
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 02 July 2024
Sec. Marine Molecular Biology and Ecology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1389708
This article is part of the Research Topic Spiralian Genomics in a Changing World View all 5 articles
One of the most salient features of marine bryozoans is their well-calcified skeleton, and many species in this phylum are important reef-builders. To date, the molecular machinery responsible for skeletal formation in these key animals remains unknown. In this study we performed de novo transcriptome assembly from RNA from Cellaria immersa colonies collected in New Zealand, the first from the family Cellariidae. The assembly resulted in a set of 125,750 transcripts and was estimated to be 97.2% complete when compared to BUSCO core gene sets. A function was predicted for 61,442 (48.8%) of the translated proteins, using similarity searches against a range of databases using BLAST and InterProScan. Cellaria species form erect, heavily calcified arborescent colonies, which when abundant can create micro-forests or meadows on the ocean floor. RNA was extracted separately from younger distal and older proximal parts of the colonies, aiming to identify the key genes involved in biomineralization as the young zooids (at the distal growth margin) of the colony are more likely to be actively growing and calcifying compared to the old zooids of the proximal parts of the colony. Differential expression analysis was carried out to identify differential expression between the distal and proximal parts of the colonies. This showed that 506 (2.2%) of the transcripts were expressed more highly in the young zooids and 4,676 (20.4%) were expressed more highly in the old zooids. Over 50 protein families were identified as candidates involved in biomineralization in C. immersa based on the functional annotation, their expression pattern and literature. Transcripts encoding 24 such protein families were more highly expressed in the young zooids. This is the first such study on a heavily calcified species from the phylum Bryozoa, increasing the amount of ‘omics’ data available for C. immersa and the phylum. These data provide a resource for current and future studies of heavily calcified bryozoans, shedding a light on the biomineralization process in this phylum.
Bryozoans are colonial marine invertebrates and are globally important reef-builders. To date, the molecular machinery responsible for their most salient characteristic, their calcified skeletal architecture, remains unknown. Only a few studies discuss bryozoan transcriptomes (Treibergs and Giribet, 2020; Kumar et al., 2020a), and available data on genome sequencing is also limited for this phylum (Rayko et al., 2020; Bishop et al., 2023; Wood et al., 2023).
Bryozoan colonies are made of tiny individual clonal units (zooids) which grow through budding at the growth margin to form complex colonies (Ryland, 2005). As seen in other biomineralizers, the skeleton is secreted on the cuticle by underlying epithelial cells at the growing edge of the colony (Tavener-Smith and Williams, 1972). Although the zooids are independent to some extent, the colonies exhibit coordinated physiology and behavior (McKinney and Jackson, 1991). Their colonial integration is also evident in skeleton formation. While the young zooids of the colony are actively growing and depositing primary calcification, the old zooids of the colony often continue to biomineralize to a lesser degree, depositing a secondary calcification layer to strengthen the colony, and to act as a basis for regeneration in case of breakage or predation (e.g., Smith and Girvan, 2010). In addition to building their skeletons, some bryozoan species can also remove carbonate through skeletal resorption via controlled dissolution of calcified material (Batson et al., 2020).
Invertebrate biomineralization processes have been best described in corals, molluscs, and echinoderms (Clark, 2020). A “basic evolutionary toolkit” has been suggested based on the molluscan skeleton, which could be extended to other biomineralizers; it includes proteins such as Tyrosinase, Carbonic anhydrase (CA), those with chitin-binding domains, and Von Willebrand factor-A/domains (Arivalagan et al., 2016; Clark, 2020). The role of these proteins in biomineralization has been well established (Robinson and King, 1963; Miyamoto et al., 1996; Shen and Jacobs-Lorena, 1999; Inoue et al., 2003; Weiss et al., 2006; Schönitzer and Weiss, 2007; Furuhashi et al., 2009; Suzuki et al., 2009; Joubert et al., 2010; Chang and Evans, 2015; Kintsu et al., 2017; Li et al., 2017b). More specifically, CA has a long evolutionary history, and it is thought to have a single ancestral origin for all metazoans, later differentiated in separate animal lineages (Le Roy et al., 2014; Arivalagan et al., 2016; Clark, 2020; Murdock, 2020). Other common proteins involved in biomineralization include transmembrane channels such as the calcium ATPase, sodium/calcium exchange, sodium/potassium ATPase and V-type proton ATPase families (Malachowicz and Wenne, 2019; Clark, 2020). Collagen proteins are present in other animals and their role in extracellular matrix and skeletogenesis is well established (Kimura et al., 2008; Germer et al., 2015; Luo et al., 2015; Flores and Livingston, 2017; Malachowicz and Wenne, 2019; Le Roy et al., 2021). Immunity-associated domains are also commonly found in the shell matrix in molluscs, but their role in mineralization is still uncertain (Ramos-Silva et al., 2013; Arivalagan et al., 2016; Malachowicz and Wenne, 2019; Song et al., 2019). The list of potential proteins associated with biomineralization is long (Malachowicz and Wenne, 2019), but further studies will likely be able to highlight common processes.
Despite the numerous studies published regarding biomineralization in other phyla, few discuss this process in bryozoans (Taylor et al., 2015; Wernström et al., 2022; Lombardi et al., 2023). A recent study showed that the skeletal matrix in bryozoans comprises a mixture of proteins and polysaccharides, with strong lectin reactivity, specifically related to chitin-binding lectins (Lombardi et al., 2023). Further, soft tissue anatomy and skeletal ultrastructure suggest that mechanisms involved in biomineralization in bryozoans could be closely related to those of Brachiopoda and Mollusca (Taylor and Kuklinski, 2011; Taylor et al., 2015). However, the key genes responsible for biomineralization in Mollusca and Brachiopoda were not found in Bryozoa and its sister clade Phoronida, suggesting that mineralized skeletons evolved secondarily in the stem lineage leading to some bryozoan subclades (Wernström et al., 2022).
Here we used Cellaria immersa (Image: Figure 1A), one of the main habitat-forming bryozoan species in New Zealand (Wood et al., 2012), as a case study for biomineralization. Cellaria immersa forms tree-like colonies, where the branches are formed by calcified internodes separated by organic joints (i.e., rhizooids). The colonies can grow up to 40 cm in height, and when abundant they create a miniature “forest” on the ocean floor. Cellaria species can colonize soft substrates due to the presence of rhizoids at the proximal part of the colony, contrasting with other heavily calcified species that often require hard substrate to settle (Achilleos et al., 2019). Cellaria belongs to the class Gymnolaemata, Order Cheilostomata and family Cellariidae, closely grouped together with its sister family Calescharidae (Figure 1B; Orr et al., 2021). Cellaria skeletal carbonate is always calcite (Smith et al., 2006; Loxton et al., 2018), generally with low Mg content, with some individuals adding subdominant high-Mg calcite as a discrete mineral (Smith and Lawton, 2010). Several Cellaria species deposit secondary calcification at a later stage during the biomineralization process (Achilleos et al., 2020). The high-Mg calcite is possibly added as a secondary strengthening layer, as in confamilial Melicerita chathamensis (Smith and Lawton, 2010), although it has been recently suggested that the more soluble high-Mg in Cellaria species could be potentially added at the newly formed branching internodes (Batson et al., 2020).
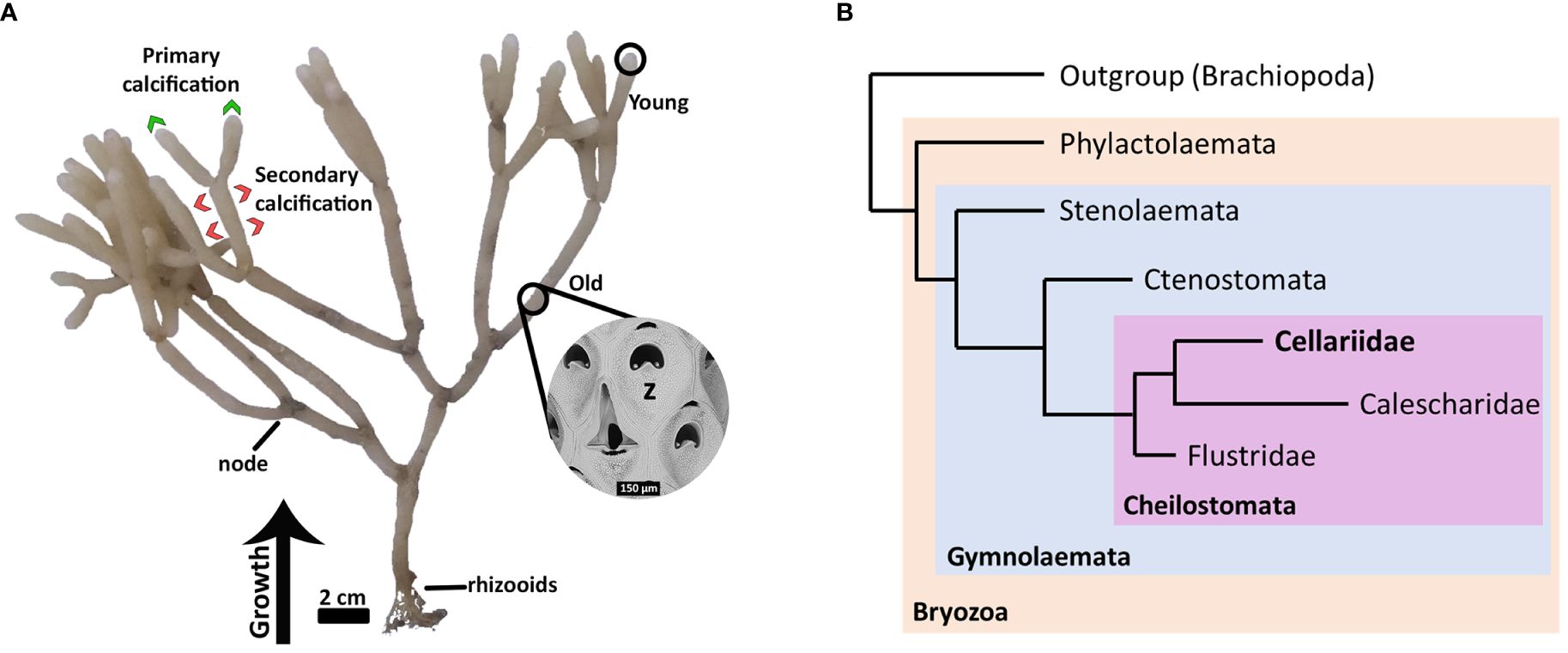
Figure 1 Cellaria immersa biology. (A) A colony of Cellaria immersa and the different colony parts (young and old) used for RNA extraction and downstream analysis. Also indicated are the areas of the colony associated with primary calcification (by young zooids) and secondary calcification (by older zooids). (B) Diagrammatic cladogram illustrating the phylogeny of Cellariidae (after Orr et al., 2021). Cellaria immersa is a member of the family Cellariidae, which belongs in the order Cheilostomata and class Gymnolaemata. z, zooid.
This study represents the first transcriptomic investigation of this family, and the first to focus on biomineralization by profiling the gene expression of young (those at the colony growth margin) and older adult zooids in C. immersa colonies. We provide the first evidence of genes likely related to skeletal resorption, and discuss possible mechanisms involved in the mineral nucleation and deposition in C. immersa. This study will be foundational for understanding biomineralization in this phylum, and of interest to those working on this essential process more broadly across the animal tree of life.
Cellaria immersa colonies were collected in 2017 and 2020 from the Otago shelf at 90 m water depth, at the same sampling location (45.7955°S, 170.921°E). Upon collection healthy, unfouled colonies were selected and flash frozen on dry ice, and subsequently held at -80°C for long-term storage.
Ten colonies were used for RNA extraction. Distal young (distal ~1 mm length of the colony) and proximal old zooids of the colonies were separated (Figure 1) for the subsequent steps, as the young zooids are more likely to be actively growing and calcifying compared to the old zooids of the colony. The ten total samples for each stage were separated in two, with five colonies pooled. The pooled approach was followed to ensure sufficient tissue for the RNA extractions, especially for the young zooids. This was due to limitations in acquiring enough specimens, given that this species is found in deeper water and is not readily available for recollection. Further, the heavily calcified nature of the skeleton overestimates the true tissue weight used for the RNA extraction. The total weight of pooled tissue, including the skeleton, used for the RNA extractions were as follow: C. immersa (1), 0.68 g (young) and 2.42 g (old); C. immersa (2), 0.71 g (young) and 0.41 g (old).
RNA was extracted using an RNeasy mini kit (Qiagen). Due to the heavily calcified skeleton, a few modifications were applied to the protocol to ensure the lysis step was successful. RLT buffer (350–700 μl) was added together with liquid nitrogen during the homogenization step. The powder mixture (specimen + RLT buffer) was then transferred into 1.5 mL collection tubes, centrifuged for 15 s and left at room temperature for about 15 min, or until the collection tubes were not cold to touch, to ensure activation of the RLT buffer. No further modifications were made to the protocol, and the subsequent steps were followed as indicated by the RNeasy mini kit. The libraries were constructed according to the Illumina TruSeq Stranded mRNA protocol.
Sequencing was done in two separate runs. The Illumina HiSeq 2000 platform was used for paired-end sequencing at 125 bp, and the MiSeq platform with a V2 kit was used for paired-end read sequencing at 2x250 bp.
FastQC v0.11.5 (Andrews, 2010) was used to perform quality assessment of all reads. The sequencing runs were treated separately prior to downstream analysis. For Illumina HiSeq 2000 libraries (replicate 1 for both old and young samples), the reads were cleaned using Trimmomatic 0.36 with the following settings: ILLUMINACLIP:2:30:10, SLIDINGWINDOW:5:20 and MINLEN:36. The adaptors file was customized to include the specific adaptor sequences for each read pair. For Illumina MiSeq libraries (replicate 2 from both old and young samples), the reads were cleaned using the same settings as HiSeq 2000 libraries. Additionally, for MiSeq libraries, the paired-end reads were merged using FLASH v2.2.00 to eliminate possible read overlaps.
The SortMeRNA tool (Kopylova et al., 2012) was used to assess and remove rRNA contaminants, as it includes SILVA databases for 16S and 23S (bacteria and archaea), 18S and 28S (eukaryotes) and Rfam database for 5S and 5.8S. The reads were also aligned against the general human genome using HISAT2 to identify possible human sequences (Supplementary File 3). The small proportion of reads flagged as contamination were removed prior the de novo assembly.
The reads were combined for the transcriptome assembly. De novo assembly of RNAseq data was performed using Trinity v2.11.0 (Grabherr et al., 2011), using default parameters. Bowtie2 (Langmead and Salzberg, 2012) was used to align the raw reads to the assembly, to assay assembly completeness. Basic numerical metrics relating to the assembly were recovered using the scripts within the Trinity suite. The de novo assembly of the quality processed reads resulted in 240,305 transcript isoforms, corresponding to 202,353 Trinity ‘genes’ (Supplementary File 3).
To assess the content of the assembly, Blobtools (Laetsch and Blaxter, 2017) was used together with Diamond (Buchfink et al., 2014) to run BLASTx searches for assignment of taxon identity against NCBI nr database (E value cutoff: 1e-5, with best hit retained). Further cleaning of the assembly was performed using the blobtools filter parameter provided by Blobtoolkit. Of the 240,305 transcripts 19,206 were assigned to the phylum Foraminifera and were removed using Blobtools by filtering based on phylum (–param bestsumorder_phylum–Keys=Foraminifera). The sequences assigned to the phylum Foraminifera can be seen in Supplementary File 1. The presence of Foraminifera is not surprising, as microscopic benthic Foraminifera often settle on the bryozoan colonies (pers. observation). BUSCO v. 3.0.2 (Simão et al., 2015) was used to evaluate the completeness of the assembly when compared to the “Metazoa” OrthoDB database v10. No other significant contamination was observed in the assembly (<1% for bacteria and human (Supplementary File 3) under the ‘other’ category as shown by Blobtools (Figure 2A).
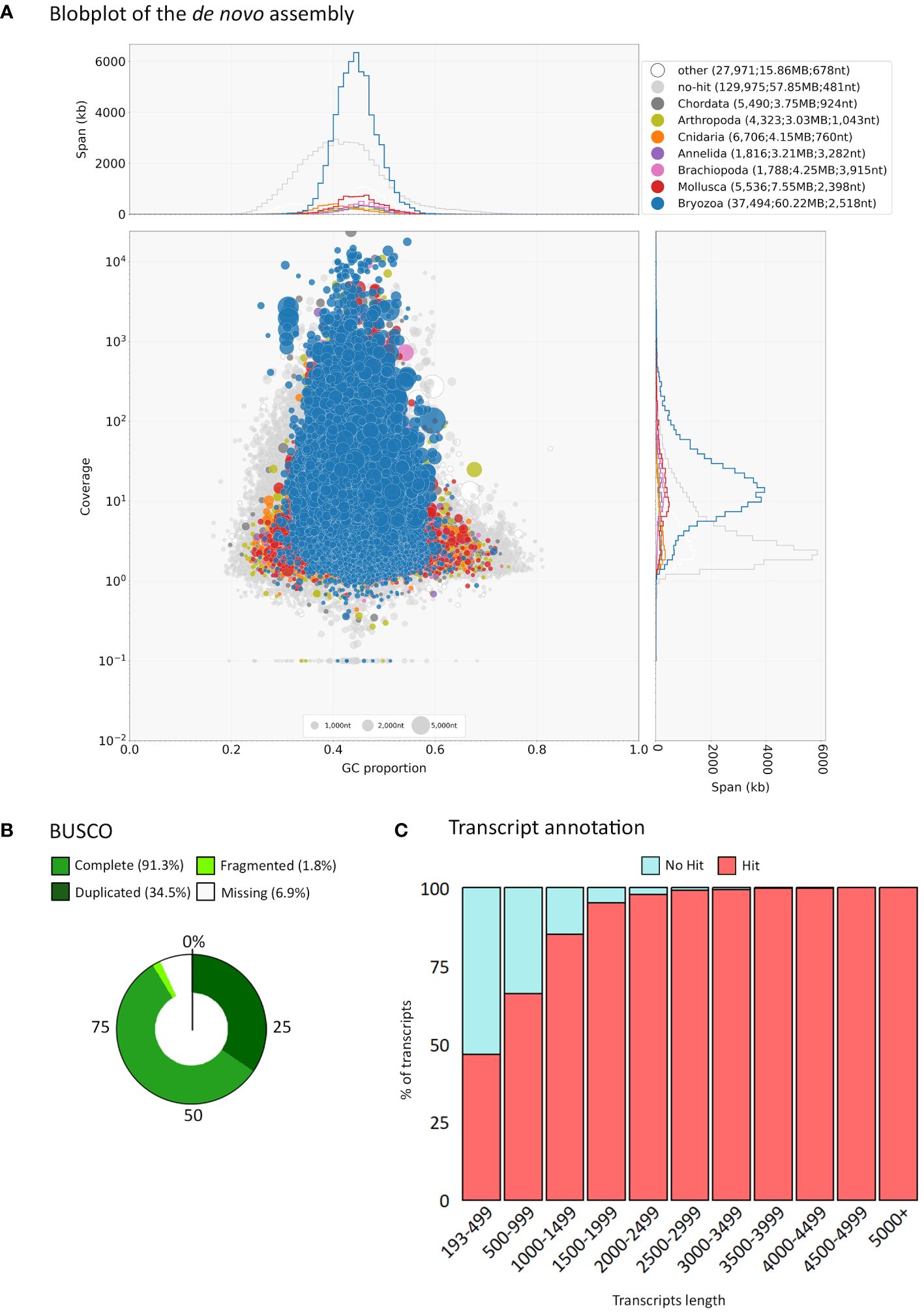
Figure 2 Summary of assembly data. (A) Blobplot showing distribution of the de novo assembly according to GC content and coverage within the final transcriptome assembly. Summary statistics are also shown at the top of the panel. (B) BUSCO gene completeness after low read support removal (cutoff of min_expr_any = 3). (C) Transcript length distribution and annotation after low read support removal (cutoff of min_expr_any = 3).
Transcript expression quantification was performed through Trinity’s align_and_estimate_abundance.pl script using Bowtie2 for alignment and RSEM to estimate abundance (as transcripts per million, TPM). Transcripts with low read support were filtered out using the filter_low_expr_transcripts.pl script within the Trinity pipeline, with a cutoff of min_expr_any = 3. Transcript expression quantification was performed once more for the filtered assembly and the resulting files used in downstream analyses. BUSCO v. 3.0.2 (Simão et al., 2015) was used to evaluate the completeness of the filtered assembly (Figure 2B).
Functional annotation analysis was first performed using OmicsBox after using the standalone BLASTx annotation (E value cutoff: 1e-5, with best hit retained, vs the nr database), including InterProScan and all default protein databases. Gene Ontology (GO) was also retrieved by OmicsBox under the categories: biological processes (BP), molecular function (MF), and cellular component (CP). Transcripts of 1000 bp and longer (22,976 transcripts) were subsampled and used for downstream analyses as they represented the majority of total annotations (Figure 2C). Further functional annotation was performed by BLASTx against a custom ‘biomineralization database’ extracted from Swissprot/Uniprot (E value cutoff: 1e-5, with best hit retained). This included all entries in Swissprot/Uniprot with the keyword ‘biomineralization’. TransDecoder was used to find coding regions within the transcripts, and InterProScan v5.51–85.0 to annotate the proteins found by TransDecoder by identifying signatures from the InterPro member databases including TMHMM and Phobius for the protein topology and signal peptide predictor.
A principal component analysis (PCA) and differential expression was performed in R (Supplementary File 2). PCA was used to assess the replicates’ relationship based on their transcript expression. A batch effect was observed which was removed by using the function removeBatchEffect (edgeR package). The dispersion value was also estimated by using the function estimateDisp (edgeR package). A differential expression analysis was performed using edgeR and the batch effect was corrected-for in the design of the model for the DE analysis. edgeR was run with adjusted p-value (BH) cut off 0.05, a log-fold change of 2 and a dispersion value of 0.36.
The results from the functional annotation (Supplementary File 12) and differential expression analyses (Supplementary File 9) were combined with known literature to compile the suite of candidate genes involved in biomineralization in this species (Supplementary File 14). Genes that were not expressed in one of the two young replicates were excluded from the final list even if marked as differentially expressed by the analysis. Similarly, the results from TMHMM and Phobius were combined with known literature for the localization of the encoding proteins.
Read assessment with FASTQC indicated high sequence quality, with more than 80% of the input read pairs surviving after cleaning, with GC content of 44%, 45%, 45% and 46% for the four samples (Supplementary File 3; Supplementary Table S1 and Supplementary Figure S1).
The final assembly consisted of 221,099 transcript isoforms corresponding to 185,188 trinity ‘genes’ and N50 of 1,139 bp. The assembly includes 1.2 times as many ‘isoforms’ as in ‘genes’, suggesting that the Trinity assembler may have recovered splice or allelic variation in the coding sequences, which is unsurprising given the pooled approach which included several colonies.
Of the 221,099 transcripts 37,494 (17%) were assigned to the phylum Bryozoa while 129, 975 (59%) did not recover a blast hit at a reasonable threshold (Figure 2A). This relatively low percentage of sequences with a match of known identity is likely due to the low representation of bryozoans in the GenBank nr database. The BUSCO metazoan set of genes was used to assess the completeness and quality of our assembly. Of the 954 genes in the metazoan BUSCO cassette, 97.2% (928) were found to be represented by complete transcripts, 1% (10) were fragmented and 1.8% (16) were missing. This result show that our de novo assembly recovers the majority of the expected genes found in metazoans. Metrics related to the transcriptome assembly and BUSCO completeness can be seen in Supplementary File 3; Supplementary Table S2.
Transcripts with low read support (min_expr_any = 3) were removed from the library as they often contain errors and obscure the detection of differentially expressed transcripts. The filtered assembly resulted in 125,750 transcripts corresponding to 107,584 Trinity ‘genes’, with a N50 of 1,472 bp (Supplementary File 3; Supplementary Table S2). Of the 954 genes in the metazoan BUSCO set of genes, 91.3% (871) were found to be represented by complete transcripts, 1.8% (17) were fragmented and 6.9% (66) were missing (Figure 2B), indicating that our assembly subset is still of sufficient quality to recover over 90% of the expected metazoan genes, and to be used for downstream analysis. The low number (<2%) of fragmented genes in the final sets indicates most genes were represented by complete transcripts (Supplementary File 3).
Functional annotation was performed on the assembly subset using OmicsBox, against the InterPro member databases and GO terms (Supplementary File 4). About half of the transcripts were annotated with InterProScan (61,442; 48.8%), and of those 32,841 (53%) were assigned to GO terms (Figure 2C). This percentage of annotated sequences is comparable to a similar bryozoan study conducted by Treibergs and Giribet (2020) on Bugulina stolonifera, where from the total of 1,097 genes 67% were annotated when blasting using the NCBI’s nr database, and 39% were assigned to GO terms. It is likely that the limited data available for bryozoans in publicly available databases restricted the recovery rate.
Of the 61,442 annotated transcripts 57% (34,785) were assigned to a blast hit from the NCBI nr database (also in Supplementary File 5), and of these 40% (13,815) were assigned to the phylum Bryozoa. Unsurprisingly, the species with the highest number of best blast hits was Bugula neritina 38% (13,388), the most well studied species in the phylum Bryozoa, followed by the cnidarian Hydra vulgaris, 4% (1,434). This is more likely due to the slow rate of protein evolution in Hydra, rather than cnidarian contamination, as Hydra is a freshwater species. Further, seven enzyme classes were assigned to 7,472 sequences in our dataset, of which Hydrolases (39%) and Transferases (28%) were the most common.
Of the 125,750 transcripts, 22,976 (≥ 1000 bp) were used for downstream analysis as they represented the majority of the annotated transcripts as shown by OmicsBox. Of the 22,976 transcripts, 39.5% (9,093) were assigned to a blast hit from the ‘biomineralization database’ by SwissProt/UniProt, and of these 64.5% were assigned to the phylum Brachiopoda, and the species Lingula unguis (Supplementary File 6). It is worth noting that the ‘biomineralization database’ by uniprot does not include the phylum Bryozoa. TransDecoder recovered 23,689 proteins (Supplementary File 7). The higher number of proteins compared to the number of the transcripts indicates that multiple coding regions occur within some of the transcripts in the assembly. Of the 23,689 proteins 96% (22,853) were annotated by InterProScan.
At ‘gene’ level PCA showed that by far the largest proportion of the variation, 84.02%, was between young and old parts of the colony, while a significantly smaller fraction (around 16% of the variance, in PC2) was between-replicate variation. At ‘isoform’ level PCA showed that the largest proportion of the variation, 76.8%, was once more between the young and old parts of the colony, while a smaller fraction (23% in PC2) was between-replicate variation (Supplementary File 8). This discrepancy suggests a slight batch effect between the sequencing runs for replicate 1 and 2, but one with markedly less influence on transcriptome content than the age of the samples. The variation observed between replicates might also partly reflect the natural biological variation as each replicate is a pool sample of five colonies.
DE analysis was performed to identify which transcripts are expressed at a higher level in the young and old zooids of the colony. Young zooids (distal part of the colony) are more likely to be actively calcifying at a higher rate compared to the rest of the colony, and therefore likely to express the key genes involved in primary calcification. The analysis showed that 506 (2.2%) of the Trinity transcript isoforms were more highly expressed in the young zooids (logFoldChange > 2, p-adjusted < 0.05), 4,676 (20.4%) were more highly expressed in the old zooids (logFoldChange < -2, p-adjusted < 0.05), and 17,794 (77.4%) Trinity transcript isoforms were equally expressed between the young and the old zooids of the colonies (Supplementary Files 8, 9).
This study represents the first transcriptomic resource for this family and the first in the phylum to investigate the process of biomineralization at this level. More, we discuss the process of skeletal resorption and remodeling and possible mechanisms of the mineral nucleation by matrix vesicles and providing a list of candidate genes and their encoded proteins that may be involved in these processes.
Skeletal formation in bryozoans occurs in a similar way to other metazoans, where the mineralized part of the skeleton is secreted by the epithelium in contact with the growing skeleton (Tavener-Smith and Williams, 1972). Based on the differences between young and old zooids in the colony, we expected to find genes expressed that encode proteins involved in growth and skeletal formation, such as those associated with extracellular matrix and transmembrane transport channels, in young compared to the old zooids. As a low level of calcification occurs in old zooids, we also anticipate that some genes involved in biomineralization process may be equally expressed in both young and old zooids in the colony. We have therefore catalogued genes expressed in young and old Cellaria. We discuss the differences between the young and old zooids, but also include the genes that were equally expressed throughout the colony when compiling the suite of genes potentially involved in biomineralization and skeletal resorption in this species.
We analyzed a range of genes related to transmembrane transporters, signaling, and secretory proteins in our data, which may be involved in the skeletal formation. Overall, genes encoding over 50 protein families were identified as candidates which could be involved in the biomineralization process. Many of the proteins are characterized by low complexity regions (e.g., consensus disorder prediction regions) in our annotation. Further, the topology prediction of the proteins is shown by the annotation of cytoplasmic, transmembrane and non-cytoplasmic regions by InterProScan. The annotation of signal peptides also indicate that the proteins are processed through the secretory pathway. All annotation is shown in Supplementary Files 10–12. Thirty-eight transcript isoforms (24 protein families) were significantly more highly expressed in the young zooids and are discussed in more detail below (Figure 3, logFoldChange < -2 or not expressed in old zooids, p-adjusted < 0.05, values in Supplementary File 13). The rest of the transcripts were equally expressed in both young and old zooids.

Figure 3 Candidate genes significantly more highly expressed in young zooids encoding proteins involved in biomineralization. O, Old; Y, Young.
Members of the suite of genes identified in C. immersa are also found in other bryozoans (Rayko et al., 2020; Kumar et al., 2020b; Lombardi et al., 2023), and biomineralizers such as Mollusca, Brachiopoda, Echinodermata and Cnidaria (Livingston et al., 2006; Ramos-Silva et al., 2013; Luo et al., 2015; Malachowicz and Wenne, 2019), including most of the proteins from the “basic evolutionary toolkit” suggested for molluscs (Arivalagan et al., 2016; Clark, 2020).
We discuss their potential function below, based on their expression in older or younger portions of the colony and the published literature on other metazoans. A schematic illustration of candidate genes involved in C. immersa biomineralization identified in the present study, and their hypothesized localization can be seen in Figure 4. All candidate proteins are listed in Supplementary File 14.
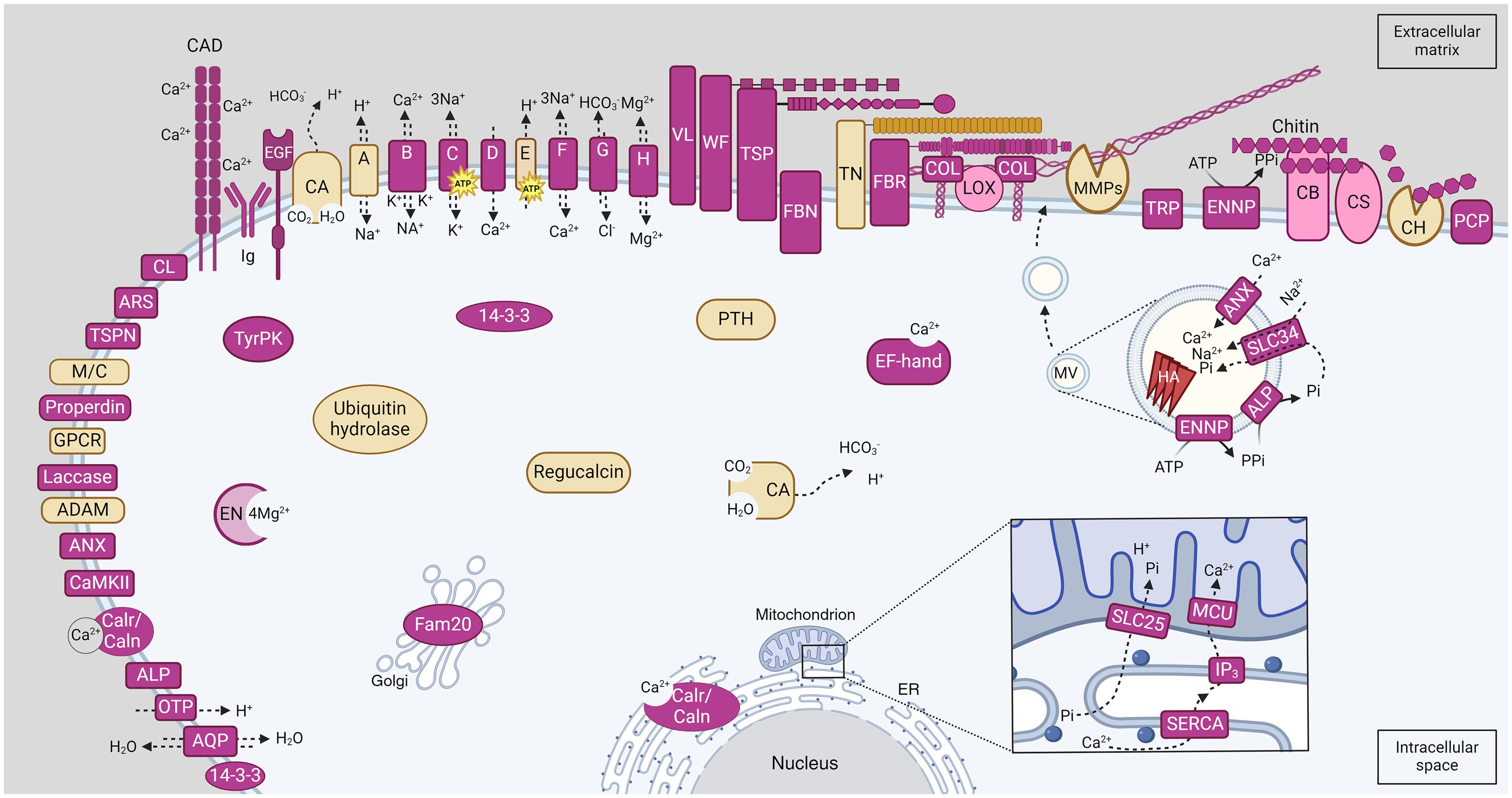
Figure 4 Schematic illustration of candidate proteins involved in C. immersa biomineralization identified in the present study. Proteins location is based on their topology as predicted by InterProScan annotation and known literature. Those predicted with a combination of a signal peptide, a cytoplasmic, transmembrane, and non-cytoplasmic region are located on the membrane. Pink colour indicates proteins involved in primary and secondary calcification, and yellow indicates proteins involved in skeletal remodeling and resorption. The square detail at bottom right shows the flow of the ions from the intracellular space into the mitochondrial matrix, according to the biogenesis mechanism for the exosome-like MV. The circular inset image at centre-right shows the mineral nucleation inside the MV. A, Sodium/Hydrogen exchanger; ADAM, Disintegrin and Metalloprotease; ALP, Alkaline Phosphatase; ANX, Annexins; AQP, Aquaporin; ARS, Arylsulfatase; B, Sodium/potassium/calcium exchanger; C, Sodium/potassium-dependent ATPase; CA, Carbonic Anhydrase; CAD, Cadherin-like; Calr/Caln, Calreticulin/Calnexin; CaMKII, Calcium/calmodulin-dependent protein kinase II, association-domain; CB, Chitin binding protein; CH, Chitinase; CL, C-type lectin-like; CS, Chitin synthase; COL, Collagenase; D, Voltage-dependent calcium channel; E, V-type proton ATPase; EN, Enolase; ENNP, Ectonucleotide pyrophosphatase/phosphodiesterase; EGF, EGF-like domain-containing protein; ER, Endoplasmic reticulum; F, Sodium/calcium exchanger; FBN, Fibrinogen related proteins; FBR, Fribronectins; G, Bicarbonate transporter; GPCR, G protein-coupled receptors; H, Magnesium transporter; HA, hydroxyapatite; Ig, Immunoglobulin; IP3, Inositol 1,4,5-trisphosphate; LOX, Lysyl oxidase-like-related; M/C, Macroglobulin/Complement; MMP, Matrix Metalloproteinase; MCU, Calcium uniporter protein; OTP, Otopetrin; PIC2, Mitochondrial phosphate carrier protein Pic2/Mir1-like; PCP, Purple acid phosphatase; PTH, Parathyroid hormone-responsive B1; TN, Tenascin; TyrPK, Tyrosine proteine kinase; TRP, Transient receptor potential channel; TSP, Thrombospondins; TSPN, Tetraspanin; SERCA, P-type ATPase, subfamily IIA, SERCA-type; SLC25; Solute carrier family 25; SLC34, Solute carrier family 34; VL, Vitellogenin; WF, von Willebrand factors.
Carbonic anhydrase (CA) is a metalloenzyme which catalyzes the reverse hydration of carbon dioxide during calcium carbonate formation, and it is a highly conserved enzyme throughout the metazoan clade (Le Roy et al., 2014; Murdock, 2020). Its role in biomineralization is well established throughout the Metazoa and it is part of the “basic evolutionary toolkit” (Robinson and King, 1963; Livingston et al., 2006; Drake et al., 2013; Ramos-Silva et al., 2013; Germer et al., 2015; Luo et al., 2015; Rose-Martel et al., 2015; Li et al., 2016; Arivalagan et al., 2017). Two genes encoding Alpha carbonic anhydrases were more highly expressed in young zooids (G1-G2 in Figure 3; Supplementary File 13). Specifically, one of the CA (G2) is predicted to have a low complexity region, a signal peptide and a non-cytoplasmic domain (Supplementary File 15). Multicopper oxidases and cupredoxin domains have been identified in the skeletal proteome of corals, and they are mainly involved in iron efflux (Ramos-Silva et al., 2013; Takeuchi et al., 2016). Laccase is a multicopper oxidase with cupredoxin domains. Two transcripts encoding Laccase 2, Isoform Gs were more expressed in the young compared to the old zooids (G15 in Figure 3; Supplementary File 13). One of the Laccase proteins encoded was predicted to have a signal peptide and a non-cytoplasmic region.
Tyrosine protein kinase has been identified to participate in biomineralization in several metazoans including molluscs and echinoderms (Mann et al., 2010; Malachowicz and Wenne, 2019), and it is involved in signal transduction. Two genes encoding tyrosine protein kinase were more highly expressed in the young zooids (G35-G36 in Figure 3 and Supplementary File 13). Ubiquitin may also play a role in biomineralization as has been detected in molluscs (Fang et al., 2012; Čadež et al., 2017). Specifically, one gene encoding ubiquitin carboxyl-terminal hydrolase 32-like protein was more highly expressed in the young zooids (G37 in Figure 3 and Supplementary File 13). This enzyme is responsible for removing the ubiquitin molecules from the peptides. Ubiquitin carboxyl-terminal hydrolases are also thought to participate in bone reformation (Guo et al., 2018). It is therefore unclear whether their role extends skeletal resorption and remodeling in this species.
Calcium binding domains such as Calcium/calmodulin and EF-hand proteins play a role in sustaining the calcium signal and are often found at the biomineralizing front of other marine invertebrates (Sun et al., 2015; Le Roy et al., 2021). Here, one gene encoding a Calcium/calmodulin-dependent protein kinase II (CaMKII), and one a EF-hand calcium-binding domain-containing protein 7 and were highly expressed in the young zooids (G4, G9 in Figure 3; Supplementary File 13).
Ion transport is a key part of the biomineralization process. Ion transportation between the intracellular and the extracellular space can be either passive or active, through transmembrane proteins (Sillanpää et al., 2020). The high expression of one gene encoding a Bicarbonate transporter, eukaryotic (G3 in Figure 3 and Supplementary File 13) suggests that the young zooids of C. immersa actively regulate the concentration of bicarbonate ions (HCO3-) in the extracellular space as it has also been observed in molluscs (Yarra et al., 2021). After the HCO3- is secreted in the extracellular space, it reacts with calcium ions to form calcium carbonate (Clark, 2020). However, the role of this transporter may extend further than that by also neutralizing the acidic environment created during calcium carbonate formation (Hu et al., 2018). Two genes encoding a sodium/potassium-dependent ATPase beta subunit were also more highly expressed in the young zooids (G25-G26 in Figure 3; Supplementary File 13), and may play a key role in ion regulation, which participates in skeletal formation by moving K+ against the ion gradient (Kahil et al., 2021; Yarra et al., 2021).
Immunity associated domains have been reported as part of the skeletal formation in several metazoans (Zhao et al., 2012; Ramos-Silva et al., 2013; Arivalagan et al., 2017; Malachowicz and Wenne, 2019; Song et al., 2019; Schwaner et al., 2022). One gene encoding members of the Immunoglobulin were only expressed in the young zooids (G12 in Figure 3; Supplementary File 13). Transcripts for other immunity associated proteins such as macroglobulin/complement were also more highly expressed in the young zooids (G17-G18 in Figure 3; Supplementary File 13). Alpha-2-macroglobulin is largely known for inhibiting the catalytic activity of peptidases and therefore its role could be related to both skeletal matrix formation and immune defense (Murphy and Nagase, 2009; Flores and Livingston, 2017; Le Roy et al., 2021). One encoded macroglobulin/complement (G18) had a signal peptide, and non-cytoplasmic and low complexity regions indicating its topology as a transmembrane protein. The second protein was also predicted with low complexity regions, and it is likely to have the same topology. Properdin is a glycoprotein of the complement system of the immune system (Lesher et al., 2013). Three genes encoding Properdin were more highly expressed in the young zooids and were predicted to have a signal peptide and non-cytoplasmic region (G20-G22 in Figure 3; Supplementary File 13). Acid phosphatases have been identified at the biomineralization site in molluscs, indicating a role in defense against pathogens (Malachowicz and Wenne, 2019; Zhang et al., 2019). However, in the pearl oyster Pinctada fucata acid phosphatase had an inhibitory activity on calcium carbonate precipitation (Zhang et al., 2019). One gene encoding a purple acid phosphatase was only expressed in the young zooids while the rest were expressed in both young and old zooids (G23 in Figure 3; Supplementary File 13). The encoded protein was predicted to have a cytoplasmic, transmembrane and a non-cytoplasmic region. Further studies will be required to determine if this enzyme also has a similar dual role in C. immersa.
Chitin and chitin-binding domains are also part of the “basic evolutionary toolkit” and have often been reported in metazoans. They appear to play a key role in providing an organic framework and controlling the growth of bio-crystals (Shen and Jacobs-Lorena, 1999; Inoue et al., 2003; Marin et al., 2013; Luo et al., 2015; Li et al., 2016; Arivalagan et al., 2017; Feng et al., 2017; Arroyo-Loranca et al., 2020; Fan et al., 2020; Sun et al., 2020). Chitin has been identified in other marine and freshwater bryozoans (Schafer et al., 2006; Kaya et al., 2015; Lombardi et al., 2023), indicating that chitin is a major structural component of the skeletal matrix in this phylum. Schafer et al. (2006) suggested that the rhizoids in Cellaria species consist mainly of chitin, which could also be the case for C. immersa. One gene encoding an invertebrate chitin-binding protein (PS50940, type-2 profile) was more highly expressed in the young zooids (G14 in Figure 3; Supplementary File 13) and were equally expressed in both young and old zooids. All chitin related proteins were predicted as transmembrane by our annotation.
Collagens are some of the most common proteins found among animals and they are involved in the organic matrix network in all metazoans (Domart-Coulon et al., 2004; Livingston et al., 2006; Drake et al., 2013; Ramos-Silva et al., 2013; Germer et al., 2015; Luo et al., 2015; Rose-Martel et al., 2015; Flores and Livingston, 2017; Malachowicz and Wenne, 2019; Mao et al., 2019; Le Roy et al., 2021). Four genes encoding collagen related proteins were highly expressed in the young zooids (G5-G8 in Figure 3; Supplementary File 13). Other collagen family members were equally well-expressed in young and old zooids. The strength of the collagen network relies on the formation of the cross-links between fibrils, formed by the enzyme lysyl oxidase (Rodriguez-Pascual and Slatter, 2016) which was also expressed in both young and old zooids. While all the encoded collagens were predicted to have low complexity regions, two (G5, G8) were also predicted with a transmembrane region.
Other large and complex extracellular glycoproteins participating in fibril formation (a common aspect of biomineralization) are encoded by our transcriptome. Fibrinogen related proteins (fibrinogen/tenascin/angiopoietin) and fibronectin are common components of the extracellular matrix in other metazoans (Ramos-Silva et al., 2013; Germer et al., 2015; Rose-Martel et al., 2015; Takeuchi et al., 2016; Arivalagan et al., 2017; Feng et al., 2017; Flores and Livingston, 2017; Li et al., 2017a; Malachowicz and Wenne, 2019; Shimizu et al., 2020; Le Roy et al., 2021). Both fibronectins and fibrinogen have a significant role in matrix assembly by binding to other extracellular proteins such as Collagen, Thrombospondin type 1 and von Willebrand factor domains (Halper and Kjaer, 2014). One gene encoding a Fibrinogen, alpha/beta/gamma chain domain with low complexity regions was more highly expressed in the young zooids (G11 in Figure 3; Supplementary File 13). This fibrinogen domain was predicted as transmembrane with a large cytoplasmic region. Other proteins participating in the fibril formation such as fibronectins and Willebrand factor proteins were expressed in both young and old zooids. Tenascin is a large oligomeric extracellular matrix glycoprotein found in the skeletal matrix (Gorski, 2011; Hemond et al., 2014; Arivalagan et al., 2017; Flores and Livingston, 2017; Malachowicz and Wenne, 2019). Members of this protein family are known to modulate adhesion and protein interaction and may also have a regulatory effect on fibronectins (Halper and Kjaer, 2014). The expression of tenascin in adult tissues is generally low, and it is often upregulated during tissue repair (Halper and Kjaer, 2014). Here, one gene annotated as encoding Tenascin with EGF-like domain was more highly expressed in the young zooids (G28 in Figure 3; Supplementary File 13).
Tetraspanins are membrane organizers and have been identified in the skeletal matrix of echinoderms (Matranga et al., 2013; Flores and Livingston, 2017). Among the proteins they regulate are the Disintegrin and Metalloprotease family (Charrin et al., 2014; Termini and Gillette, 2017). Here, one gene encoding Tetraspanin-31 was only expressed in the young zooids, and it was indicated as a transmembrane protein by our annotation (G29 in Figure 3; Supplementary File 13). Thrombospondins are considered the “adhesion-modulating” components of the extracellular matrix, and they participate in collagen assembly as well as interacting with other structural components (Halper and Kjaer, 2014; Le Roy et al., 2021). The involvement of thrombospondin in skeletal formation is well established throughout the metazoan clade (Domart-Coulon et al., 2004; Ramos-Silva et al., 2013; Takeuchi et al., 2016; Flores and Livingston, 2017; Malachowicz and Wenne, 2019; Le Roy et al., 2021). Five genes encoding Thrombospondin type 1 family members were highly expressed in the young zooids compared to older zooids (G30-G34 in Figure 3; Supplementary File 13). Topology prediction indicates their location to be transmembrane. Epidermal growth factors (EGFs) have been detected at the biomineralization site in other metazoans (Takeuchi et al., 2016; Flores and Livingston, 2017; Malachowicz and Wenne, 2019; Le Roy et al., 2021). One gene encoding multiple EGF like domains 6 was more highly expressed in the young compared to the old zooids (G10 in Figure 3; Supplementary File 13).
Skeletal resorption is the controlled dissolution and removal of skeletal parts by an animal on itself. It occurs in all biomineralizers, and although it was documented for bryozoans as early as the 19th century, it has only been reported in detail in this phylum by Batson et al. (2020). The authors suggested that the formation of the nodes in Cellaria species occurs by resorption (i.e., abscission) oriented perpendicular to the skeleton. Skeletal resorption or remodeling likely takes place throughout the colony, such as the fragmentation of the extracellular matrix at the mineralization site in the young zooids for the incorporation of the minerals, or the formation of the brooding chambers (i.e., ovicells) in the old zooids by window resorption (Anderson, 2003; Batson et al., 2020). Here, we identified transcripts encoding twelve proteins that may be involved in skeletal resorption and remodeling in C. immersa, which can be found in Supplementary File 7.
Skeletal resorption likely occurs through a combination of mechanisms that involve several biochemical mechanisms. (i) Ion regulation: Secretion of protons at the extracellular space can lead to the acidification of the area and carbonate dissolution (Batson et al., 2020; Everts et al., 2022). Proton production and transportation may occur through the activity of carbonic anhydrase and proton transmembrane channels identified in our transcriptome. For example, V-type proton ATPases are thought to play a role in skeletal resorption as they can increase the acidity of the environment in the extracellular space (Batson et al., 2020). Here, one gene encoding a V-type ATPase 116kDa subunit family was only expressed in the young zooids while the rest were expressed throughout the colony (G38 in Figure 3; Supplementary File 13). It is likely that G38 has a role in skeletal remodeling during primary calcification. Also, one gene annotated as encoding a Sodium/Hydrogen Exchanger 8, was also more highly expressed in the young zooids compared to the old zooids (G24 in Figure 3; Supplementary File 13). This exchanger is known to be involved in pH regulation in molluscs (Ramesh et al., 2020). The freed calcium ions can then be transported from the area by calcium binding proteins such as Regucalcin (Yamaguchi, 2014). (ii) Extracellular matrix degradation: This step is the process of the matrix fragmentation mostly through enzymatic activity. Metallopeptidases and Chitinase are common enzymes involved in matrix degradation (Krane and Inada, 2008; Batson et al., 2020; Everts et al., 2022). Here, two genes encoding matrix metalloproteinases (MMPs) with collagenase catalytic domain (G18-G19 in Figure 3; Supplementary File 13) were found to be more highly expressed in the young zooids, likely involved in the remodeling of the growing margin during primary calcification. MMP-28 was predicted to have a signal peptide, a non-cytoplasmic region and a low complexity region. MMP-21 was predicted with a low complexity region by InterProScan, and it is likely to have the same topology as MMP-28. Genes encoding chitinase were expressed equally in both growing and old zooids of the colony suggesting its role in this process. Alpha-2-macroglobulin’s regulatory role related to endopeptidases could also potentially be associated with skeletal resorption control (Murphy and Nagase, 2009). Tenascin has been commonly found in skeletal remodeling sites with matrix metallopeptidases, participating in the fragmentation of the matrix (Halper and Kjaer, 2014). (iii) Hormonal regulation: Parathyroid hormone (PTH) is one of the major regulators of mineral metabolism and biomineralization and does this by controlling calcium levels (Arnold et al., 2021). Collagenase is also known to be linked to PTH activity (Zhao et al., 1999). G-protein coupled receptors are part of the endocrine process regulating biomineralization and may have a regulatory effect on PTH in C. immersa (Cardoso et al., 2020).
The characterization of the epithelial cells in bryozoans has been greatly neglected. The skeletal cells of bryozoans may vary between species but are characterized by a nucleus, a large nucleolus, heterochromatin and a Golgi body or Golgi complexes that dominate the cell (Tavener-Smith and Williams, 1972; Tamberg et al., 2022). However, the cells are mainly characterized by endoplasmic reticulum in both cheilostomata and cyclostomata (Tavener-Smith and Williams, 1972; Tamberg et al., 2022), and mitochondria and small secretory vesicles in cheilostomata (Tavener-Smith and Williams, 1972). The location of “the mineral nucleation site” in bryozoans and how it is embedded in the extracellular matrix remains unknown. Here, we discuss two possible pathways for the biogenesis of matrix vesicles based on mechanisms proposed in other metazoans, and the gene annotation in our transcriptome (Figure 4; Supplementary File 7).
Matrix vesicles (MVs) are naturally secreted from the cells and selectively transported to the mineralization site (Anderson, 2003; Ansari et al., 2021). The biogenesis mechanism of these matrix vesicles remains a challenge, but the similarities between MVs and the vesicles released by non-skeletal cells suggests that the mechanism of their biogenesis may be in part conserved (Anderson, 2003; Ansari et al., 2021). There are two possible mechanisms of MVs biogenesis that may be involved in the mineral nucleation and deposition in C. immersa:
1) Exosome-Like MVs, are formed intracellularly through an exosome-like mechanism likely involving mitochondria and are believed to participate in primary and secondary calcification (Ansari et al., 2021; Yan et al., 2023). Calcium influx in the endoplasmic reticulum (ER) through sarco-endoplasmic reticulum ATPase and efflux through inositol 1,4,5-trisphosphate (IP3; G13 in Figure 3 and Supplementary File 13) eventually reaches the mitochondrial matrix through the calcium uniporter protein (Yan et al., 2023). Phosphate (Pi) is transported through the ER and enters the mitochondria through the solute carrier family 25, a mitochondrial phosphate carrier (SLC25; G27 in Figure 3; Supplementary File 13), which also promotes the influx of protons in the mitochondria (Yan et al., 2023). The accumulation of these ions in the mitochondria likely initiates the pre-nucleation of mineral complexes, resulting in the mitochondria becoming swollen and rendered dysfunctional, eventually fusing with a lysosome which breaks down the mitochondria leaving the mineral complexes in the MV (Ansari et al., 2021; Yan et al., 2023). During this process, mineral nucleation continues through enzymes and transporters on their plasma membrane. Such proteins include Alkaline Phosphatase which hydrolyses pyrophosphate (PPi) to Pi, Annexins for Ca2+ transportation and a sodium-dependent phosphate symporter (solute carrier family 34) for the Pi transportation inside the vesicle (Anderson, 2003; Yan et al., 2023). It is unclear if these vesicles are secreted from the cells as such and transported to the extracellular matrix, or whether the mineral complexes are simply discharged via exocytosis to the extracellular space and assimilated into the matrix within the collagen gaps (Yan et al., 2023). In the case that the vesicle is selectively transported to the matrix, mineral nucleation may continue, resulting in needle-like hydroxyapatite which disrupts the MV’s membrane, and is embedded in the extracellular matrix by the activity of metallopeptidases (Anderson, 2003; Ansari et al., 2021; Yan et al., 2023). Ectonucleotide pyrophosphatase/phosphodiesterase (ENNP) is also likely to be involved by generating PPi locally (Yan et al., 2023).
2) Ectosome-Like MVs are formed by budding and pinching off from the plasma membrane of skeletal cells and are likely involved in secondary calcification (Ansari et al., 2021). Similarly, to exosome-Like MVs, these vesicles accumulate calcium and phosphate ions through Alkaline phosphatase, Annexins and Sodium-dependent phosphate symporter proteins, which are located on their plasma membrane (Ansari et al., 2021). Mineral nucleation continues until the formation of needle-like hydroxyapatite, disruption of the MV membrane and incorporation in the extracellular matrix (Anderson, 2003; Ansari et al., 2021).
Alkaline phosphatase is largely involved with the formation of calcium phosphate (Anderson et al., 2005), but bryozoan skeletons are generally calcitic, aragonitic or bimineralic with a variety of levels of Mg (Smith et al., 2006; Smith and Lawton, 2010; Loxton et al., 2018). Specifically, C. immersa’s skeletal carbonate is calcite with low Mg content (Smith et al., 2006). However, calcium carbonate depositors may manipulate aspects of their biomineralization pathways with phosphate (Kahil et al., 2021). In the European abalone Haliotis tuberculata, phosphate is responsible for the stability of the transient amorphous calcium carbonate (Ajili et al., 2022). While the formation of hydroxyapatite was predominant at the very early stages of mineralization, the same hydroxyapatite crystals vanished at later stages with the conversion of calcium phosphate into carbonate (Ajili et al., 2022). This shift occurred through the activity of carbonic anhydrase (Ajili et al., 2022). A similar mineral shift may happen in C. immersa if the mineral nucleation occurs through exosome-like or ectosome-like MVs. Alternatively, another role of alkaline phosphatase may simply be to hydrolyse PPi, as increase of PPi inhibits biomineralization (Golub, 2009).
The mechanisms discussed here for the formation of the matrix vesicles and the mineral nucleation in C. immersa should only be considered a hypothesis at this stage. Most of the transcripts encoding proteins involved in these pathways, excluding IP3 and SLC25, were not highly expressed in the young zooids but were equally expressed throughout the colony. This could be due to old zooids using the same pathways for secondary calcification, or as these proteins are involved in other secretory pathways not related to the biomineralization process. Nevertheless, this catalogue will be of utility as a hypothesis generation tool and for comparison with novel resources in the future.
Bryozoans are among the primary marine biomineralizers and are important ecosystem engineers, especially in the Southern Hemisphere. However, the molecular machinery responsible for the formation of their skeleton has to date remained unknown and largely limited to proteomic data from a few studies. The assembly of the complete transcriptome in the marine bryozoan C. immersa has added considerably to the transcriptome resources available for Bryozoa and provides for the first time an extensive list of potential proteins involved in biomineralization. Transcripts encoding over 50 proteins were identified as candidates which could be involved in biomineralization in this species, including transmembrane transporters, signaling, and secretory proteins. The proteins predicted in C. immersa are similar to those seen in other metazoans, particularly molluscs and brachiopods. This similarity between bryozoans and molluscs may indicate that they calcify in similar ways, or it may simply indicate the amount of information we have for molluscs, compared to less-well studied animals. Candidate proteins involved in skeletal resorption and remodeling are also predicted here for the first time for this phylum. Finally, the mechanisms for the biogenesis of matrix vesicles and mineral nucleation are also discussed for the first time for this phylum, but these represent hypotheses to be investigated in future studies. The journey to learn about the bryozoan “biomineralization toolkit” is still in its infancy, but alongside further transcriptome studies, especially of other heavily calcified bryozoans, this work will provide the basis for comparative analysis.
Raw reads have been uploaded to NCBI Short Read Archive, with accession PRJNA1077468. The final assembly is available to download in full from Figshare, doi: 10.6084/m9.figshare.25237483.v2 and TSA, accession: GKVQ01000000.
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because no ethics were required for this phylum.
KA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. AS: Resources, Supervision, Writing – review & editing. NK: Methodology, Writing – review & editing. CB: Methodology, Resources, Supervision, Writing – review & editing, Formal analysis.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Doctoral Scholarship, University of Otago, to KA and research funding from the Departments of Biochemistry and Marine Science, University of Otago. Health Science Career Development Postdoc, University of Otago (while preparing this manuscript for publication). NK: Supported by Rutherford Discovery Fellowship MFP-UOO2109, Royal Society Te Apārangi.
We thank Bill Dickson and Evan Kenton for their help on board RV Polaris II during the collection of the specimens. We are very grateful to Dr. Kim Currie (NIWA/University of Otago Research Centre for Oceanography) for allowing to KA to be on board during her ‘Munida Time Series’ fieldwork to collect specimens. KA would like to thank Dr. Chun Shen Lim (Department of Biochemistry) for his help during the initial RNA extractions and discussions. Otago Genomics Facility, University of Otago (Dunedin, New Zealand) for Illumina MiSeq.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material for this article is available at Figshare: 10.6084/m9.figshare.25237483.v2.
Supplementary File 1 | Foraminifera-sequences.fasta.
Supplementary File 2 | DE_analysis_and_graph (R script).
Supplementary File 3 | Transcriptomes assembly overview.
Supplementary File 4 | Functional annotation by OmicsBox.
Supplementary File 5 | NCBI nr blastx results.
Supplementary File 6 | SwissprotUniprot Blastx results.
Supplementary File 7 | Protein sequences by Transdecoder.
Supplementary File 8 | PCA and Volcano plot.
Supplementary File 9 | Differential Expression analysis results.
Supplementary File 10 | InterProScan annotation.
Supplementary File 11 | InterProScan.gff3.
Supplementary File 12 | Functional annotation combined.
Supplementary File 13 | Candidate genes in young zooids.
Supplementary File 14 | Candidate biomineralization proteins.
Supplementary File 15 | Alpha Carbonic Anydrase visualization.
Achilleos K., Gordon D. P., Smith A. M. (2020). Cellaria (Bryozoa, Cheilostomata) from the deep: new species from the southern Zealandian region. Zootaxa 4801, 201–236. doi: 10.11646/zootaxa.4801.2.1
Achilleos K., Smith A. M., Gordon D. P. (2019). The articulated bryozoan genus Cellaria in the southern Zealandian Region: distribution and associated fauna. Mar. Biodiversity 49, 2801–2812. doi: 10.1007/s12526-019-01009-y
Ajili W., Tovani C. B., Fouassier J., de Frutos M., Laurent G. P., Bertani P., et al. (2022). Inorganic phosphate in growing calcium carbonate abalone shell suggests a shared mineral ancestral precursor. Nat. Commun. 13, 1496. doi: 10.1038/s41467-022-29169-9
Anderson C. H. (2003). Matrix vesicles and calcification. Curr. Rheumatol Rep. 5, 222–226. doi: 10.1007/s11926-003-0071-z
Anderson C. H., Garimella R., Tague S. E. (2005). The role of matrix vesicles in growth plate development and biomineralization. Front. Bioscience 10, 822–837. doi: 10.2741/1576
Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Ansari S., de Wildt B. W. M., Vis M. A. M., de Korte C. E., Ito K., Hofmann S., et al. (2021). Matrix vesicles: Role in bone mineralization and potential use as therapeutics. Pharmaceuticals 14, 289. doi: 10.3390/ph14040289
Arivalagan J., Marie B., Sleight V. A., Clark M. S., Berland S., Marie A. (2016). Shell matrix proteins of the clam, Mya truncata: Roles beyond shell formation through proteomic study. Mar. Genomics 27, 69–74. doi: 10.1016/j.margen.2016.03.005
Arivalagan J., Yarra T., Marie B., Sleight V. A., Duvernois-Berthet E., Clark M. S., et al. (2017). Insights from the shell proteome: Biomineralization to adaptation. Mol. Biol. Evol. 34, 66–77. doi: 10.1093/molbev/msw219
Arnold A., Dennison E., Kovacs C. S., Mannstadt M., Rizzoli R., Brandi M. L., et al. (2021). Hormonal regulation of biomineralization. Nat. Rev. Endocrinol. 17, 261–275. doi: 10.1038/s41574–021-00477–2
Arroyo-Loranca R. G., Hernandez-Saavedra N. Y., Hernandez-Adame L., Rivera-Perez C. (2020). Ps19, a novel chitin binding protein from Pteria sterna capable to mineralize aragonite plates in vitro. PloS One 15, e0230431. doi: 10.1371/journal.pone.0230431
Batson P. B., Tamberg Y., Taylor P. D., Gordon D. P., Smith A. M. (2020). Skeletal resorption in bryozoans: occurrence, function and recognition. Biol. Rev. 95, 1341–1371. doi: 10.1111/brv.12613
Bishop J., Wood C., Adkins P., Jenkins H. (2023). The genome sequence of an encrusting bryozoan, Cryptosula pallasiana (Moll 1803). Wellcome Open Res. 8, 128. doi: 10.12688/wellcomeopenres.19100.1
Buchfink B., Xie C., Huson D. H. (2014). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Čadež V., Škapin S. D., Leonardi A., Križaj I., Kazazić S., Salopek-Sondi B., et al. (2017). Formation and morphogenesis of a cuttlebone’s aragonite biomineral structures for the common cuttlefish (Sepia officinalis) on the nanoscale: Revisited. J. Colloid Interface Sci. 508, 95–104. doi: 10.1016/j.jcis.2017.08.028
Cardoso J. C. R., Félix R. C., Ferreira V., Peng M. X., Zhang X., Power D. M. (2020). The calcitonin-like system is an ancient regulatory system of biomineralization. Sci. Rep. 10, 7581. doi: 10.1038/s41598–020-64118-w
Chang E. P., Evans J. S. (2015). Pif97, a von willebrand and peritrophin biomineralization protein, organizes mineral nanoparticles and creates intracrystalline nanochambers. Biochemistry 54, 5348–5355. doi: 10.1021/acs.biochem.5b00842
Charrin S., Jouannet S., Boucheix C., Rubinstein E. (2014). Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648. doi: 10.1242/jcs.154906
Clark M. S. (2020). Molecular mechanisms of biomineralization in marine invertebrates. J. Exp. Biol. 223, jeb206961. doi: 10.1242/jeb.206961
Domart-Coulon I., Tambutté S., Tambutté E., Allemand D. (2004). Short term viability of soft tissue detached from the skeleton of reef-building corals. J. Exp. Mar. Biol. Ecol. 309, 199–217. doi: 10.1016/j.jembe.2004.03.021
Drake J. L., Mass T., Haramaty L., Zelzion E., Bhattacharya D., Falkowski P. G. (2013). Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc. Natl. Acad. Sci. U.S.A. 110, 3788–3793. doi: 10.1073/pnas.1301419110
Everts V., Jansen I. D. C., de Vries T. J. (2022). Mechanisms of bone resorption. Bone 163, 116499. doi: 10.1016/j.bone.2022.116499
Fan S., Zheng Z., Hao R., Du X., Jiao Y., Huang R. (2020). PmCBP, a novel poly (chitin-binding domain) gene, participates in nacreous layer formation of Pinctada fucata martensii. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 240, 110374. doi: 10.1016/j.cbpb.2019.110374
Fang D., Pan C., Lin H., Lin Y., Xu G., Zhang G., et al. (2012). Ubiquitylation functions in the calcium carbonate biomineralization in the extracellular matrix. PloS One 7. doi: 10.1371/journal.pone.0035715
Feng D., Li Q., Yu H., Kong L., Du S. (2017). Identification of conserved proteins from diverse shell matrix proteome in Crassostrea gigas: Characterization of genetic bases regulating shell formation. Sci. Rep. 7, 45754. doi: 10.1038/srep45754
Flores R. L., Livingston B. T. (2017). The skeletal proteome of the sea star Patiria miniata and evolution of biomineralization in echinoderms. BMC Evol. Biol. 17, 125. doi: 10.1186/s12862-017-0978-z
Furuhashi T., Beran A., Blazso M., Czegeny Z., Schwarzinger C., Steiner G. (2009). Pyrolysis GC/MS and IR spectroscopy in chitin analysis of molluscan shells. Biosci. Biotechnol. Biochem. 73, 93–103. doi: 10.1271/bbb.80498
Germer J., Mann K., Wörheide G., Jackson D. J. (2015). The skeleton forming proteome of an early branching metazoan: A molecular survey of the biomineralization components employed by the coralline sponge Vaceletia sp. PloS One 10, e0140100. doi: 10.1371/journal.pone.0140100
Golub E. E. (2009). Role of matrix vesicles in biomineralization. Biochim. Biophys. Acta Gen. Subj. 1790, 1592–1598. doi: 10.1016/j.bbagen.2009.09.006
Gorski J. P. (2011). Biomineralization of bone: a fresh view of the roles of non-collagenous proteins. Front. bioscience 16, 2598. doi: 10.2741/3875
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo Y. C., Zhang S. W., Yuan Q. (2018). Deubiquitinating enzymes and bone remodeling. Stem Cells Int., 3712083. doi: 10.1155/2018/3712083
Halper J., Kjaer M. (2014). “Basic components of connective tissues and extracellular matrix: elastin, fibrillin, fibulins, fibrinogen, fibronectin, laminin, tenascins and thrombospondins,” in Progress in Heritable Soft Connective Tissue Diseases. Ed. Halper J. (Springer Dordrecht Heidelberg, New York London), 31–47.
Hemond E. M., Kaluziak S. T., Vollmer S. V. (2014). The genetics of colony form and function in Caribbean Acropora corals. BMC Genomics 15, 1133. doi: 10.1186/1471-2164-15-1133
Hu M. Y., Yan J.-J., Petersen I., Himmerkus N., Bleich M., Stumpp M. (2018). A SLC4 family bicarbonate transporter is critical for intracellular pH regulation and biomineralization in sea urchin embryos. Elife 7, e36600. doi: 10.7554/eLife.36600.019
Inoue H., Ohira T., Ozaki N., Nagasawa H. (2003). Cloning and expression of a cDNA encoding a matrix peptide associated with calcification in the exoskeleton of the crayfish. Comp Biochem Physiol B Biochem Mol Biol. 136, 755–65. doi: 10.1016/s1096-4959(03)00210-0
Joubert C., Piquemal D., Marie B., Manchon L., Pierrat F., Zanella-Cléon I., et al. (2010). Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: Focus on biomineralization. BMC Genomics 11, 613. doi: 10.1186/1471-2164-11-613
Kahil K., Weiner S., Addadi L., Gal A. (2021). Ion pathways in biomineralization: Perspectives on uptake, transport, and deposition of calcium, carbonate, and phosphate. J. Am. Chem. Soc. 143, 21100–21112. doi: 10.1021/jacs.1c09174
Kaya M., Baublys V., Šatkauskiene I., Akyuz B., Bulut E., Tubelyte V. (2015). First chitin extraction from Plumatella repens (Bryozoa) with comparison to chitins of insect and fungal origin. Int. J. Biol. Macromol 79, 126–132. doi: 10.1016/j.ijbiomac.2015.04.066
Kimura H., Kwan K. M., Zhang Z., Deng J. M., Darnay B. G., Behringer R. R., et al. (2008). Cthrc1 is a positive regulator of osteoblastic bone formation. PloS One 3, e3174. doi: 10.1371/journal.pone.0003174
Kintsu H., Okumura T., Negishi L., Ifuku S., Kogure T., Sakuda S., et al. (2017). Crystal defects induced by chitin and chitinolytic enzymes in the prismatic layer of Pinctada fucata. Biochem. Biophys. Res. Commun. 489, 89–95. doi: 10.1016/j.bbrc.2017.05.088
Kopylova E., Noé L., Touzet H. (2012). SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217. doi: 10.1093/bioinformatics/bts611
Krane S. M., Inada M. (2008). Matrix metalloproteinases and bone. Bone. 43, 7–18. doi: 10.1016/j.bone.2008.03.020
Kumar G., Ertl R., Bartholomew J. L., El-Matbouli M. (2020a). First transcriptome analysis of bryozoan Fredericella sultana, the primary host of myxozoan parasite Tetracapsuloides bryosalmonae. PeerJ 8, e9027. doi: 10.7717/peerj.9027
Kumar G., Ertl R., Bartholomew J. L., El-Matbouli M. (2020b). Transcriptome analysis elucidates the key responses of bryozoan Fredericella sultana during the development of Tetracapsuloides bryosalmonae (Myxozoa). Int. J. Mol. Sci. 21, 1–16. doi: 10.3390/ijms21165910
Laetsch D. R., Blaxter M. L. (2017). BlobTools: Interrogation of genome assemblies. F1000Res 6, 1287. doi: 10.12688/f1000research.12232.1
Langmead B., Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Le Roy N., Ganot P., Aranda M., Allemand D., Tambutté S. (2021). The skeletome of the red coral Corallium rubrum indicates an independent evolution of biomineralization process in octocorals. BMC Ecol. Evol. 21, 1. doi: 10.1186/s12862–020-01734–0
Le Roy N., Jackson D. J., Marie B., Ramos-Silva P., Marin F. (2014). The evolution of metazoan aα-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front. Zool 11. doi: 10.1186/s12983–014-0075–8
Lesher A. M., Nilsson B., Song W. C. (2013). Properdin in complement activation and tissue injury. Mol. Immunol. 56, 191–198. doi: 10.1016/j.molimm.2013.06.002
Li H., Liu B., Huang G., Fan S., Zhang B., Su J., et al. (2017a). Characterization of transcriptome and identification of biomineralization genes in winged pearl oyster (Pteria penguin) mantle tissue. Comp. Biochem. Physiol. Part D Genomics Proteomics 21, 67–76. doi: 10.1016/j.cbd.2016.12.002
Li S., Liu C., Huang J., Liu Y., Zhang S., Zheng G., et al. (2016). Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO2 and temperature. Sci. Rep. 6. doi: 10.1038/srep18943
Li H., Wang D., Deng Z., Huang G., Fan S., Zhou D., et al. (2017b). Molecular characterization and expression analysis of chitinase from the pearl oyster Pinctada fucata. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 203, 141–148. doi: 10.1016/j.cbpb.2016.10.007
Livingston B. T., Killian C. E., Wilt F., Cameron A., Landrum M. J., Ermolaeva O., et al. (2006). A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 300, 335–348. doi: 10.1016/j.ydbio.2006.07.047
Lombardi C., Kuklinski P., Spirandelli E., Bruzzone G., Raiteri G., Bordone A., et al. (2023). Antarctic bioconstructional bryozoans from Terra Nova bay (Ross sea): morphology, skeletal structures and biomineralization. Minerals 13, 246. doi: 10.3390/min13020246
Loxton J., Jones M. S., Najorka J., Smith A. M., Porter J. S. (2018). Skeletal carbonate mineralogy of Scottish bryozoans. PloS One 13, e0197533. doi: 10.1371/journal.pone.0197533
Luo Y. J., Takeuchi T., Koyanagi R., Yamada L., Kanda M., Khalturina M., et al. (2015). The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat. Commun. 6, 8301. doi: 10.1038/ncomms9301
Malachowicz M., Wenne R. (2019). Mantle transcriptome sequencing of Mytilus spp. and identification of putative biomineralization genes. PeerJ 6, e6245. doi: 10.7717/peerj.6245
Mann K., Poustka A. J., Mann M. (2010). Phosphoproteomes of Strongylocentrotus purpuratus shell and tooth matrix: identification of a major acidic sea urchin tooth phosphoprotein, phosphodontin. Proteome Sci 8, 6. doi: 10.1186/1477-5956-8-6
Mao J., Zhang W., Wang X., Song J., Yin D., Tian Y., et al. (2019). Histological and Expression Differences Among Different Mantle Regions of the Yesso Scallop (Patinopecten yessoensis) Provide Insights into the Molecular Mechanisms of Biomineralization and Pigmentation. Mar. Biotechnol. 21, 683–696. doi: 10.1007/s10126-019-09913-x
Marin F., Marie B., Hamada S., Silva P., Le Roy N., Guichard N., et al. (2013). ‘Shellome’: proteins involved in mollusc shell biomineralization-diversity, functions in Recent Advances in Pearl Research, pp 149–166 Eds., Watabe S., Maeyama K., Nagasawa H., TERRAPUB.
Matranga V., Pinsino A., Bonaventura R., Costa C., Karakostis K., Martino C., et al. (2013). Cellular and molecular bases of biomineralization in sea urchin embryos. Cah. Biol. Mar. 54, 467–478.
McKinney F. K., Jackson J. B. C. (1991). Bryozoans as modular machines. , in: Bryozoan Evolution. Univ. Chicago Press, Chicago, pp, 1–13.
Miyamoto H., Miyashita T., Okushimat M., Nakanoi S., Morita T., Matsushiro A. (1996). A carbonic anhydrase from the nacreous layer in oyster pearls. Natl. Institutes Health 93, 9657–9660. doi: 10.1073/pnas.93.18.9657
Murdock D. J. E. (2020). The ‘biomineralization toolkit’ and the origin of animal skeletons. Biol. Rev. 95, 1372–1392. doi: 10.1111/brv.12614
Murphy G., Nagase H. (2009). Progress in matrix metalloproteinase research. Mol. Aspects Med. 29, 290–308. doi: 10.1016/j.mam.2008.05.002
Orr R. J. S., Di Martino E., Gordon D. P., Ramsfjell M. H., Mello H. L., Smith A. M., et al. (2021). A broadly resolved molecular phylogeny of New Zealand cheilostome bryozoans as a framework for hypotheses of morphological evolution. Mol. Phylogenet Evol. 161, 107172. doi: 10.1016/j.ympev.2021.107172
Ramesh K., Hu M. Y., Melzner F., Bleich M., Himmerkus N. (2020). Intracellular pH regulation in mantle epithelial cells of the Pacific oyster, Crassostrea gigas. J. Comp. Physiol. B 190, 691–700. doi: 10.1007/s00360-020-01303-3
Ramos-Silva P., Kaandorp J., Huisman L., Marie B., Zanella-Cléon I., Guichard N., et al. (2013). The skeletal proteome of the coral Acropora millepora: The evolution of calcification by co-option and domain shuffling. Mol. Biol. Evol. 30, 2099–2112. doi: 10.1093/molbev/mst109
Rayko M., Komissarov A., Kwan J. C., Lim-Fong G., Rhodes A. C., Kliver S., et al. (2020). Draft genome of Bugula neritina, a colonial animal packing powerful symbionts and potential medicines. Sci. Data 7, 356. doi: 10.1038/s41597–020-00684-y
Robinson D. S., King N. R. (1963). Carbonic anhydrase and formation of the hen’s egg shell. Nature 199, 497–498. doi: 10.1038/199497a0
Rodriguez-Pascual F., Slatter D. A. (2016). Collagen cross-linking: Insights on the evolution of metazoan extracellular matrix. Sci. Rep. 6, 37374. doi: 10.1038/srep37374
Rose-Martel M., Smiley S., Hincke M. T. (2015). Novel identification of matrix proteins involved in calcitic biomineralization. J. Proteomics 116, 81–96. doi: 10.1016/j.jprot.2015.01.002
Schafer P., Bader B., Blaschek H. (2006). Morphology and function of the flexible nodes in the cheilostome bryozoan Cellaria sinuosa (Hassall). Courier Forschungsinstitut Senckenberg 257, 119–129.
Schönitzer V., Weiss I. M. (2007). The structure of mollusc larval shells formed in the presence of the chitin synthase inhibitor Nikkomycin Z. BMC Struct. Biol. 7, 71. doi: 10.1186/1472-6807-7-71
Schwaner C., Farhat S., Haley J., Pales Espinosa E., Allam B. (2022). Transcriptomic, proteomic, and functional assays underline the dual role of extrapallial hemocytes in immunity and biomineralization in the hard clam Mercenaria mercenaria. Front. Immunol. 13. doi: 10.3389/fimmu.2022.838530
Shen Z., Jacobs-Lorena M. (1999). Evolution of chitin-binding proteins in invertebrates. J. Mol. Evol. 48, 341–347. doi: 10.1007/PL00006478
Shimizu K., Kintsu H., Awaji M., Matumoto T., Suzuki M. (2020). Evolution of biomineralization genes in the prismatic layer of the pen shell Atrina pectinata. J. Mol. Evol. 88, 742–758. doi: 10.1007/s00239-020-09977-7
Sillanpää J. K., Cardoso J. C., dos R., Félix R. C., Anjos L., Power D. M., et al. (2020). Dilution of seawater affects the Ca2+ Transport in the outer mantle epithelium of Crassostrea gigas. Front. Physiol. 11. doi: 10.3389/fphys.2020.00001
Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Smith A. M., Girvan E. (2010). Understanding a bimineralic bryozoan: Skeletal structure and carbonate mineralogy of Odontionella cyclops (Foveolariidae: Cheilostomata: Bryozoa) in New Zealand. Palaeogeogr Palaeoclimatol Palaeoecol 289, 113–122. doi: 10.1016/j.palaeo.2010.02.022
Smith A. M., Key M. M., Gordon D. P. (2006). Skeletal mineralogy of bryozoans: Taxonomic and temporal patterns. Earth Sci. Rev. 78, 287–306. doi: 10.1016/j.earscirev.2006.06.001
Smith A. M., Lawton E. I. (2010). Growing up in the temperate zone: Age, growth, calcification and carbonate mineralogy of Melicerita chathamensis (Bryozoa) in southern New Zealand. Palaeogeogr Palaeoclimatol Palaeoecol 298, 271–277. doi: 10.1016/j.palaeo.2010.09.033
Song X., Liu Z., Wang L., Song L. (2019). Recent advances of shell matrix proteins and cellular orchestration in marine molluscan shell biomineralization. Front. Mar. Sci. 6. doi: 10.3389/fmars.2019.00041
Sun J., Chen C., Miyamoto N., Li R., Sigwart J. D., Xu T., et al. (2020). The Scaly-foot Snail genome and implications for the origins of biomineralised armour. Nat. Commun. 11, 1657. doi: 10.1038/s41467-020-15522-3
Sun X., Yang A., Wu B., Zhou L., Liu Z. (2015). Characterization of the mantle transcriptome of Yesso scallop (Patinopecten yessoensis): Identification of genes potentially involved in biomineralization and pigmentation. PloS One 10, e0122967. doi: 10.1371/journal.pone.0122967
Suzuki M., Saruwatari K., Kogure T., Yamamoto Y., Nishimura T., Kato T., et al. (2009). An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Sci. (1979) 325, 1388–1390. doi: 10.1126/science.1173793
Takeuchi T., Yamada L., Shinzato C., Sawada H., Satoh N. (2016). Stepwise evolution of coral biomineralization revealed with genome-wide proteomics and transcriptomics. PloS One 11, e0156424. doi: 10.1371/journal.pone.0156424
Tamberg Y., Batson P. B., Smith A. M. (2022). The epithelial layers of the body wall in hornerid bryozoans (Stenolaemata: Cyclostomatida). J. Morphol 283, 406–427. doi: 10.1002/jmor.21451
Tavener-Smith R., Williams A. (1972). The secretion and structure of the skeleton of living and fossil Bryozoa. Phil. Trans. R. Soc. Lond. B., 26497–160. doi: 10.1098/rstb.1972.0010
Taylor P. D., Kuklinski P. (2011). Seawater chemistry and biomineralization: Did trepostome bryozoans become hypercalcified in the “calcite sea” of the Ordovician? Paleobiodivers Paleoenviron 91, 185–195. doi: 10.1007/s12549–011-0054–4
Taylor P. D., Lombardi C., Cocito S. (2015). Biomineralization in bryozoans: Present, past and future. Biol. Rev. 90, 1118–1150. doi: 10.1111/brv.12148
Termini C. M., Gillette J. M. (2017). Tetraspanins function as regulators of cellular signaling. Front. Cell Dev. Biol. 5. doi: 10.3389/fcell.2017.00034
Treibergs K. A., Giribet G. (2020). Differential gene expression between polymorphic zooids of the marine bryozoan Bugulina stolonifera. G3: Genes Genomes Genet. 10, 3843–3857. doi: 10.1534/g3.120.401348
Weiss I. M., Schönitzer V., Eichner N., Sumper M. (2006). The chitin synthase involved in marine bivalve mollusk shell formation contains a myosin domain. FEBS Lett. 580, 1846–1852. doi: 10.1016/j.febslet.2006.02.044
Wernström J. V., Gąsiorowski L., Hejnol A. (2022). Brachiopod and mollusc biomineralisation is a conserved process that was lost in the phoronid–bryozoan stem lineage. Evodevo 13, 17. doi: 10.1186/s13227–022-00202–8
Wood C., Bishop J., Adkins P., Jenkins H. (2023). The genome sequence of an erect bryozoan, Bugulina stolonifera (Ryland 1960). Wellcome Open Res. 8, 26. doi: 10.12688/wellcomeopenres.18775.1
Wood A. C. L., Probert P. K., Rowden A. A., Smith A. M. (2012). Complex habitat generated by marine bryozoans: A review of its distribution, structure, diversity, threats and conservation. Aquat Conserv. 22, 547–563. doi: 10.1002/aqc.2236
Yamaguchi M. (2014). The role of regucalcin in bone homeostasis: Involvement as a novel cytokine. Integr. Biol. (United Kingdom). 6, 258–266. doi: 10.1039/c3ib40217g
Yan X., Zhang Q., Ma X., Zhong Y., Tang H., Mai S. (2023). The mechanism of biomineralization: Progress in mineralization from intracellular generation to extracellular deposition. Japanese Dental Sci. Rev. 59, 181–190. doi: 10.1016/j.jdsr.2023.06.005
Yarra T., Blaxter M., Clark M. S. (2021). A bivalve biomineralization toolbox. Mol. Biol. Evol. 38, 4043–4055. doi: 10.1093/molbev/msab153
Zhang R., Xie L., Yan Z. (2019). “The study on enzymes related to biomineralization of Pinctada fucata,” in Biomineralization Mechanism of the Pearl Oyster, Pinctada Fucata (Singapore: Springer), 445–507. doi: 10.1007/978–981-13–1459-9_4
Zhao W., Byrne M. H., Boyce B. F., Krane S. M. (1999). Bone resorption induced by parathyroid hormone is strikingly diminished in collagenase-resistant mutant mice. J. Clin. Invest. 103, 517–524. doi: 10.1172/JCI5481
Keywords: Cellaria immersa, Bryozoa, transcriptomics, biomineralization, skeletal resorption, extracellular matrix
Citation: Achilleos K, Smith AM, Kenny NJ and Brown CM (2024) A new transcriptome resource for Cellaria immersa (Phylum: Bryozoa) reveals candidate genes and proteins related to biomineralization. Front. Mar. Sci. 11:1389708. doi: 10.3389/fmars.2024.1389708
Received: 22 February 2024; Accepted: 07 June 2024;
Published: 02 July 2024.
Edited by:
Tali Mass, University of Haifa, IsraelReviewed by:
Jeana Drake, University of California, Los Angeles, United StatesCopyright © 2024 Achilleos, Smith, Kenny and Brown. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katerina Achilleos, a2F0ZXJpbmEuYWNoaWxsZW9zQG90YWdvLmFjLm56; Chris M. Brown, Y2hyaXMuYnJvd25Ab3RhZ28uYWMubno=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.