
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 02 April 2024
Sec. Marine Megafauna
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1358695
This article is part of the Research Topic Population Structure and Connectivity of Marine Turtles in the Pacific and Indian Oceans View all 14 articles
 Lalu M. Iqbal Sani1,2*
Lalu M. Iqbal Sani1,2* Jamaludin3
Jamaludin3 Gunawan Hadiko3
Gunawan Hadiko3 Ema Herma3
Ema Herma3 Emi Inoguchi4
Emi Inoguchi4 Michael P. Jensen5,6
Michael P. Jensen5,6 Christine A. Madden5
Christine A. Madden5 Hideaki Nishizawa7
Hideaki Nishizawa7 Leni Maryani1
Leni Maryani1 Achmad Farajallah8
Achmad Farajallah8 Beginer Subhan1
Beginer Subhan1 Dietriech G. Bengen1
Dietriech G. Bengen1 Hawis Madduppa1,2
Hawis Madduppa1,2Indonesia is an archipelagic country that provides important nesting and foraging habitats for the critically endangered hawksbill turtle (Eretmochelys imbricata). Although many studies have investigated this migratory species globally, there is a lack of information on the population structure and geographic boundary of this species in Indonesia. This study aims to investigate the genetic diversity and population structure of six nesting sites in the Java Sea region of Indonesia. The control region (d-loop) sequence (818 bp) was obtained from 152 individuals, resulting in 20 haplotypes. This study revealed 13 new haplotypes, 12 of which were rare and observed only in a single sampling location. Results showed that the Indonesian population shares haplotypes with rookeries from Peninsular Malaysia and Australia, as indicated by EiIP08 and EiIP49. The haplotype diversity (h) was highest at the more northern rookery sites (Segama Besar Island, Kimar Island, East Belitung, Penambun Island) (h: 0.6033 - 0.9167; 4 - 9 haplotypes) compared to the other two rookeries located in the Seribu Archipelago (Tidung Island and Harapan Island) (h: 0.3354 - 0.5965; 3 - 6 haplotypes). Furthermore, population structure analysis showed a pan-mixed population between Tidung Island and Harapan Island (FST: 0.003, P > 0.05) but significant population structure across all other rookery sites (FST: 0.083 - 0.486, P < 0.05), resulting in five newly identified Management Units (MUs) in this area. This study showed the presence of a complex population structure with local haplotypes and narrow population boundaries for rookeries in relatively close proximity within the Java Sea, prompting local management and highlighting the need for more research on hawksbill turtle populations in Indonesia.
Indonesia, an archipelagic country, is home to many critical foraging and nesting habitats for at least four species of sea turtles: hawksbill turtle (Eretmochelys imbricata), green turtle (Chelonia mydas), olive ridley (Lepidochelys olivacea), and leatherback sea turtle (Dermochelys coriacea) (Halim et al., 2001; Wiadnyana, 2003; Hutomo and Moosa, 2005; Asaad et al., 2018). In Southeast Asia, sea turtles face numerous threats, ranging from habitat destruction, marine heatwaves to illegal trade and bycatch (Parsons, 1972; Limpus, 1993; Persoon et al., 1996; Meylan and Donnelly, 1999; da Silva et al., 2016; Konsta et al., 2022; Jeethvendra et al., 2023), leading to a worrying decline in many populations (e.g., Malaysia, Indonesia, Myanmar, Thailand) (Groombridge and Luxmoore, 1989; Sutanto-S et al., 1989; Kitchener, 1996; Meylan and Donnelly, 1999; Pilcher and Ali, 1999; Nishizawa et al., 2016; Hemelikova et al., 2021). This is particularly true for the critically endangered hawksbill turtle, listed on the International Union for Conservation of Nature (IUCN) Red List (Mortimer and Donnelly, 2008). Indonesia is known for its large number of hawksbill turtle nesting beaches (Meylan and Donnelly, 1999; Putrawidjaja, 2000; Wiadnyana, 2003; Zainudin et al., 2007; Tapilatu, 2017; Harahap et al., 2020), but there is a striking gap in our understanding of these populations, particularly in relation to their population structure. This lack of knowledge about the connectivity between nesting and foraging areas could be a significant barrier to effective conservation efforts (Vargas et al., 2016; Madden Hof et al., 2023).
In general, adult hawksbill turtles have been documented migrating to over 1,600 km from their nesting sites to foraging areas (Nietschmann, 1981; Miller et al., 1998; Hawkes et al., 2012; Marcovaldi et al., 2012; Pilcher et al., 2019), though in some regions, hawksbill turtles appear to remain near their natal nesting sites (Gaos et al., 2017). Indonesia plays an important role in preserving coral reef ecosystems, which serves as the primary habitat for hawksbill turtles (León and Bjorndal, 2002; Hutomo and Moosa, 2005; Ismuranty, 2006; Bustard, 2016; Hemelikova et al., 2021). Thus, the complex relationship between scattered nesting sites and abundant foraging grounds highlights the difficulty in managing and conserving these species. Understanding the connectivity between these areas is crucial for effective conservation (Palsbøll et al., 2007; Wallace et al., 2010). Female turtles may exhibit nesting behavior across several sites, but it’s important to identify areas with restricted genetic exchange to identify a single genetic stock based on the degree of female philopatry to particular nesting sites (Reece et al., 2005; Vargas et al., 2016).
Genetic tools provide a way to elucidate movement between nesting sites and foraging grounds (Pella and Masuda, 2001; Bolker et al., 2007), as well as gene flow between nesting sites (Jensen et al., 2019). However, the critically endangered status of the hawksbill turtle, combined with the logistical challenges of sampling across scattered and remote areas, has limited research for this species in Indonesia. While genetic studies on hawksbill turtle populations have been extensive globally (Broderick et al., 1994; Reece et al., 2005; Bowen et al., 2007; FitzSimmons and Limpus, 2014; Gaos et al., 2016; Nishizawa et al., 2016; Vargas et al., 2016; Bell and Jensen, 2018; FitzSimmons et al., 2020; Madden Hof et al., 2023), there remains a critical gap in our understanding of the genetic structure of hawksbill turtles in the Indo-Pacific region, particularly Indonesia. This lack of baseline genetic information hinders our ability to accurately determine the natal origin of foraging hawksbills or to trace the origin of illegally harvested tortoiseshell (LaCasella et al., 2021). With Indonesia known for hosting regionally large hawksbill nesting populations (Salm and Halim, 1984; Schulz, 1987; Limpus, 1993; Meylan and Donnelly, 1999), obtaining genetic information from these nesting sites is urgently needed. A previous study has indicated a loss of genetic variation in some nesting sites, possibly due to population decline (Nishizawa et al., 2016). Thus, it is vital to investigate the genetic diversity and population structure of nesting hawksbill turtles in Indonesia.
This study specifically aims to reveal the population genetic structure of Indonesian hawksbill turtles in the Java Sea by analyzing the control region (d-loop) of mitochondrial DNA (mtDNA); this region has been utilized to define management units (MUs) based on genetic differentiation (Moritz, 1994), and has been proven to effectively differentiate sea turtle nesting populations (Bowen et al., 2005; FitzSimmons and Limpus, 2014; Gaos et al., 2016; Nishizawa et al., 2016; Vargas et al., 2016; Bell and Jensen, 2018; Madden Hof et al., 2023). To date, nine MUs have been identified across the Indo-Pacific (Madden Hof et al., 2023). Many populations of hawksbill turtles in the Java Sea experienced a substantial population decline during the 20th century (Meylan and Donnely, 1999; Mortimer and Donnelly, 2008). At least 13 nesting sites have been recorded across seven provinces (i.e., Bangka Belitung, Jakarta, Jawa Tengah, Jawa Timur, Kalimantan Barat, Kalimantan Selatan, Lampung) (Suganuma et al., 1999; Meylan and Donnely, 1999). Conservation efforts have led to population increase in some of these nesting sites (https://seaturtle.or.id/). However, it is crucial to determine the connectivity between these populations to carry out effective management actions in the future. Our research investigates the genetic diversity, population structure, and connectivity among hawksbill turtles from six nesting sites in this region. Our study provides important baseline data on the genetics of hawksbill turtles in the Java Sea, which will have significant implications for their conservation and management in Indonesia.
A total of 152 tissue samples were collected from nesting female hawksbill turtles at six locations in the Java Sea of Indonesia between 2002 to 2021 year-round. A small piece of skin tissue of the hindlimb was collected from the different individuals and put into a prefilled 2 ml cryotube with 96% ethanol. The specific locations and number of samples from each location are as follows: Tidung Island (n=26), Harapan Island (n=19), Segama Island (n=25), Kimar Island (n=24), East Belitung (n=30), and Penambun Island (n=28) (Figure 1; Table 1). The samples collected from East Belitung consisted of two locations, Momparang Island (n=8) and Pesemut Island (n=22), which are approximately 3.5 km apart. Therefore, these two sites were considered one nesting site based on tagging data observations between 1999 and 2020 (personal communication to ELNA https://www.elna.or.jp/ and YPLI http://seaturtle.or.id/).
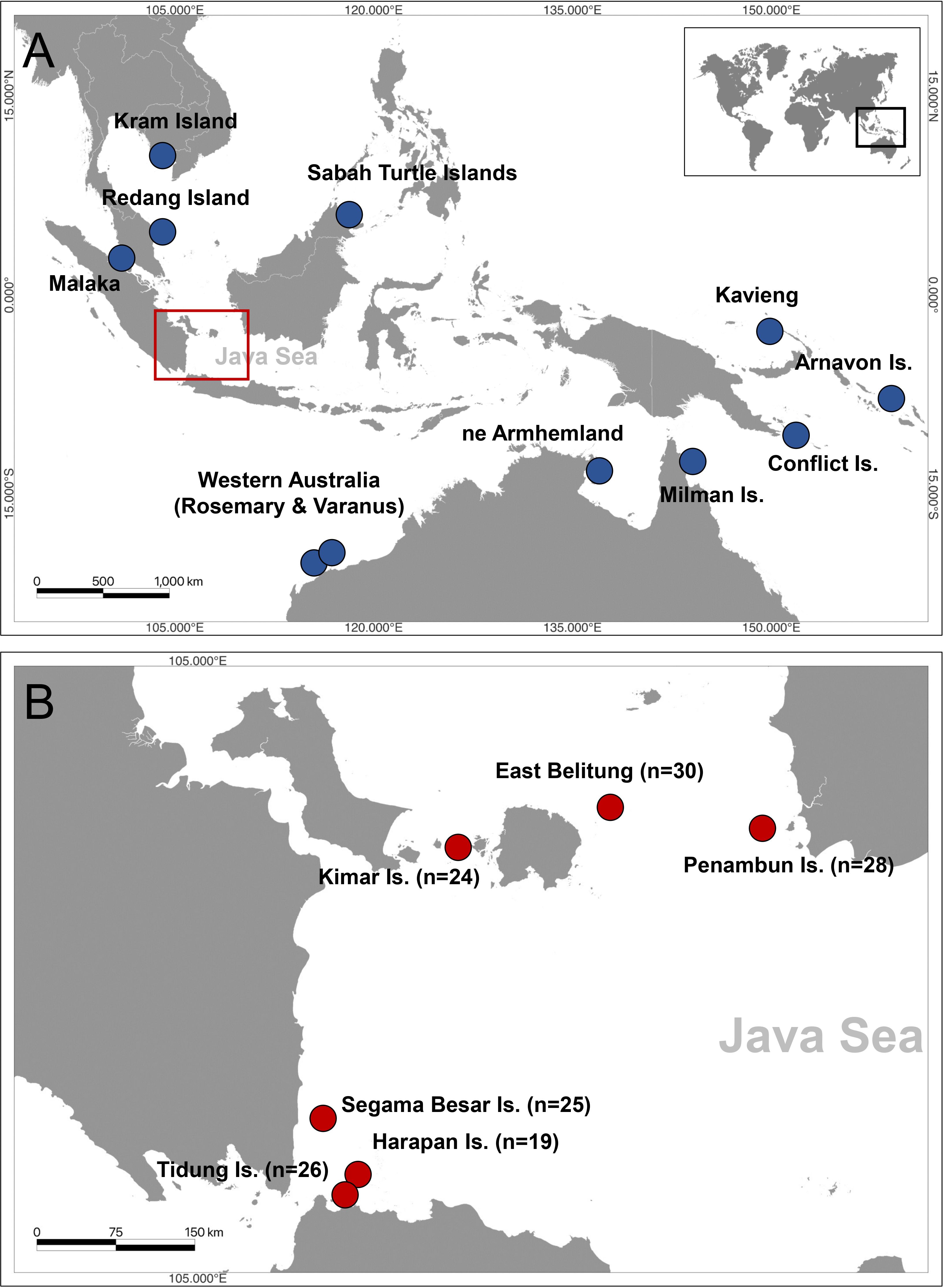
Figure 1 Map showing hawksbill turtle (Eretmochelys imbricata) genetic studies in (A) Indo-Pacific region (blue) including Malaysia, Thailand, Australia, Papua New Guinea, and Solomon Islands, (B) 152 tissue samples used from six sites in the Java Sea region of Indonesia (Red) including Tidung Island, Harapan Island, Segama Besar Island, Kimar Island, Momparang and Pesemut Island of East Belitung, and Penambun Island.
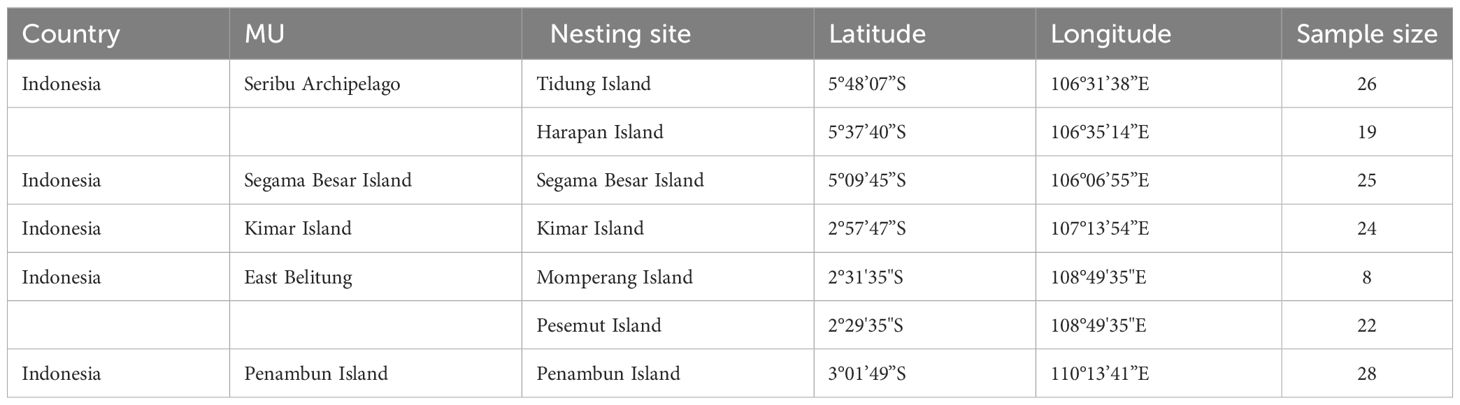
Table 1 Detailed information on hawksbill turtle (Eretmochelys imbricata) samples used in this study.
All samples were transported and stored at Marine Biodiversity and Biosystematic Laboratory (Biodivsi), Department of Marine and Science and Technology, IPB University in Bogor, Indonesia. In the laboratory, the samples were stored in a -24° C freezer to ensure long-term preservation of the genetic material.
Genomic DNA was extracted from the collected tissue samples using Qiagen Dneasy Blood and Tissue Kits. The targeted ~800 bp of mitochondrial DNA control region (d-loop) was amplified by PCR using the primers LCM15382 and H950 (Abreu-Grobois et al., 2006), the same primers used in other population genetics studies of hawksbill turtles (Reece et al., 2005; Bowen et al., 2007; FitzSimmons and Limpus, 2014; Gaos et al., 2016; Nishizawa et al., 2016; Vargas et al., 2016; Bell and Jensen, 2018; Madden Hof et al., 2023). DNA amplification was performed with 13 μL MyFi 2x hotstart polymerase (Bioline), 1 μL forward and 1 μL reverse primers, 9 μL ddH2O, and 3 μL DNA templates (3 μL ddH2O was used for the negative control). The DNA amplification processes were run with an initial denaturation at 95°C for 8 minutes, followed by 35 cycles of denaturation at 95°C for 45 seconds, annealing at 52°C for 45 seconds, and extension at 72°C for 45 seconds, then final extension at 72°C for 5 minutes (Vargas et al., 2016). All PCR products were then visualized on 2% agarose gel electrophoresis and sequenced at DNA Sanger sequencing facility in Apical Scientific, Malaysia.
Raw sequences of 152 samples collected from six sites in Indonesia were imported and edited in MEGA11 (Tamura et al., 2021). All sequences were checked to avoid double peak chromatograms of the nucleotide sequence, and then processed to identify the number of unique haplotypes using DnaSP V.6.12.03 (Rozas et al., 2017). The haplotypes were then identified using Basic Local Alignment Search Tool (BLAST) via National Centre Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/) and searched against the ShellBank database (www.shellbankproject.org), a global repository for marine turtle haplotype data. This comparison determined if sequences matched previously known haplotypes or were new to science. Haplotype sequences that were not 100% identical to previously published haplotypes were classified as new and assigned a unique name following the standardized nomenclature for Indo-Pacific haplotypes, prefixed with EiIP and the next sequential number. Haplotype identification was done strictly by double check from different people. These new haplotypes were then submitted to NCBI GenBank (accession no OR961305 - OR961456) and ShellBank for inclusion in the global genetic database.
To visualize the relation between newly identified haplotypes and existing haplotypes of Indo-Pacific hawksbill turtles, a haplotype network was constructed using the median-joining network (Bandelt et al., 1999) in PopArt (Leigh and Briant, 2015). This network provides a visual representation of the genetic relationship between haplotypes (EiIP) of hawksbill turtles identified in this study. The haplotype number, haplotype diversity (h), and nucleotide diversity (π) were calculated in the Arlequin v.3.5 program (Excoffier and Lischer, 2010) for each site. The analysis of population structure was based on conventional FST measures based on haplotype frequencies. The significance level was determined using 10,000 permutation replicates, with a threshold of p < 0.05 for statistical significance. Pairwise FST was visualized using pheatmap package (Kolde and Kolde, 2015), and Mantel test was carried out based on Spearman correlation with 10,000 permutation test to evaluate the correlation between FST value and geographical distance using vegan package (Oksanen et al., 2007) in R, using Rstudio (RStudio Team, 2013).
To provide a regional perspective on the genetic diversity and structure of hawksbill turtle populations, haplotype frequencies were visualized on a map along with previously published hawksbill turtle rookeries in the region using rookery baseline data from the ShellBank database. This included published data from nesting hawksbill turtles in Malaysia at Malaka, Redang Island, and Sabah Turtle Islands (Wahidah and Syed Abdullah, 2009; Nishizawa et al., 2016; Vargas et al., 2016), Thailand at Kram Island (Wahidah and Syed Abdullah, 2009); Australia at Western Australia, northeast Arnhemland in the Northern Territory and North Queensland (Vargas et al., 2016; Bell and Jensen, 2018; LaCasella et al., 2021), in the Solomon Islands (Vargas et al., 2016; LaCasella et al., 2021), Conflict Islands, Milne Bay, and Kavieng, New Ireland (Madden Hof et al., 2023).
The analysis of the mtDNA control region sequences from 152 individuals across six sites in the Java Sea identified a total of 20 haplotypes. Among these, 13 were new haplotypes, indicating a significant discovery in the genetic diversity of hawksbill turtles in this region (Figure 2). The distribution of both new and previously identified haplotypes varied across the sites. Previously identified haplotypes found in our study were EiIP08 (23%), EiIP49 (19%), EiIP67 (18%), EiIP122 (8%), EiIP53 (5%), EiIP50 (3%), and EiIP123 (1%) (Figure 3A). Haplotype EiIP67 was particularly widespread and found in all sites except Penambun Island, and EiP49 was found in all sites except Segama Besar Island and Tidung Island. In contrast, EiIP08 was only found in Tidung Island, Harapan Island, and Segama Besar Island. For the newly identified haplotypes, EiIP153 was dominant (7%), distributed on Kimar Island and Penambun Island. The remaining 12 newly identified haplotypes (EiIP) were rare and site-specific (0.66-6.58%). The haplotype network suggested that most of the newly identified haplotypes likely mutated from either EiP53 (EiIP152, EiIP153, EiIP154, EiIP140, EiIP157, EiIP160), or EiP08 (EiIP155, EiIP158, EiIP159).
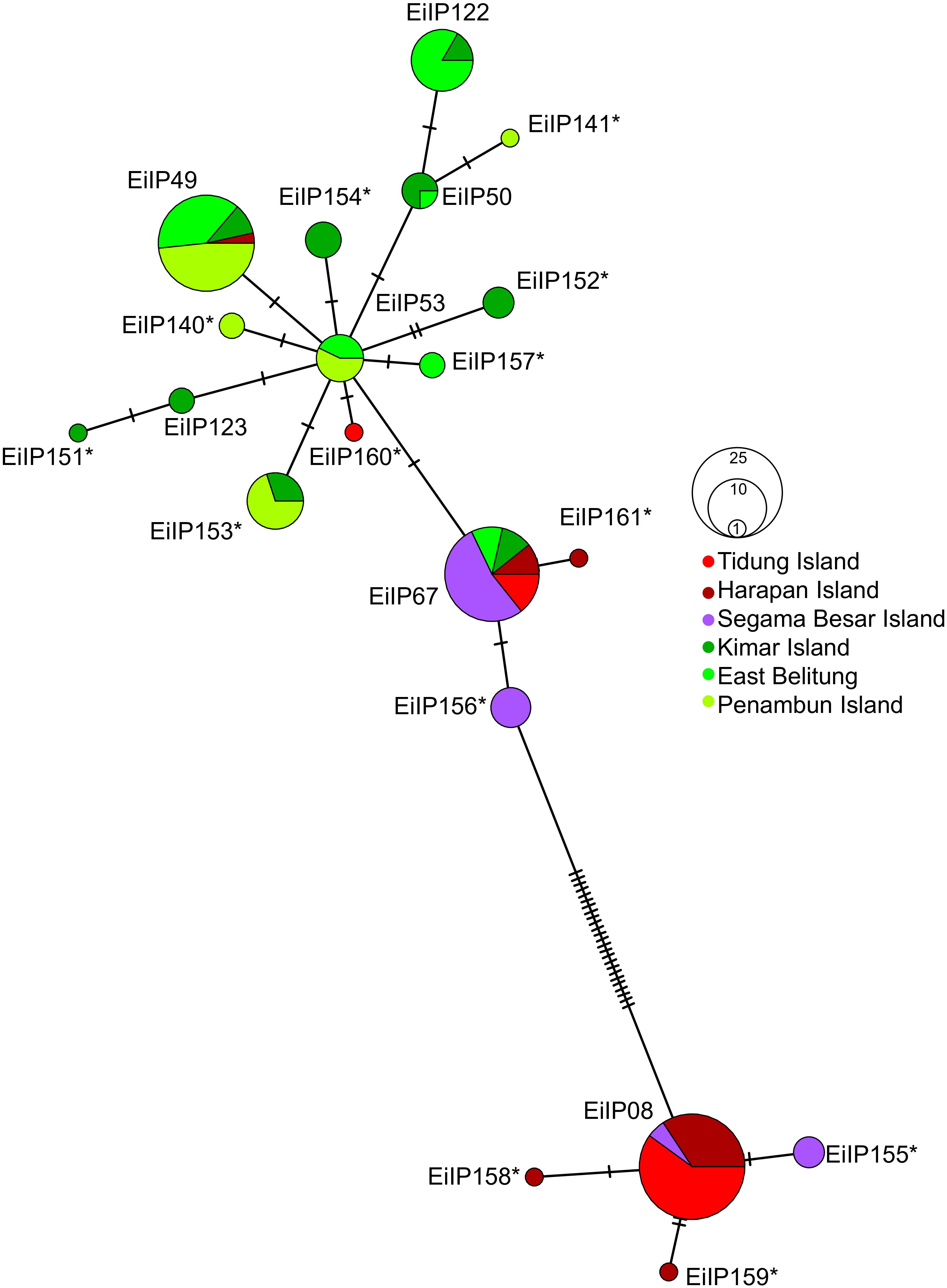
Figure 2 Haplotype network of hawksbill turtles (Eretmochelys imbricata) in the Java Sea region generated based on median-joining network. Each line between points represents a single mutational step. Asterisk symbol (*) indicates new identified haplotypes in the Indo-Pacific.
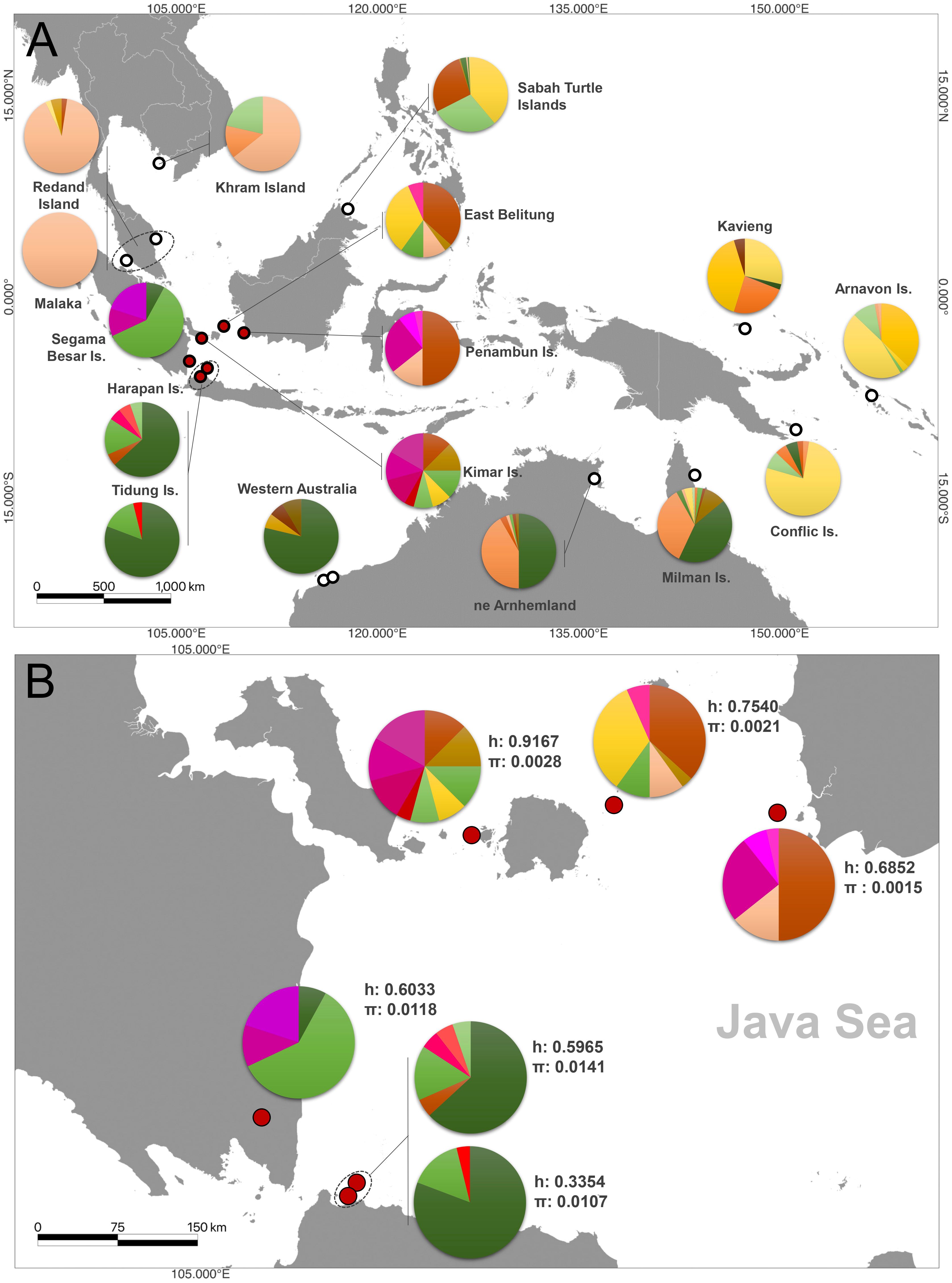
Figure 3 Haplotype distributions and frequencies of hawksbill turtles (Eretmochelys imbricata) (A) across Indo-Pacific, (B) Genetic diversity information of six nesting sites in the Java Sea region of Indonesia.
The haplotype diversity was the highest in Kimar Island (h: 0.9167), followed by East Belitung (h: 0.7540), Penambun Island (h: 0.6852), and Segama Besar Island (h: 0.6033), Harapan Island (h: 0.5965), and Tidung Island (h: 0.3354) (Figure 3B; Table 2). Nucleotide diversity was highest at Harapan Island (π: 0.0141), Segama Besar Island (π: 0.0118), and Tidung Island (π: 0.0107). These sites also exhibited a high number of polymorphic sites (28 to 32) compared to other with only 5 to 10 polymorphic sites (Table 2).
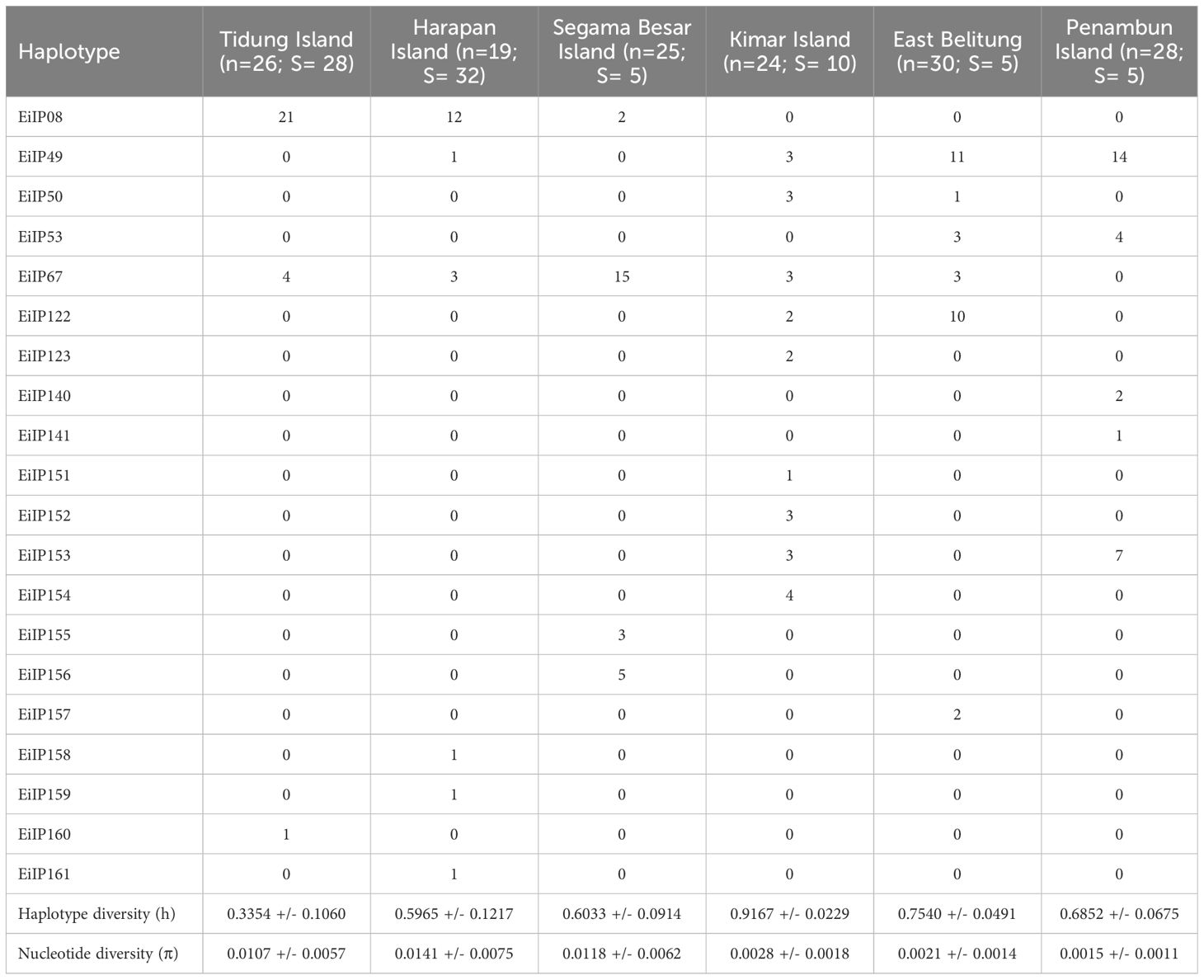
Table 2 Genetic diversity of hawksbill turtle (Eretmochelys imbricata) in the Java Sea of Indonesia (n: sample size, h: haplotype diversity, π: nucleotide diversity, S: polymorphic sites).
The results indicate substantial genetic segregation, suggesting the existence of at least five demographically independent genetic stocks (or MUs) within the Java Sea. The only exception to this pattern was observed between Tidung Island and Harapan Island, located in Seribu Archipelago (FST: 0.003, P > 0.05), suggesting a close genetic relationship between these two sites (Figure 4). The other genetic stocks showing clear differentiation include Kimar Island, Segama Besar Island, East Belitung, and Penambun Island. The Mantel test results suggest positive correlations between FST or population structure with the geographical distance (r: 0.525, P < 0.05).
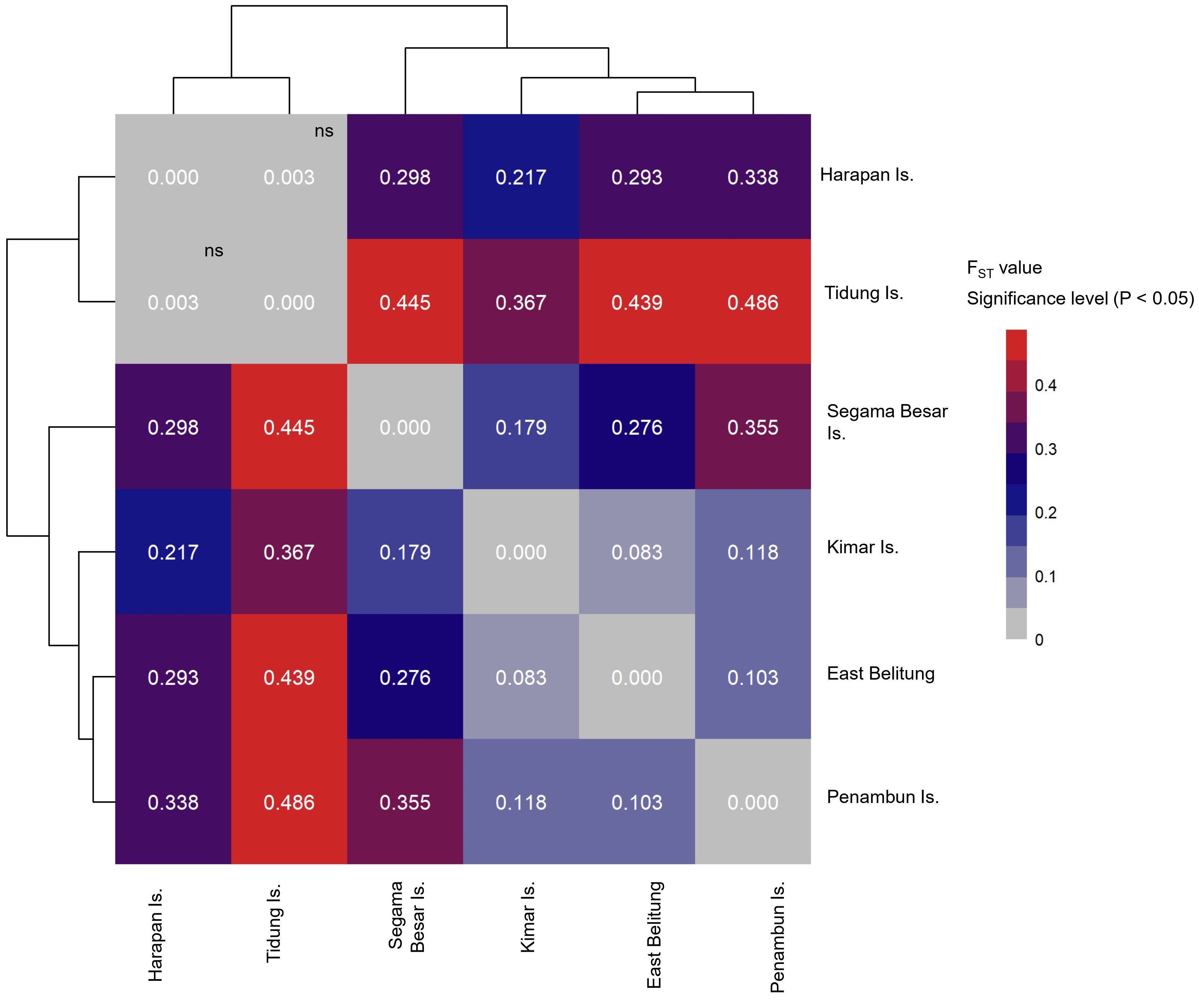
Figure 4 Heatmap of pairwise FST value of of hawksbill turtle (Eretmochelys imbricata) of six nesting sites in the Java Sea region of Indonesia (ns: not significant or P > 0.05).
The hawksbill turtle, classified as critically endangered, faces several threats, particularly illegal trade and bycatch (Meylan and Donnelly, 1999; Bourjea et al., 2008; da Silva et al., 2016), requiring effective conservation strategies, including the establishment of MUs (Broderick et al., 1994; FitzSimmons, 2020; Tabib et al., 2011, Tabib et al., 2014; Vargas et al., 2016; Bell and Jensen, 2018; Madden Hof et al., 2023). While MUs have been widely recognized as essential for sea turtle conservation, there is a significant knowledge gap in the genetic analysis of hawksbill turtle rookeries in the Indo-Pacific region, particularly across Indonesia (Vargas et al., 2016; LaCasella et al., 2021; Madden Hof et al., 2023). For sea turtle conservation, the genetic analysis contributes to determining an adequate scale for demographic (Jensen et al., 2013; Dutton et al., 2014; Reid et al., 2017; Jensen et al., 2019; FitzSimmons et al., 2020; Vilaça et al., 2022) and genetic diversity (Jensen et al., 2016; Madduppa et al., 2021), as well as identifying the likely sources of recruits for population expansion (Komoroske et al., 2017). Identifying five distinct MUs populations within the Java Sea area brings the total number of MUs in the Indo-Pacific to 14, with Indonesia contributing at least five, and likely many more. The population structure revealed in the Java Sea also brings a new perspective on how demographic variations may exist in very small areas. Therefore, it is crucial to acknowledge that many gaps still exist across the region. Identifying more MUs in this region, which is notably rich in coral reef ecosystems and a primary habitat for hawksbill turtles, could aid in shaping conservation policies at the regional level for numerous countries in Southeast Asia.
Our genetic analysis demonstrates significant population differences even across very small distances (~75 to 500 km), challenging previous assumptions about hawksbill turtle dispersal and fine-scale population structure (Vargas et al., 2016; Arantes et al., 2020). We observed significant genetic differentiation between Segama Besar Island and Harapan Island, despite their proximity (approximately 75 km). This pattern of isolation over short distances is remarkable and contrasts with previous observations in the Indo-Pacific, where differentiation is generally noted between rookeries several hundred kilometers apart (Gaos et al., 2016; Nishizawa et al., 2016; Vargas et al., 2016; Gaos et al., 2017). Our findings align more closely with studies in the Atlantic, where genetic differentiation between rookeries has been noted separated by just 30 km (Browne et al., 2010). While hawksbill turtles are known to have migrated 1,600 km (Nietschmann, 1981; Miller et al., 1998), significant differentiations among nearby nesting sites indicate a high degree of natal philopatry, where turtles return to their birthplace to nest (Gaos et al., 2017). This species has been observed to have extremely strong nesting site fidelity (Horne et al., 2023), which makes it possible to have genetic differentiations occur on a small scale area. In the Java Sea, recaptured records on different places are only between Pesemut Island and Momperang Island (~3.5 km apart), revealed by YPLI and ELNA that conducted tagging programs for nesting females from 1996 to 2024 in 4 sites (Segama Besar Island, Kimar Island and Penambun Island, and Pesemut Island and Momperang Island).
In addition to a high degree of natal philopatry that results in isolation by distance (IBD) supported by correlation between genetic differentiation and geographic distances, the distribution of coral reef foraging habitats may facilitate strong nest site fidelity (Tanabe et al., 2023). Patchy coral reef habitats could restrict movement, aggregation, and mating of hawksbill turtles, facilitating the population differentiation. Restricted movement patterns of this species have been reported in other areas (Berube et al., 2012; Martinez-Estevez et al., 2021), with dependency on the reef habitats. Utilizing different coral reef systems near the birthplace may lead IBD in the Java Sea on a relatively small geographical scale. Our result showed the genetic differentiation is likely dependent on distance between nesting sites located on different complex reef areas, while the same genetic stock occurs in one complex reef area. The Momparang Island group of East Belitung consists of two nesting sites (i.e., Momparang Island and Pesemut Island) resulting in one genetic stock. Seribu Archipelago, known as a complex reef area (Djohani, 1994; Cleary et al., 2006) also revealed a pan-mixed population indicated by Tidung Island and Harapan Island. As species that are generally known to have very dependency on coral reef areas (León and Bjorndal, 2002; Blumenthal et al., 2009; Goatley et al., 2012), the large number of coral reefs present in Indonesia (https://allencoralatlas.org/) may allowing many genetic stock to occur in many places, not only in the Java Sea. We speculate that the same pattern of genetic differentiation dependency can be observed in other parts of Indonesia (such as the Coral Triangle area). Moreover, habitat use of this species is not only limited to coral reefs (i.e., mangrove estuaries), that could be another potential to influence the genetic differentiation as has been previously reported (Gaos et al., 2016).
The discovery of 13 new haplotypes specific to the Java Sea area further highlights the region’s high genetic diversity. This high haplotype diversity is likely a result of the haplotypes at these sites belonging to two divergent clades, suggesting a complex evolutionary history. The northern Java Sea rookeries (Kimar Island, East Belitung, and Penambun Island), were characterized by haplotypes belonging to one clade that includes EiIP49 and EiIP53, mainly observed from rookeries in Sabah and the Malay Peninsula, respectively (Nishizawa et al., 2016). The clade contains haplotype EiIP67 that was shared in five sites except Penambun Island. In contrast, the southern rookeries (Segama Besar Island, Tidung Island, and Harapan Island), showed haplotypes from two distinct clades, one of which contained EiIP08 that was reported from rookeries in northern Australia (Vargas et al., 2016; Bell and Jensen, 2018). In addition, haplotypes EiIP122 and EiIP123, previously reported from a foraging ground in the South China Sea at Tiga Island near Sabah (Nishizawa et al., 2016), were identified for the first time at a nesting site in this study. We considered hawksbills turtles in the northern MUs of Java Sea potentially moving out to the northern area (i.e., Sabah, Malaysia), meanwhile, the southern Java Sea MUs are restricted or resident. This finding further supports a complex phylogeographic pattern for hawksbill turtles, indicating multiple colonization events in the Indo-Pacific (Vargas et al., 2016).
As an area that has strong evidence to have complex evolutionary lineages of the Indo-Pacific, Java Sea hawksbill population has highest genetic diversity compared to other regions (Vargas et al., 2016; Gaos et al., 2016; Bell and Jensen, 2018). For instance, the Java Sea of Indonesia certainly has a total haplotype nearly three times that exists in the Eastern Pacific, the region that has four genetic stocks with seven haplotypes (Gaos et al., 2016). For many cases, the differentiated genetic stocks measured from haplotype frequency regionally appears with low genetic diversity; three genetic stocks in Melanesia with 14 haplotypes (Madden Hof et al., 2023), three genetic stocks in Australasia with 15 haplotypes (Vargas et al., 2016). These regions have larger coverage areas compared to the Java Sea, the highest known high genetic diversity and genetic stocks in the Indo-Pacific to date. The Java Sea area itself was also recognized to have a phylogeographical break pattern as observed by restricted gene flow, which is greatly influenced by ocean currents (Carpenter et al., 2011; Kool et al., 2011). Previously, Jensen et al. (2020) reported that genetic stock differences of marine turtle (i.e., green turtle) are related to environmental factors (i.e., regional ocean current patterns). To have better understanding of hawksbill turtle distribution across Java Sea, satellite telemetry work (Chaloupka et al., 2004; Pilcher et al., 2019; Madde Hof et al., 2023) or mixed stock analysis (Bowen et al., 2007; Limpus et al., 2009; Jensen, 2010; Jones et al., 2018; Piovano et al., 2019) could bring in-depth information of main foraging areas, and also stock contributions between different rookeries and foraging areas.
Small islands play a crucial role in preserving hawksbill turtle habitats due to their isolation from human activity compared to those that nest on the mainland. It is expected that the mainland coastal area will continue to be under increased stress, especially in relation to land-based pollution from anthropogenic activity (Adyasari et al., 2021). The existence of small islands, particularly those in the Java Sea, has a major impact on preventing the number of hawksbill turtles from declining populations and also maintaining genetic diversity. More importantly, strong nesting site fidelity has been recognized for this species (Horne et al., 2023). Site-specific haplotypes were found on small islands in the Java Sea, highlighting the island’s important role in the preservation of the genetic pool. In regions such as Indo-Pasific, a large contribution of haplotypes comes from small islands such as Sabah Turtle Island, Redang Island (Malaysia), Khram Island (Thailand), Varanus Island, Rosemary Island, Groote Eylandt, Milman Island (Australia), Arvanond Island (Solomon Islands), and other 15 Islands in Papua New Guinea (Wahidah and Syed Abdullah, 2009; Nishizawa et al., 2016; Vargas et al., 2016; LaCasella et al., 2021; Madden Hof et al., 2023).
This initial study on the genetic diversity and connectivity of hawksbill turtle population from nesting sites in Indonesia (particularly from the Java Sea area), has revealed overall high genetic diversity among major rookeries (i.e., Jakarta, Lampung, Bangka Belitung, West Kalimantan) (Meylan and Donnelly, 1999; Suganuma et al., 1999). Contrary to observations in Malaysia (Nishizawa et al., 2016), our findings do not indicate reduced genetic diversity, due to past population decline. The five MUs identified here form the foundation of the national conservation management strategy under the Turtle Conservation National Action Plan (RAN) for 2022-2024, as outlined by the Indonesian Government. Despite regulations banning the utilization of six sea turtle species including hawksbill turtle (e.g., Minister of Environment and Forestry Regulation No.7/1999; No.526/MEN-KP/VIII/2015; and P.106/SETJEN/KUM.1/12/2018), their critically endangered status persists, and illegal turtle trade including tortoiseshell harvesting remains a challenge in Southeast Asia and elsewhere (Hemelikova et al., 2021; Jeethvendra et al., 2023).
The nesting sites in our study are typically small islands that have their waters (i.e., reef habitat), and all of the MUs identified in this study are covered by the conservation areas or Marine Protected Areas (MPAs). In a conservation context, MPAs play an important role in protecting marine resources, including sea turtles (Dobbs et al., 2007; Scott et al., 2012; Schofield et al., 2013; Pendoley et al., 2014). Tidung Island and Harapan Island are situated within the Seribu Islands Marine National Park, which is located ~45 km north of Jakarta and covers an area of 107,489 ha (KEPMEN HUT 6310/Kpts-II/2002); Segama Besar Island is covered under 14,878 Ha of Batang Island and Segama Island conservation area (KEPMEN KP no 125/KEPMEN-KP/2023); Kimar Island is covered under 391,820.23 Ha of Belitung Water conservation area in Kepulauan Bangka Belitung province (KEPMEN KP no 94/KEPMEN-KP/2020); Momparang Island and Pesemut Island of East Belitung are protected under the MPA of the Momparang Island group and its surroundings covering a total area of 124,320 ha (KEPMEN KP no 52/KEPMEN-KP/2017); Penambun Island is covered under 164,595 Ha of Kendawangan coastal and small islands conservation area and surrounding waters in West Kalimantan province (KEPMEN KP no 91/KEPMEN-KP/2020). It is possible to achieve the goals of conservation and sustainable management of the hawksbill turtle population with the existence of a conservation area designated for MUs revealed in this study. Notably, when considering that one of Southeast Asia's largest hawksbill populations is in Indonesia.
Given the distinct genetic structure we observed between nearby nesting sites, it is crucial to extend genetic studies to foraging grounds surrounding the Java Sea, and to other Indonesian nesting sites beyond the Java Sea. A comprehensive understanding of genetic composition in rookeries is important for accurately estimating the origin of hawksbill turtle foraging aggregations and illegally traded turtle parts and products. The genetic approach offers a powerful tool against illegal trade (Naro-Maciel et al., 2010; Foran and Ray, 2016), potentially originating from various regions within Indonesia (LaCasella et al., 2021). Enhanced forensic techniques and updated DNA databases could significantly aid conservation efforts by tracing illegal turtles, parts and products back to their source, a method proven effective in prosecuting illegal activities in marine environments (van Oppen and Coleman, 2022).
In addition, hawksbill turtle foraging aggregations in Indonesia should be further studied. Sporadic reports of foraging hawksbill turtles in Southeast Asia (Nishizawa et al., 2016; Joseph et al., 2017; Nishizawa et al., 2024) highlight gaps in our knowledge of their life history, habitat use, and movement. The presence of haplotypes EiIP122 and EiIP123 in Java Sea rookeries, previously reported from foraging ground in South China Sea (e.g., Tiga Island) (Nishizawa et al., 2016), suggests a potential migratory connection that warrants further exploration. This further indicates the possibility of connectivity patterns between hawksbill turtles in the Java Sea and foraging areas in Sabah. Previous satellite documentation (Pilcher et al., 2019) showed the movement of hawksbill turtles from Sarawak, ~560-630 km apart from the location where EiIP122 and EiIP123 were found in the Java Sea (e.g., Kimar Island and East Belitung), moving towards Sabah. This supports previous suggestion that hawksbill turtles from various rookeries have migrated to Southeast Asia’s foraging regions (Nishizawa et al., 2016). A comprehensive understanding of both foraging aggregations and nesting rookeries is essential for accurately estimating population dynamics and conservation needs. By involving numerous parties with open information (e.g., local communities), it will be highly useful in developing and evaluating conservation plans for this endangered species (Gaos and Yañez, 2012). Collaborative efforts among diverse institutions and local communities, including Universities, Non-Government Organizations or NGOs and Governments will become more important for understanding hawksbill turtles throughout Indonesia, as done in olive ridley turtles and leatherback turtles in Madduppa et al. (2019, 2021).
The data presented in the study are deposited in the GenBank repository, accession number OR961305-OR961456.
The animal study was approved by Secretariat of Scientific Authority for Biodiversity of National Research and Innovation Agency (BRIN) of Indonesia. The study was conducted in accordance with the local legislation and institutional requirements. Sample collection for this study was approved by the Ministry of Environment and Forestry of the Republic of Indonesia, under permission numbers SK.85/KSDAE/SET.3/KSA.3/3/2022.
LMIS: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. J: Data curation, Writing – review & editing. GH: Data curation, Writing – review & editing. EH: Data curation, Resources, Writing – review & editing. EI: Data curation, Resources, Writing – review & editing. MPJ: Conceptualization, Methodology, Resources, Supervision, Writing – review & editing. CAM: Conceptualization, Funding acquisition, Project administration, Resources, Validation, Writing – review & editing. HN: Conceptualization, Methodology, Resources, Supervision, Writing – review & editing. LM: Data curation, Project administration, Writing – review & editing. AF: Supervision, Writing – review & editing. BS: Data curation, Supervision, Writing – review & editing. DGB: Supervision, Validation, Writing – review & editing. HM: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was mainly funded under ShellBank and a collaboration between World Wide Fund (WWF) Australia and Oceanogen under the no of contract P0160-RCLSHB-PT01.
The authors would like to say thank you to Yayasan Penyu Laut Indonesia (YPLI), Everlasting Nature of Asia (ELNA), the Laboratory of Marine Biodiversity and Biosystematics IPB University for supporting the sample collection. The authors also want to honor the memory of our respected and much-loved co-author and friend HM who passed away on 10th May 2022. HM was an associate professor and head department in marine science and technology at IPB University who also participated actively in several initiatives aimed at improving marine and fisheries research and management in Indonesia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1358695/full#supplementary-material
Abreu-Grobois F. A., Horrocks J. A., Formia A., Dutton P. H., LeRoux R., Vélez-Zuazo X., et al. (2006). “New mtDNA D-loop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analysis,” in Book of abstracts. Twenty sixth annual symposium on sea turtle biology and conservation, Eds. Frick M., Panagopoulou A., Rees A. F., Williams K. (Athens, Greece: International Sea Turtle Society), 376.
Adyasari D., Pratama M. A., Teguh N. A., Sabdaningsih A., Kusumaningtyas M. A., Dimova N. (2021). Anthropogenic impact on Indonesian coastal water and ecosystems: Current status and future opportunities. Mar. pollut. Bull. 171, 112689. doi: 10.1016/j.marpolbul.2021.112689
Arantes L. S., Vargas S. M., Santos F. R. (2020). Global phylogeography of the critically endangered hawksbill turtle (Eretmochelys imbricata). Genet. Mol. Biol. 43, e20190264. doi: 10.1590/1678-4685-gmb-2019-0264
Asaad I., Lundquist C. J., Erdmann M. V., Costello M. J. (2018). Delineating priority areas for marine biodiversity conservation in the Coral Triangle. Biol. Conserv. 222, 198–211. doi: 10.1016/j.biocon.2018.03.037
Bandelt H. J., Forster P., Röhl A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Bell I., Jensen M. P. (2018). Multinational genetic connectivity identified in western Pacific hawksbill turtles, Eretmochelys imbricata. Wildlife Res. 45, 307–315. doi: 10.1071/WR17089
Berube M. D., Dunbar S. G., Rützler K., Hayes W. K. (2012). Home range and foraging ecology of juvenile hawksbill sea turtles (Eretmochelys imbricata) on inshore reefs of Honduras. Chelonian Conserv. Biol. 11, 33–43. doi: 10.2744/CCB-0898.1
Blumenthal J. M., Austin T. J., Bothwell J. B., Broderick A. C., Ebanks-Petrie G., Olynik J. R., et al. (2009). Diving behavior and movements of juvenile hawksbill turtles eretmochelys imbricata on a caribbean coral reef. Coral Reefs 28, 55–65. doi: 10.1007/s00338-008-0416-1
Bolker B. M., Okuyama T., Bjorndal K. A., Bolten A. B. (2007). Incorporating multiple mixed stocks in mixed stock analysis: ‘many-to-many’ analyses. Mol. Ecol. 16, 685–695. doi: 10.1111/j.1365-294X.2006.03161.x
Bourjea J., Nel R., Jiddawi N. S., Koonjul M. S., Bianchi G. (2008). Sea turtle bycatch in the West Indian Ocean: review, recommendations and research priorities. Western Indian Ocean J. Mar. Sci. 7, 137–150. doi: 10.4236/jep.2023.144020
Bowen B. W., Bass A. L., Soares L., Toonen R. J. (2005). Conservation implications of complex population structure: lessons from the loggerhead turtle (Caretta caretta). Mol. Ecol. 14, 2389–2402. doi: 10.1111/j.1365-294X.2005.02598.x
Bowen B. W., Grant W. S., Hillis-Starr Z., Shaver D. J., Bjorndal K. A., Bolten A. B., et al. (2007). Mixed-stock analysis reveals the migrations of juvenile hawksbill turtles (Eretmochelys imbricata) in the Caribbean Sea. Mol. Ecol. 16, 49–60. doi: 10.1111/j.1365-294X.2006.03096.x
Broderick D., Moritz C., Miller J. D., Guinea M., Prince R. I. T., Limpus C. J. (1994). Genetic studies of the hawksbill turtle Eretmochelys imbricata: evidence for multiple stocks in Australian waters. Pacific Conserv. Biol. 1, 123–131. doi: 10.1071/PC940123
Browne D. C., Horrocks J. A., Abreu-Grobois F. A. (2010). Population subdivision in hawksbill turtles nesting on Barbados, West Indies, determined from mitochondrial DNA control region sequences. Conserv. Genet. 11, 1541–1546. doi: 10.1007/s10592-009-9883-3
Bustard H. R. (2016). The hawksbill turtle (Eretmochelys imbricata): conservation research. Testudo 8 (3), 50–63.
Carpenter K. E., Barber P. H., Crandall E. D., Ablan-Lagman M. C. A., Mahardika G. N., Manjaji-Matsumoto B. M., et al. (2011). Comparative phylogeography of the Coral Triangle and implications for marine management. J. Mar. Sci. 2011. doi: 10.1155/2011/396982
Chaloupka M., Parker D., Balazs G. (2004). Modelling post-release mortality of loggerhead sea turtles exposed to the Hawaii-based pelagic longline fishery. Mar. Ecol. Prog. Series. 280, 285–293. doi: 10.3354/meps280285
Cleary D. F., Suharsono, Hoeksema B. W. (2006). “Coral diversity across a disturbance gradient in the Pulau Seribu reef complex off Jakarta, Indonesia,” in eds. Hawksworth D. L., Bull A. T. Marine, Freshwater, and Wetlands Biodiversity Conservation, vol 4. (Dordrecht: Springer), 285–306. doi: 10.1007/978-1-4020-5734-2_19
da Silva V. R., Mitraud S. F., Ferraz M. L., Lima E. H., Melo M. T. D., Santos A. J., et al. (2016). Adaptive threat management framework: integrating people and turtles. Environment Dev. sustainability 18, 1541–1558. doi: 10.1007/s10668-015-9716-0
Djohani R. H. (1994). “Patterns of spatial distribution, diversity and cover of corals in Pulau Seribu National Park: implications for the design of core coral sanctuaries,” in Proc IOC-WESTPAC 3rd Int Sci Symp. (Bali, Indonesia: Indonesian Institute of Science), 265–279.
Dobbs K., Fernandes L., Slegers S., Jago B., Thompson L., Hall J., et al. (2007). Incorporating marine turtle habitats into the marine protected area design for the Great Barrier Reef Marine Park, Queensland, Australia. Pacific Conserv. Biol. 13, 293–302. doi: 10.1071/PC070293
Dutton P. H., Jensen M. P., Frutchey K., Frey A., LaCasella E., Balazs G. H., et al. (2014). Genetic stock structure of green turtle (Chelonia mydas) nesting populations across the Pacific islands. Pacific Sci. 68, 451–464. doi: 10.2984/68.4.1
Excoffier L., Lischer H. E. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
FitzSimmons N. N., Limpus C. J. (2014). Marine turtle genetic stocks of the Indo-Pacific: identifying boundaries and knowledge gaps. Indian Ocean Turtle Newslett. 20, 2–18. doi: 10.1002/aqc.3270
FitzSimmons N. N., Pittard S. D., McIntyre N., Jensen M. P., Guinea M., Hamann M., et al. (2020). Phylogeography, genetic stocks, and conservation implications for an australian endemic marine turtle. Aquat. Conservation: Mar. Freshw. Ecosyst. 30, 440–460.
Foran D. R., Ray R. L. (2016). Mitochondrial DNA profiling of illegal tortoiseshell products derived from hawksbill sea turtles. J. forensic Sci. 61, 1062–1066. doi: 10.1111/1556-4029.13062
Gaos A. R., Yañez I. L.. (2012). “Saving the eastern Pacific hawksbill from extinction: last chance or chance lost?,“ in Sea Turtles of the Eastern Pacific: Advances in Research and Conservation, eds Seminoff J. A., Wallace B. P. (Tuscon, AZ: University of Arizona Press), 244–262.
Gaos A. R., Lewison R. L., Jensen M. P., Liles M. J., Henriquez A., Chavarria S., et al. (2017). Natal foraging philopatry in eastern Pacific hawksbill turtles. R. Soc. Open Sci. 4, p.170153. doi: 10.1098/rsos.170153
Gaos A. R., Lewison R. L., Liles M. J., Gadea V., Altamirano E., Henríquez A. V., et al. (2016). Hawksbill turtle terra incognita: conservation genetics of eastern Pacific rookeries. Ecol. Evol. 6, 1251–1264. doi: 10.1002/ece3.1897
Groombridge B., Luxmoore R. A. (1989). The green turtle and hawksbill (Reptilia: Cheloniidae): world status, exploitation and trade. Secretariat of the Convention on International Trade in Endangered Species of Wild Fauna and Flora.
Goatley C. H., Hoey A. S., Bellwood D. R. (2012). The role of turtles as coral reef macroherbivores. PloS one. 7(6),e39979 337–343. doi: 10.1371/journal.pone.0039979
Halim M. H., Silalahi S., Sugarjito J. (2001). Conservation and utilization trend of marine turtles in Indonesia. Tigerpaper (FAO) 28, 10–18.
Harahap S. A., Prihadi D. J., Virando G. E. (2020). Spatial characteristics of the Hawksbill (Eretmochelys imbricate Linnaeus 1766) nesting beach on Kepayang Island, Belitung-Indonesia. World Sci. News 146), 152–169.
Hawkes L. A., Tomás J., Revuelta O., León Y. M., Blumenthal J. M., Broderick A. C., et al. (2012). Migratory patterns in hawksbill turtles described by satellite tracking. Mar. Ecol. Prog. Ser. 461, 223–232. doi: 10.3354/meps09778
Hemelikova A., Zoubek P., Ouhel T., Awaluddin A., Ferasyi T. R. (2021). “Conservation of hawksbill turtle (Eretmochelys imbricata) in Indonesia,” in 2nd International Conference on Veterinary, Animal, and Environmental Sciences (ICVAES 2020). (Banda Aceh, Indonesia: Atlantis Press), 44–47. doi: 10.2991/absr.k.210420.01
Horne J. B., Frey A., Gaos A. R., Martin S., Dutton P. H. (2023). Non-random mating within an Island rookery of Hawaiian hawksbill turtles: demographic discontinuity at a small coastline scale. R. Soc. Open Sci. 10, p.221547. doi: 10.1098/rsos.221547
Hutomo M., Moosa M. K. (2005). Indonesian marine and coastal biodiversity: Present status. Indian J. Mar. Sci. 34 (1), 87–97.
Ismuranty C. (2006). “The challenges for sea turtle conservation: lessons from the Derawan islands-Indonesia,” in OF THE TWENTY-THIRD ANNUAL SYMPOSIUM ON SEA TURTLE BIOLOGY AND CONSERVATION. (Kuala Lumpur, Malaysia: National Marine Fisheries Service), 58.
Jeethvendra K., Nishizawa H., Alin J., Muin H., Joseph J. (2023). Illegal tortoiseshell harvest of hawksbill turtles (Eretmochelys Imbricata) In Southeast Asia: evidence from Baturua reef, Semporna, Sabah, Malaysia. J. Sustainability Sci. Manage. 18, 54–67. doi: 10.46754/jssm
Jensen M. P. (2010). Assessing the composition of green turtle (Chelonia mydas) foraging grounds in Australasia using mixed stock analyses. Canberra, Australia: Doctoral dissertation, University of Canberra. doi: 10.26191/3fvq-2v21
Jensen M. P., Bell I., Limpus C. J., Hamann M., Ambar S., Whap T., et al. (2016). Spatial and temporal genetic variation among size classes of green turtles (Chelonia mydas) provides information on oceanic dispersal and population dynamics. Mar. Ecol. Prog. Ser. 543, 241–256. doi: 10.3354/meps11521
Jensen M. P., Dalleau M., Gaspar P., Lalire M., Jean C., Ciccione S., et al. (2020). Seascape genetics and the spatial ecology of juvenile green turtles. Genes 11, p.278. doi: 10.3390/genes11030278
Jensen M. P., FitzSimmons N. N., Bourjea J., Hamabata T., Reece J., Dutton P. H. (2019). The evolutionary history and global phylogeography of the green turtle (Chelonia mydas). J. Biogeography 46, 860–870. doi: 10.1111/jbi.13483
Jensen M. P., Limpus C. J., Whiting S. D., Guinea M., Prince R. I., Dethmers K. E., et al. (2013). Defining olive ridley turtle Lepidochelys olivacea management units in Australia and assessing the potential impact of mortality in ghost nets. Endangered Species Res. 21, 241–253. doi: 10.3354/esr00521
Jones K., Jensen M., Burgess G., Leonhardt J., van Herwerden L., Hazel J., et al. (2018). Closing the gap: mixed stock analysis of three foraging populations of green turtles (Chelonia mydas) on the Great Barrier Reef. PeerJ 6, e5651. doi: 10.7717/peerj.5651
Joseph J., Nishizawa H., Hassan M., Zakariah M. I., Jaaman S. A., Zhang X. (2017). Utilization of Brunei Bay (Malaysia) as a developmental and foraging habitat for hawksbill turtle (Eretmochelys imbricata). Regional Stud. Mar. Sci. 16, 304–307. doi: 10.1016/j.rsma.2017.09.012
Kitchener D. J. (1996). “The status of Green and Hawksbill Turtle rookeries in Nusa Tenggara and Maluku Tenggara, Eastern Indonesia–with observations on other marine turtles in the region,” in Proceedings of the First International Conference on Eastern Indonesian-Australian Vertebrate Fauna, (Indonesia: Western Australian Museum for Lembaga Ilmu Pengetahuan). 97.
Komoroske L. M., Jensen M. P., Stewart K. R., Shamblin B. M., Dutton P. H. (2017). Advances in the application of genetics in marine turtle biology and conservation. Front. Mar. Sci. 4, 156. doi: 10.3389/fmars.2017.00156
Konsta A., Chatzimentor A., Lin M., Dimitriadis C., Kyprioti A., Liu M., et al. (2022). Marine heatwaves threaten key foraging grounds of sea turtles in Southeast Asian Seas. Regional Environ. Change 22, 97. doi: 10.1007/s10113-022-01952-w
Kool J. T., Paris C. B., Barber P. H., Cowen R. K. (2011). Connectivity and the development of population genetic structure in Indo-West Pacific coral reef communities. Global Ecol. Biogeography 20, 695–706. doi: 10.1111/geb.2011.20.issue-5
LaCasella E. L., Jensen M. P., Madden Hof C. A., Bell I. P., Frey A., Dutton P. H. (2021). Mitochondrial DNA profiling to combat the illegal trade in tortoiseshell products. Front. Mar. Sci. 7, 595853. doi: 10.3389/fmars.2020.595853
Leigh J. W., Bryant D. (2015). POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116. doi: 10.1111/2041-210X.12410
León Y. M., Bjorndal K. A. (2002). Selective feeding in the hawksbill turtle, an important predator in coral reef ecosystems. Mar. Ecol. Prog. Ser. 245, 249–258. doi: 10.3354/meps245249
Limpus C. J. (1993). “The worldwide status of marine turtle conservation,” in Proc. of the First ASEAN Symposium Workshop on Marine Turtle Conservation. (Manila, Philippines: World Wide Fund), 43–63.
Limpus C. J., Bell I., Miller J. D. (2009). Mixed stocks of green turtles foraging on Clack Reef, northern Great Barrier Reef identified from long term tagging studies. Mar. Turtle Newslett. 123, 3–5.
Madden Hof C. A., Desbiens A., Kinch J., Fitzsimmons N., Versace H., Amon A., et al. (2023). From rookeries to foraging grounds: understanding regional connectivity and genetic diversity in hawksbill turtles. Front. Mar. Sci. 10, 1–12. doi: 10.3389/fmars.2023.1201009
Madduppa H., Bahri S., Ghozali A. T., Atmadipoera A. S., Subhan B., Santoso P., et al. (2021). Population genetic structure of Olive ridley (Lepidochelys olivacea) across Indonesian archipelago revealed by mitochondrial DNA: Implication for management. Regional Stud. Mar. Sci. 41, 101600. doi: 10.1016/j.rsma.2020.101600
Madduppa H. H., Bahri S., Subhan B., Anggraini N. P., Ohoiulun H., Abdillah T., et al. (2019). “DNA barcoding of sea turtles (Dermochelyidae and Cheloniidae) and its protocol using different tissues quality: implication to conservation managers,” in The 3rd EMBRIO International Workshop on Marine Biodiversity: Understanding, Utilization, Conservation, 9–10 October 2018, Bogor, Indonesia. Vol. 278, 012041. doi: 10.1088/1755-1315/278/1/012041
Marcovaldi M.Â., Lopez G. G., Soares L. S., López-Mendilaharsu M. (2012). Satellite tracking of hawksbill turtles Eretmochelys imbricata nesting in northern Bahia, Brazil: turtle movements and foraging destinations. Endangered Species Res. 17, 123–132. doi: 10.3354/esr00421
Martinez-Estevez L., Amador J. P. C., Amador F. C., Zilliacus K. M., Pacheco A. M., Seminoff J. A., et al. (2021). Spatial ecology of hawksbill sea turtles (Eretmochelys imbricata) in foraging habitats of the Gulf of California, Mexico. Global Ecol. Conserv. 27, e01540. doi: 10.1016/j.gecco.2021.e01540
Meylan A. B., Donnelly M. (1999). Status justification for listing the hawksbill turtle (Eretmochelys imbricata) as critically endangered on the 1996 IUCN Red List of Threatened Animals. Chelonian Conserv. Biol. 3, 200–224.
Miller J. D., Dobbs K. A., Limpus C. J., Mattocks N., Landry A. M. Jr. (1998). Long-distance migrations by the hawksbill turtle, Eretmochelys imbricata, from north-eastern Australia. Wildlife Res. 25, 89–95. doi: 10.1071/WR96086
Moritz C. (1994). Applications of mitochondrial DNA analysis in conservation: a critical review. Mol. Ecol. 3, 401–411. doi: 10.1111/j.1365-294X.1994.tb00080.x
Mortimer J. A., Donnelly M. (2008). Eretmochelys imbricata (The IUCN Red List of Threatened Species 2008), e.T8005A12881238. doi: 10.2305/IUCN.UK.2008.RLTS.T8005A12881238.en
Naro-Maciel E.U.G.E.N.I.A., Reid B., Fitzsimmons N. N., Le M., Desalle R. O. B., Amato G. (2010). DNA barcodes for globally threatened marine turtles: a registry approach to documenting biodiversity. Mol. Ecol. Resour. 10, 252–263. doi: 10.1111/j.1755-0998.2009.02747.x
Nietschmann B. (1981). Following the underwater trail of a vanishing species: the hawksbill turtle. Natl. Geographic Soc. 13, 459–480.
Nishizawa H., Jolis G., Joseph J., Isnain I., Muin H., Johari S. (2024). Variations in body condition of green turtles (Chelonia mydas) in two nearby foraging grounds indicate their sensitivity to foraging habitats. Aquat. Conservation: Mar. Freshw. Ecosyst. 34, e4038. doi: 10.1002/aqc.4038
Nishizawa H., Joseph J., Chong Y. K. (2016). Spatio-temporal patterns of mitochondrial DNA variation in hawksbill turtles (Eretmochelys imbricata) in Southeast Asia. J. Exp. Mar. Biol. Ecol. 474, 164–170. doi: 10.1016/j.jembe.2015.10.015
Oksanen J., Kindt R., Legendre P., O’Hara B., Stevens M. H. H., Oksanen M. J., et al. (2007). The vegan package. Community Ecol. Package 10, 719.
Palsbøll P. J., Berube M., Allendorf F. W. (2007). Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16. doi: 10.1016/j.tree.2006.09.003
Parsons J. J. (1972). The hawksbill turtle and the tortoise shell trade. In: Etudes de Geographie Tropicale Offertes a Peirre Gourou. Mouton Paris La Haye. pp. 45–60. doi: 10.1515/9783111342733-005
Pella J., Masuda M. (2001). Bayesian methods for analysis of stock mixtures from genetic characters. Fish. Bull. 99, 151–167. doi: 10.1139/F08-169
Pendoley K. L., Schofield G., Whittock P. A., Ierodiaconou D., Hays G. C. (2014). Protected species use of a coastal marine migratory corridor connecting marine protected areas. Mar. Biol. 161, 1455–1466. doi: 10.1007/s00227-014-2433-7
Persoon G., de Iongh H., Wenno B. (1996). Exploitation, management and conservation of marine resources: the context of the Aru Tenggara Marine Reserve (Moluccas, Indonesia). Ocean Coast. Manage. 32, 97–122. doi: 10.1016/S0964-5691(96)00025-7
Pilcher N. J., Ali L. (1999). Reproductive biology of the hawksbill turtle, Eretmochelys imbricata, in Sabah, Malaysia. Chelonian Conserv. Biol. 3, 330–336.
Pilcher N. J., Bali J., Buis J., Heng C. E., Devadasan A., Isnain I., et al. (2019). A review of sea turtle satellite tracking in Malaysia. Indian Ocean Turtle Newsletter 29, 11–22.
Piovano S., Batibasaga A., Ciriyawa A., LaCasella E. L., Dutton P. H. (2019). Mixed stock analysis of juvenile green turtles aggregating at two foraging grounds in Fiji reveals major contribution from the American Samoa Management Unit. Sci. Rep. 9, 3150. doi: 10.1038/s41598-019-39475-w
Reece J. S., Castoe T. A., Parkinson C. L. (2005). Historical perspectives on population genetics and conservation of three marine turtle species. Conserv. Genet. 6, 235–251. doi: 10.1007/s10592-004-7821-y
Reid B. N., Mladenoff D. J., Peery M. Z. (2017). Genetic effects of landscape, habitat preference and demography on three co-occurring turtle species. Mol. Ecol. 26, 781–798. doi: 10.1111/mec.13962
Rozas J., Ferrer-Mata A., Sánchez-DelBarrio J. C., Guirao-Rico S., Librado P., Ramos-Onsins S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Salm R., Halim M. H. (1984). “Marine conservation data atlas Indonesia,” in Planning for the survival of Indonesia’s seas and coasts. IUCN/WWF Project, 3108. Bogor, Indonesia: Directorate General of Forest Protection and Nature Conservation.
Schofield G., Scott R., Dimadi A., Fossette S., Katselidis K. A., Koutsoubas D., et al. (2013). Evidence-based marine protected area planning for a highly mobile endangered marine vertebrate. Biol. Conserv. 161, 101–109. doi: 10.1016/j.biocon.2013.03.004
Schulz J. P. (1987). Status of and trade in Chelonia mydas and Eretmochelys imbricata in Indonesia (Cambridge, United Kingdom: IUCN Conservation Monitoring Centre).
Scott R., Hodgson D. J., Witt M. J., Coyne M. S., Adnyana W., Blumenthal J. M., et al. (2012). Global analysis of satellite tracking data shows that adult green turtles are significantly aggregated in Marine Protected Areas. Global Ecol. Biogeography 21, 1053–1061. doi: 10.1111/j.1466-8238.2011.00757.x
Suganuma H., Kamezaki N., Yusuf A. (1999). Current status of nesting populations of the hawksbill turtle (Eretmochelys imbricata) in the Java Sea, Indonesia. Chelonian Conserv. Biol. 3, 337–343.
Sutanto-S I., Somantri A., Schulz J. P. (1989). The status of sea turtle in Indonesia. Media Konservasi (Indonesia) 2, 25–30.
Tabib M., Frootan F., Hesni M. A. (2014). Genetic diversity and phylogeography of hawksbill turtle in the Persian Gulf. J. Biodivers Environ. Sci. 4, 51–57.
Tabib M., Zolgharnein H., Mohammadi M., Salari-Aliabadi M. A., Qasemi A., Roshani S., et al. (2011). mtDNA variation of the critically endangered hawksbill turtle (Eretmochelys imbricata) nesting on Iranian islands of the Persian Gulf. Genet. Mol. Res. 10, 1499–1503. doi: 10.4238/vol10-3gmr1148
Tamura K., Stecher G., Kumar S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tanabe L. K., Cochran J. E., Williams C. T., Garzon F., Langner U., Hardenstine R. S., et al. (2023). Case report: tracking data from foraging hawksbill turtles in the northern Red Sea. Anim. Biotelemetry 11, 1–8. doi: 10.1186/s40317-022-00314-x
Tapilatu R. F., Wona H., Batubara P. P. (2017). Status of sea turtle populations and its conservation at Birds Head Seascape, Western Papua, Indonesia. Biodiversitas 18, 129–136. doi: 10.13057/biodiv/d180118
van Oppen M. J., Coleman M. A. (2022). Advancing the protection of marine life through genomics. PloS Biol. 20, e3001801. doi: 10.1371/journal.pbio.3001801
Vargas S. M., Jensen M. P., Ho S. Y., Mobaraki A., Broderick D., Mortimer J. A., et al. (2016). Phylogeography, genetic diversity, and management units of hawksbill turtles in the Indo-Pacific. J. Heredity 107, 199–213. doi: 10.1093/jhered/esv091
Vilaça S. T., Hahn A. T., Naro-Maciel E., Abreu-Grobois F. A., Bowen B. W., Castilhos J. C., et al. (2022). Global phylogeography of ridley sea turtles (Lepidochelys spp.): evolution, demography, connectivity, and conservation. Conserv. Genet. 23, 995–1010. doi: 10.1007/s10592-022-01465-3
Wahidah M. A., Syed Abdullah S. A. K. (2009). “Identification of the stock/population of green and hawksbill turtles in Southeast Asian region,” in Report on the third regional technical consultation on research for stock enhancement of Sea turtles (Japanese trust fund IV program). Ed. Syed Abdullah bin Syed Abdul Kadir O. A. (Kuala Lumpur, Malaysia: Southeast Asian Fisheries Development Center), 125–135.
Wallace B. P., DiMatteo A. D., Hurley B. J., Finkbeiner E. M., Bolten A. B., Chaloupka M. Y., et al. (2010). Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PloS One 5, e15465. doi: 10.1371/journal.pone.0015465
Wiadnyana N. N. (2003). “Study on population and distribution of two common sea turtles, green turtle and hawksbill turtle in Indonesia,” in Proceedings on the 4th SEASTAR2000 Workshop, Graduate School of Informatics, Kyoto University, Bangkok, Thailand. 44–46.
Zainudin I. M., Pet-Soede L., Hitipeuw C., Adnyana W. (2007). Interaction of Sea Turtles with Indonesian fisheries–preliminary findings (Karnataka, India: Indian Ocean Turt. Newsl). Available at: https://www.iotn.org/iotn06-01-interaction-of-sea-turtles-with-Indonesian-fisheries-preliminaryfindings.
Keywords: conservation genetics, Java Sea, management units, mtDNA, population genetics, mitochondrial DNA, nesting colonies, haplotype diversity
Citation: Sani LMI, Jamaludin, Hadiko G, Herma E, Inoguchi E, Jensen MP, Madden CA, Nishizawa H, Maryani L, Farajallah A, Subhan B, Bengen DG and Madduppa H (2024) Unraveling fine-scale genetic structure in endangered hawksbill turtle (Eretmochelys imbricata) in Indonesia: implications for management strategies. Front. Mar. Sci. 11:1358695. doi: 10.3389/fmars.2024.1358695
Received: 20 December 2023; Accepted: 23 February 2024;
Published: 02 April 2024.
Edited by:
Eduardo Cuevas, Autonomous University of Baja California, MexicoReviewed by:
Elizabeth Labastida, National Autonomous University of Mexico, MexicoCopyright © 2024 Sani, Jamaludin, Hadiko, Herma, Inoguchi, Jensen, Madden, Nishizawa, Maryani, Farajallah, Subhan, Bengen and Madduppa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lalu M. Iqbal Sani, aXFiYWxzYW5pQGFwcHMuaXBiLmFjLmlk
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.