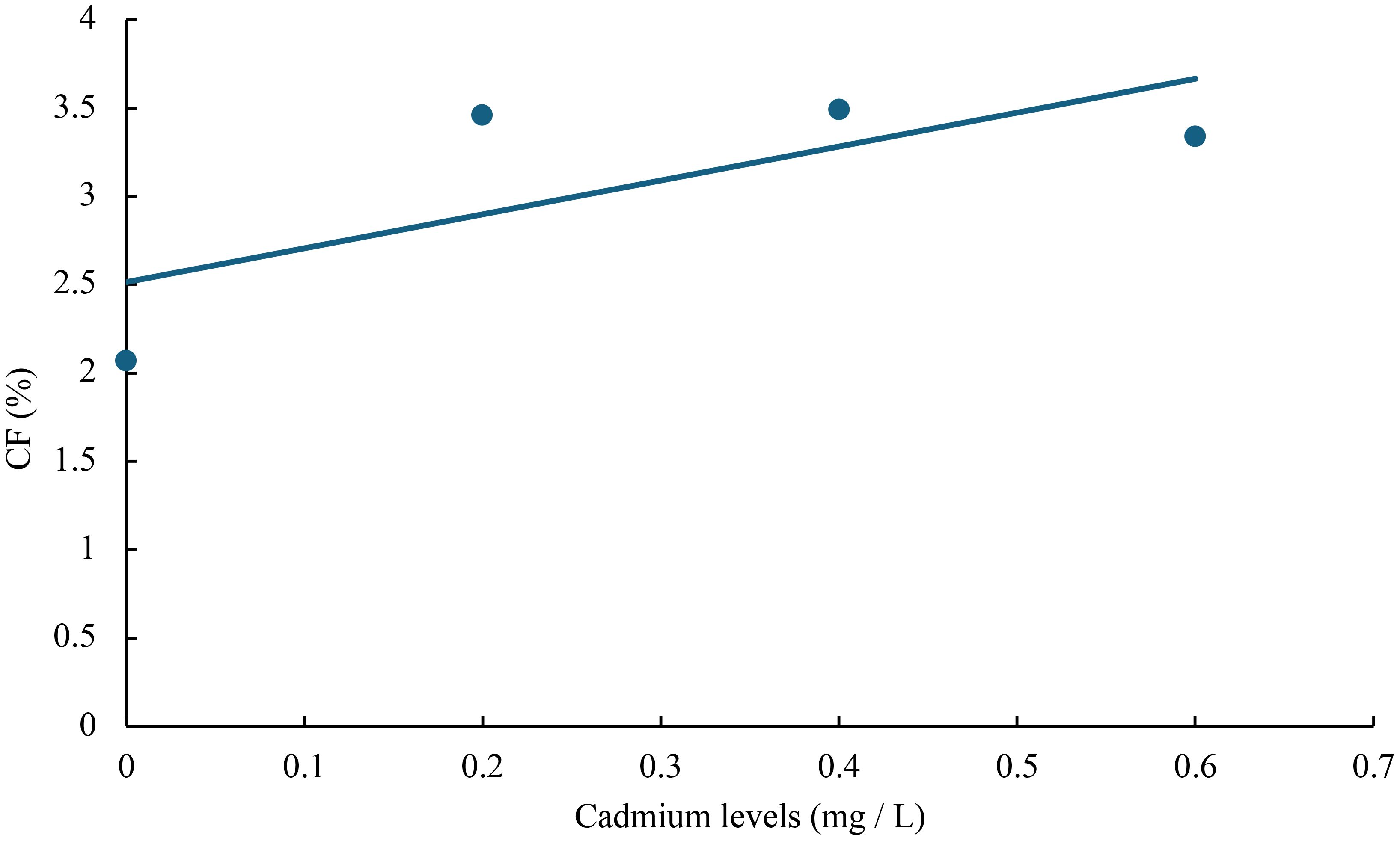
Figure 6 Relationship between cadmium levels and CF of juvenile GIFT based on a linear regression analysis, in which Y = 1.92X + 2.514, R2 = 0.5267.
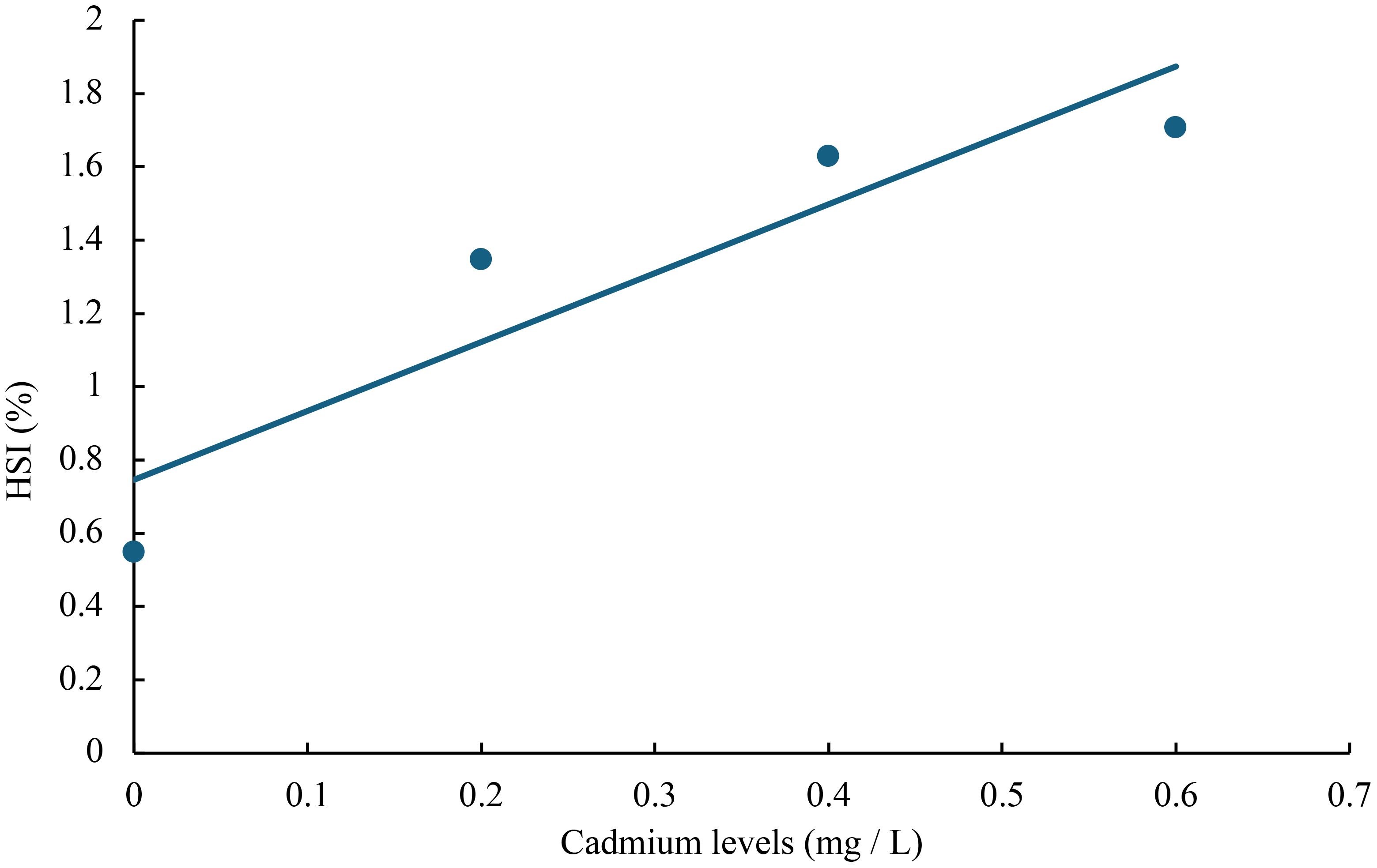
Figure 7 Relationship between cadmium levels and HSI of juvenile GIFT based on a linear regression analysis, in which Y = 1.88X + 0.746, R2 = 0.8399.
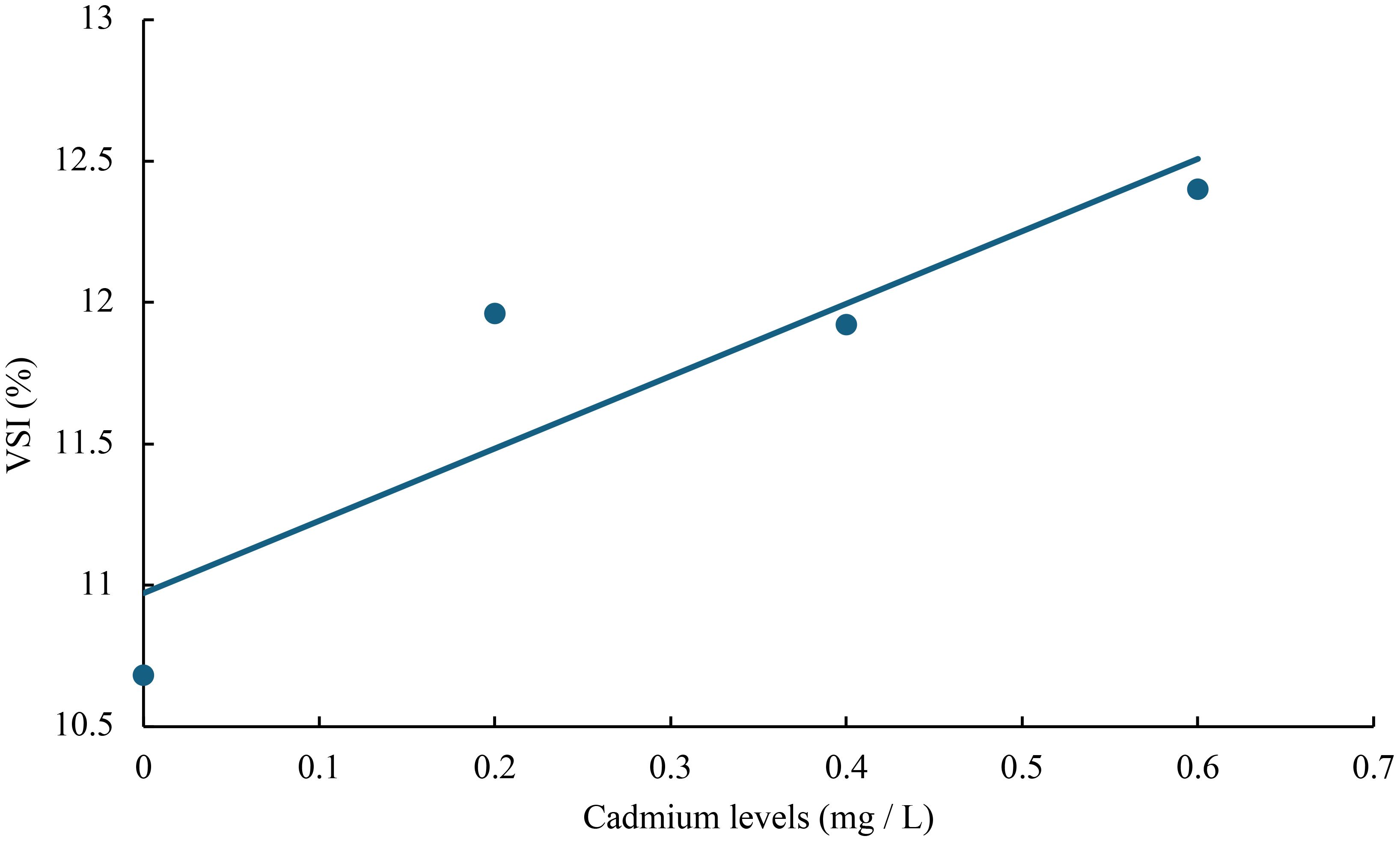
Figure 8 Relationship between cadmium levels and VSI of juvenile GIFT based on a linear regression analysis, in which Y = 2.56X + 10.972, R2 = 0.7992.
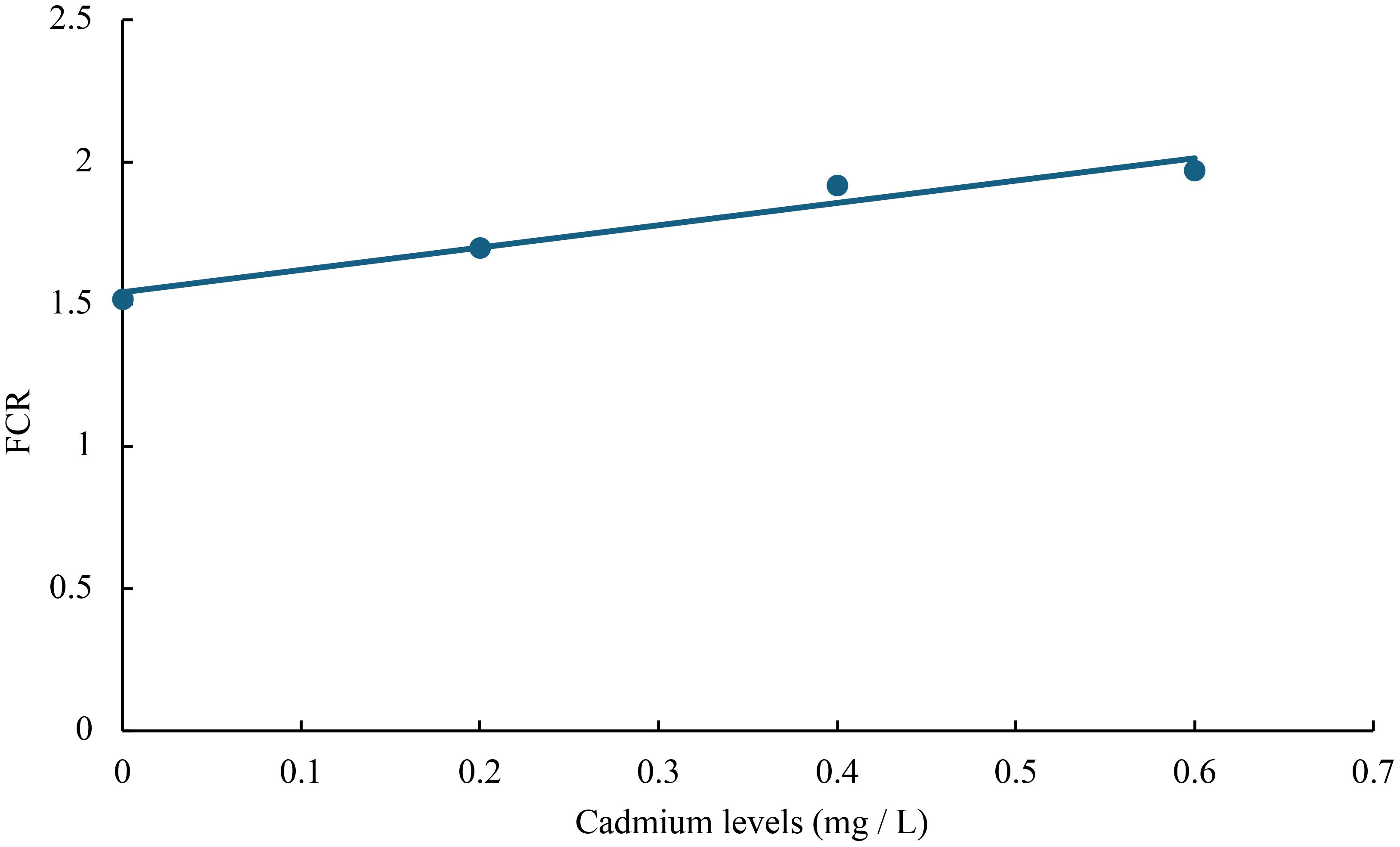
Figure 9 Relationship between cadmium levels and FCR of juvenile GIFT based on a linear regression analysis, in which Y = 0.785X + 1.542, R2 = 0.9504.
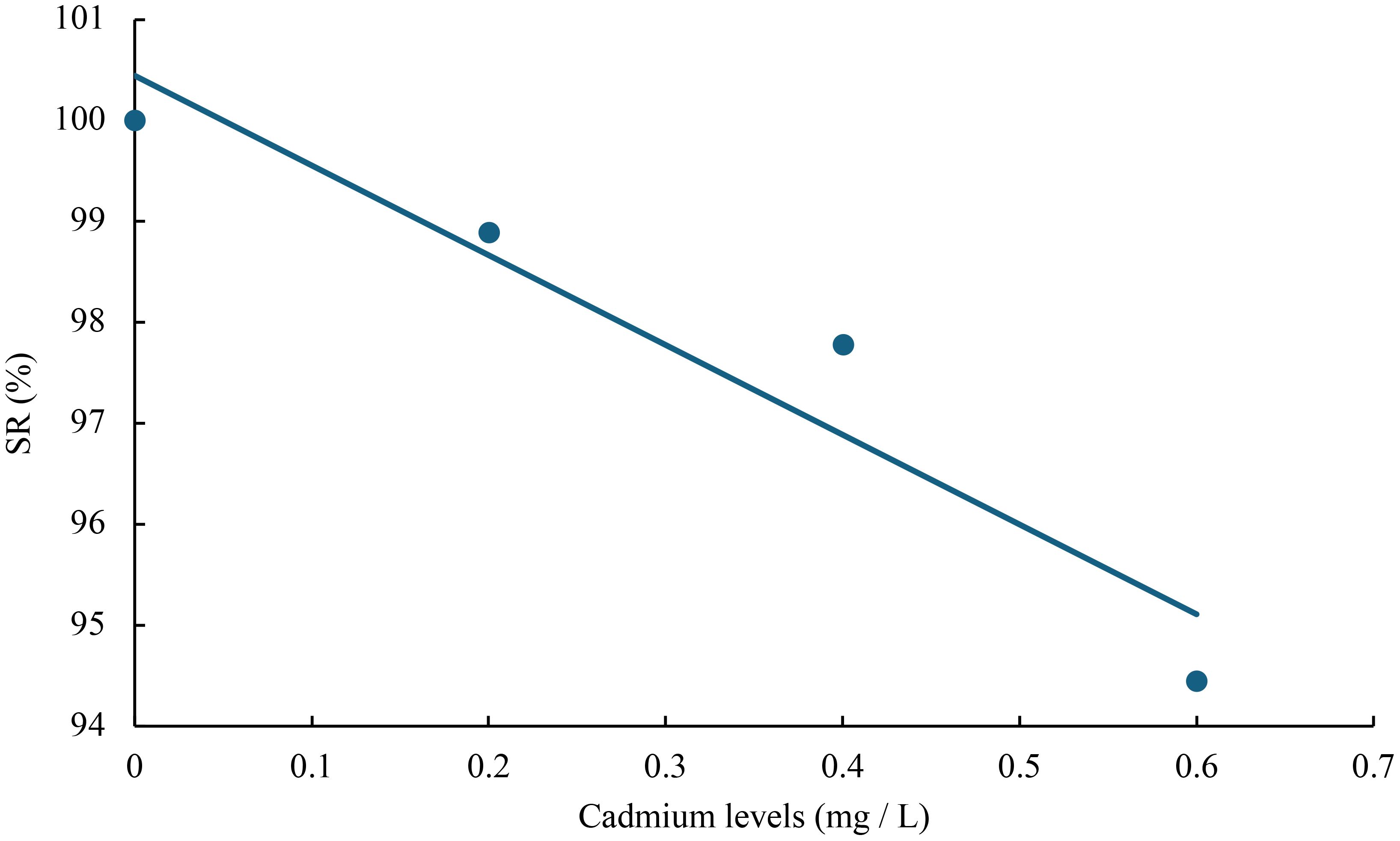
Figure 10 Relationship between cadmium levels and FCR of juvenile GIFT based on a linear regression analysis, in which Y = -8.895X + 100.45, R2 = 0.9138.
3.2 Effects of cadmium on the muscle composition of juvenile GIFT
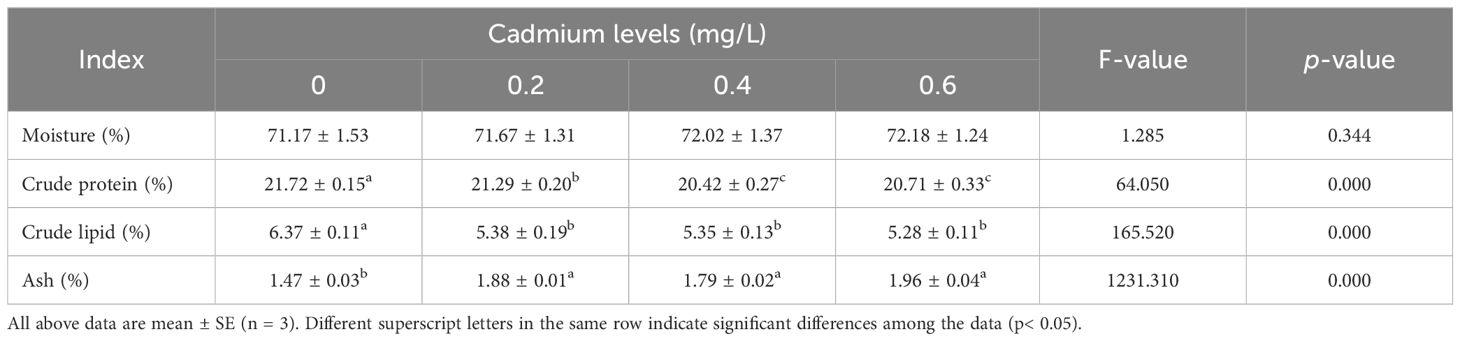
Compared to the control group (0 mg/L Cd2+), the content of crude protein and crude lipid in the muscle of juvenile GIFT significantly decreased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.4 mg/L Cd2+ group exhibited the lowest crude protein content, which was significantly lower (p< 0.05) than that in the 0.2 mg/L Cd2+ group, but no significant difference (p > 0.05) was observed compared to 0.6 mg/L Cd2+ group. The 0.6 mg/L Cd2+ group exhibited the lowest crude lipid content, but no significant difference (p > 0.05) was observed compared to 0.2 and 0.4 mg/L Cd2+ groups, as shown in Table 3.
Compared to the control group (0 mg/L Cd2+), the content of ash in the muscle of juvenile GIFT significantly decreased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the highest ash content, but no significant difference (p > 0.05) was observed compared to 0.2 and 0.4 mg/L Cd2+ groups, as shown in Table 3.
However, there was no significant difference (p > 0.05) between 0, 0.2, 0.4, and 0.6 mg/L Cd2+ groups on the moisture content in the muscle of juvenile GIFT, as shown in Table 3.
3.3 Effects of cadmium on the digestive enzyme activity of juvenile GIFT
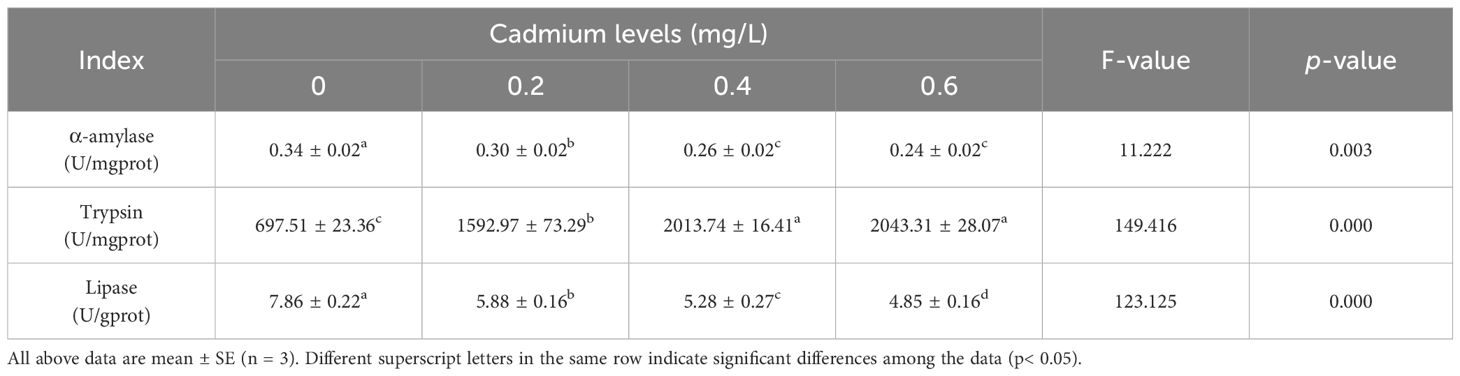
Compared to the control group (0 mg/L Cd2+), the α-amylase and lipase activity in the intestinal of juvenile GIFT significantly decreased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the lowest α-amylase activity, which was significantly lower (p< 0.05) than that in the 0.2 mg/L Cd2+ group, but no significant difference (p > 0.05) was observed compared to 0.4 mg/L Cd2+ group. The 0.6 mg/L Cd2+ group exhibited the lowest lipase activity, which was significantly lower (p< 0.05) than that in the 0.2 and 0.4 mg/L Cd2+ groups, as shown in Table 4.
Compared to the control group (0 mg/L Cd2+), the trypsin activity in the intestinal of juvenile GIFT significantly increased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the highest trypsin activity, which was significantly higher (p< 0.05) than that in the 0.2 mg/L Cd2+ group, but no significant difference (p > 0.05) was observed compared to 0.4 mg/L Cd2+ group, as shown in Table 4.
3.4 Effects of cadmium on the gene expression of lipid metabolism of juvenile GIFT
Compared to the control group (0 mg/L Cd2+), the expression levels of peroxisome proliferator-activated receptor α (pparα), malate dehydrogenase (mdh), carnitine palmityl transferase 1 (cpt-1), lipoprotein lipase (lpl), leptin (lep), hormone-sensitive lipase (hsl), peroxisome proliferator-activated receptor γ (pparγ), fatty acid synthetase (fas), cholesterol response element binding protein 1 (srebp1), stearoyl-CoA desaturase (scd), and squalene cyclooxygenase (sqle) genes in the liver of juvenile GIFT significantly decreased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the lowest expression levels of cpt-1, pparα, mdh, pparγ, hsl, lep, fas, srebp1, sqle, and scd, which were significantly lower (p< 0.05) than those in the 0.2 and 0.4 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the lowest expression levels of lpl, which was significantly lower (p< 0.05) than that in the 0.2 mg/L Cd2+ group, but no significant difference (p > 0.05) was observed compared to 0.4 mg/L Cd2+ group, as shown in Figure 11.
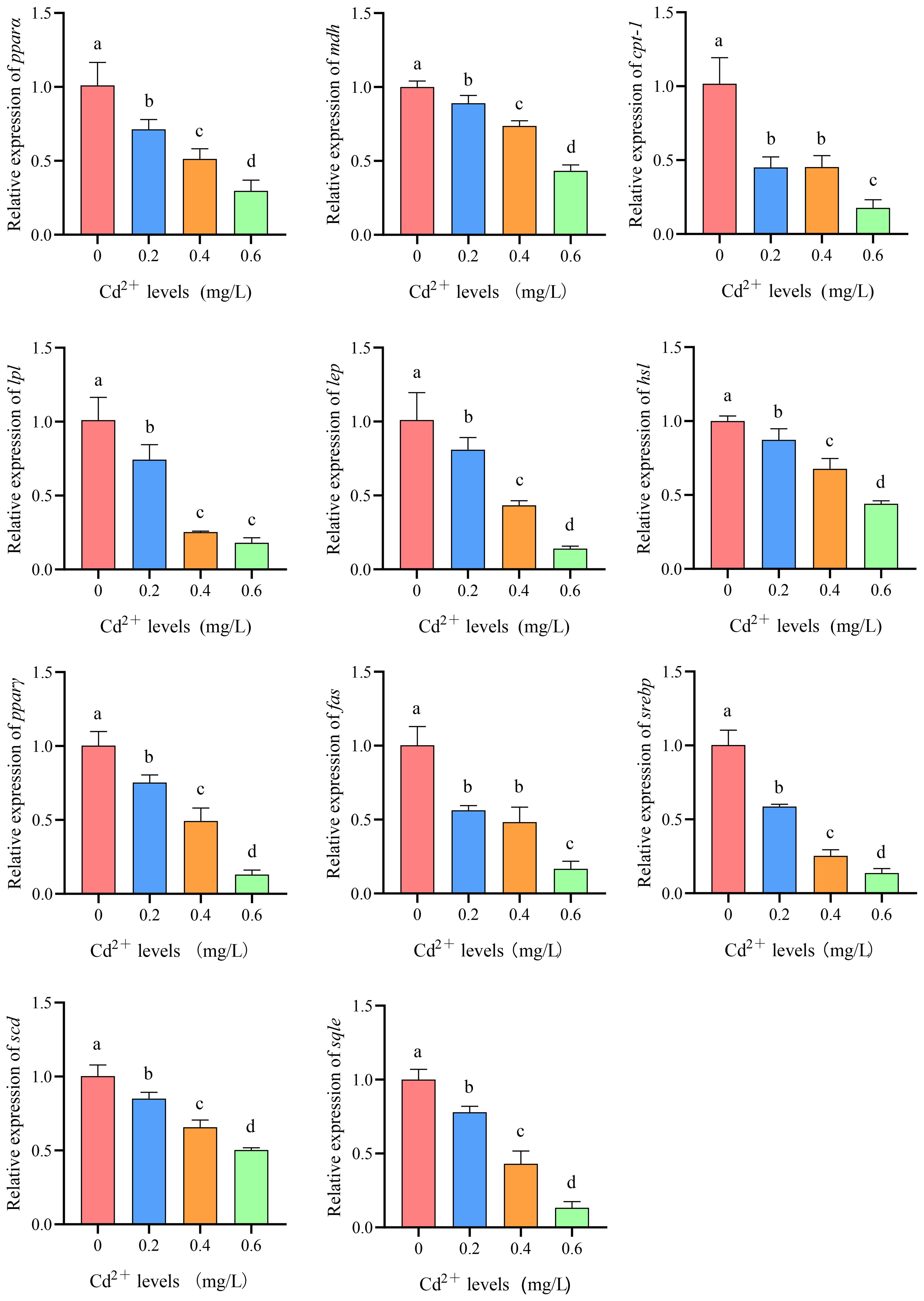
Figure 11 Effects of cadmium on the expression levels of peroxisome proliferator-activated receptor α (pparα), malate dehydrogenase (mdh), carnitine palmityl transferase 1 (cpt-1), lipoprotein lipase (lpl), leptin (lep), hormone-sensitive lipase (hsl), peroxisome proliferator-activated receptor γ (pparγ), fatty acid synthetase (fas), cholesterol response element binding protein 1 (srebp1), stearoyl-CoA desaturase (scd), and squalene cyclooxygenase (sqle) genes in the liver of juvenile GIFT. All the above data are mean ± SE (n = 3). Different superscript letters in the figure indicate significant differences among the data (p< 0.05).
3.5 Effects of cadmium on the gene expression of antioxidant of juvenile GIFT
Compared to the control group (0 mg/L Cd2+), the expression levels of catalase (cat) and superoxide dismutase (sod) genes in the liver and gills of juvenile GIFT significantly decreased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the lowest expression levels of cat and sod, which were significantly lower (p< 0.05) than those in the 0.2 and 0.4 mg/L Cd2+ group, as shown in Figure 12.
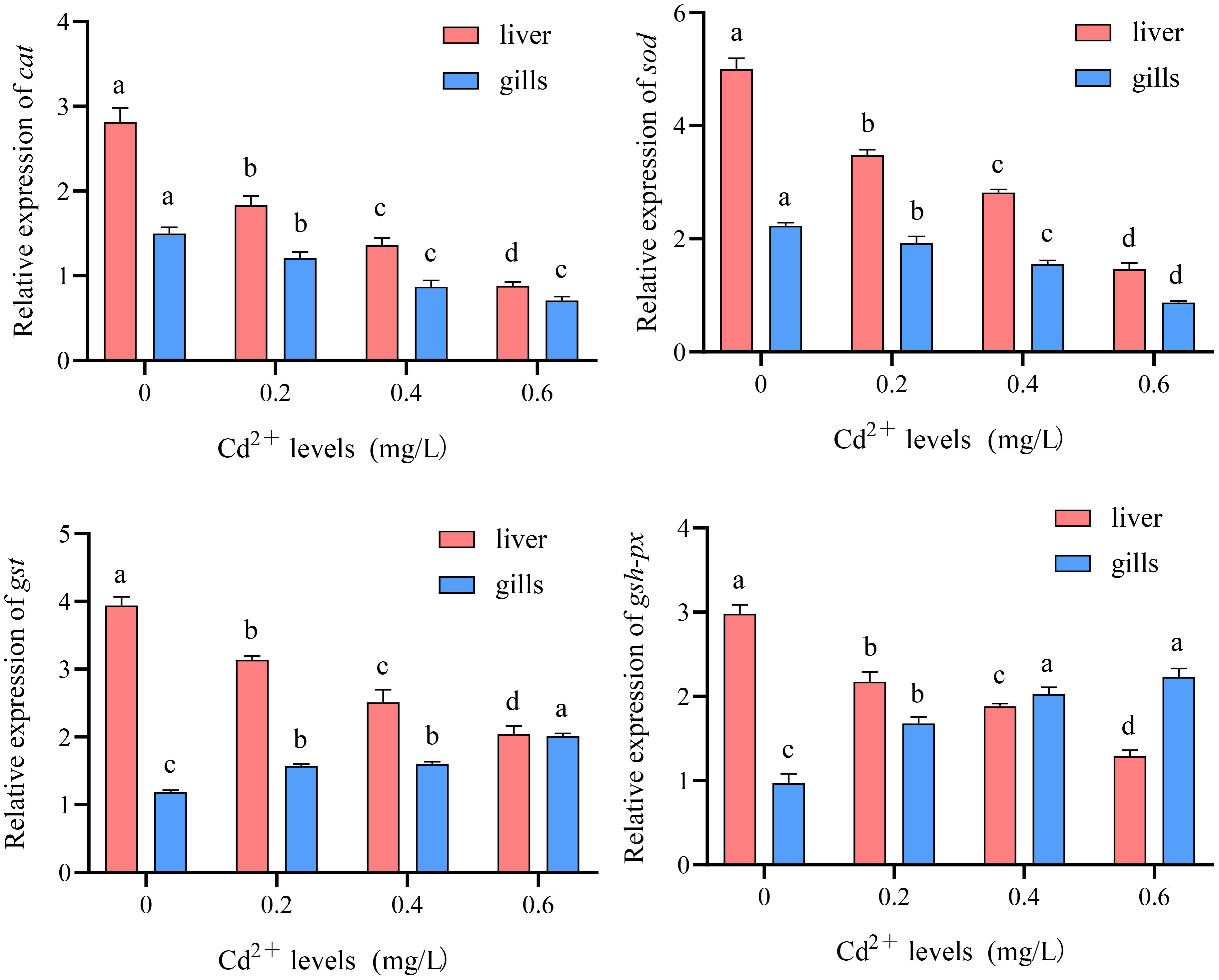
Figure 12 Effects of cadmium on the expression levels of catalase (cat), superoxide dismutase (sod), glutathione S-transferase (gst), and glutathione peroxidase (gsh-px) genes in the liver and gills of juvenile GIFT. All the above data are mean ± SE (n = 3). Different superscript letters in the figure indicate significant differences among the data (p< 0.05).
Compared to the control group (0 mg/L Cd2+), the expression levels of glutathione S-transferase (gst) and glutathione peroxidase (gsh-px) genes in the liver of juvenile GIFT significantly decreased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the lowest expression levels of cat and sod, which were significantly lower (p< 0.05) than those in the 0.2 and 0.4 mg/L Cd2+ group, as shown in Figure 12.
Compared to the control group (0 mg/L Cd2+), the expression levels of gst and gsh-px genes in the gills of juvenile GIFT significantly increased (p< 0.05) in the 0.2, 0.4, and 0.6 mg/L Cd2+ groups. The 0.6 mg/L Cd2+ group exhibited the highest expression levels of gst, which was significantly higher (p< 0.05) than that in the 0.2 and 0.4 mg/L Cd2+ group. The 0.6 mg/L Cd2+ group exhibited the highest expression levels of gsh-px, which was significantly higher (p< 0.05) than that in the 0.2 mg/L Cd2+ group, but no significant difference (p > 0.05) was observed compared to 0.4 mg/L Cd2+ group, as shown in Figure 12.
4 Discussion
The growth performance is a crucial indicator that reflects the health and growth status of fish (Zhang et al., 2023). The results of this study demonstrated that cadmium could reduce the growth performance of juvenile GIFT. This may be because: First, the cadmium in aquaculture water leads to a reduction in fish appetite and feed intake, resulting in a decrease in the final body weight, specific growth rate, and weight gain rate of fish (Khanh et al., 2022). Heydarnejad et al. conducted an experiment where rainbow trout (Oncorhynchus mykiss) were subjected to sub-lethal concentrations (1 and 3 μg/L) of cadmium, and it was observed that the rainbow trout exhibited a decrease in appetite and a significant decline in feeding activity as the concentration of cadmium increased (Heydarnejad et al., 2013). The study of Drąg-Kozak et al. revealed the presence of anorexia in Prussian carp (Carassius gibelio Bloch) when exposed to cadmium (Drąg-Kozak et al., 2021). Second, cadmium stress could induce inflammation in the viscera of fish, lesions in liver tissue, enlargement of liver cells, and dilation of capillaries. As a result, there is a significant increase in the hepatosomatic index and viscera index of fish (Abdel-Tawwab et al., 2024). Ahmed et al. discovered that exposure to heavy metals, such as lead and cadmium, during their investigation of the histological lesions of freshwater climbing perch (Anabas testudineus), resulted in hepatomegaly, obesity, and a pronounced inflammatory response (Ahmed et al., 2014). Third, cadmium stress could induce severe intestinal inflammation and reduce the activity of digestive enzymes, thereby impairing the absorption and utilization of feed by fish (Heydarnejad et al., 2013; Ayyat et al., 2017). The study of Fazio et al. revealed that catfish (Mystus seenghala) exhibited a significant reduction in feed absorption and an increase in feed conversion ratio under cadmium stress conditions (Fazio et al., 2022). In addition, cadmium stress could affect the health of fish, resulting in decreased disease resistance and stress resistance, and significantly reduced the survival rate (Hansen et al., 2002).
In this study, cadmium stress significantly reduced the content of crude protein and crude lipid, and significantly increased the content of ash in the muscle of juvenile GIFT, indicating that cadmium stress could affect the muscle composition of fish. This may be because: First, under cadmium stress, juvenile tilapia could generate a substantial quantity of reactive oxygen species (ROS), including anions, hydroxyl radicals, and hydrogen peroxide (Hu et al., 2021). Cadmium could induce oxidative stress in the fish via ROS, leading to heightened physical energy expenditure, increased respiratory rate, and accelerated energy metabolism rate in the fish. Consequently, this diminished nutrient absorption and utilization by the fish (Abdel-Tawwab et al., 2012). The content of crude protein and crude lipid was significantly reduced. Similar results were observed in yellow catfish (Pelteobagrus fulvidraco) and Nile tilapia (Oreochromis niloticus), where exposure to cadmium significantly decreased the content of crude lipid and crude protein of the fish (Abdel Tawwab and Wafeek, 2010; Tan et al., 2010). Second, the increase in heavy metal content in water leads to the excessive absorption of heavy metal elements by fish bodies (Pandey and Madhuri, 2014). However, these heavy metal elements could not be effectively metabolized within the fish bodies, resulting in an elevation of inorganic salt content and subsequently leading to an increase in ash content (Tan et al., 2010). Similar studies have demonstrated that the presence of cadmium in aquatic environments leads to an increase in the ash content of tilapia (Abdel Tawwab and Wafeek, 2010). The ash content of juvenile tilapia exposed to heavy metals exhibited a significantly higher level compared to that of juvenile tilapia cultured in unpolluted aquatic environments (Abdel-Tawwab et al., 2017).
The activities of trypsin, α-amylase, and lipase in the intestine can serve as indicators of the digestive efficiency of fish (Zaki et al., 2023). The findings of this study demonstrated a positive correlation between cadmium concentration in the water and trypsin activity in the intestinal of juvenile GIFT, while α-amylase and lipase activities exhibited a significant decrease. These results indicated that cadmium stress could significantly impact the digestive efficiency of the fish. This may be because: First, the binding of cadmium to thiol groups in the active center of digestive enzymes, specifically cysteine residues, leads to inhibition of lipase and amylase activities (Wu et al., 2014). Second, cadmium stress could induce oxidative stress within the intestinal of fish, leading to impairment of both the antioxidant and immune systems. This subsequently resulted in necrosis of the intestinal tissue structure, ultimately causing a downregulation in digestive enzyme activity (Ferain et al., 2021). Attia et al. investigated the barrier effect of cadmium on intestinal tissues and observed that cadmium disrupted the metabolic processes in these tissues, leading to structural damage within the intestines (Attia et al., 2022). Third, in cadmium-stressed environments, trypsin activity in fish was upregulated to facilitate food digestion and provide energy (Wu et al., 2013). Moreover, trypsin could activate protease-activated receptor 2 (PAR-2) through specific small interfering RNA (siRNA), thereby inducing the release of the inflammatory factor interleukin 8 (IL-8) and causing intestinal inflammation in fish (Larsen et al., 2008). Similar studies have demonstrated that the long-term exposure of three-spined stickleback (Gasterosteus aculeatus) to cadmium significantly decreased the activity of digestive enzymes in the intestine (Hani et al., 2018). When investigating the impact of dietary cadmium on the aquatic bird (Cairina moschata) lipid metabolism and storage, Lucia et al. discovered that cadmium effectively hindered fatty acid synthesis, leading to a reduction in lipase activity (Lucia et al., 2010). The study of Silvestre et al. investigated the differential protein expression of Chinese mitten crab (Eriocheir sinensis) in response to cadmium exposure, revealing an upregulation of trypsin levels in Eriocheir sinensis following prolonged cadmium exposure (Silvestre et al., 2006).
In this study, compared with the control group, the expression of lipid anabolism and catabolism-related genes in the liver of juvenile GIFT decreased significantly with the increase of cadmium concentration, indicating that cadmium could inhibit the nutrient metabolism and energy metabolism of the fish. This may be because: First, the deposition of lipids was inhibited under cadmium stress due to the inhibition of both de novo synthesis and β-oxidation of fatty acids (Pan et al., 2018). Ferain et al. observed that the relative expression levels of genes involved in lipid metabolism in the liver of rainbow trout, including carnitine palmitoyl transferase 1 (cpt-1), hormone-sensitive lipase (HSL), and lipoprotein lipase (lpl), significantly decreased under cadmium stress (Ferain et al., 2021). Guo et al. demonstrated that environmental cadmium stress had a significant impact on the triglyceride and lipid metabolism pathway of carp, specifically highlighting its disruptive effect on the β-oxidation pathway (Guo et al., 2022). Second, cadmium stress-induced oxidative stress in the liver of fish, resulting in the disruption of liver tissue structure and downregulation of genes associated with lipid metabolism (Shi et al., 2018). Wang et al. demonstrated that cadmium could cause oxidative damage and apoptosis in clam (Meretrix meretrix), and mitochondria were the key cell organelles affected by cadmium toxicity (Wang et al., 2021). Third, cadmium could cause oxidative stress in the liver, and then activate the AMPK/PPAR-γ/NF-κB pathway, resulting in the downregulation of peroxisome proliferator-activated receptor α (pparα) and pparγ gene expression, and finally induce autophagy and apoptosis (Wang et al., 2021). Zhu et al. investigated the effect of cadmium exposure on hepatic lipid metabolism in mice, and they observed that such exposure resulted in dysregulation of the transcriptome associated with lipid metabolism, particularly affecting the PPAR signaling pathway-mediated lipid metabolism pathway, thereby inducing disorders in lipid metabolism (Zhu et al., 2024). Fourth, the presence of Cd2+ could compete with Ca2+ or other bivalent metal cations, thereby inducing ion imbalances, which could alter oxidative stress levels and lead to mitochondrial dysfunction, subsequently resulting in dysregulated disruption of lipid metabolic pathways and downregulation of lipid metabolism-related genes (Liu X. et al., 2023). The effects of low-level cadmium exposure on lipid metabolic pathways in mice were investigated by Go et al., who discovered that cadmium regulated lipid synthesis, oxidation, and mitochondrial oxidative phosphorylation in mice exposed to low-level cadmium (Go et al., 2015). Dai et al. discovered that cadmium exposure induced hepatic oxidative stress in carp (Procypris merus), resulting in excessive autophagy. Additionally, cadmium competed with bivalent metal ions, leading to mitochondrial dysfunction and downregulation of genes related to lipid metabolism (Dai et al., 2020).
In this study, there was a significant decrease in the relative expression levels of catalase (cat) and superoxide dismutase (sod) genes in the liver and gills of juvenile GIFT with increasing cadmium concentrations. However, there was a significant decrease in the relative expression levels of glutathione S transferase (gst) and glutathione peroxidase (gsh-px) genes in the liver, while a significant increase was observed in their relative expression levels in the gills. The results showed that different tissues of tilapia had different responses to cadmium stress. This may be because: First, after prolonged exposure to cadmium stress, fish had a significant increase in the production of ROS that surpassed the body’s endogenous antioxidant defense system. Consequently, this leads to profound physiological distress, resulting in irreversible tissue damage and cellular necrosis while concurrently suppressing the expression of antioxidant genes (Renu et al., 2021; Mukherjee et al., 2022). Second, cadmium stress induced Nrf2 binding and dissociated Nrf2 from Keap1, leading to the translocation of Nrf2 into the nucleus and subsequent suppression of antioxidant gene expression. Simultaneously, the levels of gsh-px and gst regulated by the Nrf2 signaling pathway were significantly diminished, indicating inhibition of Nrf2 and thereby demonstrating impairment in fish’s antioxidant system (Al-sawafi et al., 2017; Ahmatjan et al., 2023). Similar studies showed that: Hu et al. demonstrated a significant down-regulation of the relative expressions of sod and cat genes in zebrafish (Danio rerio) exposed to cadmium stress (Hu et al., 2022). Woo et al. investigated the effect of heavy metals on the antioxidant system of Javanese medaka (Oryzias javanicus), and they observed a significant decline in the levels of SOD upon exposure to stressors such as Zn2+ and Cd2+ (Woo et al., 2009). Wang et al. observed that Cd could alter the molecular conformation of SOD (Wang et al., 2015). Third, the upregulation of gsh-px and gst in gills may be attributed to the toxic excitatory effect caused by cadmium stress, leading to the emergence of an overcompensation mechanism. This mechanism could trigger the upregulation of antioxidant gene expression to eliminate reactive oxygen species in vivo and mitigate oxidative damage (Boyes et al., 2012). A similar study has shown that cadmium stress could trigger an upregulation of gsh-px and gst levels in the gills of olive flounder (Paralichthys olivaceus), which facilitates the elimination of reactive oxygen species (ROS) generated under cadmium stress (Lee et al., 2022).
5 Conclusions
In conclusion, under the experimental conditions, the growth performance, body composition, digestive enzyme activity, expression of lipid metabolism, and antioxidant genes of juvenile GIFT were significantly affected by cadmium stress. Therefore, it is crucial to prioritize water quality protection efforts during this stage of juvenile fish development. Furthermore, the assessment of antioxidant responses and lipid metabolism in tilapia could potentially serve as a biomarker for heavy metal pollution such as cadmium. This finding offers valuable insights for water pollution monitoring.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Biomedical Ethics Committee of Guangxi Minzu University, Nanning, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. YX: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Supervision, Validation, Visualization, Software, Writing – original draft. RQ: Conceptualization, Investigation, Validation, Visualization, Writing – original draft. EH: Conceptualization, Investigation, Validation, Visualization, Writing – original draft. ZZ: Conceptualization, Investigation, Validation, Visualization, Writing – review & editing. JZ: Conceptualization, Investigation, Validation, Visualization, Writing – review & editing. DL: Conceptualization, Investigation, Validation, Visualization, Writing – review & editing. LM: Conceptualization, Investigation, Validation, Visualization, Writing – review & editing. YL: Conceptualization, Investigation, Validation, Visualization, Writing – review & editing, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision. TT: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Grants from the Scientific Research Foundation for the Introduced Talents of Guangxi Minzu University (2018KJQD14 and 2021KJQD17). Joint Funds of the National Natural Science Foundation of China (U20A2064). Shandong Provincial Key Research and Development Programs (2019JZZY020710). Innovation-driven Development Special Fund Project of Guangxi (AA17204044).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdel-Tawwab M., El-Sayed G. O., Monier M. N., Shady S. H. (2017). Dietary edta supplementation improved growth performance, biochemical variables, antioxidant response, and resistance of nile tilapia, oreochromis niloticus (l.) To environmental heavy metals exposure. Aquaculture 473, 478–486. doi: 10.1016/j.aquaculture.2017.03.006
Abdel-Tawwab M., El-Sayed G. O., Shady S. H. (2012). Effects of dietary protein levels and environmental zinc exposure on the growth, feed utilization, and biochemical variables of nile tilapia, oreochromis niloticus (l.). Toxicol. Environ. Chem. 94, 1368–1382. doi: 10.1080/02772248.2012.703202
Abdel-Tawwab M., Khalil R. H., Younis N. A., Abo Selema T. A., Saad A. H., El-Werwary S. O., et al. (2024). Saccharomyces cerevisiae supplemented diets mitigate the effects of waterborne cadmium toxicity on gilthead seabream (sparus aurata l.): Growth performance, haemato-biochemical, stress biomarkers, and histopathological investigations. Vet. Res. Commun. 48, 69–84. doi: 10.1007/s11259-023-10176-0
Abdel Tawwab M., Wafeek M. (2010). Response of nile tilapia, oreochromis niloticus (l.) To environmental cadmium toxicity during organic selenium supplementation. J. World Aquacult. Soc. 41, 106–114. doi: 10.1111/jwas.2010.41.issue-1
Ahmatjan B., Ruotian L., Rahman A., Bin M., Heng D., Yi H., et al. (2023). Klotho inhibits the formation of calcium oxalate stones by regulating the keap1-nrf2-are signaling pathway. Int. Urol. Nephrol. 55, 263–276. doi: 10.1007/s11255-022-03398-9
Ahmed M. K., Parvin E., Islam M. M., Akter M. S., Khan S., Al-Mamun M. H. (2014). Lead-and cadmium-induced histopathological changes in gill, kidney and liver tissue of freshwater climbing perch anabas testudineus (bloch 1792). Chem. Ecol. 30, 532–540. doi: 10.1080/02757540.2014.889123
Al-sawafi A. A., Wang L., Yan Y. (2017). Cadmium accumulation and its histological effect on brain and skeletal muscle of zebrafish. J. Heavy Met. Toxic. Dis. 02, 1. doi: 10.21767/2473-6457.100017
Association of Official Analytical Chemists (AOAC). (2005). Official Methods of Analysis of the Association of Official Analytical Chemists (Hoboken, NJ, USA: John Wiley & Sons, Inc).
Atli G., Canli E. G., Eroglu A., Canli M. (2016). Characterization of antioxidant system parameters in four freshwater fish species. Ecotox. Environ. Safe. 126, 30–37. doi: 10.1016/j.ecoenv.2015.12.012
Attia S. M., Varadharajan K., Shanmugakonar M., Das S. C., Al-Naemi H. A. (2022). Cadmium: an emerging role in adipose tissue dysfunction. Expo. Health 14, 171–183. doi: 10.1007/s12403-021-00427-3
Ayyat M. S., Mahmoud H. K., El-Hais A. E. M., Abd El-Latif K. M. (2017). The role of some feed additives in fish fed on diets contaminated with cadmium. Environ. Sci. pollut. Res. 24, 23636–23645. doi: 10.1007/s11356-017-9986-1
Boyes W. K., Bingham E., Cohrssen B., Powell C. H. (2012). “Neurotoxicology and behavior,” in Patty’s Toxicology, 5th (John Wiley & Sons Inc., New York, NY), 35–74.
Chellaiah E. R. (2018). Cadmium (heavy metals) bioremediation by pseudomonas aeruginosa: a minireview. Appl. Water Sci. 8, 154. doi: 10.1007/s13201-018-0796-5
Cheung K. C., Leung H. M., Wong M. H. (2008). Metal concentrations of common freshwater and marine fish from the pearl river delta, south China. Arch. Environ. Contam. Toxicol. 54, 705–715. doi: 10.1007/s00244-007-9064-7
Da Rocha A. M., De Freitas D. S., Burns M., Vieira J. P., de la Torre F. R., Monserrat J. M. (2009). Seasonal and organ variations in antioxidant capacity, detoxifying competence and oxidative damage in freshwater and estuarine fishes from southern Brazil. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 150, 512–520. doi: 10.1016/j.cbpc.2009.07.012
Dai Z., Cheng J., Bao L., Zhu X., Li H., Chen X., et al. (2020). Exposure to waterborne cadmium induce oxidative stress, autophagy and mitochondrial dysfunction in the liver of procypris merus. Ecotox. Environ. Safe. 204, 111051. doi: 10.1016/j.ecoenv.2020.111051
Drąg-Kozak E., Łuszczek-Trojnar E., Socha M. (2021). Cadmium accumulation and depuration in the muscle of pRussian carp (carassius gibelio bloch) after sub-chronic cadmium exposure: ameliorating effect of melatonin. Animals 11, 2454. doi: 10.3390/ani11082454
Fazio F., Habib S. S., Naz S., Hashmi M. A. H., Saoca C., Ullah M. (2022). Cadmium sub-lethal concentration effect on growth, haematological and biochemical parameters of mystus seenghala (sykes 1839). Biol. Trace Elem. Res. 200, 2432–2438. doi: 10.1007/s12011-021-02852-6
Ferain A., Delbecque E., Neefs I., Dailly H., De Saeyer N., Van Larebeke M., et al. (2021). Interplay between dietary lipids and cadmium exposure in rainbow trout liver: influence on fatty acid metabolism, metal accumulation and stress response. Aquat. Toxicol. 231, 105676. doi: 10.1016/j.aquatox.2020.105676
Go Y., Sutliff R. L., Chandler J. D., Khalidur R., Kang B., Anania F. A., et al. (2015). Low-dose cadmium causes metabolic and genetic dysregulation associated with fatty liver disease in mice. Toxicol. Sci. 147, 524–534. doi: 10.1093/toxsci/kfv149
Guo W., Gao B., Zhang X., Ren Q., Xie D., Liang J., et al. (2022). Distinct responses from triglyceride and cholesterol metabolism in common carp (cyprinus carpio) upon environmental cadmium exposure. Aquat. Toxicol. 249, 106239. doi: 10.1016/j.aquatox.2022.106239
Hamouda A. H., Abd Alkareem O. M. (2021). Insight into the correlation between parasitic infestation and heavy metal concentrations in tilapia species inhabiting lake nasser, Egypt. Aquac. Res. 52, 3425–3437. doi: 10.1111/are.15187
Hani Y. M. I., Turies C., Palluel O., Delahaut L., Gaillet V., Bado-Nilles A., et al. (2018). Effects of chronic exposure to cadmium and temperature, alone or combined, on the threespine stickleback (gasterosteus aculeatus): interest of digestive enzymes as biomarkers. Aquat. Toxicol. 199, 252–262. doi: 10.1016/j.aquatox.2018.04.006
Hansen J. A., Welsh P. G., Lipton J., Suedkamp M. J. (2002). The effects of long-term cadmium exposure on the growth and survival of juvenile bull trout (salvelinus confluentus). Aquat. Toxicol. 58, 165–174. doi: 10.1016/S0166-445X(01)00233-8
Heydarnejad M. S., Khosravian-Hemamai M., Nematollahi A. (2013). Effects of cadmium at sub-lethal concentration on growth and biochemical parameters in rainbow trout (oncorhynchus mykiss). Irish Vet. J. 66, 11. doi: 10.1186/2046-0481-66-11
Hu F., Yin L., Dong F., Zheng M., Zhao Y., Fu S., et al. (2021). Effects of long-term cadmium exposure on growth, antioxidant defense and dna methylation in juvenile nile tilapia (oreochromis niloticus). Aquat. Toxicol. 241, 106014. doi: 10.1016/j.aquatox.2021.106014
Hu W., Zhu Q., Zheng J., Wen Z. (2022). Cadmium induced oxidative stress, endoplasmic reticulum (er) stress and apoptosis with compensative responses towards the up-regulation of ribosome, protein processing in the er, and protein export pathways in the liver of zebrafish. Aquat. Toxicol. 242, 106023. doi: 10.1016/j.aquatox.2021.106023
Ji L. L., Yeo D. (2021). Oxidative stress: an evolving definition. Faculty Rev. 10, 13. doi: 10.12703/r/10-13
Jiang D., Hu Z., Liu F., Zhang R., Duo B., Fu J., et al. (2014). Heavy metals levels in fish from aquaculture farms and risk assessment in lhasa, tibetan autonomous region of China. Ecotoxicology 23, 577–583. doi: 10.1007/s10646-014-1229-3
Johri N., Jacquillet G., Unwin R. (2010). Heavy metal poisoning: the effects of cadmium on the kidney. Biometals 23, 783–792. doi: 10.1007/s10534-010-9328-y
Joseph P. (2009). Mechanisms of cadmium carcinogenesis. Toxicol. Appl. Pharmacol. 238, 272–279. doi: 10.1016/j.taap.2009.01.011
Khanh D. N. N., Vy N. T. T., Phuong T. H., Nhi P. T., Thang N. Q., Sy D. T., et al. (2022). Effects of cadmium and lead on muscle and liver glycogen levels of climbing perch (anabas testudineus). Bull. Environ. Contam. Toxicol. 168, 854–860. doi: 10.1007/s00128-021-03384-4
Larsen A. K., Seternes O., Larsen M., Aasmoe L., Bang B. (2008). Salmon trypsin stimulates the expression of interleukin-8 via protease-activated receptor-2. Toxicol. Appl. Pharmacol. 230, 276–282. doi: 10.1016/j.taap.2008.02.019
Lee D., Choi Y. J., Kim J. (2022). Toxic effects of waterborne cadmium exposure on hematological parameters, oxidative stress, neurotoxicity, and heat shock protein 70 in juvenile olive flounder, paralichthys olivaceus. Fish Shellfish Immunol. 122, 476–483. doi: 10.1016/j.fsi.2022.02.022
Lee J., Jo A., Lee D., Choi C. Y., Kang J., Kim J. (2023). Review of cadmium toxicity effects on fish: oxidative stress and immune responses. Environ. Res. 236, 116600. doi: 10.1016/j.envres.2023.116600
Leung H. M., Leung A., Wang H. S., Ma K. K., Liang Y., Ho K. C., et al. (2014). Assessment of heavy metals/metalloid (as, pb, cd, ni, zn, cr, cu, mn) concentrations in edible fish species tissue in the pearl river delta (prd), China. Mar. pollut. Bull. 78, 235–245. doi: 10.1016/j.marpolbul.2013.10.028
Liu X., Pang X., Jin L., Pu D., Wang Z., Zhang Y. (2023). Exposure to acute waterborne cadmium caused severe damage on lipid metabolism of freshwater fish, revealed by nuclear lipid droplet deposition in hepatocytes of rare minnow. Aquat. Toxicol. 257, 106433. doi: 10.1016/j.aquatox.2023.106433
Liu Y., Huang E., Xie Y., Meng L., Liu D., Zhang Z., et al. (2023). The effect of dietary lipid supplementation on the serum biochemistry, antioxidant responses, initial immunity, and mtor pathway of juvenile tilapia (oreochromis niloticus). Fishes 8, 535. doi: 10.3390/fishes8110535
Lu K., Xu W., Li X., Liu W., Wang L., Zhang C. (2013). Hepatic triacylglycerol secretion, lipid transport and tissue lipid uptake in blunt snout bream (megalobrama amblycephala) fed high-fat diet. Aquaculture 408, 160–168. doi: 10.1016/j.aquaculture.2013.06.003
Lucia M., André J., Gonzalez P., Baudrimont M., Bernadet M., Gontier K., et al. (2010). Effect of dietary cadmium on lipid metabolism and storage of aquatic bird cairina moschata. Ecotoxicology 19, 163–170. doi: 10.1007/s10646-009-0401-7
Mukherjee A. G., Wanjari U. R., Renu K., Vellingiri B., Gopalakrishnan A. V. (2022). Heavy metal and metalloid-induced reproductive toxicity. Environ. Toxicol. Pharmacol. 92, 103859. doi: 10.1016/j.etap.2022.103859
Munguti J. M., Nairuti R., Iteba J. O., Obiero K. O., Kyule D., Opiyo M. A., et al. (2022). Nile tilapia (oreochromis niloticus linnaeus 1758) culture in Kenya: emerging production technologies and socio-economic impacts on local livelihoods. Aquacul. Fish Fisheries 2, 265–276. doi: 10.1002/aff2.58
Pan Y., Luo Z., Zhuo M., Wei C., Chen G., Song Y. (2018). Oxidative stress and mitochondrial dysfunction mediated cd-induced hepatic lipid accumulation in zebrafish danio rerio. Aquat. Toxicol. 199, 12–20. doi: 10.1016/j.aquatox.2018.03.017
Pandey G., Madhuri S. (2014). Heavy metals causing toxicity in animals and fishes. Res. J. Animal Vet. Fishery Sci. 2, 17–23. Available at: https://isca.in/AVFS/Archive/v2/i2/4.ISCA-RJAVFS-2014-002.pdf.
Renu K., Chakraborty R., Myakala H., Koti R., Famurewa A. C., Madhyastha H., et al. (2021). Molecular mechanism of heavy metals (lead, chromium, arsenic, mercury, nickel and cadmium)-induced hepatotoxicity–a review. Chemosphere 271, 129735. doi: 10.1016/j.chemosphere.2021.129735
Saito T., Whatmore P., Taylor J. F., Fernandes J. M., Adam A., Tocher D. R., et al. (2021). Micronutrient supplementation affects transcriptional and epigenetic regulation of lipid metabolism in a dose-dependent manner. Epigenetics 16, 1217–1234. doi: 10.1080/15592294.2020.1859867
Schmittgen T. D., Livak K. J. (2008). Analyzing real-time pcr data by the comparative ct method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shi Q., Sun N., Kou H., Wang H., Zhao H. (2018). Chronic effects of mercury on bufo gargarizans larvae: thyroid disruption, liver damage, oxidative stress and lipid metabolism disorder. Ecotox. Environ. Safe. 164, 500–509. doi: 10.1016/j.ecoenv.2018.08.058
Silvestre F., Dierick J., Dumont V., Dieu M., Raes M., Devos P. (2006). Differential protein expression profiles in anterior gills of eriocheir sinensis during acclimation to cadmium. Aquat. Toxicol. 76, 46–58. doi: 10.1016/j.aquatox.2005.09.006
Tan X. Y., Luo Z., Zhang G. Y., Liu X. J., Jiang M. (2010). Effect of dietary cadmium level on the growth, body composition and several hepatic enzymatic activities of juvenile yellow catfish, pelteobagrus fulvidraco. Aquac. Res. 41, 1022–1029. doi: 10.1111/j.1365-2109.2009.02386.x
Turchini G. M., Torstensen B. E., Ng W. K. (2009). Fish oil replacement in finfish nutrition. Rev. Aquac. 1, 10–57. doi: 10.1111/j.1753-5131.2008.01001.x
Wang J., Deng W., Zou T., Bai B., Chang A. K., Ying X. (2021). Cadmium-induced oxidative stress in meretrix meretrix gills leads to mitochondria-mediated apoptosis. Ecotoxicology 30, 2011–2023. doi: 10.1007/s10646-021-02465-8
Wang J., Han S., Li L., Lu D., Limbu S. M., Li D., et al. (2018). Lipophagy is essential for lipid metabolism in fish. Sci. Bull. 63, 879–882. doi: 10.1016/j.scib.2018.05.026
Wang J., Zhang H., Zhang T., Zhang R., Liu R., Chen Y. (2015). Molecular mechanism on cadmium-induced activity changes of catalase and superoxide dismutase. Int. J. Biol. Macromol. 77, 59–67. doi: 10.1016/j.ijbiomac.2015.02.037
Woo S., Yum S., Park H., Lee T., Ryu J. (2009). Effects of heavy metals on antioxidants and stress-responsive gene expression in javanese medaka (oryzias javanicus). Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 149, 289–299. doi: 10.1016/j.cbpc.2008.08.002
Wu H., Xuan R., Li Y., Zhang X., Jing W., Wang L. (2014). Biochemical, histological and ultrastructural alterations of the alimentary system in the freshwater crab sinopotamon henanense subchronically exposed to cadmium. Ecotoxicology 23, 65–75. doi: 10.1007/s10646-013-1152-z
Wu H., Xuan R., Li Y., Zhang X., Wang Q., Wang L. (2013). Effects of cadmium exposure on digestive enzymes, antioxidant enzymes, and lipid peroxidation in the freshwater crab sinopotamon henanense. Environ. Sci. pollut. Res. 20, 4085–4092. doi: 10.1007/s11356-012-1362-6
Younis N. A., Laban S. E., Al-Mokaddem A. K., Attia M. M. (2020). Immunological status and histopathological appraisal of farmed oreochromis niloticus exposed to parasitic infections and heavy metal toxicity. Aquac. Int. 28, 2247–2262. doi: 10.1007/s10499-020-00589-y
Zaki M. A., Khalil H. S., Allam B. W., Khalil R. H., Basuini M. F. E., Nour A. E. M., et al. (2023). Assessment of zootechnical parameters, intestinal digestive enzymes, haemato-immune responses, and hepatic antioxidant status of pangasianodon hypophthalmus fingerlings reared under different stocking densities. Aquac. Int. 31, 2451–2474. doi: 10.1007/s10499-023-01092-w
Zhang Q., Li F., Guo M., Qin M., Wang J., Yu H., et al. (2023). Growth performance, antioxidant and immunity capacity were significantly affected by feeding fermented soybean meal in juvenile coho salmon (oncorhynchus kisutch). Animals 13, 945. doi: 10.3390/ani13050945
Zhang Q., Xie Y., Zhang Y., Huang E., Meng L., Liu Y., et al. (2024). Effects of dietary supplementation with chitosan on the muscle composition, digestion, lipid metabolism, and stress resistance of juvenile tilapia (oreochromis niloticus) exposed to cadmium-induced stress. Animals 14, 541. doi: 10.3390/ani14040541
Keywords: growth, antioxidant, lipid metabolism, cadmium, tilapia
Citation: Zhang Q, Xie Y, Qin R, Huang E, Zhang Z, Zhou J, Liu D, Meng L, Liu Y and Tong T (2024) Effects of cadmium on the growth, muscle composition, digestion, gene expression of antioxidant and lipid metabolism in juvenile tilapia (Oreochromis niloticus). Front. Mar. Sci. 11:1443484. doi: 10.3389/fmars.2024.1443484
Received: 04 June 2024; Accepted: 03 July 2024;
Published: 31 July 2024.
Edited by:
Gisele Cristina Favero, Universidade Federal de Minas Gerais, BrazilCopyright © 2024 Zhang, Xie, Qin, Huang, Zhang, Zhou, Liu, Meng, Liu and Tong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqiang Liu, bGl1eW9uZ3FpYW5nQGd4bXp1LmVkdS5jbg==; Tong Tong, dG9uZ3RvbmdAZ3htenUuZWR1LmNu
†These authors have contributed equally to this work
 Qin Zhang
Qin Zhang Yi Xie†
Yi Xie† Yongqiang Liu
Yongqiang Liu