- 1Faculty of Bioengineering and Veterinary Medicine, Don State Technical University, Rostov-on-Don, Russia
- 2Centre for Healthy Eating & Food Innovation (HEFI), Maastricht University—Campus Venlo, Venlo, Netherlands
- 3Division of Immunobiology and Biomedicine, Center of Genetics and Life Sciences, Sirius University of Science and Technology, Federal Territory Sirius, Sochi, Russia
- 4Center for Oceanography and Marine Biology “Moskvarium”, Moscow, Russia
- 5Division of Nanobiomedicine, Center of Genetics and Life Sciences, Sirius University of Science and Technology, Federal Territory Sirius, Sochi, Russia
Dolphins are aquatic mammals, whose role in microbial ecology is not fully understood. It is critical to study the microbiota of dolphins kept in captivity, as close contact with humans could unpredictably affect both human and animal health. In this study, we investigated the gut microbiota of 15 captive common bottlenose dolphins, and the relationship of microbial diversity and composition with animals’ age, sex, birthplace (dolphinarium or wildlife), and direct contacts with dolphinarium visitors using high-throughput V3-V4 16S rRNA amplicon sequencing. Based on acquired sequencing data we calculated and compared alpha and beta diversity indexes, as well as conducted differential abundance analysis of identified bacterial taxa to find multivariable associations with dolphins’ age period, sex, birthplace, and type of contact with humans with linear models. There was no association between gut microbiota composition and diversity and dolphins’ age period, sex, and birthplace. However, there were significant differences (q-value ≤ 0.00018) in the relative abundance of Edwardsiella genus in dolphins that were in contact only with dolphinarium staff in comparison to dolphins that apart from dolphinarium staff also had contact with dolphinarium visitors. These results show that dolphins born in wildlife and dolphinariums do not have significant differences in gut microbiota composition and the microbiota of captive dolphins can be affected by their contact with humans.
1 Introduction
The concept of global microbial ecology emphasizes the importance of microorganisms for the environment and the health of all living beings, which in turn intersects the ideas of this concept with the One Health approach (Lemke and DeSalle, 2023; Law et al., 2024). Microbes are involved in various biological processes: from metabolizing anthropological wastes in the oceans to modulating the immune systems of humans and animals (Zhang, 2021; Zhai et al., 2023). It is important to keep in mind that any human intervention in nature causes unpredictable alterations in any environment, including microbial communities, which in turn could cause further erratic outcomes for humans. A case in point is the domestication of cats, which increased the occurrence of schizophrenia in cat owners (Hakami et al., 2022; McGrath et al., 2023). The key role in increasing the rates of schizophrenia in cat owners is plays\ed by Toxoplasma gondii, which is greatly abundant in the Felidae microbiota (Kaushik et al., 2014). This is not the only example of emerging diseases caused by human intervention in natural environments and animal habitats, as the emergence of tuberculosis, borreliosis, SARS, and other bacterial, viral, fungal, and protozoan infections is also directly or indirectly induced by anthropogenic activities (Vouga and Greub, 2016; Donnik et al., 2021; Carpouron et al., 2022; Tunali and Korkmaz, 2023). The constant threat of emerging infections justifies the need for microbial ecology studies of different environments. Considering the degree of knowledge of microbial communities of humans, domestic animals, and terrestrial environments, the microbiomes of marine environments and wild animals, especially marine species, is little studied (Sehnal et al., 2021; Abreu et al., 2022). This, in turn, emphasizes the need of investigation of microbial communities of marine animals that are in close contact with humans and are under the process of domestication, especially the ones from zoos and oceanariums, as in comparison to animals domesticated thousands of years ago nothing is known about their microbiota, which leads to a degree of uncertainty for assessing the microbiological risks (Bai et al., 2021).
Common bottlenose dolphins (Tursiops truncatus) are the most common and well-known marine mammals of the family Delphinidae (Cetacea) (Villagra-Blanco et al., 2017). The history of domestication and training of this marine species started in 1938 when the first captive long-term colony of common bottlenose dolphins was established with the opening of Marine Studios (later Marineland) in St. Augustine, Florida (Wells, 2009). Starting from this date, many dolphinariums and aquariums featuring common bottlenose dolphins were opened all over the world, which in turn brought the need to develop veterinary healthcare management, including infectious disease control, including zoonic ones, as captive dolphins are in close contact with their trainers and sometimes with the visitors of dolphinariums (Barratclough et al., 2019; Segawa et al., 2023).
The gut microbiota of mammals plays a vital role in their health. Some researchers even state that gut microbiota should be considered as a separate organ within the body (Borgo et al., 2018) Although this statement is debatable, the role of gut microbiota in a host’s well-being is undeniable, as its composition and diversity directly affect and reflect the host’s health conditions (Bensch et al., 2023; Popov et al., 2023a). Gut microbial communities in mammals are influenced by various factors, such as age, sex, and diet of the host, and also the environment, where the host lives (de Jonge et al., 2022; Dillard et al., 2022; Kim, 2022; Xu et al., 2023; Popov et al., 2023b). The studies of gut microbiota of common bottlenose dolphins are limited, which does not allow to come to a consensus on developing healthcare guidelines and assessing the effects of the aforementioned factors on dolphins’ intestinal microbiome. In this study, we investigated gut microbiota composition and diversity of clinically healthy captive common bottlenose dolphins (Tursiops truncatus gilli) of different ages, sexes, and birthplaces, as well as with different contacts with humans (only with dolphinarium staff or also with dolphinarium visitors).
2 Materials and methods
2.1 Sampling
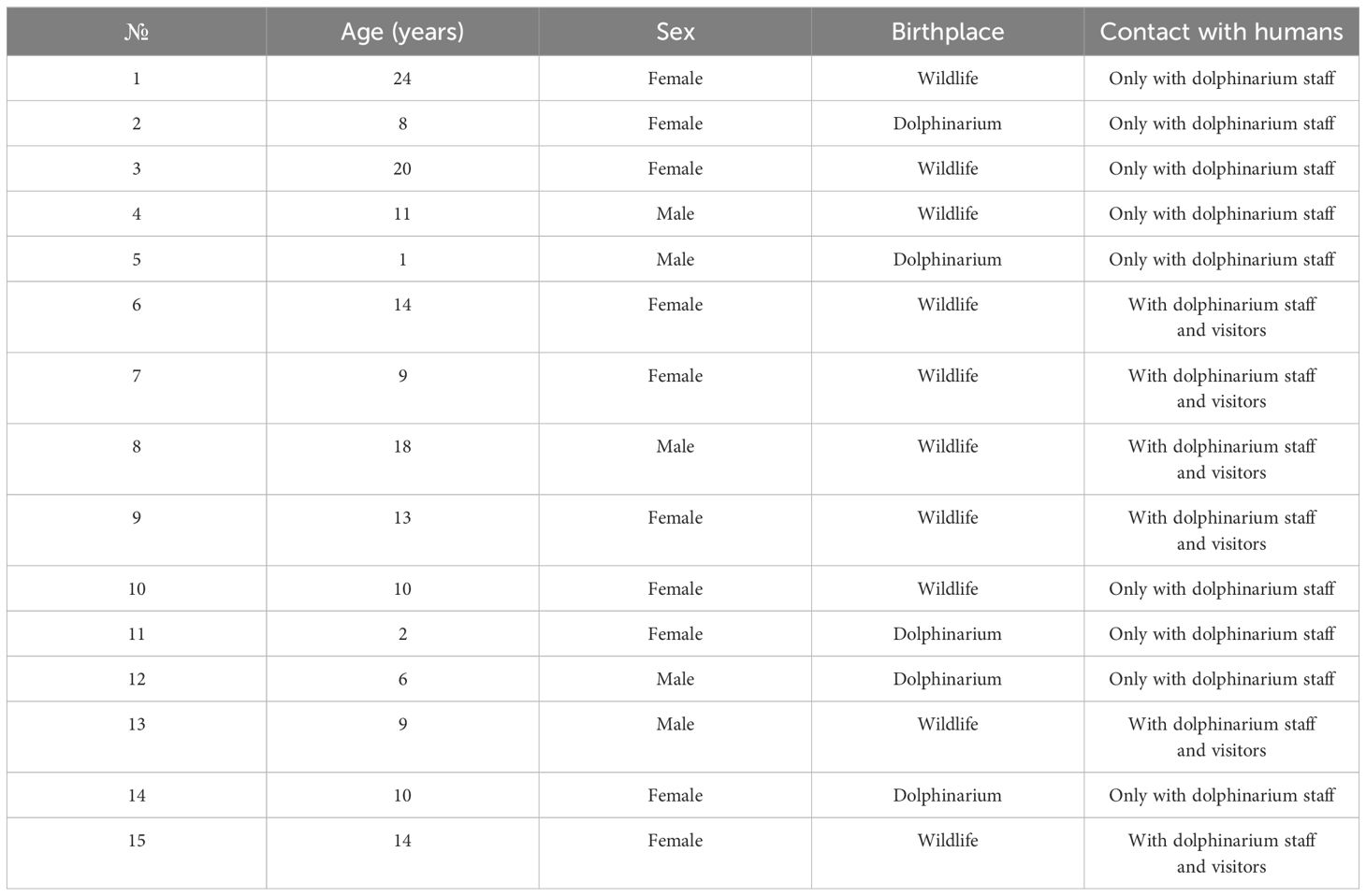
In this study, we collected fecal samples from 15 common bottlenose dolphins (T. truncatus gilli) from the Center for Oceanography and Marine Biology “Moskvarium”, Moscow. Age, sex, birthplace, and information regarding contact with dolphinarium staff and visitors are provided in Table 1. Dolphins included in this study were clinically healthy at least for six months before the analyses according to daily clinical veterinary care and monthly hematological and biochemical analyses of blood samples. The diet of all dolphins consisted of herring, pink salmon, char, greenling, and squid. The one-year-old dolphin was partially breastfed.
To collect fecal samples, each bottlenose dolphin voluntarily, on command, lay belly up in such a way that at the same time, the genital opening of the animal was below the surface of the water. The anal area was rubbed with a sterile cloth moistened with antiseptics. Then a sterile silicone gastric tube Fr16 was inserted into the rectum of the animal to a depth of about 30 cm, and the feces was gently aspirated using a Janet’s syringe attached to the probe (Figure 1). All samples were taken during the routine clinical care of dolphins after 3 hours from feeding. Collected fecal samples were placed in sterile tubes with DNA/RNA Shield (Zymo Research, USA) and transported to the laboratory at room temperature.
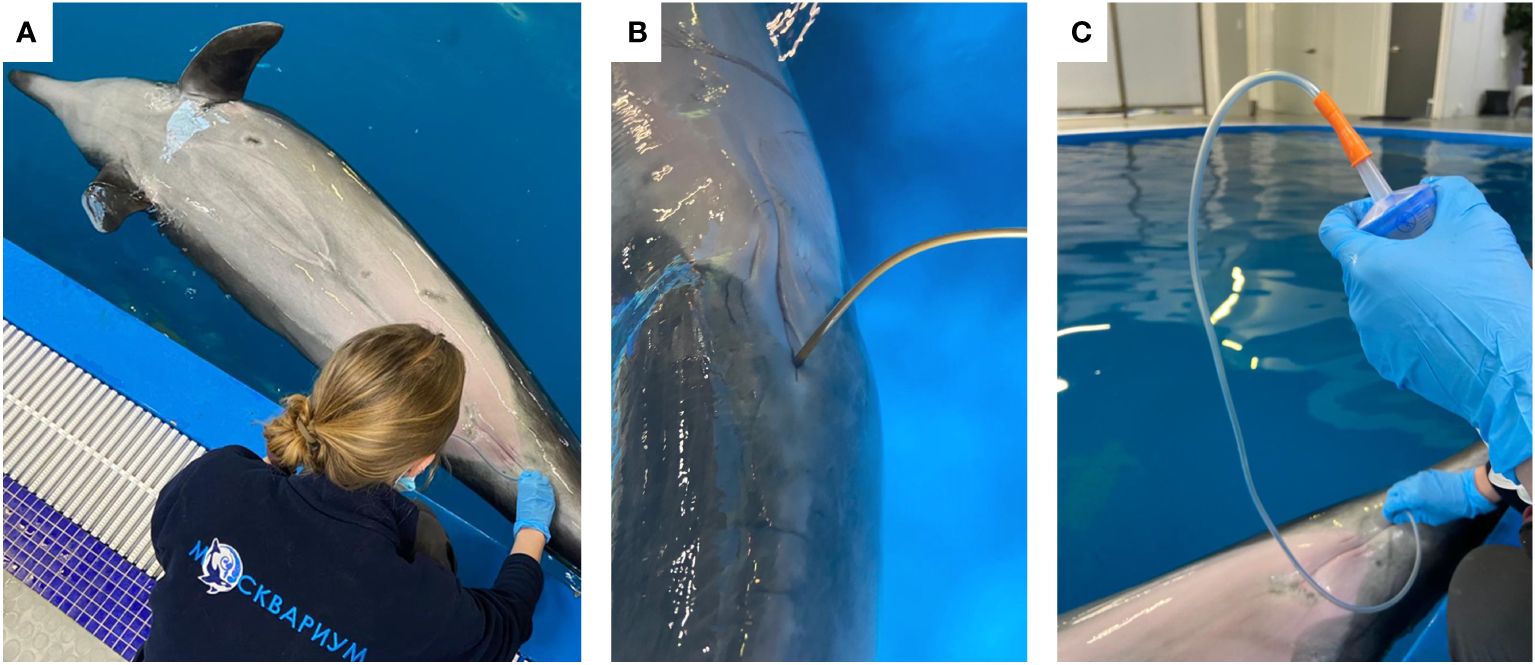
Figure 1 Photos representing the sampling process: bottlenose dolphin voluntarily, on command, lay belly up (A); sterile silicone gastric tube Fr16 inserted into the rectum of the animal (B); Janet’s syringe attached to the probe and used for the aspiration of feces (C).
2.2 Gut microbiota composition
Sequencing of the V3-V4 region of the 16S rRNA gene was performed to determine microbiota composition. In short, from the fecal samples, DNA was isolated with QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany), amplified with barcoding, pooled, and subsequently sequenced with the Illumina MiSeq sequencing system according to the manufacturer’s instructions (Illumina, USA). The Binary Base Call text-based format for storing biological sequence and corresponding quality scores pipeline (BCL2FASTQ, version 1.8.3, Illumina, USA) was used to convert the sequences into a text-based format for storing biological sequence and corresponding quality score (FASTQ) files after quality checking. Subsequently, Quantitative Insights Into Microbial Ecology 2 (QIIME-2, version 2023.5) software was used to analyze the results (Bolyen et al., 2019). Classification of the sequences into amplicon sequence variants was performed using DADA2 (version 1.26.0) software (Callahan et al., 2016). Taxonomic identification of amplicon sequence variants was conducted with the Silva database (version 138 (available online: https://www.arb-silva.de/documentation/release-138/accessed on 27 July 2023) as a reference 16S rRNA database (Quast et al., 2013; Yilmaz et al., 2014).
2.3 Statistical analysis
The software package R v4.2.3 (R Foundation for Statistical Computing, Vienna, Austria) was used for statistical analyses. “MaAsLin2” (version 1.7.3) package (Mallick et al., 2021) was used for differential abundance analysis of gut microbial composition in dolphins with different age periods, sex, birthplace, and contacts with humans. Old/senior dolphins (>18 years) were used as a reference for the linear model analysis. Age was used as a categorical variable according to T. truncatus age periods: calves – 1–2 years, adults – 6–14 years, and old/senior dolphins – 18+ years (Tezanos Pinto, 2009). Neonates, juveniles, and 30+-year-old dolphins were not included in this study. Before differential abundance analysis, we performed filtering by applying a minimal 10% threshold for the prevalence of the taxa to get more robust results (Nearing et al., 2022). Based on acquired counts, the relative abundance (RA) of taxa was calculated. Taxa with a RA higher than 1% were used for the differential abundance analysis. For the alpha and beta diversity indexes calculations, the “vegan” (version 2.6–6.1) package was used (Dixon, 2003). For the alpha diversity analysis, we used Shannon (Shannon, 1948) and Chao1 (Chao, 1984) indexes, as well as the number of observed features (amplicon sequence variants) (Chiarello et al., 2022), while for the beta diversity Bray-Curtis similarity (Sorensen, 1948) and Jaccard dissimilarity (Jaccard, 1908) indexes were used. Non-parametric Kruskal–Wallis and Mann–Whitney tests were performed to determine differences in the alpha diversity. Permutational multivariate analysis of variance (PERMANOVA) with the adonis function from the “vegan” package was used to determine differences in beta diversity distances (the number of permutations was set to 999). Multiple comparisons were performed with the non-parametric Mann–Whitney test and the results were adjusted with the Benjamini–Hochberg false discovery rate. Adjusted p-values (q-values) were considered significant at q < 0.05.
2.4 Ethical statement
Experimental procedures for this report did not include any in vivo studies. Fecal samples for high-throughput sequencing were taken during the routine clinical care of dolphins (Moscvarium veterinary care protocol no. 25/162–3).
3 Results
3.1 Gut microbiota composition
On the phylum level, we observed Pseudomonadota, Bacillota, Fusobacteriota, and Bacteroidota with a mean RA among all studied dolphins of 37%, 24%, 24%, and 11% respectively (Supplementary Figure 1A). On the class level, Gammaproteobacteria (37%), Fusobacteriia (24%), Bacilli (16%), and Clostridia (8%) were among the abundant classes (Supplementary Figure 1B). On the order level (Supplementary Figure 1C), the major abundant taxa that were observed in the bottlenose dolphins were Enterobacterales (35%), Fusobacteriales (24%), Bacteroidales (11%), Lactobacillales (11%), Mycoplasmatales (4%), Oscillospirales (3%), and Lachnospirales (3%). Enterobacteriaceae (26%), Fusobacteriaceae (24%), Enterococcaceae (9%), Bacteroidaceae (7%), Hafniaceae (4%), Mycoplasmataceae (4%), Lachnospiraceae (3%), Prevotellaceae (3%), Ruminococcaceae (2%), Vibrionaceae (2%), Morganellaceae (2%), Pasteurellaceae (1%), Oscillospiraceae (1%) were among majorly abundant families in bottlenose dolphins from Moscvarium (Supplementary Figure 1D). On the genus level (Supplementary Figure 1E), there were 12 taxa, which mean RA among all dolphins were more than 1%: Cetobacterium (23%), Escherichia-Shigella (14%), Plesimonas (11%), Enterococcus (9%), Bacteroides (7%), Edwardsiella (4%), Ureaplasma (4%), Prevotella 9 (2%), Vibrio (2%), Actinobacillus (1%), Morganella (1%), and Fusobacterium (1%). The bacterial gut composition in each studied dolphin is visualized in Figure 2; Supplementary Figures 2-6.
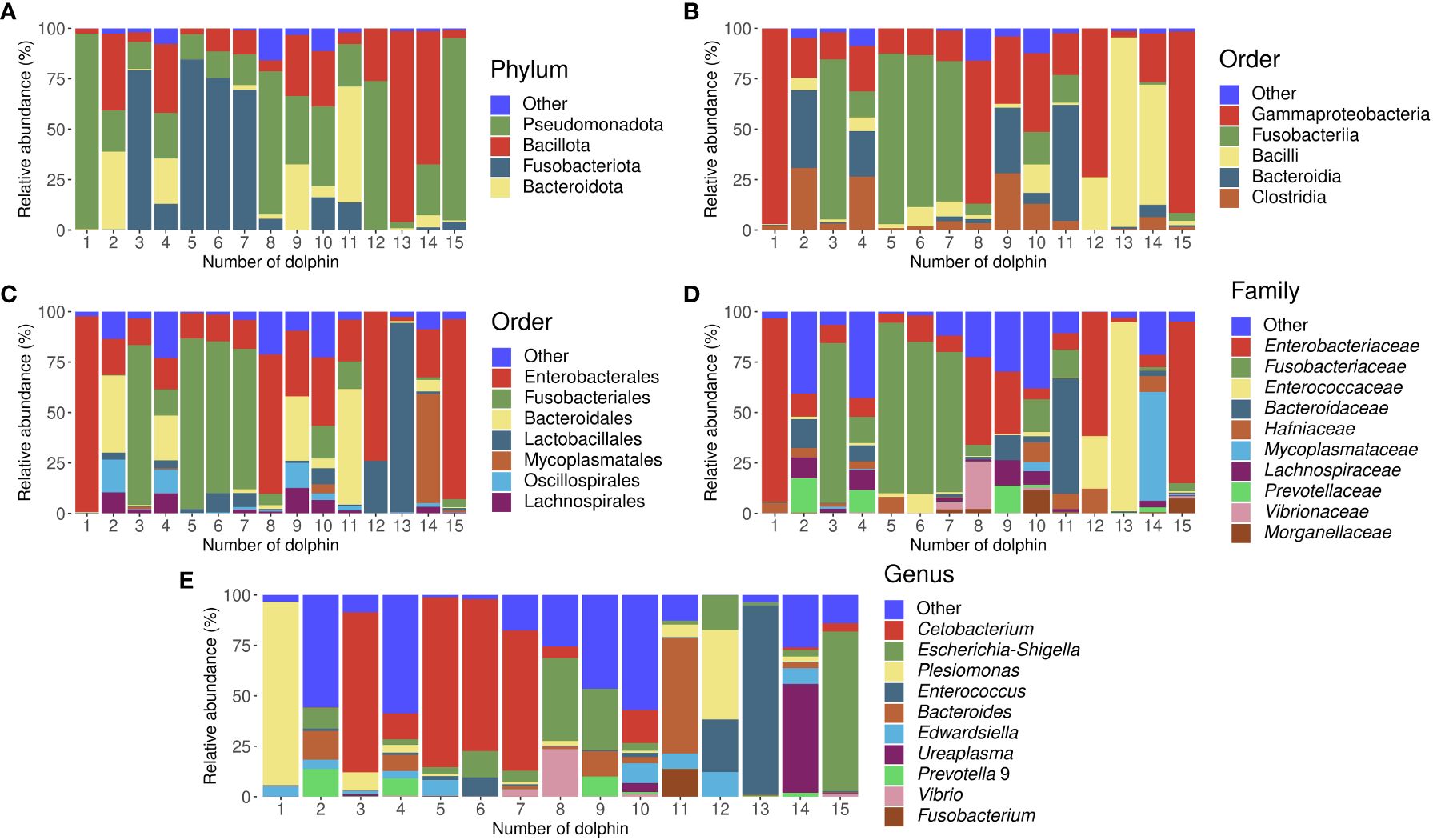
Figure 2 Stacked bar charts representing gut microbiota composition of all studied dolphins separately in the relative abundances of bacterial taxa on phylum (A), class (B), order (C), family (D), and genus (E) levels. Dolphins’ numbers correspond to Table 1.
After conducting differential abundance analysis, we did not observe any significant associations between bacterial RA and age period (q-value>0.05), sex (q-value>0.05), and birthplace (q-value>0.05). However, there was a significantly greater (q-value<0.0001) abundance of the Edwardsiella genus in dolphins, who had contact with only dolphinarium staff in comparison to animals, who also were in contact with dolphinarium visitors as well. Figure 3 shows the results of differential abundance analysis and a direct comparison of gut microbial composition in dolphins with different types of contact with humans.
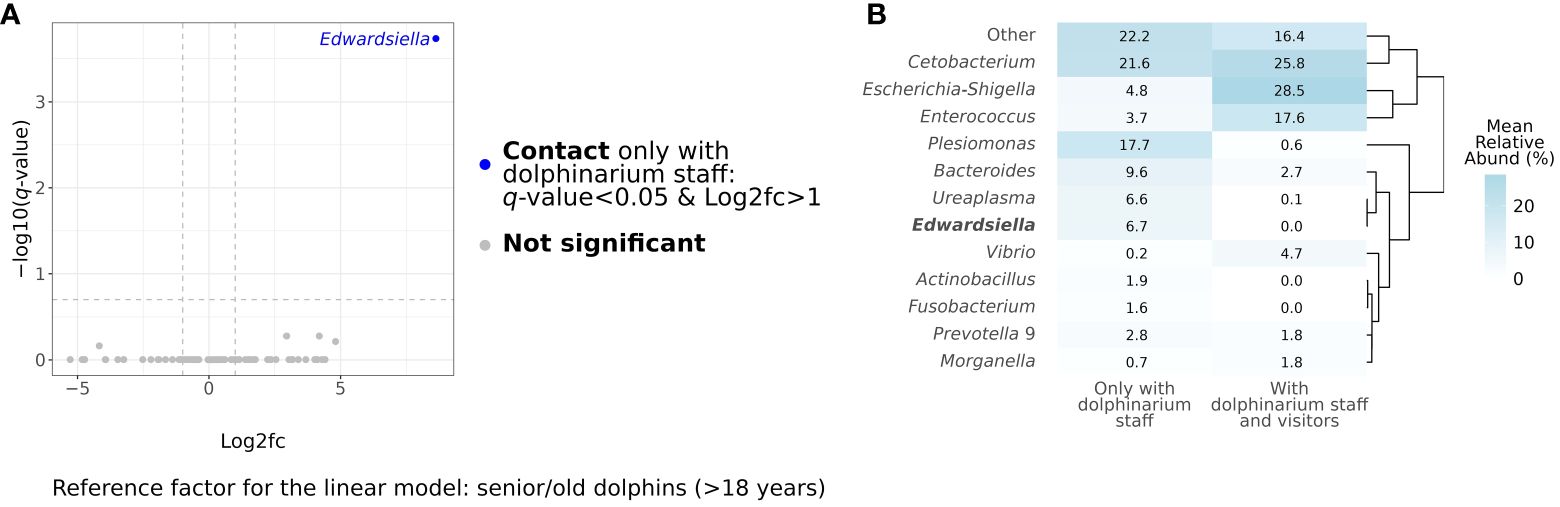
Figure 3 Results of differential abundance analysis at the genus level using MaAsLin2 (Mallick et al., 2021). The blue dot in the volcano plot represents a taxon, whose relative abundance statistically significantly differed in dolphins, which contact visitors and dolphinarium staff, in comparison to dolphins, which contact only dolphinarium staff (A). The heatmap represents the direct comparison of bacterial mean relative abundances in gut microbiota in captive bottlenose with different contacts with humans at the genus levels. Genus, whose relative abundance has significant differences, is marked in bold (B). Log2fc, log2 fold change.
3.2 Gut microbiota diversity
According to the multiple comparisons testing of alpha diversity index values, there were no statistically significant differences between the captive bottlenose dolphins of different sexes, age periods, and birthplaces, as well as between dolphins that contacted only with dolphinarium staff and the ones that also contacted dolphinarium visitors (Figure 4). Generally, the alpha diversity was more variable in adult dolphins (Figures 4A–C), and dolphins born in the wild (Figures 4J–L).
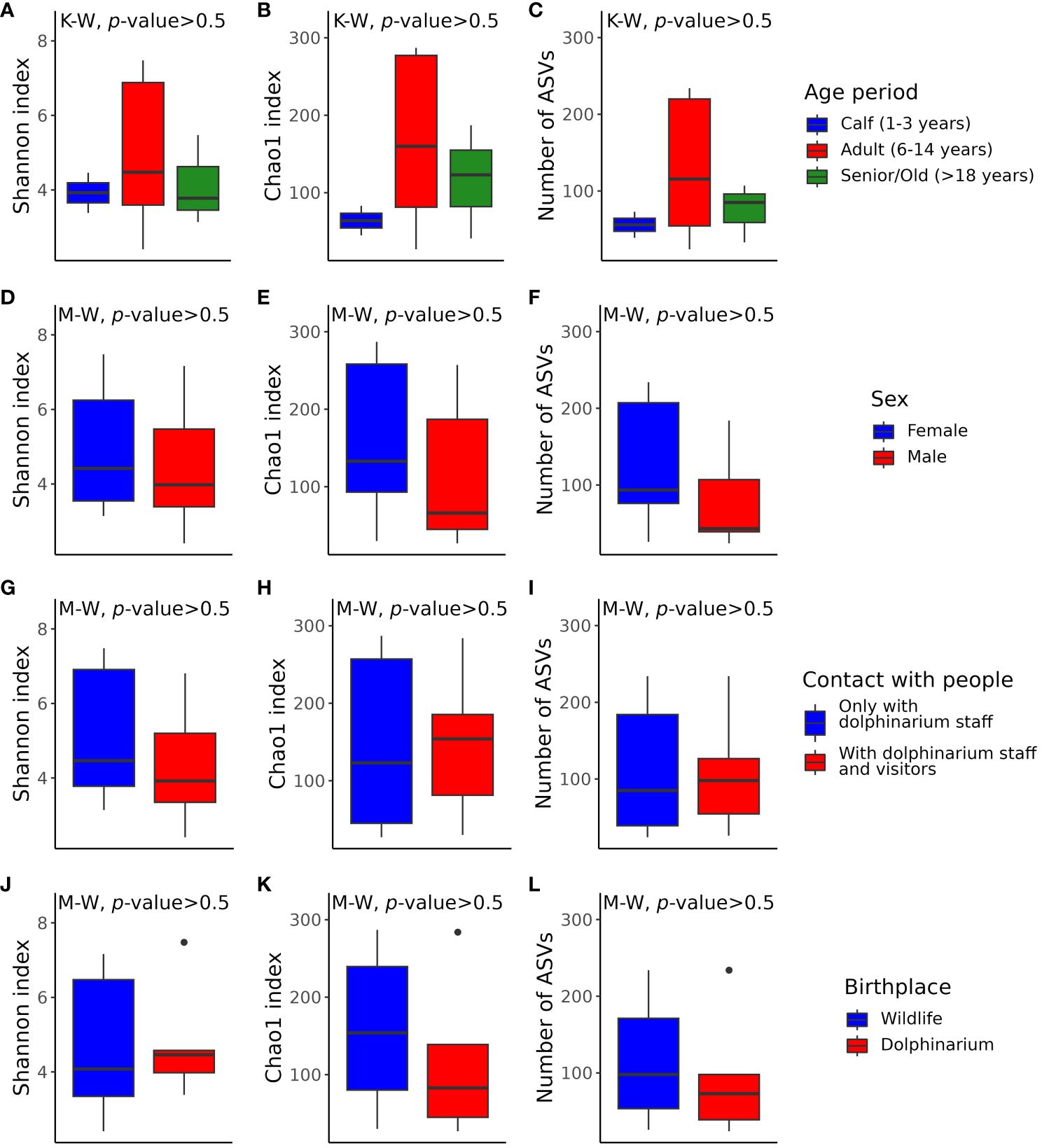
Figure 4 Box plots illustrating alpha diversity in samples acquired from dolphins of different age periods (A–C), sex (D–F), with different frequencies of contact with various humans (G–I), and birthplace (J–L). Plots (A, D, G, J) show Shannon’s indexes; plots (B, E, H), and K show Chao1 indexes; plots (C, F, I, L) show the number of observed features. The p-values were calculated with the Kruskal–Wallis (K-W) and Mann-Whitney (M-W) tests. ASVs, amplicon sequence variants.
Also, we did not find any significant differences between the studied dolphins of different age periods, sexes, and birthplaces and between dolphins that did and did not have contact with dolphinarium visitors with respect to beta diversity (Figure 5).
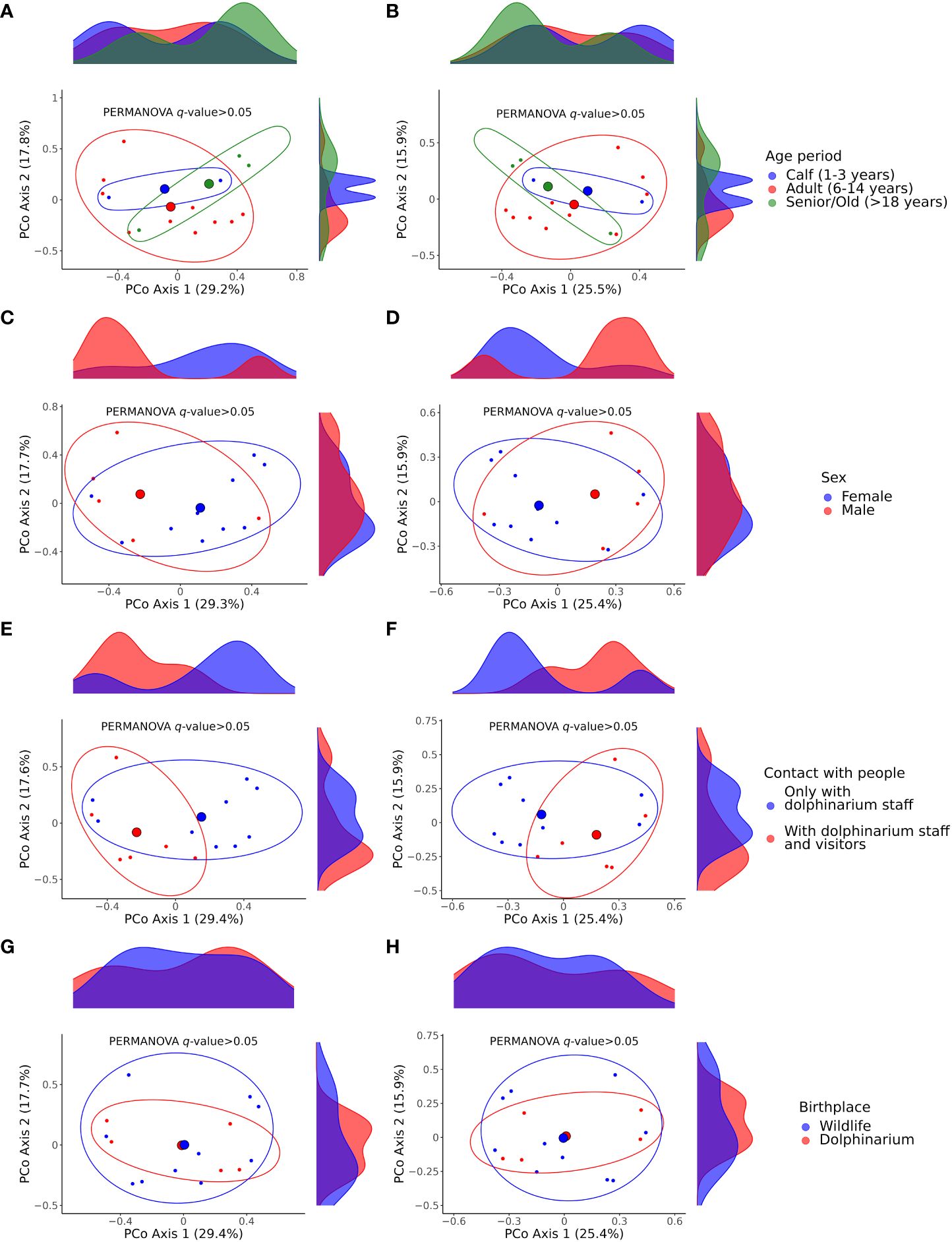
Figure 5 Principal coordinate analysis (PCoA) plots illustrating differences in beta diversity among samples acquired from dolphins of different age periods (A, B), sex (C, D), with different contacts with humans regarding dolphinarium visitors (E, F), and birthplace (G, H). Plots (A, C, E, G) show Bray–Curtis dissimilarity distances; plots (B, D, F, H) show Jaccard similarity distances. The p-values were calculated with the PERMANOVA test; the number of permutations was set to 999. The large colored spheres represent the centroids of the groups. PERMANOVA, permutational multivariate analysis of variance.
4 Discussion
In this research, we studied gut microbiota composition and diversity of captive common bottlenose dolphins T. truncatus gilli and compared these variables among dolphins of different ages, sexes, and birthplaces, and also between dolphins that did and did not have contact with dolphinarium visitors. T. truncatus gilli previously was considered a subspecies of T. truncatus, as the skull morphological features, such as the size of the mandibular condyles and the position of the optic canal of T. truncatus gilli differed from the T. truncatus (True, 1889), but recent comparative morphological studies support the evidence that T. truncatus gilli is junior synonym of T. truncatus, as the cranial morphology features had no significant differences (Costa et al., 2023). From our point of view, to reach a consensus on this question molecular studies should be conducted to detect all subspecies of T. truncatus, and conclude the status of T. truncatus gilli. In this article, we use T. truncatus gilli term for the definition of studied dolphins, as all the animals included in this study were identified by Moskvarium zoologists and veterinarians as T. truncatus gilli due to the presence of distinctive cranial morphological features. However, we do not exclude the fact that the study’s findings on gut microbiota features could also be applied to T. truncatus species in general.
Previously, the gut microbiota composition and diversity of common bottlenose dolphins were studied by Soverini et al. in Italy (Soverini et al., 2016), Suzuki et al. in Japan (Suzuki et al., 2019), Bai et al. in China (Bai et al., 2021), and Bik et al., Robles-Malagamba et al., Olmstead et al., and Linnehan et al. in USA (Bik et al., 2016; Robles-Malagamba et al., 2020; Olmstead et al., 2023; Linnehan et al., 2024). Thus, our study is the first report from Eastern Europe, Russia, on common bottlenose dolphins’ gut microbiota composition and diversity.
Soverini et al. studied the gut microbiota of ten captive common bottlenose dolphins (nine adults and one calf) from an Italian dolphinarium with V3-V4 16S rRNA sequencing of metagenomic DNA extracted from fecal samples. In that study, on the phylum level, the most abundant taxa were Bacillota and Pseudomonadota with mean relative abundances of 56% and 27% respectively. Among subdominant phyla were Actinobacteria (5%), Bacteroidetes (3%), Fusobacteria (4%) and Tenericutes (3%). Among the most abundant families were Clostridiaceae (16%), Vibrionaceae (12%), Staphylococcaceae (6%), Lactobacillaceae (7%), Peptostreptococcaceae (7%), Ruminococcaceae (5%), Fusobacteriaceae (4%) and Pasteurellaceae (4%) (Soverini et al., 2016). Suzuki et al. compared the gut microbiota of sixteen bottlenose dolphins from three different aquaria in Japan using high-throughput V3-V4 16S rRNA amplicon sequencing. In this study, they reported three majorly abundant phyla: Pseudomonadota (21–70%), Bacillota (3–50%), and Fusobacteriota (8–38%); although their RA significantly differed between aquariums. On the family level, among largely abundant taxa were Fusobacteriaceae, Peptostreptococcaceae, Lachnospiraceae, Vibrionaceae, Rhodobacteraceae, Pasteurellaceae, Brucellaceae, Clostridiaceae, Enterobacteriaceae, and others. But in comparison to the phylum level the RA of bacterial families differed even more dramatically between dolphins from the different aquariums. These results are supported by the outcomes of alpha and beta diversity analyses, where significant differences were also seen between studied aquariums. The outcomes of this study show that the aquatic environment strongly affects the gut microbiota of captive bottlenose dolphins, as all three aquariums had different rearing conditions (Suzuki et al., 2019). Bai et al. conducted a comparative study of the gut microbial communities among four different marine mammal species with one common bottlenose dolphin included from an aquarium in China using V4 16S rRNA sequencing. In the gut microbiota of the studied bottlenose dolphin, there were two dominant phyla: Pseudomonadota and Bacillota. Among dominant taxa on the family level, there were Enterobacteriaceae, Clostridiaceae 1, Peptostreptococcaceae, and Vibrionaceae, while on the genus level, there were Vibrio, Morganella, and Clostridium sensu stricto 1 (Bai et al., 2021). Bik et al. studied the microbiota of captive sea lions and bottlenose dolphins, as well as free-ranging dolphins of the same species using 16S rRNA sequencing. In this study, Bacillota and Pseudomonadota were dominant phyla in the gut microbiota of captive and free-ranging common bottlenose dolphins. Also, significant differences were observed in beta diversity between captive and free-ranging bottlenose dolphins, which additionally shows that the gut microbiota of these animals is strongly dependent on the environment (Bik et al., 2016). Robles-Malagamba et al. studied bacterial communities of 14 free-ranging common bottlenose dolphins (five calves and nine adults) from Florida, USA, also using V3-V4 16S rRNA high-throughput sequencing. Intestinal microbial communities on the phylum level were characterized by highly abundant Bacillota, Pseudomonadota, and Fusobacteriota, as in previously discussed studies. Interestingly, on the genus level, the most dominant bacterial lineage in the intestinal microbiota of free-ranging bottlenose dolphins was Cetobacterium (71%) with Clostridium XI (15%), and Clostridium sensu stricto (4%) occupying almost the entire remaining niche (Robles-Malagamba et al., 2020). Olmstead et al. used high-throughput V4 16S rRNA amplicon sequencing to investigate gut microbial communities in unusual mortality events of bottlenose dolphins associated with Morbillivirus. In this study, on the phylum level, the most abundant taxa were Bacillota (42%), Fusobacteriota (38%), Pseudomonadota (17%), and Bacteroidota (2%). The most dominant taxa on the genus level were Cetobacterium (31%), Clostridium sensu stricto 1 (24%), Paeniclostridium (10%), Photobacterium (6%), Fusobacterium (5%), Edwardsiella (3%), Escherichia-Shigella (3%), Peptostreptococcus (2%), Actinobacillus (2%), Bacteroides (2%), and Actinobacillus (1%). One of the most important outcomes of this study was the definition of the relationship between the detection of Morbillivirus in bottlenose dolphins and Actinobacillus delphinicola RA in their gut microbiota, which makes this bacteria a potential biomarker for health monitoring for this dolphin species (Olmstead et al., 2023). The recent study by Linnehan et al. was aimed at the safety and effectiveness evaluation of fecal microbiota transplantations in bottlenose dolphins using metagenomic sequencing. In this study, Bacillota and Pseudomonadota were also dominant taxa on the phylum level, as their total RA exceeded 80%. The most important outcome of this study was the successful fecal microbiota transplantation, as there were improvements in clinical signs, increases in alpha diversity, and most importantly engraftment of novel microbial species from donor in all four recipients (Linnehan et al., 2024).
In our study, the dominant bacterial taxa on the phylum level were Pseudomonadota (37%), Bacillota (24%), Fusobacteriota (24%), and Bacteroidota (11%), which generally corresponds to previous studies, although their RA differs from the above-discussed studies. On the genus level, there was a remarkable dominance of Cetobacterium (23%) which is consistent with the results of Robles-Malagamba et al. and Olmstead et al. studies, where this genus was the most dominant with the RA of 71% and 31% respectively (Robles-Malagamba et al., 2020; Olmstead et al., 2023). Cetobacterium is an important component of the aquatic animals’ microbiome (Godoy-Vitorino et al., 2017; Zeng et al., 2017; Ramírez et al., 2018; Zamora-Briseño et al., 2020; Ofek et al., 2021). Some studies even show that bacteria of this genus are involved in the host’s metabolism and have probiotic properties (Wang et al., 2021; Colorado Gómez et al., 2023; Qi et al., 2023; Zhao et al., 2023). Suzuki et al. isolated Cetobacterium ceti from the intestinal microbiota of captive common bottlenose dolphin and revealed vitamin B12 biosynthesis, which makes this bacteria important for supplying myoglobin and hemoglobin to dolphins (Suzuki et al., 2022). The second dominant taxon on the genus level in the gut microbiota of studied dolphins was Escherichia-Shigella, which also was observed in the intestinal microbiota of bottlenose dolphins in the Olmstead et al. study (Olmstead et al., 2023). Bacteria belonging to Escherichia and Shigella genera commonly inhabit the vertebrate intestinal tract, and also were found in marine mammals of other species (Bai et al., 2021; Gross et al., 2022; Watkins et al., 2022). Other bacterial genera identified in the intestinal microbiota of studied bottlenose dolphins, such as Plesimonas, Enterococcus, Bacteroides, and others, were also found in dolphins and other marine mammals (Bik et al., 2016; Janda et al., 2016; Medeiros et al., 2017; Bai et al., 2021; Olmstead et al., 2023).
We studied gut microbiota in clinically healthy captive dolphins, which had constant veterinary control and care. Although the bacterial composition and RA of identified taxa on the genus level did not fully correspond to other studies, the differences between the gut microbiota of dolphins in this study and others could be explained by different rearing environments and living conditions, which is corroborated by the study of Suzuki et al., where gut microbiota of dolphins significantly differed between different aquariums (Suzuki et al., 2019). We have to note that the most notable differences in the gut microbiota composition of bottlenose dolphins are between captive and free-ranging animals, for Cetobacterium abundance in particular. In free-ranging dolphins, the RA of this genus was around 70%, while in captive dolphins this value did not exceed 35% in above discussed studies, including ours (Robles-Malagamba et al., 2020; Olmstead et al., 2023). We speculate that Cetobacterium abundance could be a valuable health marker for captive bottlenose dolphins, and veterinary care protocols could be developed regarding this, which could include enrichment of the aquatic environment with this bacteria or special diets aimed at the promotion of its growth. Also, it is important to note that we did not detect potential pathogenic bacteria belonging to Brucella, Erysipelothrix, Streptococcus, Staphylococcus, and Salmonella genera (Venn-Watson et al., 2008; Bik et al., 2016; Marangi et al., 2022).
In this study, we investigated the relationships between the composition and diversity of the gut microbiota of dolphins and their sex, age period, birthplace, and also the influence of contact with dolphinarium visitors. We did not observe significant differences in gut microbiota between dolphins of different age periods, male and female dolphins, as well as between dolphins born in the wildlife and dolphinarium. The majority of studies show that the sex of marine mammals does not strongly affect their gut microbiota (Bates et al., 2023). However, some studies show sex-related differences in the intestinal microbiota of marine mammals, which could be related to the different physiology of males and females (Pacheco-Sandoval et al., 2022). The absence of differences in microbiota between dolphins with different birthplaces could additionally indicate the importance of aquatic rearing environments on gut microbes in dolphins in addition to the discussion above. Although we did not observe significant differences in gut bacterial diversity and composition between dolphins of different age periods, this does not mean that could not be age-related alterations in the gut microbiota of dolphins. Age-related shifts in gut microbiota composition were observed previously in marine mammals, for example in wild harbor seals (Phoca vitulina) and captive spotted seals (Phoca largha) (Tian et al., 2020; Pacheco-Sandoval et al., 2022). The physiology of immune, endocrine, and gastrointestinal systems in mammals alters with age, which results in different digestion and host-microbiome interactions affecting gut microbial composition and diversity (Coman and Vodnar, 2020; Sadoughi et al., 2022; Chen et al., 2023; Xu et al., 2023). The notable outcome of this study is the difference in Edwardsiella abundance, which was absent in dolphins who were in contact with staff and visitors, while in dolphins, that had contact with only dolphinarium staff its RA was 6.7%. Bacteria belonging to the Edwardsiella genus are commonly found in aquatic environments and animals, as well as the gut microbiota of humans, and could be both pathogenic and non-pathogenic (Sakai et al., 2007; Nakamura et al., 2013). As all dolphins included in this study were clinically healthy, we speculate that detected Edwardsiella bacteria could be non-pathogenic, although this needs to be proven in further studies. Most importantly, these finding show that the type of contact with humans could affect the microbiota of captive dolphins, as the dolphins who contact dolphinarium visitors obviously have more frequent contact with various people in comparison to dolphins who contact only with dolphinarium staff. In other studies of zoo animals’ microbiota, a gain of human-like microbiota in captive animals over time has been observed (Clayton et al., 2016). We hypothesize that close contact with various human individuals can alter the microbiota of captive wild animals with a direct decrease in their natural bacterial abundance or even its substitution, which also should be addressed in veterinary care protocols for zoo animals.
The main limitation of this study is the sample size of 15 dolphins, although in most previous studies the sample size of captive common bottlenose was similar (Soverini et al., 2016; Suzuki et al., 2019; Robles-Malagamba et al., 2020). In our study, the sample size was objectively limited by the number of available animals in the dolphinarium. Also, another limitation linked to the limited sample size could be grouping animals by age period, where we marked animals older than 18 years as “old/senior”. The results of the Jaakkola et al. study showed that the median life expectancy for captive bottlenose dolphins during the 2003–2012 years time period was 29.2 years (Jaakkola and Willis, 2019), while in the Venn-Watson et al. study the median age of death of captive dolphins for the 2009–2013 years time period was 32 years (Venn-Watson et al., 2015). There are also reports that the maximum lifespan of bottlenose dolphins exceeded 50 years (Venn-Watson et al., 2015; Buddhachat et al., 2021; Barratclough et al., 2023). Taking into account various data on the lifespan of dolphins, marking animals older than 18 years old as old or senior could be conditional. Also, having only three dolphins of this age period could influence the results of statistical analyses. We hypothesize that a larger sample size would allow for revealing alterations in the gut microbiota of dolphins associated with age and contact with humans. For this study, we used one of the most robust methods for differential abundance analysis according to the bioinformatical benchmark study by Nearing et al., which included filtering the least abundant taxa, combined with subsequent identification of multivariable associations with linear models using the Maaslin2 package (Nearing et al., 2022). Further studies should be based on a larger sample size by including animals from different dolphinariums and aquariums and free-ranging animals, which is essential for direct comparison of captive and wild dolphins’ microbiota. Also, other methods for studying microbial communities should be considered, as high-throughput 16S rRNA amplicon sequencing allows for composition and diversity analyses, while other methods would allow for the investigation of other features and properties, including functional features, of gut bacteria (Durazzi et al., 2021; Regueira-Iglesias et al., 2023).
5 Conclusions
In conclusion, we investigated the bacterial composition and diversity of intestinal microbiota of captive bottlenose dolphins from a dolphinarium (Moscvarium, Moscow, Russia). The gut bacterial composition generally corresponds to the previous studies of captive bottlenose dolphins’ intestinal microbiota, although the bacterial relative abundance on the genus level slightly differed, which can be associated with the different rearing conditions. There was no association between gut microbiota composition and diversity and dolphins’ age, sex, and birthplace, but there were some significant differences in microbial abundances between dolphins that contact dolphinarium staff and visitors compared to animals that contact only dolphinarium staff, which should be addressed in developing veterinary care protocols for dolphins in the future.
Data availability statement
All sequences associated with this study have been deposited into the National Center for Biotechnological Information’s Short Read Archive and are available under BioProject ID PRJNA1087733.
Ethics statement
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because experimental procedures for this report did not include any in vivo studies. Fecal samples for metagenomic sequencing were taken during the routine clinical care of dolphins (Moscvarium veterinary care protocol no. 25/162-3).
Author contributions
IP: Writing – original draft, Conceptualization, Formal analysis, Funding acquisition, Investigation, Software, Visualization. IS: Investigation, Methodology, Resources, Writing – review & editing. AK: Investigation, Writing – original draft. IC: Formal analysis, Writing – original draft. IT: Software, Writing – original draft. KV: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing. AE: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research has been made possible with the support of the Dutch Province of Limburg with a grant (HEFI-1) to the Centre for Healthy Eating & Food Innovation of Maastricht University -campus Venlo.
Acknowledgments
IP acknowledges the support of the Ministry of Science and Higher Education of the Russian Federation (order No. 486 of 15 June 2021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1410769/full#supplementary-material
References
Abreu A., Bourgois E., Gristwood A., Troublé R., Acinas S. G., Bork P., et al. (2022). Priorities for ocean microbiome research. Nat. Microbiol. 7, 937–947. doi: 10.1038/s41564-022-01145-5
Bai S., Zhang P., Zhang C., Du J., Du X., Zhu C., et al. (2021). Comparative study of the gut microbiota among four different marine mammals in an aquarium. Front. Microbiol. 12, 769012. doi: 10.3389/fmicb.2021.769012
Barratclough A., McFee W. E., Stolen M., Hohn A. A., Lovewell G. N., Gomez F. M., et al. (2023). How to estimate age of old bottlenose dolphins (Tursiops truncatus); by tooth or pectoral flipper? Front. Mar. Sci. 10, 364. doi: 10.3389/fmars.2023.1135521
Barratclough A., Wells R. S., Schwacke L. H., Rowles T. K., Gomez F. M., Fauquier D. A., et al. (2019). Health assessments of common bottlenose dolphins (Tursiops truncatus): past, present, and potential conservation applications. Front. Veterinary Sci. 6, 444. doi: 10.3389/fvets.2019.00444
Bates K. A., Higgins C., Neiman M., King K. C. (2023). Turning the tide on sex and the microbiota in aquatic animals. Hydrobiologia. 850, 3823–3835. doi: 10.1007/s10750-022-04862-4
Bensch H. M., Tolf C., Waldenström J., Lundin D., Zöttl M. (2023). Bacteroidetes to Firmicutes: captivity changes the gut microbiota composition and diversity in a social subterranean rodent. Anim. Microbiome. 5, 9. doi: 10.1186/s42523-023-00231-1
Bik E. M., Costello E. K., Switzer A. D., Callahan B. J., Holmes S. P., Wells R. S., et al. (2016). Marine mammals harbor unique microbiotas shaped by and yet distinct from the sea. Nat. Commun. 7, 10516. doi: 10.1038/ncomms10516
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Borgo F., Garbossa S., Riva A., Severgnini M., Luigiano C., Benetti A., et al. (2018). Body mass index and sex affect diverse microbial niches within the gut. Front. Microbiol. 9, 334465. doi: 10.3389/fmicb.2018.00213
Buddhachat K., Brown J. L., Kaewkool M., Poommouang A., Kaewmong P., Kittiwattanawong K., et al. (2021). Life expectancy in marine mammals is unrelated to telomere length but is associated with body size. Front. Genet. 12, 737860. doi: 10.3389/fgene.2021.737860
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carpouron J. E., de Hoog S., Gentekaki E., Hyde K. D. (2022). Emerging animal-associated fungal diseases. J. Fungi. 8, 611. doi: 10.3390/jof8060611
Chao A. (1984). Nonparametric estimation of the number of classes in a population. Scandinavian J. Stat 11 (4), 265–270.
Chen J., Lv W., Zhang X., Zhang T., Dong J., Wang Z., et al. (2023). Animal age affects the gut microbiota and immune system in captive koalas (Phascolarctos cinereus). Microbiol. Spectr. 11, e04101–e04122. doi: 10.1128/spectrum.04101-22
Chiarello M., McCauley M., Villéger S., Jackson C. R. (2022). Ranking the biases: The choice of OTUs vs. ASVs in 16S rRNA amplicon data analysis has stronger effects on diversity measures than rarefaction and OTU identity threshold. PloS One. 17, e0264443. doi: 10.1371%2Fjournal.pone.0264443
Clayton J. B., Vangay P., Huang H., Ward T., Hillmann B. M., Al-Ghalith G. A., et al. (2016). Captivity humanizes the primate microbiome. Proc. Natl. Acad. Sci. 113, 10376–10381. doi: 10.1073/pnas.1521835113
Colorado Gómez M. A., Melo-Bolívar J. F., Ruíz Pardo R. Y., Rodriguez J. A., Villamil L. M. (2023). Unveiling the Probiotic Potential of the Anaerobic Bacterium Cetobacterium sp. nov. C33 for Enhancing Nile Tilapia (Oreochromis niloticus) Cultures. Microorganisms. 11, 2922. doi: 10.3390/microorganisms11122922
Coman V., Vodnar D. C. (2020). Gut microbiota and old age: Modulating factors and interventions for healthy longevity. Exp. Gerontol. 141, 111095. doi: 10.1016/j.exger.2020.111095
Costa A., Archer F., Rosel P., Perrin W. (2023). Tursiops truncatus nuuanu, a new subspecies of the common bottlenose dolphin from the eastern tropical Pacific. J. Mamm. Evol. 30, 213–229. doi: 10.1007/s10914-022-09641-5
de Jonge N., Carlsen B., Pertoldi C., Nielsen J. L. (2022). The gut microbiome of 54 mammalian species. Front. Microbiol. 13, 886252. doi: 10.3389/fmicb.2022.886252
Dillard B. A., Chung A. K., Gunderson A. R., Campbell-Staton S. C., Moeller A. H. (2022). Humanization of wildlife gut microbiota in urban environments. Elife. 11, e76381. doi: 10.7554/eLife.76381.sa2
Dixon P. (2003). VEGAN, a package of R functions for community ecology. J. vegetation Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Donnik I., Popov I. V., Sereda S., Popov I. V., Chikindas M., Ermakov A. (2021). Coronavirus infections of animals: Future risks to humans. Biol. Bull. 48, 26–37. doi: 10.1134/S1062359021010052
Durazzi F., Sala C., Castellani G., Manfreda G., Remondini D., De Cesare A. (2021). Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 11, 3030. doi: 10.1038/s41598-021-82726-y
Godoy-Vitorino F., Rodriguez-Hilario A., Alves A. L., Gonçalves F., Cabrera-Colon B., Mesquita C. S., et al. (2017). The microbiome of a striped dolphin (Stenella coeruleoalba) stranded in Portugal. Res. Microbiol. 168, 85–93. doi: 10.1016/j.resmic.2016.08.004
Gross S., Müller A., Seinige D., Wohlsein P., Oliveira M., Steinhagen D., et al. (2022). Occurrence of antimicrobial-resistant Escherichia coli in marine mammals of the north and baltic seas: Sentinels for human health. Antibiotics. 11, 1248. doi: 10.3390/antibiotics11091248
Hakami R. M., Alnaami A. A., Shbeeli K. A., Al Suhaym A. Y., Khormi B. H., Faqihi I. H., et al. (2022). Association between childhood exposure to pet cats and later diagnosis of schizophrenia: A case-control study in Saudi Arabia. Cureus. 14, e32401. doi: 10.7759/cureus.32401
Jaakkola K., Willis K. (2019). How long do dolphins live? Survival rates and life expectancies for bottlenose dolphins in zoological facilities vs. wild populations. Mar. Mammal Sci. 35, 1418–1437. doi: 10.1111/mms.12601
Jaccard P. (1908). Nouvelles recherches sur la distribution florale. Bull. Soc Vaud. Sci. Nat. 44, 223–270. doi: 10.5169/seals-268384
Janda J. M., Abbott S. L., McIver C. J. (2016). Plesiomonas shigelloides revisited. Clin. Microbiol. Rev. 29, 349–374. doi: 10.1128/CMR.00103-15
Kaushik M., Knowles S., Webster J. (2014). What makes a feline fatal in Toxoplasma gondii’s fatal feline attraction? Infected rats choose wild cats. Am. Zoologist. 54, 118–128. doi: 10.1093/icb/icu060
Kim N. (2022). Sex difference of gut microbiota. Sex/gender-specific Med. Gastrointestinal Dis., 363–377. doi: 10.1007/978-981-19-0120-1
Law S. R., Mathes F., Paten A. M., Alexandre P., Regmi R., Reid C., et al. (2024). Life at the borderlands: microbiomes of interfaces critical to One Health. FEMS Microbiol. Rev. 48 (2), fuae008. doi: 10.1093/femsre/fuae008
Lemke M., DeSalle R. (2023). The next generation of microbial ecology and its importance in environmental sustainability. Microbial Ecol. 85, 781–795. doi: 10.1007/s00248-023-02185-y
Linnehan B. K., Kodera S. M., Allard S. M., Brodie E. C., Allaband C., Knight R., et al. (2024). Evaluation of the safety and efficacy of fecal microbiota transplantations in bottlenose dolphins (Tursiops truncatus) using metagenomic sequencing. J. Appl. Microbiol. 135, lxae026. doi: 10.1093/jambio/lxae026
Mallick H., Rahnavard A., McIver L. J., Ma S., Zhang Y., Nguyen L. H., et al. (2021). Multivariable association discovery in population-scale meta-omics studies. PloS Comput. Biol. 17, e1009442. doi: 10.1371/journal.pcbi.1009442
Marangi M., Carlucci R., Carlino P., Fanizza C., Cirelli G., Maglietta R., et al. (2022). Dolphins and sea turtles may host zoonotic parasites and pathogenic bacteria as indicators of anthropic pressure in the Gulf of Taranto (Northern Ionian Sea, Central-Eastern Mediterranean Sea). Veterinary Res. Commun. 46, 1157–1166. doi: 10.1007/s11259-022-10011-y
McGrath J. J., Lim C. C., Saha S. (2023). Cat ownership and schizophrenia-related disorders and psychotic-like experiences: A systematic review and meta-analysis. Schizophr. Bull. 50 (3), sbad168. doi: 10.1093/schbul/sbad168
Medeiros A. W., Blaese Amorim D., Tavares M., de Moura T. M., Franco A. C., d’Azevedo P. A., et al. (2017). Enterococcus species diversity in fecal samples of wild marine species as determined by real-time PCR. Can. J. Microbiol. 63, 129–136. doi: 10.1139/cjm-2016-0427
Nakamura Y., Takano T., Yasuike M., Sakai T., Matsuyama T., Sano M. (2013). Comparative genomics reveals that a fish pathogenic bacterium Edwardsiella tarda has acquired the locus of enterocyte effacement (LEE) through horizontal gene transfer. BMC Genomics. 14, 1–14. doi: 10.1186/1471-2164-14-642
Nearing J. T., Douglas G. M., Hayes M. G., MacDonald J., Desai D. K., Allward N., et al. (2022). Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 13, 342. doi: 10.1038/s41467-022-28034-z
Ofek T., Lalzar M., Laviad-Shitrit S., Izhaki I., Halpern M. (2021). Comparative study of intestinal microbiota composition of six edible fish species. Front. Microbiol. 12, 760266. doi: 10.3389/fmicb.2021.760266
Olmstead A. R., Mathieson O. L., McLellan W. A., Pabst D. A., Keenan T. F., Goldstein T., et al. (2023). Gut bacterial communities in Atlantic bottlenose dolphins (Tursiops truncatus) throughout a disease-driven (Morbillivirus) unusual mortality event. FEMS Microbiol. Ecol. 99, fiad097. doi: 10.1093/femsec/fiad097
Pacheco-Sandoval A., Lago-Lestón A., Abadía-Cardoso A., Solana-Arellano E., Schramm Y. (2022). Age as a primary driver of the gut microbial composition and function in wild harbor seals. Sci. Rep. 12, 14641. doi: 10.1038/s41598-022-18565-2
Popov I. V., Berezinskaia I. S., Popov I. V., Martiusheva I. B., Tkacheva E. V., Gorobets V. E., et al. (2023a). Cultivable gut microbiota in synanthropic bats: shifts of its composition and diversity associated with hibernation. Animals. 13, 3658. doi: 10.3390/ani13233658
Popov I. V., Einhardt Manzke N., Sost M. M., Verhoeven J., Verbruggen S., Chebotareva I. P., et al. (2023b). Modulation of swine gut microbiota by phytogenic blends and high concentrations of casein in a validated swine large intestinal in vitro model. Veterinary Sci. 10, 677. doi: 10.3390/vetsci10120677
Qi X., Zhang Y., Zhang Y., Luo F., Song K., Wang G., et al. (2023). Vitamin B12 produced by Cetobacterium somerae improves host resistance against pathogen infection through strengthening the interactions within gut microbiota. Microbiome. 11, 135. doi: 10.1186/s40168-023-01574-2
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramírez C., Coronado J., Silva A., Romero J. (2018). Cetobacterium is a major component of the microbiome of giant Amazonian fish (Arapaima gigas) in Ecuador. Animals. 8, 189. doi: 10.3390/ani8110189
Regueira-Iglesias A., Balsa-Castro C., Blanco-Pintos T., Tomás I. (2023). Critical review of 16S rRNA gene sequencing workflow in microbiome studies: From primer selection to advanced data analysis. Mol. Oral. Microbiol. 38, 347–399. doi: 10.1111/omi.12434
Robles-Malagamba M. J., Walsh M. T., Ahasan M. S., Thompson P., Wells R. S., Jobin C., et al. (2020). Characterization of the bacterial microbiome among free-ranging bottlenose dolphins (Tursiops truncatus). Heliyon. 6, e03944. doi: 10.1016/j.heliyon.2020.e03944
Sadoughi B., Schneider D., Daniel R., Schülke O., Ostner J. (2022). Aging gut microbiota of wild macaques are equally diverse, less stable, but progressively personalized. Microbiome. 10, 95. doi: 10.1186/s40168-022-01283-2
Sakai T., Iida T., Osatomi K., Kanai K. (2007). Detection of type 1 fimbrial genes in fish pathogenic and non-pathogenic Edwardsiella tarda strains by PCR. Fish Pathol. 42, 115–117. doi: 10.3147/jsfp.42.115
Segawa T., Ohno Y., Kurita M., Ishibashi T., Yoshioka M. (2023). Helicobacter delphinicola infection and the risk of gastric disease in common bottlenose dolphin. Dis. Aquat. Organisms. 155, 187–192. doi: 10.3354/dao03751
Sehnal L., Brammer-Robbins E., Wormington A. M., Blaha L., Bisesi J., Larkin I., et al. (2021). Microbiome composition and function in aquatic vertebrates: small organisms making big impacts on aquatic animal health. Front. Microbiol. 12, 567408. doi: 10.3389/fmicb.2021.567408
Shannon C. E. (1948). A mathematical theory of communication. Bell system Tech. J. 27, 379–423. doi: 10.1002/bltj.1948.27.issue-3
Sorensen T. (1948). A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biologiske Skrifter. 5, 1–34.
Soverini M., Quercia S., Biancani B., Furlati S., Turroni S., Biagi E., et al. (2016). The bottlenose dolphin (Tursiops truncatus) faecal microbiota. FEMS Microbiol. Ecol. 92, fiw055. doi: 10.1093/femsec/fiw055
Suzuki A., Segawa T., Sawa S., Nishitani C., Ueda K., Itou T., et al. (2019). Comparison of the gut microbiota of captive common bottlenose dolphins Tursiops truncatus in three aquaria. J. Appl. Microbiol. 126, 31–39. doi: 10.1111/jam.14109
Suzuki A., Shirakata C., Anzai H., Sumiyama D., Suzuki M. (2022). Vitamin B12 biosynthesis of Cetobacterium ceti isolated from the intestinal content of captive common bottlenose dolphins (Tursiops truncatus). Microbiology. 168, 001244. doi: 10.1099/mic.0.001244
Tezanos Pinto G. (2009). Population structure, abundance and reproductive parameters of bottlenose dolphins (Tursiops truncatus) in the Bay of Islands (Northland, New Zealand). ResearchSpace@ Auckland.
Tian J., Du J., Han J., Song X., Lu Z. (2020). Age-related differences in gut microbial community composition of captive spotted seals (Phoca largha). Mar. Mammal Sci. 36, 1231–1240. doi: 10.1111/mms.12728
True F. W. (1889). Contributions to the natural history of the cetaceans: A review of the family Delphinidae. Washington: Govt. Print. Off. doi: 10.5962/bhl.title.16278
Tunali V., Korkmaz M. (2023). Emerging and re-emerging parasitic infections of the central nervous system (CNS) in europe. Infect. Dis. Rep. 15, 679–699. doi: 10.3390/idr15060062
Venn-Watson S. K., Jensen E. D., Smith C. R., Xitco M., Ridgway S. H. (2015). Evaluation of annual survival and mortality rates and longevity of bottlenose dolphins (Tursiops truncatus) at the United States Navy Marine Mammal Program from 2004 through 2013. J. Am. Veterinary Med. Assoc. 246, 893–898. doi: 10.2460/javma.246.8.893
Venn-Watson S., Smith C. R., Jensen E. D. (2008). Primary bacterial pathogens in bottlenose dolphins Tursiops truncatus: needles in haystacks of commensal and environmental microbes. Dis. Aquat. Organisms. 79, 87–93. doi: 10.3354/dao01895
Villagra-Blanco R., Silva L., Aguilella-Segura A., Arcenillas-Hernández I., Martínez-Carrasco C., Seipp A., et al. (2017). Bottlenose dolphins (Tursiops truncatus) do also cast neutrophil extracellular traps against the apicomplexan parasite Neospora caninum. Int. J. Parasitology: Parasites Wildlife. 6, 287–294. doi: 10.1016/j.ijppaw.2017.09.002
Vouga M., Greub G. (2016). Emerging bacterial pathogens: the past and beyond. Clin. Microbiol. Infect. 22, 12–21. doi: 10.1016/j.cmi.2015.10.010
Wang A., Zhang Z., Ding Q., Yang Y., Bindelle J., Ran C., et al. (2021). Intestinal Cetobacterium and acetate modify glucose homeostasis via parasympathetic activation in zebrafish. Gut Microbes. 13, 1–15. doi: 10.1080/19490976.2021.1900996
Watkins C. A., Gaines T., Strathdee F., Baily J. L., Watson E., Hall A. J., et al. (2022). A comparative study of the fecal microbiota of gray seal pups and yearlings-a marine mammal sentinel species. Microbiologyopen. 11, e1281. doi: 10.1002/mbo3.1281
Wells R. S. (2009). Learning from nature: bottlenose dolphin care and husbandry. Zoo Biol. 28, 635–651. doi: 10.1002/zoo.20252
Xu X., Li G., Zhang D., Zhu H., Liu G. H., Zhang Z. (2023). Gut microbiota is associated with aging-related processes of a small mammal species under high-density crowding stress. Advanced Sci. 10, 2205346. doi: 10.1002/advs.202205346
Yilmaz P., Parfrey L. W., Yarza P., Gerken J., Pruesse E., Quast C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zamora-Briseño J. A., Cerqueda-García D., Hernández-Velázquez I. M., Rivera-Bustamante R., Huchín-Mian J. P., Briones-Fourzán P., et al. (2020). Alterations in the gut-associated microbiota of juvenile Caribbean spiny lobsters Panulirus argus (Latreille 1804) infected with pav1. J. Invertebrate Pathol. 176, 107457. doi: 10.1016/j.jip.2020.107457
Zeng S., Huang Z., Hou D., Liu J., Weng S., He J. (2017). Composition, diversity and function of intestinal microbiota in pacific white shrimp (Litopenaeus vannamei) at different culture stages. PeerJ. 5, e3986. doi: 10.7717/peerj.3986
Zhai X., Zhang X.-H., Yu M. (2023). Microbial colonization and degradation of marine microplastics in the plastisphere: A review. Front. Microbiol. 14, 1127308. doi: 10.3389/fmicb.2023.1127308
Zhang A. (2021). From immunity to collaboration: microbes, waste, and antitoxic politics. Curr. Anthropology. 62, S298–S310. doi: 10.1086/715542
Zhao Y., Li S., Lessing D. J., Guo L., Chu W. (2023). Characterization of Cetobacterium somerae CPU-CS01 isolated from the intestine of healthy crucian carp (Carassius auratus) as potential probiotics against Aeromonas hydrophila infection. Microbial Pathogenesis. 180, 106148. doi: 10.1016/j.micpath.2023.106148
Keywords: 16S rRNA, dolphins, microbial ecology, bacterial diversity, gut microbiota
Citation: Popov IV, Suvorova IV, Krikunova AA, Chebotareva IP, Tikhmeneva IA, Venema K and Ermakov AM (2024) Gut microbiota of captive common bottlenose dolphins Tursiops truncatus gilli: influence of age, sex, birthplace, and contacts with dolphinarium visitors. Front. Mar. Sci. 11:1410769. doi: 10.3389/fmars.2024.1410769
Received: 01 April 2024; Accepted: 10 June 2024;
Published: 27 June 2024.
Edited by:
Lifeng Zhu, Nanjing University of Chinese Medicine, ChinaReviewed by:
Xiaoling Wan, Chinese Academy of Sciences (CAS), ChinaShijie Bai, Chinese Academy of Sciences (CAS), China
Copyright © 2024 Popov, Suvorova, Krikunova, Chebotareva, Tikhmeneva, Venema and Ermakov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Igor V. Popov, aS5wb3BvdkBtYWFzdHJpY2h0dW5pdmVyc2l0eS5ubA==
†Present address: Koen Venema, Wageningen Food and Biobased Research, Wageningen University & Research, Wageningen, Netherlands
‡These authors have contributed equally to this work
 Igor V. Popov
Igor V. Popov