
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 24 July 2024
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1424999
This article is part of the Research Topic Towards an Expansion of Sustainable Global Marine Aquaculture View all 13 articles
The starter diet for Japanese eel (Anguilla japonica) has always been a difficult problem for the realization of total artificial reproduction. Therefore, this research analyzed the nutritional composition of artificially fertilized eggs, and transcriptome of samples from early hatchlings of fry to better understand nutrients requirements. The composition of crude lipid and crude protein in fertilized eggs was 7.24% ± 0.32% and 10.56% ± 0.41%, respectively. Seven kinds of essential amino acids (EAA) were detected but took a comparable lower content (3.19%) than other marine fish eggs. We randomly assembled 265.74 million clean reads and identified 1751 differentially expressed genes (DEGs) (P < 0.01) from pre-leptocephalus larvae. A total of 23 KEGG pathways related to the digestive and metabolic system were detected. Genes related to the secretion pathway of saliva, pancreatic juice and other digestive juices were significantly changed. Transcriptome analysis showed that as larvae aged, glycolytic metabolism and the transcription level of hexokinase (HK) increased significantly (day 0 to 12). This study will facilitate future studies on the nutrition of A. japonica larvae and other biological traits to reproductive research.
Japanese eel (Anguilla japonica) is an important cultured species in East Asia with a mysterious life cycle that involving spawning in the ocean and migrating to freshwater (Tsukamoto, 1992; Tanaka, 2015; Kuan-Mei et al., 2018). However, its populations have suffered a severe reduction due to overfishing, habitat loss, and environmental deterioration (Matsushige et al., 2019; Pike et al., 2020). Eel farming, which relies on wild-caught glass eels, is also experiencing a sharp decrease in annual harvests (Dekker and Casselman, 2014; Koh et al., 2017; Feunteun and Prouzet, 2020). Although the complete life cycle of A. japonica was firstly closed in the lab in 2010 (Tanaka et al., 2001; Masuda et al., 2012), commercialization remains challenging due to the low survival rate of pre-leptocephalus larvae (Okamura et al., 2014).
The low survival rate of newly hatched larvae in lab is likely due to nutritional deficiencies in their artificial diets. Despite efforts to include shark egg yolk, Antarctic krill, and rotifer paste, additional nutritional improvements are still necessary (Tanaka et al., 2003; Okamura et al., 2013; Liu et al., 2017; Okamura et al., 2019). Pousão-Ferreira et al. (1999) successfully used gilthead seabream (Sparus aurata) eggs to feed S. aurata larvae, determining nutritional requirements during the larval rearing based on egg composition. Mourente and Rosa (1996) confirmed the fatty acids levels for unfed Senegal sole (Solea senegalensis) larvae. Ohkubo et al. (2008) highlighted the importance of understanding egg nutritional composition for developing starter diets by describing nutrient changes from fertilized eggs to yolk-sac larvae of A. japonica.
The digestive system function in fish larvae depends on both genetically pre-programmed and extrinsically influenced factors (Politis et al., 2018). The expression of several selected pancreatic enzyme genes indicated that A. japonica larvae acquire full function by the onset of exogenous feeding at 8-12 dph (Okamura et al., 2019). However, other digestive enzyme genes and nutrient transporters in A. japonica are not well studied, and few studies address food digestion and nutrient absorption during the preleptocephali stage. The advent of next-generation sequencing technology has advanced genome and transcriptome analysis in aquatic organisms, providing insight into gene expression during developmental stages (Kleppe et al., 2014). Changes in the quantity and quality of transcriptome data can reflect the expression status of related genes in specific circumstances, which can effectively improve the quality of basic research. RNA-Seq, in partic3ular, is a powerful tool for profiling and quantifying RNA transcripts, enhancing the quality of basic research (Wang et al., 2010). It provides fundamental insights into biological processes and applications such as gene expression levels in developmental stages, combining advantages of high throughput, low background noise and high sensitivity (Ozsolak and Milos, 2010; Churcher et al., 2015; Hsu et al., 2015).
With the aim to explore the dietary requirements for the preleptocephali of A. japonica during the critical early life history stage, the nutritional composition of eggs was determined and the RNA of both embryos and preleptocephali was sequenced. It is hope to reveal the molecular events that affect the nutrient digestion and absorption, and to provide a basis for the future research on nutrient requirements of artificially A. japonica hatchlings.
The wild-caught broodstock A. japonica were cultured in the experimental facility of the Marine and Fisheries Research Institute of Ningbo (China), where they were kept in 8 m × 4 m × 1.7 m pond and the natural seawater was maintained at 20 ± 4°C and adjusted to 31 ± 5 psu salinity using Red Sea® Salts. Fish were not fed during the experiment and around 75% of the pond area was shaded by the black net. At the onset of experiments, all experimental broodstock A. japonica were anaesthetized (MS-222) and then tagged with a passive integrated transponder.
The A. japonica were artificially induced to mature using the method detailed in Jiang et al. (2012). Five female broodstocks were euthanized after stripping to collect eggs, which were pooled at equal ratios before being stored at -80°C until nutritional analyses. Natural spawning happened around 12 h after the remaining females and males were put together, then fertilized eggs were collected and reared at 24°C in seawater (31 psu) in darkness. Hence, 50 fertilized eggs were collected as 0 d sample, and 30 pre-leptocephalus samples were taken every three days after hatching until the 12th day. All samples were preserved by RNAlater (Tiangen Biotech, Beijing, China) and then stored at -80°C until RNA extraction.
The proximate composition analysis of eggs was performed using official methods of analysis of AOAC (1996). Moisture content was determined by drying 0.2g eggs’ sample in an oven at 110°C to a constant weight. Ash content of eggs was determined by heating 0.2g eggs’ sample in a muffle furnace for about 10-12 h at 550°C and weighting it after cooling. Crude protein content was measured by the Kjeldahl method (N content × 6.25) using a Kjeltech system (Kjeltec 2300, Foss, Sweden). The extraction of lipids from 0.2g eggs’ sample was carried out with a mixture of chloroform and methanol (2:1, v/v) containing 0.01% BHT and determination of crude lipids was performed according to Folch et al. (1957).
Fatty acid methyl esters (FAMEs) were esterified with 14% BF3 in methanol and the FAMEs extracted with hexane (Morrison and Smith, 1964). FAMEs were separated and detected by an Agilent 7890 gas chromatograph (Agilent Technologies, CA, USA) equipped with a flame ionization detector instrument. The 37-FAME Mix (Supelco, Bellefonte, PA, USA) was used to identify the FAMEs, and the fatty acid C19:0 (nonadecanoic acid, Sigma) was used as an internal standard for fatty acid quantification. All measurements were performed in triplicates, the fatty acids content was expressed as the percentage of each FA to the total FAs. The amino acid composition of the freeze-dried eggs digested with hydrochloric acid was determined with a HITACHI L-8900 amino acid automatic analyzer (Hitachi Limited, Tokyo, Japan), and the peak areas were recorded (Gilani and Peace, 2005). Standard curves were plotted with amino acid peak area as the ordinate and amino acid concentration as the abscissa.
The theoretical demand of each nutrient was calculated according to previous research (Pousão-Ferreira et al., 1999; Li et al., 2016). The crude lipid and crude protein contents of the fresh weight base were converted into dry weight base, which was the theoretical requirement for fat and protein of larvae eels. The requirement of each amino acid can be converted according to the content of crude protein on the basis of dry weight. Since the amino acids in this study were tested on the basis of dry weight, the detected value is the theoretical requirement of amino acids in the feed of larvae eels. Similarly, according to the crude fat content based on dry weight, the theoretical requirement of fatty acids in the feed can be calculated.
The total RNA of the whole fish was extracted using TRIzol® reagent (Invitrogen, California, USA) according to the manufacturer’s instructions. Purified RNA was quantified by Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). From each pooled sample, 5μg mRNA was isolated from total RNA using oligo (dT) magnetic beads (Invitrogen). Then five sequencing libraries, one for each time point, were constructed using Truseq™ RNA sample prep Kit (Illumina, San Diego, USA) according to the instruction. The mRNA was interrupted into~200 bp short fragments using the fragmentation buffer. It was then transcribed into the first-strand cDNA using reverse transcriptase and random hexamer primers, followed by second-strand cDNA synthesis. The double-stranded cDNA was subjected to end repair, phosphorylation, a-tailed and indexed adapters were ligated. Suitable fragments were selected and enriched by PCR to create the final cDNA library. The paired-end cDNA library was sequenced on an Illumina HiSeq™ 4000 platform (Majorbio Biotech Co., Ltd., Shanghai, China).
The raw RNA-seq data were processed to discard the dirty reads that include reads with adaptors, reads with more than 10% Q<20 bases. The low complexity reads were removed by Seqprep and Sickle program. Clean and high-quality reads from the five samples were then assembled using the Trinity program, meanwhile, the counts of transcripts and the N50 were calculated. The assembled unigenes of five samples were used for BlastX search and annotation against the NR, Swissprot, COG (Clusters of Orthologous Groups of proteins), KEGG (Kyoto Encyclopedia of Genes and Genomes) and GO (Gene Ontology) databases with a cut-off E-value < 10−5. Based on Nr-matched unigenes, the annotation of GO was obtained by Web Gene Ontology Annotation Plot (WEGO; http://wego.genomics.org.cn/) program. The transcripts were also blasted against the Pfam database to identify specific protein domains and acquire GO annotations.
All clean sequence reads from each of the five libraries (0, 3, 6, 9, 12 days) were mapped to reference sequences (unigenes from the assembled transcriptome data) using Bowtie2 software with default setting (Langmead, 2012). Subsequently, RSEM (http://deweylab.biostat.wisc.edu/rsem/) was used to calculate the FPKM (Fragments per kilobase of transcripts per million fragments mapped values) of the assembled transcripts (Li and Dewey, 2011). Identifying differentially expressed genes (DEGs) among five groups was performed using the R package WGCNA (Robinson et al., 2010). Because we have a single sample from each time point, pair-wise comparisons between time points are not feasible. We decided to examine time-dependent transcriptional changes using a linear model where time points are the independent variable and expression levels are the dependent variable. The false discovery rate (FDR) method was introduced to determine the threshold P-value in multiple tests. If FDR was smaller than 0.05 and FPKM values showing at least twofold difference two groups, this unigene was considered as significant time-dependent DEGs. DEGs among the samples were further annotated by GO and KEGG pathway analysis.
In the experiment, the data of each group were presented as mean ± standard deviation (SD) and analyzed by Excel 2010 statistical software and SPSS version 17 (Michigan Avenue, Chicago, IL, USA). The original transcriptomic data were analyzed by a linear model implemented in the WGCNA package. With day-age as the independent variable and gene expression as the dependent variable, genes significantly changed with day-age were identified. We applied a WGCNA module significance filter of P< 0.01 and DEG FDR < 0.05 for the final DEG gene set.
The published fatty acid composition of different marine fish eggs was summarized in Table 1. Proximate composition of unfertilized eggs is shown in Figure 1. The composition of crude lipid and crude protein was 7.24% ± 0.32% and 10.56% ± 0.41%, respectively. A total of nine saturated fatty acids (SFA) were detected in the eggs, together with 8 monounsaturated fatty acids (MUFA), and 10 polyunsaturated fatty acids (PUFA), which took 41.14% ± 1.61%, 18.19% ± 0.69%, and 40.69% ± 1.11% of total fatty acids. The n-3 PUFA takes up about 34.03% ± 0.50% of total fatty acids, while the n-6 PUFA took around 5.04% ± 0.48% in total. The EPA (C20:5n3) and DHA (C22:6n3) accounted for 4.27% ± 0.24% and 25.98% ± 0.49% of total fatty acids, respectively, in which EPA was lower than most marine fish, and DHA was higher than most marine fish (Figure 1 and Table 1). Moreover, the total EPA and DHA content is 30.25% is medium level, comparing to other marine fish. The SFA of A. japonica eggs accounted for 41.14% ± 1.61% of the total fatty acids, which was higher than most marine fishes listed in Table 1, among which the content of C16:0 was the highest. Simultaneously, compared to the contents of fatty acids in other marine fish eggs, it was found that eel eggs were rich in fatty acids, which provided a good foundation for the development of eel larvae (Table 1 and Figure 1). The theoretical fat demand of starter diet was 40.86% in the current study, that the demand of C16:0 and DHA was the highest (Table 2).
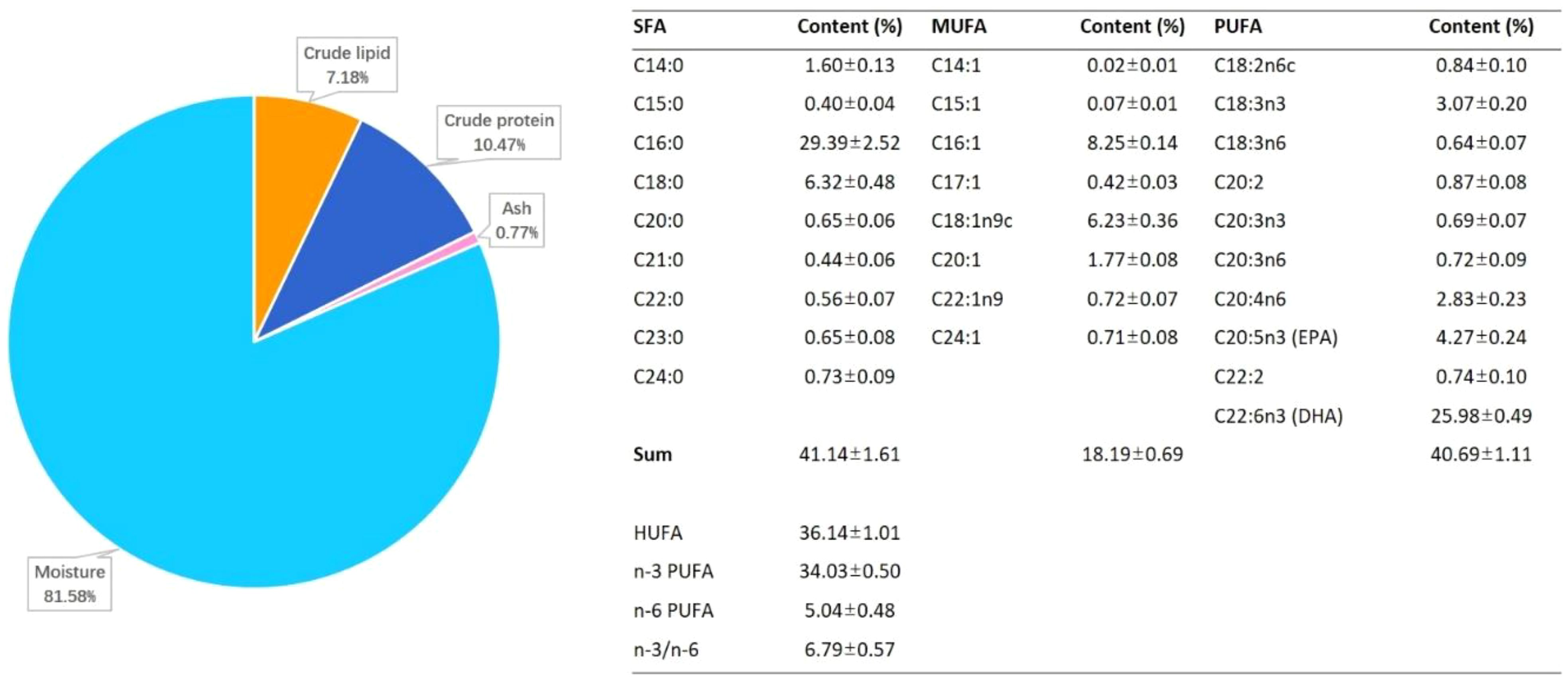
Figure 1 The proximate composition (% wet weight basis) and fatty acid composition (% total FA) in unfertilized eggs of Anguilla japonica.
A total of 17 amino acids were detected, including 7 kinds of essential amino acids (EAA) and 10 nonessential amino acids (NEAA, Figure 2). Leucine (0.69%) took a higher percentage compare to other EAAs, and the glutamic acid was the highest (0.98%) among NEAAs. The ratio of EAA/NEAA was 65.37%, and the EAA made up about 39.53% of the total amino acids (Figure 2). Compared with other amino acids, the contents of glutamic acid (0.98%), alanine (0.82%), leucine (0.69%), and lysine (0.63%) in artificial breeding eel eggs are higher, and glutamate is the highest, which is similar to most marine fish. Still, the content of each amino acid is far lower than that of other marine fish eggs (Table 3). The theoretical demand for protein in the diet of the larvae was 59.59%, the theoretical demand for glutamate was the highest, and the theoretical demand for cysteine was the lowest (Table 2).
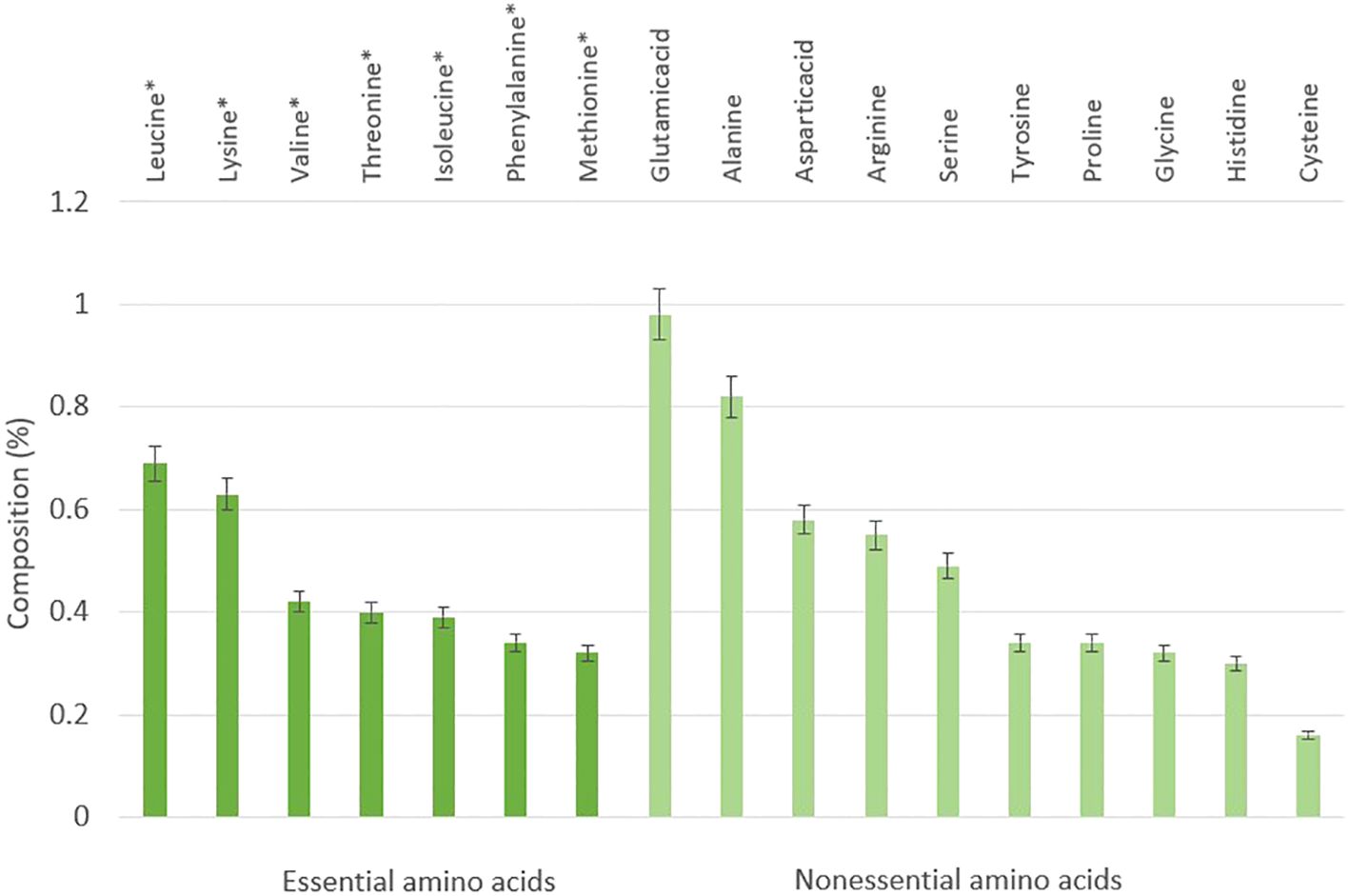
Figure 2 Composition of essential amino acids and nonessential amino acids in unfertilized eggs of Anguilla japonica. * This means essential amino acid.
The RNA sequencing generated a total of 271,423,952 raw reads, and 265,743,784 (97.9%) clean reads were obtained after removing SeqPrep adapter and low-quality reads (Table 4). Thereafter, we got a total of 102941 unigenes with a Q20 percentage over 98%. The contig N50 was 1398 bps. The average length of it was 856.6 bps (Table 5). For the functional annotation, the unigenes were aligned with sequences from major databases including Pfam, Swiss-Prot, KEGG, GO and Nr. The statistics of overall functional annotations were shown in Table 6.
A summary of unigenes classified to each term at GO level 2 is shown in Figure 3. A total of 1664 terms in the biological process category were produced, the most dominant subcategories were cellular process (17087; 21.14%), single-organism process (12101; 14.97%), metabolic process (11794; 14.59%), biological regulation (7441; 9.21%), and regulation of biological process (6915; 8.56%). In cellular component functions, we got 774 terms. And there were 15712 (19.89%) of unigenes that were assigned to the cell, followed by cell part 15469 (19.58%), membrane 11852 (15.00%), organelle 9858 (12.48%), and macromolecular complex 5782 (7.32%). In the molecular function category, 422 terms were produced, and the five most dominant subcategories were binding (20754; 43.91%), catalytic activity (15442; 32.67%), transporter activity (2474; 5.23%), signal transducer activity (2054; 4.35%) and structural molecule activity (1971; 4.17%).
By running the WGCMA to perform a linear regression of time-dependent gene expression on the original transcriptome data, genes significantly changed with day-age were identified. A total of 1,751 genes were selected that changed significantly with age, contained in WGCNA module with P < 0.01 and gene DEG FDR < 0.05. By comparing and analyzing the screened genes with KEGG database, 23 pathways related to the digestive and metabolic system were detected. With the change of age, 120 genes showed a significant increase in gene expression, and 18 genes showed a significant decrease in gene expression. Among them, genes related to the secretion pathway of saliva, pancreatic juice and other digestive juices were significantly changed. The genes of carbohydrate metabolism, glycerolipid metabolism, glycerophospholipid metabolism, and other metabolic pathways were up-regulated significantly with age (Table 7).
Statistical overrepresentation test was performed for genes whose gene expression significantly increased with age after screening. A total of 20 significantly enriched Go-slim molecular function pathways were detected. Through analysis, it is found that among the genes enriched in the transmembrane transporter activity pathway, the TRPC1 gene and CFTR gene are related to the secretion and transport of digestive enzymes such as saliva, pancreatic juice, and bile (Table 8).
Studies have demonstrated that the knowledge of nutrient composition of fish eggs will not only provide key insights into requirements of nutrient, but also support formulating the starter diet (Pousão-Ferreira et al., 1999; Yang et al., 2014). Meanwhile, the transcriptomic analysis could reveal the changes of food digestion and nutrient absorption during the preleptocephali stage to eliminating the hurdle to complete full artificial reproduction.
Protein is essential for cell and tissues function, crucial for growth and life maintenance. The protein content data from eel eggs emphasize the nutritional requirements for first larval feeding (Pousão-Ferreira et al., 1999). The theoretical protein demand of starter diet of A. japonica was 59.59% that calculated depending on the crude protein composition (10.56%) of eel eggs in present study, aligning with previous findings (Xiong et al., 1996; de Souza Romaneli et al., 2021). However, the EAA content (3.19%) was significantly lower compared to other marine fish, potentially leading to malnutrition and high mortality before feeding.
Lysine, the first restrictive amino acid, enhances the utilization of other EAAs, prevents nitrogen loss, and promotes growth (Zhou et al., 2008). Therefore, a higher lysine content in fish egg is beneficial to both fish growth and survival. Although the content of lysine (0.69%) was one of the highest amino acid in the eel eggs of this study, a comparable lower rate to other marine fish eggs still implies that a limited lysine content could be another cause for the mass death in the artificial reproduction. Fatty acids are one of the main energy sources for fish (Cejas et al., 2004; Bennett et al., 2007). The analysis shows that the SFA of A. japonica eggs accounts for 41.14% of the total fatty acids, among which the content of C16:0 was the richest that is similar to Alosa sapidissima (Yanes-Roca et al., 2009), Sardina pilchardus (Liu et al., 2018), and Centropomus undecimalis (Guedes et al., 2020). PUFA has been proved to have a significant effect in promoting fish growth and development, as well as the fish immunity and survival rate (Xu et al., 2010). A 40.69% of the total fatty acids’ PUFA was detected in this study, and a similar high proportion was found in other marine fish eggs as well (Liu et al., 2018; Tang et al., 2020; Zheng et al., 2020).
In this study, the transcription levels of various digestive enzymes and carbohydrate metabolism in the larvae were comparably high with the changes of the larvae’s age. The metabolic capability for low molecular carbohydrate, especially in galactose, fructose, and sucrose metabolism pathways, showed increasing with the day-age growing. The hyaluronic acid, a disaccharide substance, is found to be the main body composition of leptocephali, highlights the importance of carbohydrates for growth (Pfeiler, 1999). The marine snow, a most likely starter diet of A. japonica, happens to be a collection of different carbohydrates also supports the demands of low molecular carbohydrate during the early life (Pfeiler, 1999). Okamura et al. (2020) found that dietary supplementation with chitin hydrolysates including mono-, di- and trimers of N-acetylglucosamine, supporting the growth and survival of eel larvae. It demonstrates that feeding low-molecular bait such as glucose sugar and maltose to larvae eel is beneficial to improve the survival time of larvae (Skoog et al., 2008; Jang et al., 2022), which is consistent with our experimental results. During the early stage of post-hatching, the larvae consume endogenous yolk protein to supply the growth needs (Nobuyuki et al., 2008). It implies that the consumption of yolk nutrient continued after the larvae hatch, and the body carbohydrates kept strengthen in order to meet the energy demand for its growth. Our previous study indicated that the teeth began to form on the 6th day of membrane emergence, and the oil globule completely disappeared from the day 8, which marked the transition from endogenous nutrition to exogenous nutrition of larvae eel (Liu et al., 2017).
Amino acids play essential role in fish metabolism. They act as signal molecules for physiological regulation and can be oxidized to provide energy through gluconeogenesis during hunger or malnutrition (Aragão et al., 2004; Hamidoghli et al., 2019). Transcriptome analysis revealed that lysine metabolism increased steadily with age. The relatively high lysine content in fish eggs supports early larval growth. As a key ketogenic amino acid, lysine contributes to ketone body and glucose metabolism, serving as a vital energy source when other energy supplies are insufficient (Huang et al., 2021).
Transcriptome analysis showed a significant increase in glycolytic metabolism in larvae as they aged, particularly in the transcription level of hexokinase (HK), a key enzyme in glycolysis (Table 7). This suggests that larvae convert glucose and other sugars into energy to meet their growth needs. Additionally, the transcription level of gluconeogenesis and the metabolism of glycerides and glycerophospholipids increased with age. The metabolism and transcription levels of glyceride and glycerophospholipid were enhanced. This indicates that larvae may convert non-carbohydrate substances into glucose or glycogen to supplement their energy requirements due to insufficient carbohydrate reserves.
A previous study presented that the growth rate of the farmed leptocephali (which feeds on shark eggs) is lower than that of the wild leptocephali (Ishikawa et al., 2001). As one of the commonly used artificial starter diet components, the Acanthias shark egg-based diet are composed with protein (26.3%), lipids (17.5%), carbohydrates (0.1%) and moisture (54.4%) (Okamura et al., 2014). Our results demonstrate an age-dependent increase in carbohydrate conversion in larvae, suggesting a potential correlation between the sluggish growth of cultured preleptocephali larvae and insufficient carbohydrate supplementation in the bait. Okamura et al. (2020) presented that dietary supplementation with high sugar content such as N-acetylglucosamine, glucose and maltose could significantly improve the growth rate of larvae. Furthermore, our study suggests that the egg itself provides sufficient fatty acids to meet the nutritional requirements of the preleptocephali stage. Consequently, optimizing the carbohydrate content and supplementing essential amino acids like lysine in the diet of artificially bred A. japonica larvae may enhance growth, although the specific mechanisms of digestion and metabolism necessitate further experimental validation. Notably, our analysis of significantly enriched GO-Slim pathways indicates a predominance of pathways associated with ion transport, suggesting potential implications for early nutrient metabolism and transport, possibly influenced by water salinity, warranting deeper investigation.
In present study, we examined the nutritional composition and transcriptome of the artificially fertilized eggs and pre-leptocephalus larvae of A. japonica. Seven kinds of essential amino acids (EAA) were detected, with A. japonica exhibiting lower levels compared to other marine fish eggs. Through random assembly of 265.74 million clean reads, we identified 1751 differentially expressed genes. Notably, genes associated with carbohydrate metabolism, glycerolipid metabolism and glycerophospholipid metabolism showed significant up-regulation with larval growth. These findings lay a foundation for future studies into the nutrient requirements and digestive functions of newly hatched A. japonica, thereby advancing the field of artificial reproduction of eels.
The original contributions presented in the study are deposited in the NCBI repository, accession number: PRJNA1133914.
The animal study was approved by Animal Care and Use Ethics Committee of the Shanghai Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
KL: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. YL: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. TL: Data curation, Investigation, Methodology, Writing – original draft. RC: Methodology, Writing – review & editing. LL: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (No: 32072994); the Shanghai Agriculture Applied Technology Development Program, China (2020-02-08-00-10-F01471); and Startup Foundation for Young Teachers of Shanghai Ocean University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AOAC (1996). Official methods of analysis of the association of official analytical Chemists (Arlington (U.S.A: Association of Official Analytical Chemists).
Aragão C., Conceição L. E., Dinis M. T., Fyhn H. J. (2004). Amino acid pools of rotifers and Artemia under different conditions: nutritional implications for fish larvae. Aquaculture 234, 429–445. doi: 10.1016/j.aquaculture.2004.01.025
Bennett P. M., Weber L. P., Janz D. M. (2007). Comparison of chloroform–methanol-extracted and solvent-free triglyceride determinations in four fish species. J. Aquat. Anim. Health 19, 179–185. doi: 10.1577/H06-044.1
Bransden M. P., Battaglene S. C., Goldsmid R. M., Dunstan G. A., Nichols P. D. (2007). Broodstock condition, egg morphology and lipid content and composition during the spawning season of captive striped trumpeter, Latris lineata. Aquaculture 268 (1–4), 2–12.
Cejas J. R., Almansa E., Jérez S., Bolaños A., Felipe B., Lorenzo A. (2004). Changes in lipid class and fatty acid composition during development in white seabream (Diplodus sargus) eggs and larvae. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 139, 209–216. doi: 10.1016/j.cbpc.2004.07.010
Chen J. M., Liu X. F., Leng K. L., Gao H., Miao J. K., Wang S. (2016). Analysis and evaluation of nutritional components of Gadus macrocephalus roe. Journal of Qingdao University (National science edition) 29 (2), 57–62. (in Chinese).
Churcher A. M., Hubbard P. C., Marques J. P., Canario A. V., Huertas M. (2015). Deep sequencing of the olfactory epithelium reveals specific chemosensory receptors are expressed at sexual maturity in the European eel Anguilla Anguilla. Mol. Ecol. 24, 822–834. doi: 10.1111/mec.13065
Dayal J. S., Ali S. A., Thirunavukkarasu A. R., Kailasam M., Subburaj R. (2003). Nutrient and amino acid profiles of egg and larvae of Asian seabass, Lates calcarifer (Bloch). Fish Physiology and Biochemistry 29, 141–147.
Dekker W., Casselman J. M. (2014). The 2003 Québec declaration of concern about eel declines-11 years later: are eels climbing back up the slippery slope? Fisheries 39, 613–614. doi: 10.1080/03632415.2014.979342
de Souza Romaneli R., do Nascimento T. M. T., Gous R. M., de Paula Reis M., Mansano C. F. M., Khan K. U., et al. (2021). Response of Nile tilapia (Oreochromis niloticus) to lysine: Performance, body composition, maintenance and efficiency of utilization. Aquaculture 538, 736522. doi: 10.1016/j.aquaculture.2021.736522
Faulk C. K., Holt G. J. (2008). Biochemical composition and quality of captive-spawned cobia Rachycentron canadum eggs. Aquaculture 279, 70–76.
Feunteun E., Prouzet P. (2020). “Forty years of decline and 10 years of management plan: Are European eels (Anguilla Anguilla) recovering?,” in Coast bordeaux symposium and of the 17th french-Japanese oceanography symposium (Cham, Switzerland: Springer International Publishing), 269–295.
Folch J., Lees M., Sloane Stanley G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509. doi: 10.1016/S0021-9258(18)64849-5
Gilani G. S., Peace R. W. (2005). Chromatographic determination of amino acids in foods. J. AOAC Int. 88, 877–887. doi: 10.1093/jaoac/88.3.877
Guedes M., Costa-Pinto A. R., Gonçalves V. M., Moreira-Silva J., Tiritan M. E., Reis R. L., et al. (2020). Sardine roe as a source of lipids to produce liposomes. ACS Biomaterials Sci. Eng. 6, 1017–1029. doi: 10.1021/acsbiomaterials.9b01462
Hamidoghli A., Bae J., Won S., Lee S., Kim D. J., Bai S. C. (2019). A review on Japanese eel (Anguilla japonica) aquaculture, with special emphasis on nutrition. Rev. Fisheries Sci. Aquaculture 27, 226–241. doi: 10.1080/23308249.2019.1583165
Hsu H. Y., Chen S. H., Cha Y. R., Tsukamoto K., Lin C. Y., Han Y. S. (2015). De novo assembly of the whole transcriptome of the wild embryo, preleptocephalus, leptocephalus, and glass eel of Anguilla japonica and deciphering the digestive and absorptive capacities during early development. PloS One 10, e0139105. doi: 10.1371/journal.pone.0139105
Huang D., Liang H., Ren M., Ge X., Ji K., Yu H., et al. (2021). Effects of dietary lysine levels on growth performance, whole body composition and gene expression related to glycometabolism and lipid metabolism in grass carp, Ctenopharyngodon idellus fry. Aquaculture 530, 735806. doi: 10.1016/j.aquaculture.2020.735806
Ishikawa S., Suzuki K., Inagaki T., Watanabe S., Kimura Y., Okamura A., et al. (2001). Spawning time and place of the Japanese eel Anguilla japonica in the North Equatorial Current of the western North Pacific Ocean. Fisheries Sci. 67, 1097–1103. doi: 10.1046/j.1444-2906.2001.00366.x
Jang W. J., Kim S. K., Lee S. J., Kim H., Ryu Y. W., Shin M. G., et al. (2022). Effect of Bacillus sp. supplementation diet on survival rate and microbiota composition in artificially produced eel larvae (Anguilla japonica). Front. Microbiol. 13, 891070. doi: 10.3389/fmicb.2022.891070
Jiang T. B., Liu L. P., Gao X. Y., Chen W. Y., Wu J. M. (2012). The changes of serum biochemical components during carp pituitary extract and HCG induced maturation of the female Japanese eel (Anguilla japonica) (in Chinese). J. Fisheries China 36, 893–899. doi: 10.3724/SP.J.1231.2012.27821
Kleppe L., Edvardsen R. B., Furmanek T., Taranger G. L., Wargelius A. (2014). Global transcriptome analysis identifies regulated transcripts and pathways activated during oogenesis and early embryogenesis in Atlantic cod. Mol. Reprod. Dev. 81, 619–635. doi: 10.1002/mrd.22328
Koh I. C. C., Hamada D., Tsuji Y., Okuda D., Nomura K., Tanaka H., et al. (2017). Sperm cryopreservation of Japanese eel, Anguilla japonica. Aquaculture 473, 487–492. doi: 10.1016/j.aquaculture.2017.03.011
Kuan-Mei H., Shingo K., Yu-San H., Aigo T., Yoshiyuki I., Caroline D. (2018). Effect of ENSO events on larval and juvenile duration and transport of Japanese eel (Anguilla japonica). PloS One 13, e0195544. doi: 10.1371/journal.pone.0195544
Lanes C. F. C., Bizuayehu T. T., Bolla S., Martins C., de Oliveira Fernandes J. M., Bianchini A., et al. (2012). Biochemical composition and performance of Atlantic cod (Gadus morhua L.) eggs and larvae obtained from farmed and wild broodstocks. Aquaculture 324, 267–275.
Langmead B. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li C., Cheng X. F., Hong B., Chen X. Y., Wu Y. N., Li H. (2016). Nutritional analysis and evaluation on eggs of Spinibarbus caldwelli. Chin. J. Anim. Nutr. 28, 2204–2212. Available at: http://www.chinajan.com/CN/abstract/html/20160726.htm.
Li B., Dewey C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 1–16. doi: 10.1186/1471-2105-12-323
Liu Z. F., Gao X. Q., Yu J. X., Wang Y. H., Guo Z. L. (2018). Changes of protein and lipid contents, amino acid and fatty acid compositions in eggs and yolk-sac larvae of American shad (Alosa sapidissima). J. Ocean Univ. China 17, 209–215. doi: 10.1007/s11802-018-3403-3
Liu L. P., Liu D. P., Pu J. C., Du L., Chen T. Y., Chen W. Y., et al. (2017). Effects of different initial diets on the survival and behavior characteristics of the larvae of Japanese eel (Anguilla japonica) (in Chinese). J. Fisheries China 41, 703–710. doi: 10.11964/jfc.20160910542
Ma S., Hao S. X., Li L. H., Yang X. Q., Huang Y., Wei Y., et al. (2019). Comparative analysis of nutritional components of several roes. South China Fisheries Science 15 (4), 113–121.
Masuda Y., Imaizumi H., Usuki H., Oda K., Hashimoto H., Teruya K. (2012). Artificial completion of the Japanese eel, Anguilla japonica, life cycle: challenge to mass production. Bull. Fisheries Res. Agency 35, 111–117.
Matsushige K., Yasutake Y., Mochioka N. (2019). Spatial distribution and habitat preferences of the Japanese eel, Anguilla japonica, at the reach and channel-unit scales in four rivers of Kagoshima Prefecture, Japan. Ichthyological Res. 67, 68–80. doi: 10.1007/s10228-019-00704-x
Morrison W. R., Smith L. M. (1964). Preparation of fatty acid methyl esters and dimethylacetals from lipids with boron fluoride-methanol. J. Lipid Res. 5, 600–608. doi: 10.1016/S0022-2275(20)40190-7
Mourente G., Rosa V. (1996). Changes in the content of total lipid, lipid classes and their fatty acids of developing eggs and unfed larvae of the Senegal sole, Solea Senegalensis kaup. Fish Physiol. Biochem. 15, 221–235. doi: 10.1007/BF01875573
Ohkubo N., Sawaguchi S., Nomura K., Tanaka H., Matsubara T. (2008). Utilization of free amino acids, yolk protein and lipids in developing eggs and yolk-sac larvae of Japanese eel Anguilla japonica. Aquaculture 282, 130–137. doi: 10.1016/j.aquaculture.2008.06.017
Okamura A., Horie N., Mikawa N., Yamada Y., Tsukamoto K. (2014). Recent advances in artificial production of glass eels for conservation of anguillid eel populations. Ecol. Freshw. Fish 23, 95–110. doi: 10.1111/eff.12086
Okamura A., Yamada Y., Horie N., Mikawa N., Tanaka S., Kobayashi H., et al. (2013). Hen egg yolk and skinned krill as possible foods for rearing leptocephalus larvae of Anguilla japonica Temminck & Schlegel. Aquaculture Res. 44, 1531–1538. doi: 10.1111/are.2013.44.issue-10
Okamura A., Yamada Y., Horie N., Mikawa N., Tsukamoto K. (2019). Long-term rearing of Japanese eel larvae using a liquid-type diet: food intake, survival and growth. Fisheries Sci. 85, 687–694. doi: 10.1007/s12562-019-01316-0
Okamura A., Yamada Y., Mikawa N., Horie N., Tsukamoto K. (2020). Dietary supplementation with chitin hydrolysates for Anguilla japonica leptocephali. Fisheries Sci. 86, 685–692. doi: 10.1007/s12562-020-01440-2
Ozsolak F., Milos P. M. (2010). RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12, 87–98. doi: 10.1038/nrg2934
Pfeiler E. (1999). Developmental physiology of elopomorph leptocephali. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 123, 113–128. doi: 10.1016/S1095-6433(99)00028-8
Pike C., Crook V., Gollock M. (2020). Anguilla Anguilla (The IUCN Red List of Threatened Species). Available online at: https://dx.doi.org/10.2305/IUCN.UK.2020-2.RLTS.T60344A152845178.en (Accessed 2024 12 June). 2020.
Politis S. N., Sørensen S. R., Mazurais D., Servili A., Zambonino-Infante J. L., Miest J. J., et al. (2018). Molecular ontogeny of first-feeding European eel larvae. Front. Physiol. 9. doi: 10.3389/fphys.2018.01477
Pousão-Ferreira P., Morais S., Dores E., Narciso L. (1999). Eggs of gilthead seabream Sparus aurata L. as a potential enrichment product of Brachionus sp. in the larval rearing of gilthead seabream Sparus aurata L. Aquaculture Res. 30, 751–758. doi: 10.1046/j.1365-2109.1999.00394.x
Robinson M. D., Mccarthy D. J., Smyth G. K. (2010). EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Shi Y. H., Xu J. B., Xie Y. D., Liu Y. S., Shui C., Lu G. H., et al. (2020). Nutritional composition analysis and evaluation of eggs in American Shad Alosa sapidissima. Fisheries Science 39 (3), 407–413. (in Chinese).
Skoog A., Alldredge A., Passow U., Dunne J., Murray J. (2008). Neutral aldoses as source indicators for marine snow. Mar. Chem. 108, 195–206. doi: 10.1016/j.marchem.2007.11.008
Tanaka H. (2015). Progression in artificial seedling production of Japanese eel Anguilla japonica. Fisheries Sci. 81, 11–19. doi: 10.1007/s12562-014-0821-z
Tanaka H., Kagawa H., Ohta H. (2001). Production of leptocephali of Japanese eel (Anguilla japonica) in captivity. Aquaculture 201, 51–60. doi: 10.1016/S0044-8486(01)00553-1
Tanaka H., Kagawa H., Ohta H., Unuma T., Nomura K. (2003). The first production of glass eel in captivity: fish reproductive physiology facilitates great progress in aquaculture. Fish Physiol. Biochem. 28, 493–497. doi: 10.1023/B:FISH.0000030638.56031.ed
Tang L., Xiao X., Shi H., Chen J., Han C., Huang H., et al. (2020). Induction of oocyte maturation and changes in the biochemical composition, physiology and molecular biology of oocytes during maturation and hydration in the orange-spotted grouper (Epinephelus coioides). Aquaculture 522, 735115. doi: 10.1016/j.aquaculture.2020.735115
Tong X. H., Tang X. H., Bao C. M., Yang X. L., Chen Q. (2016). Relationship between embryonic growth and nucleic acids of Scophthalmus maximus. Acta Agri. Zhejiangensis. 28 (3), 428–434. (in Chinese).
Tsukamoto K. (1992). Discovery of the spawning area for Japanese eel. Nature 356, 789–791. doi: 10.1038/356789a0
Wang B., Wu H. W., Li L. Y. (2020). Analysis and evaluation of the nutritional compositions of skipjack tuna (Katsuwonus pelamis) roes. Journal of Guangdong Ocean University 40 (2), 111–116. (in Chinese).
Wang Z., Gerstein M., Snyder M. (2010). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Xiong B. X., Long L. Q., Su X., Chang Q. (1996). Crude protein and essential amino acid countens among eel feeds made in China (in Chinese). J. Huazhong Agric. Univ. 15, 60–63.
Xu Y., Li W., Ding Z. (2010). Effects of polyunsaturated fatty acids on immunity and survival of fish and their mechanisms (in Chinese). Chin. J. Anim. Nutr. 23, 551–556.
Yanes-Roca C., Rhody N., Nystrom M., Main K. L. (2009). Effects of fatty acid composition and spawning season patterns on egg quality and larval survival in common snook (Centropomus undecimalis). Aquaculture 287, 335–340. doi: 10.1016/j.aquaculture.2008.10.043
Yang J. J., Jiang Z. Q., Zuo R. T., Wang S. Y., Wen S. H., Sun H. (2014). Nutritional analysis and evaluation on eggs of Hemitripterus villosus (in Chinese). Chin. J. Anim. Nutr. 26, 1103–1110.
Zheng T. T., Zhou J., Weng X., Chen L. J., Cheng W. J., Pang J., et al. (2020). Analysis and evaluation of nutritional components of four types of marine aquatic roes. Food Fermentation Industries 46, 244–249. doi: 10.13995/j.cnki.11-1802/ts.023891
Keywords: Anguilla japonica, embryo, larvae, transcriptome analysis, nutritional components
Citation: Li K, Li Y, Li T, Cui R and Liu L (2024) Nutritional composition and transcriptome analysis of the newly hatched Anguilla japonica from embryo to preleptocephali obtained from artificial reproduction. Front. Mar. Sci. 11:1424999. doi: 10.3389/fmars.2024.1424999
Received: 29 April 2024; Accepted: 17 June 2024;
Published: 24 July 2024.
Edited by:
Yngvar Olsen, NTNU, NorwayReviewed by:
Chunyan Zhao, Qingdao Agricultural University, ChinaCopyright © 2024 Li, Li, Li, Cui and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liping Liu, bHAtbGl1QHNob3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.