
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 24 April 2024
Sec. Aquatic Microbiology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1396306
Oxygen minimum zones (OMZs) in the ocean are areas with dissolved oxygen (DO) concentrations below critical thresholds that impact marine ecosystems and biogeochemical cycling. In the northern Indian Ocean (NIO), OMZs exhibit a tendency to expand in mesopelagic waters and contribute significantly to global nitrogen loss and climate change. However, the microbial drivers of OMZ expansion in the NIO remain understudied. Here, we characterized bacterial communities across DO gradients in the NIO using high-throughput 16S rRNA gene sequencing. We found that Marinimicrobia, Chloroflexi, and the SAR324 clade were enriched in both oxygen-deficient and low oxygen mesopelagic waters. Furthermore, Marinimicrobia, Chloroflexi, and the SAR324 clade exhibited a significant negative correlation with DO (P < 0.01), suggesting that they were well-adapted to the oxygen-deficient OMZ habitat. Functional predictions revealed heightened nitrogen metabolism in OMZs, particularly nitrate reduction, suggesting its pivotal role in nitrogen loss. These findings underscore the importance of microbial communities in driving OMZ expansion in the NIO and highlight their implications in global biogeochemical cycles and climate change.
Due to the current high levels of ocean oxygenation, most marine life forms exist aerobically. However, oxygen minimum zones (OMZs), oceanic water bodies with dissolved oxygen (DO) levels below a limited concentration, are also prevalent (Cline and Richards, 1972; Stramma et al., 2008; Paulmier and Ruiz-Pino, 2009). However, there is no universal agreement on the upper threshold of the DO concentration to define an OMZ. Concentrations ranging from 2 μM to 90 μM were adopted in different studies (Lam and Kuypers, 2011). Most aquatic organisms begin to suffer in a habitat with an oxygen concentration below 63 µM (hypoxia). Microbial hypoxia refers to a water body with a DO concentration below 20 µM (Lam and Kuypers, 2011; Rixen et al., 2020). According to the data from the World Ocean Atlas, the total volume of water bodies characterized as microbial hypoxia in the global ocean occupies as much as 1.5×1016 m3 (v/v, 1.13%).
Approximately 8% of the global oceanic subsurface is covered by permanent OMZs (Paulmier and Ruiz-Pino, 2009). They are notably present in key regions, such as the eastern tropical Atlantic, the eastern tropical Pacific, and the northern Indian Ocean (NIO). The NIO makes up about 21% of the global permanent OMZs. Therefore, it contributes significantly to global climate change via nitrogen loss mediated by microbial activity. Oceanic nitrogen limitation is common (Moore et al., 2013). The nitrogen loss occurred in OMZs would intensify nitrogen limitation on marine photosynthesis, then reduce the fluxes of carbon fixed by phytoplankton (Field et al., 1998; Deutsch et al., 2011). Moreover, the NIO harbors OMZs in which the oxygen concentration is beyond the detection limit by modern techniques (Lüke et al., 2016; Menezes et al., 2020). Oceanic OMZs in the NIO usually occur at depths of 100–1,000 m, underlying highly productive surface water. The formation of oceanic OMZs is most likely ascribed to oxygen consumption caused by degradation of sinking organic matter derived from high primary productivity of the euphotic zone (Fernandes et al., 2018; Rixen et al., 2020; Sarma et al., 2020). Furthermore, the semi-enclosed geographic characteristics of the NIO are disadvantageous to the ventilation of subsurface water, and thus stratification of oxygen-deficient mesopelagic waters became more severe. Most importantly, the oceanic OMZs may be expanding both vertically and horizontally as global climate change and anthropogenic activities cause enhanced oxygen demand (Gu et al., 2022).
The oceanic OMZs exhibit active biogeochemical cycling of elements, especially that of nitrogen (Canfield et al., 2010; Lam and Kuypers, 2011). OMZs contribute approximately 30–50% of the total nitrogen loss from oceans (Gruber and Sarmiento, 1997; Codispoti et al., 2000) via anaerobic ammonium oxidation (also known as anammox) (Kuypers et al., 2005; Hamersley et al., 2007) and denitrification (Jayakumar et al., 2004; Castro-González et al., 2005). Furthermore, nitrogen loss causes nutrient limitation and reduced photosynthesis in the oceans (Gruber and Sarmiento, 1997; Codispoti et al., 2000; Lam and Kuypers, 2011; Rangamaran et al., 2023). OMZs also contribute to climate change (e.g., global warming) directly as they are large sources of carbon dioxide, nitrous oxide, and other greenhouse gases (Paulmier et al., 2008). In contrast, climate change has led to variations in the extent of OMZs. For example, the Pacific Ocean suboxic zone varied in size twice, according to historical observations. Deutsch et al. (2011) pointed out that climate-driven changes in tropical and subtropical thermoclines exhibited multiplicative effects on oxygen respiration rates to enlarge the OMZs.
Anaerobic and microaerobic microbes are metabolically active in OMZs, although their aerobic large organism counterparts normally escape from OMZs (Beman and Carolan, 2013). The structure and function of microbial communities undergo a notable shift during the formation of OMZs (Paulmier and Ruiz-Pino, 2009; Deutsch et al., 2011; Beman and Carolan, 2013). This transition from a normoxic level to a hypoxic level produces a profound impact on marine productivity (Wright et al., 2012). Microorganisms inhabiting OMZs display remarkable metabolic flexibility, and engage in various biogeochemical cycles of nitrogen (Bertagnolli and Stewart, 2018). For example, studies by Walsh et al. (2009) revealed that the SUP05 clade is not only involved in denitrification and anammox, but also mediates sulfur-oxidization and carbon assimilation in oxygen-deficient oceanic waters (Canfield et al., 2010). The Arabian Sea (AS) represents a pronounced permanent OMZ in the NIO. In the scenario of global climate change, the permanent OMZs may expand in both the vertical and horizontal dimensions in the NIO (Paulmier and Ruiz-Pino, 2009; Beman and Carolan, 2013; Rangamaran et al., 2023). Therefore, it is significant to predict the possible expansion of OMZs in the NIO from the perspective of microbial oceanography.
More recently, Gu et al. (2022) examined the microbial communities of expanding low-oxygen zones in the Bay of Bengal (BoB), where the oxygen concentration was barely above suboxic levels (5 μM oxygen). They revealed that Trichodesmium may serve as an essential source of carbon and nitrogen in maintaining the BoB OMZ. Hitherto, the key microbial taxa driving the expansion of OMZ has not been investigated in the NIO. Our hypothesis is that specific groups of marine microorganisms may play a pivotal role in driving this process. In this study, we aimed to 1) characterize the profiles of bacterial communities in three different regions of the NIO using high-throughput 16S rRNA gene sequencing, and 2) gain insight into the key bacterial taxa that may drive the formation of permanent OMZs from potential OMZs.
Water samples were collected from 12 stations located in the AS, the Carlsberg Ridge (CR), and the Ninety East Ridge (NER), with different levels of DO (Figure 1). Sample collection was performed using a Seabird 911 plus conductivity, temperature, and depth (CTD) rosette (SeaBird Electronics, Bellevue, Washington, USA) on board the R/V Shen-Hai-Yi-Hao, from May 2022 to August 2022. Each water sample was prepared in three duplicates (about 2 L). The samples were filtered through a 0.22-μm polycarbonate membrane (Whatman), and stored at −80°C prior to DNA extraction.
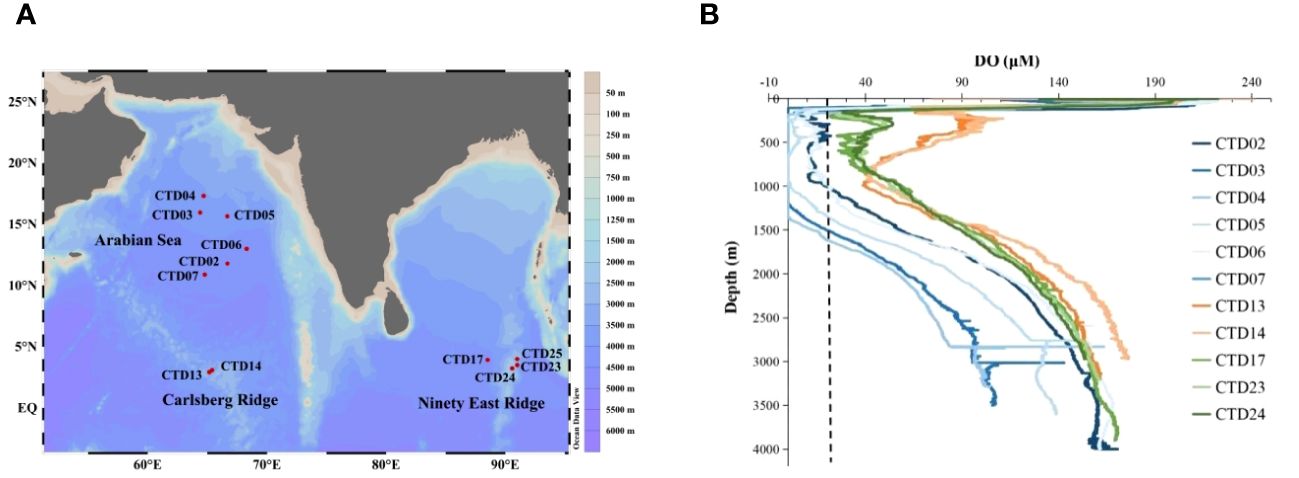
Figure 1 Investigation sites featured different levels of DO in the NIO. (A), Geographical location of the sampling stations in the NIO. (B), DO curves along the increasing depth at each station. Different colors indicate the stations located in different regions in the NIO. Blue lines indicate the AS; orange lines indicate the CR; green lines indicate the NER. The dotted line represents the threshold value of 20 μM, which defined an OMZ in this study.
The CTD rosette was used to record data, including water depth, temperature, salinity, and DO, along the water column. A total of 44 water samples were collected at various depths from different stations in the NIO and stored at −20°C. Next, dissolved nutrients (including phosphate, nitrate, nitrite, and ammonium) were analyzed using a Skalar autoanalyzer (Skalar Analytical, Netherlands) following standard methods (Strickland and Parsons, 1968).
The microbial DNA was extracted from those samples described in Section 2.1 with the E.Z.N.A. soil DNA kit (Omega Bio-tek, Norcross, GA, USA) following the manufacturer’s protocols. The quality of the extracted DNA was assessed using the 1% (w/v) agarose gel electrophoresis. The V3-V4 region of the bacterial 16S rRNA gene was amplified using the primer pair 341F (5’- CCTAYGGGRBGCASCAG -3’) and 806R (5’- GGACTACNNGGGTATCTAAT -3’) (Yu et al., 2005; Bowman et al., 2012). Thermal cycling consisted of initial denaturation at 95°C for five min, followed by 28 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s, and a final extension at 72°C for 10 min. The preparation of PCR mixtures and the purification of reaction products were performed following the method by Li et al (Li et al., 2023). Finally, the concentration and quality of purified PCR products were determined using QuantiFluor™-ST fluorometer (Promega, Madison, WI, USA).
The amplicon library was constructed using purified PCR products of the bacterial 16S rRNA gene following Illumina’s library preparation procedure. Next, the library was paired-end sequenced on an Illumina MiSeq PE250 platform (Shanghai Biozeron Biotechnology Co. Ltd., Shanghai, China) according to the standard protocol. Quality control of the raw reads was performed using Trimmomatic software (Bolger et al., 2014) based on the following criteria: (i) The 250-bp reads were truncated at any site that was assigned an average quality score <20 over a 10-bp sliding window. The truncated reads were discarded when they were shorter than 50 bp. (ii) Barcode matching was required to be exact. Two nucleotide mismatches at most were permitted in primer matching. Reads containing ambiguous characters were removed. (iii) The sequences were assembled when overlaps were longer than 10 bp. Otherwise, reads were discarded when they could not be assembled. Pair-end reads were merged and further filtered by removing primer sequences and chimeras using Vsearch (2.27.0) (Rognes et al., 2016) with the commands “–fastq_mergepairs” and “–fastx_filter”. Then, the optimized sequences were dereplicated using “–derep_fulllength”. Otherwise, reads were discarded when they could not be assembled.
The filtered sequences were clustered into operational taxonomic units (OTUs) at 97% similarity using Usearch software (version 10, http://drive5.com/uparse/). Representative sequences of the OTUs were subjected to further taxonomic assignments. Specifically, the phylogenetic affiliation of each representative sequence was analyzed using the UCLUST algorithm (v1.2.22q, http://www.drive5.com/usearch/manual/uclust_algo.html) against the SILVA (SSU138.1) database with a confidence threshold of 80% (Quast et al., 2012). The raw reads presented in the study were deposited in the NCBI Sequence Read Archive(SRA) database, accession number PRJNA1020552.
Bacterial community functions were predicted using 16S rRNA data through phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) and functional annotation of prokaryotic taxa (FAPROTAX). PICRUSt uses a computational approach to predict the functional composition of a metagenome based on biomarker genes and full genomes (Langille et al., 2013). The function predictions of all genes were assigned to the KEGG Orthology database, tier 3. Next, the differences in genes associated with metabolic processes (including nitrogen, sulfur, and methane) among groups were illustrated. FAPROTAX is well-suited to predict the biogeochemical cycle of environmental samples (Langille et al., 2013). It maps microbial taxa to microbial metabolic functions based on functional annotations of marine culturable microorganisms in a reference database (Louca et al., 2016).
In this study, the water samples described in Section 2.1 were grouped according to the water depth at each region of the NIO, including surface water (<200 m), mesopelagic water (from ≥200 m to <1,000 m), and bottom water (≥1,000 m). We also adopted a threshold value of 20 μM to define a group as OMZ or non-OMZ (Lam and Kuypers, 2011).
The alpha-diversity indices, including coverage, richness (ACE and Chao 1), and species diversity (Shannon and Simpson), were analyzed using Mothur (v.1.30.1, http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity) (Schloss et al., 2009). To assess the difference in alpha-diversity indices among different groups, a Kruskal–Wallis test was performed. Dunn’s test was performed to determine the level of differences in alpha-diversity indices between two groups. The beta diversity (non-metric multidimensional scaling, NMDS) based on the Bray–Curtis distance was calculated using the vegan package in R, and the results were visualized using the ggplot2 package. The analysis of similarities (ANOSIM) was calculated to test the results of the beta diversity. The variance inflation factor (VIF) was analyzed using SPSS (version R25.0) to characterize multi-collinearity among environmental factors (Marcoulides and Raykov, 2018), and the appropriate factors were further screened. A detrended correspondence analysis (DCA) was applied to further screen suitable ordination methods. A canonical correspondence analysis (CCA) and Pearson correlation analysis were employed to explore the influence of environmental factors on bacterial communities.
In the functional prediction, inter-group variations in metabolic pathways were analyzed using a statistical analysis of metagenomic profiles (Parks et al., 2014) with Welch’s t test at a 95% confidence interval. A Kruskal–Wallis test was conducted to characterize the differential genes between groups. Generally, a P value of <0.05 was regarded as statistically significant.
In this study, environmental parameters of 44 water samples derived from 12 stations in the NIO were recorded and analyzed. Specifically, the temperature decreased from 29.4°C to 1.4°C with increasing water depth (Supplementary Figure S1, Supplementary Table S1). Similarly, the salinity also exhibited a decreasing tendency, from 35.7 to 34.7 PSU with increasing water depth. Compared to the surface water, the DO concentration declined rapidly in the mesopelagic water of the NIO (Figure 1; Supplementary S2). Noticeably, the AS exhibited the lowest DO concentration at a mesopelagic depth (≤ 20 μM) in the NIO, suggesting the presence of an oceanic OMZ. Moreover, the OMZ in the AS distributed vertically starting from a depth of 100 m to 1500 m, indicating its enormous volume and corresponding significant impact on climate change. In contrast, an OMZ was not observed in the mesopelagic waters of the CR or NER. However, the mesopelagic layer in the NER exhibited a relatively low oxygen level (DO = 39.3 ± 11.5 μM; n=7), which was close to microbial hypoxia (DO < 20 µM).
To investigate and compare the bacterial community diversity between OMZ and non-OMZs, we divided the water samples into four groups: ASS (surface waters in the AS; DO = 142.9 ± 76.9 μM; n=7), OMZ (mesopelagic waters in the AS; DO ≤ 20 μM; n=8), ASB (bottom waters in the AS; DO = 137.4 ± 79.0 μM; n=8), and CNM (mesopelagic waters in the CR and NER; DO = 45.8 ± 19.7 μM; n=10). Both the OMZ and CNM showed a substantial difference in DO concentration compared to the normoxic samples (ASS and ASB) (Supplementary Table S2, P<0.001). There was no significant difference (P=0.47) in the DO concentration between the OMZ and CNM. A comparison of the bacterial community structure between these groups may provide insight into the key microbial taxa both in the vertical (from ASS/ASB to the OMZ) and horizontal DO transition (from CNM to the OMZ).
The Good’s coverage index approached 100% in all groups, ensuring sufficient sequencing data to accurately represent the bacterial species composition in the NIO (Figure 2A). From the perspective of the vertical DO gradient, the bacterial community in the OMZ exhibited the highest richness (Chao 1, ACE), followed by ASS, with ASB showing the lowest richness. This result aligns well with the findings of Stevens et al (Stevens and Ulloa, 2008), who observed a decline in microbial richness along the transition from the OMZ core to the oxic surface and deeper waters. Additionally, from the perspective of the horizontal DO gradient, CNM exhibited a higher richness and diversity than the OMZ did.
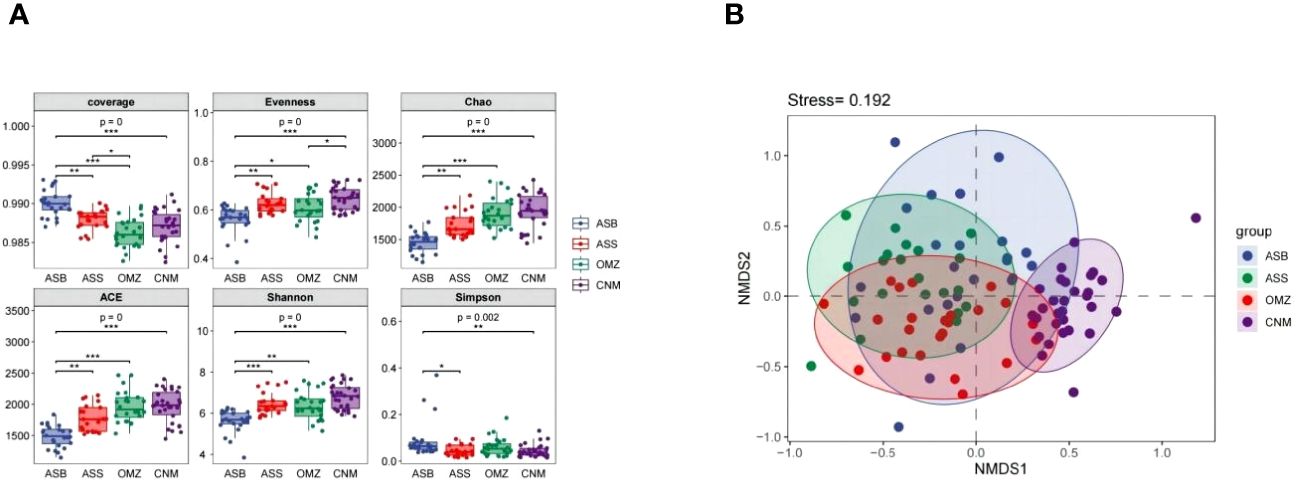
Figure 2 The diversity of bacterial communities in different groups derived from the NIO. ASS, oxic surface waters in the AS; OMZ, oxygen-deficient mesopelagic waters in the AS; ASB, oxic bottom waters in the AS; CNM, low-oxygen mesopelagic waters in the CR and NER. (A), The alpha diversity of bacterial communities. The P value represents the level of difference among the groups. The level of difference between each paired group was marked by an asterisk. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B), An NMDS profile shows the dissimilarity between bacterial community structures. Stress < 0.2 indicates that the result of the NMDS analysis was reliable. Each colored point indicates a water sample derived from a specific region in the NIO. The solid line ellipse indicates 95% confidence.
The NMDS result demonstrated that the OMZ exhibited a different bacterial community structure than did the non-OMZs (Figure 2B; Supplementary Table S3). More specifically, the OMZ exhibited a different bacterial community structure from the non-OMZ groups in the same region of the NIO (ASS and ASB), which may be ascribed to the significantly different DO levels and depth. Although there was no significant difference in DO concentrations between the OMZ and CNM, the OMZ also exhibited a bacterial community structure different from that of the CNM low-oxygen group, suggesting distinct endemic species may be present in mesopelagic waters of the different NIO regions.
The bacterial community structures in the NIO are shown at the level of phylum (Figure 3A). Proteobacteria (71.09 ± 12.37%) exhibited the highest abundance across the four groups, followed by Bacteroidota (13.97 ± 7.58%), and Actinobacteriota (4.69 ± 3.02%). Cyanobacteria exhibited the highest abundance in surface waters of AS (ASS, 1.42 ± 1.56%), where visible light favors the growth of photosynthetic microorganisms. Moreover, Firmicutes exhibited a higher abundance in bottom waters of AS (ASB, 2.56 ± 4.56%) than in the other groups (0.32 ± 0.48%). In contrast, Marinimicrobia, Chloroflexi, and the SAR324 clade exhibited a relatively higher abundance in the suboxic and low-oxygen mesopelagic waters derived from the NIO (OMZ and CNM, 1.15 ± 1.00%) than in the oxic surface and bottom waters (ASS and ASB, 0.32 ± 0.56%).
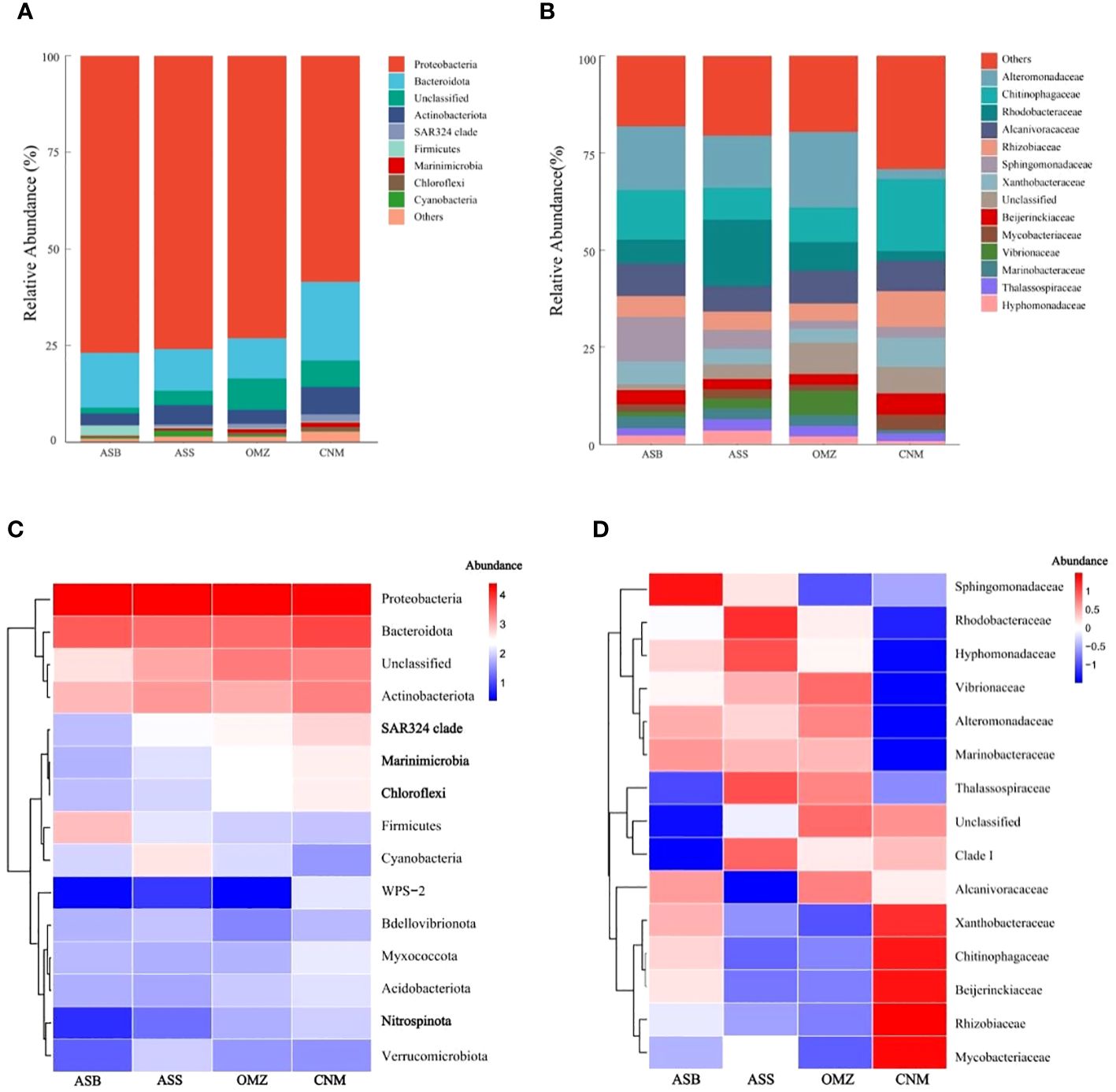
Figure 3 A comparison of the bacterial community structure between the OMZ and non-OMZs. The group explanations correspond to those in the caption of Figure 2. (A), Species composition of the bacterial communities at the phylum level. The species with a relative abundance < 0.5% were grouped as “Others”. (B), The top 15 species composition of the bacterial communities at the family level. (C), The top 15 most abundant phylum are shown in the standardized heatmap. (D), The top 15 most abundant family are shown in the standardized heatmap. The color gradient indicates the relative abundance (scaled by row) of the corresponding bacterial taxa.
We also illustrated the bacterial community structures in the NIO at the level of family (Figures 3B, D). More specifically, Rhodobacteraceae exhibited higher relative abundance in surface waters of AS (ASS, 17.09%) than that of the other groups (5.30 ± 6.64%). Sphingomonadaceae was more abundant in bottom waters of AS (ASB, 11.49%) compared to other groups (3.24 ± 2.69%). In contrast, Alteromonadaceae (19.44 ± 10.84%), Alcanivoracaceae (8.15 ± 6.90%) and Vibrionaceae (6.18 ± 9.86%) exhibited a relatively higher abundance in the OMZ group than that of the other groups. Additionally, Chitinophagaceae (18.6 ± 7.80%), Rhizobiaceae (9.14 ± 4.86%), Xanthobacteraceae (7.49 ± 3.19%), Beijerinckiaceae (5.39 ± 2.54%), and Mycobacteriaceae (3.98 ± 2.56%) were the most enriched species in the CNM group.
The standardized heatmap further demonstrated the most enriched bacterial species in each group (Figure 3C). Remarkably, Marinimicrobia, Chloroflexi, and the SAR324 clade exhibited higher relative abundances in the OMZ and CNM than in ASS and ASB, suggesting that these bacteria may adapt well in a habitat featuring suboxic or low oxygen conditions (DO = 29.17 ± 24.16 μM; n=18). Additionally, Nitrospinota (formerly Nitrospinae) also exhibited a slightly higher abundance in the OMZ and CNM than did their counterparts in oxic waters. Nitrospinota members are known as the most abundant nitrite-oxidizing bacteria (NOB) in the oceans (Mueller et al., 2021). The NOB are generally assumed to be aerobes. However, two novel NOB species in the oxygen-depleted zone (ODZ) of the eastern tropical South Pacific were discovered. This finding revealed that the novel NOB were more abundant inside ODZs than in oxic waters, indicating the versatility of NOB and the diversity of NOB niches (Sun et al., 2019).
The DCA results showed that the length of the first axis was 3.33 (Supplementary Table S4). Hence, the CCA based on a unimodal model was chosen in this study. We determined five environmental factors (DO, salinity, NO3-, NO2-, and NH4+) for a subsequent CCA based on the VIF analysis (Supplementary Table S5). The results of the CCA demonstrated that the bacterial community structures of different groups correlated to specific environmental factors (Figure 4A). Among these environmental factors, salinity, and DO emerged as the most influential in shaping the microbial community structures in the NIO (Supplementary Table S6). Notably, samples derived from different groups formed distinct clusters on the CCA plot. The bacteria in both the OMZ group and the CNM group exhibited a significant negative correlation with DO, suggesting that oxygen deficiency or low oxygen played a central role in shaping the bacterial community in the mesopelagic waters of the NIO. Additionally, salinity was highest in surface waters of the AS (Supplementary Figure S1), and displayed a positive correlation with the bacteria of the ASS group.
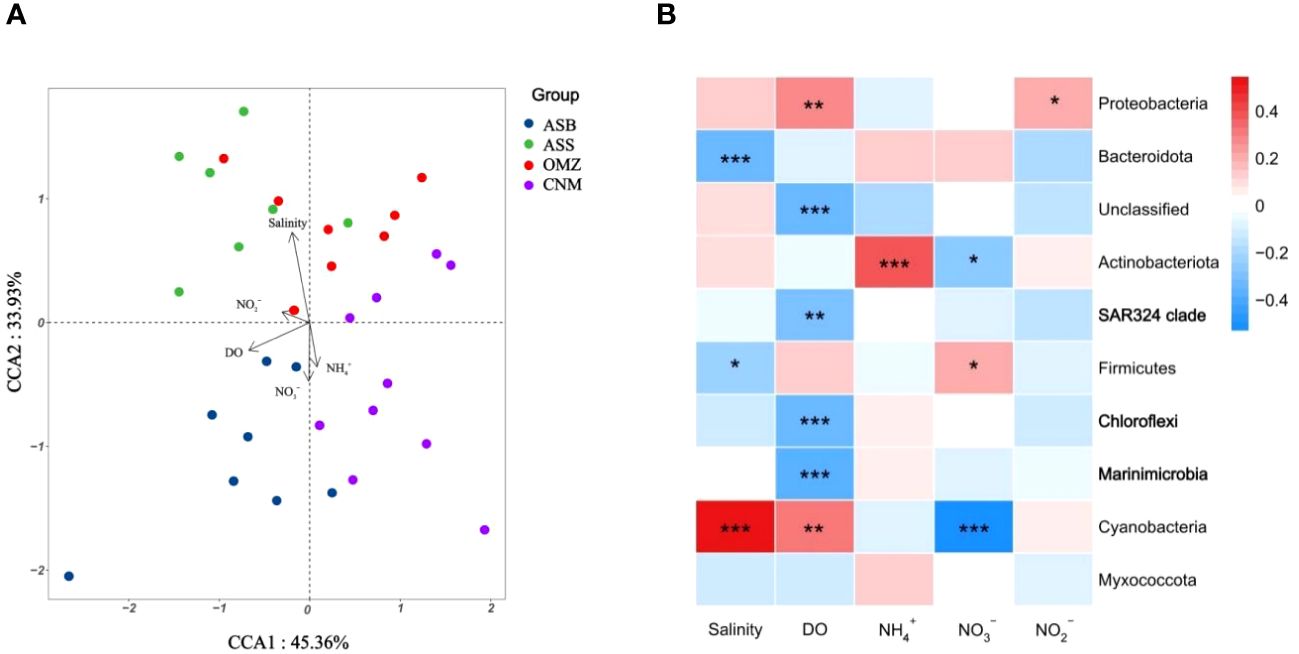
Figure 4 Correlation between microbial community structures and environmental parameters. The group descriptions correspond to those in the caption of Figure 2. (A), CCA ordination diagram illustrating the correlation between bacterial community composition and environmental factors. (B), Pearson correlation analysis between bacterial community structures and environmental parameters. In the heatmap, color gradients represent the strength of the correlation (scaled by column) between bacterial community structures and environmental parameters. Red hues indicate a positive correlation, while blue hues indicate a negative one. Additionally, the significance of the difference between the relative abundance of bacterial phyla and environmental factors is denoted with asterisks. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The Pearson correlation analysis further revealed that Marinimicrobia, Chloroflexi, and the SAR324 clade exhibited a significant negative correlation with DO (P < 0.01) (Figure 4B), suggesting that these bacterial species are well-adapted to the oxygen-deficient conditions of the OMZ habitat. Pajares et al. (2020) observed a similar trend, showing that the phyla Marinimicrobia, Chloroflexi, and the SAR324 clade were more prevalent in the OMZ core than in the euphotic zone of the Mexican Pacific.
Metabolic gene profiles in the NIO encompassing nitrogen, sulfur, and methane metabolism were assessed using PICRUSt 2 (Supplementary Figure S2). Genes associated with nitrogen metabolism were more abundant in the OMZ group (31.67%) compared to those of the non-OMZ groups (27.35–28.35%). This observation underscored the heightened activity of nitrogen metabolism under oxygen-deficient conditions in the OMZ.
Figure 5 presents the variation in metabolic processes between different groups as predicted by FAPROTAX. Compared to the OMZ, the ASB group exhibited a higher prevalence of chemoheterotrophy and nitrogen fixation (Figure 5A). Moreover, phototrophy-related metabolic processes were more prominent in the ASS group than in the OMZ group (Figure 5B). Additionally, ureolysis and nitrogen fixation exhibited a higher prevalence in the CNM group than in the OMZ (Figure 5C). In contrast, nitrate reduction was more abundant in the OMZ group than in the three other groups. Although the DO levels between the OMZ and CNM exhibited no significant difference (P=0.47; Supplementary Table S2), nitrate reduction was more prevalent in the OMZ than in CNM, suggesting that it may be a crucial and unique metabolic process in oxygen-deficient conditions.
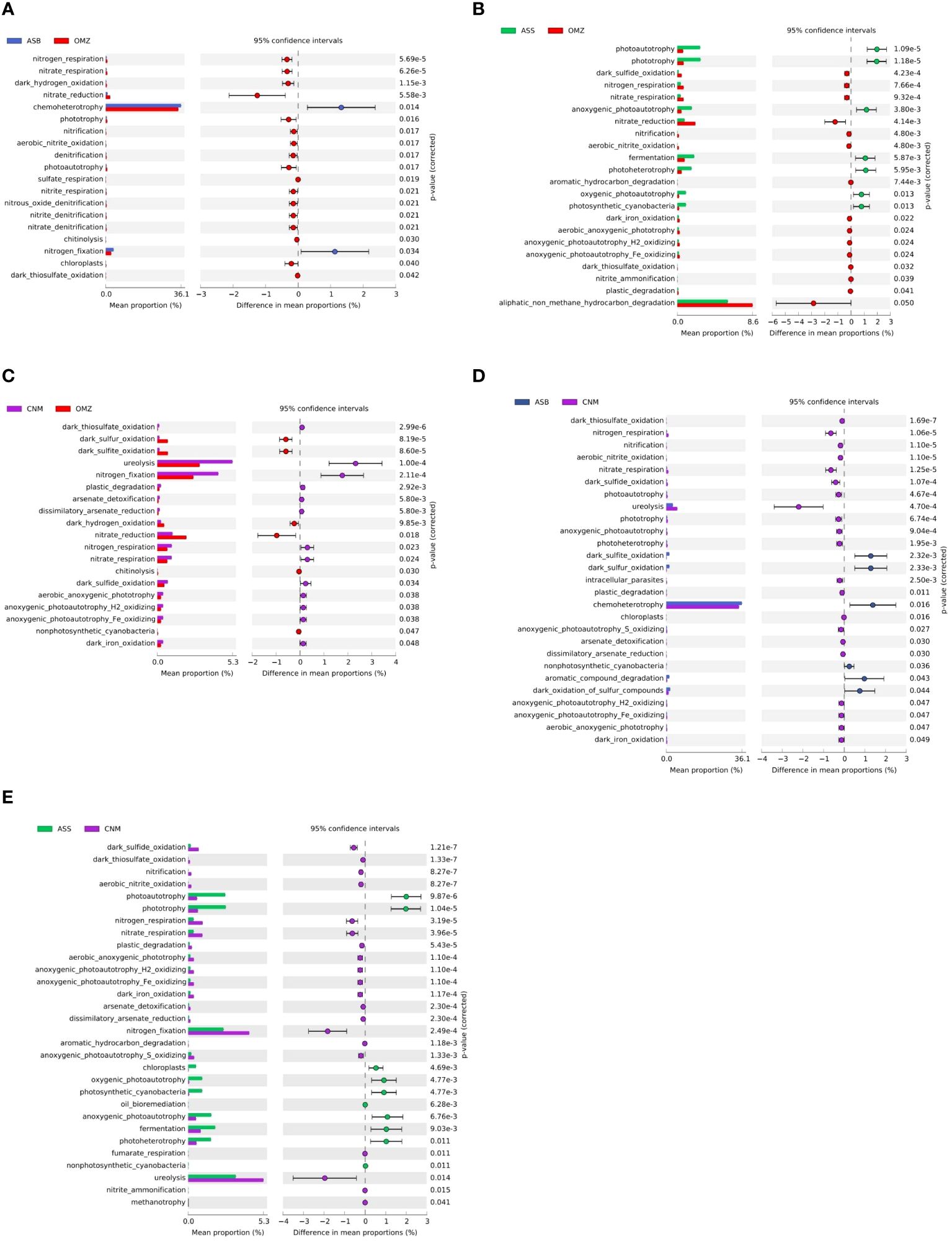
Figure 5 Variations of the abundance of metabolic processes in different groups. The group descriptions correspond to those in the caption of Figure 2. (A), Differential metabolic processes between ASB and OMZ; (B), differential metabolic processes between ASS and OMZ; (C), differential metabolic processes between CNM and OMZ; (D), differential metabolic processes between ASB and CNM; and (E), differential metabolic processes between ASS and CNM. The left panel shows the mean abundance of the metabolic processes, while the right panel presents the P values obtained from Welch’s t test.
The CNM low oxygen group exhibited a higher prevalence of nitrogen metabolism than did the oxic waters. More specifically, ureolysis, nitrate respiration, and nitrogen respiration were more prominent in the CNM group than in the ASB group (Figure 5D). Similarly, ureolysis, nitrate respiration, nitrogen respiration, and nitrogen fixation was also more prevalent in the CNM group than in the ASS group (Figure 5E). Notably, ureolysis was the predominant metabolic process in the CNM low oxygen group.
Due to different tolerances to oxygen depletion, OMZs are deficient in multicellular life forms and are instead dominated by microbial metabolism (Wishner et al., 2013; Bertagnolli and Stewart, 2018). As a result, investigation of the microbial community structure and function in the NIO is significant to predict the potential expansion of OMZs and assess the effects of expanding OMZs to ecosystem health and climate change (Beman et al., 2021). Previous studies documented that the microbial community structure of OMZs varied with changes in organic carbon availability, DO concentration, and water depth (Cavan et al., 2017; Aldunate et al., 2018; Rangamaran et al., 2023). In this study, we investigated oceanic habitats with different levels of DO, and found that the bacterial community in the NIO varied with DO and water depth. Specifically, Cyanobacteria exhibited the highest relative abundance in oxygenated surface waters of the AS (Figure 3C), and exhibited a significant positive correlation with DO (Figure 4B). Field observations have characterized an oceanic diazotrophic community dominated by Cyanobacteria (Loescher et al., 2014; Jayakumar et al., 2017; Long et al., 2021). Higher rates of nitrogen fixation often occur in the oxic waters near the surface, which exert a significant impact on the oceanic N-budget. Moreover, Firmicutes exhibited the highest relative abundance in the oxygenated bottom waters of the AS (Figure 3C). A number of previous studies identified Firmicutes as one of the most abundant phyla in the sediment underlying the OMZ (Lincy and Manohar, 2020; Amberkar et al., 2021), suggesting its preference for deep water niches. The prevalence of Firmicutes in the deep waters underlying the OMZ may be ascribed to its high hydrolytic activities to settled organic matter (Divya et al., 2010; Lin and Lin, 2022).
Marinimicrobia, Chloroflexi, and the SAR324 clade were the most enriched species in oxygen-deficient and low oxygen conditions (OMZ and CNM) compared to normoxic conditions (ASS and ASB) (Figure 3C), likely due to several factors. First, the heterotrophic bacterial phylum Marinimicrobia was abundant in the ocean, suggesting a high diversity of its metabolic functions (Hawley et al., 2017). For example, strains of Marinimicrobia have been found to participate in nitrogen and sulfur cycling, as well as fermentative amino acid degradation in methanogenic environments (Allers et al., 2013; Nobu et al., 2015; Hawley et al., 2017). Beman and Carolan (2013) reported that the abundance of Marinimicrobia increased with deoxygenation in the OMZ. Therefore, they may represent one of the key bacteria involved in nitrogen cycling processes in the oxygen-deficient and low oxygen habitats of the NIO. Second, the SAR324 clade was also ubiquitous in the ocean, and was found to be one of the most abundant bacterial groups in OMZs (Wright et al., 2012). Pajares et al. (2020) identified that this clade was significantly abundant in the OMZ core of the tropical Mexican Pacific (250 or 500 m), and mediated diverse microaerophilic or anaerobic metabolism. Moreover, this clade has been implicated in sulfur oxidation and carbon dioxide fixation, further underscoring its significance in biogeochemical cycles of elements (Swan et al., 2011; Sheik et al., 2014; Malfertheiner et al., 2022). Third, Gao et al. (2019) reported that Chloroflexi was one of the dominant bacterial taxa in the hadal zone, and emphasized its role in degrading recalcitrant dissolved organic matter. A substantial proportion (8.11%) of 16S rRNA gene amplicons from the OMZ corresponded to unclassified OTUs (Figure 3A), suggesting a reservoir of undiscovered prokaryotic diversity within this oxygen-depleted region of the AS.
The structure of microbial communities in the NIO is subject to varying degrees of influence by physicochemical factors. The influence exerted by each of the physicochemical factors depends on the biogeochemical functions and niche preferences of specific microbial taxa, as seen in previous studies (Beman et al., 2021). Our study also demonstrated a significant correlation between the bacterial community structures in the NIO and environmental factors (Figure 4).
We found that three key factors—DO, salinity, and nitrates—exhibited significant correlations with the bacterial community structures in the NIO (Supplementary Table S6). Remarkably, DO exhibited a significant correlation with most samples in the OMZ group (Figure 4A). Our study reinforced previous findings, which emphasized the strong influence of vertical oxygen gradients on microbial diversity, composition, and function within OMZs (Bertagnolli and Stewart, 2018). Furthermore, there was a significant negative correlation between DO and the abundance of bacterial species, including Marinimicrobia, Chloroflexi, and the SAR324 clade (Figure 4B), suggesting their niche preference to OMZs. Consequently, these bacteria demonstrate a higher abundance and metabolic versatility in low-oxygen and oxygen-deficient habitats of the NIO (CNM and OMZ; Figure 3C). Therefore, they may play a pivotal role as key bacterial taxa mediating biogeochemical cycles of elements, especially nitrate reduction, within OMZs of the NIO.
Previous studies have highlighted the dynamic biogeochemical processes within OMZs of the AS and BoB, showcasing variations in nitrogen and carbon recycling potential (Lincy and Manohar, 2020). In this study, we compared genes related to nitrogen, sulfur, and methane metabolism among different habitats in the NIO, in which various levels of DO were observed (Figure 1B). Notably, the OMZ group exhibited a significantly higher abundance of nitrogen metabolism genes than those of the non-OMZ groups (ASS, ASB, and CNM; Supplementary Figure S2). This result suggested that nitrogen biogeochemistry is particularly responsive to depletion of DO in the OMZ of the NIO. Furthermore, we observed that the nitrate reduction process was the most prominent metabolic process in the OMZ group, which set it apart from the non-OMZ groups (Figures 5A-C). This metabolic step represents the initiation of denitrification, and contributes significantly to nitrogen loss (Lam and Kuypers, 2011). Scholz (2018) identified nitrate reduction as a key attribute of anoxic OMZs in the water columns of the modern ocean. Additionally, Zhang et al. (2023) reported nitrate reduction was significantly enhanced by seasonal deoxygenation in the coastal marine environment. They demonstrated that nitrate reduction was sensitive to changing DO concentrations, suggesting that it might be a significant metabolic process during transition from oxic to suboxic waters.
The low-oxygen group CNM also exhibited a higher abundance of metabolic processes involving nitrogen than the oxic groups did (Figures 5D, E), which suggested a transition of microbial community function with the decline of DO (Bertagnolli and Stewart, 2018). Overall, the mesopelagic waters in the NIO (CNM and OMZ) shared certain similarities in DO levels (Supplementary Table S2), most enriched species (Figure 3C), and prominent metabolic processes (Figure 5), which implied the potential horizontal expansion of OMZs in the NIO (especially in the NER). This situation may potentially intensify the impact on global climate change via nitrogen loss in the expanding OMZ of the NIO.
In this study, we investigated the bacterial community structure and function across different regions and at various depths in the NIO. Our findings elucidate the intricate microbial ecosystems of the NIO, highlighting the features of bacterial communities within the oxygen-deficient and low oxygen mesopelagic waters of the NIO. Furthermore, our research emphasizes enhanced nitrogen metabolism, particularly nitrate reduction, as a distinct feature of the OMZ in the AS. Marinimicrobia, Chloroflexi, and the SAR324 clade may play a pivotal role in driving the biogeochemical cycling of nitrogen in oxygen-deficient and low oxygen habitats of the NIO. These revelations contribute significantly to our understanding of the ecological consequences of potential OMZ expansion in the NIO, offering valuable insights into the effects of these unique habitats on global climate change.
The raw reads presented in the study were deposited in the NCBI Sequence Read Archive (SRA) database, accession number PRJNA1020552.
JL: Writing – review & editing, Data curation, Formal analysis, Methodology, Visualization, Writing – original draft. ZC: Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Supervision. XL: Data curation, Supervision, Writing – review & editing. ZW: Supervision, Writing – review & editing. XZ: Writing – review & editing, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (42076165) and Digital Deep-sea Typical Habitats Program of China Deep Ocean Affairs Administration.
We acknowledged Hongda Wu for collecting water samples for this study. This cruise was conducted on the R/V Shen-Hai-Yi-Hao.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1396306/full#supplementary-material
Aldunate M., de la Iglesia R., Bertagnolli A. D., Ulloa O. (2018). Oxygen modulates bacterial community composition in the coastal upwelling waters off central Chile. Deep Sea Res. Part II: Topical Stud. Oceanogr. 156, 68–79. doi: 10.1016/j.dsr2.2018.02.001
Allers E., Wright J. J., Konwar K. M., Howes C. G., Beneze E., Hallam S. J., et al. (2013). Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. ISME J. 7, 256–268. doi: 10.1038/ismej.2012.108
Amberkar U., Khandeparker R., D. Menezes L., Meena R. M. (2021). Phylogenetic diversity of culturable marine bacteria from sediments underlying the oxygen minimum zone of the Arabian Sea and their role in nitrate reduction. Mar. Ecol. 42, e12646. doi: 10.1111/maec.12646
Beman J. M., Carolan M. T. (2013). Deoxygenation alters bacterial diversity and community composition in the ocean’s largest oxygen minimum zone. Nat. Commun. 4 (1), 2705. doi: 10.1038/ncomms3705
Beman J. M., Vargas S. M., Vazquez S., Wilson J. M., Yu A., Cairo A., et al. (2021). Biogeochemistry and hydrography shape microbial community assembly and activity in the eastern tropical North Pacific Ocean oxygen minimum zone. Environ. Microbiol. 23, 2765–2781. doi: 10.1111/1462-2920.15215
Bertagnolli A. D., Stewart F. J. (2018). Microbial niches in marine oxygen minimum zones. Nat. Rev. Microbiol. 16, 723–729. doi: 10.1038/s41579-018-0087-z
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowman J. S., Rasmussen S., Blom N., Deming J. W., Rysgaard S., Sicheritz-Ponten T. (2012). Microbial community structure of Arctic multiyear sea ice and surface seawater by 454 sequencing of the 16S RNA gene. ISME J. 6, 11–20. doi: 10.1038/ismej.2011.76
Canfield D. E., Stewart F. J., Thamdrup B., De Brabandere L., Dalsgaard T., Delong E. F., et al. (2010). A cryptic sulfur cycle in oxygen-minimum–zone waters off the Chilean coast. Science 330, 6009, 1375–1378. doi: 10.1126/science.1196889
Castro-González M., Braker G., Farías L., Ulloa O. (2005). Communities of nirS-type denitrifiers in the water column of the oxygen minimum zone in the eastern South Pacific. Environ. Microbiol. 7, 1298–1306. doi: 10.1111/j.1462-2920.2005.00809.x
Cavan E. L., Trimmer M., Shelley F., Sanders R. (2017). Remineralization of particulate organic carbon in an ocean oxygen minimum zone. Nat. Commun. 8, 14847. doi: 10.1038/ncomms14847
Cline J. D., Richards F. A. (1972). Oxygen deficient conditions and nitrate reduction in the eastern tropical north pacific ocean1. Limnol. Oceanogr. 17, 6, 885–900. doi: 10.4319/lo.1972.17.6.0885
Codispoti L. A., Brandes J., Christensen J. P., Devol A., Naqvi S. W. A., Paerl H., et al. (2000). The oceanic fixed nitrogen and nitrous oxide budgets: moving targets as we enter the anthropocene? Sci. Mar. 65, 85–105. doi: 10.3989/scimar.2001.65s285
Deutsch C., Brix H., Ito T., Frenzel H., Thompson L. (2011). Climate-forced variability of ocean hypoxia. Science 333, 336–339. doi: 10.1126/science.1202422
Divya B., Soumya K. V., Nair S. (2010). 16SrRNA and enzymatic diversity of culturable bacteria from the sediments of oxygen minimum zone in the Arabian Sea. Antonie Van Leeuwenhoek 98, 9–18. doi: 10.1007/s10482-010-9423-7
Fernandes S., Mazumdar A., Bhattacharya S., Peketi A., Mapder T., Roy R., et al. (2018). Enhanced carbon-sulfur cycling in the sediments of Arabian Sea oxygen minimum zone center. Sci. Rep. 8, 8665. doi: 10.1038/s41598-018-27002-2
Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. doi: 10.1126/science.281.5374.237
Gao Z. M., Huang J. M., Cui G. J., Li W. L., Li J., Wei Z. F., et al. (2019). In situ meta-omic insights into the community compositions and ecological roles of hadal microbes in the Mariana Trench. Environ. Microbiol. 21, 4092–4108. doi: 10.1111/1462-2920.14759
Gruber N., Sarmiento J. L. (1997). Global patterns of marine nitrogen fixation and denitrification. Global Biogeochem. Cycles 11, 235–266. doi: 10.1029/97GB00077
Gu B., Liu J., Cheung S., Ho N. H. E., Tan Y., Xia X. (2022). Insights into prokaryotic community and its potential functions in nitrogen metabolism in the bay of bengal, a pronounced oxygen minimum zone. Microbiol. Spectr. 10, e0089221. doi: 10.1128/spectrum.00892-21
Hamersley M. R., Lavik G., Woebken D., Rattray J. E., Lam P., Hopmans E. C., et al. (2007). Anaerobic ammonium oxidation in the Peruvian oxygen minimum zone. Limnol. Oceanogr. 52, 3, 923–933. doi: 10.4319/lo.2007.52.3.0923
Hawley A. K., Nobu M. K., Wright J. J., Durno W. E., Morgan-Lang C., Sage B., et al. (2017). Diverse Marinimicrobia bacteria may mediate coupled biogeochemical cycles along eco-thermodynamic gradients. Nat. Commun. 8, 1507. doi: 10.1038/s41467-017-01376-9
Jayakumar A., Chang B. X., Widner B., Bernhardt P., Mulholland M. R., Ward B. B. (2017). Biological nitrogen fixation in the oxygen-minimum region of the eastern tropical North Pacific ocean. ISME J. 11, 2356–2367. doi: 10.1038/ismej.2017.97
Jayakumar A., Francis C. A., Naqvi S. W. A., Ward B. (2004). Diversity of nitrite reductase genes (nirS) in the denitrifying water column of the coastal Arabian Sea. Aquat. Microbial. Ecol. 34, 69–78. doi: 10.3354/ame034069
Kuypers M. M. M., Lavik G., Woebken D., Schmid M., Fuchs B. M., Amann R., et al. (2005). Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Natl. Acad. Sci. 102, 18, 6478–6483. doi: 10.1073/pnas.0502088102
Lam P., Kuypers M. M. (2011). Microbial nitrogen cycling processes in oxygen minimum zones. Ann. Rev. Mar. Sci. 3, 317–345. doi: 10.1146/annurev-marine-120709-142814
Langille M. G. I., Zaneveld J., Caporaso J. G., McDonald D., Knights D., Reyes J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Li Y., Cui Z., Luan X., Bian X., Li G., Hao T., et al. (2023). Degradation potential and pathways of methylcyclohexane by bacteria derived from Antarctic surface water. Chemosphere 329, 138647. doi: 10.1016/j.chemosphere.2023.138647
Lin G., Lin X. (2022). Bait input altered microbial community structure and increased greenhouse gases production in coastal wetland sediment. Water Res. 218, 118520. doi: 10.1016/j.watres.2022.118520
Lincy J., Manohar C. S. (2020). A comparison of bacterial communities from OMZ sediments in the Arabian Sea and the Bay of Bengal reveals major differences in nitrogen turnover and carbon recycling potential. Mar. Biol. Res. 16, 656–673. doi: 10.1080/17451000.2020.1840593
Loescher C. R., Großkopf T., Desai F. D., Gill D., Schunck H., Croot P. L., et al. (2014). Facets of diazotrophy in the oxygen minimum zone waters off Peru. ISME J. 8, 2180–2192. doi: 10.1038/ismej.2014.71
Long A. M., Jurgensen S. K., Petchel A. R., Savoie E. R., Brum J. R. (2021). Microbial ecology of oxygen minimum zones amidst ocean deoxygenation. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.748961
Louca S., Parfrey L. W., Doebeli M. (2016). Decoupling function and taxonomy in the global ocean microbiome. Science 353, 6305, 1272–1277. doi: 10.1126/science.aaf4507
Lüke C., Speth D. R., Kox M. A. R., Villanueva L., Jetten M. S. M. (2016). Metagenomic analysis of nitrogen and methane cycling in the Arabian Sea oxygen minimum zone. PeerJ 4, e1924. doi: 10.7717/peerj.1924
Malfertheiner L., Martínez-Pérez C., Zhao Z., Herndl G. J., Baltar F. (2022). Phylogeny and metabolic potential of the candidate phylum SAR324. Biol. (Basel) 11, 599. doi: 10.3390/biology11040599
Marcoulides K. M., Raykov T. (2018). Evaluation of variance inflation factors in regression models using latent variable modeling methods. Educ. psychol. Measure. 79, 874–882. doi: 10.1177/0013164418817803
Menezes L. D., Fernandes G. L., Mulla A. B., Meena R. M., Damare S. R. (2020). Diversity of culturable Sulphur-oxidising bacteria in the oxygen minimum zones of the northern Indian Ocean. J. Mar. Syst. 209, 103085. doi: 10.1016/j.jmarsys.2018.05.007
Moore C. M., Mills M. M., Arrigo K. R., Berman-Frank I., Bopp L., Boyd P. W., et al. (2013). Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 6, 701–710. doi: 10.1038/ngeo1765
Mueller A. J., Jung M.-Y., Strachan C. R., Herbold C. W., Kirkegaard R. H., Wagner M., et al. (2021). Genomic and kinetic analysis of novel Nitrospinae enriched by cell sorting. ISME J. 15, 732–745. doi: 10.1038/s41396-020-00809-6
Nobu M. K., Narihiro T., Rinke C., Kamagata Y., Tringe S. G., Woyke T., et al. (2015). Microbial dark matter ecogenomics reveals complex synergistic networks in a methanogenic bioreactor. ISME J. 9, 1710–1722. doi: 10.1038/ismej.2014.256
Pajares S., Varona-Cordero F., Hernández-Becerril D. U. (2020). Spatial distribution patterns of bacterioplankton in the oxygen minimum zone of the tropical Mexican pacific. Microbial. Ecol. 80, 519–536. doi: 10.1007/s00248-020-01508-7
Parks D. H., Tyson G. W., Hugenholtz P., Beiko R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Paulmier A., Ruiz-Pino D. (2009). Oxygen minimum zones (OMZs) in the modern ocean. Prog. Oceanogr. 80, 113–128. doi: 10.1016/j.pocean.2008.08.001
Paulmier A., Ruiz-Pino D., Garcon V. (2008). The oxygen minimum zone (OMZ) off Chile as intense source of CO2 and N2O. Continent. Shelf Res. 28, 2746–2756. doi: 10.1016/j.csr.2008.09.012
Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–6. doi: 10.1093/nar/gks1219
Rangamaran V. R., Sankara Subramanian S. H., Balachandran K. R. S., Gopal D. (2023). Vertical microbial profiling of Arabian sea oxygen minimal zone reveals complex bacterial communities and distinct functional implications. Microbial. Ecol. 85, 357–371. doi: 10.1007/s00248-021-01952-z
Rixen T., Cowie G., Gaye B., Goes J., do Rosário Gomes H., Hood R. R., et al. (2020). Reviews and syntheses: Present, past, and future of the oxygen minimum zone in the northern Indian Ocean. Biogeosciences 17, 6051–6080. doi: 10.5194/bg-17-6051-2020
Rognes T., Flouri T., Nichols B., Quince C., Mahé F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Sarma V. V. S. S., Bhaskar T. V. S. U., Kumar J. P., Chakraborty K. (2020). Potential mechanisms responsible for occurrence of core oxygen minimum zone in the north-eastern Arabian Sea. Deep Sea Res. Part I: Oceanogr. Res. Pap. 165, 103393. doi: 10.1016/j.dsr.2020.103393
Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Scholz F. (2018). Identifying oxygen minimum zone-type biogeochemical cycling in Earth history using inorganic geochemical proxies. Earth-Sci. Rev. 184, 29–45. doi: 10.1016/j.earscirev.2018.08.002
Sheik C. S., Jain S., Dick G. J. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ. Microbiol. 16, 304–317. doi: 10.1111/1462-2920.12165
Stevens H., Ulloa O. (2008). Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. 10, 1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x
Stramma L., Johnson G. C., Sprintall J., Mohrholz V. (2008). Expanding oxygen-minimum zones in the tropical oceans. Science 320, 655–658. doi: 10.1126/science.1153847
Strickland J. D. H., Parsons T. R. (1968). A practical handbook of seawater analysis. Ottawa, Canada: Fisheries Research Board of Canada.
Sun X., Kop L. F. M., Lau M. C. Y., Frank J., Jayakumar A., Lücker S., et al. (2019). Uncultured Nitrospina-like species are major nitrite oxidizing bacteria in oxygen minimum zones. ISME J. 13, 2391–2402. doi: 10.1038/s41396-019-0443-7
Swan B. K., Martinez-Garcia M., Preston C. M., Sczyrba A., Woyke T., Lamy D., et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333, 1296–1300. doi: 10.1126/science.1203690
Walsh D. A., Zaikova E., Howes C. G., Song Y. C., Wright J. J., Tringe S. G., et al. (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326, 578–582. doi: 10.1126/science.1175309
Wishner K. F., Outram D. M., Seibel B. A., Daly K. L., Williams R. L. (2013). Zooplankton in the eastern tropical north Pacific: Boundary effects of oxygen minimum zone expansion. Deep Sea Res. Part I: Oceanogr. Res. Pap. 79, 122–140. doi: 10.1016/j.dsr.2013.05.012
Wright J. J., Konwar K. M., Hallam S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394. doi: 10.1038/nrmicro2778
Yu Y., Lee C., Kim J., Hwang S. (2005). Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679. doi: 10.1002/bit.20347
Zhang X., Yao C., Zhang B., Tan W., Gong J., Wang G.-y., et al. (2023). Dynamics of benthic nitrate reduction pathways and associated microbial communities responding to the development of seasonal deoxygenation in a coastal mariculture zone. Environ. Sci. Technol. 57, 15014–15025. doi: 10.1021/acs.est.3c03994
Keywords: oxygen deficient, denitrification, nitrogen loss, Arabian Sea, bacterial community structure
Citation: Liu J, Cui Z, Luan X, Wang Z and Zhang X (2024) Expanding oxygen minimum zones in the northern Indian Ocean predicted by hypoxia-related bacteria. Front. Mar. Sci. 11:1396306. doi: 10.3389/fmars.2024.1396306
Received: 05 March 2024; Accepted: 09 April 2024;
Published: 24 April 2024.
Edited by:
Xianbiao Lin, Ocean University of China, ChinaCopyright © 2024 Liu, Cui, Luan, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhisong Cui, Y3pzQGZpby5vcmcuY24=; Xuelei Zhang, emhhbmd4bEBmaW8ub3JnLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.