- 1Misaki Marine Biological Station, School of Science, The University of Tokyo, Miura, Kanagawa, Japan
- 2Tateyama Marine Laboratory, Marine and Coastal Research Center, Ochanomizu University, Tateyama, Chiba, Japan
- 3Department of Biology, Keio University, Yokohama, Japan
Many species of sea cucumbers (Echinodermata, Holothuroidea) perform evisceration, i.e., ejection of internal organs including digestive tracts when responding to environmental stresses. After evisceration, they also show a high regenerative capacity, in which all the eviscerated organs regenerate. Especially in anterior evisceration species, the oral complex structure consisting of multiple organs, including the digestive tract and central nervous system, are reconstructed. The detailed developmental mechanisms underlying the process remains unclear, and therefore, in this study, focusing on Eupentacta quinquesemita, gene expression analyses in the regenerating tissues were carried out. For the formation of the gut tube, genes involved in mesenchymal-epithelial transition were upregulated consistently with the histological changes. Upregulation of Hox and Parahox genes along the anterior-posterior axis was observed, implying the involvement of these genes in the spatial differentiation of the digestive tract. In addition, the expression of otx, six and pax, i.e., transcription factors patterning anterior nervous tissues, was upregulated during the regeneration of the central nervous system. Taken together, these results suggest that conserved genes are co-opted to the internal organ regeneration after evisceration in sea cucumbers.
Introduction
Sea cucumbers possess high capacity of regeneration that enables them to reconstruct whole internal organs, including the digestive tract. They perform evisceration and eject internal organs as a type of autotomy in response to various extrinsic stimuli such as attacks by predators (Byrne, 2001). A small sea cucumber, Eupentacta quinquesemita, the species used in this study, is known to perform seasonal evisceration in addition to the response to physical stimulation (Byrne, 1985). Patterns of evisceration in sea cucumbers are classified into two major types, i.e., anterior and posterior evisceration (Mashanov and García-Arrarás, 2011). In contrast to posterior evisceration, in which only the digestive tract between the esophagus and cloacal stump is discarded, anterior evisceration seen in Dendrochirotida species (including E. quinquesemita) ejects the entire anterior structures, including the oral complex (Mashanov and García-Arrarás, 2011). The oral complex contains the nerve ring that is thought to function as a part of the central nervous system (CNS) in echinoderms (Mashanov et al., 2009).
Intense studies have been conducted for years, revealing the histological changes and molecular mechanisms of drastic regeneration in holothurians after evisceration. Both in anterior and posterior evisceration species, spatial and temporal dynamics of cellular processes such as mitosis, apoptosis and cell migration have been described in detail based on histological research (reviewed in Quispe-Parra et al., 2021b) and gene expression analyses suggested that remodeling of extracellular matrix (ECM) has key roles in the regeneration of eviscerated tissues (e.g., Ortiz-Pineda et al., 2009). In addition, RNA-seq analyses implied the involvement of various signaling factors in regeneration process (e.g., Sun et al., 2013). However, in contrast to the extensive knowledge about the cellular basis of intestinal regeneration in holothurians, the molecular mechanisms of larger scale, i.e., mechanisms which organize the drastic and complicated reconstruction of multiple organ complexes, are not fully understood.
During the regeneration, genetic programs used in embryonic development are often re-employed for reconstruction and repatterning of organs (Goldman and Poss, 2020). Thus, for understanding the regulation of complicated regeneration in holothurians, it should be important to investigate to what extent the conserved molecular mechanisms of embryonic development are co-opted in the fundamental process of regeneration, i.e., the reconstruction of the tubular structure of the digestive tract, regional differentiation of the digestive tract along the anterior-posterior (AP) axis, and regeneration of the central nervous system in the oral complex.
Regarding the reconstruction of the tubular structure of the digestive tract, histological observations in. E. quinquesemita (Figure 1A) suggested that the regeneration of the digestive tract after evisceration involves mesenchymal-epithelial transition (MET) in the anterior regenerating tissues (Figure 1B, Thiery and Sleeman, 2006; Chaffer et al., 2007; Okada and Kondo, 2019). MET refers to a phenomenon in which mesenchymal cells change their adhesiveness to acquire epithelial characteristics (Pei et al., 2019), and is histologically the opposite of the epithelial-mesenchymal transition (EMT) (Figure 2A). MET and EMT are known to contribute to epithelial regeneration and embryonic development (Ekblom, 1989; Kalluri and Weinberg, 2009; Rousselle et al., 2019). In both EMT and MET, a similar molecular mechanism is suggested to be activated, in which E-cadherin plays the major roles (Liu et al., 2016). In this mechanism, two key transcription factors, snail and twist, are known to be involved (Kang and Massagué, 2004; Barrallo-Gimeno and Nieto, 2005; Lamouille et al., 2014).
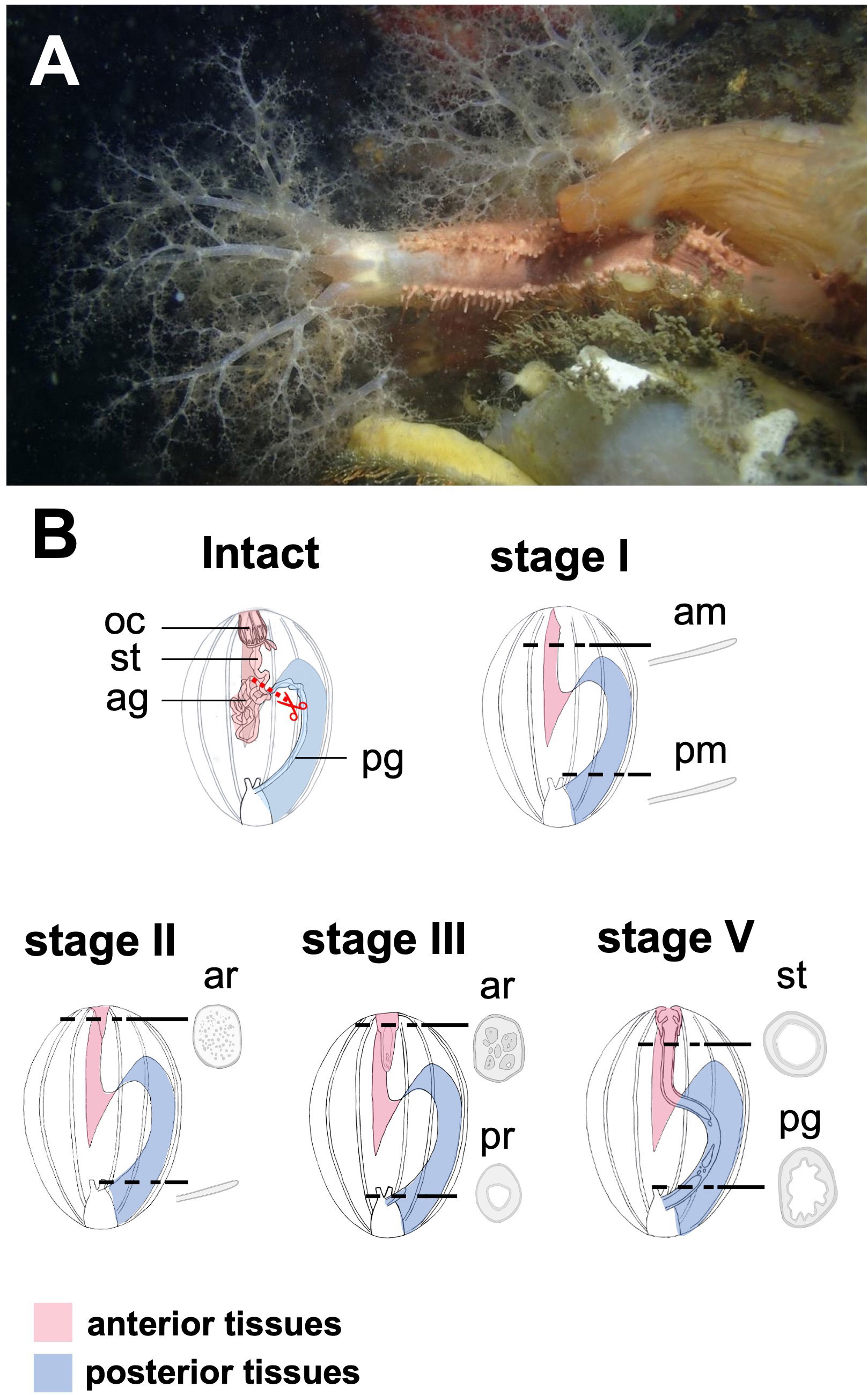
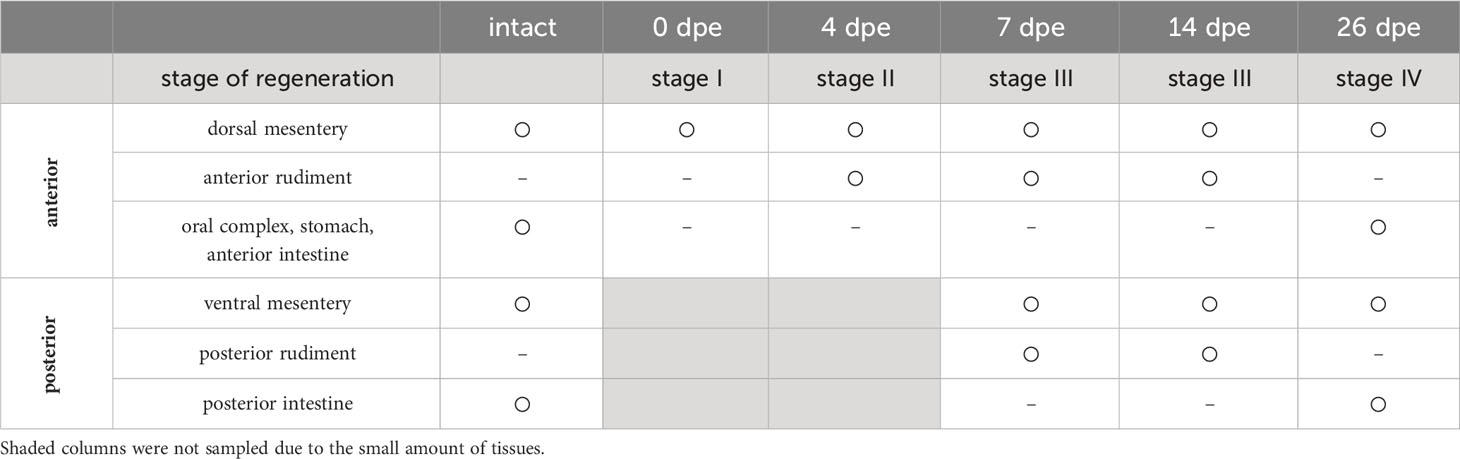
Figure 1 Regeneration process of digestive tract in a sea cucumber, Eupentacta quinquesemita, (A) The material species focused on in this study, an anterior-eviscerating sea cucumber, E. quinquesemita. (B) Schematic diagram of regeneration of the digestive tract during stages I and IV based on Okada and Kondo, 2019. MET occurs during stages II and III, and a tubular structure consisting of epithelium is newly formed inside mesenchyme. Ag, anterior gut; am, anterior mesentery; ar, anterior rudiment; oc, oral complex; pg, posterior gut; pm, posterior mesentery; pr, posterior rudiment; st, stomach.
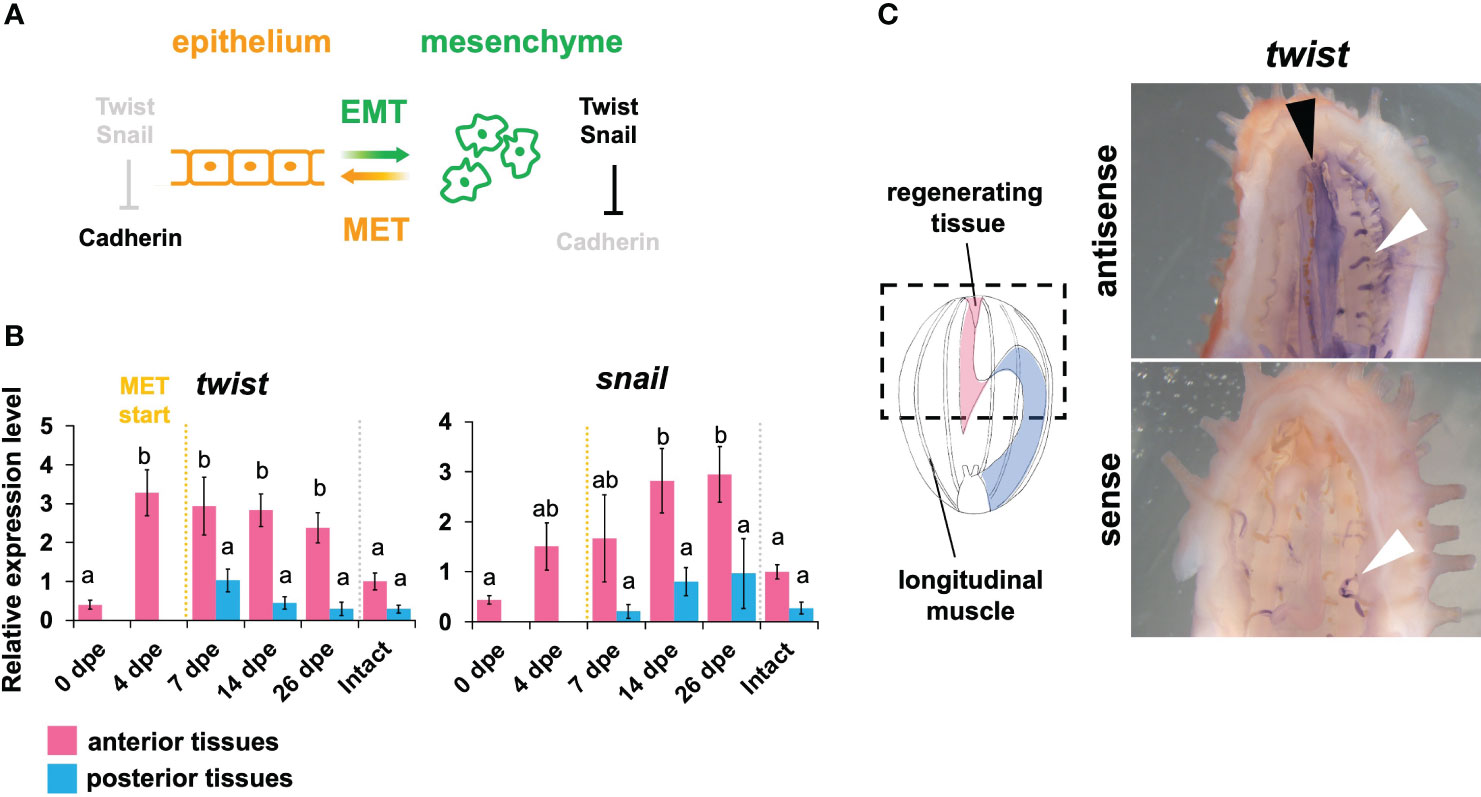
Figure 2 Expression pattern of MET- related genes in the regeneration of the digestive tract. (A) Schematic illustration of MET (mesenchymal-epithelial transition) and EMT (epithelial-mesenchymal transition) pathway. Two important transcription factors, i.e., Twist and Snail, play important roles in the regulation of Cadherin expression. At the time during which MET occurs and lasts, it is considered that the twist and snail expressions are down-regulated, while the cadherin expression is up-regulated. Based on Foroni et al. (2012). (B) Expression patterns of MET-related genes, i.e., twist and snail. Relative expression levels (mean ± S.D., n = 3) to the mean expression were calibrated using an internal control (EF1α). Horizontal axes show days post evisceration (dpe). The results were subjected to one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons tests (p < 0.05). Different letters above the bars indicate significant differences between groups. (C) in situ hybridization of twist at 4 dpe. Dotted line in schematic illustration indicates the region shown in right panels. The black arrowhead indicates signal detected in dorsal mesentery of the anterior regenerating part. Sense probes were used as controls. The white arrowheads indicate the ampullae of the water vascular system and they were also stained in the sense control.
A holothurian individual possesses a digestive tract differentiated into specific parts such as the esophagus, stomach and intestine along the A-P axis (Hyman, 1955). In the sea cucumber Apostichopus japonicus (Holothuroidea), during embryogenesis, Hox genes are suggested to provide the spatial information, leading to the differentiation of digestive tract parts (Kikuchi et al., 2015). The expression patterns of Hox and Parahox genes during development have been investigated in some species of echinoderms and shown to have a spatially colinear pattern along the A-P axis, indicative of their involvement in regional differentiation (reviewed in Byrne et al., 2016). In addition, RNA-seq analysis revealed that some Hox genes are upregulated during intestinal regeneration in a posterior eviscerating sea cucumber, Holothuria glaberrima (Quispe-Parra et al., 2021a), although the detailed spatial and temporal expression patterns of Hox and Parahox genes during the intestinal regeneration remain unclear.
In the case of anterior evisceration, not only the digestive tract but also the nerve ring, which constitutes the central nervous system in sea cucumbers, together with radial nerves in the body wall, is ejected (Byrne, 2001; Mashanov et al., 2005, 2009). The nerve ring is then regenerated at the same time as the digestive tract (Dolmatov, 1992). In various echinoderms, genes such as otx, six3/6, and pax6, which are involved in anterior neuronal patterning in deuterostomes (Hirth and Reichert, 1999), are expressed in the nerve ring and oral nervous system during development (e.g., Morris and Byrne, 2005). These three genes are known to play important roles in anterior neural patterning and sensory-organ formation. In addition, conserved neural factors such as elav, mushasi and neuroD are involved in the neurogenesis in the sea cucumber Holothuria glaberrima (Mashanov et al., 2015). However, the expression dynamics of these factors during the regeneration of the nerve ring have not yet been reported.
In this study, therefore, we conducted spatial and temporal expression analyses during regeneration after anterior evisceration using E. quinquesemita, focusing on MET-related genes, Hox and Parahox genes and neurogenesis- related genes in situ. We investigated the anterior regenerating tissues and the posterior regenerating tissues separately to clarify the spatial expression patterns, since the regeneration occurs independently in the anterior part and the posterior part, which then fuse to complete the regeneration (Figure 1B, Okada and Kondo, 2019). To obtain orthologous gene sequences in E. quinquesemita, transcriptome analysis was carried out to construct a gene database. In addition, in order to reveal the regeneration process of the nerve ring in this species, histological observations focusing on the nerve-ring regeneration were also carried out.
Materials and methods
Collection of E. quinquesemita, acclimatization, and experimental treatment
The sea cucumber, E. quinquesemita, was collected by diving at the depth of about 3 m in Tokyo Bay, near the pier of Hakkeijima Sea Paradise in Yokohama, Kanagawa Prefecture, or at the depth of about 5–10 m near Aquamarine Fukushima in Onahama, Fukushima Prefecture. Animals were kept in tanks until used for experiments. Methods for induction of evisceration and regeneration were based on Okada and Kondo, 2019. The day of evisceration was designated as 0 days post evisceration (dpe). Briefly, evisceration was induced by injection of approximately 100 μL of 0.45 M KCl into the coelom and the eviscerated animals were kept in aquaria with sea water at 13–18°C without feeding, up to 26 dpe. Embryos were collected after spontaneous spawning in laboratory tanks and kept in 2 L of filtered sea water in a plastic container at 20°C without feeding.
RNA-sequencing analysis
To identify MET-related genes, Hox and Parahox genes and neuronal patterning genes in the focal sea cucumber species, RNA sequencing was performed. Total RNAs were extracted from 2 embryonic stages (early and late gastrula) and juveniles of E. quinquesemita using QIAzol lysis reagent (Qiagen), and subsequently the RNA samples were pooled together. The pooled sample was then subjected to library preparation for RNA sequencing and sequenced on a Hiseq 4000 platform (Illumina) at a commercial service provider, Eurofins Genomics (Tokyo). The resulting 100-bp (base pairs) paired-end reads were deposited in the Sequence Read Archive of the DNA Data Bank of Japan (DDBJ) under the accession number DRA017482. Adapter and low-quality sequences were removed from the paired-end reads using Trimmomatic v0.39 (Bolger et al., 2014) with the following options: ILLUMINACLIP : TruSeq3-PE.fa:2:30:10, LEADING:20, TRAILING:20, SLIDINGWINDOW:4:20, MINLEN:25. Using the filtered reads, de novo transcriptome assembly was performed using Trinity v2.8.5 (Grabherr et al., 2011) with default options. To obtain the target orthologous genes, BLAST searches against the assembly were carried out with target gene orthologs in Strongylocentrotus purpuratus and Apostichopus japonicus (Tu et al., 2012; Zhang et al., 2017). The top-hit sequences from the E. quinquesemita transcriptome database were defined as putative orthologs of the target genes. To confirm the orthologs, reciprocal BLAST searches and phylogenetic analyses were performed (Supplementary Figures 1–7).
Real time quantitative PCR analysis
To analyze gene expression patterns by quantitative PCR, total RNAs were firstly extracted from the stages during regeneration after the evisceration in the focal sea cucumber species (Table 1). Since the regeneration requires about 3 weeks, samples were prepared at 0, 4, 7, 14, and 26 dpe. Regenerating digestive tracts were isolated by dissection and tissues from 2–4 individuals were pooled as each sample to obtain enough total RNAs. Separately from anterior and posterior digestive tracts, total RNAs were extracted by using QIAzol lysis reagent (Qiagen) and the RNA purification was performed with Agencourt AMPure XP (Beckman Coulter). Intact digestive tracts from mature individuals were also used for comparison. The extracted RNAs were reverse-transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantifications of target transcript levels were performed using Fast SYBR Green Master Mix and an ABI Prism 7500 instrument (Applied Biosystems). To evaluate endogenous control levels of constitutive expressions of putative reference genes, i.e., elongation factor 1 alpha (EF1α), tubulin beta chain (TUBB), NADH dehydrogenase (NADH) and ribosomal protein S18 (RPS18), were evaluated using geNorm (Vandesompele et al., 2002) and Normfinder (Andersen et al., 2004). The results indicated that EF1α was the most appropriate reference gene for comparisons among stages and body parts. Primers for qPCR (Supplementary Table 1) were designed using Primer Express software (ver. 3.0.0, Applied Biosystems). Data acquisition and analyses were performed using ABI Prism 7500 software ver. 2.0.4 (Applied Biosystems) with the relative standard curve method. For statistics, biological triplicates were subjected to one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons tests (p < 0.05) using R ver. 4.0.2.
In situ hybridization
To investigate the localization of target genes, whole mount in situ hybridizations (WISHs) were carried out, based on previous studies with minor modifications (Mashanov et al., 2010; Omori et al., 2011; Kikuchi et al., 2015). The PCR products were used as templates to transcribe riboprobes with a DIG RNA Labeling Kit (Roche). After dissection, the tissue samples were fixed with 4% paraformaldehyde in 0.5 M NaCl, 0.1 M 3-(N-morpholino) propanesulfonic acid (MOPS pH 7.0) overnight at 4°C. The samples were washed with phosphate buffered saline (PBS), and then decalcified with 0.5 M ethylenediaminetetraacetic acid (EDTA) (pH 8.0) in PBS for 3–4 days at room temperature. After decalcification, the samples were kept in 99.5% ethanol at –20°C until use. The samples were washed in PBS containing 0.1% Tween-20 (PBST), treated with 1/50 volume of proteinase K (TaKaRa) in PBST for 20 min at 37°C, acetylated sequentially in 0.25% and 0.5% acetic anhydride in 0.1 M triethanolamine, 5 min each. After the samples were washed twice with PBST for 5 min each, prehybridization was performed at 58°C for 2 h or longer in hybridization buffer containing 50% formamide, 5× saline-sodium citrate (SSC), 100 μg/mL yeast RNA, 5× Denhardt’s solution, 0.1% Tween 20. The riboprobes were diluted in hybridization buffer at 58°C to a final concentration of about 400 ng/ml and denatured at 80°C for 5 min. The hybridization was carried out at 58°C overnight. After hybridization, the samples were washed with 50% formamide in 5× SSC at 58°C for 20 min, 5× SSC at 58°C for 50 min, 2× SSC at 58°C for 50 min, 0.1× SSC at 58°C for 15 min and then PBST at room temperature for 15 min twice. Subsequently, the samples were incubated in 0.1% blocking reagent (Roche) in PBST (blocking buffer) at room temperature for 30 min, followed by an incubation in 1/2000 volume of anti-DIG-AP (Roche) in blocking buffer at 4°C overnight. Following a wash with PBST (15 min, 8 times), immunodetection was performed using BM purple (Roche) at room temperature. After detection, the samples were washed with PBST and then kept in 10% formalin/PBS.
Histological observations on the regeneration of nerve ring
Animals were anesthetized in 72 g/L MgCl2 in sea water for approximately 15 minutes to 1 hour before fixation. An individual body was dissected along the anterior-posterior (oral-aboral) axis of the body at the right ventral interambulacral zone, exposing the whole-body cavity, and dissected into anterior and posterior parts. The dissected bodies were fixed in Bouin’s Fixative or 4% paraformaldehyde in 0.5 M NaCl, 0.1 M 3-(N-morpholino) propanesulfonic acid (MOPS pH 7.0) overnight at 4°C. The specimens were washed with PBS, and then decalcified with 0. 5M EDTA (pH 8.0) in PBS for 3–4 days at room temperature. After decalcification, the specimens were kept in 99.5% ethanol at –20°C until embedding in paraffin (Paraplast X-TRA, SIGMA) for histological observation. Serial sections of 6- μm thickness were produced and were stained with hematoxylin-eosin (HE) or toluidine blue (TB) and observed under a light microscope (ECLIPSE TE300, Nikon). Sections were mostly made perpendicular to the anterior-posterior axis of the body. In total, 4 animals were used for histological observations.
Results
RNA-sequencing and ortholog searches
RNA-sequencing analysis of the transcriptome derived from gastrula embryos and juveniles of E. quinquesemita yielded 28,686,525 pairs of 100-bp reads. After filtering, 26,497,155 paired reads were retained. De novo transcriptome assembly of the sequence reads generated 350,742 contigs with a total of 296,553,944 bases and N50 of 1,513 bp. Using the transcriptome data, a gene database was constructed to search target genes, i.e., MET-related genes, Hox genes and neural patterning genes. As a result, the gene orthologs from the focal sea cucumber species were obtained by reciprocal BLAST searches and by constructing phylogenetic trees (Supplementary Figures 1–3). Based on the obtained gene sequences from the RNA-sequencing result, primers for qPCR were designed (Supplementary Table 1).
Expressions MET-related genes
Firstly, the expression patterns of MET-related genes, i.e., twist and snail, during the regeneration after anterior evisceration in E. quinquesemita were investigated. The results clearly showed that both of these genes were highly expressed in the anterior tissues of the regenerating digestive tract, in comparison with the posterior ones (Figure 2B). At 0 dpe, the expression level of twist in the anterior part was still low, but the twist expression became highly upregulated at 4 dpe, and was gradually decreased at the later stages. In the case of snail, the expression level was gradually upregulated, reaching the highest expression level at around 14–26 dpe, at the time when the twist expression started to decrease. The results of whole mount in situ hybridization showed that twist was expressed in anterior mesenteries (Figure 2C). On the other hand, in situ signals were not detected for snail.
Expressions of Hox genes
Anterior Hox genes such as hox1, hox5, and hox8 were highly expressed in the anterior tissues, while posterior Hox genes such as hox9/10, hox11/13/a, and hox11/13b were highly expressed in the posterior ones (Figure 3A). No difference between anterior and posterior tissues was detected in the expression levels of hox7. Temporal expression peaks varied among the Hox genes. hox5 showed the highest expression level at 0 dpe in anterior tissue, while hox 8 showed it at 4 dpe in anterior tissue. For hox1, hox9/10, hox11/13a, and hox11/13b, the temporal expression peak occurred at 14–26 dpe.
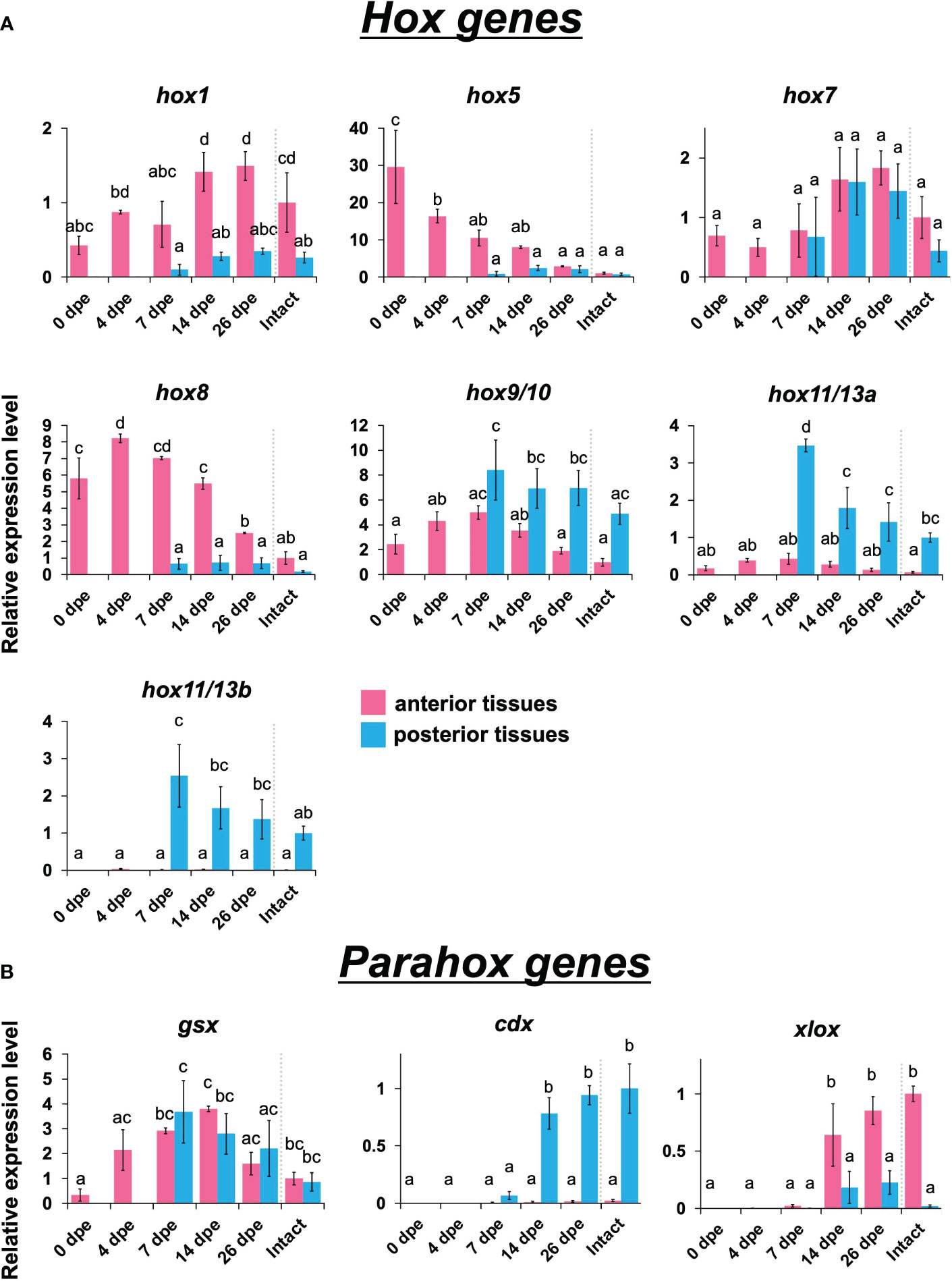
Figure 3 Expression profiles of Hox and Parahox genes in the regeneration after evisceration. Expression levels of Hox and Parahox genes were quantified by qPCR. Relative expression levels (mean ± S.D., n = 3) to the mean expression were calibrated using an internal control (EF1α). Horizontal axes show days post evisceration (dpe). The results were subjected to one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test (p < 0.05). Different letters above the bars indicate significant differences between groups. (A) Expression profiles of hox1, hox5, hox7, hox8, hox9/10, hox11/13a and hox11/13b. (B) Expression profiles of three neural patterning genes, i.e., gsx, cdx and xlox.
Expressions of Parahox genes
Like Hox genes, Parahox genes, i.e., gsx, xlox, and cdx, also showed colinear expression patterns: xlox was expressed in anterior tissues, while cdx was expressed exclusively in posterior tissues (Figure 3B). These genes were expressed at a later stage of regeneration, after 14 dpe. Among the three examined Parahox genes, gsx showed a distinctive pattern, in which no difference was seen between anterior and posterior tissues, and the expression level was highest at 7 to 14 dpe.
Regeneration of nerve ring
Histological observations clearly revealed the process of nerve ring regeneration (Figures 4B–E). At the time when anterior evisceration occurred (0 dpe), the anterior end of the sea cucumber body shrank to close the wound, so the radial nerve tissues came close to each other. At the early stage (4 dpe), neural tissues showing histological characteristics of cell proliferation extended from wounded radial nerves in the body wall, entering the regenerating tissues at the center of the body trunk (Figure 4B). At 6 and 12 dpe, the apical ends of regenerating nerves were elongated in the perpendicular direction, forming ring structures by connecting to each other (Figures 4C, D). At 12 dpe, an almost complete ring-like structure was formed by the elongated neural tissues connecting to each other (Figure 4D). At 21 dpe, the formation of the nerve ring was completed (Figure 4E). In addition, at 12 dpe, tentacular canals of the water vascular system were apparent (Figure 4D) and at 21 dpe, regenerating tentacles were observed (Figure 4E).
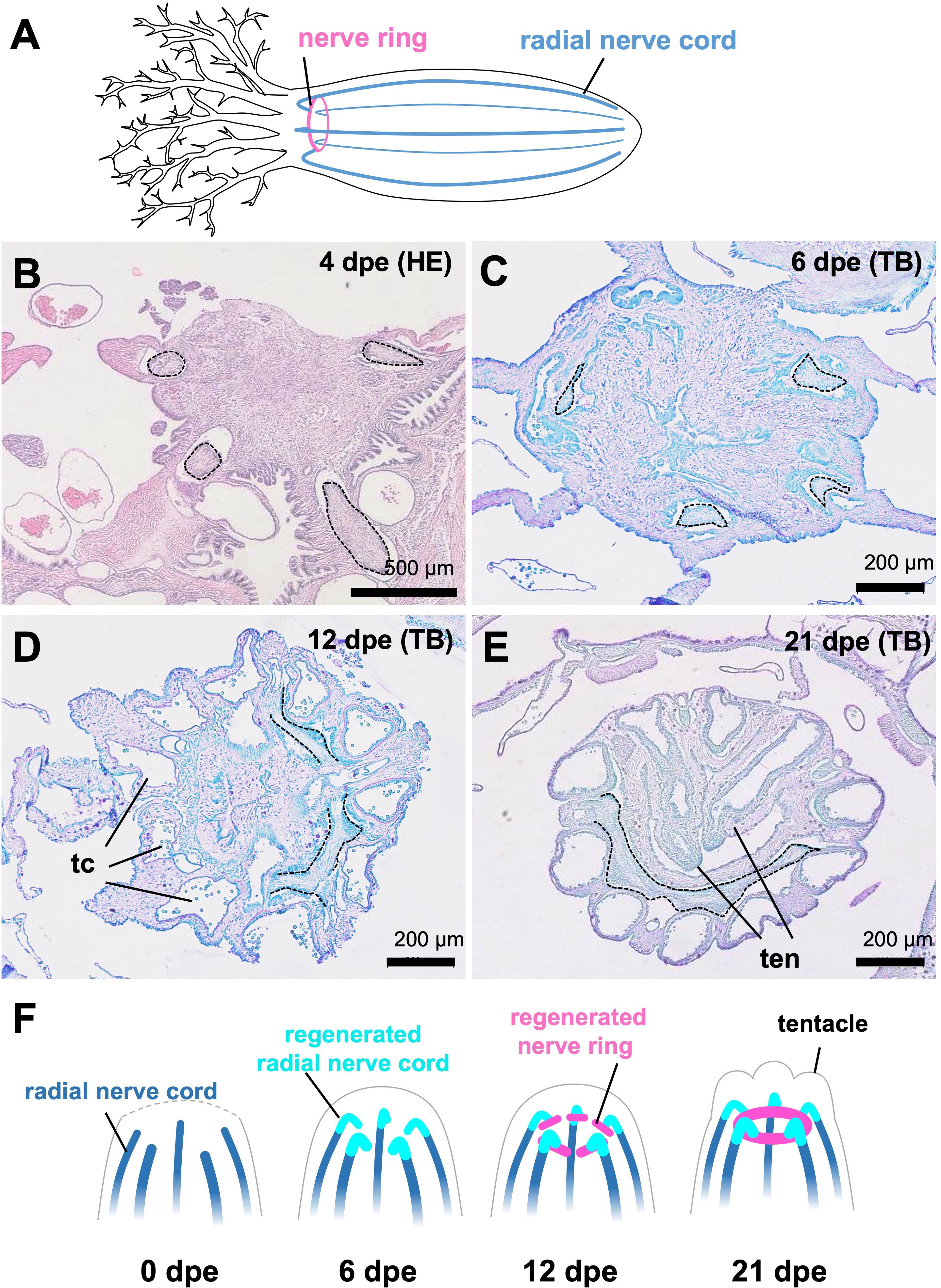
Figure 4 Regeneration process of nervous system in E. quinquwsemita. (A) Nervous system of E. quinquesemita. It consists of the nerve ring (also known as the circumoral nerve ring), tentacular nerves and radial nerve cords. (B–E) Histological images showing the regeneration process of circumoral nerve ring. Cross sections of the anterior regenerating tissues of E. quinquesemita were stained with hematoxylin-eosin (HE) or toluidine blue (TB). Dotted line indicates the outline of nervous tissue. (F) Schematic illustration of nervous system regeneration based on histological observation. Anterior parts in regenerating individuals are presented. tc, tentacular canal; ten, tentacle.
Expressions of neural patterning genes
Orthologous genes of 6 neural patterning genes, i.e., otx, six3/6, pax6, elav, musashi, and neuroD, were obtained from the gene database in E. quinquesemita (Supplementary Figures 4–7). Based on the sequences of the gene orthologs, primers for qPCR were designed (Supplementary Table 1). F or otx, six3/6, and pax6, the expression levels were higher in the anterior tissues, and almost no expression was detected in the posterior tissues (Figure 5). For otx and six3/6, the expression peaks were seen at 4 dpe, and then the expression levels were decreased. For pax6, the expression peak was seen at 26 dpe. A neural marker gene, elav, showed a strong expression level immediately after the evisceration (0 dpe) in the anterior tissue (Figures 3, 4). The expression level of musashi, which is involved in neural differentiation was slightly higher in the anterior tissues, but it was relatively constant over the entire regeneration period. For neuroD, no difference in expression levels was detected between anterior and posterior tissues, or among regeneration stages (Figure 5).
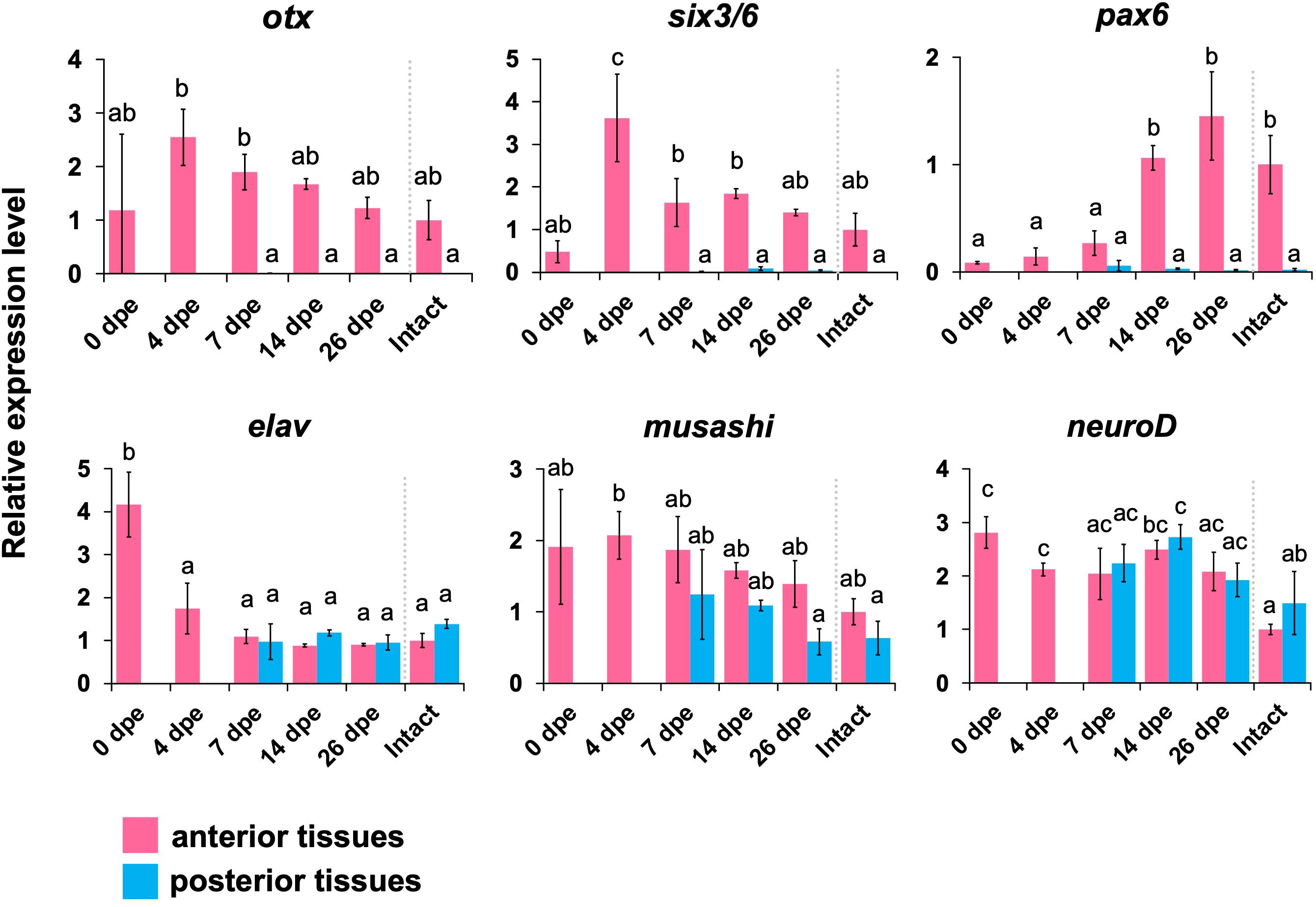
Figure 5 Expression profiles of genes that are known to play important roles in the development of the nervous system. Expression profiles of genes that are known to play important roles in the neuronal patterning (otx, six3/6, pax6) and neurogenesis (elav, musashi, neuroD) in regeneration stages, quantified by qPCR. Relative expression levels (mean ± S.D., n = 3) to the mean expression were calibrated using an internal control (EF1α). Horizontal axes show days post evisceration (dpe). The results were subjected to one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons tests (p < 0.05). Different letters above the bars indicate significant differences between groups.
Discussion
MET factors involved in digestive-tract regeneration
In general, twist and snail suppress cadherin expression (Foroni et al., 2012) and the downregulation of cadherin genes leads to the maintenance of mesenchyme (Figure 2A, Kang and Massagué, 2004; Barrallo-Gimeno and Nieto, 2005; Lamouille et al., 2014; Pei et al., 2019). Thus, our results suggest that the expression level of cadherin could be low in anterior regenerating tissues, that is, the number of mesenchyme cells should be increased, along with the upregulation of twist and snail. This process is consistent with the histological observations showing that mesenchymal cells proliferated at the earlier stages of digestive-tract regeneration in this species (Okada and Kondo, 2019). In addition, the timing of twist expression was earlier than that of snail expression (Figure 2B), which is congruent with a report showing that snail expression is regulated downstream of twist (Smit et al., 2009). Therefore, this time difference between the expressions of these two factors is suggested to reflect the epistatic relationship between the two factors.
Recent RNA-seq analysis in the related species Eupentacta Fraudatrix, which also shows anterior evisceration, revealed the upregulation of EMT-related factors such as snai2 and id2 during regeneration after evisceration (Boyko et al., 2020). It was also suggested in a posterior eviscerating species that EMT was involved in the intestinal regeneration (García-Arrarás et al., 2011). Therefore, it is possible that induction of EMT at the onset of intestinal regeneration via regulators such as twist and snail is widely shared among holothurians.
On the other hand, histological observations suggest that MET is involved in the digestive tract regeneration of anterior tissues of E. quinquesemita, in which epithelial tissues appeared among mesenchymal tissues (Okada and Kondo, 2019). Although the qPCR results in this study suggest that EMT occurs in anterior regenerating tissues as a whole, down regulation of twist and snail should occur locally at the tissues where the epithelial formation occurs after 7 dpe. Histological investigation suggested that MET does not occur in the anterior regenerating tissues of E. fraudatrix (Mashanov and Dolmatov, 2001, Mashanov et al., 2005), so the dynamics of gene expression related to MET might be different between species.
Expressions of Hox/Parahox genes during the digestive-tract regeneration
The results of this study revealed a tendency for anterior Hox genes to be expressed anteriorly while posterior ones were expressed posteriorly in regenerating tissues (Figure 3A). It is known that, in sea urchin larvae, xlox is expressed at the border between stomach and gut (Arnone et al., 2006). This is consistent with our result in which xlox was expressed in the anterior region, since the border in E. quinquesemita is located at a relatively anterior portion of the digestive tract (Byrne, 2001; Okada and Kondo, 2019). As shown by histological observations, the developmental speed in posterior regenerating tissues is a bit slower than that in anterior tissues (Okada and Kondo, 2019). Therefore, the expression of cdx, which is required for formation of the posterior digestive tract, was also seen at later stages of the regeneration process (Figure 3B). The expression of another Parahox gene, gsx, is known to be localized at a part of the neural tube in the development of amphioxus (Brooke et al., 1998; Arnone et al., 2006). It is generally believed that, in ancestral animals, gsx is expressed at the anterior (or oral) side of the digestive tract (Samadi and Steiner, 2010), although there are many animal species that do not exhibit gsx expression in their digestive tracts (Wollesen et al., 2015). In this study, gsx expression was shown to peak during the middle period of regeneration (Figure 3B), suggesting that this gene shows diverse expression patterns depending on the animal species.
Overall, our results suggest that Hox and Parahox genes provide spatial information along the A-P axis for digestive-tract differentiation. Time differences of the expression peaks among Hox/Parahox genes indicate that the timepoints for determination and differentiation vary among digestive-tract parts. Upregulation of Hox genes in the regenerating intestine has been reported in several holothurians (e.g., Méndez et al., 2000), warranting further investigation of the conservation and diversification of detailed gene expression patterns and histology among species, especially anterior eviscerating species and posterior eviscerating species.
Regeneration of central nervous system
Our histological observations focusing on the nervous tissue regeneration showed that the nerves were regenerated from the anterior ends of the remaining radial nerve cords, and later connected to each other to form the ring structure (Figure 4F). A similar regeneration pattern is known in the closely related species E. fraudatrix (Dolmatov, 1992). Since seasonal evisceration is known in E. quinquesemita (Byrne, 1985), the complete renewal of the nerve ring observed in these species might play a key role in their lifecycle.
Gene expression analyses by real time qPCR showed that the neural patterning genes otx, six3/6 and pax6 were upregulated in the anterior body part during the regeneration process. In particular, otx and six3/6 were upregulated at a relatively early stage of regeneration (4 dpe, Figure 5). The timing of upregulation corresponds to the timing when the regenerating nerve tissues start to enter the regenerating tissues composed of mesenchymal cells that later form the digestive tract. This may suggest that otx and six3/6 are required for the differentiation and elongation of nerve tissues around the digestive tract. It was previously shown that, in the echinoderm species Holopneustes purpurescens (Echinoidea), the otx gene is expressed in the nerve ring during larval stages (Morris et al., 2004; Morris and Byrne, 2005). In a feather star, Anneissia japonica, otx and six3 are expressed in oral nervous tissues (Omori et al., 2020). Therefore, it is suggested that neural patterning genes such as otx and six3/6 are co-opted from normal development to the re-formation of the nerve ring after anterior evisceration.
On the other hand, pax6 was upregulated later in the regeneration process (14–26 dpe). Around at 12 dpe, histological observations showed that the morphological characters of tentacles became apparent (Figures 4D, E). As it is known that pax6 is expressed in tentacles in adults of the sea cucumber Apostichopus japonicus (Liu et al., 2020), the upregulation of pax6 associated with the tentacular formation is consistent with previous knowledge. In other holothurians, RNA-seq analyses suggest that conserved paracrine factors involved in embryonic development such as wnt6, wnt9, and bmp1 are upregulated during intestinal regeneration (Ortiz-Pineda et al., 2009; Sun et al., 2013; Yuan et al., 2019; Auger et al., 2023) and notch is upregulated in nerve cord regeneration (Mashanov et al., 2014). It is known that, in regeneration processes in vertebrates, expressions of transcription factors such as pax, six and hox genes are regulated downstream of the signals such as the bmp and wnt pathways (e.g., Grogg et al., 2005; Patel et al., 2022), so similar epistatic relationships are also suggested in the regeneration in echinoderms.
Conclusions
Based on our results, it is suggested that the regeneration after anterior evisceration in E. quinquesemita employs three conserved mechanisms: the digestive tract formation via EMT and MET under the regulation of twist and snail, spatial differentiation of the digestive tract along the A-P axis by Hox and Parahox genes, and reconstruction of the nerve ring by recruiting the conserved neuronal patterning genes (Figure 6). Understanding how these mechanisms relate to the upstream regulators will be important in order to clarify the whole regulatory system in the regeneration after evisceration in holothurians. Since the loss of the digestive tract or central nervous system is lethal in many vertebrates, understanding the rules that regulate the regeneration which reconstructs multiple organs in holothurians should offer new insights into deuterostome regeneration and regenerative medicine for indispensable organs.
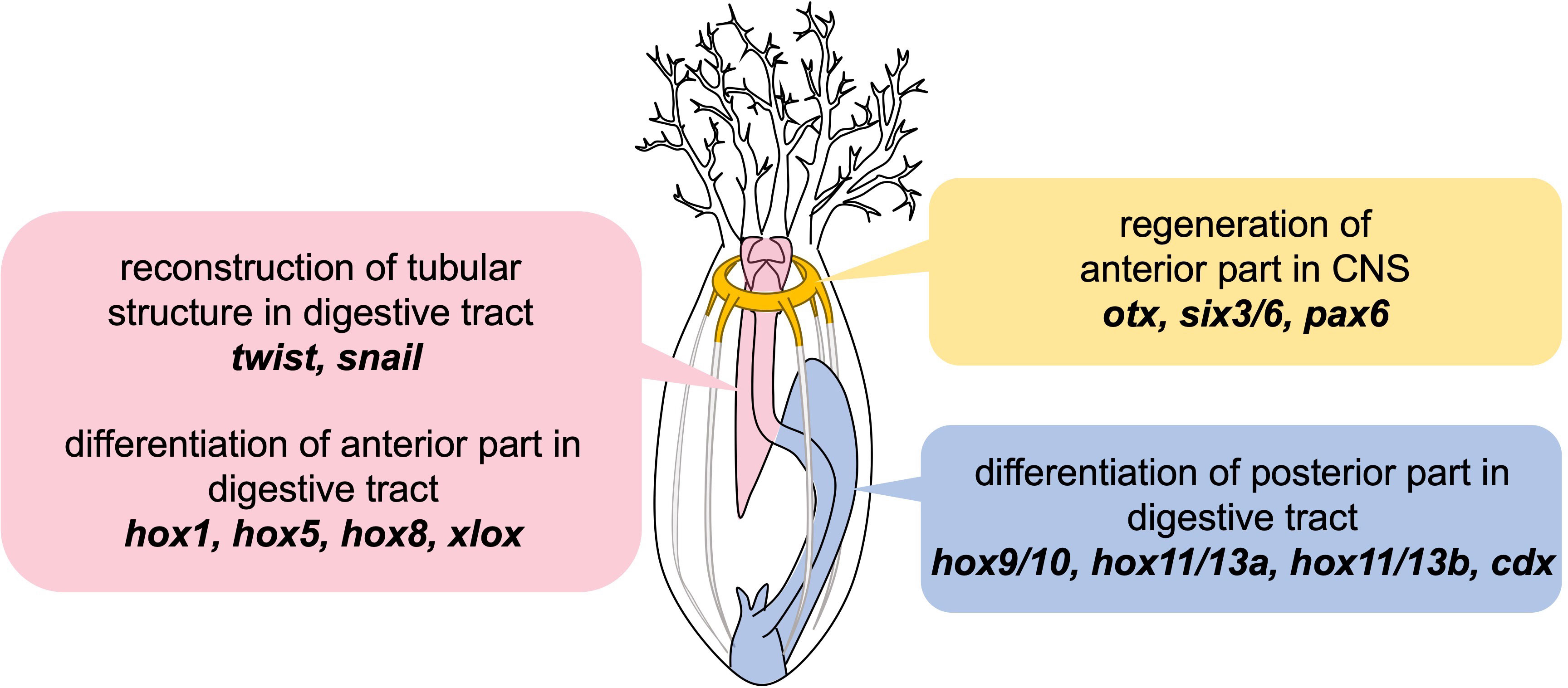
Figure 6 Schematic illustration of results in this study. Presented genes are suggested to be involved in the individual steps during regeneration after anterior evisceration in E. quinquesemita.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here in the Sequence Read Archive of the DNA Data Bank of Japan (DDBJ) under the accession number DRA017482.
Ethics statement
The animal study was approved by Life Science Research Ethics and Safety, The University of Tokyo. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
AO: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Visualization, Writing – original draft, Writing – review & editing. SU: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. HK: Conceptualization, Investigation, Resources, Writing – review & editing. YH: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. TM: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a Grant-in-Aid for Scientific Research A (No. 18H04006) and a Grant-in-Aid for Challenging Research (Pioneering, No. 21K18240) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Acknowledgments
We would like to express our gratitude to Y. Harumoto and the staff of Aquamarine Fukushima, Fukushima Ocean Science Museum, S. Abe and the staff of Yokohama Hakkeijima Sea Paradise, and M. Sekifuji, M. Kawabata and M. Kyokuwa for assisting us at the time of sampling. M. Yoshida, D. Kurokawa, M. Kondo, M. Okanishi, K.Oguchi and C. Umatani provided us with technical support and a number of constructive comments on the current study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1346172/full#supplementary-material
References
Andersen C. L., Jensen J. L., Orntoft T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496.
Arnone M. I., Rizzo F., Annunciata R., Cameron R. A., Peterson K. J., Martínez P. (2006). Genetic organization and embryonic expression of the ParaHox genes in the sea urchin S. purpuratus: insights into the relationship between clustering and collinearity. Dev. Biol. 300, 63–73. doi: 10.1016/j.ydbio.2006.07.037.
Auger N. A., Medina-Feliciano J. G., Quispe-Parra D. J., Colón-Marrero S., Ortiz-Zuazaga H., García-Arrarás J. E. (2023). Characterization and expression of holothurian wnt signaling genes during adult intestinal organogenesis. Genes 14 (2), 309. doi: 10.3390/genes14020309
Barrallo-Gimeno A., Nieto M. A. (2005). The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 32, 3151–3161. doi: 10.1242/dev.01907.
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170.
Boyko A. V., Girich A. S., Tkacheva E. S., Dolmatov I. Y. (2020). The Eupentacta fraudatrix transcriptome provides insights into regulation of cell transdifferentiation. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-58470-0
Brooke N., Garcia-Fernàndez J., Holland P. (1998). The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922. doi: 10.1038/31933.
Byrne M. (1985). Evisceration behavior and the seasonal incidence of evisceration in the holothurian Eupentacta quinquesemita (Selenka). Ophelia 24, 75–90. doi: 10.1080/00785236.1985.10426621
Byrne M. (2001). The morphology of autotomy structures in the sea cucumber Eupentacta quinquesemita before and during evisceration. J. Exp. Biol. 204, 849–863. doi: 10.1242/jeb.204.5.849.
Byrne M., Martinez P., Morris V. (2016). Evolution of a pentameral body plan was not linked to translocation of anterior Hox genes: The echinoderm HOX cluster revisited. Evol. Dev. 18, 137–143. doi: 10.1111/ede.12172
Chaffer C. L., Thompson E. W., Williams E. D. (2007). Mesenchymal to epithelial transition in development and disease. Cells Tissues Organs 185, 7–19. doi: 10.1159/000101298.
Dolmatov I. Y. (1992). Regeneration of the aquapharyngeal complex in the holothurian eupentacta fraudatrix (Holothuroidea, dendrochirota), keys for regeneration: monogr. Dev. Biol. Basel: Karger 23, 40–50.
Ekblom P. (1989). Developmentally regulated conversion of mesenchyme to epithelium. FASEB J. 3, 2141–2150. doi: 10.1096/fasebj.3.10.2666230.
Foroni C., Broggini M., Generali D., Damia G. (2012). Epithelial-mesenchymal transition and breast cancer: Role, molecular mechanisms and clinical impact. Cancer Treat Rev. 38, 689–697. doi: 10.1016/j.ctrv.2011.11.001
García-Arrarás J. E., Valentín-Tirado G., Flores J. E., Rosa R. J., Rivera-Cruz A., San Miguel-Ruiz J. E., et al. (2011). Cell dedifferentiation and epithelial to mesenchymal transitions during intestinal regeneration in H. glaberrima. BMC Dev. Biol. 11, 61. doi: 10.1186/1471-213X-11-61
Goldman J. A., Poss K. D. (2020). Gene regulatory programs of tissue regeneration. Nat. Rev. Genet. 21, 511–525. doi: 10.1038/s41576-020-0239-7
Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883.
Grogg M. W., Call M. K., Okamoto M., Vergara M. N., Del Rio-Tsonis K., Tsonis P. A. (2005). BMP inhibition-driven regulation of six-3 underlies induction of newt lens regeneration. Nature 438, 858–862. doi: 10.1038/nature04175
Hirth F., Reichert H. (1999). Conserved genetic programs in insect and mammalian brain development. BioEssays 21, 677–684. doi: 10.1002/(SICI)1521-1878(199908)21:8<677::AID-BIES7>3.0.CO;2-8
Kalluri R., Weinberg R. A. (2009). The basics of epithelial-mesenchymal transition. J. Clin. Investig. 119, 1420–1428. doi: 10.1172/JCI39104.
Kang Y., Massagué J. (2004). Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 118, 277–279. doi: 10.1016/j.cell.2004.07.011.
Kikuchi M., Omori A., Kurokawa D., Akasaka K. (2015). Patterning of anteroposterior body axis displayed in the expression of Hox genes in sea cucumber Apostichopus japonicus. Dev. Genes Evol. 225, 275–286. doi: 10.1007/s00427-015-0510-7
Lamouille S., Xu J., Derynck R. (2014). Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. doi: 10.1038/nrm3758.
Liu X., Chenggang L., Sun L., Liu S., Sun J., Yang H. (2020). Behavioral response of different epithelial tissues of sea cucumber (Apostichopus japonicus) to light and differential expression of the light-related gene Pax6. Mar. Freshw. Behav. Physiol. 53, 73–85. doi: 10.1080/10236244.2020.1767505.
Liu F., Gu L. N., Shan B. E., Geng C. Z., Sang M. X. (2016). Biomarkers for EMT and MET in breast cancer: An update (Review). Oncol. Lett. 12, 4869–4876. doi: 10.3892/ol.2016.5369.
Mashanov V. S., Dolmatov I. Y. (2001). Regeneration of digestive tract in the pentactulae of the far-eastern holothurian Eupentacta fraudatrix (Holothuroidea, Dendrochirota). Invertebr. Reprod. Dev. 39 (2), 143–151. doi: 10.1080/07924259.2001.9652477
Mashanov V. S., Dolmatov I. Y., Heinzeller T. (2005). Transdifferentiation in holothurian gut regeneration. Biol. Bull. 209, 184–193. doi: 10.2307/3593108.
Mashanov V. S., García-Arrarás J. E. (2011). Gut regeneration in Holothurians: a snapshot of recent developments. Biol. Bull. 221, 93–109. doi: 10.1086/BBLv221n1p93.
Mashanov V. S., Zueva O. R., García-Arrarás J. E. (2014). Transcriptomic changes during regeneration of the central nervous system in an echinoderm. BMC Genomics 15, 1–21. doi: 10.1186/1471-2164-15-357
Mashanov V. S., Zueva O. R., García-Arrarás J. E. (2015). Heterogeneous generation of new cells in the adult echinoderm nervous system. Front. Neuroanat. 9, 123. doi: 10.3389/fnana.2015.00123.
Mashanov V. S., Zueva O. R., Heinzeller T., Aschauer B., Naumann W. W., Grondona J. M., et al. (2009). The central nervous system of sea cucumber (Echinodermata: Holothuroidea) shows positive immunostaining for a chordate glial secretion. Front. Zool 6, 11. doi: 10.1186/1742-9994-6-11.
Mashanov V. S., Zueva O. R., Rojas-Catagena C., Garcia-Arraras J. E. (2010). Visceral regeneration in a sea cucumber involves extensive expression of survivin and mortalin homologs in the mesothelium. BMC Dev. Biol. 10. doi: 10.1186/1471-213X-10-117
Méndez A. T., Roig-Löpez J. L., Santiago P., Santiago C., García-Arrarás J. E. (2000). Identification of Hox gene sequences in the sea cucumber Holothuria glaberrima selenka (Holothuroidea: Echinodermata). Mar. Biotechnol. 2, 231–240. doi: 10.1007/s101269900027
Morris V. B., Byrne M. (2005). Involvement of two Hox genes and Otx in echinoderm body-plan morphogenesis in the sea urchin Holopneustes purpurescens. J. Exp. Zool 304B, 456–467. doi: 10.1002/(ISSN)1552-5015.
Morris V. B., Zhao J. T., Shearman D. C. A., Byrne M., Frommer M. (2004). Expression of an Otx gene in the adult rudiment and the developing central nervous system in the vestibula larva of the sea urchin Holopneustes purpurescens. Int. J. Dev. Biol. 48 (1), 17–22. doi: 10.1387/ijdb.15005570
Okada A., Kondo M. (2019). Regeneration of the digestive tract of an anterior-eviscerating sea cucumber, Eupentacta quinquesemita, and the involvement of mesenchymal-epithelial transition in digestive tube formation. Zoological Lett. 5, 1–13. doi: 10.1186/s40851-019-0133-3
Omori A., Akasaka K., Kurokawa D., Amemiya S. (2011). Gene expression analysis of Six3, Pax6, and Otx in the early development of the stalked crinoid Metacrinus rotundus. Gene Expression Patterns 11, 48–56. doi: 10.1016/j.gep.2010.09.002
Omori A., Shibata T. F., Akasaka K. (2020). Gene expression analysis of three homeobox genes throughout early and late development of a feather star Anneissia japonica. Dev. Genes Evol. 230, 305–314. doi: 10.1007/s00427-020-00665-6
Ortiz-Pineda P. A., Ramírez-Gómez F., Pérez-Ortiz J., González-Díaz S., Santiago-De Jesús F., Hernández-Pasos J., et al. (2009). Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genomics 10, 1–21. doi: 10.1186/1471-2164-10-262
Patel J. H., Schattinger P. A., Takayoshi E. E., Wills A. E. (2022). Hif1α and Wnt are required for posterior gene expression during Xenopus tropicalis tail regeneration. Dev. Biol. 483, 157–168. doi: 10.1016/j.ydbio.2022.01.007
Pei D., Shu X., Gassama-Diagne A., Thiery J. P. (2019). Mesenchymal-epithelial transition in development and reprogramming. Nat. Cell Biol. 21, 44–53. doi: 10.1038/s41556-018-0195-z.
Quispe-Parra D. J., Medina-Feliciano J. G., Cruz-González S., Ortiz-Zuazaga H., García-Arrarás J. E. (2021a). Transcriptomic analysis of early stages of intestinal regeneration in Holothuria glaberrima. Sci. Rep. 11, 1–14. doi: 10.1038/s41598-020-79436-2
Quispe-Parra D., Valentín G., García-Arrarás J. E. (2021b). A roadmap for intestinal regeneration. Int. J. Dev. Biol. 65, 427–437. doi: 10.1387/ijdb.200227dq
Rousselle P., Braye F., Dayan G. (2019). Re-epithelialization of adult skin wounds: Cellular mechanisms and therapeutic strategies. Adv. Drug Delivery Rev. 146, 344–365. doi: 10.1016/j.addr.2018.06.019.
Samadi L., Steiner G. (2010). Conservation of ParaHox ‘genes’ function in patterning of the digestive tract of the marine gastropod Gibbula varia. BMC Dev. Biol. 10, 74. doi: 10.1186/1471-213X-10-74.
Smit M. A., Geiger T. R., Song J.-Y., Gitelman I., Peeper D. S. (2009). A twist-snail axis critical for trkB-induced epithelial-mesenchymal transition-like transformation, anoikis resistance, and metastasis. Mol. Cell. Biol. 29, 3722–3737. doi: 10.1128/MCB.01164-08
Sun L., Yang H., Chen M., Ma D., Lin C. (2013). RNA-seq reveals dynamic changes of gene expression in key stages of intestine regeneration in the sea cucumber apostichopus japonicas. PloS One 8 (8), e69441. doi: 10.1371/journal.pone.0069441
Thiery J. P., Sleeman J. P. (2006). Complex networks orchestrate epithelial–mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142. doi: 10.1038/nrm1835.
Tu Q., Cameron R. A., Worley K. C., Gibbs R. A., Davidson E. H. (2012). Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Res. 22, 2079–2087. doi: 10.1101/gr.139170.112
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034. doi: 10.1186/gb-2002-3-7-research0034.
Wollesen T., Rodríguez Monje S. V., McDougall C., Degnan B. M., Wanninger A. (2015). The ParaHox gene Gsx patterns the apical organ and central nervous system but not the foregut in scaphopod and cephalopod mollusks. EvoDevo 6, 41. doi: 10.1186/s13227-015-0037-z.
Yuan J., Gao Y., Sun L., Jin S., Zhang X., Liu C., et al. (2019). Wnt signaling pathway linked to intestinal regeneration via evolutionary patterns and gene expression in the sea cucumber Apostichopus japonicus. Front. Genet. 10. doi: 10.3389/fgene.2019.00112
Keywords: sea cucumber, evisceration, regeneration, digestive tract, central nervous system, mesenchymal-epithelial transition
Citation: Okada A, Udagawa S, Kohtsuka H, Hayashi Y and Miura T (2024) Gene-expression patterns during regeneration of the multi-organ complex after evisceration in the sea cucumber Eupentacta quinquesemita. Front. Mar. Sci. 11:1346172. doi: 10.3389/fmars.2024.1346172
Received: 30 November 2023; Accepted: 12 February 2024;
Published: 07 March 2024.
Edited by:
Ana Pombo, Center for Marine and Environmental Sciences (MARE- IPLeiria), PortugalReviewed by:
Chryssanthi Antoniadou, Aristotle University of Thessaloniki, GreeceAndy Suhrbier, Pacific Shellfish Institute, United States
Copyright © 2024 Okada, Udagawa, Kohtsuka, Hayashi and Miura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toru Miura, bWl1QG1tYnMucy51LXRva3lvLmFjLmpw
 Akari Okada1
Akari Okada1 Yoshinobu Hayashi
Yoshinobu Hayashi Toru Miura
Toru Miura