- 1Northwest Fisheries Science Center, National Oceanic & Atmospheric Administration (NOAA) Fisheries, Seattle, WA, United States
- 2Biología Departamento Universitario, Universidad Autónoma de Madrid – Unidad de Genética, Madrid, Spain
- 3Ocean Ecology and Biogeochemistry, College of Earth, Ocean, and Atmospheric Sciences, Oregon State University, Corvallis, OR, United States
- 4Pacific Marine Environmental Laboratory, National Oceanic & Atmospheric Administration (NOAA), Seattle, WA, United States
A survey of marine pelagic coastal microbial communities was conducted over a large geographic latitude range, from Cape Mendocino in northern California USA to Queen Charlotte Sound in British Columbia Canada, during the spring to summer transition. DNA metabarcoding and flow cytometry were used to characterize microbial communities. Physical and chemical oceanography indicated moderate conditions during the survey with no widespread upwelling, marine heat wave, or other extreme conditions. However, four locations displayed features approaching acidified conditions: Heceta Head, Newport, Copalis Beach, and Cape Flattery. Although bacterial and archaeal communities at the Juan de Fuca canyon and northward had high similarity, those south of the Juan de Fuca canyon were well differentiated from each other. In contrast, eukaryotic microbial communities exhibited stronger geographic differentiation than bacterial and archaeal communities across the extent of the survey. Seawater parameters that were best predictors of bacterial and archaeal community structure were temperature, pH, and dissolved inorganic nutrients (nitrate, phosphate, silicate), while those that were best predictors of eukaryotic microbial community structure were salinity, dissolved oxygen, total alkalinity, and dissolved inorganic nutrients (nitrite, silicate). Although five bacterial and archaeal indicators for potentially corrosive waters were identified (Colwellia, Nitrosopumilus, Nitrosopelagicus, Sup05 cluster, Sva0996 marine group), no eukaryotic microbial indicators were found. Potentially pathogenic taxa detected in the survey included four disease-causing bacteria for mammals, finfish, and/or shellfish (Coxiella, Flavobacterium, Francisella, Tenacibaculum), sixteen genera of microalgae capable of producing biotoxins, and fifteen parasitic species. This study demonstrates the value of coordinating microbial sampling and analysis with broad-scale oceanographic surveys to generate insights into community structures of these important pelagic trophic levels.
Introduction
The eastern North Pacific Ocean has a dynamic marine ecosystem along the west coast of the United States and Canada. The major ocean currents flow parallel to the coastline, with the Alaska Current flowing northward and the California Current flowing southward from southern British Columbia in Canada (Thomson, 1981; Hickey and Banas, 2003). Each major current generates multiple smaller features including described processes that cause nearshore upwelling as well as other changes in regional conditions (Hickey, 1989; Hickey and Royer, 2001; Hickey and Banas, 2003). Both the Alaska and California Currents are characterized by high productivity and fisheries economic values, and their biological resources are subject to long-term monitoring and forecasting efforts (e.g., Thompson et al., 2022). As one of the major eastern boundary upwelling systems in the world, the California Current system also experiences the phenomenon of ocean acidification (OA) resulting from elevated dissolved inorganic carbon (DIC) and lowered pH (Feely et al., 2016) which can maintain spatial persistence (Chan et al., 2017) and affect biogenic minerals such as aragonite with anticipated consequences for exposed organisms (e.g., Kroeker et al., 2013; Waldbusser and Salisbury, 2014; Dutkiewicz et al., 2015; Mostofa et al., 2016).
In marine systems, the pelagic food web is heavily dependent upon lower trophic levels for producing fixed carbon and nitrogen and for remineralization of dissolved nutrients (Azam et al., 1983; Calbet and Landry, 2004). These trophic levels, which include phytoplankton, bacteria, archaea, protists, and other eukaryotic microbes, can undergo changes in community structures in response to physical and chemical features of seawater with a wide temporal range, from hours to seasons (reviewed in Fuhrman et al., 2015). For example, Table 1 displays a simplified summary of known metabolic and ecological roles associated with several high abundance groups of microbes.
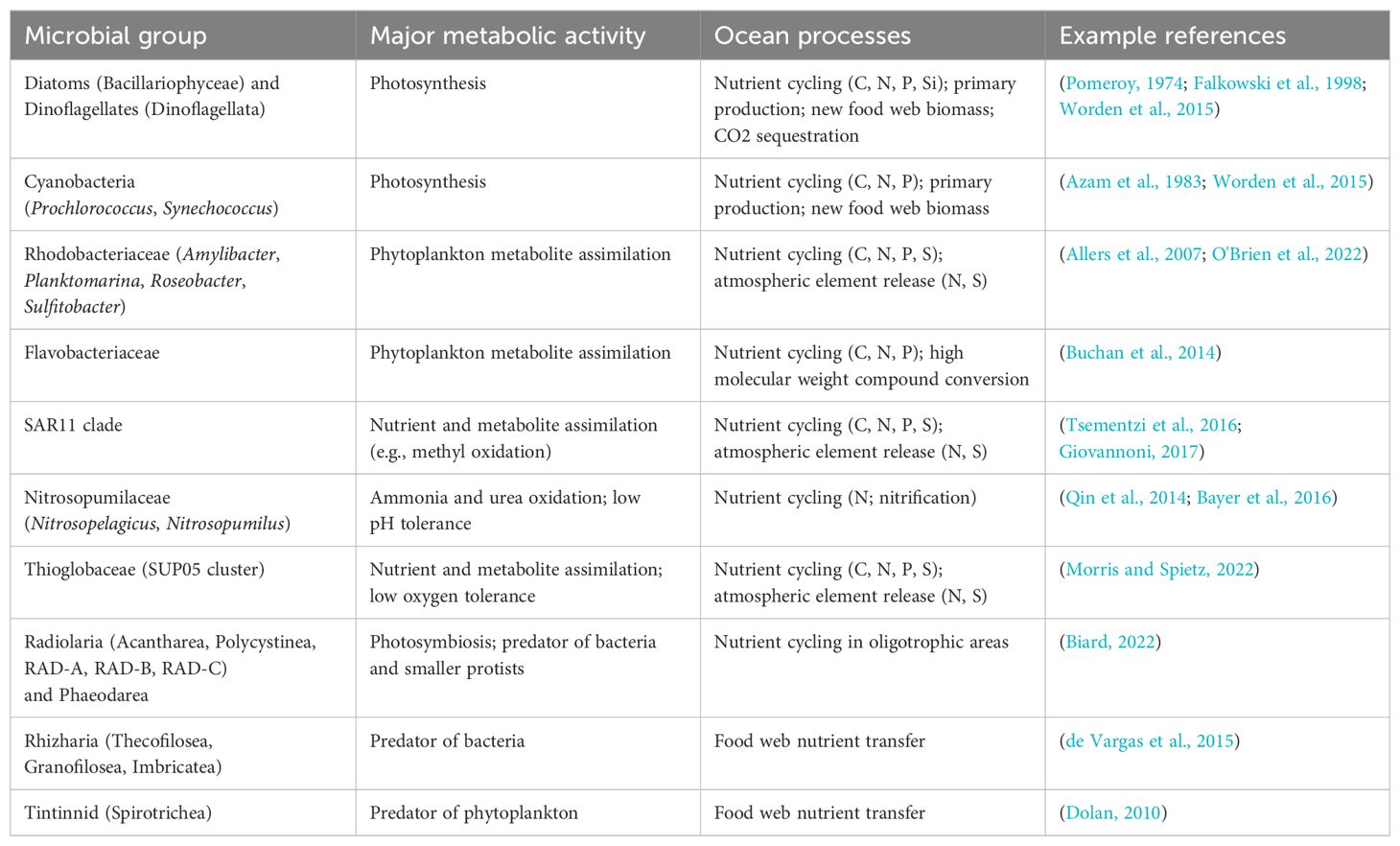
Table 1. Examples of several high abundance microbial groups with known metabolic and ecological roles.
The reliance of marine food webs on lower trophic dynamics is well acknowledged, but direct assessments of microorganisms are not usually conducted in surveys. Instead, easily measured proxies such as chlorophyll α concentrations are typically used to represent a lower trophic tier such as primary producers. However, these proxies provide no information about the underlying microbial structures, which limits both the understanding of biological processes and predicting changes associated with ocean conditions. The advent of metabarcoding technologies and highly curated sequence databases allows taxonomic identifications and descriptions of microbial community structures. When combined with matched physical and chemical seawater parameters, it is feasible to begin building better knowledge of these lower trophic levels.
This study provides a set of observations on microbial communities from a geographically broad survey along the Pacific Northwest nearshore coast, extending from the Queen Charlotte Sound of Canada to Cape Mendocino in northern California of the U.S. It leveraged the accessibility of water samples collected for ocean chemistry assessment conducted by NOAA’s Pacific Marine Environmental Laboratory (PMEL), and co-collection of biological and chemical sampling permitted direct correspondence of results. These community characterizations and evaluations provide a glimpse of the microbial communities that are present in the nearshore coastal areas and can serve as a basis for further comparisons associated with changing ocean conditions.
Methods and materials
Site descriptions and sample collections
The West Coast Ocean Acidification (WCOA) cruise is a periodic assessment of ocean chemistry led and conducted by NOAA’s Pacific Marine Environmental Laboratory (PMEL), and information about the 2016 WCOA cruise can be obtained from PMEL’s Carbon Program website1. Some of the 2016 sites reported here were coincident with sites used in prior cruises (2007, 2011, 2012, 2013) and with California Cooperative Oceanic Fisheries Investigations (CalCOFI) sites, which were originally chosen due to their significant oceanographic features. For this study, a total of 29 nearshore stations along the northwest coast of the U.S. and Canada (Figure 1) were sampled by deployment of a rosette containing a Seabird CTD and an array of Niskin bottles. Water samples were collected from Niskin bottles from vertical profiles during the northern leg of NOAA’s 2016 WCOA cruise. Temperature, salinity, pressure, and dissolved oxygen were measured during profiling with a Sea-Bird 9plus CTD. Samples for chemical analyses were processed immediately for shipboard analyses. Names of sample groups shown in Figure 1 are listed in Table 2.
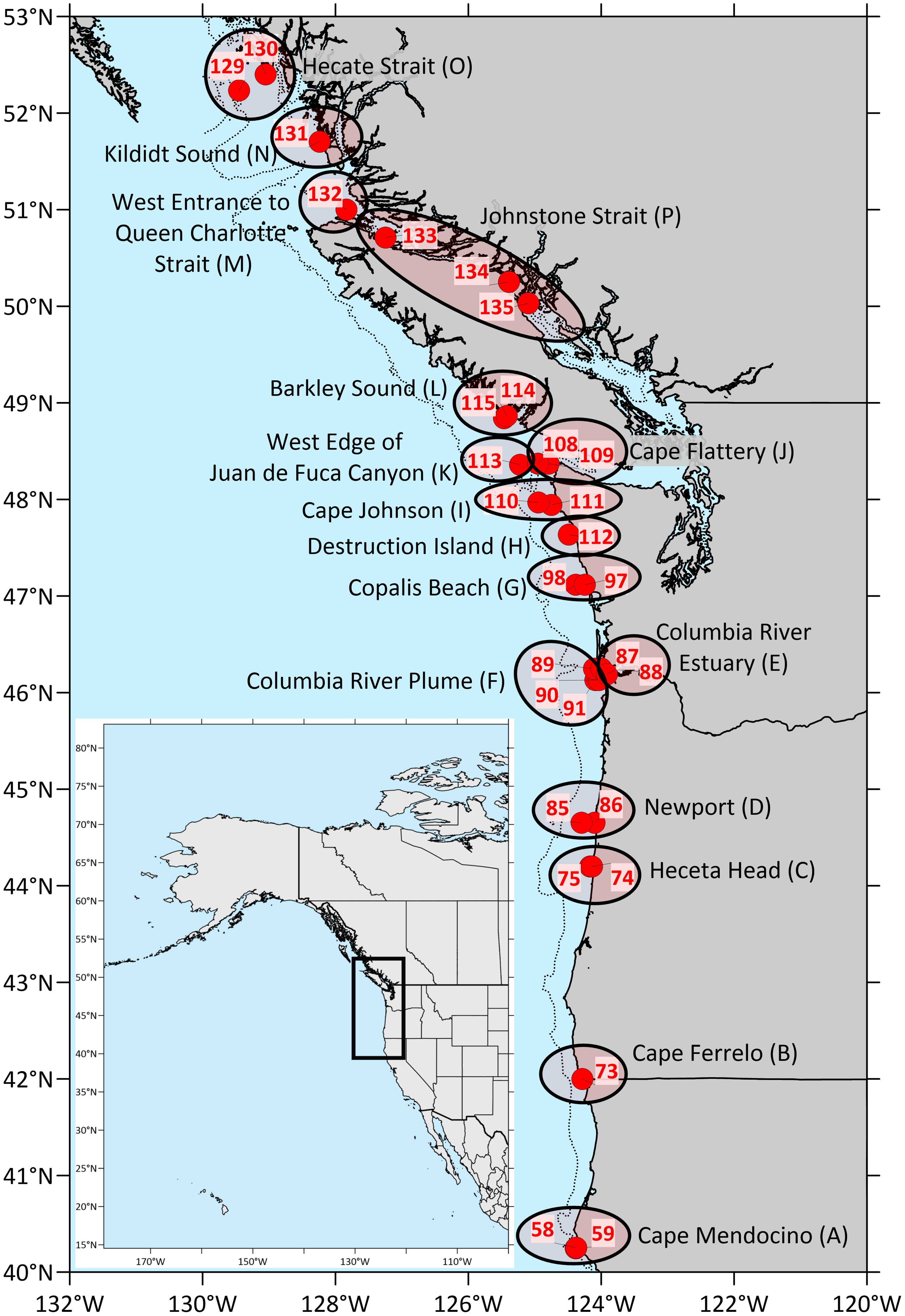
Figure 1. Maps of cruise coverage, stations, and geographic groups sampled for this study. The inset shows the area covered by the cruise in the western part of North America. Closed red circles indicate the locations of sampling stations, while the black ovals associated with place names indicate geographic groups reported in this study. Letters in parentheses denote the group code assigned to each geographic group.
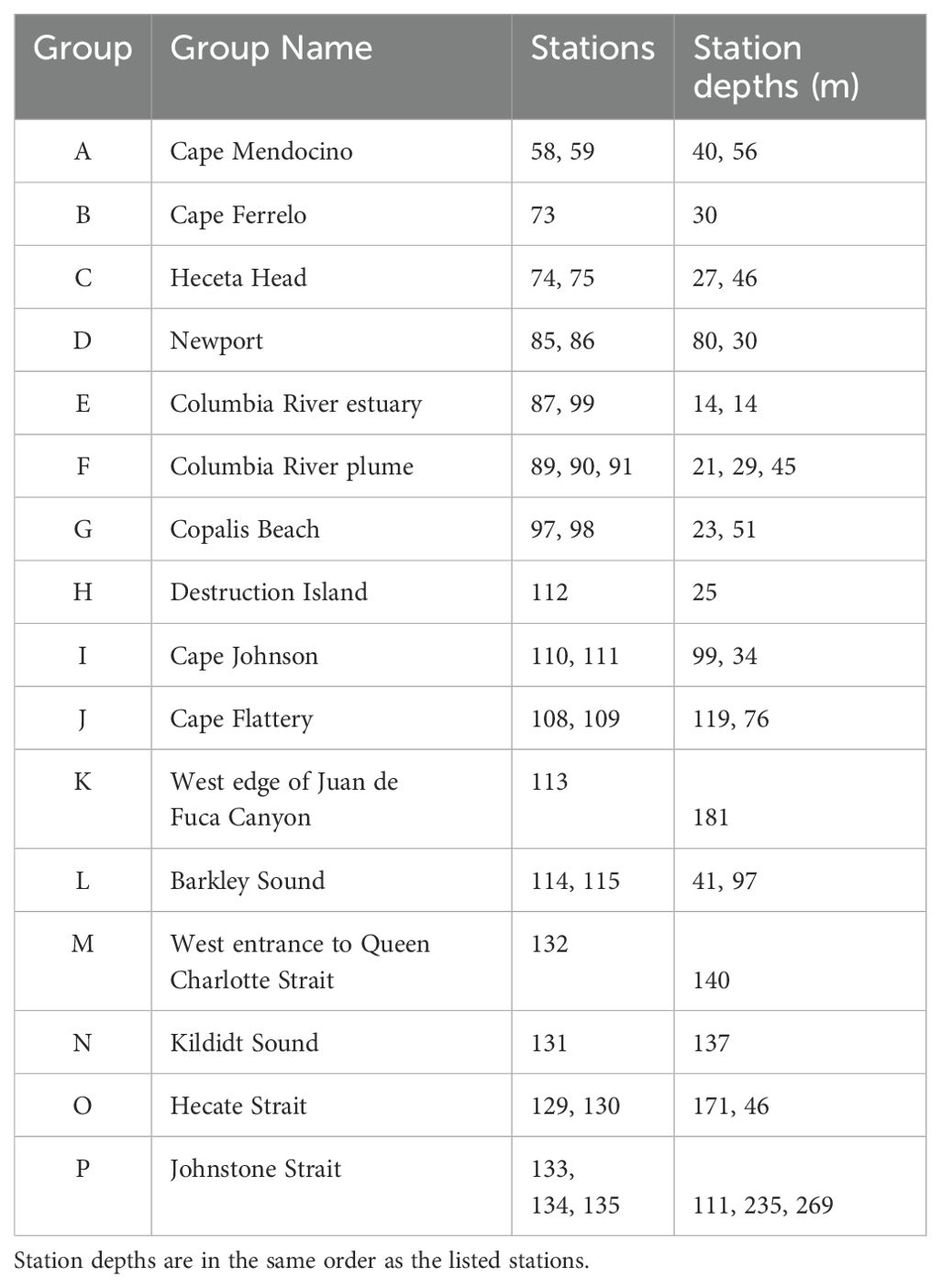
Table 2. List of geographic groups, group names, individual stations within each group, and maximum depths for each station.
Samples for biological analyses (> 2 L) were co-collected with those used for chemical analyses. Biological sampling depths were within 3 m of the surface, within 3 m of the bottom, and two to four approximately equally spaced intervals throughout the water column, depending on maximum depth. They were briefly held on ice in the dark and processed for longer term storage within an hour of collection. Whole water flow cytometry samples were preserved in 0.2% paraformaldehyde, flash-frozen in liquid nitrogen, and stored at ≤ −80°C until analysis. Duplicate 1 L samples for microbial taxonomic analyses were vacuum-filtered through 0.2 µm polyethersulfone filters (Sterlitech Corp, Kent WA), flash-frozen in liquid nitrogen, and stored at ≤ −80°C until extraction and analysis.
Sample analyses
Measurements for dissolved inorganic nutrients (nitrate, nitrite, ammonium, phosphate, silicate), dissolved inorganic carbon (DIC), and total alkalinity (TA) content were analyzed following the methods of Feely et al. (2016) and in detailed metadata included with the cruise dataset at the National Centers for Environmental Information (Alin et al., 2017). All measurements are reported in units of micromoles per kilogram (µmol kg−1). Additional carbonate system parameters—pH (on the total scale, pHT, unitless), carbonate ion content ([CO32−], µmol kg−1), and carbon dioxide fugacity (fCO2, µatm)—were calculated from inputs of DIC and TA using CO2SYS (Pelletier et al., 2007). Within CO2SYS, we used the Lueker et al. (2000) method for K1 and K2 dissociation constant formulations the Dickson (1990) method for KSO4 the Uppström (1974) method for total boron, and the seacarb option for Kf (i.e., Perez and Fraga, 1987, when temperature is 9–33°C and salinity is 10–40 and Dickson and Goyet, 1994 otherwise).
Microorganism abundances were enumerated by a BD FACSCAlibur 2-laser 4-color flow cytometer operated by the Kavanaugh Laboratory at Oregon State University. Bacteria and archaea were enumerated using the method of Longnecker et al. (2006). Coccoid cyanobacteria (Synechococcus, Prochlorococcus), eukaryotic nano- and picophytoplankton, and Cryptophytes were enumerated using the method of Sherr et al. (2005).
DNA for taxonomic analyses was extracted from frozen filters by the methods of Green and Sambrook (2018), and used to prepare amplicon libraries for Illumina sequencing. The bacterial and archaeal library was amplified using the 16S V4 primers from the Earth Microbiome Project (https://earthmicrobiome.org/protocols-and-standards/16s/):
Forward 515F (Parada): 5’ GTGYCAGCMGCCGCGGTAA-3’
Reverse 806R (Apprill): 5’ GGACTACNVGGGTWTCTAAT-3’
For the eukaryotic library, the following 18S primers from Stoeck et al. (2010) were used for amplification:
Forward 1391F: 5’-GTACACACCGCCCGTC-3’
Reverse EukBr: 5’-TGATCCTTCTGCAGGTTCACCTAC-3’
Individual samples amplified with HotStarTaq Plus Master Mix (Qiagen, USA) with the following conditions: denaturation at 95°C (5 minutes); 30 amplification cycles of 95°C (5 seconds), 53°C (40 seconds), 72°C (1 minute); elongation at 72°C (10 minutes). Products were uniquely dual indexed with Nextera adapters, purified with calibrated Ampure XP beads, and size-checked and quantified in 2% agarose gel for normalization and pooling. Library preparation and sequence analysis was performed using the MiSeq reagent kit v3 (600 cycles) on a MiSeq sequencer (Illumina, San Diego CA) following manufacturer’s guidelines by Molecular Research LP (MR DNA, Shallowater, TX USA).
Dereplicated sequence reads from the bacterial and archaeal results were quality trimmed using Trimmomatic (Bolger et al., 2014), and paired ends assembled using PANDAseq (Masella et al., 2012). Additional sequence filtering removed sequences with lengths less than 250 base pairs (bp), containing homopolymers, or ambiguous bases greater than 7 bp in length. Highly similar sequences (≥ 97% identity) were grouped into operational taxonomic units (OTUs), which were treated as the highest resolution taxon for some community analyses. OTUs were identified using QIIME2 (Bolyen et al., 2019) and taxonomy determined with the SILVA SSU database (release 132; Pruesse et al., 2012; Quast et al., 2012; Yilmaz et al., 2013). Based on analysis of a mock community of equimolar amounts of genomic DNA from 14 known bacterial species, OTUs with a frequency of < 21 in any one sample were discarded as sequencing errors. After sequencing, there were a total of 52,098,424 raw sequence reads for all station and depth combinations (113 samples), with an average count of 461,048 reads per sample for the V4 region of the 16S SSU rRNA (Supplementary Table 1). After trimming and joining paired reads followed by quality processing, 17,936,177 reads remained with an average count of 158,727 reads per sample.
For 18S eukaryotic sequence results, primer sequences were removed from demultiplexed paired-end reads using CUTADAPT version 3.0 (Martin, 2011). The R package DADA2 (version 1.2.0, Callahan et al., 2016) was used to quality filter, denoise, and trim reads, specifying a trimming length of 110 and a maximum number of expected errors (MaxEE) of 2. DADA2 was also used to remove chimeric sequences and export a table of amplicon sequence variants (ASVs). Taxonomic assignments were performed using the IDtaxa function in the R package DECIPHER (Murali et al., 2018) and the Protist Ribosomal Database (PR2; Guillou et al., 2012) using a confidence level of 70. Classifications were adjusted for consistency with the World Register of Marine Species (Ahyong et al., 2023). Eukaryotic phyla that did not contain marine protists were excluded from further analyses. A total of 17,936,177 reads remained after filtering, clustering and removal of chimeras, with an average of 138,727 reads per sample. For the V9 region of the 18S SSU rRNA, there were 16,132,082 reads from 112 station/depth combinations with an average of 144,045 reads per sample (Supplementary Table 2). After QC, clustering and denoising, 12,709,078 sequences were kept for 6763 ASVs, with an average coverage of 113,474 reads per station/depth combination. The 18S reads were filtered further to only include protist taxa resulting in 4,425,842 reads in 1976 ASVs, with an average coverage of 39,516 reads per station/depth combination.
Data are deposited in publicly accessible repositories. Physical and chemical seawater data are archived at NOAA’s National Centers for Environmental Information2 (Alin et al., 2017). Sequencing reads are archived at the National Center for Biotechnology Information under BioProject PRJNA10189553. Nonsequence biological data are archived at NOAA’s National Centers for Environmental Information (Accession number 0265154)4.
Data exploration and statistical analyses were performed using PRIMER7 (Anderson et al., 2008; Clarke and Gorley, 2015), R statistical package 4.3.2 (R Core Team, 2021; Oksanen et al., 2022), and Stata 18 (StataCorp, 2023). When appropriate, data were transformed to satisfy parametric statistical assumptions, applying a log(1+x) transform to raw sequence reads and standardizing by sample for Bray-Curtis similarity/dissimilarity matrices. Univariate metrics (species richness [d], Shannon diversity index [H’(loge)], Pielou’s evenness [J]) were calculated based on OTUs and ASVs (PRIMER7, Diverse; R, vegan). Principal components analysis (PCA) was performed to reduce variable datasets to influential subsets and summary indices (PRIMER7, PCA; Stata 18, Factor & principal components analysis), and canonical analysis of principal coordinates (CAP) was used to ordinate samples based on sequencing results (PRIMER7, CAP). Microbial community data were examined by permutational multifactorial and multivariate analysis of variance (PRIMER7, PERMANOVA; Stata 18, MANOVA; R, vegan). Network analysis was performed and displayed using R 4.3.2 (network, igraph, ggraph). For analyses utilizing depth intervals, they are: 1, surface to 10 m; 2, 11 to 20 m; 3, 21 to 30 m; 4, 31 to 40 m; 5, 41 m to 50 m; 6, 51 to 100 m; 7, >100 m.
Results
Seawater analyses
Seawater parameter characteristics by depth and geography
Up to twelve seawater parameters were measured for each sample, and principal components (PC) analysis of all parameters revealed strong alignment of samples with PC axis 1 which was best defined by fCO2 (fugacity of carbon dioxide), dissolved nitrate, dissolved inorganic phosphate (negative PC coefficients) and pHT (positive PC coefficients; Figure 2). Samples from all geographic groups, except those associated with Columbia River, were widely distributed across PC axis 1, indicating that a range of those parameters occurred within each group (Figure 2A). Instead, sample depth exhibited a relationship with PC axis 1, with deeper samples associated with higher fCO2, nitrate, and phosphate, and shallower samples associated with higher pHT (Figure 2B). Overall sample distribution was narrower along PC axis 2, which was best defined by total alkalinity, seawater density, and salinity (positive PC coefficients; Figure 2). Columbia River-associated samples (E and F) were most broadly distributed along PC axis 2, consistent with the properties of fresh water on these three parameters (Figure 2A).
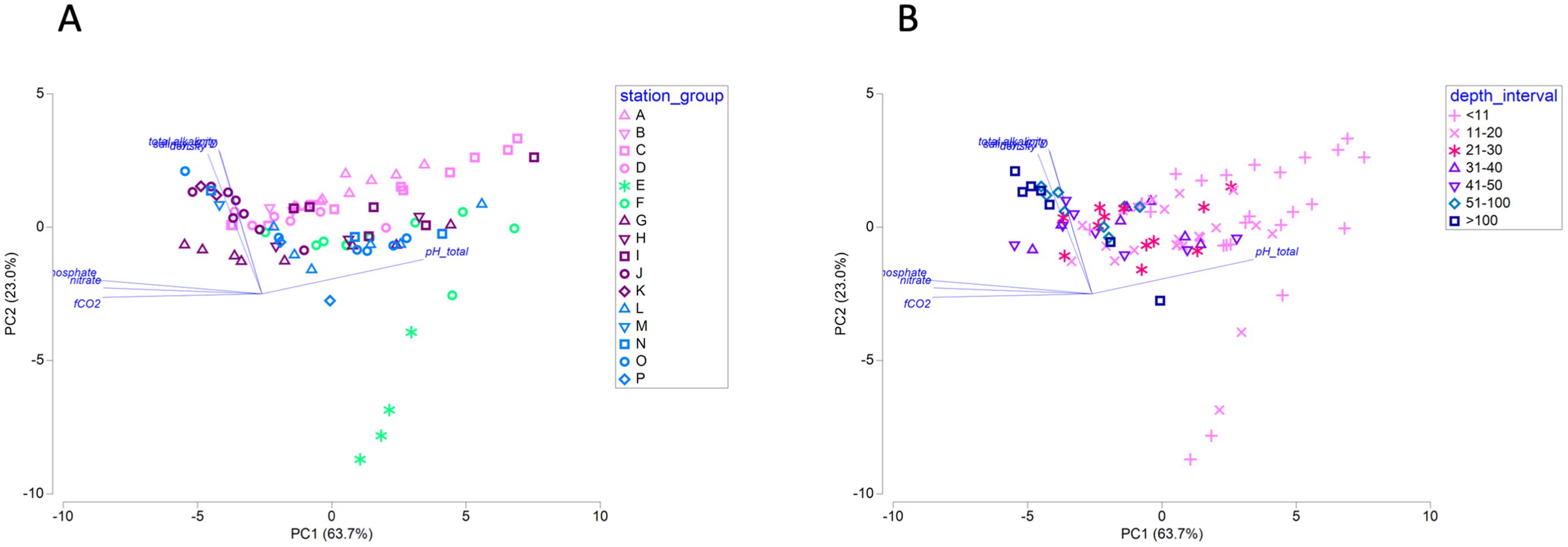
Figure 2. Principal components analysis (PCA) of twelve seawater parameters. PCA model is plotted by geographic group (A) or by depth interval (B). Parameters with PC coefficients greater than |0.3| in the first two axes are overlaid as vectors. Total variation explained by the first two PC axes is 86.6%; N = 89.
Individual seawater parameters by geography
Depth, temperature and salinity
The maximum depth of the sampling locations was typically < 50 m for the southern stations, from Cape Mendocino through Cape Johnson (A through I; Table 2 and Figure 3A), with the shallowest locations in the Columbia River estuary (11 m; E) and Destruction Island (20 m; H). Maximum depths for locations from Cape Johnson and northward (I through P) ranged between 50 and 150 m, with the deepest locations in Johnstone Strait (250 m; P).
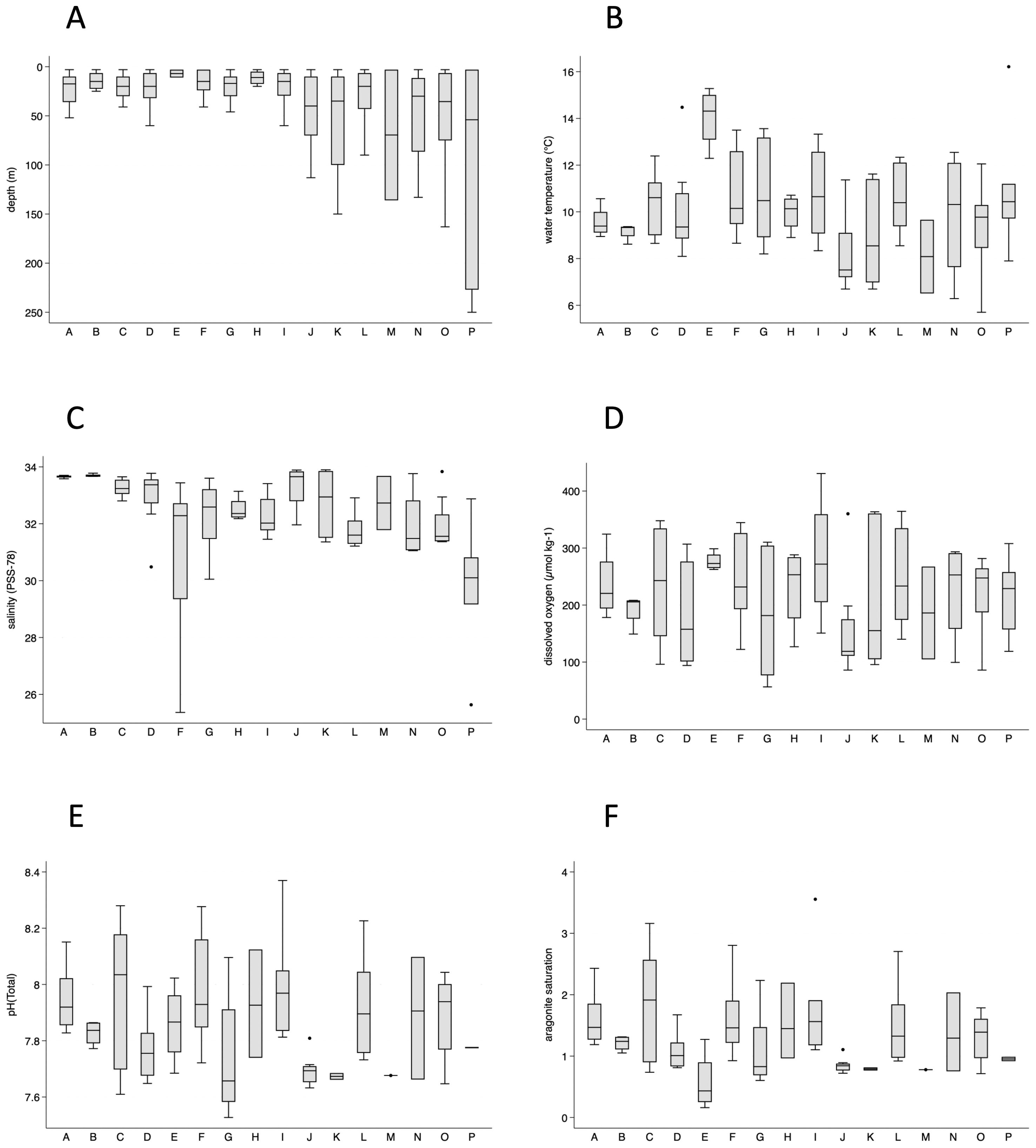
Figure 3. Box plots of sampling depths (A), temperatures (B), salinities (C), dissolved oxygen (D), pHT (E), and aragonite saturation (F) by geographic group. The Columbia River estuary group (E) had low salinities (1.9–23.1) and is not included in the salinity graph (C).
Except for samples from the Columbia River estuary (E), median temperatures ranged from 7.5°C (Cape Flattery, J) to 10.6°C (Heceta Head and Cape Ferrelo, C and I, respectively; Figure 3B). Samples from Cape Mendocino (A) and Cape Ferrelo (B) exhibited the narrowest temperature ranges (8.9–10.6°C and 8.6–9.4°C, respectively).
Salinity varied widely among the groups, with lowest values in the Columbia River estuary (1.9-23.1; not shown), and wide variation in the Columbia River plume (F; Figure 3C). Other locations with lowered salinities were Copalis Beach (G) and Johnstone Strait (P). Cape Mendocino (A) and Cape Ferrelo (B) displayed the highest values and lowest variation in salinity (Figure 3C).
Dissolved oxygen
Dissolved oxygen content of seawater ranged between 56.5 and 430.7 µmol kg−1 and varied widely across the geographic groups (Figure 3D). Thirty-two of the 113 samples had dissolved oxygen concentrations considered low (62 to 160 µmol kg−1), and one had a concentration considered hypoxic (< 62 µmol kg−1, Supplementary Figure 1A). The deepest sample from most of the stations had low dissolved oxygen, even if the depth was only 20 m, and in many cases low oxygen extended upward 10–20 m. At Cape Flattery (J) low dissolved oxygen extended to within 10 m of the surface. The three lowest values were from two stations near Copalis Beach (G). The station closer to the shore was nearly hypoxic for the bottom sample (19 m) but was highly oxic for the shallower parts of the profile. The station farther from shore was hypoxic for the bottom sample (46 m), low oxygen to 30 m, then oxic for shallower samples. No surface samples (< 10 m) had low dissolved oxygen.
pH on the total scale and aragonite saturation
The values for pH on the total scale (pHT) ranged from 7.53 to 8.37 (Figure 3E), with most of the values below the surface open ocean average value of 8.1 (Supplementary Figure 1B). Twenty-five percent (22 of 89) had values below 7.7, which is considered low (Feely et al., 2010) and a level with negative physiological impacts for multiple marine species from microbes to invertebrates (e.g., Manno et al., 2016; Nelson et al., 2020). None of the samples had pHT values at or below 7.5, which is a threshold value for biological effects such as bivalve calcification (e.g., Gazeau et al., 2007). Among the samples with values below 7.7, more than half were from Copalis Beach (G) and Cape Flattery (J), and Heceta Head (C), and most samples were collected deeper than 30 m (Supplementary Figure 1B).
Changes in carbonate saturation can result from uptake of CO2 by seawater, causing biological consequences for calcifying organisms (e.g., bivalves and pteropods) such as impeding shell formation and dissolution of existing shells (e.g., Bednaršek et al., 2014; Waldbusser et al., 2015). Aragonite saturation (Ωaragonite) is a widely used measure of carbonate ion concentration and, consequently, ocean acidification. In this study, aragonite saturation ranged from 0.16 in the Columbia River estuary to 3.55 at Cape Johnson (Figure 3F). Over a third of samples exhibited a calculated aragonite saturation (Ωaragonite) value below the aragonite saturation horizon (i.e., Ωaragonite < 1; Supplementary Figure 2). Samples from Copalis Beach (G) and Cape Flattery (J) were undersaturated throughout the depth profile. In contrast, undersaturated samples from most other locations were deep or at the bottom (Supplementary Figure 2). Higher aragonite saturation values (i.e., Ωaragonite > 2) occurred in surface and shallow depth samples, except in the Columbia River estuary (Supplementary Figure 2).
Nitrate, phosphate, nitrite, ammonium, silicate content
Dissolved nitrate content for all samples were either within the oligotrophic range (0 to 16.5 µmol kg−1) or mesotrophic range (16.5 to 82.6 µmol kg−1). Nutrient content generally followed the expectation of increase with increasing depth (Supplementary Figure 3A).
In contrast to nitrate levels, many samples had eutrophic levels of dissolved inorganic phosphate (> 1.08 µmol kg−1). Eighteen samples had mesotrophic levels, and only five samples were at oligotrophic levels (< 0.32 µmol kg−1), which is limiting for diatom growth (Supplementary Figure 3B). All other samples, including shallower ones from Cape Ferrelo (B) and Cape Flattery (J) contained eutrophic levels of phosphate.
The patterns of low nitrate and high phosphate imply that nitrate to phosphate ratios would be low, and only two of the samples approached or exceeded the theoretical Redfield ratio of 16 to 1 (Supplementary Figure 3C). Both of these samples were from the Columbia River estuary (E). Samples with low nitrate to phosphate ratios (< 2 µmol kg−1) were most frequently from Heceta Head (C), the Columbia River plume (F), Copalis Beach (G), and Cape Johnson (I).
Nitrite, a transient and intermediate product in microbial nitrification, was less than 0.5 µmol kg−1 for all samples except for deeper samples from Copalis Beach (G) and one deeper sample from Destruction Island (H). Examination of individual groups revealed a variety of profile patterns (Supplementary Figure 4A). Several locations showed an increase in nitrite with depth to a certain depth, then little change with greater depth (Cape Mendocino and Heceta Head; A and C, respectively), while one location (Cape Flattery, J) showed no pattern of nitrite content with depth (Supplementary Figure 4A). Multiple locations exhibited subsurface maxima (Columbia River plume, Cape Johnson, Barkley Sound, and Hecata Strait; F, I, L, and O, respectively). Cape Flattery (J) displayed variations in content along the depth profile, while Copalis Beach (G) had a striking linear decrease in nitrite with depth (Supplementary Figure 4A).
Ammonium can often display subsurface maxima close to the nitrite maxima. Although a preferred nitrogen source by phytoplankton, ammonium levels are typically low in seawater. Ammonium patterns varied by location, frequently displaying a subsurface maximum (Heceta Head, Columbia River plume, Cape Johnson, Cape Flattery, Barkley Sound, and Hecate Strait; C, F, I, J, L, and O, respectively; Supplementary Figure 4B). For Copalis Beach (G), the ammonium depth pattern was similar to the nitrite pattern, i.e., a nearly linear decrease with depth (Supplementary Figure 4B).
Silicate ranged widely, from 0.96 to 172.11 µmol kg−1. Four samples were close or below 2 µmol kg−1 which is considered to be limiting for diatom growth (Supplementary Figure 4C). The lowest three values were from Heceta Head (C), and the fourth lowest value was from Cape Johnson (I). The three highest values, which exceeded 100 µmol kg−1, were from the Columbia River estuary (not shown on Supplementary Figure 4C).
Microbial analyses
Cellular abundances by geography
Total bacterial and archaeal cellular abundance, measured by flow cytometry enumeration, followed an expected pattern of lower abundances with depth (data not shown), but there was wide variation in abundances among geographic groups (Figure 4A). Cape Flattery (J) samples had the lowest bacterial and archaeal abundances at all depths, where counts decreased from low surface counts (0.5–0.8 cells ml−1) to lower counts at all other depths (0.2–0.3 cells ml−1). Copalis Beach (G), also had low surface abundances (data not shown). Locations with higher abundances at the surface and across the depths included Heceta Head (C), Newport (D), Kildidt Sound (N), and Hecate Strait (O).
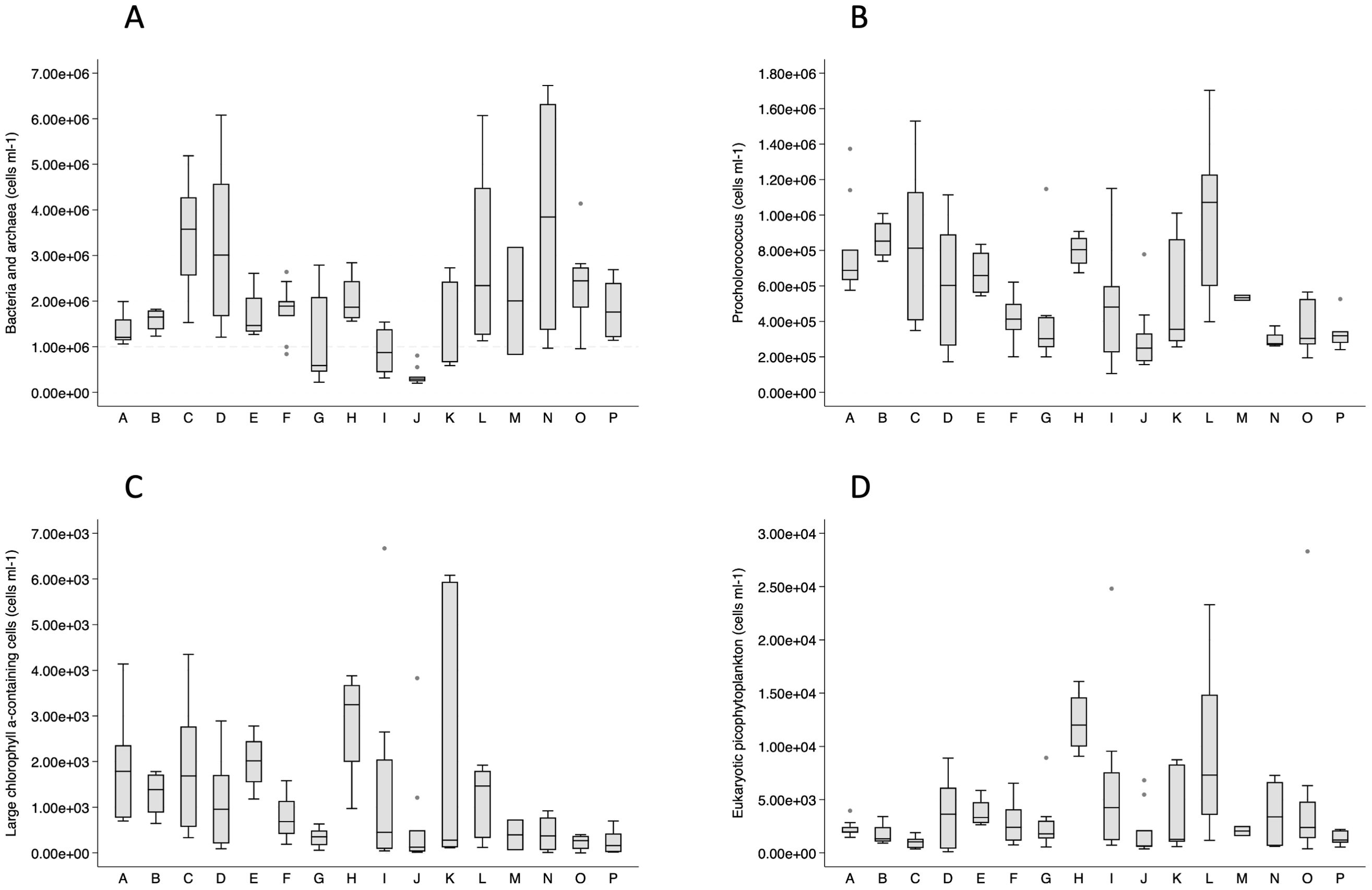
Figure 4. Box plots of flow cytometry-based cell abundances for bacteria and archaea (A), Prochlorococcus (B), large chlorophyll α-containing cells (C), and eukaryotic nano- and picophytoplankton (D) by geographic group.
Phytoplankton, including cyanobacteria, were also enumerated by flow cytometry. Synechococcus cellular abundances were low (mean < 2,500 cells ml−1) in all geographic groups except at Cape Johnson (I; mean = 4,341 cells ml−1), Barkley Sound (L; mean = 6,174 cells ml−1), Kildidt Sound (N; mean = 20,882 cells ml−1), and Hecate Strait (O; mean = 39,649 cells ml−1). In contrast, Prochlorococcus cellular abundances were high (minimum 100,000 cells ml−1) and varied geographically (Figure 4B). Cryptophyte abundances were extremely low (mean < 7 cells ml−1), with highest values in the Columbia River estuary (range 11–14 cells ml−1). Large chlorophyll α-containing cells, presumed to be primarily diatoms, were highly varied in abundance between the Columbia River (E, F) and Barkley Sound (L), and north of Barkley Sound abundances were low (Figure 4C). Eukaryotic nano- and picophytoplankton exhibited abundances intermediate to Synechococcus and Prochlorococcus (Figure 4D). Among the geographic groups, Destruction Island (H) displayed the highest consistent concentrations of Prochlorococcus, large chlorophyll α-containing cells, and eukaryotic nano- and picophytoplankton (Figures 4B–D).
Overall, there were significant correlations among microbial cell abundances (Table 3). Bacterial and archaeal abundances and eukaryotic nano- and picophytoplankton abundances were positively correlated with all other microbial categories, and a large correlation existed between Prochlorococcus and cells containing chlorophyll α, which are presumed to be primarily diatoms (Table 3). Among the geographic groups, Barkley Sound (L) had large correlations (Spearman’s r > 0.95) between Prochlorococcus, Synechococcus, chlorophyll α-containing cells, and eukaryotic nano- and picophytoplankton.
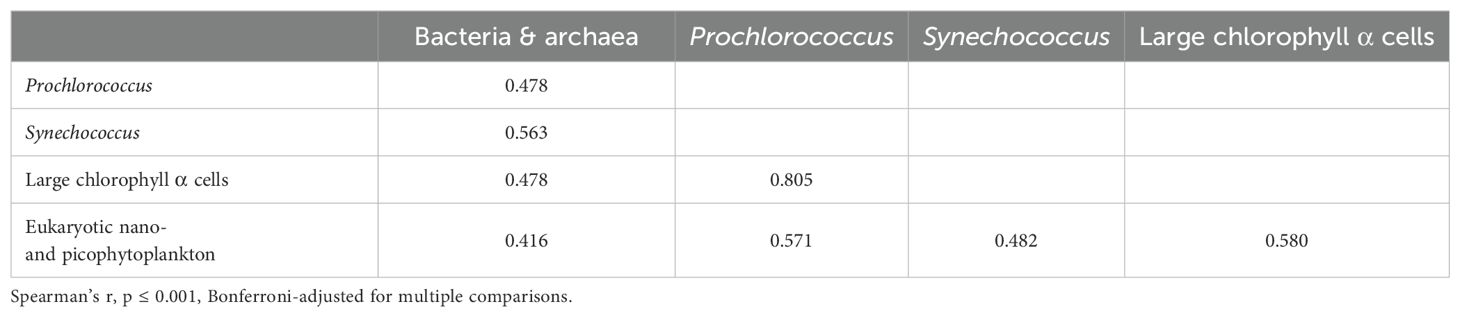
Table 3. Significant correlations of abundances for categories of microbes based on flow cytometry for all geographic groups combined.
Microbial community univariate indices
Principal components analysis (PCA) of indices for richness, evenness, and diversity for bacterial and archaeal OTUs revealed geographic separation of southern and northern stations, primarily due to richness with high explanatory power (98.5%) of the first two axes (Supplementary Figure 5A). Stations at Copalis Beach and southward (A–G) had lower OTU richness, while stations at Destruction Island and northward (H-P), except at Cape Flattery (J), had greater OTU richness (Supplementary Figure 6A). This pattern was consistent for higher resolution taxon assignments such as family and genus (data not shown). Samples from Cape Ferrelo (B) and the Columbia River estuary (E) tended toward lower diversity than most of the other samples (Supplementary Figure 6A).
PCA analysis based on diversity indices of eukaryotic ASVs showed a pattern different from bacterial and archaeal OTUs with good explanatory power (99.8%) for the first two PCA axes (Supplementary Figure 5B). ASV diversity and evenness increased north of the two southern groups (A and B) and remained elevated north of the Columbia River plume (F to P), except at Destruction Island (H; Supplementary Figures 6B, C).
Potential relationships between seawater parameters and univariate indices for bacterial and archaeal OTUs and for microbial eukaryotic ASVs were examined, but no consistent and significant correlations were found (data not shown).
Community structures by geography and depth
Nonmetric multidimensional scaling (NMDS) of bacterial and archaeal communities based on OTUs revealed strong separation between stations southward from Newport (A-D) & stations northward of Juan de Fuca Strait (L-P). The Washington coastal communities (F-K) overlapped with both the northern and southern groups, while communities within the Columbia River estuary (E) diverged from all other stations (Supplementary Figure 7A). Vectors for eight parameters that explain ~ 85% of the seawater principal components analysis aligned parallel to the main axis of the communities in the NMDS plot. The NMDS plot using the depth interval for each sample revealed that shallower samples (≤ 30 m) aligned with higher dissolved oxygen, pHT, and carbonate concentrations and deeper samples (> 30 m) aligned with higher dissolved nitrate and phosphate concentrations (Supplementary Figure 7B).
NMDS analyses of microbial eukaryotic ASVs revealed weak differentiation of community structure among geographic groups, primarily due to the greater differences of the two southern groups (Cape Mendocino, Cape Ferrelo) from all other groups (Supplementary Figure 7C). When plotted by depth interval, a similar pattern emerged of shallower samples aligned with higher dissolved oxygen, pHT, and carbonate and deeper samples aligned with higher dissolved nitrate and phosphate concentrations (Supplementary Figure 7D).
Canonical analysis of principal coordinates (CAP) of microbial communities allows ordination of the samples using the sequencing data, so that samples with stronger similarities in community structure are ordered closer together in multidimensional space (Anderson and Willis, 2003). Assignment of samples to geographic groups using a discriminant analysis method provides a cross-validation of the CAP model. CAP identified strong differences in community structure related to geographic group, and the canonical correlations for the first two axes exceeded 0.9 (Figure 5). For bacterial and archaeal OTUs, the overall ability to assign a sample to the correct geographic group was 77% and exceeded 75% for samples from Cape Mendocino (A), Cape Ferrelo (B), Heceta Head (C), the Columbia River estuary (E), the Columbia River plume (F), Copalis Beach (G), Destruction Island (H), and Cape Flattery (J). Sample assignment was modest (50–75%) for Cape Johnson (I), Juan de Fuca Canyon (K), Barkley Sound (L), Hecate Strait (O), and Johnstone Strait (P). In contrast, assignment was poor (<50%) for Newport (D), Queen Charlotte Strait (M), and Kildidt Sound (N). Associated seawater parameters revealed higher total alkalinity, salinity, and dissolved inorganic carbon within the two southern groups (A and B), and higher temperatures, silicate, nitrite, and ammonium at Copalis Beach (G) and Columbia River stations (E, F; Figure 5A).
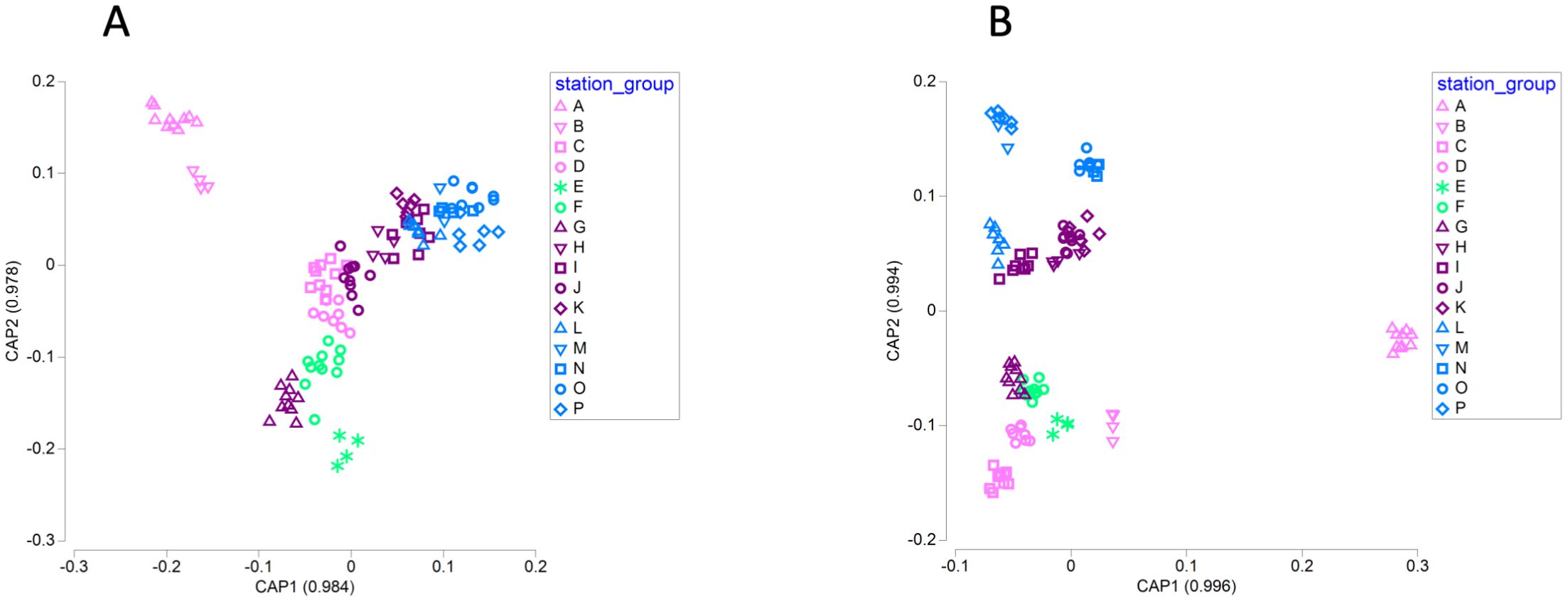
Figure 5. Plots of microbial communities for bacterial and archaeal OTUs (A) and microbial eukaryotic ASVs (B) based on canonical analysis of principal coordinates by geographic group.
CAP analysis revealed even greater distinctions among geographic groups for microbial eukaryotic ASVs (Figure 5B). Overall ability of CAP to assign samples to the correct geographic group was 79.5%. It had 100% correct classifications for samples from Cape Mendocino (A), Cape Ferrelo (B), Heceta Head (C), Copalis Beach (G), Barkley Sound (L), and Kildidt Sound (N), and had high accuracy for the Columbia River plume (F; 90.9%), Newport (D, 87.5%), Hecate Strait (O; 87.5%), the Columbia River estuary (E; 75.0%), and Destruction Island (H; 75.0%). CAP assignment accuracy was poor for samples from Cape Flattery (J; 50%), Juan de Fuca canyon (K; 50%), Johnstone Strait (P; 50%), and Cape Johnson (I; 37.5%), and CAP was unable to make any correct sample assignments for Queen Charlotte Strait (M; 0%).
Permutational multivariate analysis of variance and pairwise comparison for OTU and ASV communities revealed significant differences and similarities among the geographic groups (PERMANOVA, p ≤ 0.05). OTU communities in the three southernmost groups (Cape Mendocino [A], Cape Ferrelo [B], Heceta Head [C]) and Columbia River estuary (E) were statistically distinct from each other and from all other groups. Along much of the Washington coast, OTU communities tended to be different from other sites, but similar to proximal locations (Supplementary Figure 8A). For example, the Columbia River plume (F) was different from other groups except flanking locations of Copalis Beach (G) and Newport (D), while Destruction Island (H) was different from other groups except Cape Johnson (I). In contrast, Cape Johnson (I), the Juan de Fuca Canyon (K) and groups north of the Strait of Juan de Fuca (Barkley Sound [L], Queen Charlotte Strait [M], Kildidt Sound [N], Hecate Strait [O], Johnstone Strait [P]) were statistically similar to each other (Supplementary Figure 8A). Pairwise comparison of ASV communities found fewer statistically similar communities, and those similarities occurred primarily between groups northward of Destruction Island (H; Supplementary Figure 8B). Surprisingly, Queen Charlotte Strait (M) had ASV community similarity to a wide range of geographic groups (Supplementary Figure 8B).
Depth of sample was anticipated to be important in community structure, but the depth-based canonical correlations were modest (0.756 and 0.261 for axis 1 and 2, respectively for OTUs; 0.756 and 0.562 for axis 1 and 2, respectively for ASVs; data not shown). The ability of the depth-based CAP model to correctly assign samples was weak overall (50.4% for OTUs, 55.4% for ASVs). The depth models had poor predictive ability for OTUs (< 15%) and mediocre predictive ability for ASVs (14–43%) at intermediate depth intervals (21–50 m). Conversely, the depth models had best predictive ability for OTUs and ASVs at the depths ≤ 10 m (70.0% and 75.0%, respectively) and at depths > 50 m (76.1% and 62.5%, respectively).
Multivariate correlations between seawater parameters and OTUs or ASVs examined the relationship between microbial community structures and environmental factors. Due to the strong freshwater influence of the Columbia River, separate multivariate correlations were performed for samples associated with the river (E, F). The best fit between seawater parameters and OTUs using within-geographic group analysis identified five significant seawater parameters: temperature, pHT, nitrate, phosphate, and silicate (Bio-Env stepwise correlation, rho = 0.734). Correlation analysis for just the Columbia River-associated groups (E, F) identified five slightly different seawater parameters: temperature, salinity, dissolved inorganic carbon, nitrate, and ammonium (Bio-Env stepwise correlation, rho = 0.876). The best fit between seawater parameters and ASVs identified five significant seawater parameters: salinity, dissolved oxygen, total alkalinity, silicate, and nitrite (Bio-Env stepwise correlation, rho = 0.750). The ASV analysis using only Columbia River samples identified five significant seawater parameters: temperature, salinity, dissolved oxygen, nitrate, and ammonium (Bio-Env stepwise correlation, rho = 0.792).
Microbial phyla distribution
Proteobacteria was a dominant phylum occurring across most of the geographic groups across all depths, with relative abundances spanning 20–60% (Supplementary Figure 9). Samples from the Columbia River estuary (E) were notable exceptions, where Cyanobacteria was more abundant at all depths. The next most prominent phylum was Bacteroidetes, which was relatively abundant (~10–60%) for all geographic groups at depths up to 50 m; at depths > 50 m, Bacteroidetes occurred at < 10%. Cyanobacteria was the third most prominent phylum at depths < 50 m, especially at the southern locations (Cape Mendocino, A; Cape Ferrelo, B), around the Columbia River (Columbia River estuary, E; Columbia River plume, F), Copalis Beach (G), and Juan de Fuca Canyon (K).
Archaeal phyla were not as abundant as Proteobacteria, Bacteroidetes, and Cyanobacteria, but were frequently more abundant than most of the other bacterial phyla (Supplementary Figure 9). Euryarchaeota exhibited the most cosmopolitan geographic and depth distribution of the Archaea. Thaumarchaeota occurred at all locations, but exhibited a clear pattern of increasing relative abundance with depth. Nanoarchaeota were the least abundant of the Archaea, and occurred only at depths > 30 m.
Low relative abundance bacterial phyla that were detected at all of the locations included Actinobacteria, Marinimicrobia (SAR406 clade), Planctomycetes, and Verrucomicrobia. Although these phyla occurred at all depths, Marinimicrobia and Planctomycetes exhibited increased relative abundances with depth. Among the other low relative abundance bacterial phyla, some occurred predominantly in shallower samples (Epsilonbacteraeota, Firmicutes, Patescibacteria) while others tended to occur in deeper samples (AncK6, Chloroflexi, Gemmatimonadetes, Nitrospinae, PAUC34f).
Among eukaryotic microbes, Dinoflagellata and Bacillariophyta were dominant across all geographic groups and at all depths, although Bacillariophyta relative abundances greatly declined at depths > 50 m at geographic locations north of the Columbia River (Supplementary Figure 9). Ciliophora was the next most abundant phylum among groups north of Heceta Head (C), especially at depths > 50 m. Cercozoa were not highly abundant, but they occurred consistently across all of the geographic groups at depths < 50 m. The relative abundances of Radiolaria and Apicomplexa were low, but similar to Ciliophora, increased with depth.
Examination of co-occurrences between bacterial and archaeal phyla with eukaryotic microbial phyla revealed two major groupings across the samples. A larger group containing most of the highly abundant bacterial and archaeal phyla (e.g., Proteobacteria, Cyanobacteria) and eukaryotic microbial phyla (e.g., Ochrophyta, Dinoflagellata) displayed many strong associations (≥80% co-occurrence among samples; Figure 6A). A second, separate group consisting of less abundant bacterial and archaeal phyla (e.g., Chloroflexi, Nitrospinae, Nanoarcheaeota) were strongly associated with a few eukaryotic microbial phyla, such as Rhodophyta, Perkinsea, and Foraminifera (Figure 6A). Based on their distribution by depth, phyla in the first group occur across all depth intervals, while the second group is representative of associations that tend to occur at greater depths (Figure 6B).
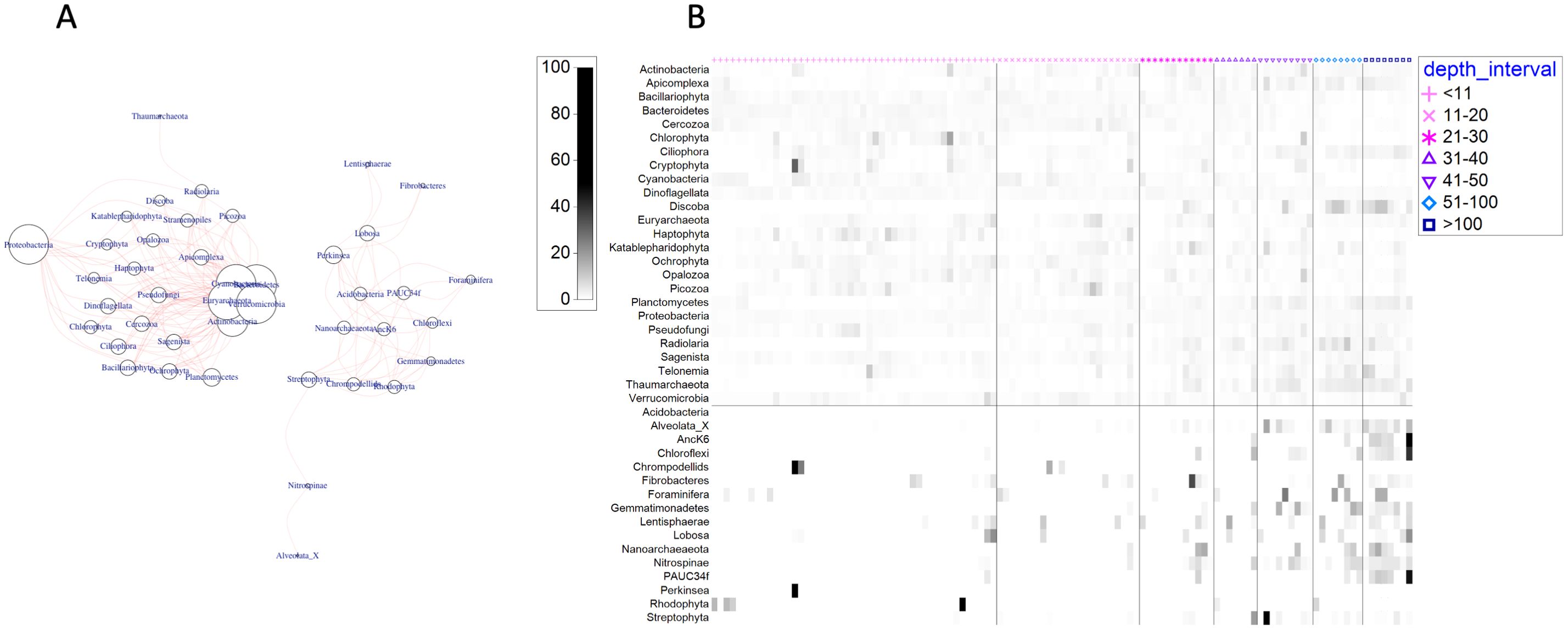
Figure 6. Network of co-occurrences between 16S and 18S phyla occurring in ≥80% of the samples (A) and shade plot of phyla relative abundances by depth interval (B). Circles at nodes in (A) are proportional to the number of associations (i.e., larger circles have more associations), and co-occurring taxa are connected by pink lines. Abundances in (B) are double standardized (by sample and by phylum) and arranged from shallow (left) to deep (right), with vertical lines between adjacent depth intervals. Horizontal line in (B) separates the network clusters displayed in (A).
Abundance and depth patterns of microbial taxa
The patterns of family abundances across the geographic locations and depths provides finer resolution both spatially and taxonomically. The most abundant bacterial and archaeal families across all geographic groups were Flavobacteriaceae and Rhodobacteriaceae (Supplementary Figure 10). Detected Flavobacteriaceae genera included Flavobacterium, Formosa, Lutibacter, NS2b marine group, NS3a marine group, NS4 marine group, NS5 marine group, Pseudofulvibacter, Tenacibaculum, and Ulvibacter. Tenacibaculum was the most highly abundant genus of this family. Detected Rhobacteriaceae genera included Amylibacter, Loktanella, and Planktomarina, and Amylibacter was the most abundant genus of this family. The next most abundant bacterial and archaeal families present in all geographic groups were Cryomorphaceae, NS9 marine group, Porticoccocaceae (SAR92 clade was most abundant), Halieaceae (Halioglobus was the most abundant), Thioglobaceae (SUP05 cluster was most abundant), and SAR11 clade I (Supplementary Figure 10).
Some bacterial and archaeal families exhibited patterns associated with depth. Although Nitrosopumilaceae occurred in all groups (except in the Columbia River estuary), the family displayed a pattern of increasing abundance with depth. Microtrichaceae also exhibited a depth-associated pattern, but with lower relative abundance than Nitrosopumilaceae (Supplementary Figure 10).
Other bacterial and archaeal families displayed patterns associated with geographic locations. Nitrosococcaceae was not detected at Destruction Island and groups south of Destruction Island (A–H), but was detected at Cape Johnson and all groups northward (I–P). Burkholderiaceae was found everywhere except the two southernmost groups, Cape Mendocino (A) and Cape Ferrelo (B). Hyphomicrobiaceae were only detected at Cape Mendocino (A) and Cape Flattery (J). Coxiellaceae, which includes the pathogenic Coxiella genus, were detected only at Cape Johnson (I) and groups north of the Strait of Juan de Fuca (L–P). In contrast, Francisellaceae, which includes the pathogenic Francisella genus, was detected in all groups except the two southernmost (Cape Mendocino [A], Cape Ferrelo [B]) and Queen Charlotte Strait (M). Chitinophagaceae, Sphingobacteriaceae, and Spirosomaceae were detected only at locations associated with the Columbia River (E, F).
Among eukaryotic phytoplankton, twelve taxa were relatively abundant across the entire cruise and at all depths: diatoms (Bacillariophyceae, Bacillariales, Chaetocerotales, Coscinodiscales, Thalassiosirales), dinoflagellates (Dino-Group-I, Dino-Group-II, Gonyaulacales, Gymnodiniales, Peridiniales), and microalgae (Cryptomonadales, Prymnesiales; Supplementary Figure 11). Rhizosoleniales were abundant from Cape Ferrelo through Barkley Sound (B through L), and Noctilucales, and Hermiaulales were abundant across the depth profiles from Cape Ferrelo to Copalis Beach (B though G). Some phytoplankton taxa occurred primarily at certain locations (e.g., Chloropicales and Pseudoscourfieldiales at Cape Mendocino, A; Cryptophyceae, Melosirales, and Aulacoseirales around the Columbia River, E and F). Several dinoflagellate taxa, such as Dino-Group-IV and Dino-Group-V, occurred primarily at lower depths (Supplementary Figure 11).
Among zooplankton classes, three taxa were common across the entire cruise and depth profile: Litostomatea (ciliates), Filosa-Thecofilosea (amoebic flagellates), and Spirotrichea (ciliates; Supplementary Figure 12). Polycistinea (radiolarians) and Plagiophylea had broad geographic distribution but were more prevalent at subsurface depths. Samples associated with the Columbia River (E, F) had a wide variety of zooplankton, although some classes (e.g., CONTH_4, CONTH_5, Perkinsida, Colpodea, Tubulinea) occurred nearly exclusively in the estuary (Supplementary Figure 12). The radiolarians (RAD-A, RAD-B, RAD-C, Acantharea, Polycistinea) and amoeboid Phaeodarea tended to occur at subsurface depths (Supplementary Figure 12).
Eukaryotic microbes other than zooplankton and the most abundant phytoplankton were also detected with 18S metabarcoding. Among these groups are free-living and parasitic protists (e.g., Apicomplexa, Gregarinomorphea, Labyrinthulomycetes), saprotrophs (e.g., Opalozoa, Oomycota), and algae (e.g., Picozoa, Chrysophyceae). Classes abundantly present in all geographic groups and by depth included Chrysophyceae, Dictyochophyceae, Gregarinomorphea, Picozoa, Labyrinthulomycetes, Telonemia, and six classes collectively called MAST (MArine STramenopiles; Massana et al., 2014) belonging to the saproptrophic phyla Pseudofungi, Opalozoa, and Sagenista (Supplementary Figure 13). Several classes limited or nearly limited to the Columbia River groups (E, F) included algae with chloroplasts (Ulvophyceae, Chlorophyceae, Synurophyceae, Eustigmatophyceae). Certain green algae (Trebouxiophyceae) and brown algae (Phaeophyceae) were highly abundant at the southernmost group (A), but occurred only sporadically at other locations. Several classes were found primarily in deeper water, including the crustacean ectoparasitic Ellobiopsidae and MOCH-4 aplastidic algae class (Supplementary Figure 13).
Indicator taxa
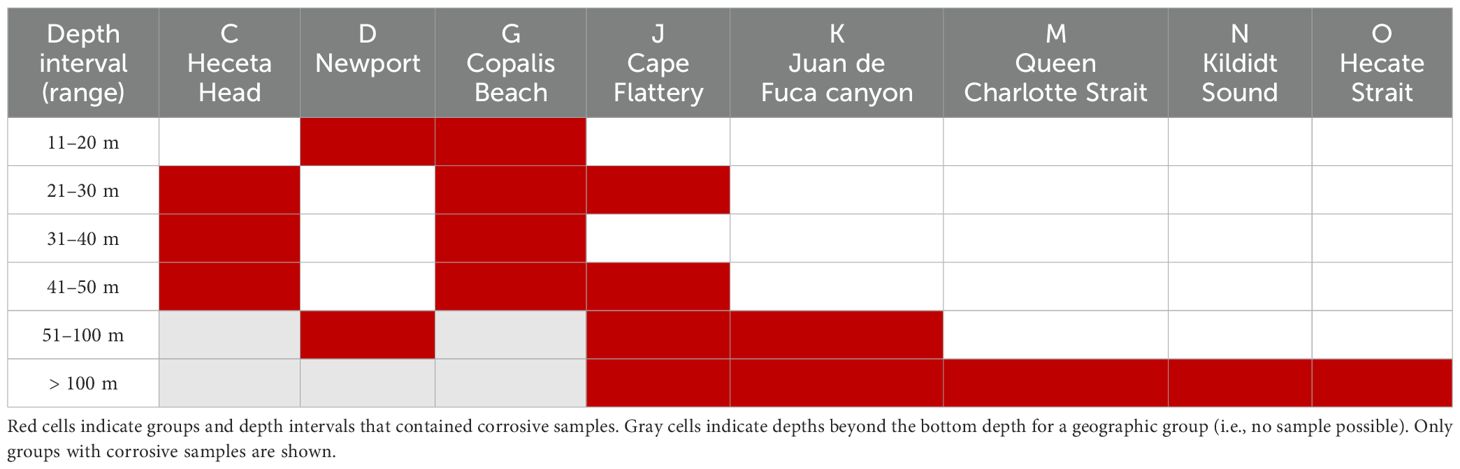
Feely et al. (2016) described “acidified, corrosive, CO2-rich waters” as having a pH < 7.75, Ωaragonite < 1.0, and dissolved inorganic carbon > 2190 µmol kg−1, and a subset of seawater samples fulfilled those criteria. Most of the samples were from three locations: Copalis Beach (G), Heceta Head (C), and Cape Flattery (J). Corrosive samples from more northern locations (i.e., Juan de Fuca Canyon (K), Queen Charlotte Strait (M), Kildidt Sound (N), Hecate Strait (O)) were at the maximum depths for the location (Table 4).
Five bacterial and archaeal genera were identified as potential indicator taxa for corrosive waters: Colwellia (Colwelliaceae), Nitrosopumilus (Nitrosopumilaceae), Nitrosopelagicus (Nitrosopumilaceae), Sup05 cluster (Thioglobaceae), and Sva0996 marine group (Microtrichiaceae). A comparison of these genera show that they can occur in noncorrosive waters, but their relative abundances are much higher in corrosive waters (Figure 7). Comparable assessment of 18S genera failed to identify any significant eukaryotic taxa as indicators of corrosive waters (data not shown).
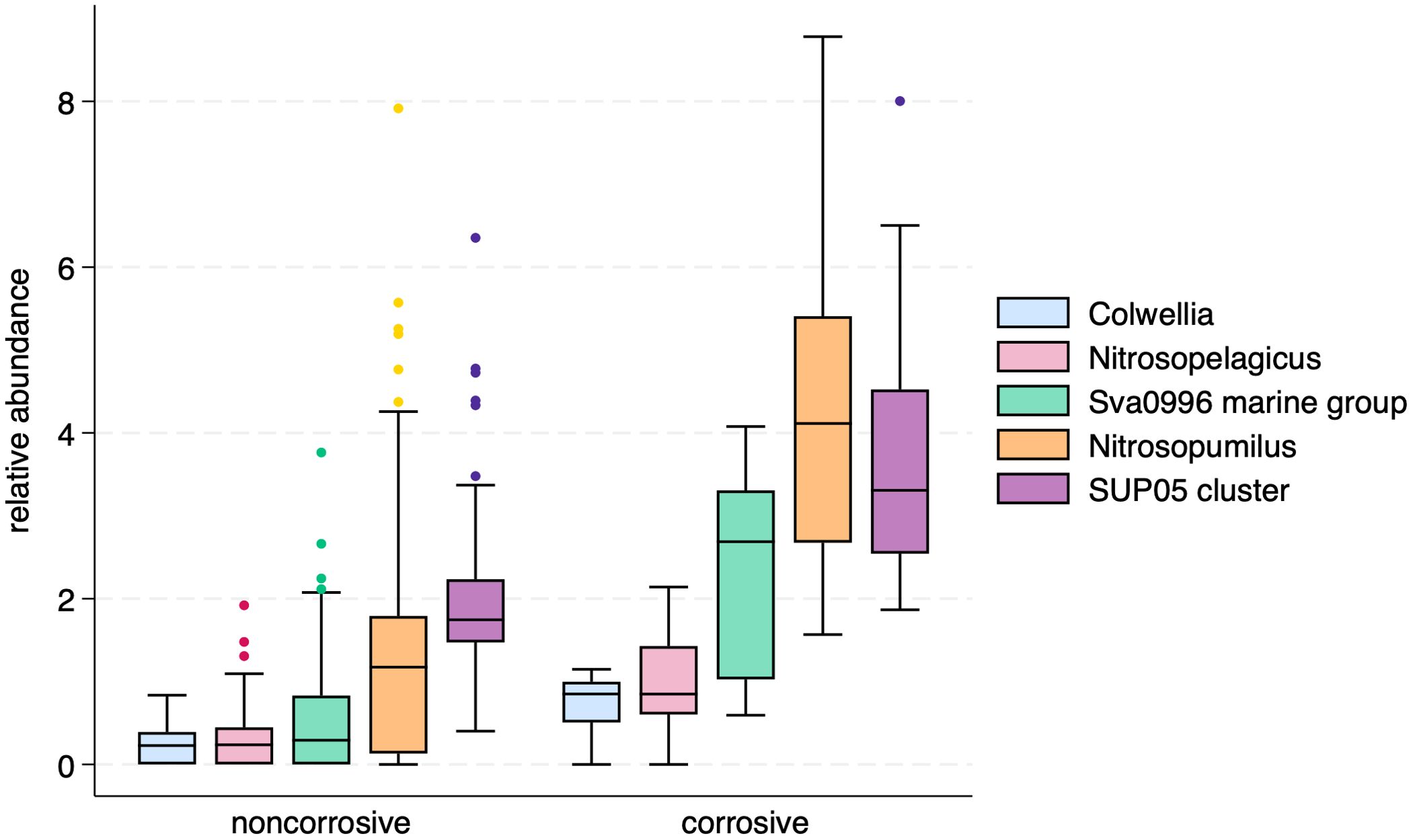
Figure 7. Box plots of relative abundances of five bacterial and archaeal indicator genera for corrosive waters.
Analysis for bacterial and archaeal genera that could be indicators for specific geographic locations found potential taxa for Cape Ferrelo (B), inside the Columbia River entrance (E), and Hecate Strait (O). The bacterial genus Thalassotalea (family Colwelliaceae) and the bacterial genus Idiomarina (family Idiomarinaceae) are potential indicator taxa for Cape Ferrelo (B) and Hecate Strait (O), respectively. For locations inside of the Columbia River entrance (E), five potential indicator taxa were identified: Sediminibacterium (family Chitinophagaceae), Pseudarcicella (family Spirosomaceae), Solitalea (family Sphingobacteriaceae), Algoriphagus (family Cyclobacteriaceae), and Flavobacterium (family Flavobacteriaceae).
Potentially pathogenic taxa
Bacterial and archaeal classification to genus revealed several potentially pathogenic bacterial taxa, and these genera included Coxiella, Francisella, Flavobacterium, and Tenacibaculum. The genus Coxiella was detected at Cape Johnson (I) and all groups north of the Strait of Juan de Fuca (L–P). Detections occurred primarily at depths > 20 m, but Coxiella was found in shallow waters (< 10 m) in the Johnstone Strait group (P). The genus Francisella had a slightly wider geographic distribution, from Heceta Head (C) and all groups northward except at Queen Charlotte Strait (C–L, N–P), and was more frequently detected at depths < 50 m. Flavobacterium was detected only in shallow waters (< 10 m) in the Columbia River estuary (E). In contrast, Tenacibaculum was detected across at all geographic groups and at all depth intervals and exhibited a decreasing gradient of abundance from southern to northern latitudes.
Sixteen eukaryotic genera considered as harmful algal bloom (HAB) taxa were detected across the geographic extent of the survey. The most frequently detected HAB taxa were Alexandrium, Gyrodinium, and Pseudo-nitzschia (Figures 8A, B). Cryptomonas occurred only in the Columbia River estuary, an observation similar to the flow cytometry results. Heterocapsa, which affects shellfish and small crustaceans, was most abundant around the western end of the Strait of Juan de Fuca (J, K), and gradually declined northward. Gynmodinium was most abundant at northern Canadian sites (M, N, P), while Dinophysis was most abundant in Johnstone Strait (P).
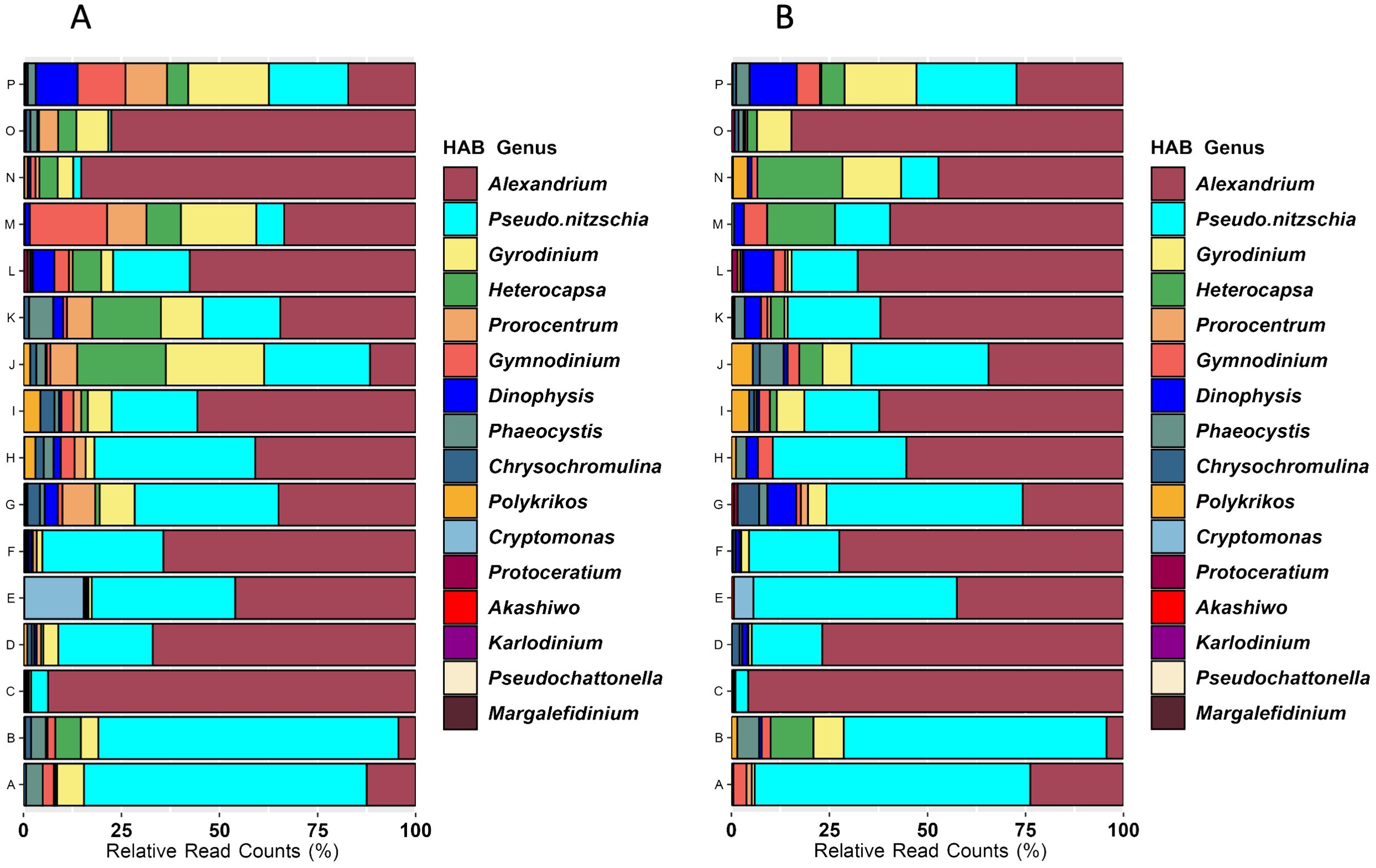
Figure 8. Average relative abundances of HAB genera detected by 18S metabarcoding by geographic group for samples from all depths (A) or from only surface samples (B). HAB genera are ordered in the legend by their abundance in the entire dataset (i.e., Alexandrium were the most abundant and Marglefidinium were the least abundant).
Among the remaining eukaryotic taxa, a number of potential parasites and parasitoids were detected (Table 5). Some parasites and pathogens exhibited limited geographic distribution (Amoebophrya ceratii, Fusiforma themisticola, Pseudochatonella spp., Thiriotia pugettiae), while others were common at most locations (e.g., Heliospora caprellae). Most of the detected parasites and parasitoids affect diatoms, dinoflagellates, or invertebrate species (Table 5).
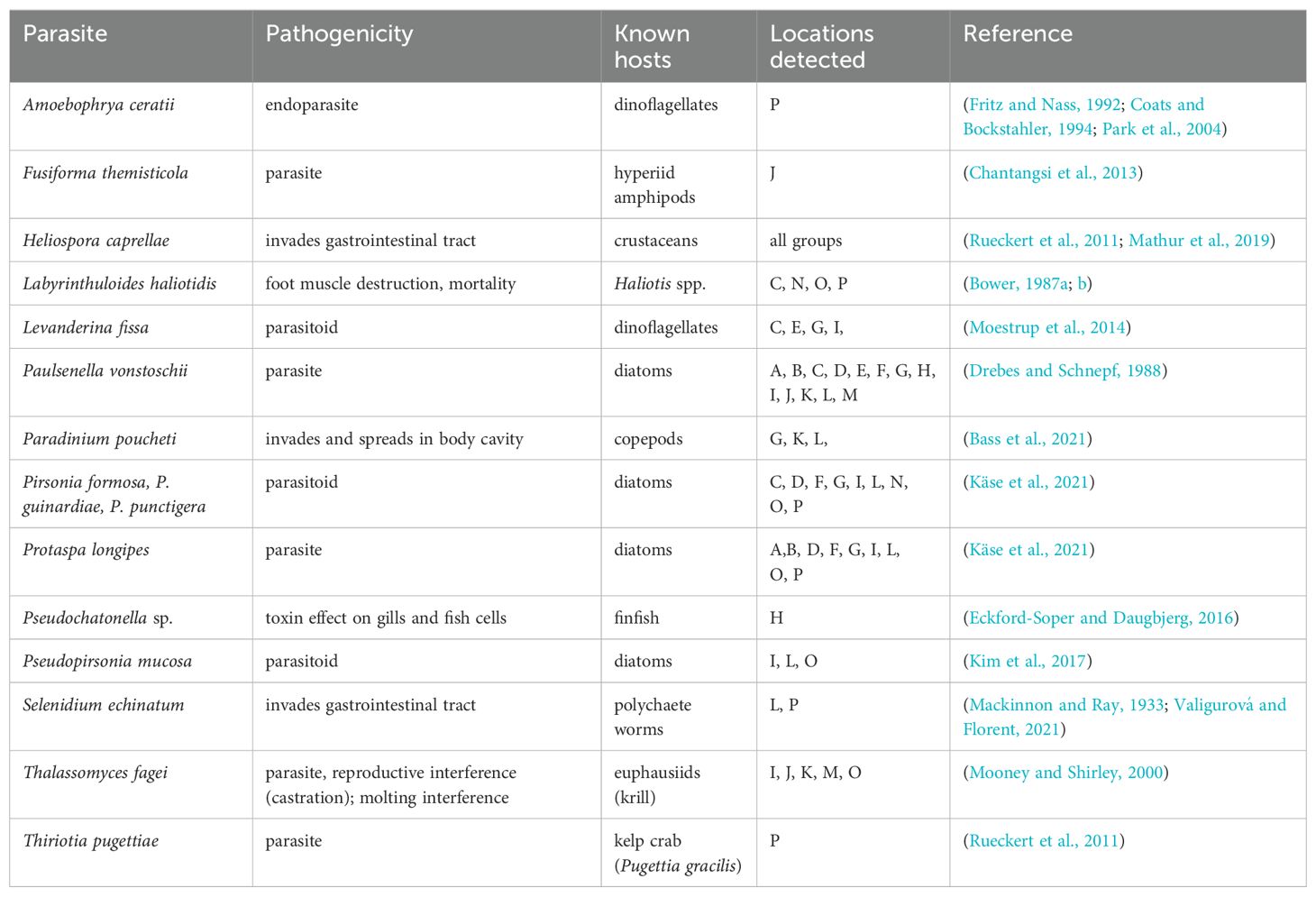
Table 5. Parasites and parasitoids detected with 18S metabarcoding, associated pathogenicity, known hosts, and geographic groups where detected.
Discussion
Seawater and microbial characteristics
The West Coast Ocean Acidification cruises are a subset of oceanographic surveys conducted since 2011 along the western coastline of the contiguous United States and portions of the Canadian west coast. Sampling for this microbial assessment was conducted in the northern portion of the cruise, ranging between latitudes 40.25°N and 52.40°N, encompassing the coastlines of northern California, Oregon, Washington, and southern British Columbia. Collecting microbial samples from the same water grabs used for analytical chemistry allows direct comparisons with microbial metabarcoding information without assumptions or adjustments for spatial or temporal differences in sampling. This direct correspondence is important for organisms with short replication periods and rapid responses to environmental conditions, such as bacteria and phytoplankton.
Overall, few physical and chemical measurements were extreme, and there was a considerable overlap in seawater profiles among locations, in spite of the large geographic range (Figure 1). Samples collected from deeper than 10 m were modestly differentiated from each other, suggesting that although seasonal stratification was in early stages, vertical mixing was still prevalent at the time of the survey (late May to early June). An El Niño was in place during the survey5, which would have weakened typical upwelling along the coast. There were no extreme temperatures, with those in shallower waters (≤ 20 m) spanning 8.9–16.2°C and waters deeper than 20 m spanning 5.7–10.6°C. Only one sample was hypoxic (< 62 µmol kg−1), and none were at eutrophic concentrations for dissolved inorganic nitrate (≥ 82.6 µmol kg−1). For shallow samples, no phosphorous or silicate limitations for diatom or other phytoplankton growth were detected. There was no evidence of primary nitrite and ammonium maxima, which often coincide and are located at the base of the euphotic zone. Instead, dissolved nitrite and ammonium concentrations were distributed throughout the water column and increased with depth, a pattern typically observed at higher latitudes where light-limited primary production coexists with high levels of nitrification (Zakem et al., 2018). Nonetheless, there were depth-dependent variations in nutrients and carbon chemistry. Deeper samples exhibited higher dissolved inorganic nitrate, phosphate, and silicate, while shallower samples exhibited higher pHT and dissolved oxygen (Figure 2; Supplementary Figures 1, 3). In general, seawater conditions across the survey sites could be considered moderate, and not under conditions associated with strong upwelling, marine heatwaves, or hypoxia.
Commonalities in bacterial and archaeal communities occurred across the geographic span of the survey. Proteobacteria were dominant at all locations and at all depths, and Bacteroidetes and Cyanobacteria were highly abundant at depths ≤ 50 m. The high abundances for Proteobacteria were due primarily to the families Rhodobacteriaceae and SAR11 clades, and for Bacteroidetes were due to the family Flavobacteriaceae. These families occur at high abundances in coastal northwest Atlantic waters, with Flavobacteriaceae associated with the particle-associated fraction and Rhodobacteriaceae and SAR11 clades associated with the free-living fraction (Zorz et al., 2019). They are also dominant in offshore northeast Pacific waters at depths < 50 m, and were identified as indicator taxa for the years preceding the marine heatwave of 2014–2015 (Traving et al., 2021). Flavobacteriaceae is considered central in maintaining multiple and variable microbial relationships as surface waters transition through seasonal temperature fluxes in northwest Pacific coastal waters (Chun et al., 2021). In our study, there was a strong association between Flavobacteriaceae and Rhodobacteriaceae in waters shallower than 50 m, with Cryomorphaceae (another Flavobacteriales family) added to the association in the midwater depths, and subsequent replacement of Flavobacteriaceae by Cryomorphaceae and Halieaceae at depths > 50 m. Cryomorphaceae are highly abundant across the seasons in temperate surface waters (< 10 m; Korlević et al., 2022), and nutrient requirements of cultured isolates suggest the genus relies on enriched organic sources, likely contributing to secondary production (Bowman, 2014). Halieaceae (also known as NOR5/OM60 clade) have a world-wide coastal distribution, are highly abundant at the surfaces of intertidal sediments (Yan et al., 2009), and based on analysis of cultured isolates, are capable of chemotrophic or heterotrophic metabolism including utilization of polysaccharides from phytoplankton blooms (Li et al., 2023).
At depths > 100 m, the second most abundant phylum was the archaeal Thaumarchaeota (also known as Marine Group I archaea), composed of the family Nitrosopumilaceae and two genera, Nitrosopumilus and Nitrosopelagicus. Both genera are characterized as chemolithoautotrophic ammonia oxidizers with the ability to fix inorganic carbon (such as bicarbonate), broad salinity tolerance, and pH optima ≤ 7.3 (Könneke et al., 2005; Santoro et al., 2015; Qin et al., 2017). Originally considered to inhabit extreme environments such as oxygen minimum zones, Nitrosopumilus and Nitrosopelagicus isolates have been cultured from coastal and open ocean habitats and from photic zone to ocean depths, indicating a more cosmopolitan distribution (Qin et al., 2014; Santoro et al., 2015; Qin et al., 2017). The high metabolic activity, flexible physiology (i.e., mixotrophy), and tolerance of low pH and low dissolved oxygen may position these archaea to be winners under conditions of ocean acidification, and we identified both genera as indicator taxa for potentially corrosive seawater conditions (pH ≤ 7.75, DIC > 2190 µmol kg−1).
Among phytoplankton, Prochlorococcus cell abundance exceeded Synechococcus and eukaryotic nano- and picophytoplankton abundances by at least an order of magnitude (Figure 4). This pattern was contrary to the generalized expectation of greater Prochlorococcus abundance in oligotrophic waters and greater Synechococcus abundance in higher nutrient, cooler coastal waters (Pierella Karlusich et al., 2020), and the absence of obvious upwelling conditions during the cruise may have favored the former cyanobacterium. Prochlorococcus cells were well distributed across depth and in water temperatures as low as 6°C, reminiscent of the HLI and LLI ecotypes that are adapted to high and low light levels, respectively, and that occur at higher latitudes (Partensky and Garczarek, 2010). Ecogenomic analysis of Prochlorococcus is an emerging research area with potential for discovering strains with previously undescribed physiological tolerances (Partensky and Garczarek, 2010).
Among zooplankton and non-phytoplanktonic eukaryotic microbes, several taxa were present at high relative abundances across the geographic extent of the cruise and in the depth profiles, suggesting these are important components of community structures. The taxa Chrysophyceae (golden algae), Litostomatea and Spirotrichea (ciliates), and Filosa-Thecofilosea (flagellates) span size ranges from 0.4 µm to greater than 20 µm and are dominant in Arctic melt pond communities (Xu et al., 2020). The depth associations of certain eukaryotic microbes in this survey were supported by known physiology. For example, the ciliated protozoans Trimyema (class Plagiopylea) detected in deeper water are known anaerobes with mitochondrial modifications to produce H2 which is consumed by Trimyema’s endosymbiotic methanogens (Lewis et al., 2018). Some eukaryotic microbes with saprotrophic lifestyles, such as Labyrinthulomycetes, were abundant throughout the geographic extent of the survey. Temporal studies of this eukaryotic class found time-lagged correlations with highly abundant bacterial phylum Flavobacteriaceae, cyanobacteria Synechococcus, algae, and fungi (Xie et al., 2021), emphasizing the multiple processes they can play in moving nutrients by heterotrophy and remineralization through marine food webs. One abundant taxon of eukaryotic microbes across all survey locations was the gregarines (Gregarinomorphea; Supplementary Figure 13), well known apicomplexans that parasitize invertebrates. Assessments of the effects of gregarines on their host (e.g., arthropod, mollusks, worms) reveal that there is a spectrum of relationships, from beneficial to harmful, which may contribute to their widespread occurrence and high diversity (Rueckert et al., 2019).
Assessment for co-occurrences among 16S and 18S phyla identified two networks of interactions, one involving highly abundant taxa that occurred across the depth profiles and one involving less abundant taxa that were more abundant in deeper waters (Figure 6). For the former network, bacteria and archaea were strong nodes for associations, with one group of phyla (Actinobacteria, Bacteroidetes, Cyanobacteria, Euryarchaeota, Verrucomicrobia) forming a tightly associated group and Proteobacteria serving as a separate strong node. A translatitudinal survey of the Atlantic Ocean using 16S and 18S metabarcoding found that Proteobacteria were a highly associated node for the free-living fraction, and Bacteroidetes and Verrucomicrobia were highly associated node for particle-associated fractions (Milici et al., 2016), indicating these taxa are central to microbial networks in both nearshore and open ocean waters. Interestingly, dominance of the nearshore network by bacteria and archaea is more similar to the Southern Ocean waters than the other open oceanographic provinces (Lima-Mendez et al., 2015), which may be a reflection of seasonal high productivity in temperate nearshore waters and in the Southern Ocean. The strongest nodes in the second network were the eukaryotic Perkinsea, Lobosa, and Streptophyta and the bacteria Acidobacteria and PAUC34f. Perkinsea include well described parasitic protists that can infect dinoflagellates, mollusks, and finfish, while Lobosa are mostly free-living amoeba that move by pseudopodial extensions. Streptophyta include some green algae and land plants, the latter presumably transported into the ocean by terrestrial runoff and rivers. The absence of an understandable functional association among these taxa suggests that co-occurrences might be driven primarily by physical factors, rather than biological interactions.
Geographic patterns
Eukaryotic phytoplankton exhibited stronger differentiation across the geographic groups than bacteria and archaea (Figure 5). The patterns in eukaryotic phytoplankton community structures can result from physical factors such as wind stress and ocean currents operating in combination with mesoscale oceanographic features such as canyons and promontories (Hickey and Banas, 2003). Submarine banks and canyons contribute to the well documented retentive features of the Juan de Fuca eddy, located near the coast of Washington State and Vancouver Island, and Heceta Head, located near the central coast of Oregon State. For example, eukaryotic phytoplankton communities from Destruction Island through Barkley Sound (groups H through L, Figure 1) displayed similarity with each other, suggesting a retentive influence by the gently sloping coastal bathymetry and the Juan de Fuca eddy (Figure 5B). Terrestrial inputs such as river plumes, and especially the Columbia River, also influence phytoplankton community structure via nutrient input, hydraulically trapping cells near the coast, or acting as a conduit for community transport (Hickey et al., 2013). The potential significance of physical transport for both nutrients and particles is reinforced by the pattern of community similarity among sites proximal to each other, such as the Columbia River plume and Copalis Beach (F and G), Kildidt Sound and Hecate Strait (N and O), entrance to Queen Charlotte Strait and Johnstone Strait (M and P); Figure 5B). Another contributing factor to the observed patterns of differentiation could be latitudinal variation in light intensity and cloud cover. The structure of ocean seascapes, which are analogous to terrestrial landscapes, can also influence the composition of microbial communities in localized areas (Kavanaugh et al., 2016).
The northern locations (groups L through P) exhibited shared microbial features, in spite of the large spatial separation among sites and geomorphic differences in adjacent coastal uplands. Bacterial and archaeal taxon richness was consistently and significantly higher at northern locations (Supplementary Figure 6A), and there was high similarity in bacterial and archaeal community structures (Figure 5A). The shortest water-based distance between Barkley Sound (L) and Hecate Strait (O) is more than 500 km, and the sites leading to and within Johnstone Strait (P) are closely bounded by land rather than exposed to open ocean locations (L through O). Although the microbial community similarity might be driven by similarity in ocean conditions, this was not reflected in the principal components analysis of the seawater parameters (Figure 2), suggesting unmeasured factors (biotic and/or abiotic) may have contributed to community homogeneity. Studies of oligotrophic open ocean communities using drifters to track changes associated with moving water parcels have found community homogeneity over distances ranging from a few km up to approximately 50 km (Hewson et al., 2006). The stretch of water from Barkley Sound to Hecate Strait is east of the division of the North Pacific Current into the Alaska Current (turning north) and the California Current (turning south), potentially placing this region beyond the main influence of these large ocean currents or even the Haida Eddies that aggregate and transport coastal water and plankton (Peterson et al., 2011). The oceanography of Hecate Strait and Queen Charlotte Strait indicates complex horizontal recirculation of waters at multiple depths (Crawford et al., 1995), which could result in transport, mixing, and possible retention of water masses. Given the single timepoint nature of this survey, the observed homogeneity may have been serendipitous, and seasonal transitions are expected to induce greater microbial heterogeneity.
In contrast to shared patterns in bacterial and archaeal communities among the northern locations, there was stronger geographic differentiation for southern locations. Communities at the two southernmost locations (Cape Mendocino and Cape Ferrelo, A and B, respectively) were strongly different from each other and all of the other locations (Figure 5). Both of these locations displayed narrow variations in temperature and salinity (Figures 3B, C), and Cape Ferrelo had low taxon diversity and evenness (Supplementary Figures 6B, C). Thalassotalea (Colwelliaceae) was identified as an indicator taxon for Cape Ferrelo, and the genus occurred nearly exclusively at the four southernmost locations (A–D) at depths > 10 m. Thalassotalea emerged as an important component of petroleum degradation during the 2010 Deepwater Horizon incident (Noirungsee et al., 2020), and genome analysis indicates that the genus has good capacity for metabolizing complex organic molecules, such as macroalgal-derived polysaccharides, and likely plays an important role in nutrient cycling (Kim et al., 2020).
Microbial communities from the Columbia River plume (F) and Copalis Beach (G) resembled each other but were distinctive from other locations (Figure 5). These two locations are approximately 100 km from each other, and each is influenced by substantial fresh water outflows from the Columbia River (average ~7,500 m3 s−1) and from the Chehalis (average ~180 m3 s−1) and Copalis Rivers (average ~80 m3 s−1), respectively. Historically and as recently as 2021, nearshore waters between the Columbia River and Copalis Beach are subject to seasonal hypoxia, driven by upwelling, variable water sources, and/or biochemical oxygen removal (Connolly et al., 2010; Peterson et al., 2013; Barth et al., 2024). Although their microbial communities were similar, the seawater features of these two locations were different, with Copalis Beach distinctive from all other locations (Figure 2A). Its samples contained high levels of dissolved nitrite and ammonium, relative to other locations (Supplementary Figure 4), as well as high levels of dissolved inorganic phosphate, low pHT, and low dissolved oxygen. The elevated concentrations of nitrite and ammonium in the presence of low dissolved oxygen suggests suitable conditions for anaerobic ammonium oxidation, or anammox, at this location (Zehr and Ward, 2002), but relative abundances of ammonia-oxidizing archaea (e.g., Thaumarchaeota) were low, and no ammonia-oxiding bacteria were identified. Elevated nitrite and ammonium concentrations can be due to local river inputs (e.g., agricultural runoff), atmospheric deposition (e.g., combustion byproduct), elevated rates of remineralization of organic matter, altered rates of nitrification and/or denitrification (e.g., due to hypoxia), or combinations of these factors (Voss et al., 2011). The adjacent uplands are forested areas with no significant agricultural activities or urbanization, and there is no obvious source or cause for the elevated ammonium and nitrite at this location.
Cape Flattery (J) was another geographic group that was distinctive: low levels of dissolved oxygen (Figure 3D); low pHT/high DIC seawater (Figure 3E); low absolute abundances of bacteria, archaea, and phytoplankton (Figure 4); and greater abundances of bacterial and archaea indicators for potentially corrosive waters (Table 4). Cape Flattery is at the intersection of water flows from the Strait of Juan de Fuca and the along-coast Davidson and California currents, and it is located at the periphery of the Juan de Fuca (or Tully) eddy. Modest upwelling can occur there (Hickey and Banas, 2003) which may have contributed to the observed seawater chemistry, but the relatively low abundances of microorganisms suggest low water retention, perhaps due to rapid or frequent movement in water masses.
The survey included several sampling locations within the Columbia River estuary (E), and, not surprisingly, the community compositions were strikingly different from other locations (Figure 5; Supplementary Figure 7). An extensive spatiotemporal and depth assessment by Fortunato et al. (2012) along the Columbia River through the estuary and plume and to the shelf edge detected distinctive spatial groups that obscured temporal variability (Fortunato et al., 2012). Our analysis found five genera that were indicators for the Columbia River estuary locations, and these taxa have a variety of geographic preferences and metabolic lifestyles. Sediminibacterium (family Chitinophagaceae), Pseudarcicella (family Spirosomaceae), and Algoriphagus (family Cyclobacteriaceae) are members of families associated with freshwater sediments and soils, and more recently, with microplastics (Miao et al., 2019; Pinnell and Turner, 2020; Rogers et al., 2020; Wright et al., 2021), and their presence is most probably linked to the estimated 5 million tons of sediment annually transported to the Pacific Ocean by the river6. The genus Solitalea (family Sphingobacteriaceae), which was detected only within the Columbia River estuary (E) and the adjacent coastal plume (F), may play a role in ammonium regulation, which is useful for wastewater treatment (Pang et al., 2022). The genus Flavobacterium (family Flavobacteriaceae) was detected only within the estuary, and potential significance is discussed in the pathogens section below.
Archaea were a prominent component of most communities, with Euryarchaeota and Thaumarchaeota each comprising ~5.5% of the overall sequence abundances, while Nanoarchaeota contributed < 0.1%. All Euryarchaeota were classified as Marine Group II, and this taxon was absent or at low relative abundance south of Newport (D) and detected at up to nearly 40% at depths < 20 m. In contrast to Thaumarchaeota, Marine Group II archaea are well distributed in photic zones and abundant in metabolically active fractions of the microbial community, strongly indicating a role in pelagic biogeochemical cycling (Zhang et al., 2015; Wemheuer et al., 2019). All Thaumarchaeota we detected were classified as Nitrosopumilaceae, including the genera Nitrosopumilus and Nitrosopelagicus, and exhibited a depth-related distribution. This family, well characterized as ammonia-oxiding archaea, is commonly associated with deeper waters and sediments, and capable of thriving under hypoxic or low pH conditions (Qin et al., 2017; Campbell et al., 2019). The prominent position of archaea in this survey without environmental extremes supports their wider role in nutrient cycle and ready availability when conditions change, such as upwelling events.
Potential pathogens and harmful algae
Although Flavobacteriaceae occurred at all locations and ten genera were identified, the genus Flavobacterium was detected only at locations in the Columbia River estuary (E). This genus is found in a wide variety of habitats and temperature ranges from freshwater to seawater (Loch and Faisal, 2015). Several species of Flavobacterium (F. columnare, F. psychrophilum, F. branchiophilum) are well characterized pathogens of finfish, including salmon (Starliper, 2011; Declercq et al., 2013). Over the past 20 years (2002–2022), an annual average of ~1.5 million adult salmon and ~7.9 million juvenile salmon migrate through the Columbia River estuary (Columbia River DART, 2023). At the time of our sampling (May 29, 2016), ~151,000 adult salmon and ~6.5 million juvenile salmon had passed into the estuary during that year (Columbia River DART, 2023). It is likely that the high numbers of salmon contributed to the Flavobacterium abundance at this location, and conversely, the estuary could represent a location where horizontal transmission of Flavobacterium among salmon could occur.
In contrast to Flavobacterium, the genus Tenacibaculum was detected in every sample and at every depth, with highest relative abundances at Destruction Island (H). Multiple species of Tenacibaculum cause disease (called tenacibaculosis) in a wide variety of finfish including pelagic (e.g., salmon, amberjack, sea bream) and epibenthic (e.g., sole, flounder; (Avendaño-Herrera et al., 2006; Fernández-Álvarez and Santos, 2018) fish. Although there are species of Tenacibaculum that do not have demonstrated disease-causing abilities, genomic analyses show that pathogenic and non-pathogenic strains are closely intertwined and there may be homologous recombination between species (Habib et al., 2014). For marine aquaculture, the genetic evidence for endemic colonization by Tenacibaculum, rather than introduction by long distance fish movements (Habib et al., 2014), suggests their ubiquity could pose a persistent challenge during growing operations.
The Francisella genus includes several species, including isolates from diseased marine shellfish and finfish that were originally classified as subspecies of known zoonotic agents (Brevik et al., 2011; Colquhoun and Duodu, 2011; Ramirez-Paredes et al., 2020). Although isolates from marine infections belong to the genus containing well-known human pathogens F. tularensis and F. philomiragia, the risk of zoonotic disease has been considered low (Birkbeck et al., 2011; Colquhoun and Duodu, 2011). One feature of the fish pathogen F. noatunensis is the ability to enter a low metabolic but persistent state termed viable but not culturable (VBNC; Duodu and Colquhoun, 2010). Induction of the VBNC state is usually in response to stressors such as starvation or unfavorable temperatures, and many pathogens such as Vibrio spp. adopt this strategy, allowing longer-term persistence in the environment (Oliver, 2010). Similar to Tenacibaculum, Francisella was detected at all depths and at all locations except the southernmost locations (Cape Mendocino, A; Cape Ferrelo, B) and Queen Charlotte Strait, M). The highest abundances occurred at Johnstone Strait (P), Destruction Island (H), Barkley Sound (L), and Cape Johnson (I).
In addition to fish pathogens, one potentially zoonotic genus was detected: Coxiella. The single species of Coxiella, C. burnetii, is a strictly intracellular bacterium and the causative agent of Q fever, which can result in abortions and still births in animals and humans (Eldin et al., 2016). C. burnetii infects a wide range of organisms (arthropods, reptiles, birds, and mammals including humans), and occurs in terrestrial, freshwater, and marine environments (Eldin et al., 2016). Among marine mammals, both pinnipeds and cetaceans can be infected, and population surveys found seroprevalence ranging from 24–50% from the outer coasts of central Oregon and southern Washington and the inland waters of the Salish Sea (Kersh et al., 2012), and as high as 80% among Alaskan pinnipeds (Minor et al., 2013). Infections among terrestrial agriculture animals are common (Eldin et al., 2016), and birds could serve as vectors through feeding on infected placenta (Gardner et al., 2023). In our survey, Coxiella was not detected south of Cape Johnson (I), with highest abundances in Canadian waters in Kildidt Sound (N), Hecate Strait (O), and Johnstone Strait (P). Recent efforts to employ genotyping to differentiate terrestrial from marine mammal-associated strains may enable a better understanding of the sources of Coxiella in marine environments (Gardner et al., 2022).
Microalgae capable of producing biotoxins or causing harm under bloom conditions were widely detected both geographically and across the depth profiles, and the identified taxa are well documented as problematic for seafood safety, marine organisms (including aquacultured and fisheries harvested species), and the marine ecosystem (Bates et al., 2020; Anderson et al., 2021; McKenzie et al., 2021). Alexandrium was a frequently detected HAB, with high abundances at northern Canadian locations (Kildidt Sound (N), Hecate Strait (O)), at the Columbia River plume (F), and at Heceta Head (C). As a dinoflagellate that includes a benthic resting cyst stage in its life history and vertical mobility of the vegetative stage, Alexandrium detection and distribution is likely to be greatly affected by temperature (controlling excystment and growth) and salinity (for horizontal transport; Anderson, 1997). Pseudo-nitzschia was also a dominant HAB detected in the survey, and eleven of the fifteen known toxic species are problematic along the west coast of the U.S. and Canada (Bates et al., 2020; Anderson et al., 2021; McKenzie et al., 2021). Our survey was conducted approximately eight months after the end of a severe bloom of Pseudo-nitzschia spp. extending from northern British Columbia, Canada to California in the U.S (McCabe et al., 2016). Fifteen years of intense documentation of Pseudo-nitzschia in the Southern California Bight was able to identify general oceanographic patterns favorable for bloom formation, but factors affecting distribution, magnitude, and toxin production are not yet known (Smith et al., 2018). Although the Heceta Bank eddy (C) and the Juan de Fuca eddy (K) are known “hot spots” for Pseudo-nitzschia blooms (Lewitus et al., 2012), the diatom genus was not at high relative abundance at those locations at the time of this survey (Figure 8). Because Pseudo-nitzschia impacts the entire west coast of North America (Lewitus et al., 2012), targeted studies of bloom vs favorable but non-bloom conditions at such known “hot spots”, including associated eukaryotic microbial communities, is likely to yield better information about drivers of this diatom’s dynamics. Gyrodinium, a dinoflagellate capable of mass mortalities in shellfish and finfish, was a prominent HAB in a quarter of the geographic groups, around Cape Flattery and the Juan de Fuca canyon (J, K), Queen Charlotte Strait (M), and Johnstone Strait (P; Figure 8). Other HAB taxa with moderate abundances such as Heterocapsa, Gymnodinium, Chrysochomulina are known hazards for marine finfish and shellfish, especially those in aquaculture facilities (Karlson et al., 2021).
Conclusion
Nearshore marine environments are highly dynamic due to the influence of oceanic factors such as upwelling and terrestrial forces such as river inputs. Microbial communities in these nearshore areas can reflect ocean conditions due to their tight physiological connections with physical and chemical environmental parameters. The synoptic view of the 2016 West Coast Ocean Acidification survey provided a wide geographic perspective within a limited time frame (i.e., late spring-early summer) when upwelling is likely to occur. Seawater parameters revealed no widespread, extreme ocean conditions across the geographic extent of the survey, although some evidence of local upwelling was found at discrete locations along the Oregon and Washington coasts. Characterizations of microbial communities show a more common community structure for bacteria and archaea in more northern locations (i.e., north of latitude 48°N), but more differentiated community structures across the entire survey for eukaryotic microbes. Indicator taxa for potentially corrosive seawater were found among bacteria and archaea that are typical for deeper ocean waters, but no indicator taxa were identified among the eukaryotic microbes. Potentially pathogenic microbes, such as harmful microalgae, pathogenic bacteria, and microbial parasites were detected, but these were not associated with potentially corrosive seawater conditions. Surveys with closely linked chemical and biological sampling, such as this one, offer insights into the alignment of lower trophic levels with seawater parameters and link the ecology of microbes to impending changes in ocean conditions.
Data availability statement
The datasets presented in this study can be found in the following online repositories. Physical and chemical seawater data are archived at NOAA’s National Centers for Environmental Information (NCEI Accession 0169412; https://www.ncei.noaa.gov/access/metadata/landing-page/bin/iso?id=gov.noaa.nodc:0169412). Sequencing reads are archived at the National Center for Biotechnology Information (NCBI BioProject PRJNA1018955; https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1018955). Nonsequence biological data are archived at NOAA’s National Centers for Environmental Information (NCEI Accession number 0265154; https://www.ncei.noaa.gov/access/metadata/landing-page/bin/iso?id=gov.noaa.nodc:0265154).
Author contributions
LR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. NA: Data curation, Formal analysis, Investigation, Project administration, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. RGS: Formal analysis, Software, Validation, Visualization, Writing – review & editing. MK: Data curation, Formal analysis, Investigation, Resources, Validation, Writing – review & editing, Methodology. SA: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing. RF: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Northwest Fisheries Science Center (NOAA Fisheries) provided funding and resource support, and NOAA’s Ocean Acidification Program (NOAA; project 21405, ROR # 02bfn4816) provided funding support. The NOAA PMEL contribution number is 5615.
Acknowledgments
The authors are grateful for the contributions of Dana Greeley (PMEL/NOAA) for seawater data management; William Nilsson (NOAA Fisheries) for assistance in sample collection; and Kelly George (CEOAS/Oregon State University) for flow cytometry data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the edits, and the reviewers. The use of trade, firm, or corporation names is the for information of the reader, and does not constitute an official endorsement or approval by the U.S. Government
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1430930/full#supplementary-material
Footnotes
- ^ https://www.pmel.noaa.gov/co2/story/2016+West+Coast+Ocean+Acidification+Cruise
- ^ https://www.ncei.noaa.gov/access/metadata/landing-page/bin/iso?id=gov.noaa.nodc:0169412
- ^ https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1018955
- ^ https://www.ncei.noaa.gov/access/metadata/landing-page/bin/iso?id=gov.noaa.nodc:0265154
- ^ https://psl.noaa.gov/enso/dashboard.html
- ^ https://www.usgs.gov/programs/cmhrp/news/new-research-informs-dredging-efforts-columbia-river-mouth-conserving-valuable
References
Ahyong S., Boyko C. B., Bailly N., Bernot J., Bieler R., Brandão S. N., et al. (2023). World Register of Marine Species (WoRMS) (Ostend, Belgium: WoRMS Editorial Board).
Alin S. R., Feely R. A., Hales B., Byrne R. H., Cochlan W., Liu X., et al. (2017). Dissolved inorganic carbon, total alkalinity, pH on total scale, and other variables collected from profile and discrete sample observations using CTD, Niskin bottle, and other instruments from NOAA Ship Ronald H. Brown in the U.S. West Coast California Current System from 2016-05-08 to 2016-06-06 (NCEI Accession 0169412) (Silver Spring, Maryland, USA: NOAA National Centers for Environmental Information). doi: 10.7289/v5v40shg
Allers E., Gómez-Consarnau L., Pinhassi J., Gasol J. M., Šimek K., Pernthaler J. (2007). Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ. Microbiol. 9, 2417–2429. doi: 10.1111/j.1462-2920.2007.01360.x
Anderson D. M. (1997). Bloom dynamics of toxic Alexandrium species in the northeastern U.S. Limnology Oceanography 42, 1009–1022. doi: 10.4319/lo.1997.42.5_part_2.1009
Anderson D. M., Fensin E., Gobler C. J., Hoeglund A. E., Hubbard K. A., Kulis D. M., et al. (2021). Marine harmful algal blooms (HABs) in the United States: History, current status and future trends. Harmful Algae 102, 101975. doi: 10.1016/j.hal.2021.101975
Anderson M. J., Gorley R. N., Clarke K. R. (2008). PERMANOVA+ for PRIMER: guide to software and statistical methods (Plymouth, UK: PRIMER-E).
Anderson M. J., Willis T. J. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84, 511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2
Avendaño-Herrera R., Toranzo A. E., Magariños B. (2006). Tenacibaculosis infection in marine fish caused by Tenacibaculum maritimum: a review. Dis. Aquat. Organisms 71, 255–266. doi: 10.3354/dao071255
Azam F., Fenchel T. M., Field J. G., Gray J. S., Meyer-Reil L.-A., Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Barth J. A., Pierce S. D., Carter B. R., Chan F., Erofeev A. Y., Fisher J. L., et al. (2024). Widespread and increasing near-bottom hypoxia in the coastal ocean off the United States Pacific Northwest. Sci. Rep. 14, 3798. doi: 10.1038/s41598-024-54476-0
Bass D., Rueckert S., Stern R., Cleary A. C., Taylor J. D., Ward G. M., et al. (2021). Parasites, pathogens, and other symbionts of copepods. Trends Parasitol. 37, 875–889. doi: 10.1016/j.pt.2021.05.006
Bates S. S., Beach D. G., Comeau L. A., Haigh N., Lewis N. I., Locke A., et al. (2020). “Marine harmful algal blooms and phycotoxins of concern to Canada,” in Canadian Technical Report of Fisheries and Aquatic Sciences (Fisheries and Oceans Canada, Moncton, New Brunswick, Canada).
Bayer B., Vojvoda J., Offre P., Alves R. J. E., Elisabeth N. H., Garcia J. A. L., et al. (2016). Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J. 10, 1051–1063. doi: 10.1038/ismej.2015.200
Bednaršek N., Tarling G. A., Bakker D. C. E., Fielding S., Feely R. A. (2014). Dissolution dominating calcification process in polar pteropods close to the point of aragonite undersaturation. PLoS One 9, e109183. doi: 10.1371/journal.pone.0109183
Biard T. (2022). Diversity and ecology of Radiolaria in modern oceans. Environ. Microbiol. 24, 2179–2200. doi: 10.1111/1462-2920.16004
Birkbeck T. H., Feist S. W., Verner-Jeffreys D. W. (2011). Francisella infections in fish and shellfish. J. Fish Dis. 34, 173–187. doi: 10.1111/j.1365-2761.2010.01226.x
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bower S. M. (1987a). Artificial culture of Labyrinthuloides haliotidis (Protozoa: Labyrinthomorpha), a pathogenic parasite of abalone. Can. J. Zoology 65, 2013–2020. doi: 10.1139/z87-306
Bower S. M. (1987b). Labyrinthuloides haliotidis n.sp. (Protozoa: Labyrinthomorpha), a pathogenic parasite of small juvenile abalone in a British Columbia mariculture facility. Can. J. Zoology 65, 1996–2007. doi: 10.1139/z87-304
Bowman J. (2014). “The family cryomorphaceae,” in The Prokaryotes: Other Major Lineages of Bacteria and The Archaea, (Heidelberg, Germany: Springer Link) 539–550. doi: 10.1007/978-3-642-38954-2_135
Brevik O. J., Ottem K. F., Kamaishi T., Watanabe K., Nylund A. (2011). Francisella halioticida sp. nov., a pathogen of farmed giant abalone (Haliotis gigantea) in Japan. J. Appl. Microbiol. 111, 1044–1056. doi: 10.1111/j.1365-2672.2011.05133.x
Buchan A., LeCleir G. R., Gulvik C. A., González J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Calbet A., Landry M. R. (2004). Phytoplankton growth, microzooplankton grazing, and carbon cycling in marine systems. Limnology Oceanography 49, 51–57. doi: 10.4319/lo.2004.49.1.0051
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Campbell L. G., Thrash J. C., Rabalais N. N., Mason O. U. (2019). Extent of the annual Gulf of Mexico hypoxic zone influences microbial community structure. PLoS One 14, e0209055. doi: 10.1371/journal.pone.0209055
Chan F., Barth J. A., Blanchette C. A., Byrne R. H., Chavez F., Cheriton O., et al. (2017). Persistent spatial structuring of coastal ocean acidification in the California Current System. Sci. Rep. 7, 2526. doi: 10.1038/s41598-017-02777-y
Chantangsi C., Lynn D. H., Rueckert S., Prokopowicz A. J., Panha S., Leander B. S. (2013). Fusiforma themisticola n. gen., n. sp., a new genus and species of apostome ciliate infecting the hyperiid amphipod Themisto libellula in the Canadian Beaufort Sea (Arctic Ocean), and establishment of the Pseudocolliniidae (Ciliophora, Apostomatia). Protist 164, 793–810. doi: 10.1016/j.protis.2013.09.001
Chun S.-J., Cui Y., Baek S. H., Ahn C.-Y., Oh H.-M. (2021). Seasonal succession of microbes in different size-fractions and their modular structures determined by both macro- and micro-environmental filtering in dynamic coastal waters. Sci. Total Environ. 784, 147046. doi: 10.1016/j.scitotenv.2021.147046
Coats D. W., Bockstahler K. R. (1994). Occurrence of the Parasitic Dinoflagellate Amoebophrya ceratii in Chesapeake Bay Populations of Gymnodinium sanguineum. J. Eukaryotic Microbiol. 41, 586–593. doi: 10.1111/j.1550-7408.1994.tb01520.x
Colquhoun D. J., Duodu S. (2011). Francisella infections in farmed and wild aquatic organisms. Vet. Res. 42, 47. doi: 10.1186/1297-9716-42-47
Columbia River DART (2023). Adult Passage Counts Graphics & Text; Smolt Index Graphics & Text (Seattle, Washington, USA: Columbia Basin Research, University of Washington).
Connolly T. P., Hickey B. M., Geier S. L., Cochlan W. P. (2010). Processes influencing seasonal hypoxia in the northern California Current System. J. Geophys Res. 115, C03021. doi: 10.1029/2009jc005283
Crawford W. R., Woodward M. J., Foreman M. G. G., Thomson R. E. (1995). Oceanographic features of hecate strait and queen charlotte sound in summer. Atmosphere-Ocean 33, 639–681. doi: 10.1080/07055900.1995.9649549
Declercq A. M., Haesebrouck F., Van den Broeck W., Bossier P., Decostere A. (2013). Columnaris disease in fish: a review with emphasis on bacterium-host interactions. Veterinary Res. 44, 27. doi: 10.1186/1297-9716-44-27
de Vargas C., Audic S., Henry N., Decelle J., Mahé F., Logares R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605. doi: 10.1126/science.1261605
Dickson A. G. (1990). Standard potential of the reaction: AgCl(s) + 12H2(g) = Ag(s) + HCl(aq), and and the standard acidity constant of the ion HSO4– in synthetic sea water from 273.15 to 318.15 K. J. Chem. Thermodynamics 22, 113–127. doi: 10.1016/0021-9614(90)90074-Z
Dickson A. G., Goyet C. (1994). Handbook of methods for the analysis of the various parameters of the carbon dioxide system in sea water. Version 2 (Washington, D.C., USA: Oak Ridge National Lab (ORNL).
Dolan J. (2010). Morphology and ecology in tintinnid ciliates of the marine plankton: Correlates of lorica dimensions. Acta Protozoologica 49, 235–244.
Drebes G., Schnepf E. (1988). Paulsenella Chatton (Dinophyta), ectoparasites of marine diatoms: development and taxonomy. Helgoländer Meeresuntersuchungen 42, 563–581. doi: 10.1007/BF02365627
Duodu S., Colquhoun D. (2010). Monitoring the survival of fish-pathogenic Francisella in water microcosms. FEMS Microbiol. Ecol. 74, 534–541. doi: 10.1111/j.1574-6941.2010.00973.x
Dutkiewicz S., Morris J. J., Follows M. J., Scott J., Levitan O., Dyhrman S. T., et al. (2015). Impact of ocean acidification on the structure of future phytoplankton communities. Nat. Climate Change 5, 1002–1006. doi: 10.1038/nclimate2722
Eckford-Soper L., Daugbjerg N. (2016). The ichthyotoxic genus Pseudochattonella (Dictyochophyceae): Distribution, toxicity, enumeration, ecological impact, succession and life history - A review. Harmful Algae 58, 51–58. doi: 10.1016/j.hal.2016.08.002
Eldin C., Mélenotte C., Mediannikov O., Ghigo E., Million M., Edouard S., et al. (2016). From Q fever to coxiella burnetii infection: a paradigm change. Clin. Microbiol. Rev. 30, 115–190. doi: 10.1128/cmr.00045-16
Falkowski P. G., Barber R. T., Smetacek V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206. doi: 10.1126/science.281.5374.200
Feely R. A., Alin S. R., Newton J., Sabine C. L., Warner M., Devol A., et al. (2010). The combined effects of ocean acidification, mixing, and respiration on pH and carbonate saturation in an urbanized estuary. Estuarine, Coastal and Shelf Science 88, 442-449. doi: 10.1016/j.ecss.2010.05.004
Feely R. A., Alin S. R., Carter B., Bednaršek N., Hales B., Chan F., et al. (2016). Chemical and biological impacts of ocean acidification along the west coast of North America. Estuarine Coast. Shelf Sci. 183, 260–270. doi: 10.1016/j.ecss.2016.08.043
Fernández-Álvarez C., Santos Y. (2018). Identification and typing of fish pathogenic species of the genus Tenacibaculum. Appl. Microbiol. Biotechnol. 102, 9973–9989. doi: 10.1007/s00253-018-9370-1
Fortunato C. S., Herfort L., Zuber P., Baptista A. M., Crump B. C. (2012). Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. Isme J. 6, 554–563. doi: 10.1038/ismej.2011.135
Fritz L., Nass M. (1992). Development of the endoparasitic dinoflagellate Amoebophrya ceratii within host dinoflagellate species. J. Phycology 28, 312–320. doi: 10.1111/j.0022-3646.1992.00312.x
Fuhrman J. A., Cram J. A., Needham D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13, 133–146. doi: 10.1038/nrmicro3417
Gardner B. R., Arnould J. P. Y., Hufschmid J., McIntosh R. R., Fromant A., Tadepalli M., et al. (2022). Understanding the zoonotic pathogen, Coxiella burnetii in Australian fur seal breeding colonies through environmental DNA and genotyping. Wildlife Res. 50, 840-848. doi: 10.1071/WR22136
Gardner B. R., Hufschmid J., Stenos J., Tadepalli M., Sutton G., Fromant A., et al. (2023). Pacific Gulls (Larus pacificus) as Potential Vectors of Coxiella burnetii in an Australian Fur Seal Breeding Colony. Pathogens 12, 122. doi: 10.3390/pathogens12010122
Gazeau F., Quiblier C., Jansen J. M., Gattuso J.-P., Middelburg J. J., Heip C. H. R. (2007). Impact of elevated CO2 on shellfish calcification. Geophysical Res. Lett. 34, L07603. doi: 10.1029/2006GL028554
Giovannoni S. J. (2017). SAR11 bacteria: the most abundant plankton in the oceans. Annu. Rev. Mar. Sci. 9, 231–255. doi: 10.1146/annurev-marine-010814-015934
Green M. R., Sambrook J. (2018). Isolation of high-molecular-weight DNA from suspension cultures of mammalian cells using proteinase K and phenol. Cold Spring Harb. Protoc. 2018, 356-359. doi: 10.1101/pdb.prot093476
Guillou L., Bachar D., Audic S., Bass D., Berney C., Bittner L., et al. (2012). The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Habib C., Houel A., Lunazzi A., Bernardet J. F., Olsen A. B., Nilsen H., et al. (2014). Multilocus sequence analysis of the marine bacterial genus Tenacibaculum suggests parallel evolution of fish pathogenicity and endemic colonization of aquaculture systems. Appl. Environ. Microbiol. 80, 5503–5514. doi: 10.1128/aem.01177-14
Hewson I., Steele J., Fuhrman J. (2006). Temporal and spatial scales of variation in bacterioplankton assemblages of oligotrophic surface waters. Mar. Ecol. Prog. Ser. 311, 67–77. doi: 10.3354/meps311067
Hickey B. M. (1989). “Chapter 2 patterns and processes of circulation over the washington continental shelf and slope,”. in Elsevier Oceanography Series Eds. Landry M. R., Hickey B. M. (Elsevier Oceanography Series:Cambridge, Massachusetts, USA: Elsevier), 41–115.
Hickey B. M., Banas N. S. (2003). Oceanography of the U.S. Pacific Northwest Coastal Ocean and estuaries with application to coastal ecology. Estuaries 26, 1010–1031. doi: 10.1007/BF02803360
Hickey B. M., Royer T. (2001). “California and alaskan currents,” in Encyclopedia of Ocean Sciences. Eds. Steele J. H., Thorpe S. A., Turekian K. A. (Academic Press, San Diego, California), 368–379.
Hickey B. M., Trainer V. L., Michael Kosro P., Adams N. G., Connolly T. P., Kachel N. B., et al. (2013). A springtime source of toxic Pseudo-nitzschia cells on razor clam beaches in the Pacific Northwest. Harmful Algae 25, 1–14. doi: 10.1016/j.hal.2013.01.006
Karlson B., Andersen P., Arneborg L., Cembella A., Eikrem W., John U., et al. (2021). Harmful algal blooms and their effects in coastal seas of Northern Europe. Harmful Algae 102, 101989. doi: 10.1016/j.hal.2021.101989
Käse L., Metfies K., Neuhaus S., Boersma M., Wiltshire K. H., Kraberg A. C. (2021). Host-parasitoid associations in marine planktonic time series: Can metabarcoding help reveal them? PLoS One 16, e0244817. doi: 10.1371/journal.pone.0244817
Kavanaugh M. T., Oliver M. J., Chavez F. P., Letelier R. M., Muller-Karger F. E., Doney S. C. (2016). Seascapes as a new vernacular for pelagic ocean monitoring, management and conservation. ICES J. Mar. Sci. 73, 1839–1850. doi: 10.1093/icesjms/fsw086
Kersh G. J., Lambourn D. M., Raverty S. A., Fitzpatrick K. A., Self J. S., Akmajian A. M., et al. (2012). Coxiella burnetii infection of marine mammals in the Pacific Northwest 1997–2010. J. Wildlife Dis. 48, 201–206. doi: 10.7589/0090-3558-48.1.201
Kim M., Cha I.-T., Lee K.-E., Lee E.-Y., Park S.-J. (2020). Genomics reveals the metabolic potential and functions in the redistribution of dissolved organic matter in marine environments of the genus thalassotalea. Microorganisms 8, 1412. doi: 10.3390/microorganisms8091412
Kim S., Jeon C. B., Park M. G. (2017). Morphological observations and phylogenetic position of the parasitoid nanoflagellate Pseudopirsonia sp. (Cercozoa) infecting the marine diatom Coscinodiscus wailesii (Bacillariophyta). Algae 32, 181–187. doi: 10.4490/algae.2017.32.7.28
Könneke M., Bernhard A. E., de la Torre J. R., Walker C. B., Waterbury J. B., Stahl D. A. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437, 543–546. doi: 10.1038/nature03911
Korlević M., Markovski M., Herndl G. J., Najdek M. (2022). Temporal variation in the prokaryotic community of a nearshore marine environment. Sci. Rep. 12, 16859. doi: 10.1038/s41598-022-20954-6
Kroeker K. J., Kordas R. L., Crim R., Hendriks I. E., Ramajo L., Singh G. S., et al. (2013). Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob Chang Biol. 19, 1884–1896. doi: 10.1111/gcb.12179
Lewis W. H., Sendra K. M., Embley T. M., Esteban G. F. (2018). Morphology and Phylogeny of a New Species of Anaerobic Ciliate, Trimyema finlayi n. sp., with Endosymbiotic Methanogens. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00140
Lewitus A. J., Horner R. A., Caron D. A., Garcia-Mendoza E., Hickey B. M., Hunter M., et al. (2012). Harmful algal blooms along the North American west coast region: History, trends, causes, and impacts. Harmful Algae 19, 133–159. doi: 10.1016/j.hal.2012.06.009
Li S.-H., Kang I., Cho J.-C. (2023). Metabolic versatility of the family halieaceae revealed by the genomics of novel cultured isolates. Microbiol. Spectr. 11, e03879–e03822. doi: 10.1128/spectrum.03879-22
Lima-Mendez G., Faust K., Henry N., Decelle J., Colin S., Carcillo F., et al. (2015). Determinants of community structure in the global plankton interactome. Science 348, 1262073. doi: 10.1126/science.1262073
Loch T. P., Faisal M. (2015). Emerging flavobacterial infections in fish: A review. J. Advanced Res. 6, 283–300. doi: 10.1016/j.jare.2014.10.009
Longnecker K., Sherr B. F., Sherr E. B. (2006). Variation in cell-specific rates of leucine and thymidine incorporation by marine bacteria with high and with low nucleic acid content off the Oregon coast. Aquat. Microbial Ecol. 43, 113–125. doi: 10.3354/Ame043113
Lueker T. J., Dickson A. G., Keeling C. D. (2000). Ocean pCO2 calculated from dissolved inorganic carbon, alkalinity, and equations for K1 and K2: validation based on laboratory measurements of CO2 in gas and seawater at equilibrium. Mar. Chem. 70, 105–119. doi: 10.1016/S0304-4203(00)00022-0
Mackinnon D. L., Ray H. N. (1933). The Life cycle of two Species of “Selenidium” from the Polychaete Worm Potamilla reniformis. Parasitology 25, 143–162. doi: 10.1017/S003118200001934X
Manno C., Peck V. L., Tarling G. A. (2016). Pteropod eggs released at high pCO2 lack resilience to ocean acidification. Sci. Rep. 6, 25752. doi: 10.1038/srep25752
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011 17, 3. doi: 10.14806/ej.17.1.200
Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., Neufeld J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 13, 31. doi: 10.1186/1471-2105-13-31
Massana R., del Campo J., Sieracki M. E., Audic S., Logares R. (2014). Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. Isme J. 8, 854–866. doi: 10.1038/ismej.2013.204
Mathur V., Kolísko M., Hehenberger E., Irwin N. A. T., Leander B. S., Kristmundsson Á., et al. (2019). Multiple independent origins of apicomplexan-like parasites. Curr. Biol. 29, 2936–2941.e2935. doi: 10.1016/j.cub.2019.07.019
McCabe R. M., Hickey B. M., Kudela R. M., Lefebvre K. A., Adams N. G., Bill B. D., et al. (2016). An unprecedented coastwide toxic algal bloom linked to anomalous ocean conditions. Geophysical Res. Lett. 43, 10,366–310,376. doi: 10.1002/2016GL070023
McKenzie C. H., Bates S. S., Martin J. L., Haigh N., Howland K. L., Lewis N. I., et al. (2021). Three decades of Canadian marine harmful algal events: Phytoplankton and phycotoxins of concern to human and ecosystem health. Harmful Algae 102, 101852. doi: 10.1016/j.hal.2020.101852
Miao L., Wang P., Hou J., Yao Y., Liu Z., Liu S., et al. (2019). Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci. Total Environ. 650, 2395–2402. doi: 10.1016/j.scitotenv.2018.09.378
Milici M., Deng Z.-L., Tomasch J., Decelle J., Wos-Oxley M. L., Wang H., et al. (2016). Co-occurrence analysis of microbial taxa in the Atlantic ocean reveals high connectivity in the free-living bacterioplankton. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00649
Minor C., Kersh G. J., Gelatt T., Kondas A. V., Pabilonia K. L., Weller C. B., et al. (2013). Coxiella burnetii in northern fur seals and Steller sea lions of Alaska. J. Wildl Dis. 49, 441–446. doi: 10.7589/2012-09-226
Moestrup Ø., Hakanen P., Hansen G., Daugbjerg N., Ellegaard M. (2014). On Levanderina fissa gen. & comb. nov. (Dinophyceae) (syn. Gymnodinium fissum, Gyrodinium instriatum, Gyr. uncatenum), a dinoflagellate with a very unusual sulcus. Phycologia 53, 265–292. doi: 10.2216/13-254.1
Mooney J. R., Shirley T. C. (2000). New hosts, prevalence, and density of the ellobiopsid parasite thalassomyces fagei on euphausiids in prince william sound, Alaska. J. Crustacean Biol. 20, 320–325. doi: 10.1163/20021975-99990043
Morris R. M., Spietz R. L. (2022). The physiology and biogeochemistry of SUP05. Annu. Rev. Mar. Sci. 14, 261–275. doi: 10.1146/annurev-marine-010419-010814
Mostofa K. M. G., Liu C. Q., Zhai W., Minella M., Vione D., Gao K., et al. (2016). Reviews and Syntheses: Ocean acidification and its potential impacts on marine ecosystems. Biogeosciences 13, 1767–1786. doi: 10.5194/bg-13-1767-2016
Murali A., Bhargava A., Wright E. S. (2018). IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6, 140. doi: 10.1186/s40168-018-0521-5
Nelson K. S., Baltar F., Lamare M. D., Morales S. E. (2020). Ocean acidification affects microbial community and invertebrate settlement on biofilms. Sci. Rep. 10, 3274. doi: 10.1038/s41598-020-60023-4
Noirungsee N., Hackbusch S., Viamonte J., Bubenheim P., Liese A., Müller R. (2020). Influence of oil, dispersant, and pressure on microbial communities from the Gulf of Mexico. Sci. Rep. 10, 7079. doi: 10.1038/s41598-020-63190-6
O'Brien J., McParland E. L., Bramucci A. R., Siboni N., Ostrowski M., Kahlke T., et al. (2022). Biogeographical and seasonal dynamics of the marine Roseobacter community and ecological links to DMSP-producing phytoplankton. ISME Commun. 2, 16. doi: 10.1038/s43705-022-00099-3
Oksanen J., Simpson G., Blanchet F., Kindt R., Legendre P., Minchin P., et al. (2022). Vegan: Community Ecology Package version 2.6-4. (Vienna, Austria: The Comprehensive R Archive Network).
Oliver J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34, 415–425. doi: 10.1111/j.1574-6976.2009.00200.x
Pang Q., Xu W., He F., Peng F., Zhu X., Xu B., et al. (2022). Functional genera for efficient nitrogen removal under low C/N ratio influent at low temperatures in a two-stage tidal flow constructed wetland. Sci. Total Environ. 804, 150142. doi: 10.1016/j.scitotenv.2021.150142
Park M. G., Yih W., Coats D. W. (2004). Parasites and phytoplankton, with special emphasis on dinoflagellate infections1. J. Eukaryotic Microbiol. 51, 145–155. doi: 10.1111/j.1550-7408.2004.tb00539.x
Partensky F., Garczarek L. (2010). Prochlorococcus: advantages and limits of minimalism. Annu. Rev. Mar. Sci. 2, 305–331. doi: 10.1146/annurev-marine-120308-081034
Pelletier G., Lewis E., Wallace D. (2007). A calculator for the CO2 system in seawater for Microsoft Excel/VBA (Olympia, Washington/Upton, New York, USA: Washington State Department of Ecology).
Perez F. F., Fraga F. (1987). Association constant of fluoride and hydrogen ions in seawater. Mar. Chem. 21, 161–168. doi: 10.1016/0304-4203(87)90036-3
Peterson T. D., Crawford D. W., Harrison P. J. (2011). Evolution of the phytoplankton assemblage in a long-lived mesoscale eddy in the eastern Gulf of Alaska. Mar. Ecol. Prog. Ser. 424, 53–73. doi: 10.3354/meps08943
Peterson J. O., Morgan C. A., Peterson W. T., Lorenzo E. D. (2013). Seasonal and interannual variation in the extent of hypoxia in the northern California Current from 1998–2012. Limnology Oceanography 58, 2279–2292. doi: 10.4319/lo.2013.58.6.2279
Pierella Karlusich J. J., Ibarbalz F. M., Bowler C. (2020). Phytoplankton in the tara ocean. Ann. Rev. Mar. Sci. 12, 233–265. doi: 10.1146/annurev-marine-010419-010706
Pinnell L. J., Turner J. W. (2020). Temporal changes in water temperature and salinity drive the formation of a reversible plastic-specific microbial community. FEMS Microbiol. Ecol. 96, fiaa230. doi: 10.1093/femsec/fiaa230
Pomeroy L. R. (1974). The ocean's food web, A changing paradigm. BioScience 24, 499–504. doi: 10.2307/1296885
Pruesse E., Peplies J., Glöckner F. O. (2012). SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Qin W., Amin S. A., Martens-Habbena W., Walker C. B., Urakawa H., Devol A. H., et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc. Natl. Acad. Sci. 111, 12504–12509. doi: 10.1073/pnas.1324115111
Qin W., Heal K. R., Ramdasi R., Kobelt J. N., Martens-Habbena W., Bertagnolli A. D., et al. (2017). Nitrosopumilus maritimus gen. nov., sp. nov., Nitrosopumilus cobalaminigenes sp. nov., Nitrosopumilus oxyclinae sp. nov., and Nitrosopumilus ureiphilus sp. nov., four marine ammonia-oxidizing archaea of the phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 67, 5067–5079. doi: 10.1099/ijsem.0.002416
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramirez-Paredes J. G., Larsson P., Thompson K. D., Penman D. J., Busse H. J., Öhrman C., et al. (2020). Reclassification of Francisella noatunensis subsp. orientalis Ottem et al. 2009 as Francisella orientalis sp. nov., Francisella noatunensis subsp. Chilensis subsp. nov. and emended description of Francisella noatunensis. Int. J. Syst. Evol. Microbiol. 70, 2034–2048. doi: 10.1099/ijsem.0.004009
R Core Team (2021). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing).
Rogers K. L., Carreres-Calabuig J. A., Gorokhova E., Posth N. R. (2020). Micro-by-micro interactions: How microorganisms influence the fate of marine microplastics. Limnology Oceanography Lett. 5, 18–36. doi: 10.1002/lol2.10136
Rueckert S., Betts E. L., Tsaousis A. D. (2019). The symbiotic spectrum: Where do the gregarines fit? Trends Parasitol. 35, 687–694. doi: 10.1016/j.pt.2019.06.013
Rueckert S., Simdyanov T. G., Aleoshin V. V., Leander B. S. (2011). Identification of a divergent environmental DNA sequence clade using the phylogeny of gregarine parasites (Apicomplexa) from crustacean hosts. PLoS One 6, e18163. doi: 10.1371/journal.pone.0018163
Santoro A. E., Dupont C. L., Richter R. A., Craig M. T., Carini P., McIlvin M. R., et al. (2015). Genomic and proteomic characterization of “Candidatus Nitrosopelagicus brevis”: An ammonia-oxidizing archaeon from the open ocean. Proc. Natl. Acad. Sci. 112, 1173–1178. doi: 10.1073/pnas.1416223112
Sherr E. B., Sherr B. F., Wheeler P. A. (2005). Distribution of coccoid cyanobacteria and small eukaryotic phytoplankton in the upwelling ecosystem off the Oregon coast during 2001 and 2002. Deep-Sea Res. Part Ii-Topical Stud. Oceanography 52, 317–330. doi: 10.1016/J.Dsr2.2004.09.020
Smith J., Connell P., Evans R. H., Gellene A. G., Howard M. D. A., Jones B. H., et al. (2018). A decade and a half of Pseudo-nitzschia spp. and domoic acid along the coast of southern California. Harmful Algae 79, 87–104. doi: 10.1016/j.hal.2018.07.007
Starliper C. E. (2011). Bacterial coldwater disease of fishes caused by Flavobacterium psychrophilum. J. Advanced Res. 2, 97–108. doi: 10.1016/j.jare.2010.04.001
Stoeck T., Bass D., Nebel M., Christen R., Jones M. D. M., Breiner H.-W., et al. (2010). Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 19, 21–31. doi: 10.1111/j.1365-294X.2009.04480.x
Thompson A. R., Bjorkstedt E. P., Bograd, Steven J., Fisher J. L., Hazen E. L., et al. (2022). State of the California current ecosystem in 2021: Winter is coming? Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.958727
Thomson R. E. (1981). Oceanography of the British Columbia Coast (Ottawa, Ontario, Canada: Canadian Special Publication of Fisheries and Aquatic Sciences). C.S.P.o.F.a.A. Sciences.
Traving S. J., Kellogg C. T. E., Ross T., McLaughlin R., Kieft B., Ho G. Y., et al. (2021). Prokaryotic responses to a warm temperature anomaly in northeast subarctic Pacific waters. Commun. Biol. 4, 1217. doi: 10.1038/s42003-021-02731-9
Tsementzi D., Wu J., Deutsch S., Nath S., Rodriguez R. L., Burns A. S., et al. (2016). SAR11 bacteria linked to ocean anoxia and nitrogen loss. Nature 536, 179–183. doi: 10.1038/nature19068
Uppström L. R. (1974). The boron/chlorinity ratio of deep-sea water from the Pacific Ocean. Deep Sea Res. Oceanographic Abstracts 21, 161–162. doi: 10.1016/0011-7471(74)90074-6
Valigurová A., Florent I. (2021). Nutrient acquisition and attachment strategies in basal lineages: A tough nut to crack in the evolutionary puzzle of apicomplexa. Microorganisms 9, 1430. doi: 10.3390/microorganisms9071430
Voss M., Baker A., Bange H. W., Conley D., Cornell S., Deutsch B., et al. (2011). “Nitrogen processes in coastal and marine ecosystems,” in The European Nitrogen Assessment. Eds. Sutton M. A., Howard C. M., Erisman J. W., Billen G., Bleeker A., Grennfelt P., van Grinsven H., Grizzetti B. (Cambridge University Press, New York, NY, uSA), 147–176.
Waldbusser G. G., Hales B., Langdon C. J., Haley B. A., Schrader P., Brunner E. L., et al. (2015). Saturation-state sensitivity of marine bivalve larvae to ocean acidification. Nat. Climate Change 5, 273–280. doi: 10.1038/nclimate2479
Waldbusser G. G., Salisbury J. E. (2014). Ocean acidification in the coastal zone from an organism's perspective: multiple system parameters, frequency domains, and habitats. Annu. Rev. Mar. Sci. 6, 221–247. doi: 10.1146/annurev-marine-121211-172238
Wemheuer F., von Hoyningen-Huene A. J. E., Pohlner M., Degenhardt J., Engelen B., Daniel R., et al. (2019). Primary production in the water column as major structuring element of the biogeographical distribution and function of archaea in deep-sea sediments of the central pacific ocean. Archaea 2019, 3717239. doi: 10.1155/2019/3717239
Worden A. Z., Follows M. J., Giovannoni S. J., Wilken S., Zimmerman A. E., Keeling P. J. (2015). Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 347, 1257594. doi: 10.1126/science.1257594
Wright R. J., Langille M. G. I., Walker T. R. (2021). Food or just a free ride? A meta-analysis reveals the global diversity of the Plastisphere. ISME J. 15, 789–806. doi: 10.1038/s41396-020-00814-9
Xie N., Hunt D. E., Johnson Z. I., He Y., Wang G. (2021). Annual partitioning patterns of labyrinthulomycetes protists reveal their multifaceted role in marine microbial food webs. Appl. Environ. Microbiol. 87, e01652-20. doi: 10.1128/aem.01652-20
Xu D., Kong H., Yang E.-J., Li X., Jiao N., Warren A., et al. (2020). Contrasting community composition of active microbial eukaryotes in melt ponds and sea water of the arctic ocean revealed by high throughput sequencing. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01170
Yan S., Fuchs B. M., Lenk S., Harder J., Wulf J., Jiao N.-Z., et al. (2009). Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria. Systematic Appl. Microbiol. 32, 124–139. doi: 10.1016/j.syapm.2008.12.001
Yilmaz P., Parfrey L. W., Yarza P., Gerken J., Pruesse E., Quast C., et al. (2013). The SILVA and “All-species Living Tree Project (LTP)“ taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zakem E. J., Al-Haj A., Church M. J., van Dijken G. L., Dutkiewicz S., Foster S. Q., et al. (2018). Ecological control of nitrite in the upper ocean. Nat. Commun. 9, 1206. doi: 10.1038/s41467-018-03553-w
Zehr J. P., Ward B. B. (2002). Nitrogen cycling in the ocean: New perspectives on processes and paradigms. Appl. Environ. Microbiol. 68, 1015–1024. doi: 10.1128/AEM.68.3.1015-1024.2002
Zhang C. L., Xie W., Martin-Cuadrado A.-B., Rodriguez-Valera F. (2015). Marine Group II Archaea, potentially important players in the global ocean carbon cycle. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01108
Keywords: nearshore microbial communities, bacteria and archaea, eukaryotic phytoplankton, 16S and 18S rRNA metabarcoding, Northeastern Pacific Ocean
Citation: Rhodes LD, Adams NG, Gallego Simon R, Kavanaugh MT, Alin SR and Feely RA (2024) Nearshore microbial communities of the Pacific Northwest coasts of Canada and the U.S.. Front. Mar. Sci. 11:1430930. doi: 10.3389/fmars.2024.1430930
Received: 11 June 2024; Accepted: 15 August 2024;
Published: 05 September 2024.
Edited by:
Ana R. Díaz-Marrero, Spanish National Research Council (CSIC), SpainReviewed by:
Cesar Cardona-Felix, National Polytechnic Institute (IPN), MexicoCarolina Pérez Reyes, University of La Laguna, Spain
Copyright © 2024 Rhodes, Adams, Gallego Simon, Kavanaugh, Alin and Feely. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda D. Rhodes, aXNsYW5kLnJlc2VhcmNoLndoaWRiZXlAZ21haWwuY29t
†Present address: Linda D. Rhodes, Island Research, PO Box 249, Greenbank, WA, United States
 Linda D. Rhodes
Linda D. Rhodes Nicolaus G. Adams
Nicolaus G. Adams Ramon Gallego Simon
Ramon Gallego Simon Maria T. Kavanaugh
Maria T. Kavanaugh Simone R. Alin
Simone R. Alin Richard A. Feely
Richard A. Feely