
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 07 May 2024
Sec. Marine Biotechnology and Bioproducts
Volume 11 - 2024 | https://doi.org/10.3389/fmars.2024.1343363
This article is part of the Research Topic Marine Biotechnology and Bioproducts session of the 13th Asia-Pacific Marine Biotechnology Conference (APMBC) and 5th Australia New Zealand Marine Biotechnology Society Conference (ANZMBS) View all 6 articles
 Yanling Guo1,2,3†
Yanling Guo1,2,3† Jing Chang1,4†
Jing Chang1,4† Yan Xu1,2,3
Yan Xu1,2,3 Yanqing Liao1,2,3
Yanqing Liao1,2,3 Dehua Ji1,2,3
Dehua Ji1,2,3 Changsheng Chen1,2,3
Changsheng Chen1,2,3 Wenlei Wang1,2,3*
Wenlei Wang1,2,3* Chaotian Xie1,2,3*
Chaotian Xie1,2,3*Pigmentation-related mutations can be utilized to distinguish between differentially colored sectors of chimeric thalli, thereby facilitating the efficient breeding of economically valuable Pyropia/Porphyra seaweed species. However, the specific trait loci and alleles responsible for Pyropia/Porphyra coloration have yet to be identified, which limits the applicability of coloration mutants for breeding and genetic analyses. In this study, to preserve the genetic integrity of the population, only four-colored thalli were considered when constructing the doubled haploid (DH) Pyropia haitanensis population, which consisted of 480 homozygous offspring lines (representing the largest DH Pyropia/Porphyra population). The offspring lines in the DH population exhibited both wild-type colored and orange sectors, with a segregation ratio of approximately 1:1, indicating that the orange coloration was controlled by a single nuclear gene. Through BSA-seq analysis (99% confidence interval), a candidate region of 0.5 Mb was identified in the P. haitanensis genome. Additionally, a non-synonymous SNP [A/G] was detected at base-pair position 481 in the coding region of PhcpcC, which encodes a phycocyanin-associated rod linker protein. This SNP locus was verified in both DH and natural populations, with the wild-type colored lines having an A base and the orange lines having a G base at this locus. Therefore, PhcpcC may be the gene associated with the orange coloration of P. haitanensis. The molecular marker developed in this study can be employed to exploit pigmentation mutants for breeding and genetic analyses of Pyropia/Porphyra species.
Pyropia/Porphyra species (Bangiales, Rhodophyta) are among the most economically and ecologically important macroalgae. They possess the highest commercial value per unit mass (approximately $523 per wet metric ton) compared to other cultivated seaweed species (FAO, 2017). Because of the growing recognition of their nutritional benefits and sustainability, as well as the globalization of their production and trade, the consumption of Pyropia/Porphyra seaweed species has witnessed a recent worldwide surge, extending beyond limited regions in Asia (Cho and Rhee, 2019). The global value of Pyropia/Porphyra production has risen from $945.1 billion in 1987 to $2,319.7 billion in 2017 (FAO, 2019). However, the lack of improved varieties has become a significant impediment to the sustainable development of the Pyropia/Porphyra industry as the scale of cultivation continues to expand.
One of the main difficulties associated with breeding new Pyropia/Porphyra varieties is the fact that these species possess chimeric thalli. Specifically, their life cycle is characterized by a heteromorphic life cycle, consisting of a haploid macroscopic gametophytic phase and a diploid microscopic conchocelis phase. Meiosis in Pyropia/Porphyra cells occurs during the first two cell divisions of germinating conchospores, and the initial four cells of a developing conchosporeling form a linear genetic tetrad, resulting in the formation of chimeric blades (Zeng, 1985). This chimeric characteristic hinders the rapid breeding of homozygous strains. Once pigmentation mutants are identified, the variation in thallus colors can be used to develop genetic markers that can distinguish between different sectors of a chimeric thallus. Consequently, the color of pigmentation mutants serves as an excellent marker trait for breeding new varieties (Niwa et al., 2009; Sano et al., 2020; Zhao et al., 2022). Unfortunately, the qualitive trait loci (QTLs) and alleles responsible for Pyropia/Porphyra coloration have yet to be identified, thereby limiting the utility of coloration mutants for breeding and genetic analyses (Yu et al., 2020).
Bulked segregant analysis (BSA) was developed as a rapid method for detecting molecular markers linked to target traits in mapping populations. It is based on sequencing major gene mutants and pooling genome-wide association study data for extreme variants (Michelmore et al., 1991; Zou et al., 2016). The success of bulked segregant analysis (BSA) relies on carefully selecting individuals with extreme phenotypes for the target trait from a segregating population, which are then used to construct an extreme mixing pool. Advancements in technology and the increased affordability of next-generation sequencing (NGS) have allowed researchers to combine whole-genome resequencing and BSA (i.e., BSA-seq). This approach has accelerated the identification of closely linked markers for important traits and improved the resolution of maps for gene identification and QTL mapping. BSA-seq has been effectively employed to identify and map genes in various land plants, including Arabidopsis thaliana (Schneeberger, 2014), Benincasa hispida (Ma et al., 2021), and Oryza sativa (Takagi et al., 2013). In a recent study utilizing BSA-seq, the genes responsible for the red coloration of Pyropia yezoensis, an economically valuable seaweed, were identified and mapped (Yu et al., 2020). However, the limited size of P. yezoensis population poses challenges in identifying useful molecular markers and performing out-of-population validations.
This study focused on Pyropia haitanensis, which accounts for approximately 70% of the total annual output of Pyropia/Porphyra seaweeds in China (Fishery Department of China, 2021). The objective was to develop single nucleotide polymorphism (SNP) markers associated with the orange coloration of P. haitanensis. To achieve this, two DNA pools were generated for the orange-colored individuals and another for the wild-type individuals, from the doubled haploid (DH) population comprising 480 homozygous offspring lines. The BSA-seq method was employed to identify molecular markers associated with the orange coloration of P. haitanensis using the BSA-seq method. Consequently, phycocyanin-associated rod linker protein (PhcpcC) was identified as the gene associated with regulating the orange coloration of P. haitanensis.
Two genetically pure P. haitanensis lines, namely the wild-type colored W28 and orange DH115-2, were utilized in this study. The DH115-2 line was obtained through hybridization between ‘YSIII’ and RTPM (Xu et al., 2015), the W28 line is a new strain obtained through four generations of purification after the mutagenesis of the new variety Z-26 (Zhao et al., 2022). A DH P. haitanensis population (N = 480) was developed by crossing DH115-2 (male parent) with W28 (female parent) (Zhao et al., 2022) as described by (Xu et al., 2012) (Figure 1). Briefly, to preserve the genetic integrity of the population, 150 mosaic blades with four-colored sectors were selected to construct the DH population. Each colored sector was converted to protoplasts through the enzyme-mediated isolation of single somatic cells. Subsequently, the protoplasts were induced to develop into F1 thalli. The F1 thalli were enzymatically converted to protoplasts, which were then induced to develop into a homozygous F2 conchocelis. In the process of constructing the DH population, each colored sector of 150 four-colored mosaic blades with normal development was initially cut and separated from each other. Subsequently, they were cultured individually to obtain pure filaments from each colored sector. However, it should be noted that some of the four-colored mosaic thalli were unable to yield filaments from the colored sector, or the filaments obtained could not be purified due to contamination with heteroalgae. As a result, the remaining 125 four-colored mosaic thalli were successfully cultured to obtain 500 pure filaments. Out of a total of 500 pure filaments obtained, 480 were capable of releasing conchospores and forming thallus, thus constituting the present DH population.
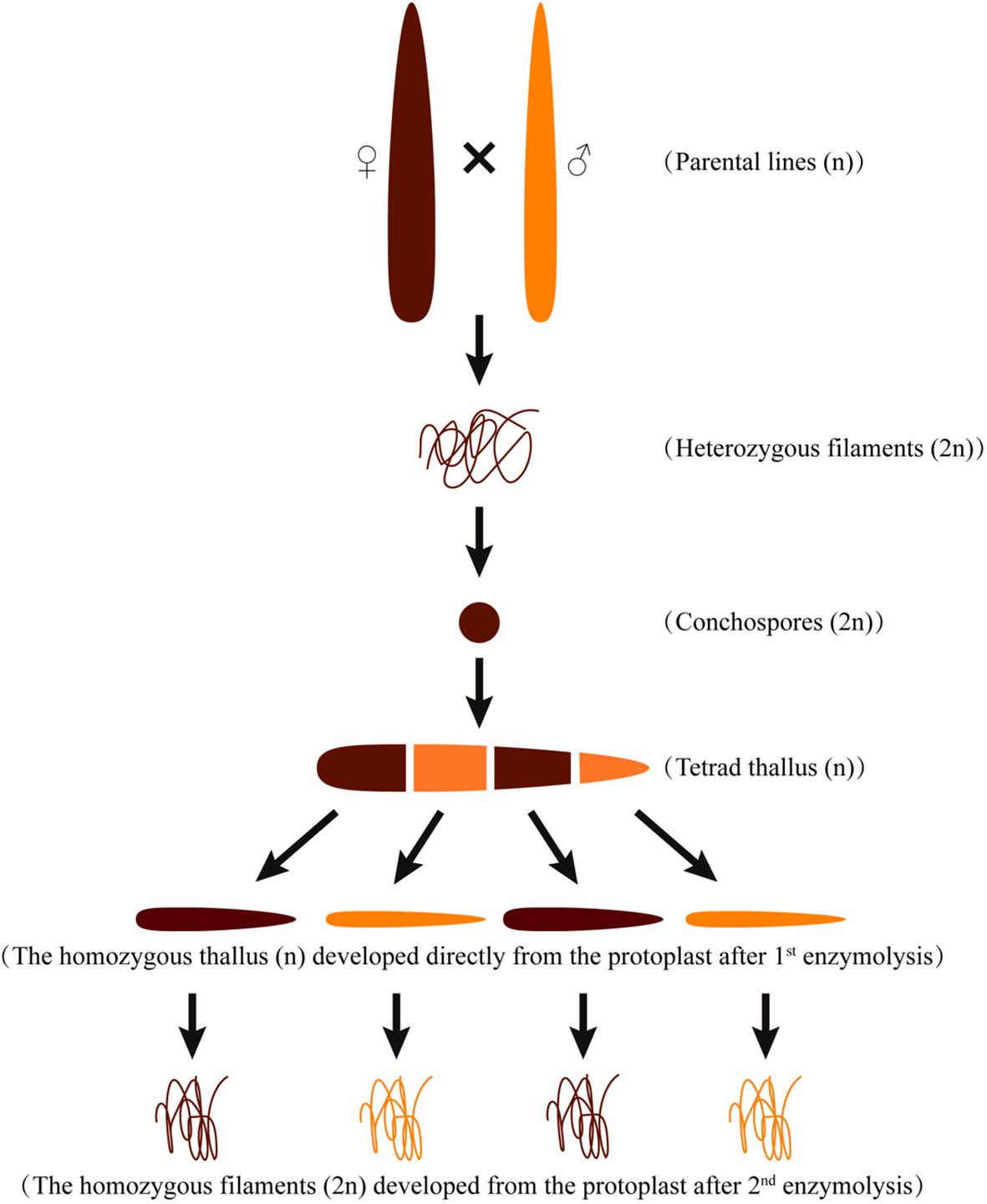
Figure 1 Schematic diagram of the cross between the maternal (W28) and paternal (DH115-2) P. haitanensis lines.
The gametophytic thalli were cultured in bubbling natural seawater supplemented with Provasoli’s enrichment solution, under cool-white fluorescent lamps (50–60 µmol photons m−2 s−1) at a temperature of 21 ± 1°C, following a 12-h light:12-h dark cycle. The free-living conchocelis (filamentous sporophyte) was maintained at 21 ± 1°C under cool-white fluorescent lamps (10 ± 1 µmol photons m−2 s−1), with a 12-h light:12-h dark cycle. The culture medium (MES) was renewed every 30 days.
For the BSA-seq analysis, F2 individuals with orange or wild-type colored conchocelis were selected. The wild-type colored (W) pool consisted of 20 strains exhibiting wild color, while the orange (O) pool comprised 20 strains displaying an orange color. Genomic DNA was extracted from the free-living conchocelis of the parents and the F2 populations. The concentrations and quality of DNA samples were assessed by 1.2% agarose gel electrophoresis and the UV-1990i UV-Vis spectrophotometer (Shimadzu, Kyoto, Japan). The DNA samples were stored at −80°C for subsequent analyses.
Genomic DNA (at least 3 µg) extracted from the above-mentioned bulks was utilized to construct paired-end libraries with an insert size of 500 bp using the Paired-End DNA Sample Prep kit (Illumina Inc., San Diego, CA, USA). These libraries were sequenced on the HiSeq X10 NGS platform (Illumina Inc.). The resequencing data for the parental materials and gene pools were aligned to the reference genome (CNP0003571) of Pyropia haitanensis (Bangiales, Rhodophyta) using the Burrows Wheeler Aligner (Li and Durbin, 2009). The genome sequencing materials included pure paternal line DH115-2, pure maternal line W28, and pure hybrid offspring WO57-1-1, WO57-1-2, WO57-1-3, and WO57-1-4 for genome sequencing (Wang et al, 2024, in revising1). The SAMtools program was employed to identify SNPs and insertions/deletions (InDels) (Li et al., 2009). The identified SNPs and InDels underwent filtration using the GATK VariantFiltration program according to appropriate standards. SNPs and InDels associated with distorted segregation or containing sequencing errors were discarded. The ANNOVAR software (Wang et al., 2010) was used to align and annotate SNPs and InDels for determining their physical positions.
A custom reference sequence was developed to calculate the SNP-index by replacing the genotype of the reference genome with homozygous SNPs from W28 (Takagi et al., 2013). Only homozygous SNP loci that segregated between the parents were selected, and their SNP-index was calculated for the BSA mapping. The SNP-index represents the frequencies of the parental alleles (DH115-2) in the bulks. The following standard pipeline was applied for filtering SNPs and InDels: 1) markers with a read depth <5× in each parent were excluded; 2) markers with missing genotype were excluded; 3) markers with a read depth <10× or >500× in each bulk were excluded to eliminate low-confidence markers due to low coverage or markers that may be in repetitive regions (i.e., inflated read depth); 4) markers with a SNP-index <0.3 or >0.7 in both bulks were excluded. The Δ(SNP-index) was calculated for the W and O pools. The average Δ(SNP-index) and SNP-index for each bulk were calculated using a 100-kb sliding window with a step size of 10 kb. Windows with <10 SNPs/InDels were discarded. Windows with an average Δ(SNP-index) greater than the 99th percentile of the genome-wide average Δ(SNP-index) were designated as significant windows. Significant windows that were overlapping or adjacent were merged into large significant genomic region. The genes within these regions were identified as candidate genes.
To analyze the differences between the candidate genes in the mapping region between the parents, non-synonymous mutations were identified by aligning the parental resequencing data. The SNP loci were amplified by PCR and the reliability of the SNP loci was validated by sequencing the PCR products of 20 F2 segregating populations and 13 differentially colored conchocelis samples from the natural population. PCR amplification for SNP genotyping was performed in a 20 μL volume containing 2 μL template DNA, 6 μL sterilized ddH2O, 10 μL Mix (TakaRa, Dalian, China), and 1 μL each primer. The PCR program was as follows: initial denaturation at 94°C for 5 min; 40 cycles of melting at 94°C for 30 s, annealing at a temperature determined by the primer sequences for 30 s, and extension at 72°C for 30 s; final extension at 72°C for 10 min. The PCR products were separated on 1.0% agarose gels and sequenced in both directions.
Chlorophyll a (Chl a) was extracted by immersing samples (approximately 0.05 g fresh weight) in 90% acetone for 24 h at 4°C in darkness. The samples were then centrifuged (5,000 × g, 4°C, 20 min), and the optical density of the supernatant was measured (400 to 800 nm) using the UV-1900i spectrophotometer (Shimadzu, Japan). The Chl a concentration was calculated in terms of mg/g dry weight (DW) as described by (Xu et al., 2020). To determine the phycobiliprotein (PBP) contents, including phycoerythrin (PE), phycocyanin (PC), and allophycocyanin (APC), approximately 0.01 g (DW) thalli was thoroughly homogenized in 50 mL sodium phosphate buffer (0.05 mol/L, pH 6.8) at 4°C. After at least six rounds of freezing (−21°C) and thawing (room temperature), the extracts were incubated in darkness for 12 h at 4°C. The extracts were then centrifuged (5,000 × g, 4°C, 20 min). The PBP concentrations were calculated according to absorbance values as described by (Hongfeng, 1993), with slight modifications as described by Wang et al. (2019).
All experiments were repeated three times. The standard deviation of the mean was used to determine the precision of the mean. The significance of any differences between samples was assessed by a one-way ANOVA and a post hoc LSD test using SPSS 19.0 (SPSS Inc., USA). The threshold for significance was set at P < 0.05.
The coloration and pigment contents exhibited variations between the parental lines, W28 and DH115-2 (Figures 2A, B). The hybridization between W28 and DH115-2 resulted in the production of heterozygous carpospores with wild-type coloration. These carpospores developed into conchocelis and were induced to release conchospores. The presence of pigmentation chimeras confirmed the successful implementation of the hybridization process. In addition, the differentially colored sectors in the F1 heterozygous thalli were utilized to distinguish the types of ordered tetrads (Figures 2C, D). The filaments derived from protoplasts of each colored sector after the second enzymolysis exhibited orange and wild-type colors (Figure 2E). Statistical analysis revealed a segregation ratio of 238:242 for the wild-type colored sectors and orange sectors in the F1 filaments.
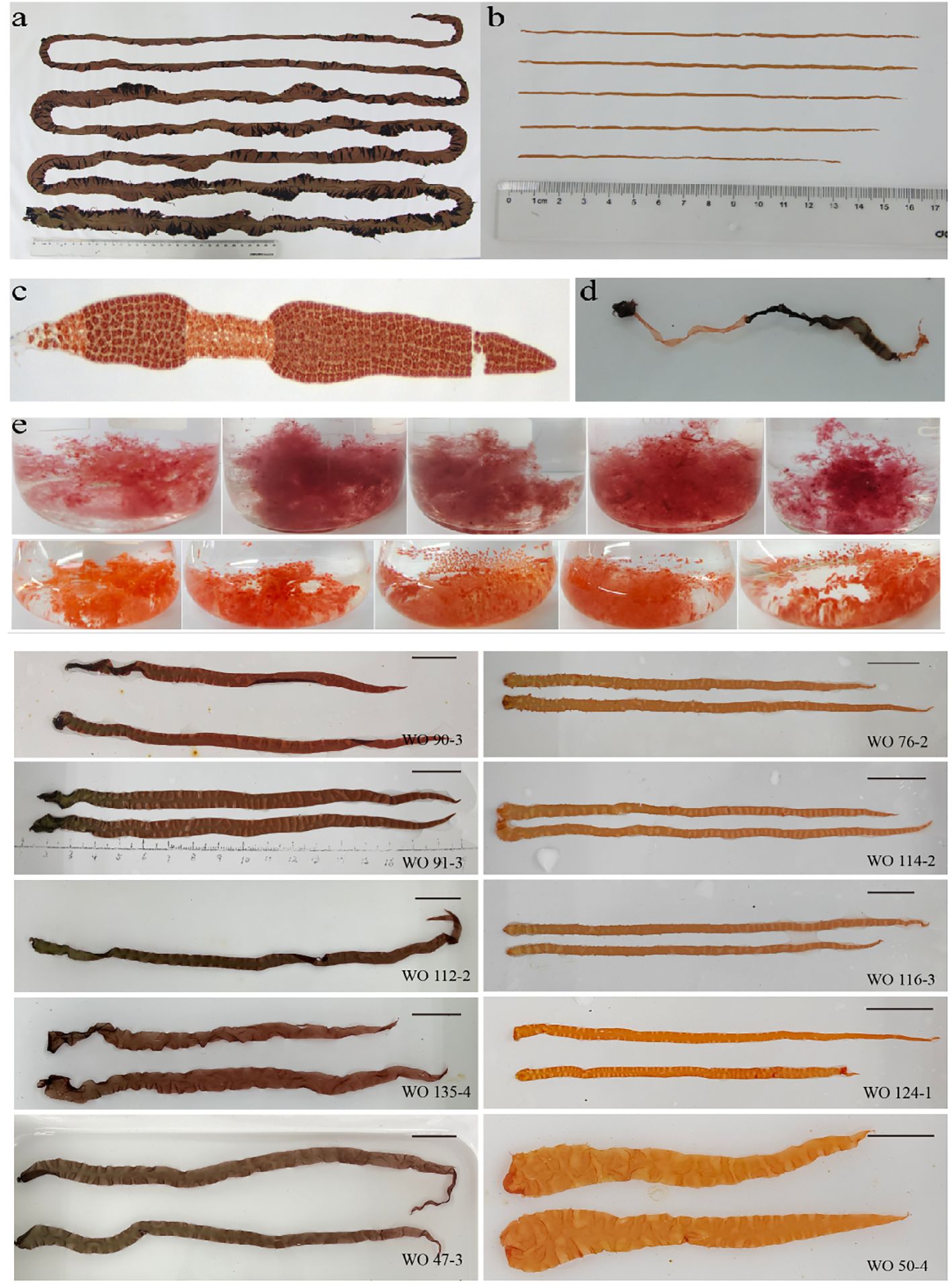
Figure 2 Contrasting colors of the parents and offspring. (A) Thalli of the maternal line. (B) Thalli of the paternal line. (C, D) Microscopic image and photograph of the F1 chimeric thalli after crossbreeding, respectively. (E) Coloration of selected homozygous F1 filaments and F2 thallus.
There were significant differences in the content of PE and PC between W28 and DH115-2 (P ≤ 0.001). Additionally, the content of Chl a in W28 was significantly higher compared to DH115-2 (P < 0.05; Figure 3A). When examining the pigment ratios, it was observed that PC/APC in W28 was significantly higher than in DH115-2 (P ≤ 0.001). Furthermore, PC/Chl a (P < 0.01) and PE/APC (P < 0.05) were significantly higher in W28 compared to DH115-2. Conversely, in DH115-2, PE/PC (P < 0.05) was significantly higher than in W28 (Figure 3B).
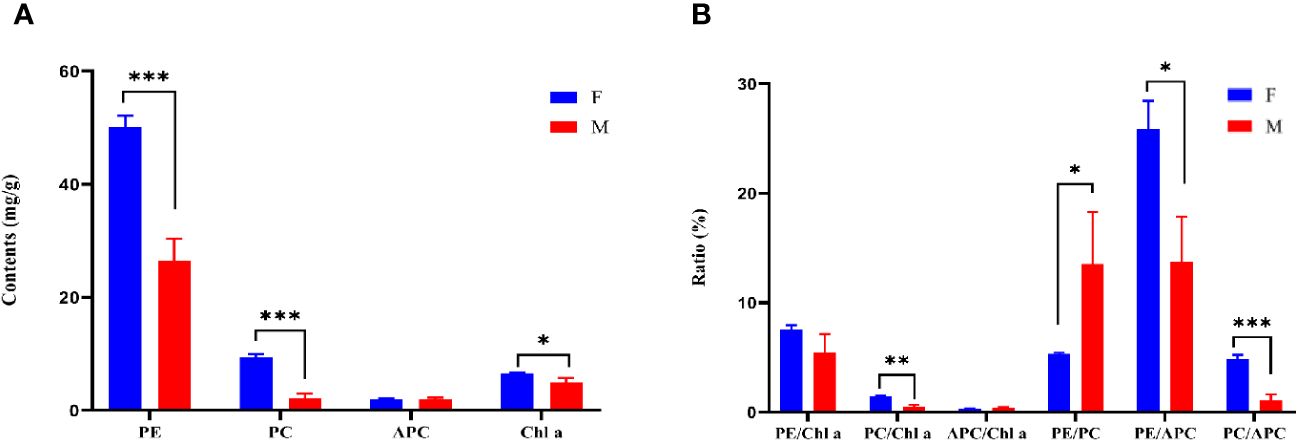
Figure 3 Pigment protein content and ratio. (A) Pigment protein contents in parental lines. (B) The ratio of pigment proteins in the parental line. W28 (F) and DH115-2 (M). Significant differences between samples are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P ≤ 0.001).
The sequencing data and the results of the alignment to the reference genome are summarized in Supplementary Tables S2 and S3. A total of 60,088 SNPs and 2,772 InDels, which were polymorphic between the parents, were selected. For all the obtained SNPs, association intervals for the orange coloration of P. haitanensis were determined using the Δ(SNP-Index), Euclidean Distance (ED), G-statistic, and Fisher’s exact test with association thresholds of 95% and 99%. Notably, a significantly larger peak-value was obviously obtained at the 99% significance level compared to the 95% significance level. The highest peak-value was observed on chromosome 2 in all four algorithms (Figure 4). At the 99% significance level, the ED algorithm identified the smallest QTL interval (0-0.5 M), while the Δ(SNP-Index), G-statistic and Fisher’s exact test algorithms identified QTL interval of 2.41 M (0-2.41 M), 0.66 M (0.03-0.15 M, 0.15-0.5 M, and 0.65-0.84 M), and 1.81 M (0-1.42M, 1.52-1.66 M, 1.79-1.90 M, and 2.03-2.17 M), respectively. By integrating abovementioned four algorithms, a QTL region of 0.03-0.5 M with the highest peak on chromosome 2 was identified, which was further analyzed to identify candidate genes.
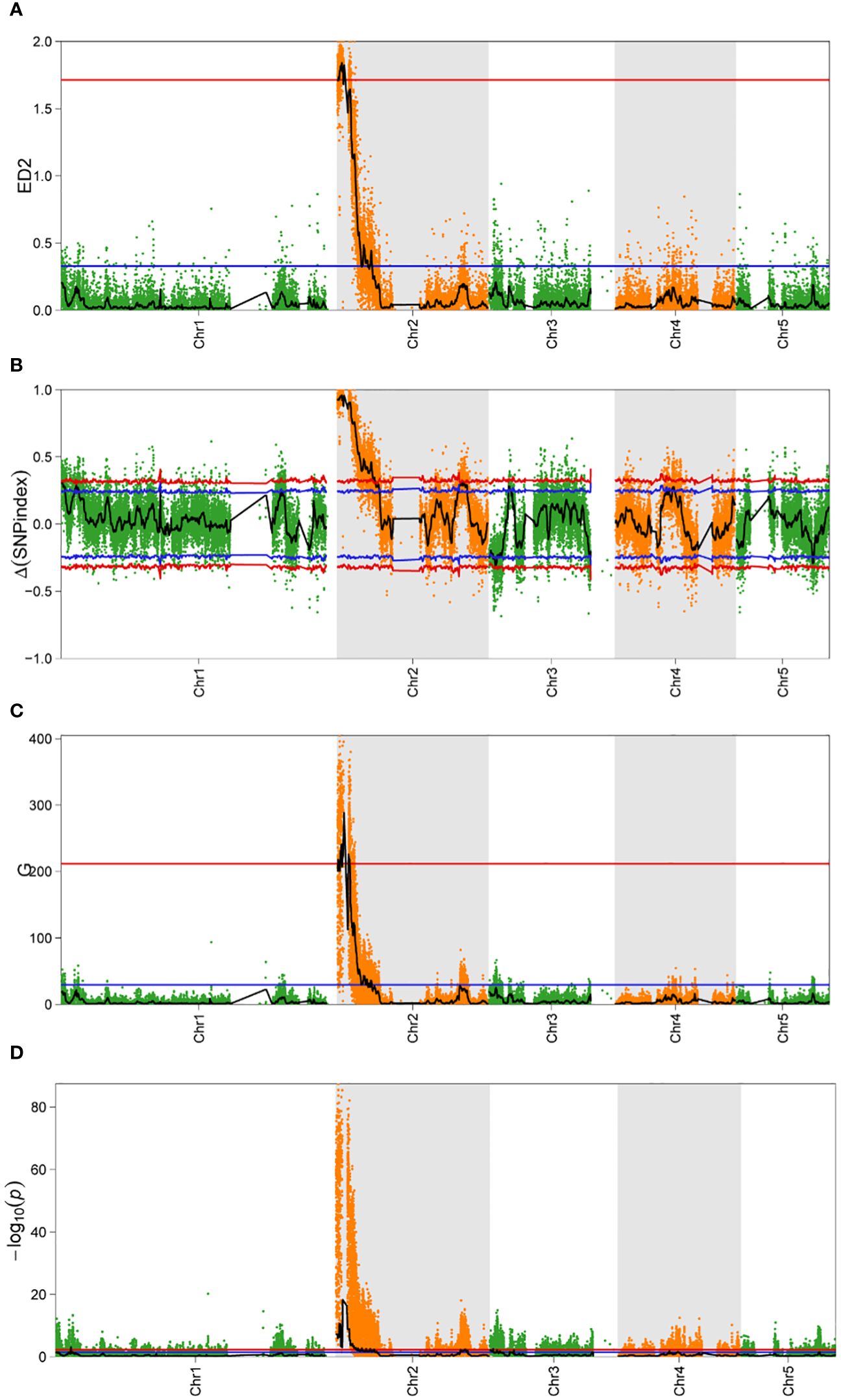
Figure 4 Bulked segregant analysis of candidate intervals associated with the coloration of P. haitanensis conchocelis. (A–D) represent Euclidean distance (ED), Δ(SNP-index), G-statistic (G), and Fisher’s exact test value for the QTL sequencing analysis, respectively. The x-axis presents the physical positions of P. haitanensis chromosomes. Red and blue lines indicate P < 0.01 and P < 0.05, respectively.
The candidate region (0.03-0.5 M) contained 57 annotated genes (Supplementary Table S4). Subsequent analysis revealed that four candidate genes associated with pigment biosynthesis and assembly, with SNP variation sites, were identified within the candidate interval (Table 1). Further examination indicated that the SNPs in the upstream and downstream regulatory regions of Porphobilinogen synthase and UPF0187 chloroplastic, as well as the synonymous mutation in the exonic region of UPF0187 chloroplastic, were considered unrelated to the orange coloration of P. haitanensis A nonsynonymous mutations (T/C) in Chlorophyll a synthase (CHLG) and a nonsynonymous SNP [A/G] at base-pair position 481 in the phycocyanin-associated rod linker protein (cpcC) coding region was detected in the final candidate region (Table 1).
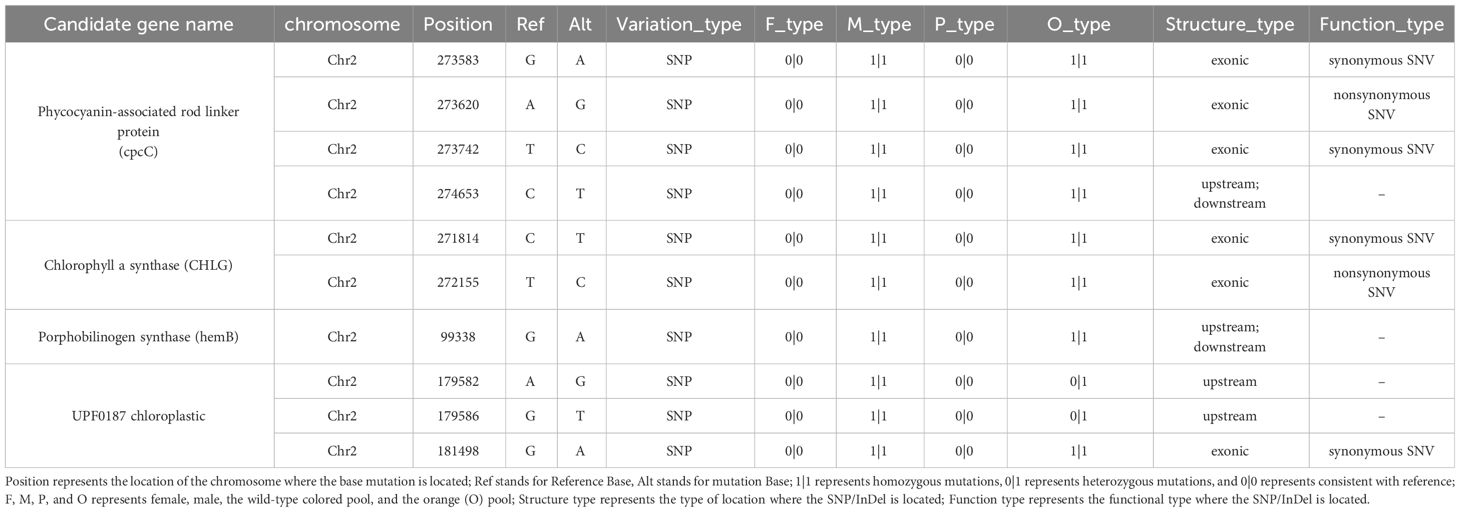
Table 1 Predicted candidate genes associated with photosynthetic pigment synthesis within the mapped region based on 99% confidence intervals.
To evaluate the association between the SNP in cpcC and the orange coloration of P. haitanensis, 20 randomly selected F1 filament lines from the DH population were analyzed through Sanger sequencing. The results demonstrated that the mutation was linked to the orange coloration of the P. haitanensis conchocelis (Supplementary Figure S1). Specifically, a base (A) in the wild-type colored lines was mutated (G) in the orange lines. To further verify that the mutation in cpcC was related to the orange coloration phenotype, 13 P. haitanensis lines with different coloration (8 orange and 5 wild-type colored lines) were selected from the P. haitanensis germplasm resource library of Fujian Province. These lines encompassed a range of germplasm resources, including wild-type lines, mutant lines, hybrid offspring lines, and breeding lines. Analysis of these 13 lines confirmed that the mutation was closely associated with the orange coloration of filaments. More specifically, a base (A) observed in all wild-type colored lines was mutated (G) in orange lines (Table 2).
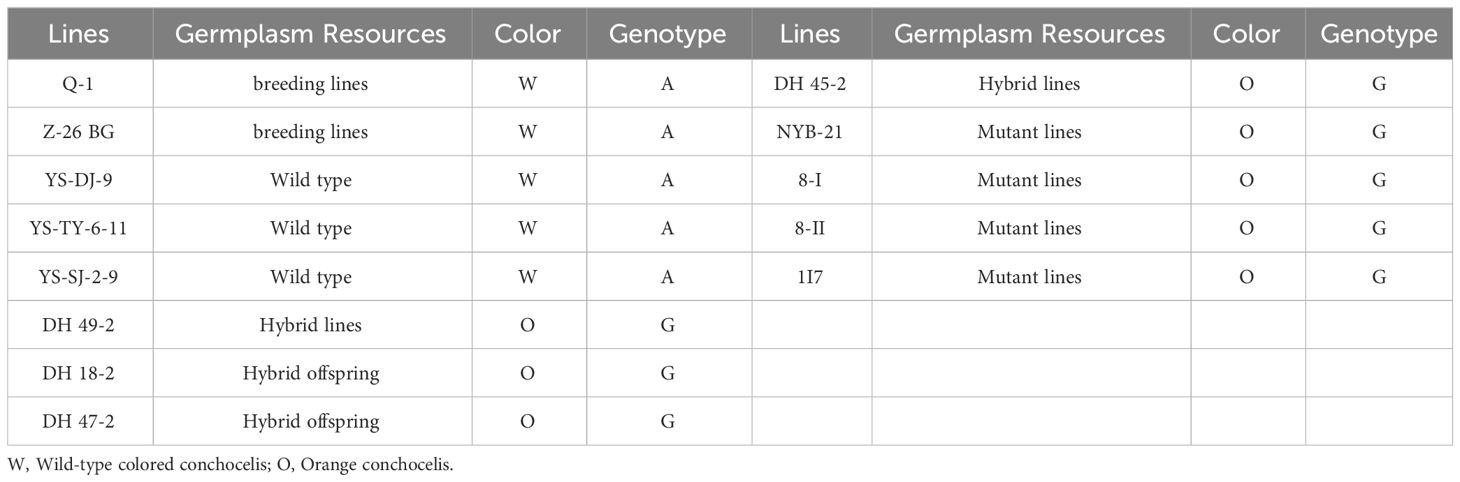
Table 2 Genotypes of base-pair position 481 in PhcpcC for the conchocelis coloration in 13 P. haitanensis germplasm resources.
A mapping population is a prerequisite for conducting QTL mapping. In Pyropia/Porphyra species, the process of meiosis occurs in the first and second cell divisions of germinated conchospores, resulting in the fusion of the four cells produced by meiosis. Due to homologous recombination during meiosis, the genotype of each sector in the tetrad may vary (Ohme and Miura, 1988; Yan et al., 2005). Therefore, the conventional protocols used for constructing mapping populations are inappropriate for Pyropia/Porphyra species. DH populations can be employed to rapidly generate homozygous breeding materials, thereby expediting the production of new varieties and improving selection efficiency (Forster and Thomas, 2005). In the present study, conchospores released from heterozygous conchocelis develop into chimeras composed of 2–4 sectors. To maintain the genetic integrity of the population, only four-colored thalli were considered during the construction of the DH population. Consequently, the largest Pyropia/Porphyra DH population to date was constructed, consisting of 480 pure offspring lines. This population size is sufficient for breeding new varieties and identifying QTLs associated with economically valuable traits. The genetic region linked to the orange coloration of P. haitanensis has been narrowed down to a 0.5 Mb interval (Figure 4), which is smaller than the 3.3 Mb region linked to the red coloration of N. yezoensis (Yu et al., 2020). This may be attributed to the presence of numerous segregating populations with tightly linked mutated regions.
Previous studies suggested that the inheritance patterns of mutated coloration phenotypes in Pyropia/Porphyra species follow Mendel’s laws, implying that abnormal coloration is caused a single nuclear gene (Niwa, 2010). In this study, the homozygous conchocelis of the parental line DH115-2 exhibited an orange coloration, which was in contrast to wild-type colored heterozygous conchocelis. Additionally, the segregation ratio of the wild-type colored sectors to orange sectors in the heterozygous conchocelis was approximately 1:1, indicating the recessive nuclear inheritance of the DH115-2 orange coloration trait. The color of Pyropia/Porphyra species is primarily determined by the contents and proportions of specific pigments, including PE, PC, APC, and Chl a (Yan et al., 2000). In the current study, the orange P. haitanensis lines exhibited significantly lower levels of PE, PC and Chl a as compared to the wild-type colored P. haitanensis lines (Figure 3A). Consequently, to identify candidate genes associated with the orange coloration, we screened for pigment-related genes within the candidate interval. On the basis of BSA, four candidate genes related to pigment biosynthesis and assembly were identified in the candidate region (Table 1). Further analyses showed that a non-synonymous SNP [A/G] at 481 bp position in PhcpcC was linked to the orange coloration phenotype (Supplementary Figure S1, Supplementary Table 1).
Phycobilisome (PBS) is composed of phycobiliproteins (PBPs) and linker polypeptides. Trimers comprising various PBPs are organized into a highly ordered supramolecular complex with the assistance of the linker proteins (Grossman, 1994) (Zhang et al., 2017). reported the cryo-electron microscopic structure of a 14.7-megadalton phycobilisome from the red alga G. pacifica and resolved the structures of all linker proteins, including four linkers in the core, 16 rod–core linkers, and 52 rod linkers, in the functionally assembled state. Linkers proteins have three proposed roles in the PBS: (i) organization and stabilization of the separate PBP subcomponents in the entire PBS complex, (ii) determination of the direction and termination of the rods and method of attachment of the rods onto the core, and (iii) modification of the spectral properties of the PBPs, by modifying the individual chromophore environments (David et al., 2011). Such as, the presence of a 40 kDa linker polypeptides between PEs and PCs enhances the energy transfer between PEs and PCs in the rod of phycobilisomes (Zhao et al., 2019), while mutations in linker polypeptides often lead to phycobilisome structures and functions alterations in phycobilisome structures (Liu et al., 2005). In the present study, a mutation was observed in the PhcpcC coding region at base-pair position 481 (A to G) in the orange paternal line DH115-2. This mutation was confirmed in the DH mapping population as well as in another population consisting of various germplasm resources, such as wild-type lines, mutant lines, hybrid offspring lines, and breeding lines (Table 2). Interestingly, the content of PE, PC in the orange-colored paternal parent was significantly lower than that in the wild-type-colored maternal parent. This observation suggests that the mutation in PhcpcC may induce structural and functional changes in the phycobilisomes, thereby affecting the content and assembly of important phycobiliproteins involved in regulating algal coloration, ultimately influencing the coloration of P. haitanensis thalli.
Transgenic and gene editing technologies have the potential to significantly facilitate breeding processes by enabling rapid, precise, and versatile customization. The expression systems of various genes, including the GUS gene (Shin et al., 2016; Kong et al., 2017), PyLHCI gene (Zheng et al., 2021), carbonic anhydrase γCAL1 gene (Shao et al., 2022), and other genes have been gradually established in P. yezoensis. These advancements provide technical support for further improvement of transgenic techniques in macroalgae and the exploration of key gene functions in Pyropia/Porphyra species. Despite the successful development of a genetic transformation system in P. yezoensis, transgenic strains of P. yezoensis can only be propagated through asexual reproduction using monospore germination. However, this method is not applicable to P. haitanensis, making it challenging to obtain stable transformed cells for this species. Additionally, the currently research progress in transgenic studies of P. yezoensis reported so far has also faced challenges in obtaining desirable phenotypes. Pigment mutations can easily cause significant changes in the color of macroalgae, thus, the genes involved in pigment synthesis and assembly can serve as target genes for genetic modification. In terms of gene editing, CRISPR/LbCas12a gene editing has been accomplished in Gracilariopsis lemaneiformis through direct transformation of protein-nucleic acid complexes using microparticle bombardment (Zhang et al., 2023). Single-strand conformation polymorphism analysis has proven to be useful in screening editing results in a large number of wild-type cells. This study serves as an important reference for the development of gene-editing systems in macroalgae. In Pyropia/Porphyra spp., both transgenic technology and gene editing technology are still in the early stages of exploration. Accordingly, there is an urgent need to establish a stable transgenic or gene editing system specifically for Pyropia/Porphyra species.
In this study, we constructed mixed pools of orange and wild-type colored individuals using the DH population of P. haitanensis to conduct a QTL sequencing analysis. The gene responsible for the orange coloration of this seaweed species was identified within an approximately 0.5 Mb region on chromosome 2. A non-synonymous SNP [A/G] was detected at base-pair position 481 in the coding region of PhcpcC, which encodes a phycocyanin-associated rod linker protein. Further investigation of natural populations confirmed PhcpcC as the associated gene for the orange coloration of P. haitanensis. The findings of this study provide breeders with molecular markers that can be valuable for exploiting orange-pigmented mutants to breed novel seaweed lines.
The data presented in the study are deposited in the NCBI SRA repository, accession number PRJNA1045300.
YG: Writing – original draft. JC: Writing – review & editing. YX: Writing – review & editing. YL: Writing – review & editing. DJ: Writing – review & editing. CC: Writing – review & editing. WW: Project administration, Writing – review & editing. CX: Project administration, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (U21A20265), the Fujian Province Science and Technology Major Project (2019NZ08003), the Young Elite Scientists Sponsorship Program by the China Association of Science and Technology (grant number: 2021QNRC001), and the China Agriculture Research System of MOF and MARA (CARS-50).
We thank Liwen Bianji (Edanz) (https://www.liwenbianji.cn) for editing the language of a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2024.1343363/full#supplementary-material
Supplementary Figure 1 | Mutation in the cpcC gene sequence.
Supplementary Table 4 | Gene function annotation results in significant intervals.
DH, doubled haploid; BSA, bulked segregant analysis; NGS, next-generation sequencing; SNP, single nucleotide polymorphism; O, orange; W, wild-type colored; PE, phycoerythrin; PC, phycocyanin; APC, allophycocyanin; PBP, phycobiliprotein; Chl a, chlorophyll a; cpcC, phycocyanin-associated rod linker protein.
Cho T. J., Rhee M. S. (2019). Health functionality and quality control of laver (Porphyra, Pyropia): current issues and future perspectives as an edible seaweed. Mar. Drugs 18, 14. doi: 10.3390/md18010014
David L., Marx A., Adir N. (2011). High-resolution crystal structures of trimeric and rod phycocyanin. J. Mol. Biol. 405, 201–213. doi: 10.1016/j.jmb.2010.10.036
FAO. (2017). Fisheries & aquaculture-fisheries and aquaculture fact sheets. Available at: http://www.fao.org/fishery/factsheets/en (Accessed on 2 August 2017).
FAO. (2019). Fishery and aquaculture statistics. Global aquaculture production. In FAO Fisheries and Aquaculture Department. Rome, Italy: FAO. Available at: http://www.fao.org/fishery/ (Accessed on 18 September 2019).
Fishery Department of China. (2021). China Fishery Bureau, Fishery Production, China Fishery Statistical Yearbook (in Chinese), Chinese Agriculture Express, 2021.
Forster B. P., Thomas W. T. (2005). Doubled haploids in genetics and plant breeding. Plant Breed Rev. 25, 57–88. doi: 10.1002/9780470650301.ch3
Grossman A. R. (1994). Phycobilisome and phycobiliprotein structure. Mol. Biol. Cyanobacteria, 139–216.
Hongfeng G. (1993). The variation in the contents of phycobilipro-teins from Porphyra haitanensis collected in different growing stages. Oceanologia Et Limnologia Sin. 24, 645–648.
Kong F., Zhao H., Liu W., Li N., Mao Y. (2017). Construction of plastid expression vector and development of genetic transformation system for the seaweed Pyropia yezoensis. Mar. Biotechnol. 19, 147–156. doi: 10.1007/s10126-017-9736-x
Li H., Durbin R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu L., Chen X., Zhang Y., Zhou B. (2005). Characterization, structure and function of linker polypeptides in phycobilisomes of cyanobacteria and red algae: an overview. Biochim. Et Biophys. Acta (Bba)-Bioenergetics 1708, 133–142. doi: 10.1016/j.bbabio.2005.04.001
Ma L., Liu Z., Cheng Z., Gou J., Chen J., Yu W., et al. (2021). Identification and application of bhaprr2 controlling peel colour in wax gourd (Benincasa hispida). Front. Plant Sci. 12, 716772. doi: 10.3389/fpls.2021.716772
Michelmore R. W., Paran I., Kesseli R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Niwa K. (2010). Genetic analysis of artificial green and red mutants of Porphyra yezoensis Ueda (Bangiales, Rhodophyta). Aquaculture 308, 6–12. doi: 10.1016/j.aquaculture.2010.08.007
Niwa K., Hayashi Y., Abe T., Aruga Y. (2009). Induction and isolation of pigmentation mutants of Porphyra yezoensis (Bangiales, Rhodophyta) by heavy-ion beam irradiation. Phycol. Res. 57, 194–202. doi: 10.1111/j.1440-1835.2009.00539.x
Ohme M., Miura A. (1988). Tetrad analysis in conchospore germlings of Porphyra yezoensis (Rhodophyta, Bangiales). Plant Sci. 57, 135–140. doi: 10.1016/0168-9452(88)90079-9
Sano F., Murata K., Niwa K. (2020). Identification, growth, and pigment content of a spontaneous green mutant of Pyropia kinositae (Bangiales, Rhodophyta). J. Appl. Phycol. 32, 1983–1994. doi: 10.1007/s10811-020-02085-5
Schneeberger K. (2014). Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 15, 662–676. doi: 10.1038/nrg3745
Shao Z., Xie X., Liu X., Zheng Z., Huan L., Zhang B., et al. (2022). Overexpression of mitochondrial γcal1 reveals a unique photoprotection mechanism in intertidal resurrection red algae through decreasing photorespiration. Algal Res. 66, 102766. doi: 10.1016/j.algal.2022.102766
Shin Y. J., Lim J., Park J. H., Choi D., Hwang M. S., Park E., et al. (2016). Characterization of pygus gene silencing in the red macroalga, Pyropia yezoensis. Plant Biotechnol. Rep. 10, 359–367. doi: 10.1007/s11816-016-0408-5
Takagi H., Abe A., Yoshida K., Kosugi S., Natsume S., Mitsuoka C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of dna from two bulked populations. Plant J. 74, 174–183. doi: 10.1111/tpj.12105
Wang K., Li M., Hakonarson H. (2010). Annovar: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Wang Y., Xu K., Wang W., Xu Y., Ji D., Chen C., et al. (2019). Physiological differences in photosynthetic inorganic carbon utilization between gametophytes and sporophytes of the economically important red algae Pyropia haitanensis. Algal Res. 39, 101436. doi: 10.1016/j.algal.2019.101436
Xu N., Xu K., Wang W., Xu Y., Ji D., Chen C., et al. (2020). Nutrient enrichment improves growth and food quality of two strains of the economic seaweed Pyropia haitanensis. Front. Mar. Sci. 7, 544582. doi: 10.3389/fmars.2020.544582
Xu Y., Huang L., Ji D., Chen C., Zheng H., Xie C. (2015). Construction of a dense genetic linkage map and mapping quantitative trait loci for economic traits of a doubled haploid population of Pyropia haitanensis (Bangiales, Rhodophyta). BMC Plant Biol. 15, 1–11. doi: 10.1186/s12870-015-0604-4
Xu Y., Xie C., Chen C., Ji D., Gao Y. (2012). Genetic analyses of six quantitative traits of a doubled haploid population of Porphyra haitanensis chang et zheng (Bangiales, Rhodophyta). J. Appl. Phycol. 24, 89–96. doi: 10.1007/s10811-011-9653-8
Yan X., Fujita Y., Aruga Y. (2000). Induction and characterization of pigmentation mutants in Porphyra yezoensis (Bangiales, Rhodophyta). J. Appl. Phycol. 12, 69–81. doi: 10.1023/A:1008129119065
Yan X., Li L., Aruga Y. (2005). Genetic analysis of the position of meiosis in Porphyra haitanensis chang et zheng (Bangiales, Rhodophyta). J. Appl. Phycol. 17, 467–473. doi: 10.1007/s10811-005-2752-7
Yu X., Wang L., Xu K., Kong F., Wang D., Tang X., et al. (2020). Fine mapping to identify the functional genetic locus for red coloration in Pyropia yezoensis thallus. Front. Plant Sci. 11, 867. doi: 10.3389/fpls.2020.00867
Zhang J., Ma J., Liu D., Qin S., Sun S., Zhao J., et al. (2017). Structure of phycobilisome from the red alga Griffithsia pacifica. Nature 551, 57–63. doi: 10.1038/nature24278
Zhang J., Wu Q., Eléouët M., Chen R., Chen H., Zhang N., et al. (2023). Crispr/lbcas12a-mediated targeted mutation of Gracilariopsis lemaneiformis (Rhodophyta). Plant Biotechnol. J. 21, 235. doi: 10.1111/pbi.13949
Zhao M., Sun L., Fu X., Chen M. (2019). Phycoerythrin-phycocyanin aggregates and phycoerythrin aggregates from phycobilisomes of the marine red alga Polysiphonia urceolata. Int. J. Biol. Macromol. 126, 685–696. doi: 10.1016/j.ijbiomac.2018.12.109
Zhao R., Xu Y., Xu K., Ji D., Chen C., Wang W., et al. (2022). Evaluation of the main economic characteristics of a narrow-thallus strain of Neoporphyra haitanensis. Aquaculture 558, 738395. doi: 10.1016/j.aquaculture.2022.738395
Zheng Z., He B., Xie X., Wang G. (2021). Co-suppression in Pyropia yezoensis (Rhodophyta) reveals the role of pylhci in light harvesting and generation switch. J. Phycol. 57, 160–171. doi: 10.1111/jpy.13073
Keywords: chimera, pigmentation marker, doubled haploid population, QTL, Pyropia haitanensis
Citation: Guo Y, Chang J, Xu Y, Liao Y, Ji D, Chen C, Wang W and Xie C (2024) Identification of orange color-related gene, PhcpcC, in Pyropia haitanensis. Front. Mar. Sci. 11:1343363. doi: 10.3389/fmars.2024.1343363
Received: 23 November 2023; Accepted: 19 April 2024;
Published: 07 May 2024.
Edited by:
Jianguang Qin, Flinders University, AustraliaReviewed by:
Guanpin Yang, Ocean University of China, ChinaCopyright © 2024 Guo, Chang, Xu, Liao, Ji, Chen, Wang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaotian Xie, Y3R4aWVAam11LmVkdS5jbg==; Wenlei Wang, d2x3YW5nQGptdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.