- Population Genomics Laboratory, School of Biological Sciences, College of Natural Sciences, Seoul National University, Seoul, Republic of Korea
Introduction: This study undertakes a detailed examination of the host range and distribution patterns of Rhizocephala, a group of specialized parasitic barnacles, within Korean marine environments. It aims to expand the understanding of their biodiversity and ecological impact.
Methods: Employing both morphological observation and mitochondrial DNA sequencing, the study identifies the variety of rhizocephalan species infecting decapod hosts in Korean waters, aiming to document their diversity and distribution.
Results: The investigation identifies 26 rhizocephalan species parasitizing 28 decapod hosts, including the discovery of three new parasite-host relationships not previously reported. It was observed that while a few rhizocephalan species infect multiple hosts, most have a narrow host range. Additionally, the study maps out regional differences in Rhizocephala diversity across three Korean marine ecoregions, noting the highest diversity in the warmer East China Sea ecoregion and lower diversity in the colder Yellow Sea and East Sea ecoregions.
Discussion: The findings underscore the necessity for continued taxonomic research on Rhizocephala and their host species to better understand and manage these parasites, which hold significant ecological and economic importance. The study suggests that geographical variations in host-parasite relationships are influenced by regional climatic conditions, particularly winter water temperatures.
1 Introduction
Rhizocephala is a taxon of specialized parasitic barnacles that infect a variety of crustacean hosts. They are mainly parasites on decapods, and some rhizocephalans infect commercial decapod species such as king crabs and swimming crabs (Noever et al., 2016; Yang et al., 2018). Rhizocephala has the potential to act as an invasive species and harm the ecological function of native decapod hosts. One of the rhizocephalans, Loxothylacus panopaei (Gissler, 1884), native to the Gulf of Mexico and Florida, has already invaded Cheshire Peak Bay and New York and is harming the ecological function of its host, Eurypanopeus depressus (Smith, 1869), which exerts top-down control in oyster reefs (Hines et al., 1997; O'Shaughnessy et al., 2014). Many previous studies have reported biological characteristics of Rhizocephala such as their complex life cycle, the body size correlation between the parasite and the host, and biological effects on their hosts (feminization, growth and survival rate, etc.) (Ritchie & Høeg, 1981; Alvarez et al., 1995; Kristensen et al., 2012; Nagler et al., 2017). A comprehensive understanding of the host range and distribution of Rhizocephala is fundamental to effective control of their prevalence on commercial decapod species and invaded areas and the biological control of invasive and harmful decapods through their parasitic barnacles (Boyko & Williams, 2016).
Nevertheless, the distribution and host relationships of rhizocephalans are largely unknown. Recently, a few studies have contributed to elucidating the distribution and host range of Rhizocephala in Japan and Korea (Yoshida et al., 2014; Jung et al., 2019, 2021). However, these studies, while pioneering, were constrained by their limited examination of specific areas or a few of the host taxa. Yoshida et al. (2014) focused on the limited paguroids (hermit crab) hosts in the limited area of southern Honshu (Japan). While Jung et al. (2019) expanded this research to the entire Korean coast, however, the host taxon in this study was also limited to Paguroidea. Jung et al. (2021) investigated the interaction of Korean Rhizocephala with Brachyura (crab) and Gebiidea (mud shrimp) hosts, however, this study has a limitation in that it mostly focused on the southern Korean Peninsula. As the host specificity of Rhizocephala is expected to vary depending on geographic location (Jung et al., 2019; 2021), a more expansive and inclusive study is required to fully elucidate the relationship between Rhizocephala and their hosts.
To fill the gap in our knowledge, an extensive examination of three host taxa—Gebiidea, Anomura, and Brachyura—sampled from 28 locations representing three marine ecoregions of (Kim et al., 2020; 2022) was performed, building upon previous work of Jung et al. (2019; 2021)1. Due to the highly degenerated morphological features of rhizocephalan species (Høeg, 1992; Høeg & Lützen, 1995; Øksnebjerg, 2000) that often make species identification very challenging, the author also determined mtDNA cox1 sequences as molecular evidence of their correct identification. Afterward, the decapod host and distribution of each identified Rhizocephala were confirmed based on the three Korean coast ecoregions, i.e., Yellow Sea, East China Sea, and East Sea (Kim et al., 2020; 2022). The author analyzed the relationship between Rhizocephala and its host and the diversity of Rhizocephala in each marine ecoregion to confirm trends and special features of the results. Through this integrative approach, this study concluded that the relationship with its host and the distribution of Rhizocephala might have been influenced by differences in geographical factors such as water temperature.
2 Materials and methods
2.1 Taxon sampling, morphological and molecular identifications
The author examined 173 Korean Rhizocephalans from 18 host decapod species collected from 21 sampling sites in Korea (Figure 1A). Voucher specimens were deposited in the National Institute of Biological Resources (NIBR), the National Marine Biodiversity Institute of Korea (MABIK), and the Honam National Institute of Biological Resources (HNIBR). All rhizocephalan specimens were fixed in 70-95% ethanol and subjected to morphological examination and molecular analysis. For morphological analysis, the externa and mantle were examined using an MZ8 dissection microscope (Leica, Wetzlar, Germany).
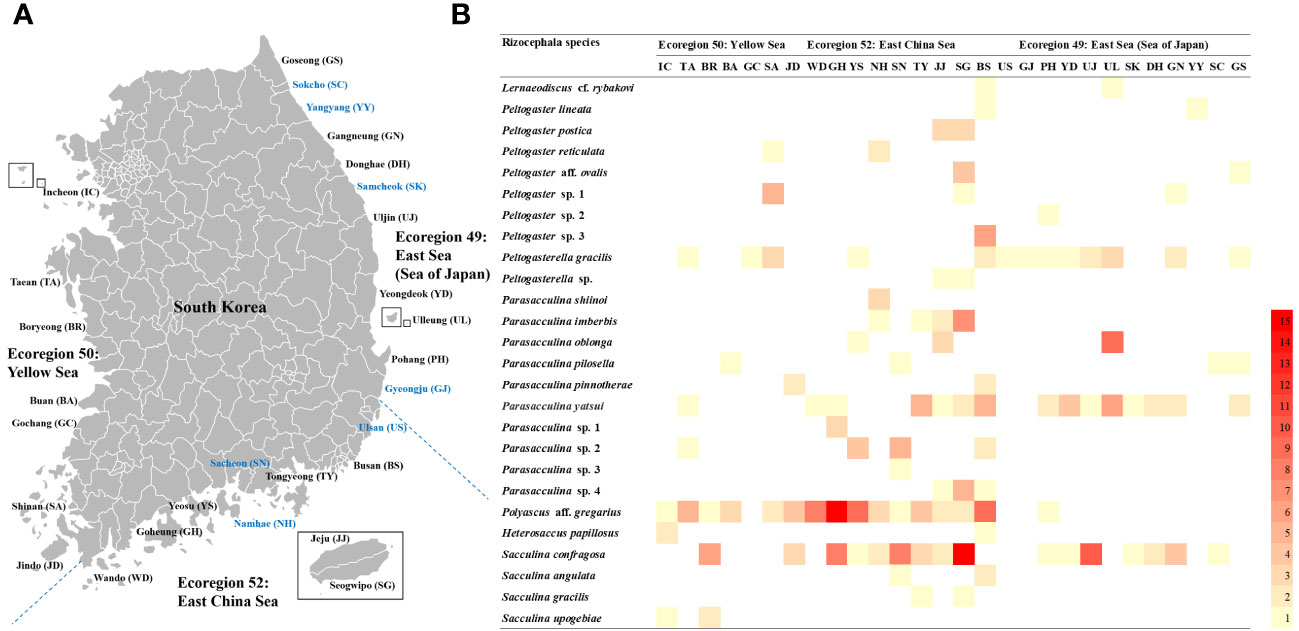
Figure 1 Map and heatmap showing the collection sites and density of the Korean rhizocephalan species. (A) the map showing 28 collection sites of the Korean rhizocephalans modified from Jung et al. (2019; 2021)1, 21 sites in black letters: investigated in this study, 7 sites in blue letters: investigated only in previous studies; (B) the heatmap showing the density of 26 Korean rhizocephalans per each 28 collection sites.
For molecular analysis, the lateral end of the externa tissue of each rhizocephalan specimen was excised for total genomic DNA extraction using the QIAamp DNA Micro Kit (QIAGEN, Hilden, Germany). Universal primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) were used to amplify a fragment of mitochondrial cytochrome c oxidase subunit I (cox1) (Folmer et al., 1994). Polymerase chain reaction (PCR) was performed in reaction volumes of 50 µL that included 2 µL DNA template, 5 µL 10 x Ex Taq Buffer, 2 µL of each primer (10 µM), 0.25 µL Go Taq DNA polymerase (Promega, Madison City, WI, USA), 2.5 µL dNTP mix (10 mM), and 35.75 µL distilled H2O. PCR amplification was performed using the following steps: 5 min denaturation at 94°C followed by 35 cycles of 30 sec at 94°C, 1 min at 52°C, 1 min at 72°C, and a final extension of 7 min at 72°C. PCR products were visualized on 1% agarose gels and sequenced with an ABI PRISM 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). Nucleotide sequences of mtDNA cox1 fragments were analyzed and edited using Geneious v. 9.1.8 (Kearse et al., 2012) and aligned using ClustalW in the MEGA10 program (Kumar et al., 2018). The mtDNA cox1 sequences of the Korean rhizocephalan species were deposited on GenBank (OR481955-OR482064). Thirty-six cox1 and 16S rRNA rhizocephalan sequences were downloaded from GenBank and included in further phylogenetic analyses (Supplementary Table 1).
Phylogenetic relationships among rhizocephalan species were inferred using maximum likelihood of RaxML version 8 (Stamatakis, 2014) and Bayesian inference of MrBayes version 3.2.6 (Ronquist et al., 2012), respectively. Maximum likelihood analyses of cox1 sequences were performed using the general time reversible substitution model (Tavaré, 1986), with a gamma distribution (+G) and invariable sites (+I) rate categories based on Bayesian Information Criterion (BIC) scores model using the Model Selection option of MEGA10 (Kumar et al., 2018). The Bayesian phylogenetic analysis was performed using the Markov chain Monte Carlo (MCMC) method, with two independent runs of 1 × 106 generations with six chains, sampling every 100 generations. Bayesian posterior probability (BPP) values were estimated after discarding the 2 × 103 initial trees as burn-in that reached a stationary stage of the parameters. The branch support in the ML trees was assessed by analysis of 1,000 bootstrap replicates. Interspecific and intraspecific sequence divergences were estimated based on the Kimura 2-parameter (K2P) distance matrix in MEGA10.
2.2 Analysis of host-parasite relationships (decapod-rhizocephalan species) and their distribution
To elucidate the ecological characteristics of Korean Rhizocephala, especially their distribution and relationships between Rhizocephala and their hosts, the author analyzed data of rhizocephalan species from 28 species and 11,016 individuals of decapod host across 28 sampling sites. This dataset includes both current study data and information from previous studies in Korea (Jung et al., 2019; 20211). Vertical bar charts were generated to depict the frequency of decapod hosts infected by rhizocephalans. Furthermore, the author investigated the correlation between the distribution of rhizocephalans and their hosts, focusing on biogeographic patterns in the three Korean coast ecoregions, i.e., Yellow Sea, East China Sea, and East Sea (Kim et al., 2020; 2022).
3 Results
3.1 Sequence divergence and phylogenetic relationships among Korean rhizocephalan species
Because this sampling targeted the 4 families (Peltogasterellidae, Peltogastridae, Polyascidae, and Sacculinidae) which can cover the majority of whole Korean Rhizocephalan fauna, phylogenetic analysis in this study was focused on relationships among rhizocephalan species belonging to these four families. A total of 110 mtDNA cox1 partial sequences (497 bp) were used for phylogenetic analysis, and the resulting phylogenies from maximum likelihood and Bayesian methods were generally consistent with each other in that four rhizocephalan families were monophyletic respectively, except Sacculina gracilis Boschma, 1931 is positioned in the Polyascidae clade (Figure 2). In the inferred phylogenetic tree, the sequences of Korean rhizocephalan species were nested and/or clustered with their congeneric species retrieved from GenBank (Figure 2). The individual sequence variations within rhizocephalan species were relatively low ranging from 0% (in S. gracilis, Parasacculina sp. 1) to 1.4% (in Sacculina confragosa Boschma, 1933), compared to interspecific differences [a minimum sequence divergence of 4.6% found between Polyascus aff. gregarius and Polyascus planus (Boschma, 1933)] (Supplementary Table 2). One notable exception to low within-species variation was found in P. aff. gregarius (Figure 2) in which sequence variation ranged from 4.6% to 5.7% between Korean and Chinese samples. From this phylogenetic analysis, the author discovered unidentified species (Peltogasterella sp., Peltogaster sp. 1, and Parasacculina sp. 1) of previous study (Jung and Park, in review) were separated from their congeneric species (Figure 2), such as Peltogasterella gracilis (Boschma, 1927), Peltogaster postica Yoshida & Osawa in Yoshida, Osawa, Hirose & Hirose, 2011, and Parasacculina yatsui (Boschma, 1936) and P. aff. gregarius having the same family of hosts and similar morphological features (Figure 2). Interspecific sequence differences of these three unidentified species from their congeneric species ranged from 18.5 to 35.8%, more or less comparable to an average distance (23.9%) of the congeneric comparison. By contrast, intraspecific variations of unidentified species were very low, from 0% (Parasacculina sp. 1) to 1.2% (in Peltogaster sp.). Unexpectedly, the phylogenetic tree depicted the family Sacculinidae as non-monophyletic because S. gracillis sequences (OR482063, OR482064) were not grouped with all the other Sacculinidae species (including Sacculina species), but instead with some Parasacculina species (including Parasacculina shiinoi (Lützen, Itani, Jespersen, Hong, Rees & Glenner, 2016), Parasacculina sp. 1, and Parasacculina sp. 3), (Figure 2A). Note that relationships at the deep branches (i.e., higher than generic level) are not strongly supported by bootstrap values (mostly ≤50%). These unresolved relationships at higher taxonomic levels are likely due to the poor resolution of mtDNA cox1 partial sequences as a molecular marker for deeper node Rhizocepala phylogeny. In the maximum likelihood phylogenetic tree of Rhizocephala combining mtDNA cox1 and 16S rRNA, deep branches were strongly supported by high bootstrap values (Figure 2B). Indeed, sequence divergences between S. gracillis and Parasacculina species belonging to different families, ranging from 21.0% to 31.5%, are not noticeably higher than that detected between congeneric species (S. gracillis and other Sacculina species) ranging from 28.1% to 32.4%.
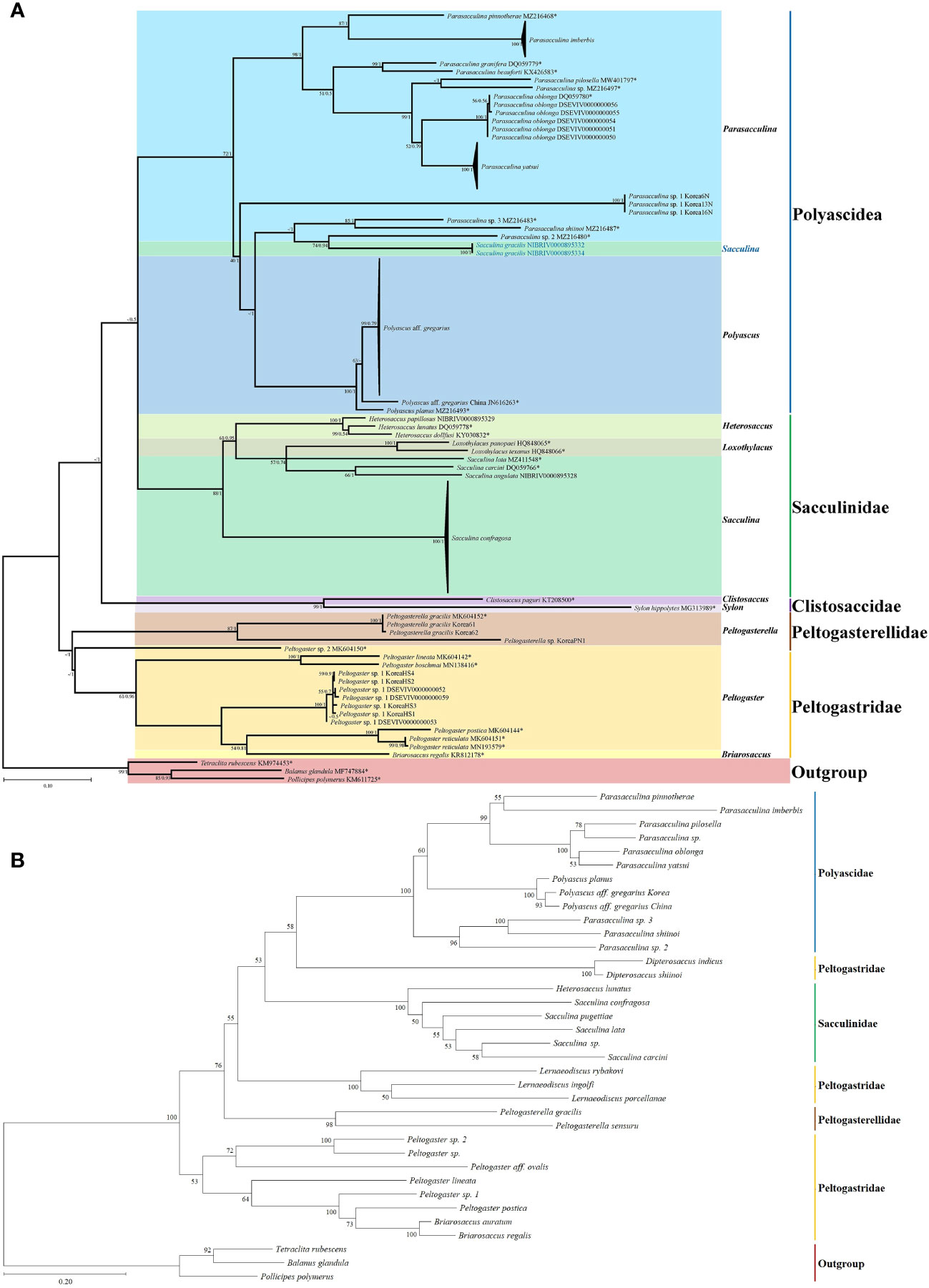
Figure 2 Phylogenetic relationships among Korean rhizocephalan species. Maximum likelihood and Bayesian inference methods were used to reconstruct phylogenetic tree using the mtDNA sequence data. Numerical values on each node indicate maximum likelihood bootstrap support/Bayesian posterior probability. Multiple sequences representing Parasacculina imberbis, Parasacculina yatsui, Polyascus aff. gregarius, and Sacculina confragosa, are presented as black triangles. Sequences of Sacculina gracilis that assumed to be belonging to the genus Parasacculina are indicated in blue. *: sequences derived from GenBank. (A) cox1 maximum likelihood and Bayesian inference tree; (B) cox1 and 16S rDNA maximum likelihood tree.
3.2 Host-parasite relationships between rhizocephalans, decapod hosts, and their distribution
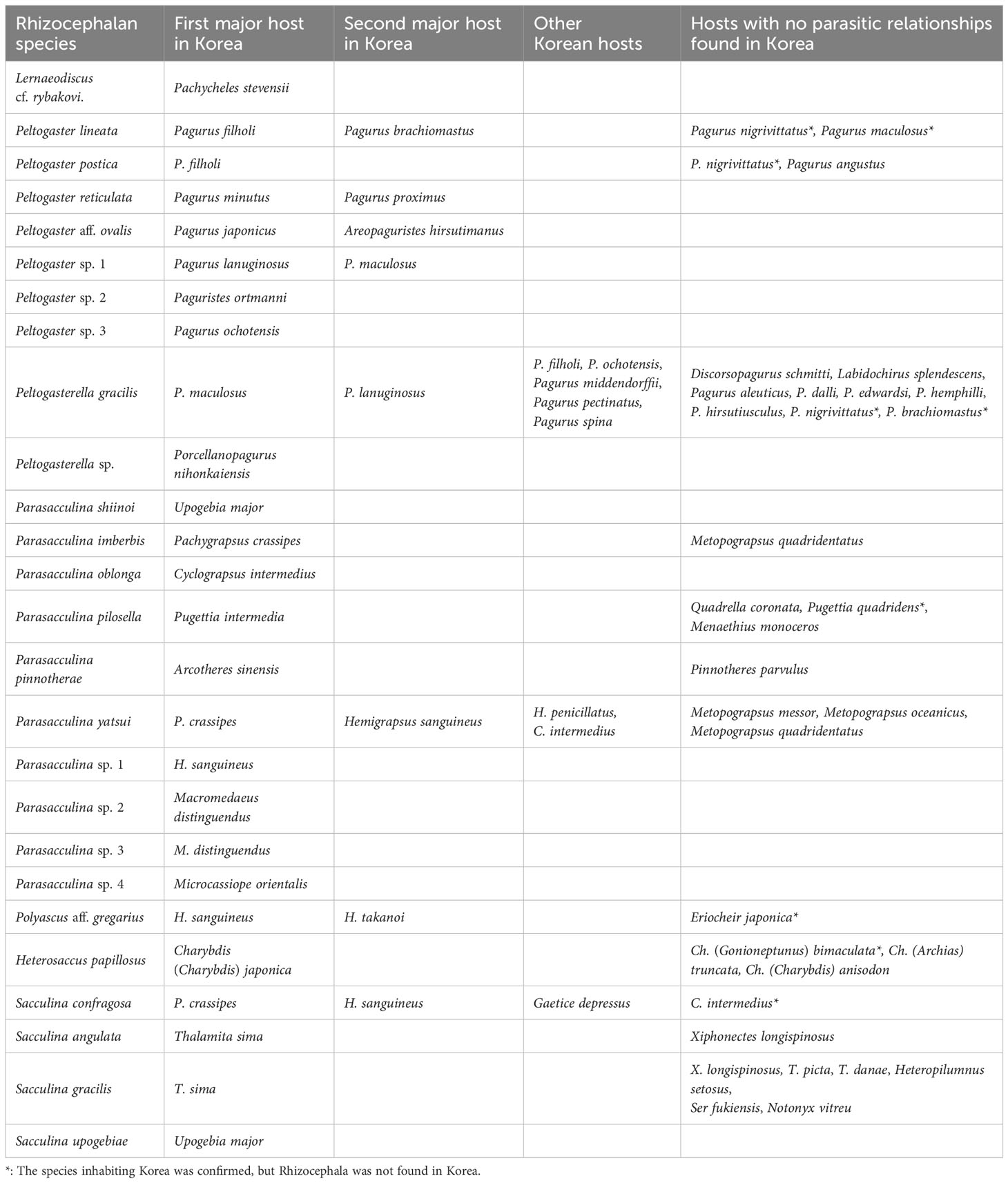
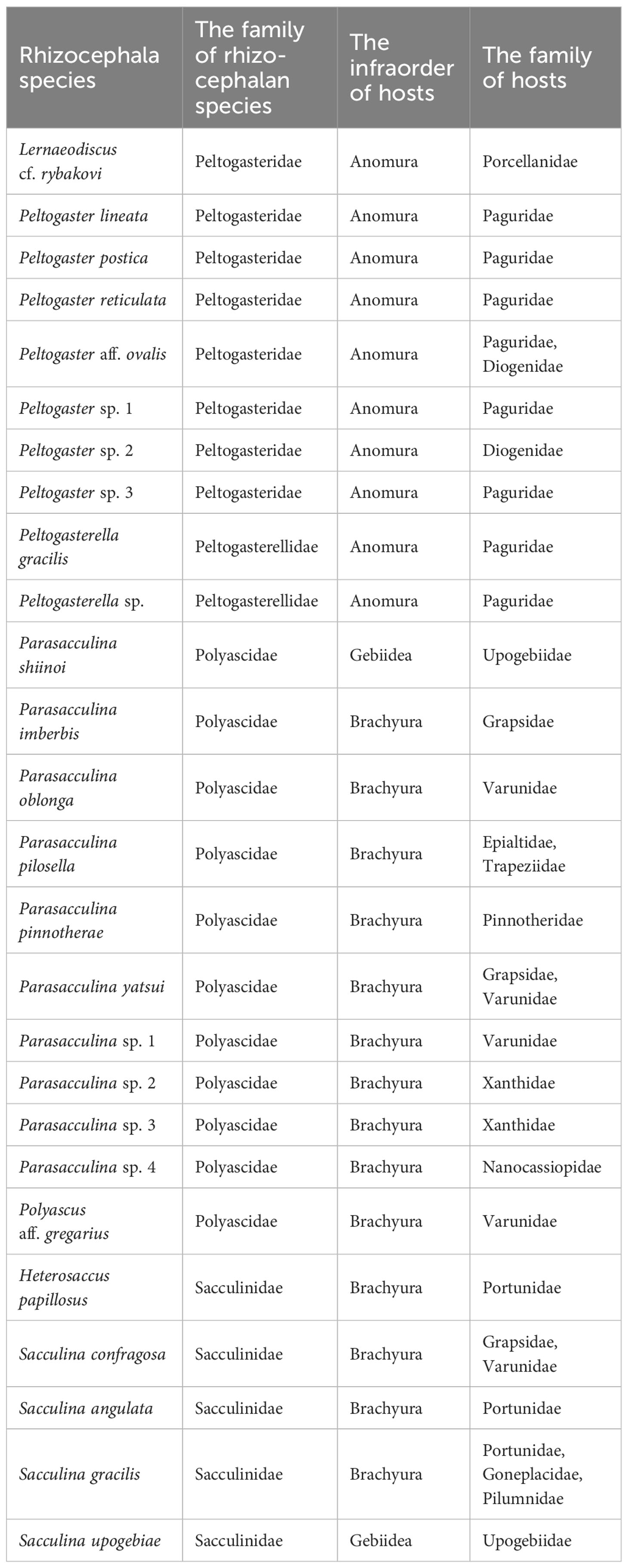
Among all 28 species and 11,016 individuals of the investigated decapod host, 2.69% (296 individuals) were infected by Rhizocephala (Table 1). Combining the data on host-parasite association of the present study with results from the previous study (Jung et al., 2019; 2021)1 provides updated, synthetic information on the relationships between 26 parasitic barnacle species and 28 decapod host species discovered in Korea (Figure 3, Table 2). From this, the author newly found four host-parasite relationships that were not previously reported from Korean rhizocephalan species. Microcassiope orientalis Takeda & Miyake, 1969 was infected by Parasacculina sp. 4, Pachycheles stevensii Stimpson, 1858 was infected by Lernaeodiscus cf. rybakovi, and Areopaguristes hirsutimanus (Kobjakova, 1971) was infected by Peltogaster aff. ovalis. Pagurus ochotensis Brandt, 1851 was found infected by P. gracilis and an unknown Peltogaster sp. 3 (Figure 3A, Table 2). Based on the host-parasite associations updated from this study, it turns out that three rhizocephalan species (P. gracillis, P. yatsui, and S. confragosa) were found from more than three host species, consistent with previous studies (Boschma, 1955b; Tsuchida et al., 2006; Yoshida et al., 2014) and 23 species were found in one or two host species (Figure 3A, Table 2). Among these 23 rhizocephalans, five species [Peltogaster lineata Shiino, 1943, P. postica, Parasacculina pilosella (Van Kampen & Boschma, 1925), Heterosaccus papillosus (Boschma, 1933), and S. gracilis] were previously reported to use more than 3 host species in Thailand, Singapore, Indonesia, Philippines, China, and Japan (Boschma, 1955a; b; Yoshida et al., 2014). The majority of host hermit crabs in Korea were infected by one or two rhizocephalan species, while Pagurus filholi (De Man, 1887), Hemigrapsus sanguineus (De Haan, 1835), and Pachygrapsus crassipes Randall, 1840 were infected by more than three rhizocephalan species (Figure 3B). Extending the host-parasite association to higher taxonomic level, the host range of the rhizocephalan families is correlated with the infraorder level of host hermit crabs: The family Peltogastridae and Peltogasterellidae rhizocephalan species parasitize the Anomura species and the Sacculinidae and Polyascidae species were found in the Brachyura and Gebiidea species (Table 3).
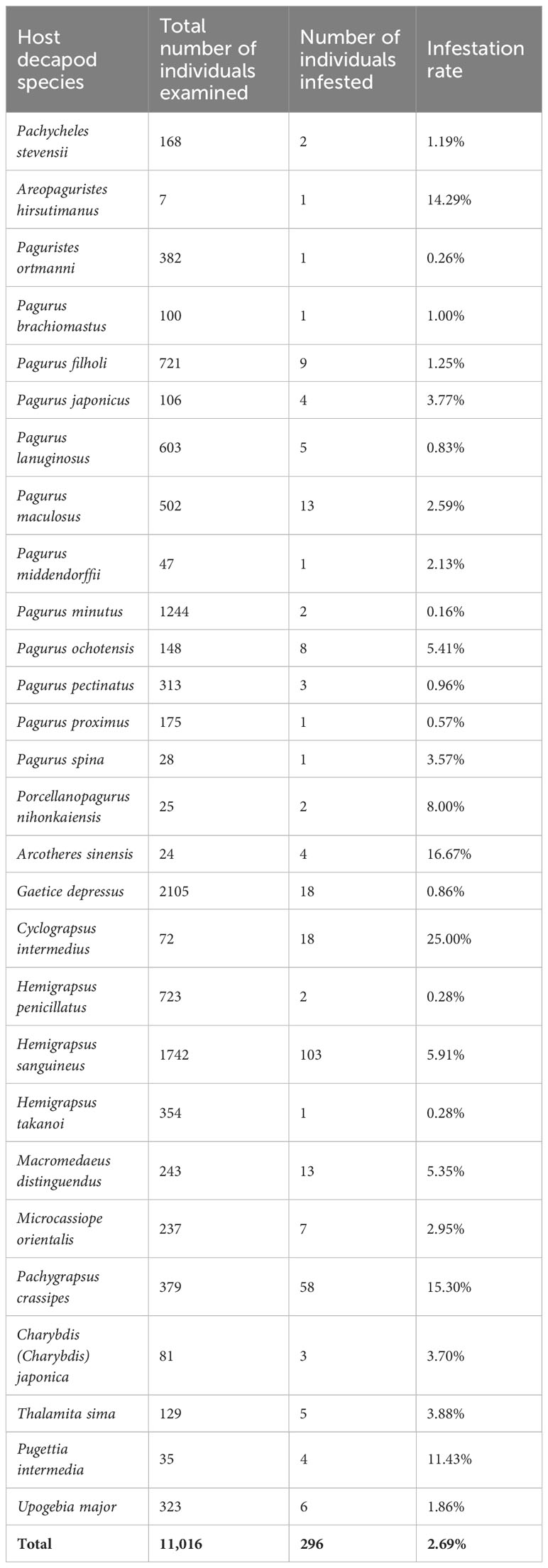
Table 1 Number of individuals and infestation rate (%) of 28 Korean decapod hosts by Rhizocephala as examined in this study and previous studies (Jung et al., 2019; 2021)1.

Figure 3 A vertical bar chart showing the correlation of 26 Korean rhizocephalan species and their 28 decapod hosts. (A) decapod host per Rhizocephala; (B) Rhizocephala per decapod host.
To analyze the frequency of occurrence of rhizocephalan species and their host species in different locations and marine ecoregions in Korea, a total of 26 rhizocephalan species and 28 host species were examined in 28 locations and three marine ecoregions (Supplementary Table 3). In the Yellow Sea ecoregion, 46 individuals of 11 rhizocephalan species were found from 10 decapod host species. Notably, Sacculina upogebiae Shiino, 1943 was exclusively found in this ecoregion. In the East China Sea ecoregion, 182 individuals representing 24 species of Rhizocephala were found from 21 host species. Among them, 10 rhizocephalan species were found solely in this region. In the East Sea ecoregion, 68 individuals of 11 rhizocephalan species were found to infect 17 host species. Peltogaster sp. 2 was specifically found only in the East Sea ecoregion. Out of the 26 rhizocephalan species, six species were found in common from three ecoregions. Notably, P. gracilis, P. yatsui, P. aff. gregarius, and S. confragosa had the highest occurrence rate among these species (Figure 1B). However, P. aff. gregarius was not observed in the upper East Sea ecoregion. Furthermore, the author examined the relationship between Rhizocephala and host species that appeared in only one ecoregion, differentiating it from the case of the rhizocephalan species that appeared in two or more ecoregions. As a result, a specific relationship was found in one instance in the Yellow Sea ecoregion, five instances in the East China Sea ecoregion, and seven instances in the East Sea ecoregion (Table 4). Despite being present in multiple ecoregions, six decapod host species were found to be infected by six or more individuals of a single Rhizocephalan species in only one specific ecoregion (Table 4). For instance, Gaetice depressus (De Haan, 1833) was found to be infected with S. confragosa exclusively in the East China Sea ecoregion, despite that G. depressus is common in all three marine ecoregions of Korea.
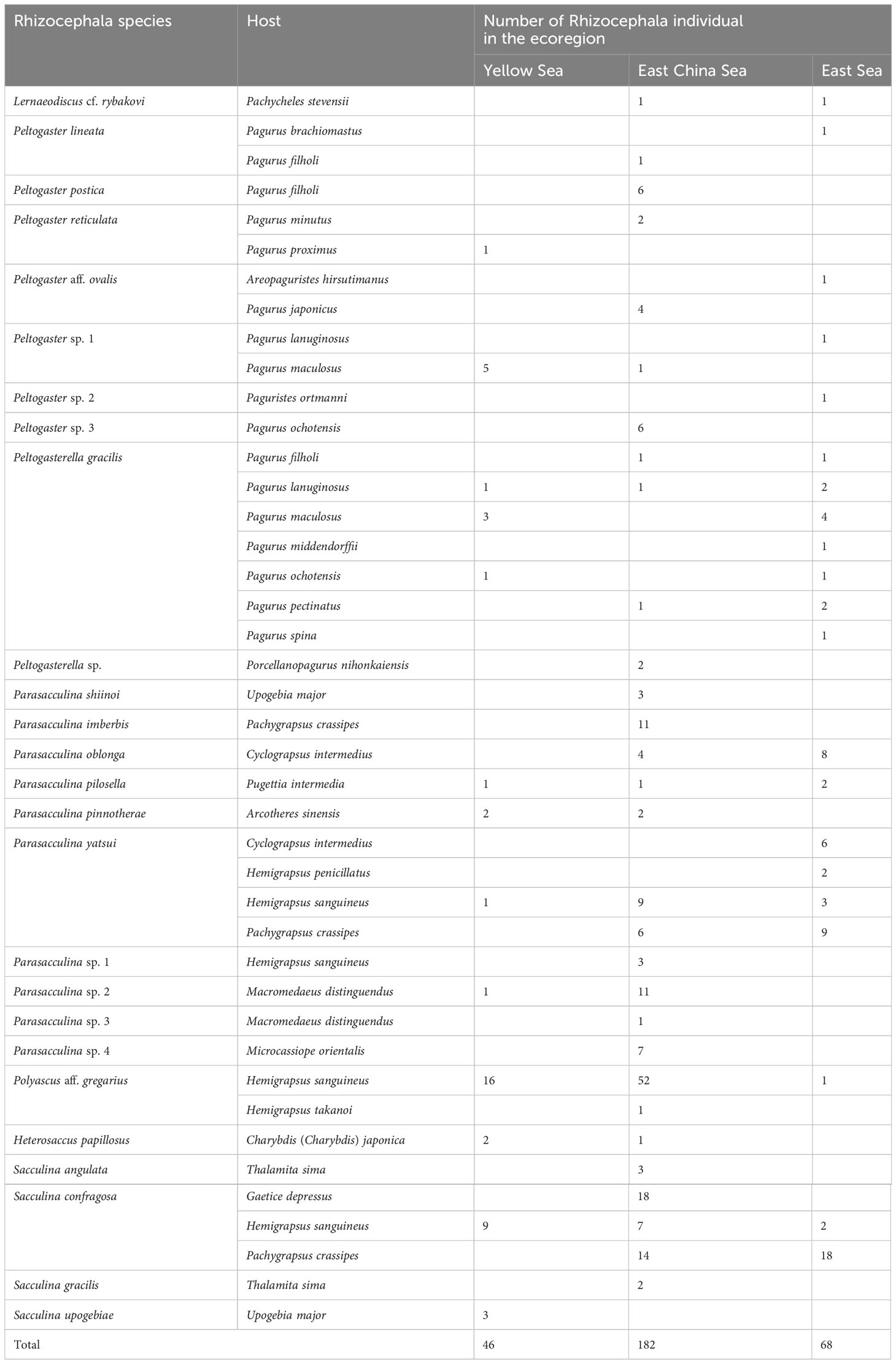
Table 4 Relationships between Rhizocephala species and host appear in each three Korean marine ecoregions.
4 Discussion
4.1 The overview of the taxonomy and phylogenetic relationships of Korean rhizocephalan
Morphological and molecular analyses provided insights into the taxonomy and phylogenetic relationships of Korean rhizocephalan species in this study. Phylogenetic trees recognized four rhizocephalan families, i.e., Polyascidae, Sacculinidae, Peltogastridae, and Peltogasterellidae, consistent with previous molecular analysis (Høeg et al., 2019; Jung et al., 2019; 2021). Three unidentified species (Lernaeodiscus cf. rybakovi, Peltogaster sp. 3, and Parasacculina sp. 4) were distinguished from their congeneric species based on their hosts and their morphological characteristics. In addition, the author assumed that P. aff. gregarius with different geographic origins (i.e., Korea and China) might represent two different cryptic species because they have different host species and high sequence divergence ranging from 4.6% to 5.7% in molecular analysis (Supplementary Table 2, Figure 2).
4.2 The characteristics of host-parasite relationships between Korean rhizocephalan and decapod hosts
From a combined analysis of host-parasite association of 26 rhizocephalans and 28 host decapods including previous studies, this study revealed that Korean rhizocephalan species have different patterns in their host range, compared to the previously reported in Japan (Tsuchida et al., 2006; Yoshida et al., 2014). Three unreported relationships between Rhizocephala and the host, i.e., M. orientalis and Parasacculina sp. 4, A. hirsutimanus and P. aff. ovalis, and P. ochotensis and Peltogaster sp. 3, were found in this study. The author also found 23 rhizocephalan species that show high host specificity, parasitizing only one or two host species in Korea (Figure 3A, Table 2). In general, parasites are highly dependent on the availability and suitability of their hosts which is chance-dependent in their early establishment of host-parasite association. In case of S. gracilis, compared to other geographic areas (Thailand, Indonesia, Singapore, Philippines, and China; see Boschma, 1955a) where relatively rich data resources of host candidates have ever known [e.g., Xiphonectes longispinosus (Dana, 1852), Thalamita picta Stimpson, 1858, Thalamita sima H. Milne Edwards, 1834, Thalamita danae Stimpson, 1858, Heteropilumnus setosus (A. Milne-Edwards, 1873), Ser fukiensis Rathbun, 1931, and Notonyx vitreu Alcock, 1900], only one host candidate T. sima was thus far reported to occur in Korea. Therefore, it is reasonable to assume that T. sima can only be utilized as a host for Korean populations of S. gracilis. However, P. lineata and P. postica were found in Pagurus brachiomastus (Thallwitz, 1891) and P. filholi, but none of these rhizocephalans were found from Pagurus nigrivittatus Komai, 2003 (examined 47 individuals) and Pagurus maculosus Komai & Imafuku, 1996 (examined 502 individuals) (Table 4) even though these hermit crab species inhabit Korea and were reported as a host of P. lineata or P. postica in Japan (Yoshida et al., 2014).
In contrast, this study provides additional information on the extensive range of host-parasite association at the higher taxonomic level between rhizocephalans and their host hermit crabs, i.e., between the family of rhizocephalans and the infraorder of the hermit crab hosts. Unlike host-parasite association at the species level which is host resource-dependent (availability and abundance of host resource), their relationships at the higher taxonomic level of host and parasite are relatively relaxed (Table 3) as previously documented in other metazoan groups (Williams, 2008; Sweet et al., 2018). Further characterization of host-parasite relationships based on extensive taxon samplings is needed to fill the gap in our current understanding of their relaxed relationship, host ranges, and preferences in diverse parasitic barnacle species.
4.3 The influence of geographical factors on the host range and distribution pattern of Korean rhizocephalans
In this study, the author compared species diversity and abundance of parasitic barnacles across the Korean sea coast that was characterized into three ecoregions (Kim et al., 2020; 2022). From this, the author found that rhizocephalan species diversity and abundance are higher in the East China Sea ecoregion (182 individuals and 24 species) than in the Yellow Sea (46 individuals and 10 species) and the East Sea ecoregions (68 individuals and 11 species) (Figure 1). Relatively lower species richness and a scanty of rhizocephalans in the Yellow Sea and the East Sea ecoregions are assumed relevant to the lower survival rate of their larva as surface water temperature decreases. Kashenko and Korn (2002; 2003) reported that most larvae of three rhizocephalan species, i.e., P. gracilis, Peltogaster reticulata Shiino, 1943, and Polyascus polygeneus (Lützen & Takahashi, 1997), were not able to survive lower than 12°C. The lowest water temperature of the Yellow Sea and the East Sea ecoregions in winter often drops below 10°C. On the other hand, the lowest water temperature in the East China Sea ecoregion in winter is approximately 11-12°C (Ma et al., 2006; Kim et al., 2020; Park et al., 2020). It is assumed that Rhizocephala larvae in the Yellow Sea and the East Sea ecoregions with relatively lower surface water temperature during the winter season might have a lower survival rate, leading to relatively lower species richness of rhizocephalans compared to the East China Sea ecoregions. However, P. gracilis exhibits high densities with a 20% infection rate in Pagurus middendorffii Brandt, 1851 in Vostok Bay (Kornienko et al., 2018), where winter water temperatures can drop below zero (Fayman et al., 2019), which suggests that low temperatures may not universally limit Rhizocephalan diversity. Therefore, this necessitates further study to elucidate the specific impact of minimum winter water temperatures on the diversity of Rhizocephalans.
This research unveiled distinct patterns in the specificity of Rhizocephala-host relationships across three Korean ecoregions (the East China Sea, the East Sea, and the Yellow Sea). In the East China Sea ecoregion, the author identified a remarkable diversity, with 10 ecoregion-specific rhizocephalan species and 5 unique host-parasite relationships that are not found in other ecoregions (Figure 3). This contrasts starkly with the East Sea ecoregion, where the author found only one ecoregion-specific rhizocephalan species, alongside 7 unique host-parasite relationships. The Yellow Sea ecoregion showed a lower specificity with a simpler pattern of rhizocephalan fauna represented by only one ecoregion-specific rhizocephalan species and a single unique host-parasite relationship. These disparities appear to be linked to the environmental characteristics of each ecoregion, particularly the lowest water temperatures in winter and the host species diversity. In the East China Sea ecoregion, the relatively higher water temperatures in winter (>11°C) and a greater abundance of host species (21 species, Table 4) might foster an environment likely to increase the diversity of Rhizocephala, might lead to the settlement of specialist Rhizocephala parasitizing one or two hosts. Conversely, in the East Sea ecoregion, despite the secondary high diversity of hosts (17 species, Table 4), the lower water temperatures in winter (<10°C) might prevent the settlement of specialist Rhizocephala. This may result in the generalist Rhizocephala parasitizing more than three hosts might have adapted to parasitize unique hosts. In the Yellow Sea ecoregion, both the lower minimum sea temperatures and a limited host species pool seem to constrain the development of unique Rhizocephala-host relationships. The hypothesis receives limited support, as evidenced by the discovery of 6 Rhizocephalan species in southern Honshu, Japan, with a minimum winter water temperature above 11°C and 7 host Paguroidea species (Yoshida et al., 2014), compared to only 3 Rhizocephalan species in Vostok Bay, Russia, where the minimum winter water temperature falls below zero and hosts 5 Paguroidea species (Kornienko et al., 2018).
In the present investigation, these findings reveal a geographically constrained parasitism of Rhizocephala on specific hosts. Notably, the parasitism of S. confragosa on G. depressus is exclusive to Namhae and its vicinities, i.e., Yeosu, Tongyeong, Sacheon, and Goheung, within the East China Sea ecoregion, despite the extensive prevalence of G. depressus along the entire Korean coastline. Similarly, Parasacculina imberbis Shiino, 1943 exhibits a unique parasitic relationship with P. crassipes solely in the southern extents of the East China Sea ecoregion, such as Jeju, Seogwipo, Namhae, and Tongyeong, similar to the distribution of Japanese P. imberbis limited to southern Honshu (Tsuchida et al., 2006). These observations resonate with the patterns documented by Boschma (1955b), wherein Heterosaccus occidentalis (Boschma, 1928) demonstrates a limited distribution relative to the wider distribution of its three hosts: Mithraculus forceps A. Milne-Edwards, 1875, Omalacantha bicornuta (Latreille, 1825), and Pitho lherminieri (Desbonne in Desbonne & Schramm, 1867). These results may be related to the gap in ecological parameters of each location, especially the difference in optimal water temperature and optimal salinity promote the development and survival of each larvae of rhizocephalan species (Kashenko and Korn, 2002; 2003; Tu et al., 2009), which can influence the inhabit of Rhizocephala. These factors could serve as ecological barriers that delineate the range of these parasitic entities. This hypothesis warrants further investigation to unravel the complexities underlying these specialized parasitic relationships.
5 Conclusions
The present study integrates multiple sources of information about the molecular phylogeny of Korean rhizocephalans and their distribution and host range based on recent collections of parasitic barnacles. Through this molecular analysis, the unidentified species of Korean Rhizocephala in the previous studies are different from other parasitic barnacles. In addition, each Rhizocephala family correlated with the infraorder of their host. The author found that the Korean rhizocephalan species show different patterns of host-parasite association from those previously documented in other geographic regions, which might be due to different host species resources depending on geographical origins. Meanwhile, in this study, the distribution of Rhizocephala in three Korean marine ecoregions was confirmed, and as a result, the East China Sea had higher diversity than other ecoregions, which might be related to the highest water temperature of the East China Sea in winter. An in-depth analysis from further extensive taxon samplings of rhizocephalans and crustacean host species is needed to complete our understanding of host-parasite association and distribution of Rhizocephala for their control and utilization.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
JJ: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1A6A3A01086374, 2021R1A6A1A10039823).
Acknowledgments
The author appreciates Dr. Joong-Ki Park (Ewha Womans University) who supervised many parts of this manuscript and Dr. Jongwoo Jung, Ms. Sung-Hyun Park (Ewha Womans University), and Dr. Hyunsoon Kim (Seoul National University) who helped sample collection of the materials used in this study.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1359503/full#supplementary-material
Footnotes
- ^ Jung, J., and Park, J. K. (2024). The fauna of parasitic barnacles (Cirripedia: Rhizocephala) in Korea. Zool. Stud 63, 01. in review.
References
Alvarez F., Hines A. H., Reaka-Kudla M. L. (1995). The effects of parasitism by the barnacle Loxothylacus panopaei (Gissler) (Cirripedia: Rhizocephala) on growth and survival of the host crab Rhithropanopeus harrisii (Gould) (Brachyura: Xanthidae). J. Exp. Mar. Biol. Ecol. 192, 221–232. doi: 10.1016/0022-0981(95)00068-3
Boschma H. (1955a). Rhizocephalan parasites of the crab Pugettia brevirostris, with notes on Sacculina gracilis. Zool. Meded. 33, 237–249.
Boyko C. B., Williams J. D. (2016). “Crustacean parasites as phylogenetic indicators in decapod evolution,” in Decapod crustacean phylogenetics (CRC Press, Boca Raton, FA), 209–232.
Fayman P., Ostrovskii A., Lobanov V., Park J. H., Park Y. G., Sergeev A. (2019). Submesoscale eddies in Peter the Great Bay of the Japan/East Sea in winter. Ocean Dynamics 69, 443–462. doi: 10.1007/s10236-019-01252-8
Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Hines A. H., Alvarez F., Reed S. A. (1997). Introduced and native populations of a marine parasitic castrator: variation in prevalence of the rhizocephalan Loxothylacus panopaei in xanthid crabs. Bull. Mar. Sci. 61, 197–214.
Høeg J. T. (1992). “Rhizocephala,” in Microscopic anatomy of invertebrates, vol. 9. Eds. Harrison F. W., Humes A. G. (Wiley, New York, NY), 313–345.
Høeg J. T., Lützen J. (1995). Life cycle and reproduction in the Cirripedia Rhizocephala. Oceanogr. Mar. Biol. 33, 427–485.
Høeg J. T., Noever C., Rees D. A., Crandall K. A., Glenner H. (2019). A new molecular phylogeny-based taxonomy of parasitic barnacles (Crustacea: Cirripedia: Rhizocephala). Zool. J. Linn. Soc 190, 632–653. doi: 10.1093/zoolinnean/zlz140
Jung J., Yoshida R., Kim W. (2019). Diversity of parasitic Peltogastrid barnacles (Crustacea: Cirripedia: Rhizocephala) on hermit crabs in Korea. Zool. Stud. 58, 33. doi: 10.6620/ZS.2019.58-33
Jung J., Yoshida R., Lee D., Park J. K. (2021). Morphological and molecular analyses of parasitic barnacles (Crustacea: Cirripedia: Rhizocephala) in Korea: preliminary data for the taxonomy and host ranges of Korean species. PeerJ 9, e12281. doi: 10.7717/peerj.12281
Kashenko S. D., Korn O. M. (2002). Effects of temperature and salinity on the larvae of two species of rhizocephalan (Crustacea: Cirripedia). Invertebr. Reprod. Dev. 41, 179–184. doi: 10.1080/07924259.2002.9652750
Kashenko S. D., Korn O. M. (2003). Combined effects of seawater temperature and salinity on development of the larvae of the rhizocephalan Peltogaster reticulatus (Crustacea: Cirripedia). Russ. J. Mar. Biol. 29, 150–155. doi: 10.1023/A:1024664631438
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analyses of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim H. K., Chan B. K. K., Lee S. K., Kim W. (2020). Biogeography of intertidal and subtidal native and invasive barnacles in Korea in relation to oceanographic current ecoregions and global climatic changes. J. Mar. Biol. Assoc. U. K. 100, 1079–1091. doi: 10.1017/S0025315420001009
Kim H. K., Chan B. K. K., Yi C., Kim I. H., Choi Y. N. (2022). Barnacle epibiosis on sea turtles in Korea: A west Pacific Region with low occurrence and intensity of Chelonibia testudinaria (Cirripedia: Chelonibiidae). Front. Ecol. Evol. 10. doi: 10.3389/fevo.2022.785692
Kornienko E. S., Korn O. M., Selin N. I. (2018). The parasitic fauna of common species of hermit crabs of Vostok Bay (Sea of Japan). Russian J. Mar. Biol. 44, 94–99. doi: 10.1134/S1063074018020062
Kristensen T., Nielsen A. I., Jørgensen A. I., Mouritsen K. N., Glenner H., Christensen J. T., et al. (2012). The selective advantage of host feminization: a case study of the green crab Carcinus maenas and the parasitic barnacle Sacculina carcini. Mar. Biol. 159, 2015–2023. doi: 10.1007/s00227-012-1988-4
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Ma J., Qiao F., Xia C., Kim C. S. (2006). Effects of the Yellow Sea Warm Current on the winter temperature distribution in a numerical model. J. Geophys. Res. Ocean 111, C11S04. doi: 10.1029/2005JC003171
Nagler C., Hörnig M. K., Haug J. T., Noever C., Høeg J. T., Glenner H. (2017). The bigger, the better? Volume measurements of parasites and hosts: Parasitic barnacles (Cirripedia, Rhizocephala) and their decapod hosts. PloS One 12, e0179958. doi: 10.1371/journal.pone.0179958
Noever C., Olson A., Glenner H. (2016). Two new cryptic and sympatric species of the king crab parasite Briarosaccus (Cirripedia: Rhizocephala) in the North Pacific. Zool. J. Linn. Soc 176, 3–14. doi: 10.1560/RCLC-NM2U-HV5L-6Q52
O'Shaughnessy K. A., Harding J. M., Burge E. J. (2014). Ecological effects of the invasive parasite Loxothylacus panopaei on the flatback mud crab Eurypanopeus depressus with implications for estuarine communities. Bull. Mar. Sci. 90, 611–621. doi: 10.5343/bms.2013.1060
Øksnebjerg B. (2000). The Rhizocephala (Crustacea: Cirripedia) of the Mediterranean and Black Seas: taxonomy, biogeography, and ecology. Isr. J. Zool. 46, 1–102. doi: 10.1560/RCLC-NM2U-HV5L-6Q52
Park S. H., Song S. K., Park H. S. (2020). Temporal and spatial variations of marine meteorological elements and characteristics of sea fog occurrence in Korean coastal waters during 2013-2017. J. Environ. Sci. Int. 29, 257–272. doi: 10.5322/JESI.2020.29.3.257
Ritchie L. E., Høeg J. T. (1981). The life history of Lernaeodiscus porcellanae (Cirripedia: Rhizocephala) and co-evolution with its porcellanid host. J. Crustac. Biol. 1, 334–347. doi: 10.2307/1547966
Ronquist F., Teslenko M., van der Mark P., Ayres D., Darling A., Höhna S., et al. (2012). MRBAYES 3.2: efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sweet A. D., Bush S. E., Gustafsson D. R., Allen J. M., DiBlasi E., Skeen H. R., et al. (2018). Host and parasite morphology influence congruence between host and parasite phylogenies. Int. J. Parasitol. 48, 641–648. doi: 10.1016/j.ijpara.2018.01.007
Tavaré S. (1986). Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math. Life Sci. 17, 57–86.
Tsuchida K., Lützen J., Nishida M. (2006). Sympatric three-species infection by Sacculina parasites (Cirripedia: Rhizocephala: Sacculinidae) of an intertidal grapsoid crab. J. Crustac. Biol. 26, 474–479. doi: 10.1651/S-2682.1
Tu T. H., Chan B. K. K., Jeng M. S. (2009). Larval development and sex ratio variation of Polyascus plana (Cirripedia: Rhizocephala), a parasite of the crab Grapsus albolineatus, in Taiwan. Bull. Mar. Sci. 84, 331–349.
Williams P. H. (2008). Do the parasitic Psithyrus resemble their host bumblebees in colour pattern? Apidologie 39, 637–649. doi: 10.1051/apido:2008048
Yang C. P., Sun D. R., Qiu Y. S., Liu Y., Shan B., Li T., et al. (2018). The parasitic barnacle Sacculina lata Boschma 1933 (Cirripedia, Rhizocephala, Sacculinidae) infecting the commercial swimming crab Charybdis miles (De Haan 1835) (Decapoda, Portunidae) in the Beibu Gulf, South China Sea. Crustaceana 91, 577–590. doi: 10.1163/15685403-00003787
Keywords: Rhizocephala, Decapoda, parasite, infestation rate, host resources, biogeography, ecoregion, water temperature
Citation: Jung J (2024) The host range and distribution pattern of rhizocephalan parasitic barnacles in Korean coasts and their relationship with geographical factors. Front. Mar. Sci. 11:1359503. doi: 10.3389/fmars.2024.1359503
Received: 21 December 2023; Accepted: 02 February 2024;
Published: 22 February 2024.
Edited by:
Jin Sun, Ocean University of China, ChinaReviewed by:
Wong Yue Him, Shenzhen University, ChinaOlga Korn, National Scientific Center of Marine Biology, Far East Branch (RAS), Russia
Copyright © 2024 Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jibom Jung, YXBvY2l2QG5hdmVyLmNvbQ==
 Jibom Jung
Jibom Jung