
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 11 March 2024
Sec. Aquatic Microbiology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1359610
Thalassia testudinum has undergone die-offs in the past century due to seagrass wasting disease caused by Labyrinthula sp. Little is known about how seagrasses resist Labyrinthula infections, but metabolites that inhibit Labyrinthula were previously extracted from seagrass leaves. Furthermore, leaf fungal endophytes from seagrasses possess antipathogenic potential, but their activity against Labyrinthula is unknown. Here, we aimed to identify whether fungal endophytes of T. testudinum can aid in disease defense against Labyrinthula. Through Illumina amplicon sequencing of the leaves’ mycobiome, we identified fungi that are known to produce antimicrobials. We also isolated and extracted organic compounds from endophytes to test their anti-Labyrinthula potential using disk diffusion assays. There were 22 isolates that inhibited Labyrinthula, from which two isolates, Trichoderma sp. P1a and Diaporthe sp. M14, displayed strong inhibition. LC-HRMS/MS analysis determined the likely bioactive compounds of Trichoderma as peptaibols and of Diaporthe as cytosporone B. Cytosporone B was confirmed bioactive against Labyrinthula via disk diffusion assays. While these organisms are low in abundance in the mycobiome, this study demonstrates that seagrass endophytes have the potential to play an important role in defense against Labyrinthula.
Within the phylum Labyrinthulomycota, the genus Labyrinthula contains colorless or yellowish protists that form slime nets containing an ectoplasmic network of hairlike structures in which they glide (reviewed in Bennett et al., 2017). Labyrinthula spp. are generally saprotrophs and have been described as either commensals, mutualists, or parasites (reviewed in Bennett et al., 2017) and include the pathogenic species that causes seagrass wasting disease, Labyrinthula zosterae (Martin et al., 2016). Labyrinthula spp. degrade the cell walls of the host using extracellular enzymes (Muehlstein, 1992), and seagrass wasting disease is characterized by splotching and dark streaking on the leaves (Renn, 1935; Duffin et al., 2020). Several consequences arise from Labyrinthula infections in seagrasses, including carbon deficiency, increased respiration, and smaller biomass, making the seagrass susceptible to sulfide, hypoxia, and possible secondary infections (Renn, 1936; Durako and Kuss, 1994; Trevathan-Tackett et al., 2013; Lohan et al., 2020). Die-offs of Thalassia testudinum in Florida Bay have been attributed to wasting disease in the past (Robblee et al., 1991; Durako, 1994), and it is important to understand how the T. testudinum holobiont responds to Labyrinthula infections.
The holobiont of a plant is composed of the plant host and its microbiome, including the phyllosphere and rhizosphere, and these factors act as a biological unit for the survival of the whole (Jones et al., 2019). The microbiomes of plants are known to provide several benefits to the plant host, including improved environmental stress tolerance, plant growth, and pathogen resistance (reviewed in Classen et al., 2015; Berg and Koskella, 2018; Jones et al., 2019). In terrestrial plants like Arabidopsis, pathogenic infections can trigger an immune response like the exudation of jasmonic acid, which can affect the abundance and/or composition of the microbial community (Carvalhais et al., 2015) and also support microbes that may have beneficial effects on the plant, including increased biofilm formation that can protect against pathogens (Berendsen et al., 2018). Furthermore, terrestrial plants can “precondition” their soil into a “protective” soil after pathogenic infections, so that new, uninfected plants of their species can obtain heightened resistance against pathogens, likely provided by soil microbial communities (Berendsen et al., 2018). Studies in which the microbiome can provide disease defense to the plant host have been reported for some aquatic plants. For example, bacterial isolates from Vallisneria americana, which is closely related to T. testudinum, were shown to inhibit the growth of the pathogenic oomycete, Pythium aphanidermatum (Kurtz et al., 2003). Although the seagrass microbiome has not been clearly shown to play a role in disease defense, studies have suggested that the microbiome can provide many benefits to the host. For instance, gene expression studies on the seagrass microbiomes show that they have the potential to detoxify harmful compounds such as sulfide, methanol, and ethanol, as well as fix nitrogen, and produce plant-growth hormones and agarase, which might inhibit algal competition (Crump et al., 2018). Additionally, seagrass endophytic isolates of Aspergillus spp (Notarte et al., 2018), Trichoderma spp., Penicillium spp., Fusarium spp., and Stephanonetria spp (Supaphon et al., 2013). have shown antipathogenic potential against human pathogens, but it is unknown whether this activity applies to seagrass pathogens.
Endophytic fungi are usually transmitted to plants via either vertical transmission to the seeds from the female plant or horizontal transmission to aboveground portions of the plant via biotic or abiotic dispersal, spores, or hyphae (Yan et al., 2019). For many years, it has been known that fungal endophytes provide many benefits to the plant host, including increased resistance to abiotic stressors, like high salinity, drought, and heavy metal exposure, and biotic factors such as herbivores and pathogens (reviewed in Yan et al., 2019 and Qin et al., 2023). Fungal endophytes can release secondary metabolites that can alter the host plant’s gene expression and biochemical pathways as a way to increase pathogen resistance (Qin et al., 2023). For example, the fungal endophyte Acrophialophora jodhpurensis was able to increase the activity of several immune responses on tomato plants against Alternaria alternata infections, including phenolic and ROS production, among other responses (Daroodi et al., 2021). Endophytic fungi also produce several antimicrobial compounds including antibiotics such as penicillin, trichodermin, and cell wall-degrading enzymes like cellulases, lipases, and proteases, all of which can offer defense against plant pathogens (Ghorbanpour et al., 2018; Fadiji and Babalola, 2020).
Few studies have examined the effects of endophytic marine fungi against diseases in marine plants; however, a few studies show that metabolite extracts from marine plants can potentially deter marine pathogens. A study on the seaweed Agarophyton vermiculophyllum demonstrated that seaweed metabolites can deter opportunistic pathogens in vitro while attracting bacteria that displayed defensive properties against disease-causing bacteria (Saha and Weinberger, 2019). Additionally, seagrasses are known to produce many secondary metabolites that have antimicrobial activity against medically and agriculturally important pathogens (reviewed in Zidorn, 2016), and antifungal activity (Ross et al., 2008). Anti-labyrinthulid activity has also been observed from phenolic acids and an unknown small molecule that was isolated from T. testudinum (Steele et al., 2005; Trevathan-Tackett et al., 2015). Furthermore, a study by Castro-González et al. (2022) showed that there are differences in the metabolic profiles of healthy vs. Labyrinthula-infected seagrasses, meaning that the pathogen infection changes the metabolic profile of the plant. Although these studies suggest that the secondary metabolites arise from the seagrass plant, the endophytic fungal secondary metabolites must be considered, due to their presence within the leaves and potential antipathogenic activity (Supaphon et al., 2013; Notarte et al., 2018). To date, no studies have determined the effects of the metabolic extracts from T. testudinum fungal endophytes on the growth of Labyrinthula spp. The aim of this paper is to determine whether fungal endophytes of T. testudinum have the potential to produce anti-labyrinthulid compounds. We first examined the potential of the fungal community to produce antipathogenic compounds based on the taxonomy of the endophytic fungi (within the leaves) and epiphytic fungi (on the leaf surface). We then cultured fungal endophytes from apparently healthy leaves, and their extracted organic compounds were tested for their activity against a strain of Labyrinthula. Finally, we identified the bioactive compounds in the two most bioactive extracts.
There were 50 healthy-appearing leaves of T. testudinum that were collected from Hobie Island Beach Park from Virginia Key, Miami, FL, USA, in November 2022. The leaves were placed in a ziplock bag with seawater while transferring to the lab. The leaves were rinsed with seawater to remove loosely attached sediment and scraped with a sterile blade on both sides of the leaf to collect the epiphyte samples, which were transferred to a 1.5-mL Eppendorf tube and stored in −80°C until DNA extractions. The leaves, which were considered surface sterilized, were then stored at −80°C overnight, and afterward transferred to a lyophilizer for 3 days. The freeze-dried leaves were crushed to almost powder and transferred to a 1.5-mL Eppendorf tube for DNA extractions. The crushed leaves were considered the endosphere samples. DNA was extracted using the Qiagen DNeasy PowerLyzer Soil Kit (Qiagen, Hilden, Germany) following manufacturer-recommended protocols, including a 10-min incubation step at 70°C after step 3 to increase DNA yield. The DNA was sent to Novogene (Beijing, China) for amplicon sequencing of the fungal ITS1-1F region (primers: ITS1-1F-F CTTGGTCATTTAGAGGAAGTAA and ITS-1F-R GCTGCGTTCTTCATCGATGC).
The preprocessed sequences were run through dada2 in R (v 4.2.2; [R Core Team, 2013]), using default settings. The amplicon sequence variants are available in the supplementary material as a fasta file. Taxonomy was assigned using the assignTaxonomy function in dada2 (v 4.2.2; [R Core Team, 2013]), with the UNITE database (version 9.0, Abarenkov et al., 2022) as reference. The R packages phyloseq (McMurdie and Holmes, 2013), vegan (Oksanen et al., 2018), ggplot2 (Wickham, 2009), dplyr (Wickham et al., 2023), ape (Paradis and Schliep, 2019), and microbiome (Lahti et al., 2017) were used for visualizations and statistical analysis including alpha (Shannon and Chao1) and beta diversity (UniFrac PCoA) plots to compare the endophytic and epiphytic fungal communities. Permutational multivariate analysis of variance (PERMANOVA) with 9,999 permutations using the Adonis command in the base package in R (v 4.2.2; [R Core Team, 2013]) was used to determine significant differences in the alpha and beta diversity between the epiphyte and endosphere samples. Phyla labeled “incertae_sedis” were removed from the dataset, as the purpose of this approach was to identify known producers of bioactive compounds. The top 20 most abundant genera were plotted using the plot_bar function in phyloseq after normalizing and filtering out “NA” genera.
Samples of Thalassia testudinum leaves were collected off the shores of Hobie Island Beach Park in Virginia Key in Miami, FL, USA, in late June 2021. Only green leaves that contained no apparent lesions were selected for isolation of potentially beneficial fungal endophytes. The leaves were collected haphazardly during low tide, with gloved hands, and were placed in ziplock bags containing seawater. The ziplock bags were then placed in a cooler with ice during transportation to the lab to be processed within 4 h.
The protocols of Mata and Cebrián (2013); Bibi et al. (2018), and Ettinger and Eisen (2020) were modified to isolate endophytic fungi. Briefly, 48 leaves were swabbed with sterile cotton swabs to remove ectosymbionts and to surface sterilize the leaves. Half of the leaves (n = 24) were then further sterilized by submersion in 0.5% NaOCl for 5 min and then rinsed with sterile salt water (Instant Ocean, VA, USA) for 5 min. All leaves were then cut into 1–2-cm pieces and plated on potato dextrose agar (PDA) and malt extract agar (MEA) media plates, one leaf per plate. This resulted in 24 plates of only scraped leaves (12 in PDA and 12 in MEA) and 24 plates of scraped and bleached leaves (12 in PDA and 12 in MEA). PDA plates were prepared with 39 g/L of PDA in DI water, and, after autoclaving, the addition of 25 mL/L of penicillin–streptomycin (10,000 units penicillin and 10 mg streptomycin per mL; Millipore Sigma). MEA plates were made with 30 g/L of malt extract and 15 g/L of agar in DI water, followed by adding 25 mL/L of penicillin–streptomycin (10,000 units penicillin and 10 mg streptomycin per mL; Millipore Sigma) after autoclaving. Once the leaves were plated, the plates were parafilmed and incubated at room temperature (~23°C), in the dark for 3–4 weeks, checking for new growth weekly. Plugs of emerging fungi were plated in the center of their respective fresh media plates (either PDA or MEA) and allowed to grow until the plate was covered. Plugs were taken from each plate until colonies were isolated.
DNA was extracted from each fungal isolate using the Qiagen PowerLyzer Microbial Kit (Qiagen, Hilden, Germany) following the manufacturer-recommended protocols with slight modification. Briefly, a sterile needle was used to collect mycelia, tissue, and/or spores from each fungus and to place them in the bead beating tubes (provided by the kit) containing the first buffer and solution SL. A 10-min incubation step at 70°C was added to increase DNA yield. Bead-beating was performed at 4.0 m/s for 20 s, twice, with a FastPrep-24 bead beater (MP Biomedicals). The ITS-28S region was amplified with primers ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG; White et al., 1990) and LR3 (5′-GGTCCGTGTTTCAAGAC; Vilgalys and Hester, 1990), as used in Ettinger and Eisen (2020). The PCR program was as follows: 95°C for 5 min, 35 cycles of 94°C for 30 s, 52°C for 15 s, 72°C for 1 min, and a final extension at 72°C for 8 min (Ettinger and Eisen, 2020).
All PCR products were sent for cleanup and Sanger sequencing with the same primer set, ITS5 and LR3 (MCLAB, San Francisco, CA, USA). Forward and reverse sequences were merged for all samples using the MegAlign program from Lasergene (v.17.4, DNASTAR). Sequences are available in GenBank (accession numbers OR355682–OR355766). NCBI BLAST was used for initial identification of all cultures. The ITS sequences obtained from the Sanger sequencing run of the most bioactive fungi were trimmed to match the ITS1-1F amplicons using Geneious v.R8.1.9 (Biomatters, Auckland, New Zealand). Amplicon sequences were used as a custom BLAST database in Geneious v.R8.1.9 and were blasted against the trimmed ITS sequences to find 100% matches, confirming the presence of the strains in our amplicon dataset. Furthermore, ASVs that matched the taxonomical assignment of the isolates that showed the most bioactivity against Labyrinthula were subset from our datasets using the subset_taxa function in phyloseq to determine their relative abundance (McMurdie and Holmes, 2013). Where appropriate, a standard two-sample T-test was performed on log-transformed data to compare the abundance of the most bioactive fungi between epiphytic and endophytic samples using the base package in R (v 4.2.2; [R Core Team, 2013]).
Fungi were grouped based on sequence data and colony morphology. A representative from each group was selected for metabolite extractions. If a culture belonged to the same taxon based on sequence data but differed in colony morphology, it was also selected for extractions. The protocols from Supaphon et al. (2013) and Notarte et al. (2018) were followed with modifications to extract metabolites from the fungal endophytes. To prepare the cultures for metabolite extractions, 17 plugs (5 mm wide) of each representative strain were placed in 125-mL flasks containing 50 mL of malt extract media (30 g/L in DI water). Malt extract media were used for growing both MEA- and PDA-derived fungal isolates because of its better yield (Supplementary Table 1). Cultures were placed in incubated mini shakers (VWR International, LLC.) set to 28°C and 130 rpm. Cultures were allowed to grow for a week, and if no growth was observed, the cultures were moved to static conditions at 28°C. If the cultures remained in the incubated shakers, they were left for an additional week, and if they were in static conditions, they were left for an additional 2 weeks.
The cultures were filtered into a 50-mL filtration unit using a GF/F Whatman filter (47-mm diameter; ©Cytiva). All glassware was rinsed with HPLC Plus grade ethyl acetate (Sigma-Aldrich, Darmstadt, Germany) between samples. For the liquid–liquid extractions, a total of 150 mL of HPLC Plus grade ethyl acetate was used to extract metabolites from the culture filtrate, three times. For the first extraction, 50 mL of ethyl acetate was mixed with the filtrate (~50 mL) and shaken vigorously for 5 min. The second and third extractions were also done with 50 mL of ethyl acetate and shaken vigorously for 2 min each. For the biomass extractions, the cultures were scraped off the GF/F Whatman filter and stored at −80°C overnight. The biomass was then lyophilized for 3 days and stored at −80°C until further processing. The biomass was then crushed with a mortar and pestle, placed in a glass beaker with 50 mL of ethyl acetate, and homogenized with a tissue homogenizer (TH Tissue Homogenizer, Omni International, USA). After 30 min in ethyl acetate, intermittently shaking the beaker, the crushed biomass was filtered through a ceramic funnel lined with a Whatman filter paper (ashless, 150 mm) shaped to fit the funnel, into a glass flask attached to a vacuum pump. The extracts derived from the supernatant and biomass were dried in vacuum using a Rotavapor (RE111, Büchi, Switzerland) at 46°C, until the ethyl acetate mostly evaporated. When the volume was near 5 mL, the extracts were transferred to scintillation vials and further evaporated on the Rotavapor. Samples were left to airdry overnight and then stored at 4°C until assayed (between 1 and 2 weeks).
The protocols of Martin et al. (2009) and Sullivan et al. (2017) were modified to culture Labyrinthula spp. from field-collected leaves. Briefly, green leaves that contained lesions were collected during low tide and placed in ziplock bags containing seawater. Upon arrival to the lab, the leaves were rinsed with seawater and swabbed with sterile cotton swabs to remove ectosymbionts. The leaf lesions were then cut into 1–2-cm pieces and placed in serum seawater agar plates (SSA, Muehlstein et al., 1988), which were prepared with modifications as follows. Twelve grams of agar was added to Instant Ocean (Spectrum Brands, Blacksburg, VA, USA; prepared according to manufacturer-recommended protocols) per liter of SSA media. After autoclaving, 10 mL of horse serum (Gibco), 25 mL of penicillin–streptomycin (10,000 units penicillin/mL, 10 mg streptomycin/mL; Millipore Sigma), and 2 mL of a germanium dioxide solution (1.5 mg/mL in DI water; filter-sterilized) were added per liter of SSA media. After isolating the strains, they were maintained in Enhanced SSA (ESSA) media (Martin et al., 2016). ESSA was prepared with modification as follows: 12 g of agar, 1 g of glucose, 0.1 g of peptone, and 0.1 g of yeast extract per liter of ESSA were autoclaved in Instant Ocean. After autoclaving and cooling, 10 mL of horse serum (Gibco) was added per liter of ESSA. After Labyrinthula colonies were isolated, they were grown in 10 mL of liquid SSA media in sterile petri dishes for 14 days in preparation for DNA extractions using the Qiagen PowerLyzer Microbial Kit (Qiagen, Hilden, Germany) following the manufacturer-recommended protocols. PCRs of the 18S rRNA gene were then performed using primer sets A, R3 and F2, B (Martin et al., 2016). PCRs were sent for Sanger sequencing using the same primers (MCLAB, San Francisco, CA, USA). Sequences were assembled using the MegAlign program from Lasergene (v.17.4, DNASTAR).
The Labyrinthula strain that grew the best was L 7.2 (GenBank accession OR356117). To determine the pathogenic probability of this strain based on Martin et al. (2016), who defined different phylogenetic clades for the pathogenic and non-pathogenic strains of Labyrinthula spp., 26 near full-length 18S rRNA gene sequences representing three distinct Labyrinthula clades (pathogenic, non-pathogenic, and terrestrial) were retrieved from the GenBank nr database. These sequences (including two outgroup sequences) were aligned using MAFFT v7.271 (Katoh and Standley, 2013) with default parameters, and gaps were masked using trimAl v1.4 (Capella-Gutiérrez et al., 2009). Phylogenetic inference was made by Maximum Likelihood methods implemented in RAxML v8.0. 0 (Stamatakis, 2014) under gamma-corrected GTR model of evolution with 1,000 bootstrap replicates based on 935 homologous positions, and the final tree was produced with FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree).
Several ESSA plates were inoculated with a plug (5-mm diameter) of Labyrinthula strain L7.2. When the Labyrinthula colonies grew at least 5 mm beyond the plugs, they were considered ready for assay. Only colonies that were actively growing were chosen. The metabolite extracts were resuspended in 400 µL methanol, from which 20 µL was pipetted onto sterile Whatman filter disks, cut with a hole puncher (6-mm diameter), in triplicates. 20 µL of methanol was used for control. The disks were airdried overnight within a closed biosafety cabinet. The disks were then placed on the edge of the Labyrinthula colony. For each replicate, both the liquid extract and the biomass extract disks from the same culture were placed at the edge of the Labyrinthula colony. The plates were checked every other day for 15 days for zones of inhibition or any visible changes to the colony, which were assessed each time. Due to the irregular and asymmetrical nature of Labyrinthula colony growth on agar media (Martin et al., 2009), measurements of colony growth can be difficult to compare between colonies; therefore, we rated bioactivity levels as strong (the colony was pushed away from the disk), moderate (the colony ceased growth), or none (the colony grew over the filter disk). The fungal extracts with the strongest bioactivity levels were selected for further analysis.
Dried fungal extracts were resuspended in 100% MeCN and diluted (1:100) in LC-HRMS starting conditions, and experiments were conducted on an Agilent 1290 Infinity II series UPLC coupled to an Agilent 6546 QTOF mass spectrometer with an electrospray ionization (ESI) source (Agilent Technologies, Santa Clara, CA, USA). Chromatography was performed using a Kinetex® C18 column (50 × 2.1 mm, 2.6, Phenomenex, Torrance, CA, USA), oven temperature was set to 40°C, and the sample injection volume was 1 µL. A binary gradient consisting of MeCN (eluent A) and water (eluent B) (both + 0.1% formic acid (FA)) at a constant flow rate of 400 μL/min was used. The gradient was applied as follows: 0 min–0.25 min, 10% A; 10.00 min, 95% A; 13.00 min, 95% A; 13.1 min, 10% A; 15.00 min 10% A. The MassHunter software (Agilent Technologies, Santa Clara, CA, USA) was used for data acquisition and subsequent qualitative and quantitative analysis. All parameters regarding the QTOF are listed in Supplementary Table 2.
Cytosporone B was identified in extracts from colony M14 (Diaporthales sp.) and was purchased as a chemical standard to confirm its bioactivity against Labyrinthula. Around 3 mg of cytosporone B (5 mg, Santa Cruz Biotechnology, Inc., USA) was resuspended with 60 µL of methanol, from which 20 µL was aliquoted onto three Whatman filter disks (6 mm diameter), totaling around 1 mg each. The filter disks were left to air-dry overnight before placing them at the edge of Labyrinthula colonies. A filter disk with 20 µL of methanol was used for control. The assays were monitored every other day for 15 days for any changes in the colonies.
The endophytic samples had a higher average alpha diversity compared with the epiphytic samples in both Shannon and Chao1 diversity indices (PERMANOVA, p = 0.001, p = 0.007, respectively; Supplementary Figure 1). The communities also differed in beta diversity, with clear, separate clusters shown by the UniFrac distance matrices (PERMANOVA, p < 0.0001; Supplementary Figure 2). In general, the top 20 most abundant fungi of the epiphytic samples differed from the endophytic samples (Figure 1). Several of the fungal genera that were isolated and cultured from the endosphere were present in the amplicons: Aspergillus, Cladosporium, and Trichoderma in both the endophytic and epiphytic samples, and Penicillium and Sarocladium in the endophytic samples (Figure 1, Supplementary Table 3). Many of the most abundant genera of the endosphere, compared with only a few of the abundant genera in the epiphytes, included taxa that have previously been described to produce antipathogenic compounds (Supplementary Table 3).
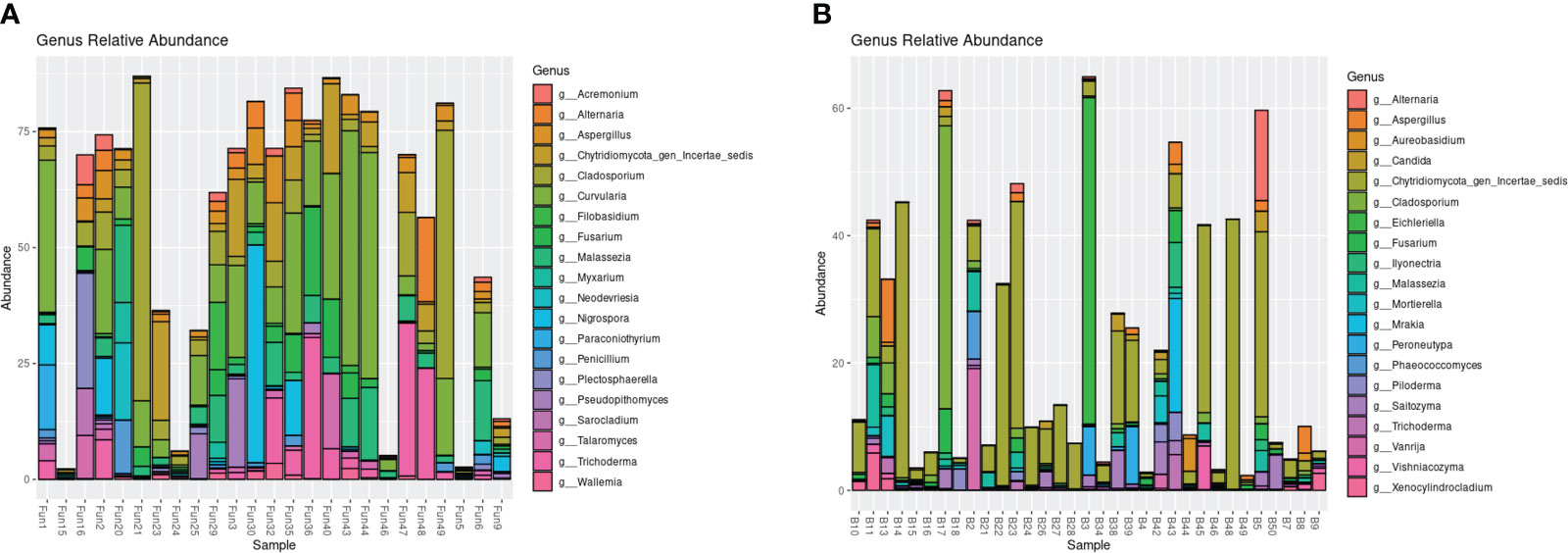
Figure 1 Relative abundance of the top 20 most abundant genera based on ITS1 amplicon sequencing of the endophytic (A) and epiphytic (B) samples.
A total of 104 fungal isolates were obtained (43 from PDA and 61 from MEA) from T. testudinum leaves. A total of 56 isolates from MEA and 30 from PDA could be successfully identified using sequencing of the ITS-28S rRNA region. The remaining cultures either had poor DNA quality after extractions or poor sequence quality after Sanger sequencing. Many sequences were identical or very similar; therefore, isolates were clustered into taxa. On MEA, there were cultures from 15 taxa: Aspergillus, Cladosporium, Diaporthales, Dothideomycetes, Eutypella, Microsphaeropsis, Myrothecium, Ohleria, Paraphaeosphaeria, Parastagonospora, Penicillium, Pestalotiopsis, Phoma, Pleosporales, and Trichoderma. On PDA, strains from nine taxa were isolated: Aspergillus, Cladosporium, Diaporthe, Dothichiza, Dothideomycetes, Microsphaeropsis, Myrothecium, Parascedosporium, and Trichoderma. In addition to NCBI’s GenBank, the UNITE database was also used to determine the taxonomy of the sequences (Supplementary Table 1). Although most of the cultures were assigned to at least the same genus on both databases, 29 of the 86 cultures had different taxonomic assignments from each database. Although the assigned taxonomy differed in each database, the taxa belonged to at least the same family, order, or class for 26 of the 29 cultures (Supplementary Table 1). Because NCBI’s GenBank yielded better %ID matches to many of the isolates compared with the UNITE database, we assigned taxonomy based on the BLAST results.
Labyrinthula strain L 7.2 was determined to be potentially pathogenic based on the similarity of its 18S rRNA sequence to the strains that were identified as pathogenic in Martin et al. (2016) (Figure 2). This strain was selected for the antagonism assays due to its faster growth rate and its potential pathogenicity.
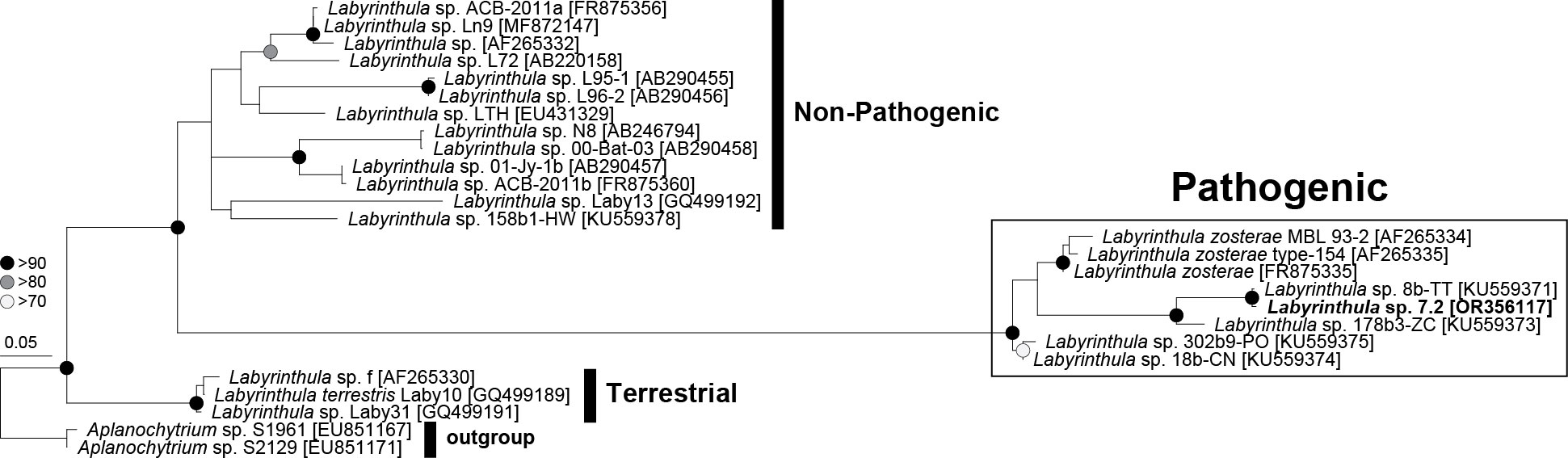
Figure 2 Phylogenetic tree of Labyrinthula sp. based on near full-length 18S rRNA sequences representing three distinct clades: pathogenic, non-pathogenic, and terrestrial. The isolated Labyrinthula colony L7.2 is in bold.
Out of 50 fungal strains tested, 22 strains showed activity against Labyrinthula. Two strains showed strong activity, meaning the Labyrinthula colonies were cleared away from the disks (Supplementary Figure 3; Table 1), whereas 20 strains showed moderate activity, meaning the Labyrinthula colonies neither grew over the disk nor cleared away from the disk (Table 1). The strain that showed the most activity against Labyrinthula, in both extracts of biomass and supernatant, was isolate M14, which belongs to the order Diaporthales per NCBI blast (97.56%; Table 1; Supplementary Table 1). Colony P1a, which belongs to the genus Trichoderma with 99.64% ID per NCBI blast, had strong activity only in the liquid extract (Table 1).
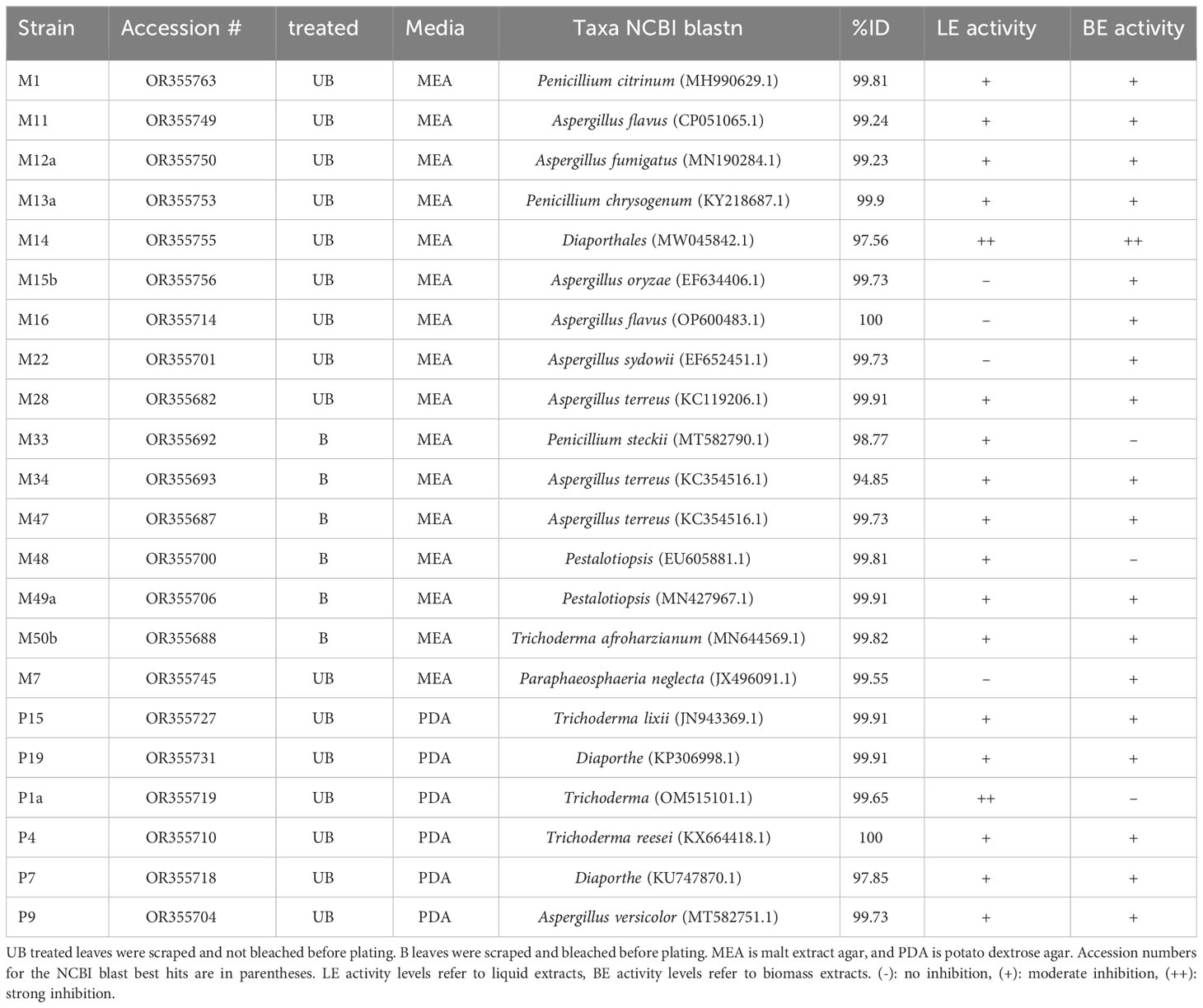
Table 1 List of bioactive fungal strains isolated from T. testudinum leaves and their predicted taxonomy.
Fungi belonging to the genera Trichoderma, Aspergillus, Penicillium, Pestalotiopsis, order Pleosporales, and class Sordariomycetes (including Diaporthales), showed moderate activity against Labyrinthula. There were 28 of the strains that showed no activity (Supplementary Table 1).
The extracts from the two fungal strains that showed the most activity against Labyrinthula spp. (M14 and P1a) were analyzed by LC-HRMS/MS. The M14 extract showed a major component and the detection of the accurate m/z 323.1855 (Δppm 0.62) suggested the molecular formula of C18H26O5. Literature revealed cytosporone B as a fit for the latter molecular formula (Chepkirui and Stadler, 2017; van Santen et al., 2019; Xu et al., 2021). Comparison of the fragmentation spectrum (MS2) with in-silico prediction software (Wang et al., 2021) suggested cytosporone B as the respective metabolite. Furthermore, the presence was unequivocally confirmed with a purchased reference standard of cytosporone B (Figure 3). Cytosporones are signature compounds described for the genus Diaporthe (Brady et al., 2000).
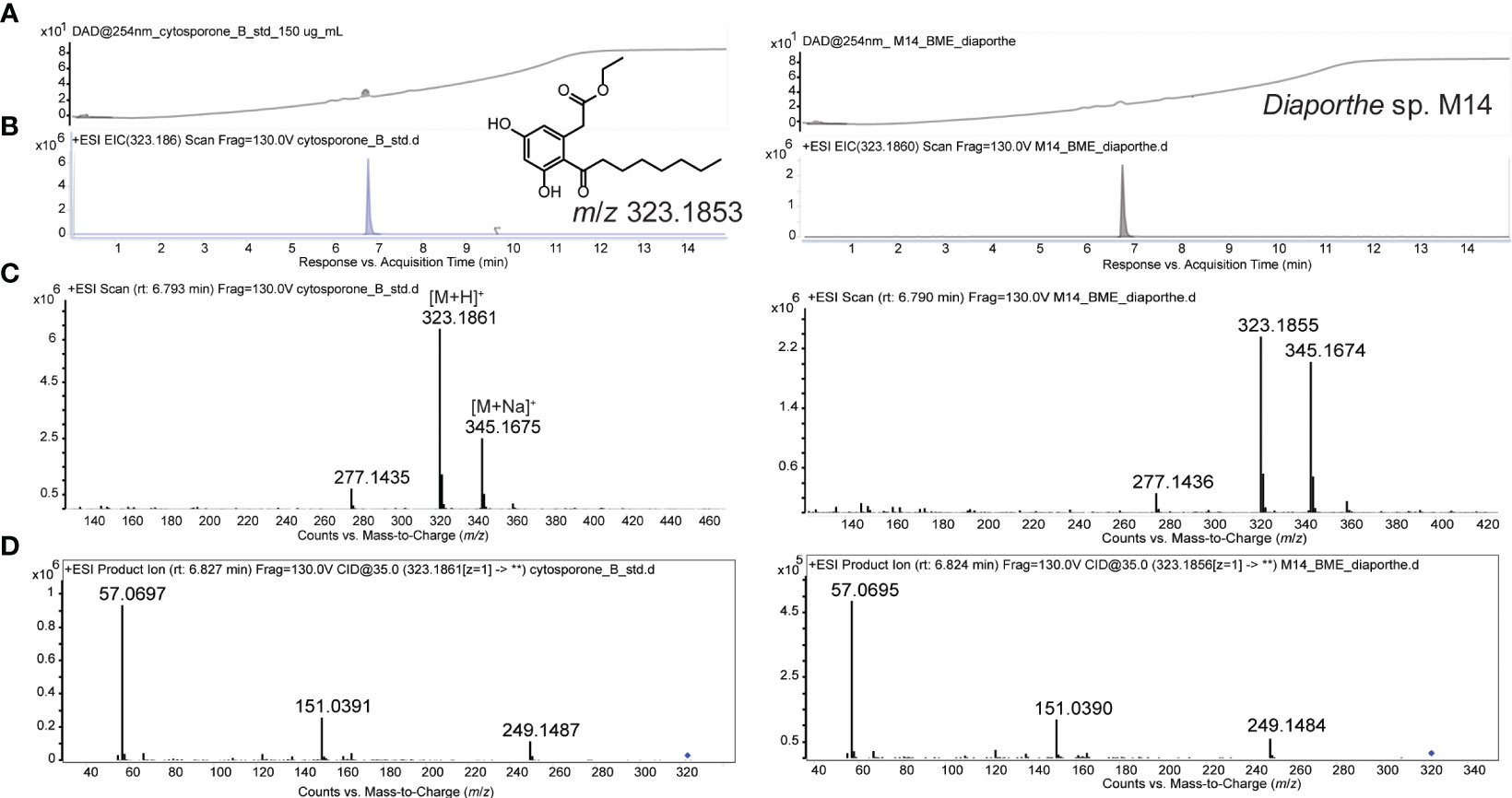
Figure 3 UV-chromatogram (254 nm; A), extracted ion chromatogram (EIC; B), MS1 spectrum (C), and MS2 spectrum (D) of cytosporone B reference (150 µg/mL) LEFT, and Diaporthe sp. (M14) extract RIGHT. Structure of cytosporone B and m/z for observed M+H ion is provided in A.
The extract of P1a showed a mixture of peptaibol-like compounds (m/z 1,108–1,120, data not shown) due to characteristic neutral losses of aminoisobutyric acid (Aib, 85.059 Da). The mixture was not further characterized since both the NCBI BLAST and UNITE database analyses indicated that P1a is in the genus Trichoderma, which produces a vast variety of peptaibols that are known to exhibit a wide range of biological activities, including antimicrobial and cytotoxic effects (Daniel and Rodrigues Filho, 2007; Hou et al., 2022).
Cytosporone B was found to be a major component in M14 extracts. As cytosporone B is commercially available, an assay was performed with 1 µg of per disk in triplicates. After 15 days of running the assay, the results showed that cytosporone B displayed moderate activity, meaning that the Labyrinthula colonies neither grew over the disk nor receded further away from them (Supplementary Figure 4).
Exact matches for the M14 and P1a sequences were not found in the amplicon sequencing dataset of the ITS1 region. The highest match for M14 was 97.6% to ASV2969, and the highest match for P1a was 99.2% to ASV4147. The relative abundance was determined to be mostly low for both taxa that were classified as the Trichoderma and Diaporthe for P1a and M14, respectively. Diaporthe spp. were only present in three samples: two epiphytic samples (B11 and B15) and one endophytic sample (FUN1), with relative abundances less than 1% in all three (Supplementary Table 4). Trichoderma spp. were present in 49 samples: 27 epiphytic samples, and 22 endophytic samples, with a higher average relative abundance in the endophytic samples (2.022% ± 4.05) compared with epiphytic samples (0.16% ± 0.17; Supplementary Table 5). Trichoderma spp. was considered rare (relative abundance <0.01%) in four epiphytic samples and one endophytic sample, moderately rare (relative abundance between 0.01% and 1%) in 23 epiphytic and 15 endophytic samples, and abundant (relative abundance >1%) in six endophytic samples. Trichoderma spp. were more abundant in endophytic samples than the epiphytic samples, although this difference was not highly significant (p = 0.062; Supplementary Table 6; Supplementary Table 7).
In the past decade, the advancement of amplicon sequencing and other shotgun sequencing methods have made it easier to study the bacterial communities of seagrasses; however, the fungal communities of seagrasses are only recently gaining attention. Recent studies on the endophytic fungi of seagrasses have shown that, like the bacterial communities, they mostly differ between sample types, at least for Z. marina (Ettinger and Eisen, 2019; Ettinger et al., 2021). Studies have also shown that there are differences in the fungal communities of the leaf endosphere compared with the leaf surface (Ettinger and Eisen, 2019). In this study, the epiphytic and endophytic fungi of T. testudinum leaves also differed in both alpha and beta diversity, with a higher diversity found in the endophytic samples. Overall, the fungal classes that have been associated with seagrasses in previous studies include Dothideomycetes, Eurotiomycetes, Sordariomycetes (Supaphon et al., 2013, 2017; Ettinger and Eisen, 2019, 2020; Ettinger et al., 2021). Several of the endophytes that occur in seagrasses have the potential to deter several pathogens through antimicrobial activities, although these were only tested on human pathogens (Supaphon et al., 2013; Notarte et al., 2018). Only a few studies have examined the effects of endophytic marine fungi against diseases in marine plants, but they found that metabolite extracts from marine plants have the potential to deter marine pathogens (Saha and Weinberger, 2019). In this study, we aimed to identify fungal taxa within the leaves that can have the potential to deter or prevent Labyrinthula infections.
Among the top 20 most abundant fungi based on ITS1 amplicon sequencing of T. testudinum leaves, we identified several fungi that can potentially provide antipathogenic benefits to the host. Five of the most abundant genera that were present as both the epiphytes and endophytes have all been previously isolated from seagrasses and shown to produce bioactive compounds against pathogens: Aspergillus (Hu et al., 2022, 2023), Cladosporium (Akmal et al., 2021; Ettinger et al., 2021), Fusarium (Supaphon et al., 2013; Alfattani et al., 2021), Malassezia (Gaitanis et al., 2019; Ettinger et al., 2021), and Trichoderma (Supaphon et al., 2014; Khan et al., 2020). Interestingly, many of the most abundant epiphytic taxa have previously been associated with either marine environments or terrestrial plants, but little is known about their potential in pathogen defense and plant benefits. On the other hand, many of the most abundant endophytic fungi have previously been isolated from sessile marine animals and plants (terrestrial and marine) and were shown to produce antipathogenic compounds (Supplementary Table 3). It is likely that the epiphytic fungal communities are mostly composed of commensal fungi that adhere to the leaves from surrounding environments, and these are usually removed during sterilization for endophyte analysis. Furthermore, four of the fungal genera that displayed bioactivity in the disk diffusion assays (Aspergillus, Trichoderma, Penicillium, and Sarocladium) were part of the most abundant genera in the leaf fungal communities (Supplementary Table 3; Figure 1). This suggests that the fungi that displayed activity against Labyrinthula in vitro might partly control Labyrinthula infections in the field.
Several of the fungal taxa isolated from T. testudinum in this study have previously been isolated from T. testudinum, like Penicillium, Cladosporium, Pestalotiopsis, Trichoderma, and Phoma spp (Mata and Cebrián, 2013). In other seagrass species, such as Zostera marina, similar taxa have been isolated, including members of the order Pleosporales and class Sordariomycetes, and the genera Trichoderma, Penicillium, and Cladosporium (Ettinger and Eisen, 2020). Terrestrial fungal endophytes have been more widely studied for their potential in pharmaceutical and agricultural industries (Aly et al., 2011) and include many of the same taxa identified and isolated here, like Aspergillus, Penicillium, and Phoma spp., which are known to produce antifungal compounds, and Trichoderma which can increase immunity against pathogens (reviewed in Yan et al., 2019). In this study, these taxa, along with Pestalotiopsis, showed moderate activity against Labyrinthula, suggesting that they may also assist in disease defense in marine systems, or at least in seagrasses. Other endophytes of marine origin that displayed activity against protistan pathogens include Penicillium, Phaeosphaeria, Chaetomium, and Phoma spp (Vallet et al., 2018), of which most were found in our dataset, although not all showed bioactivity against Labyrinthula (Supplementary Table 1). Although only a few studies are available on the benefits of fungal endophytes of marine plants and macroalgae, it is becoming clear that, like in terrestrial systems, the fungal communities of seagrasses can play a role in disease defense, and possibly even stress response to abiotic factors.
The two cultures that displayed the most bioactivity against Labyrinthula were M14 and P1a. Based on the results of NCBI BLAST and UNITE blastn, along with their chemotype, P1a is likely Trichoderma and M14 is likely Diaporthe, both of which are well known to have bioactivity against pathogens. Trichoderma spp. are known to sense and overcome other potentially pathogenic fungi and also improve plant health, growth, and nutrient absorption through excreted compounds, which has made them useful in agricultural and horticultural crops such as cucumbers, chrysanthemums, tomatoes, and chickpeas (López-Bucio et al., 2015). In addition to improving plant health, Trichoderma spp. can also provide pathogen protection, including inhibiting the growth of several Pythium spp., which are known oomycete plant pathogens, both in vitro and in planta for both tobacco and cucumber plants (Liu et al., 2022). Trichoderma spp. also produce several secondary metabolites that can inhibit phytopathogenic fungi (reviewed in Khan et al., 2020), and in our case, the isolate P1a (Trichoderma spp.) produced peptaibols, which were likely the bioactive compounds against Labyrinthula. Peptaibols isolated from Trichoderma spp. that presented antipathogenic activity include trichorzins, which displayed antifungal and antibacterial properties (Goulard et al., 1995), and trichorzianines, which inhibit spore germination and hyphal elongation of phytopathogenic fungi (Schirmböck et al., 1994). Other peptaibols isolated from Trichoderma spp. include trichokonin VI, which exhibited activity against the oomycete plant pathogen, Phytophthora parasitica, in addition to pathogenic fungi and bacteria that infect plants (Shi et al., 2012; Khan et al., 2020).
Diaporthe spp. are also known to provide many benefits to the host plant, including producing a variety of antipathogenic compounds and promoting growth and nutrient uptake (reviewed in Hilário and Gonçalves, 2022). Diaporthe spp. (along with its asexual state previously known as Phomopsis spp.) can also potentially deter the oomycete pathogen, Plasmopara viticola, in Endodesmia calphylloides (Talontsi et al., 2012; Hilário and Gonçalves, 2022). This genus also produces diverse secondary metabolites that seem to mostly differ between terrestrial and marine environments (reviewed in Wei et al., 2023). From marine environments, several compounds displayed medicinal potential with antibacterial (e.g., cytochalasin H, isochromopilones, penicilazaphilone D), antifungal (e.g., diaporthelactone), cytotoxic (e.g., phomoxanthone A, diaporisoindole A, phomopsin F, fusaristatin A), anti-inflammatory (e.g., diaporindenes, diaporisoindoles, diaporpenoids), antiviral (e.g., pestalotiopsones), anti-osteoclastogenic (e.g., phomopsichins), antiangiogenic (e.g., cordysinin A), and enzymatic activity (e.g., longidiacid A, longichalasin B; reviewed in Wei et al., 2023). In our dataset, M14 produced cytosporone B, which is likely the main compound inhibiting Labyrinthula spp., as was shown in the assay with commercially purchased cytosporone B. In previous studies, cytosporones E and P, isolated from a marine source (i.e., mangroves), showed activity against the protist, Plasmodium falciparum, which causes malaria (Kornsakulkarn et al., 2015; Xu et al., 2021). Although Plasmodium spp. and Labyrinthula spp. are only distantly related protists, the fact that marine-isolated endophytes can produce anti-protistan compounds is interesting, considering that protists can cause disease in several marine macroalgae and plants (reviewed in Schwelm et al., 2018). Cytosporone B has great potential in the pharmaceutical and medicinal industries as a compound that has shown anticancer activity (i.e., colorectal cancer; Ismaiel et al., 2021), attenuation of tissue grafts (Ding et al., 2022), antiviral activity (i.e., influenza; Egarnes et al., 2017), and inhibition of bacterial pathogens, such as Salmonella enterica (Jianfang et al., 2013). Cytosporone B and derivatives have previously been isolated from endophytes of marine plants, such as mangroves (Huang et al., 2008; Mei et al., 2021; Dankyira et al., 2022), and shown to have antifungal activity against the plant pathogens, Fusarium oxysporum (Huang et al., 2008) and Geotrichum candidum var. citri-aurantii (Yin et al., 2019), as well as other fungi such as Candida albicans (Huang et al., 2008; Dankyira et al., 2022). Cytosporone B can now also be considered an anti-Labyrinthula compound and possibly be useful when investigating potential strategies to mitigate seagrass wasting disease.
The relative abundance of the two most bioactive genera (Diaporthe and Trichoderma) was relatively low in the field samples (less than 1% in both epiphytic and endophytic samples), with the exception of Trichoderma in six samples, where it reached up to 17% (Supplementary Table 7). Furthermore, an exact 100% ID match to the ITS sequence of our strains, M14 and P1a, was not found in our amplicons. Although the bioactive strains were not represented in the ITS amplicon datasets, we cannot say that these bioactive strains are not at all present in the field, but rather we can conclude that these two strains are either rare (relative abundance <0.01%) or moderately rare (relative abundance between 0.01% and 1%; Liang et al., 2020). Moreover, fungi can have varying lengths of ITS regions in their DNA and amplicon sequencing of these regions may be biased against longer sequences (Weig et al., 2013; Bakker, 2018). Nevertheless, low abundance microbes have been recognized as important drivers of microbiome microbial community compositions in several organisms (Benjamino et al., 2018; Compant et al., 2019), and despite the potential rarity of the two most active strains against Labyrinthula growth, we cannot disregard their possible importance in the endospheres of T. testudinum.
Although we determined that certain fungi have the potential to deter Labyrinthula spp. in vitro, it is also important to study the potential of these fungi and their secondary metabolites in planta. Future studies should also consider the impact of abiotic stressors, such as temperature, light, salinity, and nutrient availability, especially because Labyrinthula spp. are opportunistic pathogens that cause seagrass wasting disease when seagrasses are under stress (Jakobsson-Thor et al., 2020).
This is the first study to investigate the relationship between fungal endophytes of T. testudinum and Labyrinthula spp. Through amplicon sequencing, we identified several strains that are known to produce antipathogenic compounds that could potentially aid in the defense against seagrass wasting disease. Through culturing and metabolite extractions, we further identified two fungal strains that had strong activity and 20 strains that had moderate activity against our lab-isolated strain of Labyrinthula. Furthermore, we also determined that four of the top 20 abundant genera showed activity against Labyrinthula in the disk diffusion assays. Although we only tested 50 strains of endophytes, it is important to consider the many fungi that might be present, but were not cultured, might be involved in disease defense against Labyrinthula.
The datasets presented in the study were deposited in GenBank, accession numbers OR355682–OR355766, OR356117, and the NCBI Sequence Read Archive, accession number PRJNA 1001931.
KU: Conceptualization, Methodology, Data curation, Formal analysis, Investigation, Visualization, Writing – original draft. AJ: Formal analysis, Methodology, Visualization, Writing – review & editing. CC: Data curation, Formal analysis, Methodology, Software, Supervision, Writing – review & editing. SL: Writing – review & editing, Conceptualization, Project administration, Supervision. US: Conceptualization, Project administration, Supervision, Writing – review & editing, Methodology, Resources.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the ForEverglades Patrick Lee Fellowship from the Patrick Lee Foundation and the Everglades Foundation. This work was also supported by NSF CH-2020110 (to S.L.) and the German Research Foundation, DFG project number 452319713 (to A.J.).
We thank our interns, Salomon Jean, Kristina Mueller, and Melissa Padron for their assistance with collecting field samples for isolation of the endophytes and Labyrinthula spp., and for culture maintenance. We thank Dr. Seemanti Chakrabarti and Dr. Braham Dhillon for their valuable fungal culturing and DNA extracting advice. We also thank Dr. David Berthold and Dr. Dail Laughinghouse for instruction and guidance on extracting metabolites.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1359610/full#supplementary-material
Abarenkov K., Zirk A., Piirmann T., Pöhönen R., Ivanov F., Nilsson R. H., et al. (2022). UNITE general FASTA release for eukaryotes (UNITE Community). doi: 10.15156/BIO/2483913
Akmal M., Khattak S., Azam N., Shuaib M. (2021). Antifungal Potential of Endophytic Cladosporium sphaerospermum L. isolated from Citrus medica L. Biosci. Res. 18, 805–812.
Alfattani A., Marcourt L., Hofstetter V., Queiroz E. F., Leoni S., Allard P.-M., et al. (2021). Combination of pseudo-LC-NMR and HRMS/MS-based molecular networking for the rapid identification of antimicrobial metabolites from fusarium petroliphilum. Front. Mol. Biosci. 8. doi: 10.3389/fmolb.2021.725691
Aly A. H., Debbab A., Proksch P. (2011). Fungal endophytes: unique plant inhabitants with great promises. Appl. Microbiol. Biotechnol. 90, 1829–1845. doi: 10.1007/s00253-011-3270-y
Bakker M. G. (2018). A fungal mock community control for amplicon sequencing experiments. Mol. Ecol. Resour. 18, 541–556. doi: 10.1111/1755-0998.12760
Benjamino J., Lincoln S., Srivastava R., Graf J. (2018). Low-abundant bacteria drive compositional changes in the gut microbiota after dietary alteration. Microbiome 6, 86. doi: 10.1186/s40168-018-0469-5
Bennett R. M., Honda D., Beakes G. W., Thines M. (2017). Labyrinthulomycota BT - Handbook of the Protists. Eds. Archibald J. M., Simpson A. G. B., Slamovits C. H., Margulis L., Melkonian M., Chapman D. J., et al (Cham: Springer International Publishing), 1–36. doi: 10.1007/978-3-319-32669-6_25-1
Berendsen R. L., Vismans G., Yu K., Song Y., de Jonge R., Burgman W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Berg M., Koskella B. (2018). Nutrient- and dose-dependent microbiome-mediated protection against a plant pathogen. Curr. Biol. 28, 2487–2492.e3. doi: 10.1016/j.cub.2018.05.085
Bibi F., Naseer M. I., Hassan A. M., Yasir M., Al-Ghamdi A. A. K., Azhar E. I. (2018). Diversity and antagonistic potential of bacteria isolated from marine grass Halodule uninervis. 3 Biotech. 8, 48. doi: 10.1007/s13205-017-1066-1
Brady S. F., Wagenaar M. M., Singh M. P., Janso J. E., Clardy J. (2000). The cytosporones, new octaketide antibiotics isolated from an endophytic fungus. Org. Lett. 2, 4043–4046. doi: 10.1021/ol006680s
Capella-Gutiérrez S., Silla-Martínez J., Gabaldón T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carvalhais L. C., Dennis P. G., Badri D. V., Kidd B. N., Vivanco J. M., Schenk P. M. (2015). Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol. Plant-Microbe Interact. 28, 1049–1058. doi: 10.1094/MPMI-01-15-0016-R
Castro-González M., Gómez López D. I., Sánchez Valencia L., Acosta Chaparro A., Coy-Barrera E. (2022). Wasting disease in Thalassia testudinum (Hydrocharitaceae) meadows and its relationship with metabolic profile. Rev. Biol. Trop. 70, 149–172. doi: 10.15517/rev.biol.trop.v70i1.46183
Chepkirui C., Stadler M. (2017). The genus Diaporthe: a rich source of diverse and bioactive metabolites. Mycol. Prog. 16, 477–494. doi: 10.1007/s11557-017-1288-y
Classen A. T., Sundqvist M. K., Henning J. A., Newman G. S., Moore J. A. M., Cregger M. A., et al. (2015). Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: What lies ahead? Ecosphere 6, art130. doi: 10.1890/ES15-00217.1
Compant S., Samad A., Faist H., Sessitsch A. (2019). A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Crump B. C., Wojahn J. M., Tomas F., Mueller R. S. (2018). Metatranscriptomics and amplicon sequencing reveal mutualisms in seagrass microbiomes. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00388
Daniel J.F.de S., Rodrigues Filho E. (2007). Peptaibols of trichoderma. Nat. Prod. Rep. 24, 1128–1141. doi: 10.1039/b618086h
Dankyira D. O., Wang B., Cai J., Zeng W., Huang G.-L., Zheng C.-J. (2022). Antifungal cytosporone derivatives from the mangrove-derived fungus dothiorella sp. ML002. Chem. Nat. Compd. 58, 842–844. doi: 10.1007/s10600-022-03812-9
Daroodi Z., Taheri P., Tarighi S. (2021). Endophytic fungus Acrophialophora jodhpurensis induced resistance against tomato early blight via interplay of reactive oxygen species, iron and antioxidants. Physiol. Mol. Plant Pathol. 115, 101681. doi: 10.1016/j.pmpp.2021.101681
Ding X., Le S., Wang K., Su Y., Chen S., Wu C., et al. (2022). Cytosporone B (Csn-B), an NR4A1 agonist, attenuates acute cardiac allograft rejection by inducing differential apoptosis of CD4+T cells. Int. Immunopharmacol. 104, 108521. doi: 10.1016/j.intimp.2022.108521
Duffin P., Martin D. L., Pagenkopp Lohan K. M., Ross C. (2020). Integrating host immune status, Labyrinthula spp. load and environmental stress in a seagrass pathosystem: Assessing immune markers and scope of a new qPCR primer set. PloS One 15, e0230108. doi: 10.1371/journal.pone.0230108
Durako M. J. (1994). Seagrass die-off in Florida Bay(USA) - Changes in Shoot Demographic Characteristics and Population-Dynamics in Thalassia testudinum. Mar. Ecol. Prog. Ser. 110, 59–66. doi: 10.3354/meps110059
Durako M. J., Kuss K. M. (1994). Effects of labyrinthula infection on the photosynthetic capacity of thalassia testudinum. Bull. Mar. Sci. 54, 727–732.
Egarnes B., Blanchet M.-R., Gosselin J. (2017). Treatment with the NR4A1 agonist cytosporone B controls influenza virus infection and improves pulmonary function in infected mice. PloS One 12, e0186639. doi: 10.1371/journal.pone.0186639
Ettinger C. L., Eisen J. A. (2019). Characterization of the mycobiome of the seagrass, zostera marina, reveals putative associations with marine chytrids. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02476
Ettinger C. L., Eisen J. A. (2020). Fungi, bacteria and oomycota opportunistically isolated from the seagrass, Zostera marina. PloS One 15, e0236135. doi: 10.1371/journal.pone.0236135
Ettinger C., Vann L., Eisen J. (2021). Global diversity and biogeography of the zostera marina mycobiome. Appl. Environ. Microbiol. 87, e02795–e02720. doi: 10.1128/AEM.02795-20
Fadiji A. E., Babalola O. O. (2020). Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioeng. Biotechnol. 8. doi: 10.3389/fbioe.2020.00467
Gaitanis G., Magiatis P., Mexia N., Melliou E., Efstratiou M. A., Bassukas I. D., et al. (2019). Antifungal activity of selected Malassezia indolic compounds detected in culture. Mycoses 62, 597–603. doi: 10.1111/myc.12893
Ghorbanpour M., Omidvari M., Abbaszadeh-Dahaji P., Omidvar R., Kariman K. (2018). Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 117, 147–157. doi: 10.1016/j.biocontrol.2017.11.006
Goulard C., Hlimi S., Rebuffat S., Bodo B. (1995). Trichorzins HA and MA, antibiotic peptides from trichoderma harzianum. J. Antibiot. (Tokyo). 48, 1248–1253. doi: 10.7164/antibiotics.48.1248
Hilário S., Gonçalves M. F. M. (2022). Endophytic diaporthe as promising leads for the development of biopesticides and biofertilizers for a sustainable agriculture. Microorganisms 10. doi: 10.3390/microorganisms10122453
Hou X., Sun R., Feng Y., Zhang R., Zhu T., Che Q., et al. (2022). Peptaibols: Diversity, bioactivity, and biosynthesis. Eng. Microbiol. 2, 100026. doi: 10.1016/j.engmic.2022.100026
Hu Z., Zhu Y., Chen J., Chen J., Li C., Gao Z., et al. (2022). Sesquiterpenoids with phytotoxic and antifungal activities from a pathogenic fungus aspergillus alabamensis. J. Agric. Food Chem. 70, 12065–12073. doi: 10.1021/acs.jafc.2c05703
Hu Z., Zhu Y., Chen J., Chen J., Li C., Gao Z., et al. (2023). Discovery of Novel Bactericides from Aspergillus alabamensis and Their Antibacterial Activity against Fish Pathogens. J. Agric. Food Chem. 71, 4298–4305. doi: 10.1021/acs.jafc.2c09141
Huang Z., Cai X., Shao C., She Z., Xia X., Chen Y., et al. (2008). Chemistry and weak antimicrobial activities of phomopsins produced by mangrove endophytic fungus Phomopsis sp. ZSU-H76. Phytochemistry 69, 1604–1608. doi: 10.1016/j.phytochem.2008.02.002
Ismaiel M., Murphy B., Aldhafiri S., Giffney H. E., Thornton K., Mukhopadhya A., et al. (2021). The NR4A agonist, Cytosporone B, attenuates pro-inflammatory mediators in human colorectal cancer tissue ex vivo. Biochem. Biophys. Res. Commun. 554, 179–185. doi: 10.1016/j.bbrc.2021.03.110
Jakobsson-Thor S., Brakel J., Toth G. B., Pavia H. (2020). Complex interactions of temperature, light and tissue damage on seagrass wasting disease in zostera marina. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.575183
Jianfang L., Chao L., Weiyang S., Zhenyu L., Xiaowei H., Yaoyao L., et al. (2013). Cytosporone B, an inhibitor of the type III secretion system of salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 57, 2191–2198. doi: 10.1128/AAC.02421-12
Jones P., Garcia B. J., Furches A., Tuskan G. A., Jacobson D. (2019). Plant host-associated mechanisms for microbial selection. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00862
Katoh K., Standley D. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Khan R. A., Najeeb S., Hussain S., Xie B., Li Y. (2020). Bioactive Secondary Metabolites from Trichoderma spp. against Phytopathogenic Fungi. Microorganisms 8. doi: 10.3390/microorganisms8060817
Kornsakulkarn J., Somyong W., Supothina S., Boonyuen N., Thongpanchang C. (2015). Bioactive oxygen-bridged cyclooctadienes from endophytic fungus Phomopsis sp. BCC 45011. Tetrahedron 71, 9112–9116. doi: 10.1016/j.tet.2015.10.015
Kurtz J. C., Yates D. F., Macauley J. M., Quarles R. L., Genthner F. J., Chancy C. A., et al. (2003). Effects of light reduction on growth of the submerged macrophyte Vallisneria americana and the community of root-associated heterotrophic bacteria. J. Exp. Mar. Bio. Ecol. 291, 199–218. doi: 10.1016/S0022-0981(03)00120-5
Lahti L., Shetty S., et al. (2017) Tools for microbiome analysis in R. Available online at: http://microbiome.github.com/microbiome.
Liang Y., Xiao X., Nuccio E. E., Yuan M., Zhang N., Xue K., et al. (2020). Differentiation strategies of soil rare and abundant microbial taxa in response to changing climatic regimes. Environ. Microbiol. 22, 1327–1340. doi: 10.1111/1462-2920.14945
Liu Y., He P., He P., Munir S., Ahmed A., Wu Y., et al. (2022). Potential biocontrol efficiency of Trichoderma species against oomycete pathogens. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.974024
Lohan K. M. P., DiMaria R., Martin D., Ross C., Ruiz G. M. (2020). Diversity and microhabitat associations of Labyrinthula spp. in the Indian River Lagoon System. Dis. Aquat. Organ. 137, 145–157. doi: 10.3354/dao03431
López-Bucio J., Pelagio-Flores R., Herrera-Estrella A. (2015). Trichoderma as biostimulant: exploiting the multilevel properties of a plant beneficial fungus. Sci. Hortic. (Amsterdam). 196, 109–123. doi: 10.1016/j.scienta.2015.08.043
Martin D. L., Boone E., Caldwell M. M., Major K. M., Boettcher A. A. (2009). Liquid culture and growth quantification of the seagrass pathogen Labyrinthula spp. Mycologia 101, 632–635. doi: 10.3852/08-171
Martin D. L., Chiari Y., Boone E., Sherman T. D., Ross C., Wyllie-Echeverria S., et al. (2016). Functional, phylogenetic and host-geographic signatures of labyrinthula spp. Provide for putative species delimitation and a global-scale view of seagrass wasting disease. Estuaries Coasts 39, 1403–1421. doi: 10.1007/s12237-016-0087-z
Mata J. L., Cebrián J. (2013). Fungal endophytes of the seagrasses Halodule wrightii and Thalassia testudinum in the north-central Gulf of Mexico. Bot. Mar. 56, 541–545. doi: 10.1515/bot-2013-0047
McMurdie P. J., Holmes S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Mei R.-Q., Nong X.-H., Wang B., Sun X.-P., Huang G.-L., Luo Y.-P., et al. (2021). A new phenol derivative isolated from mangrove-derived fungus Eupenicillium sp. HJ002. Nat. Prod. Res. 35, 4051–4057. doi: 10.1080/14786419.2020.1712388
Muehlstein L. K. (1992). The host-pathogen interaction in the wasting disease of eelgrass, zostera-marina. Can. J. Bot. Can. Bot. 70, 2081–2088. doi: 10.1139/b92-258
Muehlstein L. K., Porter D., Short F. T. (1988). Labyrinthula sp., a marine slime mold producing the symptoms of wasting disease in eelgrass, Zostera marina. Mar Biol. 99, 465–472. doi: 10.1007/BF00392553
Notarte K. I., Yaguchi T., Suganuma K., dela Cruz T. E. (2018). Antibacterial, cytotoxic and trypanocidal activities of marine-derived fungi isolated from Phillippine macroalgae and seagrasses. Acta Bot. Croat. 77, 141–151. doi: 10.2478/botcro-2018-0016
Oksanen J., Blanchet F., Guillaume Friendly M., Kindt R., Legendre P., McGlinn D., et al. (2018) vegan: Community Ecology Package. Available online at: http://cran.r-project.org/package=vegan.
Paradis E., Schliep K. (2019). ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528. doi: 10.1093/bioinformatics/bty633
Qin X., Xu J., An X., Yang J., Wang Y., Dou M., et al. (2023). Insight of endophytic fungi promoting the growth and development of woody plants. Crit. Rev. Biotechnol. 44(1), 1–22. doi: 10.1080/07388551.2022.2129579
R Core Team (2013) R: A language and environment for statistical computing. Available online at: http://www.r-project.org/.
Renn C. E. (1935). A mycetozoan parasite of zostera marina. Nature 135, 544–545. doi: 10.1038/135544b0
Renn C. E. (1936). The wasting disease of zostera marina I. A phytological investigation of the diseased plant. Biol. Bull. 70, 148–158. doi: 10.2307/1537320
Robblee M. B., Barber T. R., Carlson P. R., Durako M. J., Fourqurean J. W., Muehlstein L. K., et al. (1991). Mass mortality of the tropical seagrass thalassia-testudinum in florida bay (USA). Mar. Ecol. Prog. Ser. 71, 297–299. doi: 10.3354/meps071297
Ross C., Puglisi M. P., Paul V. J. (2008). Antifungal defenses of seagrasses from the Indian River Lagoon, Florida. Aquat. Bot. 88, 134–141. doi: 10.1016/j.aquabot.2007.09.003
Saha M., Weinberger F. (2019). Microbial “gardening” by a seaweed holobiont: Surface metabolites attract protective and deter pathogenic epibacterial settlement. J. Ecol. 107, 2255–2265. doi: 10.1111/1365-2745.13193
Schirmböck M., Lorito M., Wang Y. L., Hayes C. K., Arisan-Atac I., Scala F., et al. (1994). Parallel formation and synergism of hydrolytic enzymes and peptaibol antibiotics, molecular mechanisms involved in the antagonistic action of Trichoderma harzianum against phytopathogenic fungi. Appl. Environ. Microbiol. 60, 4364–4370. doi: 10.1128/aem.60.12.4364-4370.1994
Schwelm A., Badstöber J., Bulman S., Desoignies N., Etemadi M., Falloon R. E., et al. (2018). Not in your usual Top 10: protists that infect plants and algae. Mol. Plant Pathol. 19, 1029–1044. doi: 10.1111/mpp.12580
Shi M., Chen L., Wang X.-W., Zhang T., Zhao P.-B., Song X.-Y., et al. (2012). Antimicrobial peptaibols from Trichoderma pseudokoningii induce programmed cell death in plant fungal pathogens. Microbiology 158, 166–175. doi: 10.1099/mic.0.052670-0
Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steele L., Caldwell M., Boettcher A. (2005). Seagrass-pathogen interactions: “pseudo-induction” of turtlegrass phenolics near wasting disease lesions. Mar. Ecol. Prog. Ser. 303, 123–131. doi: 10.3354/meps303123
Sullivan B. K., Robinson K. L., Trevathan-Tackett S. M., Lilje E. S., Gleason F. H., Lilje O. (2017). The first isolation and characterisation of the protist labyrinthula sp. in southeastern Australia. J. Eukaryot. Microbiol. 64, 504–513. doi: 10.1111/jeu.12387
Supaphon P., Phongpaichit S., Rukachaisirikul V., Sakayaroj J. (2013). Antimicrobial Potential of Endophytic Fungi Derived from Three Seagrass Species: Cymodocea serrulata, Halophila ovalis and Thalassia hemprichii. PloS One 8, e72520. doi: 10.1371/journal.pone.0072520
Supaphon P., Phongpaichit S., Rukachaisirikul V., Sakayaroj J. (2014). Diversity and antimicrobial activity of endophytic fungi isolated from the seagrass Enhalus acoroides. Indian J. Geo-Marine Sci. 43, 785–797.
Supaphon P., Phongpaichit S., Sakayaroj J., Rukachaisirikul V., Kobmoo N., Spatafora J. W. (2017). Phylogenetic community structure of fungal endophytes in seagrass species. Bot. Mar. 60, 489–501. doi: 10.1515/bot-2016-0089
Talontsi F. M., Islam M. T., Facey P., Douanla-Meli C., von Tiedemann A., Laatsch H. (2012). Depsidones and other constituents from Phomopsis sp. CAFT69 and its host plant Endodesmia calophylloides with potent inhibitory effect on motility of zoospores of grapevine pathogen Plasmopara viticola. Phytochem. Lett. 5, 657–664. doi: 10.1016/j.phytol.2012.06.017
Trevathan-Tackett S. M., Lane A. L., Bishop N., Ross C. (2015). Metabolites derived from the tropical seagrass Thalassia testudinum are bioactive against pathogenic Labyrinthula sp. Aquat. Bot. 122, 1–8. doi: 10.1016/j.aquabot.2014.12.005
Trevathan-Tackett S. M., Lauer N., Loucks K., Rossi A. M., Ross C. (2013). Assessing the relationship between seagrass health and habitat quality with wasting disease prevalence in the Florida Keys. J. Exp. Mar. Bio. Ecol. 449, 221–229. doi: 10.1016/j.jembe.2013.10.004
Vallet M., Strittmatter M., Murúa P., Lacoste S., Dupont J., Hubas C., et al. (2018). Chemically-mediated interactions between macroalgae, their fungal endophytes, and protistan pathogens. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03161
van Santen J. A., Jacob G., Singh A. L., Aniebok V., Balunas M. J., Bunsko D., et al. (2019). The natural products atlas: an open access knowledge base for microbial natural products discovery. ACS Cent. Sci. 5, 1824–1833. doi: 10.1021/acscentsci.9b00806
Vilgalys R., Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wang F., Liigand J., Tian S., Arndt D., Greiner R., Wishart D. S. (2021). CFM-ID 4.0: more accurate ESI-MS/MS spectral prediction and compound identification. Anal. Chem. 93, 11692–11700. doi: 10.1021/acs.analchem.1c01465
Wei W., Khan B., Dai Q., Lin J., Kang L., Rajput N. A., et al. (2023). Potential of secondary metabolites of diaporthe species associated with terrestrial and marine origins. J. Fungi 9. doi: 10.3390/jof9040453
Weig A. R., Peršoh D., Werner S., Betzlbacher A., Rambold G. (2013). Diagnostic assessment of mycodiversity in environmental samples by fungal ITS1 rDNA length polymorphism. Mycol. Prog. 12, 719–725. doi: 10.1007/s11557-012-0883-1
White T., Bruns T., Lee S., Taylor J. (1990). Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. (Academic Press, Inc). doi: 10.1016/B978-0-12-372180-8.50042-1
Wickham H. (2009). ggplot2: elegant graphics for data analysis. J. Stat. Softw 35, 65–88. doi: 10.1007/978-0-387-98141-3
Wickham H., François R., Henry D., Müller L., Vaughan K. (2023) dplyr: A Grammar of Data Manipulation. Available online at: https://dplyr.tidyverse.org.
Xu T.-C., Lu Y.-H., Wang J.-F., Song Z.-Q., Hou Y.-G., Liu S.-S., et al. (2021). Bioactive secondary metabolites of the genus diaporthe and anamorph phomopsis from terrestrial and marine habitats and endophytes: 2010–2019. Microorganisms 9. doi: 10.3390/microorganisms9020217
Yan L., Zhu J., Zhao X., Shi J., Jiang C., Shao D. (2019). Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 103, 3327–3340. doi: 10.1007/s00253-019-09713-2
Yin C., Liu H., Shan Y., Gupta V. K., Jiang Y., Zhang W., et al. (2019). Cytosporone B as a Biological Preservative: Purification, Fungicidal Activity and Mechanism of Action against Geotrichum citri-aurantii. Biomolecules 9. doi: 10.3390/biom9040125
Keywords: seagrass, Thalassia, metabolites, fungi, Labyrinthula, endophytes
Citation: Ugarelli K, Jagels A, Choi CJ, Loesgen S and Stingl U (2024) Fungal endophytes from Thalassia testudinum show bioactivity against the seagrass pathogen, Labyrinthula spp.. Front. Mar. Sci. 11:1359610. doi: 10.3389/fmars.2024.1359610
Received: 21 December 2023; Accepted: 19 February 2024;
Published: 11 March 2024.
Edited by:
Alexander Eiler, University of Oslo, NorwayReviewed by:
Brooke K. Sullivan, Northeastern University, United StatesCopyright © 2024 Ugarelli, Jagels, Choi, Loesgen and Stingl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ulrich Stingl, dXN0aW5nbEB1ZmwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.