
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 19 April 2024
Sec. Marine Biology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1372252
The increase in UV-B radiation at the Earth’s surface due to the depletion of the stratospheric ozone layer is a notable facet of contemporary climate change patterns. The macroalgae inhabiting the intertidal zone exhibit a diverse array of adaptive strategies to cope with dramatic environmental changes. In this study, we integrated physiological, transcriptomic and metabolomic data from energy metabolism perspective to elucidate the responses and recovery mechanism of N. haitanensis to UV-B radiation exposure. UV-B radiation has a harmful impact on the photosynthetic performance of N. haitanensis. However, an increase in photosynthetic performance and upregulated expression of genes related to photosynthesis were observed during recovery, suggesting that the effect of UV-B on N. haitanensis was dynamic photoinhibition. Recovery experiments revealed that most genes and metabolites related to glycolysis were significantly upregulated, suggesting that glycolysis was activated to promote energy production. In addition, the TCA cycle was also activated, as evidenced by the increase in key substances and the upregulated expression of key enzyme-encoding genes during recovery. Correspondingly, ATP was also abundantly accumulated. These results suggested that the TCA cycle provided ATP for N. haitanensis to repair UV-B damage. Meanwhile, amino acid metabolism was enhanced during recovery as a source of intermediates for the TCA cycle. Therefore, photosynthesis, glycolysis, the TCA cycle, and amino acid metabolism synergistically cooperate to provide material and energy for recovery after UV-B radiation. This study is important for understanding the adaptive strategies of intertidal macroalgae in response to UV-B radiation.
As industrialization accelerates, the excessive release of ozone-depleting substances such as chlorofluorocarbons (CFCs), hydrochlorofluorocarbons (HCFCs) and nitrous oxide (N2O) destroys the stratospheric ozone layer (Lee et al., 2021). Due to the decrease in ozone concentration, additional UV-B radiation (280–315 nm) can penetrate the stratosphere and reach the Earth’s surface (Torres et al., 2019). Various ecological disruptions, such as changes in plant growth and distribution, changes in animal behavior and reduced agricultural productivity, can result from the increase in the intensity and dose of UV-B radiation (Liaqat et al., 2023). UV-B concentrations are expected to continue to increase by 1.3% per decade after 2050, according to current UV radiation prediction models (Soni et al., 2022). In addition, UV-B radiation can penetrate water and reach depths of 20-30 m (Dahms and Lee, 2010). This means that even when submerged, organisms can still be exposed to harmful UV-B radiation in both marine and freshwater environments (Zhao et al., 2021).
The capacity of organisms to regulate UV-B radiation stress has a certain threshold; when this threshold is exceeded, plant growth and development are negatively affected (Wu et al., 2023). Previous studies have shown that excessive or high-intensity UV-B radiation causes physiological and biochemical damage to plants (Rai and Agrawal, 2017). UV-B radiation leads to reduced photosynthetic efficiency, the inactivation of photosystem II, downregulation of photosynthesis-related gene expression, and altered carbon and nitrogen metabolism (Thakur et al., 2023). To defend against the negative effects of UV-B radiation stress, plants have evolved various adaptative and repair mechanisms, which can play crucial roles in balancing growth and stress responses. These adaptations manifest themselves in terms of changes in plant morphology, physiological characteristics (Yang et al., 2004, 2005), biochemical parameters (Kumari et al., 2010; Tripathi et al., 2011) and gene expression levels (Ekhtari et al., 2019). For example, in order to cope with UV-B stress, plants produce protective compounds that remodel the metabolism pathway to serve the energy demand under stress (Sun Y et al., 2022). Plants have developed several mechanisms to cope with UV-B-induced damage, including photoreactivation, nucleotide and base excision, and repair through recombination (Apoorva et al., 2021). Recent studies have underscored the significance of energy metabolic pathways, such as glycolysis, in plant stress resistance (Zhang et al., 2023). Evidence has demonstrated that critical genes associated with glycolysis are up-regulated in response to cold stress, facilitating energy generation to enhance plant resistance under environmental stress (Zhang et al., 2023). However, the response of energy metabolism to UV-B radiation is currently still unclear.
As sessile organisms, intertidal macroalgae passively adapt to changing conditions and are always exposed to UV-B radiation (Takshak and Agrawal, 2019; Xue et al., 2022). Neoporphyra haitanensis (Bangiales, Rhodophyta), which lives in the intertidal zone, is one of the most important economic algae in China (He et al., 2017). N. haitanensis has the highest annual production of all nori species and is widely consumed as a food in Asian countries (Yang et al., 2018). Significantly, N. haitanensis exhibits substantial stress tolerance which has attracted a lot of attention from researchers (Blouin et al., 2011). Routine tidal turning periodically exposes it to the air, and it inevitably experiences the drastic changes in environmental factors such as UV-B radiation, osmotic pressure and temperature (Cao et al., 2020). At low tide during the day, N. haitanensis is directly exposed to high levels of UV-B radiation. Especially in the context of global change, enhanced UV-B radiation may be potentially harmful to N. haitanensis. Additionally, with the large number of reports on the N. haitanensis genome and its response to intertidal adversity (Cao et al., 2020), this species has attracted considerable research interest as a genetic and physiological model for analyzing the stress resistance of intertidal red seaweeds.
In our previous work, we explored the changes in photosynthetic performance, key physiological processes, and metabolic substances in N. haitanensis under different UV-B intensities and preliminarily found that enhanced UV-B radiation affects key physiological and metabolic processes and causes damage to N. haitanensis (Fu et al., 2021; Xue et al., 2022). Here, we carried out further research to systematically analyze the physiological, metabolic and gene expression changes in N. haitanensis under UV-B radiation, especially during recovery from the perspective of energy metabolism. The aims of this study were to (1) evaluate changes in the photosynthetic physiology of N. haitanensis after UV-B radiation and during recovery; (2) characterize the DEGs in energy metabolism in N. haitanensis after UV-B radiation and during recovery using transcriptomics; and (3) analyze the response characteristics of key metabolic pathways associated with energy metabolism in N. haitanensis after UV-B radiation and during recovery using targeted metabolomics. This study is of great significance in revealing the adaptation strategies of intertidal macroalgae to UV-B radiation.
Thalli of N. haitanensis were collected in December 2021 from artificial seaweed farming in Qidong (121.66°E, 31.8°N), Jiangsu Province, China. After collection, the intact algae were washed thoroughly with natural seawater to remove sediment and other debris attached to the surface of the algae. The samples were spread flat and dried in the shade to a moisture content of 10-15% and stored at 20°C until culture to ensure that biological activity was retained. Thalli were precultured prior to the start of the experiment. Thalli were gently rinsed with sterilized seawater to prevent microbial contamination and then precultured in the same way as in previous experiments (Xue et al., 2022). The preculture conditions were as follows: Provasoli enrichment solution medium (PES) in aerated seawater at 20 ± 0.5°C for 72 h. The photosynthetically active radiation intensity was 50 μmol photons·m-2·s-1 (PAR), and the photoperiod was 12 L:12 D. The seawater was changed once a day.
After preculturing, similarly shaped healthy thalli were transferred to square plastic plates containing 300 ml of culture seawater. To prevent interference from UV-C radiation, UV-B lamps (Philips, TL 40 W/12 RS, Hamburg, Germany) wrapped in cellulose diacetate film were used. The radiation level was measured using a radiometer equipped with a UV-B sensor (UV-B 297, Beijing Normal University, China). The temperature and light conditions were identical to those used for preculture.
The UV-B radiation intensity was measured with a UV-B radiometer (Photoelectric Instrument Factory of Beijing Normal University, China) by adjusting the distance between UV-B lamps and the thalli. Based on our previous work (Fu et al., 2021; Xue et al., 2022), 0 W·m-2 UV-B was set as control and the treatment groups include UV-B radiation groups (0.5 and 1 W·m-2 UV-B) and recovery after UV-B radiation groups (deprivation of 0.5 and 1 W·m-2 UV-B radiation).
The thalli samples from different treatments were selected for photosynthetic physiological analyses with three replicates per group. Thalli were subjected to 0 W·m-2 UV-B radiation, 0.5 W·m-2 UV-B radiation, and 1 W·m-2 UV-B radiation for 4 h, followed by recovery after deprivation of the UV-B radiation, with a recovery of 24 h for continuous monitoring. Chlorophyll fluorescence measurements were conducted before UV-B radiation, during UV-B radiation and recovery using an Imaging-PAM (Walz, Germany). Three replicate samples were taken from each group. Samples were collected, allowed to adapt to the dark for 30 min on water in a square dish and transferred to Imaging-PAM. The manufacturer’s software, Imaging WinGigE, was used to implement the preprogrammed settings for actinic light duration, dark periods and saturation pulse. Minimal fluorescence (Fo) was measured under a 0.15 μmol photons·m-2·s-1 weak pulse of modulating light, and maximal fluorescence (Fm) was obtained after a saturating pulse of 0.6 s at 4000 μmol m-2·s-1. The maximal PSII quantum yield was calculated using the following equation: Fv/Fm = (Fm−Fo)/Fm, where Fm is the maximum fluorescence yield of PSII after a saturating light pulse and Fo is the baseline fluorescence of dark-adapted algae. In addition, the effective PSII quantum yield (Y(II)), the regulated nonphotochemical quantum yield (Y(NPQ)) and the nonregulated nonphotochemical quantum yield (Y(NO)) were determined; these three parameters are associated with photosynthetic activity. These parameters were calculated as follows: Y(II)=(Fm’-F)/Fm’, Y(NPQ)=1-Y(II)-1/(NPQ+1+qL(Fm/Fo-1)), and Y(NO)=1/(NPQ+1+qL(Fm/Fo-1)), where F represents the fluorescence yield, Fm’ is the maximum fluorescence yield, and qL is the coefficient of photochemical quenching. By analyzing these parameters, we were able to compare the precise photosynthetic performance. The corresponding photosynthetic parameters were obtained directly using Imaging WinGigE software, facilitating a detailed and accurate analysis.
Rapid light curves (RLCs) were used to measure the electron transfer rate (ETR) with increasing light intensity. Nine incremental steps of increasing levels of actinic light generated by the Imaging-PAM were used to irradiate the algae to obtain RLCs. Finally, the RLCs were refitted using the modified nonlinear function published by Platt et al. (1980). The relative maximum electron transport rate (rETRmax), the theoretical maximum light utilization coefficient (α) and the light saturation coefficient (Ik) were calculated by refitting the RLCs.
Thalli treated with 0 W·m-2 UV-B radiation (C), 0.5 and 1 W·m-2 UV-B radiation for 4 h (0.5U and 1U) and recovery for 18 h (0.5R and 1R) were selected for transcriptomics analysis. Total RNA was extracted from N. haitanensis using TRIzol reagent according to the manufacturer’s instructions. For transcriptomic analysis, three replicate samples were taken from each group. RNA degradation and contamination were monitored on 1% agarose gels. An RNA Nano 6000 assay kit from the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used to assess RNA integrity.
The clean reads were obtained by filtering the raw data, checking the sequencing error rate and checking the GC content distribution; then, the reads were aligned to the N. haitanensis reference genome (https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_008729055.1/). The reads were mapped to the reference genome using HISAT2 (http://www.ccb.jhu.edu/software/hisat/, 2.1.0). The reads per kilobase of exons per million fragments mapped (FPKM) values were used to express the gene expression values and the calculation of FPKM was performed using featureCounts (http://subread.sourceforge.net, 1.6.1). Analysis of differential expression was performed using the DE Seq package (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html, 1.22.1) (FDR<0.05 and |log2Fold Change|≥1). Significantly enriched biological pathways were identified with a P value≤ 0.05 via KEGG analyses of DEGs (https://www.genome.jp/kegg, version 2017.08).
Thalli treated with 0 W·m-2 UV-B radiation (C), 0.5 and 1 W·m-2 UV-B radiation for 4 h (0.5U and 1U) and recovery for 18 h (0.5R and 1R) were selected for metabolomics analysis. To further measure the changes in energy metabolites, we collected three replicate samples from each treatment group for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, and 50 mg ( ± 2.5 mg) of each replicate was used to extract metabolites. The samples were mixed with alcohol or water, and after 2 centrifugations, 200 μL of the supernatant was transferred to a protein precipitation plate for LC–MS/MS analysis.
The metabolites in the samples were analyzed quantitatively and qualitatively using the self-built MetWare database (MWDB) (MetWare Company, Wuhan, China) and multiple reaction monitoring (MRM). The differentially abundant metabolites were identified according to the criteria of VIP > 1 and P value < 0.05.
The line graphs and column charts were generated with GraphPad Prism 9. The RLCs were refitted using the modified nonlinear function published by Platt et al. (1980) with GraphPad Prism 9. The bubble diagrams and principal component analysis (PCA) plots used in omics analysis were generated using the R package ggplot2 (Version 4.3.1), and the heatmaps were produced by the R package pheatmaps (Version 4.3.1).
To investigate the effects of UV-B radiation on the photodamage and the ability of the photosynthetic system in N. haitanensis to recover, the changes in the chlorophyll fluorescence parameters of N. haitanensis were determined after UV-B radiation and recovery (Figure 1). Compared with those in the control group, the Fv/Fm values in the UV-B radiation-treated groups decreased continuously with increasing radiation time—by 25.80% after 4 h of 0.5 W·m-2 UV-B radiation treatment and by 47.17% after 4 h of 1 W·m-2 UV-B radiation treatment. After 24 h of recovery, all the thalli of N. haitanensis in the radiation-treated groups had recovered to different extents, with the 0.5 W·m-2 UV-B radiation treatment group recovering 77.43% of the initial level and the 1 W·m-2 UV-B radiation treatment group recovering 70.95% of the initial level (Figure 1A). The change in Y(II) was similar to that in Fv/Fm; with increasing UV-B radiation time, there was a different degree of decline in each radiation treatment group, and the Y(II) value in each radiation treatment group increased significantly after recovery (Figure 1B). Notably, the Y(II) values in the 0.5 W·m-2 UV-B radiation treatment group and the 1 W·m-2 UV-B radiation treatment group peaked after 18 h of recovery to 85.82% and 67.40%, respectively, of the initial values. After UV-B radiation or during recovery, there was no significant change in Y(NO), and Y(NPQ) increased first and then decreased during recovery (Figures 1C, D).
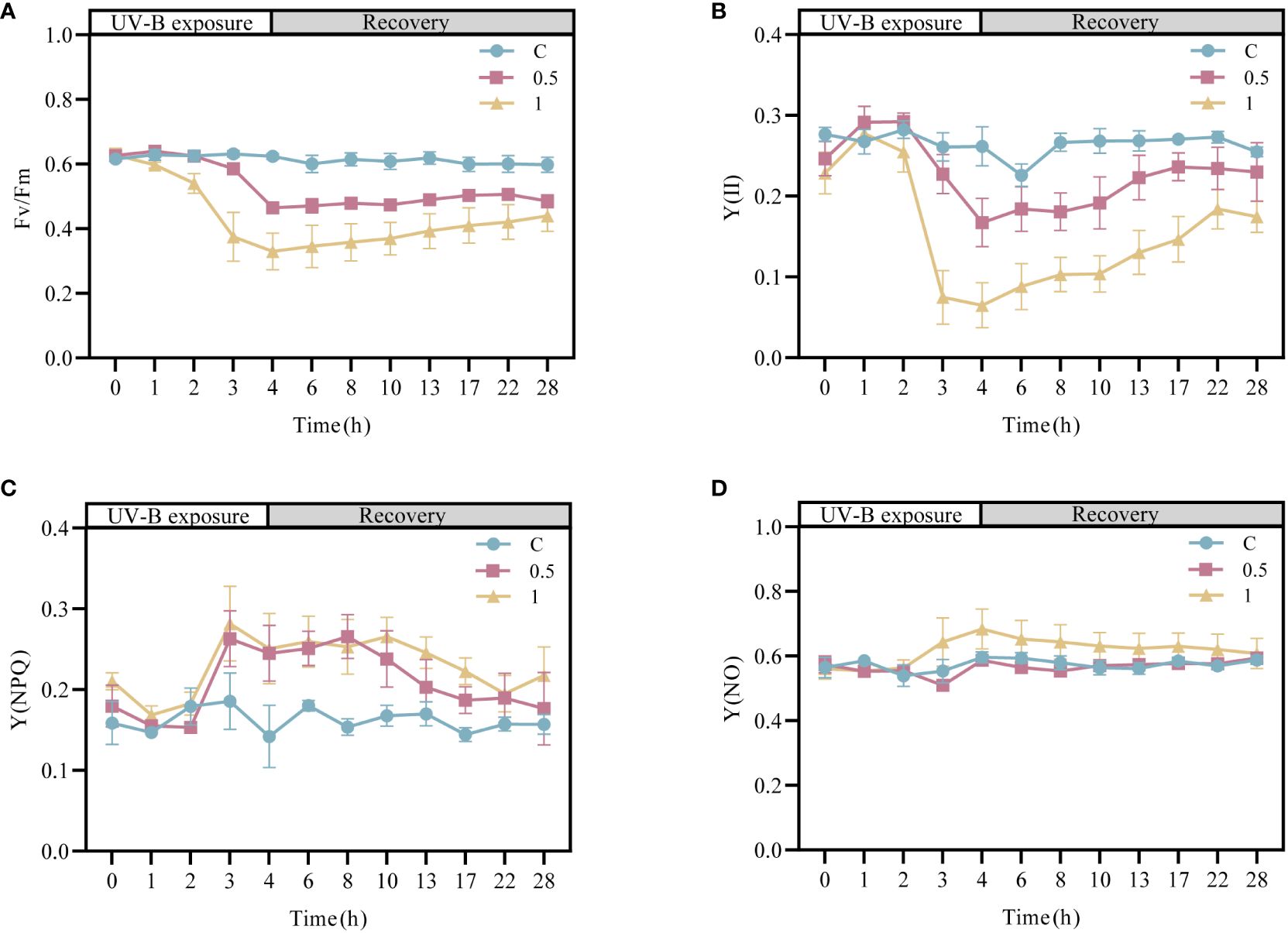
Figure 1 Changes in the photosynthetic activity of Neoporphyra haitanensis after UV-B radiation and during recovery. (A) Variations in the maximal PSII quantum yield (Fv/Fm). (B) Variations in the effective PSII quantum yield (Y(II)). (C) Variations in the quantum yield of regulated energy dissipation (Y(NPQ)). (D) Variations in the quantum yield of nonregulated energy dissipation (Y(NO)).
The RLCs were measured to further investigate the electron transport rate of PSII under UV-B radiation (Figure 2). Three parameters related to electron transfer were evaluated (Supplementary Table 1). Compared with that in the control group, α decreased by 53.33% after 4 h of 1 W·m-2 UV-B radiation, and after 18 h of recovery, α recovered to 77.70% of the initial value. Under both 0.5 W·m-2 and 1 W·m-2UV-B irradiation, both the rETRmax and IK decreased significantly; however, the rETRmax and IK recovered to 82.30% and 89.53% of the initial level after 18 h of recovery in the 0.5U group, and 33.06% and 42.55% of the initial level in the 1U group, respectively. In summary, the decrease in the electron transfer rate of N. haitanensis was more obvious in the 1 W·m-2 UV-B radiation group than in the control group, but after 18 h of recovery, there were different degrees of recovery in each group, and the recovery was more obvious in the 0.5 W·m-2 UV-B radiation group.
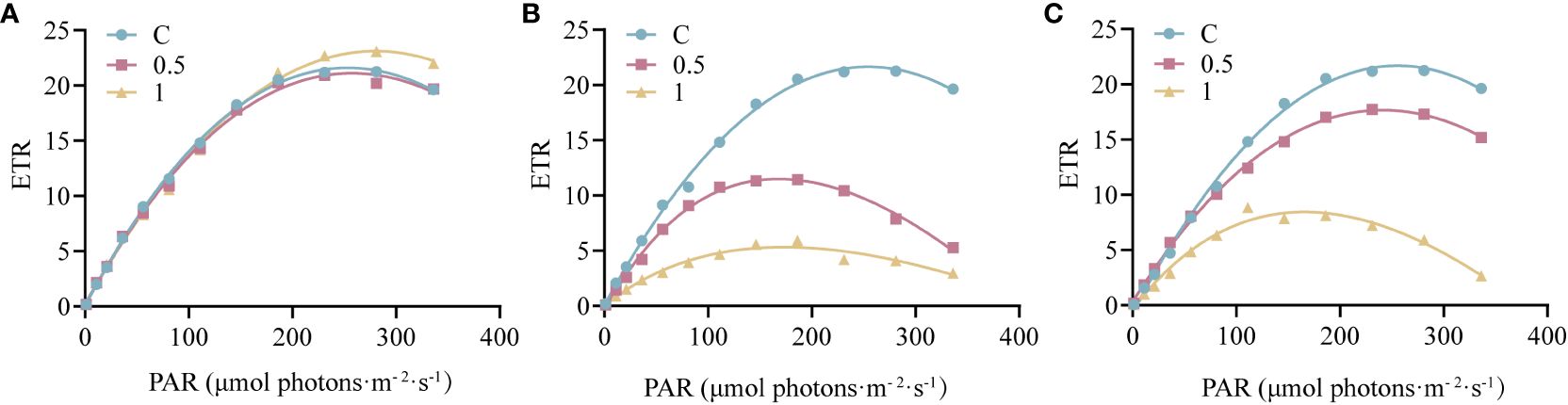
Figure 2 Changes in the rapid light curves (RLCs) of Neoporphyra haitanensis after UV-B radiation and during recovery. (A) RLCs without UV-B radiation. (B) RLCs after 4 h of UV-B radiation. (C) RLCs after 4 h of UV-B radiation and 18 h of recovery.
To explore the mechanism underlying the recovery of N. haitanensis after UV-B radiation, samples of N. haitanensis treated by different conditions were collected for transcriptome analysis. After removing the low-quality reads, a total of 112.35 G of clean reads were obtained from 15 samples. The percentages of Q30 and GC were 87.98%–90.17% and 67.54–69.62%, respectively, indicating that the quality of the transcriptome sequencing data was high (Supplementary Table 2). PCA revealed significant differences before and after UV-B radiation and recovery, with good repeatability within groups (Figure 3A). Interestingly, at both 0.5U and 1U, there were more genes with upregulated expression than genes with downregulated expression in N. haitanensis after UV-B radiation compared to controls, and there were slightly more genes with downregulated expression than genes with upregulated expression during recovery (Figure 3B). These results indicated that there were significant changes in gene expression in N. haitanensis after UV-B radiation and during recovery.
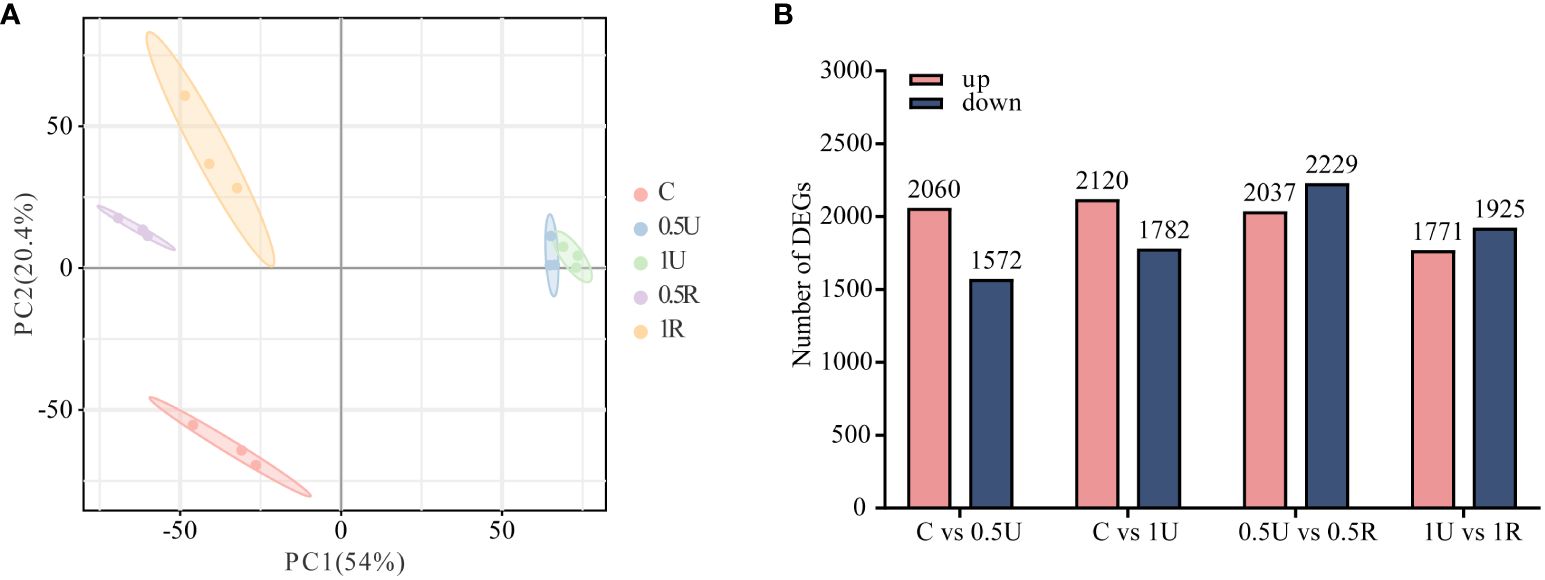
Figure 3 Transcriptomic analysis of Neoporphyra haitanensis after UV-B radiation and during recovery. (A) Principal component analysis (PCA) plot of transcriptome data obtained from samples with different levels of UV-B radiation and recovery treatments. (B) Number of up- and down-regulated genes under different treatments.
To verify the biological functions of the DEGs in N. haitanensis after UV-B radiation and during recovery, KEGG enrichment analysis of the DEGs was performed. The KEGG results showed that the DEGs were involved in DNA replication and repair, biosynthesis of secondary metabolites, ascorbate and aldarate metabolism, as well as energy-related metabolic pathways, including carbon metabolism, amino acid biosynthesis, photosynthesis and other related pathways after UV-B radiation and during recovery. In particular, the glycolytic pathway, the pentose phosphate pathway, and the TCA cycle, which are closely related to carbon metabolism, were enriched (Figure 4).
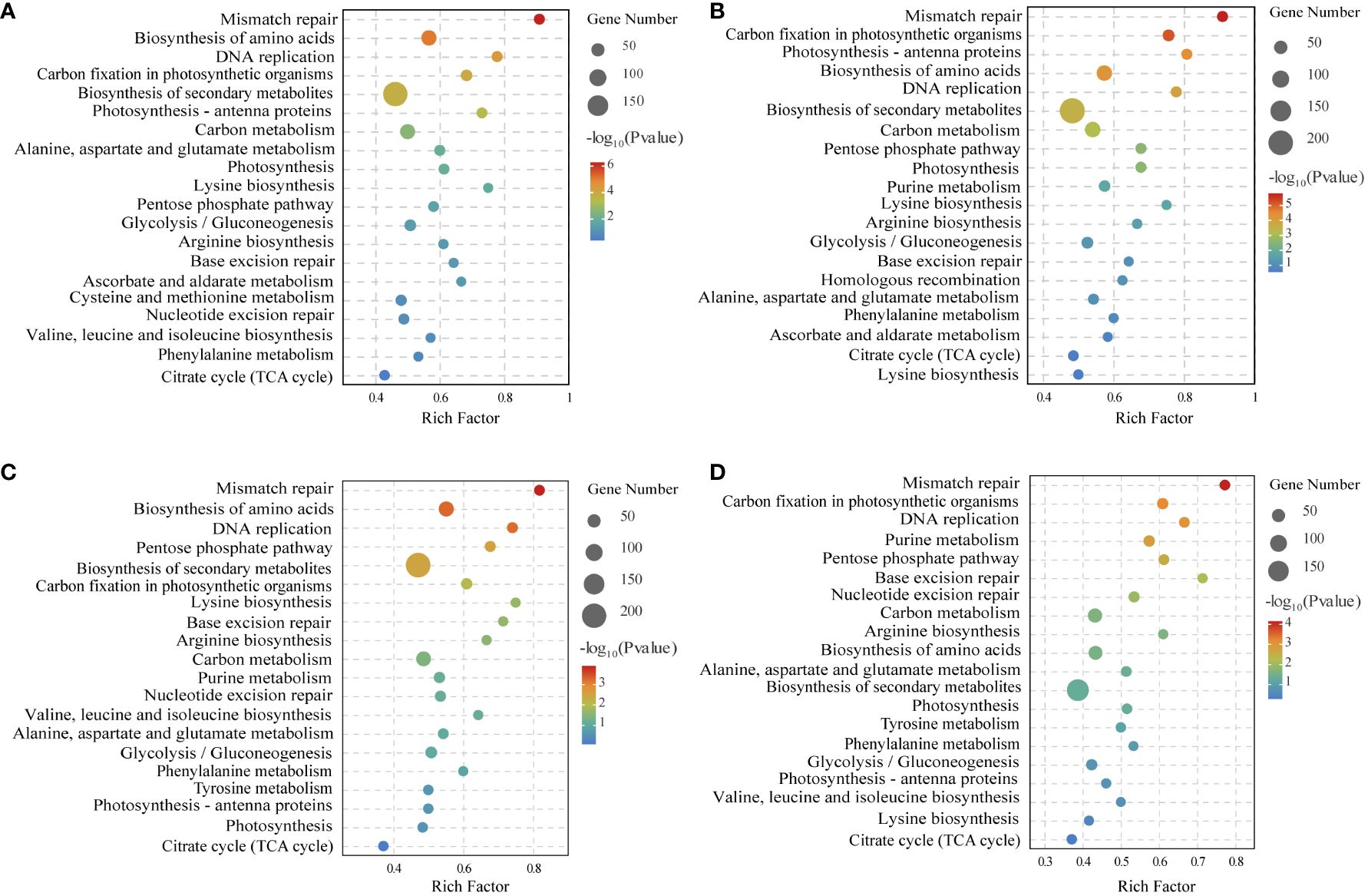
Figure 4 KEGG enrichment analysis of differentially expressed genes of comparing (A) Control (C) vs 0.5 W·m-2 UV-B radiation (0.5U), (B) Control (C) vs 1 W·m-2 UV-B radiation (1U), (C) 0.5 W·m-2 UV-B radiation (0.5U) vs Recovery after 0.5 W·m-2 UV-B radiation (0.5R), and (D) 1 W·m-2 UV-B radiation (1U) vs Recovery after 1 W·m-2 UV-B radiation (1R).
To further reveal the dynamic changes and recovery characteristics of N. haitanensis after UV-B radiation, we focused on energy metabolism and investigated metabolite changes. A total of 63 energy metabolites were identified in 15 samples (Supplementary Table 3). We observed a distinct difference in the distribution of N. haitanensis according to the PCA plot after UV-B radiation and during recovery, indicating significant variation in the metabolome of N. haitanensis given different treatments (Figure 5A). Furthermore, the number of metabolites with upregulated expression exceeded the number of metabolites with downregulated expression, especially in the recovery group (Figure 5B). Additionally, the heatmap reveals a substantial increase in the level of most substances during recovery (Figure 5C).
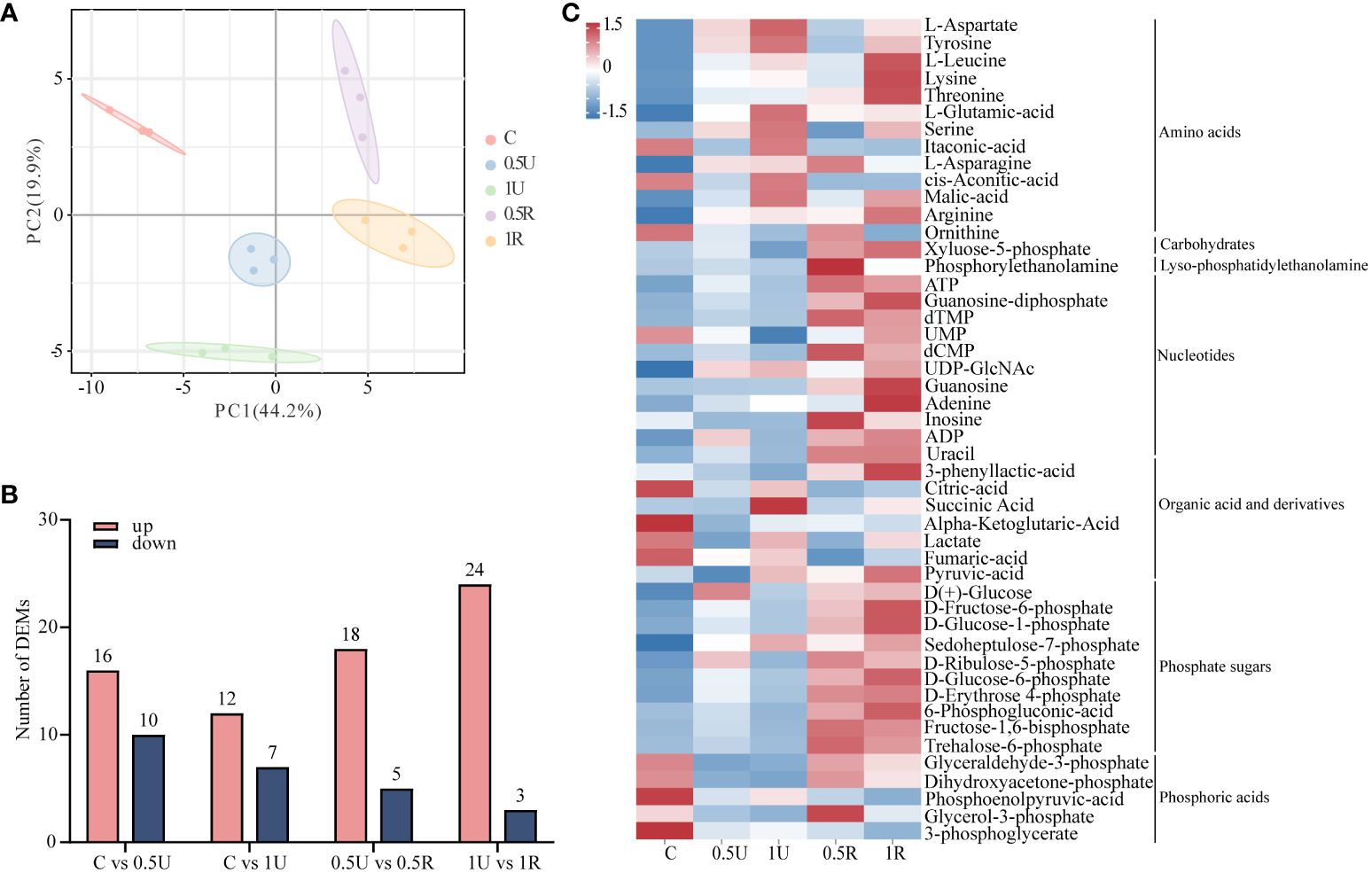
Figure 5 Metabolomic analysis of Neoporphyra haitanensis after UV-B radiation and during recovery. (A) Principal component analysis (PCA) plot of metabolome data obtained from samples with different levels of UV-B radiation and recovery treatments. (B) Number of up- and down-regulated metabolites under different treatments. (C) Classification of differentially expressed metabolites.
The levels of most genes and metabolites related to photosynthesis decreased and then increased after UV-B radiation and during recovery. The expression of genes encoding the light harvesting complex (NhLHCs) was down-regulated after UV-B radiation and remained consistently low during recovery. (Figure 6). Moreover, the expression of genes encoding the photoreaction center PSII, as well as other key components of the electron transport chain, such as Cyt b6f, Fd, FNR and ATP synthase, and the expression of genes encoding the downstream key enzymes of the Calvin cycle, such as fructose-1,6-bisphosphatase (FBP), phosphoglycerol kinase (PGK), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was also downregulated. Notably, the expression of genes encoding the photoreaction centers PSII and Cyt b6f, ATP synthase, NhFBP, NhPGK, and NhGAPDH was upregulated during recovery (Figure 6). The metabolomic results showed that the levels of glyceraldehyde-3-phosphate and ribulose-5-phosphate within the Calvin cycle also increased during recovery (Figure 6).
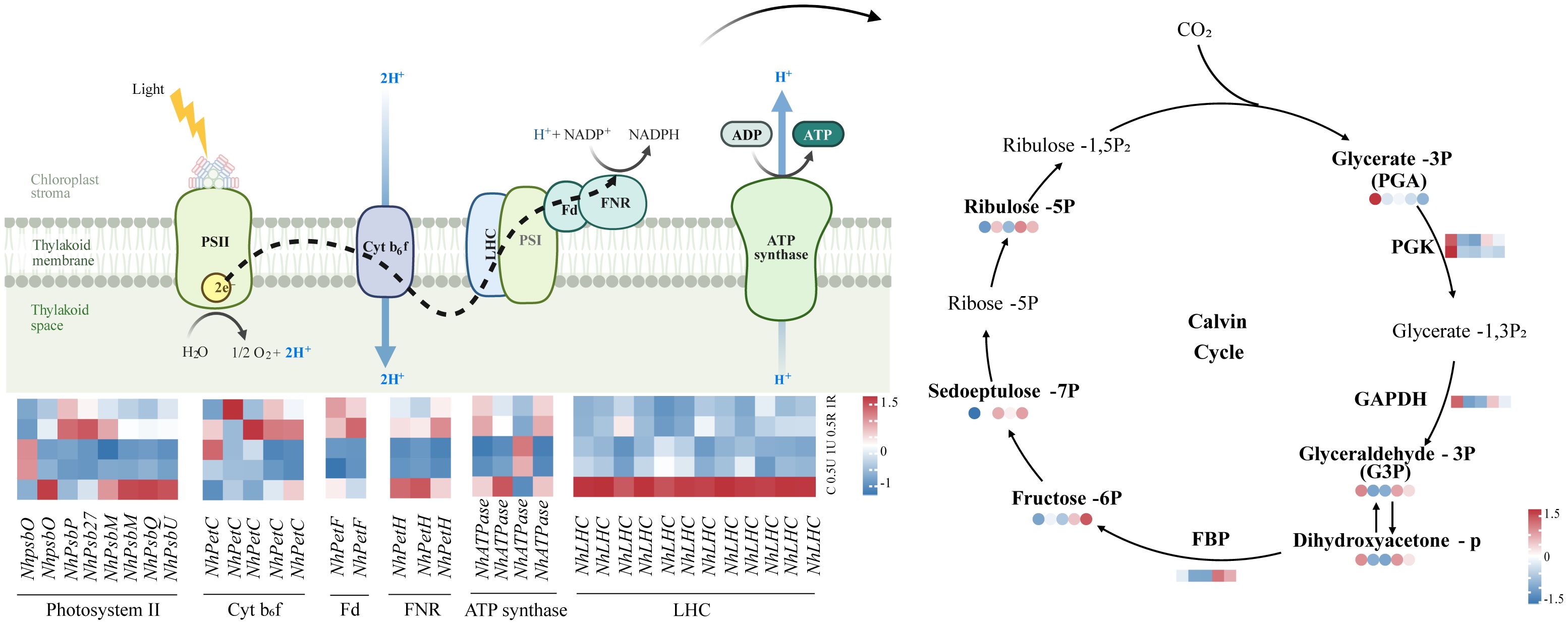
Figure 6 Differentially expressed genes and metabolites related to photosynthesis in Neoporphyra haitanensis after UV-B radiation and during recovery. The data show transcriptome and metabolome profiles from the Control (C), 0.5 W·m-2 UV-B radiation (0.5U), 1 W·m-2 UV-B radiation (1U), Recovery after 0.5 W·m-2 UV-B radiation (0.5R), Recovery after 1 W·m-2 UV-B radiation (1R) groups (from left to right). The square heatmaps show gene expression levels (normalized FPKM) (n = 5), and the circular heatmaps show the normalized peak area of metabolites (n = 5). PGK, phosphoglycerate kinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; FBP, fructose-1,6-bisphosphatase.
The expression of most genes and metabolites related to glycolysis and the pentose phosphate pathway increased during recovery in N. haitanensis (Figure 7). The expression of NhALDO, NhGAPDH and NhPGK, which are involved in the expression of fructose-bisphosphate aldolase, glyceraldehyde 3-phosphate dehydrogenase and phosphoglycerate kinase, respectively, was significantly downregulated after UV-B radiation and then upregulated during recovery. Our metabolomic results showed that the levels of 3-phosphoglyceraldehyde decreased after UV-B radiation treatment and increased again after recovery. Similarly, the expression of genes encoding phosphoglucomutase (NhPGM), phosphoglucose isomerase (NhGPI), and pyruvate kinase (NhPK) was upregulated in the recovered group, and these changes were accompanied by elevated levels of glucose-6-phosphate, fructose-6-phosphate, and pyruvate.
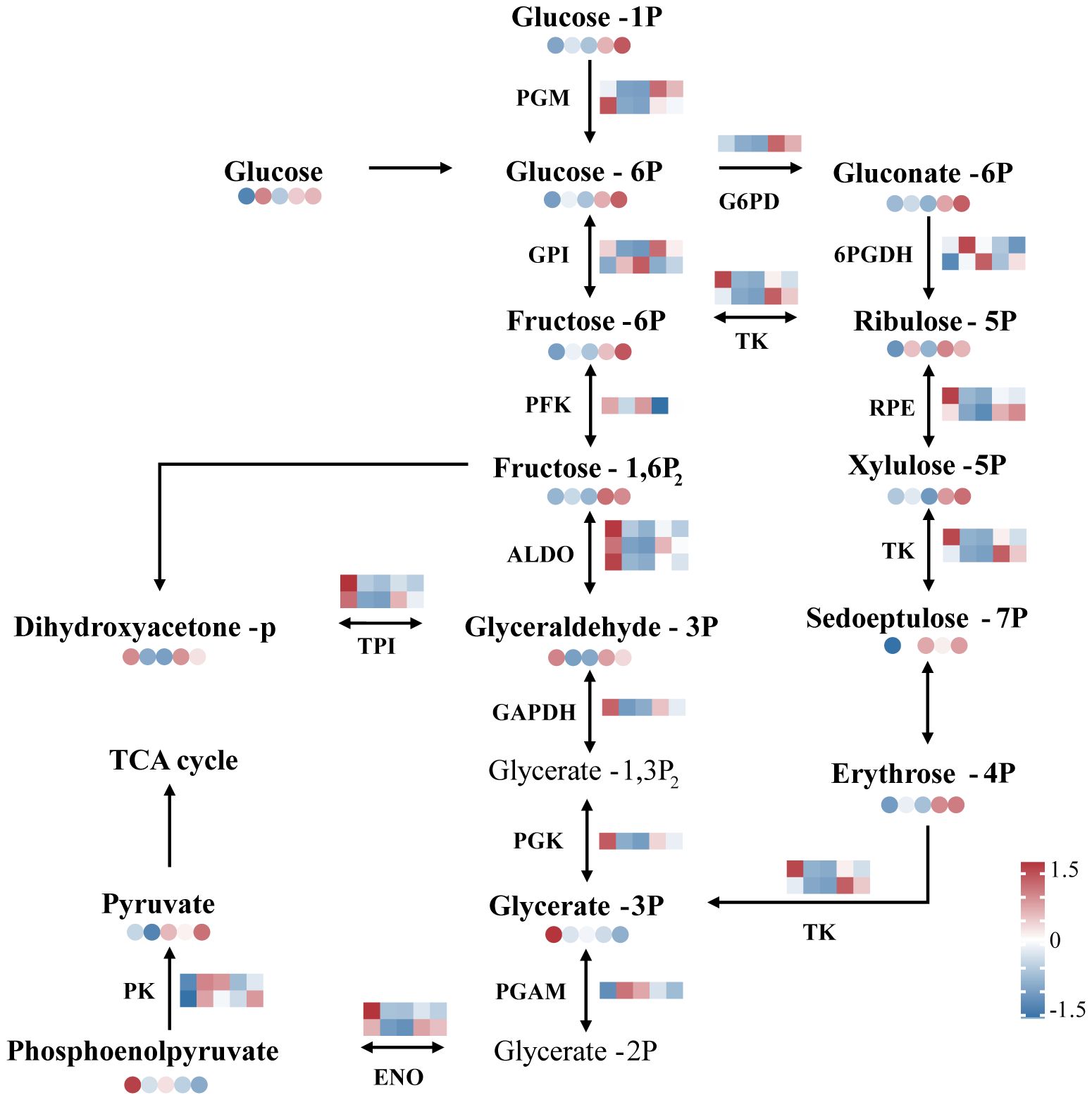
Figure 7 Overview of glycolysis and the pentose phosphate pathway of Neoporphyra haitanensis after UV-B radiation and during recovery. The data show transcriptome and metabolome profiles from the Control (C), 0.5 W·m-2 UV-B radiation (0.5U), 1 W·m-2 UV-B radiation (1U), Recovery after 0.5 W·m-2 UV-B radiation (0.5R), Recovery after 1 W·m-2 UV-B radiation (1R) groups (from left to right). The square heatmaps show gene expression levels (normalized FPKM) (n = 5), and the circular heatmaps show the normalized peak area of metabolites (n = 5). PGM, phosphoglucomutase; GPI, phosphoglucose isomerase; PFK, phosphofructokinase; ALDO, fructose-bisphosphate aldolase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; PGK, phosphoglycerate kinase; PGAM, phosphoglycerate mutase; G6PD, glucose-6-phosphate dehydrogenase; 6PGDH, 6-phosphogluconate dehydrogenase; RPE, ribulose-phosphate 3 epimerase; TK, transketolase; ENO, enolase; TPI, triosephosphate isomerase; PK, pyruvate kinase.
In the pentose phosphate pathway, the expression of genes encoding glucose-6-phosphate dehydrogenase (NhG6PD), and genes encoding 6-phosphogluconate dehydrogenase (Nh6PGDH) was significantly higher in the recovery group than in the control group, and the expression of glucose 6-phosphate, ribulose-5-phosphate, and erythrose-4-phosphate was also increased in the recovery group (Figure 7).
The transcriptomic analysis showed that the expression of genes encoding citrate synthase (NhCS), succinate dehydrogenase (NhSDH), isocitrate dehydrogenase (NhIDH), fumarate hydratase (NhFUM), and malate dehydrogenase (NhMDH) was upregulated during recovery. In contrast, the expression of genes encoding aconitase (NhACO) was downregulated. The metabolomic results showed that oxaloacetate, succinate, and malate levels were elevated during recovery. Especially, there was a sustained increase in ATP in N. haitanensis after UV-B radiation and recovery (Figure 8). In addition, a total of 9 differentially expressed amino acids were detected, including 2 acidic amino acids (Asp and Glu), 2 basic amino acids (Lys and Arg), 3 neutral amino acids (Thr, Ser and Asn), 1 aromatic family amino acid (Tyr) and 1 aliphatic amino acid (Leu). The levels of these amino acids were increased after UV-B radiation and during recovery compared with that in the controls (Figure 8).
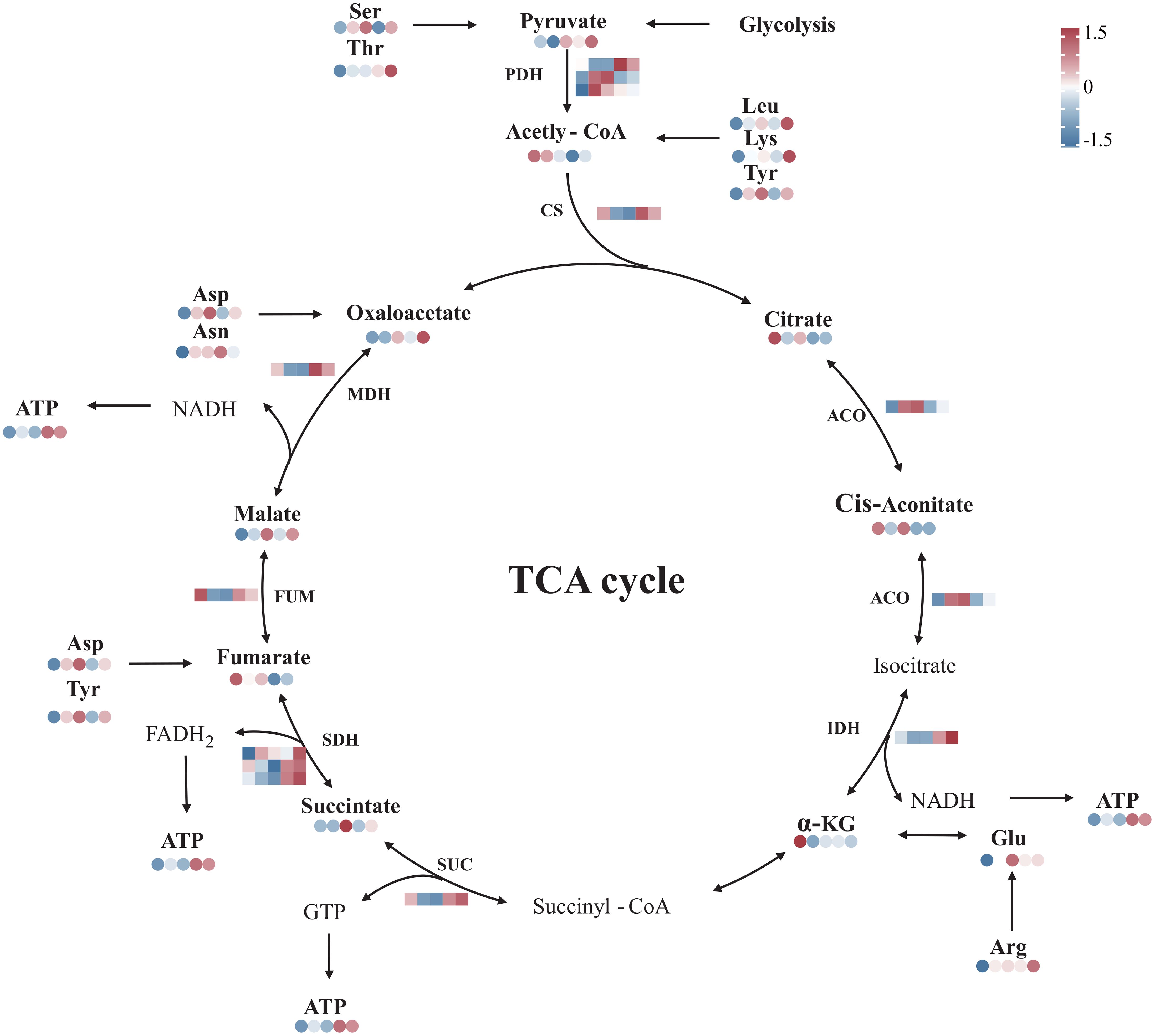
Figure 8 Overview of the TCA cycle and amino acid biosynthesis in Neoporphyra haitanensis after UV-B radiation and during recovery. The data show transcriptome and metabolome profiles from the Control (C), 0.5 W·m-2 UV-B radiation (0.5U), 1 W·m-2 UV-B radiation (1U), Recovery after 0.5 W·m-2 UV-B radiation (0.5R), Recovery after 1 W·m-2 UV-B radiation (1R) groups (from left to right). The square heatmaps show gene expression levels (normalized FPKM) (n = 5), and the circular heatmaps show the normalized peak area of metabolites (n = 5). PDH, dehydrogenase; CS, citrate synthase; ACO, aconitase; IDH, isocitrate dehydrogenase; SUC, succinyl-CoA synthetase; SDH, succinate dehydrogenase; FUM, fumarase; MDH, malate dehydrogenase.
An increase in UV-B radiation can have a detrimental effect on the rate of photosynthesis in plants (Yin et al., 2017). UV-B radiation can lead to the reversible inactivation of the light-harvesting pigment protein complex PSII, which reduces the rate of photosynthesis (Yin et al., 2017). In our study, the expression of genes encoding the light-harvesting complex (NhLHCs) decreased after UV-B radiation, and simultaneously, both the Fv/Fm ratio and the Y(II) level significantly decreased. These findings indicate that UV-B radiation has a harmful impact on the photosynthetic performance of N. haitanensis. During recovery, the expression levels of NhLHCs did not increase, reflecting the particularity of LHC in red algae. Unlike higher plants or other algae (such as green algae and brown algae), light energy capture in red algae mainly depends on phycobilisomes (Voerman et al.,2022; Li X et al., 2023). The role of LHC in red algae is far less significant than that of phycobilisomes and is not directly involved in light energy capture and electron transfer (You et al., 2023). Therefore, the persistently low expression of NhLHCs during recovery suggests that UV-B radiation can reduce the expression of NhLHCs, and this inhibition still exists after a period of recovery.
Considering the overall deleterious effect of UV-B on photosynthesis, photosynthetic performance should be greatly enhanced in the absence of UV-B during recovery (Tilbrook et al., 2016). This capacity for recovery can be explained by “dynamic photoinhibition”, which differs from photodamage and is a photoprotective mechanism that allows for the recovery of photosynthetic performance once excess radiant energy is removed (Simioni et al., 2014). In this study, the effect of UV-B on N. haitanensis presumably led to dynamic photoinhibition. Once UV-B treatment was discontinued, the photosynthetic ability of N. haitanensis was restored, as indicated by changes in heat dissipation and electron transfer. First, Y(NPQ) and Y(II) increased after 18 h of recovery and nearly reached their original levels, while Y(NO) remained relatively stable. UV-B did not cause destructive damage, and the temporary decrease in photosynthetic capacity may be a protective mechanism of N. haitanensis. This mechanism has also been found in the red alga Gelidium floridanum (Simioni et al., 2014). Second, PsbU and PsbQ are located upstream of the electron transport chain and are responsible for protecting electron donors (Zhen et al., 2021). A lack of PsbU affects electron transfer in the PSII complex (Shinde et al., 2022). The transcriptomics analysis in this study indicated the upregulated expression of key genes (NhPsbM, NhPsbU and NhPsbQ) associated with electron transport; this result provided further evidence of restored photosynthetic performance in N. haitanensis. Photosynthetic carbon fixation in algae is performed via the Calvin cycle (Kroth et al., 2008). The photosynthetic carbon fixation pathway can be promoted by an increase in ribulose content (Shi et al., 2022). In our study, an increase in ribulose 5-phosphate content during recovery indicated a rebound in the carbon sequestration capacity of N. haitanensis. Furthermore, the upregulated expression of carbon fixation-related genes, such as NhFBP, NhPGK, and NhGAPDH, promoted the production of metabolites in the carbon fixation pathway. During recovery, the enhancement of photosynthetic performance results from upstream photosynthetic capture and electron transfer, along with the accumulation of metabolites involved in downstream carbon reactions. This synergistic effect illustrates that damage to the photosynthetic system in N. haitanensis caused by UV-B radiation was reversible.
Glycolysis occurs in response to abiotic stress mainly by affecting the energy supply of plants (Zeng et al., 2019). Glycolysis can increase the production of energy by degrading carbohydrates, thereby improving the response of plants to their environment under abiotic stress conditions (Zeng et al., 2019). Under hypoxic conditions, glycolysis is activated in Sesbania cannabina to produce ATP, thereby providing energy for coping with adverse stress (Ren et al., 2017). In our study, the transcriptome and metabolome data showed that the expression of genes and metabolites related to glycolysis were significantly upregulated in the recovery group (Figure 7), suggesting that glycolysis may be activated to promote energy production during recovery.
Studies have shown that glucose is utilized as a substrate for glycolysis and is beneficial for improving plant metabolism and stress resistance (Du et al., 2022). In our study, glycolysis was active during recovery, yet the amount of glucose increased, which may be due to the increase in photosynthesis during recovery. Previous studies have shown that enhanced photosynthesis promotes the production of soluble sugars (Li Y et al., 2023). Therefore, we speculate that during recovery, the recovery of photosynthetic performance can further promote glucose production, thus providing substrates for glycolysis.
The accumulation of pyruvate in the glycolytic metabolic pathway can enhance the survival of plants under stress conditions (Tian et al., 2022). Pyruvate, the end product of glycolysis, enters the TCA cycle to undergo oxidative phosphorylation to produce ATP (Beebe et al., 2020). Pyruvate kinase (PK) and GAPDH, the key enzymes of the glycolysis pathway, play vital roles in pyruvate production (Wong et al., 2018). In particular, PK catalyzes the final and irreversible step of glycolysis to generate pyruvate and ATP (Zhong et al., 2019). In our study, the upregulation of NhPK and NhGAPDH expression was accompanied by pyruvate accumulation during recovery. We speculated that the accumulated pyruvate enters the TCA cycle to undergo oxidative phosphorylation for ATP production.
The pentose phosphate pathway (PPP) is a complementary source of cellular energy that can directly oxidize sugars and complement glycolysis (Zhu et al., 2023). Glucose-6-phosphate dehydrogenase (G6PD) is the first rate-limiting enzyme of the PPP, and the activity of this enzyme reflects the importance of the pentose phosphate pathway (Sarkar et al., 2011). For example, upregulated expression of G6PD suggests activation of the pentose phosphate pathway in wheat under high salt stress (Chang et al., 2020). In this study, the expression of NhG6PD was upregulated during recovery. 6-Phosphogluconate dehydrogenase catalyzes the conversion of 6-phosphogluconic acid to 5-phosphate ribulose and is another key enzyme in the pentose phosphate pathway (Zhao et al., 2019). In this study, the expression of Nh6PGDH was upregulated, and the content of ribulose 5-phosphate increased during recovery. Similar results were found as a result of activation of the pentose phosphate pathway during wheat seed germination (Lv et al., 2016). With the upregulated expression of genes that control key enzymes in the PPP and the increase in the level of key substances, we speculate that the PPP of N. haitanensis was activated during recovery, possibly to replenish substrates for glycolysis.
The TCA cycle is a key mechanism of ATP production in plants and plays an important role in resisting stress (Yang et al., 2020). Citrate synthetase (CS) is the first enzyme of the TCA cycle and catalyzes the reaction between acetyl coenzyme A and oxaloacetate to produce citric acid (Chaudhry et al., 2018). Changes in the activity of citrate synthase can directly affect the efficiency of the TCA cycle (Wang et al., 2018). In this study, NhCS expression was upregulated, suggesting that the efficiency of the TCA cycle was also elevated during N. haitanensis recovery. In addition, MDH and IDH can produce NADH, which can enter the oxidative respiratory chain and eventually generate ATP in plants (Qian et al., 2022). In this study, NhMDH and NhIDH expression was upregulated accompanied by a large accumulation of ATP, suggesting that ATP may have been generated for energy supply during recovery. Additionally, increased oxaloacetate levels likely stimulate the TCA cycle (Sass et al., 2019). An increase in the oxaloacetate level caused by the upregulation of NhMDH expression was also found in N. haitanensis. In addition, the expression of genes encoding succinate dehydrogenase (NhSDH) and fumarate hydratase (NhFUM) was upregulated during recovery. We speculated that the TCA cycle in N. haitanensis was activated during recovery because of the increase in key substances and the upregulated expression of key enzyme-encoding genes. Activation of the TCA cycle provided ATP for N. haitanensis to repair UV-B damage. It is noteworthy that NhACO downregulated during recovery, contrasting with the predominant trend observed in most genes. Despite this, we maintained that the TCA cycle was activated during recovery. Similar results were found in the study of the effects of UV-B radiation on Isochrysis galbana, in which genes encoding the majority of essential enzymes in the TCA cycle were predominantly upregulated. However, the gene encoding fumarase was an exception, showing no upregulation (Cao et al., 2019). This prevalent pattern suggested that the complex regulation of metabolic pathways in response to UV-B stress.
Amino acid levels are strongly correlated with the accumulation of intermediates in the TCA cycle (de la Torre et al., 2014). The decomposition of amino acids can supplement intermediates in the TCA cycle, and aspartate and asparagine can be utilized to supplement oxaloacetate (Liu et al., 2023). In our study, aspartate and asparagine levels increased during recovery. The increased aspartate and asparagine may supplement oxaloacetate. We speculated that enhanced amino acid metabolism served as a source of intermediates for the TCA cycle. Correspondingly, studies have shown that under high temperature conditions, Arabidopsis thaliana exhibited a rapid increase in amino acid levels that complement oxaloacetate and pyruvate (Kaplan et al., 2004), with concurrent upregulation of enzyme genes involved in these pathways, such as aspartate aminotransferase. This indicated a causal relationship between the variations in amino acid content and TCA cycle intermediate levels. Furthermore, the addition of certain amino acids (aspartate, asparagine, threonine, and serine) has been shown to lead to significant accumulations of pyruvate and oxaloacetate, demonstrating the role of amino acids in supplementing the TCA cycle (Zampieri et al., 2019). In addition to providing intermediates for the TCA cycle, amino acids play an important role in protecting plants against different abiotic stresses (Zhang et al., 2018). In previous studies, stress-associated defensive metabolites, including proline, leucine, isoleucine, glutamic acid and serine, were found to accumulate in plants exposed to high levels of UV-B radiation (Zhang et al., 2018; Sun Q et al., 2022). In this study, we found that UV-B radiation and recovery caused significant changes in the levels of aspartate, tyrosine, leucine, threonine, glutamic acid and serine in N. haitanensis. The levels of most amino acids increased significantly after UV-B radiation treatment and recovery, which indicated that there was a tendency to accumulate amino acids after UV-B radiation and during recovery; this result also indicated that amino acid metabolism had an important role in the response to UV-B radiation, as well as during recovery in N. haitanensis. Moreover, Lys may be involved in stress response in plants, and the accumulation of arginine is often observed in plants subjected to various types of environmental stress (Keller et al., 2018; Zhang et al., 2018). The levels of Lys and arginine increased significantly after UV-B radiation and remained high during recovery. These findings indicate that the accumulation of Lys and Arg in N. haitanensis after UV-B treatment may be associated with UV-B responses. Therefore, the accumulation of specific amino acids produced by amino acid metabolism is related to the increased UV-B radiation tolerance of N. haitanensis, as well as to the recovery process.
We explored changes in the photosynthetic physiology, gene expression and metabolite accumulation of N. haitanensis after UV-B radiation and during recovery from the perspective of energy metabolism. The results showed that UV-B radiation decreased the photosynthetic performance of N. haitanensis. Recovery experiments revealed that the synergistic effect of multiple biological processes could help N. haitanensis to repair the damage caused by UV-B radiation. The increased photosynthetic performance promoted glucose production to provide substrates for glycolysis. The upregulated genes and metabolites suggested that glycolysis was subsequently activated, facilitating energy production. Meanwhile, the TCA cycle responded positively by providing ATP for N. haitanensis to repair UV-B damage. Additionally, enhanced amino acid metabolism served as a source of intermediates for the TCA cycle.
The RNA‐Seq raw sequence data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) repository, accession number PRJNA1066830.
XW: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft. YZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Supervision, Validation, Writing – review & editing. SX: Formal analysis, Software, Validation, Writing – review & editing. SS: Formal analysis, Software, Validation, Writing – review & editing. JX: Formal analysis, Software, Validation, Writing – review & editing. LT: Writing – review & editing, Formal analysis, Software, Validation. JC: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Writing – review & editing. XT: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China (42176154, 42006144), and National Key R & D Program of China (2019YFD0901204).
The authors sincerely acknowledge Professor Yunxiang Mao from Ocean university of China for providing the genome annotation files of N. haitanensis. We also thank Zhongming Yin of Qidong Heteng Food Co., Ltd. providing experimental materials.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1372252/full#supplementary-material
Apoorva J. D., Pandey-Rai S., Agrawal S. B. (2021). Untangling the UV-B radiation-induced transcriptional network regulating plant morphogenesis and secondary metabolite production. Environ. Exp. Bot. 192, 104655. doi: 10.1016/j.envexpbot.2021.104655
Beebe K., Robins M. M., Hernandez E. J., Lam G., Horner M. A., Thummel C. S. (2020). Drosophila estrogen-related receptor directs a transcriptional switch that supports adult glycolysis and lipogenesis. Genes Dev. 34, 701–714. doi: 10.1101/gad.335281.119
Blouin N. A., Brodie J. A., Grossman A. C., Xu P., Brawley S. H. (2011). Porphyra: a marine crop shaped by stress. Trends Plant Sci. 16, 29–37. doi: 10.1016/j.tplants.2010.10.004
Cao J.-Y., Kong Z.-Y., Ye M.-W., Zhang Y.-F., Xu J.-L., Zhou C.-X., et al. (2019). Metabolomic and transcriptomic analyses reveal the effects of ultraviolet radiation deprivation on Isochrysis galbana at high temperature. Algal. Res. 38, 101424. doi: 10.1016/j.algal.2019.101424
Cao M., Xu K., Yu X., Bi G., Liu Y., Kong F., et al. (2020). A chromosome-level genome assembly of Pyropia haitanensis (Bangiales, Rhodophyta). Mol. Ecol. Resour. 20, 216–227. doi: 10.1111/1755-0998.13102
Chang B., Ma K., Lu Z., Lu J., Cui J., Wang L., et al. (2020). Physiological, transcriptomic, and metabolic responses of ginkgo biloba L. to drought, salt, and heat stresses. Biomolecules 10, 1635. doi: 10.3390/biom10121635
Chaudhry R., Kokkayil P., Ghosh A., Bahadur T., Kant K., Sagar T., et al. (2018). Bartonella henselae infection in diverse clinical conditions in a tertiary care hospital in north India. Indian J. Med. Res. 147, 189–194. doi: 10.4103/ijmr.IJMR_1932_16
Dahms H. U., Lee J. S. (2010). UV radiation in marine ectotherms: molecular effects and responses. Aquat. Toxicol. 97, 3–14. doi: 10.1016/j.aquatox.2009.12.002
de la Torre F., Cañas R. A., Pascual M. B., Avila C., Cánovas F. M. (2014). Plastidic aspartate aminotransferases and the biosynthesis of essential amino acids in plants. J. Exp. Bot. 65, 5527–5534. doi: 10.1093/jxb/eru240
Du N., Yang Q., Guo H., Xue L., Fu R., Dong X., et al. (2022). Dissection of Paenibacillus polymyxa NSY50-Induced Defense in Cucumber Roots against Fusarium oxysporum f. sp. cucumerinum by Target Metabolite Profiling. Biol. (Basel). 11, 1028. doi: 10.3390/biology11071028
Ekhtari S., Razeghi J., Hasanpur K., Kianianmomeni A. (2019). Different regulations of cell-type transcription by UV-B in multicellular green alga Volvox carteri. Plant Signal Behav. 14, 1657339. doi: 10.1080/15592324.2019.1657339
Fu S., Xue S., Chen J., Shang S., Xiao H., Zang Y., et al. (2021). Effects of different short-term UV-B radiation intensities on metabolic characteristics of porphyra haitanensis. Int. J. Mol. Sci. 22, 2180. doi: 10.3390/ijms22042180
He Y., Yan Z., Du Y., Ma Y., Shen S. (2017). Molecular cloning and expression analysis of two key genes, HDS and HDR, in the MEP pathway in Pyropia haitanensis. Sci. Rep. 7, 17499. doi: 10.1038/s41598-017-17521-9
Kaplan F., Kopka J., Haskell D. W., Zhao W., Schiller K. C., Gatzke N., et al. (2004). Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136, 4159–4168. doi: 10.1104/pp.104.052142
Keller C., Maeda J., Jayaraman D., Chakraborty S., Sussman M. R., Harris J. M., et al. (2018). Comparison of vacuum MALDI and AP-MALDI platforms for the mass spectrometry imaging of metabolites involved in salt stress in medicago truncatula. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01238
Kroth P. G., Chiovitti A., Gruber A., Martin-Jezequel V., Mock T., Parker M. S., et al. (2008). A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from comparative whole genome analysis. PloS One 3, e1426. doi: 10.1371/journal.pone.0001426
Kumari R., Singh S., Agrawal S. B. (2010). Response of ultraviolet-B induced antioxidant defense system in a medicinal plant, Acorus calamus. J. Environ. Biol. 31, 907–911.
Lee T., Kim K. D., Kim J. M., Shin I., Heo J., Jung J., et al. (2021). Genome-wide association study for ultraviolet-B resistance in soybean (Glycine max L.). Plants (Basel). 10, 1335. doi: 10.3390/plants10071335
Li X., Hou W., Lei J., Chen H., Wang Q. (2023). The unique light-harvesting system of the algal phycobilisome: structure, assembly components, and functions. Int. J. Mol. Sci. 24, 9733. doi: 10.3390/ijms24119733
Li Y., Xi K., Liu X., Han S., Han X., Li G., et al. (2023). Silica nanoparticles promote wheat growth by mediating hormones and sugar metabolism. J. Nanobiotechnol. 21, 2. doi: 10.1186/s12951-022-01753-7
Liaqat W., Altaf M. T., Barutcular C., Nawaz H., Ullah I., Basit A., et al. (2023). Ultraviolet-B radiation in relation to agriculture in the context of climate change: a review. Cereal Res. Commun. 52 (1), 1–24. doi: 10.1007/s42976-023-00375-5
Liu L., Zhao Y., Lu S., Liu Y., Xu X., Zeng M. (2023). Metabolomics investigation on the volatile and non-volatile composition in enzymatic hydrolysates of Pacific oyster (Crassostrea gigas). Food Chem. X. 17, 100569. doi: 10.1016/j.fochx.2023.100569
Lv Y., Zhang S., Wang J., Hu Y. (2016). Quantitative proteomic analysis of wheat seeds during artificial ageing and priming using the isobaric tandem mass tag labeling. PloS One 11, e0162851. doi: 10.1371/journal.pone.0162851
Platt T., Gallegos C., Harrison W. G. (1980). Photoinhibition of photosynthesis in natural assemblages of marine phytoplankton.
Qian C., Ji Z., Lin C., Zhang M., Zhang J., Kan J., et al. (2022). Nitric oxide extends the postharvest life of water bamboo shoots partly by maintaining mitochondrial structure and energy metabolism. Int. J. Mol. Sci. 23, 1607. doi: 10.3390/ijms23031607
Rai K., Agrawal S. B. (2017). Effects of UV-B radiation on morphological, physiological and biochemical aspects of plants: an overview. J. Sci. Res. 61, 87–113.
Ren C. G., Kong C. C., Yan K., Zhang H., Luo Y. M., Xie Z. H. (2017). Elucidation of the molecular responses to waterlogging in Sesbania cannabina roots by transcriptome profiling. Sci. Rep. 7, 9256. doi: 10.1038/s41598-017-07740-5
Sarkar D., Bhowmik P. C., Young In K., Shetty K. (2011). The role of proline-associated pentose phosphate pathway in cool-season turfgrasses after UV-B exposure. Environ. Exp. Bot. 70, 251–258. doi: 10.1016/j.envexpbot.2010.09.018
Sass A., Slachmuylders L., Van Acker H. (2019). Various evolutionary trajectories lead to loss of the tobramycin-potentiating activity of the quorum-sensing inhibitor baicalin hydrate in burkholderia cenocepacia biofilms. Antimicrob. Agents Chemother. 63, 10–1128. doi: 10.1128/AAC.02092-18
Shi Q., Chen C., He T., Fan J. (2022). Circadian rhythm promotes the biomass and amylose hyperaccumulation by mixotrophic cultivation of marine microalga Platymonas helgolandica. Biotechnol. Biofuels Bioprod. 15, 75. doi: 10.1186/s13068-022-02174-2
Shinde S., Singapuri S., Jiang Z., Long B., Wilcox D., Klatt C., et al. (2022). Thermodynamics contributes to high limonene productivity in cyanobacteria. Metab. Eng. Commun. 14, e00193. doi: 10.1016/j.mec.2022.e00193
Simioni C., Schmidt E. C., Felix M. R., Polo L. K., Rover T., Kreusch M., et al. (2014). Effects of ultraviolet radiation (UVA+UVB) on young gametophytes of Gelidium floridanum: growth rate, photosynthetic pigments, carotenoids, photosynthetic performance, and ultrastructure. Photochem. Photobiol. 90, 1050–1060. doi: 10.1111/php.12296
Soni S., Jha A. B., Dubey R. S., Sharma P. (2022). Application of nanoparticles for enhanced UV-B stress tolerance in plants. Plant Nano. Biol. 2, 100014. doi: 10.1016/j.plana.2022.100014
Sun Q., Liu M., Cao K., Xu H., Zhou X. (2022). UV-B irradiation to amino acids and carbohydrate metabolism in rhododendron chrysanthum leaves by coupling deep transcriptome and metabolome analysis. Plants (Basel). 11, 2730. doi: 10.3390/plants11202730
Sun Y., Wang B., Ren J., Zhou Y., Han Y., Niu S., et al. (2022). OsbZIP18, a positive regulator of serotonin biosynthesis, negatively controls the UV-B tolerance in rice. Int. J. Mol. Sci. 23, 3215. doi: 10.3390/ijms23063215
Takshak S., Agrawal S. B. (2019). Defense potential of secondary metabolites in medicinal plants under UV-B stress. J. Photochem. Photobiol. B. 193, 51–88. doi: 10.1016/j.jphotobiol.2019.02.002
Thakur K., Kumari C., Zadokar A., Sharma P., Sharma R. (2023). Physiological and omics-based insights for underpinning the molecular regulation of secondary metabolite production in medicinal plants: UV stress resilience. Plant Physiol. Biochem. 204, 108060. doi: 10.1016/j.plaphy.2023.108060
Tian Y., Peng K., Lou G., Ren Z., Sun X., Wang Z., et al. (2022). Transcriptome analysis of the winter wheat Dn1 in response to cold stress. BMC Plant Biol. 22, 277. doi: 10.1186/s12870-022-03654-1
Tilbrook K., Dubois M., Crocco C. D., Yin R., Chappuis R., Allorent G., et al. (2016). UV-B perception and acclimation in chlamydomonas reinhardtii. Plant Cell 28, 966–983. doi: 10.1105/tpc.15.00287
Torres S., Gonzalez-Ramirez M., Gavilan J., Paz C., Palfner G., Arnold N., et al. (2019). Exposure to UV-B radiation leads to increased deposition of cell wall-associated xerocomic acid in cultures of serpula himantioides. Appl. Environ. Microbiol. 85, e00870-19. doi: 10.1128/AEM.00870-19
Tripathi R., Sarkar A., Rai S. P., Agrawal S. B. (2011). Supplemental ultraviolet-B and ozone: impact on antioxidants, proteome and genome of linseed (Linum usitatissimum L. cv. Padmini). Plant Biol. (Stuttg). 13, 93–104. doi: 10.1111/j.1438-8677.2010.00323.x
Voerman S.E., Ruseckas A., Turnbull G.A., Samuel I.D., Burdett H.L. (2022). Red algae acclimate to low light by modifying phycobilisome composition to maintain efficient light harvesting. BMC biology 20 (1), 291. doi: 10.1186/s12915-022-01480-3
Wang X., Sakata K., Komatsu S. (2018). An integrated approach of proteomics and computational genetic modification effectiveness analysis to uncover the mechanisms of flood tolerance in soybeans. Int. J. Mol. Sci. 19, 1301. doi: 10.3390/ijms19051301
Wong D. C. J., Ariani P., Castellarin S., Polverari A., Vandelle E. (2018). Co-expression network analysis and cis-regulatory element enrichment determine putative functions and regulatory mechanisms of grapevine ATL E3 ubiquitin ligases. Sci. Rep. 8, 3151. doi: 10.1038/s41598-018-21377-y
Wu X., Chen B., Xiao J., Guo H. (2023). Different doses of UV-B radiation affect pigmented potatoes’ growth and quality during the whole growth period. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1101172
Xue S., Zang Y., Chen J., Shang S., Tang X. (2022). Effects of enhanced UV-B radiation on photosynthetic performance and non-photochemical quenching process of intertidal red macroalgae Neoporphyra haitanensis. Environ. Exp. Bot. 199, 104888. doi: 10.1016/j.envexpbot.2022.104888
Yang Y., Yao Y., Xu G., Li C. (2005). Growth and physiological responses to drought and elevated ultraviolet-B in two contrasting populations of Hippophae rhamnoides. Physiol. Plant. 124, 431–440. doi: 10.1111/j.1399-3054.2005.00517.x
Yang H., Zhao Z., Qiang W. Q., An L., Xu S. (2004). Effects of enhanced UV-B radiation on the hormonal content of vegetative and reproductive tissues of two tomato cultivars and their relationships with reproductive characteristics. Plant Growth Regul. 43, 251–258. doi: 10.1023/b:grow.0000046002.39513.be
Yang C., Zhao W., Wang Y., Zhang L., Huang S., Lin J. (2020). Metabolomics analysis reveals the alkali tolerance mechanism in puccinellia tenuiflora plants inoculated with arbuscular mycorrhizal fungi. Microorganisms 8, 327. doi: 10.3390/microorganisms8030327
Yang L. E., Zhou W., Hu C. M., Deng Y. Y., Xu G. P., Zhang T., et al. (2018). A molecular phylogeny of the bladed Bangiales (Rhodophyta) in China provides insights into biodiversity and biogeography of the genus Pyropia. Mol. Phylogenet. Evol. 120, 94–102. doi: 10.1016/j.ympev.2017.11.009
Yin B. F., Zhang Y. M., Lou A. R. (2017). Impacts of the removal of shrubs on the physiological and biochemical characteristics of Syntrichia caninervis Mitt: in a temperate desert. Sci. Rep. 7, 45268. doi: 10.1038/srep45268
You X., Zhang X., Cheng J., Xiao Y., Ma J., Sun S., et al. (2023). In situ structure of the red algal phycobilisome–PSII–PSI–LHC megacomplex. Nature 616, 199–206. doi: 10.1038/s41586-023-05831-0
Zampieri M., Hörl M., Hotz F., Müller N. F., Sauer U. (2019). Regulatory mechanisms underlying coordination of amino acid and glucose catabolism in Escherichia coli. Nat. Commun. 10, 3354. doi: 10.1038/s41467-019-11331-5
Zeng W., Peng Y., Zhao X., Wu B., Chen F., Ren B., et al. (2019). Comparative proteomics analysis of the seedling root response of drought-sensitive and drought-tolerant maize varieties to drought stress. Int. J. Mol. Sci. 20, 2793. doi: 10.3390/ijms20112793
Zhang X., Ding X., Ji Y., Wang S., Chen Y., Luo J., et al. (2018). Measurement of metabolite variations and analysis of related gene expression in Chinese liquorice (Glycyrrhiza uralensis) plants under UV-B irradiation. Sci. Rep. 8, 6144. doi: 10.1038/s41598-018-24284-4
Zhang N., Wang S., Zhao S., Chen D., Tian H., Li J., et al. (2023). Global crotonylatome and GWAS revealed a TaSRT1-TaPGK model regulating wheat cold tolerance through mediating pyruvate. Sci. Adv. 9, eadg1012. doi: 10.1126/sciadv.adg1012
Zhao X., Bai X., Jiang C., Li Z. (2019). Phosphoproteomic analysis of two contrasting maize inbred lines provides insights into the mechanism of salt-stress tolerance. Int. J. Mol. Sci. 20, 1886. doi: 10.3390/ijms20081886
Zhao X., Zheng W., Qu T., Zhong Y., Xu J., Jiang Y., et al. (2021). Dual roles of reactive oxygen species in intertidal macroalgae Ulva prolifera under ultraviolet-B radiation. Environ. Exp. Bot. 189, 1034. doi: 10.1016/j.envexpbot.2021.104534
Zhen Z. H., Qin S., Ren Q. M., Wang Y., Ma Y. Y., Wang Y. C. (2021). Reciprocal effect of copper and iron regulation on the proteome of synechocystis sp. PCC 6803. Front. Bioeng. Biotechnol. 9. doi: 10.3389/fbioe.2021.673402
Zhong Z., Liu S., Zhu W., Ou Y., Yamaguchi H., Hitachi K., et al. (2019). Phosphoproteomics Reveals the Biosynthesis of Secondary Metabolites in Catharanthus roseus under Ultraviolet-B Radiation. J. Proteome Res. 18, 3328–3341. doi: 10.1021/acs.jproteome.9b00267
Zhu M., Zang Y., Zhang X., Shang S., Xue S., Chen J., et al. (2023). Insights into the regulation of energy metabolism during the seed-to-seedling transition in marine angiosperm Zostera marina L.: Integrated metabolomic and transcriptomic analysis. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1130292
Keywords: UV-B radiation, recovery mechanism, intertidal macroalgae, transcriptomics, targeted metabolomics, energy metabolism
Citation: Wang X, Zang Y, Xue S, Shang S, Xin J, Tang L, Chen J and Tang X (2024) Integrated analysis of the physiological, transcriptomic and metabolomic responses of Neoporphyra haitanensis after exposure to UV-B radiation: an energy metabolism perspective. Front. Mar. Sci. 11:1372252. doi: 10.3389/fmars.2024.1372252
Received: 24 January 2024; Accepted: 02 April 2024;
Published: 19 April 2024.
Edited by:
Renato Crespo Pereira, Fluminense Federal University, BrazilCopyright © 2024 Wang, Zang, Xue, Shang, Xin, Tang, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuexi Tang, dGFuZ3h4QG91Yy5lZHUuY24=; Jun Chen, Y2hlbmp1bkBvdWMuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.