
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 18 March 2024
Sec. Marine Biology
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1358793
 Katherine Hartle-Mougiou1,2
Katherine Hartle-Mougiou1,2 Chrysoula Gubili3
Chrysoula Gubili3 Panagiota Xanthopoulou3,4
Panagiota Xanthopoulou3,4 Panagiotis Kasapidis5
Panagiotis Kasapidis5 Martha Valiadi2*†
Martha Valiadi2*† Electra Gizeli1,2*†
Electra Gizeli1,2*†The Mediterranean basin has faced an increased influx of invasive species since the Suez Canal expansion in 2015. The invasive lionfish species, Pterois miles, has rapidly established new populations in the Eastern Mediterranean Sea, impacting local fish biodiversity. Here, we have developed a new, fast (< 35 min) molecular approach to detect and quantify P. miles environmental DNA (eDNA) in combination with a portable device for field-based analysis. Using a species-specific real-time colorimetric loop-mediated isothermal amplification (qcLAMP) for the cytochrome oxidase subunit 1 (COI) gene, we demonstrate a high sensitivity with a limit of detection of 0.002 ng DNA per reaction, equivalent to only 50 copies of the COI gene. The assay is specific to the target in the presence of closely related and co-occurring species, and it is quantitative over five orders of magnitude. We validated the assay using aquarium water samples and further demonstrated its utility on natural eDNA samples collected from locations around the island of Crete where P. miles had been sighted. P. miles was indeed detected in three out of nine locations, two nature reserves and a closed bay. Lack of detection in the remaining locations suggests that populations are still at a low density. We also demonstrate the feasibility of P. miles eDNA qualitative detection directly from the filter used to collect eDNA-containing particles, completely omitting DNA extraction. Overall, we present a new approach for fast and targeted eDNA quantification. The developed LAMP assay together, with the quantitative real-time colorimetric detection approach, open new possibilities for monitoring invasive P. miles in the field.
Marine biological invasions are increasing globally, driven mostly by human-mediated transportation and elimination of natural barriers to dispersal, and further facilitated by global warming (Chan and Briski, 2017). Invasive species damage ecosystem equilibria in their new habitat, by causing new ecological pressures and benefitting from a lack of native predators (Giakoumi et al., 2019). These invasions have detrimental socio-economic impacts (Galanidi et al., 2018), and some even pose a threat to human health (Giakoumi et al., 2019). The Mediterranean Sea is one of the most vulnerable regions for biological invasions; 993 non indigenous species were reported by the end of 2021, with 751 species confirmed as established (Zenetos et al., 2022). Key environmental barriers between the Red Sea and Mediterranean Sea have been removed, facilitating species migration. Expansions of the Suez Canal, initially opened in 1869 and widened in 2015, have removed the channel’s hypersaline regions (Boudouresque, 1999) whilst the eastern Mediterranean Waters have rapidly warmed to similar temperatures as the Red Sea (Conversi et al., 2010; Pastor et al., 2020).
One of the most significant ongoing invasions in the eastern Mediterranean Sea is by the Indo-Pacific lionfish species, Pterois miles (Bennett, 1828), a fish with long venomous spines of the Family Scorpaenidae. Its indistinguishable relative, Pterois volitans, has already caused significant damage in the North West Atlantic since the 1980’s, causing decline in native fish species and affecting reef communities with associated economic impacts (Poursanidis et al., 2020). Their success is due to their high fecundity and spawning activity, lack of natural predators in invaded regions, their poisonous fins, broad diet, and ability to occupy various habitats and depths (Claydon et al., 2012; Côté and Smith, 2018; Savva et al., 2020). The invasion in the eastern Mediterranean is at an earlier stage, with reports of established populations commencing in 2015 (Phillips and Kotrschal, 2021). In the past few years, populations have primarily affected the coasts of Israel, Syria, Lebanon, Turkey and Cyprus and more recently Greece (Savva et al., 2020). Despite large-scale efforts by the EU H2020 RELIONMED programme to collect data in order to officially include this species in the EU Invasive Alien Species list and implement management strategies to intercept the invasion, this has yet to happen (Kleitou et al., 2021). Management strategies have already been tested in the Atlantic and recommendations have been made for population control by targeted removal and commercial exploitation (Ulman et al., 2022). The Mediterranean lionfish invasion front is currently situated in Greece with populations rapidly establishing in the Levantine Sea, Aegean and Ionian Seas; further spread is limited by the 15.3°C isotherm to the north and west although there have been a few isolated sightings in Sicily and Croatia (Dimitriadis et al., 2020; Poursanidis et al., 2020; Kleitou et al., 2022). The Island of Crete, in Southern Greece, has faced increasing populations in the last five years (Phillips and Kotrschal, 2021), with lionfish now regularly available at fishmongers.
Early intervention on marine invasions requires early detection of alien species, as well as information about population abundance and habitat preferences. Current methods for marine organism observation include visual surveys (snorkelling and SCUBA diving), observations by citizens, and trawl nets (Darling and Mahon, 2011; Bennett et al., 2016; Pastor et al., 2020). However, these approaches are ineffective for low density populations as, when individuals are infrequent, they can be easily missed by divers (Keller et al., 2022). Conversely, molecular detection of species from environmental DNA (eDNA) is more sensitive and cost efficient for low density populations at the initial phase of an invasion (Keller et al., 2022). eDNA is genetic material contained in free particles that have been shed from organisms in the ocean e.g., from urine, faeces, gametes, and skin (Beng and Corlett, 2020). Species-specific eDNA-based approaches (Bylemans et al., 2019) have been demonstrated for monitoring invasive fish such as carp and brook trout fish in USA freshwaters (Jerde et al., 2013; Schumer et al., 2019; Beng and Corlett, 2020). On the other hand, eDNA metabarcoding, which profiles eDNA from the whole community, might miss rare species due to insufficient sequencing depth or lack of sequences on the reference database. Additionally, this approach requires sending the sample to a dedicated laboratory and time-consuming bioinformatic analysis, increasing the cost and analysis time (Larson et al., 2020).
Species-specific molecular detection is the preferred approach for early detection and spatial tracking of specific species of concern. Quantitative PCR (qPCR) and digital droplet PCR (ddPCR) have been used in numerous quantitative eDNA studies as they have high sensitivity in detecting eDNA targets and provide quantitative estimations (Wilcox et al., 2013; Doi et al., 2015; Mauvisseau et al., 2019; Capo et al., 2021; Jerde, 2021; Yu et al., 2021). DdPCR is less prone to errors and more accurate than qPCR, especially at low eDNA concentrations, making it better suited to provide abundance and biomass estimates of the target species (Doi et al., 2015). Assays need to be carefully optimized to ensure target specificity, as eDNA samples contain genetic material from various organisms, including species that are closely related to the target organism. Quantitative data generated by these approaches enable estimates of population size, helping to identify preferred habitats, potential breeding grounds, and define modelling approaches that account for currents and life cycle history in eDNA based ecosystem studies (Doi et al., 2015; Goldberg et al., 2016; Murakami et al., 2019; Kutti et al., 2020; Keller et al., 2022). However, both aforementioned PCR-based methods are not suitable for field-based analysis as they require expensive instruments (>30k euros), the run time is long (>1 hr) and the preparation of highly purified DNA samples is also time consuming.
Given the need for intensive field surveys, there will be significant benefit from approaches that provide results quickly and at low cost. Ideally, these tests should be performed on site, to avoid the need to transport sensitive eDNA samples to a laboratory freezer in strict conditions. This approach would enable field studies with improved geographic coverage especially at remote locations. To this end, the characteristics of isothermal nucleic acid amplification enable analysis in small, simple and portable devices, as they obviate the need for energy-demanding thermal cycling as in PCR (Becherer et al., 2020; Yu et al., 2021). Isothermal amplification methods amplify DNA based on strand displacement enzymatic activity, instead of temperature cycling. For marine environments with high biodiversity, loop-mediated isothermal amplification (LAMP) stands out as a method of choice for eDNA analysis due to its high specificity. This is due to the requirement of 4-6 primers to be used to recognise 6-8 regions of the target. LAMP also has a very high sensitivity due to high product yield and is a very robust method even in the presence of complex samples (Becherer et al., 2020). Detection of LAMP amplicons can be achieved in real-time to enable target quantification by including a fluorescent DNA dye; this method is primarily lab-based and relatively expensive. An alternative approach includes the use of pH sensitive dyes and visual assessment of colour change. However, although of low cost and suitable for field testing, visual assessment suffers from low sensitivity while it is also subjected to the user’s interpretation and requires a “trained eye” for accurate monitoring (Zhang et al., 2014).
Few portable systems have been developed for marine applications so far, with the main focus being the detection of harmful algae (McPartlin et al., 2017). These are mainly based on assays targeting ribosomal genes which exist in hundreds of copies per cell to enhance the assay sensitivity. These assays are qualitative (target presence/absence only) due to a lack of portable instrumentation with enough accuracy to enable target quantification. Nevertheless, LAMP assays have been shown to achieve a high sensitivity, which is ideal for marine eDNA studies; although this is reported in non-uniform units, the Limit of Detection (LoD) is in the range of 1 cell mL-1 for LAMP with a fluorometric dye and an instrument measuring turbidity in the reaction (Fujiyoshi et al., 2021) and 0.034 pg DNA per reaction with detection using lateral flow strips (Qin et al., 2019). In our lab, we have developed a dedicated portable device to improve colorimetric measurements and enable quantitative analysis of the samples. The device can record the colour change in the LAMP reaction-tube via an embedded mini-camera; the device has been demonstrated to be suitable for fast and sensitive testing of swab samples for SARS-CoV-2 RNA and cancer genetic mutations in patients’ tissue (Papadakis et al., 2022). Overall, the method has been adapted to provide real-time results with reduced complexity and user-friendly smartphone operation, which can be used in the field.
In this study, we aimed to develop a real-time colorimetric loop-mediated isothermal amplification (qcLAMP) assay for targeted estimates of P. miles presence and population density through eDNA. This assay is implemented with the newly developed portable device to enable readiness as an on-site early detection tool when coupled with field-based DNA extraction kits. Initially, we optimised the assay for sensitivity and specificity in the lab using gold-standard lab-based DNA extraction; this allowed us to identify required optimization steps more readily at the initial assay development stage. Moreover, the assay was validated with field samples collected around the island of Crete, Greece, which now has established P. miles populations. This presumed non-uniform distribution, which should lead to variable amounts of P. miles eDNA, was the ideal setting to test our new quantitative assay. Additionally, given the ability of qcLAMP to amplify DNA from crude samples, we evaluated this approach as an alternative qualitative detection tool that can be applied on-site without the need for DNA extraction.
For the initial assay development, three P. miles individuals were obtained from local fishermen caught off the island of Dia, approximately seven nautical miles (13 km) north of Heraklion, Crete. Individuals were dissected and 1 cm2 muscle tissue samples were obtained and stored at -80°C until further analysis. Additionally, another four samples of P. miles DNA from tissues of individuals collected in Rhodes, were provided by the Hellenic Centre for Marine Research (HCMR) from Heraklion, Crete. To test the assay specificity against closely related species, one individual of each of S. scrofa, Mullus barbatus and M. surmuletus were also obtained from local fish markets in Heraklion, Crete. Further specimens of Scorpaena porcus and S. notata collected from the Thracian Sea, were provided by the Fisheries Research Institute (INALE), Nea Peramos, Kavala (Hellenic Agricultural Organization-DIMITRA).
The performance of the assay on target eDNA from seawater was initially assessed on the following samples: i) water samples were collected from the P. miles aquarium tank at CRETAquarium (Heraklion, Crete, Greece); ii) one aquarium sample from the putative P. volitans tank provided by colleagues at Dublin City University, Ireland; iii) water samples containing closely related and other local species, but lacking P. miles, from the fish tanks of the Aquaworld aquarium (Hersonissos, Crete, Greece). For each water sample, 1 L of aquarium tank water was passed through a 0.45 μm pore size polycarbonate filter (Cyclopore, Whatman, Germany) shortly after collection. Filters were stored at -80°C until further analysis.
Natural samples were collected from nine coastal sites in Crete, where P. miles had been previously sighted (sightings by fishermen, divers, and citizens recorded on gbif.org), during November 2021 and May 2022 (Figure 1; Supplementary Table S3). Sampling was carried out from either a stand-up paddle board or a small boat using a 2 L Niskin bottle (Aquatic Biotechnology, Spain). Seawater was collected from a depth of 10-20 m, and returned to the beach. Two aliquots of 0.5 L were filtered through Sterivex filter cartridges (HV 0.45 μm, Millipore, USA) using sterilized 60 ml disposable syringes, to provide technical replicates. Additionally, 1 L of bottled deionized water was filtered at each station, to serve as a negative control in the field. Following filtering, filter units were stored in individual ziplock bags on dry ice until returned to the laboratory, where they were stored at -80°C until further analysis.

Figure 1 eDNA sampling locations around Crete. Green dots denote that P. miles was detected and orange dots denote that it was not detected.
Genomic DNA was extracted from tissue samples using the DNeasy Blood & Tissue Kit (Qiagen, Germany). Fish muscle tissue was cut into small pieces and processed following the manufacturer’s instructions with a modified lysis incubation of 3 hours. The quality of extracted DNA was checked on a 1% agarose gel, purity was assessed using a Nanodrop spectrophotometer (ND-1000, Nanodrop Technologies Inc., USA), and DNA concentrations were confirmed with the Qubit 4 (Thermofisher Scientific) using the Qubit 1X dsDNA HS Assay kit (ThermoFisher Scientific).
Environmental DNA was extracted from Sterivex filters following sterile procedures in a dedicated laboratory, separate from other work involving P. miles tissues and amplicons. Sterivex filters were removed from the cartridge and the filter was cut to small pieces (approximately 0.4 cm2) using a sterile single-use scalpel and petri dish. eDNA was extracted from filters using the DNeasy Blood & Tissue Kit following the same protocol as for the tissues, with one modification. To ensure the complete disruption of particles containing eDNA, the lysate was passed through a QIAshredder (Qiagen, Germany) column as per the DNeasy plant extraction kit, prior to binding the DNA on the extraction membrane. Single use and sterile consumables were changed between samples (e.g., gloves, scalpel blades, petri dishes).
To guide the assay development and confirm the identity of target and non-target fish, we partially sequenced the mitochondrial cytochrome oxidase subunit 1 (COI) gene. For tissues excised from individual fish, we used the Universal fish PCR primers, FISH-BCL (Forward) and FISHCOIHBC targeting a 704 bp fragment of the COI gene (Baldwin et al., 2009). For P. miles aquarium water samples, which could contain DNA from other fish in the tank or from the fish food, we designed a species-specific primer; PmCOI7F was used in conjunction with primer FISHCOIHBC and the same PCR conditions to specifically amplify P. miles COI from the mixed population (Table 1); primer design was based on a 606 bp consensus sequence of 74 GenBank sequences for P. miles COI gene. The GenBank sequences were collated in Geneious Prime 2014.7.1.3 and aligned using MAFFT pairwise global alignment (Katoh and Standley, 2013). Primer PmCOI7F was combined with the universal FISHCOIHBC for species-specific amplification of a 572 bp COI gene fragment.
PCR on DNA extracted from fish tissues with primers FISH-BCL and FISHCOIHBC) as well as on eDNA extracted from aquarium water samples (with primers PmCOI7F-FISHCOIHBC) was performed in 25 μl reactions which included: 12.5 μl KAPA2G Fast HotStart mix (KAPA Biosystems, UK), 1.25 μl forward primer (10 μM), 1.25 μl reverse primer (10 μM), 9 μl molecular grade water (Sigma-Aldrich), 1 μl template (approximately 10 ng DNA). The PCR cycles were as follows: initial denaturation at 95°C for 3 min, 35 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s and extension at 72°C for 30 s, and a final extension at 72°C for 1 min. PCR amplicons were Sanger sequenced by Genewiz (Germany). Forward and reverse reads were paired in Geneious Prime 2014.7.1.3 and species identity was confirmed with the Basic Local Alignment Search Tool (BLAST) on NCBI. Sequences have been submitted to GenBank under accession numbers OR794518 - OR794524.
Primer design. Primers were designed based on the P. miles COI consensus sequence, produced from sequences obtained in this study as well as from GenBank (Supplementary Tables S1, S3). Haplotypes were generated using DnaSP 6.12.03 with the default parameters (Rozas et al., 2017). LAMP primers were designed using PrimerExplorer V5 (https://primerexplorer.jp/e/) to target a 213 bp fragment of the P. miles consensus sequence for the mitochondrial gene COI (Figure 2). This resulted in 5 LAMP primers: Forward inner primer (FIP), backward inner primer (BIP), forward outer primer (F3), backward outer primer (B3) and only a single loop primer, the backward primer (LB). A forward loop primer (LF primer) was not predicted by the software due to no suitable sequence regions. The forward primer F3 was designed with a single degeneracy due to polymorphism at the binding site. The generated primers were examined in silico in pairs for their specificity to the target by using Primer BLAST (NCBI GenBank). Sequences of the closely related P. volitans sequences contained a total of 7 SNPs in the primer binding sites that would most likely preclude amplification of this species (Supplementary Table S1; Figure 2). Potential cross-reaction and secondary structures in the primers were identified using Primer Digital (https://primerdigital.com/). The final primer sequences are shown in Table 1.
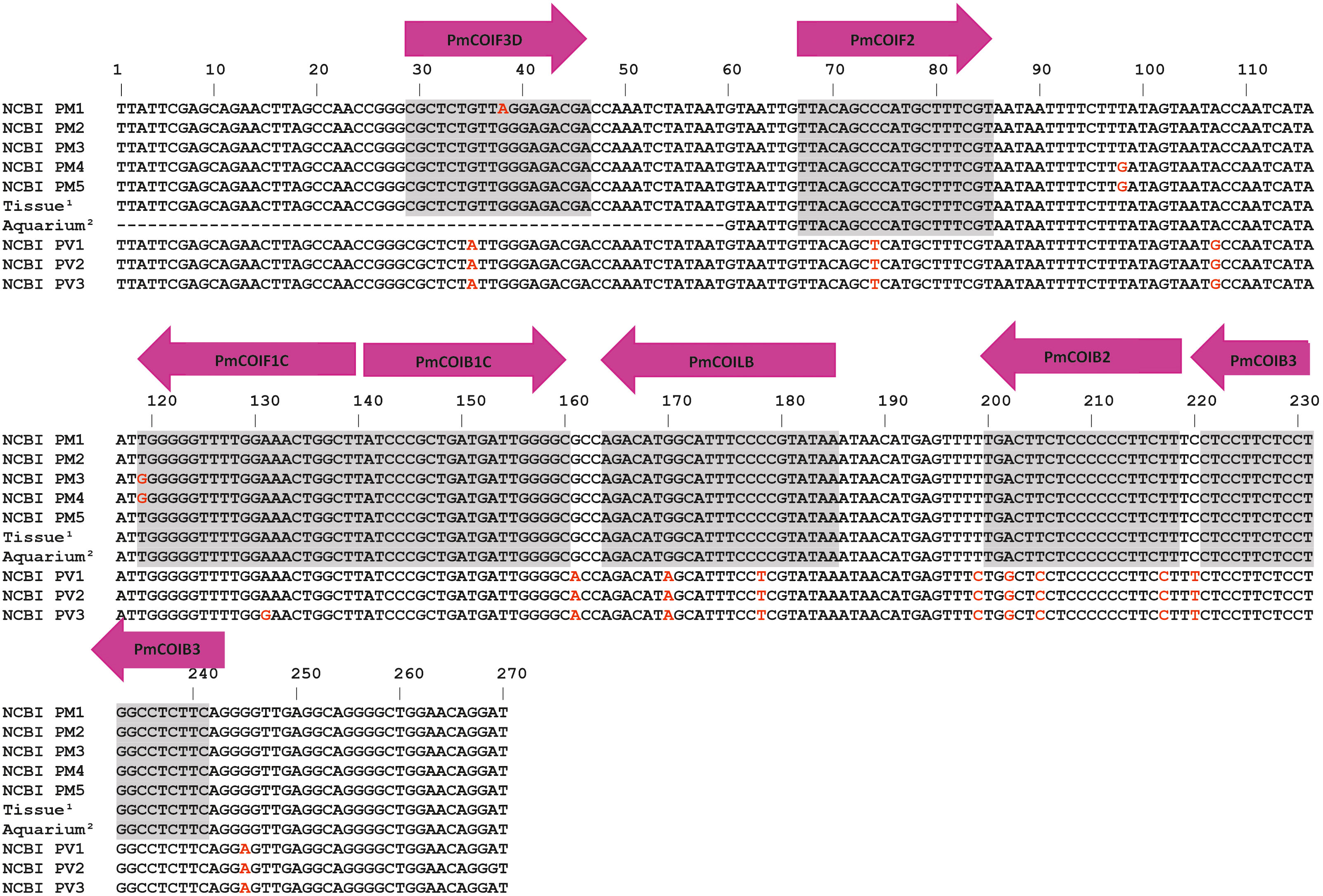
Figure 2 Alignment of the five P. miles and three P. volitans haplotypes based on GenBank sequences, and sequences obtained in this study (Supplementary Table S1). The haplotypes (NCBI PM & NCBI PV) shown in the alignment were the ones with sequence variation in the primer target region. Grey areas denote primer binding sites. Arrows show the direction and names of the LAMP primers. The naming of the binding sites follows the standard notation in LAMP primer design since a single primer binds in two positions of the sense and antisense DNA (Notomi et al., 2000). Superscripts indicate sequence accession numbers; 1: OR794524, OR794523, 2: OR794522, OR794521.
The newly developed qcLAMP device used in this work has been described in Papadakis et al. (2022). Briefly, it consists of 3D-printed mechanical and electrical parts assembled to create a light, hand-held device portable prototype device which is operated by a mobile app. It includes a heating element and a camera to measure colour change within eight reaction-tubes, resulting in a real-time amplification curve based on Colour Index. Photo-shots of the camera are collected and transferred via a bluetooth to a nearby tablet or smartphone using a custom application. For field-based detection, the commercially available version of the device and application were used (“research-based” version of the Pebble-R, sold by BIOPIX DNA Technology, GR, < 1k Euros). The Pebble-R device employs six reaction tubes and can be used together with a commercial LAMP mix or home-made reaction assays.
Initial optimization for the optimal LAMP incubation temperature was determined using fluorescence real-time LAMP following manufacturer’s instructions (WarmStart LAMP kit, New England Biolabs) and a standard laboratory qPCR thermal cycler (CFX Connect, BIO-RAD) due to higher throughput. The qcLAMP reaction contained 12.5 μl WarmStart Colorimetric LAMP 2X Master Mix with UDG to prevent amplicon contamination (New England Biolabs, USA), 2.5 μl primer mix (FIP 1.8 μM, BIP 1.4 μM, LB 0.6 μM, F3 0.3 μM, B3 0.2 μM), 4 μl DNA template and 6 μl molecular grade water (Sigma-Aldrich), to a final reaction volume of 25 μl. For both tissue and natural seawater sample, 4 μL of the extracted DNA was added to each reaction with a total volume of 25 μL. Reactions were overlayed with 15 μL mineral oil (Sigma-Aldrich) to prevent evaporation and were run in the device at 65°C for 40 min. Time to positivity (TTP) for each sample was determined by using the background values prior to the onset of amplification, as negative controls could not be included in every run due to the 8-position limit. The threshold colour index was set as the average background value plus 3 times the standard deviation (Kalsi et al., 2015) and the TTP was the time it took for the colour index to rise above this value.
Although the developed assay can be performed on a portable device, as yet, there is no validated eDNA extraction protocol that can be performed in the field. This is a bottleneck for a sample-to-result analysis at the point of sampling. Since LAMP is known to be tolerant to crude samples containing sample inhibitors (Kaneko et al., 2007; Francois et al., 2011; Yang et al., 2014), we tested whether P. miles could be detected directly from the concentrated seawater particles on the filter, without performing an extraction. We cut small pieces of the filter and added them directly into the colorimetric LAMP reactions and performed a heat lysis step at 90°C for 9 min on the same device prior to amplification. We first confirmed that a heat lysis incubation did not affect the functionality of the LAMP reagents, using P. miles tissue extracted as positive controls. Filter pieces from the P. miles aquarium tank samples were then used as simulated environmental samples. Small square filter pieces approximately 0.4 x 0.4 cm were cut from the total 4 x 4 cm filter, and a single piece was added directly to 25 μL colorimetric LAMP reactions. Following the lysis incubation, reactions were cooled down for 5 min, and re-introduced to the device for amplification to commence.
In this study, we initially designed a species-specific LAMP assay for P. miles, based on sequences from NCBI and from locally collected fish samples. We established that the assay is specific to the target species using both tissue samples and eDNA mock samples collected from aquarium tanks, with the target species present or absent. Finally, we tested the assay on field samples collected around Crete, Greece. In addition, we tested whether we could detect P. miles from crude eDNA samples without a DNA extraction step so that the whole analysis could be performed in the field. The overall approach is depicted in Figure 3. In workflow A, extracted DNA from either tissue or seawater samples was added to the qcLAMP reactions, and in workflow B, pieces of the filter were added directly without extraction of the captured particles. Target amplification in the device was monitored in real time, generating amplification curves where the faster the signal rises above the background, the more of the target gene is present.
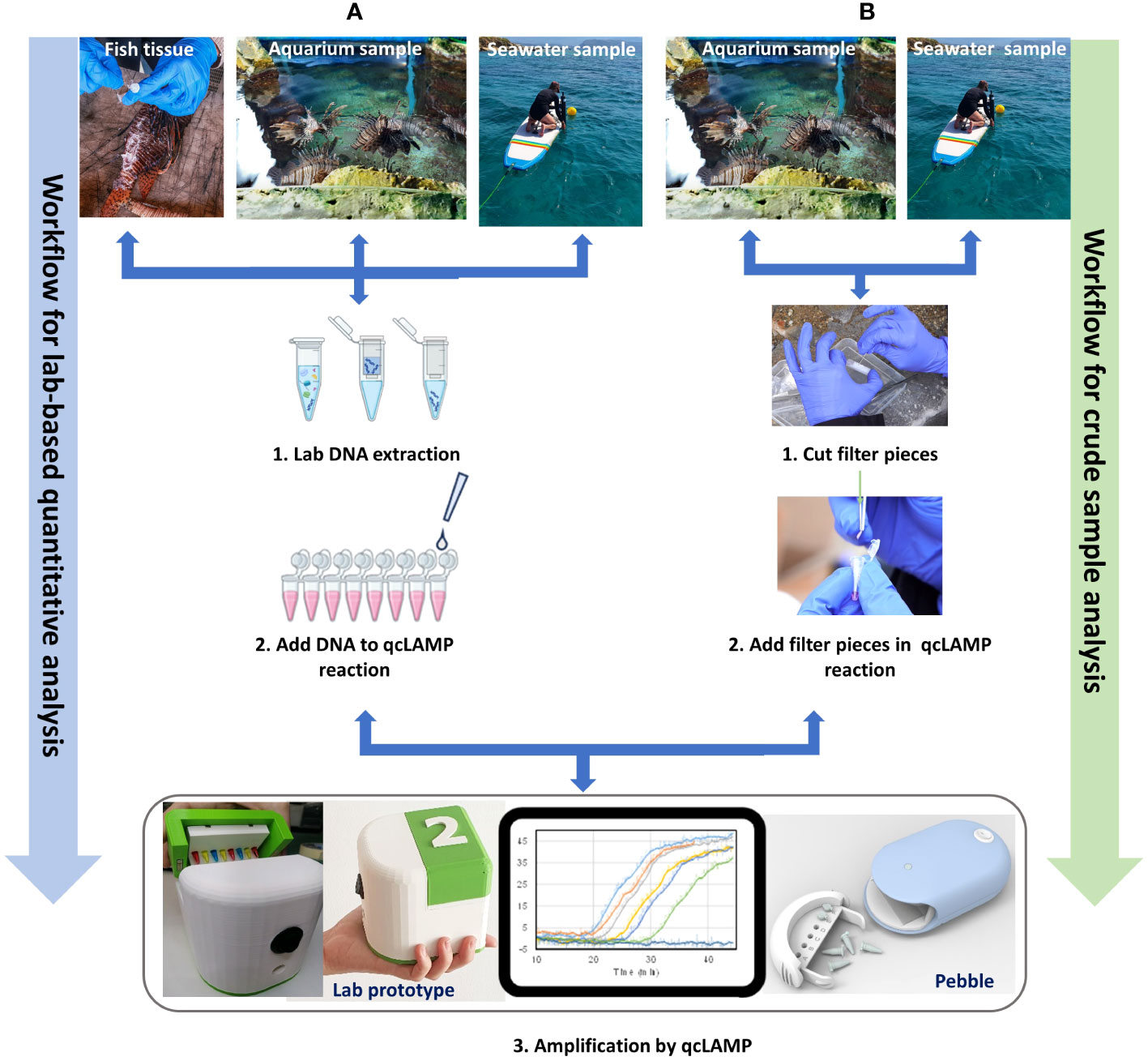
Figure 3 Assay development process used in this study. Increasing complexity of sample type, from fish tissue to aquarium and field (seawater) samples. (A). Testing the quantitative capacity of the assay with DNA from all three samples extracted in the laboratory, as well as (B) the capacity to detect the target from small pieces of the filter in the last two cases of samples (aquarium and sea).
The new LAMP assay was designed based on P. miles sequences obtained from NCBI and locally. Tissues from local fish revealed two haplotypes, DNA from the P. miles tank water of the CRETAquarium matched one of these haplotypes, and a different third haplotype was obtained from tank water of the Dublin Aquarium which was initially thought to be a P. volitans but we found it to be a P. miles (Figure 2). The assay was initially evaluated using DNA extracts from fish tissues. Primer concentrations were optimized for reaction speed and to reduce non-specific amplification. The forward primer F3 contained a two-fold degeneracy i.e., two different primer sequences at half the concentration each. Therefore, increasing the concentration of this primer by 50% relative to the standard protocol, resulted in a faster onset of amplification. Decreasing the concentration of the BIP primer resolved occasional non-specific products in the negative controls, as in silico analysis had predicted a cross-reaction between the primers BIP and LB. To assess if a second loop primer would improve the reaction speed, a forward loop primer (LF) was manually designed and tested. There was no difference in reaction speed with the inclusion of LF, therefore, this primer was omitted. All the above adjustments were adopted in our qcLAMP assay and subsequent experimentations.
The assay’s sensitivity was determined by serially diluting and amplifying P. miles tissue extracted DNA with LAMP. This test established the quantitative capacity of the assay, the LoD and the reproducibility among replicates. For initial tests, we used real-time LAMP with a fluorescence dye to obtain an assay benchmarking from an established laboratory device (i.e., the qPCR thermal cycler) prior to proceeding with colorimetric LAMP on the prototype colorimetric device. To enable direct comparisons, both tests (fluorescent LAMP and qcLAMP) were performed in parallel on the same DNA samples. These were a gDNA dilution series at the following amount of DNA per reaction: 20 ng, 2 ng, 0.2 ng, 0.02 ng, 0.002 ng, in triplicate reactions. The assay performed linearly across the whole range tested in both fluorescence and qcLAMP (Figure 4). The LoD was determined as the lowest DNA concentration at which 95% of replicates produced a positive result (Klymus et al., 2020). To ensure that the LoD was comparable to other reported studies, we estimated the number of COI copies in P. miles genomic DNA by qPCR (LoD at 0.002 ng is equivalent to 50 gene copies; Supplementary Figure S1, Supplementary Table S2). qcLAMP generally showed slower reaction times than fluorescence LAMP presumably due to device differences. At the LoD of 0.002 ng μL-1 reaction, the TTP for qcLAMP was 31.2 min, while for fluorescence LAMP it was 25.1 min, with high variability among replicates.
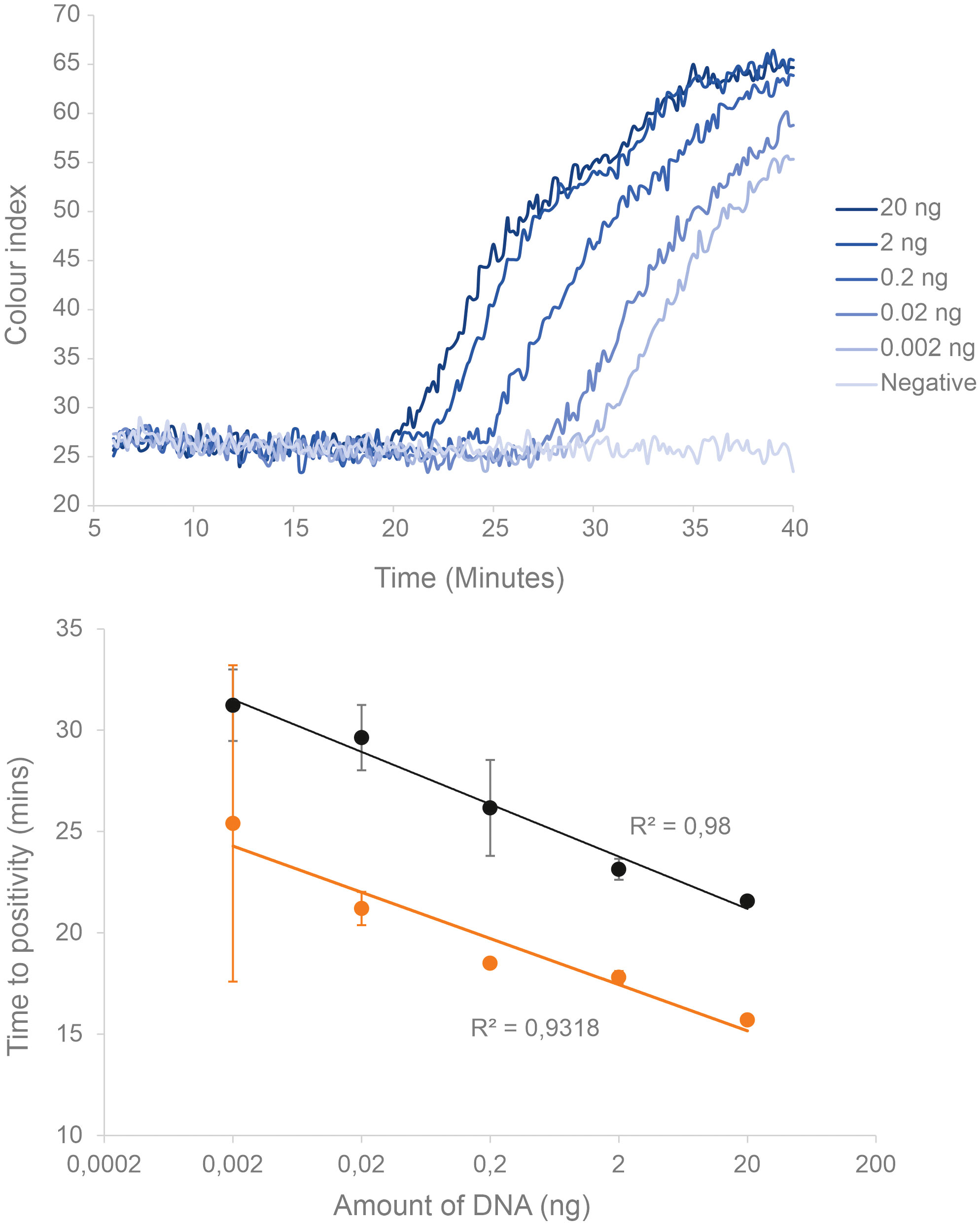
Figure 4 Sensitivity and quantitative range of the real-time P. miles LAMP assay established by serial dilutions of genomic DNA extracted from fish tissues (n=3). (Top) Real-time amplification curves for the different DNA concentrations, showing later onset of amplification for lower concentrations. (Bottom) Linear relationship of the corresponding TTP values for qcLAMP (black), as well as the equivalent experiment performed using fluorescence LAMP on a qPCR cycler (orange). Error bars indicate standard deviation.
The assay specificity was evaluated by testing against species closely related to the target as well as co-occurring species in the area. Samples were obtained both from fish tissue and from aquarium tanks containing P. miles and others that did not contain P. miles, simulating environmental negatives (Table 2). Experimental testing of the assay on the closely related P. volitans was not possible as we were unable to obtain samples. Species of the Scorpaena genus were prioritised in the tests as these are members of the Scorpenidae family and the closest relatives of P. miles after P. volitans in terms of sequence identity of the COI gene (S. porcus: 81.1%, S. scrofa: 82.3%, and S. notata: 81.4%). Specificity was further tested against fish species that typically co-occur with the P. miles in eastern Mediterranean waters. In total, 18 non-target species and six P. miles individuals were tested for primer specificity. There was no amplification from DNA of species closely related to P. miles, or DNA from aquarium tanks containing non-target species (Table 2), confirming that the assay is species specific.
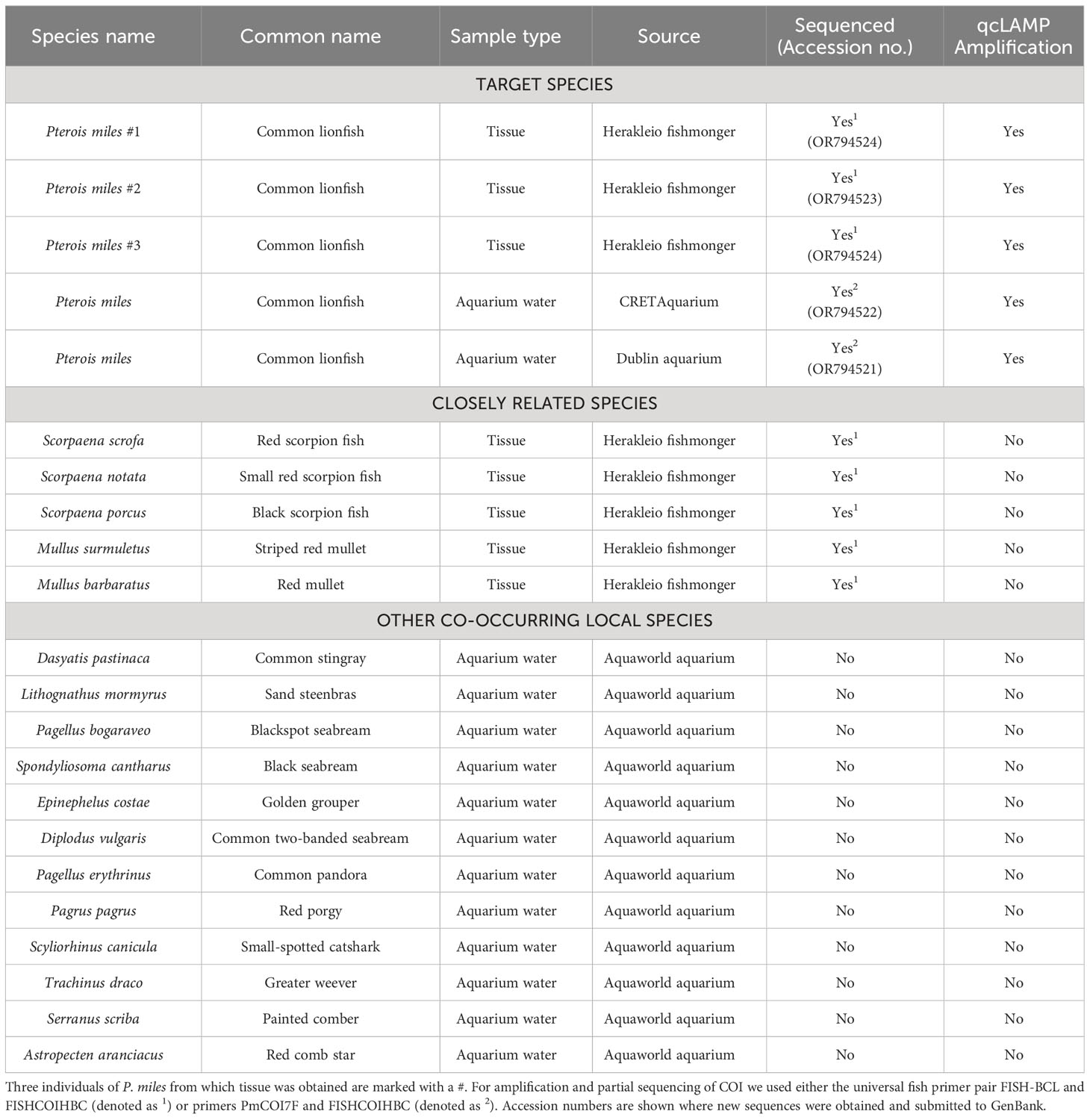
Table 2 Results of assay specificity testing showing fish species tested, including sample type and source.
The assay was validated by testing the P. miles qcLAMP protocol on seawater samples collected from nine locations around the island of Crete, Greece (Figure 1) as well as samples collected from the CRETAquarium and Dublin aquarium tanks. Following eDNA extraction, the amplification quality of the DNA was confirmed spectrophotometrically, fluorometrically and by PCR using the Universal fish primers (Supplementary Table S3). All environmental samples and negative field controls (e.g., bottled water) were analysed for P. miles by qcLAMP using the device. P. miles eDNA was detected in three of our nine field samples (Figure 1; Table 3); North Central Crete (Mononaftis beach), Eastern Crete (Vai beach), and Western Crete (Elafonisi beach), with the latter two calculated as having a considerably higher DNA concentration than the former. The target concentration in positive reactions was determined using the DNA standard curve equation. To extrapolate the DNA concentration per reaction, to the DNA concentration in the original sample in units of ng L-1, we accounted for the volume of seawater filtered per sample (Table 3), the initial elution volume of the DNA extraction (50-100 μL), and the amount of DNA added per reaction (4 μL). We calculated P. miles DNA concentrations in natural seawater ranging from ~ 0.15 – 3.19 ng L-1 and in the two aquarium samples 2.86 ng L-1 and 3.91 ng L-1 (Table 3). These concentrations are in the middle of the tested assay range.

Table 3 Details of sample volumes, TTP and calculated concentrations of eDNA recovered from natural and aquarium seawater samples.
Amplification of P. miles eDNA was feasible directly from the filter used to collect eDNA containing particles from the seawater. This was achieved with a simple heat-lysis step performed within the qcLAMP device immediately prior to amplification. We initially applied this protocol to samples collected from the CRETAquarium P. miles tank. A positive amplification was achieved with a TTP of 30 min, equivalent to 0.008 ng DNA. Extrapolating the amount of DNA from the filter piece to the equivalent for the whole filter membrane, and accounting for the volume filtered, the sample quantifications yielded a value of 2.5 ng L-1, which is similar to the 2.86 ng L-1 obtained previously for the same tank using extracted DNA samples (Table 3).
The scope of this study was to develop a simple and fast molecular assay that can accurately quantify the invasive P. miles from seawater samples, which is also applicable in the field. The LAMP method was selected since it is ideal for species-specific assays due to the six primers required along with the efficient production of amplicon. This provides the sensitivity and specificity to eliminate the risk of false positives from non-target amplification, and false negatives from low sensitivity (Rozas et al., 2017). However, several optimization steps were required to guarantee the reliability of the P. miles qcLAMP assay. An inherent limitation and risk of the LAMP method is the formation of primer dimers due to the high number of primers used (Becherer et al., 2020). We overcame this issue by recognising these interactions in silico and optimizing primer concentrations. We confirmed our assay’s ability to amplify different P. miles haplotypes to ensure its potential application on a regional scale. In silico analysis of the primers against P. volitans sequences suggests that this species is unlikely to amplify due to several SNPs in the primer binding sites. However, this should be evaluated experimentally if the assay is to be used in regions dominated by P. volitans. We also demonstrated the specificity of our assay in the presence of non-target species by testing against species of the same family (Scorpaenidae) and against additional other species that naturally occur in the regions that we conducted our field analyses. In addition to fish tissue samples, we used water samples from aquarium tanks that contained multiple local marine species to simulate a natural environmental sample.
Our assay showed a reliable and highly sensitive detection as low as 0.002 ng per reaction of the mitochondrial COI gene, comparable to other studies using LAMP as their detection method for eDNA in aquatic environments (Williams et al., 2017; Kageyama et al., 2022; Porco et al., 2022). This is approximately equivalent to 25 gene copies per reaction, which is within the usual LoD for LAMP and other quantitative DNA amplification methods, such as qPCR and qRPA. Interestingly, we noted that at this low DNA input, qcLAMP on the prototype device outperformed the standard fluorescence real-time LAMP performed on the qPCR cycler. This might be due to differences in the operation of the two devices, or the presence of inhibitors that may affect fluorophores.
Following assay design, optimization, and in vitro testing, the protocol was assessed on eDNA from natural samples collected from nine locations around Crete. The target species was detected in three out of the nine locations (Figure 1). These three locations are either semi-enclosed bays (Mononaftis) which are sheltered from onshore winds and waves, or are not exposed to prevailing winds throughout the year (Vai and Elafonissi). The highest eDNA concentrations of 3.12 ng L-1 and 3.19 ng L-1 were found at Vai and Elafonissi beaches, which were similar to those found in the aquarium water tanks. Even though these areas were sampled in different seasons (November 2021 and May 2022, respectively), both are protected nature reserves where fishing is not allowed and are therefore expected to have a higher overall fish biomass. Similar findings have been reported in Cyprus, where the highest densities of P. miles populations were recorded in Marine Protected Areas where fishing is forbidden (Kleitou et al., 2022). Mononaftis bay, which was sampled in May 2022, contained a lower amount of eDNA by one order of magnitude, even though it is SCUBA diving location due to it being rich in marine life. However, at the time of sampling the waters were still at pre-summer temperatures and only two individuals were observed by divers at the sampling location.
The lack of detection in the remaining regions could be attributed to potentially lower population densities of the target species, as well as to various biotic and abiotic factors that could affect eDNA concentration, distribution and persistence in the water column (Beng and Corlett, 2020). The effectiveness of eDNA detection depends on its persistence in the water column at the sampling point, which can, in turn, depend on its release from the target organism, temperature of water, transport across space and decay rates (Takahara et al., 2012; Klymus et al., 2015; Sansom et al., 2017; Murakami et al., 2019). Although eDNA is concentrated by filtering water samples, the relative concentration of target DNA can remain very small, thus making its detection particularly challenging (Wilcox et al., 2013). In the case of our target, its ecology and habitat preferences play a major role in the presence and distribution of its extra-organismal DNA. P. miles static lifestyle could limit the distribution of eDNA in the water column in calm waters without currents. Moreover, this species migrates to deeper waters in winter, likely to shelter from rougher weather conditions at the surface (Johnson and Swenarton, 2016). A solution could be to filter larger volumes of water instead of the 1 L filtered in this study and perform more frequent seasonal sampling.
A major bottleneck for field-based eDNA molecular assays is DNA extraction. LAMP has been demonstrated to provide sensitive target amplification from a number of crude sample matrices, including food and plant tissues (Yang et al., 2014; Kogovšek et al., 2017; Hodgetts, 2019). We tested whether we could amplify and detect P. miles eDNA directly from the filter that contained concentrated biomass, including tissue particles from P. miles. We successfully released DNA for amplification by using a portable, commercially available qcLAMP device and by directly inserting filter pieces in the LAMP reaction and preheating the reaction at 90 °C. Comparison to equivalent samples that were amplified after DNA extractions showed that LAMP was unaffected by remaining lysed particles. This agrees with previous results with LAMP using crude samples, where protozoan and bacterial DNA was amplified without a DNA extraction (Sriworarat et al., 2015; Papadakis et al., 2019; Papadakis et al., 2022). This approach is useful for on-site detection, although using only 1% of the total filter for amplification obviously reduces the overall sensitivity for field samples. Further testing is needed at locations with visually evident populations to establish the required LoD for management purposes.
In conclusion, we have developed and validated a fast, sensitive and species-specific qcLAMP assay combined with a novel prototype and commercially available qcLAMP device to detect and quantify eDNA of the marine invasive lionfish, P. miles. The assay was fully optimized for sensitivity, specificity, and quantitative capacity. We demonstrated the assay using the rapid and simple-to-use portable device which can be taken to the field for on-site analysis. We also demonstrated that on-site P. miles detection is feasible by direct amplification from the sample.
We have used our assay to detect the presence of P. miles in natural areas around Crete, where the species has been previously sighted. The presence of P. miles was detected in three out of nine locations: these are nature reserves protected from fishing and boating, and may thus have higher population densities of the target species. Our results highlight the need for further standardizing the method in the field, with targeted studies of eDNA concentration, SCUBA-based observational data and dispersal patterns in relation to species life history and hydrographic conditions so that quantitative eDNA results can be related to actual population distributions. Once fully standardized for application in the field, this will be an easy-to-use monitoring tool for the early detection of P. miles invasion routes, for collecting information about this species’ dispersal patterns and for providing timely data to policy makers to manage P. miles invasions. Our proposed qcLAMP methodology is generic and widely applicable for other species in marine and fresh waters, thus presenting a new tool for ecological monitoring.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethical approval was not required for the study involving animals in accordance with the local legislation and institutional requirements because experiments were only conducted on dead fish that were purchased from fish mongers; they were intended for human consumption.
KH-M: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. CG: Conceptualization, Formal analysis, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing. PX: Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. PK: Conceptualization, Formal analysis, Methodology, Validation, Writing – review & editing. MV: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. EG: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work (KH-M, MV and EG) was funded by the European Commission Horizon 2020 project TechOceanS under the action H2020-BG07 (GA 101000858). CG and PX were funded by the Operational Programme of Fisheries and Sea (OPFS) 2014-2020, Greece and the European Maritime and Fisheries Fund (EMFF) as part of the project “Monitoring and control of invasive alien species in Greece using innovative techniques under current and future climate conditions – INVASION” (MIS 5049543).
Dr. Aspasia Sterioti for access to the aquarium tanks at CRETAquarium, John McLaren for access to the aquarium tanks at Aquaworld Hersonissos, Philipos Marakis and Dr. Aggelos Ioannou at the Ecodive Center Heraklion for help with sampling at Mononaftis Bay and visual confirmation of lionfish presence, and Dr. Fiona Bracken at Dublin City University for aquarium samples.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1358793/full#supplementary-material
Baldwin C. C., Mounts J. H., Smith D. G., Weigt L. A. (2009). Genetic identification and color descriptions of early life-history stages of Belizean Phaeoptyx and Astrapogon (Teleostei: Apogonidae) with comments on identification of adult Phaeoptyx. Zootaxa 22, 1–22. doi: 10.11646/zootaxa.2008.1.1
Becherer L., Borst N., Bakheit M., Frischmann S., Zengerle R., Von Stetten F. (2020). Loop-mediated isothermal amplification (LAMP) - review and classification of methods for sequence-specific detection. Anal. Methods 12, 717–746. doi: 10.1039/C9AY02246E
Beng K. C., Corlett R. T. (2020). Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodiversity Conserv. 29, 2089–2121. doi: 10.1007/s10531-020-01980-0
Bennett R. H., Ellender B. R., Mäkinen T., Miya T., Pattrick P., Wasserman R. J., et al. (2016). Ethical considerations for field research on fishes. Koedoe 58, 1–15. doi: 10.4102/koedoe.v58i1.1353
Boudouresque C. F. (1999). The Red Sea-Mediterranean link: Unwanted effects of canals. In Invasive species and biodiversity management. Ed. Odd Terje Sandlund P. J. S., Åslaug Viken. Åslaug Viken: Kluwer Academic Publishers.
Bylemans J., Gleeson D. M., Duncan R. P., Hardy C. M., Furlan E. M. (2019). A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes. Environ. DNA 1, 402–414. doi: 10.1002/edn3.41
Capo E., Spong G., Koizumi S., Puts I., Olajos F., Königsson H., et al. (2021). Droplet digital PCR applied to environmental DNA, a promising method to estimate fish population abundance from humic-rich aquatic ecosystems. Environ. DNA 3, 343–352. doi: 10.1002/edn3.115
Chan F. T., Briski E. (2017). An overview of recent research in marine biological invasions. Mar. Biol. 164, 121. doi: 10.1007/s00227-017-3155-4
Claydon J. A. B., Calosso M. C., Traiger S. B. (2012). Progression of invasive lionfish in seagrass, mangrove and reef habitats. Mar. Ecol. Prog. Ser. 448, 119–129. doi: 10.3354/meps09534
Conversi A., Umani S. F., Peluso T., Molinero J. C., Santojanni A., Edwards M. (2010). The mediterranean sea regime shift at the end of the 1980s, and intriguing parallelisms with other european basins. PloS One 5. doi: 10.1371/journal.pone.0010633
Côté I. M., Smith N. S. (2018). The lionfish Pterois sp. invasion: Has the worst-case scenario come to pass. J. Fish Biol. 92, 660–689. doi: 10.1111/jfb.13544
Darling J. A., Mahon A. R. (2011). From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 111, 978–988. doi: 10.1016/j.envres.2011.02.001
Dimitriadis C., Galanidi M., Zenetos A., Corsini-Foka M., Giovos I., Karachle P., et al. (2020). Katsanevakis: Updating the occurrences of Pterois miles in the Mediterranean Sea, with considerations on thermal boundaries and future range expansion. Mediterr. Mar. Sci. 21, 62–69. doi: 10.12681/mms.21845
Doi H., Uchii K., Takahara T., Matsuhashi S., Yamanaka H., Minamoto T. (2015). Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PloS One 10, e0122763. doi: 10.1371/journal.pone.0122763
Francois P., Tangomo M., Hibbs J., Bonetti E. J., Boehme C. C., Notomi T., et al. (2011). Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 62, 41–48. doi: 10.1111/j.1574-695X.2011.00785.x
Fujiyoshi S., Yarimizu K., Miyashita Y., Rilling J., Acuña J. J., Ueki S., et al. (2021). Suitcase Lab: new, portable, and deployable equipment for rapid detection of specific harmful algae in Chilean coastal waters. Environ. Sci. pollut. Res. 28, 14144–14155. doi: 10.1007/s11356-020-11567-5
Galanidi M., Zenetos A., Bacher S. (2018). Assessing the socio-economic impacts of priority marine invasive fishes in the Mediterranean with the newly proposed SEICAT methodology. Mediterr. Mar. Sci. 107-123. doi: 10.12681/mms.15940
Giakoumi S., Katsanevakis S., Albano P. G., Azzurro E., Cardoso A. C., Cebrian E., et al. (2019). Management priorities for marine invasive species. Sci. Total Environ. 688, 976–982. doi: 10.1016/j.scitotenv.2019.06.282
Goldberg C. S., Turner C. R., Deiner K., Klymus K. E., Thomsen P. F., Murphy M. A., et al. (2016). Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 7, 1299–1307. doi: 10.1111/2041-210X.12595
Hodgetts J. (2019). Rapid sample preparation and LAMP for Phytoplasma detection. Methods Mol. Biol. 1875, 187–201. doi: 10.1007/978-1-4939-8837-2_15
Jerde C. L. (2021). Can we manage fisheries with the inherent uncertainty from eDNA? J. Fish Biol. 98, 341–353. doi: 10.1111/jfb.14218
Jerde C. L., Chadderton W. L., Mahon A. R., Renshaw M. A., Corush J., Budny M. L., et al. (2013). Detection of Asian Carp DNA as part of a Great Lakes basin-wide surveillance program. Can. J. Fisheries Aquat. Sci. 70, 522–526. doi: 10.1139/cjfas-2012-0478
Johnson E. G., Swenarton M. K. (2016). Age, growth and population structure of invasive lionfish (Pterois volitans/miles) in northeast Florida using a length-based, age-structured population model. PeerJ 2016. doi: 10.7717/peerj.2730
Kageyama S. A., Hoogland M. R., Tajjioui T., Schreier T. M., Erickson R. A., Merkes C. M. (2022). Validation of a portable eDNA detection kit for invasive carps. Fishes 7, 363. doi: 10.3390/fishes7060363
Kalsi S., Valiadi M., Tsaloglou M. N., Parry-Jones L., Jacobs A., Watson R., et al. (2015). Rapid and sensitive detection of antibiotic resistance on a programmable digital microfluidic platform. Lab. Chip 15, 3065–3075. doi: 10.1039/C5LC00462D
Kaneko H., Kawana T., Fukushima E., Suzutani T. (2007). Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 70, 499–501. doi: 10.1016/j.jbbm.2006.08.008
Katoh K., Standley D. M. (2013). MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010%JMolecularBiologyandEvolution
Keller A. G., Grason E. W., McDonald P. S., Ramón-Laca A., Kelly R. P. (2022). Tracking an invasion front with environmental DNA. Ecol. Appl. 32, e2561. doi: 10.1002/eap.2561
Kleitou P., Hall-Spencer J. M., Rees H. C., Kletou D. (2022). Guide to lionfish management in the Mediterranean (Plymouth UK: University of Plymouth).
Kleitou P., Hall-Spencer J. M., Savva I., Kletou D., Hadjistylli M., Azzurro E., et al. (2021). The case of lionfish (Pterois miles) in the mediterranean sea demonstrates limitations in EU legislation to address marine biological invasions. J. Mar. Sci. Eng. 9, 325. doi: 10.3390/jmse9030325
Klymus K. E., Merkes C. M., Allison M. J., Goldberg C. S., Helbing C. C., Hunter M. E., et al. (2020). Reporting the limits of detection and quantification for environmental DNA assays. Environ. DNA 2, 271–282. doi: 10.1002/edn3.29
Klymus K. E., Richter C. A., Chapman D. C., Paukert C. (2015). Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 183, 77–84. doi: 10.1016/j.biocon.2014.11.020
Kogovšek P., Mehle N., Pugelj A., Jakomin T., Schroers H.-J., Ravnikar M., et al. (2017). Rapid loop-mediated isothermal amplification assays for grapevine yellows phytoplasmas on crude leaf-vein homogenate has the same performance as qPCR. Eur. J. Plant Pathol. 148, 75–84. doi: 10.1007/s10658-016-1070-z
Kutti T., Johnsen I. A., Skaar K. S., Ray J. L., Husa V., Dahlgren T. G. (2020). Quantification of eDNA to map the distribution of cold-water coral reefs. Front. Mar. Sci. 7. doi: 10.3389/fmars.2020.00446
Larson E. R., Graham B. M., Achury R., Coon J. J., Daniels M. K., Gambrell D. K., et al. (2020). From eDNA to citizen science: emerging tools for the early detection of invasive species. Front. Ecol. Environ. 18, 194–202. doi: 10.1002/fee.2162
Mauvisseau Q., Davy-Bowker J., Bulling M., Brys R., Neyrinck S., Troth C., et al. (2019). Combining ddPCR and environmental DNA to improve detection capabilities of a critically endangered freshwater invertebrate. Sci. Rep. 9, 14064. doi: 10.1038/s41598-019-50571-9
McPartlin D. A., Loftus J. H., Crawley A. S., Silke J., Murphy C. S., O’Kennedy R. J. (2017). Biosensors for the monitoring of harmful algal blooms. Curr. Opin. Biotechnol. 45, 164–169. doi: 10.1016/j.copbio.2017.02.018
Murakami H., Yoon S., Kasai A., Minamoto T., Yamamoto S., Sakata M. K., et al. (2019). Dispersion and degradation of environmental DNA from caged fish in a marine environment. Fisheries Sci. 85, 327–337. doi: 10.1007/s12562-018-1282-6
Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, e63–e63. doi: 10.1093/nar/28.12.e63
Papadakis G., Pantazis A. K., Fikas N., Chatziioannidou S., Tsiakalou V., Michaelidou K., et al. (2022). Portable real-time colorimetric LAMP - device for rapid quantitative detection of nucleic acids in crude samples. Sci. Rep. 12, 1–15. doi: 10.1038/s41598-022-06632-7
Papadakis G., Pantazis A. K., Ntogka M., Parasyris K., Theodosi G. I., Kaprou G., et al. (2019). 3D-printed Point-of-Care platform for genetic testing of infectious diseases directly in human samples using acoustic sensors and a smartphone. ACS Sensors 4, 1329–1336. doi: 10.1021/acssensors.9b00264
Pastor F., Valiente J. A., Khodayar S. (2020). A warming Mediterranean: 38 years of increasing sea surface temperature. Remote Sens. 12, 1–16. doi: 10.3390/rs12172687
Phillips E. W., Kotrschal A. (2021). Where are they now Tracking the Mediterranean lionfish invasion via local dive centers. J. Environ. Manage. 298, 113354. doi: 10.1016/j.jenvman.2021.113354
Porco D., Hermant S., Purnomo C. A., Horn M., Marson G., Colling G. (2022). eDNA-based detection of the invasive crayfish Pacifastacus leniusculus in streams with a LAMP assay using dependent replicates to gain higher sensitivity. Sci. Rep. 12, 6553. doi: 10.1038/s41598-022-10545-w
Poursanidis D., Kalogirou S., Azzurro E., Parravicini V., Bariche M. (2020). Habitat suitability, niche unfilling and the potential spread of Pterois miles in the Mediterranean Sea. Mar. pollut. Bull. 154, 111054. doi: 10.1016/j.marpolbul.2020.111054
Qin Y., Chen G., Zhang C., Wang Y., Zhou J. (2019). Development of loop-mediated isothermal amplification combined with a chromatographic lateral-flow dipstick for rapid detection of Chattonella marina. Harmful Algae 89, 101666. doi: 10.1016/j.hal.2019.101666
Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248%JMolecularBiologyandEvolution
Sansom B. J., Sassoubre L. M., Environmental D. N. A. (2017). (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environ. Sci. Technol. 51, 14244–14253. doi: 10.1021/acs.est.7b05199
Savva I., Chartosia N., Antoniou C., Kleitou P., Georgiou A., Stern N., et al. (2020). They are here to stay: the biology and ecology of lionfish (Pterois miles) in the Mediterranean Sea. J. Fish Biol. 97, 148–162. doi: 10.1111/jfb.14340
Schumer G., Crowley K., Maltz E., Johnston M., Anders P., Blankenship S. (2019). Utilizing environmental DNA for fish eradication effectiveness monitoring in streams. Biol. Invasions 21, 3415–3426. doi: 10.1007/s10530-019-02056-z
Sriworarat C., Phumee A., Mungthin M., Leelayoova S., Siriyasatien P. (2015). Development of loop-mediated isothermal amplification (LAMP) for simple detection of Leishmania infection. Parasit Vectors 8, 591. doi: 10.1186/s13071-015-1202-x
Takahara T., Minamoto T., Yamanaka H., Doi H., Kawabata Z. I. (2012). Estimation of fish biomass using environmental DNA. . PloS One 7, 3–10. doi: 10.1371/journal.pone.0035868
Ulman A., Ali F. Z., Harris H. E., Adel M., Mabruk S. A. A. A., Bariche M., et al. (2022). Lessons from the western atlantic lionfish invasion to inform management in the mediterranean. Front. Mar. Sci. 9. doi: 10.3389/fmars.2022.865162
Wilcox T. M., McKelvey K. S., Young M. K., Jane S. F., Lowe W. H., Whiteley A. R., et al. (2013). Robust detection of rare species using Environmental DNA: The importance of primer specificity. PloS One 8. doi: 10.1371/journal.pone.0059520
Williams M. R., Stedtfeld R. D., Engle C., Salach P., Fakher U., Stedtfeld T., et al. (2017). Isothermal amplification of environmental DNA (eDNA) for direct field-based monitoring and laboratory confirmation of Dreissena sp. PloS One 12, e0186462. doi: 10.1371/journal.pone.0186462
Yang Q., Wang F., Prinyawiwatkul W., Ge B. (2014). Robustness of Salmonella loop-mediated isothermal amplification assays for food applications. J. Appl. Microbiol. 116, 81–88. doi: 10.1111/jam.12340
Yu D., Shen Z., Chang T., Li S., Liu H. (2021). Using environmental DNA methods to improve detectability in an endangered sturgeon (Acipenser sinensis) monitoring program. BMC Ecol. Evol. 21, 216. doi: 10.1186/s12862-021-01948-w
Zenetos A., Albano P. G., LÓPez Garcia E., Stern N. I. R., Tsiamis K., Galanidi M. (2022). Established non-indigenous species increased by 40% in 11 years in the Mediterranean Sea. Mediterr. Mar. Science 23(1). doi: 10.12681/mms.29106
Keywords: lionfish, Pterois miles, invasive species, environmental DNA, loop mediated isothermal amplification, Eastern Mediterranean
Citation: Hartle-Mougiou K, Gubili C, Xanthopoulou P, Kasapidis P, Valiadi M and Gizeli E (2024) Development of a quantitative colorimetric LAMP assay for fast and targeted molecular detection of the invasive lionfish Pterois miles from environmental DNA. Front. Mar. Sci. 11:1358793. doi: 10.3389/fmars.2024.1358793
Received: 20 December 2023; Accepted: 26 February 2024;
Published: 18 March 2024.
Edited by:
Mark Botton, Fordham University, United StatesReviewed by:
Ioannis A. Giantsis, Aristotle University of Thessaloniki, GreeceCopyright © 2024 Hartle-Mougiou, Gubili, Xanthopoulou, Kasapidis, Valiadi and Gizeli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martha Valiadi, bWFydGhhX3ZhbGlhZGlAaW1iYi5mb3J0aC5ncg==; Electra Gizeli, Z2l6ZWxpQGltYmIuZm9ydGguZ3I=
†These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.