- 1Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China
- 2Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Zhuhai, China
- 3School of Oceanography, Shanghai Jiao Tong University, Shanghai, China
A new wood-fall-associated maldanid discovered at a depth of 2,321 m in the South China Sea is here morphologically and molecularly described as Nicomache tigilli sp. nov. The new species is characterized as having 21 chaetigers, a prostomium rounded anterior, an arched cephalic keel, anterior ends of nuchal grooves curved outwards, 3–6 acicular spines on chaetigers 1–3, notopodia with simply long and narrow capillary chaetae, and an anal funnel with triangular, unequal-sized anal cirri. Our molecular analyses of the genus Nicomache, encompassing COI, 16S, 18S, and 28S genes support establishing the new species, which appear phylogenetically closely related to the other species of the genus from reduced habitats.
1 Introduction
Maldanidae, a sedentary tube-building polychaete family known as bamboo worms, are widely distributed from the intertidal to the deep sea and from sandy beaches to muddy sediments, including chemosynthetic environments (Paterson et al., 2009; De Assis et al., 2012; Kobayashi et al., 2018; He et al., 2023). Maldanidae comprise more than 270 species and 45 genera (Pamungkas et al., 2019) grouped into six subfamilies (Rhodininae Arwidsson, 1906, Lumbriclymeninae Arwidsson, 1906, Notoproctinae Detinova, 1985, Maldaninae Malmgren, 1867, Nicomachinae Arwidsson, 1906, and Euclymeninae Arwidsson, 1906). Euclymeninae are reported as paraphyletic, with members of Nicomachinae nested within them (Kobayashi et al., 2018). Nicomachinae differ from Euclymeninae in having a strongly arched head not forming a cephalic plate, versus an obliquely truncated head forming a cephalic plate with a thin rim around its margin. In turn, the pygidium is key to distinguishing the genera in Nicomachinae, so that incomplete specimens cannot be accurately identified. Within Nicomachinae, Nicomache currently includes 17 species (De Assis et al., 2007; Kongsrud and Rapp, 2012), which are characterized by a lack of cephalic plate and having a long cephalic keel on the prostomium, short and curved nuchal grooves, acicular spines on the first three neuropodia, rostrate neurochaetae arranged in one row on following chaetigers, and well-developed funnel-shaped pygidium. Petaloproctus resembles Nicomache, but has an anal plate with a reduced dorsal border (De Assis et al., 2010). Four species of Nicomachinae have been recorded to date in Chinese coastal waters, namely, Nicomache inornata (Moore, 1903), N. lumbricalis (Fabricius, 1780), N. personata Johnson 1901, and Petaloproctus terricolus Quatrefages 1866 (Yang and Sun, 1988; Liu, 2008). However, with the exception of N. lumbricalis, all other reports cannot be confirmed due to a lack of voucher specimens. The other four species of Nicomache have been described from reducing environments: N. arwidssoni (Blake, 1985) and N. venticola (Blake and Hilbig, 1990) from the Pacific Ocean, N. lokii (Kongsrud & Rapp, 2012) from an Arctic vent, and N. ohtai (Miura and Hashimoto, 1991) from Japanese cold seeps. All of them belong to the subgenus Loxochona and are characterized by having anal funnels distally oblique in lateral view. In this paper, we describe a new species of Nicomache from China, found inhabiting a peculiar chemosynthetic environment, deep-sea wood falls. Despite maldanids being frequently found in this particular habitat (Wolff, 1979; Pailleret et al., 2007), there are no previous records from Chinese waters. Deep-sea wood falls are known to serve as stepping stones for the dispersal of species inhabiting other reduced environments, such as hydrothermal vents and cold seeps (Bienhold et al., 2013; Chen and Linse, 2020), while providing a specialized habitat and energy source that contributes to trigger species evolution in the overall oligotrophic deep ocean. Therefore, we are also exploring the possible relationships of our new species with other maldanids inhabiting reducing environments.
2 Materials and methods
2.1 Specimen collection and morphological observations
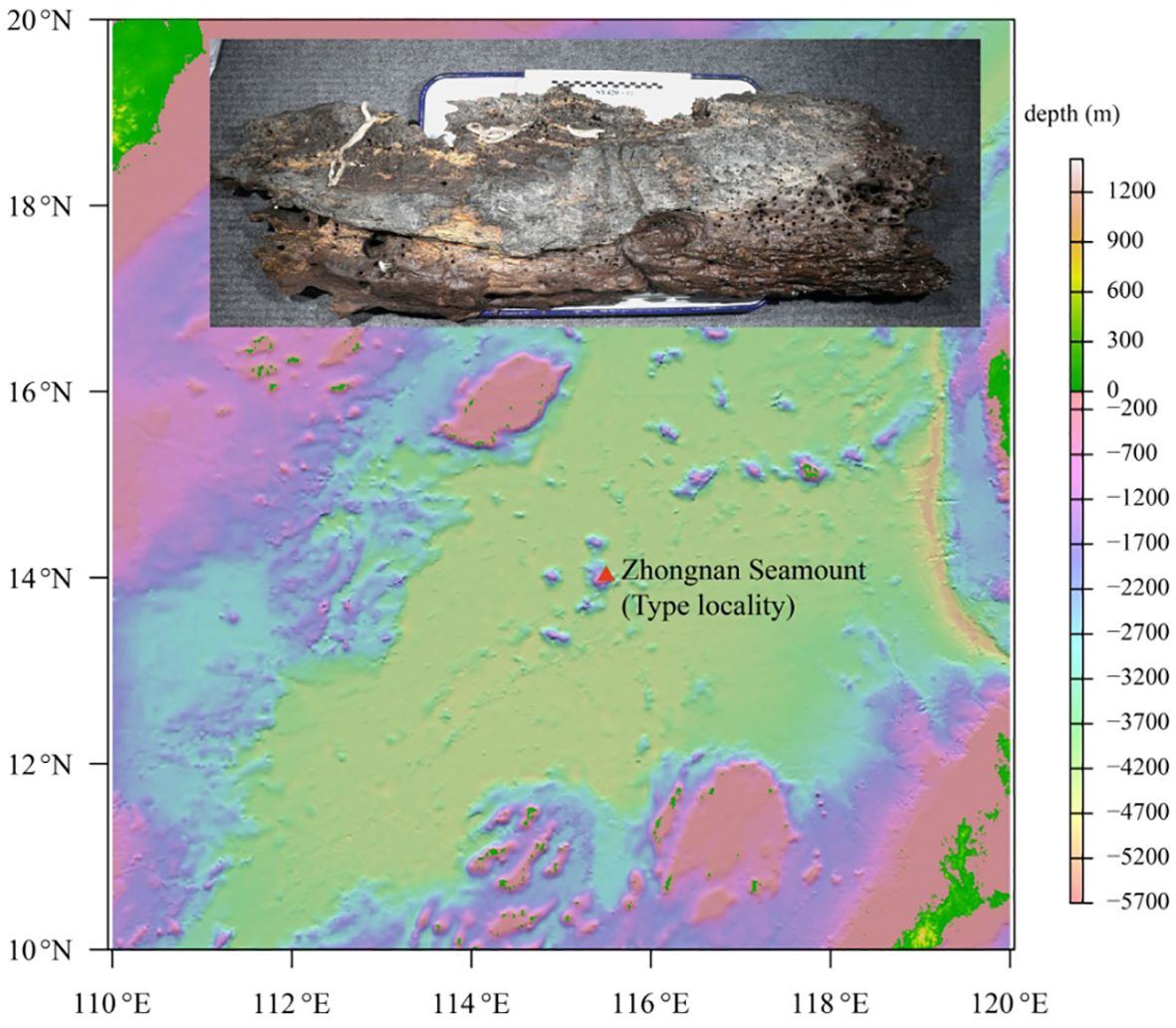
Wood falls were collected from Zhongnan Seamount in the South China Sea on 23 December 2021 during the Cruise TS2–10-2 using the submersible Shenhai Yongshi (Deep Sea Warrior). Maldanids were extracted from wood (Figures 1A, B), preserved in 75% ethanol, and deposited in the Sample Repository of the Second Institute of Oceanography (RISO, Ministry of Natural Resources, Hangzhou, China) after being studied. Morphological and morphometric observations were done under a Zeiss Stereomicroscope Discovery V20 (Germany) and a Zeiss compound microscope Axio Imager A2 (Germany). Detailed observations of key morphological traits were done under a scanning electron microscopy (SEM; HITACHI TM-1000, Japan). Morphological terminology follows De Assis et al (2010).
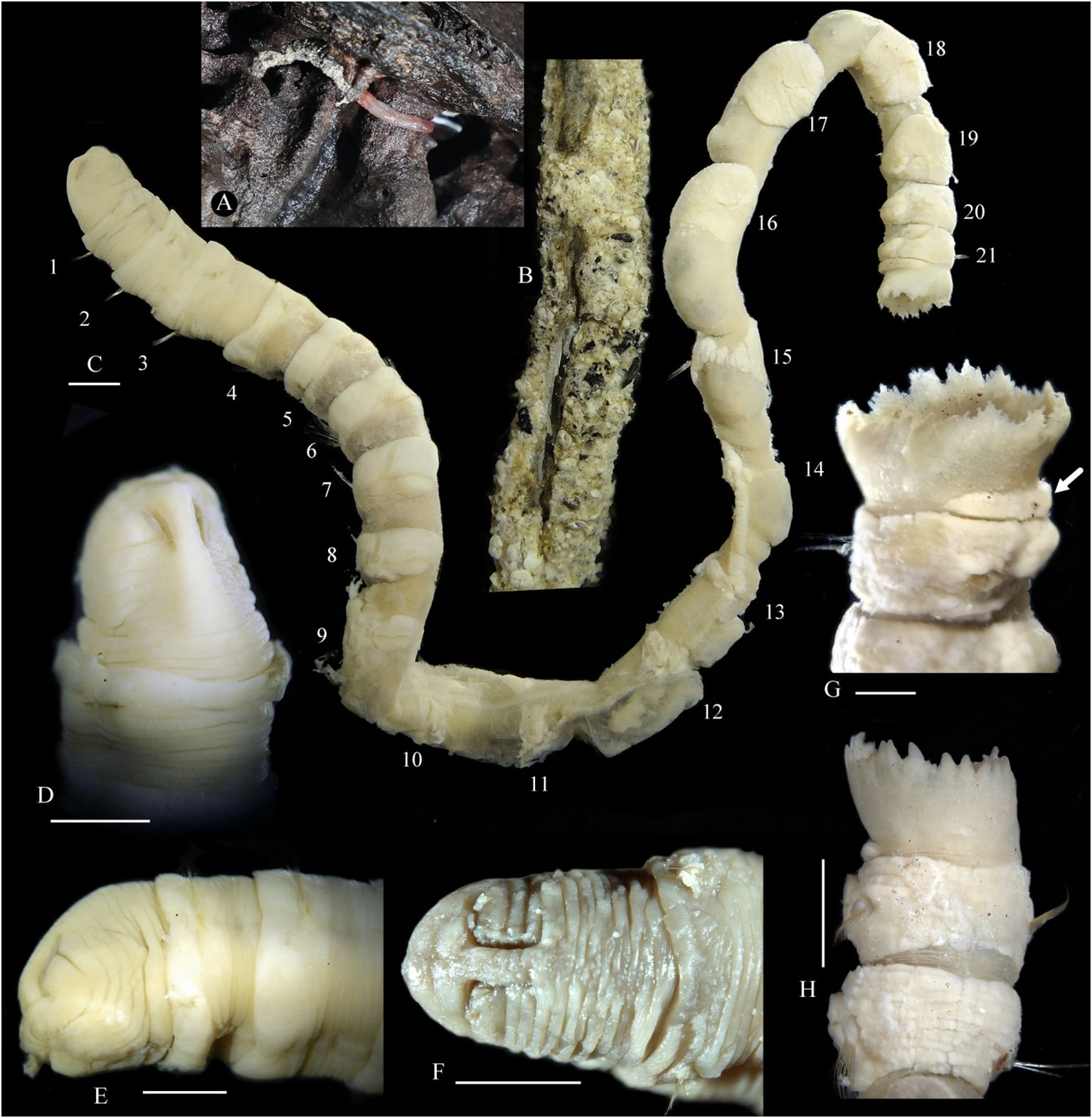
Figure 1 Nicomache tigilli sp. nov. (A) Living specimen attached to a wood fall. (B) Tube. Preserved specimens: (C) body; (D) anterior end in dorsal view; (E) anterior end in lateral view; (F) anterior end in dorsal view; (G) 21th chaetiger and anal funnel, The arrow shows the glandular pad. (H) Pygidium in dorsal view. (B-E, G) Holotype; and (F, H) paratype. Scale bar: 0.25 cm.
2.2 DNA extraction and PCR amplification
DNA was extracted from the body wall tissue using a DNeasy Tissue Kit (Qiagen) following the manufacturer’s protocol. Amplifications of specific partial fragments of the COI, 16S, and 28S genes were carried out using distinct primer pairs: LCO1490/HCO2198 for COI (Folmer et al., 1994), 16Sar/16Sbr for 16S (Palumbi, 1996), and Po28R4/28F5 for 28S (Struck et al., 2006; Passamaneck et al., 2004). The polymerase chain reaction (PCR) mixtures contained 0.5 U of polymerase (Phanta Super-Fidelity DNA Polymerase, Vazyme, China), 2.5 μL of 10 × buffer solution (supplied by the polymerase manufacturer), 0.5 μL of dNTP solution (2.5 mM), 1 μL of each primer (10 μM), 1 μL of template DNA, and ddH2O (to bring the final volume to 25 μL). The PCR amplification conditions were as follows: COI: 95°C/4 min, 35 cycles of (95°C/30 s, 45°C/45 s, and 72°C/1 min), and 72°C/7 min; 16S: 95°C/4 min, 35 cycles of (95°C/30 s, 50°C/30 s, and 72°C/1 min), and 72°C/7 min; 18S: 95°C/4 min, 35 cycles of (95°C/30 s, 50°C/1 min, and 72°C/2 min), and 72°C/10 min; 28S: 95°C/4 min, 7 cycles of (95°C/30 s, 55°C/30 s, and 72°C/2 min); and 35 cycles of (95°C/30 s, 52°C/30 s, and 72°C/2 min), and 72°C/10 min. The PCR products were purified using the QIAquick PCR purification kit (Qiagen, CA, USA) and then sequenced using the bi-directional Sanger sequencing method, which was conducted by Sangon (Shanghai, China).
2.3 Phylogenetic analyses
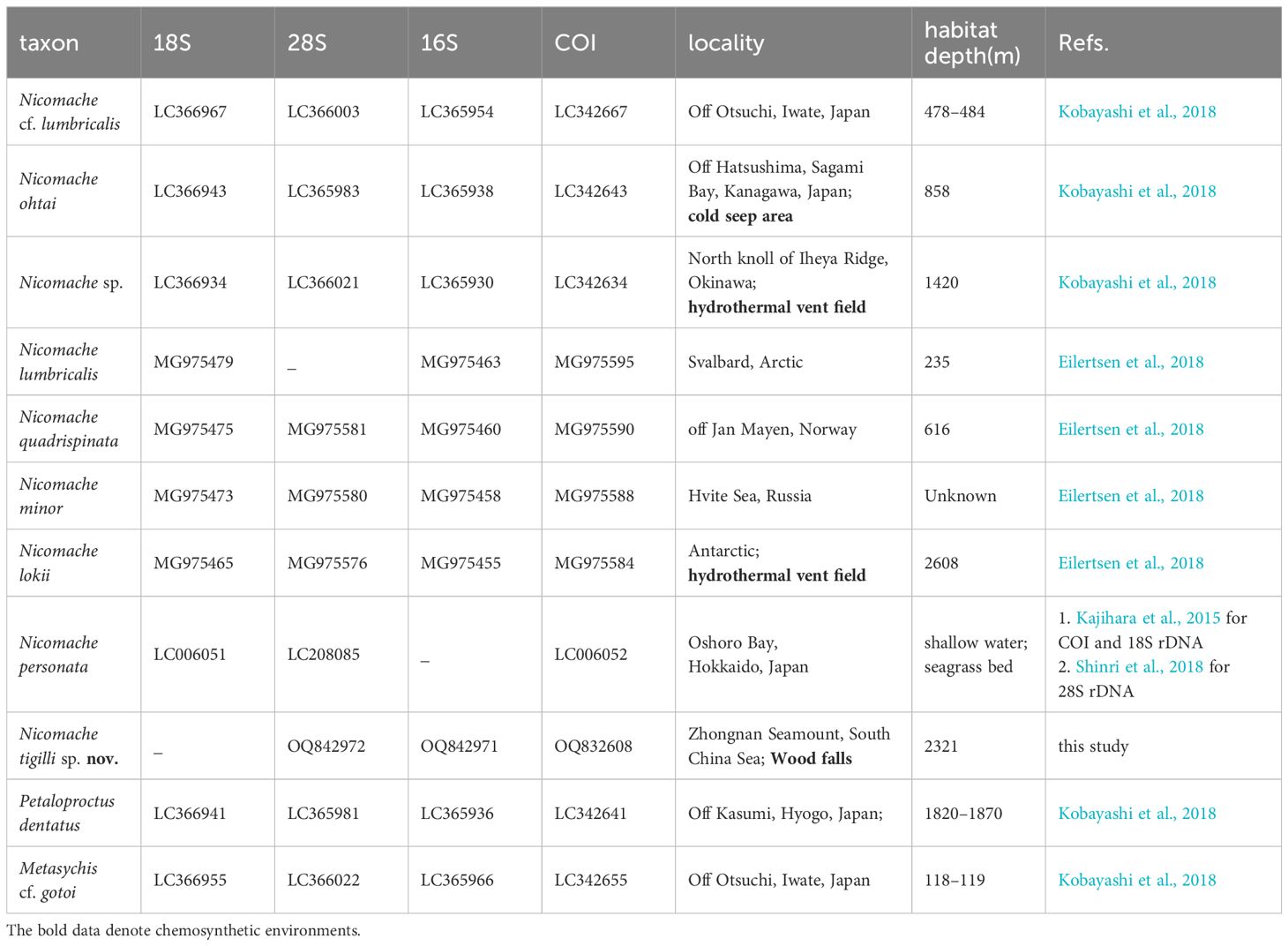
We constructed a molecular phylogenetic tree based on the four selected genes using sequences of nine species of Nicomachinae and one of Metasychis cf. gotoi (Maldaninae) from GenBank (Table 1).
The homologous sequences were normally aligned with MAFFT (Katoh and Standley, 2013), using the “—auto” strategy and concatenated using PhyloSuite v1.2.2 (Zhang et al., 2020) to select the best partitioning scheme and evolutionary models for the predefined partitions using PartitionFinder 2 (Lanfear et al., 2017), with the greedy algorithm and Corrected Akaike Information Criterion.
The phylogenies were inferred from the concatenated dataset using maximum likelihood (ML) and Bayesian inference (BI). The ML phylogenies were inferred using IQ-TREE (Nguyen et al., 2015) under the Edge-linked partition model for 5,000 standard bootstraps, and the Shimodaira–Hasegawa-like approximate likelihood-ratio test (Guindon et al., 2010). The BI phylogenies were inferred using MrBayes 3.2.6 (Ronquist et al., 2012) under a partition model (two parallel runs, 2,000,000 generations), discarding the initial 25% of the sampled data as burn-in.
3 Results
3.1 Systematics
Family: Maldanidae (Malmgren, 1865)
Subfamily: Nicomachinae (Arwidsson, 1907)
Genus: Nicomache (Malmgren, 1865)
Type of species: Sabella lumbricalis (Fabricius, 1780)
Nicomache (Loxochona) tigilli sp. nov.
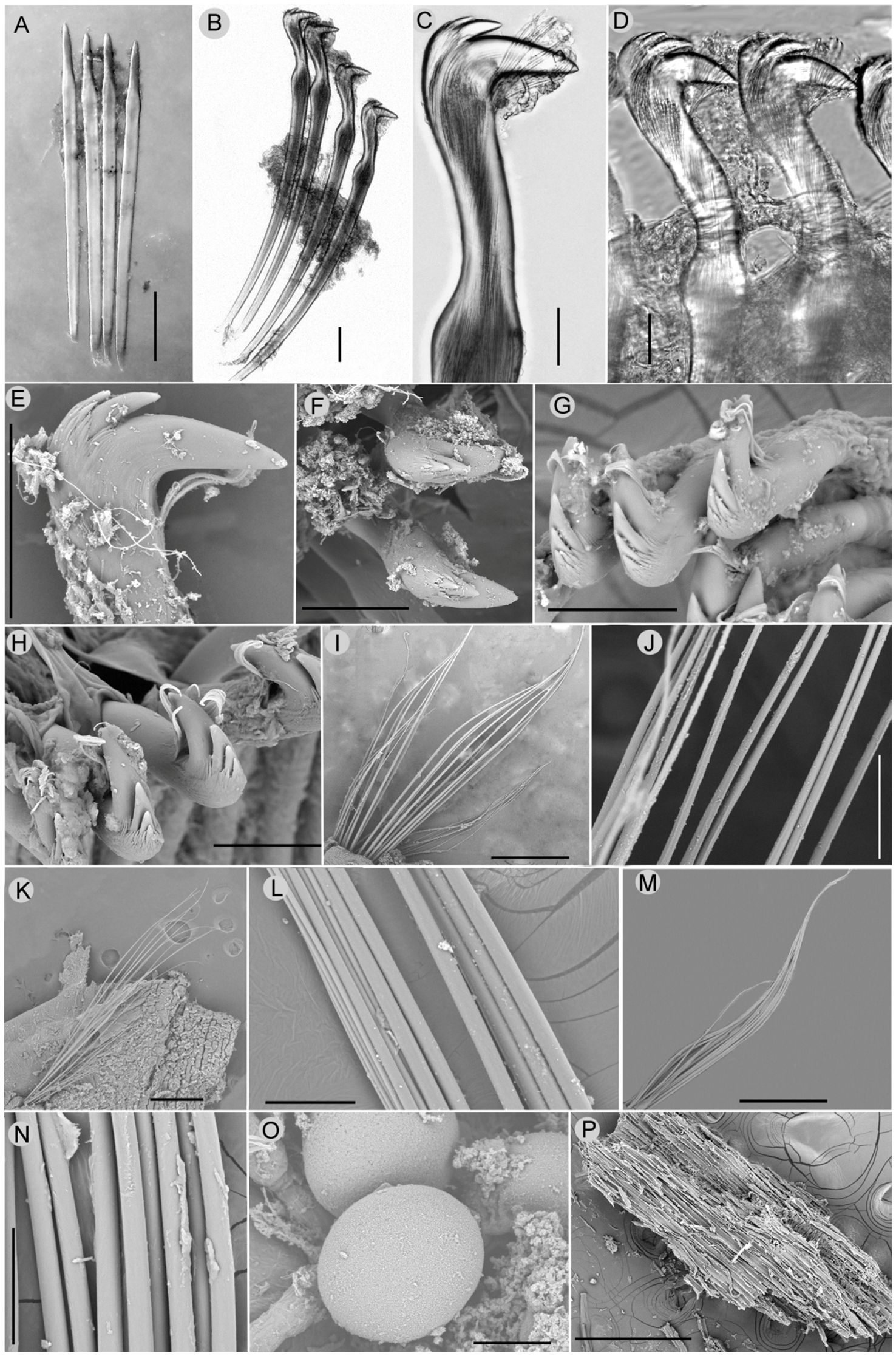
Figure 3 Nicomache tigilli sp. nov. micrographs: (A) Acicular spines from chaetiger 3. (B) Neurochaetae from chaetiger 4. (C) Capitium of uncinus. (D) neurochaetae from chaetiger 17. (E, F) Lateral and top view of the capitium of an uncinus from chaetiger 5. (G, H) Capitium of uncini from chaetiger 15 and 20, respectively. (I, J)Notochaetae from chaetiger 5 and its partial enlargement. (K, L) Notochaetae from chaetiger 15 and partial enlargement. (M, N)Notochaetae from chaetiger 20 and partial enlargement. (O) Oocytes. (P)Small wood chip extracted from gut. Scale bars: (A, B, E-H, L, N, O) = 50μm, (C, D) = 20 μm, (I, K, M, P) = 500 μm, (J) = 100 μm.
urn:lsid:zoobank.org:act:3B3930B9-D534–4105-B58D-A9FAF52CD579
3.1.1 Material examined
Holotype: B6317400001, complete specimen, ca. 110-mm long, ca. 5.0-mm wide, coll. Biao Chen, Zhongnan Seamount, South China Sea, 14.01943°N, 115.5016°E, 2,321-m depth.
Paratypes: B6317400002, one complete specimen, ca. 98.2-mm long, ca. 4.2-mm wide, same collection data as holotype.
B6317400003: One complete specimen, ca. 113-mm long, ca. 4.5-mm wide, the same collection data as the holotype, partly used for DNA sequencing and SEM.
3.1.2 Diagnosis
The body has 21 chaetigers, without achaetous pre-pygidial segment. The distal margin of the anal funnel is slightly oblique in lateral view, with the ventral part longer than the dorsal part. The prostomium is rounded anteriorly. The anterior ends of the nuchal grooves curve outwards. The neuropodia of chaetigers 1–3 with 3–6 have strongly pointed acicular spines.
3.1.3 Etymology
The specific name tigilli is the genitive form of the Latin tigillum (meaning small piece of wood), referring to the habitat of the new species.
3.1.4 Description
The body is long, cylindrical with 21 chaetigers and a pygidial funnel, without an achaetous pre-pygidial segment. Segments are shorter on the anterior and posterior body, and longer on the middle body (Figure 1C). Glands as well-developed circular bands in the anterior part of chaetigers 1–8, then present mainly on parapodial rami in the following chaetigers, and a prominent semicircular pad on the anteroventral side of the pygidial funnel (Figure 1G).
The head is well-defined, with an arched cephalic keel (Figures 1D, E). The prostomium is rounded anteriorly, with a wide prostomial palpode (anterior part). Paired nuchal grooves are long, deep, inverted J-shaped, and curved outwards in the anterior part (Figures 1E, F). Ocelli are not seen. The mouth is large, oval, and with a thick lower lip. Anterior six chaetigers are short, with its length being half of its width, the length of chaetiger 7 is approximately 1.0 -1.3 times its width; chaetiger 8 is the shortest, chaetigers 9 -19 are about twice as long as wide or longer; the last two chaetigers are shorter than wide. The pygidial funnel is short, with a straight distal margin in lateral view and an irregularly serrated border, with many small digitate papillae or triangular lobes (Figures 1G, H).
All chaetigers are biramous, with noto- and neuropodial chaetae. Neuropodia of chaetigers 1–3, with 3–6 straight acicular neuropodial spines with slender tips (Figure 3A), are replaced by a single row of rostrate uncini in the following neuropodia. There are 12–15 uncini in chaetiger 4, up to 56 in mid-body chaetigers, and 22–35 in posterior chaetigers. The capitium of uncinus has 2–4 teeth in a row, many small accessory teeth on two sides, and a tuft of subrostral bristles under the main fang (Figures 3B–H). From chaetiger 4, notochaetae are arranged in double rows (Figures 3H, K), as simple capillaries lack wings, the same in the anterior and posterior chaetigers (Figures 3I–N), but gradually changing from parallel to the neurochaetae at first few chaetigers to vertical to neurochaetae in the middle and posterior segments.
Oocytes are in anterior and middle segments, up to 108 μm in diameter (Figure 3O). Tubes are irregularly curved, covered with coarse sand and foraminifers, adhered to wood falls.
Distribution and feeding: known only from type locality; associated to wood falls (Figures 1A, B); and dissected gut containing many small wood chips (Figure 3P).
Remarks: Nicomache tigilli sp. nov. resembles N. ohtai in the shape of head, acicular spines, and rostrate uncini, but differs in having 21 chaetigers (32 in N. ohtai), short thin spinulose capillaries absent (present on all notopodia in N. ohtai). Spinulose notochaetae are present on most species of Nicomache, while simple capillary notochaetae are only present in N. arwidssoni, N. canadensis, N. lokki and N. tigilli sp. nov., which can be distinguished from these three species by having more (3–6 vs. 1–2) acicular spines on chaetigers 1–3. Remarkably, a distinct semicircular pad is present on anteroventral side of pygidial funnel of N. tigilli sp. nov., which is readily stained with methyl green as the glands on the parapodia rami. Moreover, it is easily removed and the tissues beneath it are identical to those of the adjacent body wall, indicating that it is not an achaetous pre-pygidial segment.
3.1.5 Molecular analysis
The dataset of the 28S rDNA, 16S rDNA, and COI partial sequences of N. tigilli sp. nov. (B6317400003) contained 563 bp, 480 bp, and 611 bp (Accession number OQ842972, OQ842971, and OQ832608), respectively. Interspecific pairwise genetic distances between N. tigilli sp. nov. and its close relatives ranged from 9.46% to 21.41% (Table 2), with the smallest distances (9.29%–14.14%) occurring between the species from chemosynthetic environments (N. lokii, N. ohtai, and Nicomache sp. with GenBank accession number LC342634) and N. tigilli sp. nov., while those between other species of Nicomache ranged from 12.75% to 25.5%. The ML and BI combined phylogenetic trees were totally congruent and generally well supported (Figure 4). The species from Nicomache clustered into a clade with PP > 0.95, with those from chemosynthetic environments and our new species forming a monophyletic group with maximum support. Nicomache minor Arwidsson, 1906 and N. personata cluster together with high support, being a sister group to N. lumbricalis.
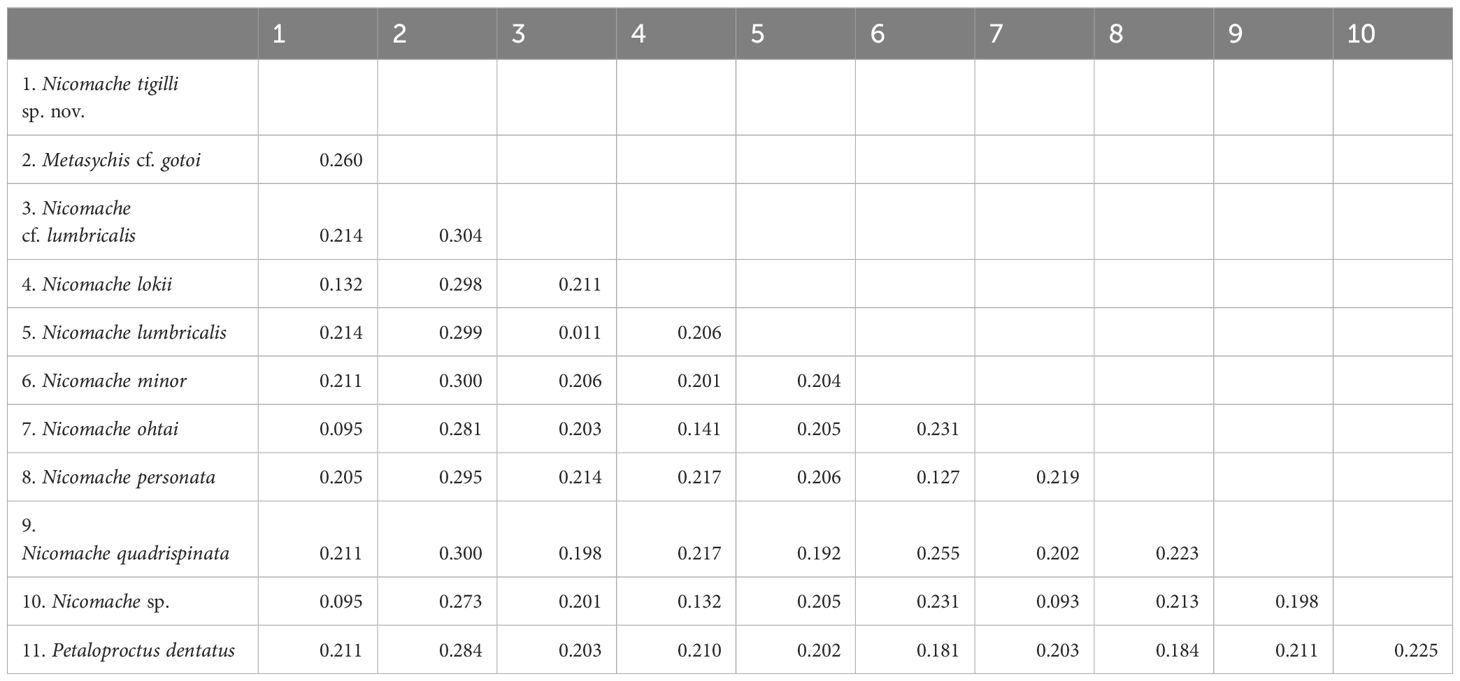
Table 2 Pairwise average Kimura 2-parameter (K2P) genetic distances for the taxa that were used in this study based on the COI gene sequences.
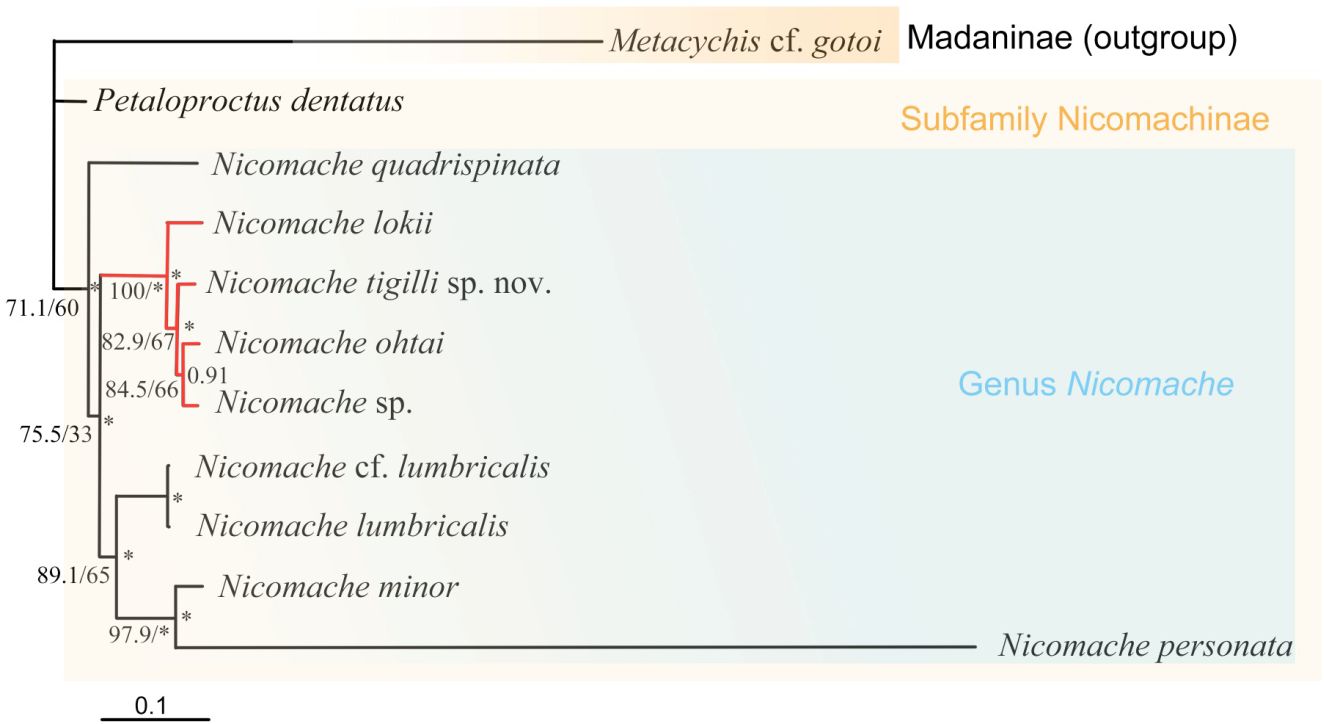
Figure 4 Phylogenetic tree obtained by the Bayesian inference based on the concatenated dataset of 16S rDNA, COI gene, 18S rDNA, and 28S rDNA sequences. The branch serves as the SH-aLRT support/standard bootstrap support (BP; left) and Bayesian posterior probabilities (PP, right) are indicated adjacent to each node. *PP > 0.95 and BP > 95%. The well-supported Nicomache clade from reducing habitats highlight with red.
4 Discussion
Nicomache tigilli sp. nov. can be clearly distinguished morphologically and molecularly from the 17 species of Nicomache accepted to date (De Assis et al., 2007; Kongsrud and Rapp, 2012), and is the first species of the genus inhabiting deep-sea wood falls, Moreover, the wood chips found in the gut strongly suggest feeding on wood-associated bacteria, in addition to organic matter from surrounding sediments, as suggested for N. lokii (Kongsrud and Rapp, 2012).
The morphological and molecular phylogenetic relationship within Nicomachinae did not reveal clear interspecific relationships, likely because a few species of Nicomache were included (De Assis and Christoffersen, 2011; Eilertsen et al., 2018; Kobayashi et al., 2018). The monophyly of Nicomachinae was suggested based on three species (N. lumbricalis, P. terriculus, and Micromaldane ornithochaeta Mesnil, 1897), with the Euclymeninae as sister clade (De Assis and Christoffersen, 2011) and later confirmed molecularly (Kobayashi et al, 2018). However, the Euclymeninae were paraphyletic because Nicomachinae were nested within. Nicomache lokii, a widespread hydrothermal vent species, showed slightly higher inter- than intra–populations when comparing its Arctic and Antarctic populations (Eilertsen et al., 2018). The phylogenetic topology in Eilertsen et al. (2018) was similar to ours, except for the absence of the chemosynthetic clade, as they included only one species associated to these environments. Conversely, our results (notably the smaller COI distances) strongly support the species of Nicomache from chemosynthetic environments to form an evolutionarily separated clade. Deep-sea chemosynthetic environments, such as hydrothermal vents, cold seeps, and organic falls have discontinuous and extremely patchy distributions that may constitute dispersal barriers likely promoting speciation. However, wood falls, like other deep-sea organic-matter-enriched environments, are known to serve as stepping stones, not only for wood associated species (Romano et al., 2020), but for seep species, such as the gastropod Cordesia provannoides Warén and Bouchet, 2009 or the siboglinid tubeworms (Dando et al., 1992). Nicomache tigilli sp. nov. is phylogenetically closer to vent/seep species of Nicomache than to those from non-reducing environments, independently of depth. Therefore, we hypothesize that sunken wood provides a specialized habitat facilitation dispersal, but also promotes speciation, to the species of Nicomache living in a wide range of chemosynthetic environments.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The article presents research on animals that do not require ethical approval for their study.
Author contributions
YW: Writing – review & editing, Writing – original draft, Methodology, Formal analysis. YZ: Writing – review & editing, Resources, Investigation, Data curation. DZ: Writing – review & editing, Visualization, Methodology, Funding acquisition. CW: Writing – review & editing, Supervision, Resources, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National Natural Science Foundation of China under contract No. 41806179, the Innovation Group Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), Grant Number: SML2022005.
Acknowledgments
We would like to thank the pilots and technical team of the HOV Shenhaiyongshi. We also thank Dr. Biao Chen for his help on sample collection, Prof. Xiaotong Peng and Prof. Kefu Yu for their support during the cruise, and Dr. Hong Cheng for assisting with the molecular analysis. We thank the reviewers for helpful comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arwidsson I. (1906). Studien über die skandinavischen und arktischen Maldaniden nebst Zusammenstellung der übrigen bisher bekannten Arten dieser Familie. Inaugural-Dissertation zur erlangung der Doktorwürde. Der Mathematisch-Naturwissenschaftlichen Sektion der Philosophischen Fakultät zu Upsala, Upsala Universitet, Upsala. 1–308.
Arwidsson I. (1907). Studien über die Skandinavischen und Arktischen Maldaniden nebst zusammenstellung der übrigen bisher bekannten Arten dieser Familie. Zoologische Jahrbcher. Supplement 9, 1–308.
Bienhold C., Pop Ristova P., Wenzhöfer F., Dittmar T., Boetius A. (2013). How deep-sea wood falls sustain chemosynthetic life. PloS One 8, e53590–e53590. doi: 10.1371/journal.pone.0053590
Blake J. A. (1985). Polychaeta from the vicinity of deep-sea geothermal vents in the eastern Pacific. I. Euphrosinidae, Phyllodocidae, Hesionidae, Nereididae, Glyceridae, Dorvilleidae, Orbiniidae, and Maldanidae. Bull. Biol. Soc. Wash 6, 67–101.
Blake J. A., Hilbig B. (1990). Polychaeta from the vicinity of deep-sea hydrothermal vents in the Eastern Pacific Ocean and records from the Juan de Fuca and Explorer Ridge systems. Pac Sci. 44, 219–253.
Chen C., Linse K. (2020). From wood to vent: first cocculinid limpet associated with hydrothermal activity discovered in the Weddell Sea. Antarct Sci. 32, 354–366. doi: 10.1017/S095410202000022X
Dando P. R., Southward A. F., Southward E. C., Dixon D. R., Crawford A., Crawford (1992). Shipwrecked tube worms. Nature. 356, 667–667. doi: 10.1038/356667a0
De Assis J. E., Alonso C., Christoffersen M. L. (2007). A catalogue and taxonomic keys of the Subfamily NicomaChinae (Polychaeta : Maldanidae) of the world. Zootaxa. 1657), 41–55.
De Assis J. E., Bleidorn C., Christoffersen M. L. (2012). “"Maldanidae malmgre". in Handbook of Zoology, A natural history of the phyla of the animal kingdom,” in Annelida. Ed. Schmidt-Rhaesa A. (De Gruyter, Osnabrück), 1–12.
De Assis J. E., Christoffersen M. L. (2011). Phylogenetic relationships within Maldanidae (Capitellida, Annelida), based on morphological characters. Syst. Biodivers 9, 233–245. doi: 10.1080/14772000.2011.604358
De Assis J. E., Christoffersen M. L., Lana P. D. C. (2010). Phylogenetic analysis of Petaloproctus (Maldanidae: Polychaeta), with description of a new species from southeastern Brazil. Sci. Mar 74, 111–120. doi: 10.3989/scimar.2010.74n1
Detinova N. N. (1985). Taxonomy, composition and distribution of polychaetes subfamily lumbriclymeninae (Maldanidae). Explor. Fauna Seas. 34 (42), 25–29.
Eilertsen M. H., Georgieva M. N., Kongsrud J. A., Linse K., Wiklund H., Glover A. G., et al. (2018). Genetic connectivity from the Arctic to the Antarctic: Sclerolinum contortum and Nicomache lokii (Annelida) are both widespread in reducing environments. Sci. Rep 8, 4810. doi: 10.1038/s41598-018-23076-0
Fabricius O. (1780). Fauna groenlandica, systematice sistens animalia groenlandiae occidentalis hactenus indagata, quoad nomen specificium, triviale, vernaculumque, synonyma auctorum plurimum, descriptionem, locum, victum, generationem, mores, usum capturamque singuli, pro ut detegendi occasio fuit, maximaque parte secundum proprias observationes (Ioannis Gottlob Rothe: Hafniae & Lipsiae), 1–452.
Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit i from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3 (5), 294–299.
Guindon S., Dufayard J. F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
He X., Xu T., Chen C., Liu X., Li Y., Zhong Z., et al. (2023). Same (sea) bed different dreams: Biological community structure of the Haima seep reveals distinct biogeographic affinities. Innovation Geoscience. 1, 100019. doi: 10.59717/j.xinn-geo.2023.100019
Kajihara H., Tomioka S., Kakui K., Iseto T. (2015). Phylogenetic position of the queer, backward-bent entoproct loxosoma axisadversum (Entoprocta: solitaria: loxosomatidae). Species diversity. 20, 83–88. doi: 10.12782/sd.20.1.083
Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kobayashi G., Goto R., Takano T., Kojima S. (2018). Molecular phylogeny of Maldanidae (Annelida): Multiple losses of tube-capping plates and evolutionary shifts in habitat depth. Mol. Phylogenet Evol, 127332–127344. doi: 10.1016/j.ympev.2018.04.036
Kongsrud J. A., Rapp H. T. (2012). Nicomache (Loxochona) lokii sp. nov.(Annelida: Polychaeta: Maldanidae) from the Loki’s Castle vent field: an important structure builder in an Arctic vent system. Polar Biol. 35, 161–170. doi: 10.1007/s00300-011-1048-4
Lanfear R., Frandsen P. B., Wright A. M., Senfeld T., Calcott B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution formolecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Malmgren A. J. (1865). Nordiska hafs-annulater. Öfversigt af Königlich Vetenskapsakademiens förhandlingar Stockholm. 22, 181–192.
Malmgren A. J. (1867). Annulata polychaeta spetsbergiae, grönlandiae, islandiae et scandinaviae hactenus cognita. Öfversigt Af Kongliga Vetenskaps-Akademiens Förhandlingar Stockholm. 24, 127–235.
Miura T., Hashimoto J. (1991). Nicomache ohtai, new species (Polychaeta: Maldanidae) collected from the Hatsushima cold-seep in Sagami Bay. Proc. Biol. Soc. Wash. 104, 159–165.
Moore J. P. (1903). Polychaeta from the coastal slope of japan and from kamchatka and bering sea. Proc. Acad. Nat. Sci. Phila. 55, 401–490.
Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Pailleret M., Haga T., Petit P. A., Privé-Gill C., Saedlou N., Gaill F., et al. (2007). Sunken wood from the Vanuatu Islands: identification of wood substrates and preliminary description of associated fauna. Mar. Ecology., 28233–28241. doi: 10.1111/j.1439-0485.2006.00149.x
Pamungkas J., Glasby C. J., Read G. B., Wilson S. P., Costello M. J. (2019). Progress and perspectives in the discovery of polychaete worms (Annelida) of the world. Helgol Mar. Res. 73. doi: 10.1186/s10152-019-0524-z
Passamaneck Y. J., Schander C., Halanych K. M. (2004). Investigation of molluscan phylogeny using large-subunit and small-subunit nuclear rRNA sequences. Mol. Phylogenet. Evol. 32 (1), 25–38. doi: 10.1016/j.ympev.2003.12.016
Paterson G. L., Glover A. G., Froján C. B., Whitaker A., Budaeva N., Chimonides J., et al. (2009). A census of abyssal polychaetes. Deep Sea Res. 2 Top. Stud. Oceanogr. 56, 1739–1746. doi: 10.1016/j.dsr2.2009.05.018
Romano C., Nunes-Jorge A., Le Bris N., Rouse G. W., Martin D., Borowski C. (2020). Wooden stepping stones: Diversity and biogeography of deep-sea wood boring Xylophagaidae (Mollusca: Bivalvia) in the North-East Atlantic Ocean, with the description of a new genus. Frot Mar. Sci. 7, 579959. doi: 10.3389/fmars.2020.579959
Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Höhna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shinri T., Keiichi K., Hiroshi K. (2018). Molecular phylogeny of the family capitellidae (Annelida). Zoolog Sci. 35, 436–445. doi: 10.2108/zs180009
Struck T. H., Pursche G., Halanych K. M. (2006). Phylogeny of eunicida (Annelida) and exploring data congruence using a partition addition bootstrap alteration (PABA) approach. Syst. Biol. 55 (1), 1–20. doi: 10.1080/10635150500354910
Warén A., Bouchet P. (2009). New gastropods from deep-sea hydrocarbon seeps off West Africa. Deep Sea Res. 2 Top. Stud. Oceanogr. 56, 2326–2349. doi: 10.1016/j.dsr2.2009.04.013
Wolff T. (1979). Magrofaunal utilization of plant remains in the deep sea. Sarsia 64, 117–143. doi: 10.1080/00364827.1979.10411373
Yang D., Sun R. (1988). Polychaetous annelids commonly seen from China coastal waters (in chinese) (Beijing: China Agriculture Press), 64–265.
Keywords: new species, South China Sea, deep-sea, polychaetes, Maldanidae
Citation: Wang Y, Zhou Y, Zhang D and Wang C (2024) Are maldanids from deep-sea reduced habitats closely related? Implications of a new wood-fall species of Nicomache from the South China Sea. Front. Mar. Sci. 11:1401688. doi: 10.3389/fmars.2024.1401688
Received: 15 March 2024; Accepted: 23 May 2024;
Published: 20 June 2024.
Edited by:
Daniel Martin, Spanish National Research Council (CSIC), SpainReviewed by:
Eriberto De Assis, Federal University of Paraíba, BrazilEkin Tilic, Senckenberg Research Institute and Natural History Museum Frankfurt, Germany
Copyright © 2024 Wang, Zhou, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Zhang, ZHN6aGFuZ0BzaW8ub3JnLmNu
 Yueyun Wang
Yueyun Wang Yadong Zhou
Yadong Zhou Dongsheng Zhang
Dongsheng Zhang Chunsheng Wang
Chunsheng Wang