- 1College of Marine Science, Ningde Normal University, Ningde, China
- 2Engineering Research Center of Mindong Aquatic Product Deep-Processing, Ningde Normal University, Ningde, China
- 3Xianghu Laboratory, Hangzhou, China
As rate-limiting enzymes of peroxisomal β-oxidation, acyl-coenzyme A oxidase (ACOXs) play vital roles in maintaining energy homeostasis and regulating reactive oxygen species (ROS) metabolism. However, there are no studies on the functions of ACOXs in crustaceans. In the present study, four full-length cDNA sequences of acoxs, namely the acox-1a (2403 bp), acox-1b (2733 bp), acox-3a (2878 bp) and acox-3b (3445 bp), were successfully isolated from mud crab Scylla paramamosain, which encoded 666, 673, 701 and 658 amino acids, respectively. Sequence analysis showed that the ACOX-1a, ACOX-1b and ACOX-3a possessed conserved structural domains like FAD-binding motif, fatty acyl CoA oxidase domain and peroxisomal targeting signal, while the ACOX-3b lacked peroxisomal targeting signal. Results of phylogenetic tree indicated that the four ACOXs of mud crab grouped gathered with their corresponding orthologues from crustaceans. The acox-1a, acox-3a and acox-3b were highly expressed in hepatopancreas, and the acox-1b was mainly distributed in muscle and hepatopancreas. Compared with feeding groups, the expression levels of acox-1a, acox-3a and acox-3b in hepatopancreas and acox-3a in muscle were markedly up-regulated in fasting groups, suggesting that the acoxs had significant effects in modulating energy balance during fasting. In addition, fasting significantly increased the transcriptional levels of nuclear factor erythroid 2-related factor (nrf2) and its downstream antioxidant genes (catalase (cat), glutathione peroxidase (gpx) and glutathione S-transferase (gst)) to improve antioxidant capacity for removing excessive ROS produced by ACOX-mediated peroxisomal β-oxidation. These results would be conducive to providing new insights into evolutionary characteristics and functions of acoxs in crustaceans.
1 Introduction
Mitochondria and peroxisomes are two important organelles for β-oxidation of fatty acids, and both play irreplaceable roles in the utilization of fatty acids (Houten and Wanders, 2010; Ding et al., 2021). The substrates, enzymes and final products involved in the specific reaction of fatty acid β-oxidation are different in mitochondria and peroxisomes. Mitochondria primarily oxidize short-chain (< C8) and medium-long (C8-C20) fatty acids (Reddy and Hashimoto, 2001). By contrast, the substrates of peroxisomes are more complex than the mitochondria, which mainly oxidizes long-chain fatty acids (> C20), branched-chain fatty acids, leukotrienes, prostaglandin and so on (Oaxaca-Castillo et al., 2007; Sun et al., 2020). The rate-limiting enzyme of β-oxidation in mitochondria is carnitine palmitoyltransferases (CPTs), while the peroxisomes are acyl-coenzyme A oxidases (ACOXs) (Adeva-Andany et al., 2019; He et al., 2021). In addition, β-oxidation of mitochondria can completely oxidize fatty acids into water and carbon dioxide (CO2) as well as release energy to synthesize adenosine triphosphate (ATP) (Panov et al., 2022). However, the β-oxidation of peroxisomes is halfway, which only can oxidize fatty acids into short or medium-chain fatty acyl-coenzyme A (CoA) providing substrates for β-oxidation of mitochondria, and meanwhile this process is accompanied with the production of hydrogen peroxide (H2O2) (Ding et al., 2021; He et al., 2021). Currently, the studies about physiological functions and regulatory mechanisms of fatty acid β-oxidation have been more focused on mitochondria (Verma et al., 2022; Monzel et al., 2023; Huang et al., 2024), while the attention on fatty acid β-oxidation in peroxisomes was far too limited, especially in aquatic animals.
Peroxisomes are a single membrane-enclosed and dynamic organelle existed in almost all eukaryotic cells, and its quantity, morphology and location can alter with the changes of environmental and nutritional status (Huybrechts et al., 2009). In addition to β-oxidation of fatty acids, emerging studies have suggested that the peroxisomes participate in multiple metabolic processes like generation and elimination of reactive oxygen species (ROS), synthesis of bile acids and ether lipid as well as catabolism of polyamines and D-amino acids (Lodhi and Semenkovich, 2014). As rate-limiting enzymes, the ACOXs catalyzed the first step of fatty acid β-oxidation in peroxisomes, and there were three genes named acox-1, acox-2 and acox-3 found in peroxisomes of mammals, which encoded ACOX-1, ACOX-2 and ACOX-3 proteins, respectively (Li et al., 2022). The ACOX-1 mainly oxidizes saturated and unsaturated straight-chain fatty acids and dicarboxylic acids, and the ACOX-2 primarily acts on 2-methyl branched fatty acids (Nöhammer et al., 2001). Additionally, the substrates of ACOX-3 overlap with that of ACOX-1 and ACOX-2, which can oxidize both methyl-branched and straight-chain fatty acids (Van Veldhoven, 2010). In comparison, the acox-2 was not observed in teleost fish, and only found the acox-1 and acox-3 (He et al., 2014; Madureira et al., 2016; Sun et al., 2020; Kong et al., 2022; Wang et al., 2022). For examples, two acox-1 isoforms were found in both Oreochromis niloticus and Ctenopharyngodon idella and highly expressed in liver (He et al., 2014; Sun et al., 2020). Madureira et al. (2016) showed that the Salmo trutta possessed an acox-3 gene, which was mainly distributed in ovary, heart, muscle, liver and stomach. Besides, six acox genes were observed in Chlamys farreri and Patinopecten yessoensis including four acox-1, an acox-2 and an acox-3, indicating the acox-2 gene was not absent in bivalves (molluscs), and these acox genes were primarily expressed in kidneys and digestive glands (Li et al., 2022). However, to the best of our knowledge, the acox genes in crustaceans have not been reported yet.
In addition to metabolic functions, increasing studies have indicated that the peroxisomes are an important sensor of stress, which played critical roles in adapting to environmental and metabolic stresses including viral and bacterial infections, hypoxia, oxidative stress, cold exposure and nutrient deprivation (Chrousos, 2009; He et al., 2021). Emerging evidence have suggested that modulation of peroxisomal abundance, morphology and location within cells as well as cooperation with other organelles like mitochondria and endoplasmic reticulum serve as an important strategy for peroxisomes in responding to external stresses (Chu et al., 2015; He et al., 2021). Besides, peroxisome-derived ROS and other metabolic products like ether lipids and acetyl-CoA, were crucial signaling molecules for peroxisomes to reply stresses (He et al., 2021). The molecular mechanisms of peroxisomes regulating various forms of stress are complex, and studies previously conducted have primarily focused on mammals (He et al., 2020; Ding et al., 2021, 2021). For example, in considering oxidative stress and fasting stress, ACOX-mediated peroxisomal β-oxidation produces abundant H2O2 in certain tissues of mammals (Boveris et al., 1972), and the imbalance of peroxisomal ROS generation and removal can cause oxidative stress that is closely associated with various human diseases (Cipolla and Lodhi, 2017). Studies have shown that the sirtuin 5 (SIRT5) can regulate the ACOX-1 activities by desuccinylation or succinylation indirectly modulating the ROS generation (Matsushita et al., 2011; Chen et al., 2018). Peroxisomes is noted to contain multiple antioxidant enzymes like catalase (CAT), superoxide dismutase (SOD), glutathione peroxidase (GPX) and glutathione S-transferase (GST) to eliminate higher amounts of ROS yielded by ACOX-mediated peroxisomal β-oxidation, and the peroxisomal biogenesis factor Pex5 participated in coping with oxidative stress through regulating the redistribution of peroxisomal CAT (Dubreuil et al., 2020). As for fasting stress, Kong et al. (2020) indicated that nutrient deprivation can stimulate physical coactions of lipid droplets and peroxisomes via kinesin-like protein. The peroxisomal biogenesis factor Pex5 further can recruit triglyceride lipase to lipid droplet-peroxisome contact positions in a manner dependent on protein kinase A activation, and the released fatty acids by lipolysis were provided substrates for β-oxidation of peroxisomes. Similarly, studies in aquatic animals also found that the fasting stress can promote β-oxidation of fatty acids in peroxisomes (Morais et al., 2007; He et al., 2014; Sun et al., 2020; Wang et al., 2022), while the mechanisms were still ill-defined.
As a dominant species with a higher economic returns in China, the mud crab, Scylla paramamosain, has been cultured in South and East China like Guangdong, Fujian and Zhejiang provinces with its yield reaching up to 154,661 tons in 2022 (China Fishery Statistical Yearbook, 2023). Studies have shown that excessive lipid in diets can exacerbate the abnormal deposition of lipid in aquatic animals (liver and enterocoelia), leading to metabolic disorders and inflammation, and ultimately inhibiting the growth of aquatic animals. In addition, excessive lipid deposition can also affect beauty and nutritional quality, and reduce the value of goods. Therefore, lipid utilization (β-oxidation of fatty acids) has been become a research hotspot with wide concern. Previously, we have reported three rate-limiting enzyme genes (cpt-2, cpt-1b and cpt-1a) involved in fatty acid β-oxidation of mitochondria, and made a preliminary investigation on their regulatory mechanisms (Lin et al., 2024). In the present study, we further explored the β-oxidation of fatty acids in peroxisomes, and study contents mainly included the features of four rate-limiting enzymes (acox-1a, acox-1b, acox-3a and acox-3b) of peroxisomal β-oxidation and their functions in respond to fasting. This study was the first time to report the peroxisomal β-oxidation in crustaceans, which would contribute to enhancing the knowledge of acoxs evolutionary characteristics and their functions in lipid utilization of crustaceans.
2 Materials and methods
2.1 Fasting experiment
The fasting experiment contained two treatments (fasting group and feeding group), and the feeding group was set as control group. The mud crabs used in the present study were obtained from a local crab farm in Sandu bay (Ningde, Fujian, China), and the fasting experiment was conducted in aquaculture system produced by company of Zhongkehai (Qingdao, China) in Ningde Normal University. After being acclimated, thirty-six able-bodied and vigorous crabs (64.51 ± 0.83 g) were assigned to six polypropylene buckets (height, 0.67 m; radius, 0.43; volume, 300 L). Each polypropylene bucket was placed with six crabs (male crabs: female crabs, 1: 1), and each treatment contained three polypropylene buckets (three replicate). During the four-week fasting experiment, the crabs in fasting group were in a state of starvation all the time. In addition, the crabs in feeding group were fed twice daily (time 8:30 and 18:00) with Sinonovacula constricta (a local bivalve) to apparent satiation. Waste (feces and uneaten) produced in feeding experiment was removed per day, and 30% water of each bucket was siphoned and injected fresh seawater daily. The water quality parameters were as follows: water temperature was between 15.3°C and 20.3°C, dissolved oxygen was higher than 7.0 mg L−1, and ammonia nitrogen was less than 0.05 mg L−1. At the end of the fasting experiment, samples including muscle and hepatopancreas were obtained after the crabs were dissected. The samples were immediately frozen in liquid nitrogen and stored at −80°C for further use.
2.2 Gene cloning
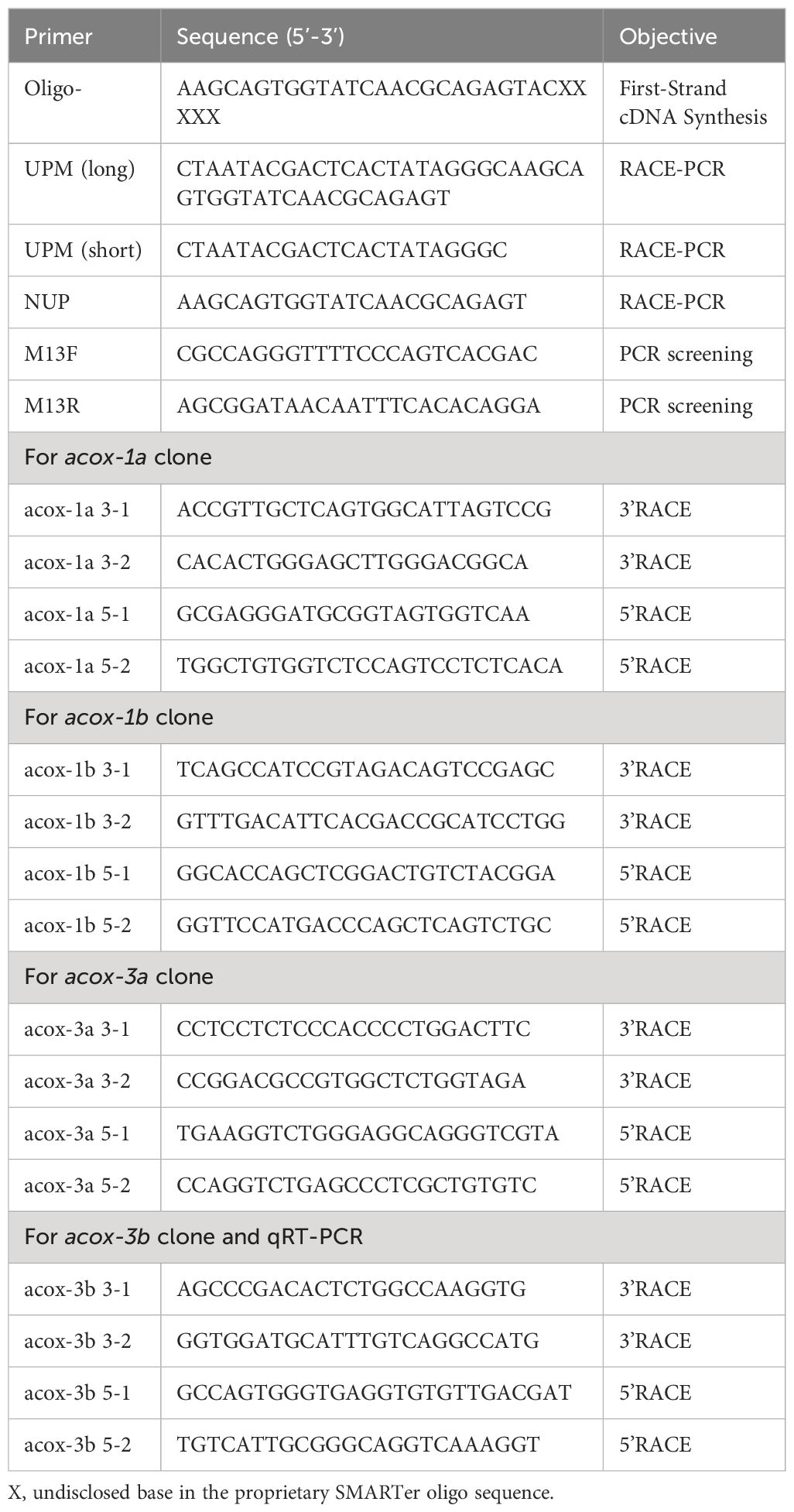
Total RNA from hepatopancreas of mud crab was extracted by using TRNzol Universal Reagent produced by company of Tiangen (Beijing, China). Total RNA above was further reverse-transcribed into clone template with SMART RACE cDNA Amplification kit (Clontech, USA) based on the kit instructions after evaluating the quality and concentration. The cDNA template was kept in -20°C for subsequent use. Four sequence fragments of acox genes, named acox-1a, acox-1b, acox-3a and acox-3b, were gotten from our previous measured transcriptome data. Subsequently, primers used for cloning the 5’ and 3’ untranslated region (UTR) of four acox genes were designed according to sequence fragments above by using software Primer Premier 5. The 5’ and 3’ UTR of four acox genes were cloned mainly through using the method of 5’ and 3’ rapid amplification of cDNA ends (RACE) respectively as well as uniting the strategies of two-round PCR ([irst round PCR (touch-down PCR) and second round PCR (nested PCR)]. The specific operation methods and steps of clone have been shown in a previous study (Lin et al., 2024). The related primes have been shown in Table 1.
2.3 Sequence and phylogenetic analysis
Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI) was chosen to identify the obtained cDNA sequences. The open reading frame prediction and multiple sequence alignment were analyzed by using ORF Finder and DNAMAN software, respectively. NPS Server and TMHMM 2.0 were selected to predict the protein secondary structure and transmembrane domain, respectively. Tertiary structure of protein was assessed by using SWISS-MODEL, and isoelectric point and molecular mass of predicted protein were counted by ExPASy ProtParam tool. SignalP5.0 Server and InterPro were used to determine the signal peptide and structural region, respectively. In addition, the phylogenetic tree was constructed by MEGA 7.0 and ClustalX softwares.
2.4 Quantitative real-time PCR
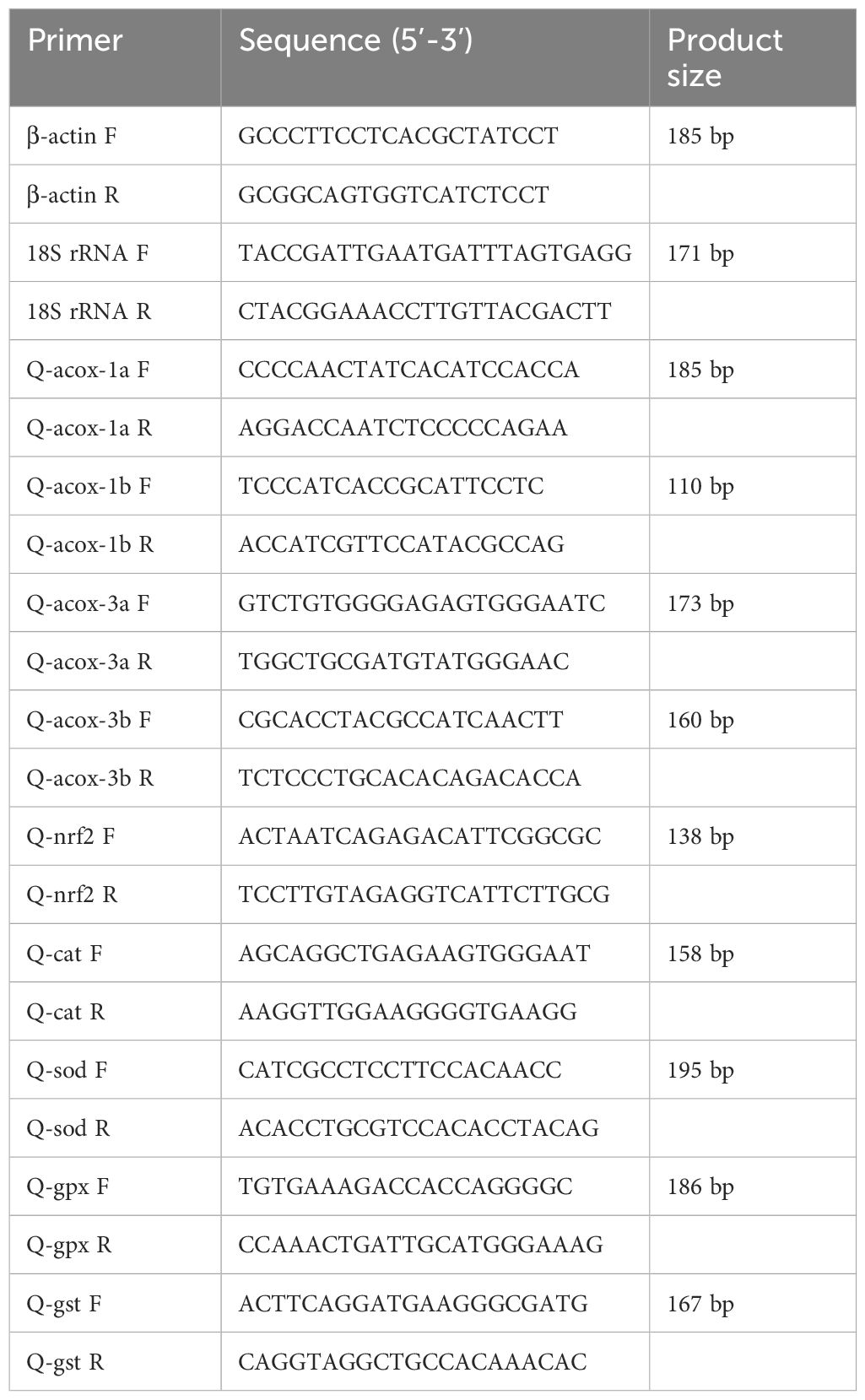
Six mud crab including three males and three females with mean mass 49.50 ± 1.18 g were selected to investigate the tissue distribution of four acox genes (acox-1a, acox-1b, acox-3a and acox-3b). The crabs were anatomized to collect the tissue samples after being anesthetized with ice including thoracic ganglia, muscle, cranial ganglia, heart, gill, eyestalk, hepatopancreas and intestine. The transcriptional levels of four acox genes [acox-1a, acox-1b, acox-3a and acox-3b) in different tissues and target genes (acox-1a, acox-1b, acox-3a, acox-3b, cat, sod, gpx, gst and nuclear factor erythroid 2-related factor 2 (nrf2)] in response to fasting stress were determined by using quantitative real-time PCR. The isolation method of total RNA was same to the gene cloning above, and 1 μg high-quality total RNA of each sample was reverse-transcribed into cDNA template using reverse transcription kit (PrimeScript™ RT reagent Kit with gDNA Eraser) produced by Takara (Dalian, China). The reaction volume of quantitative real-time PCR was 20 μL specifically comprising 10 μL 2 × ChamQ Universal SYBR qPCR Master Mix produced by Vazyme (Nanjing, China), 0.4 μL of each positive and negative primer, 1.0 μL of the four-time diluted cDNA and 8.2 μL of distilled water. The experiment was executed in QuantStudio 3 (Thermo Fisher, USA), and the program included three steps (holding stage, PCR stage and melting curve stage). The holding stage contained a 95°C for 30 s, the PCR stage was 40 cycles of 95°C for 10 s and 60°C for 30 s, and the melting curve stage included 95°C for 15 s, 60°C for 60 s and 95°C for 15 s. The 18S rRNA and β-actin were set as reference genes, and the primes used for quantitative real-time PCR have been given in Table 2. The amplification efficiency of primers used in the present study was ranged from 90% and 110%, which was calculated by formula E = 10(−1/Slope) – 1 via five different thinned cDNA samples. The expression levels of target genes were counted with the method of 2-ΔΔCt reported by Livak and Schmittgen, 2001.
2.5 Statistical analysis
The results were given in term of mean ± standard error, and all the statistical analysis was performed by using SPSS 20.0 software. Significant difference was set as P values less than 0.05, and P values lower than 0.01 was considered as highly significant difference. After checking the homogeneity and normality, one-way analysis of variance (ANOVA) followed by Duncan’s multiple comparison test was used to investigate the statistical differences of target gene expression levels in different tissues. In addition, the statistical difference of fasting experiment was determined by independent-sample T-test.
3 Results
3.1 Sequence analysis
The obtained complete cDNA sequences of acox-1a, acox-1b, acox-3a and acox-3b have been uploaded to the NCBI database, and the GenBank accession numbers were PP708699, PP708700, PP708701 and PP708702, respectively. The full-length sequence of acox-1a was 2403 bp including 29 bp of 5’ UTR, 373 bp of 3’ UTR and 2001 bp of open reading frame encoding a protein with 666 amino acids (Supplementary Figure S1). The acox-1b sequence with total length 2733 bp was comprised of 61 bp of 5’ UTR, 650 bp of 3’ UTR and 2022 bp of open reading frame encoding a protein with 673 amino acids (Supplementary Figure S2). Besides, the acox-3a and acox-3b were 2878 bp and 3445 bp, respectively. The acox-3a sequence contained 293 bp of 5’ UTR, 479 bp of 3’ UTR and 2106 bp of open reading frame, which encoded a protein with 701 amino acids (Supplementary Figure S3), and the acox-3b sequence consisted of 1977 bp of open reading frame encoding a protein with 658 amino acids, 271 bp of 5’ UTR and 1197 bp of 3’ UTR (Supplementary Figure S4). ExPASy ProtParam tool analysis showed that the theoretical isoelectric point of prediction ACOX-1a, ACOX-1b, ACOX-3a and ACOX-3b proteins were 75.14, 75.43, 78.32 and 73.04 kDa, respectively. In addition, the molecular masses of prediction ACOX-1a, ACOX-1b, ACOX-3a and ACOX-3b proteins were 7.63, 6.28, 6.60 and 6.67, respectively.
Results of TMHMM 2.0 and SignalP5.0 prediction exhibited that the ACOX-1a, ACOX-1b, ACOX-3a and ACOX-3b proteins did not possess membrane-spanning domains and signal peptides. The three-dimensional prediction models of ACOX-1a, ACOX-1b, ACOX-3a and ACOX-3b proteins have been presented in Figure 1. The secondary structure was predicted by the NPS Server, and the results indicated that the ACOX-1a, ACOX-1b, ACOX-3a and ACOX-3b proteins were mainly comprised of β-corner, random coil, extended strand and α-helix, and the α-helix occupied the largest proportion.
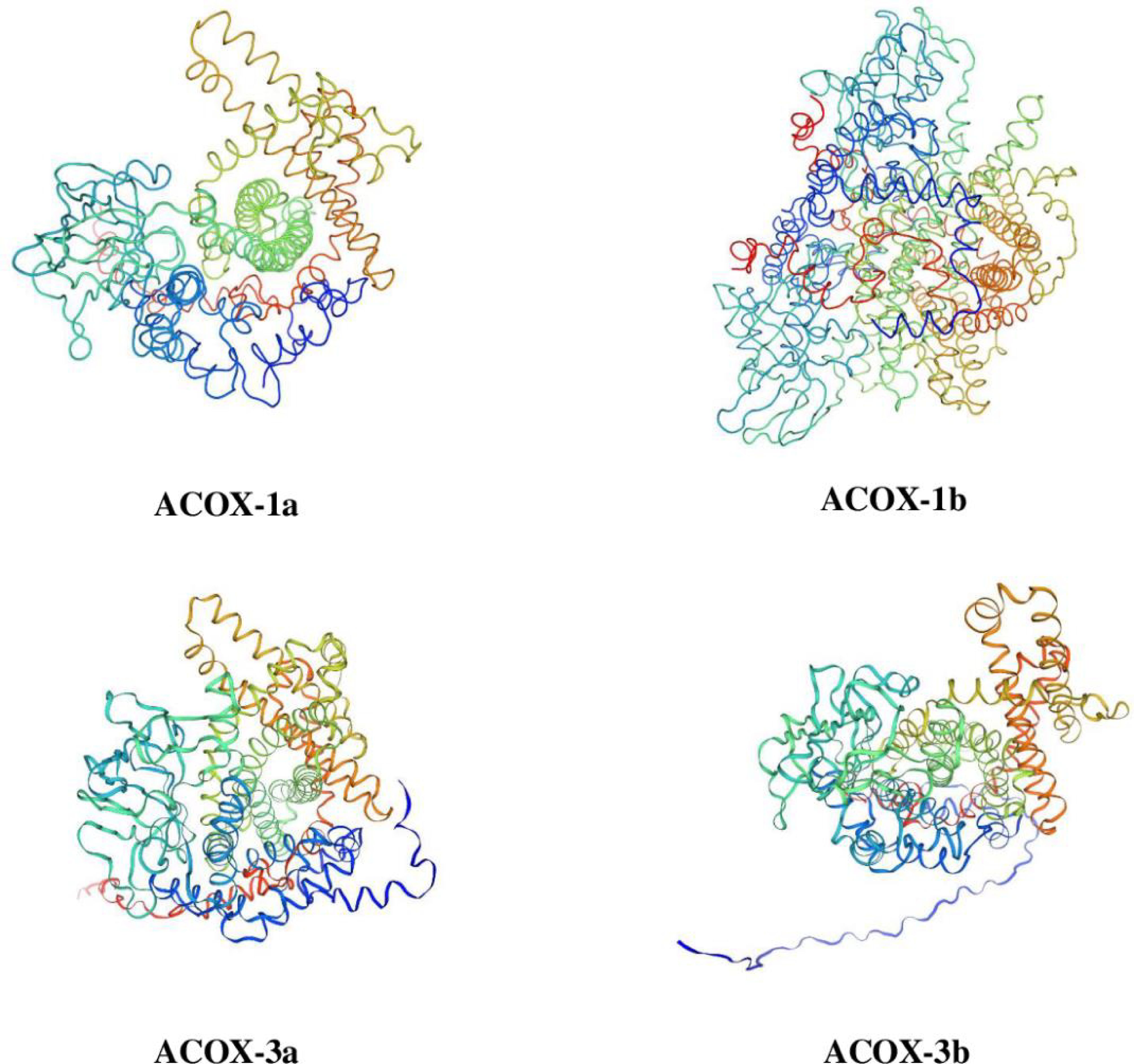
Figure 1 Three-dimensional structure prediction of acyl-coenzyme A oxidase (ACOX) proteins in Scylla paramamosain. ACOX-1a, acyl-coenzyme A oxidase 1a; ACOX-1b, acyl-coenzyme A oxidase 1b; ACOX-3a, acyl-coenzyme A oxidase 3a; ACOX-3b, acyl-coenzyme A oxidase 3b.
3.2 Conserved structural domains
Multiple sequence alignments from mud crab with mammals, teleost fish and crustaceans revealed that the mud crab ACOX proteins existed three major conserved function regions like the FAD-binding motif, peroxisomal targeting signal and fatty acyl CoA oxidase domain (Figures 2, 3). The mud crab ACOX-1a and ACOX-1b possessed typical structures of the FAD-binding motif (CGGHGY) and fatty acyl CoA oxidase domain (KWWPGG), which were highly conserved in different species. Compare with traditional highly conserved motif (SKL), the peroxisomal targeting signal of mud crab ACOX-1a and ACOX-1b has mutated and formed “SNL” and “ASL” variants, respectively. Similarly, the FAD-binding motif of mud crab ACOX-3b and the fatty acyl CoA oxidase domain of mud crab ACOX-3a and ACOX-3b also produced new variants. The FAD-binding motif of mud crab ACOX-3b was “CGGQGY”, and the fatty acyl CoA oxidase domain of mud crab ACOX-3a and ACOX-3b was “KCWAGNL” and “KYWITN”, respectively. In addition, the FAD-binding motif and peroxisomal targeting signal of mud crab ACOX-3a were highly conserved, which were “CGGHGY” and “SKL”, respectively. However, there was no peroxisomal targeting signal found in mud crab ACOX-3b (Figure 3).

Figure 2 Multiple alignments of the acyl-coenzyme A oxidase 1 (ACOX-1) amino acid sequences from Scylla paramamosain with other species. The threshold for similarity shading is set at 50%. Identical residues are shaded black. The similarity of amino acid residues more than 75% and 50% are shaded in pink and cyan, respectively. The fatty acyl CoA oxidase domain, FAD-binding motif and Peroxisomal targeting signal are marked with boxes. The sequences used for comparison are as follows: Homo sapiens (EAW89349), Mus musculus (NP_001258827), Salmo salar (XP_013986887), Danio rerio (AAH83524), Larimichthys crocea (XP_010746625), Penaeus vannamei (XP_027239044), Eriocheir sinensis (XP_050736712), Portunus trituberculatus (XP_045124550), S. paramamosain ACOX-1a (PP708699) and S. paramamosain ACOX-1b (PP708700).

Figure 3 Multiple alignments of the acyl-coenzyme A oxidase 3 (ACOX-3) amino acid sequences from Scylla paramamosain with other species. The threshold for similarity shading is set at 50%. Identical residues are shaded black. The similarity of amino acid residues more than 75% and 50% are shaded in pink and cyan, respectively. The fatty acyl CoA oxidase domain, FAD-binding motif and peroxisomal targeting signal are marked with boxes. The sequences used for comparison are as follows: Homo sapiens (AAH17053), Mus musculus (NP_109646), Salmo salar (ACN10554), Danio rerio (NP_998312), Homarus americanus (XP_042235809), Penaeus japonicas (XP_042885483), Eriocheir sinensis (XP_050713538), Portunus trituberculatus (XP_045124199), S. paramamosain ACOX-3a (PP708701) and S. paramamosain ACOX-3b (PP708702).
3.3 Phylogenetic analysis
The phylogenetic trees of mud crab ACOX-1a and ACOX-1b were given in the Figure 4. The results exhibited that the mud crab ACOX-1a and ACOX-1b grouped together with ACOX-1 from crustacean cluster, and separated from ACOX-1 of vertebrates and molluscs groups. In crustacean group, the mud crab ACOX-1a formed an independent branch with ACOX-1 from Portunus trituberculatus, and then further clustered with ACOX-1 from Eriocheir sinensis and Chionoecetes opilio, producing a bigger branch. The mud crab ACOX-1b and ACOX-1a were located in different branches. The mud crab ACOX-1b grouped most closely with ACOX-1 from Homarus americanus. The branch above was gathered with branch of Penaeus chinensis and Penaeus vannamei ACOX-1, and further clustered with Penaeus monodon and Penaeus japonicas ACOX-1 branch, finally forming a bigger branch.
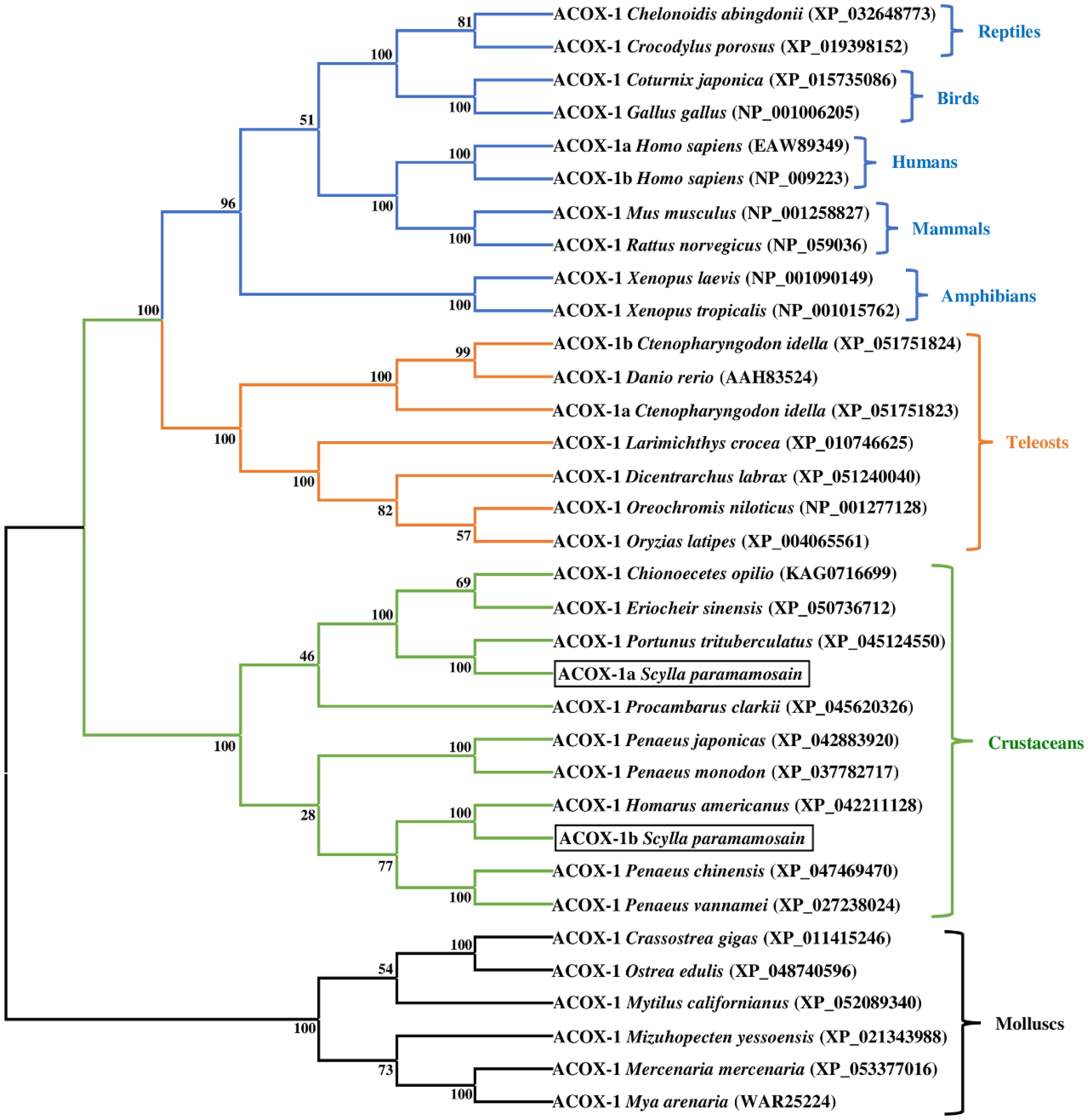
Figure 4 Neighbor-joining phylogenetic tree of representative vertebrate and invertebrate acyl-coenzyme A oxidase 1 (ACOX-1) amino acid sequences. The tree was constructed using the neighbor joining method with MEGA 7.0. The horizontal branch length is proportional to amino acid substitution rate per site. Numbers represent the frequencies with which the tree topology presented was replicated after 1000 bootstrap iterations.
The phylogenetic trees of mud crab ACOX-3a and ACOX-3b were presented in the Figure 5. The results indicated that the mud crab ACOX-3a and ACOX-3b clustered with ACOX-3 from crustacean group. Specifically, the mud crab ACOX-3a, P. trituberculatus ACOX-3 and E. sinensis ACOX-3 were gathered together into a branch, and then further clustered with ACOX-3 from other crustaceans. In addition, the mud crab ACOX-3a showed a closer genetic relationship with the P. trituberculatus ACOX-3 than the E. sinensis ACOX-3. Interestingly, the mud crab ACOX-3b gathered with ACOX-3 from C. opilio and P. vannamei, forming a relatively independent branch, which was far from the branch of the mud crab ACOX-3a. Compared with C. opilio ACOX-3, the mud crab ACOX-3b clustered in a smaller branch with P. vannamei ACOX-3, showing a more intimate genetic relationship.
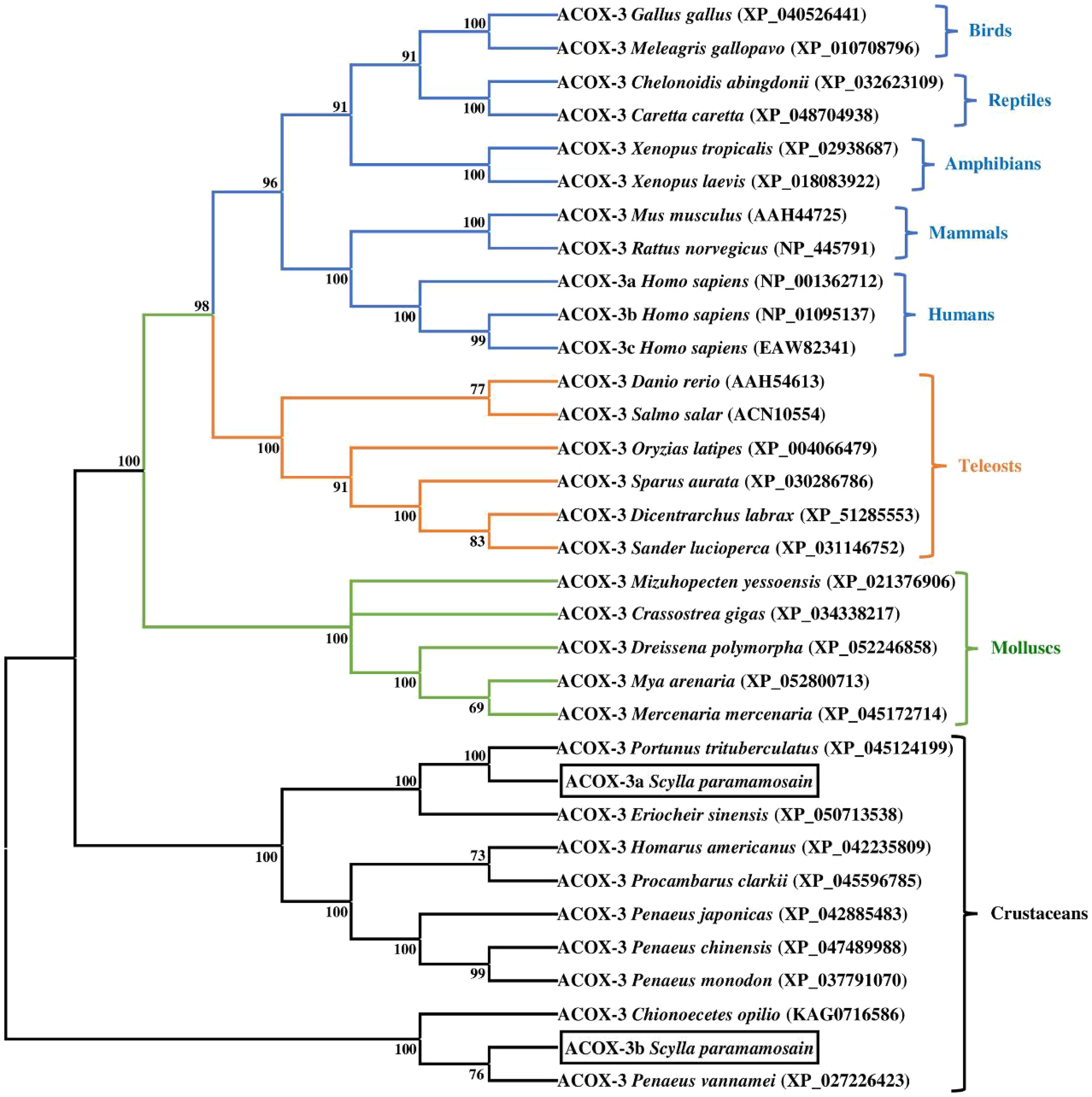
Figure 5 Neighbor-joining phylogenetic tree of representative vertebrate and invertebrate acyl-coenzyme A oxidase 3 (ACOX-3) amino acid sequences. The tree was constructed using the neighbor joining method with MEGA 7.0. The horizontal branch length is proportional to amino acid substitution rate per site. Numbers represent the frequencies with which the tree topology presented was replicated after 1000 bootstrap iterations.
3.4 Tissue distribution
The transcriptional levels of acox-1a, acox-1b, acox-3a and acox-3b in eight selected tissues were shown in Figure 6. The expression levels of acox-1a and acox-3a in hepatopancreas exhibited the highest value among all examined tissues, and were significantly higher than the cranial ganglia, intestine, thoracic ganglia, gill, heart, eyestalk and muscle (P < 0.05). In addition, there were no significant differences in acox-1a and acox-3b expressions of the cranial ganglia, intestine, thoracic ganglia, gill, heart, eyestalk and muscle (P > 0.05). Similarly, the present study also found that the highest number of acox-3b transcripts was detected in hepatopancreas among all tissues (P < 0.05), followed by the muscle and cranial ganglia. By comparison, the highest transcriptional level of acox-1b was the muscle (P < 0.05), and the acox-1b was also highly expressed in the hepatopancreas and eyestalk when compared with the cranial ganglia, heart, thoracic ganglia, intestine and gill (P < 0.05).
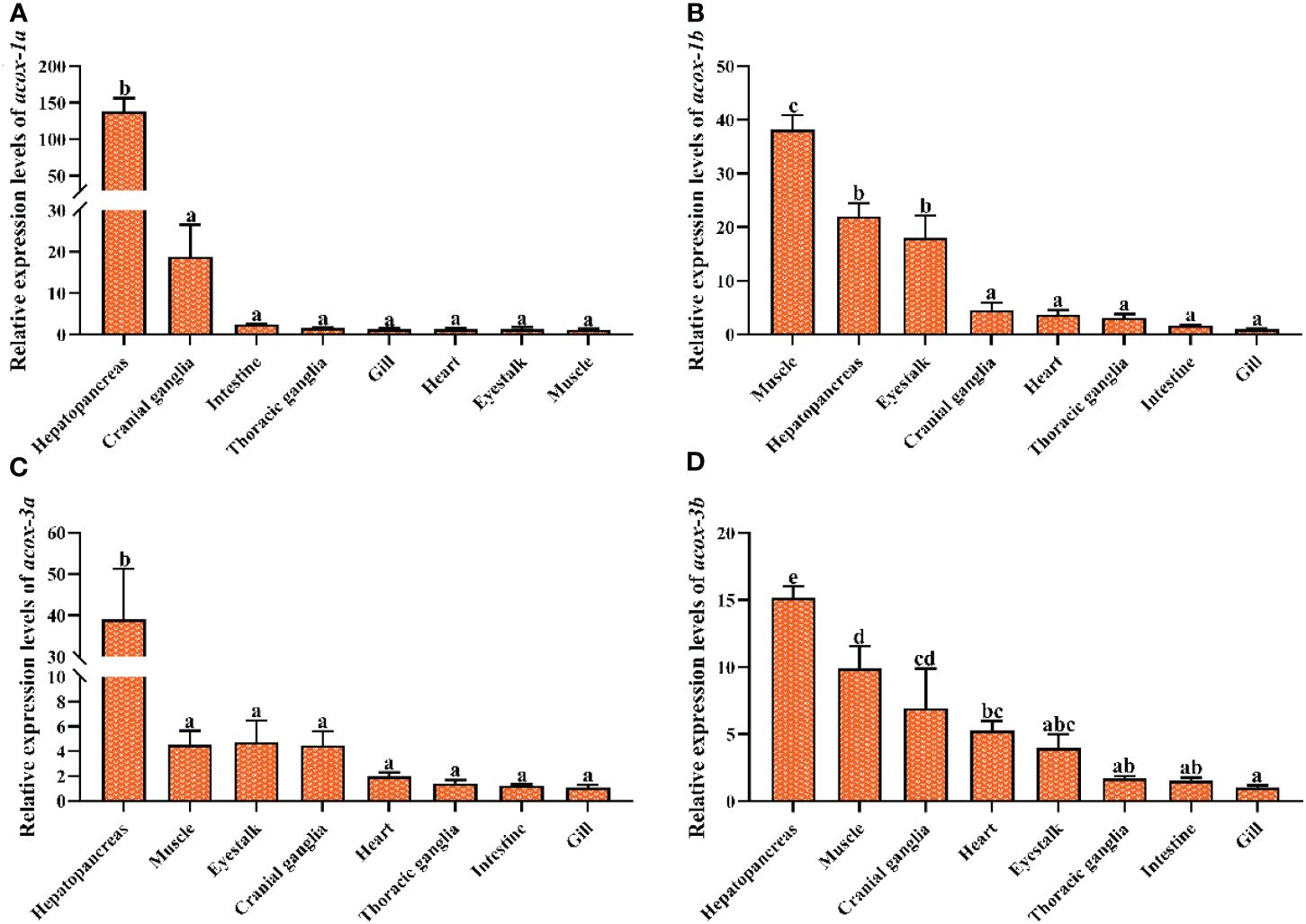
Figure 6 Relative expression levels of acyl-coenzyme A oxidase (acox) in different tissues of Scylla paramamosain. Vertical bars represented mean ± standard error of the mean (n = 6) for each tissue. acox-1a, acyl-coenzyme A oxidase 1a; acox-1b, acyl-coenzyme A oxidase 1b; acox-3a, acyl-coenzyme A oxidase 3a; acox-3b, acyl-coenzyme A oxidase 3b. (A): acox-1a expression in different tissues; (B): acox-1b expression in different tissues; (C): acox-3a expression in different tissues; (D): acox-3b expression in different tissues. Letters show significant differences (P < 0.05) among tissues as determined by one-way ANOVA followed by Duncan’s multiple comparison test.
3.5 Expression levels of acox-1a, acox-1b, acox-3a and acox-3b in reaction to fasting
The effect of fasting stress on acox-1a, acox-1b, acox-3a and acox-3b expression levels was presented in Figure 7. Compare with the feeding groups, the transcriptional levels of acox-1a, acox-3a and acox-3b in hepatopancreas were markedly up-regulated (P < 0.05), and the acox-3a exhibited a fearfully significant difference (P < 0.01). There was no significant difference in acox-1b expression of hepatopancreas (P > 0.05). In addition, the expression level of acox-3a in muscle was markedly increased in fasting group when compared with the feeding group (P < 0.05). No significant differences in transcriptional levels of acox-1a, acox-1b and acox-3b between the fasting group and feeding group (P > 0.05).
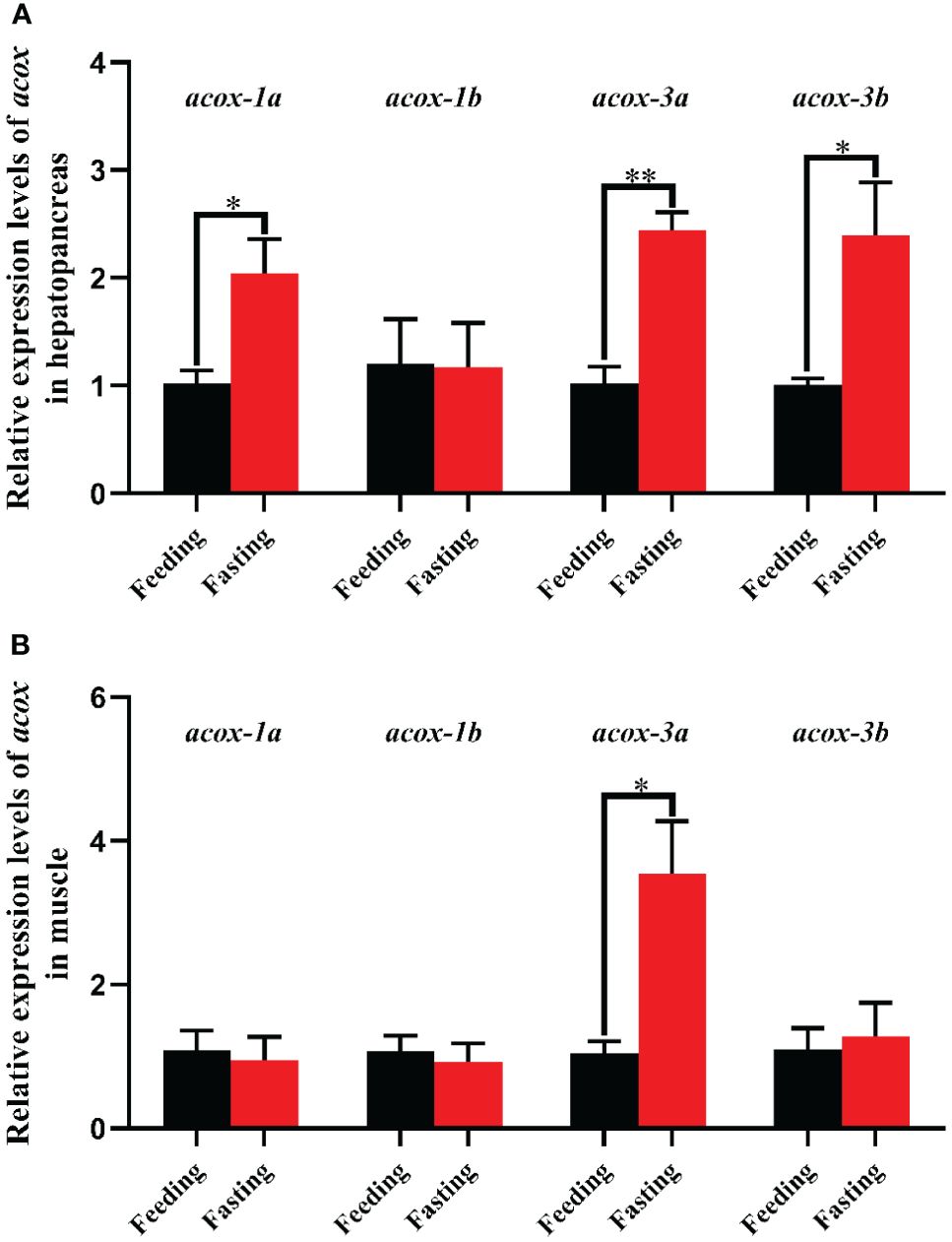
Figure 7 Relative expression levels of acyl-coenzyme A oxidase (acox) in hepatopancreas (A) and muscle (B) of Scylla paramamosain in response to fasting stress. Asterisks indicated significant differences between feeding group and fasting group (*P < 0.05 and **P < 0.01). acox-1a, acyl-coenzyme A oxidase 1a; acox-1b, acyl-coenzyme A oxidase 1b; acox-3a, acyl-coenzyme A oxidase 3a; acox-3b, acyl-coenzyme A oxidase 3b.
3.6 Expression levels of antioxidant genes in reaction to fasting
The effect of fasting stress on the expression levels of antioxidant genes was shown in Figure 8. Nrf2 is a key transcription factor, which can regulate cellular oxidative stress response. The results of present study indicated that the fasting group exhibited markedly higher transcriptional levels of nrf2 in hepatopancreas and muscle than the feeding group (P < 0.05). Meanwhile, the downstream target genes of nrf2 including cat, gpx and gst in hepatopancreas and muscle were also significantly up-regulated in fasting group when compared with the feeding group (P < 0.05), and the gpx showing highly significant difference (P < 0.01). In addition, compared with feeding group, the transcriptional level of cat in hepatopancreas was increased by fourteen times in fasting group, while the muscle only added two times.
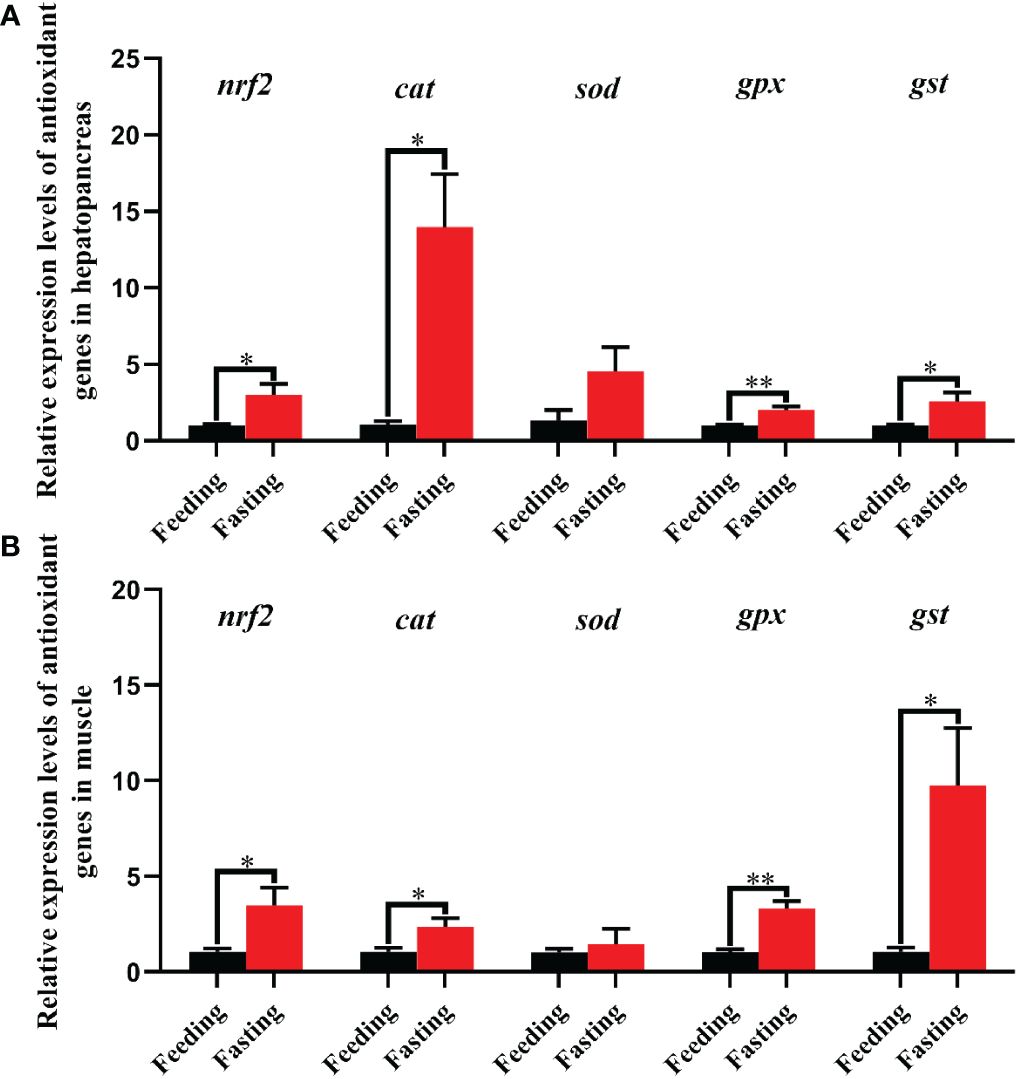
Figure 8 Relative expression levels of antioxidant genes in hepatopancreas (A) and muscle (B) of Scylla paramamosain in response to fasting stress. nrf2, nuclear factor erythroid 2-related factor 2; cat, catalase; sod, superoxide dismutase; gpx, glutathione peroxidase; gst, glutathione S-transferase. Asterisks indicated significant differences between feeding group and fasting group (*P < 0.05 and **P < 0.01).
4 Discussion
Mitochondria and peroxisomes are the main sites of β-oxidation of fatty acids in aquatic animals. However, the functions about mitochondria and peroxisomes of β-oxidation in lipid metabolism were remain unclear in aquatic animals, especially in crustaceans. Previously, Lin et al. (2024) have preliminarily investigated the β-oxidation of fatty acids in mitochondria including identification and characterization of three rate-limiting enzyme genes (cpt-2, cpt-1b and cpt-1a) and their regulatory mechanisms in mud crab. In the present study, we further explored the β-oxidation of fatty acids in peroxisomes of mud crab. Four full-length cDNA sequences of acoxs involved in peroxisomal β-oxidation were successfully obtained from the mud crab in this study, which were identified as acox-1a, acox-1b, acox-3a and acox-3b by sequence analysis. In mammals, there were three genes named acox-1, acox-2 and acox-3 found in peroxisomes, while the teleost fish was short of acox-2 and only contained acox-1 and acox-3 like O. niloticus (He et al., 2014), S. trutta (Madureira et al., 2016), C. idella (Sun et al., 2020), Larimichthys crocea (Kong et al., 2022). Consistently, the present study exhibited that there was no acox-2 observed in mud crab, which may indicate that the acox-2 gene also lacked in crustaceans. In addition, Li et al. (2022) reported that the acox-2 gene was not absent in bivalves (molluscs), and both C. farreri and P. yessoensis possessed the four acox-1, a acox-2 and a acox-3. The ACOX-1a, ACOX-1b, ACOX-3a and ACOX-3b proteins of mud crab contained 666, 673, 701 and 658 amino acids, respectively and shared high homology with crustaceans, teleost fish and mammals, suggesting that the ACOX structures were relatively conservative among different species. Past studies have exhibited that the ACOXs contained three primary conserved regions including the peroxisomal targeting signal, FAD-binding motif and fatty acyl CoA oxidase domain (Kunau et al., 1995; Hooks et al., 1999). Likewise, the mud crab ACOX-1a, ACOX-1b and ACOX-3a also possessed all the conserved structural domains above, which was in accordance with the study results from teleost fish and molluscs like L. crocea, C. idella, S. trutta, C. farreri and P. yessoensis (Madureira et al., 2016; Sun et al., 2020; Kong et al., 2022; Li et al., 2022). Besides, the mud crab ACOX-3b lacked the structure of peroxisomal targeting signal and only included the FAD-binding motif and fatty acyl CoA oxidase domain. The possible reason is that the peroxisomal targeting signal lost in the process of genome replication. The present study also found that the peroxisomal targeting signal of ACOX-1a and ACOX-1b, fatty acyl CoA oxidase domain of ACOX-3a and ACOX-3b as well as FAD-binding motif of ACOX-3b formed new variants, and the reason may be in connection with the mutation of key amino acids in genome replication. These results were in line with the study of Li et al. (2022) who reported that the conserved structural domains of ACOX-1, ACOX-2 and ACOX-3 in C. farreri and P. yessoensis contained multiple variants. Furthermore, the results of phylogenetic tree indicated that the ACOX-1 and ACOX-3 of mud crab clustered together with their homologous orthologues from crustaceans. Collectively, the results of the homology, conserved domains and genetic relationship above supported the obtained gene sequences pertained to the acox family.
Exploring tissue distribution could contribute to understanding the physiological roles of target genes. Studies in mammals have indicated that the acox-1 was abundantly expressed in liver followed by the kidney, adipose tissue and other tissues (Nöhammer et al., 2001; Vluggens et al., 2010). As for aquatic animals, the acox expression in different tissues exhibited multiple patterns because of species specificity. Kong et al. (2022) reported that the intestine and liver were the main expressed tissues of acox-1 in L. crocea, and similar results were also observed in Danio rerio (Morais et al., 2007) and Takifugu rubripes (Wang et al., 2022). Studies in S. maximus and T. rubripes also found that the acox-3 expression levels were primarily detected in liver and intestine (Wang et al., 2022). In O. niloticus and C. idellus, the acox-1 was principally distributed in kidney and liver tissues (He et al., 2014; Sun et al., 2020), and in S. maximus, except for liver, the acox-1 was also highly expressed in brain (Wang et al., 2022). Madureira et al. (2016) indicated that the acox-1 and acox-3 were most abundant in ovary followed by liver, stomach, heart and brain in S. trutta. Besides, a study from two molluscs, C. farreri and P. yessoensis, found that six acox genes including four acox-1, a acox-2 and a acox-3 were predominantly expressed in kidneys and digestive glands. From the results above, it is not difficult to find that the liver is an important site for acox gene expression despite different species existed differences. The hepatopancreas in crustaceans was analogous to liver in vertebrates, which was closely associated with lipid metabolism (Lin et al., 2024). Consistently, the present study found that the acox-1a, acox-3a and acox-3b were mainly distributed in hepatopancreas when compared with other detected tissues. Additionally, the highest number of acox-1b transcripts was observed in muscle among all tissues, and meanwhile the hepatopancreas also showed high expression level. These results may suggest that the mud crab acoxs primarily played a crucial role in peroxisomal β-oxidation of hepatopancreas and muscle, especially in hepatopancreas.
As rate-limiting enzymes of peroxisomal β-oxidation, increasing studies in mammals have suggested that the ACOXs played indispensable roles in maintaining energy homeostasis, regulating inflammatory response as well as adapting to environmental and metabolic stresses (like nutrient deprivation, oxidative stress, hypoxia and cold exposure) (Houten and Wanders, 2010; Ding et al., 2021; He et al., 2021). However, the studies on the functions of ACOXs were still at the start stage in aquatic animals, especially in crustaceans. Wang et al. (2022) reported that both high-lipid diet and fasting could markedly increase the acox transcriptional levels in liver of T. rubripe. Consistently, the acox-1 exhibited a significant up-regulation in C. idella adipocytes at 12 hours after fasting (Sun et al., 2020), and the acox-3 expression was observably up-regulated in liver of S. maximus after 10 days fasting (Wang et al., 2022). In addition, He et al. (2014) observed that refeeding significantly suppressed the expression of acox-1 in liver and kidney after starvation. Similarly, the results of present study found that the expression levels of acox-1a, acox-3a and acox-3b in hepatopancreas and acox-3a in muscle were markedly up-regulated after fasting, suggesting that the acoxs also possess important function in energy homeostasis for mud crab during fasting. Besides, previously, we have investigated the mitochondrial β-oxidation in response to fasting in same species, which observed that the cpt-1a, cpt-1b and cpt-2 (rate-limiting enzymes of mitochondrial β-oxidation) in hepatopancreas and muscle were significantly up-regulated after fasting and their fold changes were more obvious than the acoxs (Lin et al., 2024). These results showed that the β-oxidation of mitochondria and peroxisomes existed a mutual collaboration in response to starvation stress for maintaining energy balance, and the mitochondrial β oxidation might dominate during starvation stress. As an important transcription factor, nrf2 participates in regulating cellular oxidative stress responses, and activated nrf2 can induce the expression of a series of downstream antioxidant genes like cat, sod, gpx and gst (Kong et al., 2022). Studies have demonstrated that ACOX-mediated peroxisomal β-oxidation can induce the ROS production, which was an important source of intracellular ROS (Ding et al., 2021; He et al., 2021; Kong et al., 2022). For example, Kong et al. (2022) indicated that palmitate caused an increase ROS generation though mediating the acox-1 expression in L. crocea, and meanwhile up-regulated the expression of antioxidant genes (nrf2, sod and gpx) for resisting oxidative stress. The results of present study found that expression levels of nrf2 and its target genes (cat, gpx and gst) in hepatopancreas and muscle were markedly up-regulated in fasting group compared with the feeding group. The possible explanation is that fasting resulted in ROS generation via the ACOX-mediated peroxisomal β-oxidation, and then further induced expression of antioxidant genes to eliminate excessive ROS. Furthermore, the present study also found that the increased multiple of cat relative expression level in hepatopancreas was more obvious than other antioxidant genes, which may be in connection with H2O2 as a main byproduct of ACOX-mediated peroxisomal β-oxidation. Similar result was also observed in L. crocea reported by Kong et al., 2022. These results above suggested that ACOX-mediated peroxisomal β-oxidation participated in maintaining energy homeostasis after fasting, and meanwhile this process caused the ROS generation and further induced expression of antioxidant genes for relieving oxidative stress.
5 Conclusion
In the present study, four completed cDNA sequences of acoxs (acox-1a, acox-1b, acox-3a and acox-3b) were cloned from the mud crab, which was the first time to report the key genes involved in regulating peroxisomal β-oxidation in crustaceans. Subsequently, the present study analyzed the homology, conserved domains, evolutionary characteristics of ACOX-1a, ACOX-1b, ACOX-3a and ACOX-3b as well as acox-1a, acox-1b, acox-3a and acox-3b expression levels in eight selected tissues. Lastly, we further explored the roles of ACOX-mediated peroxisomal β-oxidation and oxidation states under the starvation stress. These results would contribute to enhancing the cognition of acoxs evolutionary characteristics and ACOX-mediated peroxisomal β-oxidation in crustaceans.
Data availability statement
The data presented in the study are deposited in the the National Center for Biotechnology Information (NCBI) repository, accession numbers: PP708699, PP708700, PP708701 and PP708702. The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Ethics Committee of Ningde Normal University (NDNU-LL-202308). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LZ: Conceptualization, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing. LJ: Data curation, Formal analysis, Methodology, Validation, Writing – original draft. LH: Formal analysis, Methodology, Resources, Supervision, Validation, Visualization, Writing – original draft. HC: Data curation, Formal analysis, Methodology, Supervision, Validation, Writing – original draft. ZM: Data curation, Formal analysis, Supervision, Writing – original draft. HQ: Conceptualization, Data curation, Investigation, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the Scientific Research Foundation of Ningde Normal University (Grant number 2022ZDK01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1442810/full#supplementary-material
References
Adeva-Andany M. M., Carneiro-Freire N., Seco-Filgueira M., Fernández-Fernández C., Mouriño-Bayolo D. (2019). Mitochondrial β-oxidation of saturated fatty acids in humans. Mitochondrion. 46, 73–90. doi: 10.1016/j.mito.2018.02.009
Boveris A., Oshino N., Chance B. (1972). The cellular production of hydrogen peroxide. Biochem. J. 128, 617–630. doi: 10.1042/bj1280617
Chen X. F., Tian M. X., Sun R. Q., Zhang M. L., Zhou L. S., Jin L., et al. (2018). SIRT 5 inhibits peroxisomal ACOX 1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. 19, e45124. doi: 10.15252/embr.201745124
China Fishery Statistical Yearbook (2023). China fishery statistical yearbook Vol. 28 (Beijing, China: China Agriculture Press).
Chrousos G. P. (2009). Stress and disorders of the stress system. Nat. Rev. Endocrinol. 5, 374–381. doi: 10.1038/nrendo.2009.106
Chu B. B., Liao Y. C., Qi W., Xie C., Du X., Wang J., et al. (2015). Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 161, 291–306. doi: 10.1016/j.cell.2015.02.019
Cipolla C. M., Lodhi I. J. (2017). Peroxisomal dysfunction in age-related diseases. Trends Endocrin. Met. 28, 297–308. doi: 10.1016/j.tem.2016.12.003
Ding L., Sun W., Balaz M., He A., Klug M., Wieland S., et al. (2021). Peroxisomal β-oxidation acts as a sensor for intracellular fatty acids and regulates lipolysis. Nat. Metab. 3, 1648–1661. doi: 10.1038/s42255-021-00489-2
Dubreuil M. M., Morgens D. W., Okumoto K., Honsho M., Contrepois K., Lee-McMullen B., et al. (2020). Systematic Identification of regulators of oxidative stress reveals non-canonical roles for peroxisomal import and the pentose phosphate pathway. Cell Rep. 30, 1417–1433. doi: 10.1016/j.celrep.2020.01.013
He A., Dean J. M., Lodhi I. J. (2021). Peroxisomes as cellular adaptors to metabolic and environmental stress. Trends Cell Biol. 31, 656–670. doi: 10.1016/j.tcb.2021.02.005
He A., Dean J. M., Lu D., Chen Y., Lodhi I. J. (2020). Hepatic peroxisomal β-oxidation suppresses lipophagy via RPTOR acetylation and MTOR activation. Autophagy. 16, 1727–1728. doi: 10.1080/15548627.2020.1797288
He A. Y., Liu C. Z., Chen L. Q., Ning L. J., Zhang M. L., Li E. C., et al. (2014). Identification, characterization and nutritional regulation of two isoforms of acyl-coenzyme A oxidase 1 gene in Nile tilapia (Oreochromis niloticus). Gene 545, 30–35. doi: 10.1016/j.gene.2014.05.010
Hooks M. A., Kellas F., Graham I. A. (1999). Long-chain acyl-CoA oxidases of Arabidopsis. Plant J. 20, 1–13. doi: 10.1046/j.1365-313X.1999.00559.x
Houten S. M., Wanders R. J. A. (2010). A general introduction to the biochemistry of mitochondrial fatty acid β-oxidation. J. Inherit. Metab. Dis. 33, 469–477. doi: 10.1007/s10545-010-9061-2
Huang R., Zhang C., Xiang Z., Lin T., Ling J., Hu H. (2024). Role of mitochondria in renal ischemia-reperfusion injury. FEBS J. doi: 10.1111/febs.17130
Huybrechts S. J., Van Veldhoven P. P., Brees C., Mannaerts G. P., Los G. V., Fransen M. (2009). Peroxisome dynamics in cultured mammalian cells. Traffic. 10, 1722–1733. doi: 10.1111/j.1600-0854.2009.00970.x
Kong J., Ji Y., Jeon Y. G., Han J. S., Han K. H., Lee J. H., et al. (2020). Spatiotemporal contact between peroxisomes and lipid droplets regulates fasting-induced lipolysis via PEX5. Nat. Commun. 11, 578. doi: 10.1038/s41467-019-14176-0
Kong A., Xu D., Hao T., Liu Q., Zhan R., Mai K., et al. (2022). Role of acyl-coenzyme A oxidase 1 (ACOX1) on palmitate-induced inflammation and ROS production of macrophages in large yellow croaker (Larimichthys crocea). Dev. Comp. Immunol. 136, 104501. doi: 10.1016/j.dci.2022.104501
Kunau W. H., Dommes V., Schulz H. (1995). β-Oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog. Lipid Res. 34, 267–342. doi: 10.1016/0163-7827(95)00011-9
Li M., Wang Y., Tang Z., Wang H., Hu J., Bao Z., et al. (2022). Expression plasticity of peroxisomal acyl-coenzyme A oxidase genes implies their involvement in redox regulation in scallops exposed to PST-producing alexandrium. Mar. Drugs 20, 472. doi: 10.3390/md20080472
Lin Z., Huang C., Zhuo Z., Xie J., Lan H., Hu B., et al. (2024). Molecular cloning and characterization of three carnitine palmitoyltransferase (cpt) isoforms from mud crab (Scylla paramamosain) and their roles in respond to fasting and ambient salinity stress. Front. Mar. Sci. 11. doi: 10.3389/fmars.2024.1381263
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Lodhi I. J., Semenkovich C. F. (2014). Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19, 380–392. doi: 10.1016/j.cmet.2014.01.002
Madureira T. V., Castro L. F. C., Rocha E. (2016). Acyl-coenzyme A oxidases 1 and 3 in brown trout (Salmo trutta f. fario): Can peroxisomal fatty acid β-oxidation be regulated by estrogen signaling? Fish Physiol. Biochem. 42, 389–401. doi: 10.1007/s10695-015-0146-6
Matsushita N., Yonashiro R., Ogata Y., Sugiura A., Nagashima S., Fukuda T., et al. (2011). Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells 16, 190–202. doi: 10.1111/gtc.2011.16.issue-2
Monzel A. S., Enríquez J. A., Picard M. (2023). Multifaceted mitochondria: moving mitochondrial science beyond function and dysfunction. Nat. Metab. 5, 546–562. doi: 10.1038/s42255-023-00783-1
Morais S., Knoll-Gellida A., André M., Barthe C., Babin P. J. (2007). Conserved expression of alternative splicing variants of peroxisomalacyl-CoA oxidase 1 in vertebrates and developmental and nutritional regulation in fish. Physiol. Genomics 28, 239–252. doi: 10.1152/physiolgenomics.00136.2006
Nöhammer C., El-Shabrawi Y., Schauer S., Hiden M., Berger J., Forss-Petter S., et al. (2001). cDNA cloning and analysis of tissue-specific expression of mouse peroxisomal straight-chain acyl-CoA oxidase. FEBS J. 267, 1254–1260. doi: 10.1046/j.1432-1327.2000.01128.x
Oaxaca-Castillo D., Andreoletti P., Vluggens A., Yu S., van Veldhoven P. P., Reddy J. K., et al. (2007). Biochemical characterization of two functional human liver acyl-CoA oxidase isoforms 1a and 1b encoded by a single gene. Biochem. Bioph. Res. Co. 360, 314–319. doi: 10.1016/j.bbrc.2007.06.059
Panov A., Mayorov V. I., Dikalov S. (2022). Metabolic syndrome and β-oxidation of long-chain fatty acids in the brain, heart, and kidney mitochondria. Int. J. Mol. Sci. 23, 4047. doi: 10.3390/ijms23074047
Reddy J. K., Hashimoto T. (2001). Peroxisomal β-oxidation and peroxisome proliferator-activated receptor α: an adaptive metabolic system. Annu. Rev. Nutr. 21, 193–230. doi: 10.1146/annurev.nutr.21.1.193
Sun J., Li H., Luo X., Lu R., Ji H. (2020). Identification and characterization of two isoforms of acyl-coenzyme A oxidase 1 gene and their expression in fasting-induced grass carp Ctenopharyngodon idella adipocyte lipolysis. Fish Physiol. Biochem. 46, 1645–1652. doi: 10.1007/s10695-020-00816-6
Van Veldhoven P. P. (2010). Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J. Lipid Res. 51, 2863–2895. doi: 10.1194/jlr.R005959
Verma M., Lizama B. N., Chu C. T. (2022). Excitotoxicity, calcium and mitochondria: a triad in synaptic neurodegeneration. Transl. Neurodegener. 11, 3. doi: 10.1186/s40035-021-00278-7
Vluggens A., Andreoletti P., Viswakarma N., Jia Y., Matsumoto K., Kulik W., et al. (2010). Functional significance of the two ACOX1 isoforms and their crosstalks with PPARα and RXRα. Lab. Invest. 90, 696–708. doi: 10.1038/labinvest.2010.46
Wang D., Ma Q., Liao Z., Bi Q., Wei Y., Liang M., et al. (2022). Tissue distribution and nutritional regulation of peroxisomal fatty acid β-oxidation genes: A preliminary study in two marine teleosts, turbot (Scophthalmus maximus) and tiger puffer (Takifugu rubripes). Aquac. Res. 53, 5434–5443. doi: 10.1111/are.16022
Keywords: acyl-coenzyme A oxidase, antioxidant capacity, peroxisomal β-oxidation, starvation stress, Scylla paramamosain
Citation: Zhideng L, Jinjie L, Huangbin L, Chaoyang H, Mingyao Z and Qincheng H (2024) Molecular cloning, tissue distribution and nutritional regulation of four acyl-coenzyme A oxidase (acox) isoforms in Scylla paramamosain. Front. Mar. Sci. 11:1442810. doi: 10.3389/fmars.2024.1442810
Received: 03 June 2024; Accepted: 15 July 2024;
Published: 01 August 2024.
Edited by:
Songlin Li, Shanghai Ocean University, ChinaCopyright © 2024 Zhideng, Jinjie, Huangbin, Chaoyang, Mingyao and Qincheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huang Qincheng, cWluY2hlbmdoMDgxNEAxNjMuY29t
 Lin Zhideng
Lin Zhideng Lan Jinjie1
Lan Jinjie1